Abstract
Circulating microRNAs (miRNAs) are promising biomarkers for cancer detection. However, multiethnic and multicentric studies of non-small-cell lung cancer (NSCLC) are lacking. We recruited 221 NSCLC patients, 161 controls and 56 benign nodules from both China and America. Initial miRNA screening was performed using the TaqMan Low Density Array followed by confirming individually by RT-qPCR in Chinese cohorts. Finally, we performed a blind trial from an American cohort to validate our findings. RT-qPCR confirmed that miR-483-5p, miR-193a-3p, miR-25, miR-214 and miR-7 were significantly elevated in patients compared to controls. The areas under the curve (AUCs) of the ROC curve of this five-serum miRNA panel were 0.976 (95% CI, 0.939–1.0; P < 0.0001) and 0.823 (95% CI, 0.75–0.896; P < 0.0001) for the two confirmation sets, respectively. In the blind trial, the panel correctly classified 95% NSCLC cases and 84% controls from the American cohort. Most importantly, the panel was capable of distinguishing NSCLC from benign nodules with an AUC of 0.979 (95% CI, 0.959–1.0) in the American cohort and allowed correct prediction of 86% and 95% stage I–II tumors in the Chinese and American cohorts, respectively. This serum miRNA panel holds the potential for diagnosing ethnically diverse NSCLC patients.
Abbreviations: miRNA, microRNA; NSCLC, non-small-cell lung cancer; RT-qPCR, quantitative reverse transcription polymerase chain reaction; TLDA, TaqMan Low-Density Array; Cq, quantification cycle; AUC, the area under the ROC curve; TNM, tumor–node–metastasis
Keywords: Non-small-cell lung cancer, Multiethnic, Multicentric, Serum microRNAs, Biomarker, Diagnosis
Graphical abstract
Highlights
-
•
A multiethnic, multicentric, case–control study was conducted to identify serum miRNA signature for diagnosing NSCLC.
-
•
A five-miRNA-based classifier for accurate diagnosis of NSCLC among patients of Chinese and America was constructed.
-
•
The five-miRNA panel allowed accurate detection of NSCLC especially stage I–II cases from normal and benign nodule subjects.
Wang et al. constructed a five-miRNA-based classifier which provides the potential for accurate diagnosis of NSCLC among patients of Chinese and America from a multiethnic, multicentric, case–control study. More importantly, the five-miRNA panel allowed accurate detection of NSCLC cases especially stage I–II cases from normal and benign nodule subjects. These results suggest that the five-miRNA signature might be a useful biomarker for diagnosing NSCLC in ethnically diverse patients and help discriminate early stage NSCLC from normal and benign nodule subjects, which may benefit personalized therapy of NSCLC patients in the future.
1. Introduction
Tumors of the lung are one of the highest incidence of cancers and the leading cause of cancer deaths worldwide. According to the 2008 WHO epidemiological statistics, the incidence of lung cancer is more than 130 per 100,000 individuals, which accounts for 13% (1.6 million) of the total cancer cases and 18% (1.4 million) of cancer-related mortality; even worse, the number of new cases is expected to double worldwide in next decades (Jemal et al., 2011). The vast majority (85–90%) of lung cancer cases are non-small-cell lung cancer (NSCLC) (D'Addario et al., 2010). Early-stage NSCLCs are symptom-free and difficult to detect; therefore, the disease is often diagnosed at a locally advanced or metastatic stage (stage III or stage IV), at which point few clinical treatment options remain. Despite numerous developments relating to NSCLC therapies in recent years, the overall survival rate of NSCLC patients has only improved by approximately 15% over the last few decades (Spiro and Silvestri, 2005, Siegel et al., 2012). In comparison, the five-year survival rate is approximately 33% for cases discovered at early stages (stage I or II) (Little et al., 2005). At present, diagnosis largely relies to a large extent on imaging methods, such as chest X-rays, CT scans, or PET–CT scans, followed by bronchoscopy and biopsy (Oken et al., 2005, International Early Lung Cancer Action Program Investigators et al., 2006). While no blood marker for NSCLC is available at present, some blood-based protein markers such as carcinoembryonic antigen and squamous cell carcinoma antigen have been applied as candidate diagnostic markers for NSCLC in clinics. Unfortunately, the low sensitivity and specificity of these antigens limit the diagnostic accuracy and utility. For these reasons, the development of sensitive and reliable biomarkers remains a major challenge for researchers.
MicroRNAs (miRNAs) are a family of small, single-stranded non-coding RNAs that are critical regulators of numerous diseases, including cancer, and their expression patterns have the potential to diagnose various types of cancer (Esquela-Kerscher and Slack, 2006, Kong et al., 2012, Calin and Croce, 2006). Recent studies from our group and others have demonstrated that human body fluid such as serum/plasma contains numerous stable miRNAs, potentially paving the way toward a novel class of cancer biomarkers (Chen et al., 2008, Mitchell et al., 2008, Eichelser et al., 2013). A number of studies have reported that circulating miRNA expression patterns differ between NSCLC patients and healthy controls, and some dysregulated serum miRNAs hold potential as promising biomarkers for the diagnosis and prognosis of NSCLC (Chen et al., 2008, Hu et al., 2010, Bianchi et al., 2011, Chen et al., 2012a, Heegaard et al., 2012, Wang et al., 2013). However, data produced by different groups on this topic are quite variable. Therefore, a systematical analysis of serum miRNA expression profiles from multiethnic and multicentric NSCLC cases is urgently needed.
In the present study, we used a high-throughput TaqMan Low-Density Array (TLDA) (Applied Biosystems) scanning combined with an individual quantitative reverse transcription polymerase chain reaction PCR (RT-qPCR) confirmation to establish a panel of significantly upregulated serum miRNAs in NSCLC patients using samples from three independent Chinese cohorts. A single-blind trial was then performed to assess the ability of the panel to diagnose NSCLC in an American cohort and to function as a reliable diagnostic indicator of NSCLC in patients of different ethnicities.
2. Materials and Methods
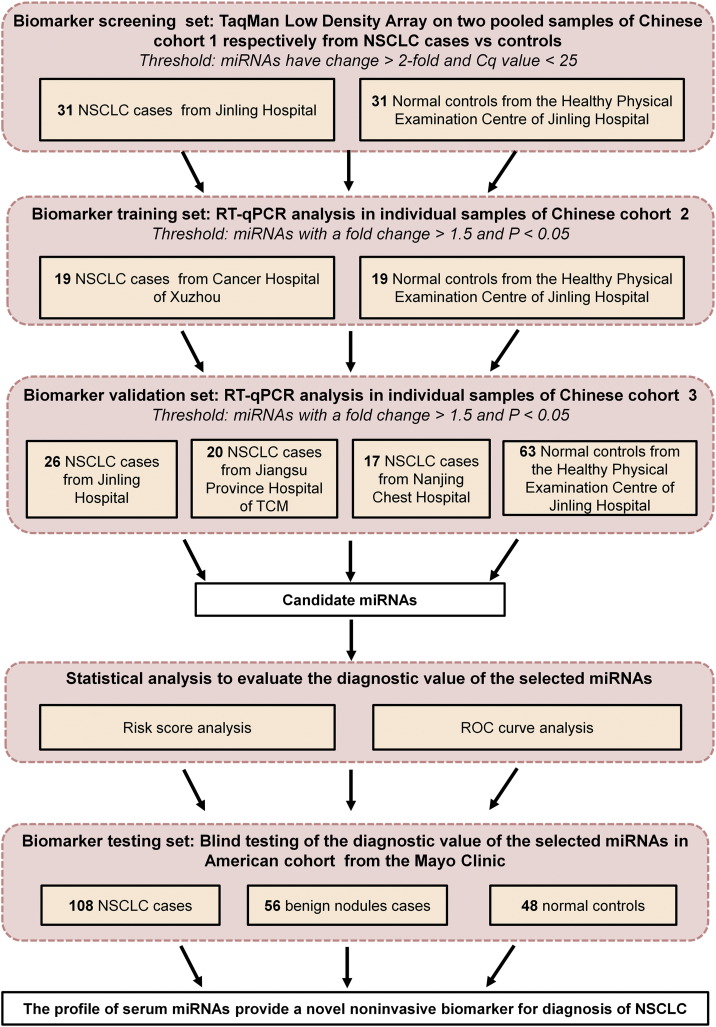
2.1. Participants and Study Design
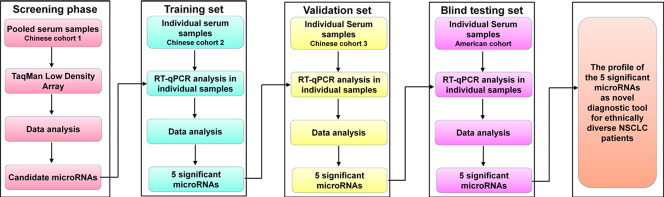
We totally enrolled 438 participants from both China and America, including 221 NSCLC patients, 161 normal controls and 56 benign nodules. To identify a miRNA signature associated with NSCLC, we first used a discovery cohort consisting of 62 individuals (31 patients with NSCLC and 31 normal controls) from Jinling Hospital, China, to find serum miRNAs as surrogate markers. Subsequently, verification was conducted using individual RT-qPCR assays to refine the number of serum miRNAs included in the NSCLC signature by a 2-phase experimental procedure from two additionally independent, randomized Chinese cohorts: one consisted of 19 lung cancer cases (from Cancer Hospital of Xuzhou) and 19 normal controls (from Jinling Hospital) and the other consisted of 63 case samples (from Jinling Hospital, Jiangsu Province Hospital of TCM and Nanjing Chest Hospital) and 63 normal controls (from Jinling Hospital). Finally, we evaluated the serum samples from an American cohort, which included 108 NSCLC patients, 48 normal controls and 56 benign nodules (as non-cancer controls). The American cohort was selected from the Mayo Clinic (Mayo Validation Support Services, 3050 Superior Drive, NW, Rochester, Minnesota) in a blinded fashion (the investigators performing the analysis on the blood samples were blinded to the patients' clinical diagnosis and the medical sample providers conducted the data analysis) to validate the predictive power of the identified miRNA signature for NSCLC. The overall study design is shown in Fig. 1. The protocol was approved by the ethics committee board at each participating institution, and written informed consent was obtained from all participants.
Fig. 1.
Overall study design and numbers of patients with NSCLC included in the discovery, training, validation and testing sets.
2.2. Eligibility, Exclusion Criteria and Processing of Serum
Histopathological analysis of the tumors was performed according to the WHO criteria. Patients were eligible if they had been diagnosed with a pathological NSCLC that met histological or cytological criteria. The tumor stage at the time of diagnosis was assessed according to the American Joint Committee on Cancer guidelines (http://www.cancerstaging.org/). Eligibility criteria for NSCLC patients both China cohorts and American cohort included: 1) if eighteen years of age old or older; 2) confirmed diagnosis of NSCLC; 3) not previously suffered from lung cancer and other types of cancers; 4) not previously with chemotherapy or radiotherapy; 5) and free of synchronous multiple cancers. For patients who underwent surgery, definitive tumor stage was determined based on the operative findings. For those patients unsuitable for surgical therapy, tumors were assessed using histopathology or imaging technology such as bronchoscopy, dynamic computed tomography (CT), magnetic resonance imaging, and/or endoscopic ultrasonography. Eligibility criteria for benign nodules enrolled from American cohort (Mayo Clinic) included: 1) nodules detected on CT scan; 2) biopsy performed and confirmed negative for cancer or biopsy performed and nodules detected and were confirmed benign based on CT scan trial algorithm; 3) five years of follow-up confirming no diagnosis of lung cancer; and 4) no previous history of cancer prior to enrollment. Individuals who showed no evidence of disease during the medical checkup were selected as normal controls. Eligibility criteria for normal controls in American cohort included: 1) no nodules detected on CT scan; 2) five years of following up confirming no diagnosis of lung cancer; and 3) no previous history of cancer prior to enrollment. The recruitment of normal subjects in Chinese cohorts was conducted in the Healthy Physical Examination Center of the Jinling Hospital (Nanjing, China). The finally selected controls underwent routine laboratory and imaging tests such as complete blood cell counts, sera biochemical profile, baseline electrocardiogram, tumor markers (CEA and AFP), a type-B ultrasound for the abdomen and pelvis, and a chest X-ray. All the healthy individuals did not have any pulmonary pathology or any constitutional symptoms after consulting the physical examination records. Peripheral venous blood (3–4 ml) was collected in serum vacutainer tubes (Becton, Dickinson and Company, Franklin Lakes, New Jersey) with clot activator from participants 12 h overnight fasting, and serum was immediately separated by a 10-min centrifugation at 1500 g. Supernatant serum samples were then carefully transferred to a new Eppendorf tube and stored at − 80 °C until analyze. In addition, we checked the hemolysis status of all the samples and only the qualified samples were used in our study.
The demographics and clinical features of all NSCLC patients and controls are shown in Table 1. All blood samples from patients were gathered before treatments, including surgery, chemotherapy, radiotherapy, and tissue manipulation (such as biopsy, bronchoscopy, or fine needle aspiration).
Table 1.
Demographic and clinical features of the NSCLC patients and controls of the Chinese cohorts and American cohort.
| Variable | Chinese cohorts |
American cohort |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Cohort 1 |
Cohort 2 |
Cohort 3 |
Normal control |
Benign nodules |
NSCLC |
||||
| Normal control |
NSCLC |
Normal control |
NSCLC |
Normal control |
NSCLC |
||||
| n = 31 | n = 31 | n = 19 | n = 19 | n = 63 | n = 63 | n = 48 | n = 56 | n = 108 | |
| Age — yra | 59.8 ± 8.5 | 59.6 ± 11.5 | 62.1 ± 9.4 | 61.8 ± 12.7 | 59.7 ± 5.7 | 61.9 ± 9.5 | 58.5 ± 5.1 | 63.7 ± 6.7 | 67.2 ± 10.2 |
| ≤ 59 | 20 (64.5) | 13 (41.9) | 10 (52.6) | 8 (42.1) | 36 (57.1) | 25 (39.7) | 36 (75.0) | 15 (26.8) | 21 (19.4) |
| > 59 | 11 (35.5) | 18 (58.1) | 9 (47.4) | 11 (57.9) | 27 (42.9) | 38 (60.3) | 12 (25.0) | 40 (71.4) | 87 (80.6) |
| Unknown | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (1.8) | 0 (0) |
| Sex — no. (%) | |||||||||
| Male | 25 (80.6) | 23 (74.2) | 13 (68.4) | 11 (57.9) | 37 (58.7) | 49 (77.8) | 20 (41.7) | 25 (44.6) | 52 (48.1) |
| Female | 6 (19.4) | 8 (25.8) | 6 (31.6) | 8 (42.1) | 26 (41.3) | 14 (22.2) | 28 (58.3) | 30 (53.6) | 56 (51.9) |
| Unknown | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (1.8) | 0 (0) |
| Smoking status — no. (%) | |||||||||
| Ever and current | 12 (38.7) | 16 (51.6) | 7 (36.8) | 8 (42.1) | 18 (28.6) | 39 (61.9) | 48 (100) | 55 (98.2) | 103 (95.4) |
| Never | 19 (61.3) | 15 (48.4) | 12 (63.2) | 11 (57.9) | 45 (71.4) | 24 (38.1) | 0 (0) | 1 (1.8) | 5 (4.6) |
| Tumor subtype — no. (%) | |||||||||
| Adenocarcinoma | – | 12 (38.7) | – | 9 (47.3) | – | 26 (41.3) | – | – | 52 (48.1) |
| Squamous cell carcinoma | 9 (29.0) | – | 4 (21.1) | – | 26 (41.3) | – | – | 27 (25.0) | |
| Other | – | 10 (32.3) | – | 6 (31.6) | – | 11 (17.4) | – | – | 29 (26.9) |
| TNM stage — no. (%) | |||||||||
| I | – | 7 (22.6) | – | 1 (5.3) | – | 3 (4.7) | – | – | 43 (39.8) |
| II | – | 4 (12.9) | – | 2 (10.5) | – | 8 (12.7) | – | – | 15 (13.9) |
| III | – | 9 (29.0) | – | 2 (10.5) | – | 9 (14.3) | – | – | 29 (26.9) |
| IV | – | 11 (35.5) | – | 12 (63.2) | – | 41 (65.1) | – | – | 17 (15.7) |
| Missing data | – | 0 | – | 2 (10.5) | – | 2 (3.2) | – | – | 4 (3.7) |
The data are expressed as the mean (SD).
2.3. RNA Isolation, TLDA and RT-qPCR Assays
The serum expression profiling of miRNA was scanned using TLDA on an ABI PRISM 7900HT (Luo et al., 2013). A hydrolysis probe-based RT-qPCR assay was then used to validate candidate miRNAs following the manufacturer's instructions as described previously (Luo et al., 2013). The concentrations of serum miRNAs were normalized to let-7d/g/i trio and relative levels were evaluated using the comparative quantification cycle (Cq) method (Chen et al., 2013). The details of RNA isolation, TLDA and RT-qPCR assays are provided in the Supplemental Methods.
2.4. Statistical Analysis
The serum miRNA concentrations were represented as the mean (SE), and the other variables were expressed as the mean (SD). The nonparametric Mann–Whitney U-test was performed to compare miRNA concentrations between the groups. The differences in other variables between the two groups were compared using Student's t-test or two-sided χ2 test. A P-value < 0.05 was considered statistically significant. We performed a risk score analysis to evaluate the associations between the concentrations of the serum miRNAs and NSCLC. Briefly, the risk score of each miRNA, denoted as s, was set to 1 if its concentration was higher than the upper 95% reference interval for the corresponding miRNA concentration in controls and to 0 otherwise. A risk score function to predict NSCLC was defined according to a linear combination of concentration for each miRNA. For example, the RSF for sample i using information from five miRNAs was: rsfi = ∑5j − 1Wj · sij. In the above equation, sij is the risk score for miRNA j on sample i, and Wj is the weight of the risk score of miRNA j. To determine the Ws, five univariate logistic regression models were fitted using the disease status with each of the risk scores. The regression coefficient of each risk score was used as the weight to show the contribution of each miRNA to the RSF. We then constructed the ROC curves to evaluate the specificity and sensitivity of NSCLC prediction for each miRNA individually and for the combination of miRNAs. All analyses above were performed with the use of SPSS statistical software (version 16.0). Additional details regarding statistical methods are provided in the Supplemental Methods.
2.5. Funding
This work was supported by the grants from the National Basic Research Program of China (973 Program) (2014CB542300), the Research Special Fund for Public Welfare Industry of Health of China (201302018), the National Natural Science Foundation of China (81171661, 81472021, 81401257 and 81301511), the Natural Science Foundation of Jiangsu Province (BK2011013, BK20140730), the Medical Scientific Research Foundation of Nanjing Military Command (12Z28), and the Research Funds of the Jinling Hospital (2014043). The funding agency has no role in the study design, date analysis, interpretation of results or writing of this manuscript.
3. Results
3.1. Serum miRNA Screening by TLDA
We globally analyzed the miRNA expression patterns in two pooled serum samples from 31 NSCLC patients and 31 normal controls in Chinese cohort 1 using TLDA approach. A miRNA was considered upregulated if its Cq value was < 25 in the NSCLC sample and its concentration showed at least a 2-fold increase in the NSCLC sample compared to the control sample. Class-comparison analysis of all 754 miRNAs revealed that 38 miRNAs (5%) were upregulated in the NSCLC sample (Supplemental Table 1), 16 of the upregulated miRNAs which have been reported to be conserved in lung tissue and involved in cell proliferation, migration, immune related or known as “oncomir” were subsequently validated in the individual patients using RT-qPCR, while the newly identified miRNAs and miRNA-star miRNAs were excluded for further confirmation.
3.2. Increases in Serum miRNA Concentrations Confirmed by RT-qPCR
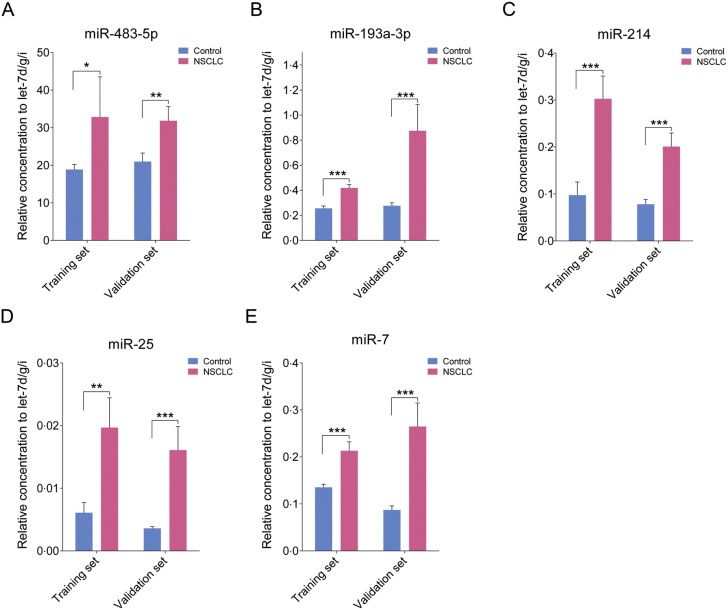
The 16 miRNAs selected from the TLDA results were first confirmed in a training set consisting of an independent Chinese cohort including 19 cases and 19 controls (the training set). Nine of the 16 miRNAs were found to be markedly dysregulated in sera from NSCLC cases. Of these 9 miRNAs, five upregulated miRNAs, including miR-483-5p, miR-193a-3p, miR-214, miR-25, and miR-7, were chosen for further analysis because the difference between cases and controls was most significant for this set (exhibiting a fold change > 1.5 and a P < 0.05) (Supplemental Table 2 and Fig. 2). The five miRNAs were further examined in another Chinese cohort including 63 cases and 63 controls (the validation set). The changes in the levels of these five miRNAs in the NSCLC group were consistent with those from the training set (fold change > 1.5 and P < 0.05) (Fig. 2), thus confirming stability of the production pattern of these miRNAs.
Fig. 2.
The relative contents of the selected five miRNAs in the sera from patient with NSCLC in the training set and validation set.
The asterisks indicate significant differences from normal controls (P < 0.05). ⁎P < 0.05; ⁎⁎P < 0.01; ⁎⁎⁎P < 0.001.
3.3. Assessment of Risk Score in Chinese Cohorts
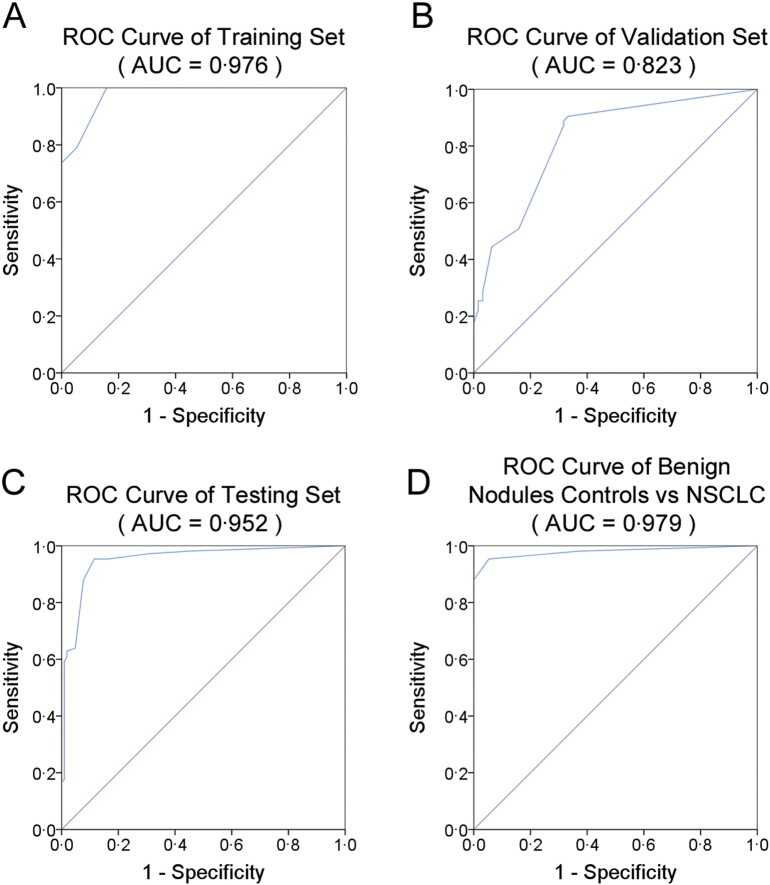
Next, we conducted ROC curve analyses on each of the individual five serum miRNAs to assess the diagnostic value for discriminating between NSCLCs and normal subjects in the training set. The AUCs for these miRNAs ranged from 0.609 to 0.900 (Supplemental Table 3). To clarify the diagnostic capability of the combination of the five miRNAs, we conducted a risk score analysis on the data set and used the results to predict NSCLC case and normal status. When the optimal cutoff value was 2.617, all of the NSCLC samples (100%) had a risk score > 2.617, whereas only 3 of the 19 controls exhibited a risk score > 2.617 (Table 2). The ROC curve for the panel demonstrated a remarkable accuracy of diagnosis, evidenced by an AUC of 0.976 (95% CI, 0.939–1.0; P < 0.0001), which was much larger than the AUC values for each of the five individual miRNAs (Supplemental Table 3 and Fig. 3A). Furthermore, at the optimal cutoff value, the diagnostic sensitivity and specificity of the five-miRNA panel for NSCLC diagnosis were 100% and 84%, respectively (Fig. 3A).
Table 2.
Risk score analysis of NSCLC cases in Chinese cohorts and American cohort.
| Risk score | 0–2.617 | > 2.617–13.820 | Prediction accuracy (%) |
|---|---|---|---|
| Chinese cohorts | |||
| Controls | 59 | 23 | 72 |
| NSCLC | 8 | 74 | 90 |
| Stage I–II NSCLC | 2 | 12 | 87 |
| Training set | |||
| Controls | 16 | 3 | 84 |
| NSCLC | 0 | 19 | 100 |
| Stage I–II NSCLC | 0 | 3 | 100 |
| Validation set | |||
| Controls | 43 | 20 | 68 |
| NSCLC | 8 | 55 | 87 |
| Stage I–II NSCLC | 2 | 9 | 82 |
| American cohort | |||
| Testing (single-blind) set | |||
| Controlsa | 87 | 17 | 84 |
| NSCLC | 5 | 103 | 95 |
| Stage I–II NSCLC | 3 | 55 | 95 |
Including normal controls and non-cancer controls (benign nodules).
Fig. 3.
Sensitivity and specificity of the five-miRNA panel in the training, validation and testing sets.
(A), ROC curve for the five-miRNA panel to discern 19 NSCLC cases from 19 normal controls in the training set. (B), ROC curve for the five-miRNA panel to discern 63 NSCLC cases from 63 normal controls in the validation set. (C), ROC curve for the five-miRNA panel to discern NSCLC cases from normal controls in the testing set. (D), ROC curve for the five-miRNA panel to discern 108 NSCLC cases from 56 benign nodules in the testing set.
We further evaluated the diagnostic values of the five individual miRNAs and the five-miRNA panel in the validation set. As shown in Supplemental Table 4, the AUCs for each of the five miRNAs ranged from 0.663 to 0.835. At the same cutoff value (2.617), 55 of the 63 NSCLC samples (87%) had a risk score > 2.617 (Table 2). And the discriminate sensitivity and specificity of the five-miRNA panel for NSCLC were 89% and 68%, respectively, and the AUC was 0.823 (95% CI, 0.750–0.896; P < 0.0001) (Supplemental Table 4 and Fig. 3B).
3.4. Evaluation of the Predictive Value of the Five-miRNA Panel in an American Cohort by Blinded Trial
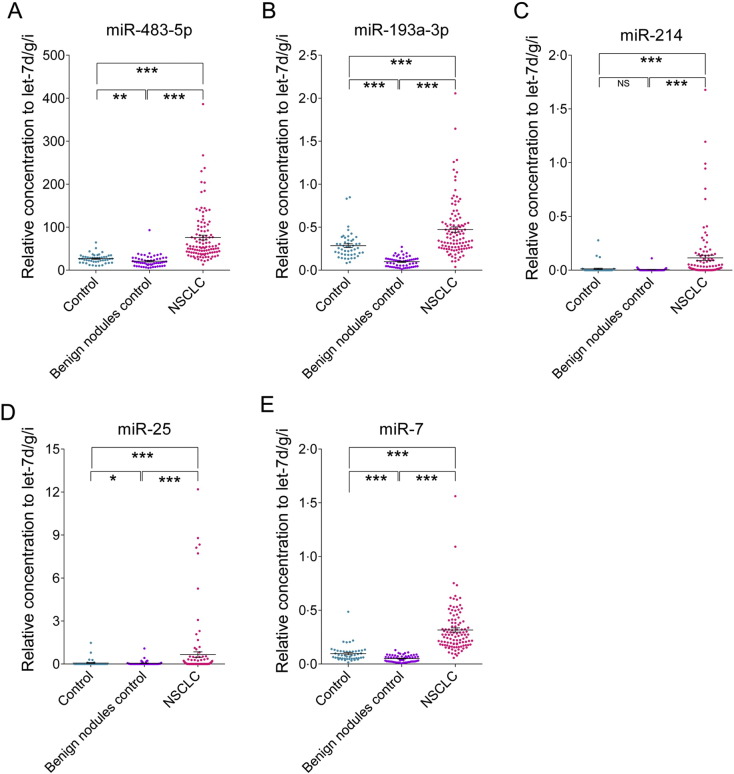
We tested an additional 212 serum samples from an American cohort in a blind trial using the RT-qPCR assay. We aimed to further confirm the diagnostic accuracy of the five serum miRNA-based biomarker for different ethnicities. First, we analyzed the difference in serum concentrations of miRNAs between the different ethnicities. We observed from RT-qPCR data that the Cq values of serum miRNAs in samples from the American cohort were generally higher than in the Chinese cohort by approximately > 3 cycles. However, the relative concentrations of four miRNAs, including miR-483-5p, miR-193a-3p, miR-25 and miR-7, were comparable between the Chinese and American cohorts except miR-214, since the average Cq value of the internal control let-7d/g/i in the American cohort with a mean (SD) of 24.59 (0.85) was also approximately 3 cycles higher than that in Chinese cohort [22.03(0.92)]. These results suggest that the relative concentrations of the five selected miRNAs correlate well between different races when let-7d/g/i is used as an internal reference for the RT-qPCR assay. Subsequently, we analyzed the relative levels of the five miRNAs from the American cohort and classified them on the basis of the risk score model of the five-miRNA panel, with a cutoff value of 2.617. As a result, 103 of 108 NSCLC samples (95%) and 87 of 104 control samples (84%) were classified correctly (Table 2). Of the 104 control samples, 48 were from healthy individuals, and the other 56 were from patients who had benign lung nodules detected by CT scan and did not develop lung cancer after a five-year period of follow-up. The relative concentrations of the five serum miRNAs were all significantly higher in NSCLC patients than in normal controls and patients with benign nodules (at least P < 0.001) (Fig. 4).
Fig. 4.
The relative contents of the selected five miRNAs in the sera from patient with NSCLC in the testing set.
The asterisks indicate significant differences from normal controls (P < 0.05). ⁎P < 0.05; ⁎⁎P < 0.01; ⁎⁎⁎P < 0.001.
In the blind American cohort, the AUCs for the five miRNAs ranged from 0.696 to 0.964 (Supplemental Table 5). The five-miRNA panel was significantly reliable when applied to the testing set, exhibiting a sensitivity of 95%, a specificity of 84% and an AUC of 0.952 (95% CI, 0.922–0.981) (Fig. 3C). Moreover, the risk score was capable of distinguishing NSCLC cases from benign nodules with an AUC of 0.979 (95% CI, 0.959–1.000) and sensitivity and specificity of 95% (Fig. 3D). Taken together, the data suggest that the five-miRNA panel is also an accurate diagnostic marker for American patients with NSCLC.
3.5. Assessment of Risk Score in Early-stage NSCLC
We further evaluated the potential of the five-miRNA panel to diagnose early-stage cancer, for which surgery is most effective. Our training set and validation set were enriched with advanced cancers (stages III and IV), while the testing set contained a relatively even proportion of early-stage patients (stages I and II) (Table 1). The mean risk score of NSCLC cases at early stages was not markedly different from that at later stages (P > 0.05). In Chinese cohorts, 12 of 14 stage I–II tumors had a risk score > 2.617 and an accuracy of 86%. In the testing set, using the cutoff value of 2.617 allowed correct prediction of 55 of 58 (95%) stage I–II tumors, demonstrating a positive performance for detecting early stages of NSCLCs (Table 2). The results indicate that the five-miRNA panel also functions as a strong predictor of early-stage cancer.
3.6. Logistic Regression Analysis of the Five Selected miRNAs and Their Panel
Finally, we conducted a forward stepwise binary logistic regression analysis to determine whether the diagnostic usefulness of the five-miRNA panel is affected by clinical features of NSCLC cases, such as TNM stage, tumor subtype, sex, smoking history or age. Sensitivity analyses revealed that the five-miRNA panel remained a strong predictor of risk of NSCLC regardless of almost all subgroupings of the patients considered in both Chinese and American cohorts (Supplemental Table 6).
To further examine whether the five altered miRNAs and their panel as well as other clinical characteristics are independent potent diagnostic markers for NSCLC, we next performed a forward stepwise univariate logistic regression using the control group as the reference category. Consequently, the odds ratios (ORs) for all of the five identified miRNAs (including miR-483-5p, miR-193a-3p, miR-214, miR-25 and miR-7) were statistically significant in the NSCLC cases in Chinese cohorts, while four of the five miRNAs (including miR-483-5p, miR-193a-3p, miR-25 and miR-7) were independently correlated with NSCLC patients in American cohort. Moreover, the panel of the five miRNAs was also significant in the NSCLC patients (P < 0.001) when the cutoff value for the panel was 2.617 in both the Chinese cohorts and American cohort (Supplementary Table 7). These results demonstrated that the 5 miRNAs and their panel are independent potent diagnostic markers for NSCLC. Subsequently, we also performed a multivariate logistic regression analysis to further determine the influence of five miRNAs' levels and clinical features on NSCLC. After adjusting for age, gender and smoking status, we found that miR-25 (OR = 12.3, 95% CI = 1.5–101.3, P = 0.02) and the five-miRNA panel (OR = 20.0, 95% CI = 7.9–50.7, P < 0.001) were still independently associated with NSCLC in Chinese cohorts; Furthermore, miR-25 (OR = 7.1, 95% CI = 2.1–23.9, P = 0.001), miR-483-5p (OR = 28.6, 95% CI = 8.1–100.5, P < 0.001) and miR-7 (OR = 91.0, 95% CI = 22.3–371.1, P < 0.001) were the independent risk factors for NSCLC in American cohort (Supplementary Table 7). Taken together, these results indicated that the 5 identified miRNAs enable the detection of NSCLC and may potentially serve as independent risk factor for NSCLC patients.
4. Discussion
To our knowledge, this study is the first multiethnic, multicentric, single-blind global analysis of miRNA expression patterns in the sera from patients with NSCLC. We identified a new five-miRNA panel in three independent cohorts from four centers in China and then validated the miRNA panel in a large cohort from America in a blind sensitivity trial. Our results reveal a five-miRNA panel which can serve as a potential biomarker for diagnosing NSCLC in patients of different races, even in the early stages of the cancer. The panel can also differentiate between malignant lesions and benign nodules that are frequently found by CT scans in high-risk populations. Our findings expand upon the results of previous studies of circulating-miRNA signatures in patients with NSCLC.
Numerous circulating-miRNA signatures have been reported for the detection of NSCLC, but the miRNAs identified by different groups vary from one another (Hu et al., 2010, Bianchi et al., 2011, Chen et al., 2012a, Heegaard et al., 2012, Wang et al., 2013). This inconsistency, we suspect, may be attributed to the differences in study design, disease type, sample size, methodology and especially in the method of data normalization. Improper normalization methods may obscure real changes and make artificial changes. In these previous studies on NSCLC, several different normalization methods were applied in circulating-miRNA expression analysis, including normalizing miRNA concentrations to serum volume, or using reference genes that include small RNAs, endogenous miRNAs or external non-human miRNAs. In the present study, we utilized a combination of let-7d, let-7g and let-7i as an internal reference for the normalization of serum miRNA concentrations. In a recent study, we comprehensively assessed let-7d/g/i and demonstrated the statistical superiority of this stable reference gene, as it corrects for experimental variations more effectively and achieves more accurate identification than alternative existing methods (Chen et al., 2013). The use of this appropriate reference for normalization of miRNAs in serum improves the reproducibility and sensitivity of the results and guarantees reliable biomarker discovery.
To realize true translational relevance and bring circulating miRNAs into routine diagnostics, multiethnic, multicentric, and reproducibly validated studies are indispensable. In the present study, we selected candidate serum miRNAs in three Chinese cohorts first and then validated the wide applicability of this biomarker in a large American cohort in a blind fashion. We found that a panel of five serum miRNAs, including miR-483-5p, miR-193a-3p, miR-25, miR-214 and miR-7, were significantly elevated in Chinese patients with NSCLC compared with control groups. The AUCs of ROC curve of this panel were 0.976 and 0.823 for the two Chinese cohorts, respectively. Furthermore, in a single-blind sensitivity trial, using a proper cutoff value of 2.617, the miRNA panel correctly classified 103 of 108 (95%) NSCLC cases and 87 of 104 (84%) controls from an American cohort. Our results demonstrate that the effectiveness of the five-miRNA panel is not limited to Chinese patients; rather, the biomarker also has high sensitivity and specificity for the diagnosis of NSCLC in American patients. Our results suggest that this miRNA panel has potential utility as a biomarker for detecting NSCLC in persons of different races. In addition, we observed that the Cq values of all serum miRNAs examined in American cohort (both patients and controls) were generally higher (approximately 3 cycles) than in the corresponding population in Chinese cohort. This result is consistent with a previous report that profiles of circulating miRNAs differed between African Americans and European Americans, suggesting that levels of circulating miRNAs can differ between races (Heegaard et al., 2012). Therefore, it is critical to select a suitable internal reference for quantification of miRNAs in serum. The endogenous internal reference miRNA let-7-d/g/i trio has similar characteristics to other serum miRNAs in that its average Cq value was also approximately 3 cycles higher in American cohort than in Chinese cohorts. Therefore, when ΔCq was calculated by subtracting the Cq values of let-7-d/g/i from the Cq values of the target miRNAs, the ΔCq values for the target miRNAs are comparable between the Chinese American cohorts. These results further demonstrate that a set of internal reference miRNA let-7-d/g/i may give highly consistent results across populations and races.
One of the weaknesses in our present study is the use of pooled samples for initially global miRNA screening since results generated from pooled samples might yield some inaccurate information and missing useful information due to individual difference, and therefore decreases the specificity of the test. Fortunately, by using RT-qPCR validation in a large number of individual serum samples arranged in multiple training and validation sets, we successfully identified the five-miRNA panel and additional bind trial test further confirmed our findings. Nevertheless, TLDA screening in individual sample should still be preferred and recommended in a similar study in the future. Another notably issue of the present study is that the fold changes of the five altered miRNAs in TLDA were not matched well with RT-qPCR analysis. The exact reason for this inconsistence, we suspected, is because a 12-cycle pre-amplification was made in TLDA, and the fold-change of miRNA expression between NSCLC patients and control subjects might be amplified. Therefore, the TLDA results must be validated using RT-qPCR assay performed at an individual level of serum samples.
The diverse, complex molecular events involved in the initiation and development of malignant neoplasm require functional changes not only in the tumor cell growth-related genes and/or tissue-specific genes but also in the body's immune response-related genes. Accordingly, there should be multiple miRNAs targeting those genes involved in tumorigenesis. Circulating miRNAs can derive from the tumor itself or from host responses to the tumor (Pritchard et al., 2012, Chen et al., 2012b). Therefore, functionally dysregulated miRNAs in circulation can be classified into three groups: (i) tissue-specific, (ii) tumor cell growth/cycle-related, and (iii) immune response-related. Of the five members of our predictive biomarker, miR-7 is a tissue-specific miRNA, because it has been found to be directly involved in NSCLC (Chou et al., 2010). miR-25, miR-483-5p and miR-193a-3p are tumor cell growth/cycle-related miRNAs (Zhang et al., 2013, Soon et al., 2009, Yu et al., 2015, Uhlmann et al., 2012). Finally, miR-214 may be categorized as an immune response-related miRNA because it has been found to play a dominant role in endothelial cell function, angiogenesis and exosome-mediated communication between endothelial cells (van Balkom et al., 2013, Ghalali et al., 2014, Schwarzenbach et al., 2012). Therefore, the combination of the identified five serum miRNAs in our study should be more reliable and specific than the single miRNA for the detection of NSCLC.
In conclusion, we have identified a new five-serum miRNA panel associated with NSCLC. Specifically, we have demonstrated that this group of serum miRNAs can potentially be used as an accurate biomarker for diagnosing NSCLC patients of different races. Further research is necessary to explore whether this panel of miRNAs is also effective for detection of NSCLC patients with other races and how much this panel adds to serum carcinoembryonic antigen.
Conflicts of Interest
The authors have no competing interests to declare.
Author Contributions
C-Y.Z., C.Z., X.C., M.X. and K.Z. designed the study. C.W., M.D., M.X., N.W., A.VL., R.S-G., X.C., J.W., W.G., X.W. and Y.Z. collected the data. C.W., M.D., A.VL., R.S-G. and S.C. did the data analysis and interpretation. C.Z., C.W. and C-Y.Z. wrote the manuscript. K.Z., M.X. and A.VL. revised the report. S.C., C.W. and X.C. did the statistical analysis. All authors reviewed the report and approved the final version.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2015.07.034.
Contributor Information
Ke Zen, Email: kzen@nju.edu.cn.
Xi Chen, Email: xichen@nju.edu.cn.
Chunni Zhang, Email: zchunni27@hotmail.com.
Chen-Yu Zhang, Email: cyzhang@nju.edu.cn.
Appendix A. Supplementary data
Supplementary material.
References
- Jemal A., Bray F., Center M.M., Ferlay J., Ward E., Forman D. Global cancer statistics. CA Cancer J. Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- D'Addario G., Früh M., Reck M., Baumann P., Klepetko W., Felip E. Metastatic non-small-cell lung cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2010;21(Suppl. 5):v116–v119. doi: 10.1093/annonc/mdq189. [DOI] [PubMed] [Google Scholar]
- Spiro S.G., Silvestri G.A. One hundred years of lung cancer. Am. J. Respir. Crit. Care Med. 2005;172:523–529. doi: 10.1164/rccm.200504-531OE. [DOI] [PubMed] [Google Scholar]
- Siegel R., Naishadham D., Jemal A. Cancer statistics, 2012. CA Cancer J. Clin. 2012;62:10–29. doi: 10.3322/caac.20138. [DOI] [PubMed] [Google Scholar]
- Little A.G., Rusch V.W., Bonner J.A., Gaspar L.E., Green M.R., Webb W.R. Patterns of surgical care of lung cancer patients. Ann. Thorac. Surg. 2005;80:2051–2056. doi: 10.1016/j.athoracsur.2005.06.071. [DOI] [PubMed] [Google Scholar]
- Oken M.M., Marcus P.M., Hu P., Beck T.M., Hocking W., Kvale P.A. Baseline chest radiograph for lung cancer detection in the randomized prostate, lung, colorectal and ovarian cancer screening trial. J. Natl. Cancer Inst. 2005;97:1832–1839. doi: 10.1093/jnci/dji430. [DOI] [PubMed] [Google Scholar]
- International Early Lung Cancer Action Program Investigators, Henschke C.I., Yankelevitz D.F., Libby D.M., Pasmantier M.W., Smith J.P. Survival of patients with stage I lung cancer detected on CT screening. N. Engl. J. Med. 2006;355:1763–1771. doi: 10.1056/NEJMoa060476. [DOI] [PubMed] [Google Scholar]
- Esquela-Kerscher A., Slack F.J. Oncomirs-microRNAs with a role in cancer. Nat. Rev. Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- Kong Y.W., Ferland-McCollough D., Jackson T.J., Bushell M. MicroRNAs in cancer management. Lancet Oncol. 2012;13:e249–e258. doi: 10.1016/S1470-2045(12)70073-6. [DOI] [PubMed] [Google Scholar]
- Calin G.A., Croce C.M. MicroRNA signatures in human cancers. Nat. Rev. Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- Chen X., Ba Y., Ma L., Cai X., Yin Y., Wang K. Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases. Cell Res. 2008;18:997–1006. doi: 10.1038/cr.2008.282. [DOI] [PubMed] [Google Scholar]
- Mitchell P.S., Parkin R.K., Kroh E.M., Fritz B.R., Wyman S.K., Pogosova-Agadjanyan E.L. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U. S. A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichelser C., Flesch-Janys D., Chang-Claude J., Pantel K., Schwarzenbach H. Deregulated serum concentrations of circulating cell-free microRNAs miR-17, miR-34a, miR-155, and miR-373 in human breast cancer development and progression. Clin. Chem. 2013;59:1489–1496. doi: 10.1373/clinchem.2013.205161. [DOI] [PubMed] [Google Scholar]
- Hu Z., Chen X., Zhao Y. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J. Clin. Oncol. 2010;28:1721–1726. doi: 10.1200/JCO.2009.24.9342. [DOI] [PubMed] [Google Scholar]
- Bianchi F., Nicassio F., Marzi M., Belloni E., Dall'olio V., Bernard L. A serum circulating miRNA diagnostic test to identify asymptomatic high-risk individuals with early stage lung cancer. EMBO Mol. Med. 2011;3:495–503. doi: 10.1002/emmm.201100154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Hu Z., Wang W., Ba Y., Ma L., Zhang C. Identification of ten serum microRNAs from a genome-wide serum microRNA expression profile as novel noninvasive biomarkers for nonsmall cell lung cancer diagnosis. Int. J. Cancer. 2012;130:1620–1628. doi: 10.1002/ijc.26177. [DOI] [PubMed] [Google Scholar]
- Heegaard N.H., Schetter A.J., Welsh J.A., Yoneda M., Bowman E.D., Harris C.C. Circulating micro-RNA expression profiles in early stage nonsmall cell lung cancer. Int. J. Cancer. 2012;130:1378–1386. doi: 10.1002/ijc.26153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Gu J., Roth J.A., Hildebrandt M.A., Lippman S.M., Ye Y. Pathway-based serum microRNA profiling and survival in patients with advanced stage non-small cell lung cancer. Cancer Res. 2013;73:4801–4809. doi: 10.1158/0008-5472.CAN-12-3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo Y., Wang C., Chen X., Zhong T., Cai X., Chen S. Increased serum and urinary microRNAs in children with idiopathic nephrotic syndrome. Clin. Chem. 2013;59:658–666. doi: 10.1373/clinchem.2012.195297. [DOI] [PubMed] [Google Scholar]
- Chen X., Liang H., Guan D., Wang C., Hu X., Cui L. A combination of Let-7d, Let-7g and Let-7i serves as a stable reference for normalization of serum microRNAs. PLoS One. 2013;8:e79652. doi: 10.1371/journal.pone.0079652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard C.C., Kroh E., Wood B., Arroyo J.D., Dougherty K.J., Miyaji M.M. Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prev. Res. (Phila.) 2012;5:492–497. doi: 10.1158/1940-6207.CAPR-11-0370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Liang H., Zhang J., Zen K., Zhang C.Y. Secreted microRNAs: a new form of intercellular communication. Trends Cell Biol. 2012;22:125–132. doi: 10.1016/j.tcb.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Chou Y.T., Lin H.H., Lien Y.C., Wang Y.H., Hong C.F., Kao Y.R. EGFR promotes lung tumorigenesis by activating miR-7 through a Ras/ERK/Myc pathway that targets the Ets2 transcriptional repressor EFR. Cancer Res. 2010;70:8822–8831. doi: 10.1158/0008-5472.CAN-10-0638. [DOI] [PubMed] [Google Scholar]
- Zhang S., Chen H., Zhao X., Cao J., Tong J., Lu J. REV3L 3′UTR 460 T > C polymorphism in microRNA target sites contributes to lung cancer susceptibility. Oncogene. 2013;32:242–250. doi: 10.1038/onc.2012.32. [DOI] [PubMed] [Google Scholar]
- Soon P.S., Tacon L.J., Gill A.J., Bambach C.P., Sywak M.S., Campbell P.R. miR-195 and miR-483-5p identified as predictors of poor prognosis in adrenocortical cancer. Clin. Cancer Res. 2009;15:7684–7692. doi: 10.1158/1078-0432.CCR-09-1587. [DOI] [PubMed] [Google Scholar]
- Yu T., Li J., Yan M., Liu L., Lin H., Zhao F. MicroRNA-193a-3p and -5p suppress the metastasis of human non-small-cell lung cancer by downregulating the ERBB4/PIK3R3/mTOR/S6K2 signaling pathway. Oncogene. 2015;34:413–423. doi: 10.1038/onc.2013.574. [DOI] [PubMed] [Google Scholar]
- Uhlmann S., Mannsperger H., Zhang J.D., Horvat E.Á., Schmidt C., Küblbeck M. Global microRNA level regulation of EGFR-driven cell-cycle protein network in breast cancer. Mol. Syst. Biol. 2012;8:570. doi: 10.1038/msb.2011.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Balkom B.W., de Jong O.G., Smits M., Brummelman J., den Ouden K., de Bree P.M. Endothelial cells require miR-214 to secrete exosomes that suppress senescence and induce angiogenesis in human and mouse endothelial cells. Blood. 2013;121:3997–4006. doi: 10.1182/blood-2013-02-478925. [DOI] [PubMed] [Google Scholar]
- Ghalali A., Ye Z.W., Högberg J., Stenius U. Phosphatase and tensin homolog deleted on chromosome 10 (PTEN) and PH domain and leucine-rich repeat phosphatase cross-talk (PHLPP) in cancer cells and in transforming growth factor β-activated stem cells. J. Biol. Chem. 2014;289:11601–11615. doi: 10.1074/jbc.M113.537241. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Schwarzenbach H., Milde-Langosch K., Steinbach B., Muller V., Pantel K. Diagnostic potential of PTEN-targeting miR-214 in the blood of breast cancer patients. Breast Cancer Res. Treat. 2012;134:933–941. doi: 10.1007/s10549-012-1988-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.