Phosphatidylinositol 3-phosphate 5-kinase is essential for the endosome maturation and cortical microtubule associations for PIN protein trafficking in young root cortical and stele cells.
Abstract
Phosphatidylinositol 3,5-bisphosphate [PtdIns(3,5)P2] is an important lipid in membrane trafficking in animal and yeast systems; however, its role is still largely obscure in plants. Here, we demonstrate that the phosphatidylinositol 3-phosphate 5-kinase, formation of aploid and binucleate cells1 (FAB1)/FYVE finger-containing phosphoinositide kinase (PIKfyve), and its product, PtdIns(3,5)P2, are essential for the maturation process of endosomes to mediate cortical microtubule association of endosomes, thereby controlling proper PIN-FORMED protein trafficking in young cortical and stele cells of root. We found that FAB1 predominantly localizes on the Sorting Nexin1 (SNX1)-residing late endosomes, and a loss of FAB1 function causes the release of late endosomal proteins, Ara7, and SNX1 from the endosome membrane, indicating that FAB1, or its product PtdIns(3,5)P2, mediates the maturation process of the late endosomes. We also found that loss of FAB1 function causes the release of endosomes from cortical microtubules and disturbs proper cortical microtubule organization.
Phosphoinositides play an important role in various cellular processes, including determination of organelle identity and mediating signal transduction by recruiting effector molecules to various organelles (Balla, 2013). Among those, D3-phosphorylated phosphoinositides, phosphatidylinositol 3-phosphate (PtdIns3P) and phosphatidylinositol 3,5-bisphosphate [PtdIns(3,5)P2], play essential roles in the endosomal trafficking and the vacuolar sorting. PtdIns3P is produced from phosphatidylinositol by class III PI3-kinase, vacuolar protein sorting34 (VPS34). In animal cells, PtdIns3P predominantly localizes to the early endosomes and controls endosome maturation, recycling, and degradation of cargo proteins coordinated with Rab5 GTPases (Jean and Kiger, 2012). In Arabidopsis (Arabidopsis thaliana), PtdIns3P mainly resides on the late endosomes and the prevacuolar membrane (Vermeer et al., 2006; Simon et al., 2014). Dysfunction of AtVPS34 resulted in a defect in growth (Welters et al., 1994), root hair elongation (Lee et al., 2008a), and pollen development (Lee et al., 2008b), indicating an important role for AtVPS34 and its product PtdIns3P in plant development. VPS34-mediated PtdIns3P synthesis at the endosomes recruits phosphatidylinositol 3-phosphate 5-kinase formation of aploid and binucleate cells1 (FAB1)/FYVE finger-containing phosphoinositide kinase (PIKfyve), then FAB1/PIKfyve produces PtdIns(3,5)P2 from PtdIns3P to mediate late endosome maturation in yeast (Saccharomyces cerevisiae) and animals (Ho et al., 2012; Jean and Kiger, 2012). PtdIns(3,5)P2 has crucial roles in the maintenance of lysosome/vacuole morphology and acidification, membrane trafficking of proteins, autophagy, and signaling mediation in response to various stresses (Shisheva, 2008).
FAB1 was discovered in yeast, where mutations were found to result in the formation of aploid and binucleate cells (hence its name FAB). In addition, a loss of Fab1p function causes defects in vacuole function and morphology, cell surface integrity, and cell growth (Yamamoto et al., 1995). In mammalian cells, this kinase is called PIKfyve (FYVE is a PI3P-binding domain). FAB1/PIKfyve forms a protein complex with an adaptor-like protein, Vacuole14 (Bonangelino et al., 1997) and PtdIns(3,5)P2 5-phosphatase (Fig. 4; Gary et al., 2002), indicating that the FAB1 complex catalyzes both PtdIns(3,5)P2 synthesis and turnover simultaneously. In mammalian cells, interference of FAB1/PIKfyve function causes severe defects during embryogenesis, resulting in embryonic lethality in Drosophila spp., Caenorhabditis elegans, and mice (Nicot et al., 2006; Rusten et al., 2006; Ikonomov et al., 2011; Takasuga et al., 2013). Whereas most genomes from human to yeast contain a single-copy gene, the Arabidopsis genome codes for four FAB1 genes (FAB1A–D), of which only FAB1A and FAB1B contain a FYVE domain (Mueller-Roeber and Pical, 2002), and fab1a/fab1b double mutant reveals male gametophyte lethality phenotype in Arabidopsis (Whitley et al., 2009). The mutant pollen shows severe defects in vacuolar reorganization following the first mitotic division of development, suggesting an important role of FAB1 and PtdIns(3,5)P2 in vacuolar rearrangement for pollen development (Whitley et al., 2009).
Figure 4.
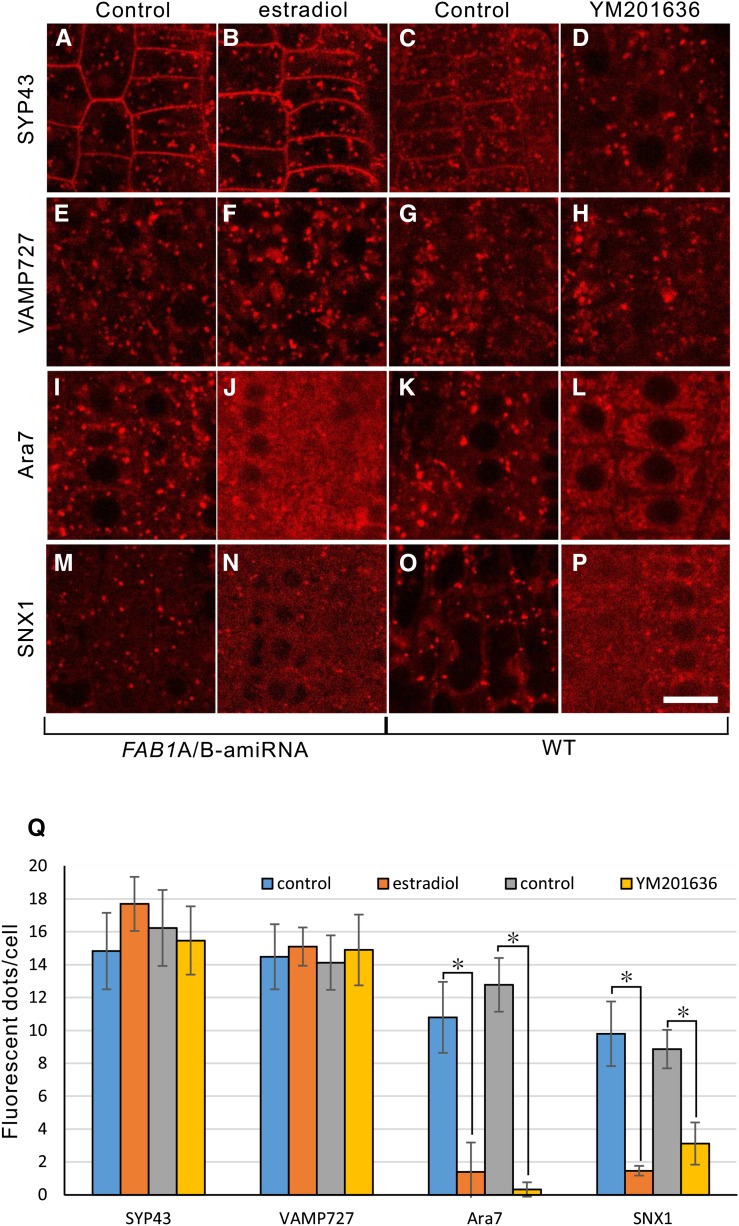
Localization of endosomal markers upon down-regulation of FAB1A/B or inhibition of PtdIns(3,5)P2 synthesis in young root cortical cells. Localization of mRFP-SYP43, mRFP-vesicle-associated membrane protein (VAMP727), mRFP-ARA7, and SNX1-mRFP without estradiol (A, E, I, and M) or with estradiol (B, F, J, and N) in the FAB1A/B-amiRNA line, or wild-type (WT) plants without YM201636 (C, G, K, and O) or with YM201636 (D, H, L, and P). Bar = 10 μm. Measurement of fluorescent dot structures (Q). Data represent fluorescent dots per cell (mean ± sd). *, P < 0.001 (Student’s t test).
We previously developed a transgenic Arabidopsis line that is able to conditionally down-regulate FAB1A and FAB1B expression simultaneously, and demonstrated that a loss of FAB1 function causes various abnormal phenotypes, including growth inhibition, hypersensitivity to exogenous auxin, disturbance of root gravitropism, and floral organ abnormalities (Hirano et al., 2011). In addition, we found that down-regulation of FAB1A/B expression impaired endomembrane homeostasis, including endocytosis, vacuole formation, and vacuolar acidification, likely causing pleiotropic developmental phenotypes that mostly related to the auxin signaling in Arabidopsis (Hirano et al., 2011; Hirano and Sato, 2011). In plants, auxin is a crucial phytohormone that has a wide variety of physiological roles associated with growth, development, and tropic responses (Zhao, 2010). The polar cell-to-cell transport of auxin is mediated by auxin transporters localized on the plasma membrane (PM), such as PIN-FORMED (PIN) proteins (Vieten et al., 2007; Feraru and Friml, 2008). PINs are used as model molecules for polarity establishment on the PM in Arabidopsis. The establishment of PIN polarity is accomplished by the recycling of PINs between the PM and endosomal compartments comprising the trans-Golgi network/early endosomes (TGN/EEs) and the late endosomes (LEs)/prevacuolar compartments. The PIN-recycling pathway is mediated by multiple endosomal regulatory proteins, such as Rab family GTPases and Sorting Nexin (SNX; Jaillais et al., 2006; Park and Jürgens, 2011).
Rab proteins function as molecular switches to regulate the tethering and fusion step of transport vesicles to target membranes. Rab5 members of the Rab GTPases have various functions in the endocytic pathway in eukaryotes. The maturation of the early-to-late endosomes is regulated by Rab5-to-Rab7 conversion, which is regulated by the Mon1/Sand-1-Ccz1 complex (Nordmann et al., 2010; Poteryaev et al., 2010). In plants, Rab5-family proteins, Ara6 and Ara7, and Rha1 play important roles in Rab5-mediated endosomal trafficking including the vacuolar trafficking pathway, thereby regulating of the polar transport of auxin and responses to environmental conditions (Ebine et al., 2011; Inoue et al., 2013).
SNXs are composed of two conserved domains: the PHOX domain, involved in the interaction with the phosphoinositides, PtdIns3P and PtdIns (3,5)P2, in the endosomal membrane in animals (Cozier et al., 2002), and the BAR domain, mediating dimerization and binding to curved membranes (Peter et al., 2004). Loss of SNX function disrupts the stable association of the retromer subcomplex, VPS26-Vps29-Vps35, with endosomal membranes, and thus results in retromer dysfunction, indicating that SNXs have a crucial role in the assembly and maintenance of the core retromer function (Teasdale et al., 2001; Cullen and Korswagen, 2012). The first plant SNX was identified as a protein that interacts with various receptor kinases in Brassica oleracea (Vanoosthuyse et al., 2003), and then three SNX genes (SNX1, SNX2a, and SNX2b) were identified in Arabidopsis. The snx1 null mutant exhibits a semidwarf phenotype with other subtle developmental defects (Pourcher et al., 2010). SNX1 is localized to the late endosome and is involved in PIN2 recycling between endosomes and the PM (Jaillais et al., 2006). SNX1 has been reported to interact with cortical microtubules via the microtubule-associated protein Cytoplasmic Linker Associated Protein (CLASP), and the clasp1 null mutant displays aberrant SNX1 endosomes and enhanced PIN2 degradation in the lytic vacuoles, suggesting that an association of SNX1 endosomes and CLASP is important for recycling of PIN transporters (Ambrose et al., 2013).
Although many analyses of FAB1/PIKfyve, Rab5 family GTPases, SNXs, and microtubles have been reported, and there are significant similarities in endosomal trafficking, a functional relationship between them is still largely obscure.
In this study, we demonstrate that FAB1 produced PtdIns(3,5)P2 in Arabidopsis, and knockdown of FAB1 expression or inhibition of FAB1 activity with a FAB1/PIKfyve inhibitor, YM201636, decreased PtdIns(3,5)P2 content. We also found that FAB1 and its product PtdIns(3,5)P2 mediate the late endosome maturation by recruiting endosomal effector molecules, Ara7 and SNX1, onto endosomes to establish endosome-cortical microtubule interaction. Subsequently, the basal polarity of PIN2 in young cortical cells and PIN1 in stele cells is achieved.
RESULTS
YM201636 Is a Selective Inhibitor for the Synthesis of PtdIns(3,5)P2 in Arabidopsis
YM201636 is a selective inhibitor for PIKfyve, blocking PtdIns(3,5)P2 production and regulating a number of intracellular membrane trafficking pathways without disturbing other PtdInsP kinases and protein kinase B functions in mammals (Jefferies et al., 2008). In Arabidopsis, YM201636 treatment reduces vacuolar acidification, convolution of guard cell, and delayed the stomatal closure in response to abscisic acid (Bak et al., 2013). However, whether YM201636 functions as a selective inhibitor of PtdIns(3,5)P2 synthesis is unknown in plants. To investigate whether YM201636 treatment could affect PtdIns(3,5)P2 levels in Arabidopsis, we used a transgenic line that moderately expressed FAB1A-GFP under the control of its native promoter (Hirano et al., 2011a), because only trace amounts of PtdIns(3,5)P2 could be detected in wild-type Arabidopsis (DeWald et al., 2001). We measured the level of PtdIns(3,5)P2 from 5-d-old seedlings of the wild-type or FAB1A-GFP expression line after overnight 32Pi labeling, with or without YM201636. 32Pi labeling and subsequent phospholipid analysis using thin-layer chromatography (TLC; Meijer et al., 2001; Munnik and Zarza, 2013) indicated that the ectopic expression of FAB1A-GFP increased the PtdIns(3,5)P2 content 5-fold compared with the wild type, whereas YM201636 effectively inhibited the production of PtdIns(3,5)P2 in the FAB1A-GFP expressing line (Fig. 1A) without significantly affecting any of the other phospholipids (Fig. 1, B–E). These results revealed that FAB1A-GFP has an ability to produce PtdIns(3,5)P2, and YM201636 functions as a specific inhibitor of FAB1 activity in Arabidopsis.
Figure 1.
TLC measurement of phospholipids in wild-type (WT) and FAB1A-GFP-expressing plants. Five-day-old seedlings were labeled overnight with 32Pi, after which they were further incubated for 2 h in the presence (YM) or absence (DMSO) of 1 μM YM201636. After extraction, lipids were separated by TLC and quantified by phosphoimaging. Data are expressed as the percentage of total 32P-phospholipids. The levels of PtdIns(3,5)P2 (A), PtdIns(4,5)P2 (B), PtdIns3P (C), phosphatidic acid (PA; D), phosphatidylinositol (PI), phosphatidylcholine (PC), phosphatidylethanolamine (PE), and phosphatidylglycerol (PG; E) are shown. Three independent samples containing three seedlings each were used. *, Significant differences between seedlings treated with or without YM201636 are indicated by (Student’s t test, P < 0.001). Data represent the mean ± sd. Experiments were repeated three times and gave similar results. DMSO, Dimethyl sulfoxide; PL, phospholipid.
Loss of FAB1 Function Impairs Gravistimulation-Dependent PIN2 Degradation in the Root Epidermal Cells
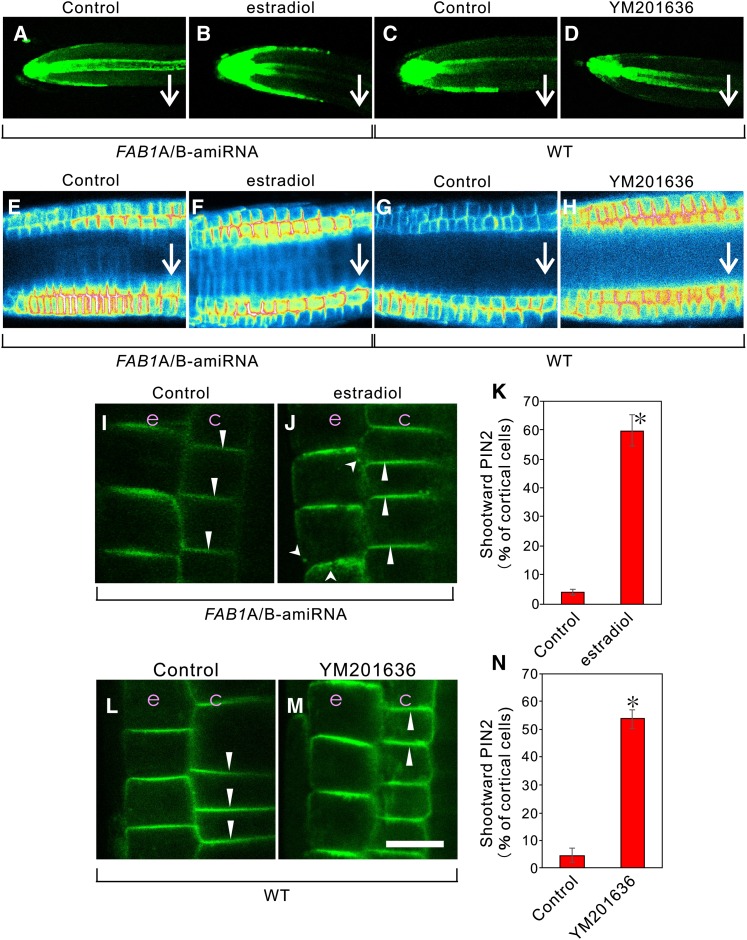
We previously generated artificial microRNA (amiRNA)-mediated knockdown plants to conditionally down-regulate FAB1A and FAB1B expression simultaneously by the addition of estradiol, and showed that the conditional knockdown of FAB1A/B expression severely impaired the gravitropic response in the root (Hirano et al., 2011). In the presence of 1 μm estradiol in the medium, FAB1A and FAB1B expression was reduced to 0.43- and 0.31-fold, respectively, compared with the control condition (Supplemental Fig. S1). Next, we examined how down-regulation of FAB1A/B or YM201636 treatment affects the auxin distribution in the root cells by crossing lines of the DR5rev::GFP (Friml et al., 2003) and the FAB1A/B-amiRNA (Hirano et al., 2011). The resultant DR5rev::GFP/FAB1A/B-amiRNA plants were subject to 180-min gravistimulation, during which the plants were vertically rotated 90°. After gravistimulation, auxin was predominantly distributed on the lower side of the root epidermal cells in the control plants (Fig. 2, A and C), whereas in estradiol (+; Fig. 2B) or YM201636 (+; Fig. 2D) plants, auxin was equally distributed on the lower and upper sides of the root epidermal cells. We then investigated the gravity-dependent degradation of PIN2 in root epidermal cells (Kleine-Vehn et al., 2008b) and observed that, in the absence of estradiol or YM201636, PIN2 was specifically degraded in the upper epidermal cell files, whereas the gravity-dependent degradation of PIN2 in the upper cell files was completely inhibited when FAB1A/B expression was down-regulated or FAB1 activity was inhibited (Fig. 2, E–H).
Figure 2.
Changes in auxin distribution and alteration of PIN2 localization by down-regulation of FAB1A/B or inhibition of PtdIns(3,5)P2 synthesis. DR5rev::GFP signal in the estradiol-inducible amiRNA-mediated conditional knockdown FAB1A/B line (FAB1A/B-amiRNA; A and B) and in the wild type (WT; C and D). PIN2::PIN2-GFP signal in the FAB1A/B-amiRNA line (E, F, I, and J) and in the wild type (G, H, L, and M). A to D, GFP fluorescence from the root tip of DR5::GFP plants after gravistimulation for 180 min with (B) or without (A) estradiol and with (D) or without (C) YM201636. E to H, Intensity of PIN2 fluorescence on the PM in the root tip after gravistimulation for 180 min with (F) or without (E) estradiol and with (H) or without (G) YM201636. I to K, PIN2 localization with (J) or without (I) estradiol and the percentage of apical PIN2 localization in young cortical cells (K; n > 22 roots). L to N, PIN2 localization with (M) or without (L) YM201636 and the percentage of apical PIN2 localization in cortical cells (N; n > 20 roots). Arrows indicate the orientation of PIN2 on the PM of young cortical cells. Arrowheads depict PIN2 localized to the endosomes, and triangles depict PIN2 polarity. c, Cortex; e, epidermis. Green signal represents GFP fluorescence. Bar = 10 μm. Data in K and N are shown as the mean ± sd. *, P < 0.001 (Student’s t test).
FAB1 Knockdown Causes Relocation of the Basal Polarity of PIN2 in Young Root Cortical Cells
We examined whether PIN2 localization is affected by the loss of FAB1 function in root cells. In the wild type, PIN2::PIN2-GFP normally shows shootward (apical) localization in the epidermis and rootward (basal) localization in the cortex of the root meristematic zone (Kleine-Vehn et al., 2008a; Rahman et al., 2010). We observed that FAB1A/B knockdown induced a shift toward the apical localization of PIN2 in young cortical cells of the root meristematic zone: about one-half of the PIN2 relocated to the apical orientation (Fig. 2, I–K). Note that the presence of cytoplasmic punctate structures and the longitudinal face localization of PIN2 in the root epidermal cells were also significantly increased in the presence of estradiol (Fig. 2, I–K; Supplemental Figs. S2 and S3). Likewise, the treatment of YM201636 induced the same effects on PIN2 localization in the young cortical and epidermal cells in the root (Fig. 2, L–N; Supplemental Figs. S2 and S3). Immunofluorescence detection of PIN2 using anti-PIN2 antibody also demonstrated that the basal localization of PIN2 was severely impaired when FAB1A/B expression was down-regulated (Supplemental Fig. S4). We also observed that the basal PIN1 localization in stele cells was severely disrupted; the apical and lateral mistargeting of PIN1 was observed in FAB1A/B-amiRNA plants expressing PIN1-GFP in the presence of estradiol (Supplemental Fig. S5) or by YM201636 treatment (Supplemental Fig. S5). These results indicated that impairment of FAB1A/B function disrupts the basal localization of both PIN1 in stele cells and PIN2 in young cortical cells, thereby possibly disturbing proper auxin flow in the root.
FAB1 Predominantly Localizes to the SNX1-Positive Endosomes
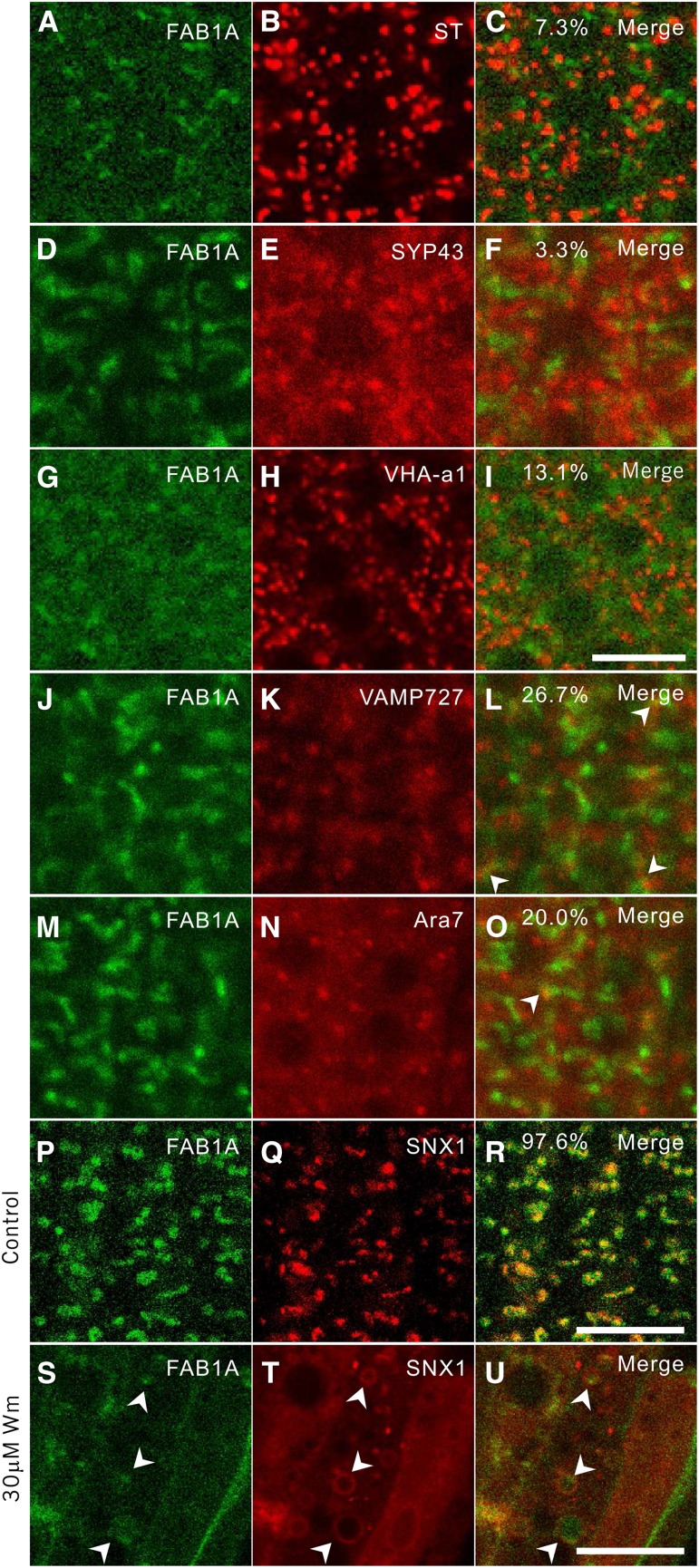
Given that the establishment of PIN polarity is accomplished by the recycling of PIN between the PM and endosomal compartments (Kleine-Vehn and Friml, 2008), the basal localization of PIN2 in the young cortical cells may be established by recycling through FAB1-positive endosomes. We therefore investigated to which endosomal compartment FAB1 localizes. We crossed the FAB1A-GFP expression line with various endosomal marker lines that express mRFP-tagged proteins targeted to different endosomal compartments. We observed that FAB1A-GFP rarely colocalized with Golgi marker ST-mRFP (7.3%) and TGN/EE markers mRFP-SYP43 (3.3%) and V-ATPase V0 subunit a1 (VHA-a1)-mRFP (13.1%) in root young cortical cells (Fig. 3, A–F). Fluorescence from FAB1A-GFP and the late endosomal markers, mRFP-VAMP727 (26.7%) and mRFP-Ara7 (20.0%), was partially colocalized (Fig. 3, G–O). In contrast, FAB1A-GFP predominantly colocalized with SNX1-mRFP (Fig. 3, P–R). Wortmannin is a selective inhibitor of PI3 kinase, and treatment with this inhibitor induces abnormal late endosomal structures in Arabidopsis (Jaillais et al., 2006; Vermeer et al., 2006). The FAB1- and SNX1-labeled endosomal compartments were apparently enlarged by treatment with wortmannin (Fig. 3, S–U). Under the wortmannin-treated condition, the SNX1-mRFP fluorescence exhibited typical bubble-like structures (Fig. 3T; Supplemental Fig. S6). Intriguingly, most of the FAB1A-GFP fluorescence was observed as dot-like structures, surrounded by the SNX-mRFP bubble-like structures. Note that a faint fluorescence of FAB1A-GFP and SNX1-mRFP in the cytosol could be observed after wortmannin treatment (Fig. 3, S–U; Supplemental Fig. S6), indicating that part of FAB1 and SNX1 was dissociated from the endosome membrane, ending up in the cytosol. Taken together, we concluded that FAB1A-GFP localized to the SNX1-positive late endosomes.
Figure 3.
Colocalization of FAB1A-GFP to the SNX1-positive late endosomes in young root cortical cells. A to O, Colocalization of young root cortical cells of FAB1A-GFP with each endosomal marker, sialyl transferase (ST)-monomeric Red Fluorescent Protein (mRFP; B and C), mRFP-Syntaxin of Plant Protein43 (SYP43; E and F), VHA-a1-mRFP (H and I), and mRFP-VAMP727 (K and L), and mRFP-ARA7 (N and O) observed by confocal microscopy. Green and red signals represent fluorescence of GFP and mRFP, respectively. P to U, Colocalization of FAB1A-GFP and SNX1-mRFP without (P–R) or with 30 μm wortmannin (S–U) for 90 min. Percentages in merged images represent colocalization rates of two fluorescent proteins captured from at least five images of five plants. Bar = 10 μm.
PtdIns(3,5)P2 Is Required for Recruitment of Late Endosomal Effector Proteins on Endosomes
In animals and yeasts, PtdIns(3,5)P2 binds and recruits various effectors to the endosomal membrane to establish endosome identity (Ho et al., 2012; McCartney et al., 2014). Similarly, we assumed that the late endosome identity would be established by assembly of effector molecules on the early endosomal membrane by binding of PtdIns(3,5)P2 in Arabidopsis. We therefore investigated whether localization of the late endosome-residing regulatory molecules is affected by FAB1 down-regulation or by inhibition of PtdIns(3,5)P2 synthesis in Arabidopsis. Neither FAB1A/B down-regulation nor treatment of the wild type with YM201636 altered the localization of mRFP-SYP43 or mRFP-VAMP727 (Fig. 4, A–H), whereas the punctate structures of mRFP-Ara7 and SNX1-mRFP were decreased, and the fluorescence from mRFP-Ara7 and SNX1-mRFP was dispersed throughout the cytosol (Fig. 4, I–P). No changes in the localization of mRFP-Ara7 or SNX1-mRFP was observed in the presence of estradiol in wild-type background (Supplemental Fig. S7).
Association of SNX1 with PtdIns(3,5)P2 and FAB1
Because human SNX binds both PtdIns3P and PtdIns(3,5)P2 in vitro (Cozier et al., 2002), we investigated whether Arabidopsis SNX1 could associate with these phosphoinositides. SNX1 tightly binds PtdIns3P and PtdIns(3,5)P2 but not PtdIns or PtdIns(4,5)P2 in vitro (Fig. 5A). Note that the full-length GST-SNX1 bound more tightly to the two phosphoinositides than the truncated one. Coexistence of FAB1 and SNX1 on FAB1/SNX1 endosomes (Fig. 3, P–R) suggests that the proteins could form a protein complex. To test this possibility, we performed a BiFC experiment. Using particle bombardment, the N-terminal fragment of yellow fluorescent protein (YFP)-fused FAB1A (NYFP-FAB1A) and the C-terminal fragment of YFP-fused SNX1 (CYFP-SNX1) were transiently introduced into vein cells of rosette leaves of wild-type and SNX1-mRFP-expressing Arabidopsis plants and onion (Allium cepa) epidermal cells. A negative control NYFP/CYFP combination yielded no fluorescence (Fig. 5, B and C), whereas full-length NYFP-SNX1 and CYFP-FAB1 showed clustered, punctate fluorescent structures in the vein cells of a rosette leaf (Fig. 5, E–G) and epidermal cells of onion (Supplemental Fig. S8). The YFP signals of the BiFC assay were completely merged with the fluorescence of SNX1-mRFP (Supplemental Fig. S9). In addition, a coimmunoprecipitation experiment using an anti-FAB1A antibody revealed the coexistence of SNX1-GFP with FAB1A (Fig. 5H). From these results, we concluded that SNX1 localizes to the late endosomes by forming a complex both with FAB1 and its product, PtdIns(3,5)P2, on endosome membranes, although we could not determine whether the association between FAB1 and SNX1 is direct or indirect via lipid components.
Figure 5.
Interaction between SNX1 and PtdIns(3,5)P2 and FAB1. A, Detection of SNX1 binding with phosphoinositides [PtdIns3P, PtdIns(3,5)P2, and PtdIns(4,5)P2] by western blotting using anti-glutathione S-transferase (anti-GST) antibody. Dried down phosphatidyl-Ser, phosphatidylethanolamine, and phosphatidylcholine (each at 28.5% [w/w]) was supplemented with 14.5% (w/w) of the relevant phosphoinositide [PtdIns, PtdIns3P, PtdIns(3,5)P2, or PtdIns(4,5)P2; total lipid content of 70 μm]. Bimolecular fluorescent complementation (BiFC) assay in vein cells of Arabidopsis leaf, a negative control NYFP and CYFP (B–D), and full-length NYFP-FAB1 and CYFP-SNX1 (E–G). SNX1 and FAB1 were fused to the C terminus of NYFP and CYFP, respectively. H, Coimmunoprecipitation of SNX1-GFP with FAB1A. Immunoprecipitation was performed with anti-FAB1A peptide antibody, and the protein was extracted from 5-d-old seedlings of the wild type (WT) and SNX1-GFP-expressing line and detected with anti-GFP antibody. Top, FAB1A blots; bottom, GFP blots. Beads, using the lysis buffer instead of lysate. I to Q, Five-day-old seedlings of the wild type (I and M), snx1-1 mutant (J and N), and overexpressing FAB1A-GFP in snx1-1 (K, L, O, and P) grown on the Suc-deficient one-half-strength Murashige and Skoog (MS) medium (M–P) or 1/2 MS medium with 1% Suc (I–L). Q, Root length of the wild type, snx1-1 mutant, and snx1-1 overexpressing FAB1A-GFP (lines #1 and #2; n > 31). Data represent the mean ± sd. *, P < 0.001 (Student’s t test). IP, Immunoprecipitation; WB, western blotting.
Ectopic Expression of FAB1A Complements the snx1-1 Phenotype
Given that the presence of FAB1 is essential for SNX1 localization on the endosomes, a mutation in SNX1 could affect FAB1 localization. To test this possibility, GFP-FAB1A was ectopically expressed in the snx1-1 mutant. As shown in Supplemental Fig. S10, the subcellular localization of FAB1A-GFP was unchanged in the snx1-1 mutant background. Next, we investigated whether FAB1 expression could complement a snx1-1 phenotype. Seedlings of snx1-1 exhibited a pronounced growth arrest after cotyledon formation on Suc-depleted medium. The growth arrest of Suc-depleted snx1-1 seedlings was conditional and could be fully rescued by Suc application (Kleine-Vehn et al., 2008b). As shown in Figure 5, I–Q, ectopic expression of FAB1A-GFP completely rescued the Suc-dependent seedling growth inhibition phenotype of the snx1-1 mutant. Under these conditions, FAB1A expression increased up to 28-fold in FAB1A-GFP/snx1-1 compared with the wild type in the Suc-depleted medium (Supplemental Figure S11). Together, these results indicated that FAB1 controls SNX1 localization and function in Arabidopsis. It is likely that the complementation capability of FAB1 to the snx1-1 phenotype might be due to the existence of other SNX family proteins, such as SNX2a and SNX2b, in Arabidopsis (Pourcher et al., 2010).
FAB1 Endosomes Communicate with Cortical Microtubules
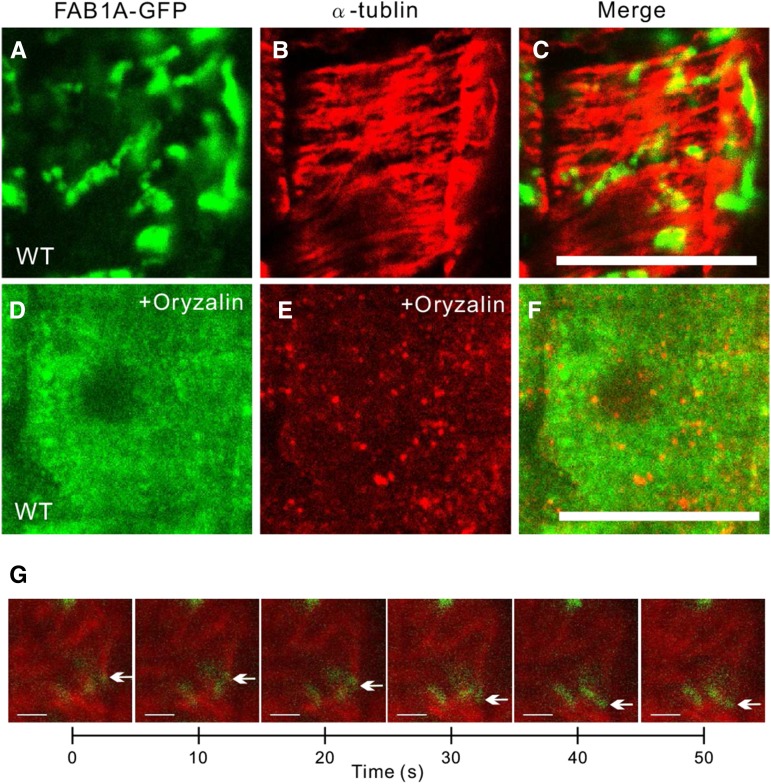
Similar to the structure of SNX1 endosomes (Ambrose et al., 2013), FAB1A-GFP-residing organelles formed tubule-like vesicular clusters that seemed to be partly associated with and slowly moving along cortical microtubules in wild-type cortical cells (Fig. 6, A–C and G; Supplemental Fig. S1; Supplemental Movie S1). In contrast, FAB1A-GFP was completely dispersed throughout the cytosol in plants treated with a microtubule polymerization inhibitor, oryzalin (Fig. 6, D–F). These results suggested that proper formation of the FAB1-poitive endosomes depends on the association with and the structure of the cortical microtubules.
Figure 6.
The structure of FAB1-positive endosomes is impaired by oryzalin treatment. Immunofluorescence detection of FAB1A-GFP and α-tubulin in the wild type (WT; A–C) and wild type with oryzalin treatment (D–F). G, Time lapse series of mRFP-β-tubilin (TUB6)-labeled microtubules (MTs; red) and FAB1A-GFP-positive endosomes (green). Arrows depict clustered endosomes along with cortical microtubules. (See also Supplemental Movie S1.) Green and red signals represent FAB1A-GFP and β-tubilin, respectively.
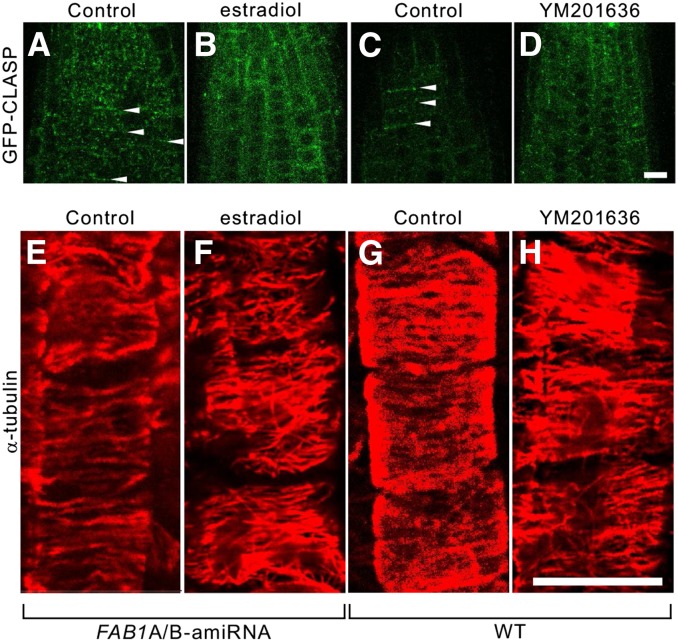
Notably, we found that the granular structures of GFP-CLASP along the apical and basal margins of cells were relocated to the lateral margins of the cells, or randomly scattered in the cytosol, probably due to knockdown of FAB1 function (Fig. 7, A and B) or YM201636 treatment (Fig. 7, C and D). In addition, transverse arrays of cortical microtubules along the longitudinal faces of growing cells were disrupted and randomly relocated upon FAB1 down-regulation (Fig. 7, E and F) or the inhibition of PtdIns(3,5)P2 synthesis (Fig. 7, G and H).
Figure 7.
Changes in the localization of GFP-CLASP and orientation of cortical microtubules by down-regulation of FAB1A/B or inhibition of PtdIns(3,5)P2 synthesis. A to D, Localization of GFP-CLASP with (B) or without (A) estradiol, and with (D) or without (C) YM201636. Orientation of cortical microtubules with (F) or without (E) estradiol in the FAB1A/B-amiRNA line, and with (H) or without (G) YM201636 in wild-type (WT) plants.
Together, these results suggested that the functional interaction between FAB1-positive endosomes and cortical microtubules is essential not only for maintaining the structure of FAB1 endosomes but also for proper organization of cortical microtubules in Arabidopsis root cells.
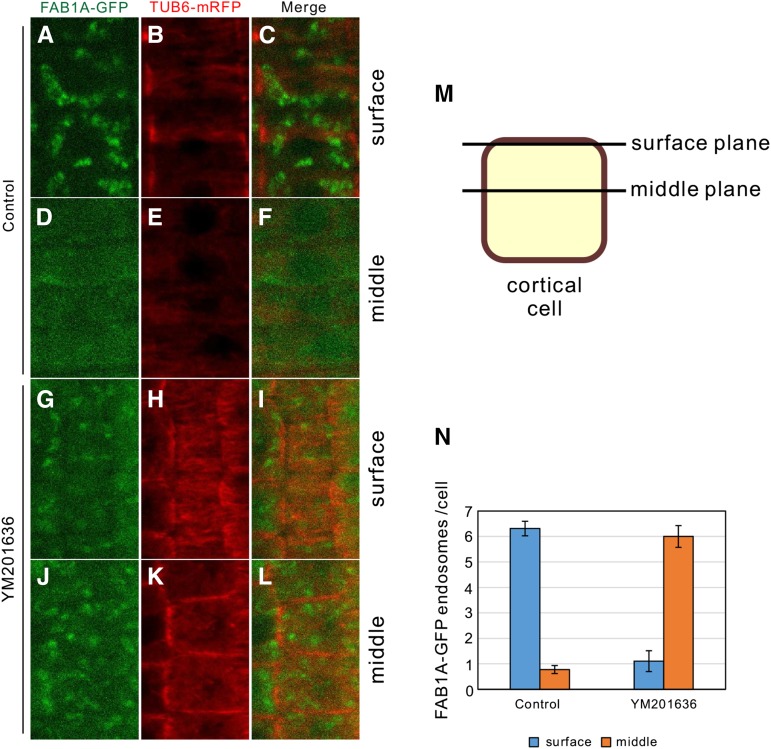
To investigate how FAB1 endosomes and cortical microtubules associate, we crossed the transgenic Arabidopsis lines expressing FAB1A-GFP and TUB6-mRFP and analyzed their association using confocal microscopy. In the absence of YM201636, FAB1 endosomes localized beneath the cortical microtubule plane (Fig. 8, A–C and N), but FAB1 endosomes were rarely observed in the middle plane of the cortical cells (Fig. 8, D–F and N). However, after YM201636 treatment, the FAB1 endosomes observed on the surface plane were decreased (Fig. 8, G–I and M), whereas FAB1 endosomes in the middle plane of the cell were significantly increased (Fig. 8, J–L and N). Taken together, we concluded that the late endosomes are associated with cortical microtubules after endosome maturation mediated by PtdIns(3,5)P2, generated by FAB1 function.
Figure 8.
Localization of FAB1 endosomes on surface layer requires their maturation by FAB1. A to F, Localization of FAB1A-GFP and α-tubulin on the surface plane (A–C) or on the middle plane (D–F) in the control. G to L, Localization of FAB1A-GFP and α-tubulin on the surface plane (G–I) or on the middle plane (J–L) with YM201636. M, Schematic drawing of surface plane and middle plane of the cortical cell. N, Number of FAB1A-GFP endosomes per one cell on the surface or middle planes (control, n = 80; YM201636, n = 19).
DISCUSSION
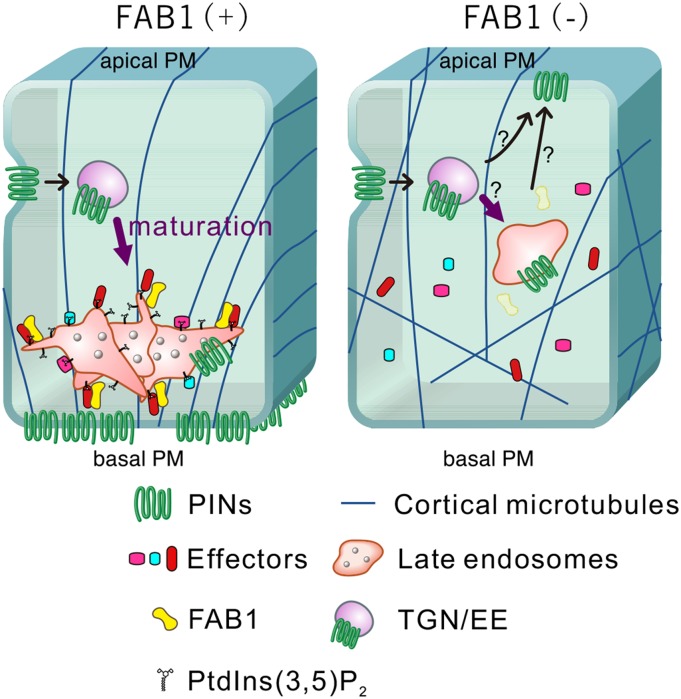
In this study, we demonstrated that FAB1 produces PtdIns(3,5)P2 from PtdIns3P on endosomes, to construct a scaffold to bind late endosomal effector proteins, such as Ara7 and SNX1, for endosome maturation. As a result, the identity and function of late endosome necessary for the proper PIN trafficking are established. If the FAB1 function is compromised, the production of PtdIns(3,5)P2 is impaired, and the endosome maturation is inhibited at the early stage of the maturation process due to lack of late endosomal effector proteins on the endosomal membrane (Fig. 9).
Figure 9.
A possible model of the function of FAB1 and its product PtdIns(3,5)P2 on late endosome maturation in young root cortical and stele cells in Arabidopsis. PIN proteins are recycled via the TGN/EE and FAB1/SNX1-positive endosomes to the basal PM in young root cortical and stele cells. Mature FAB1/SNX1-positive endosomes are partly attached and move along cortical microtubules with late endosomal effector proteins and microtubule-associated proteins. If the FAB1 function is compromised, the late endosomal effector proteins could not be accumulated due to the lack of the scaffold molecule, PtdIns(3,5)P2; consequently, endosome maturation is inhibited at the early stage of the process.
FAB1 Mediates Endosome Maturation by Recruiting Endosomal Effector Molecules
We observed that the endosomal proteins, ARA7 and SNX1, localized to the endosomal membrane in a FAB1- and PtdIns(3,5)P2-dependent manner (Fig. 3, I–P). Phosphoinositides are well known to have multiple important functions, including signal transduction, organization of the cytoskeleton, membrane trafficking, and determination of organelle identity, by recruiting various effector proteins to the host organelle membranes (Di Paolo and De Camilli, 2006; Balla 2013). Although PtdIns(3,5)P2 is a very low-abundant phosphoinositide that is predominantly found in the vacuole of yeasts and in endolysosomes of animals (McCartney et al., 2014), PtdIns(3,5)P2 is known to bind various effector molecules that contain lipid-binding domains, such as Autophagy18 (Atg18), Atg21, Hvs2, Tup1, Raptor (WD40), SNXs (PX domain), Cti6 (PHD domain), clavesin (Sec14 domain), and class II formins (PTEN domain; McCartney et al., 2014). Human and Arabidopsis SNX1 show an ability to bind both PtdIns3P and PtdIns(3,5)P2 in vitro (Cozier et al., 2002; Fig. 4A). However, the inhibition of PtdIns(3,5)P2 synthesis or FAB1A/B knockdown induced the dissociation of SNX1 from endosomal membrane (Figs. 4, M–P, and 5, A–H), suggesting that the association of SNX1 with the endosomal membrane requires PtdIns(3,5)P2 but not PtdIns3P in vivo in Arabidopsis.
We observed that an Arabidopsis Rab5 homolog, Ara7, dissociated from endosomal membranes with the loss of FAB1 function (Fig. 3, I and J) or inhibition of PtdIns(3,5)P2 production (Fig. 3, K and L). The small GTPases Rab5 and Rab7 are key determinants of early and late endosomes, respectively, and Rab5-to-Rab7 conversion on the endosome membrane is required for early-to-late endosome maturation in eukaryotic cells (Rink et al., 2005). The endosomal maturation step involves a SAND-1/MON1 family protein mediating Rab5-to-Rab7 conversion (Poteryaev et al., 2010; Cui et al., 2014). In Arabidopsis, SAND protein is not required for early-to-late endosomal maturation, but mediates the Rab5-to-Rab7 conversion process (Singh et al., 2014). If Ara7 functions in endosome maturation as a conventional Rab5 molecule, FAB1 and/or PtdIns(3,5)P2 might be required at an early stage of the transition from early to late endosomes to recruit various late-endosomal effector proteins, including Ara7, to the endosomal membrane. Considering these results, we concluded that a loss of FAB1 function prevents the binding of SNX1 and Ara7 to the early endosome membrane due to lack of PtdIns(3,5)P2 as a scaffolding molecule on the endosome membrane. As a result, no maturation of the endosomes occurs due to the lack of the late endosomal effector proteins.
FAB1 Dysfunction Impairs Proper Trafficking of PIN Proteins
Ambrose et al. (2013) have reported that the microtubule-associated protein CLASP interacts with SNX1, and SNX1 endosomes were disturbed and PIN2 degradation in the lytic vacuoles was enhanced in the clasp-1 null mutant. Similarly, we demonstrated that FAB1-positive endosomes are partly attached to cortical microtubules (Fig. 6, A–C), and that disturbance of microtubule stability by oryzalin treatment induced the disruption in FAB1-positive endosomes (Fig. 6, D–F). The abnormal FAB1-positive endosome structure induced by microtubule instability might be because FAB1 and SNX1 localize mostly to the same late endosomal compartments connecting with cortical microtubules (Fig. 3, P–R), implying that the FAB1/SNX1-carrying endosomes are associated with cortical microtubules via the SNX1-CLASP interaction. However, enhanced PIN2 degradation phenotype in snx1-1 (Kleine-Vehn et al., 2008b) and clasp-1 (Ambrose et al., 2013) mutants in root epidermal cells was not observed with FAB1 dysfunction or YM201636 treatment. Besides, the basal-to-apical relocation of PIN proteins was never observed in both snx1-1 and clasp-1 mutants (Jaillais et al., 2006; Kleine-Vehn et al., 2008b; Ambrose et al., 2013). These discrepancies in PIN localization and degradation phenotypes suggest that these phenotypes caused by loss of FAB1 function are independent of SNX1 and CLASP function. Thus, changes in PIN2 localization under various treatments or genetic backgrounds that might not always be directly associated with the polar targeting in young cortical cells.
Brefeldin A treatment or mutation in GNOM or GNOM-LIKE1 (GNL1) induces the basal-to-apical relocation of PIN1 in stele and PIN2 in young cortical cells (Geldner et al., 2003; Doyle et al., 2015). These findings indicate that the basal localization of PIN1 in stele and PIN2 in young cortical cells is mediated by a brefeldin A-sensitive ADP-ribosylation factor-Guanine nucleotide exchange factor-, GNOM-, and/or GNL1-dependent recycling pathway, whereas in epidermal and mature cortical cells, the apical PIN2 localization is GNOM independent (Kleine-Vehn et al., 2008a; Kleine-Vehn and Friml 2008; Rahman et al., 2010; Doyle et al., 2015). The basal-to-apical relocation of PIN1 in stele and PIN2 in young cortical cells is also induced by treatment with a microtubule polymerization inhibitor, oryzalin (Kleine-Vehn et al., 2008a).
In this study, we demonstrated that the basal-to-apical relocation of PIN1 in stele and PIN2 in young cortical cells was induced by the loss of FAB1 function and the inhibition of PtdIns(3,5)P2 synthesis (Fig. 2, I–N; Supplemental Figs. S2–S5). This observation is the phenocopy of gnom and/or gnl1 mutants (Geldner et al., 2003; Doyle et al., 2015). Furthermore, disruption of cortical microtubule structure by oryzalin causes the disappearance of FAB1-positive endosomes (Fig. 6, D–F). Hence, we reasoned that the basal-to-apical relocation of PIN1 in stele and PIN2 in young cortical cells by oryzalin treatment might be due to the impairment of the function of FAB1 endosomes. These data suggest that the endosome maturation process necessary for the proper basal localization of PIN proteins is inhibited at the early stage in the TGN/EEs-to-LEs transition, or the LE function is impaired by the disruption of endosome-cortical microtubule association.
We also observed the gravistimulation-dependent PIN2 degradation in root epidermal cells (Fig. 2, A–G). Given that the endocytosis of FM4-64 is severely delayed by FAB1 dysfunction (Hirano and Sato, 2011) and PIN2 is specifically degraded in the lytic vacuole of the upper epidermal cell files after gravistimulation (Kleine-Vehn et al., 2008b), we reasoned that the inhibition of PIN2 degradation in the upper cell files after gravistimulation was due to defective PIN2 endocytosis and subsequent degradation in the lytic vacuole by loss of PtdIns(3,5)P2 synthesis in root epidermal cells. However, note that the changes in auxin distribution after gravitropic stimuli occur within minutes (Band et al., 2012), whereas the gravistimulation-dependent PIN2 degradation takes place in the later stage in root gravitropic response (Kleine-Vehn et al., 2008). Therefore, it is likely that the other proteins related to regulation of auxin distribution (i.e. PINOID kinase, etc.) might be influenced faster than PIN2 degradation by gravity stimuli in roots when FAB1 function is compromised.
Considering these results, we concluded that FAB1 regulates PIN protein localization and degradation in a tissue-specific manner. Namely, FAB1 controls the basal localization of PIN1 in stele and PIN2 in young cortical cells, and the gravistimulation-dependent PIN2 degradation in root epidermal cells, simultaneously. How FAB1 regulates PIN protein localization differently in distinct types of cells is largely unknown, and we cannot yet rule out the possibility that FAB1 or PtdIns(3,5)P2 indirectly controls PIN polarity and degradation to mediate the changes in cellular auxin levels. Further studies are needed to resolve this problem.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Columbia was used in all experiments. Plants were grown under white light with a 16-h/8-h (light/dark) photoperiod at 22°C. The Arabidopsis mutants estradiol-inducible FAB1A/B-amiRNA, snx1-1 (Jaillais et al., 2006), and transgenic marker lines mRFP-SYP43 (Ebine et al., 2008), mRFP-VAMP727 (Ueda et al., 2004), ST-mRFP (Uemura et al., 2012), VHA-a1-mRFP (Uemura et al., 2012), mRFP-ARA7 (Ueda et al., 2004), SNX1-mRFP (Jaillais et al., 2006), SNX1-GFP (Jaillais et al., 2006), GFP-CLASP and TUB6-mRFP (Ambrose et al., 2013), PIN2::PIN2-GFP, and PIN1::PIN1-GFP (Friml et al., 2003) have been described previously. Transgenic marker lines in the FAB1A/B-amiRNA background (Hirano et al., 2011) and in the FAB1A-GFP background (Hirano et al., 2011) were generated by cross pollination. SNX1 and FAB1A were created by assembling entry vectors (pENTR) that were recombined into the destination vector of 35S::NYFP and 35S::CYFP. Homozygous mutant plants were then selected from the F2 segregating population based on genotyping and marker fluorescence. All seeds were germinated on one-half-strength MS medium. All experiments were performed with at least three independent biological replicates.
Confocal Microscopy
GFP and RFP fluorescence signals and differential interference contrast images were obtained using a Nikon ECLIPSE E600 laser scanning microscope equipped with the C1si ready confocal system with a 60× oil immersion lens (Nikon) or a Zeiss 710 confocal microscope equipped with a 63× water immersion lens. The captured images were processed using Nikon EZ-C1 software or Zeiss ZEN 2012 blue software.
Quantification of PIN Polarity
For PIN polarity quantification, the cells within the root meristematic zone were considered. Basal PIN2 percentages were quantified by counting the number of cells with apical PIN2 proportional to the total number of cortical cells in the root meristematic zone.
Total RNA Isolation and Complementary DNA Synthesis
The cloning of FAB1 complementary DNA (cDNA) into pENTR has been described previously. Total RNA was extracted from 6-d-old wild-type and clasp-1 seedlings using the RNeasy kit (Qiagen) following the manufacturer’s instructions. For cDNA synthesis, 1 μg of total RNA was reverse transcribed using the ReverTra Ace qPCR RT Master Mix (Toyobo). The full-length SNX1 was amplified by PCR and cloned into the pENTR entry plasmid using GATEWAY technology (Invitrogen) following the manufacturer’s protocol. For synthesis of SNX1 cDNA, the primers caccATGGAGAGCACGGAGCAGCCGAGGAA and TTAGACAGAATAAGAAGCTTCAAGTTTGGG were used.
Quantitative Reverse Transcription-PCR
Total RNA was extracted from wild-type and transgenic lines by using the RNeasy kit (Qiagen) following the manufacturer’s instructions. For cDNA synthesis, 1 μg of total RNA was reverse transcribed using ReverTra Ace qPCR RT Master Mix (Toyobo). FAB1A, FAB1B, and ubiqutin10 (UBQ10) cDNAs were amplified with an annealing temperature of 55°C during 40 cycles. Primer pairs used in the quantitative reverse transcription-PCR reaction were as follows: FAB1A-F (5′-AAGCCAGATACAAGTAAAAGTGGAG-3′) and FAB1A-R (5′-AAACAACTCCTTTCACGACCA-3′) for FAB1A, FAB1B-F (5′-TGGATCAAAACTTGATTGAAGC-3′) and FAB1B-R (5′-ATCCATCACATCACCCAATG-3′) for FAB1B, and UBQ10-F (5′-GGCCTTGTATAATCCCTGATGAATAAG-3′) and UBQ10-R (5′-AAAGAGATAACAGGAACGGAAACATAGT-3′) for UBQ10.
BiFC Analysis
Full-length SNX1 and FAB1 were recombined using the LR reaction into the BiFC destination plasmids pUGW0-nYFP (AB626694) and pUGW0-cYFP (AB626696), respectively. The binary plasmids were then transformed by the biolistic (particle bombardment) method. Plant leaves were examined using a Nikon confocal microscope. YFP signals were detected using a 540-nm emission filter. The typical scan time was 5 s, and the slice thickness was 0.5 μm.
Suc-Loaded Liposome Assay
Lipid mixtures of phosphatidylethanolamine, phosphatidylcholine, and phosphatidyl-Ser were supplemented with the required ratio of phosphatidylinositol and the relevant diC16 phosphoinositide (Olbracht Serdary Research Laboratories, Jena Bioscience) prior to being dried to form a film in a 0.5-mL tube. The subsequent formation of Suc-loaded liposomes and the analysis of recombinant GST-SNX1 binding were performed using an anti-GST antibody (1:2,500; Nakarai Tesque) as previously described (Cozier et al., 2002).
Gravity Stimulation
For gravity stimulation, 5-d-old seedlings were rotated 90° anticlockwise for 3 h. Plates were photographed before and after reorientation using a digital camera (Pentax K-5 IIs).
Coimmunoprecipitation and Western Blotting
Five-day-old seedlings of wild-type and SNX1-GFP lines were frozen in liquid nitrogen and ground into powder. The protein fraction was eluted in lysis buffer (50 mm Tris-Cl [pH 7.4], 150 mm NaCl, 1 mm EDTA, 1 mm CaCl2, 1% Triton X-100). For immunoprecipitation, 50 μL of Protein G Sepharose (GE Healthcare) was used in accordance with the manufacturer’s instructions. In brief, an anti-FAB1 peptide antibody was added to the beads. The beads with the antibody were rotated top to bottom overnight at 4°C. After rotation, the beads were washed gently and suspended in SDS loading dye. The sample was heated at 65°C for 5 min, and the isolated proteins were analyzed by western blotting. The primary antibodies used for western blotting were anti-GFP (1:2,500, Nakarai Tesque; 1:2,500, Abcam). Anti-FAB1 antibody was raised in rabbits against a short synthetic peptide, (NH2−) CTAGPSPRPKMNPRASRRVS (−COOH), from the N-terminal FAB1A fragment. The secondary antibodies used were goat anti-rabbit IgG-horseradish peroxidase (1:5,000, GE Healthcare).
Immunofluorescence
Arabidopsis seedlings grown on vertical agar plates were fixed for 1 h with 4% (w/v) formaldehyde in MTSB (50 mm PIPES, 5 mm MgSO4, 5 mm EGTA, pH 7.0), rinsed, and digested with 2% driselase in MTSB for 30 min. After rinsing, the specimens were washed five times for 5 min in MTSB supplemented with 0.1% Triton X-100, and incubated in 10% dimethyl sulfoxide and 3% Nonidet P-40 for 1 h. The specimens were then treated with 3% bovine serum albumin for 1 h and then with anti-α-tubulin antibody (1:400; NeoMarkers) or anti-GFP antibody (1:400; Nakarai Tesque) overnight. After rinsing, the specimens were washed five times for 5 min in MTSB supplemented with 0.1% Triton X-100. The secondary antibodies used were Alexa 488 anti-rabbit IgG and Alexa 555 anti-mouse IgG (1:1,000; Invitrogen).
Immunostaining of PIN2
Immunolocalization assay of PIN2 of primary roots was performed as described (Sauer et al., 2006). The anti-PIN2 antibody (1:500; Agrisera) and the secondary anti-chicken antibody coupled to Alexa 555 were used in dilutions of 1:1,000 and 1:500, respectively. In brief, 4-d-old seedlings were fixed in 4% paraformaldehyde and 0.25% glutaraldehyde in buffer A (20 mm PIPES [pH 6.8], 137 mm NaCl, and 2.7 mm KCl) under vacuum conditions for 45 min, and then cells were permeabilized by incubation with 2% driselase at room temperature for 30 min and a solution of buffer A containing 50 mm NH4Cl for 40 min. After a preincubation with buffer A containing 3% bovine serum albumin (BSA), the samples were incubated for 16 h in the primary antibodies diluted in buffer A containing 3% BSA at 37°C, and after washing the samples in buffer A, they were incubated for 3 h in Alexa 555-coupled secondary antibodies (Sigma-Aldrich) diluted 1:500 in buffer A and 3% BSA.
YM201636 Treatment and Phosphoinositide Analysis
Polyphosphoinositide levels were measured according to Munnik and Zarza (2013). In brief, radioactive PtdInsP2 levels were measured by labeling 5-d-old seedlings (three per tube in triplicate) of the wild-type or FAB1A-overexpressing line with 10 μCi of carrier-free 32P-labeled orthophosphate overnight (approximately 16 h). The next day, seedlings were treated with or without 1 μm YM201636 for 2 h, after which lipids were extracted, separated by TLC (Meijer et al., 1999), and quantified by phosphoimaging.
The Arabidopsis Genome Initiative locus identifiers for the genes mentioned in this manuscript are as follows: FAB1A (At4g33240), SYP43 (At3g05710), VAMP727 (At3g54300), ARA7 (At4g19640), SNX1 (At5g06140), CLASP (At2g20190), PIN1 (At1g73590), PIN2 (At5g57090), and UBQ10 (At4g05320).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Conditional knockdown of FAB1A and FAB1B expression in transgenic plant expressing PIN2-GFP under the FAB1A/B-amiRNA background.
Supplemental Figure S2. Alteration of PIN2 localization by down-regulation of FAB1A/B or inhibition of PtdIns(3,5)P2 synthesis.
Supplemental Figure S3. Large-field images of root of PIN2-GFP/FAB1A/B-amiRNA and PIN2-GFP/wild type.
Supplemental Figure S4. Immunolocalization of PIN2 in the FAB1A/B-amiRNA line.
Supplemental Figure S5. Alteration of PIN1 localization by down-regulation of FAB1A/B or inhibition of PtdIns(3,5)P2 synthesis.
Supplemental Figure S6. Changes in shape of FAB1A-GFP and SNX1-mRFP fluorescence by wortmannin treatment.
Supplemental Figure S7. Localization of mRFP-ARA7 and SNX1-mRFP is unchanged by the addition of estradiol in the wild-type background.
Supplemental Figure S8. BiFC assay of SNX1 and FAB1A in onion epidermal cells.
Supplemental Figure S9. BiFC assay of SNX1 and FAB1 in the transgenic Arabidopsis expressing SNX1-mRFP.
Supplemental Figure S10. FAB1A localization is not affected by snx1-1 mutation.
Supplemental Figure S11. Quantitative reverse transcription-PCR analysis of FAB1A-GFP overexpression in the snx1-1 mutant background.
Supplemental Figure S12. Fluorescence imaging of transgenic Arabidopsis expressing FAB1A-GFP and TUB6-mRFP.
Supplemental Movie S1. FAB1A-GFP associates with mRFP-TUB6-labeled microtubules.
Acknowledgments
We thank Tsuyoshi Nakagawa (Shimane University) for providing pUGW0-nYFP and pUGW0-cYFP; Tomohiro Uemura and Takashi Ueda (University of Tokyo) for providing mRFP-VAMP727, mRFP-SYP43, mRFP-ARA7, ST-mRFP, and VHA-a1-mRFP; Jiri Friml (Ghent University) for providing PIN2-GFP and PIN1-GFP; Thierry Gaude (Centre National de la Recherche Scientifique) for providing SNX1-GFP, SNX1-mRFP expression lines, and snx-1-1 mutant; and Geoffrey O. Wasteneys (University of British Columbia) for providing GFP-CLASP line and fruitful discussions. TUB6-mRFP was obtained from Arabidopsis Biological Resource Center, and the DR5rev::GFP line was provided by Jiri Friml (Institute of Science and Technology). We also thank Kentaro Tamura and Ikuko-Hara Nishimura (Kyoto University) for confocal microscopic analyses and fruitful discussions and Yumi Hirano for preparing the schematic illustration of the mode.
Glossary
- TGN/EE
trans-Golgi network/early endosome
- LE
late endosomes
- TLC
thin-layer chromatography
- amiRNA
artificial microRNA
- GST
glutathione S-transferase
- BiFC
bimolecular fluorescence complementation
- MS
Murashige and Skoog
- cDNA
complementary DNA
- BSA
bovine serum albumin
Footnotes
This work was supported by the Ministry of Education, Culture, Sports, Science and Technology Japan, a Grant-in-Aid for Scientific Research on Innovative Areas (grant no. 25119720), and the Strategic Research Funds of Kyoto Prefectural University. The authors declare no competing financial interests.
Articles can be viewed without a subscription.
References
- Ambrose C, Ruan Y, Gardiner J, Tamblyn LM, Catching A, Kirik V, Marc J, Overall R, Wasteneys GO (2013) CLASP interacts with sorting nexin 1 to link microtubules and auxin transport via PIN2 recycling in Arabidopsis thaliana. Dev Cell 24: 649–659 [DOI] [PubMed] [Google Scholar]
- Bak G, Lee EJ, Lee Y, Kato M, Segami S, Sze H, Maeshima M, Hwang JU, Lee Y (2013) Rapid structural changes and acidification of guard cell vacuoles during stomatal closure require phosphatidylinositol 3,5-bisphosphate. Plant Cell 25: 2202–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T. (2013) Phosphoinositides: tiny lipids with giant impact on cell regulation. Physiol Rev 93: 1019–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Band LR, Wells DM, Larrieu A, Sun J, Middleton AM, French AP, Brunoud G, Sato EM, Wilson MH, Péret B, et al. (2012) Root gravitropism is regulated by a transient lateral auxin gradient controlled by a tipping-point mechanism. Proc Natl Acad Sci USA 109: 4668–4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonangelino CJ, Catlett NL, Weisman LS (1997) Vac7p, a novel vacuolar protein, is required for normal vacuole inheritance and morphology. Mol Cell Biol 17: 6847–6858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozier GE, Carlton J, McGregor AH, Gleeson PA, Teasdale RD, Mellor H, Cullen PJ (2002) The phox homology (PX) domain-dependent, 3-phosphoinositide-mediated association of sorting nexin-1 with an early sorting endosomal compartment is required for its ability to regulate epidermal growth factor receptor degradation. J Biol Chem 277: 48730–48736 [DOI] [PubMed] [Google Scholar]
- Cui Y, Zhao Q, Gao C, Ding Y, Zeng Y, Ueda T, Nakano A, Jiang L (2014) Activation of the Rab7 GTPase by the MON1-CCZ1 complex is essential for PVC-to-vacuole trafficking and plant growth in Arabidopsis. Plant Cell 26: 2080–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen PJ, Korswagen HC (2012) Sorting nexins provide diversity for retromer-dependent trafficking events. Nat Cell Biol 14: 29–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeWald DB, Torabinejad J, Jones CA, Shope JC, Cangelosi AR, Thompson JE, Prestwich GD, Hama H (2001) Rapid accumulation of phosphatidylinositol 4,5-bisphosphate and inositol 1,4,5-trisphosphate correlates with calcium mobilization in salt-stressed arabidopsis. Plant Physiol 126: 759–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo G, De Camilli P (2006) Phosphoinositides in cell regulation and membrane dynamics. Nature 443: 651–657 [DOI] [PubMed] [Google Scholar]
- Doyle SM, Haeger A, Vain T, Rigal A, Viotti C, Łangowska M, Ma Q, Friml J, Raikhel NV, Hicks GR, et al. (2015) An early secretory pathway mediated by GNOM-LIKE 1 and GNOM is essential for basal polarity establishment in Arabidopsis thaliana. Proc Natl Acad Sci USA 112: E806–E815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebine K, Fujimoto M, Okatani Y, Nishiyama T, Goh T, Ito E, Dainobu T, Nishitani A, Uemura T, Sato MH, et al. (2011) A membrane trafficking pathway regulated by the plant-specific RAB GTPase ARA6. Nat Cell Biol 13: 853–859 [DOI] [PubMed] [Google Scholar]
- Ebine K, Okatani Y, Uemura T, Goh T, Shoda K, Niihama M, Morita MT, Spitzer C, Otegui MS, Nakano A, et al. (2008) A SNARE complex unique to seed plants is required for protein storage vacuole biogenesis and seed development of Arabidopsis thaliana. Plant Cell 20: 3006–3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feraru E, Friml J (2008) PIN polar targeting. Plant Physiol 147: 1553–1559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friml J, Vieten A, Sauer M, Weijers D, Schwarz H, Hamann T, Offringa R, Jürgens G (2003) Efflux-dependent auxin gradients establish the apical-basal axis of Arabidopsis. Nature 426: 147–153 [DOI] [PubMed] [Google Scholar]
- Gary JD, Sato TK, Stefan CJ, Bonangelino CJ, Weisman LS, Emr SD (2002) Regulation of Fab1 phosphatidylinositol 3-phosphate 5-kinase pathway by Vac7 protein and Fig4, a polyphosphoinositide phosphatase family member. Mol Biol Cell 13: 1238–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geldner N, Anders N, Wolters H, Keicher J, Kornberger W, Muller P, Delbarre A, Ueda T, Nakano A, Jürgens G (2003) The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 24: 219–230 [DOI] [PubMed] [Google Scholar]
- Hirano T, Matsuzawa T, Takegawa K, Sato MH (2011) Loss-of-function and gain-of-function mutations in FAB1A/B impair endomembrane homeostasis, conferring pleiotropic developmental abnormalities in Arabidopsis. Plant Physiol 155: 797–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T, Sato MH (2011) Arabidopsis FAB1A/B is possibly involved in the recycling of auxin transporters. Plant Signal Behav 6: 583–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CY, Alghamdi TA, Botelho RJ (2012) Phosphatidylinositol-3,5-bisphosphate: no longer the poor PIP2. Traffic 13: 1–8 [DOI] [PubMed] [Google Scholar]
- Ikonomov OC, Sbrissa D, Delvecchio K, Xie Y, Jin J-P, Rappolee D, Shisheva A (2011) The phosphoinositide kinase PIKfyve is vital in early embryonic development: preimplantation lethality of PIKfyve-/- embryos but normality of PIKfyve+/- mice. J Biol Chem 286: 13404–13413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue T, Kondo Y, Naramoto S, Nakano A, Ueda T (2013) RAB5 activation is required for multiple steps in Arabidopsis thaliana root development. Plant Cell Physiol 54: 1648–1659 [DOI] [PubMed] [Google Scholar]
- Jaillais Y, Fobis-Loisy I, Miège C, Rollin C, Gaude T (2006) AtSNX1 defines an endosome for auxin-carrier trafficking in Arabidopsis. Nature 443: 106–109 [DOI] [PubMed] [Google Scholar]
- Jean S, Kiger AA (2012) Coordination between RAB GTPase and phosphoinositide regulation and functions. Nat Rev Mol Cell Biol 13: 463–470 [DOI] [PubMed] [Google Scholar]
- Jefferies HBJ, Cooke FT, Jat P, Boucheron C, Koizumi T, Hayakawa M, Kaizawa H, Ohishi T, Workman P, Waterfield MD, et al. (2008) A selective PIKfyve inhibitor blocks PtdIns(3,5)P(2) production and disrupts endomembrane transport and retroviral budding. EMBO Rep 9: 164–170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine-Vehn J, Friml J (2008) Polar targeting and endocytic recycling in auxin-dependent plant development. Annu Rev Cell Dev Biol 24: 447–473 [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J, Langowski L, Wisniewska J, Dhonukshe P, Brewer PB, Friml J (2008a) Cellular and molecular requirements for polar PIN targeting and transcytosis in plants. Mol Plant 1: 1056–1066 [DOI] [PubMed] [Google Scholar]
- Kleine-Vehn J, Leitner J, Zwiewka M, Sauer M, Abas L, Luschnig C, Friml J (2008b) Differential degradation of PIN2 auxin efflux carrier by retromer-dependent vacuolar targeting. Proc Natl Acad Sci USA 105: 17812–17817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Bak G, Choi Y, Chuang WI, Cho HT, Lee Y (2008a) Roles of phosphatidylinositol 3-kinase in root hair growth. Plant Physiol 147: 624–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y, Kim ES, Choi Y, Hwang I, Staiger CJ, Chung YY, Lee Y (2008b) The Arabidopsis phosphatidylinositol 3-kinase is important for pollen development. Plant Physiol 147: 1886–1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney AJ, Zhang Y, Weisman LS (2014) Phosphatidylinositol 3,5-bisphosphate: low abundance, high significance. BioEssays 36: 52–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meijer HJ, Arisz SA, Van Himbergen JA, Musgrave A, Munnik T (2001) Hyperosmotic stress rapidly generates lyso-phosphatidic acid in Chlamydomonas. Plant J 25: 541–548 [DOI] [PubMed] [Google Scholar]
- Meijer HJG, Divecha N, van den Ende H, Musgrave A, Munnik T (1999) Hyperosmotic stress induces rapid synthesis of phosphatidyl-D-inositol 3,5-bisphosphate in plant cells. Planta 208: 294–298 [Google Scholar]
- Mueller-Roeber B, Pical C (2002) Inositol phospholipid metabolism in Arabidopsis: characterized and putative isoforms of inositol phospholipid kinase and phosphoinositide-specific phospholipase C. Plant Physiol 130: 22–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munnik T, Zarza X (2013) Analyzing plant signaling phospholipids through 32Pi-labeling and TLC. Methods Mol Biol 1009: 3–15 [DOI] [PubMed] [Google Scholar]
- Nicot AS, Fares H, Payrastre B, Chisholm AD, Labouesse M, Laporte J (2006) The phosphoinositide kinase PIKfyve/Fab1p regulates terminal lysosome maturation in Caenorhabditis elegans. Mol Biol Cell 17: 3062–3074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordmann M, Cabrera M, Perz A, Bröcker C, Ostrowicz C, Engelbrecht-Vandré S, Ungermann C (2010) The Mon1-Ccz1 complex is the GEF of the late endosomal Rab7 homolog Ypt7. Curr Biol 20: 1654–1659 [DOI] [PubMed] [Google Scholar]
- Park M, Jürgens G (2011) Membrane traffic and fusion at post-Golgi compartments. Front Plant Sci 2: 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter BJ, Kent HM, Mills IG, Vallis Y, Butler PJG, Evans PR, McMahon HT (2004) BAR domains as sensors of membrane curvature: the amphiphysin BAR structure. Science 303: 495–499 [DOI] [PubMed] [Google Scholar]
- Poteryaev D, Datta S, Ackema K, Zerial M, Spang A (2010) Identification of the switch in early-to-late endosome transition. Cell 141: 497–508 [DOI] [PubMed] [Google Scholar]
- Pourcher M, Santambrogio M, Thazar N, Thierry AM, Fobis-Loisy I, Miège C, Jaillais Y, Gaude T (2010) Analyses of sorting nexins reveal distinct retromer-subcomplex functions in development and protein sorting in Arabidopsis thaliana. Plant Cell 22: 3980–3991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman A, Takahashi M, Shibasaki K, Wu S, Inaba T, Tsurumi S, Baskin TI (2010) Gravitropism of Arabidopsis thaliana roots requires the polarization of PIN2 toward the root tip in meristematic cortical cells. Plant Cell 22: 1762–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rink J, Ghigo E, Kalaidzidis Y, Zerial M (2005) Rab conversion as a mechanism of progression from early to late endosomes. Cell 122: 735–749 [DOI] [PubMed] [Google Scholar]
- Rusten TE, Rodahl LMW, Pattni K, Englund C, Samakovlis C, Dove S, Brech A, Stenmark H (2006) Fab1 phosphatidylinositol 3-phosphate 5-kinase controls trafficking but not silencing of endocytosed receptors. Mol Biol Cell 17: 3989–4001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer M, Paciorek T, Benková E, Friml J (2006) Immunocytochemical techniques for whole-mount in situ protein localization in plants. Nat Protoc 1: 98–103 [DOI] [PubMed] [Google Scholar]
- Shisheva A. (2008) PIKfyve: Partners, significance, debates and paradoxes. Cell Biol Int 32: 591–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon ML, Platre MP, Assil S, van Wijk R, Chen WY, Chory J, Dreux M, Munnik T, Jaillais Y (2014) A multi-colour/multi-affinity marker set to visualize phosphoinositide dynamics in Arabidopsis. Plant J 77: 322–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh MK, Krüger F, Beckmann H, Brumm S, Vermeer JE, Munnik T, Mayer U, Stierhof YD, Grefen C, Schumacher K, et al. (2014) Protein delivery to vacuole requires SAND protein-dependent Rab GTPase conversion for MVB-vacuole fusion. Curr Biol 24: 1383–1389 [DOI] [PubMed] [Google Scholar]
- Takasuga S, Horie Y, Sasaki J, Sun-Wada GH, Kawamura N, Iizuka R, Mizuno K, Eguchi S, Kofuji S, Kimura H, et al. (2013) Critical roles of type III phosphatidylinositol phosphate kinase in murine embryonic visceral endoderm and adult intestine. Proc Natl Acad Sci USA 110: 1726–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teasdale RD, Loci D, Houghton F, Karlsson L, Gleeson PA (2001) A large family of endosome-localized proteins related to sorting nexin 1. Biochem J 358: 7–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueda T, Uemura T, Sato MH, Nakano A (2004) Functional differentiation of endosomes in Arabidopsis cells. Plant J 40: 783–789 [DOI] [PubMed] [Google Scholar]
- Uemura T, Kim H, Saito C, Ebine K, Ueda T, Schulze-Lefert P, Nakano A (2012) Qa-SNAREs localized to the trans-Golgi network regulate multiple transport pathways and extracellular disease resistance in plants. Proc Natl Acad Sci USA 109: 1784–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanoosthuyse V, Tichtinsky G, Dumas C, Gaude T, Cock JM (2003) Interaction of calmodulin, a sorting nexin and kinase-associated protein phosphatase with the Brassica oleracea S locus receptor kinase. Plant Physiol 133: 919–929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeer JE, van Leeuwen W, Tobeña-Santamaria R, Laxalt AM, Jones DR, Divecha N, Gadella TW Jr, Munnik T (2006) Visualization of PtdIns3P dynamics in living plant cells. Plant J 47: 687–700 [DOI] [PubMed] [Google Scholar]
- Vieten A, Sauer M, Brewer PB, Friml J (2007) Molecular and cellular aspects of auxin-transport-mediated development. Trends Plant Sci 12: 160–168 [DOI] [PubMed] [Google Scholar]
- Welters P, Takegawa K, Emr SD, Chrispeels MJ (1994) AtVPS34, a phosphatidylinositol 3-kinase of Arabidopsis thaliana, is an essential protein with homology to a calcium-dependent lipid binding domain. Proc Natl Acad Sci USA 91: 11398–11402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitley P, Hinz S, Doughty J (2009) Arabidopsis FAB1/PIKfyve proteins are essential for development of viable pollen. Plant Physiol 151: 1812–1822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto A, DeWald DB, Boronenkov IV, Anderson RA, Emr SD, Koshland D (1995) Novel PI(4)P 5-kinase homologue, Fab1p, essential for normal vacuole function and morphology in yeast. Mol Biol Cell 6: 525–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. (2010) Auxin biosynthesis and its role in plant development. Annu Rev Plant Biol 61: 49–64 [DOI] [PMC free article] [PubMed] [Google Scholar]