Abstract
Background
Characterization of the human respiratory syncytial virus (HRSV) season at the local level has important implications for appropriate decisions on the time period for administration of specific prophylaxis.
Objectives
(1) To describe five consecutive epidemic periods of HRSV in an equatorial city of Brazil and (2) to show preliminary data on genomic diversity of circulating HRSV.
Patients/Methods
Nasopharyngeal aspirates of 2885 children attending the emergency room and wards of a public hospital were collected and screened by indirect immunofluorescence for HRSV infections during five consecutive years (from January 2004 to December 2008). In addition, the genetic and antigenic variability of the HRSV strains isolated was evaluated by partial nucleotide sequencing of the protein G gene.
Results
HRSV was detected in 15·8% of the analyzed samples. HRSV seasons occurred in a restricted period of each year. The onset of each HRSV season was variable (February to May), but the end always occurred in July. From the 456 HRSV infections found, 86 cases with bronchiolitis were genotyped. Both HRSV subgroups (A and B) cocirculated during the five epidemic periods. The 58 HRSV‐A strains grouped into two clades, GA2 and GA5. In respect of the HRSV‐B strains, the 28 samples grouped into two clades: GB3 and BA.
Conclusions
HRSV accounts for a substantial proportion of ARI in the study population. As in temperate countries, HRSV infections in this equatorial area of Brazil also cause seasonal yearly epidemics, and this has implications for prophylaxis strategies. The city of Fortaleza follows the same worldwide trend of circulation of genotypes of HRSV.
Keywords: Antigenic groups and genomic diversity, human respiratory syncytial virus
Introduction
Human respiratory syncytial virus (HRSV) causes a large part of the burden of viral respiratory infections, especially that involving young children.1 Much is known about the seasonality of the HRSV in temperate areas, but there is much to learn about HRSV's behavior in equatorial areas.2 Knowledge of HRSV's seasonality can be used by clinicians and public health officials to determine when to consider HRSV as a cause of acute respiratory illnesses and when to provide HRSV immunoprophylaxis to children at high risk of serious disease3 HRSV is a member of the Paramyxoviridae family and Pneumovirinae subfamily, with a genome of RNA of approximately 15 200 nucleotides that encode the synthesis of at least 11 proteins.4 HRSV strains have been classified in two broad subgroups, A and B. A wide variety of genotypes or lineages within each subgroup have been identified by nucleotide sequencing several genes. The variability in the G protein is greater than that in other proteins both between and within the groups.5 The aims of the present study are (i) to describe epidemiological aspects of respiratory infections associated with HRSV, which can be useful for correct use of immunoprophylaxis in children at high risk of severe HRSV infection, and (ii) to show the first data on molecular epidemiology of HRSV circulating in an equatorial area of Brazil.
Materials and methods
The study population consisted of children and teenagers (0–16 years old) who were diagnosed with upper or lower acute respiratory infection (ARI) at the emergency rooms and pediatric ward of Hospital Infantil Albert Sabin (HIAS) in Fortaleza, Ceará (Northeast Brazil), from January 2004 to December 2008. Patients were included in the study if they had one or more of the following symptoms: cough, coryza, sore throat, earache, breathing difficulty, stridor, and or wheezing within seven days of onset, and if their parents or legal guardians provided signed informed consent for inclusion of the minors in the study. Demographic and clinical information on each child was recorded in standardized form. The established diagnostic in each case met clinical criteria.6 HIAS is a public and teaching hospital where care is provided to children from low‐income families living in Fortaleza and other cities in the state of Ceará. Fortaleza, the state capital, has ~2 400 000 inhabitants and is located on the coast, 4° south of the Equator. It has two distinct seasons: a rainy season that occurs in the first half of each year usually from February to June and a dry season during the rest of the year. Fortaleza presents little variation in humidity and temperature over the course of the year. Between 2004 an 2008, the absolute maximum and minimum temperatures were 34°C and 20·5°C, respectively. In the same period, the relative air humidity ranged from 75% to 78%.7 This study was approved by the Ethics Committee of HIAS (resolution 011/08).
To analyze the seasonality of HRSV in Fortaleza during the study period, we adopted the following criteria of the National Respiratory and Enteric Virus Surveillance System laboratories of the USA:8 The onset of the HRSV season was the first of two consecutive weeks during which the mean percentage of positive specimens for HRSV antigen was ≥10%. HRSV season conclusion was the last of two consecutive weeks during which the mean percentage of positive specimens was ≥10%. Season duration comprised the number of weeks lasting from the first to the last week of each HRSV season. Significant HRSV activity was defined by the detection of HRSV in ≥10% of all samples collected in the week.
Samples positive to HRSV antigens were identified using a commercial immunofluorescence assay (IFA), the Respiratory I Viral Screening and Identification kit (Chemicon International, Inc., Temucula, CA, USA) following the manufacturer's instructions. Grouping characterization of HRSV was performed in step slides of samples collected in cases of any diagnostic, except bronchiolitis, with monoclonal antibodies for subgroup A (92‐11c) and subgroup B (102‐10b), as previously described.9 Only 282 slides were available for this procedure.
Due to limited financial resources, only samples HRSV positive in cases diagnosed as bronchiolitis were selected for analysis by RT‐PCR and sequencing. Viral RNA was extracted with the Qiamp Viral RNA Mini Kit (QIAGEN, Hilden, Germany), according to the manufacturer's instructions. cDNA was synthesized using the High‐Capacity cDNA kit (Applied BiosystemsTM, Foster City, CA, USA) according to the manufacturer's instructions. The second hypervariable region of the G protein gene was the target for the external and nested PCR. External PCR was carried out with the primers Gr5 (5′‐CTGGCAATGATAATCTCAACTTC‐3′)10 and FV (5′‐GTTATGACACTGGTATACCAACC‐3′).11 PCR was carried out in a 10‐μl mixture containing 5 μl of 10X PCR buffer, 2·5 mm of each dNTP, 25 pmol of each primer, and 1·5 U of Taq DNA Polymerase (DNA polymerase, BIOTOOLS B & M Labe, S.A.) at a final volume of 50 μl. The amplification was performed in a GeneAmp PCR System 9700 thermocycler (Applied Biosystems, Inc.). A second step of nested PCR was carried out using the forward primer GAB (5′‐ YCAYTTTGAAGTGTTCAACTT‐3′) corresponding to bases 504‐524 of the G gene and F1AB (5′‐ CAACTCCATTGTTATTTGCC‐3′) corresponding to bases 3–22 of the F gene.12 The first and second PCR steps were performed with the following program: 94°C for 5 minutes, followed by 35 cycles, each composed of 1 minute at 94°C, 1 minute at 55°C, and 1 minute at 72°C, and finally 7 minutes of extension at 72°C. The amplified products were analyzed by agarose gel electrophoresis and visualized under UV light after staining with ethidium bromide. Both cDNA synthesis and PCR followed strict procedures to prevent contamination, including redundant negative controls and segregated environments for pre‐ and post‐amplification procedures. The amplified products of gene G of 490 bp were purified by precipitation with ethanol and sodium acetate and submitted to a cycle sequencing reaction using a fluorescent dye terminator kit (Applied Biosystems) and both GAB and F1AB primers in a 3100 DNA Sequencer (Applied Biosystems, Inc., USA). Both strands of each amplicon were sequenced at least twice. Sequence editing, alignments, and phylogenetic analyses were performed as described previously.13 Seventy‐one published sequences from subgroup A and 63 from subgroup B were downloaded from GenBank as reference of different lineages and genotypes. Descriptive statistics (mean, standard deviation, Student's t‐test) were used for univariate analysis. Comparisons were carried out using the Fisher's exact test and Pearson's χ2 test for categorical variables. All P values were considered significant if ≤0·05. Spearman's correlation test used to correlate the total number of ARI cases, the number of HRSV cases, and monthly rainfall.
Results
A nasopharyngeal aspirate was collected from each of the 2885 children included in the study all of whom were tested by IFA for HRSV antigens. A total of 456 samples tested positive. No deaths occurred among HRSV‐infected inpatients. Patients who were not admitted after emergency care were not followed for outcome.
The results of this study show a statistically significant association of HRSV infections with children in their first two years of life (P < 0·001, Student's t‐test). There was also a strong statistical association between HRSV and infection of the lower respiratory tract (P < 0·001, Pearson's chi‐square test), mainly represented by pneumonia and bronchiolitis (Table 1). More than one ARI peak was observed in all years of study with the first and major peak always occurring in association with the rainy season (P < 0·0001, Spearman's correlation). Human respiratory syncytial virus epidemic periods occurred within this major peak of occurrence of ARI (P < 0·0001, Spearman's correlation) (Figure 1). During the five‐year study, a considerable variation was noted in the number of HRSV cases detected at the onset as well as the duration of the HRSV epidemic periods (Figure 2) There was a difference in the number of HRSV infections during the five epidemic periods, with a maximum of 102 cases in 2004 and a minimum of 79 in 2006. The beginning of HRSV season varied, occurring as early as week 6 in 2004 and as late as week 18 in 2005. The end of HRSV season was more constant ranging from week 27 in 2006 to week 30 in 2008. The HRSV seasons lasted from 12 weeks in 2005 to 23 weeks in 2004. In some years, samples positive for HRSV were identified prior to start of the HRSV epidemic period. No HRSV cases were detected after the 30th week in any of the five years.
Table 1.
Characteristics of the population of study
| Characteristics | HRSV positive N (%) | HRSV negative N (%) |
|---|---|---|
| Gender | ||
| Male | 258 (56·5) | 1385 (57) |
| Female | 198 (43·5) | 1044 (43) |
| Local of attending | ||
| Emergency room | 384 (84·2) | 2185 (90) |
| Ward | 72(15·8) | 244 (10) |
| Age (months) | ||
| 0–12 | 278 (61) | 1123 (46·3) |
| 13–24 | 104 (22·8) | 673 (27·2) |
| 25–36 | 43 (9·5) | 281 (11·5) |
| 37–48 | 14 (3) | 130 (5·4) |
| >48 | 17 (3·7) | 222 (9·1) |
| Diagnostic | ||
| URTI | 161 (35·3) | 1249 (51·5) |
| Pneumonia | 108 (23·7) | 403 (16·5) |
| Bronchiolitis | 105 (23) | 197 (8·1) |
| Bronchial hyper‐reactivity | 59 (13) | 448 (18·5) |
| Bronchitis | 17(3·7) | 105 (4·3) |
| Other | 6 (1·3) | 27 (1·1) |
URTI, upper respiratory tract infection.
Figure 1.
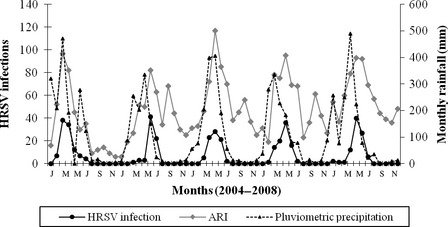
Monthly occurrence of total acute respiratory infections, HRSV infections, and rainfall in Fortaleza, Brazil, from January 2004 to December 2008.
Figure 2.
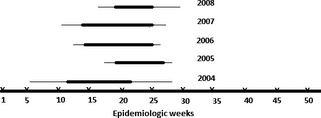
Distribution of human respiratory syncytial virus epidemic periods in Fortaleza by epidemiologic weeks (1–52) during the five years of study. The line in bold corresponds to the peak of each epidemic period.
A flow chart of the methodology used for detection, antigenic and genomic characterization, and their results is shown in Figure 3. Using IFA and nucleotide sequencing of the G2 region, it was possible determine the HRSV subgroups in 344 samples (Table 2); 215 (62·5%) of which were further classified into HRSV‐A and 129 into HRSV‐B (37·5%). Of the 105 cases of children HRSV positive with bronchiolitis analyzed by sequencing, 92 samples had sufficient volume for RNA extraction. Nested PCR was positive in 86 cases providing 86 sequences. Phylogenetic analysis of these 86 strains revealed that 58 and 28 were HRSV‐A and HRSV‐B, respectively. Phylogenetic tree was created with 27 unique partial G sequences of HRSV‐A strains from Fortaleza, clustered in two branches, and classified in genotypes GA2 (38%) and GA5(62%) with bootstrap values of 65–98% (Figure 4). The analysis of subgroup B included 19 unique partial G sequences from Fortaleza, and the sequences were grouped in two distinct clusters, previously identified as GB3 (29%) and BA (71%), with bootstrap values of 68–99% (Figure 5). HRSV‐A strains of both genotypes GA2 and GA5 were found in all years of study, except 2008, when there was exclusive circulation of GA2. HRSV‐B genotype BA was detected on 2004 and in three consecutive seasons from 2006 to 2008. Genotype GB3 cocirculated with genotype B in 2006 and was the only genotype detected in 2005. Sequences are available online in GenBank with accession numbers: JQ806027–JQ806051 and JQ814896–JQ814949.
Figure 3.
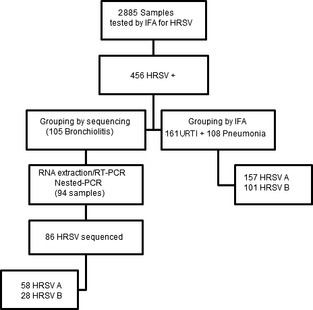
Strategies used for detection, antigenic and genomic characterization of HRSV strains circulating in Fortaleza.
Table 2.
Antigenic and genomic characteristics of HRSV strains analyzed in the study
| Year | Total of samples | HRSV positive by IFA N (%) | HRSV subgroups | Genotypes (A) | Genotypes (B) | |||
|---|---|---|---|---|---|---|---|---|
| A | B | GA2 N (%) | GA5 N (%) | GB3 | BA | |||
| 2004 | 411 | 102 (24·8) | 57 (55·8) | 33 (32·3) | 3 (37·5) | 5 (62·5) | – | 4 (100) |
| 2005 | 492 | 97 (19·7) | 40 (41·2) | 23(23·7) | 2 (9·5) | 19 (90·5) | 5 (100) | – |
| 2006 | 653 | 79 (12) | 34(43) | 17(21·5) | 3 (30) | 7 (70) | 3 (42·8) | 4 (57·2) |
| 2007 | 623 | 89 (14·3) | 45(50·5) | 24(27) | 6 (60) | 5 (40) | – | 7 (100) |
| 2008 | 706 | 89 (12·6) | 39(43·8) | 28(31·4) | 8 (100) | – | – | 5 (100) |
| Total | 2885 | 456 (15·8) | 215 (47·1) | 129 (28·2) | 22 (38) | 36 (62) | 8 (28·5) | 20 (71·5) |
IFA, immunofluorescence asssay.
Figure 4.
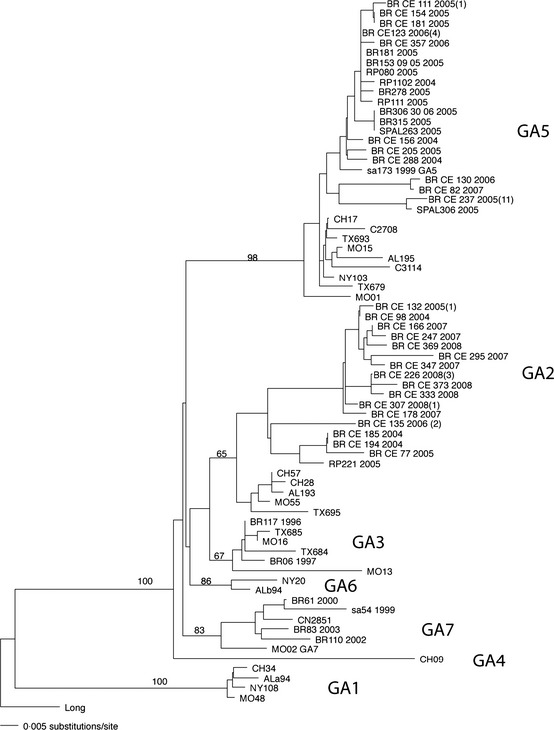
HRSV subgroup A phylogenetic tree. Sequences of 270 nucleotides of the variable region of G protein gene were compared with published sequences from GenBank. HRSV strains from Fortaleza were labeled according to the following format: country, state, number of sample, and year of collection. The number of strains with identical sequences is shown in parenthesis to the right of one representative strain.
Figure 5.
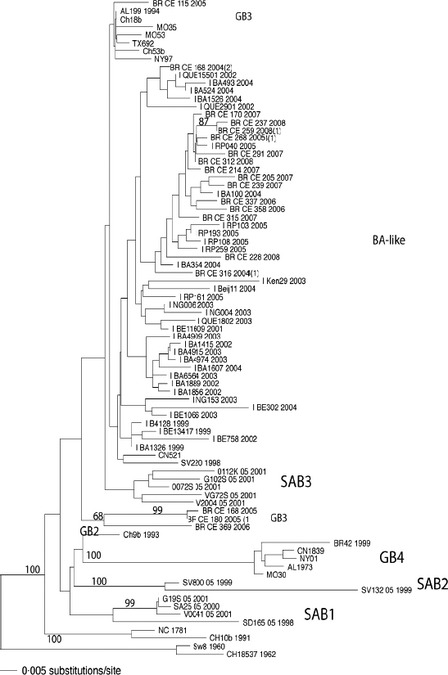
HRSV subgroup B phylogenetic tree. Sequences of 270 nucleotides of the variable region of G protein gene were compared with published sequences from GenBank. HRSV strains were labeled as in the Figure 4. The number of strains with identical sequences is shown in parenthesis to the right of one representative strain.
Discussion
The HRSV detection rate in the present study is similar to that reported in studies using detection by immunofluorescence.2, 14 In spite of the lower sensitivity of immunofluorescence compared with o‐polymerase chain reaction for detection of several viruses, including HRSV, this method continues to be used for surveillance systems of respiratory viruses in some countries, including Brazil.2, 15 Fortaleza is a city with constant high humidity, little variation in temperature, where the rain is the main climatic variable. As seen here, the major peak of ARI and the activity of HRSV in Fortaleza occur in strong association with the rainy season. This association, already demonstrated in another study, is not reported in studies conducted in cities of the South and Southeast regions of Brazil, where the association has been made with periods of lower temperatures.16, 17, 18 Interestingly, after the 30th epidemiological week of each year, no HRSV was detected. This highlights the temporal limitation of HRSV activity in Fortaleza, in contrast to what has been reported in studies conducted in cities in southeastern Brazil where HRSV activity has been observed outside the epidemic period.17, 18 Variation in the beginning and end and the duration of each HRSV season in Fortaleza are not unique, and similar variations have been reported in the USA.2 In the absence of a vaccine, palivizumab is the only licensed product currently available for prophylaxis of HRSV infections in high‐risk children. Data on seasonality and epidemic pattern of HRSV are necessary to define the timing of the start of palivizumab prophylaxis. In the USA, the American Academy of Pediatrics recommends giving a maximum of 3 to 5 monthly doses of palivizumab to infants at high risk of more severe HRSV infections, and the first dose should be administrated before the onset of community HRSV activity.19 It is known that communities in the southern USA and especially in the state of Florida are exposed earlier to the HRSV.2 Based on this regional characteristic, the first dose of palivizumab should be given earlier than in other regions of the country.19 The high costs of using palivizumab and lack of routine laboratory diagnosis to confirm the etiology of viral respiratory infections are factors that limit the use of that drug in Brazil. The regularity and well‐defined temporal extent of HRSV epidemic periods in Fortaleza shown here facilitate the implementation of prevention strategies and the provision of hospital beds for the periods of greatest impact of the virus.
Important epidemiological data are generated by analysis of antigenic and genomic characterization of circulating HRSV in a particular region. There is limited information regarding the molecular epidemiology of HRSV in Brazil, and only one published study has analyzed HRSV strains circulating in a city in the country's Northeast region.13, 20, 21, 22, 23, 24, 25, 26 Limited financial resources led to the small number of HRSV strains analyzed by sequencing in the present study. Nevertheless, the results of this analysis represent the first data on molecular epidemiology of HRSV in a Brazilian equatorial region. In Fortaleza from 2004 to 2008, both subgroups cocirculated in all epidemic periods always with predominance of group A. A similar period of predominance of HRSV‐A over HRSV‐B occurred in Uruguay, from 1995 to 1998.27 Predominance of HRSV‐B over HRSV‐A, or even exclusive circulation of HRSV‐B, may also occur in some epidemic periods.5, 27 In Fortaleza, during the five epidemic periods, genotypes GA2 and GA5 of HRSV‐A occur in varying proportions. Brazilian and international studies have shown a high diversity of genotypes of HRSV with GA2 and GA5 as the predominant circulating genotypes.24, 25, 28, 29 The small number of strains analyzed should not be the only reason for the low diversity of genotypes of HRSV‐A detected in Fortaleza. Similar analysis of a smaller number of strains of both subgroups (13 of subgroup A and 4 of subgroup B) detected in the city of Salvador (also in Northeast region of Brazil) during a single epidemic period showed HRSV‐A strains clustered in three genotypes (GA2, GA5, and GA7), while the HRSV‐B clustered in two genotypes (GB3 and SAB3).22 A new genotype of HRSV‐B was first isolated in 1999 among strains of HRSV that circulated in Buenos Aires, Argentina. This genotype, named BA, has a duplication of 60 nucleotides.30 Since 1999, the BA genotype has spread globally and in some places now predominates or has replaced genotypes that circulated for several years.29, 31 In Fortaleza, BA strains were detected in 2004 but in 2005, when there was a predominance of HRSV‐A, all five strains of HRSV‐B corresponded to genotype GB3. Strains GB3 and BA cocirculated in 2006, but only the BA genotype was found in the following two epidemic periods. This suggests that Fortaleza follows the global trend of replacement of B genotypes by BA genotype.
In summary, HRSV accounts for a substantial proportion of ARI in the population studied. The present findings also show that the circulation of HRSV can be strongly seasonal even in equatorial regions. The regularity of HRSV epidemic seasons in Fortaleza allows adoption of strategies to prevent severe HRSV infections in high‐risk children. Additionally, we generated the first data on the molecular epidemiology of HRSV‐related acute respiratory infections in children living in this city.
Addendum
Fernanda EA Moura, Marilda M Siqueira, and Edison L Durigon conceived and designed the experiments. Anne CB Perdigão, Danielle BL Oliveira, Luciano M Thomazelli, and Raquel N Caldeira performed the experiments. Fernanda EA Moura, Viviane F Botosso, Luciano M Thomazelli, and Edison L Durigon analyzed the data. Fernanda EA Moura, Marilda M Siqueira, Viviane F Botosso, and Edison L Durigon contributed reagents/materials/analysis tools. Anne CB Perdigão, Joyce F Ribeiro, Caroline MGD Florencio, Francisco MS Oliveira, and Samuel AR Pereira collected the samples.
Acknowledgments
Fernanda EA Moura received a fellowship from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq). Anne CB Perdigão and Joyce F Ribeiro received fellowship from the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES).
Moura et al (2013) Respiratory syncytial virus epidemic periods in an equatorial city of Brazil. Influenza and Other Respiratory Viruses 7(6), 1128–1135.
References
- 1. Khor C‐H, Sam I‐C, Hooi P‐S, Quek K‐F, Chan Y‐I. Epidemiology and seasonality of viral infections in hospitalized children in Kuala‐Lumpur, Malaysia: a retrospective study of 27 years. BMC Pediatrics 2012; 12:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Center for Disease Control Prevention . Respiratory Syncytial Virus‐ United States, July 2007‐June 2011. Morb Mortal Wkly Rep 2011; 60:1203–1206. [PubMed] [Google Scholar]
- 3. Goddard NL, Cooke MC, Gupta RK, Nguyen VT. Timing of monoclonal antibody for seasonal RSV prophylaxis in the United Kingdom. Epidemiol Infect 2007; 135:159–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Collins PL Jr, Crowe JE. Respiratory syncytial virus and metapneumovirus; in Knipe DM, Howley PM. (eds): Fields Virology. Philadelphia: Lippincott, 2007; 1601–1646. [Google Scholar]
- 5. Sullender WM. Respiratory syncytial virus genetic and antigenic diversity. Clin Microbiol Rev 2000; 13:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. World Health Organization . Pocket Book of Hospital Care for Children: Guidelines for the Management of Common Illnesses with Limited Resources. Geneva: WHO, 2005. [PubMed] [Google Scholar]
- 7. Governo do Estado do Ceará . Anuário Estatístico do Ceará_2010. Principais Observações Meteorológicas em Fortaleza‐2004‐2009. Available at http://www2.ipece.ce.gov/publicações/anuário/anuario2010/fisiografia/recursos.htm. (Accessed 11 January 2012).
- 8. Center for Disease Control Prevention . Respiratory syncytial virus activity‐United States, July 2008‐December 2009. Morb Mortal Wkly Rep 2010; 59:230–233. [PubMed] [Google Scholar]
- 9. Siqueira MM, Nascimento JP, Anderson LJ. Antigenic characterization of respiratory syncytial virus group A and B isolates in Rio de Janeiro, Brazil. J Clin Microbiol 1991; 29:557–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Sanz MC, Kew OM, Anderson LJ. Genetic heterogeneity of the attachment glycoprotein among group A respiratory syncytial viruses. Virus Res 1994; 33:203–217. [DOI] [PubMed] [Google Scholar]
- 11. Zheng H, Peret TC, Randolph VB, Crowlwy JC, Anderson LJ. Strain‐specific reverse transcriptase PCR assay: means to distinguish candidate vaccine from wild‐type strains of respiratory syncytial virus. J Clin Microbiol 1996; 34:334–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Peret TC, Hall CB, Scnnabel KC, Golub JA, Anderson LJ. Circulation pattern of genetically distinct groups A and B strain of human respiratory syncytial virus in community. J Gen Virol 1998; 79:2221–2229. [DOI] [PubMed] [Google Scholar]
- 13. Botosso VF, Zanotto PMA, Ueda M et al Positive selection results in frequent reversible amino acid replacements in the G protein gene of human respiratory syncytial virus. PLoS Pathog 2009; 5:e1000254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jokela P, Piiparinen H, Luiro K, Lappalainen M. Detection of human metapneumovirus and respiratory syncytial virus by duplex real‐time RT‐PCR assay um comparison with direct fluorescent assay. Clin Microbiol Infect 2010; 16:1568–1573. [DOI] [PubMed] [Google Scholar]
- 15. Secretaria de Vigilância em Saúde . Vigilância de Síndrome respiratória aguda grave (SRAG), de síndrome gripal e de internações por CID J09 a J18. Informe Técnico de Influenza 2012; 1:1–15. Available at http://portal.saude.gov.br/portal/arquivos/pdf/info_tecn_influenza_31_01_2012_28novo_29.pdf (Accessed 13 March 2012). [Google Scholar]
- 16. Tsuchiya LR, Costa LM, Raboni SM et al Viral respiratory infection in Curitiba, Southern Brazil. J Infect 2005; 51:401–407. [DOI] [PubMed] [Google Scholar]
- 17. Pecchini R, Berezin EN, Felício MC, Passos SD, Souza MC. Incidence and clinical characteristics of the infection by the respiratory syncytial virus in children admitted in Santa Casa de São Paulo Hospital. Braz J Infec Dis 2008; 12:476–479. [DOI] [PubMed] [Google Scholar]
- 18. Checon RE, Siqueira MM, Lugon AK, Portes S, Dietze R. Seasonal pattern of respiratory syncytial vírus in a region with tropical climate in Southeastern Brazil. Am J Trop Med Hyg 2002; 67:490–491. [DOI] [PubMed] [Google Scholar]
- 19. Committee on Infectious Diseases . Modified recommendations for use of Palivizumab for prevention of respiratory syncytial virus infections. Pediatrics 2009; 124:1694–1701. [DOI] [PubMed] [Google Scholar]
- 20. Lima HN, Botosso VF, Oliveira DB et al Molecular epidemiology of the SH (small hydrophobic) gene of human respiratory syncytial virus (HRSV) over 2 consecutive years. Virus Res 2012; 163:82–86. [DOI] [PubMed] [Google Scholar]
- 21. Bosso PA, Candeias JMG, Paduan KS et al Human respiratory syncytial virus detection in children admitted at a community hospital in Botucatu, SP, Brazil. Braz J Microbiol 2004; 35:348–352. [Google Scholar]
- 22. Moura FEA, Blanc A, Frabasile S et al Genetic diversity of respiratory syncytial virus isolated during an epidemic period from children of Northeastern Brazil. J Med Virol 2004; 74:156–160. [DOI] [PubMed] [Google Scholar]
- 23. Campos ACA, Durigon EL, Leal AL et al Comparison between ectodomain and G2 region of G glycoprotein for genotyping of HRSV. Braz. J. Microbiol 2007; 38:413–416. [Google Scholar]
- 24. da Silva LH, Spilki FR, Ricetto AG et al Genetic variability in the G protein gene of the human respiratory syncytial virus isolated from the Campinas metropolitan region, Brazil. J Med Virol 2008; 80:1653–1660. [DOI] [PubMed] [Google Scholar]
- 25. Ricetto AG, Silva LH, Spilk FR, Morcillo AM, Arns CW, Baracat EC. Genotypes and clinical data of respiratory syncytial virus and metapneumovirus in brazilian infants: a new perspective. Braz J Infect Dis 2009; 13:35–39. [DOI] [PubMed] [Google Scholar]
- 26. Machado AF, Sallum MA, Vilas Boas LS, Tateno AF, Machado CM. Molecular characterization of strains of respiratory syncytial virus identified in a hematopoietic stem cell transplant outpatient unit over 2 years: community or nosocomial infection? Biol Blood Marrow Transplant 2008; 14:1348–1355. [DOI] [PubMed] [Google Scholar]
- 27. Arbiza J, Delfraro A, Frabasile S. Molecular epidemiology of human respiratory syncytial virus in Uruguay: 1985‐2001‐a review. Mem Inst Oswaldo Cruz 2005; 100:221–230. [DOI] [PubMed] [Google Scholar]
- 28. Reiche J, Schweiger B. Genetic variability of group A human RSV strains circulating in Germany from 1998 to 2007. J Clin Microbiol 2009; 47:1800–1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sovero M, Garcia J, Kochel T et al Circulating strains of human respiratory syncytial virus in Central and South America. PLoS ONE 2011; 6:e2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Trento A, Galiano M, Videla GC et al Major changes in the G protein of respiratory syncytial virus isolates introduced by a duplication of 60 nucleotides. J Gen Virol 2003; 84:3115–3120. [DOI] [PubMed] [Google Scholar]
- 31. van Nierk S, Venter M. Replacement of previously circulating respiratory syncytial virus subtype B strains with the BA genotype in South Africa. J Virol 2011; 85:8789–8797. [DOI] [PMC free article] [PubMed] [Google Scholar]


