Sesquiterpene synthases for 1,8-cineole, cadinene, and geranyllinalool appear early in angiosperm lineage.
Abstract
Bay laurel (Laurus nobilis) is an agriculturally and economically important dioecious tree in the basal dicot family Lauraceae used in food and drugs and in the cosmetics industry. Bay leaves, with their abundant monoterpenes and sesquiterpenes, are used to impart flavor and aroma to food, and have also drawn attention in recent years because of their potential pharmaceutical applications. To identify terpene synthases (TPSs) involved in the production of these volatile terpenes, we performed RNA sequencing to profile the transcriptome of L. nobilis leaves. Bioinformatic analysis led to the identification of eight TPS complementary DNAs. We characterized the enzymes encoded by three of these complementary DNAs: a monoterpene synthase that belongs to the TPS-b clade catalyzes the formation of mostly 1,8-cineole; a sesquiterpene synthase belonging to the TPS-a clade catalyzes the formation of mainly cadinenes; and a diterpene synthase of the TPS-e/f clade catalyzes the formation of geranyllinalool. Comparison of the sequences of these three TPSs indicated that the TPS-a and TPS-b clades of the TPS gene family evolved early in the evolution of the angiosperm lineage, and that geranyllinalool synthase activity is the likely ancestral function in angiosperms of genes belonging to an ancient TPS-e/f subclade that diverged from the kaurene synthase gene lineages before the split of angiosperms and gymnosperms.
The aromatic Laurus nobilis, commonly known as bay laurel, is an evergreen tree originally from the Mediterranean region that is currently cultivated in many warm regions of the world. L. nobilis belongs to the family Lauraceae, which comprises 32 genera and approximately 2,000 to 2,500 species (Julianti et al., 2012). Lauraceae are in the order Laurales, which together with its sister order Magnoliales is placed among the basal dicots close to the origin of the angiosperms (Bremer et al., 2003, 2009). Fresh or dried bay leaves and bay leaf essential oils are used extensively in the food industry for flavoring soups, meats, fish, and beverages (Julianti et al., 2012) and as a food preservative, since they possess antimicrobial and insecticidal activities (Saim and Meloan, 1986). The essential oil is also used as a folk medicine, especially for rheumatism and dermatitis. In addition, it is used by the cosmetic industry in creams, perfumes, and soaps (Hafizoǧlu and Reunanen, 1993). It was reported to be used in the preparation of hair lotions for its antidandruff activity and for the external treatment of psoriasis (Hafizoǧlu and Reunanen, 1993).
The volatile constituents of various L. nobilis tissues such as leaves, flower, fruits, and buds (but not roots) have been previously examined, and they constitute mostly mono- and sesquiterpenes (Kilic et al., 2004). In fresh L. nobilis leaves, the monoterpene 1,8-cineole is the main volatile compound, and α-terpinyl acetate, terpinene-4-ol, α- and β-pinene, sabinene, and linalool have been reported to occur in appreciable levels (Kilic et al., 2004; Derwich et al., 2009; Marzouki et al., 2009).
Terpenes constitute the largest class of plant specialized (secondary) metabolites (Dudareva et al., 2004). Low-molecular terpenes such as the 10-carbon monoterpenes and 15-carbon sesquiterpenes easily volatilize at room temperature and are found as major components of floral scents and essential oils of herbs, vegetables, and fruits (Dudareva et al., 2004; Dudareva and Pichersky, 2008). All plant terpenes are made from the 5-carbon precursors isopentenyl diphosphate and its isomer, dimethylallyl diphosphate, both of which are derived from two alternative pathways, the mevalonate pathway in the cytosol or the methylerythritol phosphate pathway in plastids. Condensation of the C5 precursors leads to the formation of C10, C15, and C20 trans- or cis-prenyl diphosphate intermediates that are converted into monoterpenes, sesquiterpenes, and diterpenes, respectively, by the activity of terpene synthase (TPS) enzymes (Tholl, 2006). An unusual feature of many TPS enzymes is their ability to produce multiple terpene products from a single substrate, leading to the formation of mixtures of structurally diverse compounds (Cunningham and Gantt, 1998; Degenhardt et al., 2009).
TPSs are encoded by a gene family in all angiosperm and gymnosperm genomes so far examined (Chen et al., 2011). The ancestral gene at the root of this gene family is likely a bifunctional copalyl diphosphate synthase/kaurene synthase, which gave rise by duplication and divergence to several family clades, some of which are unique to angiosperm species (clades a, b, and g) and gymnosperm species (clade d; Chen et al., 2011). On the other hand, clade e/f, which contains the monofunctional kaurene synthases found in angiosperms and gymnosperms, also contains a subclade that was recently identified as encoding the diterpene synthase geranyllinalool synthase. Although it was inferred from sequence comparisons that the geranyllinalool synthase subclade diverged from the rest of the TPS genes in clade e/f before the split of the gymnosperm and angiosperm lineages, enzymes encoded by genes in this subclade have only been characterized from eudicot species (Falara et al., 2014). No genes belonging to this subclade have yet been found in gymnosperms, and although the recent genome sequence of the basal dicot Amborella trichopoda has revealed a gene belonging to this subclade, the enzyme it encodes has not yet been characterized (Falara et al., 2014).
Previous studies in the basal dicots have identified a few TPS genes encoding monoterpene synthases in Litsea cubeba (Lauraceae; Chang and Chu, 2011) and monoterpene and sesquiterpene synthases in Magnolia grandiflora (Lee and Chappell, 2008). These studies have shown that the monoterpene synthases fell into clade b, to which most monoterpenes from the eudicot species belong, suggesting that clade b diverged from the rest of the TPS gene family at the base of the angiosperms. Likewise, the sesquiterpene synthase from M. grandiflora fell into clade a, which contains most sesquiterpene synthases from eudicots, indicating that this clade of the TPS family also diverged from the rest of the TPS family at the time of the origin of the angiosperm lineage.
The studies of TPSs in M. grandiflora and L. cubeba involved the isolation of complementary DNAs (cDNAs), in vitro characterization of the recombinant enzymes produced in Escherichia coli, and examination of the tissues in which the genes were expressed, although they did not include the analysis of the terpenes produced in the individual plants examined. Because L. nobilis produces so many different terpenes in its various organs, because it is dioecious (having male and female plants), and because of its position at the base of the angiosperm lineage, we have begun an investigation of the TPS gene family in this species. Here, we show that male and female L. nobilis trees have distinct profiles of terpene volatiles in each of the different organs examined, including roots. Consistent with previous findings, we show that a monoterpene synthase from L. nobilis falls into TPS clade b, and that a sesquiterpene synthase from L. nobilis falls into clade a. In addition, we show that L. nobilis also has a gene belonging to the geranyllinalool synthase subclade of TPS clade e/f and demonstrate that the enzyme encoded by this gene indeed has geranyllinalool synthase activity. This result extends the conservation of this function of the genes in this subclade to the basal dicots, further supporting the hypothesis that geranyllinalool was one of the first specialized metabolites to have evolved from early stages in the evolution of the TPS gene family in plants.
RESULTS
Volatile Profiles in Leaves, Flowers, Fruits, and Roots in Male and Female L. nobilis plants
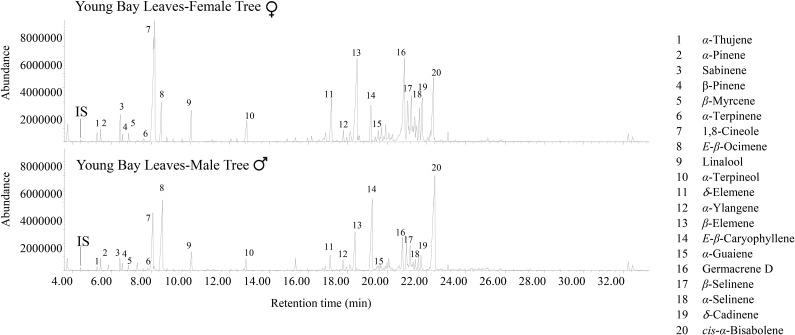
We examined the volatile compound composition of freshly harvested tissue from different organs of both male and female mature bay trees growing in our experimental station. The gas chromatography (GC)-mass spectrometry (MS) analysis of leaves, roots, flowers, and fruits (Figs. 1 and 2; Supplemental Tables S1 and S2) indicated that volatile compounds in these organs consist mainly of mono- and sesquiterpene hydrocarbons, with traces of the diterpene geranyllinalool, and small amounts of the C16-homoterpene 4,8,12-trimethyltridacan-1,3,7,11-tetraene (TMTT), which is derived from geranyllinalool, was also found in most bay tissues investigated in this study (Supplemental Table S1). Some fatty acid-derived volatiles such as 2-E-hexenal and phenylpropanoids such as eugenol and methyleugenol were also observed (Supplemental Table S2).
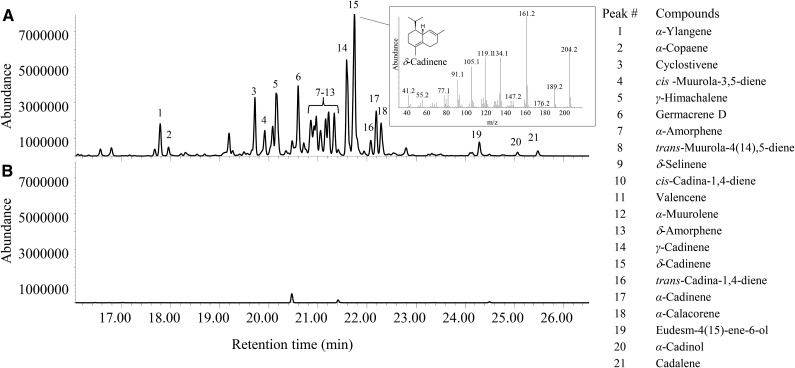
Figure 1.
GC analysis of volatiles present in young leaves of female and male L. nobilis trees.
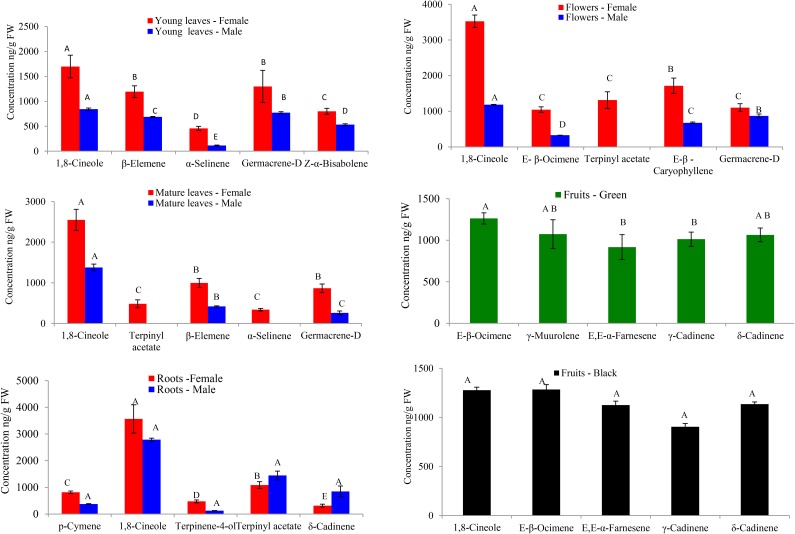
Figure 2.
The levels of the five terpene compounds with the highest concentration in female organs of L. nobilis, and comparisons with the concentrations of the corresponding compounds in male organs and in organs present in both sexes. Bars labeled with different letters indicate the significant differences as determined by JMP statistical software (P ≤ 0.05; Tukey-Kramer honestly significant difference test). All analyses were performed using three biological replicates. FW, Fresh weight.
There were significant differences in volatile composition between different organs and between male and female plants. For example, the monoterpene 1,8-cineole was the most abundant volatile in all male female organs examined except green fruit, where it was still quite abundant (Fig. 2; Supplemental Table S1). However, male organs had a consistently lower concentration of 1,8-cineole in a given organ than female plants. On the other hand, male leaves and flowers had considerably more δ-elemene than did the corresponding female organs (Supplemental Table S1). The biggest gender differences were observed in the flowers, where female flowers have predominantly monoterpenes as well as eugenol and methyleugenol, whereas male flowers contain mostly sesquiterpenes and benzaldehyde (Fig. 2; Supplemental Tables S1 and S2). It was also noteworthy that the fruits on the female plants contain high levels of two sesquiterpenes, γ-cadinene and δ-cadinene, that were found in much lower levels elsewhere in the plant (Fig. 2; Supplemental Table S1).
Analysis of Sequences of Terpene Synthases from L. nobilis
To begin analysis of the TPS gene family in L. nobilis, next-generation Illumina sequencing was performed on RNA samples extracted from young leaves of a female tree, yielding up to approximately 42.3 million 100-bp paired-end reads. A total of a 41.2 quality and adapter-filtered paired-end reads were de novo assembled and produced 63,316 contigs with an average size of 563 bp and an N50 of 752 bp. For annotation, the sequences were searched against the National Center for Biotechnology Information (NCBI) nonredundant (NR) protein database using BLASTX, the Uniref90 protein clusters from the UniProt and The Arabidopsis Information Resource databases with a cutoff value of E ≤ 1 × 10−5, and the top five hits from each database were returned. BLASTX searches resulted in at least one significant hit for 44,891 out of the 63,316 contigs (71%). Gene ontology terms were assigned using the Blast2GO search program (Conesa and Götz, 2008), which annotates high-score BLAST matches to sequences in the NCBI-NR proteins database. The results of this analysis are presented in Supplemental Figure S1.
Based on this analysis, we were able to identify eight contigs with sequence similarity to TPS genes, which we designated as LnTPS1, LnTPS2, up to LnTPS8 (Supplemental Table S3). From the sequence comparison with previously characterized TPSs from other species, the sequences of LnTPS1 and LnTPS2 appear to contain complete open reading frames (ORFs; Supplemental Fig. S2), whereas the others were incomplete. A complete ORF of LnTPS3 was obtained (Supplemental Fig. S2) as described in “Materials and Methods.”
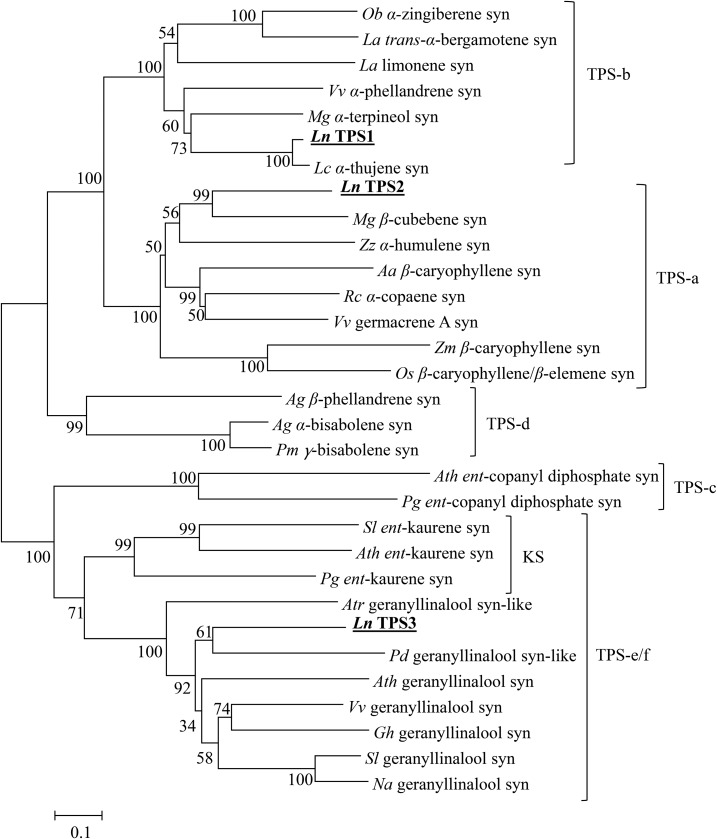
The protein encoded by LnTPS1 consists of 582 amino acids with a calculated molecular mass of 67.4 kD. The putative amino-terminal extension of 45 amino acids of the LnTPS1 upstream of the RRx8W motif and sequence analysis using ChloroP 1.1 software (http://www.cbs.dtu.dk/services/ChloroP/) predicted a plastidic localization for the protein. A phylogenetic tree based on protein sequence comparisons with representative TPS sequences from other plant species indicated that the LnTPS1 protein belongs to the TPS-b clade (Fig. 3), in which the majority of angiospermous monoterpene synthases reside (Chen et al., 2011). The TPS sequences most similar to LnTPS1 are those of α-thujene synthase from L. cubeba (Lauraceae; Chang and Chu, 2011), with 92% identity, and α-terpineol synthase from M. grandiflora (Lee and Chappell, 2008), with 57% identity.
Figure 3.
An unrooted neighbor-joining tree based on protein sequences of LnTPS1, LnTPS2, and LnTPS3 and selected plant TPS sequences. The tree was generated using Phylogeny Analysis MEGA5.1 program (Tamura et al., 2013). The resulting tree was bootstrap analyzed with 1,000 replicates. The subdivisions (clades) of the TPS gene family, designated TPS-a to TPS-e/f, are according to Chen et al. (2011). The black bold underline indicates the L. nobilis LnTPS1, LnTPS2, and LnTPS3 sequences identified in this study. Ocimum basilicum (Ob) α-zingiberene synthase (syn), accession number Q5SBP4; Lavandula angustifolia (La) transα-bergamotene syn, Q2XSC4; La limonene syn, Q2XSC6; V. vinifera (Vv) α-phellandrene syn, NP_001268167; M. grandiflora (Mg) α-terpineol syn, B3TPQ7; L. cubeba (Lc) α-thujene syn, AEJ91555; Mg β-cubebene syn, B3TPQ6; Zingiber zerumbet (Zz) α-humulene syn, B1B1U3; Artemisia annua (Aa) β-caryophyllene syn, Q8SA63; Ricinus communis (Rc) α-copaene syn, JN315864; Vv germacrene A syn, ADR66821; Zea mays (Zm) β-caryophyllene syn, ABY79213; Oryza sativa (Os) β-caryophyllene/β-elemene syn, DQ872158; Abies grandis (Ag) β-phellandrene syn, Q9M7D1; Ag α-bisabolene syn, O81086; Pseudotsuga menziesii (Pm) γ-bisabolene syn, Q4QSN4; Arabidopsis (Arabidopsis thaliana; Ath) ent-copalyl diphosphate syn, NP_192187; Picea glauca (Pg) ent-copalyl diphosphate syn, ADB55707; S. lycopersicum (Sl) ent-kaurene syn, AEP82778; Ath ent-kaurene syn, NP_178064; Pg ent-kaurene syn, ADB55708; Amborella trichopada (Atr) geranyllinalool syn-like, ERN20279; P. dactylifera (Pd) geranyllinalool syn-like, NW_008248648; Ath geranyllinalool syn, At1G61120; Vv geranyllinalool syn, NP_001268004; Grindela hirsutula (Gh) geranyllinalool syn, AGN70888; Sl geranyllinalool syn, KJ755870; Nicotiana attenuate (Na) geranyllinalool syn, KJ755868; KS, kaurene synthase.
The predicted LnTPS2 protein sequence consists of 551 amino acids, with a calculated molecular mass of 63.8 kD, and no putative chloroplast-targeting sequence identified. Comparisons with representative TPS sequences from other plant species indicated that the LnTPS2 protein belongs to the TPS-a clade (Fig. 3), in which the majority of angiospermous sesquiterpene synthases reside (Chen et al., 2011). The TPS sequence most similar to LnTPSs is β-copaene synthase (56% identity) from M. grandiflora (Lee and Chappell, 2008).
The protein encoded by LnTPS3 contains 852 amino acids, with a calculated molecular mass of 97.7 kD, and no transit peptide was predicted to be present by SignalP (http://www.cbs.dtu.dk/services/SignalP/). LnTPS3 belongs to the TPS-e/f clade, and specifically to the geranyllinalool synthase (GLS) subclade (Fig. 3), and has the highest sequence identity, 51%, to geranyllinalool synthase from Vitis vinifera (NP_001268004).
Of the remaining five TPS contigs, although incomplete, three fell into the TPS-a clade and the other two could be classified as belonging to the TPS-b clade (Supplemental Table S3). Given the large number of terpenes identified in the various organs of L. nobilis, it is likely that its genome contains additional TPS genes, and therefore the eight TPS genes that we identified from the leaf RNA sequencing (RNA-seq) data must be considered a partial list. Additional RNA-seq efforts from leaf material of different developmental stages and from other organs as well as whole genome sequencing will be required to detect more TPS genes.
Biochemical Characterization of the Enzymes Encoded by LnTPS1, LnTPS2, and LnTPS3
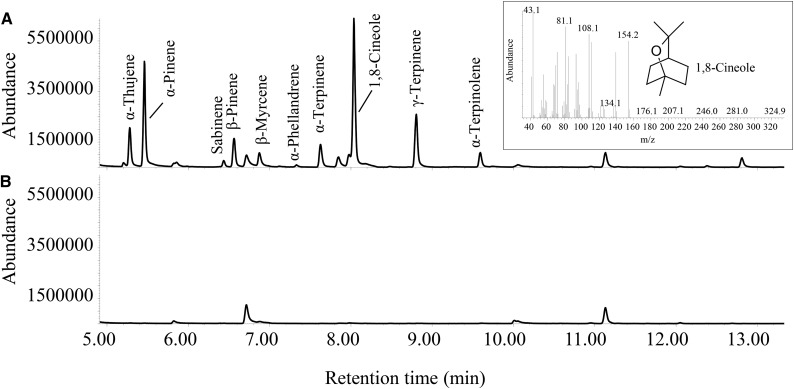
E. coli BL21 (DE3) cells expressing His-tagged LnTPS1, LnTPS2, or LnTPS3 using the expression vector pEXP5-CT/TOPO TA were harvested and lysed. LnTPS1 and LnTPS2 were purified with a nickel-agarose affinity column to a purity of >90% (Supplemental Fig. S3). However, LnTPS3 did not bind to the nickel-agarose column, and it was instead partially purified on an DE53 anion-exchange column to a purity of approximately 5% (Supplemental Fig. S3). Upon incubation with geranyl pyrophosphate (GPP) as a substrate, LnTPS1 catalyzed the formation of mostly 1,8-cineole, with α-thujene, α-pinene, β-pinene, α-terpinene, γ-terpinene, α-terpinolene, and a few other monoterpenes also produced, as detected by GC-MS analysis (Fig. 4A). The identities of these peaks were confirmed by comparisons with retention times and the mass spectra of authenticity standards. Based on the major product, LnTPS1 was designated as a 1,8-cineole synthase. Extracts prepared from E. coli (same strain) transformed with pEXP5-CT/TOPO TA lacking cDNA inserts and heat-denatured enzyme preparations served as controls for terpene formation independent of LnTPS1, and terpene products were observed in these assays (Fig. 4B).
Figure 4.
GC-MS of the products generated in vitro by recombinant LnTPS1 protein. A, Analysis of products of the reaction catalyzed by recombinant LnTPS1with GPP as the substrate. B, Analysis of products of the reaction catalyzed by boiled recombinant LnTPS1with GPP as the substrate. The insert shows the structure and the mass spectrum of the enzymatic reaction products identified as 1,8-cineole by matching the retention time and the MS pattern with an authentic standard. Identification of other products was done by GC-MS according to the retention index and by comparing the compound mass spectrum to the National Institute of Standards and Technology database.
GC-MS analysis of the reaction products catalyzed by LnTPS2 with 2E,6E-farnesyl diphosphate (eeFPP) as a substrate (Fig. 5) identified at least 21 sesquiterpenes, with δ-cadinene and γ-cadinene as two major products (peaks #15 and #14, respectively, in Fig. 5), germacrene D (#6), γ-himachalene (#5), cyclosativene (#3), α-cadinene (#17), α-ylangene (#1), α-calacorene (#18), δ-amorphene (#13), α-amorphene (#7), trans-muurola-4(14),5-diene (#8), δ-selinene (#9), cis-cadina-1,4-diene (#10), valencene (#11), α-muurolene (#12), and the minor peaks α-copaene (#2), cis-muurola-3,5-diene (#4), transcadina-1,4-diene (#16), eudesm-4(15)-ene-6-ol (#19), α-cadinol (#20), and cadalene (#21; Fig. 5A). Since δ-cadinene and γ-cadinene were the major products of the reaction, we designated LnTPS2 cadinene synthase. Extracts prepared from E. coli (same strain) transformed with pEXP5-CT/TOPO TA lacking cDNA inserts and heat-denatured enzyme preparations served as controls for terpene formation independent of LnTPS2, and no terpene products were observed in these assays (Fig. 5B).
Figure 5.
GC-MS of the products generated in vitro by recombinant LnTPS2 protein. A, Analysis of products of the reaction catalyzed by recombinant LnTPS2 with eeFPP as the substrate. B, Analysis of products of the reaction catalyzed by boiled recombinant LnTPS2 with eeFPP as the substrate. The insert shows the structure and the mass spectrum of the enzymatic reaction products identified as δ-cadinene by matching the retention time and the MS pattern with an authentic standard. Identification of other products was done by GC-MS according to the retention index and by comparing the compound mass spectrum to the National Institute of Standards and Technology database.
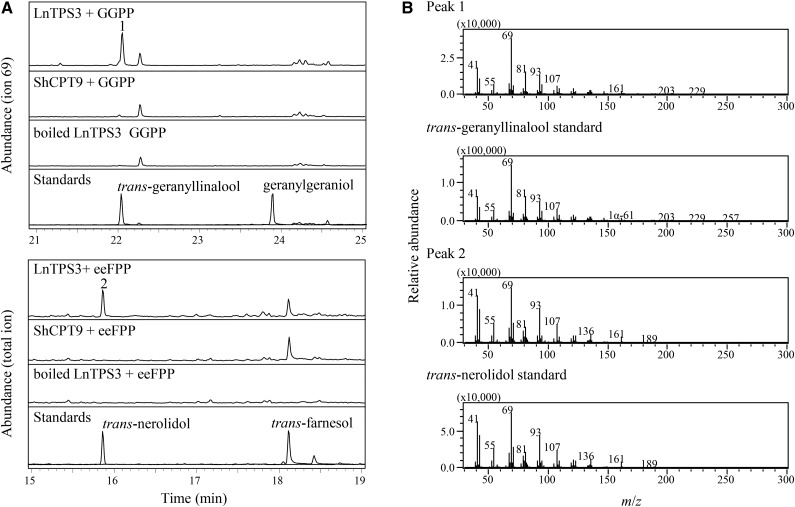
The partially purified LnTPS3 protein catalyzed the formation of geranyllinalool from geranylgeranyl diphosphate (GGPP) and trans-nerolidol from eeFPP (Fig. 6). No trans-geranyllinalool and trans-nerolidol synthase activities were detected in reactions containing crude protein extracts of E. coli cells expressing a non-TPS recombinant protein or when the LnTPS3 protein was boiled first before being added to the reaction mixture (Fig. 6). The Km and Vmax values of partially purified LnTPS3 with GGPP as the substrate were determined to be 30.5 ± 4.7 µm and 7.3 ± 0.3 pmol/ng protein·min, and the corresponding values with eeFPP were 43.4 ± 2.9 µm and 2.7 ± 0.1 pmol/ng protein·min. The Vmax/Km ratio for the geranyllinalool synthase activity (Vmax/Km = 0.24) was 4-fold higher than the ratio for transnerolidol synthase activity (Vmax/Km = 0.06). No significant TPS activity was detected using GPP as substrate.
Figure 6.
GC-MS analysis of the products from reaction assays catalyzed by recombinant LnTPS3 and containing GGPP or eeFPP as the substrate. A, Peak 1, transgeranyllinalool; peak 2, transnerolidol. B, Mass spectra comparison of peaks from enzymatic reaction products with authentic standards.
Expression Patterns of LnTPS1, LnTPS2, and LnTPS3
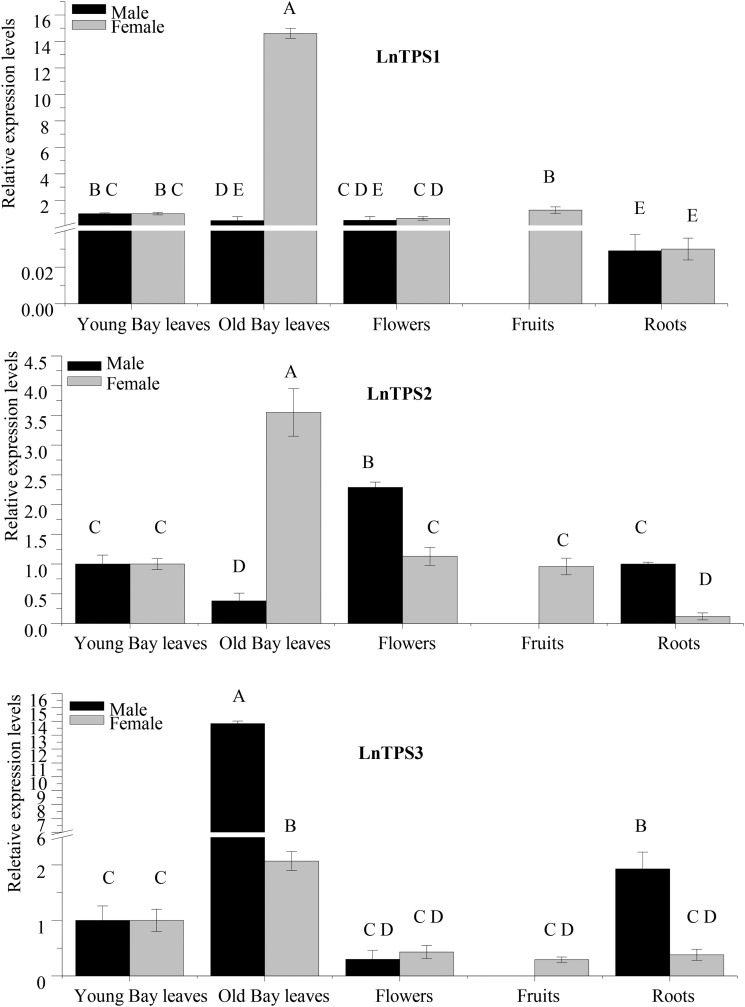
To study the TPS gene expression in the L. nobilis roots, young and mature leaves, flowers (female and male), and fruits (female), we measured the accumulation of LnTPS1, LnTPS2, and LnTPS3 transcripts in all of these tissues using quantitative real time (qRT)-PCR (Fig. 7). In female plants, all three genes showed slightly higher levels of transcripts in mature leaves than in all other tissues, with particularly high levels of LnTPS1 transcripts and low relative levels of transcripts of all three genes in roots, particularly of LnTPS1. The transcript levels of these genes in male plants showed a similar pattern to those observed in the corresponding female organs, except that in mature leaves, the levels of LnTPS3 transcripts were relatively higher than the LnTPS2 transcript levels (Fig. 7).
Figure 7.
qRT-PCR determination of transcript levels of LnTPS1, LnTPS2, and LnTPS3 in young and mature leaves, roots, flowers of male and female L. nobilis plants, and in green fruits of female plants. Quantification of LnTPS1, LnTPS2, and LnTPS3 transcript levels by real-time reverse transcription-PCR analysis normalized to equal levels of actin transcripts. All analyses were performed using three biological replicates.
DISCUSSION
L. nobilis, a Dioecious, Arboraceous Basal Dicotyledonous Species, Produces Numerous Terpenes
Although terpene production has been extensively studied in gymnosperms and eudicot, less information is available on terpenes and terpene biosynthesis in the basal dicots. Because the TPS gene family in gymnosperms has diversified independently of the TPS gene family in angiosperms (Bohlmann et al., 1998; Chen et al., 2011), the origin of terpene diversity in angiosperms needs to be investigated in the basal dicots, which are at the base of angiospermous plants (Hoot et al., 1999; Doyle and Endress, 2000; Graham and Olmstead, 2000; Bremer et al., 2003). L. nobilis is a basal dicot and therefore is a good system to understand terpene evolution in the flowering plants. Moreover, L. nobilis is a dioecious plant and thus provides an opportunity to examine gender differences in terpene production.
Both male and female plants contained high levels (several µg g−1 fresh weight of various monoterpenes and sesquiterpenes in leaves, roots (which had not been previously examined for volatiles), and flowers. The fruits on the female trees also contained similarly high levels of terpenes (Figs. 1 and 2). The monoterpene 1,8-cineole was the major terpene found in all organs examined except in green fruits, where it was present in substantial amounts nonetheless. Organs of female plants generally contained more terpenes than the corresponding male organs. Gender differences were pronounced in the flowers, but significant differences in amounts of specific terpene volatiles were also observed in other organs (Figs. 1 and 2). Fruits displayed the most divergent volatile profile from all other organs (Fig. 2).
1,8-Cineole occurs widely in plants, where it performs important ecological functions, such as to repelling insects, detering herbivores, and repressing germination in and growth of competing plants (Southwell et al., 2003; Chen et al., 2004; Franks et al., 2012). Industrially, 1,8-cineole is widely used in hygiene products, food flavors, and pharmaceutical preparation (Juergens et al., 2004; Kehrl et al., 2004; Tesche et al., 2008). The high levels of 1,8-cineole in the leaves, roots, flowers, and fruits may thus play a role in protecting L. nobilis from its enemies and increase its ability to compete belowground with neighboring plants for limited water and nutrient supplies. δ-Cadinene has been shown to provide a phytoalexin defense response in both bacterial and fungal cotton plant-pathogen interactions (Chen et al., 1995; Davis et al., 1996; Townsend et al., 2005), and δ-cadinene synthesis is induced in cotton stems infected with Verticillium dahlia (Bianchini et al., 1999; Townsend et al., 2005).
The only diterpene that we were able to detect was geranyllinalool, which was detected in trace amounts in all organs of L. nobilis trees of both sexes, except roots (Supplemental Table S1). TMTT, a compound derived from geranyllinalool by oxidation and cleavage (Boland and Gabler, 1989; Tholl et al., 2011), was also detected in these organs, with the highest concentration observed in young leaves. TMTT emission has been reported for many angiosperm plants including basal dicot and monocot plants, often together with its precursor geranyllinalool (Tholl et al., 2011). In addition to its possible role as pollinator attractant, TMTT has been implicated in attracting natural enemies of arthropod herbivores when released from damage foliage (Hopke et al., 1994; Ament et al., 2004; Tholl et al., 2011). For instance, carnivorous predatory mites (Phytoseiulus persimilis) preferred the odor source of lima bean (Phaseolus lunatus) plants attacked by spider mites (Tetranychus urticae) to the odor of plants damaged by beet armyworm (Spodoptera exigua) larvae (de Boer et al., 2004). TMTT might also play a role in indirect defense in floral tissues (Brillada et al., 2013). Still, the role of geranyllinalool in plant tissues is not well established. On the other hand, nonvolatile 17-hydroxygeranyllinalool glycosides have been shown to have a direct defense role against herbivores as toxins in leaves and flowers of Nicotiana species (Heiling et al., 2010; Jassbi et al., 2010).
It should be noted that, since we measured internal concentrations of free volatiles, the concentrations that we observed should be considered steady-state levels. In addition, we made no attempt to measure the concentration of glycosylated terpenes. However, although some monoterpene glucosides have been described from L. nobilis, geranyllinalool glycosides have not been reported to be present in this species (Kilic et al., 2005).
L. nobilis Contains TPS Genes of the a and b Clades as Well as the Geranyllinalool Subclade of the TPS-e/f Clade
After obtaining complete or partial sequences representing eight L. nobilis TPS genes by RNA-seq, we identified sequences that fall into the a, b, and e/f clades (Supplemental Table S3) and characterized one representative from each clade (Fig. 3). The L. nobilis 8-cineole synthase we characterized here produces a similar, but not identical, group of monoterpenes in addition to 1,8-cineole that previously characterized 8-cineole synthase from different plant species, and similar to them, it also belongs to the TPS-b clade (Wise et al., 1998; Chen et al., 2004; Demissie et al., 2012). For example, the Arabidopsis root-specific cineole synthase (AtTPC-Cin) produces, in addition to 1,8-cineole, (−)-(4S)-limonene and (E)-β-ocimene (Chen et al., 2004); the recombinant common sage (Salvia officinalis) cineole synthase produces 1,8-cineole as the major product along with (±)-limonene and (+)-campherene (Wise et al., 1998); and the cineole synthase from the glandular trichome of Lavandula × intermedia flowers produces, in addition to 1,8-cineole, (±)-limonene and linalool (Demissie et al., 2012). In contrast, the L. nobilis cineole synthase does not produce (±)-limonene, (E)-β-ocimene, linalool, or (+)-campherene (Fig. 4).
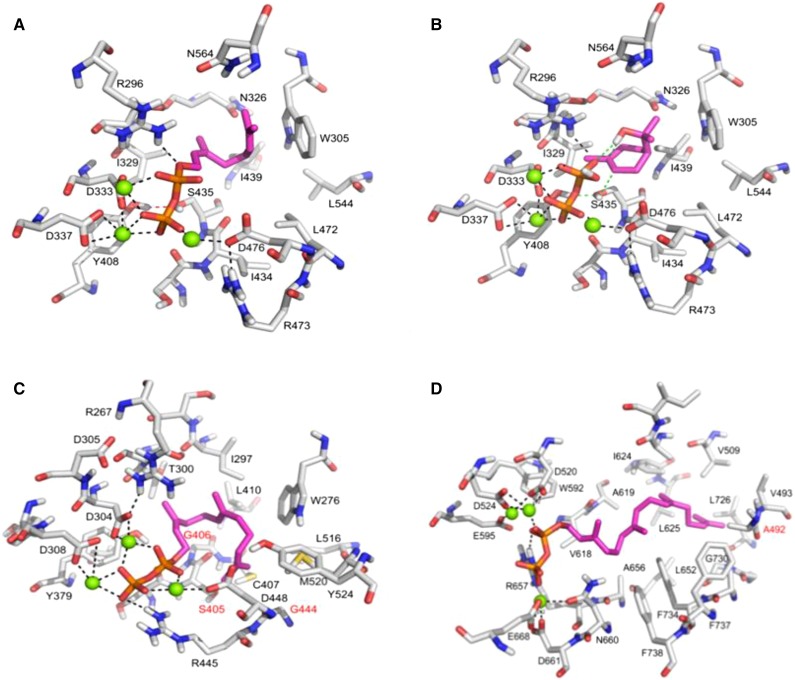
To investigate further the difference between the 1,8-cineole synthase encoded by LnTPS1 and previously characterized 1,8-cineole synthase, we created a high-quality homology model of LnTPS1 using Yet Another Scientific Artificial Reality Application (YASARA; Krieger et al., 2009; Supplemental Fig. S4). LnTPS1 forms a tertiary structure (all α-fold) very similar to other monoterpene synthases, such as (4S)-limonene synthase of spearmint (Mentha spicata; Srividya et al., 2015). The model of the active side of LnTPS1 with the bound substrate GPP (Fig. 8A) indicates that the diphosphate moiety is mainly recognized by three divalent magnesium cations complexed to three Asp side chains (D333 and D337 from the well-known DDXXD motive and D476). Furthermore, an Arg (R296) may serve as a proton donor to thermodynamically support the cleavage of the diphosphate by protonation after the first step of the reaction (Brandt et al., 2009). In agreement (e.g. with the limonene synthase), the aromatic side chain of a tryptophane (W305) stabilizes the resulting intermediate terpenyl carbocation (Fig. 8B). Based on this orientation, a water molecule activated by the diphosphate can attack the terpenyl cation to form α-terpineol (Fig. 8B). A catalytic dyad consisting of S435 and Y408 may serve as proton relay to protonate the double bond of α-terpineol, and by synchronous transfer of the proton from the hydroxyl group of terpineol to the diphosphate, the ring closure for the formation of the main product of this enzyme, 1,8-cineole, terminates the reaction. So far, the formation of the main product can be nicely explained. However, in comparison with other 1,8-cineole synthases, LnTPS1 forms no side products such as limonene, (E)-β-ocimene, or (+)-campherene. The formation of all these products needs a base in the active site serving as proton acceptor from the cation intermediates. For instance, it was shown by site-directed mutagenesis that His-579 in limonene synthase serves as the proton acceptor (Srividya et al., 2015). In LnTPS1, no such proton acceptor can be detected in the active site except the diphosphate.
Figure 8.
Models of the active sites of L. nobilis TPS enzymes. A, The active site of LnTPS1 with bound GPP (magenta carbon atoms). B, The active site of LnTPS1 with bound pyrophosphate and the intermediate terpineol (magenta carbon atoms). C, The active site of LnTPS2 with bound FPP (magenta carbon atoms). D, The active site of LnTPS3 with bound GGPP (magenta carbon atoms).
Likewise, whereas the L. nobilis cadinene synthase belongs to the TPS-a clade, as do previously characterized cadinene synthases from eudicots (the latter produce δ-cadinene as the single product; Chen et al., 1995, 1996; Davis et al., 1996; Gennadios et al., 2009), it catalyzes the formation of both δ-cadinene and γ-cadinene as well as a plethora of other sesquiterpenes from eeFPP, indicating that the L. nobilis cadinene synthase enzyme is a multiproduct cadinene synthase, a phenomenon seen with many other sesquiterpene synthases (Steele et al., 1998; Wise et al., 1998; Köllner et al., 2004). However, several terpene volatile products of LnTPS2 (e.g. cyclosativene, γ-himachalene, valencene, eudesm-4(15)-ene-6-ol, α-cadinol, and cadalene) have not actually been found to accumulate in L. nobilis tissues.
The model of the tertiary structure of LnTPS2 (Supplemental Fig. S5) and diphosphate recognition in the active site (Fig. 8C) are almost identical to those of LnTPS1. The farnesyl moiety of the substrate is also located in a mainly hydrophobic binding pockedformed by I297, I410, L516, and W305. Again, the side chain of W276 will serve to stabilize the intermediate cations. However, there are some significant differences in the active site between both enzymes. In contrast to LnTPS1, S435 (involved in the catalytic dyad) is replaced by G406 (red labeled), I434 is substituted by S405, L472 by G444, and I439 by L410. All of these alterations lead to a larger binding pocked that allows docking of FPP in many different conformations, which is not possible in LnTPS1. A comparison of the active sites of LnTPS2 with a previously characterized cadinene synthase (Protein Data Bank codes 3G4D and 3G4F; Gennadios et al., 2009) that produces δ-cadinene as the single product indicates that a Tyr might be responsible for the difference. Tyr-535 in 3G4F narrows the active site considerably, and furthermore, the conformational freedom of the bound FPP and subsequently formed cation intermediate is likely highly restricted again by being stabilized by a Trp-279. In LnTPS2, a Thr (T531) is located at a spatially identical position to Tyr-535, and this Thr does not cause a conformational restriction on FPP, leading to many different conformations of FPP and many cyclized intermediates and final products.
It was recently reported that the TPS-e/f clade branched before the split of the gymnosperm and angiosperm lineages, with one branch containing the gene for kaurene synthase of the GA hormone pathway and the other branch containing genes encoding enzymes that, in all cases examined so far, catalyze the formation of geranyllinalool from GGPP (Falara et al., 2014). However, the characterized geranyllinalool synthases have so far only come from eudicot species. LnTPS3 is the first gene from a basal dicot species that belongs to this branch of the TPS-e/f clade to be demonstrated to encode a bona fide geranyllinalool synthase. Thus, this result extends the likely function of the genes in this branch as geranyllinalool synthases to the early stages in the evolution of flowering plants. Similar genes have not been found in the genomes of the monocot grasses (order Poales); however, LnTPS3 has 48% identity to a geranyllinalool synthase-like gene (NW_008248648) from the monocot Phoenix dactylifera and a 42% identity to a gene (ERN20279) in the basal angiosperm A. trichopoda. It would be interesting to determine the biochemical function of these two genes and see how far back in plant evolution the geranyllinalool synthase activity can be traced.
The three-dimensional model of LnTPS3 (Supplemental Fig. S6) clearly shows the α, β, and γ domains of a TPS-e/f protein, and the active site of LnTPS3 (Fig. 8D) with bound GGPP (magenta carbon atoms) shows that diphosphate recognition is almost identical to that occurring in LnTPS1 and LnTPS2 (black dotted lines). However, the geranylgeranyl moiety is located in a much larger and more hydrophobic binding pocked in comparison with the two other ones. Most interestingly, the tryptophane residue (LnTPS1: W305, LnTPS2: W276) is replaced by an Ala (A492, red labeled). Therefore, cation-π stabilizing interactions are almost completely missing, which nicely explains why no cyclic terpenes are formed by this enzyme but only the rather simple hydroxylation to form geranyllinalool. The side chains of F734, F737, and F738 seem to be sterically not accessible for cation stabilization. This again shows the importance of aromatic side chains for the cation-π stabilization of cyclic terpene cations (Petersen et al., 2005; Dougherty, 2007; Brandt et al., 2009).
The Characterized L. nobilis TPS Genes Are Expressed in All Organs
The qRT-PCR analysis of the transcript levels of LnTPS1, LnTPS2, and LnTPS3 showed that they are expressed at similar levels in all organs examined, except for LnTPS1, which appears to have a lower level of transcript accumulation in the roots of both male and female trees (Fig. 7). However, although 1,8-cineole is a major compound in all L. nobilis organs and δ-cadinene and γ-cadinene are major volatiles in fruits, it is not possible with the present data to determine to what extent LnTPS1 and LnTPS2 contribute to the synthesis of these monoterpene and sesquiterpene compounds, respectively. Our RNA-seq analysis identified at least five other TPS genes (two likely monoterpene synthases and three likely sesquiterpene synthases; Supplemental Table S3), and all of these are expressed in all organs examined (Supplemental Fig. S7). Some of these genes are likely to encode TPSs that catalyze the formation of a variety of terpenes that also might include 1,8-cineole (in the case of monoterpene synthases) or cadinenes (in the case of sesquiterpene synthases). Furthermore, it is likely that the genome of L. nobilis contains additional TPS genes not identified in the RNA-seq analysis. Because terpene volatile compounds may be made by more than one TPS in the same species, and because they can accumulate to high levels in such organs, such as roots, even when the specific activity of the enzymes is relatively low, it is generally difficult to determine the direct contribution of each TPS gene to the observed mixture even when the expression levels of individual TPSs are examined in detail (Köllner et al., 2004; Tholl et al., 2005). Also, additional TPS gene transcript analysis and enzyme characterization will provide a better understanding of the individual or overlapping functions of L. nobilis TPS genes in terpene biosynthesis.
The in vitro characterization of LnTPS3 indicated that it can catalyze the formation of geranyllinalool from GGPP and nerolidol from eeFPP. The comparison of its catalytic efficiency with each substrate indicates that it is 4-fold more efficient with GGPP. A similar result has been observed in tomato (Solanum lycopersicum). SlGLS also has nerolidol synthase activities with 55-fold lower efficiency than geranyllinalool synthase activity (Falara et al., 2014). The ORF of LnTPS3, similar to those of its close relatives in this branch of the TPS-e/f clade, encodes no transit peptide, and the protein is therefore likely to be cytosolic (Herde et al., 2008; Falara et al., 2014). The cytosolic compartment contains both eeFPP and GGPP, but we could not detect any nerolidol (Supplemental Table S1). It is therefore likely that LnTPS3 is responsible for the geranyllinalool observed in L. nobilis, and ultimately TMTT biosynthesis.
MATERIALS AND METHODS
Chemicals
Chemicals, unlabeled eeFPP and GPP (1 mg mL−1), terpenoid standards, and reagents were purchased from Sigma-Aldrich unless noted otherwise. GGPP was obtained from Echelon Biosciences.
Plant Material
Laurus nobilis plants were grown in the Newe Yaar Research Center in northern Israel under standard field irrigation and fertigation conditions. Freshly harvested young and mature leaves were crushed in liquid nitrogen and stored at −80°C for terpene and transcript analysis.
Volatile Terpene Analyses
Extraction of Volatile Compounds from L. nobilis Tissues
Three biological replicates from young and old leaves, roots, flowers, and fruit of male and female L. nobilis plants (1 g) were ground into a uniform powder under liquid nitrogen with a mortar and pestle. The powder was placed in a 20-mL solid-phase microextraction (SPME) vial containing 1 g of NaCl, 7 mL of 20% (w/v) NaCl, and 0.01 µg mg−1 of 2-heptanone as an internal standard, then sealed and placed in 4°C until used. Headspace sampling was conducted immediately utilizing a 65-µm fused silica fiber coated with polydimethylsiloxane/divinylbenzene (Supelco; Yahyaa et al., 2013, 2015). After 40 min, the SPME syringe was introduced into the injector port of the GC-MS apparatus for further analysis.
Autoheadspace-SPME-GC-MS Analysis of L. nobilis Volatile Compounds
Volatile compounds were analyzed on a GC-MS apparatus (Agilent Technologies) equipped with an Rtx-5SIL MS (30-m × 0.25-mm × 0.25-µm)-fused silica capillary column, as described by Yahyaa et al. (2015).
Bay Leaves RNA Isolation and Sequencing
Since the transcriptome of L. nobilis is not known, we generated an RNA-seq database from RNA isolated from young leaves of an L. nobilis female tree, using the Spectrum Plant Total RNA Kit (Sigma-Aldrich). The cDNA library was generated from 2 μg of mRNA, and cDNA library inserts were prepared for sequencing using one pico-titer plate on the next-generation Illumina (Applied Biosystems SOLiD).
De Novo Sequence Assembly and Functional Annotation
A total of 42.3 million raw, long, 100-bp paired-end reads were cleaned from ribosomal RNA and adaptors using Silva ribosomal RNA (http://www.arb-silva.de; Quast et al., 2013) and PhiX databases. After filtering, 41.2 million paired-end reads were de novo assembled using Trinity software (Grabherr et al., 2011), with default parameters and 25-mer k-mer size, generating 63,316 sequences. The 63,316 assembled sequences were aligned using BLATSX (BLAST) algorithm with an E-value cutoff of 10−5 against the NCBI-NR (http://www.ncbi.nlm.nih.gov), the Uniref90 (Suzek et al., 2015), and Arabidopsis (The Arabidopsis Information Resource; https://www.arabidopsis.org/) protein databases. Gene ontology to three categories (molecular function, biological process, and cell component annotations) was assigned using Blast2GO (Conesa and Götz, 2008) based on an NCBI-NR search.
Isolation and Characterization of Bay Leaf TPSs
Putative bay leaves TPS-encoding genes were searched using a homology-based algorithm in the transcriptome database of L. nobilis, which has been assembled based on available GenBank sequence data entries. BLASTX searches of the assembled contigs against TPS genes in GenBank identified at least three full-length L. nobilis TPS cDNAs, of which two are predicted to be monoterpene synthases and one a sesquiterpene synthase. Two specific primers correspond to the L. nobilis monoterpene synthase (LnTPS1) coding sequence (5′-ATGTCATTTACATTGCTCACTCCATCT-3′) and the reverse primer (5′-CATAAACTTGAATGGCTCAGCCAG-3′). L. nobilis sesquiterpene synthase (LnTPS2) was obtained using a forward primer of (5′-ATGACTCCAAAAAAACTCCATCCGG-3′) and ends (5′-AACTGGAATAGGATTAACAAGCACTGAC-3′) were designed.
For transcriptome sequencing and cDNA synthesis and cloning, RNA from L. nobilis young leaves of a female tree was isolated using the Spectrum Plant Total RNA Kit (Sigma-Aldrich). For producing a cDNA clone, 5 µg of total RNA from the different L. nobilis tissues was reverse transcribed using the SuperScript One-Step Reverse Transcription-PCR (Invitrogen). The DNA molecule was then amplified using Platinum Taq DNA polymerase (Invitrogen) yielding a 1,689-bp specific fragment for LnTPS1 and a 1,746-bp fragment for LnTPS2, respectively. The cDNAs were ligated into the pEXP5-CT/TOPO TA expression vector (Invitrogen), producing pEXP-LnTPS1 and pEXP-LnTPS2, respectively, in which the LnTPS1 and the LnTPS2 coding sequences were fused with a His tag-coding extension at the C terminus and transformed into Escherichia coli Top10 cells. The constructs were verified by DNA sequencing.
The partial sequence of LnTPS3 was obtained from RNA-seq and originally assembled as contig 3. To obtain a full ORF of LnTPS3, 5′ and 3′ race templates were prepared using young leaf total RNA according manufacturer protocol (SMART RACE cDNA Amplification Kit, Clontech). 3′ end sequence of the gene was obtained by amplifying the piece of gene with the gene-specific and the poly A primers using 3′ race template. Incomplete 5′ sequence was obtained by both 5′ race with gene-specific primers and PCR with degenerate primer based on the conserved amino acid sequence of other GLSs. However, the first Met could not be obtained in this way. We therefore amplified the beginning of the ORF using a gene walking method (GenomeWalker Universal Kit, Clontech) with genomic DNA, which was digested by DraI restriction enzyme and added adaptor sequence as template. Once the DNA fragment containing the first Met was obtained in this way, the ORF of LnTPS3 was amplified with gene-specific primers using 3′ race cDNA as a template, and spliced into the pGEMTeasy vector (Supplemental Table S4).
Preparation of Bacterial Lysates
A 3-mL preculture of E. coli was grown overnight at 37°C in Luria-Bertani medium containing 100 g mL−1 ampicillin. These cultures were used to inoculate 500 mL of fresh medium to which 500 µm isopropyl-1-thio-β-d-galactopyranoside was added after 3 h to induce protein expression. Cells were further grown for 12 h at 18°C. Bacteria were lysed and soluble protein purified by nickel chromatography and assayed for purity by SDS-PAGE as previously described by Yahyaa et al. (2015).
An optimized LnTPS3 ORF for expression in E. coli was synthesized and cloned into pUC57 vector (GenScript). The ORF of the gene was amplified by PCR and subcloned into pEXP5-CT/TOPO vector (Invitrogen, C-terminal His-tag). The plasmid was transformed into BL21 (DE3) cells and the culture was grown until optical density at 600 nm = 0.8. LnTPS3 protein was inducted by adding 0.5 mm isopropyl β-d-1-thiogalactopyranoside as a final concentration and incubating at 16°C for 18 h. Cells were next collected and kept at −80°C. For control, Solanum habrochaites cis-prenyldiphosphate 9-pEXP5-CT/TOPO was introduced to BL21 (DE3) cells, and protein production was induced in the same way as for LnTPS3. Cells were resuspended in bind buffer for the DE53 anion-exchange column (Whatman) containing 50 mm Tris-HCl, 10 mm MgCl2, and 14 mm 2-mercaptoethanol, pH 7.5. After the sonication and centrifuge, the supernatant was applied to the DE53 column, which was equilibrated with the bind buffer. The column was washed with the buffer containing 50 mm Tris-HCl, 100 mm KCl, 10 mm MgCl2, and 14 mm 2-mercaptoethanol, pH 7.5. Then, elution buffer containing 50 mm Tris-HCl, 150 mm KCl, 10 mm MgCl2, and 14 mm 2-mercaptoethanol, pH 7.5, was applied to the column and the fraction was collected. This fraction was desalted and changed to buffer containing 10 mm HEPES and 5% (v/v) glycerol, pH 7.0, and used for the kinetic analysis.
Assay for L. nobilis TPS Activity
Enzyme activity assays were performed in screw-capped 2-mL GC glass vials using 1 to 500 ng of purified recontaminate protein by nickel-nitrilotriacetic acid agarose affinity chromatography, 10 µm substrate (GPP or eeFPP), 10 mm MgCl2, 10 µm MnCl2, and assay buffer 50 mm Bis-Tris, pH 7.0, in a total volume of 100 µL. The reactions were incubated for 30 min at 30°C. After incubation, the samples were analyzed by autoheadspace-SPME-GC-MS for the identification of volatile terpenes generated during the 30°C incubation.
Extracts prepared from E. coli (same strain) transformed with pEXP5-CT/TOPO TA lacking a cDNA insert and heat-denatured enzyme preparations served as controls for terpene formation independent of LnTPS1 and LnTPS2.
The enzymatic reaction mixture in a total of 50 µL containing 40 to 50 µg of either partially purified LnTPS3 protein or crude LnTPS3 E. coli protein, 20 to 200 µm substrate (GGPP or eeFPP), 50 mm HEPES, 100 mm KCl, 7.5 mm MgCl2, 5% (v/v) glycerol, and 5 mm dithiothreitol, pH 7.0, was incubated at 30°C for 20 min. As negative controls, the crude E. coli protein harboring S. habrochaites cis-prenyldiphosphate 9-pEXP5-CT/TOPO vector, instead of LnTPS3-pEXP5-CT/TOPO, and the boiled LnTPS3 protein were used. The reaction was stopped by adding 50% (v/v) methanol. The reaction compounds were extracted by 50 µL of hexane containing tetradecane as the internal standard, and 1 µL of the extract was used for the GC-MS analysis.
Estimates of Km and Vmax of LnTPS3 for eeFPP and GGPP were determined for a range of substrate concentrations from 20 to 200 µm. Values were determined using nonlinear regression of the Michaelis-Menten equation.
Samples were injected into an Rxi-5Sil MS column (30-m length, 0.25-μm film thickness, and 0.25-mm internal diameter; Restek) with splitless mode on a GC-2010 Plus coupled to a GCMS-QP2010 SE (Shimadzu). Injector temperature was 240°C, and interface temperature was 280°C. The following GC methods were used: after a 3-min isothermal hold at 60°C, the column temperature was increased by 8°C min−1 to 270°C.
LnTPS Transcript Analysis
For qRT-PCR analysis of LnTPS1 to LnTPS8, total RNA (5 µg) from young and old leaves, roots, flowers, and fruit of male and female L. nobilis plants was extracted (Spectrum Plant Total RNA Kit, Sigma-Aldrich) and reverse transcribed using an oligo primer and the SuperScript II first-strand system (Invitrogen).
qRT-PCR was performed on an Applied Biosystems StepOnePlus Real-Time PCR System (Life Technology) using ABsolute Blue qPCR SYBR Green ROX Mix (Tamar Laboratory Supplies LTD), using 5-ng reverse-translated total RNA and 100-ng primers (Supplemental Table S5). Relative quantification of gene expression was performed using as reference the housekeeping gene actin from L. nobilis with primers described in Supplemental Table S5. Differences in relative expression levels of LnTPS1 to LnTPS8 were calculated from 2-ΔΔCt values after normalization of data to actin. All analyses were performed using at least three biological replicates.
Homology Modeling of LnTPS1, LnTPS2, and LnTPS3
Homology modeling of LnTPS1, LnTPS2, and LnTPS3 was performed with YASARA (Krieger et al., 2009). For LnTPS1, 60 models (LnTPS2 96, LnTPS3 50) were created based on different alignments using more than 10 x-ray templates in each case. For LnTPS1, an isoprene synthase (Protein Data Bank code: 3N0F; Köksal et al., 2010) appeared as best suited templates from which the final model was built. For LnTPS2, the x-ray structure of a 5-epi-aristolochene synthase (Starks et al., 1997) was preferentially used, but finally a hybrid model was formed by replacing in this model some small better folded loop regions from models created from 3M02 (Noel et al., 2010), 4GAX, and 4FJQ (Li et al., 2013). For LnTPS3, an abietadiene synthase (3S9V; Zhou et al., 2012) was used as the basic template, which was slightly modified by replacement of small fragments from 4LIX (Köksal et al., 2014), 3PYA (M. Köksal, H. Hu, R.M. Coates, R.J. Peters, and D.W. Christianson, unpublished data), 3P5P (Köksal et al., 2011), and 3SAE (McAndrew et al., 2011) to create the final best scored hybrid model. The substrate of LnTPS1, GPP, was manually predocked into the active site by superposition (using the molecular modeling environment program MOE 2014.09, https://www.chemcomp.com/) with the model created from 3M00 (Noel et al., 2010), wherein an FPP derivative was located. FPP was manually modified to GPP by deleting the corresponding atoms. For LnTPS2, FPP was unmodified and overtaken from the 3M00-based model, and for LnTPS3, GGPP was formed from FPP by adding the corresponding prenyl moiety. All of these models with correct substrate for each of the enzymes were refined with a 20-cycle simulated annealing procedure (md-refine.mcr) in YASARA.
The quality of the resulting homology models were evaluated using PROCHECK (Laskowski et al., 1993) and ProSA (Sippl, 1993). All three models are of excellent quality, with more than 90% of the residues being in the most favored region of the Ramachandran plot. The ProSA energy graphs are all in the negative range, and the calculated z scores are in the range of natively folded proteins (LnTPS1 = −12.31 for 550 amino acids, LnTPS2 = −13.02, 547 amino acids, LnTPS3 = −14.55 for 773 amino acids).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers KR336614, KR336615, and KR336616.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Gene ontology assignments for the L. nobilis leaf transcriptome.
Supplemental Figure S2. Sequences of the ORF and encoded proteins of LnTPS1, LnTPS2, and LnTPS3.
Supplemental Figure S3. SDS-PAGE analysis of purified recombinant L. nobilis TPSs LnTPS1, LnTPS2, and LnTPS3.
Supplemental Figure S4. Modeling LnTPS1 structure.
Supplemental Figure S5. Modeling LnTPS2 structure.
Supplemental Figure S6. Modeling LnTPS3 structure.
Supplemental Figure S7. Expression patterns of LnTPS4 to LnTPS8 in young and mature leaves, roots, flowers, and fruits of male and female L. nobilis plants.
Supplemental Table S1. Quantification of mono- and sesquiterpene volatile compounds in different organs of L. nobilis. Volatile terpenes were measured by autoheadspace-SPME-GC-MS.
Supplemental Table S2. Levels of nonterpene volatile compounds in different organs of L. nobilis. Volatiles were measured by autoheadspace-SPME-GC-MS.
Supplemental Table S3. Classification to TPS clades of the eight L. nobilis TPS contigs found from the assembly of Illumina sequences.
Supplemental Table S4. Synthetic oligonucleotides used for isolation of full-length LnTPS3.
Supplemental Table S5. Synthetic oligonucleotides used for qRT-PCR expression of LnTPS1 to LnTPS8 in young and mature leaves, roots, flowers, and fruit of male and female L. nobilis plants.
Acknowledgments
We thank Nativ Dudai and Doya Sa'adi for taking care of the L. nobilis plants.
Glossary
- cDNA
complementary DNA
- GC
gas chromatography
- MS
mass spectrometry
- TMTT
4,8,12-trimethyltridacan-1,3,7,11-tetraene
- NCBI
National Center for Biotechnology Information
- NR
nonredundant
- ORF
open reading frame
- RNA-seq
RNA sequencing
- GGPP
geranylgeranyl diphosphate
- qRT
quantitative real time
- YASARA
Yet Another Scientific Artificial Reality Application
- SPME
solid-phase microextraction
Footnotes
This work was supported by the National Science Foundation of the United States (grant no. IOS–1025636).
Articles can be viewed without a subscription.
References
- Ament K, Kant MR, Sabelis MW, Haring MA, Schuurink RC (2004) Jasmonic acid is a key regulator of spider mite-induced volatile terpenoid and methyl salicylate emission in tomato. Plant Physiol 135: 2025–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchini GM, Stipanovic RD, Bell AA (1999) Induction of delta-cadinene synthase and sesquiterpenoid phytoalexins in cotton by Verticillium dahliae. J Agric Food Chem 47: 4403–4406 [DOI] [PubMed] [Google Scholar]
- Bohlmann J, Meyer-Gauen G, Croteau R (1998) Plant terpenoid synthases: molecular biology and phylogenetic analysis. Proc Natl Acad Sci USA 95: 4126–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland W, Gabler A (1989) Biosynthesis of homoterpenes in higher plants. Helv Chim Acta 72: 247–253 [Google Scholar]
- Brandt W, Bräuer L, Günnewich N, Kufka J, Rausch F, Schulze D, Schulze E, Weber R, Zakharova S, Wessjohann L (2009) Molecular and structural basis of metabolic diversity mediated by prenyldiphosphate converting enzymes. Phytochemistry 70: 1758–1775 [DOI] [PubMed] [Google Scholar]
- Bremer B, Bremer K, Chase MW, Fay MF, Reveal JL, Soltis DE, Soltis PS, Stevens PF, Anderberg AA, Moore MJ, et al. (2009) An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG III. Bot J Linn Soc 161: 105–121 [Google Scholar]
- Bremer B, Bremer K, Chase MW, Reveal JL, Soltis DE, Soltis PS, Stevens PF, Anderberg AA, Fay MF, Goldblatt P, et al. (2003) An update of the angiosperm phylogeny group classification for the orders and families of flowering plants: APG II. Bot J Linn Soc 141: 399–436 [Google Scholar]
- Brillada C, Nishihara M, Shimoda T, Garms S, Boland W, Maffei ME, Arimura G (2013) Metabolic engineering of the C16 homoterpene TMTT in Lotus japonicus through overexpression of (E,E)-geranyllinalool synthase attracts generalist and specialist predators in different manners. New Phytol 200: 1200–1211 [DOI] [PubMed] [Google Scholar]
- Chang YT, Chu FH (2011) Molecular cloning and characterization of monoterpene synthases from Litsea cubeba (Lour.) Persoon. Tree Genet Genomes 7: 835–844 [Google Scholar]
- Chen F, Ro DK, Petri J, Gershenzon J, Bohlmann J, Pichersky E, Tholl D (2004) Characterization of a root-specific Arabidopsis terpene synthase responsible for the formation of the volatile monoterpene 1,8-cineole. Plant Physiol 135: 1956–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F, Tholl D, Bohlmann J, Pichersky E (2011) The family of terpene synthases in plants: a mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J 66: 212–229 [DOI] [PubMed] [Google Scholar]
- Chen XY, Chen Y, Heinstein P, Davisson VJ (1995) Cloning, expression, and characterization of (+)-δ-cadinene synthase: a catalyst for cotton phytoalexin biosynthesis. Arch Biochem Biophys 324: 255–266 [DOI] [PubMed] [Google Scholar]
- Chen XY, Wang M, Chen Y, Davisson VJ, Heinstein P (1996) Cloning and heterologous expression of a second (+)-δ-cadinene synthase from Gossypium arboreum. J Nat Prod 59: 944–951 [DOI] [PubMed] [Google Scholar]
- Conesa A, Götz S (2008) Blast2GO: A comprehensive suite for functional analysis in plant genomics. Int J Plant Genomics 2008: 619832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham FX, Gantt E (1998) Genes and enzymes of carotenoid biosynthesis in plants. Annu Rev Plant Physiol Plant Mol Biol 49: 557–583 [DOI] [PubMed] [Google Scholar]
- Davis EM, Tsuji J, Davis GD, Pierce ML, Essenberg M (1996) Purification of (+)-δ-cadinene synthase, a sesquiterpene cyclase from bacteria-inoculated cotton foliar tissue. Phytochemistry 41: 1047–1055 [DOI] [PubMed] [Google Scholar]
- de Boer JG, Posthumus MA, Dicke M (2004) Identification of volatiles that are used in discrimination between plants infested with prey or nonprey herbivores by a predatory mite. J Chem Ecol 30: 2215–2230 [DOI] [PubMed] [Google Scholar]
- Degenhardt J, Köllner TG, Gershenzon J (2009) Monoterpene and sesquiterpene synthases and the origin of terpene skeletal diversity in plants. Phytochemistry 70: 1621–1637 [DOI] [PubMed] [Google Scholar]
- Demissie ZA, Cella MA, Sarker LS, Thompson TJ, Rheault MR, Mahmoud SS (2012) Cloning, functional characterization and genomic organization of 1,8-cineole synthases from Lavandula. Plant Mol Biol 79: 393–411 [DOI] [PubMed] [Google Scholar]
- Derwich E, Benziane Z, Boukir A (2009) Chemical composition and antibacterial activity of leaves essential oil of Laurus nobilis from Morocco. Aust J Basic Appl Sci 3: 3818–3824 [Google Scholar]
- Dougherty DA. (2007) Cation-π interactions involving aromatic amino acids. J Nutr 137(6, Suppl 1) 1504S–1508S, discussion 1516S–1517S [DOI] [PubMed] [Google Scholar]
- Doyle JA, Endress PK (2000) Morphological phylogenetic analysis of basal angiosperms: comparison and combination with molecular data. Int J Plant Sci 161: S121–S153 [Google Scholar]
- Dudareva N, Pichersky E (2008) Metabolic engineering of plant volatiles. Curr Opin Biotechnol 19: 181–189 [DOI] [PubMed] [Google Scholar]
- Dudareva N, Pichersky E, Gershenzon J (2004) Biochemistry of plant volatiles. Plant Physiol 135: 1893–1902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falara V, Alba JM, Kant MR, Schuurink RC, Pichersky E (2014) Geranyllinalool synthases in solanaceae and other angiosperms constitute an ancient branch of diterpene synthases involved in the synthesis of defensive compounds. Plant Physiol 166: 428–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks SJ, Wheeler GS, Goodnight C (2012) Genetic variation and evolution of secondary compounds in native and introduced populations of the invasive plant Melaleuca quinquenervia. Evolution 66: 1398–1412 [DOI] [PubMed] [Google Scholar]
- Gennadios HA, Gonzalez V, Di Costanzo L, Li A, Yu F, Miller DJ, Allemann RK, Christianson DW (2009) Crystal structure of (+)-delta-cadinene synthase from Gossypium arboreum and evolutionary divergence of metal binding motifs for catalysis. Biochemistry 48: 6175–6183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29: 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham SW, Olmstead RG (2000) Evolutionary significance of an unusual chloroplast DNA inversion found in two basal angiosperm lineages. Curr Genet 37: 183–188 [DOI] [PubMed] [Google Scholar]
- Hafizoǧlu H, Reunanen M (1993) Studies on the components of Lauras nobilis from Turkey with special reference to Laurel berry fat. Eur J Lipid Sci Technol 95: 304–308 [Google Scholar]
- Heiling S, Schuman MC, Schoettner M, Mukerjee P, Berger B, Schneider B, Jassbi AR, Baldwin IT (2010) Jasmonate and ppHsystemin regulate key malonylation steps in the biosynthesis of 17-hydroxygeranyllinalool diterpene glycosides, an abundant and effective direct defense against herbivores in Nicotiana attenuata. Plant Cell 22: 273–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herde M, Gärtner K, Köllner TG, Fode B, Boland W, Gershenzon J, Gatz C, Tholl D (2008) Identification and regulation of TPS04/GES, an Arabidopsis geranyllinalool synthase catalyzing the first step in the formation of the insect-induced volatile C16-homoterpene TMTT. Plant Cell 20: 1152–1168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoot SB, Magallon S, Crane PR (1999) Phylogeny of basal eudicots based on three molecular data sets: atpB, rbcL, and 18S nuclear ribosomal DNA sequences. Ann Mo Bot Gard 86: 1–32 [Google Scholar]
- Hopke J, Donath J, Blechert S, Boland W (1994) Herbivore-induced volatiles: the emission of acyclic homoterpenes from leaves of Phaseolus lunatus and Zea mays can be triggered by a beta-glucosidase and jasmonic acid. FEBS Lett 352: 146–150 [DOI] [PubMed] [Google Scholar]
- Jassbi AR, Zamanizadehnajari S, Baldwin IT (2010) 17-Hydroxygeranyllinalool glycosides are major resistance traits of Nicotiana obtusifolia against attack from tobacco hornworm larvae. Phytochemistry 71: 1115–1121 [DOI] [PubMed] [Google Scholar]
- Juergens UR, Engelen T, Racké K, Stöber M, Gillissen A, Vetter H (2004) Inhibitory activity of 1,8-cineol (eucalyptol) on cytokine production in cultured human lymphocytes and monocytes. Pulm Pharmacol Ther 17: 281–287 [DOI] [PubMed] [Google Scholar]
- Julianti E, Jang KH, Lee S, Lee D, Mar W, Oh KB, Shin J (2012) Sesquiterpenes from the leaves of Laurus nobilis L. Phytochemistry 80: 70–76 [DOI] [PubMed] [Google Scholar]
- Kehrl W, Sonnemann U, Dethlefsen U (2004) Therapy for acute nonpurulent rhinosinusitis with cineole: results of a double-blind, randomized, placebo-controlled trial. Laryngoscope 114: 738–742 [DOI] [PubMed] [Google Scholar]
- Kilic A, Hafizoglu H, Kollmannsberger H, Nitz S (2004) Volatile constituents and key odorants in leaves, buds, flowers, and fruits of Laurus nobilis L. J Agric Food Chem 52: 1601–1606 [DOI] [PubMed] [Google Scholar]
- Kilic A, Kollmannsberger H, Nitz S (2005) Glycosidically bound volatiles and flavor precursors in Laurus nobilis L. J Agric Food Chem 53: 2231–2235 [DOI] [PubMed] [Google Scholar]
- Köksal M, Jin Y, Coates RM, Croteau R, Christianson DW (2011) Taxadiene synthase structure and evolution of modular architecture in terpene biosynthesis. Nature 469: 116–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köksal M, Potter K, Peters RJ, Christianson DW (2014) 1.55Å-resolution structure of ent-copalyl diphosphate synthase and exploration of general acid function by site-directed mutagenesis.Biochim Biophysica Acta 1840: 184–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köksal M, Zimmer I, Schnitzler JP, Christianson DW (2010) Structure of isoprene synthase illuminates the chemical mechanism of teragram atmospheric carbon emission. J Mol Biol 402: 363–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köllner TG, Schnee C, Gershenzon J, Degenhardt J (2004) The variability of sesquiterpenes emitted from two Zea mays cultivars is controlled by allelic variation of two terpene synthase genes encoding stereoselective multiple product enzymes. Plant Cell 16: 1115–1131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieger E, Joo K, Lee J, Lee J, Raman S, Thompson J, Tyka M, Baker D, Karplus K (2009) Improving physical realism, stereochemistry, and side-chain accuracy in homology modeling: Four approaches that performed well in CASP8. Proteins 77(Suppl 9): 114–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Macarthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26: 283–291 [Google Scholar]
- Lee S, Chappell J (2008) Biochemical and genomic characterization of terpene synthases in Magnolia grandiflora. Plant Physiol 147: 1017–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JX, Fang X, Zhao Q, Ruan JX, Yang CQ, Wang LJ, Miller DJ, Faraldos JA, Allemann RK, Chen XY, et al. (2013) Rational engineering of plasticity residues of sesquiterpene synthases from Artemisia annua: product specificity and catalytic efficiency. Biochem J 451: 417–426 [DOI] [PubMed] [Google Scholar]
- Marzouki H, Piras A, Salah KBH, Medini H, Pivetta T, Bouzid S, Marongiu B, Falconieri D (2009) Essential oil composition and variability of Laurus nobilis L. growing in Tunisia, comparison and chemometric investigation of different plant organs. Nat Prod Res 23: 343–354 [DOI] [PubMed] [Google Scholar]
- McAndrew RP, Peralta-Yahya PP, DeGiovanni A, Pereira JH, Hadi MZ, Keasling JD, Adams PD (2011) Structure of a three-domain sesquiterpene synthase: a prospective target for advanced biofuels production. Structure 19: 1876–1884 [DOI] [PubMed] [Google Scholar]
- Noel JP, Dellas N, Faraldos JA, Zhao M, Hess BA Jr, Smentek L, Coates RM, O’Maille PE (2010) Structural elucidation of cisoid and transoid cyclization pathways of a sesquiterpene synthase using 2-fluorofarnesyl diphosphates. ACS Chem Biol 5: 377–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen FNR, Jensen MØ, Nielsen CH (2005) Interfacial tryptophan residues: a role for the cation-π effect? Biophys J 89: 3985–3996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, Peplies J, Glöckner FO (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41: D590–D596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saim N, Meloan CE (1986) Compounds from leaves of bay (Laurus nobilis L.) as repellents for Tribolium castaneum (Herbst) when added to wheat flour. J Stored Prod Res 22: 141–144 [Google Scholar]
- Sippl MJ. (1993) Recognition of errors in three-dimensional structures of proteins. Proteins 17: 355–362 [DOI] [PubMed] [Google Scholar]
- Southwell IA, Russell MF, Maddox CDA, Wheeler GS (2003) Differential metabolism of 1,8-cineole in insects. J Chem Ecol 29: 83–94 [DOI] [PubMed] [Google Scholar]
- Srividya N, Davis EM, Croteau RB, Lange BM (2015) Functional analysis of (4S)-limonene synthase mutants reveals determinants of catalytic outcome in a model monoterpene synthase. Proc Natl Acad Sci USA 112: 3332–3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starks CM, Back K, Chappell J, Noel JP (1997) Structural basis for cyclic terpene biosynthesis by tobacco 5-epi-aristolochene synthase. Science 277: 1815–1820 [DOI] [PubMed] [Google Scholar]
- Steele CL, Crock J, Bohlmann J, Croteau R (1998) Sesquiterpene synthases from grand fir (Abies grandis). Comparison of constitutive and wound-induced activities, and cDNA isolation, characterization, and bacterial expression of delta-selinene synthase and gamma-humulene synthase. J Biol Chem 273: 2078–2089 [DOI] [PubMed] [Google Scholar]
- Suzek BE, Wang Y, Huang H, McGarvey PB, Wu CH; UniProt Consortium (2015) UniRef clusters: a comprehensive and scalable alternative for improving sequence similarity searches. Bioinformatics 31: 926–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30: 2725–2729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesche S, Metternich F, Sonnemann U, Engelke JC, Dethlefsen U (2008) The value of herbal medicines in the treatment of acute non-purulent rhinosinusitis. Results of a double-blind, randomised, controlled trial. Eur Arch Otorhinolaryngol 265: 1355–1359 [DOI] [PubMed] [Google Scholar]
- Tholl D. (2006) Terpene synthases and the regulation, diversity and biological roles of terpene metabolism. Curr Opin Plant Biol 9: 297–304 [DOI] [PubMed] [Google Scholar]
- Tholl D, Chen F, Petri J, Gershenzon J, Pichersky E (2005) Two sesquiterpene synthases are responsible for the complex mixture of sesquiterpenes emitted from Arabidopsis flowers. Plant J 42: 757–771 [DOI] [PubMed] [Google Scholar]
- Tholl D, Sohrabi R, Huh JH, Lee S (2011) The biochemistry of homoterpenes--common constituents of floral and herbivore-induced plant volatile bouquets. Phytochemistry 72: 1635–1646 [DOI] [PubMed] [Google Scholar]
- Townsend BJ, Poole A, Blake CJ, Llewellyn DJ (2005) Antisense suppression of a (+)-<delta>-cadinene synthase gene in cotton prevents the induction of this defense response gene during bacterial blight infection but not its constitutive expression. Plant Physiol 138: 516–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wise ML, Savage TJ, Katahira E, Croteau R (1998) Monoterpene synthases from common sage (Salvia officinalis). cDNA isolation, characterization, and functional expression of (+)-sabinene synthase, 1,8-cineole synthase, and (+)-bornyl diphosphate synthase. J Biol Chem 273: 14891–14899 [DOI] [PubMed] [Google Scholar]
- Yahyaa M, Bar E, Dubey NK, Meir A, Davidovich-Rikanati R, Hirschberg J, Aly R, Tholl D, Simon PW, Tadmor Y, et al. (2013) Formation of norisoprenoid flavor compounds in carrot (Daucus carota L.) roots: characterization of a cyclic-specific carotenoid cleavage dioxygenase 1 gene. J Agric Food Chem 61: 12244–12252 [DOI] [PubMed] [Google Scholar]
- Yahyaa M, Tholl D, Cormier G, Jensen R, Simon PW, Ibdah M (2015) Identification and characterization of terpene synthases potentially involved in the formation of volatile terpenes in carrot (Daucus carota L.) roots. J Agric Food Chem 63: 4870–4878 [DOI] [PubMed] [Google Scholar]
- Zhou K, Gao Y, Hoy JA, Mann FM, Honzatko RB, Peters RJ (2012) Insights into diterpene cyclization from structure of bifunctional abietadiene synthase from Abies grandis. J Biol Chem 287: 6840–6850 [DOI] [PMC free article] [PubMed] [Google Scholar]