Abstract
Background Influenza A viruses are of major concern for public health, causing worldwide epidemics associated with high morbidity and mortality. Vaccines are critical for protection against influenza, but given the recent emergence of new strains with pandemic potential, and some limitations of the current production systems, there is a need for new approaches for vaccine development.
Objective To demonstrate the immunogenicity and protective efficacy of plant‐produced influenza antigens.
Method We engineered, using influenza A/Wyoming/3/03 (H3N2) as a model virus, the stem and globular domains of hemagglutinin (HA) produced in plants as fusions to a carrier protein and used purified antigens with and without adjuvant for ferret immunization.
Results These plant‐produced antigens were highly immunogenic and conferred complete protection against infection in the ferret challenge model. The addition of plant‐produced neuraminidase was shown to enhance the immune response in ferrets.
Conclusions Plants can be used as a production vehicle for vaccine development against influenza. Domains of HA can generate protective immune responses in ferrets.
Keywords: Influenza, launch‐vector, plant production, vaccine
Introduction
Influenza is a highly contagious disease that typically results in fever and respiratory symptoms with frequent complications that can lead to hospitalization and death, particularly in young children, adults over 65, and individuals with certain chronic underlying health conditions. 1 Annually, in the United States, there are some 30 million cases, 200 000 hospitalizations and 36 000 deaths from influenza, with an economic impact of $10 billion. 2 Outbreaks of influenza associated with type A virus subtypes H3N2 and H1N1 and type B virus occur almost annually in many countries, 3 and are caused by emerging new strains resulting from ‘antigenic drift’ in the envelope proteins hemagglutinin (HA) and neuraminidase (NA). 4 Drifted strains can evade the immune responses raised to previous infections or vaccinations and necessitate the almost annual revision of vaccine composition. 5 In addition, the periodic emergence of radically different virus strains possessing novel HA and NA antigens resulting from ‘antigenic shift’, and for which there is no prior immunity, 6 can lead to pandemics, as in 1918 caused by an H1N1 virus, when there were up to 50 million fatalities worldwide. 5 Currently, highly pathogenic H5N1 strains of avian origin are of particular public health concern and are panzootic among domestic and wild birds in Asia, Europe, and Africa. 7 Since 1997 these strains have shown the capacity to be transmitted to humans who have been in contact with infected poultry. So far, 353 cases of human H5N1 infection have been reported worldwide, with over 60% mortality. 8
Our major defense against infection with influenza viruses is immunization of individuals with an annually updated vaccine 9 that is currently produced in chicken eggs, with a global annual capacity of about 400 million doses, 10 a scale of production insufficient to combat a pandemic. Furthermore, at least 6 months is required between the identification of new virus strains to be included in the vaccine formulation and the manufacture of bulk quantities. 11 Uncertainties over the robustness of egg‐based vaccine production are intensified even further by the emergence of H5N1 strains that are highly virulent to both chickens and eggs. 7 Thus, there is a need to develop alternative vaccine production systems capable of rapid turnaround and high capacity. Recombinant subunit vaccines should circumvent some of the concerns regarding our current dependence on egg‐based production.
Antibodies to HA and NA play a key role in the immunogenicity and protective efficacy of influenza vaccines. 12 HA binds to the target cell receptor and consists of a stem domain (SD), that is relatively well conserved between strains of virus within an HA subtype, and a more variable globular domain (GD), that contains the majority of antigenic sites and epitopes that generate virus‐neutralizing antibodies. 13 NA is present at around 20% the molar equivalent of HA, 14 and has been shown to contribute to immunity. 15 Thus, HA and NA are prime candidates for influenza subunit vaccine development.
Here we report the production and evaluation of domains of HA (SD and GD) of influenza A/Wyoming/3/03 (H3N2) virus 16 expressed as fusions to the engineered thermostable enzyme, lichenase (LicKM), 17 and of NA (amino acids 38‐469) from the same virus. All vaccine targets were produced using a plant‐based transient expression system. 17 LicKM is derived from Clostridium thermocellumβ‐1,3‐1,4‐glucanase, which has previously been used as a carrier molecule for reporter gene expression in recombinant prokaryotic and eukaryotic systems. 18 When tested in ferrets, vaccine candidates containing these engineered plant‐produced influenza HA and NA antigens were highly immunogenic, and were protective against infection following challenge with homologous H3N2 virus. This plant‐based production system offers safety and capacity advantages, which, taken together with the protective efficacy data reported here, demonstrate the promise of this approach for subunit influenza vaccine development.
Materials and methods
Production of influenza antigens in plants
To evaluate the feasibility of our approach for subunit influenza vaccine development target antigens were selected from the previously epidemic A/Wyoming/3/03 (H3N2) virus 16 and engineered as fusions to LicKM. 17 The LicKM carrier molecule is based on the thermostable enzyme β‐1,3‐1,4‐glucanase (LicB) of C. thermocellum. The original sequences of HA and NA were obtained from the National Institute for Biological Standards and Control. The sequences were based on egg‐produced virus. HA and NA nucleotide sequences were optimized for expression in plants and synthesized by GENEART (Regensburg, Germany). During this optimization, no amino acid changes were introduced. Nucleotide sequences encoding amino acids 17–67 plus 294–532 of HA, which together comprise the SD, 19 were inserted into LicKM to give LicKM‐SD, and nucleotide sequences encoding amino acids 68–293 of HA, comprising the GD, 19 were similarly inserted to give LicKM‐GD. Sequence encoding the signal peptide of the Nicotiana tabacum pathogenesis‐related protein PR1a 20 was included at the N‐terminus of the fusions. Also, sequences encoding the poly‐histidine affinity purification tag (6xHis) and the endoplasmic reticulum retention signal (KDEL) were included at the C‐terminus. The LicKM fusions were introduced into the launch vector pBID4 17 allowing for viral genome transcription from the cauliflower mosaic virus 35S promoter, followed by viral replication and target sequence expression from the tobacco mosaic virus coat protein subgenomic mRNA promoter. Recombinant viral vectors were introduced into Agrobacterium tumefaciens strain GV3101 by electroporation 17 . The sequence encoding amino acids 38–469 of NA from the same influenza virus strain was introduced into pBID4, without prior fusion to LicKM. As above, the signal peptide of PR1a was included at the N‐terminus and 6xHis plus KDEL were included at the C‐terminus. Suspensions of recombinant A. tumefaciens carrying launch vectors were introduced into Nicotiana benthamiana plants, a wild variety of tobacco, by inoculating leaves 6 weeks after sowing. Plants were grown in potting soil under 12 h light/12 h dark conditions at 21°C. Leaves were harvested 4 days after inoculation with LicKM‐SD and LicKM‐GD and 7 days after inoculation with NA to assure optimum accumulation of each target. Protein extracts were prepared by grinding leaves in 50 mm sodium phosphate buffer pH 7·0, 100 mm sodium chloride, 10 mm sodium diethyldithiocarbamate, and 10 mmβ‐mercaptoethanol, and recombinant antigens were enriched by ammonium sulfate precipitation followed by immobilized metal affinity chromatography and anion exchange chromatography, with dialysis after each step, to at least 80% purity. The purity of the final product was determined on a protein basis using a Coomassie gel. The average yield of LicKM‐SD and LicKM‐GD was estimated to be 100 mg/kg of fresh plant tissue, whereas the average yield of NA was estimated to be 300 mg/kg of fresh plant tissue.
In vitro characterization of plant‐produced influenza antigens
The reactions of plant‐produced antigens with reference antisera were assessed by ELISA analysis and immunoblotting. For ELISA, 96‐well plates were coated with LicKM‐SD, LicKM‐GD or NA purified from plants, or with inactivated influenza A/Wyoming/3/03 virus, and were incubated with sheep antiserum raised against purified HA of A/Wyoming/3/03 virus, sheep antiserum raised against NIBRG‐18 (H7N2) reassorted virus or sheep antiserum raised against NIBRG‐17 (H7N1) reassorted virus. For immunoblot analysis, 100 ng of LicKM‐SD and LicKM‐GD, and inactivated influenza A/Wyoming/3/03 corresponding to 100 ng of HA, were separated by SDS‐PAGE, transferred to polyvinylidene fluoride membrane, and incubated with rabbit antiserum raised against LicKM or sheep antiserum raised against purified HA from A/Wyoming/3/03 virus. NA activity was assayed according to the standard WHO protocol WHO/CDS/CSR/NCS 2002.5 Rev.1 21 The inhibition of NA activity was assessed by pre‐incubating plant‐produced NA with sheep antiserum raised against homologous [NIBRG‐18 (H7N2)] or heterologous [NIBRG‐17 (H7N1)] reassorted virus prior to conducting the NA assay.
Assessment of immunogenicity and efficacy of plant‐produced antigens
The ferret challenge study was carried out under UK Home Office license as required by the UK Animal (Scientific Procedures) Act, 1986. Female, outbred fitch or albino ferrets, 4·5 months old, and weighing from 441 to 629 g at the initiation of the study, were maintained on high‐density ferret LabDiet 5L15 (IPS Product Supplies, London, UK). The study consisted of five groups of eight animals per group. Experimental groups received VC1 + A, VC2, or VC2 + A (Table 1) by subcutaneous injection on days 0, 14, and 28. The negative control (NC) group received alum adjuvant alone under the same dosing regimen. The positive control (PC) group were infected intranasally with 0·5 ml of influenza A/Wyoming/3/03 virus at a concentration of 105·5 TCID50 per ml on day 0 only. Following immunization, animals were monitored daily for lesions or irritation, mobility, erythema, and general activity. Animals were challenged intranasally with 0·5 ml of influenza A/Wyoming/3/03 virus at a concentration of 105·5 TCID50 per ml 10 days after the final dose. Blood samples were collected from superficial tail veins at the time of vaccination and challenge and 4 days post‐challenge. Nasal washes were collected on each of the 4 days post‐challenge. Serum HI titers were determined for homologous influenza A/Wyoming/3/03 virus and heterologous influenza A/Sydney/5/97 (H3N2), A/California/7/04 (H3N2), and A/New Caledonia/20/99 (H1N1) viruses. Hemagglutination was visually assessed following incubation with turkey red blood cells. The microneutralization assay was carried out as described by Rowe et al. 22 except that serum samples from immunized ferrets were treated with receptor‐destroying enzyme (Denka Seiken Co Ltd, Tokyo, Japan) prior to incubation with 2 × 103 TCID50 per ml of H3N2 influenza A/Wyoming/03/03 virus. Viral shedding was determined using a Madin‐Darby Canine Kidney (MDCK) cell titration on the nasal wash samples. The endpoint of the MDCK cell titration assay was determined by performing a hemagglutination assay with turkey red blood cells. The Karber calculation was used to determine log10 TCID50 per ml for each sample. The inflammatory cell response was assessed in post‐challenge nasal washes by staining with Trypan blue and counting leukocytes. Post‐challenge, animals were kept under close surveillance for 4 days for clinical evidence of influenza infection. Animals were monitored for body temperature increase and weight loss. They were also assessed for clinical signs indicative of respiratory symptoms, comprising singular sneezing or nasal rattling (1 point) or excessive sneezing (2 points), purulent discharge from the external nares (1 point), decreased alertness, spontaneous activity or play (1 point), or no activity (2 points).
Table 1.
Candidate vaccine formulations
| Vaccine candidate | Composition* |
|---|---|
| VC1 + A | 100 μg LicKM‐SD and 100 μg LicKM‐GD plus 1·3 mg alum† |
| VC2 | 100 μg LicKM‐SD, 100 μg LicKM‐GD and 50 μg NA |
| VC2 + A | 100 μg LicKM‐SD, 100 μg LicKM‐GD and 50 μg NA plus 1·3 mg alum |
*100 μg of LicKM‐SD and 100 μg of LicKM‐GD correspond to approximately 50 μg of SD and GD, respectively, which is equivalent to approximately 100 μg of full‐length HA.
†Alhydrogel was added to the vaccine candidates and incubated for 30 min on ice with agitation prior to immunization.
Results and discussion
In vitro characterization of plant‐produced influenza antigens
To evaluate the feasibility of our approach for producing immunoprotective influenza antigens we employed A. tumefaciens containing ‘launch vectors’ 17 engineered to express LicKM‐SD, LicKM‐GD, or NA. These were separately inoculated into N. benthamiana plants. Sequences encoding NA were not fused to LicKM so as to avoid potential interference with NA tetrameric structure formation and enzymatic activity that could be important for generating target‐specific immune responses. Four to seven days after inoculation, recombinant proteins were recovered and characterized. Plant‐produced LicKM‐SD and LicKM‐GD were detected by reference polyclonal sheep serum raised against HA purified from influenza A/Wyoming/3/03 virus in an ELISA (Figure 1A) and under denaturing conditions in an immunoblot (Figure 1B). In both assays LicKM‐SD was more strongly recognized by the reference serum than LicKM‐GD, although polyclonal rabbit serum raised against LicKM recognized each fusion to a similar extent (Figure 1B). This observation will be further studied. Plant‐produced NA was also recognized by reference polyclonal sheep serum raised against reassortant H7N2 virus (Figure 1C), and showed enzymatic activity that was inhibited by reference serum in a strain‐specific manner (Figure 1D).
Figure 1.
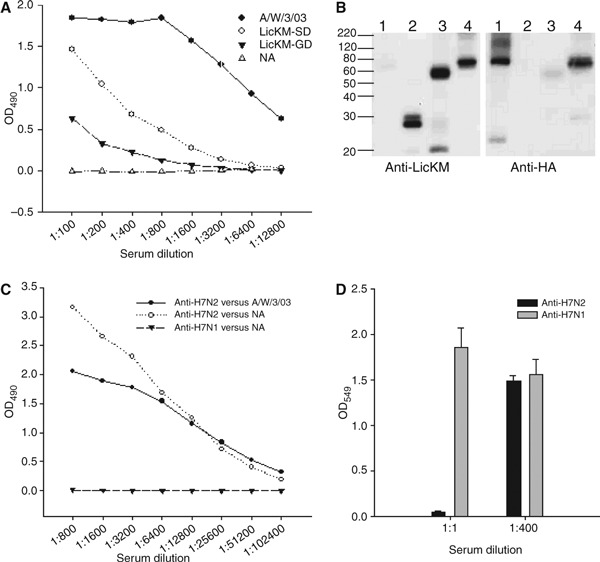
Characterization of influenza A/Wyoming/3/03 virus antigens produced in plants. (A) ELISA analysis of LicKM‐SD and LicKM‐GD using sheep serum raised against purified HA from influenza A/Wyoming/3/03 virus. Homologous virus (A/W/3/03) and plant‐produced NA were used as positive and negative controls, respectively. (B) Immunoblot analysis of LicKM‐GD (lane 3) and LicKM‐SD (lane 4) using rabbit serum raised against LicKM (Anti‐LicKM) and sheep serum raised against purified HA of influenza A/Wyoming/3/03 virus (Anti‐HA). Homologous virus (lane 1) and LicKM (lane 2) were used as controls. (C) ELISA analysis of NA using sheep sera raised against NIBRG‐18 reassorted virus (Anti‐H7N2) and NIBRG‐17 reassorted virus (Anti‐H7N1). Homologous virus (A/W/3/03) assessed using sheep serum to NIBRG‐18 (Anti‐H7N2) was used as a positive control. (D) Strain‐specific inhibition of neuraminidase activity following pre‐incubation with sheep serum raised against NIBRG‐18 (Anti‐H7N2) or NIBRG‐17 (Anti‐H7N1). Each bar represents mean enzymatic activity from three replicates with standard deviations.
Immunogenicity and protective efficacy in ferret challenge model
The ability of candidate influenza vaccines to elicit immune responses and confer protection in animal models is key to pre‐clinical evaluation. 12 We assessed the immunogenicity and protective efficacy of the plant‐produced antigens in ferrets, the accepted and well‐validated animal model for influenza. 19 , 23 It should be emphasized, however, that there is no prior report of immunizing animals with plant‐produced recombinant influenza antigens. Therefore, we adopted an immunization regimen and route of administration that would allow us to assess whether the plant‐produced influenza antigens are immunogenic. The dose of antigen chosen for this study was based on prior reports in which plant‐produced vaccine candidates induced protective immunity against relevant pathogens. 24 , 25 In the present study, three groups of eight ferrets were immunized subcutaneously by priming and boosting twice with candidate vaccine formulations (VC1 + A, VC2, and VC2 + A) containing combinations of plant‐produced influenza antigens (Table 1). NC animals received alum adjuvant alone, and PC animals were given a single intranasal dose of live influenza A/Wyoming/3/03 virus. No adverse effects were noted in any animals receiving plant‐produced vaccine candidates. Hemagglutination‐inhibition (HI) activity of sera from immunized animals is regarded as a strong correlate of protection, 12 , 25 and therefore, ferret sera were assessed for HI activity to A/Wyoming/3/03 virus. No HI activity was observed in pre‐immune sera from any animal, or in sera from NC animals (Figure 2A). However, sera from all ferrets vaccinated with VC2 + A exhibited extremely high HI titers in the range of 1:320 to 1:2560 (mean titer 1273) following the first dose (Figure 2A). These titers are much higher than 1:40, regarded as the minimum HI titer consistent with protection in humans. 12 , 26 While more experimentation is needed, the data suggest that a single dose of VC2 + A could provide protection against virus challenge. Fewer responders and lower HI titers following the first dose were observed among animals that received VC1 + A (Figure 2A), a vaccine candidate lacking NA. This suggests that NA might have modulated the immune response. Interestingly, five of the eight animals that received VC2 gave HI titers in the range of 1:160 to 1:1280 following the first dose, whereas commercial inactivated influenza vaccines in the absence of adjuvant typically induce very low HI titers. 27 , 28 , 29 Following the second dose of VC1 + A, VC2, or VC2 + A, sera from all ferrets had HI titers in the range of 1:640 to 1:2560, and these remained similarly high after the third dose (Figure 2A). Again, sera from all of these animals had HI titers well in excess of 1:40. It is of interest that HI titers in sera from ferrets receiving two or three doses of any of the plant‐produced vaccine candidates were equivalent to those in sera from intranasally infected PC animals (Figure 2A). Sera from ferrets immunized with all three vaccine candidates were also assessed for the presence of A/Wyoming/3/03 virus neutralizing (VN) antibodies using a micro‐neutralization assay. The VN titers correlated well with observed HI titers for each group (Figure 2B), and also for individual animals within the groups.
Figure 2.
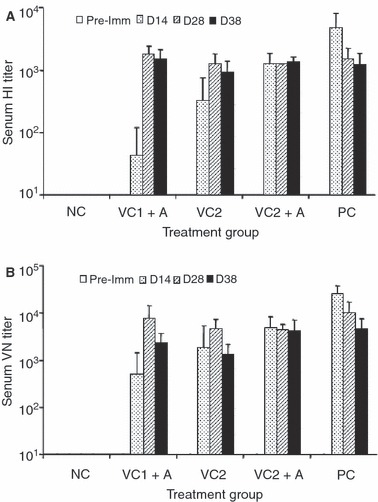
Hemagglutination inhibition titers (A) and virus neutralizing titers (B) of sera from ferrets immunized with VC1 + A, VC2, or VC2 + A. Serum samples were collected prior to the first dose (Pre‐imm), 14 days after the first dose (D14), 14 days after the second dose (D28), and 10 days after the third dose (D38). Titers were measured against A/Wyoming/3/2003. Mean titers with standard deviations are shown.
To assess the breadth of the HI response induced by the three vaccine candidates, sera from ferrets immunized with VC1 + A, VC2, or VC2 + A were tested against the heterologous H3N2 virus strains A/Sydney/5/97 and A/California/7/04. Immunization with all three candidates generated cross‐reactive serum HI titers well in excess of 1:40 (Table 2), although these titers were two‐ to 32‐fold lower than HI titers observed against homologous A/Wyoming/3/03. These results suggest that the plant‐produced vaccine candidates could provide some protective immunity against heterologous H3N2 strains. No HI activity (titers ≤10) was observed against influenza A/New Caledonia/20/99 (H1N1) (Table 2), indicating the H3 subtype specificity of the HI antibody responses generated by these vaccine candidates.
Table 2.
Serum HI antibody titers for individual animals against homologous and heterologous influenza virus strains
| Pre‐Imm | D14* | D38* | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Vaccine formulation | A/Sydney /97(H3N2) | A/Wyoming /03 (H3N2) | A/California /99 (H3N2) | A/New Caledonia /99 (H1N1) | A/Sydney /97 (H3N2) | A/Wyoming /03 (H3N2) | A/California /99 (H3N2) | A/New Caledonia /99 (H1N1) | |
| VC1+A | <10 | <10 | <10 | <10 | <10 | 80 | 2560 | 640 | <10 |
| VC2 | <10 | <10 | <10 | <10 | <10 | 40 | 320 | 160 | <10 |
| VC2+A | <10 | 80 | 1280 | 320 | <10 | 113 | 905 | 320 | <10 |
| PC† | <10 | 160 | 10240 | 1280 | <10 | 160 | 905 | 320 | <10 |
*D14 and D38 represent days at which serum samples were taken. D14 was after the first dose and D38 ten days after the third dose.
†Animals in PC group received only a single intranasal dose of egg‐produced A/Wyoming/3/03 (H3N2), although serum samples were collected on days 14 and 38 post‐infection.
The protective efficacy of the plant‐produced HA and NA antigens was assessed in the immunized ferrets by intranasal challenge with live egg‐grown influenza A/Wyoming/3/03 virus. The extent of viral infection following challenge was determined for each animal by monitoring the titer of virus shed in nasal washes for 4 days post‐challenge. Clear evidence of protection was observed for animals receiving any of the candidate vaccine formulations. Only one animal that received any of the three candidate vaccine formulations showed detectable virus shedding, and even then at less than 102 TCID50. By contrast, animals in the PC group showed a low level of virus shedding, in the range of 102 to 103 TCID50 (Figure 3A) and animals in the NC group shed virus in the range of 106 to 107 TCID50 (Figure 3A). Following the challenge, animals were also observed for weight loss, body temperature, respiratory symptoms, and leukocyte count in nasal washes of ferrets. Weight loss, an indicator of the severity of influenza infections in ferrets, was greatly reduced in ferrets that received VC1 + A, VC2 + A, or the homologous virus, compared with those in the NC group (Figure 3B). The reduction in weight loss for animals that received VC2 was less striking (Figure 3B). The febrile response following challenge was also monitored as an indicator of infection. The rise in body temperature in ferrets immunized with VC2 + A was less than that observed for animals in the NC group (Figure 3C). Furthermore, the mean peak of symptom scores, an index indicating the frequency of several influenza‐related symptoms following challenge, was significantly reduced in animals that received the candidate vaccine formulations compared with those in the NC group (Figure 3D). Similarly, counts of leukocytes in nasal washes of ferrets, taken as an indicator of upper respiratory tract infection, were significantly reduced in candidate vaccine recipients compared with animals in the NC group (Figure 3E). Overall, the challenge study clearly indicated that the plant‐produced HA and NA antigens confer a high degree of protective immunity in ferrets, showing promise for vaccine development. In future studies we will elucidate the minimum protective dose for the vaccine candidates, the protective role of LicKM‐SD and LicKM‐GD when administered individually, and the role of NA in further facilitating immune responses.
Figure 3.
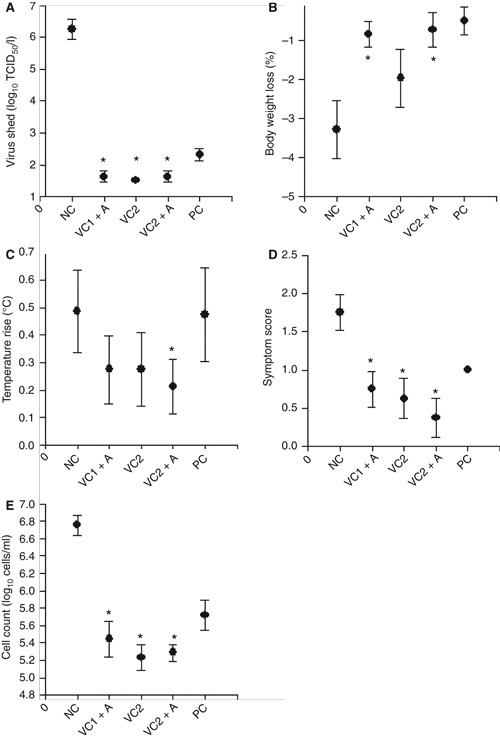
Post‐challenge monitoring of ferrets immunized with VC1 + A, VC2, or VC2 + A. Mean values with standard deviations are shown, and statistical analysis of data was conducted using anova with the Bonferroni correction for multiple testing. Groups showing statistically significant differences (P < 0·05) from the negative control group are marked with an *. (A) Peak of virus shed post‐infection. (B) Maximum weight loss post‐infection. (C) Peak temperature rise post‐infection. (D) Peak of symptom scores post‐infection. (E) Peak of total leukocyte counts per ml of nasal wash samples post‐infection.
The continual emergence of new influenza strains necessitates annual updating of the vaccine. 5 Egg‐based production has served us well for decades in providing effective and safe vaccines. 30 However, given current concerns over the transmission of highly pathogenic avian influenza type A H5N1 strains from poultry to humans, 8 several alternative approaches are being pursued for influenza vaccine production. Animal cell cultures are the most advanced in development. 31 Mammalian cells are being applied to produce target vaccine strains, either directly 31 or following reverse engineering 32 , 33 and insect cells are being utilized to produce subunit vaccine candidates expressed from baculovirus vectors. 34 In recent years plants have emerged as systems for protein expression, and are being evaluated for commercial production of vaccine antigens. Here we used a ‘launch vector’ that combines an agrobacterial binary plasmid and plant RNA viral sequences, 17 and allows for the production of target antigens within a week of plant inoculation. As the vectors are introduced into non‐genetically modified plants, there is no requirement for the development of production lines. Thus, new targets can be engineered into expression vectors, produced in plants, and purified for formulation within the time frame required for the annual influenza vaccine.
Conflict of Interest
InB contracted Retroscreen to perform aspects of the work.
Acknowledgements
We thank Caroline Nicholson and John Wood of the National Institute for Biological Standards and Control for providing target antigen sequence information and standard influenza reagents, respectively, and Dr. Jessica Chichester for critical reading of the manuscript. We also thank Dieter Hennecke, Carolyn Reifsnyder, and Jarred Lyons for assistance with protein purification, and Margaret Shillingford for care of plants. This study was supported by funds from InB Biotechnologies Inc. and Fraunhofer USA Inc.
References
- 1. Stuart‐Harris CH, Schild GC, Oxford JS. Influenza: The Viruses and the Disease. London: Edward Arnold, 1985. [Google Scholar]
- 2. Molinari NA, Ortega‐Sanchez IR, Messonnier ML et al. The annual impact of seasonal influenza in the US: measuring disease burden and costs. Vaccine 2007; 25:5086–5096. [DOI] [PubMed] [Google Scholar]
- 3. Cox NJ, Subbarao K. Global epidemiology of influenza: past and present. Annu Rev Med 2000; 51:407–421. [DOI] [PubMed] [Google Scholar]
- 4. Hampson AW. Influenza virus antigens and ‘antigenic drift’; in Potter CW. (ed): Influenza (Perspectives in Medical Virology). Amsterdam: Elsevier Science, 2002. [Google Scholar]
- 5. Ghedin E, Sengamalay NA, Shumway M et al. Large‐scale sequencing of human influenza reveals the dynamic nature of viral genome evolution. Nature 2005; 437:1162–1166. [DOI] [PubMed] [Google Scholar]
- 6. Scholtissek C. Pandemic influenza: antigenic shift; in Potter CW. (ed): Influenza (Perspectives in Medical Virology). Amsterdam: Elsevier Science, 2002. [Google Scholar]
- 7. Martin V, Sims L, Lubroth J, Pfeiffer D, Slingenbergh J, Domenech J. Epidemiology and ecology of highly pathogenic avian influenza with particular emphasis on South East Asia. Dev Biol 2006; 124:23–36. [PubMed] [Google Scholar]
- 8. World Health Organization . 2008. http://www.who.int/csr/disease/avian_influenza/en/indexhtml .
- 9. Fedson DS, Hirota Y, Shin HK et al. Influenza vaccination in 22 developed countries: an update to 1995. Vaccine 1997; 15:1506–1511. [DOI] [PubMed] [Google Scholar]
- 10. Emanuel EJ, Wertheimer A. Public health. Who should get influenza vaccine when not all can? Science 2006; 312:854–855. [DOI] [PubMed] [Google Scholar]
- 11. Gerdil C. The annual production cycle for influenza vaccine. Vaccine 2003; 21:1776–1779. [DOI] [PubMed] [Google Scholar]
- 12. Brown F, Haaheim LR, Wood JM, Schild GC (ed). Laboratory Correlates of Immunity to Influenza – A Reassessment. Basel: Karger, 2002. [Google Scholar]
- 13. Wiley DC, Wilson IA, Skehel JJ. Structural identification of the antibody‐binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature 1981; 289:373–378. [DOI] [PubMed] [Google Scholar]
- 14. Skehel JJ, Schild GC. The polypeptide composition of influenza A viruses. Virology 1971; 44:396–408. [DOI] [PubMed] [Google Scholar]
- 15. Johansson BE, Matthews JT, Kilbourne ED. Supplementation of conventional influenza A vaccine with purified viral neuraminidase results in a balanced and broadened immune response. Vaccine 1998; 16:1009–1015. [DOI] [PubMed] [Google Scholar]
- 16. Recommended composition of influenza virus vaccines for use in the 2005 influenza season Wkly Epidemiol Rec 2004; 79:369–373. [PubMed] [Google Scholar]
- 17. Musiychuk K, Stephenson N, Bi H et al. A launch vector for the production of vaccine antigens in plants. Influenza 2007; 1:19–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Goldenkova IV, Musiichuk KA, Piruzian ES. A thermostable Clostridium thermocellum lichenase‐based reporter system for studying the gene expression regulation in prokaryotic and eukaryotic cells. Mol Biol 2002; 36:868–876. [PubMed] [Google Scholar]
- 19. Sweet C, Smith H. Pathogenicity of influenza virus. Microbiol Rev 1980; 44:303–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gjetting T, Carver TL, Skot L, Lyngkjaer MF. Differential gene expression in individual papilla‐resistant and powdery mildew‐infected barley epidermal cells. Mol Plant Microbe Interact 2004; 17:729–738. [DOI] [PubMed] [Google Scholar]
- 21. World Health Organization . WHO Animal Influenza Manual. http://www.wpro.who.int/NR/rdonlyres/EFD2B9A7‐2265‐4AD0‐BC98‐97937B4FA83C/0/manualonanimalaidiagnosisandsurveillance.pdf [accessed 27 January 2008]. [Google Scholar]
- 22. Rowe T, Abernathy RA, Hu‐Primmer J et al. Detection of antibody to avian influenza A (H5N1) virus in human serum by using a combination of serologic assays. J Clin Microbiol 1999; 37:937–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Maassab HF, Kendal AP, Abrams GD, Monto AS. Evaluation of a cold‐recombinant influenza virus vaccine in ferrets. J Infect Dis 1982; 146:780–790. [DOI] [PubMed] [Google Scholar]
- 24. Chichester JA, Musiychuk K, De La Rosa P et al. Immunogenicity of a subunit vaccine against Bacillus anthracis . Vaccine 2007; 25:3111–3114. [DOI] [PubMed] [Google Scholar]
- 25. Mett V, Lyons J, Musiychuk K et al. A plant‐produced plague vaccine candidate confers protection to monkeys. Vaccine 2007; 25:3014–3017. [DOI] [PubMed] [Google Scholar]
- 26. Hobson D, Curry RL, Beare AS, Ward‐Gardner A. The role of serum haemagglutination‐inhibiting antibody in protection against challenge infection with influenza A2 and B viruses. J Hyg 1972; 70:767–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Potter CW, Shore SL, McLaren C, Stuart‐Harris C. Immunity to influenza in ferrets. II. Influence of adjuvants on immunization. Br J Exp Pathol 1972; 53:168–179. [PMC free article] [PubMed] [Google Scholar]
- 28. Potter CW, McLaren C, Shore SL. Immunity to influenza in ferrets. V. Immunization with inactivated virus in adjuvant 65. J Hyg 1973; 71:97–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Potter CW, Jennings R, McLaren C. Immunity to influenza in ferrets. VI. Immunization with adjuvanted vaccines. Arch Gesamte Virusforsch 1973; 42:285–296. [DOI] [PubMed] [Google Scholar]
- 30. Keitel WA, Couch RB. Inactivated influenza vaccines; in Potter CW. (ed): Influenza (Perspectives in Medical Virology). Amsterdam: Elsevier, 2002. [Google Scholar]
- 31. Brown F, Roberstson JS, Schild G, Wood JM (ed). Inactivated Influenza Vaccines Prepared in Cell Culture. Basel: Karger, 1997. [Google Scholar]
- 32. Ozaki H, Govorkova EA, Li C, Xiong X, Webster RG, Webby RJ. Generation of high‐yielding influenza A viruses in African green monkey kidney (Vero) cells by reverse genetics. J Virol 2004; 78:1851–1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wood JM, Robertson JS. From lethal virus to life‐saving vaccine: developing inactivated vaccines for pandemic influenza. Nat Rev 2004; 2:842–847. [DOI] [PubMed] [Google Scholar]
- 34. Treanor JJ, Schiff GM, Couch RB et al. Dose‐related safety and immunogenicity of a trivalent baculovirus‐expressed influenza‐virus hemagglutinin vaccine in elderly adults. J Infect Dis 2006; 193:1223–1228. [DOI] [PubMed] [Google Scholar]


