The functions of over 60% of plant enzymes are unknown; nearly one-half of these proteins have microbial homologs, enabling the use of cross-kingdom comparative genomics to predict their functions.
Abstract
The number of sequenced genomes is rapidly increasing, but functional annotation of the genes in these genomes lags far behind. Even in Arabidopsis (Arabidopsis thaliana), only approximately 40% of enzyme- and transporter-encoding genes have credible functional annotations, and this number is even lower in nonmodel plants. Functional characterization of unknown genes is a challenge, but various databases (e.g. for protein localization and coexpression) can be mined to provide clues. If homologous microbial genes exist—and about one-half the genes encoding unknown enzymes and transporters in Arabidopsis have microbial homologs—cross-kingdom comparative genomics can powerfully complement plant-based data. Multiple lines of evidence can strengthen predictions and warrant experimental characterization. In some cases, relatively quick tests in genetically tractable microbes can determine whether a prediction merits biochemical validation, which is costly and demands specialized skills.
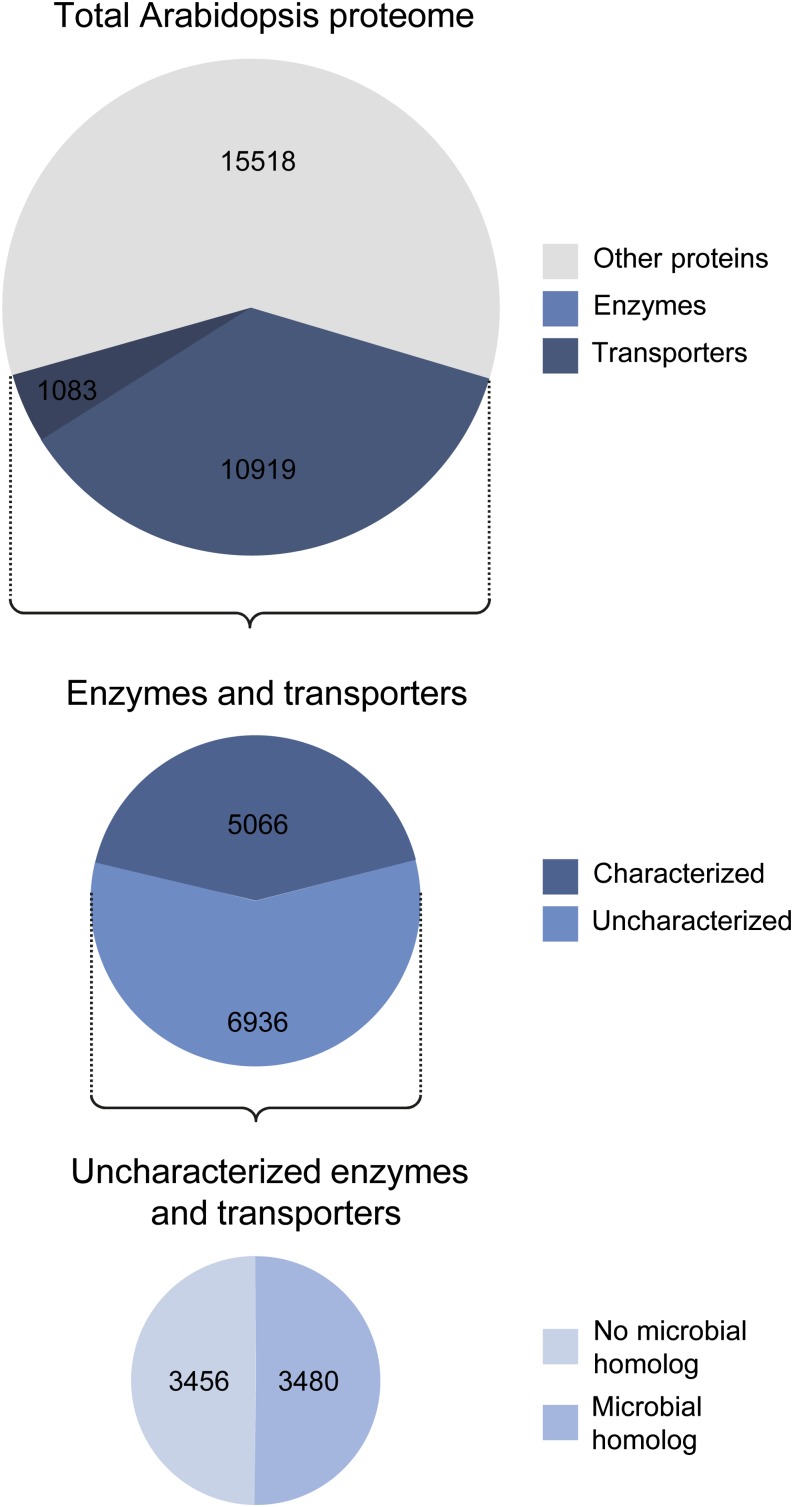
Sequencing the Arabidopsis (Arabidopsis thaliana) genome in the year 2000 (Arabidopsis Genome Initiative, 2000) has driven advances in various disciplines, including genetics, cell biology, and biochemistry. Since 2000, technological advances have dramatically reduced the time and cost of sequencing projects, resulting in a drastic increase in the number of sequenced genomes and transcriptomes. As of June 2015, almost 60,000 organisms have been sequenced, including 45,000 bacteria and 9,000 eukaryotes (www.genomesonline.org). However, functional annotation of genomes, including plant genomes, has fallen far behind the rate of genome sequencing. For example, although the Arabidopsis 2010 project sought to identify functions for all of the approximately 27,000 Arabidopsis genes by the year 2010 (Chory et al., 2000), the actual outcome of this project was far more modest. As of 2015, the functions of at least 60% of predicted Arabidopsis enzymes and transporters remain unclear or unknown (Fig. 1). If a stringent definition of function is used that includes biochemical activity, subcellular localization, and biological role based on experimental evidence, only approximately 5% of all Arabidopsis genes are characterized (Rhee and Mutwil, 2014), and this number is even lower in nonmodel plant species. The situation in microbes is not much better (Campbell et al., 2014); for instance, although approximately 80% of Escherichia coli genes have some sort of annotation (Hanson et al., 2010), experimental information is available for only 54% (Frishman, 2007). Furthermore, gene annotations are routinely propagated between genomes without experimental or genomic context-based evidence, thus misannotations are a common problem (Osterman and Overbeek, 2003; Schnoes et al., 2009). Annotations can only be confirmed by biochemical, physiological, and genetic tests, but these are expensive and time consuming and demand a high skill level (Earnshaw, 2013).
Figure 1.
Estimate of the number of Arabidopsis genes that encode uncharacterized enzymes or transporters that have good bacterial homologs. Enzymes were identified by filtering the Arabidopsis proteome (after manual removal of duplicates) for enzyme commission number and text searching for ase. The characterization status for every hundredth enzyme-encoding gene was determined using The Arabidopsis Information Resource (www.Arabidopsis.org). Repeating this subsampling approach (Politis et al., 1999) with a different set of genes gave nearly identical results. Uncharacterized enzymes were blasted against prokaryote proteins, and homologs were identified with a cutoff value of 1 × 10−15, as used previously as an appropriate threshold to detect isofunctional proteins (Woebken et al., 2007; Barchi et al., 2012). Transporters were identified from a list of Arabidopsis membrane proteins, provided by Rainer Schwacke (Forschungszentrum Jülich, Germany), by searching for terms such as transport, translocase, translocon, channel, antiporter, export, carrier, import, symport, and porin. Every 25th transporter gene was manually checked for characterization status and the occurrence of bacterial homologs as above.
With plant science funding becoming ever scarcer, it is increasingly important to leverage every possible resource to target experimental work to the most promising unknown genes. Extensive amounts of information can be obtained about a particular plant gene through online databases that enable mining of various types of plant-based data. Furthermore, about one-half of the unknown enzymes and transporters in Arabidopsis have microbial homologs (Fig. 1), and this opens the door to vast microbial databases to provide functional clues.
Well-focused use of online resources can quickly lead to solid predictions of gene function (de Crécy-Lagard and Hanson, 2007; Hanson et al., 2010; Bradbury et al., 2013). These predictions can often be tested genetically in microbes to obtain provisional evidence that they are correct before proceeding to far more costly and skill-demanding biochemical and physiological characterization. To emphasize these points, we review cases in which cross-kingdom comparative genomics led to correct prediction and subsequent validation of the function of an unidentified plant gene. We also provide a list of online databases that can be used to make predictions (Box 1) and provide a how-to guide for the powerful PubSEED database for cross-kingdom comparative genomics (Supplemental File S1).
Box 1.
A selection of online tools for identifying unknown plant enzymes.
USING CROSS-KINGDOM COMPARATIVE GENOMICS TO DISCOVER THE FUNCTION OF PLANT GENES
The following case histories illustrate how cross-kingdom comparative genomics and other data mining led to predictions of the function of unknown genes and their subsequent experimental validation. Two are drawn from B vitamin and cofactor metabolism (thiamin and riboflavin), one from chlorophyll metabolism, and one from chaperone biochemistry.
Discovering a Nudix Protein Involved in Thiamin Damage Preemption
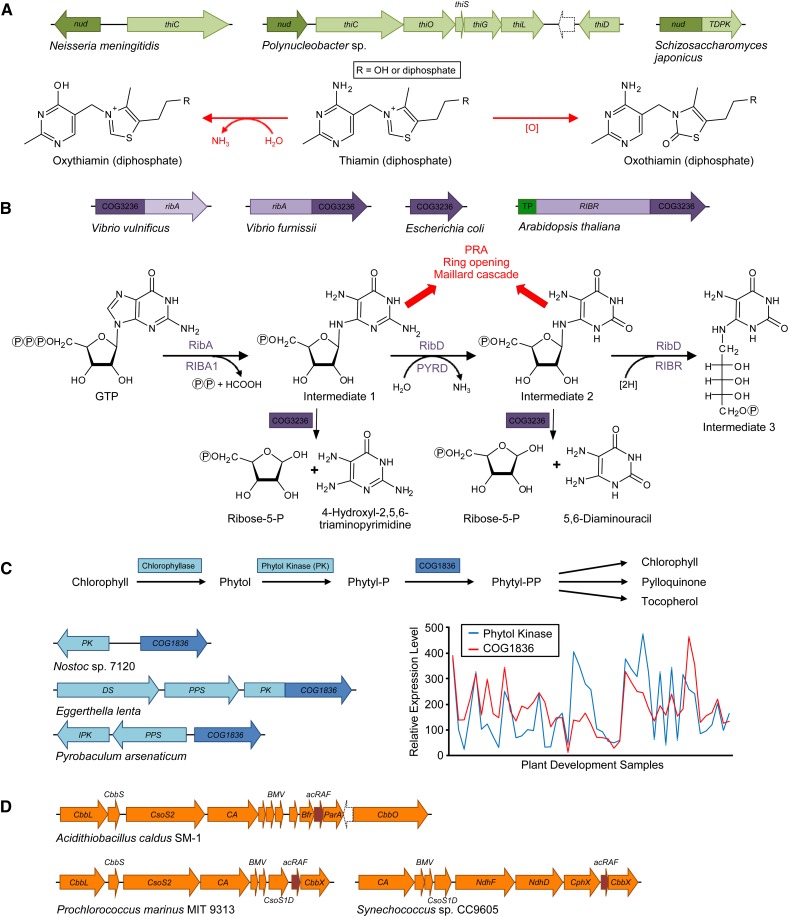
The Nudix family is a large set of proteins that typically remove a terminal phosphate from small molecules containing a diphosphate or triphosphate moiety (Bessman et al., 1996). In some fungi, a distinctive Nudix protein is fused to the enzyme thiamin diphosphokinase (TDPK; Fig. 2A), which adds a diphosphate group to thiamin (vitamin B1), yielding the active thiamin diphosphate cofactor. Orthologs of this Nudix protein occur in plants and certain bacteria and animals; some bacterial genes for these orthologs are clustered on the chromosome with various thiamin synthesis genes (Fig. 2A). Comparative genomics thus suggests that this subfamily of Nudix proteins plays a role in thiamin metabolism. In plants, the initial form of vitamin B1 synthesized is thiamin monophosphate, which must be dephosphorylated by an unidentified thiamin monophosphate phosphatase before TDPK converts it to the active cofactor. One possibility is that these Nudix proteins are the missing thiamin monophosphate phosphatase (Gerdes et al., 2012). However, the occurrence of animal orthologs suggests that these Nudix proteins are involved in thiamin metabolism rather than synthesis. Further, Nudix proteins commonly break P-O-P bonds but not C-O-P bonds, making it somewhat unlikely that the missing monophosphatase in thiamin synthesis is a Nudix protein. These inferences suggested that the thiamin-related Nudix proteins should be tested for phosphatase activity against a wide variety of thiamin-related phosphorylated compounds. Recombinant plant proteins had virtually no activity against thiamin monophosphate and only a little against thiamin diphosphate or triphosphate, but showed good activity against the thiamin damage products oxythiamine diphosphate and oxythiamin diphosphate, whose structures are shown in Figure 2A (Goyer et al., 2013). Furthermore, overexpressing an Arabidopsis protein in yeast (Saccharomyces cerevisiae) increased resistance to oxythiamin (Goyer et al., 2013). These data indicate that this Nudix subfamily is involved in removing toxic forms of thiamin diphosphate before it can cause damage by displacing the native cofactor from thiamin diphosphate-dependent enzymes, which both oxythiamin and oxothiamin diphosphate do very effectively (Tylicki et al., 2005). Note that without the cross-kingdom comparative genomics that suggested the Nudix enzymes were involved in thiamin metabolism but not synthesis, this project would probably have been jettisoned after it was discovered that these proteins had no thiamin monophosphate phosphatase activity.
Figure 2.
Clues to the function of unknown plant enzymes and transporters can come from cross-kingdom comparative genomics and plant-based data. A, The nudix (nud) gene encoding the Nudix enzyme (dark green) that preempts damage in thiamin metabolism is fused to, or clusters with, thiamin biosynthetic genes (pale green) in certain bacteria and fungi. The Nudix enzyme preferentially dephosphorylates the thiamin diphosphate analogs oxythiamin and oxothiamin diphosphate and prevents their inhibitory effects on thiamin diphosphate-utilizing enzymes. thi, Thiamin biosynthesis genes. B, The N-glycosidase COG3236 is fused to the riboflavin biosynthesis genes (lilac) ribA in some bacteria or RIBR in plants; COG3236 deglycosylates excess reactive riboflavin intermediates to fairly innocuous ribose-5-phosphate and pyrimidine moieties, preventing ring opening and Maillard reactions, which give rise to very harmful products. PRA, 5-Phosphoribosylamine; TP, targeting peptide. C, The COG1836 gene (dark blue) clusters with genes of polyprenoid metabolism (light blue) in several bacteria, and COG1836 is also fused to phytol kinase in some species. Expression of COG1836 and phytol kinase is correlated during Arabidopsis development (http://csbdb.mpimp-golm.mpg.de/csbdb/dbxp/ath/ath_xpmgq.html). DS, Phytoene desaturase; PPS, polyprenyl pyrophosphate synthetase; IPK, isopentenyl phosphate kinase. D, The pterin-4a-carbinolamine dehydratase (PCD) paralog, α-carboxysome Rubisco assembly factor (acRAF; dark orange), is associated with α-carboxysome gene clusters (orange). CbbL and CbbS, Rubisco large and small subunit; CsoS2, shell protein of unknown function; CA, carbonic anhydrase; BMV, bacterial microcompartment vertex shell proteins; Bfr, bacterioferritin family; ParA, partitioning A family; CbbO, CbbQ activase; CsoS1D, double domain shell protein; CbbX, Rubisco activase; NdhF, complex I NADH oxidoreductase chain F family protein; NdhD, complex I NADH dehydrogenase oxidoreductase M family; CphX, CO2 hydration protein.
Identifying a Riboflavin Damage Control Enzyme
Riboflavin (vitamin B2) gives rise to the cofactors FAD and FMN. The first two intermediates in riboflavin biosynthesis are reactive glycosylamines that can spontaneously break down to 5-phosphoribosylamine and Maillard products, which are highly reactive and harmful (Isbell and Frush, 1958; Foor and Brown, 1975; Fischer et al., 2004; Munanairi et al., 2007). Because riboflavin biosynthesis is not feedback regulated, intermediates 1 and 2 can potentially build up and cause harm to the cell via their decomposition (Foor and Brown, 1975; Fischer et al., 2004). Comparative genomics showed that the third enzyme of riboflavin biosynthesis in plants (RIBR) is fused to a domain of unknown function, COG3236. Strikingly, this domain also occurs as a fusion with a different enzyme of riboflavin biosynthesis (RibA) in some bacteria. In addition, COG3236 occurs as a free-standing protein in other bacteria (Fig. 2B). Furthermore, an Arabidopsis mutant lacking the COG3236 domain of RIBR shows a reduced flavin content (Ouyang et al., 2010). COG3236 is distantly related to de-ADP-ribosylation proteins that cleave N-glycosidic bonds. The fusion between COG3236 and riboflavin biosynthetic enzymes in two different kingdoms suggests that COG3236 is involved in the early riboflavin pathway. Further, based on the fact that the first two pathway intermediates are N-glycosides, it was hypothesized that COG3236 hydrolyzes these intermediates and thus prevents the harm they could cause. Biochemical characterization showed that COG3236 indeed catalyzes hydrolysis of the N-glycosidic bonds of these intermediates, yielding a relatively harmless pyrimidine moiety and ribose-5-phosphate (Fig. 2B). Deletion of COG3236 in E. coli caused a 10% to 20% reduction in flavin content, further confirming the connection between COG3236 in riboflavin biosynthesis (Frelin et al., 2015). In this case, the very demanding biochemical characterization of COG3236 could never have been justified without the extremely strong comparative genomic evidence.
Finding a Missing Phytyl-phosphate Kinase
Arabidopsis has a pathway that salvages phytol released from chlorophyll degradation (Ischebeck et al., 2006). Two kinases associated with chloroplast membranes act successively to phosphorylate phytol to phytyl-phosphate and then phytyl-diphosphate, which is a precursor to chloroplast prenyl lipids; phytol kinase had been identified, but the gene encoding phytyl-phosphate kinase was not known in plants or any other organism. A candidate membrane protein, COG1836, was discovered by comparative analysis of plant and microbial genomes (Seaver et al., 2014). Bacterial COG1836 genes cluster with genes encoding phytol kinase and other genes of polyprenyl metabolism (Fig. 2C). Further support for COG1836 as a phytyl phosphate kinase candidate comes from plant-based data: proteomics data show that Arabidopsis COG1836 localizes to the chloroplast envelope (Ferro et al., 2010), and transcriptomic data show that its expression pattern tracks that of phytol kinase (Fig. 2C). The strength of this evidence, from both cross-kingdom comparative genomics and plant-based data sets, warranted lengthy biochemical and plant genetic tests, which confirmed that Arabidopsis COG1836 indeed has phytyl-phosphate kinase activity (P. Doermann, personal communication).
Uncovering the Rubisco Chaperone Function of a PCD Paralog
PCD is involved in recycling oxidized pterin cofactors back to the reduced (tetrahydro) form and occurs in animals, protists, bacteria, and nonflowering plants. Paralogs (distant homologs) of canonical PCDs occur in bacteria and plants, including some that lack pterin-dependent enzymes. None of the PCD paralogs tested had PCD activity in genetic complementation tests (Naponelli et al., 2008) or enzyme assays (Wheatley et al., 2014). Clues to the function of these PCD paralogs in photosynthetic bacteria came from comparative genomics; the genes for these paralogs occur in clusters encoding α-carboxysomes, which are proteinaceous bacterial microcompartments for carbon fixation (Fig. 2D). Plant PCD paralogs localize to plastids (Naponelli et al., 2008), strengthening the connection to carbon fixation. The overall structure of the paralogs is similar to PCD but with major differences in the active site region. Heterologous coexpression of PCD paralogs from several photosynthetic bacteria with GroEL and Rubisco increased the amount of assembled, soluble Rubisco in E. coli, indicating a chaperone function and leading to the name α-carboxysome Rubisco assembly factor (Wheatley et al., 2014). This example again shows how valuable comparative genomics can be in making functional, and testable, hypotheses. It also emphasizes that paralogs are not necessarily just inactive copies of known enzymes but can have new and important functions of their own.
A REDEMPTIVE ROADMAP
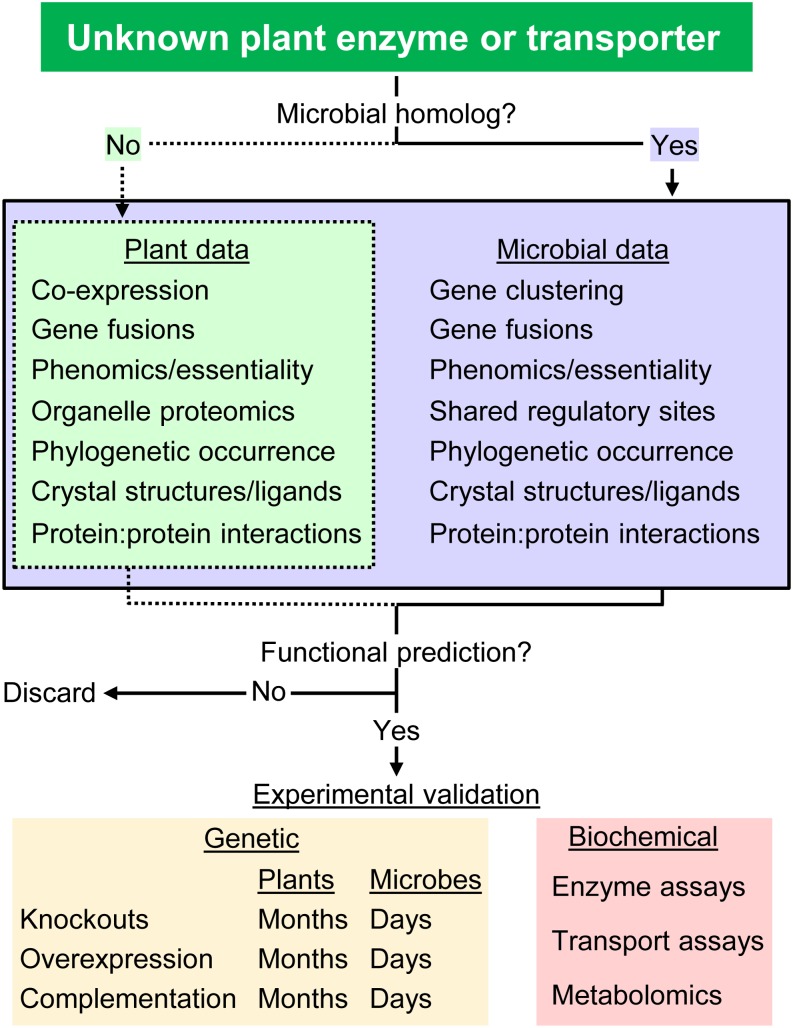
The first thing to do when beginning a scientific inquiry is to gather as much information about the topic as possible. To gain information about a particular plant gene, powerful online resources are available (Box 1), all of which collate various types of data and can provide clues to the gene’s function (Fig. 3). For plant genes that have good microbial homologs, and approximately 50% of Arabidopsis genes do, vast microbial resources can be mined for information, which greatly increases the chances of developing a functional prediction (Fig. 3). Note that, in the examples discussed above, the functional predictions were always primarily derived from microbe-based data, with plant-based data further supporting the hypothesis.
Figure 3.
A roadmap for identifying functions for uncharacterized plant genes. Various prediction methods are available for plant genes that can be complemented with microbial prediction methods if good microbial homologs exist. Functional predictions can be validated experimentally; relatively quick genetic tests in microbes may help decide if more expensive and time-consuming plant genetics and biochemical assays are worthwhile.
When microbial homologs of a plant gene exist, a good place to start a cross-kingdom comparative genomics exploration is the STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) database (Szklarczyk et al., 2015), which is intuitive, but precomputed and therefore rigid. STRING can often give useful first hints about potential functions. More in-depth analyses can be achieved by using the powerful PubSEED database (Overbeek et al., 2005). PubSEED uses the subsystem approach to genome annotation (Overbeek et al., 2005) and allows substantial user control. It contains sequenced genomes of all kingdoms and enables users to easily detect homologs and identify gene clustering patterns. A step-by-step description of how PubSEED was used to predict the function of COG1836 is given in Supplemental File S1. For Arabidopsis genes of unknown function without microbial homologs, postgenomics databases (e.g. transcriptomic, proteomic, metabolomic) can provide clues through a combination of transcriptomics, gene fusions, localization, and other data (Box 1) that lead to testable predictions (Fig. 3). The classical literature can also provide a treasure trove of useful information, and this resource is becoming more available as the older literature is digitized and text search tools become more efficient.
Testing functional predictions can sometimes be done quite quickly in microbial systems, and if warranted, can then be extended to plant systems that take much longer to yield results (Fig. 3). In the PCD paralog example above, genetic tests in microbes showed that the paralog lacked PCD activity, and subsequent coexpression of the paralog with Rubisco and GroEL in E. coli indicated that it has chaperone activity. Thus, work done in microbes confirms a link between the PCD paralog and carbon fixation and so warrants experiments to define the specific function of the paralog in plants.
Plant scientists should not be afraid to use bacterial genetics in their work. Working with microbes is often much faster and easier than working with plants. The large number of genetically tractable microbes means there are many more genetic systems available for microbes than for plants, and microbial genetics is usually much quicker and more straightforward than plant genetics. Biochemistry is almost invariably simpler, easier, and cheaper in microbes; gene cloning can be done with genomic DNA due to a lack of introns, and protein production often requires less optimization. Thus, from a practical standpoint, it is generally better to do as much as possible in microbes before moving to plants.
CONCLUSION
Despite tremendous effort in the plant science community to identify functions of all unknown plant genes, only about one-half of all Arabidopsis genes are characterized to any extent. We have highlighted the power of comparative genomics and plant omics databases to predict functions of unknown plant genes, and how quick genetic tests in microbes can be used to test these predictions. Our detailed guide on how to use PubSEED (Supplemental File S1) aims to encourage plant scientists to embrace and use comparative genomics to predict the functions of unknown plant genes.
Supplemental Data
The following supplemental materials are available.
Supplemental File S1. A tutorial showing how the PubSEED database was used to find the missing phytyl-phosphate kinase.
Supplementary Material
Acknowledgments
We thank Dr. Rainer Schwacke (Forschungszentrum Jülich, Germany) for providing a list of Arabidopsis membrane proteins and Dr. Peter Doermann (University of Bonn) for sharing data on characterizing Arabidopsis COG1836 as a phytyl-phosphate kinase.
Glossary
- PCD
pterin-4a-carbinolamine dehydratase
Footnotes
This work was supported by the National Science Foundation (grant no. MCB–1153413 to A.D.H.) and by an endowment from the C.V. Griffin Sr. Foundation.
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Barchi L, Lanteri S, Portis E, Valè G, Volante A, Pulcini L, Ciriaci T, Acciarri N, Barbierato V, Toppino L, et al. (2012) A RAD tag derived marker based eggplant linkage map and the location of QTLs determining anthocyanin pigmentation. PLoS One 7: e43740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessman MJ, Frick DN, O’Handley SF (1996) The MutT proteins or “Nudix” hydrolases, a family of versatile, widely distributed, “housecleaning” enzymes. J Biol Chem 271: 25059–25062 [DOI] [PubMed] [Google Scholar]
- Bradbury LM, Niehaus TD, Hanson AD (2013) Comparative genomics approaches to understanding and manipulating plant metabolism. Curr Opin Biotechnol 24: 278–284 [DOI] [PubMed] [Google Scholar]
- Campbell MS, Law M, Holt C, Stein JC, Moghe GD, Hufnagel DE, Lei J, Achawanantakun R, Jiao D, Lawrence CJ, et al. (2014) MAKER-P: a tool kit for the rapid creation, management, and quality control of plant genome annotations. Plant Physiol 164: 513–524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J, Ecker JR, Briggs S, Caboche M, Coruzzi GM, Cook D, Dangl J, Grant S, Guerinot ML, Henikoff S, et al. (2000) National Science Foundation-Sponsored Workshop Report: “The 2010 Project” functional genomics and the virtual plant: a blueprint for understanding how plants are built and how to improve them. Plant Physiol 123: 423–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Crécy-Lagard V, Hanson AD (2007) Finding novel metabolic genes through plant-prokaryote phylogenomics. Trends Microbiol 15: 563–570 [DOI] [PubMed] [Google Scholar]
- Earnshaw WC. (2013) Deducing protein function by forensic integrative cell biology. PLoS Biol 11: e1001742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferro M, Brugière S, Salvi D, Seigneurin-Berny D, Court M, Moyet L, Ramus C, Miras S, Mellal M, Le Gall S, et al. (2010) AT_CHLORO, a comprehensive chloroplast proteome database with subplastidial localization and curated information on envelope proteins. Mol Cell Proteomics 9: 1063–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer M, Römisch W, Saller S, Illarionov B, Richter G, Rohdich F, Eisenreich W, Bacher A (2004) Evolution of vitamin B2 biosynthesis: structural and functional similarity between pyrimidine deaminases of eubacterial and plant origin. J Biol Chem 279: 36299–36308 [DOI] [PubMed] [Google Scholar]
- Foor F, Brown GM (1975) Purification and properties of guanosine triphosphate cyclohydrolase II from Escherichia coli. J Biol Chem 250: 3545–3551 [PubMed] [Google Scholar]
- Frelin O, Huang L, Hasnain G, Jeffryes JG, Ziemak MJ, Rocca JR, Wang B, Rice J, Roje S, Yurgel SN, et al. (2015) A directed-overflow and damage-control N-glycosidase in riboflavin biosynthesis. Biochem J 466: 137–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frishman D. (2007) Protein annotation at genomic scale: the current status. Chem Rev 107: 3448–3466 [DOI] [PubMed] [Google Scholar]
- Gerdes S, Lerma-Ortiz C, Frelin O, Seaver SM, Henry CS, de Crécy-Lagard V, Hanson AD (2012) Plant B vitamin pathways and their compartmentation: a guide for the perplexed. J Exp Bot 63: 5379–5395 [DOI] [PubMed] [Google Scholar]
- Goyer A, Hasnain G, Frelin O, Ralat MA, Gregory JF III, Hanson AD (2013) A cross-kingdom Nudix enzyme that pre-empts damage in thiamin metabolism. Biochem J 454: 533–542 [DOI] [PubMed] [Google Scholar]
- Hanson AD, Pribat A, Waller JC, de Crécy-Lagard V (2010) ‘Unknown’ proteins and ‘orphan’ enzymes: the missing half of the engineering parts list--and how to find it. Biochem J 425: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isbell HS, Frush HL (1958) Mutarotation, hydrolysis, and rearrangement reactions of glycosylamines. J Org Chem 23: 1309–1319 [Google Scholar]
- Ischebeck T, Zbierzak AM, Kanwischer M, Dörmann P (2006) A salvage pathway for phytol metabolism in Arabidopsis. J Biol Chem 281: 2470–2477 [DOI] [PubMed] [Google Scholar]
- Munanairi A, O’Banion SK, Gamble R, Breuer E, Harris AW, Sandwick RK (2007) The multiple Maillard reactions of ribose and deoxyribose sugars and sugar phosphates. Carbohydr Res 342: 2575–2592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naponelli V, Noiriel A, Ziemak MJ, Beverley SM, Lye LF, Plume AM, Botella JR, Loizeau K, Ravanel S, Rébeillé F, et al. (2008) Phylogenomic and functional analysis of pterin-4a-carbinolamine dehydratase family (COG2154) proteins in plants and microorganisms. Plant Physiol 146: 1515–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterman A, Overbeek R (2003) Missing genes in metabolic pathways: a comparative genomics approach. Curr Opin Chem Biol 7: 238–251 [DOI] [PubMed] [Google Scholar]
- Ouyang M, Ma J, Zou M, Guo J, Wang L, Lu C, Zhang L (2010) The photosensitive phs1 mutant is impaired in the riboflavin biogenesis pathway. J Plant Physiol 167: 1466–1476 [DOI] [PubMed] [Google Scholar]
- Overbeek R, Begley T, Butler RM, Choudhuri JV, Chuang HY, Cohoon M, de Crécy-Lagard V, Diaz N, Disz T, Edwards R, et al. (2005) The subsystems approach to genome annotation and its use in the project to annotate 1000 genomes. Nucleic Acids Res 33: 5691–5702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Politis DN, Romano JP, Wolf M (1999) Subsampling, Ed 1 Springer Science & Business Media, New York [Google Scholar]
- Rhee SY, Mutwil M (2014) Towards revealing the functions of all genes in plants. Trends Plant Sci 19: 212–221 [DOI] [PubMed] [Google Scholar]
- Schnoes AM, Brown SD, Dodevski I, Babbitt PC (2009) Annotation error in public databases: misannotation of molecular function in enzyme superfamilies. PLoS Comput Biol 5: e1000605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seaver SMD, Gerdes S, Frelin O, Lerma-Ortiz C, Bradbury LM, Zallot R, Hasnain G, Niehaus TD, El Yacoubi B, Pasternak S, et al. (2014) High-throughput comparison, functional annotation, and metabolic modeling of plant genomes using the PlantSEED resource. Proc Natl Acad Sci USA 111: 9645–9650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, et al. (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43: D447–D452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tylicki A, Czerniecki J, Dobrzyn P, Matanowska A, Olechno A, Strumilo S (2005) Modification of thiamine pyrophosphate dependent enzyme activity by oxythiamine in Saccharomyces cerevisiae cells. Can J Microbiol 51: 833–839 [DOI] [PubMed] [Google Scholar]
- Wheatley NM, Sundberg CD, Gidaniyan SD, Cascio D, Yeates TO (2014) Structure and identification of a pterin dehydratase-like protein as a ribulose-bisphosphate carboxylase/oxygenase (RuBisCO) assembly factor in the α-carboxysome. J Biol Chem 289: 7973–7981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woebken D, Teeling H, Wecker P, Dumitriu A, Kostadinov I, Delong EF, Amann R, Glöckner FO (2007) Fosmids of novel marine Planctomycetes from the Namibian and Oregon coast upwelling systems and their cross-comparison with planctomycete genomes. ISME J 1: 419–435 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.