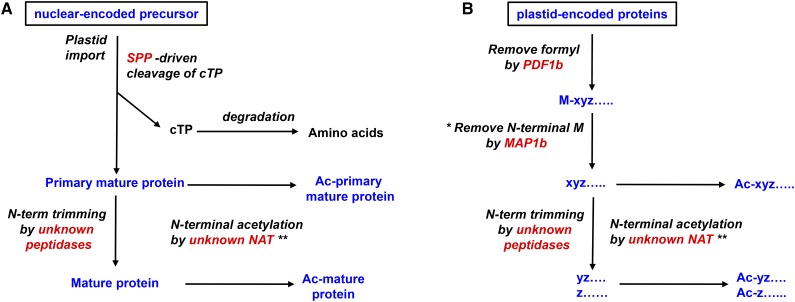
Following cleavage of chloroplast transit peptides by stromal processing peptidase, additional processing may occur, avoiding unstable or otherwise unfavorable N-terminal residues.
Abstract
Protein amino (N) termini are prone to modifications and are major determinants of protein stability in bacteria, eukaryotes, and perhaps also in chloroplasts. Most chloroplast proteins undergo N-terminal maturation, but this is poorly understood due to insufficient experimental information. Consequently, N termini of mature chloroplast proteins cannot be accurately predicted. This motivated an extensive characterization of chloroplast protein N termini in Arabidopsis (Arabidopsis thaliana) using terminal amine isotopic labeling of substrates and mass spectrometry, generating nearly 14,000 tandem mass spectrometry spectra matching to protein N termini. Many nucleus-encoded plastid proteins accumulated with two or three different N termini; we evaluated the significance of these different proteoforms. Alanine, valine, threonine (often in N-α-acetylated form), and serine were by far the most observed N-terminal residues, even after normalization for their frequency in the plastid proteome, while other residues were absent or highly underrepresented. Plastid-encoded proteins showed a comparable distribution of N-terminal residues, but with a higher frequency of methionine. Infrequent residues (e.g. isoleucine, arginine, cysteine, proline, aspartate, and glutamate) were observed for several abundant proteins (e.g. heat shock proteins 70 and 90, Rubisco large subunit, and ferredoxin-glutamate synthase), likely reflecting functional regulation through their N termini. In contrast, the thylakoid lumenal proteome showed a wide diversity of N-terminal residues, including those typically associated with instability (aspartate, glutamate, leucine, and phenylalanine). We propose that, after cleavage of the chloroplast transit peptide by stromal processing peptidase, additional processing by unidentified peptidases occurs to avoid unstable or otherwise unfavorable N-terminal residues. The possibility of a chloroplast N-end rule is discussed.
Following synthesis, most proteins undergo various N-terminal (Nt) protein modifications, including removal of the Nt Met and signal peptide, N-terminal α-acetylation (NAA), ubiquitination, and acylations. These Nt modifications play an important role in the regulation of cellular functions. The N terminus of proteins has also been shown to be a major determinant of protein stability in bacteria (Varshavsky, 2011), eukaryotes (Graciet et al., 2009), mitochondria, and perhaps in plastids/chloroplasts (Apel et al., 2010; Nishimura et al., 2013; van Wijk, 2015). The role of the N terminus in protein stability is conceptualized in the N-end rule, which states that certain amino acids, when exposed at the N terminus of a protein, act as triggers for degradation (Bachmair et al., 1986; Dougan et al., 2012; Tasaki et al., 2012; Gibbs et al., 2014).
Most of the approximately 3,000 plastid proteins are nucleus encoded (n-encoded) and are targeted to the plastid through an Nt chloroplast transit peptide (cTP). After import, the cTP is cleaved by the stromal processing peptidase (SPP; Richter and Lamppa, 1998; Trösch and Jarvis, 2011). The consensus site of cTP cleavage by SPP is only loosely defined, and the rules, mechanisms, and enzymes for possible subsequent processing, stabilization, and other posttranslational modifications (PTMs) are not well characterized (for discussion, see van Wijk, 2015). The exact N terminus is unknown for many chloroplast proteins and cannot be accurately predicted, because SPP specificity is not sufficiently understood (Emanuelsson et al., 2000; Zybailov et al., 2008) and probably also because additional Nt processing occurs for many chloroplast proteins (Fig. 1A). The approximately 85 plastid-encoded (p-encoded) proteins typically first undergo cotranslational Nt deformylation, followed by N-terminal Met excision (NME; Giglione et al., 2009; Fig. 1B); both these PTMs are required for normal plastid/chloroplast development and protein stability (Dirk et al., 2001, 2002; Giglione et al., 2003; Meinnel et al., 2006). Both n-encoded and p-encoded proteins can undergo NAA inside the plastid (Zybailov et al., 2008; Fig. 1). Postulated functions of NAA in eukaryotes include the mediation of protein location, assembly, and stability (Jones and O’Connor, 2011; Starheim et al., 2012; Hoshiyasu et al., 2013; Xu et al., 2015), thereby affecting a variety of processes, including drought tolerance in Arabidopsis (Arabidopsis thaliana; Linster et al., 2015).
Figure 1.
Conceptual illustration of Nt maturation of n-encoded and p-encoded proteins. Ac, Acetylated; MAP, Met amino peptidase; NAT, N-acetyltransferase; N-term, N-terminal; PDF, peptide deformylase. A, Nt maturation of n-encoded plastid proteins including removal of cTP by SPP and potential subsequent Nt modifications. B, Nt maturation of p-encoded proteins. *, The removal depends on the penultimate residue, generally following the N-terminal Met Excision (NME) rule; **, N-terminal acetylation typically occurs only for selected residues; “Results”).
Typical proteomics work flows generally yield only partial coverage of protein sequences, and it is often difficult to know which peptides represent the true N termini (Nti) or C termini. Systematic identification of Nti or C termini requires specific labeling and enrichment strategies, such as combined fractional diagonal chromatography, developed by Gevaert and colleagues (Staes et al., 2011), and terminal amine isotopic labeling of substrates (TAILS), developed by the group of Overall (Kleifeld et al., 2011; Lange and Overall, 2013). These strategies allow the identification of different Nt proteoforms and were recently also applied to plants (Tsiatsiani et al., 2013; Carrie et al., 2015; Kohler et al., 2015; Zhang et al., 2015) and diatoms (Huesgen et al., 2013). We previously reported on Nti of chloroplast proteins based on tandem mass spectrometry (MS/MS) analysis, but because no Nt enrichment/labeling technique was used, only those that underwent NAA could be considered bona fide Nti (Zybailov et al., 2008). Nt Edman degradation sequencing was systematically carried out for thylakoid lumen proteins (Peltier et al., 2000, 2002) but not for stromal proteins or chloroplast membrane proteins with their Nti exposed to the stroma. The Nti of thylakoid lumen proteins are mostly generated by lumenal peptidases (Hsu et al., 2011; Midorikawa et al., 2014), and the thylakoid lumen contains a different set of peptidases than the stroma; hence, rules for Nt maturation and stability are likely different than those for stroma-exposed proteins.
The objective of this study was to systematically determine the Nti of stroma-exposed chloroplast proteins of Arabidopsis (the N-terminome) and to provide a baseline for understanding Nt protein maturation and protein stability in the chloroplast stroma. To that end, we applied the TAILS technique and determined the Nti of approximately 250 chloroplast proteins by mass spectrometry (MS). We observed that many n-encoded plastid proteins accumulated with two or even three different Nt residues, in many cases both with and without NAA. The extent of accumulation of different Nt proteoforms is surprising and will be discussed. The p-encoded proteins generally showed very similar Nt residues as compared with the n-encoded proteins, with the exception of Met. Our data show that small, apolar, or hydroxylated residues (Ala, Val, Ser, and Thr) are the most frequent Nt residues of stromal proteins, whereas other residues are strictly avoided or are only present for very specific proteins likely to aid in their function. Chloroplast protein degradation products were also detected, with enrichment for peptides generated by cleavage between Arg and Thr residues. We present testable hypotheses for understanding Nt processing and maturation, stability, and a possible N-end rule in chloroplast stroma.
RESULTS
Systematic Identification of Protein Nti
To systematically identify the Nti of chloroplast proteins, we employed the TAILS method for labeling and enrichment of chloroplast protein Nti, followed by MS/MS-based identification (Kleifeld et al., 2011). The TAILS work flow removes the internal non-Nt peptides, whereas both unmodified (free) α-amino Nti and NAA Nti are retained, greatly simplifying the remaining proteome. For a general description of the TAILS method, we refer to excellent articles from Overall and colleagues (Kleifeld et al., 2011; Lange and Overall, 2013), “Materials and Methods,” and Supplemental Figure S1A. In brief, the TAILS method involves, first, the dimethyl labeling of the free Nt α-amines as well ε-amines of Lys residues. Following digestion with proteases (here trypsin or endoproteinase GluC), the unmodified peptides are removed by cross-linking to a specialized polymer matrix, allowing the collection of dimethyl-labeled peptides as well as peptides with NAA Nti. As a starting material, we used developed leaf rosettes of soil-grown Arabidopsis plants, analyzing both soluble stromal protein extracts from isolated chloroplasts (four independent preparations, 10 TAILS experiments) as well as total soluble leaf protein extracts (three independent preparations, nine TAILS experiments). Comparison of Nt sequences of these leaf extracts and chloroplast stromal extracts allowed us to consider processing of dual-targeted chloroplast proteins (i.e. also targeted to other subcellular locations, in particular mitochondria) and to identify chloroplast precursor proteins (i.e. with their cTP). Protein recovery across the labeling and enrichment steps was verified by SDS-PAGE followed by silver staining (Supplemental Fig. S1B). Dimethyl labeling efficiency and proteolytic digestion were monitored by liquid chromatography-MS/MS analysis of each sample prior to the negative selection step. This showed that more than 99% of Lys residues were dimethylated, indicating nearly quantitative labeling, which allowed a semiquantitative comparison of different Nti proteoforms.
Assessment and Filtering of Nt Sequences
All MS/MS search results were pooled and filtered to identify only Nt-labeled peptides. Of the complete set of acquired MS/MS spectra across all experiments, 13,858 spectra matched to Nt peptides (Supplemental Table S1). We then pooled the Nt peptides with the same molecular mass and sequence (irrespective of charge state), resulting in 1,037 nonredundant Nti matching to 577 proteins. Matched proteins were annotated for subcellular location to aid in the identification of subcellular Nt maturation events (Supplemental Table S2). Peptides starting with the same Nt modification and amino acid sequence, but with different C-terminal ends or different modified side chains, were merged into 894 Nti matching to 577 proteins (Supplemental Table S3). Importantly, these overlapping peptides strengthened Nt identifications. We did not condense peptide sequences with or without NAA, because these NAA sequences should be considered functionally distinct from their unmodified sequences. A total of 544 of these merged Nti matched to 250 plastid proteins, and the remaining peptides matched to proteins located in other subcellular compartments or without assigned subcellular locations (Supplemental Table S3).
The two main objectives of this study were (1) to develop a working hypothesis for cTP cleavage specificity and subsequent maturation steps and (2) to determine underrepresentation or overrepresentation of specific amino acids at the Nti for the steady-state, stroma-exposed proteins and deduce potential Nt stability rules. Chloroplast proteins with their Nti exposed to the plastid stroma, lumen, or intraenvelope space or facing the cytoplasm should be considered separately, because each of these compartments has its own set of peptidases and possibly involves different maturation steps. Hence, we carefully evaluated the intraplastid location for each identified protein. Those encoded by the plastid genome also represent a distinct set, since they undergo specific cotranslational and posttranslational processing (Fig. 1B). Sixteen of the detected n-encoded chloroplast proteins are known to be dual targeted to chloroplasts and mitochondria or cytosol (Carrie and Small, 2013; Supplemental Table S3). For most of these dual-targeted proteins, we identified a single N terminus, which appeared to represent the chloroplast-localized form. This is not surprising, because we used either protein extracts from photosynthetic leaves, in which chloroplast proteins are far more abundant than mitochondrial proteins, or isolated chloroplasts. Three dual-targeted plastid/cytosolic proteins (Arabidopsis Dynamin-like1, Glycolate oxidase1, and glutathione S-transferase phi) were only identified in their cytosolic forms and were not further considered for chloroplast N-terminome analysis. In the remaining analysis, we will focus on the n-encoded or p-encoded proteins that have stroma-exposed Nti. We note that, even if these proteins have their Nti facing the stroma, they may actually be buried within the protein structure and, thus, only truly exposed to the stroma during biogenesis or degradation.
Nt Amino Acid Frequency and Acetylation State of n-Encoded Chloroplast Proteins
For 126 plastid proteins, only a single N terminus was identified; examples are shown in Table I, scenario A. These proteins are interesting because other Nt proteoforms of these proteins must be quickly degraded, or the SPP cleaves the cTPs at only a single location, or additional peptidases trim the Nti to a single proteoform. Multiple Nti were detected for approximately 100 proteins representing three different possibilities. (1) Nt peptides well upstream of the predicted or previously documented mature N terminus (for approximately a dozen proteins; Table I, scenario B). These Nt peptides were generally found in the total leaf extracts rather than stromal extracts. This suggests that these upstream Nti were from proteins not yet imported into the chloroplast. (2) Proteins with multiple closely spaced Nti that each could represent the mature N terminus of the respective protein (Table I, scenario C). In most cases, a single N terminus had both the highest number of spectral counts (SPC; these are matched MS/MS spectra) and the most Nt residue, thus representing the most likely candidate for the N terminus of the steady-state protein. (3) Nti of degradation products. A total of 129 Nti (matching to 31 n-encoded and four p-encoded proteins) were likely degradation products (see “Accumulation of Proteolytic Products”).
Table I. Examples of experimentally determined Nt peptides for selected n-encoded proteins demonstrating three different physiological scenarios.
A number of details are provided: Nt residue position for mature protein N terminus (predicted/experimental), the residue immediately upstream of the observed Nt peptide (Prev. AA), the Nt modification (TAILS), and the number of matched MS/MS spectra (SPC). Examples include cases where the predicted N terminus is upstream, downstream, or identical to the observed N terminus. SPC values for scenario B are expressed as stroma/leaf.
| Scenario | Protein Name | N Terminus | Prev. AA | Nt Peptide | TAILS | SPC |
|---|---|---|---|---|---|---|
| Scenario Aa | ||||||
| AT1G35680 | 50S ribosomal L21 | 84/66 | F | AESVVEAEPETTDIEAVVVSDVSEVTEEKAKR | Dimethyl | 46 |
| AT4G09650 | CF1d-atpD | 49/48 | M | SATAASSYAMALADVAKR | Dimethyl | 51 |
| AT3G27830 | 50S ribosomal L12-A | 59/59 | A | AVEAPEKIEKIGSEISSLTLEEAR | Dimethyl | 161 |
| AT1G54630 | ACP3 plastid | 52/53 | C | AAKPETVDKVCAVVR | Dimethyl | 73 |
| AT4G23100 | GSH1 | 74/75 | A | ASPPTEEAVVATEPLTR | Dimethyl | 58 |
| AT5G04140 | Fd-GOGAT1 | 63/106 | A | CGVGFIANLDNIPSHGVVKDALIALGCMEHR | Dimethyl | 6 |
| Scenario Bb | ||||||
| AT1G67090 | RBCS4 | 55/2 | M | ASSMLSSATMVASPAQATMVAPFNGLKSSAAFPATR | Acetyl | 0/3 |
| 55/56 | C | MQVWPPIGKKKFETLSYLPDLTDSELAKEVDYLIR | Dimethyl | 9/3 | ||
| AT5G38410 | RBCS3B | 55/2 | M | ASSMLSSAAVVTSPAQATMVAPFTGLKSSAAFPVTR | Acetyl | 0/3 |
| 55/56 | C | MKVWPPIGKKKFETLSYLPDLSDVELAKEVDYLLR | Dimethyl | 5/0 | ||
| AT4G38970 | SFBA2 | 47/2 | M | ASTSLLKASPVLDKSEWVKGQSVLFR | Acetyl | 0/1 |
| 47/47 | R | AASSYADELVKTAKTIASPGR | Dimethyl | 0/26 | ||
| 47/48 | A | ASSYADELVKTAKTIASPGR | Dimethyl | 85/119 | ||
| Scenario Cc | ||||||
| AT3G60750 | TRANSKETOLASE1 | 66/66 | R | AAAVETVEPTTDSSIVDKSVNSIR | Dimethyl | 37 |
| 66/67 | A | AAVETVEPTTDSSIVDK | Acetyl | 4 | ||
| 66/67 | A | AAVETVEPTTDSSIVDKSVNSIR | Dimethyl | 215 | ||
| 6668 | A | AVETVEPTTDSSIVDKSVNSIR | Dimethyl | 74 | ||
| AT4G24280 | cpHSP70-1 | 93/75 | R | VVNEKVVGIDLGTTNSAVAAMEGGKPTIVTNAEGQR | Dimethyl | 16 |
| 93/78 | N | EKVVGIDLGTTNSAVAAMEGGKPTIVTNAEGQR | Dimethyl | 71 | ||
| AT4G24830 | Argininosuccinate synthase | 74/74 | R | AVLSGDGTALTTDSKEAGLR | Dimethyl | 17 |
| 74/75 | A | VLSGDGTALTTDSKEAGLR | Acetyl | 22 | ||
| 74/75 | A | VLSGDGTALTTDSKEAGLR | Dimethyl | 6 |
Scenario A is proteins for which only a single Nt peptide was identified with multiple MS/MS spectra.
The Nt amino acid frequency for all mature n-encoded chloroplast Nti was calculated (Fig. 2A; Supplemental Table S4). This demonstrates that Ala and Ser are heavily favored as Nt residues, followed by Val and Thr, whereas the remaining residues are underrepresented (in particular Asp, Tyr, and Trp each only once) or not observed at all (Pro and His; see legend to Fig. 2A). The ratio between NAA and unmodified (but dimethylated in the TAILS procedure) Nti can be approximated based on matched MS/MS spectra, in particular if a relatively high total number of MS/MS sequences (e.g. more than 50) are obtained. The NAA rates for the high-frequency residues Val, Thr, Ala, and Ser were 54%, 47%, 21%, and 19%, respectively. The few cases of Trp, Arg, Ile, and Pro were mostly in NAA form, whereas NAA was not observed for Tyr, Leu, Phe, Asp, and Cys.
Figure 2.
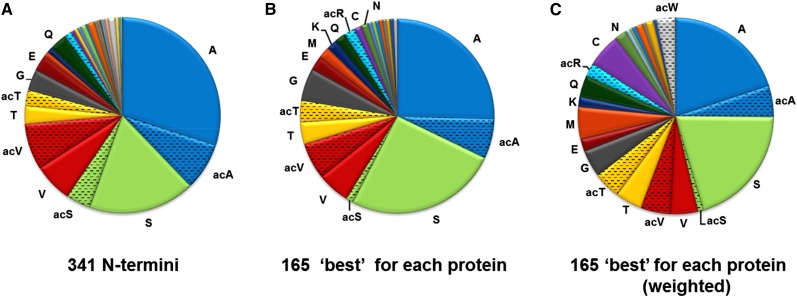
Nt amino acid frequency for stroma-exposed n-encoded chloroplast proteins. A, All detected stromal Nti (341), excluding unprocessed proteins and obvious breakdown products (Supplemental Table S4). This shows that Ala and Ser are heavily favored as Nt residues, followed by Val and Thr, while 14 residues were underrepresented (Gly, 14×; Gln, 14×; Glu, 10×; Ile, 6×; Arg, 5×; Lys, 5×; Met, 4×; Asn, 3×; Leu, 3×; Cys, 2×; Phe, 2×; Trp, 1×; Tyr, 1×; and Asp, 1×) or not observed (Pro and His). A significant portion of these highly favored residues were acetylated (Val, 54%; Thr, 47%; Ala, 21%; and Ser, 19%), whereas the acetylation rate for other residues was either 0% (Tyr, Leu, Phe, Asp, and Cys) or 100% (Trp; acetylation is indicated as ac). B, Single highest ranked N terminus per protein (165), excluding Nti with less than two SPC. Selecting a single best or highest ranked N terminus for each protein (see “Materials and Methods”) hardly influenced the Nt amino acid frequency, except that it slightly decreased the dominance of Ala, increased Ser, and reduced acetylated Ser. Less frequent residues were Gly (9×), Glu (5×), Gln (4×), Lys (4×), Arg (3×), Met (3×), Asn (2×), Cys (2×), Leu (1×), Ile (1×), Trp (1×), Tyr (1×), and Asp (1×), whereas Phe, Pro, and His were not observed. C, Single highest ranked Nti as in B but normalized (weighted) to the frequency of each amino acid in the known (from the Plant Proteome Data Base [PPDB]; 1,575 proteins) n-encoded plastid proteome with predicted cTPs removed.
Because many proteins were present as different Nt proteoforms, we ranked Nt peptides for each protein such that a single representative N terminus for each protein could be assigned. This ranking was based on the number of observed spectra, the proximity to the predicted cTP cleavage site (ChloroP), and, if available, previously published Nt sequence data (for Nt ranks and for a description of the ranking process, see Supplemental Table S4). Importantly, selecting a single best-ranked Nt for each protein hardly influenced the frequency distribution of the amino acid at the N terminus (Fig. 2B). Moreover, Ile, Leu, Try, Tyr, and Asp were each found only once as best-ranked Nt residue, whereas Pro, His, and Phe were not observed at all. Interestingly, Nt Arg (three) and Trp (one) were only found in their NAA form, perhaps suggesting that NAA is needed for stabilization.
Some amino acids are far more frequent in the known chloroplast proteome than others (Leu is the most frequent [approximately 9.5%], followed by Ala, Ser, and Val [each approximately 7.7%]), whereas His, Cys, and Trp are the least abundant (1%–2%), possibly biasing the Nt amino acid frequencies. Therefore, the frequencies of these 165 best-ranked Nti were normalized to the natural frequency of each amino acid in the known n-encoded plastid proteome (1,575 proteins; see “Materials and Methods”) with predicted cTPs removed (Fig. 2C). This showed again that Ala, Ser, and, to a lesser extent, Val and Thr (in NAA and free form) are still strongly favored, whereas Met and Cys are more prominent than before weighing (compare with Fig. 2A), and Leu is clearly avoided.
Physiological Nt Methylation
The four paralogs of Rubisco small subunit (RBCS) were the only observed n-encoded mature proteins that started with an Nt Met residue. This Nt Met of RBCS has been shown previously to be methylated at its N terminus through the activity of Rubisco methyltransferase (Houtz et al., 2008). The dimethylation reaction used in the TAILS method would mask this physiological (mono)methylation, since it generates a dimethylated N terminus. To asses if in vivo Nt methylation occurs in chloroplasts for other proteins, TAILS experiments were also performed with deuterated formaldehyde (CD2O) instead of formaldehyde (CH2O), which allowed us to differentiate between natural methylation and methylation by formaldehyde. Indeed, we observed that RBCS4 (Supplemental Fig. S2) and RBCS1b (data not shown) accumulated with in vivo monomethylated Nti Met. No other convincing cases for Nt methylation were detected, which is perhaps not surprising because we observed so few mature n-encoded proteins (only RBCS family members) that start with a Met residue. The lack of observed Nt Met in the n-encoded stroma-exposed proteome suggests very efficient NME. The lack of NME for just RBCS is likely due to the presence of a bulky residue (Lys) immediately after the Met. It should be noted that Lys methylation has been observed for several Arabidopsis chloroplast proteins downstream of their mature Nti (Zybailov et al., 2009; Alban et al., 2014). Lys-14 of RBCL has been shown in pea (Pisum sativum) to be (tri)methylated (Houtz et al., 2008). However, we found no evidence for such a modification in Arabidopsis (the detected Nt peptide of RBCL is long enough to include this Lys: SPQTETKASVGFKAGVKEY), in agreement with a recent study indicating that Arabidopsis RBCL is not naturally (tri)methylated at this position (Mininno et al., 2012).
Conservation around the cTP Cleavage Site
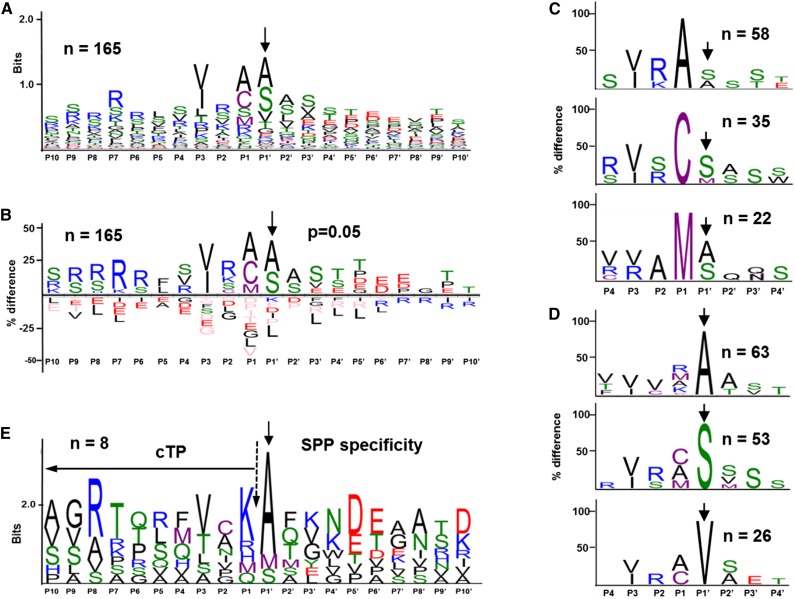
In an effort to obtain more insight into the relationship between cTP cleavage and the ultimate Nt residue/sequence, we generated a sequence logo of residues surrounding the observed mature Nti using the best-ranked N terminus for each protein (as defined above; Fig. 3A). This data set is much larger than the previously published data sets of experimental chloroplast protein Nti. Furthermore, these previous data sets were necessarily enriched for NAA Nti, since only these could be confidently identified as in vivo Nti (in the absence of Nt labeling; Zybailov et al., 2008); the dimethyl labeling in the TAILS work flow allowed us to avoid this bias. The sequence logo shows only a weak consensus around the observed N terminus (Fig. 3A); however, a (still weak) consensus motif was more clearly visualized using iceLogo (Colaert et al., 2009; Fig. 3B). The iceLogo involves weighing against the total amino acid frequency of the chloroplast proteome, thereby visualizing significantly overrepresented and underrepresented amino acids (Fig. 3B). Cys was highly enriched in the P1 position but not anywhere else. Furthermore, this showed that acidic residues were disfavored within the cTP, whereas basic residues (in particular Arg) were enriched in the cTP, but Arg was avoided within the first 10 residues of the mature protein (Fig. 3B). Small uncharged and often hydrophilic residues were favored within the first four residues of the observed proteins (P1′–P4′), whereas Leu was underrepresented in these positions. Cys, Met, and Ala were strongly enriched immediately upstream of the N terminus (P1 position). Both Cys and Met are easily oxidized, and oxidized Cys has been shown to act as a degradation signal outside of the plastid, leading to protein degradation by the proteasome (Graciet et al., 2010; Graciet and Wellmer, 2010).
Figure 3.
Analysis of amino acid conservation around experimentally determined Nti for n-encoded stroma-exposed proteins and comparison with Nti generated by in vitro SPP cleavage assays reported in the literature. As per consensus, P1′ is the observed Nt residue and P1 is the residue immediate upstream of P1′. Solid arrows indicate the experimentally determined Nt residue. For plots A to D, the best-ranked Nti of 165 plastid proteins with n-encoded stroma-exposed Nti were used. In all plots, proteins were aligned around the experimentally determined Nt residue (P1′). Color coding for residues is as follows: blue, basic residues (R, K, and H); red, acidic residues (D and E); black, apolar, or hydrophobic residues (A, V, L, I, P, F, W, and G); purple, reactive residues (M and C); and green, uncharged, polar residues (S, T, Y, Q, and N). A, Sequence Logo of the 165 stroma-exposed proteins shows a weak motif around the mature Nt. The conservation level of amino acids in this sequence alignment is represented as vertical stacks of the amino acid symbols; the stack height reflects the level of conservation. B to D, iceLogo plots of the stroma-exposed proteins in which the amino acid frequency is normalized (weighted) against the total amino acid frequency of the n-encoded chloroplast proteome (from PPDB; 1,575 proteins). Amino acid residues significantly enriched are shown above the x axis, whereas those underrepresented are shown below the x axis. Residues below the x axis colored in pink were entirely absent in this position in the experimental sequences. B, iceLogo of the 165 n-encoded stroma-exposed proteins (P = 0.05). C, iceLogo plots (P = 0.01) for n-encoded stroma-exposed proteins for which the residue immediately upstream of the experimentally determined Nti (P1) is an Ala (58 sequences), Cys (35 sequences), or Met (22 sequences). D, iceLogo plots (P = 0.01) for n-encoded stroma-exposed proteins for which the experimentally determined Nti (P1′) is an Ala (63 sequences), Ser (53 sequences), or Val (26 sequences). E, Sequence logo for eight sequences shown to be cleaved in vitro by SPP (seven using pea SPP and one using C. reinhardtii SPP), with SPP purified from chloroplasts or recombinant SPP expressed in Escherichia coli and immobilized on beads via an Nt biotin tag. Substrates are from a range of organisms (wheat [Triticum aestivum], tomato [Solanum lycopersicum], spinach [Spinacia oleracea], pea, C. reinhardtii, Arabidopsis, Saliva pratensis). Sequences and other details are provided in Supplemental Table S5.
The lack of a visible consensus cleavage site motif, despite this large and high-quality data set, suggests that SPP does not have a strict consensus cleavage motif for imported plastid proteins. Alternatively, this lack of observed motif might indicate the activity of subsequent maturation steps by additional peptidases, thereby masking the SPP cleavage site motif. Indeed, chloroplasts do possess a significant number of mostly uncharacterized aminopeptidases (Walling, 2006; van Wijk, 2015). For instance, the observation that Cys, Met, and Ala were strongly enriched immediately upstream of the N terminus (P1 position) may be explained by the activity of aminopeptidases that specifically remove these unstable residues following SPP processing.
To try and distinguish between the various scenarios and possibly reveal hidden motifs, subsets of sequences with either highly conserved residues at P1 and P1′ positions were analyzed separately by iceLogos (Fig. 3, C and D). Cys in the P1 position was preferentially flanked (in P2 and P1′) by Ser and, to a lesser extent, Arg at P2, whereas Met in the P1 position was flanked by Ala (Fig. 3C). Subsets of Nti with Ala, Ser, or Val at the N terminus (in P1′; Fig. 3D) reveal that Ser and Val Nt proteins are mostly produced by cleavage after Cys or Ala, whereas Ala Nt proteins are preceded by Arg, Lys, Ala, or Met. Furthermore, it can be observed that, for both P1-Met (Fig. 3C) and P1′-Ala (Fig. 3D), Val/Ile conservation at P3 breaks down, which could be indicative of sequential processing. These comparisons suggests that a possible cTP cleavage motif is obscured by additional processing steps.
To better understand SPP cleavage and possible subsequent maturation by other peptidases, we collected all available direct evidence for SPP cleavage site specificity (Supplemental Table S5). Such specificity has been determined for recombinant proteins using either recombinant SPP from pea (Richter and Lamppa, 2002) or semipurified SPP from isolated chloroplasts of pea or Chlamydomonas reinhardtii (Richter et al., 2005). It should be noted that these substrates are from five different plant species. Some of the substrates lack a cTP and seem less relevant to test the specificity of a processing peptidase. Using only the eight bona fide intraplastid proteins, we then generated a sequence logo of residues around the observed N terminus (Fig. 3E). This suggests cleavage primarily after basic residues (in particular Lys but also Arg and His) and upstream of Ala (Fig. 3E), which matches well with the top plot in Figure 3D. Determination of SPP cleavage specificity using a wider variety of substrates from Arabidopsis, as well as analysis of putative chloroplast aminopeptidases, are needed to improve our understanding of plastid protein maturation.
The N-Terminome of p-Encoded Proteins
The maturation process of p-encoded proteins (Fig. 1B) is very different from that of n-encoded chloroplast proteins (Fig. 1A). Moreover, the Nti of nascent p-encoded proteins are likely protected by proteins interacting with the 70S ribosome near the exit gate, such as trigger factor. Furthermore, Nt deformylation, NME, and NAA are likely cotranslational processes for p-encoded proteins (Giglione et al., 2009, 2014; Preissler and Deuerling, 2012; Sandikci et al., 2013). Hence, the Nt sensitivity to proteolytic activity may differ between p-encoded and n-encoded chloroplast proteins. The p-encoded proteins are synthesized with an Nt Met, and a subset undergoes NME. In general, the penultimate position (P1′) is the major determinant for NME, and cleavage occurs if the side chain is small (Ala, Cys, Pro, Ser, Thr, Gly, and Val; Giglione et al., 2004). Whereas p-encoded proteins generally follow this rule, there are a few outliers, and several other proteins undergo additional maturation steps (Zybailov et al., 2008, 2009; Bienvenut et al., 2012).
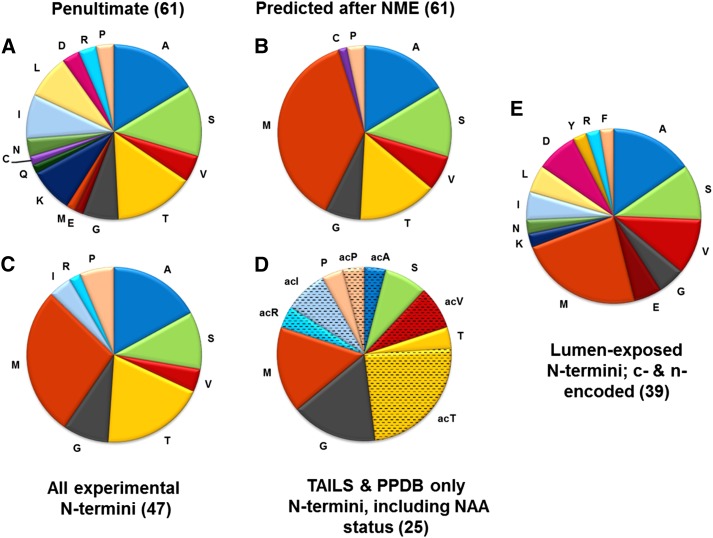
There are 88 proteins encoded by the plastid genome in Arabidopsis; 65 of these proteins have Nti in the stroma, whereas the other remaining proteins have their Nti exposed to the thylakoid lumen or their topology is currently not clear to us (Supplemental Table S6). Frequency analysis of the penultimate residues for Arabidopsis p-encoded proteins with stroma-exposed Nti showed 16 possible residues (absent are bulky His, Tyr, Trp, and Phe; Fig. 4A). Applying the general NME rule (Giglione et al., 2004) to these stroma-exposed Nti results in a simpler amino acid distribution of chloroplast Nt residues, with just eight possible amino acids (Fig. 4B).
Figure 4.
Nt amino acid frequency for stroma-exposed p-encoded proteins and comparison with all known lumenally exposed Nti (both p-encoded and n-encoded proteins). Detailed information is available in Supplemental Table S6. A, The penultimate residues (i.e. residues immediately downstream of the initiating Met) of 65 p-encoded proteins for which the N terminus is facing the stroma. This sequence information is derived from the protein sequences listed in The Arabidopsis Information Resource (TAIR; https://www.arabidopsis.org/). Within this group, there are three sets of identical homologs (ribosomal proteins S7A,B, ribosomal proteins S12A,B,C, and a full-length YCF1.2 protein and a truncated form; for details, see Supplemental Table S6). Rather than including each of these homologs, we counted each set only once, thus resulting into 61 Nti. B, The predicted Nt residues of mature proteins after application of the general NME rule for the p-encoded proteins in A. C, Experimentally determined Nt residues for p-encoded proteins for which the N terminus is facing the stroma (a total of 47 proteins). Experimental evidence was obtained from the TAILS experiments described in this study, from semitryptic or NAA Nti detected previously (Zybailov et al., 2008, 2009; Bienvenut et al., 2012), and additional data from in-house experiments in PPDB. Also included is information from Giglione et al. (2004), which were mostly based on Nt Edman sequencing data from various plant species. We note that Edman sequencing cannot sequence proteins for which the Nt is NAA; these modified Nti are blocked, preventing Edman chemistry. The experimental Nt information from these other plant species was projected onto Arabidopsis homologs if the Nti were identical. D, Experimentally determined Nt residues for 25 p-encoded proteins for which the N terminus is facing the stroma as determined by TAILS and in-house experiments in PPDB. This is a subset of the proteins in C. E, Experimentally determined Nt residues for 39 p-encoded and n-encoded proteins for which the N terminus is facing the thylakoid lumen. Experimental evidence was obtained from the TAILS experiments, previous publications (Zybailov et al., 2008, 2009), and additional data in PPDB (for details, see Supplemental Table S7).
We then combined our TAILS results with previous in-house MS/MS data for other Arabidopsis chloroplast proteome experiments in PPDB (Zybailov et al., 2008, 2009; Kim et al., 2013; Lundquist et al., 2013; Nishimura et al., 2013) as well as information from Giglione et al. (2004) that was mostly based on Nt Edman sequencing data from various plant species. The Edman sequencing method does not yield NAA state because these Nti prevent Edman chemistry (blocked Nti). The information from these other plant species was projected onto Arabidopsis homologs if the Nti were identical. The distribution of Nt residues is summarized in Figure 4C and Supplemental Table S6. We then compared Figure 4B (predicted after NME) with Figure 4C (experimental observations). This shows the presence of experimental Nti starting with Ile and Arg, which must have been due to unusual NME activity, namely that Met was removed to expose Ile (Photosystem I core subunit A [PsbA] and RPS15) or Arg (Coupling factor 1β [CF1β]); these are bulky residues that typically would prevent NME activity. It should be noted that, in all three cases, these Nt residues were acetylated, again suggesting that NAA is required for stabilization. NME did not occur for the three other observed proteins with Ile in the penultimate position (Cytochrome subunit G [PetG], NADPH dehydrogenase A [NDH-A], Photosystem I subunit J [PsaJ], and Ribosomal protein large subunit14 [RPL14]), nor was Met removed for the only other observed case with Arg in the penultimate position (PsaJ). The Nti for p-encoded YCF1.2 (Translocon inner membrane214 [TIC214]; Kikuchi et al., 2013), RBCL, and chloroplast core protein43 (CP43) did not start with Met nor with the penultimate residue, indicating that these Nti must have been generated by additional peptidase activity; however the responsible peptidases are unknown. For RBCL, the N terminus starts with the third residue, Pro (observed by 537 MS/MS spectra), and it was always in NAA form; this is in agreement with previous observations (Zybailov et al., 2008). The unprocessed YCF1.2 protein is predicted to start with formyl-Met-Met, but both Met residues were removed, resulting in an Nt Val. In the case of CP43, 12 amino acids were removed, exposing an Nt Thr, which is known to undergo NAA and reversible phosphorylation (Vener et al., 2001; van Wijk et al., 2014). We did not observe this phosphorylated form because we did not take any precautions to prevent dephosphorylation (i.e. by the addition of phosphatase inhibitors) and/or because we did not enrich for phosphopeptides, which is typically needed to observe the phosphorylated forms.
Finally, Figure 4D shows the extent of NAA for experimentally observed Nti of the stroma-faced p-encoded proteins determined only by TAILS or from previous in-house experiments listed in PPDB (25 proteins in total). This shows that Arg, Ile, Ala, and Val are always observed in their NAA form, but in the case of RPS15, Ile was also observed unmodified.
The Thylakoid Lumen-Exposed Nti Show a Wide Distribution of Amino Acids
The thylakoid lumen has its own (limited) set of proteases. We assembled all available information for p-encoded and n-encoded lumenally exposed Nti (Fig. 4E; Supplemental Table S7). In addition to the abundant Ala, Ser, Val, and Met, this shows the presence of residues essentially absent at the Nti of stroma-exposed proteins. Examples are Tyr, Asp, Glu, and Leu, indicating a far greater Nt flexibility, likely reflecting a lack of Nt-driven instability.
Accumulation of Proteolytic Products
A total of 129 Nti are likely breakdown products of chloroplast proteins (Supplemental Table S8). Interestingly, none are NAA, suggesting a short half-life and/or generation of these proteolytic products after stromal isolation and assuming that N-α-acetylases are not very active at that point. About 60% of these Nti were from the very abundant RBCL, RBCS, and Rubisco Activase (RCA), which is not surprising given that these are among the most abundant proteins. However, only a few degradation products were detected for several other highly abundant enzymes, such as transketolase, Glu-ammonia ligase, and Chaperone21. Perhaps this indicates that the RBCS/L and RCA have a shorter lifetime than other abundant stromal proteins (Recuenco-Muñoz et al., 2015). Importantly, we do note that the Nti of mature proteins are always far more frequent than the Nti of the fragments; an example is shown for the abundant stromal proteins PEPTIDYLPROLYL ISOMERASE ROC4, TRANSKETOLASE1, PHOSPHORIBULOSE KINASE2, and SEDOHEPTULOSE FRUCTOSE-BISPHOSPHATASE (SFBA; Supplemental Fig. S3). Analysis of the breakdown products revealed a strong preference for cleavage after Arg and, to a lesser extent, before Thr.
Correlation with Other Large-Scale N-Terminome Studies
In the last few months, several Arabidopsis studies were published that employed TAILS or (a variant of) combined fractional diagonal chromatography to study protein Nti in roots (Zhang et al., 2015), mitochondria (Carrie et al., 2015), and leaves of wild-type and a chloroplast import mutant (Kohler et al., 2015). Additionally, there was a large-scale study of NAA leaf proteins (Bienvenut et al., 2012) and an assessment of mitochondrial protein Nti based on classical proteomics (Huang et al., 2009). None of these studies shared the objectives of the current study; nevertheless, these studies are a good opportunity to probe the consistency with the data presented here. To that end, we systematically cross-checked the observed Nti for stroma-exposed Nti as well as Nti of mitochondrial proteins (Supplemental Table S9). Of the Nti of the 206 stroma-exposed mature proteins identified in our study, 104 matched exactly with those found by others. The observed start position for 16 other proteins observed in our data set was within five residues of that found by others. Other chloroplast proteins in our data set were either not observed by other studies or were detected with an N terminus too far downstream to represent the bona fide mature N terminus; examples are RBCS and related proteins RCA and CP12, the Calvin cycle enzymes glyceraldehyde 3-phosphate dehydrogenase A (GAP-A)/B, SFBA, and several enzymes in the methylerythritol 4-phosphate pathway.
We also detected 19 mitochondrial and 17 peroxisomal proteins (Supplemental Table S3). More than half of the mitochondrial Nti started with Ser, and the Nti were typically preceded by Met, Ser, Leu, Phe, and Tyr, in good agreement with previously described mitochondrial presequence cleavage motifs (Huang et al., 2009; Carrie et al., 2015; Supplemental Table S9). Peroxisomal proteins are targeted to the matrix by a noncleavable tripeptide at the extreme C terminus (PTS1) or a cleavable nanopeptide at the N terminus (PTS2; Hu et al., 2012). Of the 17 detected peroxisomal proteins, all except four were NAA and started either at the initiating Met or Ala in the second position, presumably because they are targeted through a PTS2 signal.
DISCUSSION
The objectives of this study were to determine the Nti of the stroma-exposed chloroplast proteome and develop a testable model for Nt processing, maturation, and stability. Through systematic TAILS analysis of soluble proteins from total leaf extracts and isolated chloroplasts, we obtained nearly 14,000 MS/MS spectra matching to protein Nti. Following condensation and curation of this data set, as well as annotation of subcellular localization, we then obtained a comprehensive set of chloroplast Nti. Comparison of this data set with previously published information for individually studied proteins and other N-terminome studies (see below) showed that our TAILS work flow provided reliable and physiologically relevant information. The parallel acquisition of N-terminomes of total leaf extract and stromal extracts from isolated chloroplasts was important for the recognition of extraplastidic proteins and chloroplast precursor proteins. This also confirmed that the accumulation of unprocessed chloroplast proteins (or cleaved cTPs) within the chloroplast is exceedingly rare, indicating a high efficiency of cTP cleavage and subsequent degradation of cleaved cTPs within the chloroplast, in agreement with Richter and Lamppa (1999) and others.
Working Hypotheses for Nt Maturation of n-Encoded Proteins
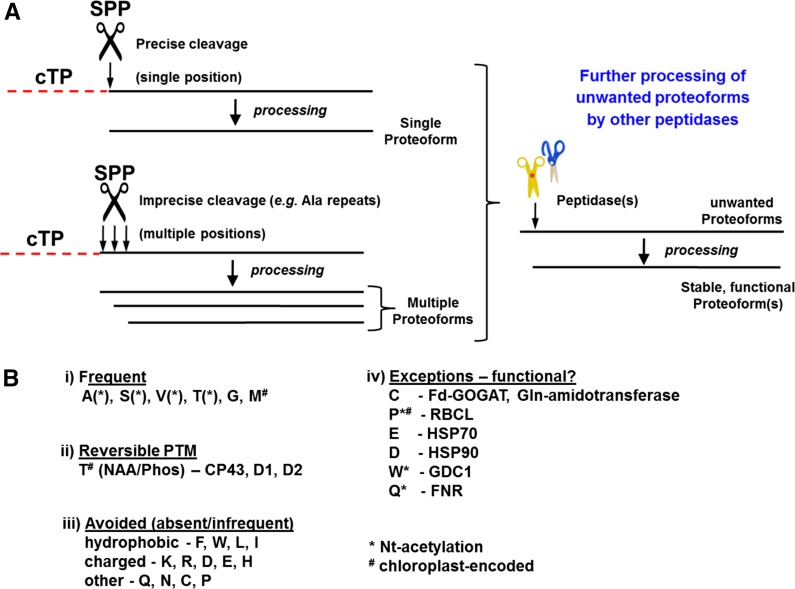
Based on the analysis of Nt amino acid frequency, sequence logos, and iceLogos, as well as published information (Richter and Lamppa, 1998; Richter et al., 2005; Zybailov et al., 2008; Bienvenut et al., 2012), we formulated a working model for Nt maturation of n-encoded proteins (Fig. 5A; for a broader discussion and many cited references, see van Wijk, 2015). Upon import into the chloroplast, the cTP is cleaved by SPP. This cleavage could either be very precise at a single position (a specific peptidyl bond; Fig. 5A, top left) or less precise, with cleavages occurring at closely spaced, multiple positions, depending on the residues neighboring the cleavage site. Additional peptidases will subsequently perhaps remove one, or in some cases two or three, residue(s) from the N terminus; this likely depends on the Nt residue and the immediate downstream sequence as well as protein fold (accessibility of the N terminus). Seven stromal amino peptidases were identified with high confidence, and their relative abundance was quantified in chloroplasts of Arabidopsis (Zybailov et al., 2008). These include the higher abundance Leu-amino peptidase (AP), Glu-AP, and Amino-AP as well as four lower abundance peptidases (Met-AP1B, Gly-AP, Pro-AP, and Ser-AP). Whereas the substrate specificity of these peptidases has generally not been characterized, they are strong candidates for performing the proposed role in Nt maturation (Fig. 5A). The combination of single and multiple SPP cleavages and activity of multiple aminopeptidases provides the most flexible scenario to arrive at the highly restricted N-terminome (i.e. high prevalence of Ala, Val, Thr, and Ser) and best accommodates all observations. Finally, a limited number of proteins likely undergo an additional downstream cleavage, as exemplified by p-encoded CP43, which accumulates with an NAA (and reversibly phosphorylated) Thr-13; we suggest that a specific peptidase (as yet unidentified) generates the N terminus of this abundant (and essential for photosynthesis) PSII core protein.
Figure 5.
Working model for Nt maturation of n-encoded proteins and the classification of different types of Nti. A, Model for the generation of mature and stable Nti of n-encoded chloroplast proteins. Upon chloroplast import, the cTPs of precursor proteins are either cleaved at a specific single site or cleaved at closely spaced multiple positions. Proteins with unwanted and/or unstable Nti are further processed by one or more stromal aminopeptidases to stabilize the proteins. B, Classification of different types of chloroplast stroma-exposed Nti and examples. We distinguish three types of amino acids: i, amino acids that are very frequent in the Nt position and that are presumably very stable in the chloroplast stroma; ii, Nti with reversible PTMs and that play a functional role; and iii, amino acids that are not or rarely observed and likely result in the destabilization of proteins in the chloroplast when these Nti are exposed to the stroma. Group iv shows examples of proteins that were observed with rare amino acids at the Nt position; these are discussed in the text.
Classification of Chloroplast Stroma-Exposed Nti Residues and Examples
Figure 5B summarizes the observed frequencies of each amino acid in the stroma-exposed Nt position for the n-encoded and p-encoded proteome. The most frequent, and perhaps the most stable, Nti start with the small polar (Ser and Thr) or apolar (Ala, Val, and Gly) residues; together, these represent approximately 75% to 80% of all Nti. Except for Gly, a significant percentage of these residues are NAA; however, the general function of NAA is poorly understood (Jones and O’Connor, 2011; Hollebeke et al., 2012; Starheim et al., 2012), but it can influence protein stability, as shown in Arabidopsis for a nod-like receptor (Xu et al., 2015). It was recently shown that reduced NAA rates trigger a drought response in Arabidopsis (Linster et al., 2015). In the case of p-encoded proteins, Met has a high frequency in the Nt position, dictated by the penultimate residue and the NME. In selected cases, such as the three PSII core proteins D1, D2, and CP43, the N terminus plays an active regulatory role through reversible phosphorylation of the (stable) NAA Thr (Fig. 5B; Vener, 2007; Rokka et al., 2011).
Whereas just six residues occupied most of the stroma-exposed Nti, other amino acids were never observed in the Nt position (His and Phe) or were observed in just one or a few cases (and sometimes only in NAA form [e.g. Trp]; Fig. 5B). We discuss a number of such cases below.
Redox-Active Cys
Cys residues are redox active, and the thiol often forms intermolecular or intramolecular disulfide bonds, participates in enzymatic reactions, and undergoes many PTMs. We observed two cases of Nt Cys residues; these are for Fd-GOGAT (AT5G04140) and GLUTAMINE PHOSPHORIBOSYL PYROPHOSPHATE AMIDOTRANSFERASE2 (AT4G34740). Surprisingly, both mature Nti start with the residues CGV as well as unusual acidic stretches upstream of the cTP cleavage site, even if these proteins are otherwise completely unrelated and have distinct functions. In both cases, these CGV Nt sequences are conserved in plants, algae, and even cyanobacteria, suggesting that these play a specific, but as yet unknown, function. Furthermore, for both cases, these proteins were only detected with these specific Cys Nt proteoforms, further suggesting that these Nt Cys residues play a functional role.
Aromatic Amino Acids Tyr, Trp, His, and Phe
These residues are destabilizing Nt residues in prokaryotes (Tyr, Trp, and Phe), where they are likely targets of the Clp protease APS system, and in the cytosol of eukaryotes (Tyr, Trp, His, and Phe), where they are targets of the proteasome (Dougan et al., 2012; Tasaki et al., 2012; Gibbs et al., 2014). His and Phe are absent as Nt residues in chloroplast stroma, whereas Tyr and Trp were each only observed once. This Trp is the N terminus of Grana-Deficient Chloroplast1 (GDC1; AT1G50900) and was observed 33 out of 35 times (always NAA); manual inspection of several of the underlying spectra confirmed the assignment. GDC1 is a mostly stroma-localized protein involved in the sorting of members of the light harvesting complex protein family and interacts with the signal recognition particle in the stroma (Cui et al., 2011; Ouyang et al., 2011). The Trp is largely conserved across land plants and is typically preceded by a Cys residue. The significance of this NAA Trp remains to be determined. It is not known if these aromatic residues confer instability to proteins in the chloroplast.
Acidic Residues Glu and Asp
The two abundant stromal chaperones cpHSP70-1 (AT4G24280) and heat shock protein90 (HSP90; AT2G04030) both start with acidic residues (Glu and Asp, respectively). The N terminus of HSP90 is generated by cleavage after Cys. In the case of HSP90, there was one other Nt proteoform, starting with an Ala one residue downstream of the Asp; however, it was only observed twice compared with 29 times for the acidic Nt proteoform. In the case of HSP70, the acidic Nt proteoform was observed 71 out of 87 times. These essential chaperones perhaps require these unusual acidic Nti to interact with their targets or partners. Another Nt Glu was found for ribosomal protein RPL13, also generated by cleavage after Cys. It seems quite logical that a stromal aminopeptidase might exist that removes these Cys residues.
Pro
We observed Nt Pro (an unreactive amino acid) with a high number of MS/MS spectra for the abundant RBCL (the Pro was always NAA), in agreement with a previous study in spinach (Mulligan et al., 1988). Because Pro is the third residue of this p-encoded protein after Met-Ser, it was likely generated by NME, followed by cleavage of the Ser by a different peptidase. We predict that a specific, as yet unknown, peptidase evolved to carry out this specific cleavage of RBCL.
Functional Significance of Nt Maturation and Nt Proteoforms
The N-terminome analysis presented in this study clearly established that many chloroplast proteins are represented by more than one Nt proteoform. Based on the number of matched MS/MS spectra presented in Supplemental Table S4, it is possible to calculate tentative abundance ratios between Nt proteoforms for these proteins. For example, in the case of inorganic phosphatase (AT5G09650.1), observed with 185 MS/MS spectra, 5% started with Ser-Ala-Ile, 27% started with Ala-Ile, and 68% started with the downstream Ile. In the case of enoyl-acyl carrier protein (ACP) reductase (AT2G05990.1), observed with 134 MS/MS spectra, two Nt proteoforms were observed starting with Ala-Met-Ser (13%) or the downstream Ser (87%), but interestingly, no Nt proteoform starting with this Met was observed. Whereas the asymmetric distribution of Nt proteoforms may relate to functional differences, very few studies exist that have looked at the significance of Nt proteoforms. One example of such studies is for thylakoid-associated Fd NADPH reductase1 (FNR1; AT5G66190) and FNR2 (AT1G20020; Lehtimäki et al., 2014), which each have two Nt variants, with Nt sequences AQVT and AQIT being observed with and without the Ala for FNR1 and FNR2, respectively (Lehtimäki et al., 2014), in agreement with our TAILS data. However, in this case, our TAILS data did not allow determination of the ratio between the two proteoforms for each FNR protein. Furthermore, NAA forms of each Nti were observed, and this modification appeared to be induced by light (Lehtimäki et al., 2014). However, these Nt variations did not influence their association with the thylakoid membrane, and the exact physiological relevance remained unclear (Lehtimäki et al., 2014). A systematic analysis to investigate the functional significance of the phenomenon of multiple proteoforms is warranted.
Differences between p-Encoded and n-Encoded Proteins
The maturation processes of n-encoded and p-encoded proteins differ from each other; it is strictly posttranslational in the case of n-encoded proteins but cotranslational for many p-encoded proteins. Furthermore, following import and cTP cleavage, most n-encoded chloroplast proteins do not start with a Met, whereas p-encoded proteins do. Except for the much higher frequency of observed Nt Met (approximately 30%), the p-encoded protein Nti are dominated by the small, uncharged residues Ala, Ser, Thr, Val, and Gly, similar to what was observed for n-encoded proteins. NAA rates appeared higher for p-encoded proteins than for n-encoded proteins, perhaps because this occurs cotranslationally rather than posttranslationally. Additionally, observation of a wide range of NAA amino acids (Ile, Pro, Arg, Trp, and Gln but not Gly) suggests that more than one Nt acetylase operates in the chloroplast (Starheim et al., 2012; Giglione et al., 2014).
Comparison with Protein Maturation in Mitochondria
Chloroplasts and mitochondria have many similarities with respect to protein biogenesis, and more than 100 proteins are dually targeted to both organelles (Carrie and Small, 2013), including several proteases such as LON1 (Daras et al., 2014), PREP1 (Kmiec et al., 2014), FTSH11 (Urantowka et al., 2005), and OOP (Kmiec et al., 2013). It is likely, therefore, that they also show similarities in protein maturation and in mechanisms of protease substrate recognition. Recent observations for plant mitochondrial Nt peptidases INTERMEDIATE CLEAVAGE PEPTIDASE55 (ICP55; Carrie et al., 2015; Huang et al., 2015) and Octapeptidyl amino peptidase1 (OCT1; Carrie et al., 2015) support a similar model for the maturation and stabilization of n-encoded proteins as the proposed model for chloroplasts (Fig. 5A). After cleavage of the Nt Mitochondrial transit peptide (mTP) by the general mitochondrial processing peptidase (the functional equivalent of chloroplast SPP), one (or sometimes two) amino acid residue (in particular Phe, Tyr, Leu, and Ile) is cleaved by ICP55 for a high portion of mitochondrial proteins. The specificity of OCT1 was not very clear, and it was suggested that OCT1 might act after the assembly of proteins, rather than immediately following mTP cleavage (Carrie et al., 2015). Based on these observations for ICP55, it was suggested that removal of specific Nt residues (in particular Phe, Tyr, and Leu) is needed to confer protein stability (Carrie et al., 2015; Huang et al., 2015). The Arabidopsis genome contains a homolog (AT4G29490) of mitochondrial ICP55, which is a candidate to play a similar function in chloroplasts.
Nt Residues, N-Degron, and the N-End Rule
The N terminus of proteins has been shown to be a major determinant of protein stability/half-life in many organisms. Early observations in yeast (Saccharomyces cerevisiae) led to the formulation of the N-end rule, which states that certain amino acids, when exposed at the N terminus of a protein, act as triggers for degradation (Bachmair et al., 1986). The N-end rule in prokaryotes is different from that in eukaryotes in part because most prokaryotes do not have a proteasome and also lack ubiquitination (Tobias et al., 1991). In bacteria, such N-end rule proteins are recognized by the adaptor ClpS, which delivers such proteins for unfolding and degradation to the Clp chaperone and protease system. Recent reviews summarize the history of these discoveries and the current understanding of the N-end rule pathway for prokaryotes and eukaryotes (Dougan et al., 2012; Tasaki et al., 2012; Gibbs et al., 2014). Whereas an N-end rule for chloroplasts/plastids in plants is not known, overexpression studies for p-encoded proteins have shown that the amino acids at the N terminus can greatly affect protein stability (Apel et al., 2010). Moreover, a plant homolog of ClpS was recently identified and characterized in chloroplasts of Arabidopsis (Nishimura et al., 2013). Recent data of the N-terminome of an Arabidopsis mitochondrial ICP55 null mutant indicated that ICP55 removes in particular the Nt residues Phe, Tyr, and Leu. These residues are generally considered unstable residues; therefore, it was suggested that plant mitochondria also utilize an N-end rule pathway (Carrie et al., 2015; Huang et al., 2015). Our study was in part motivated by the search for a possible N-end rule in the chloroplast stroma. From the observed amino acid frequencies in the stroma-exposed Nti, there appears to be a strong overlap between residues avoided in chloroplast stroma-exposed Nti and the bacterial and mitochondrial primary N-end rule residues (Trp, Tyr, Phe, and Leu). Secondary destabilizing residues Asp, Glu, Arg, and Lys in prokaryotes also are among the low-frequency amino acids in the Nt position. In contrast, Met, a secondary destabilizing residue in E. coli (but nevertheless quite frequent in prokaryotes; Bonissone et al., 2013), is clearly a very frequent and likely stable residue for p-encoded proteins in chloroplast stroma. Secondary destabilizing residues only become destabilizing upon the transfer of an amino acid to the N terminus (Dougan et al., 2012; Tasaki et al., 2012; Gibbs et al., 2014); such aminotransferase remains to be identified (or recognized) in chloroplasts. Measurements of chloroplast protein stability in dependence of their Nt residues in different peptidase, protease, and protease adaptor (e.g. ClpS1) mutant backgrounds will be needed to determine to what extent chloroplast proteostasis is governed by an N-end rule.
MATERIALS AND METHODS
Plant Growth and Generation of Protein Samples
Arabidopsis (Arabidopsis thaliana Columbia-0) was grown on soil in a temperature-controlled chamber at 150 µmol photons m−2 s−1 in a 12-h light period and harvested at developmental stages 1 to 12. Total leaf was frozen in liquid nitrogen and ground to a powder in cooled mortar and pestle. Proteins were then extracted in 50 mm HEPES-KOH, pH 8, 1 mm EDTA, 1 mm Pefabloc, and 10 µg µL−1 E64, using 1 mL of volume per 1 g fresh weight; cell debris was removed by spin columns (Friso et al., 2011). Protein concentrations were determined by the bicinchoninic acid protein assay (Thermo Fisher Scientific). Chloroplast stroma was obtained from isolated chloroplasts as described (Olinares et al., 2010).
TAILS Experiments
A TAILS strategy was employed as described (Kleifeld et al., 2011; Guryča et al., 2012) with minor modifications. Briefly, 100 to 200 μg of protein in extraction buffer (above) was mixed 1:1 with 8 m guanidine hydrochloride (GuHCl) in a single 1.6- or 2-mL tube. Dithiothreitol was added at a final concentration of 5 mm, and the solution was incubated at 65°C for 1 h. Cys residues were alkylated with iodoacetamide, at a 15 mm final concentration, for 20 min in darkness at room temperature, and the residual iodoacetamide was quenched by the addition of dithiothreitol, at a final concentration of 15 mm. For dimethylation of amines, 2 m formaldehyde (CH2O) or CD2O (heavy isotope; made fresh in distilled, deionized water) and 1 m NaCNBH3 were added to give 40 and 20 mm, respectively. The pH was lowered to 7 with 1 m HCl, and the solution was incubated at 37°C for between 8 and 16 h. The reaction was quenched with 1 m ammonium bicarbonate (NH4HCO3) at final concentration of 100 mm for 2 h at 37°C. Proteins were precipitated with between 4 and 8 volumes of ice-cold acetone and 1 volume of methanol, and the solution was kept at −80°C for 3 h. Proteins were pelleted at 14,000g for 20 min, the supernatant was removed, and the pellet was washed twice with ice-cold methanol. The pellet was resuspended in 10 to 20 μL of 8 m GuHCl or dimethyl sulfoxide followed by stepwise dilution with 50 mm HEPES, pH 8, while vortexing to give a GuHCl concentration of less than 0.8 m and a protein concentration of 1 mg mL−1. Some precipitate typically remained after resuspension.
Sequencing-grade trypsin (V5111; Promega) at a 1:100 ratio or Glu-C (V165A; Promega) at a 1:20 ratio (enzyme:protein) was added, and the solution was incubated overnight at 37°C. One aliquot (80 µg) of Glu-C-digested sample was further digested (1:20, enzyme:protein) with trypsin overnight (20 h). Alternatively, 200 μg of labeled, precipitated protein was resuspended in 1× Laemmli buffer and then resolved by SDS-PAGE with 12% (w/v) T Laemmli. The whole gel lane was then cut into four bands, and in-gel trypsin digestion was performed as described (Friso et al., 2011), except that no reduction and alkylation was performed and the digestion was performed in 50 mm HEPES-KOH, pH 8. The resulting peptide extracts were dried and then resuspended in 50 mm HEPES-KOH, pH 8. Following each digestion, a 5% (v/v) aliquot of the digested protein was reserved for testing of labeling efficiency (before the negative selection step). These aliquots were desalted using C-18 solid-phase extraction spin columns (Pierce 89870) using the manufacturer’s guidelines and subjected to MS analysis. The remaining protein digest was added to 600 μg (15- to 20-µL aliquots) of dendritic high-Mr polyglycerol-aldehyde polymer (Flintbox Innovation Network) followed immediately by 1 m NaCNBH3 to give a 40 mm final concentration. The pH was adjusted to 7 with 1 m HCl, and the sample was incubated overnight at 37°C. The reaction was quenched as above with 100 mm NH4HCO3, and the solution was filtered through a prewashed (3 × 0.5 mL of water, 2 × 100 mm NH4HCO3) Amicon 30-kD molecular mass ultrafiltration device (Millipore). The filtrate was acidified with formic acid, and the peptides were desalted as described above. The peptides were then dried in a vacuum centrifuge and resuspended in 20 μL of 2% (w/v) formic acid. SDS-PAGE was performed to ensure the recovery of protein across the labeling and precipitation steps and to ensure that digestion went to completion. Please note that the GuHCl concentration in the SDS sample buffer must be less than 0.04 m to avoid precipitate and spoiled gel separation.
Liquid Chromatography-MS/MS Analysis
All samples were analyzed by nano-liquid chromatography-MS/MS using an LTQ Orbitrap mass spectrometer (Thermo Fisher Scientific) run at 100,000 resolution in MS (Orbitrap portion) at least once prior to the negative selection step, to confirm label efficiency, and at least twice following the negative selection step. The liquid chromatography and MS tuning and acquisition conditions were as described previously (Friso et al., 2011) with some minor variations. In some cases, a reject list for the most abundant, persisting, highly concentrated RBCS and RBCL peptides was added.
Database Search/Peptide Identification
Peak lists (mgf files) were generated from Thermo Fisher Scientific raw data files using DTA Supercharge. The peak lists were searched using MASCOT 2.4 (Matrix Science) against TAIR 10, appended with all reverse sequences (Decoy) and common contaminants (71,149 sequences and 29,099,754 residues). Following an initial database search performed at 30-ppm MS tolerance and 0.8-D MS/MS tolerance, the peak list was then recalibrated as described previously (Friso et al., 2010). A semispecific enzyme search was then conducted, using semiArgC, semiGluC (V8), or semi(ArgC and GluC), allowing for two missed cleavages, 4-ppm MS tolerance, and 0.8-D MS/MS tolerance. Fixed modifications were carboxamidomethyl Cys and dimethyl Lys; variable modifications were oxidized Met, pyro-Glu Nt Gln, acetyl Nt, and dimethyl Nt (light, +28 D; heavy, +32 D). Another search including singly methylated Nt was conducted for select files in order to detect methylated Nt Pro. For samples labeled with heavy formaldehyde, a search was conducted with intermediate light and heavy (+30 D) dimethylation to account for proteins that underwent physiological (mono)methylation at the N terminus.
Additional Database Searches
To test labeling efficiency, samples taken prior to negative selection were subjected to semitryptic or semiGluC searches with only fixed carboxamidomethyl Cys and variable dimethyl Lys, dimethyl Nt, acetyl Nt, and oxidized Met. These search parameters enabled the detection of unlabeled Lys side chains and semitryptic peptides that should not be present if dimethyl labeling was complete. To detect monomethylated Pro, monomethylated Nt was added to the list of variable modifications. To detect physiological Nt monomethylation in samples labeled with heavy formaldehyde, a search was conducted with intermediate dimethylation: light/heavy (+30 D). To monitor dimethyl labeling efficiency, prenegative selection database search results were exported and the number of Lys per peptide was calculated. The number of assigned dimethylated Lys was then compared with the Lys number for each peptide as well as the number of missed cleavages (which should equal the number of Lys unless they are followed by Pro).
Filtering of MS Data
Each MASCOT result (MS/MS ion search) was filtered, P < 0.01, minimum ion score of 30. The spectra were then exported and sorted to remove any contaminant peaks such as trypsin and keratin and to ensure that the number of reverse hits gave a false discovery rate of less than 1%, with false discovery rate = 2 × FP/(FP + TP), where FP is false positive, and TP is true positive. Identified peptides from all database searches were combined and all peptides without an Nt label were removed, leaving only those with dimethyl, acetyl, or pyro-GluQ Nti. Peptides were then sorted by criteria in the following order: peptide sequence, modification 1, modification 2, and peptide score (highest to lowest). All identical peptide species were collapsed, and the number was summed to give the number of SPC for that peptide. In total, 13,858 spectra were matched using the above criteria (Supplemental Table S1), representing 1,087 Nt peptides. All pyro-GluQ Nti, downstream of Lys, Arg, or Glu (resulting from artificial trypsin or GluC cleavage), were deemed artifacts and were removed (50 peptides). Certain groups of peptides were found that matched to the same N terminus due to ragged ends at the C terminus, or alternate enzyme cleavage sites (e.g. GluC) and SPC for these peptides were grouped such that the SPC for all peptides matching the same N terminus were combined (the peptide with the most SPC is the parent of that group). If, for example, a missed cleavage led to the same NAA N terminus being found twice, each with a distinct mass, both spectra were counted toward that N terminus. Therefore, all redundant Nti that cannot be distinguished by Nt modification (148 peptides) were collapsed and their Nti added to the parent group, leaving 894 unique Nt sequences (Supplemental Table S2) matching to 577 proteins.
Validation of Nti, Terminology, and Subcellular Localization
TAILS-identified chloroplast Nti were compared with Nt information from the scattered Edman sequencing data available in the literature (Richter and Lamppa, 1998; Peltier et al., 2002; Candat et al., 2013) and with semitryptic peptides identified in PPDB. A unique Nt sequence refers to a single sequence that could be identified from different charge states of the same peptide or from separate overlapping peptides with the same modified/labeled N terminus. If the same sequence was identified with alternate modifications of the NAA amino group, these are considered unique Nti. As such, the same exact sequence can be found in three unique forms, dimethylated (free Nt in the original sample), NAA, or pyro-Glu. Proteins were annotated for subcellular location based on manually curated experimental information from PPDB (http://ppdb.tc.cornell.edu/; Sun et al., 2009).
Generation of Sequence Logos and iceLogos
Sequence logos and iceLogos were generated with iceLogo version 1.2 (http://www.proteomics.be). The sequence logos generated are identical to those made with WebLogo (http://weblogo.berkeley.edu/). For n-encoded proteins, we aligned sequences starting 10 residues upstream of the N terminus (P10 position) and ending 10 residues downstream (P10′). Sets of sequences were loaded into iceLogo along with the 1,663 known chloroplast proteins (PPDB), as a reference (background) proteome. The iceLogo results were filtered to show only residues that were significantly different from the reference proteome, with P < 0.05 for Figure 4B and P < 0.01 for Figure 4, C and D. Amino acid colors were chosen to accentuate basic (blue), acidic (red), hydrophilic or polar (green), and sulfur-containing (purple) residues. All other, generally hydrophobic, residues are in black.
Annotation of Protein Subcellular Localization
Proteins were annotated for subcellular localization based on curated information as listed in PPDB (http://ppdb.tc.cornell.edu/) updated with the most recent data from the literature and other public resources. The subcellular localization in PPDB is based on available information from the literature as well as information from specific databases, such as those by the Rolland laboratory (AtChloro), but also SUBA (from the Millar laboratory), TAIR, and the new Arabidopsis Information Portal, etc. For annotation in PPDB, all this public information is considered together with extensive in-house information to then manually assign a subcellular localization, in particular for plastids/chloroplasts. Furthermore, it also considers information from orthologs in maize (Zea mays), as described recently (Huang et al., 2013). Annotation of the orientation of Nti of chloroplast proteins (facing the stroma, lumen, or envelope intramembrane space) was based on the literature.
Availability of Mass Spectrometry Proteomics Data
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (Vizcaino et al., 2014) via the PRIDE partner repository with the data set identifier PXD002476 and 10.6019/PXD002476.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. TAILS experimental work flow, using a method adapted from Kleifeld et al. (2011).
Supplemental Figure S2. RBCS4 Nt peptide MS spectrum labeled light and heavy to distinguish physiological monomethylation status.
Supplemental Figure S3. TAILS-identified Nti of intact stromal proteins and their degradation products.
Supplemental Table S1. The 13,858 matched MS/MS spectra that provide the basis for this N-terminome analysis matching to 606 nonredundant proteins.
Supplemental Table S2. Nt peptides after merging redundant Nt peptides (i.e. with the same mass), resulting in 1,037 Nt sequences matching to 577 proteins and including degradation products.
Supplemental Table S3. Nt peptides after merging redundant and/or overlapping Nt sequences irrespective of PTMs (exception for NAA, treated as distinct) and C-terminal ends (ragged ends), resulting in 894 Nt sequences matching to 577 proteins.
Supplemental Table S4. The 344 stroma-exposed Nti of 205 n-encoded proteins used for Figures 2 and 3 and associated analysis; degradation products are removed.
Supplemental Table S5. Experimentally determined SPP cleavage sites described in the literature.
Supplemental Table S6. Nti of all 88 p-encoded proteins, their topology, predicted mature Nti based on the general NME rule, and experimental Nt information from this study, PPDB, and published literature.
Supplemental Table S7. The 39 Nti exposed to the thylakoid lumen.
Supplemental Table S8. The 129 degradation Nti of 498 stroma-exposed Nti.
Supplemental Table S9. Comparison of identified chloroplast and mitochondrial protein Nti with other published TAILS and N-terminome studies.
Glossary
- Nt
N-terminal
- NAA
N-terminal α-acetylation
- n-encoded
nucleus-encoded
- cTP
chloroplast transit peptide
- SPP
stromal processing peptidase
- PTMs
posttranslational modifications
- p-encoded
plastid-encoded
- NME
Met excision
- TAILS
terminal amino isotopic labeling of substrates
- Nti
N termini
- MS/MS
tandem mass spectrometry
- MS
mass spectrometry
- SPC
spectral counts
- GuHCl
guanidine hydrochloride
- TAIR
The Arabidopsis Information Resource
Footnotes
This work was supported by the National Science Foundation (grant nos. MCB–1021963 and IOS–1127017 to K.J.v.W.).
Articles can be viewed without a subscription.
References
- Alban C, Tardif M, Mininno M, Brugière S, Gilgen A, Ma S, Mazzoleni M, Gigarel O, Martin-Laffon J, Ferro M, et al. (2014) Uncovering the protein lysine and arginine methylation network in Arabidopsis chloroplasts. PLoS One 9: e95512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apel W, Schulze WX, Bock R (2010) Identification of protein stability determinants in chloroplasts. Plant J 63: 636–650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmair A, Finley D, Varshavsky A (1986) In vivo half-life of a protein is a function of its amino-terminal residue. Science 234: 179–186 [DOI] [PubMed] [Google Scholar]
- Bienvenut WV, Sumpton D, Martinez A, Lilla S, Espagne C, Meinnel T, Giglione C (2012) Comparative large scale characterization of plant versus mammal proteins reveals similar and idiosyncratic N-alpha-acetylation features. Mol Cell Proteomics 11: M111 015131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonissone S, Gupta N, Romine M, Bradshaw RA, Pevzner PA (2013) N-terminal protein processing: a comparative proteogenomic analysis. Mol Cell Proteomics 12: 14–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Candat A, Poupart P, Andrieu JP, Chevrollier A, Reynier P, Rogniaux H, Avelange-Macherel MH, Macherel D (2013) Experimental determination of organelle targeting-peptide cleavage sites using transient expression of green fluorescent protein translational fusions. Anal Biochem 434: 44–51 [DOI] [PubMed] [Google Scholar]
- Carrie C, Small I (2013) A reevaluation of dual-targeting of proteins to mitochondria and chloroplasts. Biochim Biophys Acta 1833: 253–259 [DOI] [PubMed] [Google Scholar]
- Carrie C, Venne AS, Zahedi RP, Soll J (2015) Identification of cleavage sites and substrate proteins for two mitochondrial intermediate peptidases in Arabidopsis thaliana. J Exp Bot 66: 2691–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colaert N, Helsens K, Martens L, Vandekerckhove J, Gevaert K (2009) Improved visualization of protein consensus sequences by iceLogo. Nat Methods 6: 786–787 [DOI] [PubMed] [Google Scholar]
- Cui YL, Jia QS, Yin QQ, Lin GN, Kong MM, Yang ZN (2011) The GDC1 gene encodes a novel ankyrin domain-containing protein that is essential for grana formation in Arabidopsis. Plant Physiol 155: 130–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daras G, Rigas S, Tsitsekian D, Zur H, Tuller T, Hatzopoulos P (2014) Alternative transcription initiation and the AUG context configuration control dual-organellar targeting and functional competence of Arabidopsis Lon1 protease. Mol Plant 7: 989–1005 [DOI] [PubMed] [Google Scholar]
- Dirk LM, Williams MA, Houtz RL (2001) Eukaryotic peptide deformylases: nuclear-encoded and chloroplast-targeted enzymes in Arabidopsis. Plant Physiol 127: 97–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dirk LM, Williams MA, Houtz RL (2002) Specificity of chloroplast-localized peptide deformylases as determined with peptide analogs of chloroplast-translated proteins. Arch Biochem Biophys 406: 135–141 [DOI] [PubMed] [Google Scholar]
- Dougan DA, Micevski D, Truscott KN (2012) The N-end rule pathway: from recognition by N-recognins, to destruction by AAA+ proteases. Biochim Biophys Acta 1823: 83–91 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, Brunak S, von Heijne G (2000) Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J Mol Biol 300: 1005–1016 [DOI] [PubMed] [Google Scholar]
- Friso G, Majeran W, Huang M, Sun Q, van Wijk KJ (2010) Reconstruction of metabolic pathways, protein expression, and homeostasis machineries across maize bundle sheath and mesophyll chloroplasts: large-scale quantitative proteomics using the first maize genome assembly. Plant Physiol 152: 1219–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friso G, Olinares PDB, van Wijk KJ (2011) The workflow for quantitative proteome analysis of chloroplast development and differentiation, chloroplast mutants, and protein interactions by spectral counting. In Jarvis RP, ed, Chloroplast Research in Arabidopsis. Humana Press, New York, pp 265–282 [DOI] [PubMed] [Google Scholar]
- Gibbs DJ, Bacardit J, Bachmair A, Holdsworth MJ (2014) The eukaryotic N-end rule pathway: conserved mechanisms and diverse functions. Trends Cell Biol 24: 603–611 [DOI] [PubMed] [Google Scholar]
- Giglione C, Boularot A, Meinnel T (2004) Protein N-terminal methionine excision. Cell Mol Life Sci 61: 1455–1474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giglione C, Fieulaine S, Meinnel T (2009) Cotranslational processing mechanisms: towards a dynamic 3D model. Trends Biochem Sci 34: 417–426 [DOI] [PubMed] [Google Scholar]
- Giglione C, Fieulaine S, Meinnel T (2014) N-terminal protein modifications: bringing back into play the ribosome. Biochimie 114: 134–146 [DOI] [PubMed] [Google Scholar]
- Giglione C, Vallon O, Meinnel T (2003) Control of protein life-span by N-terminal methionine excision. EMBO J 22: 13–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graciet E, Mesiti F, Wellmer F (2010) Structure and evolutionary conservation of the plant N-end rule pathway. Plant J 61: 741–751 [DOI] [PubMed] [Google Scholar]
- Graciet E, Walter F, Ó’Maoiléidigh DS, Pollmann S, Meyerowitz EM, Varshavsky A, Wellmer F (2009) The N-end rule pathway controls multiple functions during Arabidopsis shoot and leaf development. Proc Natl Acad Sci USA 106: 13618–13623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graciet E, Wellmer F (2010) The plant N-end rule pathway: structure and functions. Trends Plant Sci 15: 447–453 [DOI] [PubMed] [Google Scholar]
- Guryča V, Lamerz J, Ducret A, Cutler P (2012) Qualitative improvement and quantitative assessment of N-terminomics. Proteomics 12: 1207–1216 [DOI] [PubMed] [Google Scholar]
- Hollebeke J, Van Damme P, Gevaert K (2012) N-terminal acetylation and other functions of Nα-acetyltransferases. Biol Chem 393: 291–298 [DOI] [PubMed] [Google Scholar]
- Hoshiyasu S, Kohzuma K, Yoshida K, Fujiwara M, Fukao Y, Yokota A, Akashi K (2013) Potential involvement of N-terminal acetylation in the quantitative regulation of the ε subunit of chloroplast ATP synthase under drought stress. Biosci Biotechnol Biochem 77: 998–1007 [DOI] [PubMed] [Google Scholar]
- Houtz RL, Magnani R, Nayak NR, Dirk LM (2008) Co- and post-translational modifications in Rubisco: unanswered questions. J Exp Bot 59: 1635–1645 [DOI] [PubMed] [Google Scholar]
- Hsu SC, Endow JK, Ruppel NJ, Roston RL, Baldwin AJ, Inoue K (2011) Functional diversification of thylakoidal processing peptidases in Arabidopsis thaliana. PLoS One 6: e27258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Baker A, Bartel B, Linka N, Mullen RT, Reumann S, Zolman BK (2012) Plant peroxisomes: biogenesis and function. Plant Cell 24: 2279–2303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M, Friso G, Nishimura K, Qu X, Olinares PD, Majeran W, Sun Q, van Wijk KJ (2013) Construction of plastid reference proteomes for maize and Arabidopsis and evaluation of their orthologous relationships: the concept of orthoproteomics. J Proteome Res 12: 491–504 [DOI] [PubMed] [Google Scholar]
- Huang S, Nelson CJ, Li L, Taylor NL, Ströher E, Petereit J, Millar AH (2015) INTERMEDIATE CLEAVAGE PEPTIDASE55 modifies enzyme amino termini and alters protein stability in Arabidopsis mitochondria. Plant Physiol 168: 415–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S, Taylor NL, Whelan J, Millar AH (2009) Refining the definition of plant mitochondrial presequences through analysis of sorting signals, N-terminal modifications, and cleavage motifs. Plant Physiol 150: 1272–1285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huesgen PF, Alami M, Lange PF, Foster LJ, Schröder WP, Overall CM, Green BR (2013) Proteomic amino-termini profiling reveals targeting information for protein import into complex plastids. PLoS One 8: e74483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, O’Connor CD (2011) Protein acetylation in prokaryotes. Proteomics 11: 3012–3022 [DOI] [PubMed] [Google Scholar]
- Kikuchi S, Bédard J, Hirano M, Hirabayashi Y, Oishi M, Imai M, Takase M, Ide T, Nakai M (2013) Uncovering the protein translocon at the chloroplast inner envelope membrane. Science 339: 571–574 [DOI] [PubMed] [Google Scholar]
- Kim J, Olinares PD, Oh SH, Ghisaura S, Poliakov A, Ponnala L, van Wijk KJ (2013) Modified Clp protease complex in the ClpP3 null mutant and consequences for chloroplast development and function in Arabidopsis. Plant Physiol 162: 157–179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleifeld O, Doucet A, Prudova A, auf dem Keller U, Gioia M, Kizhakkedathu JN, Overall CM (2011) Identifying and quantifying proteolytic events and the natural N terminome by terminal amine isotopic labeling of substrates. Nat Protoc 6: 1578–1611 [DOI] [PubMed] [Google Scholar]
- Kmiec B, Teixeira PF, Berntsson RP, Murcha MW, Branca RM, Radomiljac JD, Regberg J, Svensson LM, Bakali A, Langel U, et al. (2013) Organellar oligopeptidase (OOP) provides a complementary pathway for targeting peptide degradation in mitochondria and chloroplasts. Proc Natl Acad Sci USA 110: E3761–E3769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kmiec B, Teixeira PF, Glaser E (2014) Phenotypical consequences of expressing the dually targeted presequence protease, AtPreP, exclusively in mitochondria. Biochimie 100: 167–170 [DOI] [PubMed] [Google Scholar]
- Köhler D, Montandon C, Hause G, Majovsky P, Kessler F, Baginsky S, Agne B (2015) Characterization of chloroplast protein import without Tic56, a component of the 1-Megadalton translocon at the inner envelope membrane of chloroplasts. Plant Physiol 167: 972–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange PF, Overall CM (2013) Protein TAILS: when termini tell tales of proteolysis and function. Curr Opin Chem Biol 17: 73–82 [DOI] [PubMed] [Google Scholar]
- Lehtimäki N, Koskela MM, Dahlström KM, Pakula E, Lintala M, Scholz M, Hippler M, Hanke GT, Rokka A, Battchikova N, et al. (2014) Posttranslational modifications of FERREDOXIN-NADP+ OXIDOREDUCTASE in Arabidopsis chloroplasts. Plant Physiol 166: 1764–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linster E, Stephan I, Bienvenut WV, Maple-Grødem J, Myklebust LM, Huber M, Reichelt M, Sticht C, Geir Møller S, Meinnel T, et al. (2015) Downregulation of N-terminal acetylation triggers ABA-mediated drought responses in Arabidopsis. Nat Commun 6: 7640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundquist P, Poliakov A, Giacomelli L, Friso G, Appel M, McQuinn RP, Krasnoff SB, Rowland O, Ponnala L, Sun Q, et al. (2013) Loss of plastoglobule kinases ABC1K1 and ABC1K3 causes conditional degreening, modified prenyl-lipids, and recruitment of the jasmonic acid pathway. Plant Cell 25: 1818–1839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meinnel T, Serero A, Giglione C (2006) Impact of the N-terminal amino acid on targeted protein degradation. Biol Chem 387: 839–851 [DOI] [PubMed] [Google Scholar]
- Midorikawa T, Endow JK, Dufour J, Zhu J, Inoue K (2014) Plastidic type I signal peptidase 1 is a redox-dependent thylakoidal processing peptidase. Plant J 80: 592–603 [DOI] [PubMed] [Google Scholar]
- Mininno M, Brugière S, Pautre V, Gilgen A, Ma S, Ferro M, Tardif M, Alban C, Ravanel S (2012) Characterization of chloroplastic fructose 1,6-bisphosphate aldolases as lysine-methylated proteins in plants. J Biol Chem 287: 21034–21044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan RM, Houtz RL, Tolbert NE (1988) Reaction-intermediate analogue binding by ribulose bisphosphate carboxylase/oxygenase causes specific changes in proteolytic sensitivity: the amino-terminal residue of the large subunit is acetylated proline. Proc Natl Acad Sci USA 85: 1513–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Asakura Y, Friso G, Kim J, Oh SH, Rutschow H, Ponnala L, van Wijk KJ (2013) ClpS1 is a conserved substrate selector for the chloroplast Clp protease system in Arabidopsis. Plant Cell 25: 2276–2301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olinares PD, Ponnala L, van Wijk KJ (2010) Megadalton complexes in the chloroplast stroma of Arabidopsis thaliana characterized by size exclusion chromatography, mass spectrometry, and hierarchical clustering. Mol Cell Proteomics 9: 1594–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang M, Li X, Ma J, Chi W, Xiao J, Zou M, Chen F, Lu C, Zhang L (2011) LTD is a protein required for sorting light-harvesting chlorophyll-binding proteins to the chloroplast SRP pathway. Nat Commun 2: 277. [DOI] [PubMed] [Google Scholar]
- Peltier JB, Emanuelsson O, Kalume DE, Ytterberg J, Friso G, Rudella A, Liberles DA, Söderberg L, Roepstorff P, von Heijne G, et al. (2002) Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. Plant Cell 14: 211–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peltier JB, Friso G, Kalume DE, Roepstorff P, Nilsson F, Adamska I, van Wijk KJ (2000) Proteomics of the chloroplast: systematic identification and targeting analysis of lumenal and peripheral thylakoid proteins. Plant Cell 12: 319–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preissler S, Deuerling E (2012) Ribosome-associated chaperones as key players in proteostasis. Trends Biochem Sci 37: 274–283 [DOI] [PubMed] [Google Scholar]
- Recuenco-Muñoz L, Offre P, Valledor L, Lyon D, Weckwerth W, Wienkoop S (2015) Targeted quantitative analysis of a diurnal RuBisCO subunit expression and translation profile in Chlamydomonas reinhardtii introducing a novel Mass Western approach. J Proteomics 113: 143–153 [DOI] [PubMed] [Google Scholar]
- Richter S, Lamppa GK (1998) A chloroplast processing enzyme functions as the general stromal processing peptidase. Proc Natl Acad Sci USA 95: 7463–7468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter S, Lamppa GK (1999) Stromal processing peptidase binds transit peptides and initiates their ATP-dependent turnover in chloroplasts. J Cell Biol 147: 33–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter S, Lamppa GK (2002) Determinants for removal and degradation of transit peptides of chloroplast precursor proteins. J Biol Chem 277: 43888–43894 [DOI] [PubMed] [Google Scholar]
- Richter S, Zhong R, Lamppa G (2005) Function of the stromal processing peptidase in the chloroplast import pathway. Physiol Plant 123: 362–368 [Google Scholar]
- Rokka A, Aro EM, Vener AV (2011) Thylakoid phosphoproteins: identification of phosphorylation sites. Methods Mol Biol 684: 171–186 [DOI] [PubMed] [Google Scholar]
- Sandikci A, Gloge F, Martinez M, Mayer MP, Wade R, Bukau B, Kramer G (2013) Dynamic enzyme docking to the ribosome coordinates N-terminal processing with polypeptide folding. Nat Struct Mol Biol 20: 843–850 [DOI] [PubMed] [Google Scholar]
- Staes A, Impens F, Van Damme P, Ruttens B, Goethals M, Demol H, Timmerman E, Vandekerckhove J, Gevaert K (2011) Selecting protein N-terminal peptides by combined fractional diagonal chromatography. Nat Protoc 6: 1130–1141 [DOI] [PubMed] [Google Scholar]
- Starheim KK, Gevaert K, Arnesen T (2012) Protein N-terminal acetyltransferases: when the start matters. Trends Biochem Sci 37: 152–161 [DOI] [PubMed] [Google Scholar]
- Sun Q, Zybailov B, Majeran W, Friso G, Olinares PD, van Wijk KJ (2009) PPDB, the Plant Proteomics Database at Cornell. Nucleic Acids Res 37: D969–D974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasaki T, Sriram SM, Park KS, Kwon YT (2012) The N-end rule pathway. Annu Rev Biochem 81: 261–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobias JW, Shrader TE, Rocap G, Varshavsky A (1991) The N-end rule in bacteria. Science 254: 1374–1377 [DOI] [PubMed] [Google Scholar]
- Trösch R, Jarvis P (2011) The stromal processing peptidase of chloroplasts is essential in Arabidopsis, with knockout mutations causing embryo arrest after the 16-cell stage. PLoS One 6: e23039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsiatsiani L, Timmerman E, De Bock PJ, Vercammen D, Stael S, van de Cotte B, Staes A, Goethals M, Beunens T, Van Damme P, et al. (2013) The Arabidopsis metacaspase9 degradome. Plant Cell 25: 2831–2847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urantowka A, Knorpp C, Olczak T, Kolodziejczak M, Janska H (2005) Plant mitochondria contain at least two i-AAA-like complexes. Plant Mol Biol 59: 239–252 [DOI] [PubMed] [Google Scholar]
- van Wijk KJ. (2015) Protein maturation and proteolysis in plant plastids, mitochondria, and peroxisomes. Annu Rev Plant Biol 66: 75–111 [DOI] [PubMed] [Google Scholar]
- van Wijk KJ, Friso G, Walther D, Schulze WX (2014) Meta-analysis of Arabidopsis thaliana phospho-proteomics data reveals compartmentalization of phosphorylation motifs. Plant Cell 26: 2367–2389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. (2011) The N-end rule pathway and regulation by proteolysis. Protein Sci 20: 1298–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vener AV. (2007) Environmentally modulated phosphorylation and dynamics of proteins in photosynthetic membranes. Biochim Biophys Acta 1767: 449–457 [DOI] [PubMed] [Google Scholar]
- Vener AV, Harms A, Sussman MR, Vierstra RD (2001) Mass spectrometric resolution of reversible protein phosphorylation in photosynthetic membranes of Arabidopsis thaliana. J Biol Chem 276: 6959–6966 [DOI] [PubMed] [Google Scholar]
- Vizcaíno JA, Deutsch EW, Wang R, Csordas A, Reisinger F, Ríos D, Dianes JA, Sun Z, Farrah T, Bandeira N, et al. (2014) ProteomeXchange provides globally coordinated proteomics data submission and dissemination. Nat Biotechnol 32: 223–226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walling LL. (2006) Recycling or regulation? The role of amino-terminal modifying enzymes. Curr Opin Plant Biol 9: 227–233 [DOI] [PubMed] [Google Scholar]
- Xu F, Huang Y, Li L, Gannon P, Linster E, Huber M, Kapos P, Bienvenut W, Polevoda B, Meinnel T, et al. (2015) Two N-terminal acetyltransferases antagonistically regulate the stability of a nod-like receptor in Arabidopsis. Plant Cell 27: 1547–1562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Deery MJ, Gannon L, Powers SJ, Lilley KS, Theodoulou FL (2015) Quantitative proteomics analysis of the Arg/N-end rule pathway of targeted degradation in Arabidopsis roots. Proteomics 15: 2447– 2457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zybailov B, Rutschow H, Friso G, Rudella A, Emanuelsson O, Sun Q, van Wijk KJ (2008) Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS One 3: e1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zybailov B, Sun Q, van Wijk KJ (2009) Workflow for large scale detection and validation of peptide modifications by RPLC-LTQ-Orbitrap: application to the Arabidopsis thaliana leaf proteome and an online modified peptide library. Anal Chem 81: 8015–8024 [DOI] [PubMed] [Google Scholar]