A Jacalin-lectin manipulates an RNA binding protein in shaping plant development through regulation of pre-mRNA processing of the key flowering repressor genes.
Abstract
Lectins selectively recognize sugars or glycans for defense in living cells, but less is known about their roles in the development process and the functional network with other factors. Here, we show that Arabidopsis (Arabidopsis thaliana) JACALIN-LECTIN LIKE1 (AtJAC1) functions in flowering time control. Loss of function of AtJAC1 leads to precocious flowering, whereas overexpression of AtJAC1 causes delayed flowering. AtJAC1 influences flowering through regulation of the key flowering repressor gene FLOWERING LOCUS C (FLC). Genetic analysis revealed that AtJAC1’s function is mostly dependent on GLYCINE-RICH RNA-BINDING PROTEIN7 (GRP7), an upstream regulator of FLC. Biochemical and cell biological data indicated that AtJAC1 interacted physically with GRP7 specifically in the cytoplasm. AtJAC1 influences the nucleocytoplasmic distribution of GRP7, with predominant nuclear localization of GRP7 when AtJAC1 function is lost but retention of GRP7 in the cytoplasm when AtJAC1 is overexpressed. A temporal inducible assay suggested that AtJAC1’s regulation of flowering could be compromised by the nuclear accumulation of GRP7. In addition, GRP7 binds to the antisense precursor messenger RNA of FLC through a conserved RNA motif. Loss of GRP7 function leads to the elevation of total FLC antisense transcripts and reduced proximal-distal polyadenylation ratio, as well as histone methylation changes in the FLC gene body region and increased total functional sense FLC transcript. Attenuating the direct binding of GRP7 with competing artificial RNAs leads to changes of FLC antisense precursor messenger RNA processing and flowering transition. Taken together, our study indicates that AtJAC1 coordinates with GRP7 in shaping plant development through the regulation of RNA processing in Arabidopsis.
Lectins are highly diverse proteins that selectively recognize specific free sugars or glycans present on glycoproteins and glycolipids (Peumans and Van Damme, 1995). They are widespread among living organisms and are involved in diverse biological processes (Moreira et al., 1991; Sharon, 2008). In animals, they are involved in self-recognition and cell-to-cell interaction in addition to acting as chaperones during glycoprotein synthesis (Rabinovich and Toscano, 2009; Lam and Ng, 2011). In plants, they serve as storage proteins and have roles in defense against pathogens and insect pests (Peumans and Van Damme, 1995; Babosha, 2008; Vandenborre et al., 2011). As a subgroup of lectins, Jacalin-lectin proteins have one or more jacalin-like lectin domains with binding specificity to Man or Gal (Kabir, 1998; Peumans et al., 2001). They are involved in multiple biological processes (Aucouturier et al., 1989; Yong et al., 2003; Jiang et al., 2006). Arabidopsis (Arabidopsis thaliana) JASMONATE ACID RESPONSE1 and wheat (Triticum aestivum) JACALIN-RELATED LECTIN LIKE1 are involved in jasmonic acid-mediated defense responses (León et al., 1998; Xiang et al., 2011). In rice (Oryza sativa), JACALIN-RELATED LECTIN1 (OsJAC1) suppresses the coleoptiles and stem elongation (Jiang et al., 2006). In wheat, a jacalin-like lectin, VER2, is induced during vernalization in young leaves and the shoot apical meristem, thus regulating flowering and spikelet development (Chong et al., 1998; Yong et al., 2003). Recently, we found that this is mainly mediated by the interaction between VER2 and proteins with O-GlcNAc modification during the course of vernalization (Xing et al., 2009; Xiao et al., 2014). To date, whether Jacalin-lectin functions in development regulation and how it cooperates with other factors are still unclear in dicotyledons such as Arabidopsis.
Flowering is an important developmental transition during the life cycle of plants. It is precisely regulated by endogenous programs and environmental cues (Fornara et al., 2010). In Arabidopsis, flowering is regulated by five major pathways: vernalization, photoperiod, GA, autonomous, and age (Amasino, 2010; Srikanth and Schmid, 2011; Song et al., 2013a; Romera-Branchat et al., 2014). Multiple genetic pathways converge to regulate the expression of FLOWERING LOCUS C (FLC), which encodes a MADS box transcription factor that plays a key role in the repression of flowering (Michaels and Amasino, 1999; Sheldon et al., 1999). During long-term cold, the vernalization pathway epigenetically silences FLC (Song et al., 2013a). In ambient temperatures, the FRIGIDA pathway up-regulates FLC expression while the autonomous pathway down-regulates FLC (Amasino, 2010; Crevillén and Dean, 2011). Chromatin modification and RNA processing are involved in the precise regulation of FLC transcription under both cold (vernalization) and ambient temperatures (He, 2012; Song et al., 2013a; Csorba et al., 2014). Autonomous pathway members, the RNA-binding proteins FCA and FPA, together with the 3′ processing factors CLEAVAGE STIMULATING FACTOR64 (CstF64), CstF77, and FY are involved in the polyadenylation site selection of a set of antisense transcripts of FLC, named COOLAIR (Swiezewski et al., 2009; Hornyik et al., 2010; Liu et al., 2010). The alternative usage of polyadenylation sites of COOLAIR is coupled with the expression state of a functional FLC sense transcript in a process requiring the histone H3K4me2 demethylase FLOWERING LOCUS D (Liu et al., 2007) and the positive transcription elongation factor b complex component CYCLIN-DEPENDENT KINASE C (Wang et al., 2014).
A member of the highly conserved small Gly-rich RNA-binding proteins (GRPs; Mangeon et al., 2010), GRP7, is also reported to regulate flowering in the autonomous pathway (Streitner et al., 2008). It contains an N-terminal RNA recognition motif and a C-terminal Gly-rich domain with nucleocytoplasmic shuttling ability (Lummer et al., 2011). GRP7 has multiple functions in RNA processing, including RNA chaperone activity (Kim et al., 2007), facilitating messenger RNA (mRNA) transport from the nucleus to the cytoplasm (Kim et al., 2008), regulating precursor messenger RNA (premRNA) splicing (Streitner et al., 2012), and processing of the primary microRNA (Köster et al., 2014). However, how GRP7 is involved in the regulation of flowering and the direct target of GRP7 in the flowering control network are unknown.
Here, we report that the Jacalin-lectin AtJAC1 functions in flowering time control in Arabidopsis. AtJAC1 interacts genetically and physically with GRP7 to regulate the FLC functional transcript level, which is coupled by influencing the RNA processing of COOLAIR through direct binding of GRP7 to FLC antisense premRNA.
RESULTS
Jacalin-Lectin AtJAC1 Regulates Flowering Time in Arabidopsis
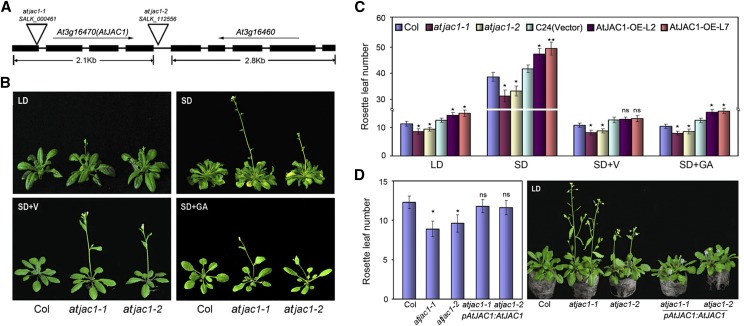
Phylogenetic analysis based on the amino acid sequence similarity of the Jacalin domain showed that there were 15 proteins in Arabidopsis that are potential orthologs of wheat Jacalin-lectin VER2 (Supplemental Fig. S1A), which is induced by cold and regulates vernalization-mediated flowering in wheat (Yong et al., 2003; Xiao et al., 2014). Among those genes, AtJAC1 (At3g16470) contained three Jacalin-lectin domains with hemagglutination activity and could bind Man, Rib, and Glc with high affinity (Supplemental Fig. S1, B–E; Supplemental Table S1). AtJAC1 is expressed in lots of tissues, with a dynamic expression level in the shoot apex during the floral transition process (Supplemental Fig. S1F), indicating its potential role in flowering time regulation.
To investigate whether AtJAC1 plays a role in the floral transition, two transfer DNA (T-DNA) insertion mutants of AtJAC1 were obtained from SALK (Alonso et al., 2003). In the null mutant atjac1-1 (SALK_000461), T-DNA was inserted in the first exon to cause the absence of the full-length mRNA of AtJAC1, whereas in atjac1-2 (SALK_112556), T-DNA was inserted into the intergenic region between At3g16470 (AtJAC1) and At3g16460, causing the down-regulation of both genes (Fig. 1A; Supplemental Fig. S2A). At3g16460 (AtJAC2) was also predicted to be a jacalin-like lectin, with four jacalin-lectin domains (Supplemental Fig. S2B). However, its expression was low in the shoot apex (Supplemental Fig. S2C). A knockdown mutant of At3g16460 (atjac2; SALK_028332) showed no change in flowering time compared with wild-type Columbia (Col; Supplemental Fig. S2, D and E). Contrarily, both atjac1-1 and atjac1-2 were early flowering, with significantly fewer rosette leaves when bolting compared with wild-type Col, under either long-day (LD) or short-day (SD) conditions (Fig. 1, B and C). Either vernalization or GA3 treatment could accelerate the flowering of both atjac1 mutants and Col, but still atjac1 mutants flower slightly earlier than Col (Fig. 1, B and C). This suggests that AtJAC1 may not function in the vernalization- or GA-regulated flowering time pathways. Meanwhile, AtJAC1 overexpression lines (AtJAC1-OE; Supplemental Fig. S2F) showed delayed flowering either under varied photoperiod conditions or with GA3 treatment as compared with the control, ecotype C24, transformed with the empty vector pBI121-GFP (Fig. 1C). By contrast, vernalization rescued the delayed flowering of AtJAC1-OE lines to the control level (Fig. 1C). Furthermore, we performed genetic complementation by introducing pAtJAC1::AtJAC1 in atjac1-1 or atjac1-2 mutants to confirm the function of AtJAC1 in flowering regulation (Fig. 1D; Supplemental Fig. S2G). As expected, the complementation lines rescued the early-flowering phenotype of both atjac1-1 and atjac1-2 (Fig. 1D). Taken together, AtJAC1 negatively regulates flowering in Arabidopsis.
Figure 1.
AtJAC1 inhibits the flowering transition in Arabidopsis. A, Schematic diagram of the T-DNA insertion sites of atjac1-1 and atjac1-2 mutants. B, atjac1 mutants flowered earlier than the wild type (Col) under SD and LD conditions, SD conditions with vernalization (SD+V), and SD conditions with GA (SD+GA). C, Rosette leaf number at bolting in atjac1 mutants, AtJAC1-OE plants, and controls (Col and C24 transformed with vector pBI121-GFP). D, Morphological phenotype and rosette leaf numbers in atjac1-1, atjac1-2, and genetic complementation lines with pAtJAC1::AtJAC1. Data are means ± sd of 15 plants for each line. Statistical analysis was done by Student’s t test in C and D; atjac1 mutants were compared with Col, whereas AtJAC1-OE lines were compared with C24 (vector). *, P < 0.05; **, P < 0.01; ns, no significant change.
AtJAC1 Influences Flowering through the Regulation of FLC Transcript Level
AtJAC1 mutants and ectopic overexpression transgenic lines retain the photoperiodic response, which is even more severe in SD conditions (Fig. 1C), which is a feature of autonomous pathway regulators (He and Amasino, 2005). In addition, vernalization fully overcomes the late-flowering phenotype of AtJAC1 overexpression lines (Fig. 1C), which is similar to the autonomous pathway late-flowering mutants (Bäurle and Dean, 2006). This hints at the possibility that AtJAC1 regulates flowering through the autonomous pathway. FLC is a key flowering repressor, integrating the inputs from autonomous pathway members (He, 2012). How the FLC response works in the floral repression effect of AtJAC1 is an attractive issue.
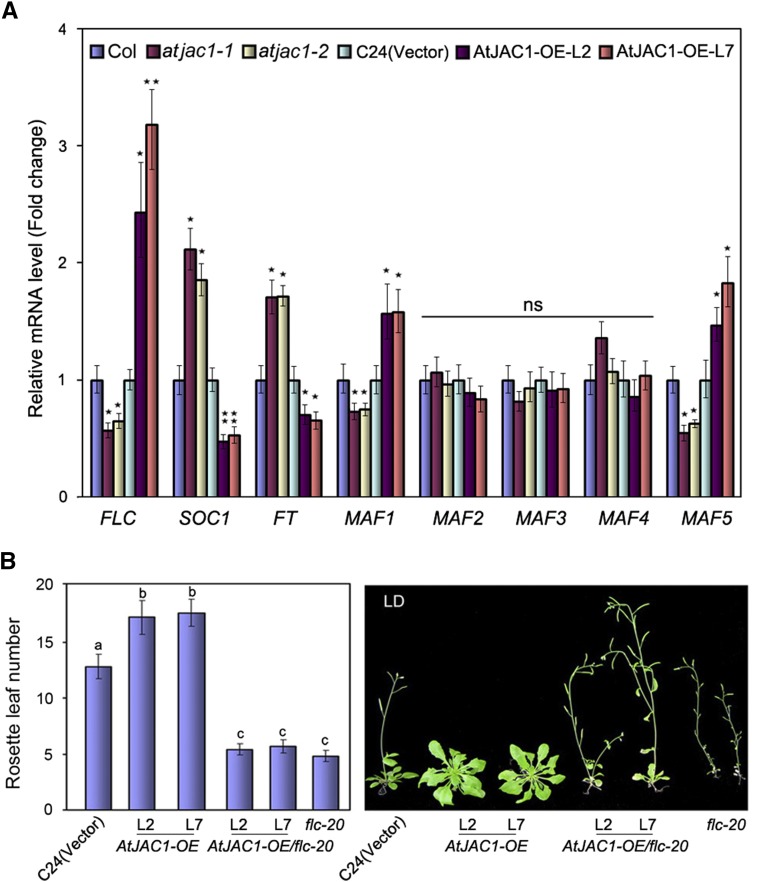
To address this question, we quantified FLC mRNA expression in AtJAC1 mutants and ectopic overexpression transgenic lines. The FLC mRNA level was reduced in atjac1-1 and atjac1-2 mutants but elevated in AtJAC1-OE lines (Fig. 2A). Concomitantly, SUPRESSOR OF OVEREXPRESSION OF CONSTANS1 (SOC1) and FLOWERING LOCUS T (FT), which are negatively controlled by direct binding of FLC, were up-regulated in atjac1 mutants but down-regulated in AtJAC1-OE lines (Fig. 2A). This suggests that AtJAC1 may influence flowering time through the regulation of FLC expression. In addition, the FLC homologs MADS AFFCTING FLOWERING1 (MAF1) and MAF5, but not MAF2, MAF3, and MAF4, showed a similar expression pattern to FLC in atjac1 mutants and AtJAC1-OE lines, although the fold change is moderate (Fig. 2A). In contrast, the mRNA levels of other flowering pathway key regulators, such as CONSTANS (CO) of the photoperiod pathway (Putterill et al., 1995) and SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) of the age pathway (Wang et al., 2009; Wu et al., 2009), were not significantly changed in either atjac1 mutants or AtJAC1-OE lines (Supplemental Fig. S3).
Figure 2.
AtJAC1 influences flowering through the regulation of FLC transcript level. A, Transcript levels of key flowering genes in atjac1 mutants and AtJAC1-OE plants (normalized to Ubiquitin C [UBC], a reference gene for quantitative PCR [qPCR]). Data are means ± sd of three parallel samples from one replicate, and three biological repeats were performed. B, Morphological phenotypes and rosette leaf numbers in AtJAC1-OE/flc-20 under LD conditions. Statistical analysis was done by Student’s t test in A; atjac1 mutants were compared with Col, whereas AtJAC1-OE lines were compared with C24 (vector). *, P < 0.05; **, P < 0.01; ns, no significant change. Tukey’s honestly significant difference (HSD) test was used for B, and different letters indicate statistically significant differences.
To study the genetic relation between FLC and AtJAC1, the late-flowering lines AtJAC1-OE-L2 and AtJAC1-OE-L7 were crossed to a null mutant, flc-20, which has a Dissociation element insertion, leading to extremely early flowering (Helliwell et al., 2002). The F2 generation, homozygous for both the flc-20 allele and AtJAC1-OE constructs, flowered almost the same time as flc-20 plants in terms of rosette leaf number when bolting (Fig. 2B) but significantly earlier than the control (ecotype C24 transformed with the empty vector). Therefore, AtJAC1 function genetically depends on FLC in the regulation of flowering.
AtJAC1 Interacts Physically with GRP7 for the Regulation of FLC Transcript Level and Flowering
We previously reported that TaGRP2, an ortholog of the autonomous pathway member GRP7 in Arabidopsis, could interact physically with Jacalin-lectin VER2 in the regulation of flowering transition in wheat (Xiao et al., 2014). This may be a clue that AtJAC1 regulates FLC transcript level by directly interacting with GRP7.
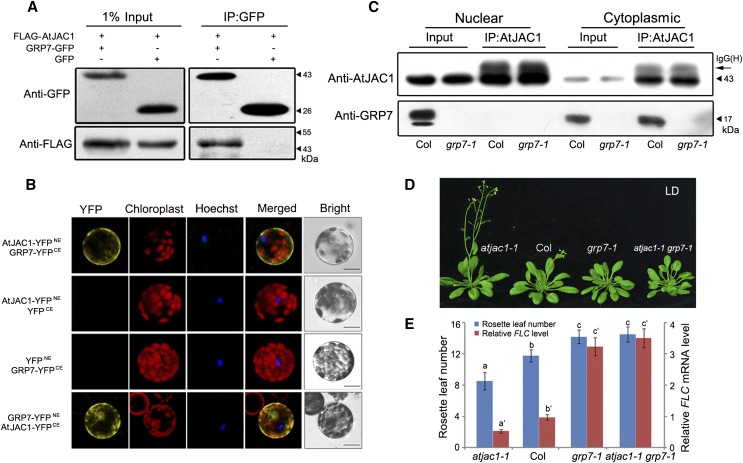
To confirm this hypothesis, a coimmunoprecipitation (CoIP) assay was performed. FLAG-AtJAC1 was coexpressed with GRP7-GFP or GFP in tobacco (Nicotiana tabacum) leaves. After immunoprecipitation (IP) with GFP antibody, FLAG-AtJAC1 was detected in the elution of the FLAG-AtJAC1/GRP7-GFP coexpression line but not in that of the FLAG-AtJAC1/GFP line, demonstrating that AtJAC1 and GRP7 interacted in vivo (Fig. 3A). Bimolecular fluorescence complementation (BiFC) assays revealed that yellow fluorescent protein (YFP) signal generated from the interaction of AtJAC1 and GRP7 was only in the cytoplasm in Arabidopsis protoplasts (Fig. 3B). However, both AtJAC1-GFP and GRP7-GFP could localize in the cytoplasm and the nucleus (Supplemental Fig. S4A). Immunoblotting of nuclear and cytoplasmic fractions indicated that AtJAC1 and GRP7 were detected in both fractions (Fig. 3C; Supplemental Fig. S4, B and C). However, the IP assay showed that GRP7 was coimmunoprecipitated with AtJAC1 only in the cytoplasmic fraction and not the nuclear fraction (Fig. 3C). This further supports the idea that AtJAC1 and GRP7 directly interact specifically in the cytoplasm.
Figure 3.
AtJAC1 interacts physically and genetically with GRP7 in the regulation of flowering. A, CoIP assay confirming the interaction between AtJAC1 and GRP7 in vivo. AtJAC1-FLAG together with GRP7-GFP or GFP were cotransfected into tobacco leaves, IP was performed with anti-GFP antibody, and immunoblotting was done with anti-FLAG antibody. B, BiFC assay showing the interaction between AtJAC1 and GRP7 in the cytoplasm in Arabidopsis protoplasts. Hoechst staining of nuclei is shown. Bars = 10 μm. C, CoIP assay of the interaction between AtJAC1 and GRP7 in the cytoplasm. Nuclear and cytoplasmic protein fractions were isolated from Col and grp7-1 plants separately. IP was performed with anti-AtJAC1 antibody, and immunoblotting was done with anti-GRP7 antibody. IgG(H), IgG H chain. D and E, AtJAC1 regulates flowering through GRP7. Morphological phenotypes (D) as well as rosette leaf numbers and FLC mRNA levels (E) are shown in Col, atjac1-1, grp7-1, and atjac1-1 grp7-1 double mutants. Tukey’s HSD test was performed for E. Letters with or without ′ represent different groups; different letters indicate statistically significant differences within groups.
To further analyze their genetic relation, we generated the double mutant atjac1-1 grp7-1 (Supplemental Fig. S5). The flowering time in each genotype was quantified by measuring the rosette leaf number when bolting. As reported previously (Streitner et al., 2008), the grp7-1 single mutant is late flowering with more rosette leaves when bolting compared with Col (Fig. 3, D and E). In the F2 generation, the homozygous double mutant atjac1-1 grp7-1 showed delayed flowering compared with Col and atjac1-1 but similar to grp7-1 (Fig. 3, D and E). As well, the FLC expression level coincides with the flowering time of different genotypes (Fig. 3E). Thus, AtJAC1 is genetically dependent on GRP7 in the regulation of FLC transcript level and flowering.
AtJAC1 Inhibits the Nuclear Accumulation of GRP7
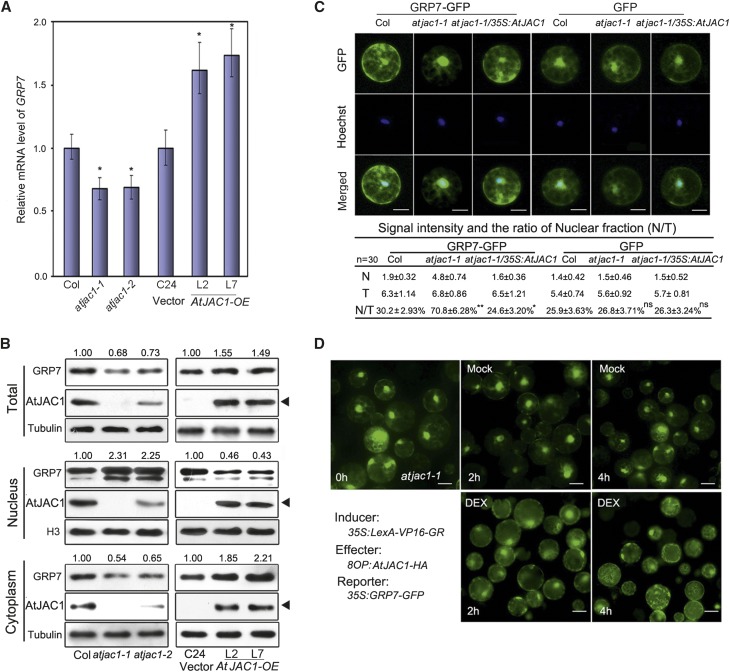
The opposite phenotype of flowering time suggested that AtJAC1 may negatively regulate GRP7 function. We then checked if this repression effect is based on the amount of mRNA and/or protein level change of GRP7 in AtJAC1 mutants and overexpression plants. The mRNA level of GRP7 was slightly down-regulated in atjac1 mutants and up-regulated in AtJAC1-OE lines (Fig. 4A). As well, the total protein levels of GRP7 decrease slightly in atjac1 mutants and increase in AtJAC1-OE lines (Fig. 4B). However, neither transcript of AtJAC1 nor protein level showed a change in either grp7-1 or GRP7-OE plants (Supplemental Fig. S6, A and B).
Figure 4.
AtJAC1 inhibits the nuclear accumulation of GRP7. A, GRP7 transcription was up-regulated in AtJAC1-OE lines and down-regulated in the atjac1 mutant. Student’s t test was performed; atjac1 mutants were compared with Col, whereas AtJAC1-OE lines were compared with C24 (vector): *, P < 0.05. B, Immunoblotting of GRP7 protein in atjac1 mutant and AtJAC1-OE transgenic lines. Tubulin is shown as a loading control for total protein and cytoplasmic fractions, and H3 shows loading for the nuclear fraction. Black triangles indicate AtJAC1-GFP (transgene) bands. Relative fold changes of GRP7 were quantified using ImageJ (http://rsb.info.nih.gov/ij). Experiments were repeated with three batches of independent materials. C, Quantification of GRP7-GFP distribution to the nucleus in protoplasts. The fluorescence intensities (106×) and the ratio between the nucleus and the total (N/T) in each type of protoplast are shown below the images. N, Nuclear signal; T, total signal. Data are means ± sd of 30 individual cells of one transformation; three different transformations were done in total. Hoechst staining indicates the nucleus area. Statistical analysis was done by Student’s t test; GRP7-GFP or GFP intensity in atjac1-1 or atjac1-1/35S:AtJAC1 were compared with Col: *, P < 0.05; **, P < 0.01; ns, no significant change. Bars = 10 μm. D, DEX-induced expression of AtJAC1 accelerated the spread of nucleus-localized GRP7-GFP to the cytoplasm in atjac1-1 protoplasts. DEX, Treatment with solution containing 10 µm DEX; Mock, treatment with solution without DEX reagent. Bars = 15 μm.
GRP7 is a nucleocytoplasmic shuttling protein and functions in regulating premRNA splicing within the nucleus (Lummer et al., 2011; Streitner et al., 2012). We further quantified the protein level of GRP7 in nuclear and cytoplasm fractions separately in loss- or gain-of-function AtJAC1 mutants and transgenic plants (Fig. 4B). The GRP7 protein level in the nuclear fraction was obviously elevated in atjac1 mutants (more than 2-fold change) and reduced in AtJAC1-OE lines (Fig. 4B). By contrast, the cytoplasm-distributed GRP7 obviously decreased in atjac1 mutants and increased in AtJAC1-OE lines (Fig. 4B). Furthermore, we used fluorescence fusion protein to visualize and quantify the distribution of GRP7-GFP in subcellular components in a transient protoplast assay. This showed that the nuclear distribution of GRP7-GFP was about 70.8% in protoplast generated from atjac1-1 but 30.2% in that of Col, regardless of the similar total protein level of GRP7-GFP in the transient expression assay (Fig. 4C; Supplemental Fig. S6C). When 35S:AtJAC1 was introduced into atjac1-1, the percentage of nucleus-distributed GRP7-GFP was reduced to 24.6%, which is even significantly lower than that in Col (Fig. 4C). By contrast, the control protein GFP alone showed no significant nucleocytoplasmic distribution differences in protoplasts among Col, atjac1-1, and atjac1-1 transformed with 35S:AtJAC1 (Fig. 4C). To gain more insight whether GRP7’s nucleocytoplasmic distribution change is directly related to AtJAC1 or secondary effects caused by constant misexpression of AtJAC1, an inducible transient expression system (Ryu et al., 2010) was used to quantitatively increase AtJAC1 protein level. A similar phenomenon was observed in that the nucleus-localized GRP7-GFP in atjac1-1 gradually decreased after dexamethasone (DEX)-induced expression of AtJAC1 (Fig. 4D; Supplemental Fig. S6D). Therefore, AtJAC1 negatively regulates the nuclear accumulation of GRP7.
Nuclear Accumulation of GRP7 Compromises AtJAC1 Function
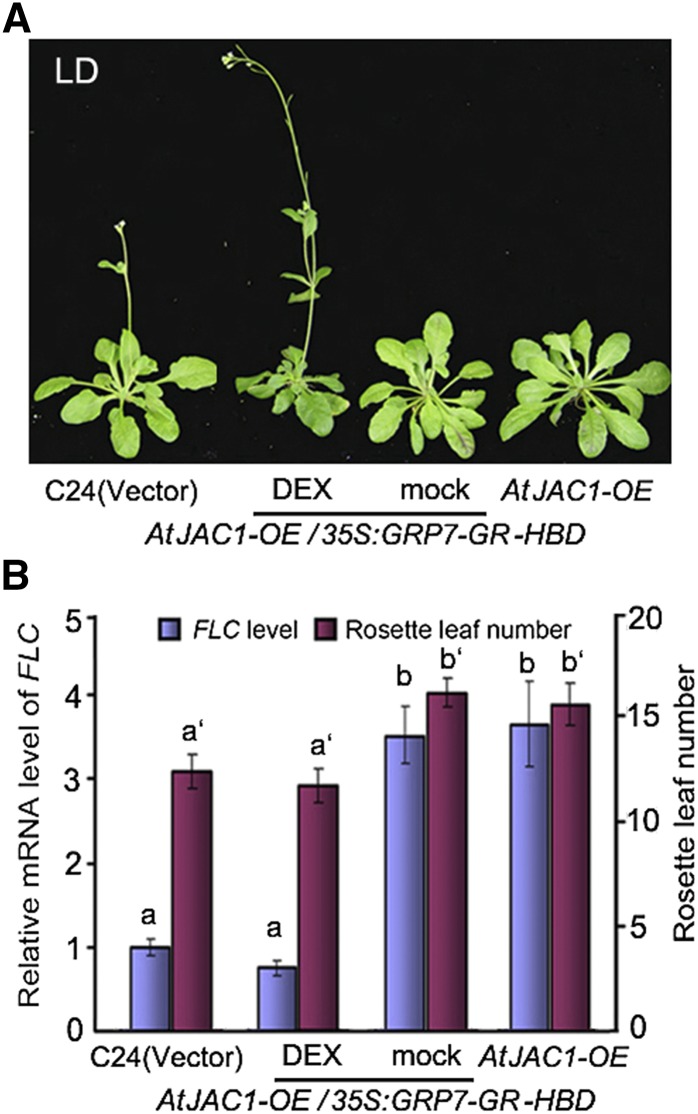
Does the nuclear accumulation level of GRP7 mediate AtJAC1’s regulation for flowering time? GRP7 was in-frame fused with the GLUCOCORTICOID RECEPTOR (GR) HORMONE-BINDING DOMAIN (HBD; Schena et al., 1991) driven by the Cauliflower mosaic virus (CaMV) 35S promoter in 35S:GRP7-GR-HBD and transformed into AtJAC1-OE lines (Supplemental Fig. S7A). With mock treatment (solution containing no DEX reagent), the GRP7-GR-HBD fusion protein was constitutively expressed but held in the cytoplasm by HEAT SHOCK PROTEIN90 (HSP90; Supplemental Fig. S7B). In the nuclear fraction, the endogenous GRP7 protein was lower compared with the control (transformed with empty vector) due to the overexpression of AtJAC1. Consistent with the low accumulation of GRP7 in the nucleus, the transgenic AtJAC1-OE35S:GRP7-GR-HBD lines showed delayed flowering with elevated FLC transcript level, similar to AtJAC1-OE, compared with control plants (Fig. 5, A and B). However, with the DEX treatment, GRP7-GR-HBD protein was released from HSP90 and a portion of the fusion protein was translocated into the nucleus, where the accumulation of GRP7 (endogenous GRP7 and GRP7-GR-HBD fusion protein) was increased significantly compared with that in AtJAC1-OE (Supplemental Fig. S7B). As expected, AtJAC1-OE35S:GRP7-GR-HSD plants rescued the delayed flowering phenotype of AtJAC1-OE, resulting in a similar rosette leaf number to the control plant when bolting. As well, FLC expression returned to the level of the control (Fig. 5, A and B). Therefore, the nuclear accumulation level of GRP7 coordinates the regulation of flowering by AtJAC1.
Figure 5.
Nuclear accumulation of GRP7 compromises AtJAC1 function. Delayed flowering in AtJAC1-OE was rescued by DEX-induced nuclear accumulation of GRP7. Morphological phenotypes (A) as well as rosette leaf numbers and FLC mRNA levels (B) in AtJAC1-OE and DEX-inducible lines are shown. Statistical analysis of multiple comparisons was done by Tukey’s HSD test. Letters with or without ′ represent different groups; different letters indicate statistically significant differences within groups.
GRP7 Directly Binds FLC Antisense premRNA through a Conserved Motif
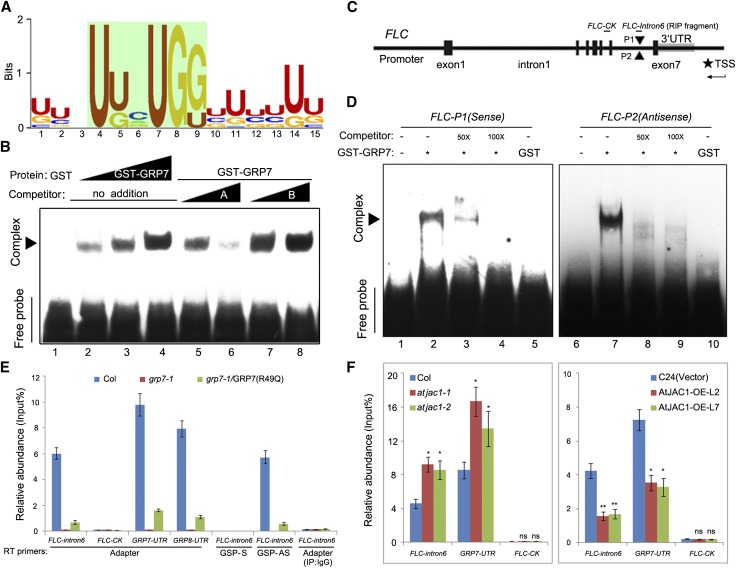
To find the direct target of GRP7, systematic evolution of ligands by exponential enrichment (SELEX; Tuerk and Gold, 1990; Xiao et al., 2014) was used to screen for the RNA-binding preference of GRP7. The most enriched RNA motif, U/G1N2N3U4U/G5N6U7G8G9N10U/G11N12N13U/G14U/G15 (E value of 7.4e-042), was calculated by MEME2 (Bailey et al., 2009) with the input sequences from the SELEX screening (Fig. 6A; Supplemental Table S2). RNA-electrophoresis mobility shift assay (EMSA) data showed that GRP7 could bind this RNA motif in vitro (Fig. 6B). Point mutation of specific nucleotides (U to A and G to A) suggests that U4U/G5 and U7G8G9 are necessary for the binding, whereas U/G1U/G11 and U/G14U/G15 contribute less (Supplemental Fig. S8A). Swapping of U to G or G to U at positions 4, 5, 7, 8, and 9 of this motif reduced the binding intensity of GRP7 in the RNA-EMSA (Supplemental Fig. S8B). This suggested that the sequence itself contributes more to the binding of GRP7 than the nucleotide composition.
Figure 6.
GRP7 directly binds FLC antisense premRNA through a conserved motif. A, The most significantly enriched RNA-binding motif of GRP7 screened by SELEX and analyzed by MEME2. B, RNA-EMSA to confirm the binding of GRP7 to the motif screened out by SELEX in vitro. Competitor A was a non-biotin-labeled GRP7-binding motif, and competitor B was a nonrelevant RNA fragment without biotin labeling. Sequences of individual oligonucleotides are listed in the supplemental sheet. C, Schematic diagram of the positions of putative GRP7-binding motifs in the FLC genomic sequence. Gene structure is as indicated. P1 and P2, Two putative GRP7-binding sites in FLC intron 6 with opposite directions; TSS, transcription start site for antisense FLC. D, GRP7 binds to FLC sense and antisense RNA probes in RNA-EMSA in vitro. Competitors are non-biotin-labeled RNA fragment FLC-P1 or FLC-P2. E, RIP followed by qPCR to confirm the binding of GRP7 to antisense FLC premRNA in vivo. FLC-intron6, FLC sense or antisense premRNA fragment containing putative GRP7-binding sites, with position indicated in C; FLC-control check fragment (FLC-CK), FLC premRNA fragment containing no GRP7-binding sites as a negative control, with position indicated in C; GSP, gene-specific primer. The GRP7/GRP8-UTR fragment served as a positive control, and grp7-1 transformed with pGRP7:GRP7(R49Q) served as a negative control for GRP7-binding activity. F, The binding of GRP7 to FLC antisense premRNA was altered in atjac1 mutants and AtJAC1-OE plants compared with controls. Adaptor primers were used for RT after determining the RNA pool with IP. Data shown in E and F are means ± sd of three parallel samples from one replicate, and three biological repeats were performed. Statistical analysis was done by Student’s t test: *, P < 0.05; **, P < 0.01; ns, no significant change.
Sequence analysis showed the two putative GRP7-binding motifs located closely in intron 6 of FLC, on opposite strands (Fig. 6C; Supplemental Table S3). RNA-EMSA indicated that GRP7 could bind both sense and antisense RNA probes in vitro (Fig. 6D). We used in vivo RNA immunoprecipitation (RIP) followed by reverse transcription (RT)-PCR to identify the binding status of GRP7 at the FLC locus in planta. Point mutation of R49Q (Arg to Gln at the 49th amino acid) in the RNA recognition motif domain of GRP7 dramatically reduced its RNA-binding ability (Schöning et al., 2007), which was used as a negative control for the RNA-binding specificity of GRP7. After IP with GRP7 antibody, the coimmunoprecipitated RNA pools were reverse transcribed with adaptor primer and further amplified with specific primers as indicated (Supplemental Fig. S9). Consistent with a previously reported EMSA in vitro (Schöning et al., 2007), the GRP7 and GRP8 untranslated region (UTR) was enriched in the IP-GRP7 sample from Col but reduced in grp7-1 transformed with pGRP7:GRP7R49Q and not detected in grp7-1, suggesting that the RIP assay works in vivo (Fig. 6E; Supplemental Fig. S10). The FLC-intron6 fragment, which contained the two putative binding motifs, was indeed enriched in Col but hard to detect in grp7-1 or grp7-1 transformed with pGRP7:GRP7R49Q, with no detection in Col using a nonrelated IgG for IP. By contrast, the FLC-CK fragment in intron 5 lacking the GRP7-binding motif was not detected in Col (Fig. 6, C and E; Supplemental Fig. S9). Primers complemented to sense or antisense strands specifically were then used to substitute for the adaptor primer when generating the first-strand complementary DNA (cDNA) from the RNA pool CoIP with GRP7 antibody. The FLC-intron6 fragment could only be detected in Col when using the primer complemented to antisense RNA (Fig. 6E; Supplemental Fig. S10). Thus, GRP7 binds directly to FLC antisense premRNA in vivo.
As expected, we find more coimmunoprecipitated FLC antisense premRNA with GRP7 in the background of atjac1 mutants compared with wild-type Col by RIP-qPCR (Fig. 6F). By contrast, less was detected in AtJAC1-OE transgenic plants as compared with the control (C24 transformed with vector; Fig. 6F).
GRP7 and AtJAC1 Influence the RNA Processing of FLC Antisense Transcripts (COOLAIR)
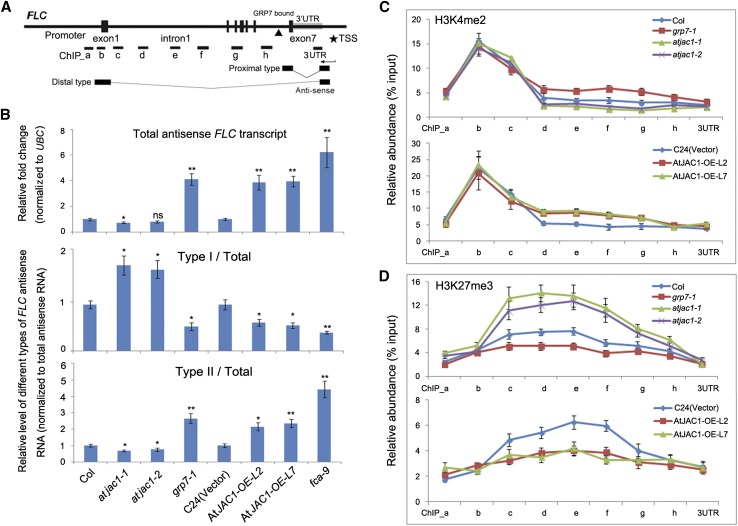
The consequence of directly binding GRP7 to FLC antisense premRNA was analyzed. Naturally occurring long noncoding RNA antisense transcripts, named COOLAIR, originate from the promoter adjacent to the FLC 3′ UTR and consist of two classes, terminating at proximal (sense intron 6) or distal (sense promoter) sites; each class contains several subclasses of alternative splicing variants (Swiezewski et al., 2009; Hornyik et al., 2010; Helliwell et al., 2011; Fig. 7A). Of note, the GRP7-binding site is close to the polyadenylation site of the proximal type COOLAIR, where the RNA-binding factor FCA and the 3′ RNA processing factors Cst64 and Cst77 are also associated (Liu et al., 2007, 2010). Therefore, we investigate whether loss of GRP7 function could affect the targeted 3′ processing and polyadenylation pattern of COOLAIR. Compared with wild-type Col, the grp7-1 mutant had elevated levels of total antisense transcripts (Fig. 7B). In addition, the relative abundance of proximal type ASI (Type I/Total) was decreased in grp7-1, whereas relative distal type ASII (Type II/Total) was increased (Fig. 7B), which also matches the previous report in fca-9 (Liu et al., 2007, 2010).
Figure 7.
GRP7 and AtJAC1 influence the RNA processing of COOLAIR and histone modification within the FLC gene body. A, Diagram of FLC gene structure and antisense transcripts. The black triangle indicates the GRP7-binding site. PCR-amplified fragments for chromatin immunoprecipitation (ChIP) in C and D are indicated as ChIP_a to 3UTR. B, Relative expression of total antisense mRNA (normalized to UBC) as well as relative ratio of proximal (type I) and distal (type II) antisense transcripts (normalized to total antisense transcripts) in atjac1, grp7-1, and fca-9 mutants and AtJAC1-OE plants compared with controls. In total, three biological repeats were performed. Statistical analysis was done by Student’s t test; atjac1 mutants, grp7-1, and fca-9 were compared with Col, whereas AtJAC1-OE lines were compared with C24 (vector): *, P < 0.05; **, P < 0.01; ns, no significant change. C and D, Histone modification change (H3K4me2 [C] and H3K27me3 [D]) at the FLC locus in atjac1, grp7-1 mutants, and AtJAC1-OE plants compared with controls. The y axis shows the relative abundance of ChIP sample compared with input. Data are means ± sd of three parallel samples from one replicate, and three biological repeats were performed.
Alternative polyadenylation of the COOLAIR transcripts has been shown to trigger changes in histone methylation status at the FLC locus (Liu et al., 2007). Therefore, we checked whether grp7 influenced H3K4 dimethylation at the FLC gene body. Mutant grp7-1 increased H3K4me2 in the body of the gene downstream of the proximal COOLAIR poly(A) site (Fig. 7C). In addition, the H3K27me3 level decreased in the gene body of FLC in the grp7-1 mutant (Fig. 7D). Moreover, relative changes of different types of FLC antisense transcripts and histone modification levels (H3K4me2 and H3K27me3) at FLC gene body regions were observed in AtJAC1-OE plants, with contrivance for atjac1 mutants (Fig. 7, B–D). It has been shown that the COOLAIR amount as well as the ratio of proximal type over distal type increased during the vernalization process and led to reductions of H3K36me3 and H3K4me3 at the FLC locus (Csorba et al., 2014; Yang et al., 2014), and we further checked these two histone modification marks. Increased H3K4me3 and H3K36me3 levels around the transcription start site of the FLC locus was observed in grp7-1 mutant and AtJAC1-OE lines, whereas reduction of those histone modifications was found in atjac1 mutants (Supplemental Fig. S11). Thus, AtJAC1 and GRP7 may be involved in FLC transcript level regulation by influencing the processing of COOLAIR and related histone modification dynamics.
Competition of GRP7 Binding Influences COOLAIR Process and Flowering
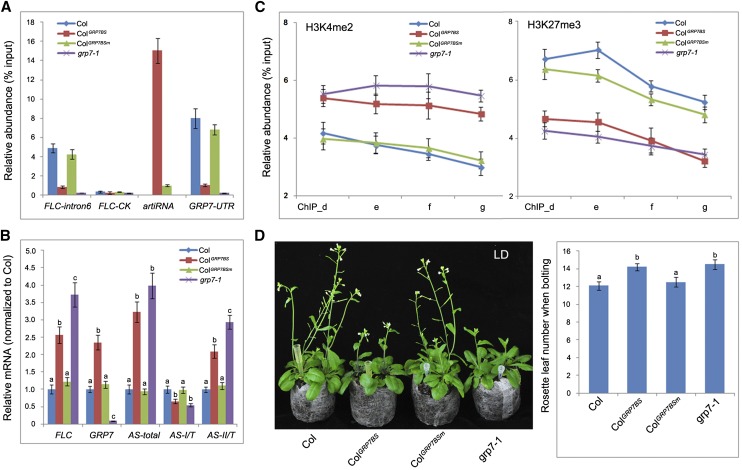
Is the RNA processing change of COOLAIR associated with direct binding of GRP7? To address this, a DNA fragment containing 12 repeats of the GRP7-binding site (GRP7BS) driven by the CaMV 35S promoter was transformed into Col, which could generate bait RNA fragments to compete the binding of endogenous GRP7 (Supplemental Fig. S12A; Supplemental Table S4). As a control, a DNA fragment of the same size with a mutation at the consensus site (U4U/G5U7G8G9) of the GRP7-binding motif (GRP7BSm) was transformed in parallel to Col (Supplemental Fig. S12B; Supplemental Table S4). RIP followed by RT-qPCR revealed that the endogenous GRP7 strongly bound to this artificial bait RNA fragment, whereas decreased binding was seen with the FLC antisense premRNA fragment (FLC-intron6) in ColGRP7BS transgenic plants compared with Col (Fig. 8A). Consistently, GRP7-UTR RNA fragments also showed a reduced enrichment in the coimmunoprecipitated RNA pool of anti-GRP7 (Fig. 8A). In contrast, GRP7 rarely bound to the GRP7BSm RNA fragments, and there was no difference between Col and ColGRP7BSm plants with respect to GRP7 binding to either FLC antisense premRNA or GRP7/8-UTR RNA fragments (Fig. 8A). The endogenous GRP7 mRNA level increased in ColGRP7BS, which is likely caused by the autonegative regulation machinery of GRP7 (Staiger et al., 2003; Fig. 8B). Consistent with the reduced binding of GRP7 to FLC antisense premRNA, the relative abundance of different poly(A) types of COOLAIR transcripts showed similar changes in ColGRP7BS transgenic plants to those in grp7-1, with increased total COOLAIR production and reduced proximal polyadenylation (type I) site usage (Fig. 8B). Meanwhile, the histone modification status of H3K4me2 and H3K27me3 at the FLC gene body also was altered in ColGRP7BS as in grp7-1 compared with Col (Fig. 8C). As expected, the total amount of FLC sense transcript level increased (Fig. 8B), with concomitant delayed flowering, in ColGRP7BS compared with Col (Fig. 8D). By contrast, in ColGRP7BSm plants, neither COOLAIR polyadenylation, histone modification, nor flowering time change was observed compared with Col (Fig. 8, B–D).
Figure 8.
Reducing GRP7 binding influences COOLAIR process and flowering. A, Competing the binding of GRP7 to its target RNA by artificial RNA fragments reduced the binding of GRP7 to FLC antisense premRNA. RIP was followed by qPCR to check the relative abundance of GRP7 binding at FLC-intron6, FLC-CK, GRP7-UTR, and artificial RNA fragments. Adaptor primers were used in RT. ColGRP7BS, Col with abundant expression of an RNA fragment containing the GRP7-binding site; ColGRP7BSm, Col with abundant expression of an RNA fragment containing a mutated GRP7-binding site. B, Relative expression level of sense FLC, GRP7, and different polyadenylated FLC antisense (AS) mRNAs in Col, artificial RNA transgenic plants, and the grp7-1 mutant. C, Histone modification change (H3K4me2 and H3K27me3) at FLC gene body regions (ChIP_d, e, f, and g as in Fig. 7A) in Col, artificial RNA transgenic plants, and the grp7-1 mutant. D, Flowering time (morphology, left; rosette leaf number when bolting, right) changes in Col, artificial RNA transgenic plants, and the grp7-1 mutant. Tukey’s HSD test was used for B and D, and different letters within the same genetic background indicate statistically significant differences.
Therefore, the binding of GRP7 to FLC antisense premRNA influences its RNA processing and, consequently, flowering time.
DISCUSSION
JACALIN-RELATED LECTIN Regulates Flowering in Both Dicot and Monocot Plants
JACALIN-RELATED LECTIN (JRL) belongs to one of the superfamily of plant lectins, with numerous members among different plant species. There are 48 JRL proteins in Arabidopsis (Nagano et al., 2008), 31 in rice (Jiang et al., 2010), and 67 in wheat (Song et al., 2013b). Beyond the classic lectins (Lannoo and Van Damme, 2010), as defined by characteristics such as high expression in storage organs, distribution in extracellular compartments, and recognition of exogenous carbohydrate or glycoprotein, increasing numbers of inducible JRLs were reported to be important in the regulation of plant development as well as biotic and abiotic stress responses (for review, see Lannoo and Van Damme, 2010). Our data here revealed that the Jacalin-lectin AtJAC1/JRL35 (Nagano et al., 2008) regulates the transition of flowering in Arabidopsis. Mutants of atjac1 were early flowering, whereas overexpression of AtJAC1 led to delayed flowering (Fig. 1). We reported previously that VER2, a Jacalin domain-containing lectin, is also involved in the regulation of the flowering transition in wheat (Yong et al., 2003; Xiao et al., 2014). In contrast to VER2, a promoter of flowering transition in wheat, AtJAC1 is a flowering repressor in Arabidopsis. VER2 is induced by cold temperature, and its function in the regulation of flowering is mainly dependent on the vernalization pathway, whereas AtJAC1 has high basal expression under normal growth conditions (Supplemental Fig. S1F) and regulates flowering through the autonomous pathway instead of vernalization (Fig. 1). VER2 is a positive regulator of TaVRN1, an AP1-like MADS box transcription factor that mediates the vernalization-accelerated flowering transition in wheat (Xiao et al., 2014). Here, we find that AtJAC1 participates in flowering control through the regulation of the transcript of FLC, an integrator of the vernalization and autonomous pathways (Fig. 2). Interestingly, FLC-like repressors were also shown in cereals to mediate vernalization-regulated flowering (Ruelens et al., 2013). To conclude, JRL could regulate the flowering transition in both monocot and dicot plants, although the exact players and regulation models in different species may vary.
Jacalin-Lectins Interact with RNA-Binding Proteins and Regulate Their Function
It is intriguing how JRLs participate in the regulation of gene expression. In wheat, the Jacalin-lectin domain-containing protein VER2 could bind to the O-GlcNAc-ylated RNA-binding protein TaGRP2 during the process of vernalization through the high-affinity association of VER2 to the GlcNAc side chain. Their interaction leads to the dissociation of TaGRP2 from TaVRN1 premRNA and releasing the inhibition of transcription (Xiao et al., 2014). It suggests an important input of O-GlcNAc signaling in shaping plant development meditated by lectin and target proteins (Liu et al., 2015). Unlike VER2, AtJAC1 shows highly specific recognition of Man, Rib, and Glc instead of N-acetyl-d-glucosamine (Supplemental Table S1; Supplemental Fig. S1E). Accordingly, it is unlikely that AtJAC1 could bind an O-GlcNAc-ylated protein as VER2. Although the specific recognition of carbohydrate is a fundamental character of lectins, other studies indicate that many plant lectins could also interact with small molecules and peptides independent of their carbohydrate-binding properties (for review, see Komath et al., 2006). Evidence from CoIP and BiFC assays indicated that AtJAC1 could also interact with GRP7 in Arabidopsis (Fig. 3). Whether this interaction is mediated by the carbohydrate-binding property of AtJAC1 needs to be analyzed further. Nevertheless, the interaction between JRL and RNA-binding proteins seems to be conserved in both monocots and dicots.
Like the inducible lectins (Lannoo and Van Damme, 2010), AtJAC1 is a nucleocytoplasmic protein (Supplemental Fig. S4A). As well, GRP7 could also localize in both nucleus and cytoplasm (Supplemental Fig. S4A). However, interaction between AtJAC1 and GRP7 occurs only in the cytoplasm, based on the BiFC and CoIP data presented here (Fig. 2). This is different from the case of VER2 and TaGRP2 in wheat, where interaction occurs in both nucleus and cytoplasm (Xiao et al., 2014). Of note, immunoblot data indicate that there are two bands of GRP7 in the nuclear fraction but only one in the cytoplasm (Figs. 3 and 4). The two bands might be related to posttranslational modification or alternative splicing and possibly contribute to the specific subcellular interaction with AtJAC1. GRP7 was shown to shuttle between nucleus and cytoplasm (Lummer et al., 2011), which may be linked to its multiple functions in RNA processing. Immunoblot data and transient fluorescence reporter assays suggest that AtJAC1 is involved in the regulation of GRP7’s distribution between nucleus and cytoplasm. With high expression of AtJAC1, GRP7 was detained in the cytoplasm, whereas reduced expression of AtJAC1 caused an accumulation of GRP7 in the nucleus (Fig. 4). In addition, a DEX-inducible assay suggests that the nuclear accumulation of GRP7 rescued the developmental defects and altered gene expression caused by the overexpression of AtJAC1 (Fig. 5). Thus, AtJAC1 could participate in the developmental program through regulating the nucleocytoplasmic distribution of GRP7. Furthermore, the subcellular distribution of GRP7 provides a novel regulation of GRP7’s function besides its well-known transcriptional autonegative feedback machinery and posttranslational modifications such as ADP-ribosylation and methylation on Arg (Staiger et al., 2003; Fu et al., 2007; Deng et al., 2010).
GRP7 Binds Directly to COOLAIR premRNA and Affects Its Processing
Although GRP7 was reported to regulate the FLC transcript level and influence flowering time (Streitner et al., 2008), the direct link between GRP7 and FLC is still unclear. Here, we identified a GRP7-binding RNA motif as U/G1N2N3U4U/G5N6U7G8G9N10U/G11N12N13U/G14U/G15, with the core consensus motif of UUNUGG. Sequence alignment showed two GRP7-binding motifs present on opposite strands of the last intron of the FLC gene, where important regulatory factors are associated (Liu et al., 2007, 2010). GRP7 could bind both RNA probes in vitro; however, only the antisense premRNA of FLC could be coimmunoprecipitated with GRP7 in vivo (Fig. 6). The data presented here provide evidence of a direct link between GRP7 and FLC transcript regulation. They also suggests that, besides the sequence conservation, other regulatory events like RNA secondary structure may also be required in facilitating the association of RNA-binding proteins to their target (Brown et al., 2009).
At the FLC locus, a group of long noncoding antisense transcripts named COOLAIR generates from the 3′ UTR (Swiezewski et al., 2009). It is induced by vernalization but also exists in warm conditions (Swiezewski et al., 2009). Although its function in the vernalization-triggered silencing of FLC expression is still under debate (Helliwell et al., 2011; Csorba et al., 2014), numerous data indicate that COOLAIR has an important role in mediating FLC expression in nonvernalized plants (Liu et al., 2010; Marquardt et al., 2014; Wang et al., 2014). COOLAIR consists of two major types with alternative polyadenylation and alternative splicing (Swiezewski et al., 2009; Hornyik et al., 2010). The relative ratio of these two types (type I with proximal polyadenylation in the last intron of FLC and type II with distal polyadenylation in the promoter of FLC) regulates the amount of FLC sense transcript in a way linked to dynamic histone modification at the FLC gene body region (Liu et al., 2007; Csorba et al., 2014). The processing of COOLAIR is regulated by RNA-binding proteins like FCA, FPA, and RNA 3′ processing factors like FY, CstF77, and CstF64 (Hornyik et al., 2010; Liu et al., 2010). In addition, the promoter of COOLAIR is regulated by the presence of an R loop, which could reduce COOLAIR expression upon stabilization by homeodomain protein (Crevillén et al., 2013; Sun et al., 2013). Here, we find that GRP7 could directly bind COOLAIR premRNA just downstream of the polyadenylation site of proximal class COOLAIRs (Fig. 6). Furthermore, the total amount of COOLAIR, as well as the relative abundance of the proximal class COOLAIR, were altered in the grp7-1 mutant (Fig. 7). The effect of GRP7 on COOLAIR processing is likely to rely on the direct binding. To support this view, we find that when overexpressing an RNA fragment containing a multimer of GRP7-binding motifs in wild-type plants, it could phenotypically mimic the grp7-1 mutant at both the morphological level (flowering time) and the molecular level (total COOLAIR and FLC transcript levels as well as the relative abundance of proximal COOLAIR transcript). In contrast, overexpression of the RNA fragment containing the mutated GRP7-binding motifs behaves like the wild type (Fig. 8).
Our data suggested that GRP7 may be involved in the regulation of COOLAIR processing, such as facilitating the poly(A) site chosen and/or the generation of COOLAIR. RNA-binding proteins frequently interact with each other in different RNA processing steps. Interestingly, there is no physical interaction between GRP7 and multiple known factors in the regulation of poly(A) site selection (Supplemental Fig. S13). GRP7 was suggested to have a function in premRNA splicing, particularly in affecting the splicing site choice (Streitner et al., 2012). Alternative splicing of both proximal and distal COOLAIR can quantitatively modulate FLC sense transcript expression through a feedback mechanism linking COOLAIR processing to FLC gene body histone demethylation (Marquardt et al., 2014). It is possible that GRP7 could participate in the alternative splicing of COOLAIR through direct binding. In addition, we find that GRP7 could interact with the nuclear exosome component RRP6 like protein1 protein in a yeast two-hybridization screening study (Supplemental Fig. S14). The latter protein has been reported to influence FLC antisense transcript generation (both COOLAIR and a novel antisense transcript, ASL; Shin and Chekanova, 2014).
Jacalin-Lectin AtJAC1 May Have Broad Function through the Regulation of GRP7-Manipulated RNA Processing
Of interest, the total COOLAIR expression level and relative proximal COOLAIR abundance also changed in atjac1 mutants and AtJAC1-OE lines, indicating that AtJAC1 could take part in RNA processing regulation through the manipulation of GRP7’s nuclear accumulation. Besides the dynamic expression profile during different developmental stages, AtJAC1 also shows elevated or reduced expression at the mRNA or protein level upon hormone treatments (JA [León et al., 1998], ethylene [Tamaoki et al., 2008], and Brassinosteroid [Tang et al., 2008]) and environmental cues (light [Piippo et al., 2006], cold [Bae et al., 2003], wounding [León et al., 1998], and pathogens [Thilmony et al., 2006]). This suggests that AtJAC1 may have broad function in plant development and stress responses. Coincidentally, GRP7 also responds to multiple endogenous and environmental cues, such as high salinity, cold, or osmotic stress (Kim et al., 2008), pathogens (Fu et al., 2007), and hormones (Cao et al., 2006; Hackmann et al., 2014). The working model we present here for AtJAC1 and GRP7 in the regulation of flowering could also be adapted to other developmental aspects and stress-response scenarios. It will be interesting to study the transcriptome profile change upon misexpression of AtJAC1 combined with the GRP7 genome-wide binding target RNA data.
MATERIALS AND METHODS
Plant Materials and Treatment
Arabidopsis (Arabidopsis thaliana) ecotypes Col and C24 were used as the genetic backgrounds of mutants and transgenic lines, respectively. Mutants atjac1-1 (SALK_000461), atjac1-2 (SALK_112556), At3g16460 (SALK_028332), grp7-1 (SALK_039556), and grp8 (SALK_007963) were ordered from the SALK T-DNA collection (Alonso et al., 2003); flc-20 was described previously (Helliwell et al., 2002). Plants were grown at 22°C under LD (16 h of light/8 h of dark) or SD (8 h of light/16 h of dark) conditions. Vernalization and GA treatments were as described (Bastow et al., 2004; Lim et al., 2004). Flowering time was measured as the number of rosette leaves when bolting. For DEX treatment, seedlings of grp7-1 and Col transformed with 35S:GRP7-GR-HBD were sprayed with a solution containing 10 μm DEX (Sigma), 0.01% (v/v) ethanol, and 0.015% (v/v) Silwet L-77 at the shoot apex region every 3 to 4 d until bolting.
Transgenic Plants
The pBI121 plasmids carrying 35S:AtJAC1-GFP and 35S:GFP were transformed into Agrobacterium tumefaciens strain GV3101 separately, and the resulting bacteria were used to transform Arabidopsis ecotype C24. The AtJAC1-OE plants were selected on one-half-strength Murashige and Skoog (MS) medium containing 40 µg mL−1 kanamycin. The pSN1301-GRP7 plasmid was transformed in the Col ecotype to generate GRP7-OE plants, with selection by 25 mg mL−1 hygromycin. The 35S:GRP7-GR construct was made by creating a translational fusion between the GRP7 cDNA and the ligand-binding domain of the rat GR under the promoter of CaMV 35S. It was transformed in AtJAC1-OE lines to test the DEX-inducible nuclear accumulation of GRP7’s effects on flowering. The transformants were selected on one-half-strength MS medium with 30 µg mL−1 Basta. DNA fragments containing the GRP7-binding motif or mutated versions of GRP7-binding motifs were synthesized, cloned into the binary vector pSN1301 plasmid, and transformed in Col in A. tumefaciens strain GV3101. The transformants were selected on one-half-strength MS medium with 20 µg mL−1 hygromycin.
Plasmids and Antibodies
All plasmids and antibodies used are described in Supplemental Text S1.
Real-Time PCR
Total RNA samples were treated with Turbo DNA-free (Ambion) to remove any contaminating genomic DNA. The first-strand cDNA was prepared using the SuperScript III First Strand Synthesis System (Invitrogen) using oligo(dT) or gene-specific primers. qPCR was performed on the Mx3000p (Agilent) using SYBR Green reagent (Toyobo). Every PCR was performed using at least three biological replicates with three technical repeats. qPCR data were first normalized to UBC as reported (Liu et al., 2010) and then normalized to either Col (for atjac1-1, atjac1-2, and grp7-1) or C24 transformed with empty vector (for AtJAC1-OE-L2/L7). For measuring different polyadenylated COOLAIRs, oligo(dT) primer was used for RT, and then specific primers for different types of COOLAIR were used for qPCR analysis as reported previously (Liu et al., 2010). qPCR was performed in triplicate for each sample. Average values were normalized to the expression of total COOLAIR. The primers used are listed in the supplemental sheet.
Subcellular Fractionation
Nuclear and cytoplasmic protein fractions were isolated as described (Xiao et al., 2014). Briefly, plant material was ground in extraction buffer and filtered through Miracloth (Calbiochem) to obtain the total extract and then centrifuged at 3,000g. The supernatant (cytoplasmic fraction) was removed and stored on ice until use. The pellets were further washed with a resuspension buffer and then reconstituted as the nuclear fraction. Immune detection of histone H3 and tubulin was used to confirm the fractionation of nucleus and cytoplasm, respectively.
Protein Interaction Analysis
CoIP assays were performed as described (Xiao et al., 2014). Briefly, FLAG-AtJAC1 was coexpressed with GRP7-GFP or GFP in tobacco (Nicotiana tabacum) leaves, and after 2 to 3 d, plant material was harvested and ground in native protein extraction buffer, filtered, and centrifuged. The supernatant was incubated with anti-GFP antibody or anti-AtJAC1 antibody coupled to protein A/G Sepharose beads (Millipore), the beads were washed four times with wash buffer, and bound proteins were eluted with elution buffer containing 2% (w/v) SDS for immunoblotting with FLAG or anti-GRP7 antibody. BiFC assays were as described (Xiao et al., 2014). Briefly, constructs for the expression of AtJAC1-YFPNE, GRP7-YFPCE, AtJAC1-YFPCE, and GRP7-YFPNE were generated and cotransformed into Arabidopsis protoplasts. YFP fluorescence was visualized with a confocal scanning microscope 40 to 48 h after infiltration.
Protoplast Transient Expression
Transient expression of 35S:GRP7-GFP, 35S:AtJAC1-GFP, 35S:GFP, 35S:VP16-GR, and 8OP:AtJAC1 in protoplasts from Col or atjac1-1 was as described (Yoo et al., 2007). Briefly, 4 × 104 mesophyll protoplasts were isolated from 4-week-old seedlings and transformed with 10 to 15 µg of purified plasmid DNA. Transformed protoplasts were incubated under weak light at 22°C. For DEX treatment, 10 µm DEX was added to the protoplasts 8 h after transfection and incubated for 2 to 4 h. GRP7-GFP, AtJAC1-GFP, or GFP signal was observed by fluorescence microscopy (Fig. 4D) or confocal microscopy (Fig. 4C; Supplemental Fig. S4A; Leica TCS SP5).
Confocal Microscopy Observation and Quantification of Fluorescent Protein Signal
GFP/YFP fluorescence was visualized using a confocal microscope (Leica TCS SP5) with an argon laser (488 nm); the autofluorescence of chloroplasts also appeared at this excitation wavelength. Hoechst fluorescence was excited by a laser at 405 nm. Images of fluorescence at different layers along the z-axis were collected and overlaid to give a full view of the GFP/YFP signal. Hoechst fluorescence and bright field were captured at their proper layers of the z-axis to show the positions of the nucleus and whole protoplast cells, respectively. Quantification of the fluorescent protein signal was performed using ImageJ (http://rsb.info.nih.gov/ij). To calculate the ratio of the nucleus to total signals of GRP7-GFP for each cell, nucleus positional area (indicated by Hoechst inflorescence) and GFP signal intensity were measured and divided by the total GFP signal of the whole cell. A representative image of each transformant is shown in Figure 4C. The numbers indicate the relative nucleus-distributed ratio of GRP7-GFP or GFP. This experiment was carried out for three biological replicates, with each sample consisting of at least 30 cells analyzed, and the signal intensity is presented as means ± sd.
RNA-EMSA
RNA probes were generated in vitro by the use of a T7 high-yield transcription kit (Thermo). Biotin was added to the 3′ end of RNA probes using the RNA 3′ End Biotinylation kit (Pierce). RNA-EMSA was performed according to the kit instructions (Pierce). Purified RNase-free GST-GRP7 was used. All probe sequences, including those for mutations, are listed in the supplemental sheet. Typically, 20 to 50 pm protein and 0.1 pm probe were used in this study.
SELEX
SELEX was performed as described previously (Xiao et al., 2014). The RNA library was reduced to 20 random nucleotides. The oligonucleotide template and primers are listed in the supplemental sheet. The in vitro-transcribed RNA library was incubated with GST-GRP7 fusion proteins bound to glutathione Sepharose 4B resin to select for the binding of RNA fragments. Then, the selected RNA fragments were reverse transcribed to DNA using adaptor primers. PCR amplification with specific primers was used to determinate the proper number of cycles to distinguish sample and negative controls. After that, PCR products were purified and used as a template to generate the next-round RNA library by in vitro transcription. The selection was repeated six times, and the final products were gel purified, ligated into pGEM-T Easy vector (Promega), and sequenced.
ChIP Assay
ChIP experiments were performed as described (Bowler et al., 2004; Xiao et al., 2013) using 2 g of young seedling tissue for each sample. Antibodies with specificity to H3K4me2, H3K4me3, H3K27me3 (Millipore), or H3K36me3 (Signalway Antibody) were used to immunoprecipitate the chromatin. The amount of immunoprecipitated FLC chromatin was determined by real-time PCR on different regions of the FLC locus as shown in Figure 7A. Primer sequences are given in the supplemental sheet. The relative abundance as a percentage of input is shown for each position checked in different genetic backgrounds. Three biological replicates were performed. One representative result is shown in Figure 7.
RIP and RT-PCR
RIP was performed as described (Xiao et al., 2014). Briefly, 3 g of Arabidopsis seedling tissue (20 d after germination growth in LD conditions) was harvested and cross-linked by 0.5% (v/v) formaldehyde. Plant material was ground and suspended in 15 mL of Honda buffer (0.44 m Suc, 1.25% [w/v] Ficoll, 2.5% [w/v] Dextran T40, 20 mm HEPES-KOH, pH 7.4, 10 mm MgCl2, 0.5% [v/v] Triton X-100, 5 mm dithiothreitol, 1 mm phenylmethylsulfonyl fluoride [PMSF], and 1× plant protease inhibitors) supplemented with 8 units mL−1 RNase inhibitor and filtered through two layers of Miracloth (Calbiochem). Centrifugation was at 3,000g for 7.5 min at 4°C. The pellet was resuspended in 500 µL of nucleus lysis buffer (50 mm Tris-HCl, pH 8, 10 mm EDTA, 1% [w/v] SDS, 1 mm PMSF, and 1× plant protease inhibitors) + 160 units mL−1 RNase inhibitor. Samples were sonicated and centrifuged, and the supernatant was transferred to the fresh tubes. DNA concentration was measured in the samples to ensure that nuclear protein concentrations were similar in all samples. Aliquots of 100 µL were prepared. The samples were then diluted 10-fold with ChIP dilution buffer (1.1% [v/v] Triton X-100, 1.2 mm EDTA, 16.7 mm Tris-HCl, pH 8, and 167 mm NaCl) + 350 units mL−1 RNase inhibitor. IP was done by adding 2 µL of GRP7 antibody (purified mouse polyclonal antibody) and 20 µL of protein A agarose/salmon sperm DNA (Upstate; 16-157) prewashed three times by binding/washing buffer (150 mm NaCl, 20 mm Tris-HCl, pH 8, 2 mm EDTA, 1% [v/v] Triton X-100, 0.1% [w/v] SDS, and 1 mm PMSF) to the solution. Preimmune mouse serum was used as a negative antibody control, grp7-1 sample served as the antigen negative control, and grp7-1 transformed with pGRP7:GRP7(R49Q) was also used as a negative control. After incubating on a rotator for 3 h at 4°C, beads were washed four times with 1 mL of binding/washing buffer + 40 units mL−1 RNase inhibitor, and the protein-RNA complex was eluted with 50 µL of RIP elution buffer (100 mm Tris-HCl, pH 8, 10 mm EDTA, and 1% [w/v] SDS) + 40 units mL−1 RNase inhibitor. Then, the protein was degraded by proteinase K, and RNA was extracted by acidic phenol/chloroform and precipitated by ethanol supplemented with 3 m NaAc, pH 5.2, and 5 mg mL−1 glycogen (Ambion). The pellet was washed with 70% (v/v) ethanol, air dried, and dissolved in RNase-free water. The RNA sample was incubated with DNase I to digest the DNA contaminant.
For identification and quantification by RT-PCR, RNA samples were polyadenylated at 37°C for 30 min with 5 units of poly(A) polymerase (Takara). Poly(A)-tailed RNA was recovered by phenol/chloroform extraction and ethanol precipitation. A 5′ adapter (GeneRacer RACE ready cDNA kit; Invitrogen) was ligated to poly(A)-tailed RNA using T4 RNA ligase (Invitrogen), and the ligation products were recovered by phenol/chloroform extraction followed by ethanol precipitation. RT was performed using GeneRacer Oligo(dT) primer (SuperScript III) with 200 units of SuperScript III reverse transcriptase (Invitrogen). The first-round PCR was carried out for 22 cycles with GeneRacer 5′/3′ primers. The PCR products were separated on a 1% (w/v) agarose gel, and DNA smear from 200 to 800 bp was recovered and diluted 20-fold to be used as template for the second-round PCR with GeneRacer nested 5′/3′ primers. The PCR products were separated on a 1% (w/v) agarose gel, and DNA smear from 150 to 800 bp was recovered and diluted 50-fold to be used as template for the detection of gene fragments with gene-specific primers. For qualification and quantification, the genomic DNA served as a positive control for PCR, and input samples (without IP) were used for equal loading.
Sequence data for genes in this article can be found in the GenBank/EMBL databases under accession numbers AtJAC1 (At3g16470), AtJAC2 (At3g16460), GRP7 (At2g21660), FLC (At5g10140), SOC1 (At2g45660), FT (At1g65480), CO (At5g15840), MAF1 (At1g77080), MAF2 (At5g65050), MAF3 (At5g65060), MAF4 (At5g65070), MAF5 (At5g65080), SPL3 (At2g33810), SPL4 (At1g53160), and SPL5 (At3g15270).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Expression pattern of AtJAC1 and its biochemical characteristics.
Supplemental Figure S2. Identification of atjac1 mutants and AtJAC1-OE plants and functional analysis of AtJAC2 in flowering.
Supplemental Figure S3. AtJAC1 does not influence transcript level of photoperiod and age pathway factors.
Supplemental Figure S4. Subcellular localization of AtJAC1 and GRP7.
Supplemental Figure S5. Identification of atjac1-1grp7 double mutant.
Supplemental Figure S6. Analysis of AtJAC1 mRNA and protein level in grp7-1 and GRP7-OE plants.
Supplemental Figure S7. DEX-induced nuclear distribution of GRP7.
Supplemental Figure S8. The sequence preference of GRP7 binding RNA motif.
Supplemental Figure S9. Workflow of RIP.
Supplemental Figure S10. Direct binding of GRP7 to FLC antisense premRNA.
Supplemental Figure S11. GRP7 and AtJAC1 influence histone modification around transcription start site of FLC locus.
Supplemental Figure S12. Diagram of generating artificial RNA fragments to compete for GRP7 binding in planta.
Supplemental Figure S13. Interaction between GRP7 and other factors involved in COOLAIR polyadenylation regulation.
Supplemental Figure S14. Interaction between GRP7 and RRP6L1.
Supplemental Table S1. Comparison of the carbohydrate-binding specificities of AtJAC1.
Supplemental Table S2. Sequences of GRP7 binding RNA fragments screened by SELEX.
Supplemental Table S3. Prediction of GRP7 binding motif in FLC gene.
Supplemental Table S4. GRP7 binding motif fragments used in transforming.
Supplemental Text S1. Plasmids, antibodies, and primers used in this study.
Supplementary Material
Acknowledgments
We thank Dr. Elizabeth Dennis (Commonwealth Scientific and Industrial Research Organization) for the flc-20 seeds; the SALK Center for the atjac1-1, atjac1-2, SALK_028332, SALK_007963, and grp7-1 seeds; Dr. Jianru Zuo (Institute of Genetics and Development, Chinese Academy of Sciences) for providing the DEX-inducible system; and Dr. Ildoo Hwang (Pohang University of Science and Technology) and Dr. Sang-Dong Yoo (Sungkyunkwan University) for the DEX-inducible transient expression system.
Glossary
- T-DNA
transfer DNA
- Col
Columbia
- LD
long-day
- SD
short-day
- CoIP
coimmunoprecipitation
- BiFC
bimolecular fluorescence complementation
- DEX
dexamethasone
- CaMV
Cauliflower mosaic virus
- SELEX
systematic evolution of ligands by exponential enrichment
- EMSA
electrophoresis mobility shift assay
- RIP
RNA immunoprecipitation
- UTR
untranslated region
- RT
reverse transcription
- qPCR
quantitative PCR
- IP
immunoprecipitation
- cDNA
complementary DNA
- MS
Murashige and Skoog
- ChIP
chromatin immunoprecipitation
- PMSF
phenylmethylsulfonyl fluoride
- HSD
honestly significant difference
- mRNA
messenger RNA
- premRNA
precursor messenger RNA
Footnotes
This work was supported by the major state basic research program of China (973 Program; grant no. 2011CB915404) and the China-Netherlands Joint Scientific Thematic Research Program (grant no. GJHZ1309).
Articles can be viewed without a subscription.
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Amasino R. (2010) Seasonal and developmental timing of flowering. Plant J 61: 1001–1013 [DOI] [PubMed] [Google Scholar]
- Aucouturier P, Pineau N, Brugier JC, Mihaesco E, Duarte F, Skvaril F, Preud’homme JL (1989) Jacalin: a new laboratory tool in immunochemistry and cellular immunology. J Clin Lab Anal 3: 244–251 [DOI] [PubMed] [Google Scholar]
- Babosha AV. (2008) Inducible lectins and plant resistance to pathogens and abiotic stress. Biochemistry (Mosc) 73: 812–825 [DOI] [PubMed] [Google Scholar]
- Bae MS, Cho EJ, Choi EY, Park OK (2003) Analysis of the Arabidopsis nuclear proteome and its response to cold stress. Plant J 36: 652–663 [DOI] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastow R, Mylne JS, Lister C, Lippman Z, Martienssen RA, Dean C (2004) Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427: 164–167 [DOI] [PubMed] [Google Scholar]
- Bäurle I, Dean C (2006) The timing of developmental transitions in plants. Cell 125: 655–664 [DOI] [PubMed] [Google Scholar]
- Bowler C, Benvenuto G, Laflamme P, Molino D, Probst AV, Tariq M, Paszkowski J (2004) Chromatin techniques for plant cells. Plant J 39: 776–789 [DOI] [PubMed] [Google Scholar]
- Brown JW, Birmingham A, Griffiths PE, Jossinet F, Kachouri-Lafond R, Knight R, Lang BF, Leontis N, Steger G, Stombaugh J, et al. (2009) The RNA structure alignment ontology. RNA 15: 1623–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S, Jiang L, Song S, Jing R, Xu G (2006) AtGRP7 is involved in the regulation of abscisic acid and stress responses in Arabidopsis. Cell Mol Biol Lett 11: 526–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong K, Bao SL, Xu T, Tan KH, Liang TB, Zeng JZ, Huang HL, Xu J, Xu ZH (1998) Functional analysis of the ver gene using antisense transgenic wheat. Physiol Plant 102: 87–92 [DOI] [PubMed] [Google Scholar]
- Crevillén P, Dean C (2011) Regulation of the floral repressor gene FLC: the complexity of transcription in a chromatin context. Curr Opin Plant Biol 14: 38–44 [DOI] [PubMed] [Google Scholar]
- Crevillén P, Sonmez C, Wu Z, Dean C (2013) A gene loop containing the floral repressor FLC is disrupted in the early phase of vernalization. EMBO J 32: 140–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csorba T, Questa JI, Sun Q, Dean C (2014) Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc Natl Acad Sci USA 111: 16160–16165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X, Gu L, Liu C, Lu T, Lu F, Lu Z, Cui P, Pei Y, Wang B, Hu S, et al. (2010) Arginine methylation mediated by the Arabidopsis homolog of PRMT5 is essential for proper pre-mRNA splicing. Proc Natl Acad Sci USA 107: 19114–19119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornara F, de Montaigu A, Coupland G (2010) SnapShot: control of flowering in Arabidopsis. Cell 141: 550–550.e2 [DOI] [PubMed] [Google Scholar]
- Fu ZQ, Guo M, Jeong BR, Tian F, Elthon TE, Cerny RL, Staiger D, Alfano JR (2007) A type III effector ADP-ribosylates RNA-binding proteins and quells plant immunity. Nature 447: 284–288 [DOI] [PubMed] [Google Scholar]
- Hackmann C, Korneli C, Kutyniok M, Köster T, Wiedenlübbert M, Müller C, Staiger D (2014) Salicylic acid-dependent and -independent impact of an RNA-binding protein on plant immunity. Plant Cell Environ 37: 696–706 [DOI] [PubMed] [Google Scholar]
- He Y. (2012) Chromatin regulation of flowering. Trends Plant Sci 17: 556–562 [DOI] [PubMed] [Google Scholar]
- He Y, Amasino RM (2005) Role of chromatin modification in flowering-time control. Trends Plant Sci 10: 30–35 [DOI] [PubMed] [Google Scholar]
- Helliwell CA, Robertson M, Finnegan EJ, Buzas DM, Dennis ES (2011) Vernalization-repression of Arabidopsis FLC requires promoter sequences but not antisense transcripts. PLoS One 6: e21513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Wesley SV, Wielopolska AJ, Waterhouse PM (2002) High-throughput vectors for efficient gene silencing in plants. Funct Plant Biol 29: 1217–1225 [DOI] [PubMed] [Google Scholar]
- Hornyik C, Terzi LC, Simpson GG (2010) The spen family protein FPA controls alternative cleavage and polyadenylation of RNA. Dev Cell 18: 203–213 [DOI] [PubMed] [Google Scholar]
- Jiang JF, Han Y, Xing LJ, Xu YY, Xu ZH, Chong K (2006) Cloning and expression of a novel cDNA encoding a mannose-specific jacalin-related lectin from Oryza sativa. Toxicon 47: 133–139 [DOI] [PubMed] [Google Scholar]
- Jiang SY, Ma Z, Ramachandran S (2010) Evolutionary history and stress regulation of the lectin superfamily in higher plants. BMC Evol Biol 10: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabir S. (1998) Jacalin: a jackfruit (Artocarpus heterophyllus) seed-derived lectin of versatile applications in immunobiological research. J Immunol Methods 212: 193–211 [DOI] [PubMed] [Google Scholar]
- Kim JS, Jung HJ, Lee HJ, Kim KA, Goh CH, Woo Y, Oh SH, Han YS, Kang H (2008) Glycine-rich RNA-binding protein 7 affects abiotic stress responses by regulating stomata opening and closing in Arabidopsis thaliana. Plant J 55: 455–466 [DOI] [PubMed] [Google Scholar]
- Kim JS, Park SJ, Kwak KJ, Kim YO, Kim JY, Song J, Jang B, Jung CH, Kang H (2007) Cold shock domain proteins and glycine-rich RNA-binding proteins from Arabidopsis thaliana can promote the cold adaptation process in Escherichia coli. Nucleic Acids Res 35: 506–516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komath SS, Kavitha M, Swamy MJ (2006) Beyond carbohydrate binding: new directions in plant lectin research. Org Biomol Chem 4: 973–988 [DOI] [PubMed] [Google Scholar]
- Köster T, Meyer K, Weinholdt C, Smith LM, Lummer M, Speth C, Grosse I, Weigel D, Staiger D (2014) Regulation of pri-miRNA processing by the hnRNP-like protein AtGRP7 in Arabidopsis. Nucleic Acids Res 42: 9925–9936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam SK, Ng TB (2011) Lectins: production and practical applications. Appl Microbiol Biotechnol 89: 45–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lannoo N, Van Damme EJ (2010) Nucleocytoplasmic plant lectins. Biochim Biophys Acta 1800: 190–201 [DOI] [PubMed] [Google Scholar]
- León J, Rojo E, Titarenko E, Sánchez-Serrano JJ (1998) Jasmonic acid-dependent and -independent wound signal transduction pathways are differentially regulated by Ca2+/calmodulin in Arabidopsis thaliana. Mol Gen Genet 258: 412–419 [DOI] [PubMed] [Google Scholar]
- Lim MH, Kim J, Kim YS, Chung KS, Seo YH, Lee I, Kim J, Hong CB, Kim HJ, Park CM (2004) A new Arabidopsis gene, FLK, encodes an RNA binding protein with K homology motifs and regulates flowering time via FLOWERING LOCUS C. Plant Cell 16: 731–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Marquardt S, Lister C, Swiezewski S, Dean C (2010) Targeted 3′ processing of antisense transcripts triggers Arabidopsis FLC chromatin silencing. Science 327: 94–97 [DOI] [PubMed] [Google Scholar]
- Liu F, Quesada V, Crevillén P, Bäurle I, Swiezewski S, Dean C (2007) The Arabidopsis RNA-binding protein FCA requires a lysine-specific demethylase 1 homolog to downregulate FLC. Mol Cell 28: 398–407 [DOI] [PubMed] [Google Scholar]
- Liu Y, Dai SJ, Xing LJ, Xu YY, Chong K (2015) O-linked β-N-acetylglucosamine modification and its biological functions. Sci Bull 60: 1055–1061 [Google Scholar]
- Lummer M, Humpert F, Steuwe C, Caesar K, Schüttpelz M, Sauer M, Staiger D (2011) Reversible photoswitchable DRONPA-s monitors nucleocytoplasmic transport of an RNA-binding protein in transgenic plants. Traffic 12: 693–702 [DOI] [PubMed] [Google Scholar]
- Mangeon A, Junqueira RM, Sachetto-Martins G (2010) Functional diversity of the plant glycine-rich proteins superfamily. Plant Signal Behav 5: 99–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt S, Raitskin O, Wu Z, Liu F, Sun Q, Dean C (2014) Functional consequences of splicing of the antisense transcript COOLAIR on FLC transcription. Mol Cell 54: 156–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels SD, Amasino RM (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11: 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreira RdeA, Ainouz IL, De Oliveira JT, Cavada BS (1991) Plant lectins, chemical and biological aspects. Mem Inst Oswaldo Cruz (Suppl 2) 86: 211–218 [DOI] [PubMed] [Google Scholar]
- Nagano AJ, Fukao Y, Fujiwara M, Nishimura M, Hara-Nishimura I (2008) Antagonistic jacalin-related lectins regulate the size of ER body-type β-glucosidase complexes in Arabidopsis thaliana. Plant Cell Physiol 49: 969–980 [DOI] [PubMed] [Google Scholar]
- Peumans WJ, Van Damme EJ (1995) Lectins as plant defense proteins. Plant Physiol 109: 347–352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peumans WJ, Van Damme EJ, Barre A, Rougé P (2001) Classification of plant lectins in families of structurally and evolutionary related proteins. Adv Exp Med Biol 491: 27–54 [DOI] [PubMed] [Google Scholar]
- Piippo M, Allahverdiyeva Y, Paakkarinen V, Suoranta UM, Battchikova N, Aro EM (2006) Chloroplast-mediated regulation of nuclear genes in Arabidopsis thaliana in the absence of light stress. Physiol Genomics 25: 142–152 [DOI] [PubMed] [Google Scholar]
- Putterill J, Robson F, Lee K, Simon R, Coupland G (1995) The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847–857 [DOI] [PubMed] [Google Scholar]
- Rabinovich GA, Toscano MA (2009) Turning ‘sweet’ on immunity: galectin-glycan interactions in immune tolerance and inflammation. Nat Rev Immunol 9: 338–352 [DOI] [PubMed] [Google Scholar]
- Romera-Branchat M, Andrés F, Coupland G (2014) Flowering responses to seasonal cues: what’s new? Curr Opin Plant Biol 21: 120–127 [DOI] [PubMed] [Google Scholar]
- Ruelens P, de Maagd RA, Proost S, Theißen G, Geuten K, Kaufmann K (2013) FLOWERING LOCUS C in monocots and the tandem origin of angiosperm-specific MADS-box genes. Nat Commun 4: 2280. [DOI] [PubMed] [Google Scholar]
- Ryu H, Cho H, Kim K, Hwang I (2010) Phosphorylation dependent nucleocytoplasmic shuttling of BES1 is a key regulatory event in brassinosteroid signaling. Mol Cells 29: 283–290 [DOI] [PubMed] [Google Scholar]
- Schena M, Lloyd AM, Davis RW (1991) A steroid-inducible gene expression system for plant cells. Proc Natl Acad Sci USA 88: 10421–10425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schöning JC, Streitner C, Page DR, Hennig S, Uchida K, Wolf E, Furuya M, Staiger D (2007) Auto-regulation of the circadian slave oscillator component AtGRP7 and regulation of its targets is impaired by a single RNA recognition motif point mutation. Plant J 52: 1119–1130 [DOI] [PubMed] [Google Scholar]
- Sharon N. (2008) Lectins: past, present and future. Biochem Soc Trans 36: 1457–1460 [DOI] [PubMed] [Google Scholar]
- Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES (1999) The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11: 445–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin JH, Chekanova JA (2014) Arabidopsis RRP6L1 and RRP6L2 function in FLOWERING LOCUS C silencing via regulation of antisense RNA synthesis. PLoS Genet 10: e1004612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song J, Irwin J, Dean C (2013a) Remembering the prolonged cold of winter. Curr Biol 23: R807–R811 [DOI] [PubMed] [Google Scholar]
- Song M, Xu W, Xiang Y, Jia H, Zhang L, Ma Z (2013b) Association of jacalin-related lectins with wheat responses to stresses revealed by transcriptional profiling. Plant Mol Biol 84: 95–110 [DOI] [PubMed] [Google Scholar]
- Srikanth A, Schmid M (2011) Regulation of flowering time: all roads lead to Rome. Cell Mol Life Sci 68: 2013–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staiger D, Zecca L, Wieczorek Kirk DA, Apel K, Eckstein L (2003) The circadian clock regulated RNA-binding protein AtGRP7 autoregulates its expression by influencing alternative splicing of its own pre-mRNA. Plant J 33: 361–371 [DOI] [PubMed] [Google Scholar]
- Streitner C, Danisman S, Wehrle F, Schöning JC, Alfano JR, Staiger D (2008) The small glycine-rich RNA binding protein AtGRP7 promotes floral transition in Arabidopsis thaliana. Plant J 56: 239–250 [DOI] [PubMed] [Google Scholar]
- Streitner C, Köster T, Simpson CG, Shaw P, Danisman S, Brown JWS, Staiger D (2012) An hnRNP-like RNA-binding protein affects alternative splicing by in vivo interaction with transcripts in Arabidopsis thaliana. Nucleic Acids Res 40: 11240–11255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Csorba T, Skourti-Stathaki K, Proudfoot NJ, Dean C (2013) R-loop stabilization represses antisense transcription at the Arabidopsis FLC locus. Science 340: 619–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swiezewski S, Liu F, Magusin A, Dean C (2009) Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462: 799–802 [DOI] [PubMed] [Google Scholar]
- Tamaoki M, Freeman JL, Pilon-Smits EA (2008) Cooperative ethylene and jasmonic acid signaling regulates selenite resistance in Arabidopsis. Plant Physiol 146: 1219–1230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Deng Z, Oses-Prieto JA, Suzuki N, Zhu S, Zhang X, Burlingame AL, Wang ZY (2008) Proteomics studies of brassinosteroid signal transduction using prefractionation and two-dimensional DIGE. Mol Cell Proteomics 7: 728–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thilmony R, Underwood W, He SY (2006) Genome-wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen Pseudomonas syringae pv. tomato DC3000 and the human pathogen Escherichia coli O157:H7. Plant J 46: 34–53 [DOI] [PubMed] [Google Scholar]
- Tuerk C, Gold L (1990) Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249: 505–510 [DOI] [PubMed] [Google Scholar]
- Vandenborre G, Smagghe G, Van Damme EJ (2011) Plant lectins as defense proteins against phytophagous insects. Phytochemistry 72: 1538–1550 [DOI] [PubMed] [Google Scholar]
- Wang JW, Czech B, Weigel D (2009) miR156-regulated SPL transcription factors define an endogenous flowering pathway in Arabidopsis thaliana. Cell 138: 738–749 [DOI] [PubMed] [Google Scholar]
- Wang ZW, Wu Z, Raitskin O, Sun Q, Dean C (2014) Antisense-mediated FLC transcriptional repression requires the P-TEFb transcription elongation factor. Proc Natl Acad Sci USA 111: 7468–7473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Park MY, Conway SR, Wang JW, Weigel D, Poethig RS (2009) The sequential action of miR156 and miR172 regulates developmental timing in Arabidopsis. Cell 138: 750–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Song M, Wei Z, Tong J, Zhang L, Xiao L, Ma Z, Wang Y (2011) A jacalin-related lectin-like gene in wheat is a component of the plant defence system. J Exp Bot 62: 5471–5483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Xu S, Li C, Xu Y, Xing L, Niu Y, Huan Q, Tang Y, Zhao C, Wagner D, et al. (2014) O-GlcNAc-mediated interaction between VER2 and TaGRP2 elicits TaVRN1 mRNA accumulation during vernalization in winter wheat. Nat Commun 5: 4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao J, Zhang H, Xing L, Xu S, Liu H, Chong K, Xu Y (2013) Requirement of histone acetyltransferases HAM1 and HAM2 for epigenetic modification of FLC in regulating flowering in Arabidopsis. J Plant Physiol 170: 444–451 [DOI] [PubMed] [Google Scholar]
- Xing L, Li J, Xu Y, Xu Z, Chong K (2009) Phosphorylation modification of wheat lectin VER2 is associated with vernalization-induced O-GlcNAc signaling and intracellular motility. PLoS One 4: e4854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Howard M, Dean C (2014) Antagonistic roles for H3K36me3 and H3K27me3 in the cold-induced epigenetic switch at Arabidopsis FLC. Curr Biol 24: 1793–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yong WD, Xu YY, Xu WZ, Wang X, Li N, Wu JS, Liang TB, Chong K, Xu ZH, Tan KH, et al. (2003) Vernalization-induced flowering in wheat is mediated by a lectin-like gene VER2. Planta 217: 261–270 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho YH, Sheen J (2007) Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc 2: 1565–1572 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.