Abstract
Background Cellula r immune responses play a critical role in providing help for the production of neutralizing antibodies to influenza virus, as well as producing anti‐viral cytokines and killing infected cells in the lung. Heterosubtypic T‐cell responses between different subtypes of influenza have been shown to exist in humans and to provide protection against morbidity and mortality associated with H5N1 infection in animal challenge models. Therefore, existing T‐cell responses induced by natural infection or vaccination in humans may provide some degree of protection from infection with H5N1 strains, or may attenuate the severity of disease.
Objectives To investigate heterosubtypic T‐cell responses to avian influenza in humans.
Methods T‐cell responses to an overlapping set of H5 HA peptides and inactivated viruses (H1N1, H3N2 and H5N1) were assessed using IFN‐γ and IL‐2 enzyme‐linked immunospot (ELISpot) assays in a cohort of adults either vaccinated against seasonal influenza in the last 3 years (n = 20) or previously infected (n = 40).
Results T‐cell responses to all three subtypes of virus were found in both infected and vaccinated individuals by IFN‐γ and IL‐2 ELISpot assays. Approximately half of the participants from each group had a positive T‐cell response to the H5 HA peptides in the IFN‐γ or IL‐2 ELISpot assay.
Conclusions Heterosubtypic T‐cell responses to H5 HA occur quite frequently in vaccinated and infected individuals. Further investigation of these responses and what role they may play upon challenge or vaccination against H5N1 may assist in vaccine design for avian influenza.
Keywords: Cytokines, haemagglutinin, heterosubtypic, human, T cell, vaccination
Introduction
Neutralizing antibodies produced by natural infection or vaccination can provide protection against influenza infection by blocking entry of the virus into cells of the respiratory epithelium. However, these antibodies are typically inadequate at protecting against serologically distinct strains. 1 , 2 Therefore, antibodies produced by prior infection or vaccination with circulating human strains of influenza are thought to offer little neutralizing capacity against H5N1 avian influenza. The cellular immune response plays a critical role in the generation of neutralizing antibodies to influenza virus; helper CD4 T cells produce cytokines and CD8 T cells mediate the killing of infected cells in the lung. 1 Although the cellular immune response in isolation does not prevent infection, it does reduce morbidity and mortality in murine models. 3 , 4 , 5 , 6 , 7
The existence of heterosubtypic T‐cell responses that cross react to epitopes on different subtypes of influenza virus has been demonstrated in mice 3 , 5 , 6 , 7 , 8 , 9 and in humans. 10 , 11 , 12 , 13 Heterosubtypic T‐cell responses induced by infection or vaccination have been shown to provide protection against morbidity and mortality from H5N1 challenge in chickens 14 and mice. 3 , 5 , 6 , 7 There is also some evidence to suggest that heterosubtypic T‐cell responses may contribute to reducing mortality following a H5N1 challenge in ferrets. 15 , 16 Human infection cases of avian influenza tend to occur in the young, 17 which may be due to greater exposure to H5N1, but more likely may indicate that adults, who will have had more exposure to influenza A strains in their lifetime, have some existing immunity against avian influenza. Therefore, existing T‐cell responses produced from natural infection or vaccination with the seasonal influenza vaccine in humans may provide some degree of protection from infection with H5N1 strains, or may attenuate the severity of disease in humans. 13 However, there has been relatively little published about the prevalence or specificity of influenza‐specific heterosubtypic T‐cell responses to H5N1 strains in humans.
T‐cell responses can be directed to any protein of influenza virus. 18 Over 150 human T‐cell epitopes of influenza have been published and nearly 300 have been published for mice. 18 Two animal studies have demonstrated that T‐cell responses to components of influenza virus other than the major surface glycoprotein, haemagglutinin (HA), are able to provide help for antibody responses to HA. 19 , 20 Therefore, whilst total T‐cell response to influenza virus is important for help in prevention or recovery from infection, the major aim of most current avian influenza vaccine candidates is to induce neutralizing antibodies to HA; hence HA is the major antigenic component of many prototype vaccines 21 , 22 , 23 and is likely to be the major antigenic component of next generation vaccines against H5N1. Therefore, when considering the heterosubtypic T‐cell response in humans to avian influenza, it is valuable to investigate T‐cell responses to H5 HA.
The enzyme‐linked immunospot (ELISpot) assay is a sensitive, valuable tool for assessing T‐cell responses to antigen and is able to detect antigen‐specific responses at the single‐cell level. 24 ELISpot assays have been widely used to assess T‐cell responses to HIV and HCV and many studies have also used this assay to examine T‐cell responses to influenza in mice 25 , 26 , 27 as well as humans. 10 , 13 , 26 , 28
The primary aim of the present study was therefore to develop IFN‐γ and IL‐2 ELISpot assays to investigate whether existing T‐cell responses in human participants induced by either prior vaccination or infection were cross‐reactive to the avian influenza H5 HA. In addition, H1, H3 and H5 HA‐specific antibody responses in serum were assessed by ELISA. We show that heterosubtypic T‐cell responses to H5 HA peptides and H1N1, H3N2 and H5N1 inactivated viruses are frequently detected in individuals previously vaccinated and/or infected with human subtypes of influenza. Cross‐reactive antibody responses to H5 HA in these individuals were also observed.
Materials and methods
Human subjects
Sixty healthy adult volunteers were recruited from the staff of the Burnet Institute and Alfred Hospital. Upon recruitment, participants consented to giving 30 ml of blood. Participant age ranged from 20 to 61 (mean: 34; median: 31) and 70% were female. Exposure to H5 strains of influenza in this cohort is not known, but is highly unlikely. Together with serology results, medical records were used to group the participants as ‘infected’ (n = 40; I1–40) for those that were seropositive and had not been vaccinated in the last 3 years, or ‘vaccinated’ (n = 20; V1–20) for those that had been vaccinated in the previous 3 years. The vaccine formulation was either Fluvax (CSL) or Vaxigrip (Sanofi Pasteur), which are both inactivated influenza vaccines containing influenza A viruses H1N1 and H3N2 and influenza B virus (15 μg HA of each per dose), of strains according to recommendations by WHO. Vaccinated individuals tended to be healthy adult healthcare workers vaccinated for reasons of potential occupational exposure (e.g. nurses) and had been vaccinated, on average, 13 months prior to participation in this study. Of the vaccinated group, 65% individuals had been vaccinated at least once prior to their most recent vaccination. The study was approved by the Alfred Human Research Ethics Committee (project number 6/06).
HA ELISA
Sera from participants were obtained by centrifugation of SSTTM Vacutainer tubes (BD, North Ryde, NSW, Australia) containing 4–8·5 ml blood at 1110 g. Nunc‐immuno MaxiSorp 96‐well plates (Thermo Fisher Scientific, Rochester, NY, USA) were coated with H1 (A/New Caledonia/20/99), H3 (A/Wyoming/3/2003) or H5 (A/Vietnam/1203/2004) purified (>90%) recombinant HA (rHA) (Protein Sciences, Meriden, CT, USA) diluted in carbonate buffer (pH 9·7) to 100 ng/ml. One hundred microlitres per well of this antigen solution was added to the plate, which was incubated overnight at 4°C. Plates were washed in between all steps using an ELx405 Bio‐Tek automated plate washer (Biotek Instruments Inc., Winooski, VT, USA), which washed the plates six times with PBS‐Tween (0·05%). Plates were blocked with 200 μl/well PBS‐Tween (0·05%) containing 2% goat serum (Millipore, North Ryde, NSW, Australia) and incubated at 37°C for 1 hour. Human serum specimens were diluted fourfold in specimen diluent buffer [PBS‐Tween (0·05%), 1% casein; Millipore] and plates were incubated at 37°C for 1 hour. Antibody binding was detected using a sheep anti‐human IgG antibody conjugated with horse radish peroxidase (IgG‐HRP; Millipore) and 3,3′,5,5′‐Tetramethylbenzidine substrate (Millipore). Colour development was stopped after 10 minutes with stop solution (0·1 m sulfuric acid; Millipore). Plates were read in a Multiskan RC plate spectrophotometer (Labsystems Helsinki, Finland) at 450–620 nm. A pool of children’s sera (kindly provided by Prof. William Rawlinson, Southern Eastern Area Laboratory Service, Sydney, NSW, Australia) negative for HA reactivity in the ELISA was used as a negative control in each assay and gave a mean maximum OD reading of 0·5 at 1:40. This value of 0·5 was arbitrarily set as the cut‐off to detect the end‐point titre of the adult sera. Using this cut‐off, we observed that 54 of the 60 participants had a titre below 2560 for H5 HA antibodies. We also observed 85% vaccinated individuals to have a titre of 2560 or above for H1 HA, whereas only 27·5% non‐vaccinated individuals had a titre of 2560 or above to H1 HA, compared to higher titres to H3 HA. This fits with the fact that H1 is included in the seasonal influenza vaccine and that in recent years, H1 has been the less common circulating subtype, compared to H3. A titre of 2560 and above was therefore considered positive for each influenza subtype.
Preparation of peripheral blood mononuclear cells
Peripheral blood mononuclear cells (PBMC) were isolated by centrifugation of lithium‐heparinized blood on a Ficoll‐paque density gradient (GE Healthcare, Rydalmere, NSW, Australia). Isolated PBMCs were cryopreserved in RPMI 1640 medium supplemented with 2 mm l‐glutamine, 100 U/ml penicillin and 10 μg/ml streptomycin (Invitrogen, Mount Waverley, VIC, Australia), 20% heat inactivated fetal calf serum (FCS; Invitrogen) and 10% DMSO (Sigma‐Aldrich Pty. Ltd., Castle Hill, NSW, Australia). PBMCs were frozen at −80°C in a controlled rate freezing unit containing isopropanol overnight before being transferred into liquid nitrogen.
Peptides
Seventy‐three peptides covering residues F8‐V522 and L554‐I568 (of 568) of the HA protein of influenza A virus A/Vietnam/1203/04 (H5N1) were synthesized (Auspep, Parkville, Vic., Australia). Peptides covering amino acids M1‐L7 and K523‐S553 could not be synthesized because of their hydrophobic nature. The peptides were 18 amino acids in length overlapping adjacent peptides by 11 amino acids. The peptides were reconstituted in 10–40 μl DMSO and then diluted up to 100 μl with RPMI 1640 supplemented with 2 mm l‐glutamine, 100 U/ml penicillin and 10 μg/ml streptomycin and pooled into eight separate pools (pools 1–8), such that each peptide was at a concentration of 100 μg/ml. The peptides were pooled in order of sequence, with pool one containing the first nine peptides, pools 2–7 containing 10 peptides each and pool eight containing the last four peptides. The peptides were used at a final concentration of 1 μg/ml in the ELISpot assays and DMSO to a final concentration of 0·8% was added to media controls.
Inactivated influenza viruses
The inactivated influenza viruses A/New Caledonia/20/99 (H1N1), A/Wellington/1/2004 (H3N2) and A/Vietnam/1194/2004 (H5N1) were used at a final concentration of 5 μg/ml in the ELISpot assays to assess total T‐cell responses. Purified, concentrated viruses were inactivated using Beta‐propiolactone (BPL; Ferak Berlin, Berlin, Germany) and the inactivation confirmed by passing dilutions of the inactivated virus through two passages in 10‐ to 12‐day‐old embryonated hen’s eggs to ensure there was no growth, as determined by the lack of agglutination of chicken RBC in the allantoic fluid. A/Vietnam/1194/2004 and A/Vietnam/1203/2004 H5 HA differ only at amino acid position 52.
ELISpot assays for IFN‐γ and IL‐2
MultiScreenHTS 96‐well plates (Millipore) were coated with 5 μg/ml of either anti‐human IFN‐γ (clone 1‐D1K) or anti‐human IL‐2 (clone IL2‐I) monoclonal antibodies (Mabtech, Nacka Strand, Sweden) and left at 4°C overnight. Plates were washed six times with 200 μl/well of 1× D‐PBS (Invitrogen) between all steps. Wells were blocked with RPMI 1640 supplemented with 2 mm l‐glutamine, 100 U/ml penicillin and 10 μg/ml streptomycin and 10% heat inactivated FCS, hereafter referred to as complete medium, for 1 hour at 37°C. The coated wells were filled with complete medium containing 1 × 105 either previously cryopreserved or freshly isolated PBMCs in triplicate. These PBMC were incubated with the HA peptides (1 μg/ml), inactivated influenza virus (5 μg/ml), or the positive controls phytohaemagglutinin (PHA, 5 μg/ml; Sigma), anti‐human CD3 monoclonal antibody (10 μg/ml; Mabtech, Nacka Strand, Sweden) or a pool of CD8 epitopes from cytomegalovirus, Epstein–Barr virus and influenza virus (CEF, 2 μg/ml; Mabtech) and incubated at 37°C for 18–24 hours. Secreted cytokine was detected using 1 μg/ml of either anti‐human IFN‐γ (clone 7‐B6‐1) or anti‐human IL‐2 (clone IL2‐II) monoclonal antibodies (Mabtech). Streptavidin‐alkaline phosphatase (1 μg/ml; Sigma‐Aldrich) was added and spots were detected using 5‐Bromo‐4‐chloro‐3‐indolyl phosphate/nitroblue tetrazolium liquid substrate (Sigma‐Aldrich). Spots were counted using AID ELISpot Reader System, Version 3·5 (Autoimmun Diagnostika GmbH, Strassberg, Germany). All tests were performed in triplicate and the mean values were calculated. A response was considered to be positive when the number of spots in the wells with antigen‐stimulated cells, after subtraction of the background (wells without antigen stimulation), was at least 50 SFC/106 PBMC. In our hands, the background was usually <20 SFC/106 PBMC. As all adults in this study had influenza‐specific responses, it was impossible to assess background for the ELISpot assay using PBMC from individuals never exposed. Therefore, we arbitrarily chose a very conservative 50 SFC/106 PBMC as a cut‐off to decrease the chance of false positives.
CD4 and CD8 depletion
Previously cryopreserved PBMC were seeded in a 24‐well plate at 2–4 × 106 cells in 2 ml complete medium per well. The amount of 1 μg/ml of each H5 HA peptide pool was added to the culture and incubated at 37°C for 7 days. After 3 days, 10 ng/ml of IL‐7 (Sigma‐Aldrich) was added to the culture. On day 7, undepleted, CD4‐depleted and CD8‐depleted populations from these cells were tested in the IFN‐γ ELISpot as described above, with exceptions being that test wells were performed in duplicate at 50 000 cells per well and responses to the rHA proteins (Protein Sciences) (1 μg/ml) were also investigated.
CD4 and CD8 MACS MicroBeads [MicroBeads conjugated to monoclonal mouse anti‐human CD4 (IgG1; clone M‐T466) or CD8 (IgG2a; clone BW135/80) antibodies; Miltenyi Biotec GmbH, Bergisch Gladbach, Germany] were used to deplete the cells of CD4+ and CD8+ cell populations respectively and depletions were performed according to the manufacturer’s instructions. Briefly, up to 107 cells were incubated in 80 μl MACS buffer [PBS supplemented with 0·5% BSA (Sigma‐Aldrich) and 2 mm EDTA] and 20 μl MicroBeads at 4°C for 15 minutes. The cells were washed with MACS buffer to remove unbound beads, then resuspended in 500 μl MACS buffer and placed through an LD column (Miltenyi Biotec) in the magnetic field of a MACS separator (Miltenyi Biotec). Negative fractions were allowed to flow through while CD4 or CD8 positive cells with MicroBeads bound to the surface remained bound in the column.
Cell populations were analysed by flow cytometry for cell surface expression of CD3, CD4 and CD8 using the following mouse IgG1κ antibodies: anti‐CD3‐PE, anti‐CD4‐PerCP and anti‐CD8‐FITC (BD, North Ryde, NSW, Australia). Data was analysed using cellquest software (BD, North Ryde, NSW, Australia). Purity of the depleted populations was 91–98% (data not shown). Enzyme‐linked immunospot assay and flow cytometry data were used together to calculate SFC/106 depleted population back to represent SFC/106 input PBMC in the undepleted population.
Haemagglutination inhibition and neutralization assay
The HI 29 and neutralization 30 assays were performed according to standard protocols. Briefly, for the haemagglutination inhibition (HI) assay, 25 μl of (4HAU) reverse genetics (RG) A/Vietnam/1203/2004 (H5N1) (St Jude Children’s Research Hospital, Memphis, TN, USA) virus was incubated at room temperature with an equal volume of receptor‐destroying enzyme (RDE)‐treated sera (RDE (II), Deka Seiken Co. Ltd, Tokyo, Japan). Sera were diluted twofold, beginning at 1:10. Following one hour incubation, 25 μl of 1% (v/v) horse red blood cells was added to each well. Haemagglutination was read after 30 minutes. For the neutralization assay, an equal volume of RDE‐treated sera (twofold dilutions of sera beginning at 1:10) and 200TCID50/100 μl RG A/Vietnam/1203/2004 virus was incubated at 35°C for 1 hour. The virus/sera mix was then added to washed, confluent (80–90%) monolayers of MDCK cells in 96‐well plates and incubated at 35°C for 2 hours. The virus/sera mix was replaced with FCS‐free tissue culture media supplemented with 4 μg/ml trypsin and the cells incubated at 35°C. Four days later, monolayers were stained with 0·036% (w/v) neutral red, washed and dye released by ethanol:PBS (1:1). Absorbance was read at 490 nm and titres were expressed as the reciprocal of the highest dilution of sera where haemagglutination or cell death was prevented.
Statistical analyses
The Wilcoxon–Rank Sum test was used to compare the distributions of serum HA antibody titres in previously vaccinated and infected individuals. This test was also used for analysing differences in the distribution of magnitude of T‐cell responses to the inactivated viruses and the H5 HA peptides between groups. The differences between the proportion of individuals with or without a positive T‐cell response to the inactivated viruses and H5 HA peptides between groups was analysed by using either the chi‐square test or the Fisher’s exact test as appropriate. Fisher’s exact test was used to measure the difference in proportion of positive anti‐H5 antibody titres between the two groups. The Spearman’s Rank Correlation test was used to analyse the correlation between antibody titre and magnitude of T‐cell responses. A P‐value < 0·05 was considered significant.
Results
H1, H3 and H5 HA antibody responses detected in both previously vaccinated and non‐vaccinated participants
To confirm previous exposure to influenza (either by infection or vaccination), serum samples from all participants were tested for anti‐HA responses in an indirect ELISA against H1, H3, and H5 HA. HA‐specific antibody responses were detected in all participants. Various patterns of antibody specificity and level against each of the three subtypes were found. Of the infected group, 19 (48%) individuals had positive antibody titres against H3 HA only, whereas 17 (85%) of vaccinated individuals had positive responses to both H1 and H3. Vaccinated individuals had a significantly higher distribution of antibody titres against H1, H5 (P‐value < 0·001) and H3 (P‐value = 0·0013) HA compared to non‐vaccinated individuals (Figure 1). The increase in antibody titre in the vaccinated group is likely to be due to the boosting of pre‐existing memory B cells, given that most individuals from the vaccinated group had been vaccinated more than once previously and are very likely to have experienced a prior influenza infection.
Figure 1.

H1, H3 and H5 HA‐specific antibody titres in previously infected and vaccinated individuals. Serum samples were obtained from both previously infected (I1–40) and vaccinated (V1–20) individuals and serially diluted out in a 96‐well plate coated with H1, H3 and H5 HA (100 ng/ml). Titres were determined by the reciprocal of the highest dilution that gave an OD reading of 0·5 or above. Titres of 2560 and above were considered positive. The distribution of the titres were significantly higher for each H1, H5 (P < 0·001) and H3 (P = 0·0031) HA in the vaccinated group (n = 20) compared to the infected individuals (n = 40). The black line represents the median.
Interestingly, five individuals in the vaccinated group had a positive H5 HA antibody titre (25%), compared to just one individual in the infected group (2·5%) (P‐value < 0·001). Ten individuals in the infected group had titres against each of H1, H3 and H5 HA below our definition of a positive response, but had detectable memory T‐cell responses to influenza virus (described below), indicating previous exposure to influenza A virus.
Serum samples positive for anti‐H5 antibodies by ELISA from the vaccinated group and samples negative for anti‐H5 antibodies by ELISA from the infected group were analysed for anti‐A/Vietnam/1203/2004 (H5N1) neutralizing and haemagglutination inhibiting antibodies by HI assay and neutralization assay. All samples tested were negative for HI antibodies and the majority was also negative in the neutralization assay. However, one of the vaccinated individuals (V3), who had an ELISA titre against H5 HA of 10240, was weakly positive in the neutralization assay with a titre of 1:20.
T‐cell responses detected to H1N1, H3N2 and H5N1 inactivated influenza viruses in both vaccinated and infected individuals by IFN‐γ and IL‐2 ELISpot
We examined the T‐cell response in vaccinated and infected individuals to inactivated influenza viruses A/New Caledonia/20/99 (H1N1), A/Wellington/1/2004 (H3N2) and A/Vietnam/1194/04 (H5N1) by IFN‐γ and IL‐2 ELISpot assay. All participants had IFN‐γ+ and/or IL‐2+ memory T‐cell responses to at least one influenza A virus. T‐cell responses to each of the three viruses were detected in both assays in both vaccinated and infected groups (Figure 2). There was no statistically significant difference between proportions of individuals with or without positive T‐cell responses to the viruses in each group, with the exception being responses to H3N2 in the IL‐2 ELISpot assay in which two (10%) vaccinated individuals compared to 12 (20%) infected individuals had IL‐2+ H3N2‐specific T‐cell responses (P = 0·034). Interestingly, 59 (98%) of the 60 participants had IFN‐γ+ H1N1‐T‐cell responses and all 60 had IFN‐γ+ H5N1‐specific T‐cell responses; only 40 (67%) had IFN‐γ+ H3N2‐specific T‐cell responses. Of all 60 participants, 38 (63%), 14 (23%) and 15 (25%) had IL‐2+ H1N1−, H3N2− and H5N1‐specific T‐cell responses respectively.
Figure 2.

T‐cell responses to H1N1, H3N2 and H5N1 inactivated influenza viruses, detected by IFN‐γ and IL‐2 ELISpot. PBMC from all 40 previously infected and 20 previously vaccinated individuals were cultured overnight with 5 μg/ml inactivated influenza virus in triplicate at 1 × 105 cells/well and IFN‐γ and IL‐2 production was measured. (A) Representative results from six individuals are shown from both previously infected (top) and vaccinated (bottom) groups. Positive T‐cell responses to each virus were detected by both assays in each group. The dashed line at 50 SFC/106 PBMC represents the positive response cut‐off. (B) Representative images of triplicate wells are shown from both IFN‐γ (left) and IL‐2 (right) ELISpot assays. Responses to the media alone (negative control), H1N1, H3N2 and H5N1 inactivated viruses and PHA (positive control) are shown. Spot counts are indicated in the top left corner of each well.
The distribution of the magnitude of responses in the IFN‐γ ELISpot did not significantly differ between groups and all 60 participants averaged at 260, 87 and 287 SFC/106 PBMC for H1N1, H3N2 and H5N1 viruses respectively. The distribution of the magnitude of IL‐2+ H3N2‐specific T‐cell responses was statistically significant between groups and averaged at 51 and 28 SFC/106 PBMC for the vaccinated and infected groups respectively (P = 0·0417). There was no statistically significant difference in the distribution of the magnitude of responses in the IL‐2 ELISpot for H1N1 and H5N1. The average magnitude of responses in the IL‐2 ELISpot for H1N1, H3N2 and H5N1 viruses were 109, 40 and 56 SFC/106 PBMC respectively. There was no statistically significant correlation between antibody titre and magnitude of T‐cell response against any of the three subtypes of virus in either IFN‐γ and IL‐2 ELISpot assays in either group of participants (P > 0·05).
T‐cell responses detected to H5 HA synthetic peptides by IFN‐γ and IL‐2 ELISpot in both previously vaccinated and infected individuals
To examine whether cross‐reactive T‐cell responses that exist in individuals either previously infected with or vaccinated against human subtypes of influenza are able to cross‐react with H5 HA, T‐cell responses to a set of peptides corresponding to the H5 HA protein were investigated using IFN‐γ and IL‐2 ELISpot assays. Seventy‐three peptides spanning H5 HA of strain A/Vietnam/1203/04 were grouped into eight pools (pools 1–8) for use in the ELISpot assays, with each peptide within the pools at a final concentration of 1 μg/ml.
T‐cell responses to the H5 HA peptides were detected in both groups of participants (Figure 3). Of the infected participants, 22 (55%) had T‐cell responses to at least one peptide pool in the IFN‐γ or IL‐2 assay. In the vaccinated group, eight (40%) individuals had T‐cell responses to at least one peptide pool in the IFN‐γ or IL‐2 ELISpot assay. The magnitude of responses ranged from 0 to 233 SFC/106 PBMC in IFN‐γ and 0 to 170 SFC/106 PBMC in the IL‐2 ELISpot (Figure 4). There was no statistically significant difference between the distribution of the magnitude of responses, or the proportion of individuals with positive responses to the peptides in each group. There was no statistically significant correlation between H5 antibody titre and magnitude of T‐cell response against the H5 HA peptides in either IFN‐γ or IL‐2 ELISpot assays in either group of participants (P > 0·05).
Figure 3.

T‐cell responses to H5 HA peptide pools (pools 1–8), detected by IFN‐γ and IL‐2 ELISpot. PBMC from all 60 participants were cultured overnight with 1 μg/ml of each H5 HA peptide pool in triplicate at 1 × 105 cells/well and IFN‐γ and IL‐2 production was measured. (A) Representative results are shown from six individuals both previously infected (top) and vaccinated (bottom) groups. Positive T‐cell responses to the peptides were detected by both assays in each group. The dashed line at 50 SFC/106 PBMC represents the positive response cut‐off. (B) Representative images of triplicate wells are shown from both IFN‐γ (left) and IL‐2 (right) ELISpot assays. Responses to media alone (negative control), H5 HA peptide pools and PHA (positive control) are shown. Spot counts are indicated in the top left corner of each well.
Figure 4.

Mean summed SFC/106 PBMC to the H5 HA peptides in IFN‐γ and IL‐2 ELISpot assays in both infected and vaccinated groups. Responses to the H5 HA peptides were summed in each individual and the average was calculated for each group; 120 and 101 SFC/106 in the IFN‐γ ELISpot assay and 144 and 84 SFC/106 in the IL‐2 ELISpot assay for the infected and vaccinated groups respectively.
CD4‐depleted and CD8‐depleted cells contribute to the anti‐influenza T‐cell response in both vaccinated and infected individuals
To determine the responding cell phenotype of the influenza‐specific T cells, CD4‐ and CD8‐depletions were performed on three individuals from each group. To increase the frequency of H5 HA‐specific T cells, PBMCs were first cultured for 7 days with 1 μg/ml of all H5 HA peptides. Following 7 days of culture, the cells underwent either CD4‐ or CD8‐depletion. Undepleted, CD4‐depleted and CD8‐depleted cells were then tested in the IFN‐γ ELISpot for influenza‐specific responses to the H5 HA peptides, recombinant HA proteins as well as the inactivated viruses. All individuals had both CD4‐ and CD8‐depleted influenza‐specific T‐cell responses. An example of the per cent contribution of each cell phenotype to the overall influenza antigen‐specific response in two individuals (I38 and V20) is shown in Figure 5. The infected individual, I38, had an even contribution of CD4‐ and CD8‐depleted T‐cell responses, whereas the vaccinated individual (V20) had predominantly CD8‐depleted (i.e. CD4+) T‐cell responses to the influenza antigens.
Figure 5.
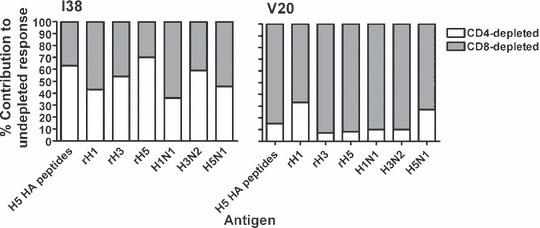
Per cent contribution to the influenza‐specific T‐cell response by CD4‐ and CD8‐depleted cell populations, as detected by IFN‐γ ELISpot. PBMCs from three vaccinated individuals and three infected individuals were cultured for 7 days with 1 μg/ml H5 HA peptides, following which the cells were depleted for CD4+ and CD8+ cells and tested in the IFN‐γ ELISpot against a pool containing all of the H5 HA peptides, recombinant HA proteins and the inactivated viruses. Together with flow cytometry data of separated populations, the ELISpot results were used to normalize the CD4‐depleted and CD8‐depleted responses as a percentage of the undepleted PBMC response. Representative results are shown from both the previously infected (I38) and vaccinated (V20) groups, demonstrating that both individuals had CD8‐depleted and CD4‐depleted T‐cell responses to the influenza antigens.
Discussion
It is possible that T‐cell responses generated from a less pathogenic strain of influenza A could provide cross‐reactive T‐cell responses that assist in the production of antibodies and lessen the severity and duration of disease and reduce viral shedding upon challenge with a highly pathogenic avian influenza (HPAI). 31 However, little is known about such cross‐reactive T‐cell responses in humans. It is also important to note that the role of T‐cell‐mediated responses in protection and recovery from seasonal strains of influenza in humans, particularly on CD4+ T‐cell responses, is not well defined. In this study, we have detected T‐cell responses that cross‐react with H5N1 inactivated virus and H5 HA peptides, as well as antibodies that cross‐react with H5 rHA, in individuals either previously infected with or vaccinated against human influenza strains.
Whilst antibody responses to influenza are often reported to be subtype and even strain‐specific, cross‐reactive antibody responses between subtypes of influenza A virus have previously been demonstrated in animal models 7 , 32 , 33 , 34 , 35 and in humans. 12 , 35 , 36 , 37 , 38 A recent study by Gioia et al. (2008), 12 reported that the seasonal influenza vaccine was able to induce neutralizing antibodies to both H5N1 as well as the influenza vaccine in some individuals. Furthermore, serological results from several H5N1 vaccine human trials have detected pre‐existing neutralizing antibodies to H5N1 in individuals never exposed to this subtype. 21 , 22 , 39 Therefore, it is not unexpected that we detected antibodies that reacted to H5 HA.
It is likely that the heterosubtypic antibody responses detected in this study are non‐neutralizing and not haemagglutination inhibiting responses, and are directed at conserved regions of the HA molecule. This is supported by the HI data, which showed samples with a positive titre in the H5 ELISA to be negative for neutralizing anti‐H5 HI antibodies. However, studies have shown the HI method to be less sensitive compared to ELISA and microneutralization, as it failed to detect HI antibodies against avian viruses in mammals, even in cases where infection was confirmed by virus isolation. 38 , 40 , 41 This is also highlighted by the fact that one individual with a positive anti‐H5 HA titre in the ELISA was also positive in the neutralization assay, but negative in the HI assay. It is unlikely that the baculovirus‐produced H5 HA is picking up antibodies in human sera that cross‐react with insect proteins because of its high purity, but rather the reactivity of human sera with H5 HA is caused by cross‐reactive epitopes common to HA proteins of different influenza subtypes that have become exposed with partial denaturation of antigen bound to a solid surface, such as an ELISA plate. 38 This assay would therefore not be useful for detection of strain‐specific or neutralizing antibodies, but rather, is useful for detecting total antibody response.
Whilst unlikely to be able to provide protection against a H5N1 challenge, non‐neutralizing, heterosubtypic antibodies directed to conserved regions of HA may provide assistance in antibody‐dependent cell‐mediated cytotoxicity function and/or clearance of antigen–antibody complexes mediated by macrophages 6 and therefore may aid in resolution of infection or reduction of morbidity and mortality. Two murine vaccination studies have demonstrated protection 6 or reduced disease severity and decreased mortality 42 following challenge with H5N1 in the absence of HI and low or undetectable levels of neutralizing antibodies, but in the presence of cross‐reactive non‐neutralizing antibody and T‐cell responses. Furthermore, a vaccine study in ferrets showed that all ferrets were protected against a lethal challenge of H5N1 in the absence of HI and virus‐neutralization antibodies, which together suggests that a low level or even the absence of neutralizing antibodies in the serum after immunization with H5 vaccines does not necessarily indicate that a vaccine is ineffective. 16 If indeed these non‐neutralizing, cross‐reactive antibodies play an important role in protection against infection or morbidity and mortality against H5N1, then these responses are worthwhile measuring in response to candidate H5N1 vaccines, rather than solely measuring neutralizing antibody responses. A memory B‐cell ELISpot assay, which has been previously used successfully to investigate influenza‐specific memory B‐cell frequency 43 , 44 could be used to further investigate these responses.
All participants had IFN‐γ+ or IL‐2+ T‐cell responses to the inactivated viruses in the ELISpot assays, indicating previous infection in the non‐vaccinated group and vaccination (as well as possible previous infection) in the vaccinated group. It is not unexpected that some individuals exhibited only IFN‐γ+ T‐cell responses, some only IL‐2+, whilst others showed both. This may correlate with the presence of CD8 and CD4 T cells respectively. The overall variability in responses and in particular the lower frequency of T‐cell responses to H3N2 may be caused by the strain(s) to which each individual has previously been exposed. Differences in sequence to the testing strain in the ELISpot may have influenced epitopes recognized and hence the magnitude of response. Other factors include time since last infection as well as the preparation of inactivated virus used, which may have contained varying amounts of each influenza protein. Alternatively, the reason behind the lower magnitude and frequency of H3N2‐specific T‐cell responses may be immunological, such as differences in immunogenicity or replication levels between the subtypes.
Detection of cross‐reactive T‐cell responses to H5N1 inactivated virus was not unexpected because the existence of such responses following vaccination or infection with human influenza subtypes has been documented previously in animal models 3 , 5 , 6 , 7 , 14 and two studies in humans. 12 , 13 Furthermore, the internal proteins are more conserved between influenza A virus strains compared to the surface glycoproteins. Therefore, much of the responses seen are likely to have been directed to internal proteins. However, it was surprising that all 60 participants had IFN‐γ+ H5N1‐specific T‐cell responses at an average magnitude of 287 SFC/106 PBMC, a higher frequency and magnitude compared to H1N1 and H3N2 responses.
Approximately 50% of participants overall had H5 HA peptide‐specific T‐cell responses in either the IFN‐γ or IL‐2 ELISpot assay, demonstrating that indeed some individuals who have been previously infected and/or vaccinated against human influenza strains have heterosubtypic T‐cell responses that are responsive to epitopes on H5 HA. The sequence similarity between A/New Caledonia/20/99 (H1N1) HA (GenPept: ABF21272) and A/Vietnam/1203/2004 (H5N1) HA (GenPept: ABP51977 45 ) is 78·2%, compared to 59·2% similarity between A/Wellington/1/2004 (H3N2) (GenPept: ABG48258) and A/Vietnam/1203/2004 (H5N1) (alignment performed using the EMBOSS Pairwise Alignment Algorithms at http://www.ebi.ac.uk/Tools/emboss/align/index.html). Therefore, whilst it is possible that these heterosubtypic responses have arisen from previous infection with H1 or H3 influenza, given the sequence similarities with H5, it is logical to assume that most of the responses have arisen from previous infection with H1. However, there was no significant difference in prevalence or magnitude of IFN‐γ+ and IL‐2+ H5 HA peptide‐specific responses between the vaccinated and infected group. The lack of difference in influenza‐specific T‐cell responses between the vaccinated and infected group fits with the idea that split, inactivated vaccine is a poor inducer of cellular immunity. 46
Whilst there are regions of homology spread throughout the H1 and H5 HA proteins that are covered by peptides from each eight peptide pools from this study, much homology between the two proteins is seen between R346 and D383, W437 and Y465, and W556 and C567 of H5 HA, which are sequences predominantly covered by peptide pools 6, 7 and 8. However, we did not detect higher responses to these three pools, which may be as a result of location of T‐cell epitopes on the HA protein, as well as the diverse HLA‐types of the participants.
It is well understood that both CD4+ and CD8+ T cells play a role in immunity against influenza virus. 1 Therefore, we expected to see both CD4+ and CD8+ T‐cell responses to the influenza antigens in the depletion experiments. It is not surprising that the non‐vaccinated individual had a higher proportion of influenza‐specific CD8+ T cells in their overall response compared to the vaccinated individual as more viral antigen is likely to be entering the MHC class I presentation pathway in an active infection and hence generating more CD8+ T‐cell responses, compared to the CD4+ responses likely to be generated by vaccination with inactivated virus. CD4‐depleted (i.e. CD8+) responses to rHA were unexpected, but may be explained by the fact that these proteins self‐assemble into particulate ‘rosettes’ of HA, which may allow access into class I presentation pathways. 47
T‐cell responses to the H5 HA peptides were detected to all eight pools of peptides and with no significant bias to any one pool, it appears that there are a number of cross‐reactive epitopes. It may be of interest to further characterize the heterosubtypic T‐cell responses to H5 HA peptides by performing epitope mapping. This will add to the growing database of T‐cell epitopes in influenza 18 and may elucidate any major cross‐reactive epitopes in H5 HA, which may provide valuable information for the design of a H5 vaccine containing HA.
Whilst much research into vaccines for avian influenza still focus on the production of neutralizing antibodies, cellular immune responses to novel vaccines for influenza have been investigated, including virus‐like particles, 43 DNA vaccines, 5 , 9 , 48 , 49 , 50 cell‐culture‐derived vaccines, 51 as well as reassortant viruses 3 and an immunostimulating complex (ISCOM), 6 which have been shown to effectively induce heterosubtypic T‐cell responses, and therefore may play a role in reduction of morbidity and mortality. Whilst a vaccine based on the induction of cellular immunity alone may not prevent infection, it could reduce the morbidity and mortality associated with lethal influenza infection, as well as reducing viral transmission. 50 New vaccine strategies that induce cross‐reactive or heterosubtypic immunity may overcome limitations in efficacy imposed by the low antigenicity of many prototype vaccines against H5N1 and the need for multiple doses.
Our study has demonstrated the existence of heterosubtypic T‐cell responses to H5 HA in healthy adults previously infected with or vaccinated against influenza. Investigation into the role of these T‐cell responses would be valuable for understanding more about what level of existing immunity, if any, we have to this virus if we face a pandemic challenge and may also assist in understanding more about responses to candidate H5N1 vaccines, and hence in vaccine design also. The efficacy of influenza vaccines may be improved with induction of heterosubtypic T cell as well as neutralizing antibody immunity.
Acknowledgements
This research was funded by the National Health and Medical Research Council of Australia (381789). The WHO Collaborating Centre for Reference and Research on Influenza receives financial and other support from the Australian Government Department of Health and Ageing. We thank Geza Paukovics for the flow cytometry analysis and all of the participants involved in this study and with thanks also to Lorena Brown for critical review of the manuscript.
References
- 1. Thomas PG, Keating R, Hulse‐Post DJ, Doherty PC. Cell‐mediated protection in influenza infection. Emerging Infect Dis 2006; 12:48–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Cox RJ, Brokstad KA, Ogra P. Influenza virus: immunity and vaccination strategies. Comparison of the immune response to inactivated and live, attenuated influenza vaccines. Scand J Immunol 2004; 59:1–15. [DOI] [PubMed] [Google Scholar]
- 3. O’Neill E, Krauss SL, Riberdy JM, Webster RG, Woodland DL. Heterologous protection against lethal A/HongKong/156/97 (H5N1) influenza virus infection in C57BL/6 mice. J Gen Virol 2000; 81:2689–2696. [DOI] [PubMed] [Google Scholar]
- 4. Graham MB, Braciale TJ. Resistance to and recovery from lethal influenza virus infection in B lymphocyte‐deficient mice. J Exp Med 1997; 186:2063–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Epstein SL, Kong W‐P, Misplon JA et al. Protection against multiple influenza A subtypes by vaccination with highly conserved nucleoprotein. Vaccine 2005; 23:5404–5410. [DOI] [PubMed] [Google Scholar]
- 6. Sambhara S, Kurichh A, Miranda R et al. Heterosubtypic immunity against human influenza A viruses, including recently emerged avian H5 and H9 viruses, induced by FLU‐ISCOM vaccine in mice requires both cytotoxic T‐lymphocyte and macrophage function. Cell Immunol 2001; 211:143–153. [DOI] [PubMed] [Google Scholar]
- 7. Tumpey TM, Renshaw M, Clements JD, Katz JM. Mucosal delivery of inactivated influenza vaccine induces B‐cell‐dependent heterosubtypic cross‐protection against lethal influenza A H5N1 virus infection. J Virol 2001; 75:5141–5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Effros RB, Doherty PC, Gerhard W, Bennink J. Generation of both cross‐reactive and virus‐specific T‐cell populations after immunization with serologically distinct influenza A viruses. J Exp Med 1977; 145:557–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fu T‐M, Friedman A, Ulmer JB, Liu MA, Donnelly JJ. Protective cellular immunity: cytotoxic T lymphocyte responses against dominant and recessive epitopes of influenza virus nucleoprotein induced by DNA immunization. J Virol 1997; 71:2715–2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jameson J, Cruz J, Ennis FA. Human cytotoxic T‐lymphocyte repertoire to influenza A viruses. J Virol 1998; 72:8682–8689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sterkers G, Michon J, Henin Y, Gomard E, Hannoun C, Levy JP. Fine specificity analysis of human influenza‐specific cloned cell lines. Cell Immunol 1985; 94:394–405. [DOI] [PubMed] [Google Scholar]
- 12. Gioia C, Castilletti C, Tempestilli M et al. Cross‐subtype immunity against avian influenza in persons recently vaccinated for influenza. Emerging Infect Dis 2008; 14:121–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jameson J, Cruz J, Terajima M, Ennis FA. Human CD8+ and CD4+ T lymphocyte memory to influenza A viruses of swine and avian species. J Immunol 1999; 162:7578–7583. [PubMed] [Google Scholar]
- 14. Seo SH, Webster RG. Cross‐reactive, cell‐mediated immunity and protection of chickens from lethal H5N1 influenza virus infection in Hong Kong poultry markets. J Virol 2001; 75:2516–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Govorkova EA, Webby RJ, Humberd J, Seiler JP, Webster RG. Immunization with reverse‐genetics‐produced H5N1 influenza vaccine protects ferrets against homologous and heterologous challenge. J Infect Dis 2006; 194:159–167. [DOI] [PubMed] [Google Scholar]
- 16. Lipatov AS, Hoffmann E, Salomon R, Yen H‐L, Webster RG. Cross‐protectiveness and immunogenicity of influenza A/Duck/Singapore/3/97(H5) vaccines against infection with A/Vietnam/1203/04(H5N1) virus in ferrets. J Infect Dis 2006; 194:1040–1043. [DOI] [PubMed] [Google Scholar]
- 17. Chotpitayasunondh T, Ungchusak K, Hanshaoworakul W et al. Human disease from influenza A (H5N1), Thailand, 2004. Emerging Infect Dis 2005; 11:201–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bui H‐H, Peters B, Assarsson E, Mbawuike I, Sette A. Ab and T cell epitopes of influenza A virus, knowledge and opportunities. PNAS 2007; 104:246–251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Scherle PA, Gerhard W. Functional analysis of influenza‐specific helper T cell clones in vivo . J Exp Med 1986; 164:1114–1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Scherle PA, Gerhard W. Differential ability of B cells specific for external vs. internal influenza virus proteins to respond to help from influenza virus‐specific T‐cell clones in vivo . PNAS 1988; 85:4446–4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bresson J‐L, Perronne C, Launay O et al. Safety and immunogenicity of an inactivated split‐virion influenza A/Vietnam/1194/2004 (H5N1) vaccine: phase I randomised trial. Lancet 2006; 367:1657–1664. [DOI] [PubMed] [Google Scholar]
- 22. Lin J, Zhang J, Dong X et al. Safety and immunogenicity of an inactivated adjuvanted whole‐virion influenza A (H5N1) vaccine: a phase I randomised control trial. Lancet 2006; 368:991–997. [DOI] [PubMed] [Google Scholar]
- 23. Stephenson I, Nicholson KG, Gluck R et al. Safety and antigenicity of whole virus and subunit influenza A/Hong Kong/1073/99 (H9N2) vaccine in healthy adults: phase I randomised trial. Lancet 2003; 362:1959–1966. [DOI] [PubMed] [Google Scholar]
- 24. Cox JH, Ferrari G, Janetzki S. Measurement of cytokine release at the single cell level using the ELISPOT assay. Methods 2006; 38:274–282. [DOI] [PubMed] [Google Scholar]
- 25. Chapman TJ, Castrucci MR, Padrick RC, Bradley LM, Topham DJ. Antigen‐specific and non‐specific CD4+ T cell recruitment and proliferation during influenza infection. Virology 2005; 340:296–306. [DOI] [PubMed] [Google Scholar]
- 26. Hu N, D’Souza C, Cheung H, Lang H, Cheuk E, Chamberlain JW. Highly conserved pattern of recognition of influenza A wild‐type and variant CD8+ CTL epitopes in HLA‐A2+ humans and transgenic HLA‐A2+ H2 class I‐deficient mice. Vaccine 2005; 23:5231–5244. [DOI] [PubMed] [Google Scholar]
- 27. Riberdy JM, Flynn KJ, Stech J, Webster RG, Altman JD, Doherty PC. Protection against a lethal avian influenza A virus in a mammalian system. J Virol 1999; 73:1453–1459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Avetisyan G, Ragnavolgyi E, Toth GT, Hassan M, Ljungman P. Cell‐mediated immune responses to influenza vaccination in healthy volunteers and allogneic stem cell transplant recipients. Bone Marrow Transplant 2005; 36:411–415. [DOI] [PubMed] [Google Scholar]
- 29. Centre for Disease Control (CDC) . Concepts and Procedures for laboratory‐based influenza surveillance. U.S. Department of Health, Public Health Service, Atlanta, Georgia: 1982. [Google Scholar]
- 30. Tannock GA, Paul JA, Herd R et al. Improved colorimetric assay for detecting influenza B virus neutralizing antibody responses to vaccination and infection. J Clin Microbiol 1989; 27:524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Booy R, Brown LE, Grohmann GS, MacIntyre CR. Pandemic vaccines: promises and pitfalls. Med J Aust 2006; 185:S62–S65. [DOI] [PubMed] [Google Scholar]
- 32. Mitchell DM, Callard RE. Fine specificity of the in vitro antibody response to influenza virus by human blood lymphocytes. J Immunol 1983; 131:1229–1233. [PubMed] [Google Scholar]
- 33. Quinnan GV, Ennis FA, Tuazon CU et al. Cytotoxic lymphocytes and antibody‐dependent complement‐mediated cytotoxicity induced by administration of influenza vaccine. Infect Immun 1980; 30:362–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Tamura M, Webster RG, Ennis FA. Subtype cross‐reactive, infection‐enhancing antibody responses to influenza A viruses. J Virol 1994; 68:3499–3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sandbulte MR, Jimenez GS, Boon ACM, Smith LR, Treanor JJ, Webby RJ. Cross‐reactive neuraminidase antibodies afford partial protection against H5N1 in mice and are present in unexposed humans. PLoS Med 2007; 4:265–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Murphy BR, Phelan MA, Nelson DL et al. Hemagglutinin‐specific enzyme‐linked immunosorbent assay for antibodies to influenza A and B viruses. J Clin Microbiol 1981; 13:554–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Burlington DB, Wright PF, Van Wyke KL, Phelan MA, Mayner RE, Murphy BR. Development of subtype‐specific and heterosubtypic antibodies to the influenza A virus hemagglutinin after primary infection in children. J Clin Microbiol 1985; 21:847–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Rowe T, Abernathy RA, Hu‐Primmer J et al. Detection of antibody to avian influenza A (H5N1) virus in human serum by using a combination of serologic assays. J Clin Microbiol 1999; 37:937–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Treanor JJ, Campbell JD, Zangwill KM, Rowe T, Wolff M Safety and immunogenicity of an inactivated subvirion influenza A (H5N1) vaccine. N Engl J Med 2006; 354:1343–1351. [DOI] [PubMed] [Google Scholar]
- 40. Beare AS, Webster RG. Replication of avian influenza viruses in humans. Arch Virol 1991; 2:37–42. [DOI] [PubMed] [Google Scholar]
- 41. Profeta ML, Palladino G. Serological evidence of human infections with avian influenza viruses. Brief report. Arch Virol 1986; 4:355–360. [DOI] [PubMed] [Google Scholar]
- 42. Lu X, Edwards LE, Desheva JA et al. Cross‐protective immunity in mice induced by live‐attenuated or inactivated vaccines against highly pathogenic influenza A (H5N1) viruses. Vaccine 2006; 10:44–46. [DOI] [PubMed] [Google Scholar]
- 43. Quan F‐S, Huang C, Compans RW, Kang S‐M. Virus‐like particle vaccine induces protective immunity against homolous and heterologous strains of influenza virus. J Virol 2007; 81:3514–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Sasaki S, Jaimes MC, Holmes TH et al. Comparison of the influenza virus‐specific effector and memory B‐cell responses to immunization of children and adults with live attenuated or inactivated influenza virus vaccines. J Virol 2006; 81:215–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. World Health Organisation Global Influenza Program Surveillance Network . Evolution of H5N1 avian influenza viruses in Asia. Emerging Infect Dis 2005; 11:1515–1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ennis FA, Cruz J, Jameson J, Klein M, Burt M, Thipphawong J. Augmentation of human influenza A virus‐specific cytotoxic T lymphocyte memory by influenza vaccine and adjuvanted carriers (ISCOMS). Virology 1999; 259:256–261. [DOI] [PubMed] [Google Scholar]
- 47. Holtz KM, Anderson DK, Cox MMJ. Production of a recombinant influenza vaccine using the baculovirus expression vector system. Bioprocess J 2005; 2:65–73. [Google Scholar]
- 48. Gao W, Soloff AC, Lu X et al. Protection of mice and poultry from lethal H5N1 influenza virus through adenovirus‐based immunization. J Virol 2006; 80:1959–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hoelscher MA, Garg S, Bangari DS et al. Development of adenoviral‐vector‐based pandemic influenza vaccine against antigenically distinct human H5N1 strains in mice. Lancet 2006; 367:475–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Laddy DJ, Yan J, Corbitt N, Kobasa D, Kobinger GP, Weiner DB. Immunogenicity of novel consensus‐based DNA vaccines against avian influenza. Vaccine 2007; 25:2984–2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kistner O, Howard MK, Spruth M et al. Cell culture (vero) derived whole virus (H5N1) vaccine based on wild‐type virus strain induces cross‐protective immune responses. Vaccine 2007; 25:6028–6036. [DOI] [PMC free article] [PubMed] [Google Scholar]


