Ecological, evolutionary, and mechanistic insights arise from the study of natural variation in plant metabolism.
Abstract
Combining quantitative genetics studies with metabolomics/metabolic profiling platforms, genomics, and transcriptomics is creating significant progress in identifying the causal genes controlling natural variation in metabolite accumulations and profiles. In this review, we discuss key mechanistic and evolutionary insights that are arising from these studies. This includes the potential role of transport and other processes in leading to a separation of the site of mechanistic causation and metabolic consequence. A reilluminated observation is the potential for genomic variation in the organelle to alter phenotypic variation alone and in epistatic interaction with the nuclear genetic variation. These studies are also highlighting new aspects of metabolic pleiotropy both in terms of the breadth of loci altering metabolic variation as well as the potential for broader effects on plant defense regulation of the metabolic variation than has previously been predicted. We also illustrate caveats that can be overlooked when translating quantitative genetics descriptors such as heritability and per-locus r2 to mechanistic or evolutionary interpretations.
The study of quantitative genetics and ecology and evolution in plants has a long history of reliance on the natural variation of metabolic traits. One of the first identified quantitative trait loci (QTLs) in any organism was for the metabolic control of seed color in Phaseolus vulgaris (Sax, 1923). This analysis helped to develop and empirically test some of the foundations of quantitative genetics. Similarly, a significant fraction of ecology and evolutionary theory has focused on the pressures leading to the diversification of plant metabolism and the contravening costs on these defenses (Ehrlich and Raven, 1964; Karban and Baldwin, 1997). These studies have used measurements of metabolite variation to make significant conceptual progress in understanding the underlying pressures without access to the underlying causal genes (Strauss and Agrawal, 1999; Agrawal, 2011; Cook-Patton et al., 2011).
Recent advances in genomics and metabolic profiling have opened new opportunities to study the natural variation of metabolic traits. These include the advent of rapid metabolomic platforms allowing the quantification of hundreds to thousands of metabolites in as many different genotypes (Fiehn, 2001; Meyer et al., 2007; Fiehn et al., 2008). In combination with the ability to sequence and measure the transcriptome of all of these same lines, there is a massive influx of studies reporting on the identification of causal genes controlling the variation in metabolites in numerous species, from crop plants like maize (Zea mays), rice (Oryza sativa), and tomato (Solanum lycopersicum) to models like Arabidopsis (Arabidopsis thaliana) and ecological models like Boechera stricta and Nicotiana attenuata (Fu and Xue, 2010; Hartings et al., 2011; Li et al., 2011, 2015; Kausch et al., 2012; Prasad et al., 2012; Matsuba et al., 2013; Chang et al., 2015; Yan et al., 2015). These studies provide new insights into the mechanistic and evolutionary structures that influence how plant metabolism functions within a broader context. Other reviews have focused on the specific genes being cloned that control metabolite variation and the approaches utilized (Saito et al., 2008; Kliebenstein, 2009, 2012; Kusano et al., 2015; Luo, 2015; Omranian et al., 2015). As such, this review will focus on the broader insights being provided by these new studies into the genetic, mechanistic, and evolutionary processes shaping plant metabolism.
GENETIC ARCHITECTURE OF METABOLOMIC VARIATION
Quantitative genetic studies typically report several descriptors of the measured phenotypes and the candidate loci. These include the heritability of the phenotype, often as broad-sense heritability, a measure of genotypic reproducibility, and the r2 of the individual locus linked to a given phenotype, often called the effect size (Lynch and Walsh, 1998). These values are often linked to general conclusions, such as metabolites have high heritability or secondary metabolite loci have higher r2 than do primary metabolite loci. However, these values have significant caveats that need to be considered when interpreting the results, which may confuse any ability to make conclusions.
Heritability
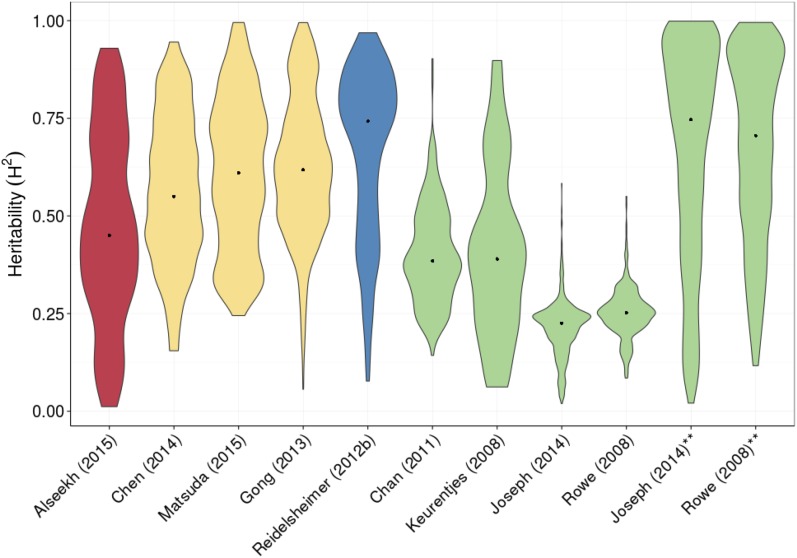
Most plant metabolomics studies that focus on natural variation report broad-sense heritability, which is a measure of the phenotype reproducibility within a set of genotypes (Lynch and Walsh, 1998). These studies have shown that the heritability of metabolomic traits displays a wide range in any given population (Fig. 1). In general, however, the heritability of maize metabolic traits appears to be higher than that for rice, with apparently lower Arabidopsis heritabilities (Keurentjes et al., 2006; Rowe et al., 2008; Chan et al., 2010a, 2010b; Yang et al., 2010; Riedelsheimer et al., 2012; Gong et al., 2013; Joseph et al., 2013a, 2013b, 2015; Li et al., 2013; Lipka et al., 2013; Chen et al., 2014; Alseekh et al., 2015; Zhang et al., 2015). These results are similar when using either structured populations like nested association mapping, recombinant inbred line (RIL), or introgression line populations or unstructured genome-wide association (GWA) populations within the same species. While it is tempting to argue that different domestication and selection processes may be influencing the difference in heritability across species, the estimation of heritability is not an absolute value and is influenced by numerous experimental, technical, and quantitative factors (Lynch and Walsh, 1998). For example, the rice analyses exclude residual error variance in the calculation of heritability, while two of the Arabidopsis studies include residual error variance. Recalculating the variance in these two Arabidopsis studies shows that they actually have a highly similar distribution of metabolite heritabilities (Fig. 1). In addition to the calculation choices, there are also biological and experimental differences among the experiments that complicate the comparison. Among three studies in rice, the growth conditions, age at harvest, and metabolite quantification methods all differed (Gong et al., 2013; Chen et al., 2014; Matsuda et al., 2015). Experimental designs range from randomized complete block design (Keurentjes et al., 2008; Chan et al., 2011; Chen et al., 2014; Alseekh et al., 2015) to α-lattice incomplete block design (Riedelsheimer et al., 2012). Thus, it is currently unclear if comparisons of heritability among these studies provide biological insight or simply reflect the technical and experimental differences. Future experiments wherein all technical and experimental differences are controlled would be required to assess if there is any biological difference in heritability among the species, potentially driven by domestication.
Figure 1.
Distribution of broad-sense heritability estimates of metabolic traits across species and methods. Red shows studies involving S. lycopersicum × S. pennellii, green shows studies using Arabidopsis, blue is for maize studies, and peach is for studies involving rice. All maize and Arabidopsis comparisons are intraspecific. Rice studies are intraspecific (Gong et al., 2013) and across subspecies (Chen et al., 2014; Matsuda et al., 2015). Heritability in the Rowe et al. (2008) and Joseph et al. (2014) data sets is shown in their original form, and heritability has been recalculated using solely environmental and genetic variance as in the other studies; the new results are indicated by asterisks.
A series of experiments did estimate the heritability of metabolic, transcriptomic, and physiological traits using the same experimental design, genotypes, and calculations to allow for direct comparison across mechanistic levels. This showed that the heritability of metabolic phenotypes is intermediate between the higher heritability of transcripts and the lower heritability of integrative traits like growth (Keurentjes et al., 2006, 2007, 2008; West et al., 2007; Rowe et al., 2008; Fu et al., 2009; Chan et al., 2010a, 2010b; Joseph et al., 2013a, 2013b, 2015). This could suggest that metabolic genetic variance is, in fact, intermediate between transcripts and integrative traits or that the integrative traits are more responsive to environmental variation leading to lower heritability. A related explanation that combines these options is that vastly more quantitative loci control integrative traits like growth of which the loci for specific metabolic traits are a subset, which would lead to diminishing heritability estimates. This model agrees with work on growth and metabolite traits that found that focusing solely on the growth QTL underestimated the link between metabolic and growth variation (Joseph et al., 2013a, 2013b). Additional studies are required to understand why mechanistically linked traits have varying heritability in the same population.
Effect Size
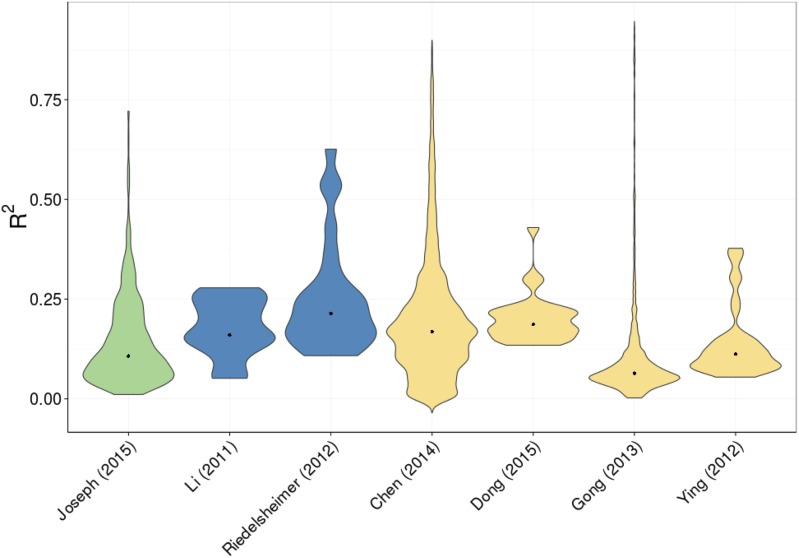
While heritability is a trait-level descriptor, most studies also provide the estimated effect size of individual loci. This is usually provided as r2 or the fraction of total phenotypic variance in a metabolite that is linked to a specific locus. This shows a wide spectrum of effect sizes, where metabolites can be under the control of few loci of large effect or numerous loci of small effect. Across Arabidopsis, rice, and maize, each locus explained on average 15% to 25% of metabolite variation (Fig. 2; Rowe et al., 2008; Ying et al., 2012; Gong et al., 2013; Chen et al., 2014; Dong et al., 2015; Matsuda et al., 2015). These averages, however, hide a wide range of individual locus variation. For example, individual rice loci have been found to explain 35% or more of an individual metabolite’s variation up to nearly 90% (Ying et al., 2012; Chen et al., 2014; Dong et al., 2015; Matsuda et al., 2015). In intraspecific studies, a single locus explained at most over 90% of metabolite variation in Arabidopsis (Rowe et al., 2008) and in rice (Gong et al., 2013) but at most only 62% in maize (Riedelsheimer et al., 2012; Fig. 2). In contrast, most loci found to control carbon and nitrogen metabolism were of small effect size in maize (Zhang et al., 2015). These studies show a wide range of effect sizes for loci linked to variation in metabolic traits.
Figure 2.
Estimates of r2 of loci controlling metabolic variation compared across species and methods. Green shows studies using Arabidopsis, blue is for maize studies, and peach is for studies involving rice. All maize and Arabidopsis comparisons are intraspecific. The rice comparisons are intraspecific (Ying et al., 2012; Gong et al., 2013; Dong et al., 2015) and across subspecies (Chen et al., 2014).
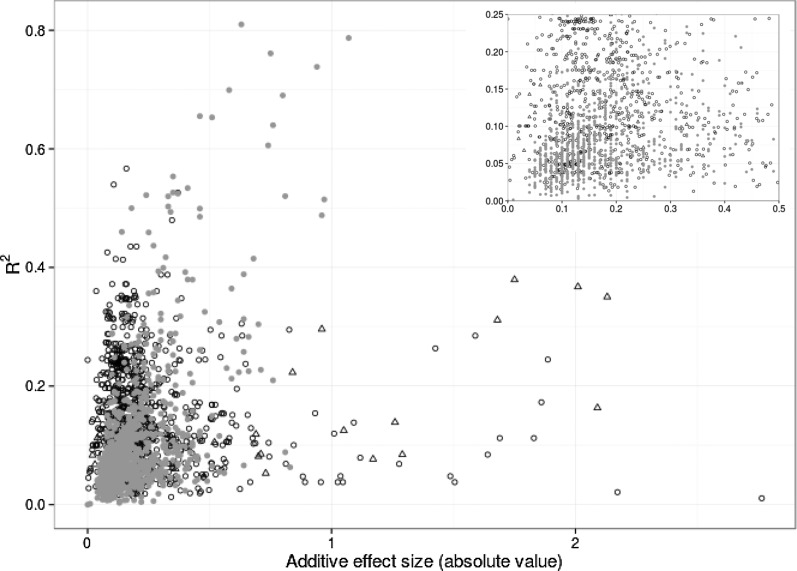
The equivalence of r2 effects per locus-to-locus effect, as commonly interpreted by mechanistic or molecular biology studies, is not straightforward. The r2 of a locus is the variance attributed to that locus divided by the total variance. As such, the calculation of per-locus r2 depends on factors that can affect the numerator (number of loci across which the variance is divided, missing loci, overestimates, etc.) as well the denominator (total variance, errors in total variance estimation, etc.). Thus, it is possible to have large-effect loci as defined by r2 that have additive effects of 10% or less in metabolite accumulation when comparing the two alleles (Fig. 3). Similarly, if the metabolite shows a large range of variation, it is possible to have small-effect loci per r2 that have additive effects of 100% difference in metabolite accumulation between the two alleles (Fig. 3). This leads to a value that is, at best, relative and of use only in that population and that can vary from differences in the number of loci identified (Beavis, 1994). The number of loci found per metabolite shows a wide range of variation across experiments and populations due to replication and statistical methods. In rice, studies have found a range of three to nine loci per metabolite, while in maize, this has ranged from five to 18 (Gong et al., 2013; Li et al., 2013; Chen et al., 2014; Dong et al., 2015; Matsuda et al., 2015; Zhang et al., 2015). While this difference will mathematically lead to the maize loci having smaller r2, because there are more loci per metabolite to share the variance, it is not clear if this is a reflection of the biological reality of the genetics controlling metabolite variation in the two species or if there are significant differences in the false-negative error rates leading to fewer detected loci in rice (Joseph et al., 2014). Thus, while r2 is a useful quantitative descriptor, it should be handled with care when making mechanistic arguments, as it does not directly scale to additive effects.
Figure 3.
Low correlation between r2 and additive effect size estimates for metabolic loci. Intraspecific analysis of nontargeted metabolites in Arabidopsis is indicated by black open circles (r2 = 0.006, P = 0.011), genetics of flavonoid abundance in a rice intraspecific comparison is indicated by gray circles (r2 = 0.38, P < 2.2e−16), and lipid genetics studied across rice subspecies (japonica and indica) is indicated by black open triangles (r2 = 0.43, P = 7.19e−06). The inset shows a closer look at loci with low effect size (<0.5) and low r2 (<0.25).
Clustering of Metabolite Loci
To summarize the identified loci, these studies frequently search for genomic hotspots that alter variation in more metabolites than expected by random chance. Every study identifies hotspots no matter what the species or population utilized, but the number and position of the hotspots can vary across tissues within a specific population, as found in rice (Gong et al., 2013). The position of hotspots can also differ depending upon the metabolite class being measured (Schauer et al., 2006, 2008; Riedelsheimer et al., 2013; Alseekh et al., 2015). The position and frequency of hotspots vary when using different populations from specific rice subspecies; in indica, hotspots were detected on chromosomes 2, 6, and 12, while japonica had hotspots on chromosomes 4 and 12 (Chen et al., 2014). Efforts to use these results to make mechanistic or evolutionary conclusions about the differences, such as arguing that the genetic architecture of metabolic variation is unique between the two rice subspecies, should proceed carefully for a couple of reasons. Different populations within Arabidopsis give different hotspots, which is likely solely associated with the different genes varying in that population regardless of whether the variation alters adaptation (Keurentjes et al., 2006; Rowe et al., 2008; Joseph et al., 2013a, 2013b). Using near-isogenic lines to assess the impact of a nonhotspot region in Arabidopsis showed that variation in this region affected more metabolites than any hotspot found using the associated RILs (Rowe et al., 2008). Thus, the difference between hotspot and nonhotspot regions could be driven by statistical rather than biological issues. As such, while hotspots may generate intriguing lines of research, any broad conclusions should wait for the cloning and characterization of the underlying causal gene(s).
Network Structure
Measuring metabolites across a set of natural genotypes allows investigations into metabolic network properties prior to conducting any locus-specific analysis. This can be done by correlating the variation in average metabolite abundance across the genotypes to look for genetic correlations. A key use of this correlation approach is to help address the significant difficulty presented by the fact that most metabolites measured have no known structure. Significant progress in identifying the structure of unknown metabolites is being made by querying for groups of metabolites that show a high genetic correlation under the assumption that they are chemically related. This approach has been used to identify sets of metabolites produced by the same biosynthetic pathway as found in rice, where using metabolite coaccumulation and structural similarity generated putative biosynthetic networks for amino acids and flavone-O-hexosides (Matsuda et al., 2012, 2015).
These approaches have been extended to other metabolites in both rice and maize to link previously unknown metabolites to each other and to new biosynthetic genes (Gong et al., 2013; Chen et al., 2014; Dong et al., 2014, 2015; Hu et al., 2014; Wen et al., 2014; Hashemi et al., 2015; Kusano et al., 2015; Luo, 2015). Similar approaches have been used to identify and expand pathways for unknown metabolite classes within nonmodel systems like N. attenuata, Barbarea vulgaris, and Capsicum spp. (Dalby-Brown et al., 2011; Wahyuni et al., 2014; Agerbirk et al., 2015; Li et al., 2015). While this approach is very powerful at identifying both new compound structures and new enzymes, it is solely reliant on the presence of genetic variation in the genes controlling the accumulation of these unknown compounds. As such, compounds with low or no genetic variation in their underlying genes will not be amenable to this approach. Fortunately, metabolites that play key roles in adaptation to biotic and abiotic stress are, by default, highly likely to be naturally variable due to both fluctuating and local variation in these selective pressures (Hancock et al., 2011; Züst et al., 2012; Brachi et al., 2015; Kerwin et al., 2015). As such, this approach may be uniquely powerful to identify and classify new metabolic pathways that play key roles in fitness in the wild.
In addition to classifying chemical pathways, it is also possible to use this approach to link metabolic traits with other yield or physiological traits (Lisec et al., 2008; Sulpice et al., 2009; Carreno-Quintero et al., 2012; Shen et al., 2013; Hill et al., 2015). Correlational analyses clustered metabolic traits into large modules, with a strong tendency toward positive correlations among metabolic and yield-associated traits in Solanum pennellii × S. lycopersicum introgression lines (Schauer et al., 2006). This grouping subdivided into three major modules: one containing whole-plant traits and metabolic intermediates, one containing all amino acids, and one including sugars and organic acids that was consistent across multiple years (Schauer et al., 2006, 2008). Similar networks were found using other tomato species (Do et al., 2010; Sauvage et al., 2014). This correlational structure was suggested to indicate network-level competition for photoassimilates that is consistent across tomato species (Schauer et al., 2006). The correlational approach has also been used to rapidly link metabolite variation to insect resistance in nonmodel systems (Kuzina et al., 2011). Thus, the use of genetic correlations has the ability to convey phenotypic insight about metabolite roles in planta.
It is also possible to use this approach to query for factors that alter metabolic network structure. For example, the correlational structure of metabolites in rice was significantly different when comparing the indica versus japonica subspecies, suggesting that the metabolic networks in these two subspecies are dissimilar, potentially in response to unique selective pressures during their separate domestication histories (Hu et al., 2014). In addition to network structure changing in response to long-term selective processes, the structure can change in response to short-term environmental perturbations. In Arabidopsis, the correlational structure shifted when the same population was harvested at different times of day (Chan et al., 2010a). In that case, there were more connections between metabolites at the dawn sampling than at the dusk sampling (Chan et al., 2010a). Thus, it is possible to learn general concepts about the structure of the metabolome using genetic correlations between metabolites even prior to mapping specific loci.
CONDITIONALITY IN METABOLIC VARIATION
The ability to make broad conclusions or identify causal genes using quantitative studies of metabolic variation is greatly influenced by the fact that metabolic abundances measured in these studies are highly conditioned on the environmental, developmental, and genetic variations present within the experiment. While most studies are conducted in a single tissue, a single environment, and without an assessment of dependency on the genetic background, insights are emerging into how these factors effect metabolic variation.
Tissue Specificity
Many metabolites display tissue or ontogenic specificity in their accumulation or synthesis (Moco et al., 2007; Kliebenstein, 2013; Moussaieff et al., 2013; Dong et al., 2015). This developmental specificity limits the feasibility of developing a simple complete picture of the genetic variation in a plant’s metabolome that can be extrapolated across tissues and developmental stages, as each tissue or stage may have completely different genetics. Within rice, only 31% of the metabolites detected in seeds and 15% of the metabolites detected in leaves were shared across both tissues (100 shared metabolites total; Gong et al., 2013). Similarly, there was a large effect of tissue specificity in tomato metabolite QTLs when comparing leaf and fruit loci (Schauer et al., 2006, 2008; Fernie and Schauer, 2009). This complicates the ability to infer when or where a gene may be working to influence a plant’s metabolism.
The tissue specificity of metabolism is partially attributable to differential transcriptional control for the underlying enzyme genes. Transcriptome analysis of tomato fruit tissues found strong spatial variation for the expression of central metabolism and secondary metabolism genes (Matas et al., 2011). Similar results were found in Arabidopsis, with tissue-specific expression of metabolic enzyme genes (Brady et al., 2007; Dinneny et al., 2008). Work with Arabidopsis glucosinolates has shown that ontogenic variation in glucosinolate hydrolysis products maps to the enzymatic loci that produce these products, suggesting that transcriptional variation in these genes controls the ontogenic variation (Wentzell et al., 2008; Wentzell and Kliebenstein, 2008). This has also been shown for the accumulation of tissue-specific metabolites in tomato and rice (Tsai et al., 2012; Gong et al., 2013). This suggests that cis-variation in the promoters of specific enzymatic loci that alters their expression may be one source for tissue specificity in metabolomic variation.
In addition to tissue-specific expression of the enzymes, transporters also play a key role in controlling when or where metabolites may accumulate, allowing the sites of synthesis and accumulation to be separated (Chen et al., 2012; Nour-Eldin et al., 2012; Andersen et al., 2013). A QTL modulating primary metabolism within Arabidopsis was cloned that displayed the potential for metabolite transport, confounding QTL interpretation with regard to tissue specificity (Li and Kliebenstein, 2014). In that study, an AT-HOOK was shown to be the basis of quantitative variation in the tricarboxylic acid cycle. Intriguingly, however, the causal gene was expressed in tissues that were distinct from where the metabolites were measured (Li and Kliebenstein, 2014). Because plant metabolism is highly interconnected across tissues by metabolite transport, it is possible for a gene with a highly localized effect to alter metabolite accumulation in a wider range of tissues. It remains to be seen how frequently cause and effect may be separated across tissues in plant metabolic variation.
Genotype × Environment Interaction
Environmental effects strongly influence metabolism, often interacting with genetic variation. This interaction limits the apparent repeatability of metabolomic variation but is a fundamental property of their biological function. For example, the induction of maize volatiles is highly specific to the attacking herbivore (McCormick et al., 2012). As such, the interaction of genotypic variation with environmental variation should instead be considered fundamental to understanding metabolic variation, and loci that vary across environments are likely important for specific environments and should be studied intensively.
Environmental effects have been studied intensively in tomato. In an interspecific cross between S. pennellii and S. lycopersicum (Eshed and Zamir, 1995), 889 metabolite QTLs were identified through gas chromatography-mass spectrometry of the fruit pericarp using three different years of harvest at a single farm site (Schauer et al., 2006). However, only 5% of the 889 metabolite QTLs were consistently identified across all three tomato harvests (Schauer et al., 2008). In volatile profiling of these same lines, significant season × line interactions were present for nine of the 23 metabolites studied in comparisons of spring and autumn harvests (Tieman et al., 2006). Furthermore, in a GWA analysis of diverse S. lycopersicum and Solanum pimpinellifolium accessions, only 47% of primary metabolic traits studied were stable across 2 years of field cultivation (Sauvage et al., 2014).
Similar environment × genotype interactions have been observed in other systems for metabolite accumulation. In rice, analysis of an indica × japonica cross indicated that only eight out of 29 QTLs associated with fatty acid abundance were consistently identified across two generations in two field trials (Ying et al., 2012). In maize GWA studies, only 17% of the metabolite-locus associations were consistently detectable across at least two of three concurrent field trials (Wen et al., 2014). A study of a high-oil maize RIL population over 2 years of field experiments showed that year and the year × genotype interactions contributed significantly to phenotypic variance (Yang et al., 2010). Identifying environment × genotype interactions does not require conducting field trials, as Arabidopsis GWA studies showed that most identified metabolite-locus associations varied across the time of day of the sampling within a single environment (Chan et al., 2010). Similarly, QTL mapping in the presence or absence of jasmonic acid and GWA studies in the presence or absence of different abiotic stresses identified different metabolic QTLs in Arabidopsis (Kliebenstein et al., 2002; Chan et al., 2011).
Thus, there is a strong interaction of environment with genetics in defining variation within the plant metabolome. One alternative is to analyze QTLs associated with relatively stable ratios between metabolites rather than single compounds (Morreel et al., 2006). Most often, the QTLs or associations that are consistent across environments are the loci followed up for study. However, given that plants evolved to adapt to constantly changing environments, it is likely that the loci that are only found in specific environments may be playing a key role in adaptation to those environments (Kerwin et al., 2015). As such, it is important to move beyond the stable loci and begin to clone and understand the mechanistic basis of environmentally conditional loci.
Epistasis: Genotype × Genotype and Genome × Genome Interactions
Another conditionality that influences the study of metabolite variation is the epistatic interaction of specific loci with the genetic background. In this article, we define epistasis as the interaction of genetic variation at two or more loci that creates a nonadditive or unpredictable change in the trait being studied. Molecular studies often consider epistasis to be evidence for a mechanistic interaction between two genes, but this is not an absolute requirement, as epistasis can also occur between genes in different pathways or even within duplicated genes in a gene family (Segrè et al., 2005; Roguev et al., 2008). In a study of the rice metabolome, 241 metabolites (53% of the total examined) exhibited 3,351 significant pairwise interactions between loci (Chen et al., 2014), and in a study of 16 phenolamides, eight significant pairwise interactions were detected between loci (Dong et al., 2015). In multiple single-nucleotide polymorphism (SNP) models of the loblolly pine (Pinus taeda) metabolome, more SNP effects were identified as dominance effects than as additive effects (Eckert et al., 2012). Twenty-four pairs of epistatic QTLs were detected in the study of high-oil maize RILs, but they accounted for 16% or less of the variance in individual oil phenotypes (Yang et al., 2010). Similarly, there are extensive epistatic interactions found for metabolomic variation in Arabidopsis in both primary and secondary metabolism (Wentzell et al., 2007; Rowe et al., 2008; Joseph et al., 2013a, 2013b; Kerwin et al., 2015). In Arabidopsis, this epistasis is largely higher order, involving the interaction of three or more loci rather than simple interactions of two loci (Wentzell et al., 2007; Rowe et al., 2008; Joseph et al., 2013a, 2013b; Kerwin et al., 2015). Cloning of the underlying loci shows that the epistatic interactions include the interaction of transcription factors with each other and with variation in enzymatic genes (Wentzell et al., 2007; Sønderby et al., 2010). It remains to be seen if higher order epistasis can be detected in other species.
Most studies on natural variation in plants limit their analysis to genetic variation within the nuclear genome. However, there is extensive evidence for genetic variation in the organelles altering adaptive phenotypes and interacting with nuclear loci (Greiner and Bock, 2013). Recent populations have been developed that are allowing quantitative assessment of the role of genetic variation in the organelle in quantitative phenotypes (McKay et al., 2008; Lovell et al., 2015). Analyzing metabolomic variation in the Arabidopsis Kas × Tsu population showed that genomic variation within the organelle altered the accumulation of nearly all metabolites (Joseph et al., 2013a, 2013b, 2015). This analysis also showed that there are extensive epistatic interactions between genetic variation in the organellar and nuclear genomes (Joseph et al., 2013a, 2013b, 2015). It remains to be tested how extensive this may be across different plant species.
Not all studies identify evidence of epistasis. A study of 342 rice metabolites found little evidence for epistasis in a comparison of metabolite abundance between parent accessions and offspring (Matsuda et al., 2015). Similarly, no significant epistatic interactions were detected in a pairwise analysis of genes associated with oil accumulation traits in maize (Li et al., 2013). It remains to be determined what differs between studies that identify epistasis in metabolic variation and those that do not. The differences could be the germplasm being used, the statistical methodologies, the environment, or some blend of these factors. Further studies will be required to quantify the extent of epistasis in plant metabolomic variation.
CAUSAL LOCI CONTROLLING METABOLIC VARIATION
A major goal of all quantitative metabolomic studies is to clone the underlying genes to understand the mechanistic basis of this variation. Recent reports have described the protocols to clone these loci, such as comparing transcriptomic and metabolic variation (Saito et al., 2008; Chan et al., 2011; Kliebenstein, 2012, 2014; Atwell and Kliebenstein, 2013). These correlational/colocalization approaches are greatly speeding up progress in cloning the causal genes, including a broad array of new enzymes, transcription factors, and other genes. This ever-increasing list of cloned loci is vastly beyond any single article’s ability to summarize. As such, we will focus on instances in which the cloned genes are illuminating new and unexpected mechanistic aspects of plant biology.
Interplay of Physiological and Metabolic Variation
A complexity in understanding plant metabolism is defining when one trait ends and another begins. Recent studies are beginning to show that metabolic variation is highly responsive to physiological variation, such as in resource availability and partitioning. A set of 126 maize SNPs associated with major carbon and nitrogen metabolic traits was overrepresented with genes linked to C4 photosynthesis and the regulation of carbon sink-source relationships (Zhang et al., 2015). Whole-plant morphology and growth conditions are also large contributors to tomato metabolic variation relative to the genetics studied (Do et al., 2010). Harvest index, a measure of fruit yield relative to biomass, was identified as the pleiotropic hub of the S. pennellii × S. lycopersicum metabolomic network linked to 46% of the metabolites (Schauer et al., 2006). Similar results were found in other tomato crosses (Prudent et al., 2009; Do et al., 2010). Overall, these results indicate that the interaction between genetics and source-sink dynamics plays a major role in defining central metabolism. Thus, metabolomic variation may often identify key physiological regulators that are important control nodes for the metabolome with pleiotropic effects.
In support of this are recent studies that cloned major metabolic variation QTLs within Arabidopsis. In one of these studies, most metabolic variation was linked to variation in the circadian clock (Kerwin et al., 2011). A key locus controlling this variation is natural variation in the EARLY FLOWERING3 gene that has been linked to altering flowering time, shade avoidance, hypocotyl elongation, circadian clock oscillation, and metabolic variation (Coluccio et al., 2011; Jimenez-Gomez et al., 2011; Kerwin et al., 2011; Undurraga et al., 2012; Anwer et al., 2014; Nieto et al., 2015). In tomato, the transcription factor APETALA2a that controls physiological pathways by regulating hormone synthesis to control fruit ripening also has impacts on phenylpropanoid and carotenoid metabolite accumulation (Karlova et al., 2011). Also in tomato, the ethylene receptor Never-Ripe controls variation in ascorbate and carotenoids but also strongly influences fruit ripening and seed production (Alba et al., 2005). In rice, the transcription factor Rc regulates flavonoid biosynthesis through abscisic acid signaling, with pleiotropic effects on seed dormancy and seed weight (Gu et al., 2011). This raises the question of whether these loci are specific metabolic regulators or are better interpreted as general regulators of physiology whose effect can be measured using metabolic variation. However, it will require analyzing the metabolic consequences of variation in other key physiological genes to tweeze apart the direct and indirect consequences on metabolite accumulation (Fukushima et al., 2009; Li and Kliebenstein, 2014).
Interplay of Metabolic and Defense Variation
The previous results suggest a pleiotropic link wherein variation in physiological or growth regulators influences metabolic traits. The cloning of genes underlying metabolic QTLs is beginning to highlight instances where variation in metabolic genes leads to unexpected effects, particularly on plant defense regulation. A major-effect QTL for aphid susceptibility in maize due to decreased levels of a benzoxazinoid, 2-hydroxy-4,7-dimethoxy-1,4-benzoxazin-3-one glucoside (HDMBOA-Glc), accumulation was caused by natural variation in transposon inactivation of a methyltransferase enzyme (Meihls et al., 2013). Inactivation of this enzyme also altered callose deposition in response to aphid infestation, suggesting a regulatory link between benzoxazinoid accumulation and callose induction (Meihls et al., 2013). A link between defense metabolite accumulation and callose regulation has been found for other methoxylated indolic metabolites with natural variation in their enzyme-encoding genes (Clay et al., 2009; Pfalz et al., 2009).
Supporting the potential for metabolic variation to influence defense regulation was a reanalysis of natural variation in a 2-oxoacid-dependent dioxygenase (AOP2) that controls the production of alkenyl glucosinolates in Arabidopsis and Brassica spp. (Kliebenstein et al., 2001; Li and Quiros, 2003). The presence or absence of this gene was found to alter the periodicity of the circadian clock as well as flowering time in both the laboratory and the field (Kerwin et al., 2011, 2015). A transcriptomic survey of genotypes with or without this enzyme showed that lines with a functional AOP2 enzyme had altered expression of both the biosynthetic and regulatory genes in the jasmonate signaling cascade, leading to decreased jasmonate sensitivity (Burow et al., 2015). As such, detailed studies of the benzoxazinoid and glucosinolate causal genes is beginning to show that metabolic loci, even enzyme-encoding ones, can have unexpected effects on other pathways, indicating potential regulatory influences for these metabolites in planta.
Diversifying Selection and Major-Effect Polymorphisms at Large-Effect Loci
A common conclusion of cloning studies is that secondary metabolite loci are controlled by large-effect presence/absence polymorphisms at the causal loci. This observation has been supported by a number of cloning studies in Arabidopsis, Brassica spp., rice, and maize (de Quiros et al., 2000; Kliebenstein et al., 2001; Li and Quiros, 2003; Hansen et al., 2008; Pfalz et al., 2009; Leckie et al., 2012; Meihls et al., 2013). However, there are also primary metabolite loci with presence/absence polymorphisms. In maize, seed starch and carotenoid contents are both controlled by large-effect polymorphisms in the causal genes (Thévenot et al., 2005; Vignesh et al., 2012; Lipka et al., 2013; Owens et al., 2014). As such, it is not accurate to make a primary versus secondary metabolism split when discussing large-effect or presence/absence polymorphisms in causal loci. Instead, it is more likely the shape of selection on the trait that is critical. In this case, all of these examples are in traits that are under either diversifying or fluctuating selection. In the case of the glucosinolates for Arabidopsis and Brassica spp., the defense traits are responding to fluctuating herbivore populations that create diversifying or balancing selective pressures that likely maintain and possibly even drive the variation (Prasad et al., 2012; Züst et al., 2012; Brachi et al., 2015; Kerwin et al., 2015). In the case of maize, carotenoid color is driven by diversified cultural preferences for white or yellow corn, and starch content is driven by diversifying selection on field corn versus popcorn. Similar to the herbivore pressure in wild Arabidopsis, the diversifying selection applied by human breeders for extremely divergent morphs of maize likely lead to the selection of large-effect presence/absence causal polymorphisms (Springer et al., 2009; Hufford et al., 2012). Thus, before classifying the type of causal polymorphisms expected when working with a metabolic trait, it is probably more important to understand the selective pressure on the metabolite rather than the type of metabolite.
What Genes Alter Variation in Metabolism?
A common question in all natural variation studies, including those on metabolism, is what are the genes that typically cause the phenotypic variation? A reading of the current literature suggests that we have an answer to this question in that we have to assume that if a gene has genetic variation that causes phenotypic variation in a trait, there will be a measurable shift in metabolism. The current set of naturally variable genes validated to impact natural variation include representatives from nearly all types of genes, from enzymes to transcription factors (Thévenot et al., 2005; Vignesh et al., 2012; Angelovici et al., 2013; Lipka et al., 2013; Meihls et al., 2013; Owens et al., 2014). As shown above, these genes can directly influence the metabolite or have potential indirect influences on the metabolite accumulation. As such, it is probably more appropriate to move beyond the question of what type of genes cause metabolic variation in natural populations and on to the more critical question of how these genes influence metabolite variation. Are they direct regulators that have immediate molecular impacts on the pathway regulation or biosynthetic potential? Or, alternatively, do these genes have more distant (sometimes thought of as indirect pleiotropic effects) links to the metabolic pathway whose output is being measured, and if so, how distant are these effects? Association mapping and other systems biology studies of glucosinolates in Arabidopsis have shown that there are dozens to hundreds of genes that can alter accumulation to a level that likely alters fitness (Chan et al., 2011; Li et al., 2014; Brachi et al., 2015; Kerwin et al., 2015). Yet, it is highly unlikely that all of these genes directly interact with the pathway in a molecular context. This generates a system whereby a metabolite’s natural variation can be altered by potentially hundreds of candidate genes, and each gene can conversely alter an array of metabolites (Angelovici et al., 2013). Within this system, the question arises of how selection can identify the proper combination of alleles to optimize fitness at the metabolic level. This mechanistic question is a key topic for the field to begin querying to understand how the metabolic system functions within an individual or species.
STATISTICAL INFLUENCES AND CHOICES
There is a large body of literature and reviews about the technical, statistical, experimental design, and population choices that are required to accomplish a quantitative analysis of natural variation in metabolism (Lynch and Walsh, 1998; Fiehn et al., 2008; Myles et al., 2009; Atwell and Kliebenstein, 2013; Luo, 2015). Thus, we will not provide a detailed step-by-step approach, as this has been done better. Instead, we will work to convey that every step has different options and there is no best choice. Rather, each option/choice introduces a different bias, and it is critical to understand what that bias is to properly interpret the results. This may be best conveyed in the choices of mapping populations available to conduct metabolomics analysis.
A highly popular approach that is gaining momentum is the use of GWA populations that are collections of random wild genotypes (Atwell et al., 2010). This population has the benefit of containing a sampling of the allelic diversity in the species. However, these populations contain significant population structures and, with even hundreds of accessions, have limited capacity to find epistasis or the effect of rare alleles, thus generating an unrecognized false-negative error rate (Chan et al., 2010a, 2010b; Long et al., 2013; Brachi et al., 2015). In contrast, the classical RIL population derived from two parents has the flaw of only having two alleles per gene, but this limitation also provides this population the greatest power to identify epistasis and small-effect loci (Falconer and Mackay, 1996; Mackay, 2014). The nested association mapping population was devised to alleviate the issue of allelic diversity in the RIL design and the rare allele issue in the GWA design. To accomplish this, it combines a set of recursive RIL populations involving multiple parents into a single combined population (Buckler et al., 2009). The true strength of this population lies in its ability to identify moderate-effect additive loci. A similar approach is the multiparent advanced generation intercross (MAGIC) population design, wherein multiple parents are admixed to create a single population (Kover et al., 2009). This population design also works well for moderate-effect, moderate-frequency additive loci, but like a GWA design, it struggles with complex epistasis or small-effect loci (Falconer and Mackay, 1996; Mackay, 2014). Thus, there is not a single population design that is suitable for all studies; instead, the population must be carefully chosen to match the strengths and weaknesses to the goal of the study.
After choosing the population, the next choice is to determine the number of lines and associated bioreplicates. This is then followed by choosing the statistical approach to link genotype with phenotype. However, these choices are highly linked. The rapid explosion of new statistical approaches for quantitative genetics is not a reflection of the flaws in the original approaches but instead a reflection of the mere fact that most experiments do not have sufficient numbers of genotypes to even fractionally sample the complete genotype-to-phenotype matrix. For example, it was recently estimated that a single RIL population would need at least 1,000 to 1,200 independent lines before it could even be determined how many more lines were needed to identify all the possible QTLs (Joseph et al., 2014). This is in a situation when most available RIL populations are maximally 200 or so lines. As such, the newer approaches, like Bayesian, multitrait, or other approaches, are simply trying to maximize the information obtained from significantly underpowered populations. Thus, rather than focusing on the optimal statistical approach, it is better to focus on maximizing the number of lines and replicates used to generate the data input into the statistical algorithms. Maximizing the power of the data will go a longer way toward optimizing the output than any particular statistical algorithm.
CONCLUSION
Plant biology over the past decades has focused largely on the qualitative assessment of small collections of genotypes within limited environments to assess the mechanistic function of one or a few genes. The future, however, will require the quantitative analysis of systematic genotype collections within a range of environments and tissues to understand the functions of entire systems. Metabolomic analysis of natural variation is well positioned to enable these very types of experiments and begin to assess questions that, until now, have been largely overlooked or inaccessible. What is the level of cell or tissue autonomy in metabolism? How many different mechanisms coordinate genetic variation in the organellar and nuclear genomes? What types of selection alter natural genetic variation in the field? All of these questions require the rapid and cheap quantitative ability that metabolomics provides in addition to the large body of enzymatic knowledge about the system. In combination, metabolomics analysis of natural variation should be a key component of future plant studies working to understand how a plant species functions in the wild or the field.
Glossary
- QTL
quantitative trait locus
- RIL
recombinant inbred line
- GWA
genome-wide association
- SNP
single-nucleotide polymorphism
Footnotes
This work was supported by the National Science Foundation (grant nos. IOS–1339125 and MCB–1330337 to D.J.K.), by the U.S. Department of Agriculture National Institute of Food and Agriculture (Hatch grant no. CA–D–PLS–7033–H to D.J.K.), and by the Danish National Research Foundation (grant no. DNRF99 to D.J.K.).
References
- Agerbirk N, Olsen CE, Heimes C, Christensen S, Bak S, Hauser TP (2015) Multiple hydroxyphenethyl glucosinolate isomers and their tandem mass spectrometric distinction in a geographically structured polymorphism in the crucifer Barbarea vulgaris. Phytochemistry 115: 130–142 [DOI] [PubMed] [Google Scholar]
- Agrawal AA. (2011) New synthesis: trade-offs in chemical ecology. J Chem Ecol 37: 230–231 [DOI] [PubMed] [Google Scholar]
- Alba R, Payton P, Fei Z, McQuinn R, Debbie P, Martin GB, Tanksley SD, Giovannoni JJ (2005) Transcriptome and selected metabolite analyses reveal multiple points of ethylene control during tomato fruit development. Plant Cell 17: 2954–2965 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alseekh S, Tohge T, Wendenberg R, Scossa F, Omranian N, Li J, Kleessen S, Giavalisco P, Pleban T, Mueller-Roeber B, et al. (2015) Identification and mode of inheritance of quantitative trait loci for secondary metabolite abundance in tomato. Plant Cell 27: 485–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen TG, Nour-Eldin HH, Fuller VL, Olsen CE, Burow M, Halkier BA (2013) Integration of biosynthesis and long-distance transport establish organ-specific glucosinolate profiles in vegetative Arabidopsis. Plant Cell 25: 3133–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angelovici R, Lipka AE, Deason N, Gonzalez-Jorge S, Lin H, Cepela J, Buell R, Gore MA, Dellapenna D (2013) Genome-wide analysis of branched-chain amino acid levels in Arabidopsis seeds. Plant Cell 25: 4827–4843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anwer MU, Boikoglou E, Herrero E, Hallstein M, Davis AM, James GV, Nagy F, Davis SJ (2014) Natural variation reveals that intracellular distribution of ELF3 protein is associated with function in the circadian clock. eLife 3: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwell S, Huang YS, Vilhjálmsson BJ, Willems G, Horton M, Li Y, Meng D, Platt A, Tarone AM, Hu TT, et al. (2010) Genome-wide association study of 107 phenotypes in Arabidopsis thaliana inbred lines. Nature 465: 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atwell S, Kliebenstein DJ (2013) Conducting genome-wide association mapping of metabolites. In Weckwerth W, Kahl G, eds, The Handbook of Plant Metabolomics, Wiley-VCH Verlag, Weinheim, Germany, pp 255–271 [Google Scholar]
- Beavis WD. (1994) The power and deceit of QTL experiments: lessons from comparative QTL studies. In Proceedings of the Forty-Ninth Annual Corn & Sorghum Industry Research Conference. American Seed Trade Association, Washington, DC, pp 250–266 [Google Scholar]
- Brachi B, Meyer CG, Villoutreix R, Platt A, Morton TC, Roux F, Bergelson J (2015) Coselected genes determine adaptive variation in herbivore resistance throughout the native range of Arabidopsis thaliana. Proc Natl Acad Sci USA 112: 4032–4037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brady SM, Orlando DA, Lee JY, Wang JY, Koch J, Dinneny JR, Mace D, Ohler U, Benfey PN (2007) A high-resolution root spatiotemporal map reveals dominant expression patterns. Science 318: 801–806 [DOI] [PubMed] [Google Scholar]
- Buckler ES, Holland JB, Bradbury PJ, Acharya CB, Brown PJ, Browne C, Ersoz E, Flint-Garcia S, Garcia A, Glaubitz JC, et al. (2009) The genetic architecture of maize flowering time. Science 325: 714–718 [DOI] [PubMed] [Google Scholar]
- Burow M, Atwell S, Fancisco-Candeiro M, Kerwin RE, Halkier BA, Kliebenstein DJ (2015) The glucosinolate biosynthetic gene AOP2 mediates feedback regulation of jasmonic acid signaling independent of its known enzymatic function. Mol Plant 8: 1201–1212 [DOI] [PubMed] [Google Scholar]
- Carreno-Quintero N, Acharjee A, Maliepaard C, Bachem CW, Mumm R, Bouwmeester H, Visser RG, Keurentjes JJ (2012) Untargeted metabolic quantitative trait loci analyses reveal a relationship between primary metabolism and potato tuber quality. Plant Physiol 158: 1306–1318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EK, Rowe HC, Corwin JA, Joseph B, Kliebenstein DJ (2011) Combining genome-wide association mapping and transcriptional networks to identify novel genes controlling glucosinolates in Arabidopsis thaliana. PLoS Biol 9: e1001125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EK, Rowe HC, Hansen BG, Kliebenstein DJ (2010a) The complex genetic architecture of the metabolome. PLoS Genet 6: e1001198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan EKF, Rowe HC, Kliebenstein DJ (2010b) Understanding the evolution of defense metabolites in Arabidopsis thaliana using genome-wide association mapping. Genetics 185: 991–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang S, Berman J, Sheng Y, Wang Y, Capell T, Shi L, Ni X, Sandmann G, Christou P, Zhu C (2015) Cloning and functional characterization of the maize (Zea mays L.) carotenoid epsilon hydroxylase gene. PLoS One 10: e0128758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen LQ, Qu XQ, Hou BH, Sosso D, Osorio S, Fernie AR, Frommer WB (2012) Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 335: 207–211 [DOI] [PubMed] [Google Scholar]
- Chen W, Gao Y, Xie W, Gong L, Lu K, Wang W, Li Y, Liu X, Zhang H, Dong H, et al. (2014) Genome-wide association analyses provide genetic and biochemical insights into natural variation in rice metabolism. Nat Genet 46: 714–721 [DOI] [PubMed] [Google Scholar]
- Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM (2009) Glucosinolate metabolites required for an Arabidopsis innate immune response. Science 323: 95–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coluccio MP, Sanchez SE, Kasulin L, Yanovsky MJ, Botto JF (2011) Genetic mapping of natural variation in a shade avoidance response: ELF3 is the candidate gene for a QTL in hypocotyl growth regulation. J Exp Bot 62: 167–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook-Patton SC, McArt SH, Parachnowitsch AL, Thaler JS, Agrawal AA (2011) A direct comparison of the consequences of plant genotypic and species diversity on communities and ecosystem function. Ecology 92: 915–923 [DOI] [PubMed] [Google Scholar]
- Dalby-Brown L, Olsen CE, Nielsen JK, Agerbirk N (2011) Polymorphism for novel tetraglycosylated flavonols in an eco-model crucifer, Barbarea vulgaris. J Agric Food Chem 59: 6947–6956 [DOI] [PubMed] [Google Scholar]
- de Quiros HC, Magrath R, McCallum D, Kroymann J, Schnabelrauch D, Mitchell-Olds T, Mithen R (2000) α-Keto acid elongation and glucosinolate biosynthesis in Arabidopsis thaliana. Theor Appl Genet 101: 429–437 [Google Scholar]
- Dinneny JR, Long TA, Wang JY, Jung JW, Mace D, Pointer S, Barron C, Brady SM, Schiefelbein J, Benfey PN (2008) Cell identity mediates the response of Arabidopsis roots to abiotic stress. Science 320: 942–945 [DOI] [PubMed] [Google Scholar]
- Do PT, Prudent M, Sulpice R, Causse M, Fernie AR (2010) The influence of fruit load on the tomato pericarp metabolome in a Solanum chmielewskii introgression line population. Plant Physiol 154: 1128–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X, Chen W, Wang W, Zhang H, Liu X, Luo J (2014) Comprehensive profiling and natural variation of flavonoids in rice. J Integr Plant Biol 56: 876–886 [DOI] [PubMed] [Google Scholar]
- Dong X, Gao Y, Chen W, Wang W, Gong L, Liu X, Luo J (2015) Spatiotemporal distribution of phenolamides and the genetics of natural variation of hydroxycinnamoyl spermidine in rice. Mol Plant 8: 111–121 [DOI] [PubMed] [Google Scholar]
- Eckert AJ, Wegrzyn JL, Cumbie WP, Goldfarb B, Huber DA, Tolstikov V, Fiehn O, Neale DB (2012) Association genetics of the loblolly pine (Pinus taeda, Pinaceae) metabolome. New Phytol 193: 890–902 [DOI] [PubMed] [Google Scholar]
- Ehrlich PR, Raven PH (1964) Butterflies and plants: a study in coevolution. Evolution 18: 586–608 [Google Scholar]
- Eshed Y, Zamir D (1995) An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141: 1147–1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falconer DS, Mackay TFC (1996) Introduction to Quantitative Genetics. Longman, Harlow, UK [Google Scholar]
- Fernie AR, Schauer N (2009) Metabolomics-assisted breeding: a viable option for crop improvement? Trends Genet 25: 39–48 [DOI] [PubMed] [Google Scholar]
- Fiehn O. (2001) Combining genomics, metabolome analysis, and biochemical modelling to understand metabolic networks. Comp Funct Genomics 2: 155–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiehn O, Wohlgemuth G, Scholz M, Kind T, Lee Y, Lu Y, Moon S, Nikolau B (2008) Quality control for plant metabolomics: reporting MSI-compliant studies. Plant J 53: 691–704 [DOI] [PubMed] [Google Scholar]
- Fu FF, Xue HW (2010) Coexpression analysis identifies Rice Starch Regulator1, a rice AP2/EREBP family transcription factor, as a novel rice starch biosynthesis regulator. Plant Physiol 154: 927–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J, Keurentjes JJB, Bouwmeester H, America T, Verstappen FWA, Ward JL, Beale MH, de Vos RCH, Dijkstra M, Scheltema RA, et al. (2009) System-wide molecular evidence for phenotypic buffering in Arabidopsis. Nat Genet 41: 166–167 [DOI] [PubMed] [Google Scholar]
- Fukushima A, Kusano M, Nakamichi N, Kobayashi M, Hayashi N, Sakakibara H, Mizuno T, Saito K (2009) Impact of clock-associated Arabidopsis pseudo-response regulators in metabolic coordination. Proc Natl Acad Sci USA 106: 7251–7256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong L, Chen W, Gao Y, Liu X, Zhang H, Xu C, Yu S, Zhang Q, Luo J (2013) Genetic analysis of the metabolome exemplified using a rice population. Proc Natl Acad Sci USA 110: 20320–20325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greiner S, Bock R (2013) Tuning a ménage à trois: co-evolution and co-adaptation of nuclear and organellar genomes in plants. BioEssays 35: 354–365 [DOI] [PubMed] [Google Scholar]
- Gu XY, Foley ME, Horvath DP, Anderson JV, Feng J, Zhang L, Mowry CR, Ye H, Suttle JC, Kadowaki K, et al. (2011) Association between seed dormancy and pericarp color is controlled by a pleiotropic gene that regulates abscisic acid and flavonoid synthesis in weedy red rice. Genetics 189: 1515–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock AM, Brachi B, Faure N, Horton MW, Jarymowycz LB, Sperone FG, Toomajian C, Roux F, Bergelson J (2011) Adaptation to climate across the Arabidopsis thaliana genome. Science 334: 83–86 [DOI] [PubMed] [Google Scholar]
- Hansen BG, Kerwin RE, Ober JA, Lambrix VM, Mitchell-Olds T, Gershenzon J, Halkier BA, Kliebenstein DJ (2008) A novel 2-oxoacid-dependent dioxygenase involved in the formation of the goiterogenic 2-hydroxybut-3-enyl glucosinolate and generalist insect resistance in Arabidopsis. Plant Physiol 148: 2096–2108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartings H, Lauria M, Lazzaroni N, Pirona R, Motto M (2011) The Zea mays mutants opaque-2 and opaque-7 disclose extensive changes in endosperm metabolism as revealed by protein, amino acid, and transcriptome-wide analyses. BMC Genomics 12: 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashemi FSG, Rafii MY, Ismail MR, Mohamed MTM, Rahim HA, Latif MA, Aslani F (2015) Application of an effective statistical technique for an accurate and powerful mining of quantitative trait loci for rice aroma trait. PLoS One 10: e0129069. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Hill CB, Taylor JD, Edwards J, Mather D, Langridge P, Bacic A, Roessner U (2015) Detection of QTL for metabolic and agronomic traits in wheat with adjustments for variation at genetic loci that affect plant phenology. Plant Sci 233: 143–154 [DOI] [PubMed] [Google Scholar]
- Hu C, Shi J, Quan S, Cui B, Kleessen S, Nikoloski Z, Tohge T, Alexander D, Guo L, Lin H, et al. (2014) Metabolic variation between japonica and indica rice cultivars as revealed by non-targeted metabolomics. Sci Rep 4: 5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hufford MB, Xu X, van Heerwaarden J, Pyhäjärvi T, Chia JM, Cartwright RA, Elshire RJ, Glaubitz JC, Guill KE, Kaeppler SM, et al. (2012) Comparative population genomics of maize domestication and improvement. Nat Genet 44: 808–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jimenez-Gomez JM, Corwin JA, Joseph B, Maloof JN, Kliebenstein DJ (2011) Genomic analysis of QTLs and genes altering natural variation in stochastic noise. PLoS Genet 7: e1002295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph B, Atwell S, Corwin JA, Li B, Kliebenstein DJ (2014) Meta-analysis of metabolome QTLs in Arabidopsis: trying to estimate the network size controlling genetic variation of the metabolome. Front Plant Sci 5: 461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph B, Corwin JA, Kliebenstein DJ (2015) Genetic variation in the nuclear and organellar genomes modulates stochastic variation in the metabolome, growth, and defense. PLoS Genet 11: e1004779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph B, Corwin JA, Li B, Atwell S, Kliebenstein DJ (2013a) Cytoplasmic genetic variation and extensive cytonuclear interactions influence natural variation in the metabolome. eLife 2: e00776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph B, Corwin JA, Züst T, Li B, Iravani M, Schaepman-Strub G, Turnbull LA, Kliebenstein DJ (2013b) Hierarchical nuclear and cytoplasmic genetic architectures for plant growth and defense within Arabidopsis. Plant Cell 25: 1929–1945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karban R, Baldwin IT (1997) Induced Responses to Herbivory. University of Chicago Press, Chicago [Google Scholar]
- Karlova R, Rosin FM, Busscher-Lange J, Parapunova V, Do PT, Fernie AR, Fraser PD, Baxter C, Angenent GC, de Maagd RA (2011) Transcriptome and metabolite profiling show that APETALA2a is a major regulator of tomato fruit ripening. Plant Cell 23: 923–941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kausch KD, Sobolev AP, Goyal RK, Fatima T, Laila-Beevi R, Saftner RA, Handa AK, Mattoo AK (2012) Methyl jasmonate deficiency alters cellular metabolome, including the aminome of tomato (Solanum lycopersicum L.) fruit. Amino Acids 42: 843–856 [DOI] [PubMed] [Google Scholar]
- Kerwin R, Feusier J, Corwin J, Rubin M, Lin C, Muok A, Larson B, Li B, Joseph B, Francisco M, et al. (2015) Natural genetic variation in Arabidopsis thaliana defense metabolism genes modulates field fitness. eLife 4: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerwin RE, Jiménez-Gomez JM, Fulop D, Harmer SL, Maloof JN, Kliebenstein DJ (2011) Network quantitative trait loci mapping of circadian clock outputs identifies metabolic pathway-to-clock linkages in Arabidopsis. Plant Cell 23: 471–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keurentjes JJB, Fu J, de Vos CHR, Lommen A, Hall RD, Bino RJ, van der Plas LHW, Jansen RC, Vreugdenhil D, Koornneef M (2006) The genetics of plant metabolism. Nat Genet 38: 842–849 [DOI] [PubMed] [Google Scholar]
- Keurentjes JJB, Fu J, Terpstra IR, Garcia JM, van den Ackerveken G, Snoek LB, Peeters AJM, Vreugdenhil D, Koornneef M, Jansen RC (2007) Regulatory network construction in Arabidopsis by using genome-wide gene expression quantitative trait loci. Proc Natl Acad Sci USA 104: 1708–1713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keurentjes JJB, Sulpice R, Gibon Y, Steinhauser MC, Fu J, Koornneef M, Stitt M, Vreugdenhil D (2008) Integrative analyses of genetic variation in enzyme activities of primary carbohydrate metabolism reveal distinct modes of regulation in Arabidopsis thaliana. Genome Biol 9: R129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein D. (2009) Advancing genetic theory and application by metabolic quantitative trait loci analysis. Plant Cell 21: 1637–1646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein D, Lambrix V, Reichelt M, Gershenzon J, Mitchell-Olds T (2001) Gene duplication and the diversification of secondary metabolism: tandem 2-oxoglutarate–dependent dioxygenases control glucosinolate biosynthesis in Arabidopsis. Plant Cell 13: 681–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ. (2012) Plant defense compounds: systems approaches to metabolic analysis. Annu Rev Phytopathol 50: 155–173 [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ. (2013) Making new molecules: evolution of structures for novel metabolites in plants. Curr Opin Plant Biol 16: 112–117 [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ. (2014) Synthetic biology of metabolism: using natural variation to reverse engineer systems. Curr Opin Plant Biol 19: 20–26 [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, Figuth A, Mitchell-Olds T (2002) Genetic architecture of plastic methyl jasmonate responses in Arabidopsis thaliana. Genetics 161: 1685–1696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kover PX, Valdar W, Trakalo J, Scarcelli N, Ehrenreich IM, Purugganan MD, Durrant C, Mott R (2009) A multiparent advanced generation inter-cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet 5: e1000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusano M, Yang Z, Okazaki Y, Nakabayashi R, Fukushima A, Saito K (2015) Using metabolomic approaches to explore chemical diversity in rice. Mol Plant 8: 58–67 [DOI] [PubMed] [Google Scholar]
- Kuzina V, Nielsen JK, Augustin JM, Torp AM, Bak S, Andersen SB (2011) Barbarea vulgaris linkage map and quantitative trait loci for saponins, glucosinolates, hairiness and resistance to the herbivore Phyllotreta nemorum. Phytochemistry 72: 188–198 [DOI] [PubMed] [Google Scholar]
- Leckie BM, De Jong DM, Mutschler MA (2012) Quantitative trait loci increasing acylsugars in tomato breeding lines and their impacts on silverleaf whiteflies. Mol Breed 30: 1621–1634 [Google Scholar]
- Li B, Gaudinier A, Tang M, Taylor-Teeples M, Nham NT, Ghaffari C, Benson DS, Steinmann M, Gray JA, Brady SM, et al. (2014) Promoter-based integration in plant defense regulation. Plant Physiol 166: 1803–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Kliebenstein DJ (2014) The AT-hook motif-encoding gene METABOLIC NETWORK MODULATOR 1 underlies natural variation in Arabidopsis primary metabolism. Front Plant Sci 5: 415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li D, Baldwin IT, Gaquerel E (2015) Navigating natural variation in herbivory-induced secondary metabolism in coyote tobacco populations using MS/MS structural analysis. Proc Natl Acad Sci USA 112: 4147–4155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Quiros CF (2003) In planta side-chain glucosinolate modification in Arabidopsis by introduction of dioxygenase Brassica homolog BoGSL-ALK. Theor Appl Genet 106: 1116–1121 [DOI] [PubMed] [Google Scholar]
- Li H, Peng Z, Yang X, Wang W, Fu J, Wang J, Han Y, Chai Y, Guo T, Yang N, et al. (2013) Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat Genet 45: 43–50 [DOI] [PubMed] [Google Scholar]
- Li L, Li H, Li Q, Yang X, Zheng D, Warburton M, Chai Y, Zhang P, Guo Y, Yan J, et al. (2011) An 11-bp insertion in Zea mays fatb reduces the palmitic acid content of fatty acids in maize grain. PLoS One 6: e24699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka AE, Gore MA, Magallanes-Lundback M, Mesberg A, Lin HN, Tiede T, Chen C, Buell CR, Buckler ES, Rocheford T, et al. (2013) Genome-wide association study and pathway-level analysis of tocochromanol levels in maize grain. G3 Genes Genomes Genetics 3: 1287–1299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec J, Meyer RC, Steinfath M, Redestig H, Becher M, Witucka-Wall H, Fiehn O, Törjék O, Selbig J, Altmann T, et al. (2008) Identification of metabolic and biomass QTL in Arabidopsis thaliana in a parallel analysis of RIL and IL populations. Plant J 53: 960–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long Q, Rabanal FA, Meng D, Huber CD, Farlow A, Platzer A, Zhang Q, Vilhjálmsson BJ, Korte A, Nizhynska V, et al. (2013) Massive genomic variation and strong selection in Arabidopsis thaliana lines from Sweden. Nat Genet 45: 884–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell JT, Mullen JL, Lowry DB, Awole K, Richards JH, Sen S, Verslues PE, Juenger TE, McKay JK (2015) Exploiting differential gene expression and epistasis to discover candidate genes for drought-associated QTLs in Arabidopsis thaliana. Plant Cell 27: 969–983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J. (2015) Metabolite-based genome-wide association studies in plants. Curr Opin Plant Biol 24: 31–38 [DOI] [PubMed] [Google Scholar]
- Lynch M, Walsh B (1998) Genetics and Analysis of Quantitative Traits. Sinauer Associates, Sunderland, MA [Google Scholar]
- Mackay TFC. (2014) Epistasis and quantitative traits: using model organisms to study gene-gene interactions. Nat Rev Genet 15: 22–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matas AJ, Yeats TH, Buda GJ, Zheng Y, Chatterjee S, Tohge T, Ponnala L, Adato A, Aharoni A, Stark R, et al. (2011) Tissue- and cell-type specific transcriptome profiling of expanding tomato fruit provides insights into metabolic and regulatory specialization and cuticle formation. Plant Cell 23: 3893–3910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuba Y, Nguyen TT, Wiegert K, Falara V, Gonzales-Vigil E, Leong B, Schäfer P, Kudrna D, Wing RA, Bolger AM, et al. (2013) Evolution of a complex locus for terpene biosynthesis in Solanum. Plant Cell 25: 2022–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda F, Nakabayashi R, Yang Z, Okazaki Y, Yonemaru J, Ebana K, Yano M, Saito K (2015) Metabolome-genome-wide association study dissects genetic architecture for generating natural variation in rice secondary metabolism. Plant J 81: 13–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda F, Okazaki Y, Oikawa A, Kusano M, Nakabayashi R, Kikuchi J, Yonemaru J, Ebana K, Yano M, Saito K (2012) Dissection of genotype-phenotype associations in rice grains using metabolome quantitative trait loci analysis. Plant J 70: 624–636 [DOI] [PubMed] [Google Scholar]
- McCormick AC, Unsicker SB, Gershenzon J (2012) The specificity of herbivore-induced plant volatiles in attracting herbivore enemies. Trends Plant Sci 17: 303–310 [DOI] [PubMed] [Google Scholar]
- McKay JK, Richards JH, Nemali KS, Sen S, Mitchell-Olds T, Boles S, Stahl EA, Wayne T, Juenger TE (2008) Genetics of drought adaptation in Arabidopsis thaliana. II. QTL analysis of a new mapping population, KAS-1 x TSU-1. Evolution 62: 3014–3026 [DOI] [PubMed] [Google Scholar]
- Meihls LN, Handrick V, Glauser G, Barbier H, Kaur H, Haribal MM, Lipka AE, Gershenzon J, Buckler ES, Erb M, et al. (2013) Natural variation in maize aphid resistance is associated with 2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one glucoside methyltransferase activity. Plant Cell 25: 2341–2355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RC, Steinfath M, Lisec J, Becher M, Witucka-Wall H, Törjék O, Fiehn O, Eckardt A, Willmitzer L, Selbig J, et al. (2007) The metabolic signature related to high plant growth rate in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 4759–4764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moco S, Capanoglu E, Tikunov Y, Bino RJ, Boyacioglu D, Hall RD, Vervoort J, De Vos RC (2007) Tissue specialization at the metabolite level is perceived during the development of tomato fruit. J Exp Bot 58: 4131–4146 [DOI] [PubMed] [Google Scholar]
- Morreel K, Goeminne G, Storme V, Sterck L, Ralph J, Coppieters W, Breyne P, Steenackers M, Georges M, Messens E, et al. (2006) Genetical metabolomics of flavonoid biosynthesis in Populus: a case study. Plant J 47: 224–237 [DOI] [PubMed] [Google Scholar]
- Moussaieff A, Rogachev I, Brodsky L, Malitsky S, Toal TW, Belcher H, Yativ M, Brady SM, Benfey PN, Aharoni A (2013) High-resolution metabolic mapping of cell types in plant roots. Proc Natl Acad Sci USA 110: E1232–E1241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myles S, Peiffer J, Brown PJ, Ersoz ES, Zhang Z, Costich DE, Buckler ES (2009) Association mapping: critical considerations shift from genotyping to experimental design. Plant Cell 21: 2194–2202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto C, López-Salmerón V, Davière JM, Prat S (2015) ELF3-PIF4 interaction regulates plant growth independently of the evening complex. Curr Biol 25: 187–193 [DOI] [PubMed] [Google Scholar]
- Nour-Eldin HH, Andersen TG, Burow M, Madsen SR, Jørgensen ME, Olsen CE, Dreyer I, Hedrich R, Geiger D, Halkier BA (2012) NRT/PTR transporters are essential for translocation of glucosinolate defence compounds to seeds. Nature 488: 531–534 [DOI] [PubMed] [Google Scholar]
- Omranian N, Kleessen S, Tohge T, Klie S, Basler G, Mueller-Roeber B, Fernie AR, Nikoloski Z (2015) Differential metabolic and coexpression networks of plant metabolism. Trends Plant Sci 20: 266–268 [DOI] [PubMed] [Google Scholar]
- Owens BF, Lipka AE, Magallanes-Lundback M, Tiede T, Diepenbrock CH, Kandianis CB, Kim E, Cepela J, Mateos-Hernandez M, Buell CR, et al. (2014) A foundation for provitamin A biofortification of maize: genome-wide association and genomic prediction models of carotenoid levels. Genetics 198: 1699–1716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfalz M, Vogel H, Kroymann J (2009) The gene controlling the Indole Glucosinolate Modifier1 quantitative trait locus alters indole glucosinolate structures and aphid resistance in Arabidopsis. Plant Cell 21: 985–999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad KVSK, Song BH, Olson-Manning C, Anderson JT, Lee CR, Schranz ME, Windsor AJ, Clauss MJ, Manzaneda AJ, Naqvi I, et al. (2012) A gain-of-function polymorphism controlling complex traits and fitness in nature. Science 337: 1081–1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prudent M, Causse M, Génard M, Tripodi P, Grandillo S, Bertin N (2009) Genetic and physiological analysis of tomato fruit weight and composition: influence of carbon availability on QTL detection. J Exp Bot 60: 923–937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedelsheimer C, Brotman Y, Méret M, Melchinger AE, Willmitzer L (2013) The maize leaf lipidome shows multilevel genetic control and high predictive value for agronomic traits. Sci Rep 3: 2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedelsheimer C, Lisec J, Czedik-Eysenberg A, Sulpice R, Flis A, Grieder C, Altmann T, Stitt M, Willmitzer L, Melchinger AE (2012) Genome-wide association mapping of leaf metabolic profiles for dissecting complex traits in maize. Proc Natl Acad Sci USA 109: 8872–8877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roguev A, Bandyopadhyay S, Zofall M, Zhang K, Fischer T, Collins SR, Qu H, Shales M, Park HO, Hayles J, et al. (2008) Conservation and rewiring of functional modules revealed by an epistasis map in fission yeast. Science 322: 405–410 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HC, Hansen BG, Halkier BA, Kliebenstein DJ (2008) Biochemical networks and epistasis shape the Arabidopsis thaliana metabolome. Plant Cell 20: 1199–1216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Hirai MY, Yonekura-Sakakibara K (2008) Decoding genes with coexpression networks and metabolomics: ‘majority report by precogs.’ Trends Plant Sci 13: 36–43 [DOI] [PubMed] [Google Scholar]
- Sauvage C, Segura V, Bauchet G, Stevens R, Do PT, Nikoloski Z, Fernie AR, Causse M (2014) Genome-wide association in tomato reveals 44 candidate loci for fruit metabolic traits. Plant Physiol 165: 1120–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sax K. (1923) The association of size differences with seed-coat pattern and pigmentation in Phaseolus vulgaris. Genetics 8: 552–560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N, Semel Y, Balbo I, Steinfath M, Repsilber D, Selbig J, Pleban T, Zamir D, Fernie AR (2008) Mode of inheritance of primary metabolic traits in tomato. Plant Cell 20: 509–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schauer N, Semel Y, Roessner U, Gur A, Balbo I, Carrari F, Pleban T, Perez-Melis A, Bruedigam C, Kopka J, et al. (2006) Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat Biotechnol 24: 447–454 [DOI] [PubMed] [Google Scholar]
- Segrè D, Deluna A, Church GM, Kishony R (2005) Modular epistasis in yeast metabolism. Nat Genet 37: 77–83 [DOI] [PubMed] [Google Scholar]
- Shen M, Broeckling CD, Chu EY, Ziegler G, Baxter IR, Prenni JE, Hoekenga OA (2013) Leveraging non-targeted metabolite profiling via statistical genomics. PLoS One 8: e57667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sønderby IE, Burow M, Rowe HC, Kliebenstein DJ, Halkier BA (2010) A complex interplay of three R2R3 MYB transcription factors determines the profile of aliphatic glucosinolates in Arabidopsis. Plant Physiol 153: 348–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer NM, Ying K, Fu Y, Ji T, Yeh CT, Jia Y, Wu W, Richmond T, Kitzman J, Rosenbaum H, et al. (2009) Maize inbreds exhibit high levels of copy number variation (CNV) and presence/absence variation (PAV) in genome content. PLoS Genet 5: e1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss SY, Agrawal AA (1999) The ecology and evolution of plant tolerance to herbivory. Trends Ecol Evol 14: 179–185 [DOI] [PubMed] [Google Scholar]
- Sulpice R, Pyl ET, Ishihara H, Trenkamp S, Steinfath M, Witucka-Wall H, Gibon Y, Usadel B, Poree F, Piques MC, et al. (2009) Starch as a major integrator in the regulation of plant growth. Proc Natl Acad Sci USA 106: 10348–10353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thévenot C, Simond-Côte E, Reyss A, Manicacci D, Trouverie J, Le Guilloux M, Ginhoux V, Sidicina F, Prioul JL (2005) QTLs for enzyme activities and soluble carbohydrates involved in starch accumulation during grain filling in maize. J Exp Bot 56: 945–958 [DOI] [PubMed] [Google Scholar]
- Tieman DM, Zeigler M, Schmelz EA, Taylor MG, Bliss P, Kirst M, Klee HJ (2006) Identification of loci affecting flavour volatile emissions in tomato fruits. J Exp Bot 57: 887–896 [DOI] [PubMed] [Google Scholar]
- Tsai YC, Weir NR, Hill K, Zhang W, Kim HJ, Shiu SH, Schaller GE, Kieber JJ (2012) Characterization of genes involved in cytokinin signaling and metabolism from rice. Plant Physiol 158: 1666–1684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Undurraga SF, Press MO, Legendre M, Bujdoso N, Bale J, Wang H, Davis SJ, Verstrepen KJ, Queitsch C (2012) Background-dependent effects of polyglutamine variation in the Arabidopsis thaliana gene ELF3. Proc Natl Acad Sci USA 109: 19363–19367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vignesh M, Hossain F, Nepolean T, Saha S, Agrawal P, Guleria S, Prasanna B, Gupta H (2012) Genetic variability for kernel β-carotene and utilization of crtRB1 3′TE gene for biofortification in maize (Zea mays L.). Indian J Genet Plant Breed 72: 189–194 [Google Scholar]
- Wahyuni Y, Stahl-Hermes V, Ballester AR, de Vos RCH, Voorrips RE, Maharijaya A, Molthoff J, Zamora MV, Sudarmonowati E, Arisi AC, et al. (2014) Genetic mapping of semi-polar metabolites in pepper fruits (Capsicum sp.): towards unravelling the molecular regulation of flavonoid quantitative trait loci. Mol Breed 33: 503–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen W, Li D, Li X, Gao Y, Li W, Li H, Liu J, Liu H, Chen W, Luo J, et al. (2014) Metabolome-based genome-wide association study of maize kernel leads to novel biochemical insights. Nat Commun 5: 3438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentzell AM, Boeye I, Zhang Z, Kliebenstein DJ (2008) Genetic networks controlling structural outcome of glucosinolate activation across development. PLoS Genet 4: e1000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentzell AM, Kliebenstein DJ (2008) Genotype, age, tissue, and environment regulate the structural outcome of glucosinolate activation. Plant Physiol 147: 415–428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wentzell AM, Rowe HC, Hansen BG, Ticconi C, Halkier BA, Kliebenstein DJ (2007) Linking metabolic QTLs with network and cis-eQTLs controlling biosynthetic pathways. PLoS Genet 3: 1687–1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- West MAL, Kim K, Kliebenstein DJ, van Leeuwen H, Michelmore RW, Doerge RW, St Clair DA (2007) Global eQTL mapping reveals the complex genetic architecture of transcript-level variation in Arabidopsis. Genetics 175: 1441–1450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Aboshi T, Teraishi M, Strickler SR, Spindel JE, Tung CW, Takata R, Matsumoto F, Maesaka Y, McCouch SR, et al. (2015) The tyrosine aminomutase TAM1 is required for β-tyrosine biosynthesis in rice. Plant Cell 27: 1265–1278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Guo Y, Yan J, Zhang J, Song T, Rocheford T, Li JS (2010) Major and minor QTL and epistasis contribute to fatty acid compositions and oil concentration in high-oil maize. Theor Appl Genet 120: 665–678 [DOI] [PubMed] [Google Scholar]
- Ying JZ, Shan JX, Gao JP, Zhu MZ, Shi M, Lin HX (2012) Identification of quantitative trait loci for lipid metabolism in rice seeds. Mol Plant 5: 865–875 [DOI] [PubMed] [Google Scholar]
- Zhang N, Gibon Y, Jason GW, Nick KL, Li P, Dedow L, Chen C, So YS, Kremling K, Bradbury P (2015) Genome-wide association of carbon and nitrogen metabolism in the maize nested association mapping population. Plant Physiol 168: 575–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Züst T, Heichinger C, Grossniklaus U, Harrington R, Kliebenstein DJ, Turnbull LA (2012) Natural enemies drive geographic variation in plant defenses. Science 338: 116–119 [DOI] [PubMed] [Google Scholar]