Genome sequencing and genomic technologies facilitate discovery and understanding of plant metabolic pathways.
Abstract
Genome-enabled discoveries are the hallmark of 21st century biology, including major discoveries in the biosynthesis and regulation of plant metabolic pathways. Access to next generation sequencing technologies has enabled research on the biosynthesis of diverse plant metabolites, especially secondary metabolites, resulting in a broader understanding of not only the structural and regulatory genes involved in metabolite biosynthesis but also in the evolution of chemical diversity in the plant kingdom. Several paradigms that govern secondary metabolism have emerged, including that (1) gene family expansion and diversification contribute to the chemical diversity found in the plant kingdom, (2) genes encoding biochemical pathway components are frequently transcriptionally coregulated, and (3) physical clustering of nonhomologous genes that encode components of secondary metabolic pathways can occur. With an increasing knowledge base that is coupled with user-friendly and inexpensive technologies, biochemists are poised to accelerate the annotation of biochemical pathways relevant to human health, agriculture, and the environment.
Arabidopsis (Arabidopsis thaliana) has been in a postgenomics era for 15 years; as a consequence, a substantial portion of our knowledge of plant metabolism is derived from this model species, including both primary and secondary metabolites. Primary metabolites are considered to be essential for plant growth and development, whereas secondary metabolites (also known as specialized metabolites) were traditionally believed to improve plant function, such as defense against abiotic and biotic stress. However, a more recent definition to distinguish between these groups of metabolites and their pathways is based on their taxonomic distribution, with primary metabolism referring to pathways universally distributed throughout the plant kingdom and secondary metabolism referring to pathways and resulting metabolites that are taxa or species specific (Pichersky and Gang, 2000).
GENOME SEQUENCING AS A MAJOR CATALYST IN UNDERSTANDING PLANT METABOLISM
The first 15 years of the 21st century have seen a revolution in genomic technologies, with advances continuing to impact all areas of plant biology, including plant metabolism, as access to the genome sequence along with associated large-scale data sets, such as expression and metabolite profiles, can empower the discovery of the biosynthetic and regulatory pathways of not only discrete but also large classes of metabolites. Indeed, generation of the genome sequence of Arabidopsis in 2000 (Arabidopsis Genome Initiative, 2000) was seminal in plant metabolism research, as not only was the full genome sequence of a plant determined but the development of accessible and robust functional genomics resources, such as full-length complementary DNA clone sets, tagged mutant lines, and a rapid and facile transformation method, collectively enabled improvements in our understanding of plant metabolism (Buell and Last, 2010). Another key attribute of completing the Arabidopsis genome was the high quality of the underlying sequence and the investment in genome annotation with publicly accessible and curated databases, including a dedicated biochemical pathway database (Mueller et al., 2003), thereby enabling broad access to the wealth of data generated by the community on this focal species.
The Arabidopsis genome was sequenced using dideoxy chain termination chemistry with a bacterial artificial chromosome (BAC)-by-BAC approach that is now outdated (Arabidopsis Genome Initiative, 2000). Current sequencing technologies utilize a sequencing-by-synthesis approach that is ultra high throughput, extremely inexpensive, and available to any researcher in the world. Collectively, these technologies are termed next generation sequencing (NGS) methods, with the Illumina platform dominating the market (http://www.illumina.com) and the Pacific Biosciences and Ion Torrent platforms employed primarily for long-read generation and amplicon sequencing, respectively (http://www.pacificbiosciences.com and https://www.lifetechnologies.com/us/en/home/brands/ion-torrent.html). Due to the rapid evolution of sequencing technologies, the technical aspects of these platforms are not discussed in this article, and the reader is referred to a review article available on these platforms (Mardis, 2013). One obvious application of NGS is the generation of de novo genome and transcriptome sequences, both of which have been highly informative across many species for understanding plant metabolism. However, NGS technologies have numerous applications outside de novo assemblies, and some emerging examples of their use in revealing complexities in plant metabolism are highlighted in this Update article.
While Arabidopsis has and will continue to serve as a model species for plant metabolism, it fails to capture the full diversity of chemistry present within the plant kingdom. In particular, given that secondary metabolites are taxa or even species specific, accessibility to the genomic resources of each target species is essential for genome-enabled pathway discovery. Recent improvements in assembly algorithms and annotation software, coupled with access to increased computational power and NGS technology, have enabled the generation of genome sequences of a wide number of plant species that can be used as a platform for secondary metabolite pathway discovery (Fig. 1). While challenges still exist in the assembly of large and/or repetitive genomes, polyploid species, and heterozygosity, these barriers can be addressed, in part, using approaches that bypass or minimize these complexities in the genome assembly process (Hirsch and Buell, 2013). Indeed, genome sequences are available for most crop species, albeit in varying degrees of quality, including a wide set of species with relevant secondary metabolism. The discovery potential of NGS in plant metabolism is unlimited, and below, we highlight how access to genome sequence(s) enabled metabolite pathway discovery in species across the plant kingdom and some interesting features in plant metabolism discovered along the way. Due to the broad application of genomics in secondary metabolism, this Update article focuses on genome-enabled discoveries in secondary rather than primary metabolism.
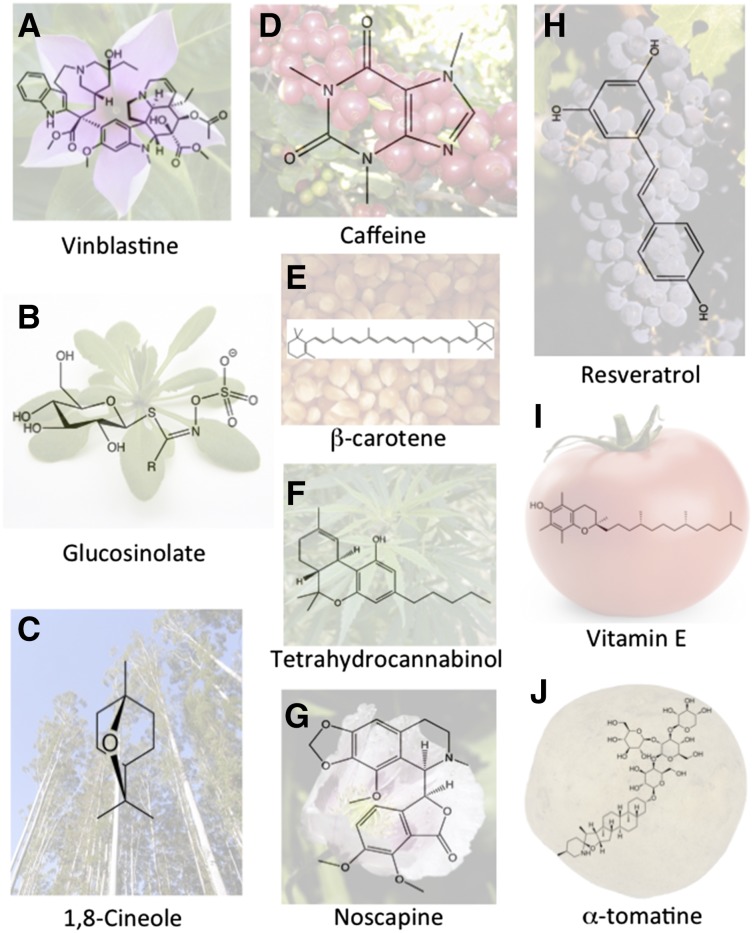
Figure 1.
Secondary metabolites for which genome sequence access facilitated gene discovery and the understanding of the evolution of secondary metabolites. A, Madagascar periwinkle (Catharanthus roseus) with the monoterpene indole alkaloid vinblastine (Kellner et al., 2015). B, Arabidopsis with the amino acid-derived secondary metabolite glucosinolate (Chan et al., 2010). C, E. grandis with the terpene 1,8-cineole (Myburg et al., 2014). D, Coffea canephora with the alkaloid caffeine (Denoeud et al., 2014). E, Maize (Zea mays) with the carotenoid β-carotene (Fu et al., 2013). F, Marijuana (Cannabis sativa) with the cannabinoid tetrahydrocannabinol (van Bakel et al., 2011). G, Poppy (Papaver somniferum) with the phthalide isoquinoline alkaloid noscapine (Winzer et al., 2012). H, Grape (Vitis vinifera) with the polyphenol resveratrol (Jaillon et al., 2007). I, Tomato with vitamin E (Quadrana et al., 2014). J, Potato with the glycoalklaloid α-tomatine (Itkin et al., 2013).
BASIC GENOMIC PRINCIPLES OF GENES INVOLVED IN SECONDARY METABOLISM: DUPLICATION, COEXPRESSION, AND PHYSICAL CLUSTERING
One complication in plant metabolism research, especially that of secondary metabolism, is that all angiosperms sequenced to date show evidence of whole-genome duplication, which has resulted in expansions of gene families leading to metabolic diversity over evolutionary time. Additional partial genome duplications, known as segmental duplications, and tandem duplications further contribute to the large gene family size observed in plant genomes. Once duplicated, genes can evolve new functions (neofunctionalization), partition function into the duplicates (subfunctionalization), or become pseudogenes (Conant et al., 2014). Examination of genome sequences for 16 species that span evolution from algae to higher plants enabled the identification of genomic signatures that distinguish plant secondary metabolism genes from primary metabolism genes (Chae et al., 2014). Features of genes involved in secondary metabolism include a tendency to be retained after gene duplication, lineage specificity, and physical clustering within the genome compared with primary metabolism genes.
A correlative approach to annotating gene function is the association of gene expression patterns and profiles with phenotype, including metabolites. Access to NGS permits the generation of rapid and inexpensive expression abundance data through RNA sequencing. From an expression atlas, in which a wide range of tissues and treatments are examined, coordinated expression analyses (coexpression) can reveal genes that are within the same regulatory network (Fig. 2). This guilt-by-association approach, when coupled with an examination of the functional annotation of genes or transcripts, has been highly successful in identifying a subset of candidate biosynthetic pathway genes for functional validation (Weber, 2015).
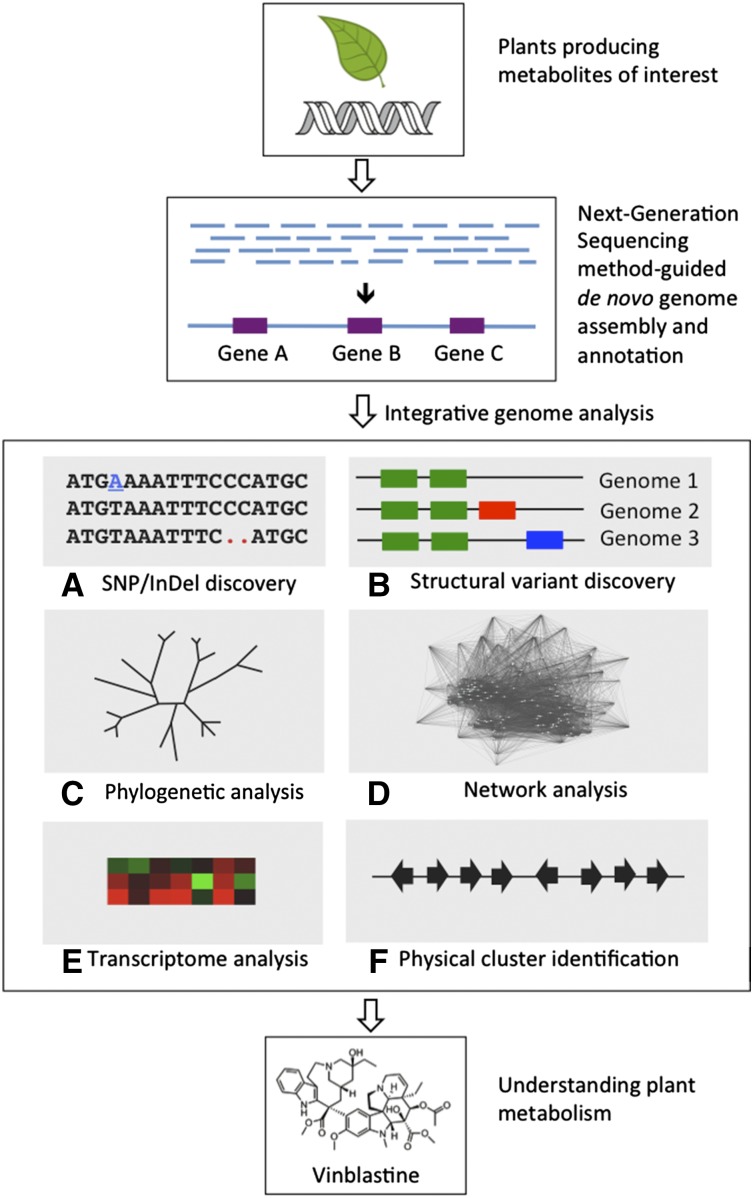
Figure 2.
Schematic work flow of NGS-guided de novo genome assembly and integrative genome analysis to understand plant metabolism. The high accuracy and coverage of NGS-derived reads allow de novo genome assembly and gene annotation. Integrative genome analyses coupled with transcriptomic and metabolic data sets enable the identification of biosynthetic and regulatory pathways controlling plant metabolism. Commonly used approaches are shown with an example of a study where these approaches were applied. A, Single-nucleotide polymorphism (SNP) and insertion/deletion (InDel) discovery (Chen et al., 2014). B, Structural variant discovery (Da Silva et al., 2013). C, Phylogenetic analysis (Denoeud et al., 2014). D, Network analysis (Itkin et al., 2013). E, Transcriptome analysis (Fu et al., 2013). F, Physical cluster identification (Winzer et al., 2012).
In bacterial genomes, genes involved in metabolic and regulatory pathways are frequently arranged in operons, which are absent in plant genomes. However, one theme that has emerged from genome analyses of secondary metabolism is that genes within specialized metabolic pathways can be physically clustered (for review, see DellaPenna and O’Connor, 2012). This nonhomologous gene clustering is hypothesized to be a consequence of evolutionary pressure to retain all components of the pathway in a discrete interval, thereby ensuring that the multigenic trait is inherited as a single locus and avoiding the buildup of toxic intermediates or an inability to synthesize the final metabolite, which may have a role in adaptation or survival. We highlight below three classes of secondary metabolites that demonstrate the extent to which gene/genome duplication and diversification, coexpression, and/or physical clustering can facilitate discovery and an improved understanding of secondary metabolic pathways.
Terpenes
Terpenes, diverse compounds derived from five-carbon isoprene units, are the largest class of plant secondary metabolites, are important in plant defense, and have health benefits for humans (Tholl and Lee, 2011). The chemical diversity of terpene metabolites is mainly governed by terpene synthases (TSs) that generate the scaffold, with structural diversity containing various numbers of carbons and a multitude of cytochrome P450-dependent monooxygenases (CYPs) that modify the scaffold. From a comparative genome analysis, 113 TSs were identified in Eucalyptus grandis compared with two in Physcomitrella patens, some of which were lineage specific to E. grandis and contributed to the diversity of terpene-derived molecules such as 1,8-cineole in E. grandis (Myburg et al., 2014).
A recent study revealed terpene diversification and the evolution of terpene biosynthetic genes in multiple plant genomes (Boutanaev et al., 2015). That study analyzed 17 sequenced plant genomes, including 12 eudicot and five monocot species, to determine the organization and evolution of terpene pathway genes. By identifying TS and CYP gene pairs and their physical location within the genome, they observed frequencies of TS/CYP gene pairs higher than that expected by chance, suggesting nonrandom association of TS and CYP gene pairs in the surveyed genomes. In addition, TSs were predominantly paired with CYP71 family genes in both eudicots and monocots, confirming previously identified TS/CYP gene clusters in Ricinus communis known to be involved in diterpene synthesis. Novel TS/CYP pairs were also observed, such as three functional TS/CYP pairs in R. communis, Arabidopsis, and Cucumis sativus. This study also revealed different assembly patterns of TS/CYP pairs in eudicots and monocots, suggesting diverged evolutionary mechanisms to assemble terpene pathways in the two major divisions of angiosperms. Based on the frequency of TS/CYP pairs and their correlations, it was hypothesized that eudicot TS/CYP gene pairs arose from a common ancestral gene pair by duplication, providing building blocks to diversify terpene biosynthetic genes. On the contrary, no correlation was observed for TS/CYP gene pairs in monocots, suggesting that they may have arisen independently as a consequence of genome rearrangements.
Monoterpene Indole Alkaloids
Madagascar periwinkle synthesizes an array of monoterpene indole alkaloids with high structural diversity, including the anticancer compounds vincristine and vinblastine (O’Connor and Maresh, 2006). Historical efforts to elucidate the pathway yielded some of the genes in the pathway using an array of slow and laborious processes. In 2010, large expression and metabolite profiling data sets were generated that enabled the discovery of a large number of genes in the vinblastine/vincristine pathway (Miettinen et al., 2014). To further accelerate pathway discovery in Madagascar periwinkle, a draft genome sequence was generated (Kellner et al., 2015) that was used to refine the expression atlas and coexpression networks and guide the discovery of additional and novel candidate genes involved in monoterpene indole alkaloid regulation, biosynthesis, and transport. The Madagascar periwinkle genome sequence revealed evidence of gene expansion coupled with the neofunctionalization of genes that encode not only the biosynthetic pathway but also regulatory components of monoterpene indole alkaloids. Physical clustering of a subset of genes involved in monoterpene indole alkaloid synthesis was also observed, showing that even with a draft genome sequence that has a limited scaffold length, new discoveries in metabolism can be readily made.
Steroidal Glycoalkaloids
Solanum spp. such as potato (Solanum tuberosum) and tomato (Solanum lycopersicum) produce steroidal glycoalkaloids (SGAs), including α-solanine, α-chaconine, and α-tomatine. SGAs are derived from alkaloids linked to a sugar moiety and are known to play crucial roles in plant defense yet have toxicity in humans (Friedman, 2006). Potato and tomato are agriculturally valuable crop species and have served as model systems for tuber and fruit development studies, respectively. De novo genome sequences are available for both species, with substantial synteny and conserved gene content between the two genomes (Potato Genome Sequencing Consortium, 2011; Tomato Genome Consortium, 2012). It was previously known that the SGA pathway entails the conversion of cholesterol to steroidal alkaloids through a series of hydroxylation, oxidation, and transamination reactions followed by decoration with various sugar moieties using UDP-glycosyltransferases such as SOLANIDINE GALACTOSYLTRANSFERASE (SGT1) and GLYCOALKALOID METABOLISM1 (GAME1) in potato and tomato, respectively. However, the pathway still remained incomplete. To discover novel genes in the SGA pathway, Itkin et al. (2013) investigated genes that were coexpressed with GAME1/SGT1 and identified 16 genes in potato and tomato, including GAME4, which encodes a cytochrome P450. They also discovered that the SGA pathway genes were physically clustered and coexpressed with GAME1 on chromosome 7 and with GAME4 on chromosome 12, with conserved synteny between tomato and potato. With functional validation of the newly identified genes, they proposed an SGA pathway starting from cholesterol and the decoration of up to four sugar moieties linked to steroidal alkaloids.
STRUCTURAL VARIATION AS A DRIVER OF METABOLITE DIVERSITY
Early sequencing efforts in bacteria revealed substantial genome diversity among isolates, leading to the concept of the core and dispensable genome, with the collective genome sequence of a species termed the pan-genome (Tettelin et al., 2005). This structural variation is in the form of copy number variation and presence/absence variation, in which genes are variable among accessions of a single species (Fig. 2). In plants, copy number variation and presence/absence variation have been reported in several species, some of which are associated with phenotypic diversity, including abiotic and biotic stress, development, and secondary metabolites (Cook et al., 2012; Maron et al., 2013); below are two examples in which structural variation is associated with the production of secondary metabolites of importance to human health.
Polyphenols
Grapes are known for the production of resveratrol, a natural polyphenol with health benefits on obesity and aging-related diseases (Baur et al., 2006), while terpene-derived metabolites in grapes contribute to the aromatic features of wine. The grapevine genome was sequenced in 2007 using the inbred Pinot Noir variety PN40024 (Jaillon et al., 2007), and access to the grapevine genome enabled the annotation of stilbene synthases and TSs that led to the synthesis of resveratrol and terpenoids, respectively. Both gene families were highly expanded relative to other sequenced plant genomes, consistent with the hypothesis that gene duplication and diversification lead to metabolic diversity. With access to NGS technologies, Da Silva et al. (2013) generated sequence data for the Uruguayan Tannat grapevine clone UY11, which accumulates high levels of polyphenols in the berry skin and seed. Comparison of the UY11 genome with the PN40024 reference genome revealed 1,873 genes that were not present in the PN40024 genome. With respect to polyphenol biosynthesis, 141 novel UY11 genes that encode 19 different enzymes involved in polyphenol biosynthesis and the expression of cultivar-specific genes associated with altered polyphenol accumulation in UY11 were identified.
Phthalide Isoquinoline Alkaloids
Poppy is well known for its production of opiates that are used as pain suppressants. Noscapine, a phthalide isoquinoline alkaloid, has cough suppressant and antitumor activities. Poppy varieties exhibit differential metabolite profiles with respect to the isoquinoline alkaloids, including high- and null-noscapine-producing varieties. Through an integrated approach using an F2 mapping population, sequencing of large insert BAC clones, and virus-induced gene silencing, a 10-gene cluster on a 221-kb segment novel to the high-noscapine-producing variety was identified for noscapine biosynthesis (Winzer et al., 2012). Based on the occurrence of small gene families (carboxylesterase, CYP82 cytochrome P450, and O-methyltransferase) and single-copy genes (short-chain dehydrogenase/reductase, acetyltransferase, and CYP719A21 cytochrome P450) in the cluster of 10 biosynthetic genes, the authors hypothesized that genome reorganization occurred before and after duplication, shaping the physical cluster of noscapine biosynthetic genes. They speculated that this cluster evolution might benefit the coinheritance of a favorable combination of alleles leading to the production of desired secondary metabolites.
GENOMICS-FACILITATED GENETIC APPROACHES FOR METABOLITE PATHWAY DISCOVERY
Classical genetic approaches to identify causal genes within a biosynthetic pathway have included forward and reverse genetic screens, positional cloning, and quantitative trait locus mapping. These classical approaches can be accelerated and performed with increased efficiency through NGS methodologies. Not only can NGS be used to readily identify sequence variants (SNPs and insertions/deletions) between accessions (Fig. 2), it can be further applied in approaches such as whole-genome resequencing in bulk segregant analyses (James et al., 2013) and k-mer-based approaches to find causal mutations (Nordström et al., 2013). Indeed, due to low cost, high coverage, and accuracy, as well as user-friendly bioinformatics pipelines, a single laboratory can now utilize NGS methods combined with genetics approaches for gene discovery. For example, access to genome sequences for multiple accessions permits association studies to link phenotype (e.g. metabolite) with genotype using either linkage mapping or a genome-wide association approach. With access to large transcriptome data sets, transcript levels can be linked with sequence variants to identify expression quantitative trait loci (eQTLs) associated with the synthesis, regulation, or transport of metabolites.
Genome-Wide Association Studies
NGS methods can enable the discovery of large numbers of SNPs for use in a genome-wide association analysis of metabolism. In rice (Oryza sativa), Chen et al. (2014) resequenced 529 diverse accessions generating 6.4 million SNPs. When combined with metabolite profiling of 840 metabolites, including amino acids, flavonoids, and terpenoids, they revealed the genetic and biochemical basis of metabolic diversity in rice by associating casual SNPs and genes with metabolite profiles. For example, an SNP located in a methyltransferase gene was associated with the levels of trigonelline, the N-methyl conjugate of nicotinic acid. In vitro assays revealed its enzymatic activity as a nicotinic acid:N-methyltransferase, and overexpression of the methyltransferase increased trigonelline accumulation in transgenic lines, showing that it is a functional enzyme in trigonelline biosynthesis. For flavonoid biosynthesis, an SNP located in a putative UDP-glucosyltransferase gene was associated with the production of a number of flavonoids and chlorogenic acid. In this study, the candidate gene was functionally annotated as a flavone 5-O-glucosyltransferase, and based on metabolite profiles of transgenic lines that overexpressed this gene, it was shown to regulate the level of flavone 5-O-glucosyltransferase.
Arabidopsis and other Brassicaceae species produce glucosinolates, which are amino acid-derived secondary metabolites known to have roles in plant defense against insects and pathogens. Glucosinolates can be categorized into three groups, aliphatic, aromatic, and indole glucosinolates, depending on the types of amino acids from which they are derived. Chan et al. (2010) performed a genome-wide association study with 96 Arabidopsis accessions, assessing the natural diversity of 43 glucosinolate phenotypes using approximately 230,000 SNPs. They identified 172 genes containing SNPs associated with glucosinolate profiles and discovered two 2-oxoglutarate-dependent dioxygenase genes ALKENYL HYDROXALKYL PRODUCING2 (AOP2) and AOP3 and METHYLTHIOALKYLMALATE SYNTHASE1 (MAM1) that control two previously known glucosinolate-related quantitative trait loci. They also detected extended linkage disequilibrium surrounding AOP2, AOP3, and MAM1 genes as a haplotype that was associated with glucosinolate chemotypes in the 96 Arabidopsis accessions, suggesting selection during evolution of the pathway. In addition to the genes directly affecting glucosinolate biosynthesis, transcriptional variation of a GluCys ligase involved in glutathione metabolism, which had been hypothesized to indirectly control glucosinolate accumulation via a regulatory and/or biosynthetic linkage, was observed to alter glucosinolate production in that study.
eQTLs
In maize, Fu et al. (2013) integrated transcriptome and metabolite profiling to identify genes involved in kernel development, including carotenoid levels. RNA sequencing was used to generate transcript abundances and SNPs from developing kernels of 368 maize inbred lines, which yielded 16,408 eQTLs for 14,375 genes. Of the 20 genes in the carotenoid metabolic pathway whose expression was associated with carotenoid levels, six genes were associated with an eQTL, including LYCOPENE EPSILON CYCLASE1 and β-CAROTENE HYDROXYLASE1. In addition, 55 genes were correlated with carotenoid production, of which 19 genes were associated with an eQTL. Coexpression analyses of genes associated with eQTLs revealed three clusters of genes that were coexpressed with known carotenoid pathway genes, providing a list of candidate genes in the pathway. This study demonstrates how an eQTL study, when combined with metabolite profiling, can discover novel genes even in well-studied biosynthetic pathways.
COMPARATIVE AND PHYLOGENETIC APPROACHES LEVERAGE EVOLUTIONARY EVENTS ACROSS TAXA TO REVEAL GENES INVOLVED IN SECONDARY METABOLISM
With the increasing number of genome sequences available, the power of comparative and phylogenetic approaches to gene discovery can be harnessed. Traditionally, phylogenetic relationships of key metabolic enzymes were performed at the single gene level with limited sequence information from a small number of species. However, with advances in NGS technology, whole-genome sequences and annotation are available not only for multiple species within a taxonomic family but also for basal angiosperm genera such as Amborella, gymnosperms, lycopods, and mosses that permit more robust queries into the mechanisms by which metabolic pathways evolved. With the availability of multiple genomes from a single or related taxonomic group, synteny (shared gene order), which reflects shared evolution and enables the prediction of the emergence of a trait and orthologous genes in a pathway between species and taxa, can be identified. With the abundance of genome sequences within some clades as well as the breadth of genome sequences across the plant kingdom, comparative analyses not only between species but also within species can be a powerful approach to reveal the evolutionary origins of biochemical pathways.
Purine Alkaloids
Caffeine belongs to a class of purine alkaloid metabolites whose synthesis is derived from purine nucleotides followed by multiple steps of N-methylation (Ziegler and Facchini, 2008). The coffee (Coffea spp.) genome, an important beverage crop with stimulating effects due to the production of caffeine, was sequenced in 2014 using a doubled haploid accession of C. canephora (Denoeud et al., 2014). Analyses of the genome revealed that N-methyltransferases (NMTs), including xanthosine methyltrasferase, theobromine synthase, and caffeine synthase, that catalyze later steps in caffeine biosynthesis were enriched in a C. canephora lineage-specific ortholog group. Phylogenetic analysis with genes from the ortholog group containing known caffeine biosynthetic genes and NMTs from tea (Camellia sinensis) and cacao (Theobroma cacao) revealed a cluster of C. canephora NMTs distinct from tea and cacao NMTs. The species-specific expansion of NMT gene families in the C. canephora genome, coupled with the convergent evolution of caffeine biosynthesis genes in coffee, cacao, and tea, highlight the power of comparative genomics in understanding the evolution of secondary metabolite pathways in plants.
Cannabinoids
The marijuana genome is an example of the successful application of NGS for metabolism study in a nonmodel system (van Bakel et al., 2011). Marijuana produces cannabinoids, a class of prenylated polyketides that are synthesized from the short-chain fatty acid hexanoate and geranyl diphosphate. To understand cannabinoid biosynthesis, the authors generated a draft genome of the marijuana-producing strain Purple Kush and identified cannabinoid biosynthetic genes such as tetrahydrocannabinol acid synthase. By resequencing a nonmarijuana strain (Finola) and comparing it with the Purple Kush sequence, they revealed a large expansion of an ACYL ACTIVATING ENZYME3 gene encoding an enzyme that may be involved in the synthesis of hexanoate, a precursor of tetrahydrocannabinol acid, in the marijuana-producing strain but not in the non-marijuana-producing strain. They extended their comparative analysis by resequencing two additional strains, USO-31 (nonmarijuana strain) and Chemdawg (marijuana strain), and generating a phylogenetic tree that revealed closer relationships within marijuana strains and nonmarijuana strains than between the two chemotypes. This approach expanded our understanding of the metabolic pathway for the production of alkaloids and revealed genetic variation associated with chemical variation within the species.
IT IS NOT JUST THE DNA SEQUENCE PER SE: EPIGENETIC MODIFICATION OF DNA LEADS TO NEW PHENOTYPES
The application of NGS technologies extends past that of determining the primary sequence of DNA and RNA molecules, and a wide range of applications permit the assessment of other features of the genome and transcriptome, including that of the epigenome. In tomato, Quadrana et al. (2014) identified epialleles that regulate vitamin E accumulation. Through promoter sequence analyses, differential methylation of a retrotransposon located in the promoter of 2-methyl-6-phytylquinol methyltransferase (VITAMIN E DEFECTIVE3 [VTE3]), which encodes the final step in the biosynthesis of γ- and α-tocopherols, was discovered. Using a combination of genomic, transcriptomic, and metabolomic approaches in different tomato genetic backgrounds, it was shown that different epialleles lead to differential expression of the VTE3 gene, causing diverse accumulation of vitamin E content in tomato fruit. This study shows the example of how advances in NGS technologies (i.e. epigenome profiling), when coupled with genetics and metabolite profiling, can enable new discoveries on the regulation of metabolism.
CONCLUSION AND FUTURE PROSPECTS
In less than a decade since the emergence of the first NGS platform, the incorporation of this technology into plant metabolism has been astounding. Certainly, improvements in the read length, quality, throughput, and cost will further integrate NGS methods into metabolic pathway discovery and reconstruction. Having complete and high-quality draft genomes with limited expenditure of effort will facilitate the discovery of physically clustered pathway components and the dissection of gene duplications associated with biochemical diversity within or between species. However, one challenge in employing a genomics approach to understand plant metabolism is that the majority of functional annotation is automated and derived from transitive annotation from other genomes or via sequence similarity. The circular nature of functional annotation leads to, at best, vague and, at worst, incorrect annotation. Coupled with the high frequency of whole-genome duplication and the resulting large gene family content in most plant genomes, the lack of high-quality functional annotation presents a challenge in accelerating plant metabolism research. Thus, the need for paradigm-changing methods to sift through a set of candidate genes with improved efficiency and effort is great, and the development of methods for high-throughput functional screening will enable more robust data mining of biochemical pathways across the plant kingdom. Another challenge is how to select an optimal computational pipeline and validate its performance and how to interpret the results properly. The application of different algorithms with arbitrary assumptions or cutoffs may lead to substantial differences in the output and affect biological interpretations; thus, knowledge of the underlying principles of the employed algorithms and associated software as well as the biology is imperative (Omranian et al., 2015). Lastly, higher spatial resolution transcriptomic and metabolomics methods need to be incorporated into genome-guided pathway discovery approaches to enable the annotation of tissue- and organelle-specific enzymatic reactions and/or intercellular or intracellular transportation of intermediates and final products, thereby minimizing the errors generated from automated genome prediction and whole-tissue expression and metabolite data (Ziegler and Facchini, 2008; Dixon and Pasinetti, 2010). Regardless of these challenges, the era of genome-enabled pathway discovery is upon us now, and the eventual rewards of this knowledge to agriculture, human health, and the environment will be many.
Glossary
- BAC
bacterial artificial chromosome
- NGS
next-generation sequencing
- TS
terpene synthase
- CYP
cytochrome P450-dependent monooxygenase
- SGA
steroidal glycoalkaloid
- SNP
single-nucleotide polymorphism
- eQTL
expression quantitative trait locus
- NMT
N-methyltransferase
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Baur JA, Pearson KJ, Price NL, Jamieson HA, Lerin C, Kalra A, Prabhu VV, Allard JS, Lopez-Lluch G, Lewis K, et al. (2006) Resveratrol improves health and survival of mice on a high-calorie diet. Nature 444: 337–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boutanaev AM, Moses T, Zi J, Nelson DR, Mugford ST, Peters RJ, Osbourn A (2015) Investigation of terpene diversification across multiple sequenced plant genomes. Proc Natl Acad Sci USA 112: E81–E88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buell CR, Last RL (2010) Twenty-first century plant biology: impacts of the Arabidopsis genome on plant biology and agriculture. Plant Physiol 154: 497–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae L, Kim T, Nilo-Poyanco R, Rhee SY (2014) Genomic signatures of specialized metabolism in plants. Science 344: 510–513 [DOI] [PubMed] [Google Scholar]
- Chan EKF, Rowe HC, Kliebenstein DJ (2010) Understanding the evolution of defense metabolites in Arabidopsis thaliana using genome-wide association mapping. Genetics 185: 991–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Gao Y, Xie W, Gong L, Lu K, Wang W, Li Y, Liu X, Zhang H, Dong H, et al. (2014) Genome-wide association analyses provide genetic and biochemical insights into natural variation in rice metabolism. Nat Genet 46: 714–721 [DOI] [PubMed] [Google Scholar]
- Conant GC, Birchler JA, Pires JC (2014) Dosage, duplication, and diploidization: clarifying the interplay of multiple models for duplicate gene evolution over time. Curr Opin Plant Biol 19: 91–98 [DOI] [PubMed] [Google Scholar]
- Cook DE, Lee TG, Guo X, Melito S, Wang K, Bayless AM, Wang J, Hughes TJ, Willis DK, Clemente TE, et al. (2012) Copy number variation of multiple genes at Rhg1 mediates nematode resistance in soybean. Science 338: 1206–1209 [DOI] [PubMed] [Google Scholar]
- Da Silva C, Zamperin G, Ferrarini A, Minio A, Dal Molin A, Venturini L, Buson G, Tononi P, Avanzato C, Zago E, et al. (2013) The high polyphenol content of grapevine cultivar Tannat berries is conferred primarily by genes that are not shared with the reference genome. Plant Cell 25: 4777–4788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DellaPenna D, O’Connor SE (2012) Plant gene clusters and opiates. Science 336: 1648–1649 [DOI] [PubMed] [Google Scholar]
- Denoeud F, Carretero-Paulet L, Dereeper A, Droc G, Guyot R, Pietrella M, Zheng C, Alberti A, Anthony F, Aprea G, et al. (2014) The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science 345: 1181–1184 [DOI] [PubMed] [Google Scholar]
- Dixon RA, Pasinetti GM (2010) Flavonoids and isoflavonoids: from plant biology to agriculture and neuroscience. Plant Physiol 154: 453–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman M. (2006) Potato glycoalkaloids and metabolites: roles in the plant and in the diet. J Agric Food Chem 54: 8655–8681 [DOI] [PubMed] [Google Scholar]
- Fu J, Cheng Y, Linghu J, Yang X, Kang L, Zhang Z, Zhang J, He C, Du X, Peng Z, et al. (2013) RNA sequencing reveals the complex regulatory network in the maize kernel. Nat Commun 4: 2832. [DOI] [PubMed] [Google Scholar]
- Hirsch CN, Buell CR (2013) Tapping the promise of genomics in species with complex, nonmodel genomes. Annu Rev Plant Biol 64: 89–110 [DOI] [PubMed] [Google Scholar]
- Itkin M, Heinig U, Tzfadia O, Bhide AJ, Shinde B, Cardenas PD, Bocobza SE, Unger T, Malitsky S, Finkers R, et al. (2013) Biosynthesis of antinutritional alkaloids in solanaceous crops is mediated by clustered genes. Science 341: 175–179 [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al. (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449: 463–467 [DOI] [PubMed] [Google Scholar]
- James GV, Patel V, Nordström KJ, Klasen JR, Salomé PA, Weigel D, Schneeberger K (2013) User guide for mapping-by-sequencing in Arabidopsis. Genome Biol 14: R61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellner F, Kim J, Clavijo BJ, Hamilton JP, Childs KL, Vaillancourt B, Cepela J, Habermann M, Steuernagel B, Clissold L, et al. (2015) Genome-guided investigation of plant natural product biosynthesis. Plant J 82: 680–692 [DOI] [PubMed] [Google Scholar]
- Mardis ER. (2013) Next-generation sequencing platforms. Annu Rev Anal Chem (Palo Alto, Calif) 6: 287–303 [DOI] [PubMed] [Google Scholar]
- Maron LG, Guimarães CT, Kirst M, Albert PS, Birchler JA, Bradbury PJ, Buckler ES, Coluccio AE, Danilova TV, Kudrna D, et al. (2013) Aluminum tolerance in maize is associated with higher MATE1 gene copy number. Proc Natl Acad Sci USA 110: 5241–5246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miettinen K, Dong L, Navrot N, Schneider T, Burlat V, Pollier J, Woittiez L, van der Krol S, Lugan R, Ilc T, et al. (2014) The seco-iridoid pathway from Catharanthus roseus. Nat Commun 5: 3606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller LA, Zhang P, Rhee SY (2003) AraCyc: a biochemical pathway database for Arabidopsis. Plant Physiol 132: 453–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myburg AA, Grattapaglia D, Tuskan GA, Hellsten U, Hayes RD, Grimwood J, Jenkins J, Lindquist E, Tice H, Bauer D, et al. (2014) The genome of Eucalyptus grandis. Nature 510: 356–362 [DOI] [PubMed] [Google Scholar]
- Nordström KJ, Albani MC, James GV, Gutjahr C, Hartwig B, Turck F, Paszkowski U, Coupland G, Schneeberger K (2013) Mutation identification by direct comparison of whole-genome sequencing data from mutant and wild-type individuals using k-mers. Nat Biotechnol 31: 325–330 [DOI] [PubMed] [Google Scholar]
- O’Connor SE, Maresh JJ (2006) Chemistry and biology of monoterpene indole alkaloid biosynthesis. Nat Prod Rep 23: 532–547 [DOI] [PubMed] [Google Scholar]
- Omranian N, Kleessen S, Tohge T, Klie S, Basler G, Mueller-Roeber B, Fernie AR, Nikoloski Z (2015) Differential metabolic and coexpression networks of plant metabolism. Trends Plant Sci 20: 266–268 [DOI] [PubMed] [Google Scholar]
- Pichersky E, Gang DR (2000) Genetics and biochemistry of secondary metabolites in plants: an evolutionary perspective. Trends Plant Sci 5: 439–445 [DOI] [PubMed] [Google Scholar]
- Potato Genome Sequencing Consortium (2011) Genome sequence and analysis of the tuber crop potato. Nature 475: 189–195 [DOI] [PubMed] [Google Scholar]
- Quadrana L, Almeida J, Asís R, Duffy T, Dominguez PG, Bermúdez L, Conti G, Corrêa da Silva JV, Peralta IE, Colot V, et al. (2014) Natural occurring epialleles determine vitamin E accumulation in tomato fruits. Nat Commun 5: 3027. [DOI] [PubMed] [Google Scholar]
- Tettelin H, Masignani V, Cieslewicz MJ, Donati C, Medini D, Ward NL, Angiuoli SV, Crabtree J, Jones AL, Durkin AS, et al. (2005) Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial “pan-genome.” Proc Natl Acad Sci USA 102: 13950–13955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tholl D, Lee S (2011) Terpene specialized metabolism in Arabidopsis thaliana. The Arabidopsis Book 9: e0143, doi/10.1199/tab.0143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomato Genome Consortium (2012) The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485: 635–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Bakel H, Stout JM, Cote AG, Tallon CM, Sharpe AG, Hughes TR, Page JE (2011) The draft genome and transcriptome of Cannabis sativa. Genome Biol 12: R102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber APM. (2015) Discovering new biology through sequencing of RNA. Plant Physiol 169: 1524–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzer T, Gazda V, He Z, Kaminski F, Kern M, Larson TR, Li Y, Meade F, Teodor R, Vaistij FE, et al. (2012) A Papaver somniferum 10-gene cluster for synthesis of the anticancer alkaloid noscapine. Science 336: 1704–1708 [DOI] [PubMed] [Google Scholar]
- Ziegler J, Facchini PJ (2008) Alkaloid biosynthesis: metabolism and trafficking. Annu Rev Plant Biol 59: 735–769 [DOI] [PubMed] [Google Scholar]