Abstract
Background
Influenza C virus can cause both upper and lower respiratory tract infections and has been reported to be prevalent in children. However, these infections have been under‐diagnosed, and epidemiological data available are limited due to the lack of convenient detection assays.
Objective
Design and validate a real‐time reverse‐transcriptase PCR (rt RT‐PCR) assay for the detection of influenza C.
Study design
Respiratory samples from two primary settings, namely, children who were hospitalized or seen in the emergency department, and respiratory outbreaks for which no other viral etiology was found were used for the detection of influenza C.
Results and Conclusions
The assay was sensitive and specific for the detection of influenza C. Eleven of 474 (2·32%) patients, all less than 10 years of age, were positive for influenza C. The strains clustered into two lineages, namely C/Kanagawa and C/Sao Paulo, based upon sequencing of the hemagglutinin‐esterase gene. Epidemiological data showed that a higher proportion of influenza C infections occur in younger children and during the winter months. This is the first report of the detection of influenza C in Alberta, Canada, and suggests that the detection of this virus should be included in respiratory virus testing panels.
Keywords: Influenza C, real‐time PCR, respiratory virus
Background
Influenza C virus remains poorly studied compared with influenza A and B, although it has been shown to cause upper respiratory tract (URT) and lower respiratory tract (LRT) infections of varying severity similar to those associated with the other respiratory viruses. Symptoms include fever, cough, rhinorrhea, and LRT illness such as pneumonia, bronchitis, and bronchiolitis.1, 2 Influenza C virus has been shown to be a significant cause of URT illness in children less than 6 years old, and the risk of complications with LRT illness is particularly high in children less than 2 years old,2, 3 resulting in more severe illness and hospitalization.4, 5 In adult volunteer studies, the disease caused by influenza C can vary from an asymptomatic to a mild upper respiratory tract infection.6 A case of acute encephalopathy associated with influenza C has also been reported.7 Influenza C virus has been documented as the etiological cause of several outbreaks in schools and the community8, 9, 10, 11, 12, 13; hence, its role in adding to the overall burden of respiratory illness should not be underestimated. High rates of seroprevalence have been reported for influenza C, suggesting that the virus is circulating widely in the population.1, 14, 15, 16
Historically, the under‐diagnosis of influenza C has resulted from the difficulty in culturing this virus and the lack of readily available monoclonal antibodies for detection by direct florescence microscopy. Although MDCK and HMV‐II cells have been used for the isolation of influenza C,17, 18, 19, 20 the rate of recovery is low. Amniotic inoculation of embryonated hen's eggs remains the most sensitive technique to isolate this virus; however, few clinical laboratories have this capability. Molecular assays for detection of this virus have been few and essentially based on end‐point or two‐step real‐time reverse‐transcriptase PCR (rt RT‐PCR).21, 22, 23, 24, 25 Here, we report on the development of a one‐step rt RT‐PCR assay and its application to the detection of influenza C in a selected panel of respiratory samples.
Methods
Clinical specimens
A subset of samples collected during hospital visits from children less than 10 years of age and from respiratory outbreaks (n = 47 from 19 outbreaks) submitted to the Provincial Laboratory for Public Health (ProvLab) for respiratory virus testing between Sept 1, 2010 and April 30, 2011 were included in the study. Pediatric samples were randomly selected for influenza C screening to include similar number of samples per month based on availability (approximately 55); multiple samples from the same patient were excluded. All specimens collected from the study patients had tested negative for influenza A and B, respiratory syncytial virus (RSV), human metapneumovirus, parainfluenza virus types 1–4, coronaviruses 229E, OC43, NL63, and HKUI, adenovirus, entero/rhinovirus using a real‐time RT‐PCR for influenza A and B26 and the respiratory viral panel (RVP) assay from Luminex Molecular Diagnostics. The numbers and types of specimens tested included: nasopharyngeal/nasal (n = 431), throat (n = 24), bronchoalveolar lavage (n = 4), sputum (n = 1), endotrachael tube (n = 2), and the collection site was not provided for 12 specimens.
Respiratory samples were extracted from an input volume of 200 μl into an elution volume of 110 μl using the easyMAG® automated extractor (bioMérieux), according to the manufacturer's instructions.
Design of primers and probes
All available matrix (M) gene sequences from GenBank were aligned to design primers (INFC‐M‐For/INFC‐M‐Rev) and a minor groove binding probe (INFC‐M‐Probe) labeled with FAM as the reporter dye to amplify and detect a 64‐bp region of the matrix gene of influenza C virus. INFC‐Clone‐For and INFC‐Clone‐Rev were designed for amplification of the M gene to generate a plasmid clone with the detection region. Additional primers were designed for sequencing the M and hemagglutinin‐esterase (HE) genes, sequences and locations of all oligos are provided in Table 1.
Table 1.
Primers and probes used for the detection and characterization of influenza C virus
| Target | Primer/Probe name | Source | Primer/Probe sequence (5′‐3′) | Nucleotide location related to GenBanka |
|---|---|---|---|---|
| Matrix | INFC‐M‐For | In‐house | TGGGAGAGATGGTGTGGAGATA | 983‐1004 |
| Matrix | INFC‐M‐Rev | In‐house | TCTTTTTCCATCGAGTCAATTTCA | 1024‐1047 |
| Matrix | INFC‐M‐Probe | In‐house | FAM‐AAAGACCACAATTATGC‐MGB | 1006‐1022 |
| Matrix | INFC‐Clone‐For | In‐house | GTTGCTCCTGAGACCAGGACAG‐ | 74‐95 |
| Matrix | INFC‐Clone‐Rev | In‐house | TGTCGGTTTCGTCAGGGGCATCC | 1084‐1106 |
| Matrix | INFC‐M‐425For | In‐house | GACTACACACCAGACATCCG | 425‐444 |
| Matrix | INFC‐M‐1110Rev | In‐house | GAGTTGTCGGTTTCGTCAG | 1092‐1110 |
| Matrix | INFC‐M‐566Rev | In‐house | CTGTGCTGGCTTTTCTTACTTC | 545‐566 |
| HE | INFC‐HE‐19F | Kimura34 | ATAATGTTTTTCTCATTACT | 19‐33 |
| HE | INFC‐HE‐1149R | In‐house | TCCCTCATTTCTTGATCTCC | 1129‐1148 |
| HE | INFC‐HE‐847F | In‐house | CCTTACACAGGGAATTCTGG | 847‐866 |
| HE | INFC‐HE‐1963R | In‐house | CAGAGATCACCAAAGCTGC | 1945‐1963 |
HE, hemagglutinin‐esterase.
Nucleotide positions for the primers and probes targeting the matrix gene are as for segment 6 of influenza C (C/Johannesburg/1/66) GenBank AM410042.1
and the hemagglutinin‐esterase gene are as for segment 4 of influenza C (C/Johannesburg/66) GenBank M17868.1.
Real‐time RT‐PCR assay
A one‐step RT‐PCR method was used for the amplification and detection of influenza C virus. The TaqMan® Fast Virus One‐Step RT‐PCR Master Mix (ABI) was used with 0·8 μm each of the sense and antisense primers and 0·2 μm of the probe. Five microliters of the extracted RNA was combined with 5 μl of the master mix, and the RT step was performed at 50°C for 5 minutes followed by incubation at 95°C for 20 seconds. Amplification included 45 cycles of denaturation at 95°C for 3 seconds, followed by annealing, extension, and data acquisition at 60°C for 30 seconds on the 7500 Fast Real‐Time PCR system (ABI).
Preparation of RNA transcripts for sensitivity studies
Primers INFC‐Clone‐For and INFC‐Clone‐Rev (Table 1) were designed to amplify 1032 bp of the M gene from influenza C‐type strain (C/Taylor/1233/47, kindly provided by Dr Yan Li, National Microbiology Laboratory, Winnipeg, Canada). The PCR products were cloned using the TOPO® TA Cloning Dual Promoter Kit (Life Technologies, California, USA). The plasmid DNA was linearized using restriction enzymes Hind III and transcribed using the T7 RiboMAX™ Express (Promega, Madison, WI, USA) to synthesize negative‐strand RNA in vitro. The transcribed RNA was spectrophotometrically quantified.
Sensitivity, specificity, and reproducibility of RT‐PCR
Ten‐fold serial dilutions ranging from 4·5 × 108 to 4·5 × 10−2 copies/reaction of quantified in vitro transcribed RNA were tested to determine assay sensitivity. End‐point sensitivity was assessed in eight replicates on three independent runs.
The specificity of the assay was determined by testing high copy number samples of common respiratory pathogens including different strains of influenza virus A and B, parainfluenza virus 1, 2, 3, 4A and 4B, RSV A and B, human coronaviruses 229E, NL63, HKU1 and OC43, human bocavirus, coxsackievirus A16 and B6, echovirus 2, human metapneumovirus, rhinovirus serotype 1B, adenovirus serotype 4, Legionella pneumophila, Mycoplasma pneumoniae, Bordetella bronchiseptica, B. holmseii, B. parapertussis, B. pertussis, Hemophilus influenzae, Neisseria meningitidis, and Streptococcus pneumoniae.
The reproducibility of the influenza C RT‐PCR was evaluated using serial dilutions of positive patient specimens made in a negative nasopharyngeal matrix at Ct values of 21·10, 27·65, and 34·08 and in negative auger suctions at Ct values of 20·36, 27·89, and 30·94. All samples were tested in triplicate on three independent runs.
Sequence analysis of influenza C positive samples
The primers described in Table 1 were used to amplify the M and HE genes from positive samples. Sequences were analyzed using SeqScape v2.6 and ClustalX Multiple Sequence Alignment Program (Version 1.81). Phylogenetic analysis was conducted using Treecon.27
A total of 1806 bases of the hemagglutinin‐esterase (HE) gene (from base pair 24 to 1830 based on numbering of C/Taylor/1233/47; GenBank M11637.1) from six influenza C positive samples were used for sequence comparison. The HE gene of influenza C viruses has been classically divided into six lineages, represented by C/Taylor/1233/47, C/Aichi/1/81, C/Sao Paulo/378/82, C/Kanagawa/1/76, C/Yamagata/26/81, and C/Mississippi/805, 11, 28, 29; these sequences in addition to representative sequences from different continents of the world and one sequence from a porcine isolate were included for comparison.
A total of 963 bases of the matrix (M) gene (from base pair 67 to 1029 based on numbering of the Influenza C/Taylor/1233/47; GenBank D26546.1) from 10 influenza C positive samples were used for sequence comparison. Influenza C viruses have been divided into three lineages based on the M gene.30 Lineage I consists of viruses with the HE gene from the C/Yamagata/26/81‐related lineage, Lineage II consists of viruses with HE gene of either C/Aichi/1/81‐ or C/Mississippi/80‐related lineage, and Lineage III consists of viruses with C/Aomori/74. The sequence of influenza C viruses belonging to the three lineages based on the M gene, six lineages based on the HE gene, and a porcine isolate were used for comparison.
All HE (JX080409‐JX080414) and M (JX133150‐JX133159) gene sequences were submitted to GenBank.
Results
Assessment of RT‐PCR assay performance
All the experimentally determined assay parameters are listed in Table 2. The assay was sensitive, specific, reproducible, and precise for the detection of a range of influenza C viral loads from patient samples.
Table 2.
RT‐PCR assay performance
| Sensitivity | Five copies of in vitro transcribed RNA/reaction |
| Specificitya | 100% |
| Intra‐assay variabilityb | 0·20–1·73% |
| Interassay variabilityb | 0·51–1·83% |
Assay parameters were assessed as described in the methods.
The assay did not amplify other viral and bacterial respiratory pathogens tested.
Six specimens with a range of viral loads were tested in triplicate on different runs. These samples gave mean crossing threshold (Ct) values of 20·36 ± 1·0, 21·10 ± 0·4, 27·65 ± 0·4, 27·89 ± 0·1, 30·94 ± 0·2, 34·08 ± 0·5.
Screening of respiratory specimens for influenza C
For the study period (Sept 1, 2010 to April 30, 2011), 427 respiratory specimens from individual patients and 47 specimens from 19 respiratory outbreaks were screened for influenza C virus using this assay. Analysis of the data shows that 11 specimens obtained from individual patients were positive for influenza C virus giving a detection rate of 2·58%. The Ct values for the positive samples ranged from 15·08 to 38·7. Influenza C virus was detected in eight nasopharyngeal and three auger suction specimens, none of the outbreak samples were positive. The age of patients ranged from 2 days to 97 years, and positive cases were detected in patients 7 months to 7 years old (Figure 1).
Figure 1.
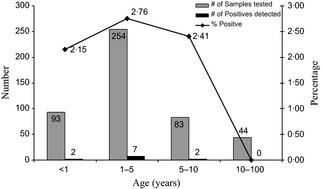
Age range of patients positive for influenza C virus. The number of samples tested and positives detected in each group is indicated. Also indicated is the percentage of positive samples detected in each age group.
Monthly distribution of influenza C positives
For the study period (Sept 1, 2010 to April 30, 2011), positives cases were detected between December and April. The seasonal distribution of the number of samples tested and positives detected is indicated in Figure 2, illustrating an increased circulation of this virus during the winter months.
Figure 2.
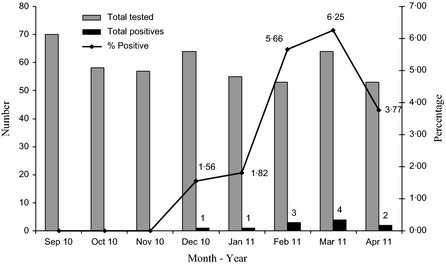
Seasonality of influenza C virus. The number of samples tested, positives detected, and percentage of positive samples in each month is indicated.
Sequence analysis of influenza C positive samples
A total of 1806 bp of the HE gene from influenza C positive isolates was compared with the six classically defined lineages described above. One sample isolated from our population clustered with the C/Sao Paulo/378/82 lineage and five samples clustered with the C/Kanagawa/1/76 lineage (Figure 3) suggesting that the two lineages were co‐circulating during the same period. Of the five sequences that clustered with the C/Kanagawa/1/76 lineage, four samples (AB‐4941‐11, AB‐2921‐11, AB‐10161‐11, and AB‐4753‐11) had greater than 99% sequence identity, sample AB‐3087‐11 was 99% identical to these sequences. The closest match for these sequences as compared with the NCBI database was C/Catalonia/1754/2009.28 Sample number AB‐3502‐11 clustered with the Sao Paulo lineage, and the closest match was C/Catalonia/1318/2009. AB‐3502‐11 was about 93% identical to the five samples belonging to the C/Kanagawa/1/76 lineage.
Figure 3.
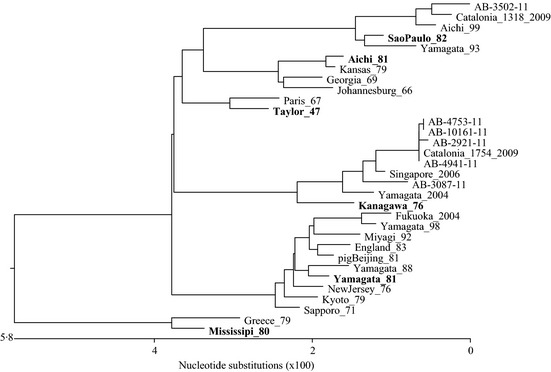
Phylogenetic tree showing the relationship between influenza C viruses based on partial hemagglutinin‐esterase (HE) gene sequence. The phylogenetic tree includes the six classically different lineages of influenza C (representatives in bold), positive samples from this study, representative sequences from different parts of the world and one sequence from a pig isolate for comparison. The Genbank numbers for sequences used in the alignment are as follows: Taylor_47 = C/Taylor/1233/47 (M11637.1), Aichi_81 = Aichi/1/81 (D28970), Sao Paulo_82 = C/Sao Paulo/378/82 (AB035364.1), Kanagawa_76 = C/Kanagawa/1/76 (D63470), and Yamagata_81 = Yamagata/26/81 (D28971.1), Aichi_99 = C/Aichi/1/99 (AB182357), Catalonia_2009 = C/Catalonia/1457/2009 (HM748633.1), England_83 = C/England/892/83 (M11642.1), Fukuoka_2004 = C/Fukuoka/2/2004 (AB252164.1), Georgia_69 = C/Georgia/1/69 (AB035359.1), Greece_79 = C/Greece/1/79 (AB035363), Johannesburg_66 = C/Johannesburg/66 (M17868), Kyoto_79 = C/Kyoto/1/79 (D63472), Miyagi_92 = C/Miyagi/3/92 (AB219076), Mississippi_80 = C/Mississippi/80 (M11640), Paris_67 = C/Paris/1/67 (AB035357), Kansas_79 = C/Kansas/2/79 (AB035361.1), Yamagata_93 = C/Yamagata/1/93 (AB035365.1), Singapore_2006 = C/Singapore/DSO‐050530/2006 (GQ853455.1), Yamagata_2004 = C/Yamagata/3/2004 (AB252153.1), Yamagata_98 = C/Yamagata/6/98 (AB064402.1), pigBeijing_81 = C/pig/Beijing/115/81 (M11644.1), Yamagata_88 = C/Yamagata/3/88 (D63473.1), New Jersey_76 = C/New Jersey/1/76 (AB035362.1), Sapporo_71 = C/Sapporo/71 (D63468.1).
A total of 963 bp of the matrix (M) gene from 10 influenza C positive isolates was compared with the classically defined lineages based on the M and HE genes. Phylogenetic clustering based on the HE and M genes was different, showing that the viruses were re‐assortants. All the positive samples from this study clustered with Miyagi/92 (D87384.1) in the M gene (Figure 4). As previously suggested, clustering in the M gene was different from that in the HE gene.30 All 10 samples were over 98% identical. The group of viruses from samples AB‐21100‐10, AB‐4753‐11, AB‐2921‐11, AB‐10161‐11, AB‐4941‐11, and AB‐2193‐11 were over 99% identical with only two base pair changes. The group of viruses from samples AB‐3502‐11, AB‐2616‐11, and AB‐4406‐11 were over 99% identical with a total of seven base pair changes. AB‐3087‐11 was slightly different with 11 base pair changes as compared with AB‐2193‐11 (98·9%) and 15 changes as compared with AB‐3502‐11 (98·4%). The predicted amino acid sequence comparison included 7 changes (2·18%) of which two changes were included in the M1 region and five in the M2 region.
Figure 4.
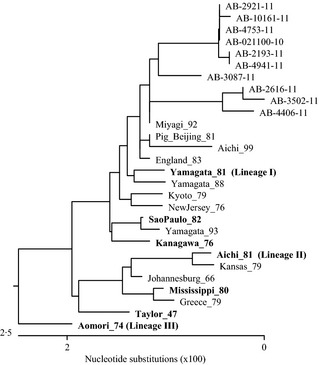
Phylogenetic tree showing the relationship between influenza C viruses based on partial M gene sequence. The phylogenetic tree compares the matrix gene from the lineages defined based on the M and hemagglutinin‐esterase (HE) genes (representatives in bold), representative sequences from different parts of the world and one sequence from a pig isolate for comparison. The Genbank numbers for sequences used in the alignment are as follows: Pig_Beijing_81 = C/pig/Beijing/115/81(AB000722.1); Mississippi_80 = C/Mississippi/80 (AB000720.1); Kyoto_79 = C/Kyoto/1/79 (AB000609.1); England_83 = C/England/83 (AB000725.1); Yamagata_81 = C/Yamagata/26/81(AB000721.1); Kanagawa_76 = C/Kanagawa/1/76 (AB000606.1); Johannesburg_66 = C/Johannesburg/1/66 (AM410041.1); Greece_79 = C/Greece/79 (AB099602.1); NewJersey_76 = C/NewJersey/76 (AB099600.1); Taylor_47 = C/Taylor/1233/47 (D26546.1); Yamagata_88 = C/Yamagata/1/88 (D16261.1); Miyagi_92 = C/Miyagi/2/92 (D87384.1); Sao Paulo_82 = C/Sao Paulo/378/82 (AB035372.1); Yamagata_93 = C/Yamagata/1/93 (AB035373.1); Aichi_81 = C/Aichi/1/81 (D16260.1); Aichi_99 = C/Aichi/1/99 (D16260.1); Kansas/1/79 = C/Kansas/1/79 (AB099601.1); Aomori_74 = C/Aomori/74 (D16259.1).
Discussion
In this study, we report the detection of influenza C in respiratory samples from Alberta, Canada. There have been no other reports of the occurrence of this virus in Canada; however, detection in respiratory samples has been previously reported from different countries.1, 5, 12, 15, 16, 23, 28 The prevalence of antibodies to influenza C has been shown to range from 60 to 100%.1, 15, 16 Influenza C has been shown to cause a spectrum of symptoms2, 3, 4, 5, suggesting that diagnosis of influenza C should be considered in the range of viral etiologies that cause respiratory illness.
As a preliminary study, our data shows that influenza C circulates in the community causing respiratory infections severe enough to require medical intervention as all the samples that tested positive for influenza C were collected during hospital visits and were also negative for all other commonly tested respiratory viruses. However, as the samples tested were restricted to hospitalized patients, the prevalence of influenza C respiratory illness in the community is unclear. Others studies have reported the presence of influenza C in non‐hospitalized patients.28 Similar to previous observations, this study detected influenza C infections primarily in children less than 10 years of age2, 3, 5, 31; however, influenza C infections in older patients have also been reported.28 Using our limited sample size, the peak of illness occurs in late winter and early spring; studies spanning more respiratory seasons will be required to understand whether influenza C infections are endemic or cyclical.
The absence of influenza C positive samples in the outbreaks is likely due to the small number of specimens available. Other studies have reported on the detection of influenza C in samples collected from outbreaks.8, 13 It will be interesting to determine whether there is a relationship between the incidence of influenza C infections in the community and the numbers of outbreaks in residential facilities caused by this virus.
As more laboratories incorporate molecular assays into their test menus, the availability of a sensitive real‐time RT‐PCR will make it easier to implement surveillance studies for influenza C. This will allow us to understand the burden of influenza C infections in the community both as single and mixed infections with other respiratory viruses. Mixed infections in children are not uncommon, with various studies showing that coinfection rates range from 5 to 65%,32 and have a higher likelihood of being admitted to a pediatric ICU.33 The participating role of influenza C as a sole or mixed infection in this vulnerable patient category deserves further study.
Another area of interest is patients with haematopoietic dysfunctions, such as stem cell transplants, who are subject to respiratory infections that can persist for a long period of time. Given the relative frequency of influenza C from studies in the literature, it is likely that some of these infections could be caused by this virus; presently the proportion is unknown. Recent studies32 show that human metapneumovirus and parainfluenza virus in this group of patients have a significantly higher rate of mortality, and defining the frequency and outcome of influenza C infections in this group of patients is worthy of study.
In our study, we found that strains from two lineages (C/Sao Paulo and C/Kanagawa) were co‐circulating, and other publications have also reported the co‐circulation of different influenza C lineages.28 While there have been few studies on the epidemiology of this virus in North America, data from Japanese studies show that up to five lineages can co‐circulate, resulting in strain replacement and frequent reassortment, allowing the virus to persist and spread in the human population.34, 35, 36 Genetic drift in the HE gene of influenza C has been shown to be independent of the year of isolation and nucleotide changes do not appear to accumulate with time. This suggests that epidemiologically dominant variants of influenza C viruses do not emerge successively.37
In summary, sensitive nucleic acid‐based assays will make it possible to study the disease burden and epidemiology of influenza C viruses. Further studies are planned to look at a number of research questions in the hospitalized and community patients.
Pabbaraju et al (2013) Detection of influenza C virus by a real‐time RT‐PCR assay. Influenza and Other Respiratory Viruses 7(6), 954–960.
References
- 1. Dykes AC, Cherry JD, Nolan CE. A clinical, epidemiologic, serologic, and virologic study of influenza C virus infection. Arch Intern Med 1980; 140:1295–1298. [PubMed] [Google Scholar]
- 2. Matsuzaki Y, Katsushima N, Nagai Y et al Clinical features of influenza C virus infection in children. J Infect Dis 2006; 193:1229–1235. [DOI] [PubMed] [Google Scholar]
- 3. Moriuchi H, Katsushima N, Nishimura H, Nakamura K, Numazaki Y. Community‐acquired influenza C virus infection in children. J Pediatr 1991; 118:235–238. [DOI] [PubMed] [Google Scholar]
- 4. Calvo C, Garcia‐Garcia ML, Centeno M, Perez‐Brena P, Casas I. Influenza C virus infection in children, Spain. Emerg Infect Dis 2006; 12:1621–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gouarin S, Vabret A, Dina J et al Study of influenza C virus infection in France. J Med Virol 2008; 80:1441–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Joosting AC, Head B, Bynoe ML, Tyrrell DA. Production of common colds in human volunteers by influenza C virus. Br Med J 1968; 4:153–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Takayanagi M, Umehara N, Watanabe H et al Acute encephalopathy associated with influenza C virus infection. Pediatr Infect Dis J 2009; 28:554. [DOI] [PubMed] [Google Scholar]
- 8. Greenbaum E, Morag A, Zakay‐Rones Z. Isolation of influenza C virus during an outbreak of influenza A and B viruses. J Clin Microbiol 1998; 36:1441–1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Katagiri S, Ohizumi A, Homma M. An outbreak of type C influenza in a children's home. J Infect Dis 1983; 148:51–56. [DOI] [PubMed] [Google Scholar]
- 10. Katagiri S, Ohizumi A, Ohyama S, Homma M. Follow‐up study of type C influenza outbreak in a children's home. Microbiol Immunol 1987; 31:337–343. [DOI] [PubMed] [Google Scholar]
- 11. Matsuzaki Y, Sugawara K, Mizuta K et al Antigenic and genetic characterization of influenza C viruses which caused two outbreaks in Yamagata City, Japan, in 1996 and 1998. J Clin Microbiol 2002; 40:422–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Matsuzaki Y, Abiko C, Mizuta K et al A nationwide epidemic of influenza C virus infection in Japan in 2004. J Clin Microbiol 2007; 45:783–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ramos AP, Herrera BA, Ramirez OV et al Detection of influenza C during an outbreak at an internal school, using a molecular tool; Havana, Cuba, September 2006. Int J Infect Dis 2008; 12:e129–e130. [DOI] [PubMed] [Google Scholar]
- 14. Manuguerra JC, Hannoun C, Aymard M. Influenza C virus infection in France. J Infect 1992; 24:91–99. [DOI] [PubMed] [Google Scholar]
- 15. Manuguerra JC, Hannoun C, Saenz MC, Villar E, Cabezas JA. Sero‐epidemiological survey of influenza C virus infection in Spain. Eur J Epidemiol 1994; 10:91–94. [DOI] [PubMed] [Google Scholar]
- 16. Motta FC, Luiz MO, Couceiro JN. Serological analysis reveals circulation of influenza C viruses, Brazil. Rev Saude Publica 2000; 34:204–205. [DOI] [PubMed] [Google Scholar]
- 17. Moriuchi H, Oshima T, Nishimura H, Nakamura K, Katsushima N, Numazaki Y. Human malignant melanoma cell line (HMV‐II) for isolation of influenza C and parainfluenza viruses. J Clin Microbiol 1990; 28:1147–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nerome K, Nakayama M, Ishida M. Established cell line sensitive to influenza C virus. J Gen Virol 1979; 43:257–259. [DOI] [PubMed] [Google Scholar]
- 19. Nishimura H, Sugawara K, Kitame F et al A human melanoma cell line highly susceptible to influenza C virus. J Gen Virol 1989; 70(Pt 7):1653–1661. [DOI] [PubMed] [Google Scholar]
- 20. Yamaoka M, Homma M, Hotta H. MDCK cell cultures supplemented with high concentrations of trypsin exhibit remarkable susceptibility to influenza C virus. Arch Virol 1995; 140:937–944. [DOI] [PubMed] [Google Scholar]
- 21. Claas EC, Sprenger MJ, Kleter GE, van BR, Quint WG, Masurel N. Type‐specific identification of influenza viruses A, B and C by the polymerase chain reaction. J Virol Methods 1992; 39:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Coiras MT, Perez‐Brena P, Garcia ML, Casas I. Simultaneous detection of influenza A, B, and C viruses, respiratory syncytial virus, and adenoviruses in clinical samples by multiplex reverse transcription nested‐PCR assay. J Med Virol 2003; 69:132–144. [DOI] [PubMed] [Google Scholar]
- 23. Hirsila M, Kauppila J, Tuomaala K et al Detection by reverse transcription‐polymerase chain reaction of influenza C in nasopharyngeal secretions of adults with a common cold. J Infect Dis 2001; 183:1269–1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zhang WD, Evans DH. Detection and identification of human influenza viruses by the polymerase chain reaction. J Virol Methods 1991; 33:165–189. [DOI] [PubMed] [Google Scholar]
- 25. Matsuzaki Y, Ikeda T, Abiko C et al Detection and quantification of influenza C virus in pediatric respiratory specimens by real‐time PCR and comparison with infectious viral counts. J Clin Virol 2012; 54:130–134. [DOI] [PubMed] [Google Scholar]
- 26. Dawood FS, Jain S, Finelli L et al Emergence of a novel swine‐origin influenza A (H1N1) virus in humans. N Engl J Med 2009; 360:2605–2615. [DOI] [PubMed] [Google Scholar]
- 27. Van de PY, De WR. Construction of evolutionary distance trees with TREECON for Windows: accounting for variation in nucleotide substitution rate among sites. Comput Appl Biosci 1997; 13:227–230. [DOI] [PubMed] [Google Scholar]
- 28. Anton A, Marcos MA, Codoner FM et al Influenza C virus surveillance during the first influenza A (H1N1) 2009 pandemic wave in Catalonia, Spain. Diagn Microbiol Infect Dis 2011; 69:419–427. [DOI] [PubMed] [Google Scholar]
- 29. Matsuzaki Y, Mizuta K, Sugawara K et al Frequent reassortment among influenza C viruses. J Virol 2003; 77:871–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tada Y, Hongo S, Muraki Y, Sugawara K, Kitame F, Nakamura K. Evolutionary analysis of influenza C virus M genes. Virus Genes 1997; 15:53–59. [DOI] [PubMed] [Google Scholar]
- 31. Troisi CL, Monto AS. Comparison of enzyme‐linked immunosorbent assay and hemagglutination inhibition in a seroepidemiological study of influenza type C infection. J Clin Microbiol 1981; 14:516–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Weigt SS, Gregson AL, Deng JC, Lynch JP III, Belperio JA. Respiratory viral infections in hematopoietic stem cell and solid organ transplant recipients. Semin Respir Crit Care Med 2011; 32:471–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Paranhos‐Baccala G, Komurian‐Pradel F, Richard N, Vernet G, Lina B, Floret D. Mixed respiratory virus infections. J Clin Virol 2008; 43:407–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kawamura H, Tashiro M, Kitame F, Homma M, Nakamura K. Genetic variation among human strains of influenza C virus isolated in Japan. Virus Res 1986; 4:275–288. [DOI] [PubMed] [Google Scholar]
- 35. Kimura H, Abiko C, Peng G et al Interspecies transmission of influenza C virus between humans and pigs. Virus Res 1997; 48:71–79. [DOI] [PubMed] [Google Scholar]
- 36. Matsuzaki Y, Takao S, Shimada S et al Characterization of antigenically and genetically similar influenza C viruses isolated in Japan during the 1999–2000 season. Epidemiol Infect 2004; 132:709–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Buonagurio DA, Nakada S, Desselberger U, Krystal M, Palese P. Noncumulative sequence changes in the hemagglutinin genes of influenza C virus isolates. Virology 1985; 146:221–232. [DOI] [PubMed] [Google Scholar]


