Antagonized functions between a heat shock and calcium-binding protein affects both preinvasion and postinvasion phases of defense responses.
Abstract
The plant immune system consists of multiple layers of responses targeting various phases of pathogen infection. Here, we provide evidence showing that two responses, one controlling stomatal closure and the other mediated by intracellular receptor proteins, can be regulated by the same proteins but in an antagonistic manner. The HEAT SHOCK COGNATE70 (HSC70), while previously known as a negative regulator of stomatal closure, is a positive regulator of immune responses mediated by the immune receptor protein SUPPRESSOR OF NPR1-1, CONSTITUTIVE1 (SNC1) as well as basal defense responses. In contrast to HSC70, a calcium-binding protein, BONZAI1 (BON1), promotes abscisic acid- and pathogen-triggered stomatal closure in addition to and independent of its previously known negative role in SNC1 regulation. BON1 likely regulates stomatal closure through activating SUPPESSOR OF THE G2 ALLELE OF SKP1 VARIANT B and inhibiting HSC70. New functions of BON1 and HSC70 identified in this study thus reveal opposite effects of each of them on immunity. The opposing roles of these regulators at different phases of plant immune responses exemplify the complexity in immunity regulation and suggest that immune receptors may guard positive regulators functioning at stomatal closure control.
The plant immune system consists of multiple layers of recognition that target different phases of pathogen infection. The two major pathogen recognition and defense-signaling branches are pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI; Dangl and Jones, 2001; Chisholm et al., 2006; Jones and Dangl, 2006; Dodds and Rathjen, 2010). PAMPs such as flagellin and elongation factor (EF)-Tu are recognized by pattern recognition receptors such as FLAGELLIN-SENSITIVE2 (FLS2) and EF-TU RECEPTOR (Monaghan and Zipfel, 2012) at the plasma membrane to initiate PTI. The short N-terminal peptides of flagellin and EF-Tu, named flg22 and elf18, respectively, are sufficient to trigger PTI (Felix et al., 1999; Kunze et al., 2004). ETI is engaged following the recognition of microbial effectors via plant intracellular immune receptors that are mostly nucleotide-binding leucine-rich repeat (NB-LRR) proteins (Chisholm et al., 2006; Jones and Dangl, 2006). Activation of NB-LRR proteins often leads to rapid and effective defense responses, including programed cell death, to restrict the growth of biotrophic pathogens.
Closure of stomata is one of the responses activated following PAMP recognition to prevent pathogen entry into plant cells (Melotto et al., 2006; Xin and He, 2013). As a gateway for water vapor and CO2 exchange between the mesophyll cells and the atmosphere, stomata pore is finely controlled in its aperture to maximize photosynthesis while preventing water loss. Stress hormone abscisic acid (ABA) and CO2 can each be perceived by their receptors and activate signaling pathways involving kinases/phosphatases and secondary messengers to modify ion channel activities that change the aperture of the stomatal pores (Kim et al., 2010b). Stomata is also the battle ground between plants and pathogens, as plants prevent entry of foliar pathogens by closing the gate upon PAMP perception, while pathogens use different strategies to open the stomata for their entries (Melotto et al., 2006). While ABA is largely responsible for abiotic stress-induced stomatal closure, the oxylipin pathway is thought to mediate biotic stress-induced closure (Montillet and Hirt, 2013). Downstream signaling events in response to biotic and abiotic signals share common components, including reactive oxygen species (ROS), calcium, and nitric oxide (Montillet et al., 2013; Sawinski et al., 2013). Protein kinases, including calcium-dependent protein kinases and mitogen-activated protein kinases, presumably transduce these signals and regulate activities of ion channels and transporters to control stomata opening (Sawinski et al., 2013).
Chaperone or cochaperone proteins such as SUPPESSOR OF THE G2 ALLELE OF SKP1 VARIANT B (SGT1b) and HEAT SHOCK PROTEIN70 (HSP70) are important regulators of plant immunity. SGT1b is part of the Skp1/Cullin/F-box (SCF) ubiquitin ligase complex that targets protein for degradation. It has multiple substrates and regulates multiple processes such as development, defense responses, and abiotic stress responses (Austin et al., 2002; Azevedo et al., 2002; Gray et al., 2003; Noël et al., 2007). SGT1b has opposing roles on NB-LRR protein regulation. On the one hand, it is required for multiple NB-LRRs to mediate defense responses, likely by assisting their proper folding and/or positively regulating their protein accumulation (Austin et al., 2002; Peart et al., 2002; Hubert et al., 2003; Leister et al., 2005; Azevedo et al., 2006). On the other hand, the SGT1b-SCF complex is implicated in coupling NB-LRR proteins to cellular degradation machinery and therefore inhibits defense responses (Liu et al., 2002; Holt et al., 2005).
HSP70 proteins are induced by a rapid temperature rise. They are generally involved in protein folding and degradation of unfolded proteins (Hartl, 1996; Park et al., 2007). Arabidopsis (Arabidopsis thaliana) has 14 HSP70s, including five cytosolic HSP70s (named HSC70s), three endoplasmic reticulum (ER)-localized luminal binding proteins (BiPs), two plastid HSP70s, and two mitochondrial HSP70s (Lin et al., 2001; Sung et al., 2001). The cytosolic HSC70 proteins are important for tolerance to abiotic stress including heat (Sung and Guy, 2003; Cazalé et al., 2009). They were recently implicated in regulating plant immunity as well, but their reported functions are contradictory from different studies. In one study, HSC70s are shown to physically interact with SGT1b, and overexpression of HSC70.1 confers susceptibility to the virulent oomycete pathogens Hyaloperonospora arabidopsidis as well as virulent and avirulent strains of Pseudomonas syringae pv tomato (Pst) DC3000 (Noël et al., 2007). This study indicates that HSC70s are negative regulators of basal resistance and NB-LRR-mediated resistance. In another study, HSC70s, especially HSC70.1 and HSC70.3, are shown to be targeted by the P. syringae effector protein HopI1, and a hsc70.1 transfer DNA (T-DNA) insertion mutant exhibits enhanced susceptibility to virulent P. syringae pv maculicola (Psm) ES4326 and type III secretion-deficient Pst DC3000 hrcC– (Jelenska et al., 2010). This study indicates that HSC70s have a positive role in plant basal defense responses. The reason for the apparent contradiction regarding the role of HSC70 in plant immunity is not known. Varying effects of HopI1 or different functions of HSC70 under different environments are suspected to contribute to varying outcomes.
The chaperone and cochaperone proteins were recently shown to be involved in stomatal control (Clément et al., 2011). HSC70.1 overexpression lines and an sgt1b mutant have delayed stomatal closure compared with the wild type under several environmental inductions. The clients of HSC70 or SGT1b in stomata regulation are not known, but they likely regulate shared components between biotic and abiotic responses.
BON1 (BONZAI1) is an intriguing regulator of NB-LRRs. It encodes a calcium-dependent phospholipid-binding copine protein that is conserved in protozoa, plants, nematodes, and mammals (Hua et al., 2001). Copine proteins in diverse species studied so far are involved in signaling, although no common processes can be readily deduced (Li et al., 2010a). In Arabidopsis, the loss-of-function (LOF) mutant bon1-1 (hereafter, bon1) has a dwarf phenotype due to constitutive defense responses triggered by the up-regulation of the NB-LRR gene SUPPRESSOR OF NPR1-1, CONSTITUTIVE1 (SNC1; Hua et al., 2001; Yang and Hua, 2004). Mutants with constitutive immune responses such as bon1 are referred to as autoimmune mutants and some of them are due to activation of NB-LRR proteins (Gou and Hua, 2012). SNC1 is highly similar to RECOGNITION OF PERONOSPORA PARASITICA4 that confers resistance to pathovars of the oomycete pathogen H. arabidopsidis (Noël et al., 1999; van der Biezen et al., 2002). The SNC1 protein is under a complex negative regulation (Gou and Hua, 2012). Although the pathogen effector that SNC1 recognizes remains elusive, SNC1 could be a minor resistance protein for the effector AvrRps4 (Kim et al., 2010a). A gain-of-function mutant of SNC1, snc1, has an autoimmune phenotype (Li et al., 2001; Zhang et al., 2003b). The BON1 gene belongs to a three-member gene family in Arabidopsis, and the knockout of all three genes (BON1, BON2, and BON3) results in lethality (Yang et al., 2006b). Genetic dissection of a differential phenotype of the bon1 bon3 double mutants in two accession backgrounds revealed that multiple NB-LRR genes are responsible for the lethality phenotype (Li et al., 2009). Therefore, BON1 is a negative regulator of multiple NB-LRR proteins in addition to the SNC1.
The BON1 protein is localized to the plasma membrane through N-terminal myristoylation; and its calcium-binding property, though not responsible for its localization, is essential for its function (Li et al., 2010a). How BON1 at the plasma membrane regulates the transcript level of the NB-LRR-coding gene SNC1 is not fully understood. Genetic studies suggest that BON1 modulates SNC1 transcript through chromatin remodeling factors HISTONE MONOUBIQUITINATION1 (HUB1) and HUB2 as well as MODIFIER OF SNC1 1 (Li et al., 2010c). It is thus likely that signaling regulated by BON1 at the plasma membrane influences expression level of SNC1. From yeast (Saccharomyces cerevisiae) two-hybrid screens, two homologous calcium-binding proteins BON1-ASSOCIATED PROTEIN1 (BAP1) and BAP2 were found to interact with the full-length BON1 and specifically the von Willebrand A domain of BON1, and these two proteins function similarly to the BON proteins in the regulation of immunity (Hua et al., 2001; Yang et al., 2006a, 2007). Another yeast two-hybrid screen identified BAK1-INTERACTING RECEPTOR-LIKE KINASE1 (BIR1) as an additional BON1 interactor, and BIR1 is also a negative regulator of defense responses (Wang et al., 2011).
To further elucidate the role of BON1 in immunity, we immunoprecipitated (IPed) BON1 and identified associated proteins by liquid chromatography (LC)-mass spectrometry (MS). Using bimolecular fluorescence complementation (BiFC) and yeast two-hybrid assays, we found that HSC70 and SGT1b are potentially BON1-interacting proteins. With the generation of a double mutant with reduced function of HSC70.1 and HSC70.3, we show that BON1, HSC70, and SGT1b have roles in NB-LRR-mediated defense responses as well as stomatal closure regulation. The opposing effects on two layers of defense responses suggest complex regulation of plant immunity as well as connection between two branches of immune responses.
RESULTS
BON1 Associates with HSC70 and SGT1b Proteins
We searched for BON1-associated proteins by immunoprecipitation (IP) of hemagglutinin (HA)-tagged BON1 protein followed by LC-MS analysis of co-IPed proteins. This BON1-HA was expressed under the strong Cauliflower mosaic virus (CaMV) 35S promoter, and it is functional, as it rescued the bon1 defect (Supplemental Fig. S1). A putative BON1-HA protein complex was stabilized by the crosslinker dithiobis(succinimidyl propionate) and then IPed with anti-HA antibodies (Supplemental Fig. S2A). The putative BON1-HA protein complex was visible after being separated on the SDS-PAGE gel after Coomassie Blue staining (Supplemental Fig. S2B). LC-MS identified three cytosolic HSC70 proteins in the top-ranking BON1-associated proteins (Supplemental Fig. S2C). We designated them as HSC70.1 (AT5G02500), HSC70.2 (AT5G02490), and HSC70.3 (AT3G09440), respectively, instead of their previous names HSC70-1, HSC70-2, and HSC70-3 (Lin et al., 2001; Noël et al., 2007), to follow the conventional usage of a dash to indicate mutant alleles.
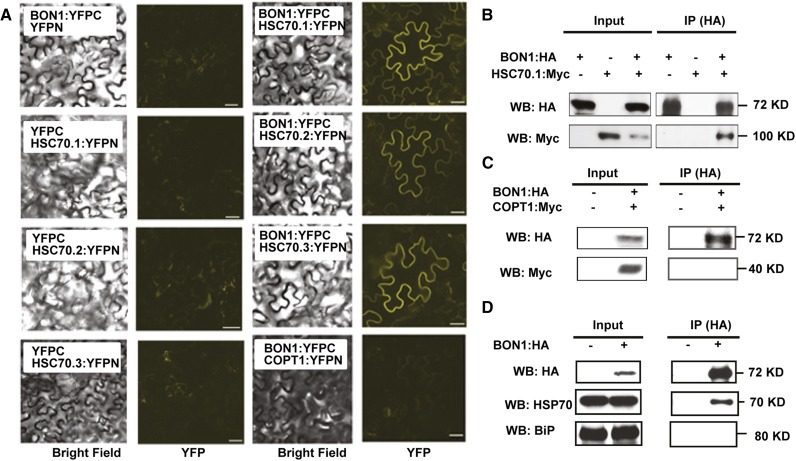
The association between BON1 and the three HSC70 proteins were subsequently tested using BiFC and co-IP methods (Schütze et al., 2009). For BiFC assays, the BON1 protein was fused to the HA tag as well as the C-terminal half of yellow fluorescence protein (YFP). For simplicity, HA:YFP-C terminus will be referred to as YFPC in the BiFC assay and as HA in the co-IP assay. The three HSC70 proteins were each fused at their C termini with a Myc tag and the N-terminal half of YFP (YFP N terminus). Again, the Myc:YFPN tag will be referred to as YFPN in the BiFC assay and as Myc in the co-IP assay. When BON1:YFPC and any one of the three HSC70:YFPN were coexpressed in Nicotiana benthamiana, YFP signals were detected (Fig. 1A). No signal was observed when BON1:YFPC or HSC70:YFPN were coexpressed with the YFPN or YFPC controls (Fig. 1A). Because BON1 is a plasma membrane-localized protein (Hua et al., 2001), proteins that are directly or indirectly attached to the plasma membrane could potentially be isolated as false-positive BON1-interacting proteins. We therefore coexpressed the YFPN fusions of the plasma membrane protein COPPER TRANSPORTER1 (COPT1) with BON1:YFPC in N. benthamiana as an additional control. While both BON1 and COPT1 proteins were expressed (Fig. 1C), no BiFC signal was detected (Fig. 1A), indicating that the BiFC signals from BON1 and HSC70 are unlikely due to physical closeness of BON1 with any proteins on the plasma membrane.
Figure 1.
Characterization of BON1 interaction with HSC70 proteins. A, BiFC assay for BON1 and HSC70 interaction. Shown are confocal images (bright field and YFP fluorescence) of N. benthamiana leaves coinfiltrated with BON1:YFPC and HSC70.1(2,3):YFPN constructs. YFPN, YFPC, or COPT1-YFPN coinfiltrated with BON1 or HSC70 constructs were used as negative controls. B, Co-IP assay for BON1 and HSC70.1 interaction in N. benthamiana leaves. Crude lysates (Input) were IPed with anti-HA antibody and then detected with anti-HA and anti-Myc antibodies for BON1:HA and HSC70.1:Myc, respectively. C, Co-IP assay for BON1 and COPT1 in N. benthamiana leaves. Blot is displayed as in B. D, Co-IP assay for BON1 and HSC70 using Arabidopsis BON1:HA/bon1 (indicated by +) transgenic plants with the wild type (indicated by –) as control. Crude lysates (Input) were IPed with anti-HA antibody and then detected with anti-HSC70 and anti-BiP antibodies for cytosolic and ER HSP70 proteins, respectively. WB, Western blot. Bar = 50 µm.
In parallel, we IPed BON1:HA using an anti-HA antibody and could detect signals of HSC70.1:Myc from coinfiltrated leaves (Fig. 1B). In contrast, no HSC70.1:Myc signal could be detected from the control sample coexpressed with the HA vector (Fig. 1B). As a control, the Myc-tagged membrane protein COPT1 could not be co-IPed with BON1:HA, indicating the specificity of BON1:HA in the co-IP assay (Fig. 1C). Therefore, BON1 specifically associates with HSC70 proteins in plants.
We further determined the interaction between BON1 and HSC70 by co-IP in BON1-HA/bon1 transgenic plants. BON1-HA was IPed by the anti-HA antibody from total protein extract without the crosslinker, and associated proteins were separated on SDS-PAGE for western blot. Positive signals could be detected by anti-HSC70 antibodies on the blot in the BON1-HA co-IP sample but not the control wild-type ecotype Columbia-0 (Col-0) sample (Fig. 1D). Furthermore, the ER-localized HSP70 family protein BiP could not be detected by the anti-BiP antibody in the BON1-HA co-IP sample (Fig. 1D). These results indicate that BON1 is associated with cytosolic-localized HSC70 proteins but not all HSP70 family proteins.
However, we did not detect a positive interaction between HSC70 and BON1 in a GAL4-based yeast two-hybrid assay. Full-length BON1 or the C-terminal von Willebrand A domain of BON1 was fused to the GAL4 transcription activation domain (AD), while HSC70.1, HSC70.2, and HSC70.3 were fused with the GAL4 DNA-binding domain (BD). Western-blot analysis indicated that all proteins were expressed at comparable levels in yeasts cotransformed with these constructs (Supplemental Fig. S3). No growth of yeast containing BON1 and HSC70 constructs was observed on the same selection media (Supplemental Fig. S3), while yeasts harboring BON1 and BAP1 constructs grew as previously reported (Hua et al., 2001). This data suggests that BON1 may not have a direct physical interaction with HSC70 proteins, although the possibility of direct interaction could not be excluded.
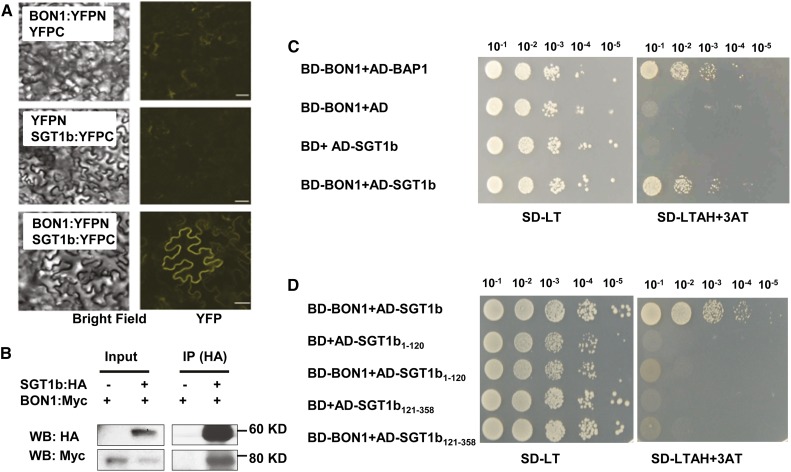
Because HSC70 is known to associate with SGT1b (Noël et al., 2007), we tested if BON1 and SGT1b can interact. Coexpression of BON1:YFPN and SGT1b:YFPC in N. benthamiana leaves led to a strong YFP signal, while coexpression of BON1:YFPN with YFPC or YFPN with SGT1b:YFPC did not (Fig. 2A). In addition, when the SGT1b:HA protein was IPed with anti-HA antibodies, BON1:Myc was detected from coinfiltrated leaves, while no signal could be detected in leaves coinfiltrated with BON1:HA and the Myc vector (Fig. 2B). The association between BON1 and SGT1b was further tested by the yeast two-hybrid assay. Yeast coexpressing BON1-BD and SGT1b-AD fusions grew on the selection medium, while yeast coexpressing BON1-AD and BD or AD and SGT1b-BD did not (Fig. 2C). Growth from coexpression of BON1 and SGT1b was at a similar extent as that of BON1 and BAP1, a previously identified BON1 interactor (Fig. 2C). These results suggest that BON1 and SGT1b proteins potentially could have a direct interaction. We subsequently assayed the interaction of BON1 with the TPR domain (amino acids 1–120) and the CS-SGS domain (amino acids 121–358), which is necessary and sufficient for SGT1b to interact with HSC70 (Azevedo et al., 2002; Li et al., 2010b). Neither of these two domains exhibited positive interaction with BON1 in the yeast two-hybrid assay (Fig. 2D), suggesting that a full-length SGT1b is required for its interaction with BON1.
Figure 2.
Characterization of BON1 interaction with SGT1b. A, BiFC assay for BON1 and SGT1b interaction. Experiments were performed similarly as in Figure 1A. B, Co-IP assay for BON1 and SGT1b interaction. Blot is displayed similarly as in Figure 1B. C to D, Yeast two-hybrid assay for BON1 and full-length SGT1b (C) and truncated SGT1b proteins (D). Yeasts were grown on synthetic dextrose (SD) plates without Leu or Trp (SD-LT) and synthetic dextrose plates without Leu, Trp, adenine, or His but with 3 mm 3-amino-1,2,4-triazole (SD-LTAH+3AT). BD-BON1 and AD-BAP1 were cotransformed as a positive control, while BD-BON1 and AD empty vector or BD empty vector and AD-SGT1b were cotransformed as negative controls. SGT1b1–120 and SGT1b121–358 denote the AD fusions of the SGT1b domains of amino acids 1 to 120 and amino acids 121 to 358, respectively. Shown is yeast growth with a serial dilution of 10–1 to 10–5 at 3 d after being spotted onto the synthetic dextrose plates. WB, Western blot. Bar = 50 µm.
The Double Mutations in HSC70.1 and HSC70.3 Partially Rescued the bon1 Growth Defects
The interaction among BON1, HSC70, and SGT1b prompted us to look at the functional involvement of HSC70 and SGT1b in bon1-triggered autoimmune responses. Because HSC70 overexpression was reported to have reduced disease resistance (Noël et al., 2007), we tested whether HSC70 overexpression can suppress the autoimmune responses in bon1. The HSC70.1 and HSC70.3 were each overexpressed by the strong CaMV 35S promoter in bon1, and 18 and 16 transgenic lines were obtained, respectively. None of the transgenic lines had reduced autoimmune phenotype compared with bon1 (Supplemental Fig. S4A). When detected with an antibody against cytosolic HSC70 proteins, a slight increase (1.1- to 1.4-fold) of total HSC70 proteins was observed in four HSC70.1 transgenic lines analyzed (Supplemental Fig. S4B). We were not able to assess expression levels of HSC70.1 due to the lack of specific antibodies, but we reason that there is a moderate increase of HSC70.1 in some transgenic lines, but that increase does not inhibit the bon1 phenotype.
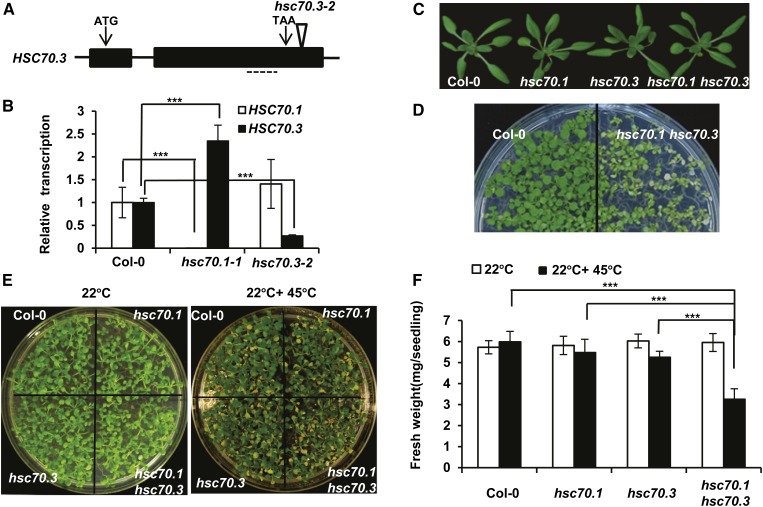
We subsequently analyzed the effect of loss of the HSC70 function on the bon1 phenotypes. T-DNA insertion mutants were isolated from the Salk collection (Alonso et al., 2003) for each of the three HSC70 genes, namely hsc70.1-1 (SALK_135531C, referred to as hsc70.1), hsc70.2-1 (SALK_085076C, referred to as hsc70.2), and hsc70.3-2 (SALK_148168, referred to as hsc70.3). Both hsc70.1 and hsc70.2 mutants were reported to be LOF mutants earlier (Noël et al., 2007). The hsc70.3 mutant allele was not characterized previously, and it has a T-DNA inserted in the 3′ untranslated region of the gene (Fig. 3A). The transcription levels of HSC70.1 and HSC70.3 in the hsc70.1 and hsc70.3 mutants were analyzed by quantitative real-time reverse transcription (qRT)-PCR. Consistent with previous reports, HSC70.1 expression was greatly decreased in hsc70.1 mutant (Noël et al., 2007). HSC70.3 expression was reduced to about 20% of the wild type in the hsc70.3 mutant (Fig. 3B), indicating that the hsc70.3 mutant is a knockdown but not a null mutant. It is also noted that HSC70.3 was significantly up-regulated in hsc70.1 (Fig. 3B), suggesting that HSC70.3 is induced perhaps to compensate for the reduction of HSC70.1 function.
Figure 3.
Characterization of the hsc70 mutants. A, Diagram of the T-DNA insertion mutant of the HSC70.3 gene. Arrows point to the translation start (ATG) and stop (TAA) codons. The triangle indicates the site of T-DNA insertion in the SALK_148168 (hsc70.3-2) mutant. Dashed lines mark the regions for qRT-PCR analysis of HSC70.3 expression. B, qRT-PCR analysis of HSC70 transcripts. HSC70.1 and HSC70.3 expression levels were assayed in the Col-0 wild type, hsc70.1-1 (referred to as hsc70.1 afterward), and hsc70.3-2 (referred to as hsc70.3 afterward) plants at 3 weeks old. Error bars represent sds (Student’s t test; ***, P < 0.001). C, Rosette leaf phenotype of hsc70.1, hsc70.3, and hsc70.1 hsc70.3 mutants compared with the wild-type Col-0 before bolting. D, Heat stress sensitivity of the hsc70 double mutants. Shown are seedlings of the wild-type Col-0 and hsc70.1 hsc70.3 after a 45°C heat shock for 20 min followed by 5-d recovery at 22°C. E, Heat stress sensitivity of the hsc70 mutants. Shown are seedlings of the wild-type Col-0, hsc70.1, hsc70.3, and hsc70.1 hsc70.3 without (left) or with (right) a 45°C heat shock for 20 min followed by 5-d recovery at 22°C. F, Fresh weights of Arabidopsis seedlings in E. The average fresh weight was calculated from seven seedlings, and error bars represent sds (Student’s t test; ***, P < 0.001).
Genetic redundancy has been reported for the HSC70 family members (Sung and Guy, 2003; Noël et al., 2007). Single mutants of individual members did not exhibit an obvious mutant phenotype, while RNA silencing of the gene family caused embryo lethality. To reduce genetic redundancy and compensation, we generated double mutants between the three HSC70 genes by crossing among the single mutants. As HSC70.1 and HSC70.2 are next to each other on the chromosome, we were not able to obtain the hsc70.1 hsc70.2 double mutant. A double null mutant of HSC70.1 and HSC70.3 genes was reported to be lethal (Noël et al., 2007), but we were able to obtain the hsc70.1 hsc70.3 double mutant, likely because the hsc70.3 allele used is a reduction-of-function allele but not a null allele.
The hsc70 single and double mutants exhibited wild-type growth phenotypes under standard growth conditions (Fig. 3C). However, the double mutant is less heat tolerant than the single mutants or the wild type. We initially noticed a drastically reduced heat tolerance in the hsc70.1 hsc70.3 double mutant compared with the wild type when very young seedlings were exposed to 45°C for 20 min (Fig. 3D). We subsequently quantified heat tolerance of single and double mutants in 10-d-old seedlings using 20 min of 45°C treatment. After recovery at 22°C for 5 d, the hsc70.1 hsc70.3 mutant plants had slightly more chlorosis than the wild-type plants or single mutants (Fig. 3E). Although the chlorosis phenotype was subtle, quantification of fresh weight revealed that the double mutant had a 40% biomass reduction with heat treatment compared with nontreatment, while the wild type and the single mutants had no or less than 10% reduction with heat treatment. Therefore, the hsc70.1 hsc70.3 double mutant we generated is more reduced in HSC70 function than the single mutants (Fig. 3F).
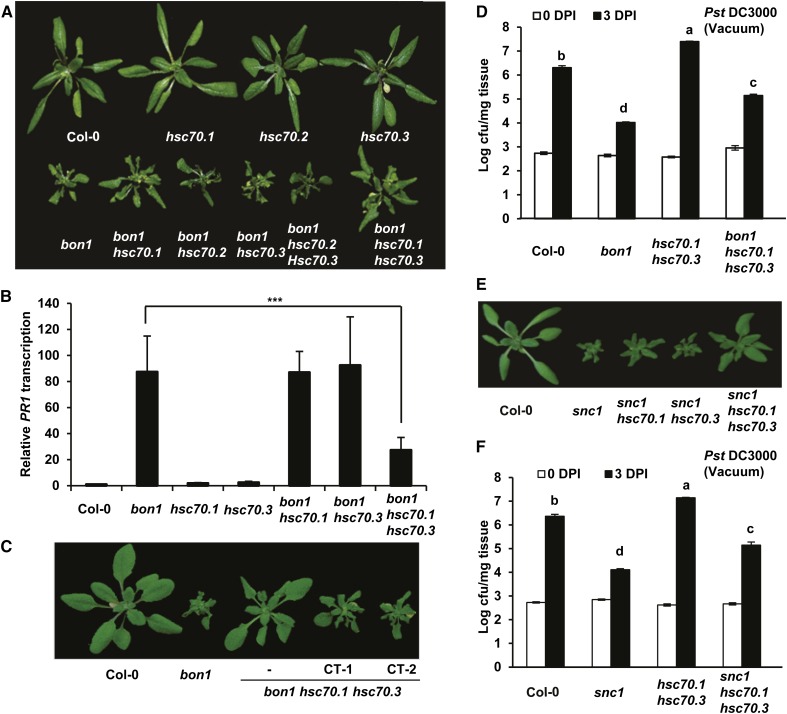
We then investigated the effect of loss of HSC70 function on the bon1 phenotype. Double mutants were constructed between bon1 and the three hsc70 single mutants. While neither bon1 hsc70.2 nor bon1 hsc70.3 had any morphological differences from the bon1 single mutant, bon1 hsc70.1 showed a milder growth defect than that of bon1 (Fig. 4A; Supplemental Fig. S5). We further generated two triple mutants bon1 hsc70.1 hsc70.2 and bon1 hsc70.1 hsc70.3. Plants of bon1 hsc70.1 hsc70.3 but not bon1 hsc70.2 hsc70.3 exhibited an even milder growth defect than the bon1 hsc70.1 plant (Fig. 4A; Supplemental Fig. S5), which was verified by biomass quantification of seedlings (Supplemental Fig. S6A). Using complementation assay, we confirmed that the inhibition of bon1 growth defect is due to the hsc70 mutations. When a wild-type HSC70.1 genomic fragment was transformed into bon1 hsc70.1 hsc70.3, 12 out of the 13 T1 transgenic plants showed a bon1-like phenotype (Fig. 4C). Therefore, the growth defect of the bon1 mutant is partially rescued by the loss of HSC70.1 function and further rescued by additional reduction of HSC70.3 function.
Figure 4.
Mutations in HSC70.1 and HSC70.3 partially suppress the bon1 and snc1 phenotypes. A, Partial rescue of the bon1 growth phenotype by the hsc70 mutations. Shown are 40-d-old plants of Col-0, hsc70.1, hsc70.2, hsc70.3, bon1, bon1 hsc70.1, bon1 hsc70.2, bon1 hsc70.3, bon1 hsc70.2 hsc70.3, and bon1 hsc70.1 hsc70.3. B, Down-regulation of PR1 gene expression in bon1 by hsc70 mutations. Shown are relative expression levels of the PR1 gene in Col-0, bon1, hsc70.1, hsc70.3, bon1 hsc70.1, bon1 hsc70.3, and bon1 hsc70.1 hsc70.3 assayed by qRT-PCR. Error bars represent sds (Student’s t test; ***, P < 0.001). C, Complementation test of bon1 hsc70.1 hsc70.3 mutant. Shown are Col-0, bon1, bon1 hsc70.1 hsc70.3, and two independent complementation transgenic (CT) T2 lines (CT-1 and CT-2) of bon1 hsc70.1 hsc70.3 transformed with the genomic fragment of HSC70.1 in the vector pMDC99 (pMDC99:HSC70.1). D, Partial suppression of resistance against the virulent bacterial strain Pst DC3000 in bon1 by the hsc70 mutations. Shown is the growth of bacteria as log value of cfu per milligram tissue in Col-0, bon1, hsc70.1 hsc70.3, and bon1 hsc70.1 hsc70.3 via vacuum inoculation at 0 and 3 DPI. Values represent averages of three biological repeats, and error bars represent sds. Letters indicate statistical difference (P < 0.001; Bonferonni posttest) of different genotypes. E, Partial rescue of the snc1 growth defect by the hsc70 mutations. Shown are wild-type Col-0, snc1, snc1 hsc70.1, snc1 hsc70.3, and snc1 hsc70.1 hsc70.3 plants before bolting. F, Partial suppression of enhanced resistance against virulent bacterial pathogen Pst DC3000 in snc1 by the hsc70 mutations. Shown are bacterial growths in Col-0, snc1, hsc70.1 hsc70.3, and snc1 hsc70.1 hsc70.3 plants at 0 and 3 DPI. Letters indicate statistical difference (P < 0.001; Bonferonni posttest) of different genotypes.
The Loss of HSC70.1 and HSC70.3 Function Compromises Immune Responses in bon1
The suppression of growth defect by the hsc70 mutations is associated with a reduced disease resistance in bon1. In bon1, salicylic acid-mediated defense responses are constitutively turned on, resulting in the highly expressed defense response marker gene PATHOGENESIS-RELATED GENE1 (PR1; Fig. 4B; Yang and Hua, 2004. In the bon1 hsc70.1 hsc70.3 triple mutant, but not the bon1 hsc70.1 or bon1 hsc70.3 double mutants, this elevated expression of PR1 in bon1 was greatly reduced, as detected by qRT-PCR (Fig. 4B). Furthermore, enhanced resistance to the virulent bacterial pathogen Pst DC3000 in bon1 was reduced by the hsc70 mutations: bacteria grew to a greater extent in bon1 hsc70.1 hsc70.3 than in bon1 (Fig. 4D).
The reduction of HSC70.1 and HSC70.3 function also inhibited the growth and defense defects in the autoactive NB-LRR SNC1 mutant snc1 (Li et al., 2001; Zhang et al., 2003b). The snc1 growth defect was significantly reduced by the hsc70.1 hsc70.3 double mutation, as supported by the biomass quantification (Fig. 4E; Supplemental Fig. S6B). The enhanced resistance to Pst DC3000 in snc1 was also reduced by mutations of HSC70.1 and HSC70.3 (Fig. 4F). Therefore, the inhibition of the bon1 autoimmune phenotype by the reduction of HSC70 activity could result from the inhibition of SNC1 activity by the loss of HSC70 activity.
Interestingly, the hsc70.1 hsc70.3 mutant displayed enhanced susceptibility to the virulent pathogen Pst DC3000 (Fig. 4D). This phenotype was unexpected because overexpression of HSC70.1 was shown previously to compromise resistance to both virulent and avirulent bacterial pathogens of Pst DC3000 (Noël et al., 2007). We analyzed disease resistance phenotypes in the hsc70.1 hsc70.3 double mutant we generated, as it has a stronger reduction of the HSC70 family than the single mutants. We monitored the growth of additional Pst strains on the hsc70.1 hsc70.3 double mutant, as single hsc70.1 and hsc70.3 mutants did not show changes in disease resistance to the virulent bacterial strain Pst DC3000 (Noël et al., 2007). At 3 d post inoculation (DPI), the mutant supported more bacterial growth for the type III secretion-deficient strain Pst DC3000 hrcU– compared with the wild type (Fig. 5A). This indicates that basal defense is compromised in the hsc70.1 hsc70.3 double mutant plants.
Figure 5.
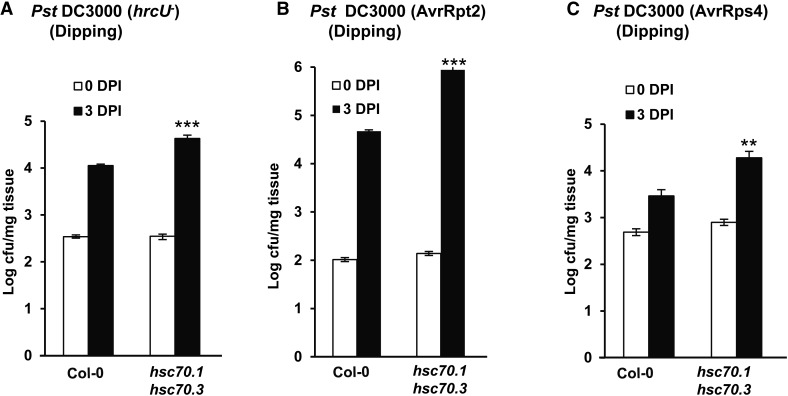
Enhanced susceptibility of the hsc70.1 hsc70.3 mutant to type III secretion-deficient and avirulent bacterial pathogens. Shown are growth of pathogen strains in the wild-type Col-0 plant and the hsc70.1 hsc70.3 double mutant at 0 and 3 DPI. Strains are type III secretion-deficient strain Pst DC3000 hrcU– (A), avirulent strain Pst DC3000 AvrRpt2 (B), and avirulent strain Pst DC3000 AvrRps4 (C). Values represent averages of three biological repeats, and error bars represent sds. A star indicates a statistical difference from the wild type at 3 DPI (Student’s t test; **, P < 0.01; and ***, P < 0.001).
The hsc70.1 hsc70.3 double mutant is also compromised in resistance conferred by NB-LRR proteins RESISTANT TO P. SYRINGAE2 (RPS2) and RPS4. When dip inoculated with the avirulent strains Pst DC3000 AvrRpt2 and Pst DC3000 AvrRps4, hsc70.1 hsc70.3 supported more bacterial growth than the wild-type Col-0 (Fig. 5, B and C). Therefore, immune responses conferred by NB-LRR proteins are compromised by the reduction of HSC70 function. Considering the dipping infection method used here, it is yet to be determined whether the positive role of HSC70s in ETI is solely due to its positive role in PTI.
Opposite Roles of HSC70 in Pre- and Postinvasion Phases of Immune Responses
Compromised basal and NB-LRR-mediated resistance was observed in the hsc70.1 hsc70.3 double mutant (this study) as well as in the HSC70.1 overexpression line (Noël et al., 2007). One scenario to explain the apparent discrepancy is that HSC70 might have opposite roles in two layers of defense: it inhibits immune responses at the stomatal closure phase, while it positively regulates immune responses after pathogen invasion. We tested this hypothesis by comparing pathogen growth in the hsc70.1 hsc70.3 double mutant from two inoculation methods: dipping and vacuum. With the dipping method, plants presumably mount both pre- and postinvasion defense responses, while with the vacuum method, only the postinvasion defense mechanism is effective.
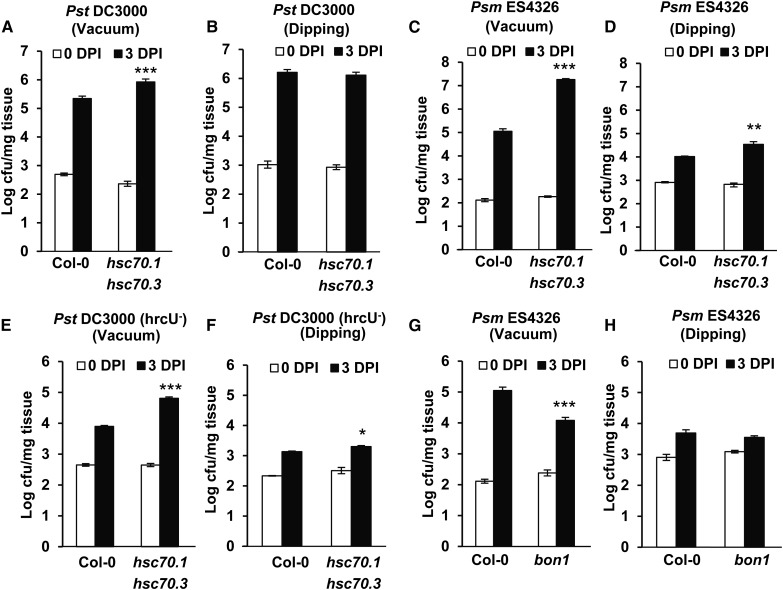
We observed differences in resistance in the hsc70.1 hsc70.3 mutant by the two methods. The virulent pathogen Pst DC3000, with vacuum infiltration, had more growth in the double mutant than the wild type by 1.1 ± 0.1, 0.8 ± 0.1, and 0.6 ± 0.1 of log (lg) value of colony forming unit (cfu) per milligram of leaf tissue in three independent experiments (Figs. 4, D and F, and 6A). With dipping inoculation, it had no more growth in the double mutant compared with the wild type: the growth difference between the mutant and the wild type was 0.1 ± 0.1, 0.1 ± 0.1, and 0.3 ± 0.2 lg cfu mg–1 in three independent experiments (Fig. 6B; Supplemental Fig. S7, A and B). For the virulent pathogen Psm ES4326, with vacuum infiltration, the differential growth in the mutant and the wild type was 2.2 ± 0.1 and 1.3 ± 0.2 lg cfu mg–1 in two independent experiments (Fig. 6C; Supplemental Fig. S7C). By dipping inoculation, the differential growth was 0.2 ± 0.1 and 0.5 ± 0.1 lg cfu mg–1 in two experiments (Fig. 6D; Supplemental Fig. S7D). For nonvirulent pathogen Pst DC3000 hrcU–, the growth increase in the double mutant compared with the wild type was 0.6 ± 0.2 and 0.9 ± 0.1 lg cfu mg–1 with vacuum (Fig. 6E; Supplemental Fig. S7E) and 0.2 ± 0.1 and 0.3 ± 0.1 lg cfu mg–1 with dipping inoculation (Fig. 6F; Supplemental Fig. S7F).
Figure 6.
Comparing pathogen growth in hsc70.1 hsc70.3 and bon1 mutants with different inoculation methods. A to F, Pathogen growth of Pst DC3000 (A and B), Psm ES4326 (C and D), and Pst DC3000 hrcU– (E and F) in Col-0 and the hsc70.1 hsc70.3 mutant by vacuum (A, C, and E) or dipping (B, D, and E) inoculations at 0 and 3 DPI. G and H, Pathogen growth of Psm ES4326 in Col-0 and the bon1-1 mutant by vacuum (G) or dipping (H) inoculations at 0 and 3 DPI. Values represent averages of three biological repeats, and error bars represent sds. A star indicates a statistical difference from the wild type (Student’s t test; *, P < 0.05; **, P < 0.01; and ***, P < 0.001).
The reduced susceptibility to pathogens in the hsc70.1 hsc70.3 double mutant with dipping inoculation compared with the vacuum inoculation indicates that the double mutant might be more resistant to pathogens at the preinvasion phase compared with the wild type. Therefore, the HSC70 genes very likely have opposing functions in two layers of immune responses during PTI and/or ETI: a positive role after the pathogen invades the apoplastic space and a negative role before invasion, likely at the level of stomatal closure.
BON1 Positively Regulates Preinvasion Resistance via Modulating Stomatal Closure
We investigated whether physical interactions of BON1 with HSC70 and SGT1b have a more direct biological relevance in stomata control. Supporting this idea that BON1 has a role in preinvasion defense, a difference in resistance level in bon1 compared with the wild type was observed with vacuum and dipping infiltration methods. With vacuum inoculation, the decrease of pathogen growth in bon1 compared with the wild type was 1.0 ± 0.2 and 1.9 ± 0.1 lg cfu mg–1 in two independent assays (Fig. 6G; Supplemental Fig. S7G). With dipping, the decrease in growth of Psm ES4326 in bon1 compared with the wild-type Col-0 was 0.1 ± 0.2 and 0.3 ± 0.2 lg cfu mg–1 at 3 d in two assays (Fig. 6H; Supplemental Fig. S7H). Therefore, the bon1 mutant is likely compromised in resistance at the preinvasion phase, indicating a positive role of BON1 in preinvasion defense response. This is in contrast to the previously known role of BON1 in negatively regulating NB-LRR genes at the postinvasion phase (Yang and Hua, 2004).
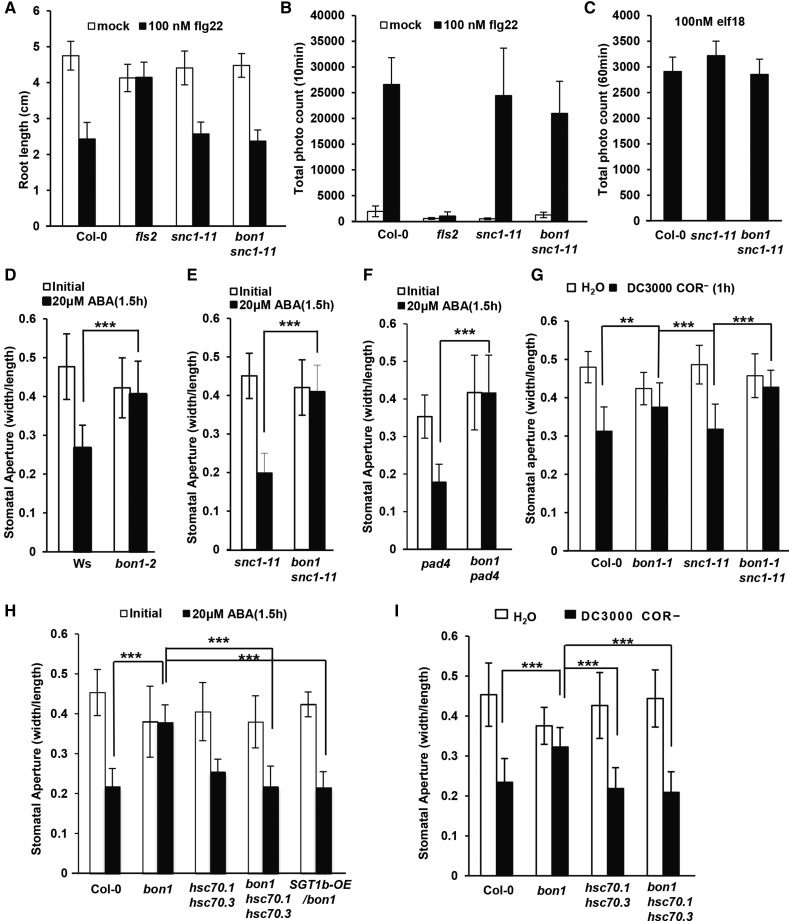
Because BON1 localizes to the plasma membrane where PAMP receptors reside and has a role in preinvasion immune response, we asked whether BON1 might function as a positive regulator of early perception of PAMPs. Responses toward different PAMP treatments were assayed in bon1 or bon1 snc1-11 (where effects from activation of SNC1 are minimized). One response to the PAMP flg22 is root growth inhibition, which is mediated by the PAMP receptor FLS2 (Zipfel et al., 2004). As expected, treatment of flg22 at 100 nm significantly inhibited root growth of Col-0 but not the fls2 mutant. Roots of bon1 snc1-11 and snc1-11 were inhibited by flg22 similarly to the wild-type Col-0 (Fig. 7A), indicating that flg22 effects are not significantly altered by the loss of BON1 function. As a more immediate response to 100 nm flg22, the production of ROS was also comparable in the bon1 snc1-11 mutant and the wild type. While the fls2 mutant had much less ROS at 10 min after treatment compared with the wild-type Col-0, bon1 snc1-11 only showed a slight reduction of ROS compared with Col-0 and snc1-11 (Fig. 7B). Similarly, in response to 100 nm elf18, another PAMP, ROS accumulation in bon1 snc1-11 and Col-0 or snc1-11 also did not exhibit significant differences (Fig. 7C). These data indicate that BON1 does not play a major role in flg22 and elf18 perception.
Figure 7.
BON1 negatively regulates stomatal closure but not PAMP perception. A, Response to flg22 assayed by root growth inhibition. Shown are root lengths of Col-0, fls2, snc1-11, and bon1 snc1-11 at 2 weeks after growth on plates with or without 100 nm flg22. B, Analysis of oxidative burst triggered by flg22 measured by luminol. Shown are averages of total photo counts of Col-0, fls2, snc1-11, and bon1 snc1-11 leaf discs at 10 min after treatment with 100 nm of flg22. C, Analysis of oxidative burst triggered by elf18 measured by luminol. Shown are averages of total photo count of Col-0, snc1-11, and bon1 snc1-11 leaf discs 60 min after treatment with 100 nm elf18. D to F, Stomatal aperture after treatment with 20 μm ABA for snc1-11 and bon1 snc1-11 (D), wild-type Ws and bon1-2 in Ws (D), and pad4 and bon1 pad4 (F) leaves. G, Stomatal aperture after treatment with Pst DC3000 COR– for Col-0, bon1, snc1-11, and bon1 snc1-11 leaves. H, Stomatal apertures for the Col-0 wild type, bon1, hsc70.1 hsc70.3, bon1 hsc70.1 hsc70.3, and SGT1b-OE/bon1 leaves before or after 20 μm ABA treatment for 1.5 h. I, Stomatal apertures for Col-0 wild-type, bon1, hsc70.1 hsc70.3, and bon1 hsc70.1 hsc70.3 leaves after treatment with Pst DC3000 COR– or water for 1 h. For D to I, values represent average apertures of at least 20 stomata, and error bars represent sds. Stars indicate statistical differences (Student’s t test; **, P < 0.01; and ***, P < 0.001). Experiments were repeated three times with similar results.
Because stomatal closure is an important defense mechanism at the preinvasion phase, we investigated whether stomatal response is altered in the bon1 mutants. We first analyzed ABA-induced closure because ABA is involved in both abiotic and PAMP-induced stomatal response (Fan et al., 2004; Melotto et al., 2006). Because activation of NB-LRR genes such as SNC1 has been shown to inhibit ABA-induced stomatal closure (Kim et al., 2011), we analyzed the effect of loss of BON1 function in two pair of genotypes with no functional SNC1 to eliminate potential secondary effects from its up-regulation. The first pair was the wild-type ecotype Wassilewskija (Ws) and a LOF bon1-2 allele in the Ws accession. No functional SNC1 exists in Ws, and therefore immune responses are not up-regulated in bon1-2 (Yang and Hua, 2004). The second pair was a SNC1 LOF mutant snc1-11 and a bon1 snc1-11 double mutant (Yang and Hua, 2004). Leaf peels made from these plants were treated with 20 μm ABA. While Ws wild-type and snc1-11 plants closed their stomata at 1.5 h after ABA treatment, no obvious stomatal closure was observed in bon1-2 or bon1 snc1-11 (Fig. 7, D and E; Supplemental Fig. S8). To exclude the possibility that this effect results from activation of genes other than SNC1 in bon1 (Li et al., 2009), we measured ABA response in a third pair of plants: the phytoalexin deficient4 (pad4) mutant where Toll/Interleukin-1 Receptor (TIR)-NB-LRR signaling in general is blocked and the bon1 pad4 double mutant. Compared with pad4, bon1 pad4 did not respond to ABA by closing stomata (Fig. 7F). Therefore, BON1 has a positive role in ABA response in stomatal closure, and this function is independent of its negative regulation of NB-LRR genes.
We further tested if the loss of BON1 function also compromises pathogen-induced stomatal closure. We used a coronatine-deficient (COR–) Pst DC3000 strain for such assay to reveal the early defense response (stomatal closure) to pathogen without the counteracting effect (stomatal opening) by coronatine from pathogens (Melotto et al., 2006). While snc1-11 and Col-0 wild-type plants closed their stomata at 1.5 h after pathogen treatment, very little stomatal closure was observed in bon1 or bon1 snc1-11 (Fig. 7G). Therefore, BON1 has a positive role in ABA and pathogen-induced stomatal closure, which likely counts toward its positive role in preinvasion defense.
The Function of BON1 at Stomata Is Related to HSC70 and SGT1b
Previous studies have demonstrated the involvement of HSC70 and SGT1b in stomatal control (Clément et al., 2011). We determined whether the function of BON1 in stomata control is related to HSC70 and SGT1b functions by analyzing the ABA response in the bon1 hsc70.1 hsc70.3 triple mutant. At 1.5 h after ABA treatment, the wild type closed its stomata, and the hsc70.1 hsc70.3 double mutant had a similar response to the wild type. While the bon1 mutant kept its stomata open, the bon1 hsc70.1 hsc70.3 closed its stomata similarly to the wild type (Fig. 7H). Similarly, overexpression of SGT1b largely suppressed the stomatal closure defect of bon1. In a representative SGT1b-OE/bon1 line where bon1 growth defect was largely suppressed by SGT1b-OE, stomata closed in response to ABA in a similar fashion as the wild type (Fig. 7H).
We further tested if the hsc70.1 and hsc70.3 double mutations could also rescue bon1’s defect in pathogen-triggered stomatal closure. At 1 h after Pst DC3000 (COR–) treatment, hsc70.1 hsc70.3 closed its stomata similarly to the wild type. While the bon1 mutant kept its stomata open, the bon1 hsc70.1 hsc70.3 closed its stomata similarly to the wild type (Fig. 7I). Therefore, BON1 positively regulates stomatal closure in response to ABA and pathogens, and this function is closely related to the function of HSC70 and SGT1b in stomata regulation.
STG1b Negatively Regulates Immune Responses in bon1
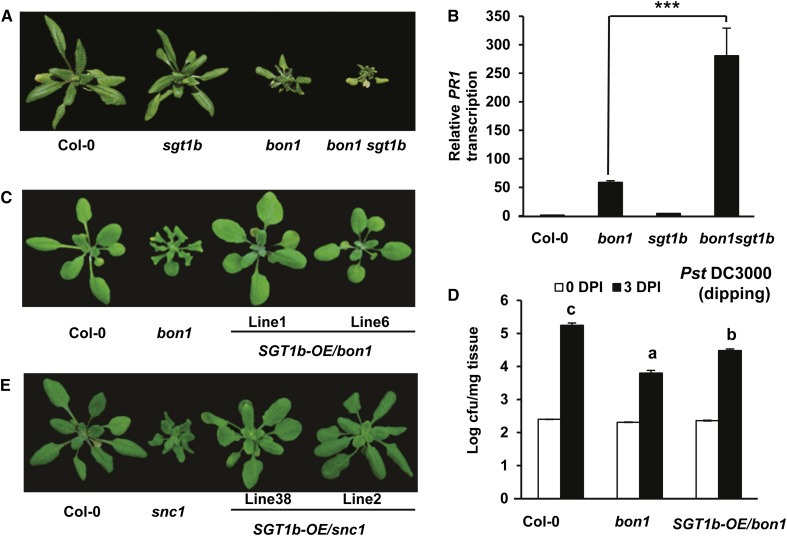
We next assessed the biological relevance of physical interaction between BON1 and SGT1b. We generated a double mutant between bon1 and the SGT1b LOF mutant allele enhanced downy mildew1.1 (edm1-1; Tör et al., 2002), which we refer to here as sgt1b. While the bon1 hsc70.1 mutant had a milder growth defect than bon1, the bon1 sgt1b double mutant had a more severe growth defect than bon1 (Fig. 8A). The visual observation was verified by biomass measurement, where fresh weight of bon1 sgt1b was significantly lower than that of bon1 (Supplemental Fig. S6C). Expression of PR1 is further up-regulated in the bon1 sgt1b double mutant compared with the bon1 single mutant as assayed by qRT-PCR (Fig. 8B), indicating that the defense responses in bon1 are enhanced by the loss of the SGT1b function.
Figure 8.
STG1b negatively regulates immune responses in bon1. A, Growth defect in bon1 is enhanced by the sgt1b mutation. Shown are plants of wild-type Col-0, sgt1b, bon1, and bon1 sgt1b at 5 weeks old. B, PR1 gene expression in bon1 is enhanced by the sgt1b mutation. Shown is the relative expression of PR1 in Col-0, bon1, sgt1b, and bon1 sgt1b plants assayed by qRT-PCR. Error bars represent sds (Student’s t test; ***P < 0.001). C, Partial rescue of bon1 growth defect by overexpressing the SGT1b gene. Shown are plants of the wild-type Col-0, bon1, and two individual T2 lines (lines 1 and 6) of SGT1b-OE/bon1. D, Partial suppression of enhanced resistance against the virulent bacterial strain Pst DC3000 in bon1 by SGT1b overexpression. Shown are bacterial growths in Col-0, bon1, and SGT1b-OE/bon1 plants via dipping inoculation at 0 and 3 DPI. Values represent averages of three biological repeats, and error bars represent sds. Letters indicate statistical difference (P < 0.001; Bonferonni posttest) of different genotypes. E, Growth phenotype of snc1 overexpressing the SGT1b gene. Shown are wild-type Col-0, snc1, and two independent lines (lines 38 and 2) of SGT1b-OE/snc1.
We subsequently overexpressed SGT1b in bon1 by the CaMV 35S promoter using the pHPT vector (Tzfira et al., 2005). All nine SGT1b-OE-independent lines showed a milder growth defect compared with bon1 (Fig. 8C), which was verified by biomass quantification (Supplemental Fig. S6C). The SGT1b transcript level was increased in two selected transgenic lines as assayed by qRT-PCR (Supplemental Fig. S9). In the progenies of the two T1 lines tested, the close-to-wild-type phenotype cosegregated with the presence of the SGT1b-OE transgene, further supporting that overexpression of SGT1b inhibited the bon1 growth phenotype. Although SGT1b-OE in the Col-0 wild type did not appear to affect disease symptom to Pst DC3000 (Uppalapati et al., 2011), it compromised the enhanced disease resistance to Pst DC3000 in bon1 (Fig. 8D). Therefore, SGT1b, opposite to HSC70, inhibits autoimmune responses in bon1.
We asked if the partial inhibition of the bon1 phenotype by SGT1b overexpression has resulted from blocking signaling leading to SNC1 transcript up-regulation or from disrupting signaling after SNC1 up-regulation. To this end, SGT1b was overexpressed in the autoimmune mutant snc1. Among 27 SGT1b-OE transgenic lines generated in snc1, 26 showed inhibition of the growth defect of snc1 (Fig. 8E), which was also verified by biomass quantification (Supplemental Fig. S6C). The SGT1b expression was higher in two representative SGT1b-OE lines than in the wild type as assayed by qRT-PCR (Supplemental Fig. S9), and the growth defect suppression was strictly correlated with the SGT1b-OE transgene. Therefore, SGT1b modulates SNC1-mediated defense responses, likely through SNC1 protein degradation, which could account for its modulation of immune responses in bon1.
STG1b Inhibits SNC1 Protein Accumulation
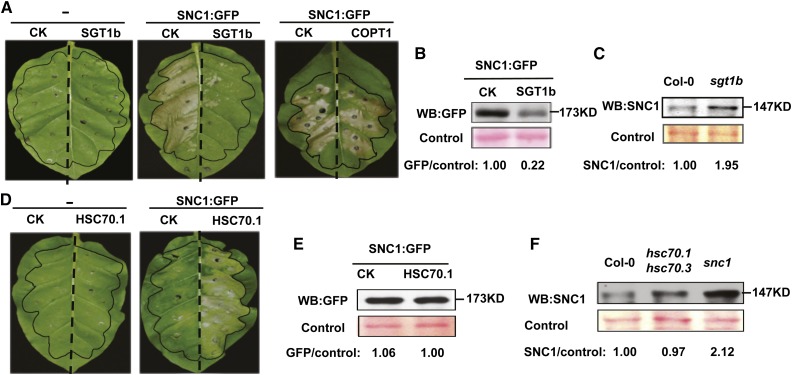
A previous study showed a requirement of SGT1b in resistance mediated by a number of NB-LRR proteins but not SNC1 (Goritschnig et al., 2007). In fact, the SNC1 protein accumulated to a higher level in the sgt1b mutant, although no increase of disease resistance was observed in the sgt1b mutant compared with the wild type (Li et al., 2010b). This prompted us to test whether the suppression of snc1 phenotype by SGT1b overexpression is due to the reduction of SNC1 protein accumulation.
To assay both SNC1 activity and SNC1 protein level, we used a previously established expression system where overexpression of a GFP-tagged SNC1 protein induced cell death in N. benthamiana (Zhu et al., 2010; Gou et al., 2012; Mang et al., 2012). SGT1b and a control gene, COPT1, were overexpressed by the CaMV 35S promoter using the vector pGWB402 (Nakagawa et al., 2007) via agroinfiltration in tobacco (Nicotiana tabacum). At 3 DPI, extensive cell death was observed in leaf areas coinfiltrated with SNC1:GFP and a pGWB402 empty vector, while neither the empty vector nor the pGWB402:SGT1b construct triggered visible cell death (Fig. 9A). When SGT1b was coinfiltrated with SNC1:GFP, cell death was greatly reduced, while coinfiltration of the control gene COPT1 with SNC1:GFP did not affect cell death (Fig. 9A). This indicates that the reduction of SNC1-triggered cell death by SGT1b was not due to a nonspecific effect from coinfiltration. This is consistent with the observation in Arabidopsis transgenic plants where overexpression of SGT1b suppressed the snc1 phenotype (Fig. 8E). We then analyzed SNC1:GFP accumulation with or without SGT1b coexpression in tobacco leaves before the onset of cell death by western blotting. Quantified with the control protein signal stained by Ponceau S, the SNC1:GFP protein level was reduced in tissues coinfiltrated with SGT1b compared with the vector control (Fig. 9B). As a control, we also detected the SNC1 level in the Arabidopsis sgt1b mutant using an anti-SNC1 antibody previously described (Cheng et al., 2011). Consistent with previous reports (Li et al., 2010b), SNC1 accumulates to a higher level in sgt1b than in the Col-0 wild type (Fig. 9C). Together, these data indicate that SGT1b inhibits SNC1 protein accumulation, which likely leads to suppression of SNC1-mediated disease resistance.
Figure 9.
Antagonistic regulation of SNC1 by SGT1b and HSC70. A, Inhibition of SNC1-triggerred cell death by transient overexpression of SGT1b in tobacco leaves. For the left leaf, each half was infiltrated with the pGWB402 vector control (CK) or pGWB402:SGT1b (SGT1b). For the middle leaf, each half was coinfiltrated with pHPTN1:SNC1 (SNC1:GFP) and pGWB402 (CK) or pGWB402:SGT1b (SGT1b). For the right leaf, each half was coinfiltrated with pHPTN1:SNC1 (SNC1:GFP) and pGWB402 (CK) or pGWB402:COPT1 (COPT1). Infiltrated areas were marked with black color. Images were taken at 3 DPI, and the experiments were repeated at least three times with similar results. B, Western-blot (WB) detection of SNC1:GFP in the coinfiltrated tobacco leaves. C, Western-blot detection of the endogenous SNC1 protein in the Arabidopsis sgt1b mutant. D, Activation of SNC1-triggerred cell death by overexpression of HSC70.1 in tobacco leaves. For the left leaf, each half was infiltrated with pMDC32 (CK) or pMDC32:HSC70.1 (HSC70.1). For the right leaf, each half was infiltrated with pHPTN1:SNC1 (SNC1:GFP) together with pMDC32 (CK) or pMDC32:HSC70.1 (HSC70.1). Images were taken at 2 DPI, and a representative image is shown here. This experiment was repeated at least three times with similar results. E, Western-blot detection of SNC1:GFP in the coinfiltrated tobacco leaves. F, Western-blot detection of the endogenous SNC1 protein in Arabidopsis. For B and E, total proteins were extracted 40 h after coinfiltration, and an anti-GFP antibody was used to detect SNC1:GFP. For C and F, total proteins were extracted from seedlings of 12-d-old plants, and an anti-SNC1 antibody was used for western blot. Proteins prominently stained by Ponceau S were used as loading controls. GFP/control or SNC1/control are the relative ratios between the GFP or SNC1 signal and the Ponceau S-stained control signal quantified by ImageJ software. The ratio of CK or the Col-0 wild type is set as 1.00.
HSC70.1 Positively Regulates SNC1 Activity But Not SNC1 Protein Accumulation
We used the same expression system to analyze the effect of HSC70 on SNC1-mediated defense responses. At 2 DPI, neither the pMDC32 empty vector (Curtis and Grossniklaus, 2003) nor HSC70.1 in pMDC32 triggered any cell death in the infiltrated leaf area (Fig. 9D). Mild cell death was observed in leaf areas coinfiltrated with SNC1:GFP and a pMDC32 empty vector. When HSC70.1 was coinfiltrated with SNC1:GFP, more extensive cell death was induced (Fig. 9D), indicating that HSC70.1 enhances SNC1-mediated defense responses.
We subsequently analyzed the effect of HSC70 on the accumulation of SNC1:GFP protein before the onset of cell death. Analyzed by the relative ratio of GFP to the control protein signal, no difference could be detected between the SNC1:GFP signal from tobacco leaves coinfiltrated with the pMDC32 vector or HSC70.1 in pMDC32 (Fig. 9E), suggesting that overexpressing HSC70, though enhancing SNC1 activity, does not increase the accumulation of the SNC1 protein. We further compared the endogenous SNC1 protein level in the wild type and the hsc70.1 hsc70.3 double mutant by western blot using anti-SNC1 antibody. No significant difference in the SNC1 protein level was observed in the hsc70.1 hsc70.3 double mutant compared with the wild type after the relative ratio of SNC1 to the control signal was calculated (Fig. 9F). As a control, an increase of SNC1 level was observed in snc1 mutant compared with the Arabidopsis wild-type Col-0 as reported earlier (Fig. 9F; Cheng et al., 2011). Therefore, HSC70 may not regulate the SNC1 protein accumulation to affect its activity.
DISCUSSION
HSC70 Genes Have Opposite Roles in Pre- and Postinvasion Phases of Immune Responses
The roles of HSC70 proteins in plant immunity were not clearly defined due to genetic redundancy and functional compensation among HSC70 genes and lethality of the knockout mutant of the gene family. In this study, we used a reduction-of-function mutant combination of HSC70.1 and HSC70.3 to sufficiently reduce the total HSC70 activity without causing lethality and consequently revealed opposite roles of HSC70 proteins in pre- and postinvasion defenses. This hsc70.1 hsc70.3 double mutant is more susceptible than the wild type to virulent and type III secretion-deficient bacterial strains (Figs. 4 and 5), indicating a positive role of HSC70 genes in regulating PTI. The hsc70.1 hsc70.3 double mutant is also more susceptible than the wild type to avirulent bacterial pathogens, and the double mutation largely suppresses the defense phenotypes conferred by the active form of SNC1 (Figs. 4 and 5). This may result from the positive role of HSC70s in PTI (as in resistance to virulent and nonvirulent pathogen), or it could be from a positive role of HSC70s in ETI as well. Overexpression of HSC70 can enhance SNC1-triggered cell death in tobacco, suggesting that HSC70 could have a positive role in ETI in addition to PTI.
In addition to revealing a positive role of HSC70s in PTI/ETI, our data support the negative role of HSC70 proteins on stomatal closure in plant immunity. The hsc70.1 hsc70.3 double mutant is more susceptible when pathogens are dip inoculated compared with when they are vacuum infiltrated (Fig. 6; Supplemental Fig. S7), suggesting that the double mutant might more effectively restrict pathogen invasion than the wild type. This is consistent with the earlier finding that HSC70 overexpression or the loss of SGT1b function confers insensitivity in stomatal closure to environmental factors (Clément et al., 2011). However, we did not observe an enhanced sensitivity of stomatal response to ABA or nonvirulent pathogen in the hsc70.1 hsc70.3 double mutant (Fig. 7), which can be due to the low detection sensitivity of stomatal measurement and/or limited time points we used in this study. In light of this study implicating opposing roles of HSC70s in two phases of immune responses, contradictory roles reported for HSC70s in disease resistance from earlier studies can now be explained by differential contribution of two layers of defenses in a particular plant-pathogen interaction as well as potential differential roles of each of the HSC70 family members in specific immune responses.
Interaction of HSC70 and SGT1b in Regulating SNC1
The role of HSC70 in SNC1-mediated defense response is opposite to that of SGT1b, which is implicated in the degradation of SNC1. The SNC1 protein accumulated to high levels in the sgt1b mutant (Li et al., 2010b), and it is targeted by the F-box protein CONSTITUTIVE EXPRESSOR OF PR GENES1 (CPR1)/CPR30 for degradation (Gou et al., 2009, 2012; Cheng et al., 2011). Here, we found that defects in bon1 or snc1 could be enhanced by the sgt1b mutation and inhibited by SGT1b overexpression (Fig. 8). SGT1b is involved in degrading the SNC1 protein, as seen with a reduced level of SNC1 and SNC1-triggered cell death in SGT1b-OE tobacco leaves (Fig. 9). By contrast, HSC70.1, while enhancing SNC1-triggered cell death when overexpressed, does not affect the protein accumulation of SNC1 (Fig. 9). As the HSC70s can assist proper folding of client protein (Hartl, 1996), they may enhance SNC1 activity by aiding the correct folding of SNC1. Alternatively, HSC70s might affect components downstream of SNC1 in defense responses and have no direct effect on the SNC1 protein itself. It is yet to be determined whether the opposite function of HSC70 and SGT1b results from direct physical interaction of these two proteins or their independent interactions with the SNC1 protein.
There is no evidence so far to support the hypothesis that BON1 regulates SNC1 primarily at the protein level, although physical interaction of BON1 and SGT1b is observed here. Elevated defense response in bon1 results from up-regulation of the NB-LRR gene SNC1 (Yang and Hua, 2004). Because hsc70.1 hsc70.3 compromised defense responses in the active snc1 mutant, and the hsc70.1 hsc70.3 mutant is more susceptible to Pst DC3000 (Fig. 4), the reduction of defense responses in bon1 by the hsc70.1 hsc70.3 mutation results from an additive effect from bon1 and hsc70.1 hsc70.3 (Fig. 4). Therefore, BON1 and HSC70 may not function strictly in a linear pathway. The suppression of bon1 by hsc70 mutations or SGT1b overexpression likely results indirectly from their effects on the SNC1 protein translated from the SNC1 transcript that is up-regulated in bon1.
Function of BON1 in Stomatal Closure
We identified, to our knowledge, a new role of BON1 in plant immunity, that is, the modulation of stomatal closure. Previous screening of BON1-interacting proteins with yeast two-hybrid assays have identified BAP1 and BIR1 proteins that are involved in negative regulation of plant defense that is dependent on SNC1 (Hua et al., 2001; Yang et al., 2006a; Wang et al., 2011). However, the link between the plasma membrane-localized BON1 and the transcriptional regulation of NB-LRR-encoding genes remained elusive. Here, we found that BON1 physically interacts with HSC70 and SGT1b, both of which are involved in regulating stomatal closure induced by environmental stimuli (Clément et al., 2011). The bon1 mutant is compromised in both ABA- and pathogen-triggered stomatal closure, and this effect is independent of SNC1 (Fig. 7). Furthermore, the defect in ABA response in the bon1 mutant is suppressed in the hsc70.1 hsc70.3 mutant and SGT1b-OE lines (Fig. 7). Thus, the BON1 protein might interact with HSC70 and SGT1b to regulate stomatal closure. How BON1 connects with the known signaling components in stomatal closure control is not known. One possibility is that BON1 interacts with receptor-like kinases in addition to BIR1, and some receptor-like kinases such as GUARD CELL HYDROGEN PEROXIDE-RESISTANT1 have been shown to mediate ABA-induced stomatal closure (Hua et al., 2012). Further research should identify the regulatory target(s) of BON1, HSC70, or SGT1b in stomata regulation and reveal how BON1 activity might be regulated by biotic and abiotic environmental signals.
The bon1 mutants do not appear to have major defects in PAMP perception (Fig. 7), and therefore BON1 may not directly regulate the activity of PAMP receptors. As BON1 is a calcium-binding protein and calcium-binding activity is critically essential for its function (Hua et al., 2001; Li et al., 2010a), it could be involved in calcium signal perception and, in turn, modulates signaling pathways or channels/transporters. Similarly, HSC70 and SGT1b were thought to function not in early signal perception but in later signaling and execution events (Clément et al., 2011).
Previous studies revealed a negative regulation of NB-LRR-encoding genes by BON1 and that this regulation likely occurs at the transcriptional level (Yang and Hua, 2004; Li et al., 2009). The positive role of BON1 in plant immunity at the preinvasion phase suggests that this function of BON1 at the stomata might be guarded by SNC1 and that the loss of BON1 function might be recognized as a manipulation by a pathogen. As there is no evidence for direct interaction of BON1 and SNC1 proteins, the guarding of BON1 by SNC1 would be indirect, similar to a NB-LRR gene SUPPRESSOR OF MKK1 MKK2 2 guarding the MAP KINASE4 activity (Zhang et al., 2012). The regulation of SNC1 by BON1 may mimic the regulation of RESISTANCE TO P. SYRINGAE PV MACULICOLA1 and RPS2 by RPM1 INTERACTING PROTEIN4 (RIN4), a classical example of guard hypothesis (Chisholm et al., 2006; Jones and Dangl, 2006). Coincidently, both RIN4 and BON1 are plasma membrane-localized proteins (Hua et al., 2001; Kim et al., 2005; Takemoto and Jones, 2005; Li et al., 2010a), and RIN4 is also involved in stomatal closure (Liu et al., 2009). It remains to be tested if BON1 is targeted by potential effectors for modification or degradation.
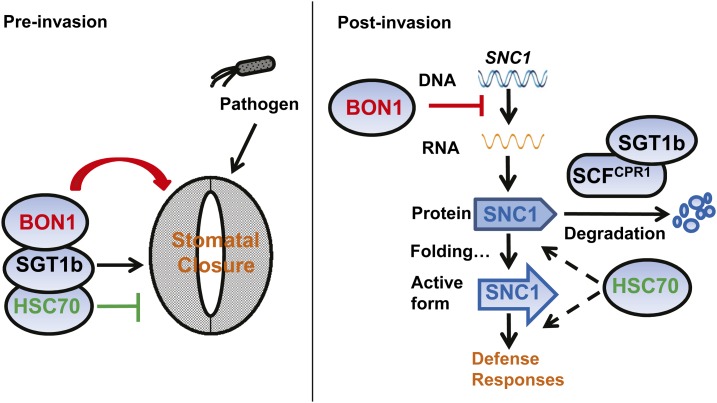
In sum, our study reveals that both BON1 and HSC70 have opposing roles in pre- and postinvasion phases of immune responses (Fig. 10). In the preinvasion phase, HSC70 negatively and SGT1b positively regulate stomatal closure, while BON1 perhaps works together with HSC70 and SGT1b to positively regulate pathogen- and ABA-induced stomatal closure. In the postinvasion phase, BON1 negatively regulates the transcript level of SNC1 through an as yet undetermined mechanism, while SGT1b and HSC70 have opposing roles in regulating SNC1 protein activity, likely by CPR1-coupled degradation, proper protein folding, or downstream signaling. The opposing roles of the same protein on plant immunity at different stages suggest that plant immunity is an evolving system resulting from targeting positive immune regulators by pathogens and guarding the immune regulators with NB-LRR genes by plants.
Figure 10.
Working model for the roles of BON1, SGT1b, and HSC70 in pre- and postinvasion defenses. In the preinvasion phase, HSC70 negatively and SGT1b positively regulate stomatal closure, while BON1 works together with HSC70 and SGT1b to positively regulate pathogen- and ABA-induced stomatal closure. In the postinvasion phase, BON1 negatively regulates the transcript level of NB-LRR gene SNC1 through a yet-to-be-determined mechanism, while SGT1b and HSC70 have opposing roles in regulating SNC1 protein activity likely by CPR1-coupled degradation, proper protein folding, or downstream signaling.
MATERIALS AND METHODS
Plant Growth
Arabidopsis (Arabidopsis thaliana) plants were grown in growth chambers either under constant light for growth phenotyping or at a 12-h/12-h photoperiod for pathogen growth tests, with light intensity at 100 µmol m–2 s–1 and relative humidity at 50% to 70%. Seeds were planted either on one-half-strength Murashige and Skoog (Sigma) medium containing 0.8% (w/v) agar and 2% (w/v) Suc or directly in soil (Metro-Mix 200; SunGro). Nicotiana benthamiana or tobacco (Nicotiana tabacum) plants were grown in the greenhouse at 24°C for 4 to 6 weeks before use for transient expression studies.
BON1 Protein Complex Purification and LC-MS Analysis
Wild-type and BON1-HA transgenic Arabidopsis plants were grown to 5 weeks old, and one-half of each was inoculated with Pst DC3000. Ten grams of leaf tissues at 2 h after inoculation was collected and combined with 10 g of tissue not inoculated with pathogen. Proteins were extracted, and the BON1 protein complex was purified following the protocol previously described (Qi and Katagiri, 2009). Protein gels for the control and BON1-HA sample were each cut into four parts for the in-gel digestion and manual extraction following a previously reported protocol (Zhang et al., 2003a). The tryptic digest was subject to Nanoscale liquid chromatographic electrospray ionization tandem mass spectrometry analysis using a LTQ-Orbitrap Velos (Thermo-Fisher Scientific) mass spectrometer equipped with a plug-and-play nano ion source device (CorSolutions). Proteins detected in the BON1-HA sample but not in the wild-type control in two biological repeats were selected as candidate BON1-interacting proteins.
BiFC Assay
The full-length complementary DNA (cDNA) fragments (without stop codon) of BON1, HSC70.1(2,3), and SGT1b(a) genes were amplified from the wild-type Col-0 cDNA using primers in Supplemental Table S1 and cloned into the Gateway entry vector pCR8 TOPO TA vector (Invitrogen). For BiFC experiments, BON1 was cloned into pSPYNE-35SGW or pSPYCE-35SGW (Walter et al., 2004; Schütze et al., 2009) using LR clonase (Invitrogen catalog no. 11791) to generate BON1:YFPC or BON1:YFPN constructs, while HSC70.1(2,3) and SGT1b were cloned similarly to generate HSC70.1(2,3):YFPN and SGT1b:YFPC constructs, respectively. A previously described protocol (Walter et al., 2004; Schütze et al., 2009) was followed to observe BiFC signals with some modification. The constructs were transformed into the Agrobacterium tumefaciens strain GV3101. Overnight cell cultures were collected and resuspended in 1 mL of AS medium (1 mL of 1 m MES-KOH, pH 5.6, 333 μL of 3 m MgCl2, and 100 μL of 150 mm acetosyringone) to optical density at 600 nm (OD600) of 0.7 to 0.8. The working suspensions were prepared by mixing, at 1:1:1 ratio, three A. tumefaciens strains respectively carrying the YFPN fusion, the YFPC fusion, and the gene-silencing inhibitor pBA-HcPro (Menke et al., 2005) and letting them stand for 2 to 4 h on a bench. The A. tumefaciens suspensions were then coinfiltrated into the abaxial surface side of 4- to 6-week-old N. benthamiana plant leaves. Fluorescence of the epidermal cell layer of the lower leaf surface was examined at 2 to 4 DPI. Images were captured by a Leica TCS SP2 Confocal Microscope with excitation wavelength at 488 and 496 nm and emission wavelength between 520 and 535 nm for YFP signals.
Co-IP and Immunoblot Analyses Using Tobacco and Arabidopsis Materials
An HA tag is present in the pSPYCE-35S GW constructs, and a Myc tag is present in the pSPYNE-35S GW constructs (Schütze et al., 2009). Tobacco leaves transiently coexpressing each of the BiFC constructs harvested after 2 to 4 DPI or 3-week-old, soil-grown Arabidopsis seedlings were ground with liquid nitrogen. Total proteins were extracted from tobacco leaves with buffer containing 50 mm Tris-HCl (pH 7.5), 2 mm EDTA, 150 mm NaCl, 10% (v/v) glycerol, 5 mm dithiothreitol, 0.25% (v/v) Triton X-100, and 1× complete protease inhibitor cocktail. After spinning protein extracts twice at 12,000g for 10 min, the supernatant was subject to desalting by passing through a Sephadex G-25 column (GE Healthcare Illustra NAP-5). For immune precipitation, 15 µL of EZviewRed Anti-HA Affinity Gel Beads (E6779, Sigma) was mixed with crude protein extracts and agitated at 4°C overnight. The affinity beads were then pelleted by centrifugation for 1 min at 1,000g and washed three times with 1 mL of IP buffer (25 mm Tris-HCl, pH7.5, 1 mm EDTA, 150 mm NaCl, 0.15% (v/v) Nonidet P-40, and 1× protease inhibitor cocktail). The bound proteins were eluted from the beads by boiling them in 50 µL of SDS-PAGE sample buffer for 5 min, and 10 µL of IPed samples were separated by SDS-PAGE gel. Immunoblot analyses were performed according to the enhanced chemiluminescence western-blotting procedure (GE Healthcare) using commercial anti-HA antibody HAII clone 16B12 (Covance), c-Myc antibody (9E10) sc-40 (Santa Cruz), anti-HPC70 antibody ADI-SPA-817 (Enzo Life Sciences), and anti-BiP antibody COP-080017 (Cosmo Bio).
Yeast Two-Hybrid Assay
The yeast (Saccharomyces cerevisiae) two-hybrid constructs were made in the pDEST-GBKT7 and pDEST-GADT7 Gateway vectors as previously described (Rossignol et al., 2007). The BON1 cDNA was cloned from the entry vector into the pDEST-GBKT7 to generate a BD-BON1 construct, while HSC70 genes and SGT1b or truncated SGT1b were cloned into pDEST-GADT7 to generate AD-HSC70 or AD-SGT1b constructs, respectively. The yeast two-hybrid assay was performed as previously described (Li et al., 2010a).
Characterization of Growth Phenotypes of bon1-, hsc70-, and sgt1b-Related Mutants
The HSC70 mutants hsc70.1 (Salk_135531C), hsc70.2 (Salk_085076C), and hsc70.3 (Salk_148168) were ordered from Arabidopsis Biological Resource Center and genotyped to obtain homozygous seeds using primers listed in Supplemental Table S1. Double and triple mutants were generated by crossing hsc70 and sgt1b (edm1-1) mutants to bon1 or snc1. Unless stated otherwise, at least 10 3-week-old seedlings grown in soil were used for growth phenotyping and biomass quantification.
Heat Shock Tolerance Assay of hsc70 Mutants
Arabidopsis seedlings were grown in one-half-strength Murashige and Skoog (one-half-strength Murashige and Skoog salts, MES, pH 5.7, and 1% [w/v] Suc) plates in a growth chamber as described above. Plates containing the 10-d-old seedlings were sealed with plastic tape and submerged in a water bath at 45°C for 30 min, and the heat-shocked seedlings were recovered at 22°C for 3 to 5 d before the photographs were taken and the biomasses were quantified.
Plant Transformation and Transgenic Plant Selection
The SGT1b cDNA from the Gateway entry vector was subcloned into the destination vector pHPTN1GW, which was modified from the binary vector previously described (Tzfira et al., 2005). pHPTN1:SGT1b was transformed into bon1 and snc1 mutants, and transgenic plants were obtained using 50 μg mL–1 hygromycin for selection on MS plates. The HSC70.1 and HSC70.3 full-length cDNAs (with stop codons) were cloned into the pDONR207 entry vector and subcloned into the pGWB402 destination vector (Nakagawa et al., 2007) to generate the overexpression constructs that were transformed into bon1. Transgenic plants were selected with 50 μg mL–1 kanamycin on MS plates. The genomic piece of HSC70.1 was amplified using primers listed in Supplemental Table S1, cloned into the pDONR222 entry vector, and subcloned into the PMDC99 Gateway binary vector. The pMDC99:HSC70.1 construct was transformed into bon1 hsc70.1 hsc70.3 for the complementation test.
Bacterial Growth Assay
Bacteria grown on plates with King’s B medium were washed and collected with 10 mL of 10 mm MgCl2. They were diluted in 10 mm MgCl2 with 0.02% (v/v) Silwet L-77 to OD600 of 0.05 for dipping inoculation and OD600 of 0.002 for vacuum infiltration. Plants were immersed in inoculums for 10 s by dipping or 2 min by vacuum. They were covered immediately to keep humidity after inoculation and uncovered 1 h later. Bacterial growth in plants was assayed at 1 h (day 0) and 3 d (day 3) after inoculation. Three whole seedlings were collected as one sample, weighed, ground in 1 mL of 10 mm MgCl2 with 0.02% (v/v) Silwet L-77, and shaken at room temperature for 1 h. Serial dilutions of the ground solution were spotted on growth media, and the number of cfu per fresh weight was determined. Three samples were analyzed for each genotype and condition combination.
qRT-PCR Analysis
Total RNAs were extracted from soil-grown, 3-week-old plants using Trizol reagent (Invitrogen) as instructed. SuperScript II Reverse Transcriptase (Invitrogen) was used to synthesize cDNA from the mRNA. qRT-PCR was performed using primers listed in Supplemental Table S1. ACTIN2 and TUBULIN2 genes were used as internal controls. SsoAdvanced Universal SYBR Green Supermix (Bio-Rad) was used for qRT-PCR.
Analysis of Responses to PAMP Signals
Seedlings were grown in sterile one-half-strength MS medium with flg22 at 10 µm for 1 to 2 weeks for root inhibition assay. ROS responses to PAMPs were carried out as previously described (Schwessinger et al., 2011). Eight leaf discs (4-mm diameter) per genotype were collected in 96-well plates and allowed to recover overnight in sterile water. The water was then removed and replaced with an eliciting solution containing 17 mg mL–1 luminol (Sigma Aldrich), 200 µg mL–1 horseradish peroxidase (Sigma Aldrich), and an appropriate concentration of the desired PAMP in water. Luminescence was recorded over a 40- to 60-min time period using a CCD camera (Photek). Peptide sequences for flg22 and elf18 have been previously described (Felix et al., 1999; Kunze et al., 2004) and were synthesized by EZBiolab.
Stomatal Assay
Stomatal closure assays were done as previously described (Zeng and He, 2010) with slight modifications. Plants were grown under a 12-h/12-h photoperiod at 22°C for ABA-induced stomata closure assay. Leaves were collected at 4 weeks after germination and placed in MES buffer (25 mm MES-KOH, pH 6.15, and 10 mm KCl) or MES buffer with 20 μm ABA in closed petri dishes for 1.5 h. For pathogen-induced stomatal closure assay, leaves from 5-week-old plants were incubated in water for 2 h before being transferred into water (control) or Pst DC3000 COR– (5 × 107 cfu mL–1) bacteria suspended in water for 1 h in closed petri dishes. Epidermis were peeled and imaged with a Leica ICC50HD microscope. At least 20 stomatal apertures were measured for each sample using ImageJ software. Each experiment as repeated at least three times.
Transient Expression of SNC1 with SGT1b or HSC70.1 in Tobacco Leaves
The pHPTN1:SNC1 construct (Zhu et al., 2010), the pMDC32:HSC70.1 construct, the pGWB402:SGT1b construct, the pMDC32 vector, and the pGWB402 vector were each transformed into the A. tumefaciens strain GV3101. Overnight cell cultures were collected and resuspended in 1 mL of AS medium (1 mL of 1 m MES-KOH, pH 5.6, 333 μL of 3 m MgCl2, and 100 μL of 150 mm acetosyringone) to OD600 of 0.7 to 0.8. The A. tumefaciens suspensions containing different constructs were combined at a 1:1 ratio and coinfiltrated into the abaxial surface side of tobacco leaves of 4- to 6-week-old plants. A picture was taken at 2 or 3 DPI for the cell death phenotype. Protein was extracted, and western blot was performed as described (Gou et al., 2012; Mang et al., 2012). The polyclonal GFP antibody A6455 (Invitrogen) was used to detect the SNC1-GFP fusion proteins.
Arabidopsis Total Protein Extraction and Western-Blot Analyses of SNC1
Arabidopsis seedlings grown on one-half-strength MS plate for 12 d were used for total protein extraction and western blot using anti-SNC1 antibody following a previously described method (Cheng et al., 2011).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Complementation of the bon1 defect by overexpression of BON1-HA.
Supplemental Figure S2. BON1 protein complex purification and MS detection of BON1-associated proteins.
Supplemental Figure S3. Yeast two-hybrid assay of interactions between BON1 and HSC70s.
Supplemental Figure S4. Characterization of HSC70.1 and HSC70.3 overexpression lines in bon1.
Supplemental Figure S5. Growth phenotypes of bon1- and hsc70-related mutants after bolting.
Supplemental Figure S6. Quantification of biomass for bon1-, hsc70-, and sgt1b-related plants.
Supplemental Figure S7. Additional repeats for growth of Pst strains in Col-0 and the hsc70.1 hsc70.3 mutant by dipping and vacuum inoculation shown in Figure 6.
Supplemental Figure S8. Stomatal closure responses in bon1 mutants upon ABA treatment.
Supplemental Figure S9. Relative expression level of SGT1b detected by qRT-PCR.
Supplemental Table S1. Primers used in this study.
Acknowledgments
We thank Dr. Sheng Zhang for help and discussion of the LC-MS analysis of the BON1 protein complex, Dr. Xin Li for the kind gift of the anti-SNC1 antibody and advice on western blot using this antibody, Dr. Tsuyoshi Nakagawa for the pGWB vectors, Dr. Alan Collmer and Dr. Shengyang He for bacterial pathogen strains, Dr. Hailei Wei and Dr. Yuan Wang for the kind help of the flg22-triggered ROS assay, and the Arabidopsis Biological Resource Center for the Arabidopsis mutants.
Glossary
- PAMP
pathogen-associated molecular pattern
- PTI
pathogen-associated molecular pattern-triggered immunity
- ETI
effector-triggered immunity
- ABA
abscisic acid
- BiP
binding protein
- Pst
Pseudomonas syringae pv tomato
- T-DNA
transfer DNA
- Psm
Pseudomonas syringae pv maculicola
- LOF
loss-of-function
- ER
endoplasmic reticulum
- LC
liquid chromatography
- MS
mass spectrometry
- BiFC
bimolecular fluorescence complementation
- IPed
immunoprecipitated
- IP
immunoprecipitation
- CaMV
Cauliflower mosaic virus
- Col-0
ecotype Columbia
- qRT
quantitative real-time reverse transcription
- DPI
days post inoculation
- lg
log
- ROS
reactive oxygen species
- Ws
ecotype Wassilewskija
- COR–
coronatine-deficient
- cDNA
complementary DNA
- OD600
optical density at 600 nm
- cfu
colony forming unit
- Col-0
ecotype Columbia-0
Footnotes
This work was supported by the National Science Foundation (grant nos. IOS–0919914 and IOS–1353738 to J.H.) and the China Scholarship Council (to Q.H.).
References
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Austin MJ, Muskett P, Kahn K, Feys BJ, Jones JD, Parker JE (2002) Regulatory role of SGT1 in early R gene-mediated plant defenses. Science 295: 2077–2080 [DOI] [PubMed] [Google Scholar]
- Azevedo C, Betsuyaku S, Peart J, Takahashi A, Noël L, Sadanandom A, Casais C, Parker J, Shirasu K (2006) Role of SGT1 in resistance protein accumulation in plant immunity. EMBO J 25: 2007–2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azevedo C, Sadanandom A, Kitagawa K, Freialdenhoven A, Shirasu K, Schulze-Lefert P (2002) The RAR1 interactor SGT1, an essential component of R gene-triggered disease resistance. Science 295: 2073–2076 [DOI] [PubMed] [Google Scholar]
- Cazalé AC, Clément M, Chiarenza S, Roncato MA, Pochon N, Creff A, Marin E, Leonhardt N, Noël LD (2009) Altered expression of cytosolic/nuclear HSC70-1 molecular chaperone affects development and abiotic stress tolerance in Arabidopsis thaliana. J Exp Bot 60: 2653–2664 [DOI] [PubMed] [Google Scholar]
- Cheng YT, Li Y, Huang S, Huang Y, Dong X, Zhang Y, Li X (2011) Stability of plant immune-receptor resistance proteins is controlled by SKP1-CULLIN1-F-box (SCF)-mediated protein degradation. Proc Natl Acad Sci USA 108: 14694–14699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chisholm ST, Coaker G, Day B, Staskawicz BJ (2006) Host-microbe interactions: shaping the evolution of the plant immune response. Cell 124: 803–814 [DOI] [PubMed] [Google Scholar]
- Clément M, Leonhardt N, Droillard MJ, Reiter I, Montillet JL, Genty B, Laurière C, Nussaume L, Noël LD (2011) The cytosolic/nuclear HSC70 and HSP90 molecular chaperones are important for stomatal closure and modulate abscisic acid-dependent physiological responses in Arabidopsis. Plant Physiol 156: 1481–1492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U (2003) A Gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dangl JL, Jones JD (2001) Plant pathogens and integrated defence responses to infection. Nature 411: 826–833 [DOI] [PubMed] [Google Scholar]
- Dodds PN, Rathjen JP (2010) Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet 11: 539–548 [DOI] [PubMed] [Google Scholar]
- Fan LM, Zhao Z, Assmann SM (2004) Guard cells: a dynamic signaling model. Curr Opin Plant Biol 7: 537–546 [DOI] [PubMed] [Google Scholar]
- Felix G, Duran JD, Volko S, Boller T (1999) Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J 18: 265–276 [DOI] [PubMed] [Google Scholar]
- Goritschnig S, Zhang Y, Li X (2007) The ubiquitin pathway is required for innate immunity in Arabidopsis. Plant J 49: 540–551 [DOI] [PubMed] [Google Scholar]
- Gou M, Hua J (2012) Complex regulation of an R gene SNC1 revealed by auto-immune mutants. Plant Signal Behav 7: 213–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou M, Shi Z, Zhu Y, Bao Z, Wang G, Hua J (2012) The F-box protein CPR1/CPR30 negatively regulates R protein SNC1 accumulation. Plant J 69: 411–420 [DOI] [PubMed] [Google Scholar]
- Gou M, Su N, Zheng J, Huai J, Wu G, Zhao J, He J, Tang D, Yang S, Wang G (2009) An F-box gene, CPR30, functions as a negative regulator of the defense response in Arabidopsis. Plant J 60: 757–770 [DOI] [PubMed] [Google Scholar]
- Gray WM, Muskett PR, Chuang HW, Parker JE (2003) Arabidopsis SGT1b is required for SCFTIR1-mediated auxin response. Plant Cell 15: 1310–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartl FU. (1996) Molecular chaperones in cellular protein folding. Nature 381: 571–579 [DOI] [PubMed] [Google Scholar]
- Holt BF III, Belkhadir Y, Dangl JL (2005) Antagonistic control of disease resistance protein stability in the plant immune system. Science 309: 929–932 [DOI] [PubMed] [Google Scholar]
- Hua D, Wang C, He J, Liao H, Duan Y, Zhu Z, Guo Y, Chen Z, Gong Z (2012) A plasma membrane receptor kinase, GHR1, mediates abscisic acid- and hydrogen peroxide-regulated stomatal movement in Arabidopsis. Plant Cell 24: 2546–2561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua J, Grisafi P, Cheng SH, Fink GR (2001) Plant growth homeostasis is controlled by the Arabidopsis BON1 and BAP1 genes. Genes Dev 15: 2263–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert DA, Tornero P, Belkhadir Y, Krishna P, Takahashi A, Shirasu K, Dangl JL (2003) Cytosolic HSP90 associates with and modulates the Arabidopsis RPM1 disease resistance protein. EMBO J 22: 5679–5689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jelenska J, van Hal JA, Greenberg JT (2010) Pseudomonas syringae hijacks plant stress chaperone machinery for virulence. Proc Natl Acad Sci USA 107: 13177–13182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL (2006) The plant immune system. Nature 444: 323–329 [DOI] [PubMed] [Google Scholar]
- Kim HS, Desveaux D, Singer AU, Patel P, Sondek J, Dangl JL (2005) The Pseudomonas syringae effector AvrRpt2 cleaves its C-terminally acylated target, RIN4, from Arabidopsis membranes to block RPM1 activation. Proc Natl Acad Sci USA 102: 6496–6501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Gao F, Bhattacharjee S, Adiasor JA, Nam JC, Gassmann W (2010a) The Arabidopsis resistance-like gene SNC1 is activated by mutations in SRFR1 and contributes to resistance to the bacterial effector AvrRps4. PLoS Pathog 6: e1001172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Böhmer M, Hu H, Nishimura N, Schroeder JI (2010b) Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu Rev Plant Biol 61: 561–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Hauser F, Ha T, Xue S, Böhmer M, Nishimura N, Munemasa S, Hubbard K, Peine N, Lee BH, et al. (2011) Chemical genetics reveals negative regulation of abscisic acid signaling by a plant immune response pathway. Curr Biol 21: 990–997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunze G, Zipfel C, Robatzek S, Niehaus K, Boller T, Felix G (2004) The N terminus of bacterial elongation factor Tu elicits innate immunity in Arabidopsis plants. Plant Cell 16: 3496–3507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leister RT, Dahlbeck D, Day B, Li Y, Chesnokova O, Staskawicz BJ (2005) Molecular genetic evidence for the role of SGT1 in the intramolecular complementation of Bs2 protein activity in Nicotiana benthamiana. Plant Cell 17: 1268–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Clarke JD, Zhang Y, Dong X (2001) Activation of an EDS1-mediated R-gene pathway in the snc1 mutant leads to constitutive, NPR1-independent pathogen resistance. Mol Plant Microbe Interact 14: 1131–1139 [DOI] [PubMed] [Google Scholar]
- Li Y, Gou M, Sun Q, Hua J (2010a) Requirement of calcium binding, myristoylation, and protein-protein interaction for the Copine BON1 function in Arabidopsis. J Biol Chem 285: 29884–29891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Li S, Bi D, Cheng YT, Li X, Zhang Y (2010b) SRFR1 negatively regulates plant NB-LRR resistance protein accumulation to prevent autoimmunity. PLoS Pathog 6: e1001111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Pennington BO, Hua J (2009) Multiple R-like genes are negatively regulated by BON1 and BON3 in Arabidopsis. Mol Plant Microbe Interact 22: 840–848 [DOI] [PubMed] [Google Scholar]
- Li Y, Tessaro MJ, Li X, Zhang Y (2010c) Regulation of the expression of plant Resistance gene SNC1 by a protein with a conserved BAT2 domain. Plant Physiol 153: 1425–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin BL, Wang JS, Liu HC, Chen RW, Meyer Y, Barakat A, Delseny M (2001) Genomic analysis of the Hsp70 superfamily in Arabidopsis thaliana. Cell Stress Chaperones 6: 201–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Elmore JM, Fuglsang AT, Palmgren MG, Staskawicz BJ, Coaker G (2009) RIN4 functions with plasma membrane H+-ATPases to regulate stomatal apertures during pathogen attack. PLoS Biol 7: e1000139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Schiff M, Serino G, Deng XW, Dinesh-Kumar SP (2002) Role of SCF ubiquitin-ligase and the COP9 signalosome in the N gene-mediated resistance response to Tobacco mosaic virus. Plant Cell 14: 1483–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mang HG, Qian W, Zhu Y, Qian J, Kang HG, Klessig DF, Hua J (2012) Abscisic acid deficiency antagonizes high-temperature inhibition of disease resistance through enhancing nuclear accumulation of resistance proteins SNC1 and RPS4 in Arabidopsis. Plant Cell 24: 1271–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M, Underwood W, Koczan J, Nomura K, He SY (2006) Plant stomata function in innate immunity against bacterial invasion. Cell 126: 969–980 [DOI] [PubMed] [Google Scholar]
- Menke FL, Kang HG, Chen Z, Park JM, Kumar D, Klessig DF (2005) Tobacco transcription factor WRKY1 is phosphorylated by the MAP kinase SIPK and mediates HR-like cell death in tobacco. Mol Plant Microbe Interact 18: 1027–1034 [DOI] [PubMed] [Google Scholar]
- Monaghan J, Zipfel C (2012) Plant pattern recognition receptor complexes at the plasma membrane. Curr Opin Plant Biol 15: 349–357 [DOI] [PubMed] [Google Scholar]
- Montillet JL, Hirt H (2013) New checkpoints in stomatal defense. Trends Plant Sci 18: 295–297 [DOI] [PubMed] [Google Scholar]
- Montillet JL, Leonhardt N, Mondy S, Tranchimand S, Rumeau D, Boudsocq M, Garcia AV, Douki T, Bigeard J, Laurière C, et al. (2013) An abscisic acid-independent oxylipin pathway controls stomatal closure and immune defense in Arabidopsis. PLoS Biol 11: e1001513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kurose T, Hino T, Tanaka K, Kawamukai M, Niwa Y, Toyooka K, Matsuoka K, Jinbo T, Kimura T (2007) Development of series of gateway binary vectors, pGWBs, for realizing efficient construction of fusion genes for plant transformation. J Biosci Bioeng 104: 34–41 [DOI] [PubMed] [Google Scholar]
- Noël L, Moores TL, van Der Biezen EA, Parniske M, Daniels MJ, Parker JE, Jones JD (1999) Pronounced intraspecific haplotype divergence at the RPP5 complex disease resistance locus of Arabidopsis. Plant Cell 11: 2099–2112 [PMC free article] [PubMed] [Google Scholar]
- Noël LD, Cagna G, Stuttmann J, Wirthmüller L, Betsuyaku S, Witte CP, Bhat R, Pochon N, Colby T, Parker JE (2007) Interaction between SGT1 and cytosolic/nuclear HSC70 chaperones regulates Arabidopsis immune responses. Plant Cell 19: 4061–4076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SH, Bolender N, Eisele F, Kostova Z, Takeuchi J, Coffino P, Wolf DH (2007) The cytoplasmic Hsp70 chaperone machinery subjects misfolded and endoplasmic reticulum import-incompetent proteins to degradation via the ubiquitin-proteasome system. Mol Biol Cell 18: 153–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peart JR, Lu R, Sadanandom A, Malcuit I, Moffett P, Brice DC, Schauser L, Jaggard DA, Xiao S, Coleman MJ, et al. (2002) Ubiquitin ligase-associated protein SGT1 is required for host and nonhost disease resistance in plants. Proc Natl Acad Sci USA 99: 10865–10869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Katagiri F (2009) Purification of low-abundance Arabidopsis plasma-membrane protein complexes and identification of candidate components. Plant J 57: 932–944 [DOI] [PubMed] [Google Scholar]
- Rossignol P, Collier S, Bush M, Shaw P, Doonan JH (2007) Arabidopsis POT1A interacts with TERT-V(I8), an N-terminal splicing variant of telomerase. J Cell Sci 120: 3678–3687 [DOI] [PubMed] [Google Scholar]
- Sawinski K, Mersmann S, Robatzek S, Böhmer M (2013) Guarding the green: pathways to stomatal immunity. Mol Plant Microbe Interact 26: 626–632 [DOI] [PubMed] [Google Scholar]
- Schütze K, Harter K, Chaban C (2009) Bimolecular fluorescence complementation (BiFC) to study protein-protein interactions in living plant cells. Methods Mol Biol 479: 189–202 [DOI] [PubMed] [Google Scholar]
- Schwessinger B, Roux M, Kadota Y, Ntoukakis V, Sklenar J, Jones A, Zipfel C (2011) Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet 7: e1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung DY, Guy CL (2003) Physiological and molecular assessment of altered expression of Hsc70-1 in Arabidopsis. Evidence for pleiotropic consequences. Plant Physiol 132: 979–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung DY, Vierling E, Guy CL (2001) Comprehensive expression profile analysis of the Arabidopsis Hsp70 gene family. Plant Physiol 126: 789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takemoto D, Jones DA (2005) Membrane release and destabilization of Arabidopsis RIN4 following cleavage by Pseudomonas syringae AvrRpt2. Mol Plant Microbe Interact 18: 1258–1268 [DOI] [PubMed] [Google Scholar]
- Tör M, Gordon P, Cuzick A, Eulgem T, Sinapidou E, Mert-Türk F, Can C, Dangl JL, Holub EB (2002) Arabidopsis SGT1b is required for defense signaling conferred by several downy mildew resistance genes. Plant Cell 14: 993–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzfira T, Tian GW, Lacroix B, Vyas S, Li J, Leitner-Dagan Y, Krichevsky A, Taylor T, Vainstein A, Citovsky V (2005) pSAT vectors: a modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol Biol 57: 503–516 [DOI] [PubMed] [Google Scholar]
- Uppalapati SR, Ishiga Y, Ryu CM, Ishiga T, Wang K, Noël LD, Parker JE, Mysore KS (2011) SGT1 contributes to coronatine signaling and Pseudomonas syringae pv. tomato disease symptom development in tomato and Arabidopsis. New Phytol 189: 83–93 [DOI] [PubMed] [Google Scholar]
- van der Biezen EA, Freddie CT, Kahn K, Parker JE, Jones JD (2002) Arabidopsis RPP4 is a member of the RPP5 multigene family of TIR-NB-LRR genes and confers downy mildew resistance through multiple signalling components. Plant J 29: 439–451 [DOI] [PubMed] [Google Scholar]
- Walter M, Chaban C, Schütze K, Batistic O, Weckermann K, Näke C, Blazevic D, Grefen C, Schumacher K, Oecking C, et al. (2004) Visualization of protein interactions in living plant cells using bimolecular fluorescence complementation. Plant J 40: 428–438 [DOI] [PubMed] [Google Scholar]
- Wang Z, Meng P, Zhang X, Ren D, Yang S (2011) BON1 interacts with the protein kinases BIR1 and BAK1 in modulation of temperature-dependent plant growth and cell death in Arabidopsis. Plant J 67: 1081–1093 [DOI] [PubMed] [Google Scholar]
- Xin XF, He SY (2013) Pseudomonas syringae pv. tomato DC3000: a model pathogen for probing disease susceptibility and hormone signaling in plants. Annu Rev Phytopathol 51: 473–498 [DOI] [PubMed] [Google Scholar]
- Yang H, Li Y, Hua J (2006a) The C2 domain protein BAP1 negatively regulates defense responses in Arabidopsis. Plant J 48: 238–248 [DOI] [PubMed] [Google Scholar]
- Yang H, Yang S, Li Y, Hua J (2007) The Arabidopsis BAP1 and BAP2 genes are general inhibitors of programmed cell death. Plant Physiol 145: 135–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Hua J (2004) A haplotype-specific Resistance gene regulated by BONZAI1 mediates temperature-dependent growth control in Arabidopsis. Plant Cell 16: 1060–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang S, Yang H, Grisafi P, Sanchatjate S, Fink GR, Sun Q, Hua J (2006b) The BON/CPN gene family represses cell death and promotes cell growth in Arabidopsis. Plant J 45: 166–179 [DOI] [PubMed] [Google Scholar]
- Zeng W, He SY (2010) A prominent role of the flagellin receptor FLAGELLIN-SENSING2 in mediating stomatal response to Pseudomonas syringae pv tomato DC3000 in Arabidopsis. Plant Physiol 153: 1188–1198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Van Pelt CK, Henion JD (2003a) Automated chip-based nanoelectrospray-mass spectrometry for rapid identification of proteins separated by two-dimensional gel electrophoresis. Electrophoresis 24: 3620–3632 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Goritschnig S, Dong X, Li X (2003b) A gain-of-function mutation in a plant disease resistance gene leads to constitutive activation of downstream signal transduction pathways in suppressor of npr1-1, constitutive 1. Plant Cell 15: 2636–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wu Y, Gao M, Zhang J, Kong Q, Liu Y, Ba H, Zhou J, Zhang Y (2012) Disruption of PAMP-induced MAP kinase cascade by a Pseudomonas syringae effector activates plant immunity mediated by the NB-LRR protein SUMM2. Cell Host Microbe 11: 253–263 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Qian W, Hua J (2010) Temperature modulates plant defense responses through NB-LRR proteins. PLoS Pathog 6: e1000844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel C, Robatzek S, Navarro L, Oakeley EJ, Jones JD, Felix G, Boller T (2004) Bacterial disease resistance in Arabidopsis through flagellin perception. Nature 428: 764–767 [DOI] [PubMed] [Google Scholar]