Protein interactions for two plasmodesmata-localized reticulon proteins suggest that these proteins, in addition to a role in endoplasmic reticulum modeling, may play important roles in linking the endoplasmic reticulum and plasma membrane.
Abstract
The endoplasmic reticulum (ER) is a ubiquitous organelle that plays roles in secretory protein production, folding, quality control, and lipid biosynthesis. The cortical ER in plants is pleomorphic and structured as a tubular network capable of morphing into flat cisternae, mainly at three-way junctions, and back to tubules. Plant reticulon family proteins (RTNLB) tubulate the ER by dimerization and oligomerization, creating localized ER membrane tensions that result in membrane curvature. Some RTNLB ER-shaping proteins are present in the plasmodesmata (PD) proteome and may contribute to the formation of the desmotubule, the axial ER-derived structure that traverses primary PD. Here, we investigate the binding partners of two PD-resident reticulon proteins, RTNLB3 and RTNLB6, that are located in primary PD at cytokinesis in tobacco (Nicotiana tabacum). Coimmunoprecipitation of green fluorescent protein-tagged RTNLB3 and RTNLB6 followed by mass spectrometry detected a high percentage of known PD-localized proteins as well as plasma membrane proteins with putative membrane-anchoring roles. Förster resonance energy transfer by fluorescence lifetime imaging microscopy assays revealed a highly significant interaction of the detected PD proteins with the bait RTNLB proteins. Our data suggest that RTNLB proteins, in addition to a role in ER modeling, may play important roles in linking the cortical ER to the plasma membrane.
The endoplasmic reticulum (ER) is a multifunctional organelle (Hawes et al., 2015) and is the site of secretory protein production, folding, and quality control (Brandizzi et al., 2003) and lipid biosynthesis (Wallis and Browse, 2010), but it is also involved in many other aspects of day-to-day plant life, including auxin regulation (Friml and Jones, 2010) and oil and protein body formation (Huang, 1996; Herman, 2008). The cortical ER network displays a remarkable polygonal arrangement of motile tubules that are capable of morphing into small cisternae, mainly at the three-way junctions of the ER network (Sparkes et al., 2009). The cortical ER network of plants has been shown to play multiple roles in protein trafficking (Palade, 1975; Vitale and Denecke, 1999) and pathogen responses (for review, see Pattison and Amtmann, 2009; Beck et al., 2012).
In plants, the protein family of reticulons (RTNLBs) contributes significantly to tubulation of the ER (Tolley et al., 2008, 2010; Chen et al., 2012). RTNLBs are integral ER membrane proteins that feature a C-terminal reticulon homology domain (RHD) that contains two major hydrophobic regions. These regions form two V-shaped transmembrane wedges joined together via a cytosolic loop, with the C and N termini of the protein facing the cytosol. RTNLBs can dimerize or oligomerize, creating localized tensions in the ER membrane, inducing varying degrees of membrane curvature (Sparkes et al., 2010). Hence, RTNLBs are considered to be essential in maintaining the tubular ER network.
The ability of RTNLBs to constrict membranes is of interest in the context of cell plate development and the formation of primary plasmodesmata (PD; Knox et al., 2015). PD formation involves extensive remodeling of the cortical ER into tightly furled tubules to form the desmotubules, axial structures that run through the PD pore (Overall and Blackman, 1996; Ehlers and Kollmann, 2001). At only 15 nm in diameter, the desmotubule is one of the most constricted membrane structures found in nature, with no animal counterparts (Tilsner et al., 2011). PD are membrane-rich structures characterized by a close association of the plasma membrane (PM) with the ER. The forces that model the ER into desmotubules, however, are poorly understood. RTNLBs are excellent candidates for this process and can constrict fluorescent protein-labeled ER membranes into extremely fine tubules (Sparkes et al., 2010). We showed recently that two of the RTNLBs present in the PD proteome, RTNLB3 and RTNLB6 (Fernandez-Calvino et al., 2011), are present in primary PD at cytokinesis (Knox et al., 2015). However, nothing is known of the proteins that interact with RTNLBs identified in the PD proteome or that may link RTNLBs to the PM. To date, the only protein shown to bind to plant RTNLBs is RHD3-LIKE2, the plant homolog of the ER tubule fusion protein ATLASTIN (Lee et al., 2013).
Here, we used a dual approach to identify interacting partners of RTNLB3 and RTNLB6 (Fernandez-Calvino et al., 2011; Knox et al.., 2015). First, we used GFP immunoprecipitation assays coupled to mass spectrometry (MS) to identify proteins potentially binding to RTNLB3 and RTNLB6. Second, from the proteins we identified, we conducted a detailed Förster resonance energy transfer by fluorescence lifetime imaging microscopy (FRET-FLIM) analysis to confirm prey-bait interactions in vivo.
The application of time-resolved fluorescence spectroscopy to imaging biological systems has allowed the design and implementation of fluorescence lifetime imaging microscopy (FLIM). The technique allows measuring and determining the space map of picosecond fluorescence decay at each pixel of the image through confocal single and multiphoton excitation. The general fluorescence or Förster resonance energy transfer (FRET) to determine the colocalization of two color chromophores can now be improved to determine physical interactions using FRET-FLIM and protein pairs tagged with appropriate GFP fluorophores and monomeric red fluorescent protein (mRFP). FRET-FLIM measures the reduction in the excited-state lifetime of GFP (donor) fluorescence in the presence of an acceptor fluorophore (e.g. mRFP) that is independent of the problems associated with steady-state intensity measurements. The observation of such a reduction is an indication that the two proteins are within a distance of 1 to 10 nm, thus indicating a direct physical interaction between the two protein fusions (Osterrieder et al., 2009; Sparkes et al., 2010; Schoberer and Botchway, 2014). It was shown previously that a reduction of as little as approximately 200 ps in the excited-state lifetime of the GFP-labeled protein represents quenching through a protein-protein interaction (Stubbs et al., 2005).
Our interaction data identified a large percentage (40%) of ER proteins, including other RTNLB family members. However, we also found a relatively large number (25%) of proteins present in the published PD proteome (Fernandez-Calvino et al., 2011) and a surprisingly high proportion (35%) of PM proteins. Of the PD-resident proteins we identified, a significant number were shown previously to be targets of viral movement proteins (MPs) or proteins present within lipid rafts, consistent with the view that PD are lipid-rich microdomains (Bayer et al., 2014). Additional proteins identified suggested roles for RTNLBs in transport and pathogen defense. We suggest that RTNLBs may play key roles in anchoring and/or signaling between the cortical ER and PM.
RESULTS
Identification of Proteins That Interact with RTNLB3 and RTNLB6
The reticulon proteins RTNLB3 and RTNLB6 are found in the PD proteome (Fernandez-Calvino et al., 2011). We showed recently that when both RTNLBs are coexpressed transiently in tobacco (Nicotiana tabacum) epidermal leaf cells with the viral MP of Tobacco mosaic virus, there is significant colocalization (Knox et al., 2015). Both these RTNLBs are located to the developing cell plate at cytokinesis and, therefore, are strong candidates for proteins that model the cortical ER into desmotubules (Knox et al., 2015).
As it is likely that RTNLBs form protein complexes with proteins in the PM and desmotubule in order to stabilize the desmotubule constriction and to allow gating in PD (see model in Knox et al., 2015), we searched for potential interacting proteins capable of carrying out these tasks.
To find interaction partners for these RTNLBs, we used Arabidopsis (Arabidopsis thaliana) plants stably expressing RTNLB3-YFP (for yellow fluorescent protein) or RTNLB6-YFP fusion proteins under the control of a 35S promoter to perform coimmunoprecipitation using GFP-Trap_A beads (Chromotek). For this approach, whole-seedling protein extracts were incubated with agarose slurry linked to anti-GFP camelid antibodies. This antibody is capable of binding the YFP tag on the reticulon proteins. The RTNLB bait, along with the proteins that bind/interact with the RTNLB, were pelleted by slow centrifugation. The resulting proteins in this pellet were identified using MS (Thermo Orbitrap Fusion; Thermo Scientific), and the data were analyzed using Scaffold (version Scaffold_4.4.1.1; Proteome Software). As a control, proteins bound to the antibody in untransformed plants were also analyzed by MS.
The MS data showed a high percentage of overlay between proteins identified with RTNLB3 and RTNLB6 as baits (Fig. 1). Of a total of 706 identified proteins, only 93 (13%) or 85 (12%) proteins were unique to RTNLB3 or RTNLB6, respectively (Table I). Proteins that were also identified in the control samples (two independent sets of wild-type Arabidopsis plants) were subtracted from the list of proteins, resulting in 146 potential interacting partners for either RTNLB3 or RTNLB6. Despite the removal of false positives, a high percentage of proteins in common were maintained for both reticulons: out of a total of 146 proteins, RTNLB3 had 135 potential interaction candidates, with only 11 proteins (8%) showing unique specificity for RTNLB3. Similarly, 126 proteins were coimmunoprecipitated with RTNLB6, with 20 proteins (17%) being unique to RTNLB6 (Table I).
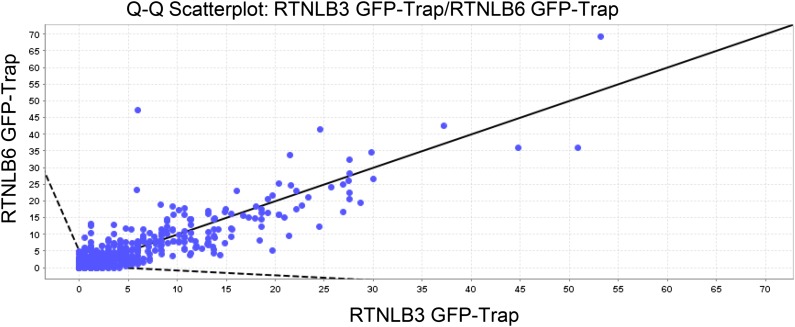
Figure 1.
Quantitative scatterplot for overlay and distribution of candidate interaction proteins for RTNLB3 and RTNLB6 (Scaffold_4.4.1.1; Proteome Software). Each protein is plotted as a point on a two-dimensional scatterplot, with the x axis showing a normalized spectral count for proteins binding to RTNLB3 and the y axis for RTNLB6. The Scaffold software shows a line with a slope of 1 on the graph. Therefore, proteins with similar abundances in both coimmunoprecipitation assays will plot as points near this line. Proteins that plot outside the dashed lines on the plot are more than 2 sd away from being the same in both coimmunoprecipitations. These proteins are considered to be differentially expressed.
Table I. Analysis steps and number of proteins derived from MS analysis.
| Analysis Step | No. of Proteins |
|---|---|
| Total (RTNLB3/RTNLB6) | 706 (613/621) |
| Minus wild-type control total (RTNLB3/RTNLB6) | 146 (135/126) |
| Proteins subjected to FRET-FLIM analysis | 17 |
These resulting protein candidates were ranked according to their percentage of the total spectra, which represents the number of spectra matching a specific protein (across all MS samples) as a percentage of the total number of spectra in the sample (Supplemental Table S1). This ranking indicates the amount of a specific protein bound to the RTLNB baits and, therefore, can be used as a measure of the reliability of each potential interaction.
FRET-FLIM Analysis to Validate Immunoprecipitation Data
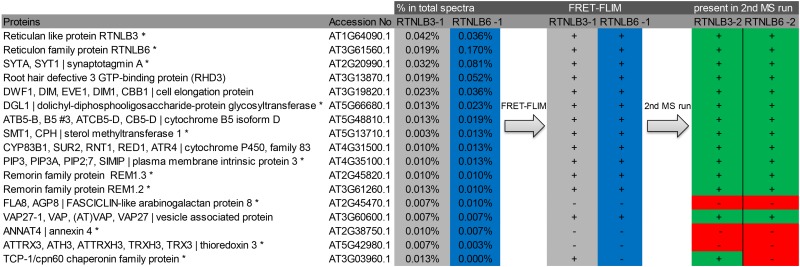
Seventeen proteins from the list of 146 potential interacting proteins were subjected to further analysis to test for interactions in vivo using a different methodology (FRET-FLIM; Tables I and II). The choice of these 17 proteins was based on four main criteria. (1) Known or expected interacting partners of RTNLBs from published work. These were used as positive controls and included RTNLB3 and RTNLB6 (RTNLB3 dimerization in Sparkes et al., 2010) and ROOT HAIR DEFECTIVE3 (RHD3)/RHD3-like2 (RL2; Lee et al., 2013). (2) Proteins present in the PD proteome, listed in Table II. (3) A selection of high- and low-abundance proteins distributed throughout the quantitative lists (Table II; Supplemental Table S1). These were used to test the hypothesis that proteins with low abundance in the GFP-trap assays were likely to represent weak or false-positive interactions. For example, THIOREDOXIN3 (TRX3; Table II) showed very low abundance relative to DWARF1 (DWF1). (4) The T-complex protein (TCP1)/chaperonin60 (cpn60) chaperonin family protein TCP1 was chosen as a further control, as this protein was found in the proteome for RTNLB3 but not for RTNLB6 (Table II; Supplemental Table S1).
Table II. Flow chart of the 17 proteins tested by FRET-FLIM.
The protein description (column 1) and accession number (column 2) indicate the percentage abundance in the total spectrum for RTNLB3 (column 3) and RTNLB6 (column 4). Proteins present in the PD proteome (Fernandez-Calvino et al., 2011) are marked with asterisks. Positive (+) and negative (−) results for interaction in the FRET-FLIM analysis with either RTNLB3 (column 5) or RTNLB6 (column 6) are shown. The reappearance of the corresponding protein in the second MS run is shown (RTNLB3-2 and RTNLB6-2; columns 7 and 8).
 |
FRET (Förster, 1948) measured by donor excited-state FLIM (Becker, 2012; Schoberer and Botchway, 2014) was used to confirm independently the interactions suggested by the GFP-trap assays. FRET-FLIM measures the reduction in the lifetime of the GFP (donor) fluorescence when an acceptor fluorophore (mRFP) is within a distance of 1 to 10 nm, thus allowing FRET to occur and indicating a physical interaction between the two protein fusions (Osterrieder et al., 2009; Sparkes et al., 2010). In the FRET-FLIM assay, each of the above 17 proteins was expressed transiently as an mRFP fusion (acceptor) in tobacco leaf epidermal cells expressing either RTNLB3-GFP or RTNLB6-GFP as donor. At least two biological samples with a minimum of three technical replicates each were used for the statistical analysis.
Due to limitations in the speed of photon counting of the FLIM apparatus, measurements were taken from high-expressing areas of ER regions with relatively low mobility, such as the ER associated with the nuclear envelope. This allowed more reliable measurements than the fast-moving cortical ER (Sparkes et al., 2010). Furthermore, to allow consistent and reliable measurements, also proteins that usually localize to PD or PM were driven to the ER by protein overexpression. FRET-FLIM interactions are shown in Table II. RTNLB3-GFP or RTNLB6-GFP expression without acceptor presence was used as a negative control, while known self-interactions between the RTNLBs (e.g. RTNLB3 against RTNLB3) or with the second RTNLB (e.g. RTNLB3 against RTNLB6) were used as positive controls and to determine the value of fluorescence that could be considered as a significantly positive interaction. Figure 2 shows a comparison of such negative and positive controls.
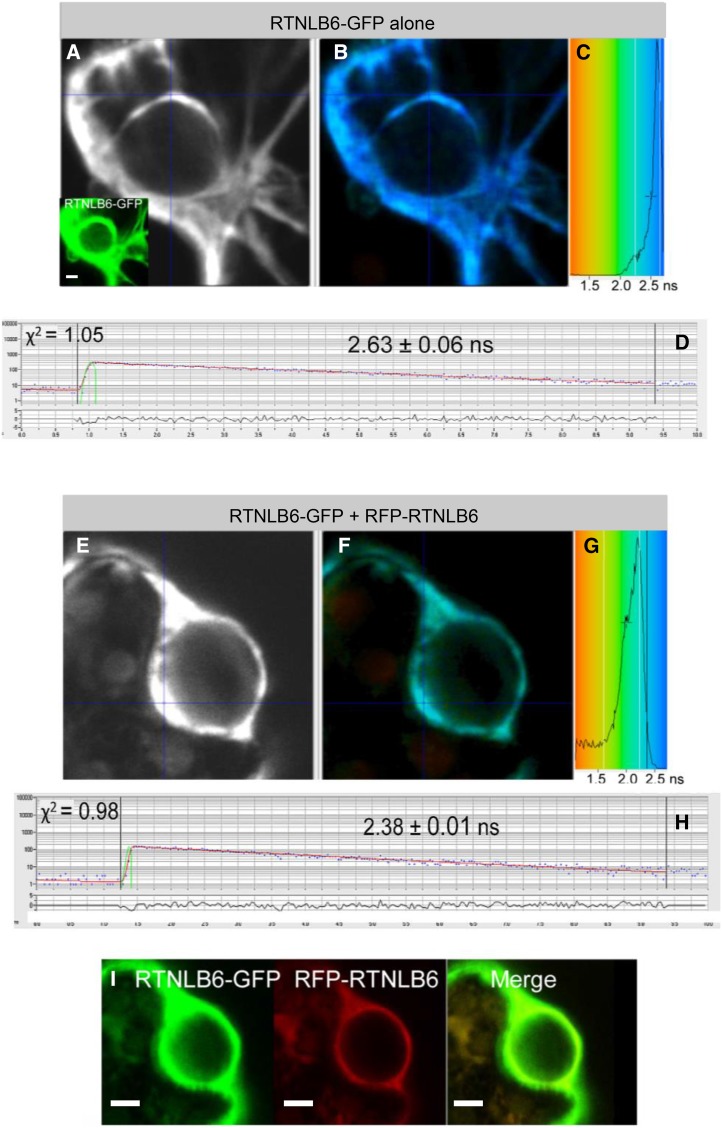
Figure 2.
FRET-FLIM analysis of RTNLB6 without an interaction partner (A–D) or RTNLB6 dimerization (E–I). A and E display the raw FRET-FLIM data. In B and F, pseudocolored lifetime maps show the lifetime values for each point within the region of interest, while the distribution of lifetimes across the entire image is shown in C and G, with blue shades representing longer GFP fluorescence lifetimes than green ones. D and H display representative decay curves of a single point with an optimal single exponential fit, where χ2 values from 0.9 to 1.2 were considered an excellent fit to the data points (binning factor of 2). The inset in A and I are the respective confocal images for the analysis, showing the GFP construct in green and the mRFP construct in red. This example of FRET-FLIM analysis shows that RTNLB6 homodimerizes, because the lifetime values for the GFP/mRFP fusion pair (H; 2.38 ± 0.01 ns) are lower than those for the GFP fusion alone (D; 2.63 ± 0.06 ns). Bars = 5 µm.
RTNLB3-GFP alone showed a fluorescence lifetime of 2.47 ± 0.05 ns, and RTNLB6-GFP alone a lifetime of 2.63 ± 0.06 ns. Excited-state lifetimes determined for RTNLB-RTNLB homomeric and heteromeric interactions varied from 2.31 to 2.38 ns (Table III), which is statistically significantly different from that of the GFP alone. Figure 2 shows the FRET-FLIM analysis steps for RTNLB6-GFP alone (Fig. 2, A–D) as a negative control and for RTNLB6-GFP interacting with RFP-RTNLB6 (Fig. 2, E–I) as a positive control. Raw FRET-FLIM images are shown in Figure 2, A and E. This analysis takes into account the lifetime values of each pixel within the image visualized by a pseudocolored lifetime map (Fig. 2, B and F). The graph shows the distribution of lifetimes within the image (Fig. 2, C and G), with blue shades representing longer GFP fluorescence lifetimes than green ones. Decay curves (Fig. 2, D and H) of a representative single pixel highlight an optimal single exponential fit, where χ2 values from 0.9 to 1.2 were considered an excellent fit to the data points (binning factor of 2). Confocal images for the region of interest showing the GFP construct in green and the mRFP construct in red are shown in Figure 2, A (inset) and I. This specific example shows that RTNLB6 homodimerizes, because the lifetime values for the GFP/mRFP fusion pair (2.38 ± 0.01 ns; Table III) are lower than those for the GFP fusion alone (2.63 ± 0.06 ns).
Table III. Fluorescence lifetimes in FRET-FLIM analysis.
Donor and acceptor protein constructs are indicated together with the average fluorescence lifetime (in ns) for the donor fluorophore and the sd for each combination. It was shown previously that a reduction in excited-state lifetime of 200 ps is indicative of energy transfer (Stubbs et al., 2005). For each combination, at least two biological samples with a minimum of three technical replicates were used for the statistical analysis.
| Donor (GFP) | Acceptor (mRFP) | Average | se |
|---|---|---|---|
| ns | |||
| RTNLB3 | – | 2.51 | 0.05 |
| RTNLB3 | RTNLB3 | 2.28 | 0.01 |
| RTNLB3 | RTNLB6 | 2.31 | 0.01 |
| RTNLB3 | SYTA | 2.29 | 0.04 |
| RTNLB3 | RHD3 | 2.30 | 0.01 |
| RTNLB3 | DWF1 | 2.28 | 0.01 |
| RTNLB3 | DGL1 | 2.37 | 0.01 |
| RTNLB3 | Cyb5D | 2.30 | 0.01 |
| RTNLB3 | SMT1 | 2.29 | 0.03 |
| RTNLB3 | SUR2 | 2.28 | 0.06 |
| RTNLB3 | PIP3 | 2.37 | 0.02 |
| RTNLB3 | REM1.3 | 2.33 | 0.02 |
| RTNLB3 | REM1.2 | 2.31 | 0.01 |
| RTNLB3 | FLA8 | 2.53 | 0.09 |
| RTNLB3 | VAP27 | 2.33 | 0.03 |
| RTNLB3 | ANNAT4 | 2.50 | 0.03 |
| RTNLB3 | TRX3 | 2.48 | 0.03 |
| RTNLB3 | TCP1 | 2.32 | 0.01 |
| RTNLB6 | – | 2.63 | 0.06 |
| RTNLB6 | RTNLB3 | 2.37 | 0.04 |
| RTNLB6 | RTNLB6 | 2.38 | 0.01 |
| RTNLB6 | SYTA | 2.46 | 0.02 |
| RTNLB6 | RHD3 | 2.34 | 0.01 |
| RTNLB6 | DWF1 | 2.32 | 0.01 |
| RTNLB6 | DGL1 | 2.37 | 0.07 |
| RTNLB6 | Cyb5D | 2.47 | 0.02 |
| RTNLB6 | SMT1 | 2.36 | 0.07 |
| RTNLB6 | SUR2 | 2.34 | 0.11 |
| RTNLB6 | PIP3 | 2.35 | 0.04 |
| RTNLB6 | REM1.3 | 2.44 | 0.05 |
| RTNLB6 | REM1.2 | 2.48 | 0.01 |
| RTNLB6 | FLA8 | 2.67 | 0.07 |
| RTNLB6 | VAP27 | 2.33 | 0.04 |
| RTNLB6 | ANNAT4 | 2.66 | 0.03 |
| RTNLB6 | TRX3 | 2.62 | 0.06 |
| RTNLB6 | TCP1 | 2.60 | 0.02 |
Next, RTNLB3-GFP and RTNLB6-GFP were coinfiltrated independently with each of the 17 chosen proteins, and the resulting lifetimes were measured (Table III; Fig. 3). Representative FRET-FLIM data are shown for each combination (Supplemental Fig. S1). As mentioned above, ER regions with relatively low mobility, such as the ER associated with the nuclear envelope, allow more reliable measurements (Sparkes et al., 2010). Therefore, to be comparable with other data, the interactions between RTNLBs and PM-localized proteins such as remorins and plasma membrane intrinsic protein3 (PIP3) were also measured in this area. These PM-bound proteins are also normally detected as they transit through the ER in transient expression experiments.
Figure 3.
Fluorescence lifetimes in FRET-FLIM interactions. The bar graphs represent average fluorescence lifetimes (ns) and the corresponding sd values for the GFP donors RTNLB3 and RTNLB6. The data show 17 candidate interaction proteins (blue bars) compared with RTNLB3-GFP or RTNLB6-GFP without interaction partners (gray bars). Lifetimes significantly lower than those of RTNLB3-GFP or RTNLB6-GFP alone (left side of the red line) indicate protein-protein interactions.
Among the RTNLB3 putative interactors, all proteins with the exceptions of FASCICLIN-LIKE ARABINOGALACTAN PROTEIN8 (FLA8), ANNEXIN4 (ANNAT4), and TRX3 showed interaction. Significantly, these proteins were not present in the second MS data set (Table II), and their lack of interaction using FRET-FLIM confirmed that these were likely to be false positives. The results for TCP1 are also significant, because TCP1 was pulled down by RTNLB3 but not by RTNLB6, and in the FRET-FLIM assays, TCP1 interacted with RTNLB3 but not with RTNLB6, confirming the results of the GFP-trap data (Table III; Fig. 3). To summarize, the proteomics data from one biological sample yielded less than 18% false negatives in the chosen selection, with the three false-negative proteins showing comparatively low peptide coverage.
MS Confirmation of Proteomics Data
For further confirmation of the data, the GFP immunoprecipitation and MS proteomics were repeated with an independent biological sample of RTNLB3-YFP and RTNLB6-YFP plants, as well as wild-type Arabidopsis and a stable Arabidopsis line expressing the ER membrane marker calnexin tagged with GFP. The ER-integral protein calnexin was used to detect false-positive interactions resulting from proteins binding to the fluorescent tag rather than the RTNLBs. Results from the wild-type and calnexin immunoprecipitations were subtracted from the proteins pulled down with RTNLB3 or RTNLB6. This second data set was then compared with data from the first experiment, and only proteins present in both data sets were compiled into a final list of interaction candidates (Table IV; Supplemental Table S2).
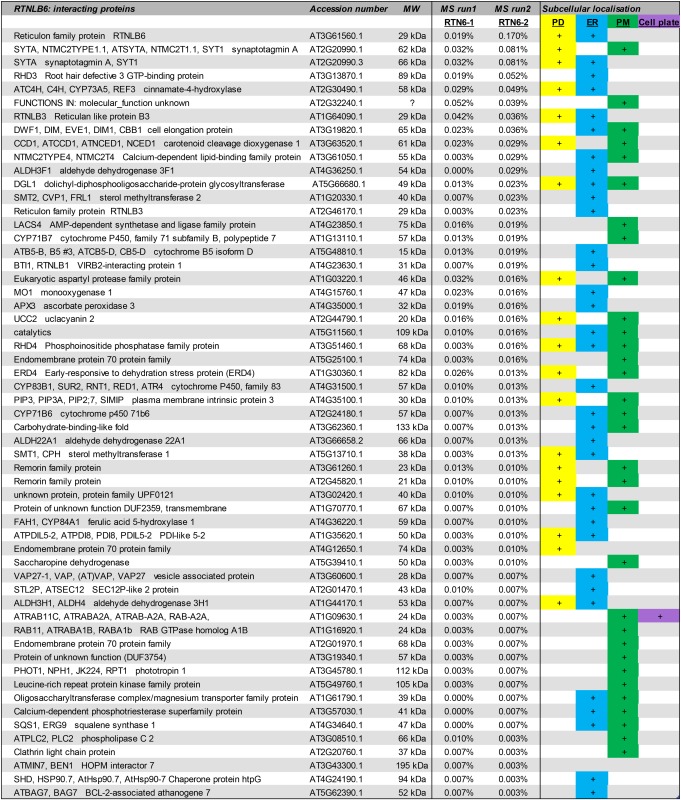
Table IV. List of interacting proteins for RTNLB3 and RTNLB6 present in both MS data sets.
The protein name, accession number, and molecular mass (MW) are given. The percentage of the total spectra in both MS data sets (MS runs 1 and 2) is given as well as known or predicted subcellular localization for PD (yellow), ER (blue), PM (green), or cell plate (purple). More detail for the interacting proteins can be found in Supplemental Table S2.
|
This resulted in 42 interaction candidates for RTNLB3 and 57 for RTNLB6. Proteins were again ranked according to the quantity of peptide present in the total spectra. Interestingly, proteins that were identified by FRET-FLIM to be false positives in the first MS run (FLA8, ANNAT4, and TRX3) were not present in the second MS data set, thereby confirming and validating the FRET-FLIM methodology (Table II). Furthermore, the final list of interaction candidates comprises a high percentage of proteins localized or predicted to be localized to PD and ER (Fig. 4).
Figure 4.
Quantitative distribution (%) of predicted or known subcellular localizations for RTNLB3 and RTNLB6 interaction candidates validated by two MS data sets and FRET-FLIM.
DISCUSSION
Validation of the Proteomics Approach
Immunoprecipitation using the camelid GFP-trap system with two of the PD proteome reticulons, RTNLB3 and RTNLB6 (Fernandez-Calvino et al., 2011; Knox et al., 2015), identified a high percentage of PD-localized proteins and also proteins that are more widely distributed over the ER and PM. These proteomics data were validated by in vivo testing with FRET-FLIM, and further rounds of immunoprecipitation using different controls confirmed the initial data and removed the few proteins that did not interact in vivo, indicating that these were most likely false positives. The following points are stressed.
(1) FRET-FLIM analysis corresponded with the second MS run. In the FRET-FLIM analysis for both RTNLB3 and RTNLB6, FLA8, ANNAT4, and TRX3 did not show a decreased florescence lifetime (Table III; Fig. 3). These proteins were absent from the second MS data set for both reticulons and, therefore, were most likely false positives in the first MS run. Additionally, FLA8, ANNAT4, and TRX3 showed low peptide abundances in the MS spectra (Table II), indicating a higher rate of false positives in the lower ranges of abundance.
(2) TCP1 was pulled down with RTNLB3 but not with RTNLB6 (Table III; Fig. 3), and indeed, FRET-FLIM analysis showed an interaction of TCP1 with RTNLB3 but not RTNLB6, validating both the proteomics data and the FRET-FLIM approach as a confirmatory method.
(3) Selectivity of reticulon protein-protein interactions. Out of 21 Arabidopsis reticulons, some of which have been shown to interact previously (Sparkes et al., 2010), only RTNLB3, RTNLB6, RTNLB5, and RTNLB1 showed up as interactors in the immunoprecipitation analysis with the bait PD reticulons RTNLB3 and RTNLB6 (Table IV; Supplemental Table S2). RTNLB5 is 84% identical at the amino acid level with RTNLB6 and, therefore, difficult to distinguish. However, the MS analysis revealed peptides unique to RTNLB5 and not RTNLB6, indicating that RTNLB5 was indeed detected. The role of this potential RTNLB5 interaction is unclear, as RTNLB5 is mainly expressed in pollen (Arabidopsis eFP Browser; Winter et al., 2007) and involved in the karrikin response (Nelson et al., 2010). Thus, it is likely that RTNLB3 and RTNLB6 interact with each other in PD and are involved in the generation of the extremely fine ER-derived desmotubule (Knox et al., 2015). RTNLB1 is ubiquitously expressed in different tissues and developmental stages (Arabidopsis eFP Browser; Winter et al., 2007). Interestingly, it has been shown that a Ser-rich region in the N-terminal tail of RTNLB1, and also RTNLB2, interacts with the FLAGELLIN-SENSITIVE2 (FLS2) receptor (Lee et al., 2011). The double mutant rtnlb1/rtnlb2, as well as an RTNLB1 overexpressor, displayed reduced FLS2-dependent signaling and enhanced susceptibility to pathogen attacks (Lee et al., 2011). RTNLB1 and RTNLB2 may regulate FLS2 transport to the PM. FLS2 is localized at the PM but also within PD (Monaghan and Zipfel, 2012) and may mediate the flg22-induced closure of PD (Faulkner et al., 2013).
(4) Preference for PD and ER localization of the interaction candidates. The immunoprecipitation experiments identified several proteins present in the PD proteome, suggesting that RTNLB3 and RTNLB6 may be part of a protein complex within PD. We also found a number of PM-specific proteins that interacted with RTNLB3 and RTNLB6. Some of these proteins have a role in anchoring the ER to the PM. For example, synaptotagmin (SYTA) is prevalent at ER-PM contact points in both animal cells (Giordano et al., 2013; Lin et al., 2014) and also in plant cells (Schapire et al., 2008; Lewis and Lazarowitz, 2010; Yamazaki et al., 2010; Uchiyama et al., 2014). Pérez-Sancho et al. (2015) have suggested that SYTA on the PM may link the PM to the cortical ER, conferring mechanotolerance at these points. However, they did not identify the interacting ER protein. Our work here suggests that RTNLBs on the cortical ER may perform such a linking function through a direct interaction with SYTA on the PM.
Another protein prevalent at ER-PM contact sites is Vesicle-associated Membrane Protein-associated Protein27 (VAP27; Wang et al., 2014), also identified here as an interacting partner of RTNLB3 and RTNLB6. Recent studies suggest that a unique complex of proteins resides at such ER-PM contacts. VAP27 can bind microtubules and RTNLBs (this study) and also NETWORKED3C (NET3C; Wang et al., 2014), a protein that links the actin cytoskeleton to the ER contacts. This protein complex may perform unique functions in anchoring and signaling between ER and PM (Wang et al., 2014). We suggest that the same complex may also function to anchor the desmotubule to the PM within or at the neck of the PD, perhaps explaining their prevalence in the PD proteome. SYTA is a Ca2+-sensitive contractile protein (Yamazaki et al., 2010) that, in the contracted form, reduces the distance between adjacent membrane bilayers to about 5 nm (Lin et al., 2014). PD closure is acutely sensitive to elevated Ca2+ levels (Tucker and Boss, 1996); therefore, SYTA emerges as a potential candidate for forcing the desmotubule and PM together upon Ca2+ influx. Significantly, like RTNLB3 and RTNLB6, SYTA appears in developing primary PD during cell plate formation and remains associated with the entrances of mature PD (Schapire et al., 2008).
A number of the PD proteins that we found to interact with RTNLB3 and RTNLB6 are also the targets of viral MPs. These include SYTA (Lewis and Lazarowitz, 2010; Uchiyama et al., 2014), VAP27 (Carette et al., 2002), and the remorin proteins REMORIN1.2 (REM1.2) and REM1.3 (Borner et al., 2005; Marín et al., 2012). A recent study (Levy et al., 2015) demonstrated that SYTA forms ER-PM junctions that are specifically recruited to PD during virus movement. Thus, proteins associated with the ER-PM contacts may be the specific targets of MPs during cell-to-cell movement. The association of these proteins with PD may provide a mechanism for targeting and concentrating viral genomes assembled on the actin-ER network and subsequently recruited to the entrances of PD (Tilsner et al., 2013; Levy et al., 2015). The PM intrinsic protein, PIP3, functions as an aquaporin and is induced by salt stress (Hachez et al., 2014b). The correct delivery of PIP3 to the PM involves specific interactions with two syntaxin proteins, SYP61 and SYP121 (Hachez et al., 2014a). PIP3 is also present in the PD proteome (Fernandez-Calvino et al., 2011) and, via an interaction with RTNLBs, may provide an additional link between the desmotubule and PM. TCP1 was pulled down only with RTNLB3 but not RTNLB6 and interacted only with RTNLB3 in FRET-FLIM assays. The TCP1 protein is part of a chaperonin complex involved in transcription factor trafficking through PD (Xu et al., 2011). One protein of this complex, Chaperonin Containing TCP1, Subunit 8 (CCT8), was shown recently to be required for KNOTTED1 trafficking through PD and is a target of the viral MP of Tobacco mosaic virus (Fichtenbauer et al., 2012). It appears that the entire chaperonin complex may be recruited for cell-to-cell trafficking (Xu et al., 2011). Conceivably, RTNLB3 provides a means of linking this complex to PD for the cell-to-cell movement of transcription factors.
In addition to proteins present at ER-PM contacts, our data reveal a number of PD proteins associated with lipid-rich domains in plants (Tapken and Murphy, 2015). This finding is in agreement with the view that PD are rich in lipid components (Naulin et al., 2014; Grison et al., 2015) and may function as unique lipid rafts (Mongrand et al., 2010), perhaps involved in receptor-mediated signaling (Faulkner et al., 2013). The PM within PD is rich in sterols and sphingolipids relative to the general PM (Naulin et al., 2014). The PD-localized protein STEROL METHYLTRANSFERASE1 (AT5G13710) controls cholesterol levels (Diener et al., 2000), while the remorin proteins that interact with RTNLB3 and RTNLB6 are components of lipid rafts (Mongrand et al., 2010). REM1.3 (AT2G45820) has been localized to PD in planta (Raffaele et al., 2009) and is differentially phosphorylated upon contact with bacterial elicitors. It may function as a scaffold protein in plant innate immunity (Benschop et al., 2007; Jarsch and Ott, 2011). The tomato (Solanum lycopersicum) REM1.3 is required for the restriction of potato virus X trafficking (Perraki et al., 2012), while the potato REM1.3 affects the ability of the TRIPLE GENE BLOCK1 MP of Potato virus X and other viral MPs to increase PD permeability (Perraki et al., 2014). Several remorins, including Arabidopsis REM1.3, form nonamyloid filamentous structures of 5.7 to 8 nm (Bariola et al., 2004; Marín et al., 2012). These remorins could be linked with the cytoskeleton in superstructures to maintain cell integrity or act as scaffold proteins for signaling and defense mechanisms (Bariola et al., 2004), a process that might occur in combination with the structural RTNLB proteins.
Additional Interacting Proteins
DWF1 (AT3G19820) is a Ca2+-dependent calmodulin-binding protein involved in the conversion of the early brassinosteroid precursor 24-methylenecholesterol to campesterol. As brassinosteroids affect cellular elongation, dwf1 mutants display a dwarf phenotype due to reduced cell expansion. SUPERROOT2 (SUR2; AT4G31500) catalyzes the conversion of indole-3-acetaldoxime to indole-3-thiohydroxymate in indole glucosinolate biosynthesis (Barlier et al., 2000; Bak et al., 2001) and was found here to interact with both RTNLB3 and RTNLB6. The biologically active degradation products of glucosinolates are formed under tissue disruption and are well known as the characteristic flavor compounds in mustard (Brassica juncea) or cabbage (Brassica capitata; for review, see Glawischnig et al., 2003). This could potentially link RTNLBs with defense mechanisms. DEFECTIVE GLYCOSYLATION (DGL1; AT5G66680) is a subunit of the ER oligosaccharyltransferase complex (Lerouxel et al., 2005). This protein complex is responsible for the transfer of N-linked glycan precursors onto Asn residues of candidate proteins in the ER. N-Glycan synthesis pathways contribute to plant development as well as defense. The mutant dgl1-1 displays developmental defects, including reduced cell elongation and differentiation defects, together with changes in the noncellulosic matrix polysaccharides (Lerouxel et al., 2005).
CONCLUSION
Our combined experimental approach of using sensitive pull-down assays coupled with FRET-FLIM provides a robust means of identifying functional interactions for reticulon proteins. The primary MS data set was validated using FRET-FLIM and showed that more than 80% of the candidate proteins were indeed interacting with the reticulons. The intermediate data set was confirmed by a second set of proteomics data for both reticulons and confirmed both the proteomics as well as, in particular, the FRET-FLIM analysis, indicating a high confidence for the final protein interactome.
Using two RTNLB proteins as bait, we have highlighted a significant number of PD proteins that interact with RTNLB3 and RTNLB6. We identified predominantly proteins associated with ER-PM contacts, proteins resident in lipid rafts, and proteins that interact with viral MPs. These interaction studies will form the basis for future research aimed at unraveling the PD interactome. It will be interesting to determine which of these interactions are significant in regulating PD functions, such as the gating response that occurs during viral infection (Oparka et al., 1997).
MATERIALS AND METHODS
Immunoprecipitation (GFP-Trap_A beads)
Tobacco (Nicotiana tabacum) plant material for immunoprecipitation with the GFP-Trap_A beads (Chromotek) was prepared according to the company’s protocol with slight modifications.
In brief, approximately 5 g of whole-seedling plant material grown for 2 weeks on Murashige and Skoog plates was ground in liquid nitrogen and in lysis buffer (10 mm Tris-HCl, pH 7.5, 150 mm NaCl, 0.5 mm EDTA, 0.5% [v/v] Nonidet P-40, 1 mm phenylmethylsulfonyl fluoride, and protease inhibitor). The extracts were incubated on ice for 30 min and then centrifuged at 10,000g for 10 min at 4°C. The supernatant (about 2–3 mL) was poured into fresh tubes via two layers of muslin cloth.
The GFP-Trap_A beads were equilibrated in 500 µL of dilution buffer (10 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 0.5 mm EDTA) and centrifuged at 2,500g for 2 min. The supernatant was discarded, and this wash was repeated twice.
A total of 100 µL of the washed beads was added to the plant extract, and the mixture was shaken on ice for 2 h. After this, tubes were centrifuged at 2,500g for 2 min at 4°C, the supernatant was discarded, and the resulting agarose pellet was washed twice with dilution buffer.
MS Analysis
Reverse-phase chromatography was used to separate tryptic peptides prior to MS analysis. Two columns were utilized, an Acclaim PepMap µ-precolumn cartridge (300-µm i.d. × 5 mm; 5 μm; 100 Å) and an Acclaim PepMap RSLC (75 µm × 50 cm; 2 µm; 100 Å; Thermo Scientific). The columns were installed on an Ultimate 3000 RSLCnano system (Dionex). Mobile phase buffer A was composed of 0.1% (v/v) aqueous formic acid, and mobile phase B was composed of acetonitrile containing 0.1% (v/v) formic acid. Samples were loaded onto the µ-precolumn equilibrated in 2% (v/v) aqueous acetonitrile containing 0.1% (v/v) trifluoroacetic acid for 8 min at 10 µL min−1, after which peptides were eluted onto the analytical column by increasing the mobile phase B concentration from 3% (v/v) B to 35% (v/v) over 87 min and then to 90% (v/v) B over 5 min, followed by a 4-min wash at 90% (v/v) B and a 15-min reequilibration at 3% (v/v) B.
Eluting peptides were converted to gas-phase ions by means of electrospray ionization and analyzed on a Thermo Orbitrap Fusion device (Q-OT-qIT; Thermo Scientific). Survey scans of peptide precursors from 350 to 1,500 mass-to-charge ratio were performed at 120K resolution (at 200 mass-to-charge ratio) with a 4 × 105 ion count target. Tandem mass spectrometry (MS/MS) was performed by isolation at 1.6 thomson using the quadrupole, higher energy collisional dissociation fragmentation with normalized collision energy of 35, and rapid-scan MS analysis in the ion trap. The MS/MS ion count target was set to 104, and the maximum injection time was 200 ms. Precursors with charge state 2 to 7 were selected and sampled for MS/MS. The dynamic exclusion duration was set to 45 s with a 10-ppm tolerance around the selected precursor and its isotopes. Monoisotopic precursor selection was turned on. The instrument was run in top speed mode with 3-s cycles.
MS Data Analysis
Raw data were processed using MSConvert in ProteoWizard Toolkit (version 3.0.5759; Kessner et al., 2008). MS/MS spectra were searched with Mascot engine (Matrix Science, version 2.4.1) Mascot was set up to search The Arabidopsis Information Resource 10 database (version 20101214; 35,508 entries) assuming the digestion enzyme trypsin. Mascot was searched with a fragment ion mass tolerance of 0.8 D and a parent ion tolerance of 20 ppm. Carbamidomethylation of Cys was specified in Mascot as a fixed modification. Oxidation of Met was specified in Mascot as a variable modification.
Scaffold (version Scaffold_4.4.1.1; Proteome Software) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at greater than 95% probability by the Scaffold Local FDR algorithm. Protein identifications were accepted if they could be established at greater than 99% probability and contained at least two identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm (Nesvizhskii et al., 2003). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
Cloning of Expression Plasmids
Primers were obtained from MWG Biotech. Q5 high-fidelity DNA polymerase (New England Biolabs) was used for all PCRs. Vectors containing the genes of interest from the proteomics data set were obtained from the Nottingham Arabidopsis Stock Centre (Scholl et al., 2000). Genes of interest were cloned into the modified binary vector pB7FWG2,0 or pB7WGR2,0, providing expression from Agrobacterium tumefaciens transfer DNA, using the cauliflower mosaic virus 35S promoter upstream of coding fusions to GFP or RFP, respectively (Karimi et al., 2005).
Tobacco Plant Material and Transient Expression in Tobacco Leaves
For A. tumefaciens-mediated transient expression, 5-week-old tobacco (‘Petit Havana’) plants grown in the greenhouse were used. Transient expression was induced and detected according to Sparkes et al. (2006). In brief, each expression vector was introduced into A. tumefaciens strain GV3101 by heat shock. Transformants were inoculated into 5 mL of YEB medium (5 g L−1 beef extract, 1 g L−1 yeast extract, 5 g L−1 Suc, and 0.5 g L−1 MgSO4∙7H2O) supplemented with the antibiotics for the vector and rifampicin to select for agrobacteria. After overnight shaking at 25°C, 1 mL of the bacterial culture was pelleted by centrifugation at 2,500g for 5 min at room temperature. The pellet was washed twice with 1 mL of infiltration medium (50 mm MES, 2 mm Na3PO4∙12H2O, 0.1 mm acetosyringone, and 5 mg mL−1 Glc) and then resuspended in 1 mL of infiltration buffer. The suspension was diluted to a final optical density at 600 nm of 0.1 and gently pressed through the stomata on the lower epidermal surface using a 1-mL syringe. Transformed plants then were incubated under normal growth conditions for 48 to 72 h. Images were taken using a Zeiss LSM510 Meta laser scanning confocal microscope with a 63× oil immersion objective. For imaging of GFP/RFP combinations, samples were excited using 488- and 543-nm laser lines in multitrack mode with line switching. Images were edited using the LSM510 image browser.
FRET-FLIM Data Acquisition
Epidermal samples of infiltrated tobacco leaves were excised, and FRET-FLIM data capture was performed according to Osterrieder et al. (2009) and Schoberer and Botchway (2014) using a two-photon microscope at the Central Laser Facility of the Rutherford Appleton Laboratory. In brief, a two-photon microscope built around a Nikon TE2000-U inverted microscope was used with a modified Nikon EC2 confocal scanning system to allow for multiphoton FLIM (Botchway et al., 2015). Laser light at a wavelength of 920 nm was produced by a mode-locked titanium sapphire laser (Mira; Coherent Lasers), producing 200-fs pulses at 76 MHz, pumped by a solid-state continuous wave 532-nm laser (Verdi V18; Coherent Laser). The laser beam was focused to a diffraction-limited spot through a water-immersion objective (Nikon VC; ×60, numerical aperture of 1.2) to illuminate specimens on the microscope stage. Fluorescence emission was collected without descanning, bypassing the scanning system, and passed through a BG39 (Comar) filter to block the near-infrared laser light. Line, frame, and pixel clock signals were generated and synchronized with an external detector in the form of a fast microchannel plate photomultiplier tube (Hamamatsu R3809U). Linking these via a time-correlated single-photon-counting PC module SPC830 (Becker and Hickl) generated the raw FLIM data. Prior to FLIM data collection, the GFP and mRFP expression levels in the plant samples within the region of interest were confirmed using a Nikon EC2 confocal microscope with excitation at 488 and 543 nm, respectively. A 633-nm interference filter was used to significantly minimize the contaminating effect of chlorophyll autofluorescence emission that would otherwise obscure the mRFP emission as well as that of GFP. Data were analyzed by obtaining excited-state lifetime values of a region of interest on the nucleus, and calculations were made using SPCImage analysis software version 5.1 (Becker and Hickl). The distribution of lifetime values within the region of interest was generated and displayed as a curve. Only values that had a χ2 between 0.9 and 1.4 were taken. The median lifetime value and minimum and maximum values for one-quarter of the median lifetime values from the curve were taken to generate the range of lifetimes per sample.
At least three nuclei from at least three independent biological samples per protein-protein combination were analyzed, and the average of the ranges was taken.
Sequence data for the genes mentioned in this article can be found in the GenBank/EMBL databases using the following accession numbers: RTNLB3, At1g64090; and RTNLB6, At3g61560. All accession numbers from the proteomic analysis can be found in the corresponding tables.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Representative FRET-FLIM data for RTNLB6 and RTNLB3, respectively, with potential interaction partners.
Supplemental Table S1. Protein interactions for RTNLB3 and RTNLB6 in the first MS data set.
Supplemental Table S2. List of interacting proteins for RTNLB3 and RTNLB6 present in both MS data sets.
Supplementary Material
Glossary
- ER
endoplasmic reticulum
- PD
plasmodesmata
- PM
plasma membrane
- FRET-FLIM
Förster resonance energy transfer by fluorescence lifetime imaging microscopy
- FLIM
fluorescence lifetime imaging microscopy
- FRET
Förster resonance energy transfer
- MP
movement protein
- MS
mass spectrometry
- MS/MS
tandem mass spectrometry
Footnotes
This work was supported by the British Biotechnology and Biological Sciences Research Council (grant no. BB/J004987/1 to K.O. and C.H.) and by the Science and Technology Facilities Council Program (grant no. 14230008 to C.H.).
References
- Bak S, Tax FE, Feldmann KA, Galbraith DW, Feyereisen R (2001) CYP83B1, a cytochrome P450 at the metabolic branch point in auxin and indole glucosinolate biosynthesis in Arabidopsis. Plant Cell 13: 101–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bariola PA, Retelska D, Stasiak A, Kammerer RA, Fleming A, Hijri M, Frank S, Farmer EE (2004) Remorins form a novel family of coiled coil-forming oligomeric and filamentous proteins associated with apical, vascular and embryonic tissues in plants. Plant Mol Biol 55: 579–594 [DOI] [PubMed] [Google Scholar]
- Barlier I, Kowalczyk M, Marchant A, Ljung K, Bhalerao R, Bennett M, Sandberg G, Bellini C (2000) The SUR2 gene of Arabidopsis thaliana encodes the cytochrome P450 CYP83B1, a modulator of auxin homeostasis. Proc Natl Acad Sci USA 97: 14819–14824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer EM, Mongrand S, Tilsner J (2014) Specialized membrane domains of plasmodesmata, plant intercellular nanopores. Front Plant Sci 5: 507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M, Heard W, Mbengue M, Robatzek S (2012) The INs and OUTs of pattern recognition receptors at the cell surface. Curr Opin Plant Biol 15: 367–374 [DOI] [PubMed] [Google Scholar]
- Becker W. (2012) Fluorescence lifetime imaging: techniques and applications. J Microsc 247: 119–136 [DOI] [PubMed] [Google Scholar]
- Benschop JJ, Mohammed S, O’Flaherty M, Heck AJ, Slijper M, Menke FL (2007) Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol Cell Proteomics 6: 1198–1214 [DOI] [PubMed] [Google Scholar]
- Borner GH, Sherrier DJ, Weimar T, Michaelson LV, Hawkins ND, Macaskill A, Napier JA, Beale MH, Lilley KS, Dupree P (2005) Analysis of detergent-resistant membranes in Arabidopsis: evidence for plasma membrane lipid rafts. Plant Physiol 137: 104–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botchway SW, Scherer KM, Hook S, Stubbs CD, Weston E, Bisby RH, Parker AW (2015) A series of flexible design adaptations to the Nikon E-C1 and E-C2 confocal microscope systems for UV, multiphoton and FLIM imaging. J Microsc 258: 68–78 [DOI] [PubMed] [Google Scholar]
- Brandizzi F, Hanton S, Pinto DaSilva LL, Boevink P, Evans D, Oparka K, Denecke J, Hawes C (2003) ER quality control can lead to retrograde transport from the ER lumen to the cytoplasm and the nucleoplasm in plants. Plant J 34: 269–281 [DOI] [PubMed] [Google Scholar]
- Carette JE, Verver J, Martens J, van Kampen T, Wellink J, van Kammen A (2002) Characterization of plant proteins that interact with cowpea mosaic virus ‘60K’ protein in the yeast two-hybrid system. J Gen Virol 83: 885–893 [DOI] [PubMed] [Google Scholar]
- Chen J, Doyle C, Qi A, Zheng H (2012) The endoplasmic reticulum: a social network in plant cells. J Integr Plant Biol 54: 840–850 [DOI] [PubMed] [Google Scholar]
- Diener AC, Li H, Zhou W, Whoriskey WJ, Nes WD, Fink GR (2000) STEROL METHYLTRANSFERASE 1 controls the level of cholesterol in plants. Plant Cell 12: 853–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers K, Kollmann R (2001) Primary and secondary plasmodesmata: structure, origin, and functioning. Protoplasma 216: 1–30 [DOI] [PubMed] [Google Scholar]
- Faulkner C, Petutschnig E, Benitez-Alfonso Y, Beck M, Robatzek S, Lipka V, Maule AJ (2013) LYM2-dependent chitin perception limits molecular flux via plasmodesmata. Proc Natl Acad Sci USA 110: 9166–9170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Calvino L, Faulkner C, Walshaw J, Saalbach G, Bayer E, Benitez-Alfonso Y, Maule A (2011) Arabidopsis plasmodesmal proteome. PLoS One 6: e18880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fichtenbauer D, Xu XM, Jackson D, Kragler F (2012) The chaperonin CCT8 facilitates spread of tobamovirus infection. Plant Signal Behav 7: 318–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förster T. (1948) Zwischenmolekulare Energiewanderung und Fluoreszenz. Ann Phys 437: 55–75 [Google Scholar]
- Friml J, Jones AR (2010) Endoplasmic reticulum: the rising compartment in auxin biology. Plant Physiol 154: 458–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giordano F, Saheki Y, Idevall-Hagren O, Colombo SF, Pirruccello M, Milosevic I, Gracheva EO, Bagriantsev SN, Borgese N, De Camilli P (2013) PI(4,5)P(2)-dependent and Ca(2+)-regulated ER-PM interactions mediated by the extended synaptotagmins. Cell 153: 1494–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glawischnig E, Mikkelsen MD, Halkier BA (2003) Glucosinolates: biosynthesis and metabolism. In Abrol Y, Ahmad A, eds, Sulphur in Plants. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 145–162 [Google Scholar]
- Grison MS, Brocard L, Fouillen L, Nicolas W, Wewer V, Dörmann P, Nacir H, Benitez-Alfonso Y, Claverol S, Germain V, et al. (2015) Specific membrane lipid composition is important for plasmodesmata function in Arabidopsis. Plant Cell 27: 1228–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachez C, Laloux T, Reinhardt H, Cavez D, Degand H, Grefen C, De Rycke R, Inzé D, Blatt MR, Russinova E, et al. (2014a) Arabidopsis SNAREs SYP61 and SYP121 coordinate the trafficking of plasma membrane aquaporin PIP2;7 to modulate the cell membrane water permeability. Plant Cell 26: 3132–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hachez C, Veljanovski V, Reinhardt H, Guillaumot D, Vanhee C, Chaumont F, Batoko H (2014b) The Arabidopsis abiotic stress-induced TSPO-related protein reduces cell-surface expression of the aquaporin PIP2;7 through protein-protein interactions and autophagic degradation. Plant Cell 26: 4974–4990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawes C, Kiviniemi P, Kriechbaumer V (2015) The endoplasmic reticulum: a dynamic and well-connected organelle. J Integr Plant Biol 57: 50–62 [DOI] [PubMed] [Google Scholar]
- Herman EM. (2008) Endoplasmic reticulum bodies: solving the insoluble. Curr Opin Plant Biol 11: 672–679 [DOI] [PubMed] [Google Scholar]
- Huang AHC. (1996) Oleosins and oil bodies in seeds and other organs. Plant Physiol 110: 1055–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarsch IK, Ott T (2011) Perspectives on remorin proteins, membrane rafts, and their role during plant-microbe interactions. Mol Plant Microbe Interact 24: 7–12 [DOI] [PubMed] [Google Scholar]
- Karimi M, De Meyer B, Hilson P (2005) Molecular cloning in plant cells. Trends Plant Sci 10: 103–105 [DOI] [PubMed] [Google Scholar]
- Kessner D, Chambers M, Burke R, Agus D, Mallick P (2008) ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics 24: 2534–2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knox K, Wang P, Kriechbaumer V, Tilsner J, Frigerio L, Sparkes I, Hawes C, Oparka KJ (2015) Putting the squeeze on plasmodesmata: a role for reticulons in primary plasmodesmata formation. Plant Physiol 168: 1563–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H, Sparkes I, Gattolin S, Dzimitrowicz N, Roberts LM, Hawes C, Frigerio L (2013) An Arabidopsis reticulon and the atlastin homologue RHD3-like2 act together in shaping the tubular endoplasmic reticulum. New Phytol 197: 481–489 [DOI] [PubMed] [Google Scholar]
- Lee HY, Bowen CH, Popescu GV, Kang HG, Kato N, Ma S, Dinesh-Kumar S, Snyder M, Popescu SC (2011) Arabidopsis RTNLB1 and RTNLB2 Reticulon-like proteins regulate intracellular trafficking and activity of the FLS2 immune receptor. Plant Cell 23: 3374–3091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerouxel O, Mouille G, Andème-Onzighi C, Bruyant MP, Séveno M, Loutelier-Bourhis C, Driouich A, Höfte H, Lerouge P (2005) Mutants in DEFECTIVE GLYCOSYLATION, an Arabidopsis homolog of an oligosaccharyltransferase complex subunit, show protein underglycosylation and defects in cell differentiation and growth. Plant J 42: 455–468 [DOI] [PubMed] [Google Scholar]
- Levy A, Zheng JY, Lazarowitz SG (2015) Synaptotagmin SYTA forms ER-plasma membrane junctions that are recruited to plasmodesmata for plant virus movement. Curr Biol 3: 2018–2025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis JD, Lazarowitz SG (2010) Arabidopsis synaptotagmin SYTA regulates endocytosis and virus movement protein cell-to-cell transport. Proc Natl Acad Sci USA 107: 2491–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CC, Seikowski J, Pérez-Lara A, Jahn R, Höbartner C, Walla PJ (2014) Control of membrane gaps by synaptotagmin-Ca2+ measured with a novel membrane distance ruler. Nat Commun 5: 5859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marín M, Thallmair V, Ott T (2012) The intrinsically disordered N-terminal region of AtREM13 remorin protein mediates protein-protein interactions. J Biol Chem 287: 39982–39991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan J, Zipfel C (2012) Plant pattern recognition receptor complexes at the plasma membrane. Curr Opin Plant Biol 15: 349–357 [DOI] [PubMed] [Google Scholar]
- Mongrand S, Stanislas T, Bayer EM, Lherminier J, Simon-Plas F (2010) Membrane rafts in plant cells. Trends Plant Sci 15: 656–663 [DOI] [PubMed] [Google Scholar]
- Naulin PA, Alveal NA, Barrera NP (2014) Toward atomic force microscopy and mass spectrometry to visualize and identify lipid rafts in plasmodesmata. Front Plant Sci 5: 234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson DC, Flematti GR, Riseborough JA, Ghisalberti EL, Dixon KW, Smith SM (2010) Karrikins enhance light responses during germination and seedling development in Arabidopsis thaliana. Proc Natl Acad Sci USA 107: 7095–7100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75: 4646–4658 [DOI] [PubMed] [Google Scholar]
- Oparka KJ, Prior DA, Santa Cruz S, Padgett HS, Beachy RN (1997) Gating of epidermal plasmodesmata is restricted to the leading edge of expanding infection sites of tobacco mosaic virus (TMV). Plant J 12: 781–789 [DOI] [PubMed] [Google Scholar]
- Osterrieder A, Carvalho CM, Latijnhouwers M, Johansen JN, Stubbs C, Botchway S, Hawes C (2009) Fluorescence lifetime imaging of interactions between Golgi tethering factors and small GTPases in plants. Traffic 10: 1034–1046 [DOI] [PubMed] [Google Scholar]
- Overall RL, Blackman LM (1996) A model of the macro-molecular structure of plasmodesmata. Trends Plant Sci 1: 307–311 [Google Scholar]
- Palade G. (1975) Intracellular aspects of the process of protein synthesis. Science 189: 347–358 [DOI] [PubMed] [Google Scholar]
- Pattison RJ, Amtmann A (2009) N-Glycan production in the endoplasmic reticulum of plants. Trends Plant Sci 14: 92–99 [DOI] [PubMed] [Google Scholar]
- Pérez-Sancho J, Vanneste S, Lee E, McFarlane HE, Esteban Del Valle A, Valpuesta V, Friml J, Botella MA, Rosado A (2015) The Arabidopsis synaptotagmin1 is enriched in endoplasmic reticulum-plasma membrane contact sites and confers cellular resistance to mechanical stresses. Plant Physiol 168: 132–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perraki A, Binaghi M, Mecchia MA, Gronnier J, German-Retana S, Mongrand S, Bayer E, Zelada AM, Germain V (2014) StRemorin13 hampers Potato virus X TGBp1 ability to increase plasmodesmata permeability, but does not interfere with its silencing suppressor activity. FEBS Lett 588: 1699–1705 [DOI] [PubMed] [Google Scholar]
- Perraki A, Cacas JL, Crowet JM, Lins L, Castroviejo M, German-Retana S, Mongrand S, Raffaele S (2012) Plasma membrane localization of Solanum tuberosum remorin from group 1, homolog 3 is mediated by conformational changes in a novel C-terminal anchor and required for the restriction of potato virus X movement. Plant Physiol 160: 624–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffaele S, Bayer E, Lafarge D, Cluzet S, German Retana S, Boubekeur T, Leborgne-Castel N, Carde JP, Lherminier J, Noirot E, et al. (2009) Remorin, a Solanaceae protein resident in membrane rafts and plasmodesmata, impairs potato virus X movement. Plant Cell 21: 1541–1555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schapire AL, Voigt B, Jasik J, Rosado A, Lopez-Cobollo R, Menzel D, Salinas J, Mancuso S, Valpuesta V, Baluska F, et al. (2008) Arabidopsis synaptotagmin 1 is required for the maintenance of plasma membrane integrity and cell viability. Plant Cell 20: 3374–3388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoberer J, Botchway SW (2014) Investigating protein-protein interactions in the plant endomembrane system using multiphoton-induced FRET-FLIM. Methods Mol Biol 1209: 81–95 [DOI] [PubMed] [Google Scholar]
- Scholl RL, May ST, Ware DH (2000) Seed and molecular resources for Arabidopsis. Plant Physiol 124: 1477–1480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes IA, Runions J, Kearns A, Hawes C (2006) Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc 1: 2019–2025 [DOI] [PubMed] [Google Scholar]
- Sparkes I, Runions J, Hawes C, Griffing L (2009) Movement and remodeling of the endoplasmic reticulum in nondividing cells of tobacco leaves. Plant Cell 21: 3937–3949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I, Tolley N, Aller I, Svozil J, Osterrieder A, Botchway S, Mueller C, Frigerio L, Hawes C (2010) Five Arabidopsis reticulon isoforms share endoplasmic reticulum location, topology, and membrane-shaping properties. Plant Cell 22: 1333–1343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs CD, Botchway SW, Slater SJ, Parker AW (2005) The use of time-resolved fluorescence imaging in the study of protein kinase C localisation in cells. BMC Cell Biol 6: 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapken W, Murphy AS (2015) Membrane nanodomains in plants: capturing form, function, and movement. J Exp Bot 66: 1573–1586 [DOI] [PubMed] [Google Scholar]
- Tilsner J, Amari K, Torrance L (2011) Plasmodesmata viewed as specialised membrane adhesion sites. Protoplasma 248: 39–60 [DOI] [PubMed] [Google Scholar]
- Tilsner J, Linnik O, Louveaux M, Roberts IM, Chapman SN, Oparka KJ (2013) Replication and trafficking of a plant virus are coupled at the entrances of plasmodesmata. J Cell Biol 201: 981–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolley N, Sparkes I, Craddock CP, Eastmond PJ, Runions J, Hawes C, Frigerio L (2010) Transmembrane domain length is responsible for the ability of a plant reticulon to shape endoplasmic reticulum tubules in vivo. Plant J 64: 411–418 [DOI] [PubMed] [Google Scholar]
- Tolley N, Sparkes IA, Hunter PR, Craddock CP, Nuttall J, Roberts LM, Hawes C, Pedrazzini E, Frigerio L (2008) Overexpression of a plant reticulon remodels the lumen of the cortical endoplasmic reticulum but does not perturb protein transport. Traffic 9: 94–102 [DOI] [PubMed] [Google Scholar]
- Tucker EB, Boss WF (1996) Mastoparan-induced intracellular Ca2+ fluxes may regulate cell-to-cell communication in plants. Plant Physiol 111: 459–467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiyama A, Shimada-Beltran H, Levy A, Zheng JY, Javia PA, Lazarowitz SG (2014) The Arabidopsis synaptotagmin SYTA regulates the cell-to-cell movement of diverse plant viruses. Front Plant Sci 5: 584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitale A, Denecke J (1999) The endoplasmic reticulum: gateway of the secretory pathway. Plant Cell 11: 615–628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallis JG, Browse J (2010) Lipid biochemists salute the genome. Plant J 61: 1092–1106 [DOI] [PubMed] [Google Scholar]
- Wang P, Hawkins TJ, Richardson C Cummins I, Deeks MJ, Sparkes I, Hawes C, Hussey PJ (2014) The plant cytoskeleton, NET3C, and VAP27 mediate the link between the plasma membrane and endoplasmic reticulum. Curr Biol 24: 1397–1405 [DOI] [PubMed] [Google Scholar]
- Winter D, Vinegar B, Nahal H, Ammar R, Wilson GV, Provart NJ (2007) An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XM, Wang J, Xuan Z, Goldshmidt A, Borrill PG, Hariharan N, Kim JY, Jackson D (2011) Chaperonins facilitate KNOTTED1 cell-to-cell trafficking and stem cell function. Science 333: 1141–1144 [DOI] [PubMed] [Google Scholar]
- Yamazaki T, Takata N, Uemura M, Kawamura Y (2010) Arabidopsis synaptotagmin SYT1, a type I signal-anchor protein, requires tandem C2 domains for delivery to the plasma membrane. J Biol Chem 285: 23165–23176 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.