Rice root curling, a response to a mechanical barrier that involves the plant hormone jasmonic acid, is modulated by an E3-ubiquitin ligase.
Abstract
Plant roots can sense and respond to a wide diversity of mechanical stimuli, including touch and gravity. However, little is known about the signal transduction pathways involved in mechanical stimuli responses in rice (Oryza sativa). This work shows that rice root responses to mechanical stimuli involve the E3-ubiquitin ligase rice HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE1 (OsHOS1), which mediates protein degradation through the proteasome complex. The morphological analysis of the roots in transgenic RNA interference::OsHOS1 and wild-type plants, exposed to a mechanical barrier, revealed that the OsHOS1 silencing plants keep a straight root in contrast to wild-type plants that exhibit root curling. Moreover, it was observed that the absence of root curling in response to touch can be reverted by jasmonic acid. The straight root phenotype of the RNA interference::OsHOS1 plants was correlated with a higher expression rice ROOT MEANDER CURLING (OsRMC), which encodes a receptor-like kinase characterized as a negative regulator of rice root curling mediated by jasmonic acid. Using the yeast two-hybrid system and bimolecular fluorescence complementation assays, we showed that OsHOS1 interacts with two ETHYLENE-RESPONSE FACTOR transcription factors, rice ETHYLENE-RESPONSIVE ELEMENT BINDING PROTEIN1 (OsEREBP1) and rice OsEREBP2, known to regulate OsRMC gene expression. In addition, we showed that OsHOS1 affects the stability of both transcription factors in a proteasome-dependent way, suggesting that this E3-ubiquitin ligase targets OsEREBP1 and OsEREBP2 for degradation. Our results highlight the function of the proteasome in rice response to mechanical stimuli and in the integration of these signals, through hormonal regulation, into plant growth and developmental programs.
During their life cycle, plants are subjected to many different stimuli from which they cannot escape. Therefore, they have evolved mechanisms to perceive environmental cues, such as gravity, temperature, light, water, and nutrients, and to adjust their growth accordingly. Plants are highly sensitive to mechanical stimuli, which include touch, wind, gravity, and herbivore attack (Chehab et al., 2009; Toyota and Gilroy, 2013). The response to mechanical stimuli can involve nastic movements that are rapid, temporary, and independent of the stimulus direction (Braam, 2005), such as the rapid folding of the leaves of Mimosa pudica in response to touch. Tropic movements, in contrast, are determined by the direction or placement of the stimulus (Braam, 2005). For example, some plants have thigmotropic tendrils in which the direction of coiling is related to the direction or point of the touch stimulus (Braam, 2005). When the mechanical stimulus is constant, plant response can result in changes in morphogenesis, growth, and/or development (Chehab et al., 2009; Kurusu et al., 2013). The sum of these modifications is termed thigmomorphogenesis, which is considered an adaptive response enhancing plant resistance to mechanical perturbations (Chehab et al., 2009).
Roots have the capacity to explore the soil and adapt to a wide diversity of mechanical stimuli (Massa et al., 2003; Falik et al., 2005). Touch and gravity sensing are mechanical signals that roots need to integrate in their growth and developmental program (Fasano et al., 2002; Massa and Gilroy, 2003a; Toyota and Gilroy, 2013). Several studies suggest that root response to a fixed obstacle involves a combination of thigmotropic and gravitropic reactions (Massa and Gilroy, 2003a; Monshausen et al., 2009). Upon touch stimulation, roots tend to bend and grow away from the obstacle, reducing the root’s ability to respond to gravity. This coordinated response to touch and gravity suggests that some perception and/or signaling mechanisms are shared by both stimuli (Massa and Gilroy, 2003a, 2003b). However, the signal transduction pathways related to mechanical stimuli responses remain largely unknown. Jaffe et al. (2002) proposed that mechanical stimulation of the cell wall can perturb the cell wall-plasma membrane-cytoskeleton network and, thus, transduce the extracellular stimulus into an intracellular signal (Fasano et al., 2002; Jaffe et al., 2002). Stretch-activated channels and mechanosensitive Ca2+-permeable channels, located in the plasma membrane, were suggested to be involved in these initial perception mechanisms (Monshausen et al., 2009; Kurusu et al., 2013). The rapid and transient increase in cytosolic Ca2+ observed upon mechanical perturbations further suggested that this ion functions as a second messenger to activate downstream transcriptional or developmental responses (Braam, 2005; Monshausen et al., 2009; Kurusu et al., 2013).
Phytohormones such as ethylene, GAs, and jasmonates have been implicated in mechanical stimuli responses (Braam, 2005; Chehab et al., 2009, 2012; Lange and Lange, 2015). Ethylene is a gaseous hormone that, when exogenously applied, can induce morphological and physiological changes similar to those occurring during thigmomorphogenesis (e.g. reduced elongation and radial swelling; Fasano et al., 2002; Chehab et al., 2009). However, only some features of touch response were shown to be regulated by this hormone, suggesting that ethylene is not the primary factor in the mechanical signal transduction pathways (Braam, 2005; Chehab et al., 2009). Nevertheless, the coordinated action of ethylene and other signaling molecules may promote the morphological responses associated with thigmomorphogenesis.
Jasmonates, such as methyl jasmonate (MeJA), 12-oxo-phytodienoic acid (12-OPDA), and jasmonic acid (JA), are lipid-derived metabolites known to be involved in the transduction of mechanical signals (Chehab et al., 2009). Increased levels of MeJA and the JA precursor 12-OPDA were reported in plants challenged by mechanical stimuli. In Bryonia dioica tendrils, MeJA levels rapidly increased upon touch stimuli, and this increase was correlated with the tendril coiling response (Weiler et al., 1993). JA signaling pathways have been extensively studied in recent years, especially in dicots such as Arabidopsis (Arabidopsis thaliana) and tomato (Solanum lycopersicum; Chico et al., 2008; Fonseca et al., 2009). In Arabidopsis, the core JA signaling module was shown to include the CORONATINE INSENSITIVE1 (COI1) protein, an F-box component of the SKIP-CULLIN-F-box (SCF)-type E3-ubiquitin ligase complex. In response to JA stimuli, the SCFCOI1 complex targets the JA response repressors JASMONATE-ZIM DOMAIN (JAZ) proteins to degradation by the ubiquitin/26S proteasome system (UPS), allowing the activation of JA-responsive genes by MYC2 (Chico et al., 2008; Fonseca et al., 2009). In rice (Oryza sativa), however, little is known regarding the JA-signaling pathway, although most of the key components of the SCF-COI1 pathway have homologs in this monocot plant (Yoshii et al., 2010). Nonetheless, JA-signaling elements different from the ones identified in Arabidopsis have been identified in rice (Riemann et al., 2008; Yoshii et al., 2010), which indicates molecular differences in the JA-signaling mechanism between monocots and dicots.
The UPS remodeling of the proteome has emerged as an important mechanism modulating abiotic and biotic stress responses (Dreher and Callis, 2007; Lee and Kim, 2011; Lyzenga and Stone, 2012). The regulation of key stress-responsive proteins by the UPS pathway underlies a rapid and precise transcriptional activation/inhibition and physiological changes, eventually leading to adaptation.
Ubiquitin is a small protein (8 kD) ubiquitous to all eukaryotic organisms. Ubiquitin covalently modifies target proteins as a single molecule at a single site (monoubiquitination), as a single molecule at multiple sites (multimonoubiquitination), or as a ubiquitin chain (polyubiquitination). The latter is the most common type of ubiquitination and often prompts the ubiquitinated target protein for degradation through the UPS machinery (Vierstra, 2009). Molecules of ubiquitin are transferred to the target protein in an ATP-dependent manner, requiring sequentially an E1-activase, an E2-conjugase, and an E3-ligase. Among these, the E3-ubiquitin ligases are the most abundant group conferring specificity to target proteins.
The rice ROOT MEANDER CURLING (OsRMC) gene encodes an apoplastic receptor-like kinase with a putative role in JA-signaling responses (Jiang et al., 2007). Knockdown lines showed enhanced JA-sensitive response and increased root curling, suggesting that OsRMC may be involved in the JA-mediated rice root curling response. OsRMC was also implicated in rice abiotic stress responses (Zhang et al., 2009; Serra et al., 2013). Enhanced tolerance to high salinity was described in knockdown lines for this gene (Zhang et al., 2009). In addition, two ETHYLENE-RESPONSE FACTOR (ERF) transcription factors (TFs) with a putative role in rice abiotic stress responses were shown to bind the promoter of OsRMC gene (Serra et al., 2013).
Root waving, skewing, and coiling/curling are touch-dependent thigmomorphogenic responses associated with growth on hard surfaces (Chen et al., 2009). This behavior is thought to facilitate root penetration in soil, promote interaction with microorganisms, and increase plant structural support (Chen et al., 2009). Here, we propose that the rice root response to mechanical stimuli is modulated by the proteasome. We show that a rice REALLY INTERESTING NEW GENE (RING)-type E3-ubiquitin ligase, HIGH EXPRESSION OF OSMOTICALLY RESPONSIVE GENE1 (OsHOS1), previously reported (Lourenço et al., 2013) as the ortholog of Arabidopsis HOS1 (Ishitani et al., 1998; Lee et al., 2001), targets the two ERF proteins involved in OsRMC transcriptional regulation (Serra et al., 2013) for degradation through the proteasome. Moreover, we demonstrate that the rice root curling phenotype depends on a mechanical barrier and is modulated by JA.
RESULTS
RNA Interference::OsHOS1 Plants Show a Straight Root Phenotype and an Up-Regulation of OsRMC Gene Expression
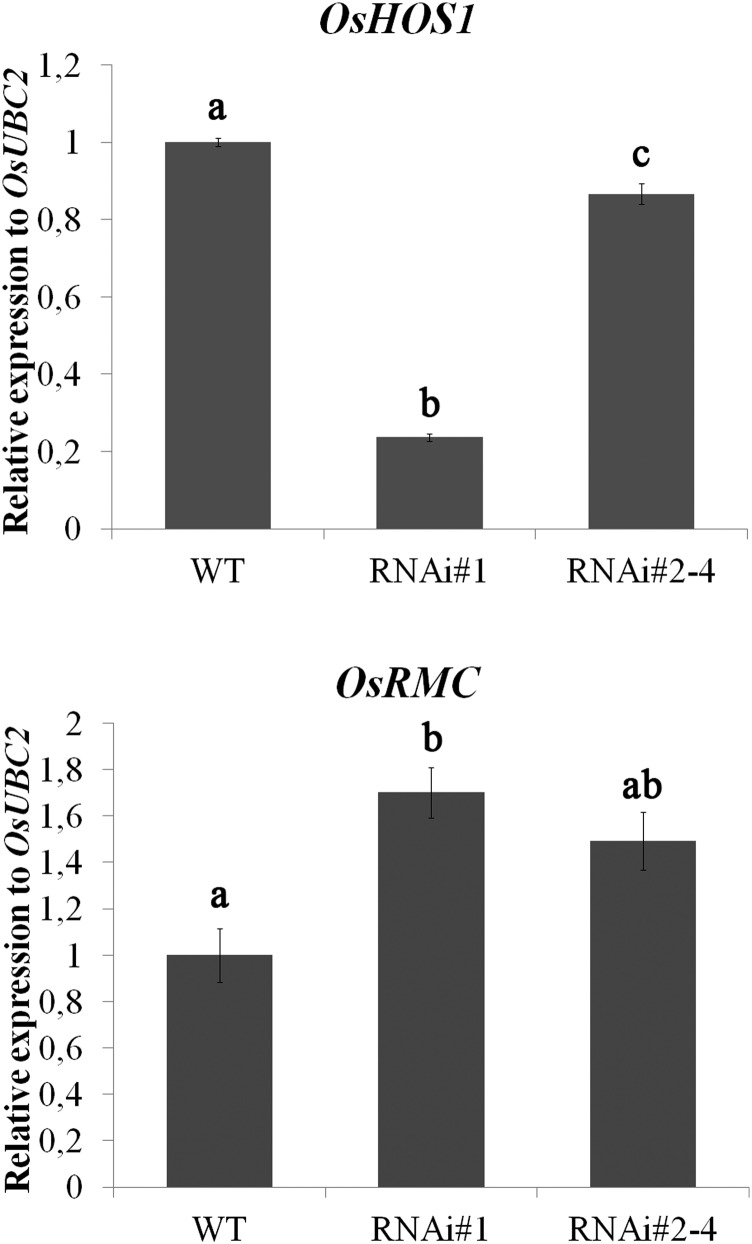
We have generated transgenic rice lines silencing the expression of OsHOS1 (RNA interference [RNAi]::OsHOS1; Lourenço et al., 2013). When rice seedlings were grown in test tubes for 10 d after germination (DAG) in liquid Yoshida’s nutritional medium (Yoshida et al., 1976), RNAi::OsHOS1 seedlings showed a straight root phenotype, whereas wild-type seedlings showed a root curling phenotype (Fig. 1). Given that OsRMC was reported to be involved in rice root development (Jiang et al., 2007), acting as a negative regulator of root curling, we hypothesized that the curled/straight root phenotype could be related to the altered expression of the OsRMC gene. Aiming to understand if this gene was indeed involved in the root phenotype observed in the RNAi::OsHOS1 roots, we performed reverse transcription (RT)-quantitative PCR (qPCR) analyses to assess OsRMC transcript levels in the RNAi::OsHOS1 and wild-type plants. Interestingly, we found an up-regulation of OsRMC gene expression in the transgenic lines as compared with wild-type plants (Fig. 2), indicating that the E3-ubiquitin ligase OsHOS1 may influence OsRMC gene expression. Furthermore, it seems that the level of OsHOS1 silencing in the transgenic lines is inversely correlated with the up-regulation of OsRMC expression (Fig. 2). To further confirm the hypothesis of OsHOS1 silencing being involved in the modulation of OsRMC expression, we used rice protoplasts to transiently express the same RNAi genetic construct (pANDA::OsHOS1). As it happened in the stable transformants, the down-regulation of OsHOS1 in protoplasts was also accompanied by an increase in OsRMC transcripts as compared with control protoplasts (Supplemental Fig. S1). Altogether, these results showed that the E3-ubiquitin ligase OsHOS1 is able to influence OsRMC gene expression and thus affect root curling phenotype.
Figure 1.
Root phenotype of the different rice plants analyzed. Rice seedlings (10 DAG) grown in liquid Yoshida’s medium in test tubes. RNAi::OsHOS1#1 (similar results for line RNAi::OsHOS1#2–4; Supplemental Fig. S2) plants show a straight root phenotype when compared with the wild type (WT; images representative of 15 plants). Arrows show root curl phenotype on wild-type plants. Scale bar = 1 cm.
Figure 2.
Gene expression analyses for OsHOS1 (JQ866627) and OsRMC (LOC_Os04g25650) were performed by RT-qPCR in wild-type (WT) and RNAi::OsHOS1 silencing lines (#1 and #2–4) under control conditions. The specific primers are described in Supplemental Table S2. The transcript level for Ubiquitin-Conjugating Enzyme E2 (OsUBC2, LOC_Os02g42314) was used to normalize the expression as an internal control for the RT-qPCR. The expression value of the wild-type sample was set to 1. Values represent means ± se (n ≥ 5 biological samples from at least three independent experiments). Different letters above the columns represent significant statistical difference between the different lines using Tukey’s analysis (P < 0.05).
Rice Root Curl Phenotype Is a Response to a Mechanical Barrier
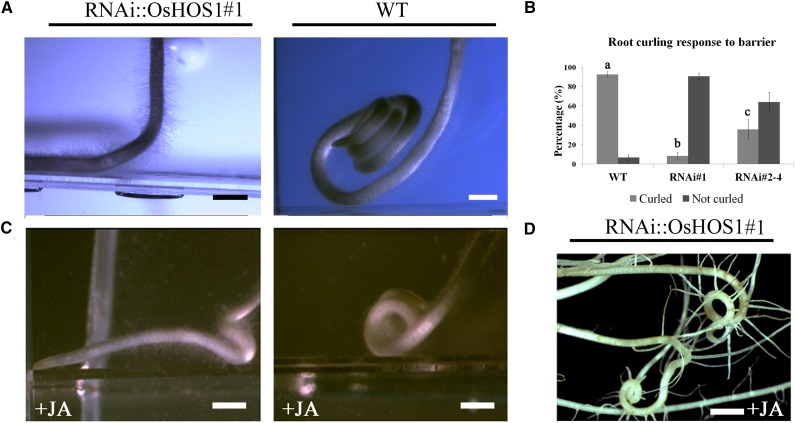
Although we could correlate the altered expression of the OsRMC with the root phenotype, we also aimed to understand the cause of the rice root curling. We hypothesized that the curled phenotype in wild-type seedlings could be due to the mechanical confinement inside the test tube to which the RNAi::OsHOS1 seedlings were, somehow, insensible. To test this hypothesis, we have grown rice seedlings (wild type and RNAi::OsHOS1#1) vertically on the surface of solidified Yoshida’s nutritional medium facing a glass barrier placed in front of the root tip. We observed that, in wild-type seedlings, the root started curling after touching the glass barrier, whereas the RNAi::OsHOS1#1 roots did not (Fig. 3A; Supplemental Table S1; Supplemental Videos S1 and S2). We further tested this phenotype by growing rice seedlings in different containers (test tubes, glass or plastic flasks of different diameters). We observed that, independent of the type of container used, the wild-type roots would curl, whereas the RNAi::OsHOS1 (lines #1 and #2–4) roots would show different curling responses dependent on the silencing level and OsRMC expression (Fig. 3B). These results confirmed the hypothesis that the curled phenotype of the wild-type roots was most likely triggered by the mechanical sensing of a barrier, and that the RNAi::OsHOS1 roots do not sense this obstacle.
Figure 3.
A, Root phenotype of rice seedlings from the wild type (WT) and RNAi::OsHOS1#1 (3 DAG) grown under control conditions on solid Yoshida’s medium and vertically oriented against a mechanical barrier. The bright field images were taken using an Olympus S2×12 stereo microscope coupled to a CCD camera QColor3 (Olympus; images are representative of at least eight plants of each line). Scale bar = 1 mm. B, Root curling response of 10-DAG seedlings of wild type and RNAi::OsHOS1 (#1 and #2–4) grown in Yoshida’s liquid medium under control conditions in test tubes. Values represent means ± se (n ≥ 4 independent assays using at least 15 seedlings per line per assay). Letters above the columns represent significant statistical difference between the different lines using Tukey’s analysis (P < 0.05). C, Root phenotype of rice seedlings (3 DAG) grown vertically against a mechanical barrier on solid Yoshida’s medium supplemented with JA (1 µm). Scale bar = 1 mm. D, Root phenotype of 10-DAG RNAi::OsHOS1#1 plants grown in test tubes for 1 week (2–10 DAG) in liquid Yoshida’s medium supplemented with 100 µm JA (images representative of eight plants). Scale bar = 4 mm.
RNAi::OsHOS1 Straight Root Phenotype Is Reverted by Exogenous Application of JA
Given that JA had already been shown to have a positive effect in the rice root curling phenotype (Jiang et al., 2007), we decided to test whether JA could induce root curling in the RNAi::OsHOS1 plants. To test this, wild-type and RNAi::OsHOS1#1 rice lines were grown vertically against a mechanical barrier, as before, in solid Yoshida’s medium supplemented with 1 µm JA. We found that JA intensified the touch-induced curling phenotype in wild-type roots, and that even the RNAi::OsHOS1#1 roots curled after hitting the obstacle, if grown with this hormone (Fig. 3C, compare with Fig. 3A for lack of curling RNAi::OsHOS1 roots without JA; Supplemental Table S1).
To investigate whether the reversion of the root phenotype could depend on the JA concentration, 2-DAG RNAi::OsHOS1 (#1 and #2–4) rice seedlings were exposed to different concentrations of JA (2 and 100 µm). After 1 week of incubation in liquid Yoshida’s medium in test tubes with the lowest concentration of JA, the RNAi::OsHOS1#2 to 4 exhibited a reversion of the straight root phenotype (Supplemental Fig. S2). However, using 2 µm JA, the RNAi::OsHOS1#1 did not curl as the wild type (data not shown). When only the highest concentration of JA (100 µm) was applied, the RNAi::OsHOS1#1 seedlings exhibited a full reversion of the straight root phenotype (Fig. 3D). With these experiments, we could confirm that the straight root phenotype of the RNAi::OsHOS1 plants could be fully reverted by increasing JA concentration. These results indicate that JA is associated with the rice root touch response through mechanisms involving OsHOS1 and OsRMC.
OsHOS1 Gene Silencing Affects JA Sensitivity But Not Its Biosynthesis
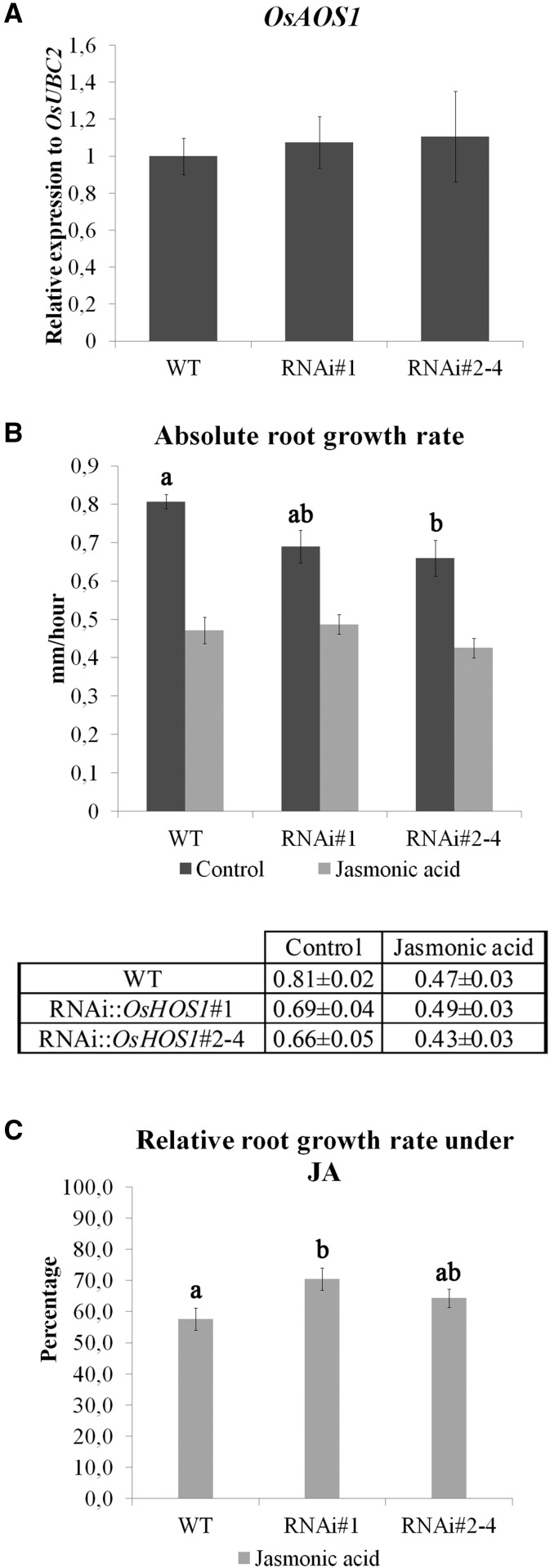
The results shown so far regarding the root curling phenotype in the RNAi::OsHOS1 suggest that these transgenic plants have either a lower JA abundance or a decreased sensitivity to the signaling pathway. To test these hypotheses, we analyzed the gene expression of the ALLENE OXIDE SYNTHASE1 (OsAOS1, LOC_Os03g55800), a key enzyme of the JA biosynthesis pathway, and used a classic assay for the JA effect (root growth inhibition) to test the effect on the JA-signaling pathway by comparing the relative root growth rate of the different lines. For the analysis of OsAOS1 expression, we used 10-DAG seedlings from the different lines grown in liquid Yoshida’s medium in test tubes. We did not observe significant variations in the expression of OsAOS1 among the different lines (Fig. 4A), indicating an absence of effect of the OsHOS1 silencing over the JA biosynthesis pathway. Regarding the sensitivity to JA, all rice lines showed a reduction in root growth rate when treated with JA, but the growth rate reduction in wild-type seedlings was higher than that observed in both RNAi::OsHOS1 transgenic lines (Fig. 4B). When treated with JA, the wild-type seedlings had a relative root growth rate of 58% (as compared with control roots; Fig. 4C). On the other hand, RNAi::OsHOS1#1 and #2-#4 retained 70% and 65% of their control root growth rate, respectively (Fig. 4C), thus showing a lower sensitivity to JA-induced root growth inhibition as compared with the wild type. Altogether, these results indicate that OsHOS1 is involved in rice root touch response phenotypes through mechanisms involving the JA-signaling pathway rather than regulating its biosynthesis.
Figure 4.
A, Gene expression analysis of OsAOS1 (LOC_Os03g55800) was performed by RT-qPCR in wild-type (WT) and RNAi::OsHOS1 silencing lines (#1 and #2–4) in control conditions. The specific primers are described in Supplemental Table S2. The transcript level for OsUBC2 (LOC_Os02g42314) was used to normalize the expression as an internal control for the RT-qPCR, and the expression value in the wild-type sample was set to 1. Values represent means ± se (n ≥ 5 biological samples from at least three independent assays). No significant statistical differences were observed between the groups using ANOVA one-way analysis (P < 0.05). B, Absolute root growth rate of wild-type and RNAi::OsHOS1 (#1 and #2–4) rice seedlings under control and JA treatment. Root growth observations started 24 h after the transfer of the seedlings to the treatment plate (time point 0). Root growth was analyzed every 24 h for 2 d (time points 1 and 2). Root growth measurements were performed using ImageJ software. The graphic shows the absolute root growth rate of wild-type and RNAi::OsHOS1 (#1 and #2–4) rice seedlings (mm h–1) under control conditions or treated with JA (2 µm). Values represent means ± se (n ≥ 10 seedlings per line per treatment from three independent assays). The different letters above the control bars represent significant statistical difference between the different groups using Tukey’s analysis (P < 0.05). No significant statistical differences were observed for the JA treatment of the three different lines. The table shows the absolute root growth rate values (mm h–1) ± se for the three lines analyzed. C, Relative root growth rate of the wild-type and RNAi::OsHOS1 (#1 and #2–4) seedlings shown in B. The bars represent the percentage of root growth rate of wild-type and RNAi::OsHOS1 (#1 and #2–4) seedlings under JA treatment relative to the mean growth rate of each line in control conditions (100%). Values represent means ± se (n ≥ 10 seedlings per line per treatment from three independent assays). The different letters above the bars represent significant statistical difference between the different groups using Tukey’s analysis (P < 0.05).
The Expression of Two Transcription Factors Binding to the OsRMC Promoter Is Identical in RNAi::OsHOS1 and Wild-Type Plants
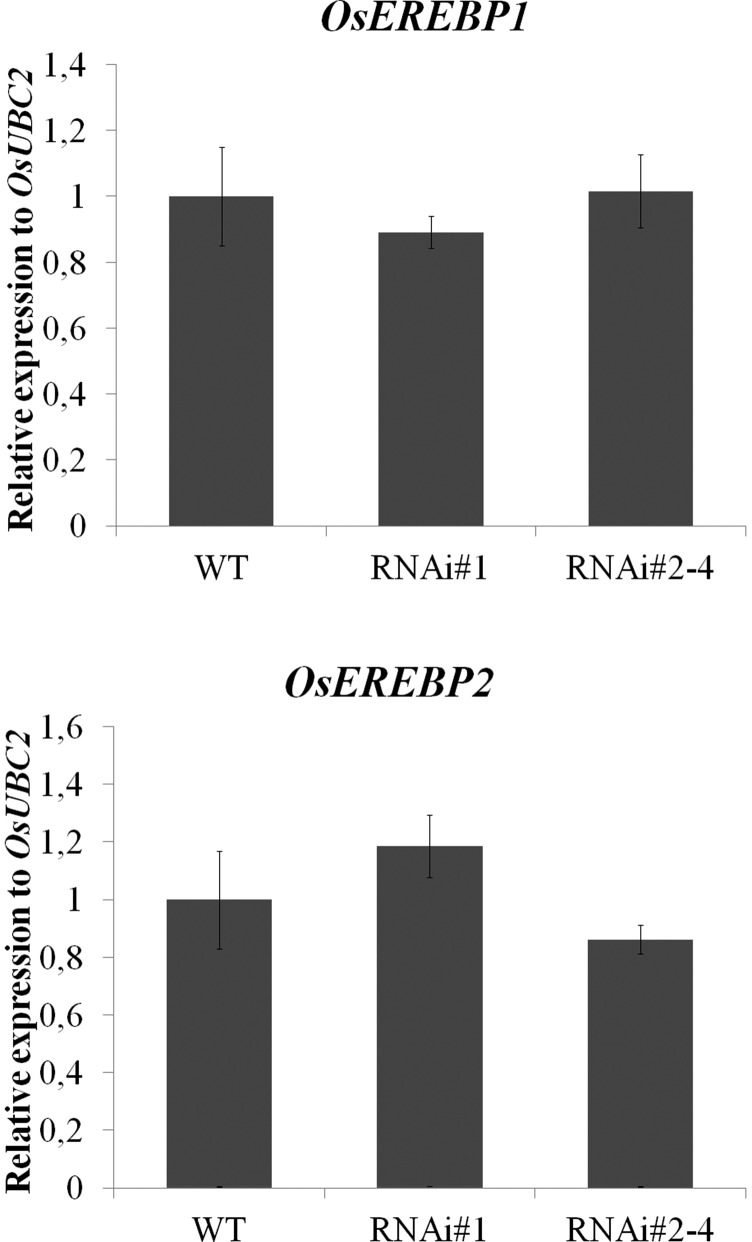
We have previously identified two TFs, OsEREBP1 and OsEREBP2, as binding to the OsRMC promoter region (Serra et al., 2013). Since the down-regulation of OsHOS1 in the transgenic line is up-regulating OsRMC gene expression, we hypothesized that altered OsEREBP1 and OsEREBP2 gene expression could contribute to the higher gene expression levels of OsRMC. However, expression analyses of OsEREBP1 and OsEREBP2 in wild-type and RNAi::OsHOS1 (#1 and #2–4) seedlings showed no significant difference between the different lines (Fig. 5). These results suggest that OsHOS1 may affect OsRMC gene expression through other TF regulators, or by modulating OsEREBP1 and OsEREBP2 protein levels.
Figure 5.
Gene expression analysis for OsEREBP1 (LOC_Os02g54160) and OsEREBP2 (LOC_Os01g64790) was performed by RT-qPCR in wild-type (WT) and RNAi::OsHOS1 silencing lines (#1 and #2–4) under control conditions. The specific primers are described in Supplemental Table S2. The transcript level for OsUBC2 (LOC_Os02g42314) was used as an internal control for the RT-qPCR. The expression value of the wild-type sample was set to 1. Values represent means ± se (n ≥ 5 biological samples from at least three independent assays). No significant statistical differences were observed within the groups using ANOVA one-way analysis (P < 0.05).
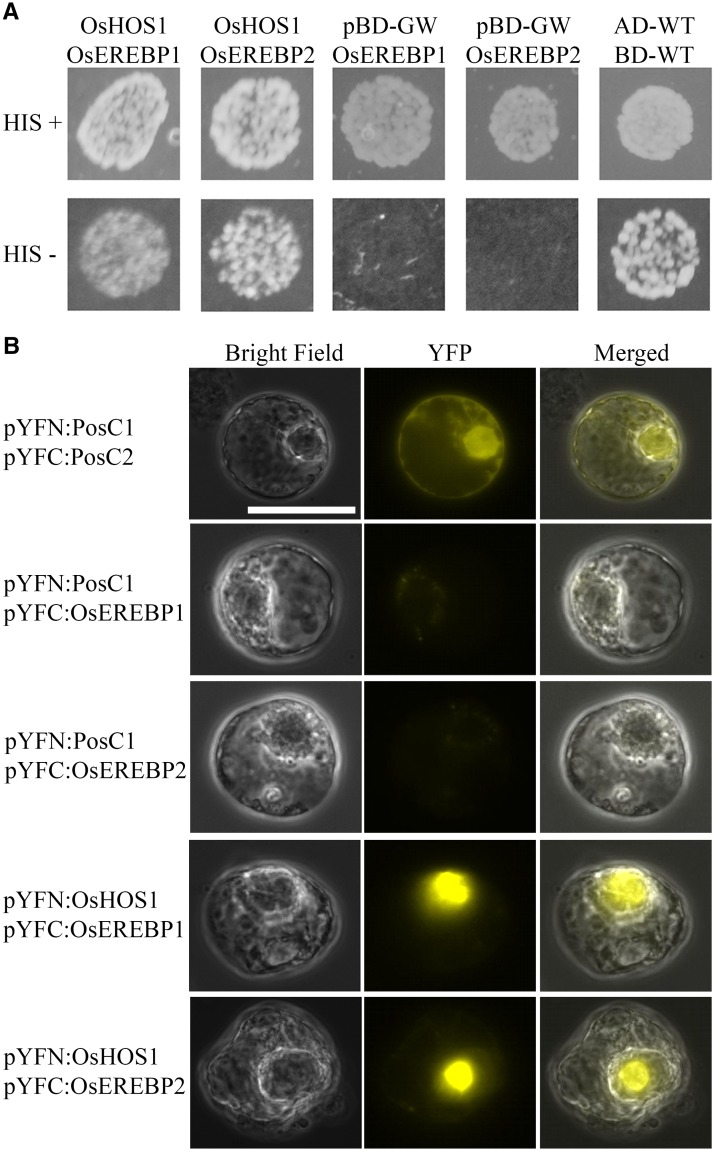
OsHOS1 Interacts with OsEREBP1 and OsEREBP2
We have previously demonstrated that OsHOS1 interacts with the rice TF INDUCER OF CBF EXPRESSION1 (OsICE1) by targeting this TF to proteasome-mediated degradation (Lourenço et al., 2013). To assess whether OsEREBP1 and OsEREBP2 could also be OsHOS1 targets, we first used a yeast two-hybrid assay. This analysis showed that, in yeast (Saccharomyces cerevisiae), OsHOS1 indeed interacts with both OsEREBP1 and OsEREBP2 (Fig. 6A). Using BiFC in Arabidopsis protoplasts, we could further assess in vivo interactions between these proteins and confirm that only the combination of OsHOS1 with the target TFs produced YFP fluorescence signal (Fig. 6B). Altogether, our results confirm the interaction of OsHOS1 with both OsEREBP1 and OsEREBP2. This observation suggests that OsHOS1 may influence the TFs' protein stability, likely targeting them for proteasome-mediated degradation and thus modulating the OsRMC gene expression.
Figure 6.
A, Yeast two-hybrid assay to analyze the interaction between OsHOS1 and OsEREBP1, and between OsHOS1 and OsEREBP2. The OsHOS1 sequence was cloned into pBD-GW (binding domain). The sequences of OsEREBP1 and OsEREBP2 fused with the activation domain (AD) were retrieved from a previous yeast one-hybrid screening (Serra et al., 2013). The plasmid combinations of pBDOsHOS1/pADOsEREBP1 and pBDOsHOS1/pADOsEREBP2 were cotransformed into the yeast strain YRG2. Transformation control plates +HIS (His) were used to assess transformation with both plasmids. The interaction was evaluated in selection complete minimum (CM) minus HIS (HIS−) plates. The interaction of pADWT and pBDWT (Stratagene) was used as a positive control. As negative controls, empty pBD-GW was cotransformed with pADOsEREBP1 and pADOsEREBP2. Each plasmid combination is representative of three independent colonies from three independent transformations. B, Bimolecular fluorescence complementation (BiFC) assay to test the interaction of OsHOS1 and the two OsEREBP proteins (1 and 2). The sequence of OsHOS1 was fused in frame with the N terminus (pYFN), whereas the sequences of OsEREBP1 and OsEREBP2 were fused in frame with the C-terminal (pYFC) part of YFP. The genetic construct pairs pYFN-OsHOS1/pYFC-OsEREBP1 and pYFN-OsHOS1/pYFC-OsEREBP2 were used to transiently transform Arabidopsis protoplasts. As a positive control (PosC), two known interacting proteins were used (pYFN-PosC1/pYFC-PosC2). As negative controls, pYFC-OsEREBP1 and pYFC-OsEREBP2 were cotransformed in pairs with a pYFN-PosC unrelated construct. YFP, Yellow fluorescent protein image; Merged, overlay of bright field and YFP images. Protoplast images are representative of four independent transformations. Scale bar = 25 µm.
OsHOS1 Modulates OsEREBP1 and OsEREBP2 Protein Stability through the 26S Proteasome
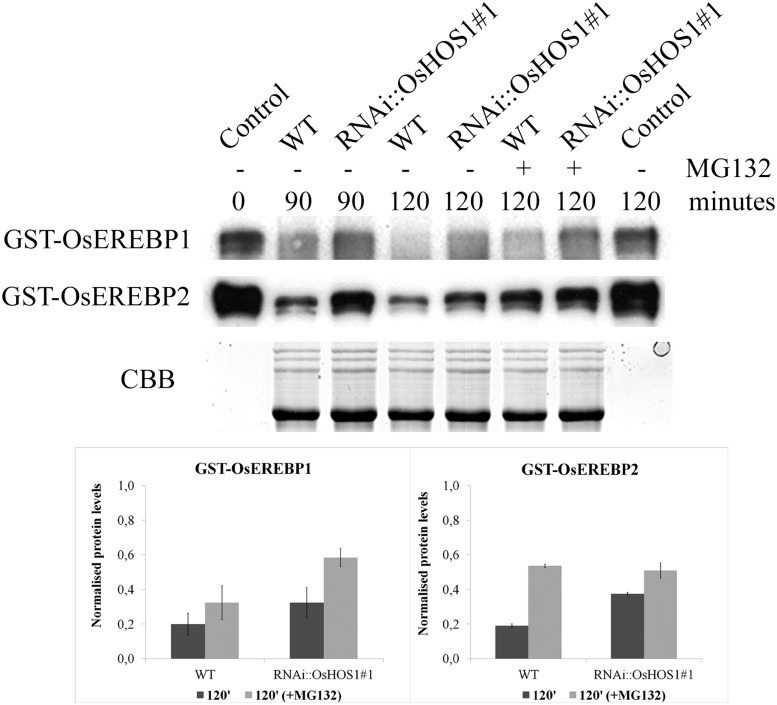
The previous results showed that OsHOS1 can interact with both OsEREBP1 and OsEREBP2. Since OsHOS1 is an E3-ubiquitin ligase, it is thus important to clarify if this interaction affected the TF protein stability. To test this, we performed in vitro degradation assays (Osterlund et al., 2000) using crude protein extracts from the RNAi::OsHOS1#1 and wild-type plants (10-DAG) and purified glutathione S-transferase (GST)-tagged TFs (Fig. 7). Assuming that a lower amount of OsHOS1 transcripts in the transgenic line (Lourenço et al., 2013) would imply less available OsHOS1 protein in these plants (as compared with the wild type), the rate of OsEREBP1 and OsEREBP2 degradation would be higher in wild-type crude extracts. This was in fact the case, since the degradation rate of GST-OsEREBP1 and GST-OsEREBP2 proved to happen faster in wild-type than in RNAi::OsHOS1#1 crude protein extracts (Fig. 7). Moreover, incubation of the crude protein extracts with MG132, a well-known proteasome inhibitor, led to slower degradation of the GST-tagged proteins (Fig. 7), further confirming the involvement of the ubiquitin/26S proteasome pathway controlling the stability of these proteins through OsHOS1.
Figure 7.
Proteasome-mediated degradation of GST-OsEREBP1 and GST-OsEREBP2. Crude protein extracts from RNAi::OsHOS1#1 and wild-type (WT) plants under control conditions were incubated at 30°C with GST-OsEREBP1 or GST-OsEREBP2, with or without MG132, for the different sampling time points shown. Control represents the GST-OsEREBP1 (or GST-OsEREBP2) input for all assays without crude protein extract at time 0 or after 120 min of incubation at 30°C. The different samples were resolved on 10% (v/v) SDS-PAGE, and the detection of GST-tagged proteins was made as described in “Materials and Methods.” The intensity of Coomassie Blue (CBB) staining was used as a loading control of the protein extracts from RNAi::OsHOS1#1 and wild-type plants. The immunoblot results are representative of two technical replicates from two independent biological replicates. The immunoblot bands (at time 120 min) were quantified using ImageJ software and normalized against Coomassie Blue total protein. Values represent means ± se.
DISCUSSION
Plants, due to their sessile nature, have evolved several mechanisms to perceive external cues and to adjust growth accordingly. Roots are an important organ for plant anchorage and uptake of water and nutrients. Therefore, roots need to integrate several external cues, including gravity, water, nutrients, and obstacles, to optimize their growth and develop a root system architecture to fulfill the plant's needs.
In this work, we have identified an interaction between a RING-type E3-ubiquitin ligase, OsHOS1 (Lourenço et al., 2013), and two TFs, OsEREBP1 and OsEREBP2, previously described as binding to the OsRMC promoter (Serra et al., 2013). OsRMC, in turn, had already been reported as being involved in rice root curling (Jiang et al., 2007). In our work, we demonstrate that the root curling phenotype modulated by OsRMC occurs upon contact with a mechanical barrier, through the ubiquitin/26 proteasome. We could also confirm that JA is involved in the curling response to this mechanical stimulus.
OsHOS1 was previously described as being involved in the rice cold response pathway, modulating the expression of rice DEHYDRATION-RESPONSIVE ELEMENT-BINDING PROTEIN 1A (OsDREB1A) gene by controlling the abundance of a key TF, OsICE1 (Lourenço et al., 2013). In recent years, the Arabidopsis HOS1 has been shown to be involved in several developmental processes (abiotic stress response, flowering time, chromatin remodeling), even beyond its proteolytic function (Jung et al., 2014; MacGregor and Penfield, 2015). In the present work, we showed that the down-regulation of OsHOS1 (RNAi::OsHOS1) led to altered root morphology in response to contact with a barrier. The involvement of genes related to abiotic stress with an effect on mechanical sensing has been previously described (Zarka et al., 2003; Figueiredo et al., 2012). In Arabidopsis, a gene from the DREB1 regulon, C-REPEAT-BINDING FACTOR2 (CBF2/DREB1C), was shown to have in its promoter cis-elements responding to cold as well as to mechanical stimulus (Zarka et al., 2003). In rice, another member of the DREB1 regulon, OsDREB1B (CBF1), also shows rapid induction in response to touch stimulus (Figueiredo et al., 2012). It is therefore possible that members of the DREB1 regulon, such as OsDREB1A and OsICE1 (both regulated by OsHOS1; Lourenço et al., 2013), are not only responsible in the response to abiotic stress but also in the response to mechanical stimuli. In rice, the ubiquitination and proteasome pathway have been implicated in root development through the modulation of rice ROOT ARCHITECTURE ASSOCIATED1 (Ge et al., 2004; Han et al., 2008). This protein has been linked to the auxin-mediated modulation of rice root development (Ge et al., 2004) as a cell cycle controller (Han et al., 2008). These studies implicate the proteasome and auxins in the regulation of rice root development. Together with our results, it is possible to speculate that the development of rice roots and their response to mechanical stimulus may be regulated by the proteasome complex by integrating different hormone-signaling pathways (such as auxin and JA).
Previously, in a yeast one-hybrid screening using the OsRMC promoter as bait, two TFs (OsEREBP1 and OsEREBP2) were identified and further shown to be negative regulators of OsRMC expression in transactivation studies (Serra et al., 2013). In the present work, we showed that these two TFs interact with OsHOS1, an E3-ubiquitin ligase, and that this interaction mediates their degradation via the 26S/proteasome system. Thus, OsEREBP1 and OsEREBP2 proteins are likely more available in the RNAi::OsHOS1 transgenic line. If these two TFs were in fact repressors of OsRMC expression, we should expect a lower expression of this gene in the RNAi::OsHOS1 plants, which was not the case. It was previously described that several TF families may form homodimers and heterodimers, implying modifications in their binding and activation/repression functions (Cutcliffe et al., 2011; Figueiredo et al., 2012). Therefore, we can hypothesize that these TFs may form heterodimers and activate the expression of OsRMC, whereas they have a repression role as single or homodimers.
In Arabidopsis, wavy root phenotypes have been correlated with touch sensing (Okada and Shimura, 1990). Arabidopsis mutants for two transmembrane proteins, MILDEW RESISTANCE LOCUS O4 (MLO4) and MLO11, located primarily at the root tip, show a tight root curve phenotype when touching a surface as compared with Columbia-0 plants. This response was dependent on touch, light, nutrients, and an intact polar auxin transport (Chen et al., 2009). In Arabidopsis, auxins are thought to integrate the signals generated in the root tip upon a touch stimulus (Ca2+, pH, reactive oxygen species), modulating root growth (Legué et al., 1997; Fasano et al., 2002; Massa and Gilroy, 2003b; Monshausen et al., 2009). Other hormones, such as cytokinins and ethylene, have also been proposed to interact with auxins in the control of root directional growth in Arabidopsis (Kushwah et al., 2011).
In rice, root curling has been associated with the down-regulation of OsRMC expression and influenced by JA (Jiang et al., 2007). JA has also been implicated in mechanical transduction. For example, the mechanically stimulated tendrils of B. dioica and Phaseolus vulgaris accumulate more 12-OPDA, a known precursor of JA biosynthesis in plants (Stelmach et al., 1998). In roots, however, not much is known regarding how this hormone may affect mechanical signal transduction. In our work, we show that rice root curling is induced by a mechanical stimulus, and that the curling is influenced by JA. Nevertheless, other factors may also play a role in root morphology, such as light, nitrogen, or high humidity, or a combination of stimuli (Shimizu et al., 2009; Wang et al., 2011). Interestingly, Shimizu et al. (2009) and Wang et al. (2011) were able to identify a root curling phenotype similar to the one we report here for wild-type plants. Both groups observed the root curling when the seedlings were grown under continuous light and supplemented with nitrogen (≥0.5 mm), although the nitrogen source alone was not able to induce root curling in the dark, nor was the curling response dependent on nitrogen concentration in the light (Shimizu et al., 2009). In our work, root curling occurred in wild-type seedlings grown under a 12-/12-h photoperiod in response to a mechanical barrier. The nutritional medium used in our experiments contains 1.5 mm NH4NO3, which is above the nitrogen threshold described to induce root curling. Together with the photoperiod, these conditions should be enough to induce root curling in wild-type seedlings. However, in our work, root curling only occurred upon sensing a mechanical barrier. In our conditions, we did not observe root curling in the wild-type seedlings grown vertically in the absence of a mechanical barrier (data not shown). This apparently different behavior may in fact be similar to that reported by Shimizu et al. (2009) and Wang et al. (2011), as they grew seedlings in glass tubes or solid medium, and possibly the curling occurred also in response to unintended contact with harder surfaces of the medium or container.
Jiang et al. (2007) observed that OsRMC down-regulation was enough to produce a root curling phenotype, and that the curling could be intensified with the addition of JA. We were able to confirm this regulation using RNAi::OsHOS1 seedlings, in which a higher OsRMC gene expression level led to a straight root phenotype. The straight root phenotype could be reverted when the roots were mechanically stimulated in the presence of JA. Although OsRMC expression level correlates with the root phenotype, it is difficult to understand the relationship between the OsHOS1, OsRMC, JA, light, and nitrogen.
OsRMC was described as a receptor-like kinase of the DOMAIN OF UNKNOWN FUNCTION26 (DUF26) family, but lacking the kinase domain (Jiang et al., 2007). Besides its involvement in rice root curling, OsRMC was linked to salt stress response (Rabbani et al., 2003; Zhang et al., 2009; Serra et al., 2013) and nutritional regulation (Torabi et al., 2009; Yang et al., 2013). Yang et al. (2013) described OsRMC as being involved in Fe nutritional uptake in rice. The overexpression of OsRMC led to a higher Fe uptake, even in Fe deficiency due to the up-regulation of rice IRON-REGULATED TRANSPORTER1 (OsIRT1; a known Fe2+ transporter) and of genes involved in the synthesis of phytosiderophores. Linking these data (Yang et al., 2013) and the work we present here shows that jasmonates may have a negative role in the fine tuning of Fe-deficiency gene expression (FERRIC REDUCTION OXIDASE2 and IRT) in Arabidopsis (Maurer et al., 2011). Recently, an EcoTILLING study using 392 rice accessions representative of the diversity of the species was used to identify allelic variants in salt-responsive genes, and found only three different protein variants in the OsRMC sequence. From the accessions studied, 99.2% had exactly the same OsRMC protein (Negrão et al., 2013). We believe that the high degree of conservation of this gene may be due to the close proximity to SHATTERING4 (SH4), a gene involved in rice domestication through reduced grain shattering (Li et al., 2006). Despite the above-mentioned evidences regarding the involvement of OsRMC in several biological processes, the role of this receptor-like kinase is still unclear and needs to be further explored.
Our RNAi::OsHOS1 lines only curled when facing a barrier in the presence of JA, in a dose-dependent manner that could be correlated to the silencing level of OsHOS1 and up-regulation of OsRMC. The exogenous addition of JA was necessary not to compensate lower JA biosynthesis levels but to overcome the lower sensibility to this hormone. This reduced sensibility may also lead to a higher up-regulation of genes involved in iron uptake, as described by Yang et al. (2013). Nevertheless, we cannot rule out the hypothesis that the reduced sensibility to JA in the down-regulated OsHOS1 lines is not a direct consequence of OsRMC action. Supporting this hypothesis is the fact that the ICE1 TFs of banana (Musa acuminata ICE1) and Arabidopsis can interact, respectively, with MaMYC2 (in banana) or JAZ repressors (in Arabidopsis; Hu et al., 2013; Zhao et al., 2013). Therefore, it can be hypothesized that the higher availability of OsICE1 in RNAi::OsHOS1 plants (Lourenço et al., 2013) may account for the different sensibility of these plants to jasmonates and not directly through, but probably in coordination with, OsRMC.
CONCLUSION
Taken together, our results prove the involvement of the proteasome (through OsHOS1) in rice root response to touch by modulating OsRMC expression (through OsEREBP1 and OsEREBP2). In addition, we confirm the influence of JA in the rice root response to touch. JA has been implicated in the mechanical response of tendrils and leaves (Chehab et al., 2012; Monshausen and Haswell, 2013), but there is no information regarding root mechanical responses. Nevertheless, the influence of light in JA-mediated root mechanosensing must also be taken into account. Different reports have shown a reduced sensibility of Arabidopsis phytochrome mutants to JA-induced root growth inhibition (Robson et al., 2010). However, it is still not clear how light, together with mechanosensing, regulates JA synthesis/signaling in the rice root. Considering the role that auxins, together with cytokinins and ethylene, play in mechanical sensing to adjust root growth in Arabidopsis, it is likely that other hormones (e.g. GAs; Lange and Lange, 2015) besides JA may also be involved in rice root touch response.
FINAL REMARK
“Nothing in Biology Makes Sense Except in the Light of Evolution” is the title of a work by Theodosius Dobzhansky (born 1900, died 1975). To that extent, we may ask the following. Why do rice roots curl when sensing a mechanical barrier? How is light involved in this process, and how does it correlate with JA? Does it have an evolutionary advantage? We may hypothesize that the curling phenotype, upon sensing a mechanical barrier, would allow the seedling to create a sort of anchor to be fixed to the upper layers of soil (where light can still penetrate). This would establish the seedling and facilitate the growth of the remaining root system, allowing the plant survival.
MATERIALS AND METHODS
Rice Root Phenotypic Analysis
The transgenic rice (Oryza sativa; RNAi::OsHOS1) plants used in this work were produced as previously described (Hiei et al., 1994; Lourenço et al., 2011, 2013). Transgenic and wild-type (rice ‘Nipponbare’) seeds were dehusked and surface sterilized with ethanol (70% [v/v]) for 1 min with gentle shaking. The seeds were then washed twice with sterile water and further sterilized with a 2% (v/v) sodium hypochlorite solution (commercial bleach solution) for 10 to 15 min with agitation. The seeds were then washed thoroughly six to seven times with sterile water and germinated in water, in the dark, at 28°C for 3 d. The germinated seedlings were then transferred to glass test tubes (height, 13.5 cm; diameter, 3 cm) with approximately 5 mL of Yoshida’s medium (Yoshida et al., 1976), capped with aluminum foil, and grown for further 10 d at 28°C with a 12-/12-h photoperiod and 500 µmol m−2 s−1 light intensity (control conditions). The Yoshida’s medium was changed every 4 to 5 d.
The presence, or absence, of a rice root curl in the different rice lines was scored after 10 DAG (Figs. 1 and 3B). The influence of JA (Duchefa) in the root curl response was also analyzed. The Yoshida’s medium from 2-DAG seedlings was changed by Yoshida’s medium plus 2 µm (or 100 µm) JA (1% [v/v] ethanol). To act as control, 2-DAG seedlings were grown with Yoshida’s medium plus 1% (v/v) ethanol. The seedlings were grown in the same temperature, photoperiod, and light intensity as above. The score of the rice root curling was performed at 10 DAG (Fig. 3D; Supplemental Fig. S2).
Vertical Growth System for Rice Root Imaging and Relative Growth Rate Measure
The rice seeds were dehusked and surface sterilized as above. The seeds were placed in square plates with solid Yoshida’s medium supplemented with 0.75% (w/v) Phytagel (Sigma-Aldrich). The plates were placed vertically in the dark for seed germination at 28°C for 3 d. After germination, the seeds from both lines were transferred to fresh Yoshida’s plates or to medium supplemented with 1 µm JA and transferred to light at 24°C to 30°C and with a 12-/12-h photoperiod for another 2 d. To facilitate root imaging, the roots were kept between two layers of solid growth medium (with or without JA). The roots grew vertically in these conditions overnight prior to imaging.
The barrier test was performed using two sterile glass coverslips placed perpendicularly in front of the growing root. The rice roots’ responses to barrier (n ≥ 8) were visualized using the ProScope digital microscope (Bodelin Technologies) with a 50× magnification, and pictures were taken every 5 min. This system was used to produce the data depicted in Figure 3, A and C, and Supplemental Table S1. Bright field images of root response to the barrier were collected using an Olympus S2×12 stereo microscope coupled with a CCD camera QColor3.
We further analyzed the relative root growth rate (wild type, RNAi::OsHOS1#1 and #2–4) using seeds prepared as above for vertical growth, with JA (2 µm) or without. Root growth observations started 24 h after the transfer of the seedlings to the treatment plate (time point 0). Root growth was analyzed every 24 h for 2 d (time points 1 and 2). The average root growth was calculated using the ImageJ software (Schneider et al., 2012; absolute and relative values in Fig. 4, B and C, respectively) from n ≥ 10 rice seedling roots per line and treatment, in three independent biological replicates.
RT-qPCR Analysis
The gene expression analysis of OsHOS1 (JQ866627), OsRMC (LOC_Os04g56430), OsAOS1 (LOC_Os03g55800), OsEREBP1 (LOC_Os02g54160), and OsEREBP2 (LOC_Os01g64790) was performed using the total RNA extracted from rice seedlings grown under control conditions (described above) for 10 DAG using the Direct-zol RNA MiniPrep kit (Zymo Research). The complementary DNA (cDNA) first strand was synthesized from 2 µg of total RNA according to the instructions from the Transcriptor High Fidelity cDNA Synthesis Kit (Roche). RT-qPCR reactions were performed using the Light Cycler 480 system (Roche) and the SYBR Green I Master mix (Roche). The equivalent of 50 ng of total RNA was used per reaction, using the gene-specific primers described in Supplemental Table S2. The PCR reaction was carried out in a volume of 20 µL with 1× SYBR Green I Master mix, cDNA, specific primers, and sterile water (for the PCR reaction of OsEREBP2, 3% [v/v] dimethyl sulfoxide was added to the reaction volume). The conditions of the PCR reaction were as follows: one cycle at 95°C for 5 min, 45 cycles of amplification at 95°C for 10 s, annealing at 58°C to 60°C (see Supplemental Table S2) for 10 s, and extension at 72°C for 10 s. A PCR product melting curve analysis was performed after the PCR reaction. The cycle threshold (CT) values were calculated from three technical replicates and normalized against the CT values of OsUBC2 (LOC_Os02g42314), used as internal control. The relative expression of transcripts was calculated with the kinetic PCR efficiency correction using the 2−ΔΔCT comparative method (Livak and Schmittgen, 2001). Three technical replicates were performed for each sample from, at least, three biological replicates (pools of two to three seedlings).
RT-Semiquantitative PCR from Protoplast Samples
Rice protoplasts were prepared from etiolated wild-type rice seedlings (7–10 DAG) using an adapted protocol (Lourenço et al., 2013) from previously described ones (Bart et al., 2006; Chen et al., 2006). The protoplasts were transformed with the RNAi::OsHOS1 construct pANDA::OsHOS1 (Miki and Shimamoto, 2004; Lourenço et al., 2013) or no-plasmid transformed (control). Total RNA extraction and cDNA synthesis (0.5 µg of total RNA) were performed as described above. RT-semiquantitative PCR was performed using gene-specific primers (Supplemental Table S2) for OsHOS1 and OsRMC, using OsUBC2 (LOC_Os02g42314) gene expression as an internal control. The gel images were analyzed using ImageJ software (Schneider et al., 2012). Two technical replicates for each sample from, at least, three independent transformations per condition were used for the analysis.
BiFC Assays
To construct the BiFC plasmids overexpressing OsEREBP1 and OsEREBP2 in fusion with the C-terminal portion of the YFP, the respective coding sequences were PCR amplified from cDNA using gene-specific primers containing Gateway adapters, GW-OsEREBP1-Fw/-Rv and GW-OsEREBP2-Fw/-Rv (Supplemental Table S2), and the Phusion high-fidelity DNA polymerase (Life Technologies). To amplify the OsEREBP2 sequence, the reaction needed to be supplemented with 1 m betaine and 2.5% (v/v) dimethyl sulfoxide. The amplified coding sequences were cloned into the vector pDONR221 according to the manufacturer’s instructions (Life Technologies). The fragments were afterward transferred to the binary Gateway-based vector pYFPC43. The coding region of the OsHOS1 was amplified with primers GW-OsHOS1-Fw/Rv and recombined from plasmid pDONR221 into the vector pYFPN43 to be in frame with the N-terminal portion of the YFP, according to Gateway technology.
Arabidopsis (Arabidopsis thaliana) protoplasts were prepared as previously described (Anthony et al., 2004). For each transformation, 3 µg of each plasmid was used. Transformed protoplasts were incubated overnight in the dark at 22°C, and protein detection was done with a fluorescence microscope (Leica DMRA2) using the YFP filter cube (excitation filter band pass, 500/20 nm; a 515-nm dichroic mirror; and 535-/30-nm emission filter). As negative control in the interaction assays, an unrelated rice TF was tested in fusion with the N-terminal portion of the YFP protein.
Yeast Two-Hybrid Assay
Yeast (Saccharomyces cerevisiae) strain YRG-2 (MATα ura3-52 his3-200 ade2-101 lys2-801 trp1-901 leu2-3 112 Gal4-542 Gal80-538 LYS2::UASGAL1-TATA GAL1-HIS3 URA3::UASGAL4 17mers[x3]-TATACYC1-lacZ) was grown in yeast peptone dextrose medium supplemented with 20 mg L−1 adenine for the preparation of competent cells through the lithium-acetate method. Plasmid DNA (pBD-OsHOS1 and pAD-OsEREBP1 or pAD-OsEREBP2) transformation was also performed using the lithium-acetate method. Yeast-transformed cells were plated in CM medium lacking Leu and Trp for plasmid transformation control and in CM lacking Leu, Trp, and His for detection of interaction. The interaction was evaluated in three individual colonies transformed with both plasmids. The interaction of pAD-wild type and pBD-wild type from the HybriZAP 2.1 kit (Stratagene) was used as a positive interaction control. As a negative interaction control, pBD-GW and pAD-OsEREBP1 or pAD-OsEREBP2 were also used.
Production of Recombinant Proteins
For recombinant protein expression in Escherichia coli, OsEREBP1 and OsEREBP2 coding sequences were amplified with the GX-OsEREBP1-Fw/-Rv and GX-OsEREBP2-Fw/-Rv primers (Supplemental Table S2), respectively, and the Phusion high-fidelity DNA polymerase (Life Technologies) according to the manufacturer’s instructions. The OsEREBP1 and OsEREBP2 fragments were cloned in frame with the GST tag as an EcoRI-XhoI fragment into the pGEX-4T-1 vector.
The E. coli Rosetta strain was transformed with the pGEX-4T-1 vector alone or pGEX-4T-1 containing the OsEREBP1 and OsEREBP2 coding sequences. Protein production and purification were performed as previously described (Serra et al., 2013).
Degradation Assays
Cell-free degradation assays were performed as previously described (Osterlund et al., 2000). Rice seeds from both wild-type and RNAi::OsHOS1#1 lines were grown for 10 DAG in a growth chamber in control conditions. The plant material was collected, immediately frozen in liquid nitrogen, and ground to a fine powder. Total protein was extracted as previously described (Osterlund et al., 2000). Five micrograms of total protein extracts from the wild type and RNAi::OsHOS1#1 was incubated with an equal amount of GST-OsEREBP1 or GST-OsEREBP2 for 90 and 120 min at 30°C. To inhibit the proteasome, we incubated the protein extracts with 200 μm MG132 (Sigma-Aldrich) at the start of the assay. The degradation reaction was stopped at each time point by adding 5× SDS-loading buffer. The reaction products were separated by SDS-PAGE and transferred onto polyvinylidene difluoride membrane (PerkinElmer). Detection of the GST proteins was carried out using an anti-GST mouse monoclonal antibody and an anti-mouse horseradish peroxidase-conjugated antibody (NBS Biologicals) according to the manufacturer’s instructions. Coomassie Blue staining was used as a loading control. Each assay was repeated twice with the same total protein extract (technical replicate) from two independent experiments (biological replicates). The degradation assay immunoblots were analyzed using ImageJ software (Schneider et al., 2012) to quantify the degradation band density.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers OsHOS1, JQ866627; OsRMC, Os04g0659300; OsAOS1, Os03g0767000; OsEREBP1, Os02g0782700; OsEREBP2, Os01g0868000; and OsUBC2, Os02g0634800.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Gene expression analyses for OsHOS1 (JQ866627) and OsRMC (LOC_Os04g25650) in rice protoplasts transiently down-regulating OsHOS1 (pANDA-OsHOS1).
Supplemental Figure S2. Root curling response of 10-DAG RNAi::OsHOS1#2 to 4 seedlings grown in liquid Yoshida’s medium on test tubes, with or without JA.
Supplemental Table S1. Analyses of rice root response to barrier with or without JA (1 µm) using a Proscope digital microscope.
Supplemental Table S2. Oligonucleotide sequences used in the cloning procedures and gene expression studies.
Supplemental Video S1. Vertical growth of wild-type rice root (cv Nipponbare) on solid medium against a glass barrier.
Supplemental Video S2. Vertical growth of RNAi::OsHOS1 rice root on solid medium against a glass barrier.
Supplementary Material
Acknowledgments
We thank Dr. Alejandro Ferrando (IBMCP) for providing the BiFC vectors.
Glossary
- MeJA
methyl jasmonate
- JA
jasmonic acid
- UPS
ubiquitin/26S proteasome system
- TF
transcription factor
- RNAi
RNA interference
- DAG
days after germination
- RT
reverse transcription
- qPCR
quantitative PCR
- CM
complete minimum
- BiFC
bimolecular fluorescence complementation
- GST
glutathione S-transferase
- cDNA
complementary DNA
- 12-OPDA
12-oxo-phytodienoic acid
Footnotes
This work was supported by the Fundação para a Ciência e a Tecnologia through projects POCI/BIA–BCM/56063/2004, PTDC/BIA_BCM/099836/2008, and research unit GREEN-iT “Bioresources for Sustainability” (UID/Multi/04551/2013), and fellowships SFRH/BPD/102872/2014 (to T.F.L.), SFRH/BD/31011/2006 (to T.S.S.), and SFRH/BD/74946/2010 (to A.M.C.); Programa Ciência 2007 and Fundação para a Ciência e a Tecnologia Investigator 2012, financed by Programa Operacional Potencial Humano/Quadro de Referência Estratégico Nacional (POPH/QREN; to N.J.M.S.); and the National Science Foundation (grant no. IOS–11213800 to S.G.) and National Aeronautics and Space Administration (grant no. NNX12AK79G to S.G.).
References
- Anthony RG, Henriques R, Helfer A, Mészáros T, Rios G, Testerink C, Munnik T, Deák M, Koncz C, Bögre L (2004) A protein kinase target of a PDK1 signalling pathway is involved in root hair growth in Arabidopsis. EMBO J 23: 572–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bart R, Chern M, Park CJ, Bartley L, Ronald PC (2006) A novel system for gene silencing using siRNAs in rice leaf and stem-derived protoplasts. Plant Methods 2: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braam J. (2005) In touch: plant responses to mechanical stimuli. New Phytol 165: 373–389 [DOI] [PubMed] [Google Scholar]
- Chehab EW, Eich E, Braam J (2009) Thigmomorphogenesis: a complex plant response to mechano-stimulation. J Exp Bot 60: 43–56 [DOI] [PubMed] [Google Scholar]
- Chehab EW, Yao C, Henderson Z, Kim S, Braam J (2012) Arabidopsis touch-induced morphogenesis is jasmonate mediated and protects against pests. Curr Biol 22: 701–706 [DOI] [PubMed] [Google Scholar]
- Chen S, Tao L, Zeng L, Vega-Sanchez ME, Umemura K, Wang GL (2006) A highly efficient transient protoplast system for analyzing defence gene expression and protein-protein interactions in rice. Mol Plant Pathol 7: 417–427 [DOI] [PubMed] [Google Scholar]
- Chen Z, Noir S, Kwaaitaal M, Hartmann HA, Wu MJ, Mudgil Y, Sukumar P, Muday G, Panstruga R, Jones AM (2009) Two seven-transmembrane domain MILDEW RESISTANCE LOCUS O proteins cofunction in Arabidopsis root thigmomorphogenesis. Plant Cell 21: 1972–1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chico JM, Chini A, Fonseca S, Solano R (2008) JAZ repressors set the rhythm in jasmonate signaling. Curr Opin Plant Biol 11: 486–494 [DOI] [PubMed] [Google Scholar]
- Cutcliffe JW, Hellmann E, Heyl A, Rashotte AM (2011) CRFs form protein-protein interactions with each other and with members of the cytokinin signalling pathway in Arabidopsis via the CRF domain. J Exp Bot 62: 4995–5002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreher K, Callis J (2007) Ubiquitin, hormones and biotic stress in plants. Ann Bot (Lond) 99: 787–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falik O, Reides P, Gersani M, Novoplansky A (2005) Root navigation by self inhibition. Plant Cell Environ 28: 562–569 [Google Scholar]
- Fasano JM, Massa GD, Gilroy S (2002) Ionic signaling in plant responses to gravity and touch. J Plant Growth Regul 21: 71–88 [DOI] [PubMed] [Google Scholar]
- Figueiredo DD, Barros PM, Cordeiro AM, Serra TS, Lourenço T, Chander S, Oliveira MM, Saibo NJM (2012) Seven zinc-finger transcription factors are novel regulators of the stress responsive gene OsDREB1B. J Exp Bot 63: 3643–3656 [DOI] [PubMed] [Google Scholar]
- Fonseca S, Chico JM, Solano R (2009) The jasmonate pathway: the ligand, the receptor and the core signalling module. Curr Opin Plant Biol 12: 539–547 [DOI] [PubMed] [Google Scholar]
- Ge L, Chen H, Jiang JF, Zhao Y, Xu ML, Xu YY, Tan KH, Xu ZH, Chong K (2004) Overexpression of OsRAA1 causes pleiotropic phenotypes in transgenic rice plants, including altered leaf, flower, and root development and root response to gravity. Plant Physiol 135: 1502–1513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Cao H, Jiang J, Xu Y, Du J, Wang X, Yuan M, Wang Z, Xu Z, Chong K (2008) Rice ROOT ARCHITECTURE ASSOCIATED1 binds the proteasome subunit RPT4 and is degraded in a D-box and proteasome-dependent manner. Plant Physiol 148: 843–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T (1994) Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J 6: 271–282 [DOI] [PubMed] [Google Scholar]
- Hu Y, Jiang L, Wang F, Yu D (2013) Jasmonate regulates the INDUCER OF CBF EXPRESSION-C-REPEAT BINDING FACTOR/DRE BINDING FACTOR1 cascade and freezing tolerance in Arabidopsis. Plant Cell 25: 2907–2924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani M, Xiong L, Lee H, Stevenson B, Zhu JK (1998) HOS1, a genetic locus involved in cold-responsive gene expression in Arabidopsis. Plant Cell 10: 1151–1161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe MJ, Leopold AC, Staples RC (2002) Thigmo responses in plants and fungi. Am J Bot 89: 375–382 [DOI] [PubMed] [Google Scholar]
- Jiang J, Li J, Xu Y, Han Y, Bai Y, Zhou G, Lou Y, Xu Z, Chong K (2007) RNAi knockdown of Oryza sativa root meander curling gene led to altered root development and coiling which were mediated by jasmonic acid signalling in rice. Plant Cell Environ 30: 690–699 [DOI] [PubMed] [Google Scholar]
- Jung JH, Lee HJ, Park MJ, Park CM (2014) Beyond ubiquitination: proteolytic and nonproteolytic roles of HOS1. Trends Plant Sci 19: 538–545 [DOI] [PubMed] [Google Scholar]
- Kurusu T, Kuchitsu K, Nakano M, Nakayama Y, Iida H (2013) Plant mechanosensing and Ca2+ transport. Trends Plant Sci 18: 227–233 [DOI] [PubMed] [Google Scholar]
- Kushwah S, Jones AM, Laxmi A (2011) Cytokinin interplay with ethylene, auxin, and glucose signaling controls Arabidopsis seedling root directional growth. Plant Physiol 156: 1851–1866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange MJP, Lange T (2015) Touch-induced changes in Arabidopsis morphology dependent on gibberellin breakdown. Nature Plants 1: 14025. [DOI] [PubMed] [Google Scholar]
- Lee H, Xiong L, Gong Z, Ishitani M, Stevenson B, Zhu JK (2001) The Arabidopsis HOS1 gene negatively regulates cold signal transduction and encodes a RING finger protein that displays cold-regulated nucleo--cytoplasmic partitioning. Genes Dev 15: 912–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Kim WT (2011) Regulation of abiotic stress signal transduction by E3 ubiquitin ligases in Arabidopsis. Mol Cells 31: 201–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legué V, Blancaflor E, Wymer C, Perbal G, Fantin D, Gilroy S (1997) Cytoplasmic free Ca2+ in Arabidopsis roots changes in response to touch but not gravity. Plant Physiol 114: 789–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Zhou A, Sang T (2006) Rice domestication by reducing shattering. Science 311: 1936–1939 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lourenço T, Saibo N, Batista R, Ricardo CP, Oliveira MM (2011) Inducible and constitutive expression of HvCBF4 in rice leads to differential gene expression and drought tolerance. Biol Plant 55: 653–663 [Google Scholar]
- Lourenço T, Sapeta H, Figueiredo DD, Rodrigues M, Cordeiro A, Abreu IA, Saibo NJM, Oliveira MM (2013) Isolation and characterization of rice (Oryza sativa L.) E3-ubiquitin ligase OsHOS1 gene in the modulation of cold stress response. Plant Mol Biol 83: 351–363 [DOI] [PubMed] [Google Scholar]
- Lyzenga WJ, Stone SL (2012) Abiotic stress tolerance mediated by protein ubiquitination. J Exp Bot 63: 599–616 [DOI] [PubMed] [Google Scholar]
- MacGregor DR, Penfield S (2015) Exploring the pleiotropy of hos1. J Exp Bot 66: 1661–1671 [DOI] [PubMed] [Google Scholar]
- Massa GD, Fasano JM, Gilroy S (2003) Ionic signaling in plant gravity and touch responses. Gravit Space Biol Bull 16: 71–82 [PubMed] [Google Scholar]
- Massa GD, Gilroy S (2003a) Touch and gravitropic set-point angle interact to modulate gravitropic growth in roots. Adv Space Res 31: 2195–2202 [DOI] [PubMed] [Google Scholar]
- Massa GD, Gilroy S (2003b) Touch modulates gravity sensing to regulate the growth of primary roots of Arabidopsis thaliana. Plant J 33: 435–445 [DOI] [PubMed] [Google Scholar]
- Maurer F, Müller S, Bauer P (2011) Suppression of Fe deficiency gene expression by jasmonate. Plant Physiol Biochem 49: 530–536 [DOI] [PubMed] [Google Scholar]
- Miki D, Shimamoto K (2004) Simple RNAi vectors for stable and transient suppression of gene function in rice. Plant Cell Physiol 45: 490–495 [DOI] [PubMed] [Google Scholar]
- Monshausen GB, Bibikova TN, Weisenseel MH, Gilroy S (2009) Ca2+ regulates reactive oxygen species production and pH during mechanosensing in Arabidopsis roots. Plant Cell 21: 2341–2356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monshausen GB, Haswell ES (2013) A force of nature: molecular mechanisms of mechanoperception in plants. J Exp Bot 64: 4663–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrão S, Almadanim MC, Pires IS, Abreu IA, Maroco J, Courtois B, Gregorio GB, McNally KL, Oliveira MM (2013) New allelic variants found in key rice salt-tolerance genes: an association study. Plant Biotechnol J 11: 87–100 [DOI] [PubMed] [Google Scholar]
- Okada K, Shimura Y (1990) Reversible root tip rotation in Arabidopsis seedlings induced by obstacle-touching stimulus. Science 250: 274–276 [DOI] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Rabbani MA, Maruyama K, Abe H, Khan MA, Katsura K, Ito Y, Yoshiwara K, Seki M, Shinozaki K, Yamaguchi-Shinozaki K (2003) Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol 133: 1755–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riemann M, Riemann M, Takano M (2008) Rice JASMONATE RESISTANT 1 is involved in phytochrome and jasmonate signalling. Plant Cell Environ 31: 783–792 [DOI] [PubMed] [Google Scholar]
- Robson F, Okamoto H, Patrick E, Harris SR, Wasternack C, Brearley C, Turner JG (2010) Jasmonate and phytochrome A signaling in Arabidopsis wound and shade responses are integrated through JAZ1 stability. Plant Cell 22: 1143–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serra TS, Figueiredo DD, Cordeiro AM, Almeida DM, Lourenço T, Abreu IA, Sebastián A, Fernandes L, Contreras-Moreira B, Oliveira MM, et al (2013) OsRMC, a negative regulator of salt stress response in rice, is regulated by two AP2/ERF transcription factors. Plant Mol Biol 82: 439–455 [DOI] [PubMed] [Google Scholar]
- Shimizu H, Tanabata T, Xie X, Inagaki N, Takano M, Shinomura T, Yamamoto KT (2009) Phytochrome-mediated growth inhibition of seminal roots in rice seedlings. Physiol Plant 137: 289–297 [DOI] [PubMed] [Google Scholar]
- Stelmach BA, Müller A, Hennig P, Laudert D, Andert L, Weiler EW (1998) Quantitation of the octadecanoid 12-oxo-phytodienoic acid, a signalling compound in plant mechanotransduction. Phytochemistry 47: 539–546 [DOI] [PubMed] [Google Scholar]
- Torabi S, Wissuwa M, Heidari M, Naghavi MR, Gilany K, Hajirezaei MR, Omidi M, Yazdi-Samadi B, Ismail AM, Salekdeh GH (2009) A comparative proteome approach to decipher the mechanism of rice adaptation to phosphorous deficiency. Proteomics 9: 159–170 [DOI] [PubMed] [Google Scholar]
- Toyota M, Gilroy S (2013) Gravitropism and mechanical signaling in plants. Am J Bot 100: 111–125 [DOI] [PubMed] [Google Scholar]
- Vierstra RD. (2009) The ubiquitin-26S proteasome system at the nexus of plant biology. Nat Rev Mol Cell Biol 10: 385–397 [DOI] [PubMed] [Google Scholar]
- Wang SJ, Ho CH, Chen HW (2011) Rice develop wavy seminal roots in response to light stimulus. Plant Cell Rep 30: 1747–1758 [DOI] [PubMed] [Google Scholar]
- Weiler EW, Albrecht T, Groth B, Xia ZQ, Luxem M, Liss H, Andert L, Spengler P (1993) Evidence for the involvement of jasmonates and their octadecanoid precursors in the tendril coiling response of bryonia dioica. Phytochemistry 32: 591–600 [Google Scholar]
- Yang A, Li Y, Xu Y, Zhang WH (2013) A receptor-like protein RMC is involved in regulation of iron acquisition in rice. J Exp Bot 64: 5009–5020; erratum Yang A, Li Y, Xu Y, Zhang WH (2013) J Exp Bot 64: 5769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Foorno DA, Cock JH, Gomez KA, editors (1976) Laboratory Manual for Physiological Studies of Rice, Ed 3. International Rice Research Institute, Los Banos, Philippines [Google Scholar]
- Yoshii M, Yamazaki M, Rakwal R, Kishi-Kaboshi M, Miyao A, Hirochika H (2010) The NAC transcription factor RIM1 of rice is a new regulator of jasmonate signaling. Plant J 61: 804–815 [DOI] [PubMed] [Google Scholar]
- Zarka DG, Vogel JT, Cook D, Thomashow MF (2003) Cold induction of Arabidopsis CBF genes involves multiple ICE (inducer of CBF expression) promoter elements and a cold-regulatory circuit that is desensitized by low temperature. Plant Physiol 133: 910–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Tian LH, Zhao JF, Song Y, Zhang CJ, Guo Y (2009) Identification of an apoplastic protein involved in the initial phase of salt stress response in rice root by two-dimensional electrophoresis. Plant Physiol 149: 916–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao ML, Wang JN, Shan W, Fan JG, Kuang JF, Wu KQ, Li XP, Chen WX, He FY, Chen JY, et al (2013) Induction of jasmonate signalling regulators MaMYC2s and their physical interactions with MaICE1 in methyl jasmonate-induced chilling tolerance in banana fruit. Plant Cell Environ 36: 30–51 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.