A transcriptomic and metabolomic profiling time course of maize foliar responses to aphid feeding identifies genes for the synthesis of benzoxazinoids, terpenes, and other induced defense metabolites.
Abstract
As a response to insect attack, maize (Zea mays) has inducible defenses that involve large changes in gene expression and metabolism. Piercing/sucking insects such as corn leaf aphid (Rhopalosiphum maidis) cause direct damage by acquiring phloem nutrients as well as indirect damage through the transmission of plant viruses. To elucidate the metabolic processes and gene expression changes involved in maize responses to aphid attack, leaves of inbred line B73 were infested with corn leaf aphids for 2 to 96 h. Analysis of infested maize leaves showed two distinct response phases, with the most significant transcriptional and metabolic changes occurring in the first few hours after the initiation of aphid feeding. After 4 d, both gene expression and metabolite profiles of aphid-infested maize reverted to being more similar to those of control plants. Although there was a predominant effect of salicylic acid regulation, gene expression changes also indicated prolonged induction of oxylipins, although not necessarily jasmonic acid, in aphid-infested maize. The role of specific metabolic pathways was confirmed using Dissociator transposon insertions in maize inbred line W22. Mutations in three benzoxazinoid biosynthesis genes, Bx1, Bx2, and Bx6, increased aphid reproduction. In contrast, progeny production was greatly decreased by a transposon insertion in the single W22 homolog of the previously uncharacterized B73 terpene synthases TPS2 and TPS3. Together, these results show that maize leaves shift to implementation of physical and chemical defenses within hours after the initiation of aphid feeding and that the production of specific metabolites can have major effects in maize-aphid interactions.
Investigation of plant defense mechanisms, which provides information about genes that are suitable for the control of agricultural pests, has been the target of extensive research involving both classical breeding and transgenic approaches (Thompson and Goggin, 2006). Under herbivore attack, gene expression responses are strongly correlated with the mode of herbivore feeding, the amount of tissue damage, the specific temporal and spatial patterns at the feeding site, as well as the host plant species (Walling, 2000; Kessler and Baldwin, 2002). A model of insect recognition by plants through herbivore-associated molecular patterns and damage-associated molecular patterns triggering plant defense responses has been proposed (Heil, 2009). Since transcriptional reprogramming underlies many plant defense responses, transcriptomic analyses of responses to aphids and other insect herbivores have been conducted using several plant species (Thompson and Goggin, 2006; Couldridge et al., 2007; Coppola et al., 2013; Appel et al., 2014; Heidel-Fischer et al., 2014). However, large-scale studies directed at investigating the interactive effects of genomic and metabolomic plant responses to aphid feeding, which are key elements to understanding these complex and dynamic interactions, are relatively uncommon (Ferry et al., 2011; Coppola et al., 2013; Guan et al., 2015).
Maize (Zea mays) is the world’s most productive grain crop, with 967 million metric tons harvested in 2013 to 2014 (National Corn Growers Association, 2014). However, biotic stress factors can severely limit maize yields, and a large percentage of production is lost due to feeding by more than 90 species of herbivorous insects (Machado et al., 2002). Among aphids, the largest group of phloem-feeding insects (Smith and Boyko, 2007), the corn leaf aphid (Rhopalosiphum maidis) is most commonly found as a pest on maize. Like most aphids, corn leaf aphids feed from vascular tissues by inserting their piercing mouthparts intercellularly to reach the phloem, thereby causing relatively limited mechanical damage to foliar tissue. Aphids also have the ability to manipulate host plant physiology by introducing effectors that influence defense signaling in prolonged interactions with the attacked plant tissue (Will et al., 2007; Hogenhout and Bos, 2011). Corn leaf aphid infestation causes direct damage to maize plants by reducing growth and yield (Bing and Guthrie, 1991). Additionally, heavy accumulation of aphid honeydew on maize tassels can block pollen shed and thereby reduce seed set (Foott and Timmins, 1973; Carena and Glogoza, 2004). Indirect damage comes from the fact that the corn leaf aphid is the most effective vector of several plant viruses, including Maize yellow dwarf virus and Barley yellow dwarf virus, which are among the most economically important diseases of cereal crops and forage grasses (El-Muadhidi et al., 2001; Hawkes and Jones, 2005; Power et al., 2011; Jarošová et al., 2013; Krueger et al., 2013). As protection against aphids and other insect pests, maize plants implement not only constitutive defenses but also inducible responses that require broad shifts in gene expression and biochemical pathways (Thompson and Goggin, 2006; Meihls et al., 2012).
Many major plant defense mechanisms require the involvement of two signaling molecules, salicylic acid and jasmonic acid, which are induced in distinct patterns by insect and pathogen damage (Pieterse and Dicke, 2007). Whereas salicylic acid is the predominant phytohormone regulating responses to pathogens and phloem-feeding insects such as aphids (Walling, 2000; Heidel and Baldwin, 2004; Mewis et al., 2006; Broekgaarden et al., 2011), plant responses to chewing herbivores are primarily regulated via jasmonic acid (Kessler and Baldwin, 2002; De Vos et al., 2005; Heidel-Fischer et al., 2014). Cross talk between these two hormone signal transduction pathways is thought to enable fine-tuning of plant responses to herbivore and pathogen attack (Thaler et al., 2012).
The benzoxazinoids are a major class of specialized metabolites found in maize and other poaceous grasses (Zuniga et al., 1983; Niemeyer, 1988; Jonczyk et al., 2008). The predominant benzoxazinoids in maize foliar tissue are 4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one glucoside (DIMBOA-Glc) and 2-hydroxy-4,7-dimethoxy-1,4-benzoxazin-3-one glucoside (HDMBOA-Glc; Frey et al., 1997, 2009). These compounds inhibit the growth of fungi, arrest insect herbivore growth, and cause allelopathic effects (Niemeyer, 2009; Ahmad et al., 2011). Benzoxazinoids are stored in a glucoside form that has reduced toxicity compared with the aglycones, which are produced upon cell disruption during pathogen or herbivore attack (Frey et al., 2009). Aphid feeding does not increase overall DIMBOA-Glc or HDMBOA-Glc accumulation in tested maize cultivars (Cambier et al., 2001; Meihls et al., 2013). However, recent studies demonstrated that DIMBOA-Glc, but not HDMBOA-Glc, is secreted into the apoplast upon aphid infestation (Ahmad et al., 2011) and may regulate the accumulation of callose as a defense against aphid feeding (Meihls et al., 2013; Betsiashvili et al., 2015).
In response to mechanical damage or herbivory, plants emit a complex blend of volatile organic compounds (Dicke, 1999; Paré and Tumlinson, 1999; Walling, 2000) that not only influence the herbivores themselves but also serve additional ecological functions, including the attraction of predators and parasitoids. This indirect defense mechanism provides natural enemies with reliable long-distance cues to recognize plants infested with their host insects. Release of terpenes, an abundant class of plant volatiles and an important component of floral odors, also is induced by herbivore feeding on vegetative tissue (Pichersky and Gershenzon, 2002; Pichersky et al., 2006; Tholl et al., 2011). Depending on the context, plant-derived terpenes can have deterrent effects, can attract predators of herbivorous insects, or can serve as host-finding cues for the herbivores themselves (Gershenzon and Dudareva, 2007).
In this study, we integrated gene expression profiling by high-throughput RNA sequencing (RNA-Seq) with metabolite profiles to reveal a dynamic and complex network of maize responses to corn leaf aphids. We exposed leaves of inbred line B73 to corn leaf aphid feeding for varying amounts of time and used statistical approaches to identify patterns in the resulting transcriptomic and metabolomic data sets. Preexisting Dissociator (Ds) transposon insertions in inbred line W22 (Vollbrecht et al., 2010) made it possible to investigate the role of specific benzoxazinoid and terpene biosynthesis genes in maize-aphid interactions.
RESULTS
Overview of the Transcriptomic and Metabolomic Data Sets
To identify transcriptomic and metabolomic changes that occur in response to aphid feeding, the second true leaves of maize inbred line B73 were infested with 10 adult corn leaf aphids for 0, 2, 4, 8, 24, 48, or 96 h. Aphid infestations were started in a staggered manner, such that all samples were harvested on the same day (Supplemental Fig. S1). Although all tissue collection occurred during the middle of the day, there is nevertheless potential for diurnal variation in the maize transcriptome and metabolome during collection of the sample sets. Therefore, individual tissue collection times were recorded, and both transcriptomic and metabolite data were normalized on the basis of uninfested control samples that were harvested at the beginning and end of the collection period.
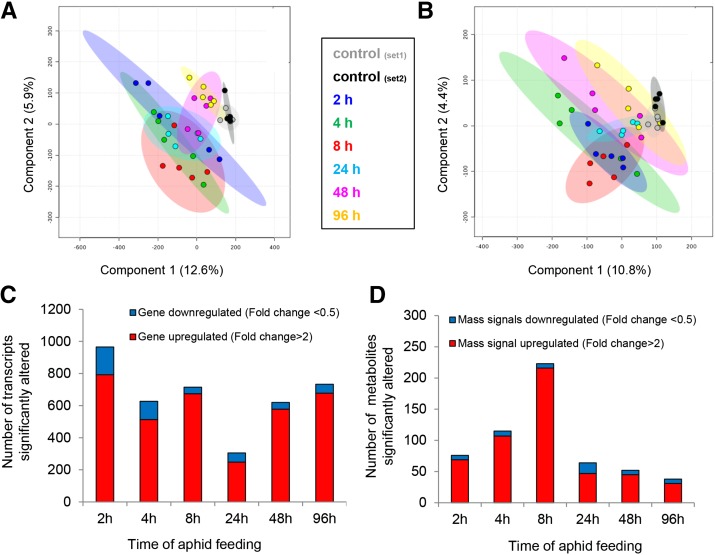
Comparison of transcriptome data (Illumina RNA-Seq) with the B73 genomic sequence (Schnable et al., 2009), which has 110,451 predicted gene models (AGPv3.20; http://www.maizegdb.org), showed approximately 41,700 unique transcripts (Supplemental Table S1). Expression patterns of selected genes were confirmed by quantitative reverse transcription (RT)-PCR using independently generated plant samples (Supplemental Fig. S2). After data filtering (see “Materials and Methods”), approximately 20,000 transcripts from the RNA-Seq data set were analyzed using edgeR (Robinson et al., 2010) for each of the six aphid-infested time points to detect genes that were differentially expressed relative to uninfested control leaves (Supplemental Table S2). Genes with significant expression differences (P ≤ 0.05, false discovery rate [FDR] adjusted) and at least 2-fold changes relative to the controls for at least one of the time points were selected, resulting in 1,607 genes that were differentially expressed (Supplemental Table S3). The gene expression levels were used to conduct a partial least-squares discriminant analysis (PLS-DA) for each of the biological replicates (Fig. 1A). In this analysis, the 4- and 8-h time points clustered farthest from the control samples, indicating the greatest changes in gene expression after the onset of aphid feeding. The 2- and 24-h aphid infestation-responsive genes clustered in the center of the plot. However, after 48 and 96 h of aphid feeding, foliar gene expression had reverted to being more similar to that of the uninfested controls.
Figure 1.
Overview of maize transcriptome and metabolome responses to aphid feeding. A, PLS-DA plots of 1,607 genes identified by transcript profiling (RNA-Seq) of inbred line B73 infested with aphids for the indicated time periods. Ovals indicate 95% confidence intervals. B, PLS-DA of 319 mass signals (negative ion mode) identified by LC-TOF-MS. Ovals indicate 95% confidence intervals. Similar data for the positive ion mode are shown in Supplemental Figure S3. C and D, Number of individual transcripts and mass spectrometry features that were significantly up- or down-regulated at each time point, respectively. P < 0.05 (FDR adjusted), and fold change greater than 2 or less than 0.5.
In a separate experiment, metabolic changes were studied in aphid-infested maize leaf samples using untargeted liquid chromatography-time of flight-mass spectrometry (LC-TOF-MS) in negative and positive ion modes (Supplemental Table S4). Mass signals with significant differences (P ≤ 0.05, FDR adjusted) and at least 2-fold changes in at least one of the time points relative to the controls were selected, resulting in 319 negative ion mode and 334 positive ion mode mass features (Supplemental Table S5). The PLS-DA clustering pattern of the untargeted metabolite analysis (negative ion mode, Fig. 1B; positive ion mode, Supplemental Fig. S3A) was similar to that of the gene expression changes observed in response to aphid herbivory (Fig. 1A): data for the 4- and 8-h time points were the most distinct, the 2- and 24-h time points were intermediate, and the 48- and 96-h time points were more similar to the control samples.
After the initiation of aphid feeding, hundreds of transcripts showed altered expression levels for each one of the time points, with up-regulated transcripts being much more abundant than down-regulated ones (Fig. 1C). Similarly, nonspecific analysis of metabolic changes induced by aphid feeding showed that many more small molecules increased significantly in abundance after aphid feeding than decreased in abundance (Fig. 1D; Supplemental Fig. S3B). Whereas changes in gene expression peaked at 2 h after the initiation of aphid feeding, the greatest number of induced metabolites was observed at 8 h after aphid feeding. The overall similarity of the transcriptomic and metabolomic data suggested that aphid-induced gene expression changes led to induced changes in the metabolome at the same or later time point in our experiments.
Clustering of Expression Patterns in the Transcriptome Data
The significantly differentially expressed genes were subjected to K-means clustering using Pearson correlation distances (TM4 software; http://www.tm4.org). Each cluster is represented by the average of the gene expression of the set of genes showing similar response patterns to aphid herbivory (Fig. 2A). The most common expression pattern is an early induction at 2 and 4 h (clusters 1 and 2) or 8 h (cluster 3), followed by a drop in the expression level for the 24-h time point and sometimes a smaller increase later during the infestation. A smaller number of genes showed an initial drop in the expression level followed by reversion to basal levels at later stages of aphid feeding (cluster 4). Finally, a subset of genes was induced by aphid feeding throughout the experiment, although the 24-h time point sometimes did not show a significant increase (clusters 5 and 6).
Figure 2.
Overview of gene expression clusters calculated by K-means clustering. A, Pearson correlation was used to identify six clusters involving a total of 1,607 transcripts with significant expression profile changes for at least one time point after the initiation of aphid feeding. The total number of transcripts in each cluster is indicated, and data for individual genes are shown in light gray. Average expression responses for each cluster are shown in red. All genes selected for this analysis have significant differences of 2-fold (up- or down-regulated), P < 0.05 (FDR adjusted). B, Overrepresentation analysis of each cluster using the MetGenMAP Web site to identify metabolic functions that are being regulated.
To elucidate the biological processes that are involved in each gene expression cluster, overrepresentation analysis was performed using MetGenMAP (Joung et al., 2009). Pathways that are enriched among the overexpressed transcripts include many that are associated with plant defense and stress responses (Fig. 2B). Genes related to jasmonic acid biosynthesis, which is commonly associated with insect defense, are expressed early in the infestation but then revert to control levels. Genes involved in the synthesis of suberin, cellulose, and coniferin, which are induced early after aphid feeding (clusters 1–3), may provide physical defense against aphid stylet penetration by leading to toughed cell walls. Similarly, the observed gene expression patterns indicate that chemical defenses such as activation of benzoxazinoids and biosynthesis of phenylpropanoids are induced early after aphid feeding but then decline at later stages. Nitrogen assimilation via nitrate reductase and Asn synthesis is reduced in the early stages of aphid infestation (cluster 4). Pathways that are induced in a more continuous manner in response to aphid feeding (clusters 5 and 6) are mostly related to primary metabolism or general stress responses (e.g. up-regulation of polyamine biosynthesis).
Differentially expressed genes from the six time points also were functionally categorized using the MapMan tool (Usadel et al., 2006; Supplemental Fig. S4). Similar to the clustering shown in Figure 2, this analysis provided evidence that jasmonic acid and salicylic acid pathways, as well as the production of defense-related secondary metabolites, are up-regulated. Other gene categories related to defense responses, including receptor kinases, phospholipase C, and different types of small molecule transporters, are also overrepresented among the transcripts that are induced by aphid feeding. Conversely, genes involved in photosynthesis, carbohydrate metabolism, gluconeogenesis, the glyoxylate cycle, nitrogen metabolism, and lipid degradation were overrepresented among the down-regulated genes and underrepresented among the up-regulated genes. Together, these observations indicate that there is a shift from primary metabolism to the production of defensive metabolites in response to corn leaf aphid feeding on maize.
Targeted Analysis of Metabolite Changes in Response to Aphid Feeding
To complement the untargeted LC-TOF-MS metabolite profiling (Supplemental Tables S4 and S5), we conducted more targeted assays by HPLC-absorbance detection, gas chromatography-mass spectrometry (GC-MS), and HPLC-mass spectrometry to identify aphid-induced changes in metabolite abundance (Supplemental Table S6). Phospholipids, including phosphatidylcholines (36:6, 18:2/18:3. 34:3, 36:4, and 16:0/18:2 lipid chains), phosphatidylglycerols (16:1/18:3, 16:0/18:3, 16:0/16:1, and 16:0/16:0 lipid chains), and phosphatidylethanolamines (16:0/18:2 lipid chain), had no significant aphid-induced changes in their abundance (data not shown). Similarly, although there were small changes in the abundance of individual amino acids, there was no consistent pattern in the abundance of free amino acids in the course of aphid feeding. In contrast, as described in more detail below, there were significant changes in the abundance of known plant signaling molecules and associated metabolic pathways.
Plant Hormone-Related Genes Induced by Aphid Feeding
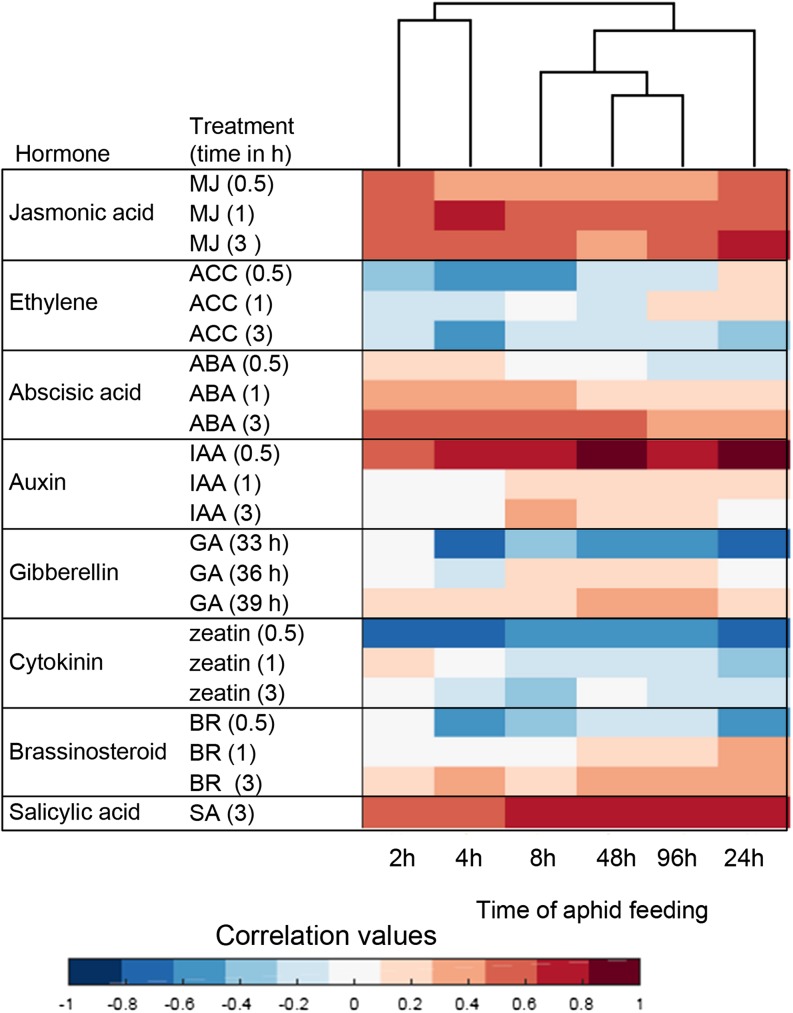
Both clustering (Fig. 2) and overrepresented gene categories (Supplemental Fig. S3) suggested that maize defense signaling pathways are differentially regulated by aphid feeding. To identify the transcriptional signatures of hormonal responses in aphid-infested maize plants, we used the Hormonometer program (http://hormonometer.weizmann.ac.il/hormonometer), which compares the variation in gene expression with indexed data sets of hormone treatments (Volodarsky et al., 2009). We evaluated the similarity in expression profiles elicited by aphid herbivory to the application of the plant hormones methyl jasmonate, 1-aminocyclopropane-1-carboxylic acid (a metabolic precursor of ethylene), abscisic acid, indole-3-acetic acid, cytokinin (zeatin), brassinosteroid, GA3, and salicylic acid. As the hormone treatments were conducted with Arabidopsis (Arabidopsis thaliana), we selected the orthologous genes from Arabidopsis and the B73 genome. Then, only the genes included in the RNA-Seq analysis after the filtering processes containing Arabidopsis probe set identifiers, which is required by Hormonometer, were kept. Finally, a total of 6,805 Arabidopsis orthologs of maize genes were used as input for the Hormonometer analysis (Supplemental Table S8).
The most common aphid-induced maize gene expression changes were associated with jasmonic acid-, salicylic acid-, and auxin-dependent signaling (Fig. 3). Additionally, there was an overall positive correlation between aphid-induced genes and those that were induced within 0.5 h after auxin treatment. Cytokinin-responsive genes showed a negative correlation with aphid feeding from maize, consistent with an approximately 40% decrease in overall cytokinin content in response to extended aphid feeding (Supplemental Fig. S5). A dendrogram analysis of the data showed that hormone-related gene expression changes in the first 4 h after aphid feeding are distinct from those observed at later time points (Fig. 3).
Figure 3.
Identification of plant hormone signatures based on transcriptomic data. The analysis was conducted using the Hormonometer program (Volodarsky et al., 2009). Red shading indicates a positive correlation between the maize aphid treatment and a particular hormone response; blue shading indicates a negative correlation. MJ, Methyl jasmonate; ACC, 1-aminocyclopropane-1-caroxylic acid (a metabolic precursor of ethylene); ABA, abscisic acid; IAA, indole-3-acetic acid, GA, GA3; BR, brassinosteroid; SA, salicylic acid.
Aphid-Induced Changes in Oxylipin Biosynthesis
Plant lipoxygenases (Lox) initiate fatty acid oxidation pathways for the synthesis of oxylipins, cyclic, or acyclic compounds that are known to have diverse functions in plant responses to herbivory (Porta and Rocha-Sosa, 2002). Up-regulation of gene expression (Fig. 2) as well as hormonal response signatures (Fig. 3) suggested that aphid feeding elicits the production of a complex array of oxylipins. Therefore, we expanded our analysis to look in more detail at genes and metabolites associated with oxylipin production (Fig. 4). Linolenic and linoleic acids, the most common substrates for plant oxylipin synthesis, were less abundant early after the initiation of aphid feeding but were increased about 4-fold after 48 and 96 h. The lipoxygenase genes GRMZM2G102760 (Lox5), GRMZM5G822593 (Lox13), GRMZM2G109056 (Lox4), GRMZM2G109130 (Lox3), and GRMZM2G156861 (Lox1) were induced at all time points of aphid feeding. The expression patterns of allene oxide synthase (AOS) genes, which encode the second step of the jasmonic acid pathway, were varied: GRMZM2G067225 was up-regulated at all time points, GRMZM2G033098 was up-regulated except at the 24-h time point, and GRMZM2G002178 was only up-regulated at 4 and 24 h after the initiation of aphid feeding. Aphid infestation generally reduced the abundance of 12-oxo-phytodienoic acid (OPDA) conjugates, trans-OPDA-ME and cis-OPDA-ME. 12-Oxophytodienoate reductase (OPR7; GRMZM2G148281) expression was slightly increased at 2 h, and trans-jasmonic acid was induced only at 24 h of aphid feeding, with an approximately 2-fold increase (Fig. 4B). Other downstream genes and metabolites associated with OPR genes were not significantly affected by aphid feeding (Supplemental Table S2). Although Lox, AOS, and OPR gene expression is induced by aphid feeding, the main product is unlikely to be jasmonic acid. Instead, oxylipins with as yet unknown structures are induced in response to aphid feeding (mass-to-charge ratio 238 and 240 in Supplemental Table S6).
Figure 4.
Effects of aphid feeding on jasmonic acid pathway gene expression and metabolites. A, Schematic diagram of the jasmonic acid biosynthesis pathway. The dashed arrow represents multiple enzymatic steps. Metabolites shaded in blue were measured. B, Transcript and metabolite abundance after aphid infestation. Values are means ± se (n = 5). *, P < 0.05 by Student’s t test relative to uninfested controls and fold change greater than 2 or less than 0.5.
Biosynthesis of Aromatic Amino Acids, Salicylic Acid, and Auxin
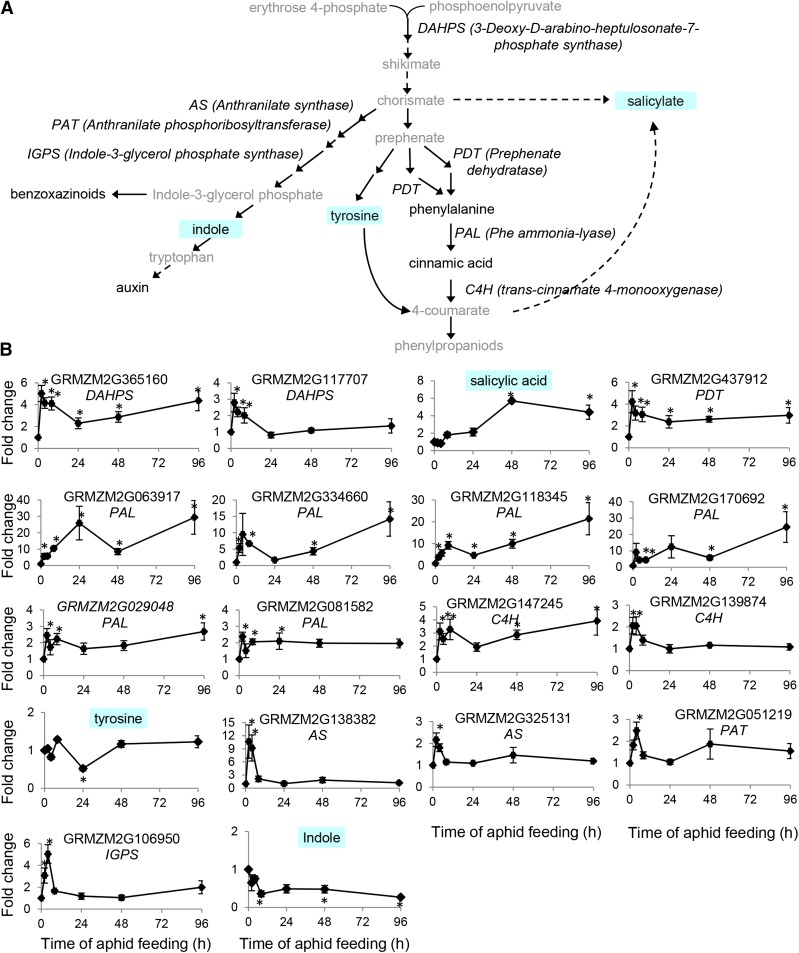
The shikimate pathway (Fig. 5A), which leads to the synthesis of Phe, Tyr, Trp, auxin, salicylic acid, lignin, and phenylpropanoids, is a major biosynthetic pathway in plants (Tzin and Galili, 2010; Vogt, 2010). Transcriptional up-regulation of the pathway is indicated by both the clustering analysis (Fig. 2) and in the hormone-regulated gene expression signature (Fig. 3). As shown in Figure 5B, two maize genes encoding deoxy-d-arabino-heptulosonate-7-phosphate synthase were up-regulated by aphid feeding, GRMZM2G365160 at all time points and GRMZM2G117707 in the first few hours. Prephenate dehydratase (GRMZM2G437912) also was induced. Six isozymes of maize Phe ammonia-lyase were up-regulated, and trans-cinnamate 4-monooxygenase gene (GRMZM2G147245) expression increased shortly after the initiation of aphid feeding. Phe and cinnamic acid accumulation remained unchanged throughout the experiment (Supplemental Table S6), and Tyr was slightly decreased at 24 h (Fig. 5B). The levels of salicylic acid, which is synthesized via either chorismate or 4-coumarate, was found to be induced by 8 h after aphid feeding and remained high for the remainder of the experiment.
Figure 5.
Effects of aphid feeding on genes and metabolites of the aromatic amino acids, salicylic acid, auxin, and other metabolites derived from the shikimate pathway. A, Pathway schematic. Measured metabolites are shaded in blue. B, Gene and metabolite abundance. Values are means ± se (n = 5). *, P < 0.05 by Student’s t test relative to uninfested controls, and fold change greater than 2 or less than 0.5.
Four genes of the Trp biosynthesis pathway, anthranilate synthases (GRMZM2G138382 and GRMZM2G325131), anthranilate phosphoribosyl transferase (GRMZM2G051219), and indole-3-glycerol phosphate synthase (GRMZM2G106950), were found to increase only transiently 2 and 4 h after aphid infestation. Concomitantly, indole showed a similar accumulation trend after aphid feeding (Fig. 5B). However, indole-3-acetic acid (auxin) abundance decreased significantly at 24 h after aphid infestation (Supplemental Table S6).
Benzoxazinoid Biosynthesis Is Involved in Herbivore Defense Mechanisms
In addition to serving as a metabolic precursor for Trp and auxin, indole is also a substrate for the biosynthesis of benzoxazinoids (Figs. 5A and 6A), a class of specialized metabolites in grasses that provide defense against insect herbivory (Frey et al., 2009; Ahmad et al., 2011). Ten enzymes (Bx1–Bx9 and Igl1) catalyze the formation of DIMBOA-Glc from indole-3-glycerol phosphate (Frey et al., 2009; Niemeyer, 2009; Fig. 6A). This is followed by methylation into HDMBOA-Glc through the activity of three Bx10 enzymes (Meihls et al., 2013; Mijares et al., 2013). Transcripts of Bx1 (GRMZM2G085381), Bx2 (GRMZM2G085661), Bx6 (GRMZM6G617209), and Bx7 (GRMZM2G441753) were significantly induced at early time points as well as at 96 h after aphid attack. Bx4 (GRMZM2G172491) expression was reduced, but only at 8 h. Expression of other genes in this pathway was either not significantly altered by aphid feeding or not detected in our transcript profiling assays. Previously, we demonstrated that the abundance of DIMBOA-Glc, HDMBOA-Glc, 2-hydroxy-7-methoxy-1,4-benzoxazin-3-one glucoside, and 2,4-dihydroxy-7,8-dimethoxy-1,4-benzoxazin-3-one glucoside was not affected by aphid feeding on B73 (Meihls et al., 2013). The abundance of 6-methoxybenzoxazolinone, an insect-deterrent breakdown product of benzoxazinoids, was reduced significantly after 8 h of aphid infestation.
Figure 6.
Effects of aphid feeding on benzoxazinoid-related genes and metabolites. A, Maize benzoxazinoid biosynthesis pathway. B, Genes and metabolites that were significantly altered by aphid feeding for at least one time point. Values are means ± se (n = 5). *, P < 0.05 by Student’s t test relative to uninfested controls and fold change greater than 2 or less than 0.5. C, DIMBOA-Glc levels in wild-type W22 and bx1::Ds, bx2::Ds, and bx6::Ds transposon knockout mutants. Values are means ± se (n = 6–11). *, P < 0.05 by Student’s t test relative to the wild type. FW, Fresh weight; ND, not detected. D, Corn leaf aphid progeny production on the same lines as in C. Values are means ± se (n = 6–11). *, P < 0.05 by Student’s t test relative to the wild type. DIM2BOA-Glc, 2,4-Dihydroxy-7,8-dimethoxy-1,4-benzoxazin-3-one glucoside; HMBOA-Glc, 2-hydroxy-7-methoxy-1,4-benzoxazin-3-one glucoside.
To investigate how the benzoxazinoid pathway affects aphid reproduction, we employed a previously identified bx1::Ds transposon insertion and confirmed new bx2::Ds and bx6::Ds mutations in the W22 genetic background by PCR-based genotyping (primers are listed in Supplemental Table S7). Whereas the bx1::Ds and bx2::Ds mutations completely eliminated DIMBOA-Glc accumulation, bx6::Ds reduced DIMBOA-Glc abundance by about 70% (Fig. 6C), suggesting that there is still residual 2,4-dihydroxy-(2H)-1,4-benzoxazin-3(4H)-one glucoside (DIBOA-Glc) dioxygenase activity in this strain. As in the case of B73 (Meihls et al., 2013), DIMBOA-Glc abundance was not significantly affected by 2 d of aphid feeding on W22 maize seedlings (Supplemental Fig. S6). Aphid reproduction was increased more than 7-fold on homozygous bx1::Ds and bx2::Ds mutant seedlings relative to the wild-type W22 (Fig. 6D). In contrast, there was only a 3-fold increase in aphid progeny production on bx6::Ds seedlings.
A Role for Terpene Synthases in Maize-Aphid Interactions
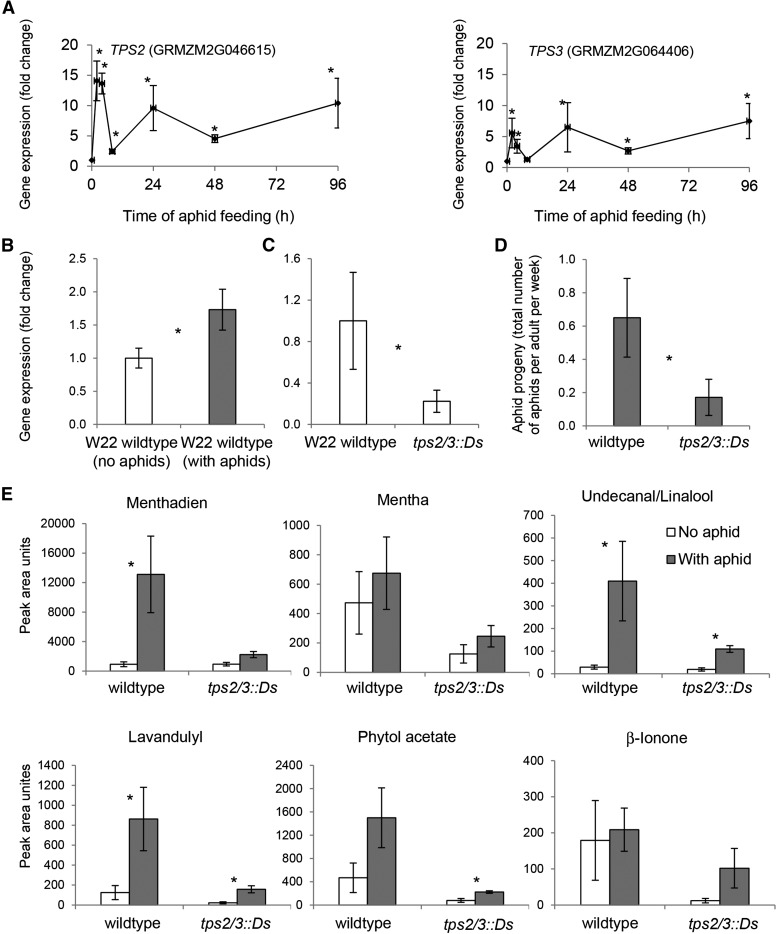
Terpenes are a class of compounds derived from isopentenyl diphosphate and its allylic isomer dimethylallyldiphosphate (Tholl et al., 2004). Terpene synthases catalyze key steps in the formation of the terpene carbon skeleton (Roberts, 2007). The aphid-induced B73 transcriptome showed that four genes encoding terpene synthases were significantly induced by aphid feeding (Supplemental Table S2; Fig. 7A). Alignment of the B73 TERPENE SYNTHASE2 (TPS2; GRMZM2G046615) and TPS3 (GRMZM2G064406) genes, which are tandem duplicated and approximately 95% identical at the DNA sequence level, to the newly sequenced W22 genome showed only a single gene in W22 (Supplemental Fig. S7). Based on the chromosomal sequence, it was not possible to unambiguously assign the W22 gene as a homolog of either B73 TPS2 or TPS3. As in the case of the B73 genes, expression of the W22 TPS2/3 gene was significantly induced by aphid feeding (Fig. 7B). Homozygous mutant and wild-type progeny of plants heterozygous for Ds transposon insertion B.S08.0585 (http://acdstagging.org/) in the W22 TPS2/3 gene were identified by PCR-based genotyping. Quantitative RT-PCR showed significantly decreased TPS2/3 transcription in the Ds insertion mutant (Fig. 7C). Aphid reproduction was decreased by approximately 73% on the mutant line relative to wild-type W22 (Fig. 7D).
Figure 7.
Effect of aphid feeding on terpene biosynthesis. A, B73 TPS2 and TPS3 gene expression in response to aphid feeding on B73. Values are means ± se (n = 5). *, P < 0.05 by Student’s t test. B, TPS2/3 gene expression in wild-type W22 with and without aphids (n = 7). C, TPS2/3 gene expression in wild-type W22 and tps2/3::Ds from a segregating population (n = 7–10). D, Aphid reproduction on wild-type W22 and tps2/3::Ds from a segregating population. *, P < 0.05 by Student’s t test (n = 7–10). E, Terpene abundance in wild-type W22 and tps2/3::Ds with and without aphid feeding. *, P < 0.05 by Student’s t test (n = 3–7).
GC-MS assays of homozygous mutant and wild-type siblings were conducted to determine whether aphid feeding and/or the tps2/3::Ds mutation were associated with altered terpene accumulation in maize seedlings. Four terpenes were significantly induced by aphid feeding: undecanal/linalool (which could not be differentiated in the assay) and lavandulyl were induced in both tps2/3::Ds and wild-type siblings, menthadiene was induced in the wild-type siblings, and phytol acetate increased only in tps2/3::Ds (Fig. 7E). In each case, the aphid-induced levels of these compounds were lower in the tps2/3::Ds mutants. The abundance of two additional terpenes, menthol (mentha) and β-ionone, also was reduced in tps2/3::Ds mutants relative to wild-type siblings but was not altered significantly by aphid feeding.
DISCUSSION
In order to investigate the dynamic plant responses to herbivore feeding, it is necessary to consider multiple time points after initiation of the interaction. With few exceptions, for example, in Arabidopsis (Couldridge et al., 2007), most prior studies of plant transcriptomic responses to aphid feeding have focused on time points of 6 h or later. However, our results show that, despite the fact that there was minimal tissue damage from the 10 aphids that were added to each feeding cage, some of the most significant transcriptional responses of maize to aphid feeding occurred at the earliest stages of aphid infestation, within 2 to 4 h after the start of our experiments (Fig. 1). Complementing this transcriptional analysis with measurement of changes in the maize metabolome improves our understanding of maize responses to aphid infestation.
Perhaps the most striking effect in the overall pattern of maize responses is that different sets of genes and metabolites are induced in the first few hours after the initiation of aphid feeding and at later time points (Figs. 1 and 2). In particular, after 48 and 96 h of aphid feeding, the transcriptomic and metabolic patterns in the maize leaves are more similar to those of control plants than at 4 and 8 h of aphid feeding. Similar, time point-specific variation in plant gene expression has been observed in other aphid-plant interactions, for instance in comparisons of significantly altered transcripts after 24, 48, and 96 h of potato aphid (Macrosiphum euphorbiae) feeding on tomato (Solanum lycopersicum; Coppola et al., 2013), 6 and 24 h of green peach aphid (Myzus persicae) and cabbage aphid (Brevicoryne brassicae) feeding on Arabidopsis (Appel et al., 2014), and 6, 12, 24, and 48 h of greenbug aphid (Schizaphis graminum) feeding on sorghum (Sorghum bicolor; Zhu-Salzman et al., 2004). In each experiment, there was limited overlap in the plant gene expression patterns at each time point, indicating dynamic shifts in plant responses to aphid feeding over time. One possible explanation is that aphids are able to suppress some plant defense responses during long-term feeding. Conversely, the shift to different long-term responses may indicate that maize and other plants are able to fine-tune their transcriptomic and metabolomic responses over time to adjust defenses for the specific type of attack that is being perceived.
Elevated jasmonic acid levels have been associated with insect resistance in several plant species (Ellis et al., 2002; Mewis et al., 2006; Shivaji et al., 2010). In maize, treatment with jasmonic acid increased the production of defense-related benzoxazinoids (Oikawa et al., 2002, 2004). However, although the jasmonic acid concentration is slightly elevated early in the aphid infestation (2-fold increased at 24 h), it drops to basal levels at later time points (Fig. 4B). Salicylic acid, which suppresses some jasmonic acid-regulated plant defenses (Zarate et al., 2007; Thaler et al., 2012), increases in abundance during later stages of aphid feeding on maize (Fig. 5B) and could account for the observed suppression of jasmonic acid signaling. Similar induction of salicylic acid-dependent gene expression has been reported in response to feeding by several other aphid species, including green peach aphid, cabbage aphid, greenbug aphid, and potato aphid (Giordanengo et al., 2010; Appel et al., 2014), suggesting that this may be a more general plant response to aphids. Aphids secrete a variety of proteins into the phloem as they are feeding (Elzinga and Jander, 2013), and some of these are known to suppress plant defense responses (Will et al., 2007; Bos et al., 2010; Elzinga et al., 2014). However, although it has been proposed that aphids actively induce salicylic acid production to misdirect plant defenses (Walling, 2008), specific salivary effectors involved in this process have not yet been identified.
In contrast to jasmonic acid and OPDA, both gene expression and metabolites that are upstream of OPDA in the oxylipin pathway (Fig. 4A) remain elevated after 48 and 96 h of aphid feeding (Fig. 4B). This indicates that other products of this metabolic pathway, including previously identified death acids (Christensen et al., 2015) and the as yet uncharacterized oxylipins with mass-to-charge ratio 238 and 240 that were detected in our GC-MS assays (Supplemental Table S6), continue to be produced in response to aphid feeding. Recent research suggests that oxylipins other than jasmonic acid can have defense signaling functions in maize (Constantino et al., 2013) or direct nutritive effects on aphids (Nalam et al., 2012). The identification of novel aphid-induced oxylipins, in combination with knockout mutations that impair their synthesis, may lead to the characterization of previously unknown maize defense responses or signaling pathways leading to aphid resistance.
A correlation between abscisic acid-induced gene expression and aphid-induced gene expression (Fig. 3) has been observed in several plant species (Zhu-Salzman et al., 2004; Park et al., 2006; Kerchev et al., 2013; Hillwig et al., 2015). Abscisic acid signaling often has been associated with osmotic stress responses in plants, leading to the hypothesis that removal of phloem content by aphids can cause wilting and thereby mimic osmotic stress. However, infiltration of aphid saliva by itself induces the characteristic expression of abscisic acid-regulated genes (De Vos and Jander, 2009), indicating that osmotic stress caused by the removal of phloem sap is not essential for this transcriptional response. It has been suggested that aphids up-regulate abscisic acid signaling, which decreases the stomatal aperture and leaf transpiration, thereby maintaining the leaf water potential that aphids need for feeding (Sun et al., 2015). Arabidopsis mutants that are defective in abscisic acid signaling are more resistant to aphids (Kerchev et al., 2013; Hillwig et al., 2015), which also indicates a more direct role in plant-aphid interactions.
In contrast to the generally positive correlation between aphid feeding and jasmonic acid-, salicylic acid-, and abscisic acid-regulated gene expression, genes regulated by cytokinins were negatively correlated with the effects of aphid feeding (Fig. 3). Cytokinins regulate plant growth and development, source/sink relations, as well as biotic and abiotic environmental interactions (Kieber and Schaller, 2014). The up-regulation of endogenous plant cytokinin production can delay the process of leaf senescence (Gan and Amasino, 1995). Thus, the overall down-regulation of cytokinin-responsive genes, as well as the decrease in leaf cytokinin content during aphid infestation of maize leaves (Supplemental Fig. S5), may be indications that the maize leaves attenuate growth processes and, instead, might be senescing.
Consistent with the observed increase in jasmonic acid content in the early stages of aphid infestation (Fig. 4B), there is an expression increase in some genes of the benzoxazinoid pathway (Fig. 6B). In particular, Bx1, which may be the rate-limiting step in the pathway (Zhang et al., 2015), shows increased expression. We previously demonstrated that a bx1::Ds mutation reduces benzoxazinoid content and increases aphid reproduction on maize inbred line W22 (Betsiashvili et al., 2015). As indole, the product of the Bx1 enzyme, is an essential component of primary metabolism and also serves as a precursor for other defensive secondary metabolites, we could not rule out the possibility that not only the absence of benzoxazinoids but perhaps other maize metabolites improves aphid growth. However, a bx2::Ds mutation has the same effect on aphid reproduction as bx1::Ds (Fig. 6D). Bx2 encodes a cytochrome P450 that uses indole as a substrate to produce indolin-3-one (Frey et al., 1997) and is thus the committing enzyme for benzoxazinoid biosynthesis. Since a bx2 mutation is less likely to affect the production of other indole-derived plant defensive metabolites (Erb et al., 2015), the improved aphid growth on this mutant line is more plausibly caused by reduced benzoxazinoid content.
Whereas maize and wheat (Triticum aestivum) accumulate primarily DIMBOA-Glc and HDMBOA-Glc, the benzoxazinoid pathway in rye (Secale cereale) and wild barley (Hordeum vulgare) ends at DIBOA-Glc (Frey et al., 2009). Therefore, a bx6 mutation, which blocks the first step in the conversion of DIBOA-Glc to DIMBOA-Glc (Fig. 6A), would be predicted to make the maize benzoxazinoid profile more like that of rye or wild barley. In addition to having direct toxic effects, DIMBOA can also lead to callose deposition as an antiaphid defense response in maize (Ahmad et al., 2011; Betsiashvili et al., 2015). It is not known whether DIBOA-Glc or its breakdown products can lead to the induction of callose formation in maize. Thus, the improved aphid reproduction on the bx6::Ds mutant (Fig. 6D) could be a result of reduced callose formation rather than the lower toxicity of DIBOA-Glc relative to DIMBOA-Glc.
Unlike bx1::Ds and bx2::Ds, which almost completely eliminated benzoxazinoid production in W22 maize seedlings, the bx6::Ds insertion reduced DIMBOA-Glc content by only about 60%. This suggests that some other enzyme in W22 has similar DIBOA-Glc dioxygenase activity to Bx6. The availability of a W22 genome sequence will make it possible to identify such genes, create knockout mutations, and eventually shift the benzoxazinoid profile of maize to resemble that of rye and wild barley. Such mutant lines, once they have been identified, will enable experiments to determine what defensive benefits the extended benzoxazinoid pathway in maize and wheat provide relative to the DIBOA-Glc pathway that is present in rye and wild barley.
Corn leaf aphid feeding significantly induced the expression of TPS2 and TPS3 in B73 (Fig. 7A) as well as the single TPS2/3 homolog found in W22 (Fig. 7B). In vitro assays with B73 TPS2 show that this enzyme uses geranyldiphosphate, farnesyldiphosphate, and geranylgeranyldiphosphate as substrates, thereby enabling the production of monoterpenes, sesquiterpenes, and diterpenes, including linalool, (E)-nerolidol, and (E,E)-geranyl linalool (Richter, 2014). Consistent with this broad substrate specificity, a Ds transposon insertion near the 5′ end of TPS2/3 significantly reduced the aphid-induced accumulation of terpenes in W22 (Fig. 7E). Given that TPS2 and TPS3 expression is induced as a B73 response to aphid feeding, it was somewhat surprising that aphid reproduction was significantly reduced by the W22 tps2/3::Ds mutation (Fig. 7D). It is possible that the attenuated TPS2/3 activity in the mutant line decreases the abundance of a terpene that attracts corn leaf aphids or serves as a feeding stimulant. Conversely, the absence of TPS2/3 activity could result in the accumulation of the geranyldiphosphate, farnesyldiphosphate, or geranylgeranyldiphosphate substrates and, thereby, the accumulation of other, as yet unidentified aphid-deterrent terpenes. Experiments with several plant-aphid combinations have demonstrated aphid-repellent effects of terpenes (Unsicker et al., 2009; Zhang et al., 2015). Further analysis of the W22 terpene content and perhaps the identification of additional mutant lines will be required to determine why the absence of TPS2/3 activity has such a strong effect on corn leaf aphid reproduction.
In addition to alterations in the production of terpenes and benzoxazinoids, aphid-induced changes in the maize transcript profile indicate the up-regulation of other defenses, including the production of suberin, cellulose, coniferin, and phenylpropanoids. Consistent with the frequently discussed tradeoff between growth and defense (Schwachtje and Baldwin, 2008), the aphid-induced transcript profiles indicate a general shift away from primary metabolism in favor of defenses in maize. However, free amino acids, which are the main source of nitrogen for phloem-feeding aphids, do not show a consistent pattern of changes (Supplemental Table S6). There may be limitations in maize physiology that do not allow significant decreases in free amino acids, which would be predicted to be detrimental to aphids. It is also possible that the leaf senescence that occurs in response to insect feeding results in an unavoidable release of free amino acids. Nevertheless, given the highly localized feeding site of aphids, we cannot rule out the possibility of changes in amino acid metabolism at a local level in the phloem sieve elements that would make nitrogen less available for aphid feeding.
CONCLUSION
Together, the results presented here provide new insight into the defense responses of maize, one of the world’s most important crop plants. By examining both transcriptional and metabolic changes in a time course after the initiation of aphid feeding, we show that maize responses in the first hours after the initiation of aphid feeding are distinct from those that occur over a period of days. Our analysis of the transcriptomic and metabolomic data has led to the discovery of a previously uncharacterized terpene synthase activity that strongly influences aphid reproduction on maize. Future research on this terpene synthase and other genes that are induced by aphid feeding in our experiments will enable the breeding of maize cultivars with enhanced resistance to insect herbivores.
MATERIALS AND METHODS
Plants and Growth Conditions
Single seeds of B73, W22, and Ds transposon insertion lines of maize (Zea mays) were planted 1.5 cm deep in 7.6- × 7.6-cm plastic pots (200 cm3) filled with moistened maize mix (produced by combining 0.16 m3 of Metro-Mix 360, 0.45 kg of finely ground lime, 0.45 kg of Peters Unimix [Griffin Greenhouse Supplies], 68 kg of Turface MVP [Banfield-Baker], 23 kg of coarse quartz sand, and 0.018 m3 of pasteurized field soil). Plants were grown in growth chambers under a 16-h-light/8-h-dark photoperiod and 180 mmol photons m−2 s−1 light intensity at constant 23°C and 60% humidity for 2 weeks (V2–V3 developmental stage).
Aphid Assays
A corn leaf aphid (Rhopalosiphum maidis) colony was maintained on B73 maize plants as described previously (Meihls et al., 2013). For the time-course bioassay, 10 adult aphids were confined on the second true leaf of 2-week-old seedlings for 2, 4, 8, 24, 48, or 96 h using clip cages. All plants received cages at the start of the experiment and the addition of aphids was staggered, so that all plant tissue for gene expression and metabolite assays was harvested at the same time (96 h after the start of the experiment; Supplemental Fig. S1). Control plants received empty cages without aphids for 96 h. For assays measuring aphid progeny reproduction, 10 adult corn leaf aphids were confined on 2-week-old plants with microperforated polypropylene bags (15 × 61 cm; PJP Marketplace; http://www.pjpmarketplace.com), and the adults and nymphs were counted 1 week later. Samples for cytokinin analysis were collected 10 d after placing aphids on the plants, when the infestation had reached a steady state.
Preparation of mRNA and Complementary DNA
Leaf material was harvested, flash frozen in liquid nitrogen, and stored at −80°C. Frozen leaf material was ground to a fine powder in nitrogen using a paint shaker (Harbil) and 3-mm steel balls. Following homogenization, RNA was extracted using TRI Reagent (Sigma) and was purified with the SV Total RNA Isolation Kit with on-column DNase treatment (Promega). RNA quality, purity, and concentration were assessed using a spectrophotometer (NanoDrop 2000c; Thermo Fisher Scientific). For complementary DNA (cDNA) synthesis, 500 ng of total RNA was reverse transcribed with SMART-MMLV Reverse Transcriptase (Clontech Laboratories) using oligo(dT) as a primer (IDT).
Quantitative RT-PCR
Gene-specific primers used for quantitative RT-PCR were designed using Primer3Plus (www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi/; Supplemental Table S7). Reactions were performed using 6.7 μL of the SYBR Green PCR master mix (Applied Biosystems), with 800 nm primers and 10 ng of cDNA, in a 7900HT instrument (Applied Biosystems). The PCR was initiated by incubation at 95°C for 10 min. For DNA amplification, the following cycle was repeated 40 times: 95°C for 15 s, 60°C for 15 s, and 72°C for 15 s. The cycle threshold values were quantified and analyzed according to the standard curve method and were normalized for differences in cDNA amount using the expression of the maize housekeeping gene ACTIN1 as an internal standard (Meihls et al., 2013). Quantitation of gene expression was calculated using the relative standard curve method (Applied Biosystems).
Transcriptome Sequencing
Total RNA was extracted from maize leaves as describe above. Tissue from five individual maize plants was combined into one experimental replicate, and five replicates were collected for each time point of the experiment. The purity of total RNA extracted was determined in NanoDrop 2000 (Thermo Scientific), and 2 to 3 μg of total RNA was used for the preparation of strand-specific RNA-Seq libraries (Zhong et al., 2011; Chen et al., 2012). The purified libraries were quantified, and 20 ng of each was used for sequencing. Samples were sequenced on an Illumina HiSeq2000 device (Illumina) at the Weill Medical College Sequencing Facility (Cornell University) with a 101-bp single-end read length. Libraries were multiplexed and sequenced in three lanes.
RNA-Seq and Metabolite Data Normalization
In order to reduce the effects of the variation in gene expression or metabolite abundance during the 4 h used for harvesting all of the samples, a multiple regression analysis was performed on the RNA-Seq and metabolite data. The actual time of sample harvest was recorded for each sample, and five replicates of no-aphid control samples were harvested at the beginning and the end of the tissue-harvesting period. Assuming a linear influence of the circadian cycle between the first and last control sample times, new artificial controls were calculated based on the value expected in the regression line between the early and late controls for each one of the replicates of the samples. Custom Ruby scripts using the module Statssample (http://ruby-statsample.rubyforge.org) were developed to calculate the artificial controls for all the samples.
RNA-Seq Data Analysis
Read quality values were checked using FASTQC. Adaptors and low-quality sequences were trimmed and removed using Fastq-mcf with a minimum length of 50 bp and a minimum quality value of 30. RNA-Seq analysis was performed following a protocol published by Anders et al. (2013), using as reference the B73 maize genome, AGPv3.20 (www.maizegdb.org). The protocol was only altered to include the counts from the artificial controls for the edgeR (Robinson et al., 2010) analysis (step 14). The counts for the early and late controls were obtained using htseq-count, and new counts were calculated for the artificial controls based on the multiple regression analysis. From the approximately 41,700 transcripts showing expression values in the maize genome sequence, the ones showing at least 1 count per million in three or more replicates for each time point (each time point has five control replicates and five treatment replicates) were kept for differentially expressed gene detection; the rest of the genes were filtered out. edgeR with a simple design was used for each one of the time points (step 14 of the protocol).
Targeted and Untargeted Metabolite Assays
For assays of maize metabolites, adult aphids were caged on the tip of the second leaf, in the same manner as for the gene expression experiments described above. Approximately 5 cm of leaf material was collected from the tip of the second leaf, including the leaf tissue contained within the aphid cage. Corresponding tissue was collected from control plants with cages but without aphids. Samples were weighed, and all data were normalized relative to the tissue fresh weight or dry weight, depending on the assay.
For nontargeted metabolite assays, separation was performed using a Dionex Ultimate 3000 UHPLC system with an Acclaim (Thermo Scientific) column, particle size 2.2 µm, 2.1 × 150 mm. Solvent A was 99.85% water + 0.1% acetonitrile + 0.05% (v/v) formic acid, and solvent B was 99.95% acetonitrile + 0.05% (v/v) formic acid. Temperature was 30°C, and the gradient was isocratic for 1 min 10% B, linear to 85% B in 24 min, and 5 min isocratic at 85% (v/v) B. Flow rate was 0.3 mL min−1. The injection volume was 2 µL (positive detection mode) or 3 µL (negative detection mode). Metabolites were detected using a quadrupole time-of-flight mass spectrometer (MicrOTOF-Q II; Bruker Daltronics) with an electrospray ionization (ESI) ion source. The instrument was run with the following parameters: scan mass-to-charge ratio of 50 to 1,400, 1.4 bar nitrogen nebulizer, dry gas as 10 L min−1 nitrogen, capillary with 4,500 V (positive) and 2,500 V (negative), end plate offset of 2500 V, funnel 1 radio frequency (RF) set at 200 volt peak-to-peak (Vpp), funnel 2 RF set at 200 Vpp, in-source collision-induced dissociation energy set at 0 V, hexapole RF set at 100 Vpp, quadrupole ion energy set at 3 eV, ion energy set at 3 eV, collision RF set at 150 Vpp, transfer time set at 70 ms, prepulse storage set at 5 ms, and spectral rate set at 1 Hz. Raw mass spectrometry data files were processed using the XCMS (http://metlin.scripps.edu/download/) and CAMERA (http://www.bioconductor.org/packages/release/bioc/html/CAMERA.html) software packages in R. Processed positive and negative ionization data sets were transferred to Microsoft Excel for further analysis.
For amino acid analysis, samples were analyzed using a Waters 2790 HPLC system as described previously (Joshi et al., 2006), with a minor modification whereby the ratio of tissue to extraction buffer was 1:3. For measuring leaf DIMBOA-Glc content, sample extraction and HPLC analysis were performed as described previously (Mijares et al., 2013). For quantification of free fatty acids, oxylipins, salicylic acid, indole, and 6-methoxybenzoxazolinone, samples were solvent extracted, methylated, collected on a polymeric adsorbent using vapor-phase extraction, and analyzed using gas chromatography-isobutane positive ion chemical ionization-mass spectrometry as described (Schmelz et al., 2011). For analysis of cytokinins, maize leaves were harvested, frozen in liquid nitrogen, and lyophilized. Dry leaf tissue was weighed and extracted with 75% methanol, and cytokinins were analyzed by mass spectrometry as described previously (Schäfer et al., 2015).
Phospholipids were extracted in methanol and analyzed using an LC/MS2010 device equipped with an LC-10ADvp pump (Shimadzu). The curved desolvation line temperature was 250°C, voltage was 1.5 kV, nebulizer gas flow was 1.5 L min−1, and the analysis mode was ESI negative, SCAN. A Capcell Pak C8 column (UG120; 50 × 2-mm i.d.; Shiseido) was eluted (0.2 mL min−1) with a gradient of 70% to 99% acetonitrile in water containing 0.05% ammonium formate over 15 min with column temperature maintained at 40°C. Quantitative analyses of phospholipids used diheptadecanoylphosphatidylcholine (500 ng mL−1) as an internal standard. Samples were also analyzed using a Prominence HPLC system coupled to an LCMS-IT-TOF detector (Shimadzu) to confirm the structures. HPLC conditions were the same as for the LC 10 ADvp pump. The mass spectrometer was operated with probe voltage of 4.5 kV, curved desolvation line temperature of 200°C, block heater temperature of 200°C, nebulizer gas flow of 1.5 L min−1, and ion accumulation time of 30 ms, and the analytical mode was ESI negative. Phospholipids were identified by mass fragmentation patterns (Han and Gross, 1996; Fang et al., 2003; Hsu et al., 2007) and comparison with authentic standards.
For terpene assays, leaf material was harvested, ground with a mortar and pestle in liquid nitrogen, and stored at −80°C until sample preparation. The volatile terpenes released from the powdered plant material was collected on a solid-phase microextraction fiber and analyzed by GC-MS (GC-2010 and GCMS-QP 2010 Plus; Shimadzu). Collection of terpenes from frozen and macerated tissue in this manner produces similar results to terpene collection from intact maize leaves (Köllner et al., 2004). To release the volatiles from the fiber, an injection temperature of 220°C was used. Hydrogen served as a carrier gas with a flow rate of 1 mL min−1. To separate the volatiles, an EC5-MS column (30-m length, 0.25-mm i.d., and 0.25-µm film; Grace) was used under the following conditions: 80°C for 3 min, first ramp of 7°C min−1 to 200°C, second ramp of 100°C min−1 to 300°C, and final 2-min hold. Compounds were identified by comparison of retention times and using the Shimadzu software GCMS Postrun Analysis with the mass spectral libraries Wiley8 (http://www.wiley.com) and Adams (Adams, 2007).
Isolation of Transposon Insertion Knockout Lines and Genomic DNA Preparation
Ds transposon insertions maintained in the W22 genetic background were identified in the following genes of interest through the Activator (Ac)/Ds tagging project Web site (http://www.acdstagging.org; Vollbrecht et al., 2010). Seed stocks for bx1::Ds (gene identifier GRMZM2G085381; Ds, B.W06.0775) and bx2::Ds (gene identifier GRMZM2G085661; Ds, I.S07.3472) are available from the Maize Genetics Cooperation Stock Center (http://maizecoop.cropsci.uiuc.edu/) as AcDs-00565 and AcDs-00212, respectively. Seed stocks for both bx6::Ds (gene identifier GRMZM6G617209; Ds, I.S07.0479) and tps2::Ds (gene identifier GRMZM2G046615; Ds, B.S08.0585) are available through the Ac/Ds tagging project Web site (http://www.acdstagging.org/seed/index.php). Primer pairs were designed to identify Ds-positive segregants using Ds-end primers in combination with Ds-flanking sequences using Primer3 software (http://www.bioinformatics.nl/cgi-bin/primer3plus/primer3plus.cgi/). Additionally, primer sets for each Ds allele were designed to amplify PCR products spanning each insertion site in an effort to identify plants carrying each homozygous or heterozygous Ds insertion allele. These primers were designed using the B73 genome browser found at MaizeGDB (http://www.maizegdb.org/; RefGen_v2 for the Bx1 gene and RefGen_v3 for Bx2, Bx6, and TPS2). For the full primer list, see Supplemental Table S9. Genomic DNA preparation was described previously (Gao et al., 2010). All PCRs were performed in a 20-µL GoTaq PCR device (Promega) with either 4% or 5% dimethyl sulfoxide added.
Statistical Analysis
Data for the PLS-DA plot were normalized as described previously (Tzin et al., 2009). Graphs were drawn using MetaboAnalyst 3.0 software (Xia et al., 2009). Overrepresentation analyses were performed using PageMan (Usadel et al., 2006). Statistical comparisons were made using JMP Pro 11 (SAS; http://www.jmp.com). The CLUSTAL Omega Multiple Sequence Alignment Tool (http://www.ebi.ac.uk/Tools/msa/clustalo/) was used to align terpene synthase chromosomal and protein sequences.
Maize RNA sequences from transcript profiling experiments were submitted to the National Center for Biotechnology Information Short Read Archive (accession no. PRJNA295410).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Design of the aphid feeding experiments.
Supplemental Figure S2. Comparison of RNA-Seq and quantitative RT-PCR gene expression data.
Supplemental Figure S3. Metabolite overview of aphids feeding on maize foliage.
Supplemental Figure S4. Enriched terms of MapMan categories of significant differentially expressed genes altered by aphid feeding.
Supplemental Figure S5. Cytokinin content in maize leaves with or without aphid feeding.
Supplemental Figure S6. DIMBOA-Glc content of wild-type W22, with and without aphid feeding.
Supplemental Figure S7. Comparison of TPS2 and TPS3 from B73 and W22.
Supplemental Table S1. Complete list of normalized transcript abundance (RNA-Seq) data for six time points using counts per million reads.
Supplemental Table S2. RNA-Seq data for six aphid feeding time points after analysis by edgeR.
Supplemental Table S3. List of differentially expressed maize genes for at least one time point (plus or minus greater than 2-fold changed) with P < 0.05 (FDR adjusted), used for PLD-SA analysis (Fig. 1A), clustering overrepresentation (Fig. 2), and PageMan analysis (Supplemental Fig. S2).
Supplemental Table S4. LC-TOF-MS data from negative and positive ion modes.
Supplemental Table S5. LC-TOF-MS mass signatures that were significantly altered for at least one aphid feeding time point, P < 0.05 (FDR adjusted), fold change less than 0.5 or greater than 2.
Supplemental Table S6. Known and putative metabolites of central and specialized metabolism, measured by multiple methods.
Supplemental Table S7. Primers used for quantitative RT-PCR analysis.
Supplemental Table S8. Orthologous Arabidopsis and maize genes used for Hormonometer analysis.
Supplemental Table S9. Primers used to screen for Ds transposon insertion knockout mutations.
Supplementary Material
Acknowledgments
We thank Meena Haribal and Rayko Halitschke for technical support related to this project.
Glossary
- DIMBOA-Glc
4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one glucoside
- HDMBOA-Glc
2-hydroxy-4,7-dimethoxy-1,4-benzoxazin-3-one glucoside
- RNA-Seq
RNA sequencing
- RT
reverse transcription
- FDR
false discovery rate
- PLS-DA
partial least-squares discriminant analysis
- LC-TOF-MS
liquid chromatography-time of flight-mass spectrometry
- GC-MS
gas chromatography-mass spectrometry
- OPDA
12-oxo-phytodienoic acid
- cDNA
complementary DNA
- ESI
electrospray ionization
Footnotes
This work was supported by the U.S. National Science Foundation (grant no. 1139329 to G.J., E.A.S., and A.H. and grant no. 1339237 to G.J.), the Alexander von Humboldt Foundation (Friedrich Wilhelm Bessel research award to G.J.), Vaadia-Binational Agricultural Research and Development Fund (postdoctoral fellowship award no. FI–471–2012 to V.T.), the Japan Science and Technology Agency (to N.M.), the U.S. Department of Agriculture (grant no. 2011–67012–30675 to L.N.M.), and the Triad Foundation (to G.J. and L.A.M.).
Articles can be viewed without a subscription.
References
- Adams RP. (2007) Identification of Essential Oil Components by Gas Chromatography/Quadruple Mass Spectrometry, Ed 4 Allured Publishing, Carol Stream, IL [Google Scholar]
- Ahmad S, Veyrat N, Gordon-Weeks R, Zhang Y, Martin J, Smart L, Glauser G, Erb M, Flors V, Frey M, et al. (2011) Benzoxazinoid metabolites regulate innate immunity against aphids and fungi in maize. Plant Physiol 157: 317–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK, Huber W, Robinson MD (2013) Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc 8: 1765–1786 [DOI] [PubMed] [Google Scholar]
- Appel HM, Fescemyer H, Ehlting J, Weston D, Rehrig E, Joshi T, Xu D, Bohlmann J, Schultz J (2014) Transcriptional responses of Arabidopsis thaliana to chewing and sucking insect herbivores. Front Plant Sci 5: 565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betsiashvili M, Ahern KR, Jander G (2015) Additive effects of two quantitative trait loci that confer Rhopalosiphum maidis (corn leaf aphid) resistance in maize inbred line Mo17. J Exp Bot 66: 571–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bing JW, Guthrie WD (1991) Generation mean analysis for resistance in maize to the corn leaf aphid (Homoptera: Aphididae). J Econ Entomol 84: 1080–1082 [Google Scholar]
- Bos JI, Prince D, Pitino M, Maffei ME, Win J, Hogenhout SA (2010) A functional genomics approach identifies candidate effectors from the aphid species Myzus persicae (green peach aphid). PLoS Genet 6: e1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broekgaarden C, Voorrips RE, Dicke M, Vosman B (2011) Transcriptional responses of Brassica nigra to feeding by specialist insects of different feeding guilds. Insect Sci 18: 259–272 [Google Scholar]
- Cambier V, Hance T, De Hoffmann E (2001) Effects of 1,4-benzoxazin-3-one derivatives from maize on survival and fecundity of Metopolophium dirhodum (Walker) on artificial diet. J Chem Ecol 27: 359–370 [DOI] [PubMed] [Google Scholar]
- Carena MJ, Glogoza P (2004) Resistance of maize to the corn leaf aphid: a review. Maydica 49: 241–254 [Google Scholar]
- Chen YR, Zheng Y, Liu B, Zhong S, Giovannoni J, Fei Z (2012) A cost-effective method for Illumina small RNA-Seq library preparation using T4 RNA ligase 1 adenylated adapters. Plant Methods 8: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christensen SA, Huffaker A, Kaplan F, Sims J, Ziemann S, Doehlemann G, Ji L, Schmitz RJ, Kolomiets MV, Alborn HT, et al. (2015) Maize death acids, 9-lipoxygenase-derived cyclopente(a)nones, display activity as cytotoxic phytoalexins and transcriptional mediators. Proc Natl Acad Sci USA 112: 11407–11412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantino NN, Mastouri F, Damarwinasis R, Borrego EJ, Moran-Diez ME, Kenerley CM, Gao X, Kolomiets MV (2013) Root-expressed maize lipoxygenase 3 negatively regulates induced systemic resistance to Colletotrichum graminicola in shoots. Front Plant Sci 4: 510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppola V, Coppola M, Rocco M, Digilio MC, D’Ambrosio C, Renzone G, Martinelli R, Scaloni A, Pennacchio F, Rao R, et al. (2013) Transcriptomic and proteomic analysis of a compatible tomato-aphid interaction reveals a predominant salicylic acid-dependent plant response. BMC Genomics 14: 515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couldridge C, Newbury HJ, Ford-Lloyd B, Bale J, Pritchard J (2007) Exploring plant responses to aphid feeding using a full Arabidopsis microarray reveals a small number of genes with significantly altered expression. Bull Entomol Res 97: 523–532 [DOI] [PubMed] [Google Scholar]
- De Vos M, Jander G (2009) Myzus persicae (green peach aphid) salivary components induce defence responses in Arabidopsis thaliana. Plant Cell Environ 32: 1548–1560 [DOI] [PubMed] [Google Scholar]
- De Vos M, Van Oosten VR, Van Poecke RM, Van Pelt JA, Pozo MJ, Mueller MJ, Buchala AJ, Métraux JP, Van Loon LC, Dicke M, et al. (2005) Signal signature and transcriptome changes of Arabidopsis during pathogen and insect attack. Mol Plant Microbe Interact 18: 923–937 [DOI] [PubMed] [Google Scholar]
- Dicke M. (1999) Are herbivore-induced plant volatiles reliable indicators of herbivore identity to foraging carnivorous arthropods? Entomol Exp Appl 91: 131–142 [Google Scholar]
- Ellis C, Karafyllidis I, Turner JG (2002) Constitutive activation of jasmonate signaling in an Arabidopsis mutant correlates with enhanced resistance to Erysiphe cichoracearum, Pseudomonas syringae, and Myzus persicae. Mol Plant Microbe Interact 15: 1025–1030 [DOI] [PubMed] [Google Scholar]
- El-Muadhidi MA, Makkouk KM, Kumari SG, Myasser J, Murad SS, Mustafa R (2001) Survey for legume and cereal viruses in Iraq. Phytopathol Mediterr 40: 224–233 [Google Scholar]
- Elzinga DA, De Vos M, Jander G (2014) Suppression of plant defenses by a Myzus persicae (green peach aphid) salivary effector protein. Mol Plant Microbe Interact 27: 747–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elzinga DA, Jander G (2013) The role of protein effectors in plant-aphid interactions. Curr Opin Plant Biol 16: 451–456 [DOI] [PubMed] [Google Scholar]
- Erb M, Veyrat N, Robert CA, Xu H, Frey M, Ton J, Turlings TC (2015) Indole is an essential herbivore-induced volatile priming signal in maize. Nat Commun 6: 6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang N, Yu S, Badger TM (2003) LC-MS/MS analysis of lysophospholipids associated with soy protein isolate. J Agric Food Chem 51: 6676–6682 [DOI] [PubMed] [Google Scholar]
- Ferry N, Stavroulakis S, Guan W, Davison GM, Bell HA, Weaver RJ, Down RE, Gatehouse JA, Gatehouse AMR (2011) Molecular interactions between wheat and cereal aphid (Sitobion avenae): analysis of changes to the wheat proteome. Proteomics 11: 1985–2002 [DOI] [PubMed] [Google Scholar]
- Foott WH, Timmins PR (1973) Effects of infestations by corn leaf aphid, Rhopalosiphum maidis (Homoptera-Aphididae), on field corn in southwestern Ontario. Can Entomol 105: 449–458 [Google Scholar]
- Frey M, Chomet P, Glawischnig E, Stettner C, Grün S, Winklmair A, Eisenreich W, Bacher A, Meeley RB, Briggs SP, et al. (1997) Analysis of a chemical plant defense mechanism in grasses. Science 277: 696–699 [DOI] [PubMed] [Google Scholar]
- Frey M, Schullehner K, Dick R, Fiesselmann A, Gierl A (2009) Benzoxazinoid biosynthesis, a model for evolution of secondary metabolic pathways in plants. Phytochemistry 70: 1645–1651 [DOI] [PubMed] [Google Scholar]
- Gan S, Amasino RM (1995) Inhibition of leaf senescence by autoregulated production of cytokinin. Science 270: 1986–1988 [DOI] [PubMed] [Google Scholar]
- Gao H, Smith J, Yang M, Jones S, Djukanovic V, Nicholson MG, West A, Bidney D, Falco SC, Jantz D, et al. (2010) Heritable targeted mutagenesis in maize using a designed endonuclease. Plant J 61: 176–187 [DOI] [PubMed] [Google Scholar]
- Gershenzon J, Dudareva N (2007) The function of terpene natural products in the natural world. Nat Chem Biol 3: 408–414 [DOI] [PubMed] [Google Scholar]
- Giordanengo P, Brunissen L, Rusterucci C, Vincent C, van Bel A, Dinant S, Girousse C, Faucher M, Bonnemain JL (2010) Compatible plant-aphid interactions: how aphids manipulate plant responses. C R Biol 333: 516–523 [DOI] [PubMed] [Google Scholar]
- Guan W, Ferry N, Edwards MG, Bell HA, Othman H, Gatehouse JA, Gatehouse AMR (2015) Proteomic analysis shows that stress response proteins are significantly up-regulated in resistant diploid wheat (Triticum monococcum) in response to attack by the grain aphid (Sitobion avenae). Mol Breed 35: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Gross RW (1996) Structural determination of lysophospholipid regioisomers by electrospray ionization tandem mass spectrometry. J Am Chem Soc 118: 451–457 [Google Scholar]
- Hawkes JR, Jones RAC (2005) Incidence and distribution of Barley yellow dwarf virus and Cereal yellow dwarf virus in over-summering grasses in a Mediterranean-type environment. Aust J Agric Res 56: 257–270 [Google Scholar]
- Heidel AJ, Baldwin IT (2004) Microarray analysis of salicylic acid- and jasmonic acid-signalling in responses of Nicotiana attenuata to attack by insects from multiple feeding guilds. Plant Cell Environ 27: 1362–1373 [Google Scholar]
- Heidel-Fischer HM, Musser RO, Vogel H (2014) Plant transcriptomic responses to herbivory. In Voelckel C, Jander G, eds, Annual Plant Reviews. John Wiley & Sons, Sussex, UK, pp 155–196 [Google Scholar]
- Heil M. (2009) Damaged-self recognition in plant herbivore defence. Trends Plant Sci 14: 356–363 [DOI] [PubMed] [Google Scholar]
- Hillwig MS, Chiozza M, Casteel CL, Lau ST, Hohenstein J, Hernández E, Jander G, MacIntosh GC (June 7, 2015) Abscisic acid deficiency increases defence responses against Myzus persicae in Arabidopsis. Mol Plant Pathol http://dx.doi.org/10.1111/mpp.12274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogenhout SA, Bos JIB (2011) Effector proteins that modulate plant-insect interactions. Curr Opin Plant Biol 14: 422–428 [DOI] [PubMed] [Google Scholar]
- Hsu FF, Turk J, Williams TD, Welti R (2007) Electrospray ionization multiple stage quadrupole ion-trap and tandem quadrupole mass spectrometric studies on phosphatidylglycerol from Arabidopsis leaves. J Am Soc Mass Spectrom 18: 783–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarošová J, Chrpová J, Šíp V, Kundu JK (2013) A comparative study of the Barley yellow dwarf virus species PAV and PAS: distribution, accumulation and host resistance. Plant Pathol 62: 436–443 [Google Scholar]
- Jonczyk R, Schmidt H, Osterrieder A, Fiesselmann A, Schullehner K, Haslbeck M, Sicker D, Hofmann D, Yalpani N, Simmons C, et al. (2008) Elucidation of the final reactions of DIMBOA-glucoside biosynthesis in maize: characterization of Bx6 and Bx7. Plant Physiol 146: 1053–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi V, Laubengayer KM, Schauer N, Fernie AR, Jander G (2006) Two Arabidopsis threonine aldolases are nonredundant and compete with threonine deaminase for a common substrate pool. Plant Cell 18: 3564–3575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joung JG, Corbett AM, Fellman SM, Tieman DM, Klee HJ, Giovannoni JJ, Fei Z (2009) Plant MetGenMAP: an integrative analysis system for plant systems biology. Plant Physiol 151: 1758–1768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerchev PI, Karpińska B, Morris JA, Hussain A, Verrall SR, Hedley PE, Fenton B, Foyer CH, Hancock RD (2013) Vitamin C and the abscisic acid-insensitive 4 transcription factor are important determinants of aphid resistance in Arabidopsis. Antioxid Redox Signal 18: 2091–2105 [DOI] [PubMed] [Google Scholar]
- Kessler A, Baldwin IT (2002) Plant responses to insect herbivory: the emerging molecular analysis. Annu Rev Plant Biol 53: 299–328 [DOI] [PubMed] [Google Scholar]
- Kieber JJ, Schaller GE (2014) Cytokinins. The Arabidopsis Book 12: e0168, doi/10.1199/tab.0168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köllner TG, Schnee C, Gershenzon J, Degenhardt J (2004) The sesquiterpene hydrocarbons of maize (Zea mays) form five groups with distinct developmental and organ-specific distributions. Phytochemistry 65: 1895–1902 [DOI] [PubMed] [Google Scholar]
- Krueger EN, Beckett RJ, Gray SM, Miller WA (2013) The complete nucleotide sequence of the genome of Barley yellow dwarf virus-RMV reveals it to be a new polerovirus distantly related to other yellow dwarf viruses. Front Microbiol 4: 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machado S, Bynum E, Archer T, Lascano R, Wilson L, Bordovsky J, Segarra E, Bronson K, Nesmith D, Xu W (2002) Spatial and temporal variability of corn growth and grain yield. Crop Sci 42: 1564–1576 [Google Scholar]
- Meihls LN, Handrick V, Glauser G, Barbier H, Kaur H, Haribal MM, Lipka AE, Gershenzon J, Buckler ES, Erb M, et al. (2013) Natural variation in maize aphid resistance is associated with 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one glucoside methyltransferase activity. Plant Cell 25: 2341–2355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meihls LN, Kaur H, Jander G (2012) Natural variation in maize defense against insect herbivores. Cold Spring Harb Symp Quant Biol 77: 269–283 [DOI] [PubMed] [Google Scholar]
- Mewis I, Tokuhisa JG, Schultz JC, Appel HM, Ulrichs C, Gershenzon J (2006) Gene expression and glucosinolate accumulation in Arabidopsis thaliana in response to generalist and specialist herbivores of different feeding guilds and the role of defense signaling pathways. Phytochemistry 67: 2450–2462 [DOI] [PubMed] [Google Scholar]
- Mijares V, Meihls LN, Jander G, Tzin V (2013) Near-isogenic lines for measuring phenotypic effects of DIMBOA-Glc methyltransferase activity in maize. Plant Signal Behav 8: e26779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalam VJ, Keeretaweep J, Sarowar S, Shah J (2012) Root-derived oxylipins promote green peach aphid performance on Arabidopsis foliage. Plant Cell 24: 1643–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Corn Growers Association (2014) World of Corn Statistics Book. National Corn Growers Association, Chesterfield, MO [Google Scholar]
- Niemeyer HM. (1988) Hydroxamic acids (4-hydroxy-1,4-benzoxazin-3-ones), defence chemicals in the Gramineae. Phytochemistry 27: 3349–3358 [Google Scholar]
- Niemeyer HM. (2009) Hydroxamic acids derived from 2-hydroxy-2H-1,4-benzoxazin-3(4H)-one: key defense chemicals of cereals. J Agric Food Chem 57: 1677–1696 [DOI] [PubMed] [Google Scholar]
- Oikawa A, Ishihara A, Iwamura H (2002) Induction of HDMBOA-Glc accumulation and DIMBOA-Glc 4-O-methyltransferase by jasmonic acid in poaceous plants. Phytochemistry 61: 331–337 [DOI] [PubMed] [Google Scholar]
- Oikawa A, Ishihara A, Tanaka C, Mori N, Tsuda M, Iwamura H (2004) Accumulation of HDMBOA-Glc is induced by biotic stresses prior to the release of MBOA in maize leaves. Phytochemistry 65: 2995–3001 [DOI] [PubMed] [Google Scholar]
- Paré PW, Tumlinson JH (1999) Plant volatiles as a defense against insect herbivores. Plant Physiol 121: 325–332 [PMC free article] [PubMed] [Google Scholar]
- Park SJ, Huang Y, Ayoubi P (2006) Identification of expression profiles of sorghum genes in response to greenbug phloem-feeding using cDNA subtraction and microarray analysis. Planta 223: 932–947 [DOI] [PubMed] [Google Scholar]
- Pichersky E, Gershenzon J (2002) The formation and function of plant volatiles: perfumes for pollinator attraction and defense. Curr Opin Plant Biol 5: 237–243 [DOI] [PubMed] [Google Scholar]
- Pichersky E, Noel JP, Dudareva N (2006) Biosynthesis of plant volatiles: nature’s diversity and ingenuity. Science 311: 808–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CM, Dicke M (2007) Plant interactions with microbes and insects: from molecular mechanisms to ecology. Trends Plant Sci 12: 564–569 [DOI] [PubMed] [Google Scholar]
- Porta H, Rocha-Sosa M (2002) Plant lipoxygenases: physiological and molecular features. Plant Physiol 130: 15–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Power AG, Borer ET, Hosseini P, Mitchell CE, Seabloom EW (2011) The community ecology of barley/cereal yellow dwarf viruses in western US grasslands. Virus Res 159: 95–100 [DOI] [PubMed] [Google Scholar]
- Richter A. (2014) Identifizierung von QTLs der Terpenbiosynthese mittels “nested association mapping” (NAM) und “genome wide association study” (GWAS). PhD thesis. Universitäts-und Landesbibliothek Sachsen-Anhalt, Halle (Saale), Germany
- Roberts SC. (2007) Production and engineering of terpenoids in plant cell culture. Nat Chem Biol 3: 387–395 [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer M, Meza-Canales ID, Navarro-Quezada A, Brütting C, Vanková R, Baldwin IT, Meldau S (2015) Cytokinin levels and signaling respond to wounding and the perception of herbivore elicitors in Nicotiana attenuata. J Integr Plant Biol 57: 198–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz EA, Kaplan F, Huffaker A, Dafoe NJ, Vaughan MM, Ni X, Rocca JR, Alborn HT, Teal PE (2011) Identity, regulation, and activity of inducible diterpenoid phytoalexins in maize. Proc Natl Acad Sci USA 108: 5455–5460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. (2009) The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115 [DOI] [PubMed] [Google Scholar]
- Schwachtje J, Baldwin IT (2008) Why does herbivore attack reconfigure primary metabolism? Plant Physiol 146: 845–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivaji R, Camas A, Ankala A, Engelberth J, Tumlinson JH, Williams WP, Wilkinson JR, Luthe DS (2010) Plants on constant alert: elevated levels of jasmonic acid and jasmonate-induced transcripts in caterpillar-resistant maize. J Chem Ecol 36: 179–191 [DOI] [PubMed] [Google Scholar]
- Smith CM, Boyko EV (2007) The molecular bases of plant resistance and defense responses to aphid feeding: current status. Entomol Exp Appl 122: 1–16 [Google Scholar]
- Sun Y, Guo H, Yuan L, Wei J, Zhang W, Ge F (2015) Plant stomatal closure improves aphid feeding under elevated CO2. Glob Change Biol 21: 2739–2748 [DOI] [PubMed] [Google Scholar]
- Thaler JS, Humphrey PT, Whiteman NK (2012) Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci 17: 260–270 [DOI] [PubMed] [Google Scholar]
- Tholl D, Chen F, Gershenzon J, Pichersky E (2004) Arabidopsis thaliana, a model system for investigating volatile terpene biosynthesis, regulation, and function. In John TR, ed, Recent Advances in Phytochemistry, Vol 38. Elsevier, Amsterdam, pp 1–18 [Google Scholar]
- Tholl D, Sohrabi R, Huh JH, Lee S (2011) The biochemistry of homoterpenes: common constituents of floral and herbivore-induced plant volatile bouquets. Phytochemistry 72: 1635–1646 [DOI] [PubMed] [Google Scholar]
- Thompson GA, Goggin FL (2006) Transcriptomics and functional genomics of plant defence induction by phloem-feeding insects. J Exp Bot 57: 755–766 [DOI] [PubMed] [Google Scholar]
- Tzin V, Galili G (2010) The biosynthetic pathways for shikimate and aromatic amino acids in Arabidopsis thaliana. The Arabidopsis Book 8: e0132, [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tzin V, Malitsky S, Aharoni A, Galili G (2009) Expression of a bacterial bi-functional chorismate mutase/prephenate dehydratase modulates primary and secondary metabolism associated with aromatic amino acids in Arabidopsis. Plant J 60: 156–167 [DOI] [PubMed] [Google Scholar]
- Unsicker SB, Kunert G, Gershenzon J (2009) Protective perfumes: the role of vegetative volatiles in plant defense against herbivores. Curr Opin Plant Biol 12: 479–485 [DOI] [PubMed] [Google Scholar]
- Usadel B, Nagel A, Steinhauser D, Gibon Y, Bläsing OE, Redestig H, Sreenivasulu N, Krall L, Hannah MA, Poree F, et al. (2006) PageMan: an interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics 7: 535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt T. (2010) Phenylpropanoid biosynthesis. Mol Plant 3: 2–20 [DOI] [PubMed] [Google Scholar]
- Vollbrecht E, Duvick J, Schares JP, Ahern KR, Deewatthanawong P, Xu L, Conrad LJ, Kikuchi K, Kubinec TA, Hall BD, et al. (2010) Genome-wide distribution of transposed Dissociation elements in maize. Plant Cell 22: 1667–1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volodarsky D, Leviatan N, Otcheretianski A, Fluhr R (2009) HORMONOMETER: a tool for discerning transcript signatures of hormone action in the Arabidopsis transcriptome. Plant Physiol 150: 1796–1805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walling LL. (2000) The myriad plant responses to herbivores. J Plant Growth Regul 19: 195–216 [DOI] [PubMed] [Google Scholar]
- Walling LL. (2008) Avoiding effective defenses: strategies employed by phloem-feeding insects. Plant Physiol 146: 859–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will T, Tjallingii WF, Thönnessen A, van Bel AJ (2007) Molecular sabotage of plant defense by aphid saliva. Proc Natl Acad Sci USA 104: 10536–10541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Psychogios N, Young N, Wishart DS (2009) MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Res 37: W652–660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarate SI, Kempema LA, Walling LL (2007) Silverleaf whitefly induces salicylic acid defenses and suppresses effectual jasmonic acid defenses. Plant Physiol 143: 866–875 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Li ZX, Yu XD, Fan J, Pickett JA, Jones HD, Zhou JJ, Birkett MA, Caulfield J, Napier JA, et al. (2015) Molecular characterization of two isoforms of a farnesyl pyrophosphate synthase gene in wheat and their roles in sesquiterpene synthesis and inducible defence against aphid infestation. New Phytol 206: 1101–1115 [DOI] [PubMed] [Google Scholar]
- Zhong S, Joung JG, Zheng Y, Chen YR, Liu B, Shao Y, Xiang JZ, Fei Z, Giovannoni JJ (2011) High-throughput Illumina strand-specific RNA sequencing library preparation. Cold Spring Harb Protoc 2011: 940–949 [DOI] [PubMed] [Google Scholar]
- Zhu-Salzman K, Salzman RA, Ahn JE, Koiwa H (2004) Transcriptional regulation of sorghum defense determinants against a phloem-feeding aphid. Plant Physiol 134: 420–431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuniga GE, Argandona VH, Niemeyer HM, Corcuera LJ (1983) Hydroxamic acid content in wild and cultivated Gramineae. Phytochemistry 22: 2665–2668 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.