Abstract
There are several evidences supporting the role of 5–10 methylenetetrahydrofolate reductase (MTHFR) gene polymorphisms in breast cancer (BC). Case control association studies on breast cancer have been repeatedly performed over the last two decades, but results are inconsistent. We performed a meta-analysis to confirm the association between MTHFR C677T polymorphism and BC risk.
The articles were retrieved by searching the PubMed, Google Scholar, and Springer Link databases. Crude odds ratios (OR) with 95% confidence intervals (CIs) was used to assess the strength of association between C677T polymorphism and BC. Publication bias was assessed by Egger's and Begg-Mazumdar tests. Meta-analysis was performed with Open Meta Analyst.
Total 75 studies with 31,315 cases and 35, 608 controls were found suitable for the inclusion in the present meta-analysis. The results of meta-analysis suggested that there were moderate significant association between C677T polymorphism and BC risk using overall comparisons in five genetic models (T vs. C: OR = 1.08, 95% CI = 1.03–1.13, p = < 0.001; TT + CT vs. CC: OR = 1.06, 95% CI = 1.02–1.09, p = < 0.001; TT vs. CC: OR = 1.17, 95% CI = 1.06–1.28, p = 0.001; CT vs. CC OR = 1.05, 95% CI = 1.01–1.08, p = 0.005; TT vs. CT + CC: OR = 1.12, 95% CI = 1.03–1.22, p = 0.005). In conclusion, results of present meta-analysis showed modest association between MTHFR C677T polymorphism with breast cancer in total studies. However, sub-group analysis results based on ethnicity showed strong significant association between TT genotype and breast cancer (TT vs. CC; OR°=°1.26; 95% CI: 1.06–1.51; p = 0.009) in Asian population but in Caucasian population such association was not observed (TT vs. CC; OR°=°1.08; 95% CI: 0.99–1.14; p = 0.05).
Keywords: MTHFR, Folic acid, Breast cancer, Meta-analysis, Polymorphism
1. Introduction
Breast cancer (BC) is a leading cause of morbidity and mortality in women in the developed countries. Global BC incidence has been increasing by more than one million new cases every year; and is significantly higher in developed countries than in developing countries (Liang et al., 2014, Sturgeon et al., 2004, Ferlay et al., 2000). The lifetime BC risk in the general population is estimated to be 10% (Yang and Lippman, 1999). Several risk factors for BC have been suggested like- age of menarche and menopause, diet, reproductive history, hormone administration and genetic factors (Langsenlehner et al., 2003, Collaborative Group on Hormonal Factors in Breast Cancer, 1997, Hulka and Stark, 1995, Kelsey, 1993). The etiology of breast cancer is not very well understood. However, it has been suggested that low-penetrance susceptibility genes combining with environmental factors may be important in the development of cancer (Zhang et al., 2010). In past decade, several common low-penetrant genes have been identified as potential breast cancer susceptibility genes, one of which is 5,10-methylenetetrahydrofolate reductase (MTHFR) gene (Zhang et al., 2010).
One carbon metabolism (OCM) and MTHFR enzyme play key roles in physiologic processes by regulating the one carbon units transfer between the DNA synthesis (nucleotide synthesis) and the DNA methylation cycle (Laanpere et al., 2010, Frankenburg, 2007). MTHFR reduces 10-methylenetetrahydrofolate (10-MTHF) to 5-methylenetetrahydrofolate (5-MTHF), which is a cofactor for the remethylation of homocysteine to convert it to S-adenosyl methionine (SAM). SAM is sole methyl group donor for DNA, RNA and protein methylation. Dysfunction of the OCM cycle has been linked to congenital abnormalities (Rai et al., 2014, Zhang et al., 2013, van der Put et al., 2001), psychiatric disorders (Rai, 2011, Gilbody et al., 2007), and different types of cancers (Rai, 2014, Zhang et al., 2012, Kim, 1999).
C677T is the most common and functional polymorphism in the MTHFR gene, which involves a cytosine-to-thymine substitution at position 677, a consequence of transformation from an alanine to a valine in the enzyme (Ala222Val) (Frosst et al., 1995). This change leads to reduced enzyme activity, and individuals heterozygous (677CT) or homozygous (677TT) for this variant had enzyme activity reduced to approximately 60% and 30%, respectively, of that of the wild type (677CC) (Ueland et al., 2001) and elevate homocysteine levels (Holmes et al., 2011, Kang et al., 1988). The genotype frequencies of the polymorphism are CC, 0.583; CT, 0.35; TT, 0.067 in Europeans and CC, 0.267; CT, 0.444; TT, 0.289 in Asians (www.hapmap.org).
MTHFR gene T allele has been widely studied as a possible low-penetrance susceptibility allele for a variety of cancers, and in particular, BC. Several studies reported significant association between C677T polymorphism and BC risk (Kakkoura et al., 2015, Lu et al., 2015, He et al., 2014, Weiwei et al., 2014, Cheng et al., 2008), however some other studies have reported no association between BC and C677T polymorphism (Singh et al., 2015, Huang et al., 2014, Wu et al., 2012, Ma et al., 2009a, Ma et al., 2009b). The variation of these results might be induced by difference in ethnicities, sample size, study design and background of patients as well as random error (Wen et al., 2013). Hence we performed a meta-analysis of published case control studies to reevaluate the association between C677T polymorphism and BC susceptibility. Meta-analysis is a technique that has proven useful in resolving discrepancies between association studies is meta-analysis (Sen et al., 2008, Lohmueller et al., 2003). Meta-analysis is a quantitative method of combining the results independent studies and synthesizing summaries and conclusions. This method increases power to distinguish between small effects and no effect.
2. Methods
2.1. Literature search and inclusion/exclusion criteria
The articles were retrieved by searching the PubMed (http://www.ncbi.nlm.nih.gov/pubmed), Google Scholar (http://scholar.google.com), and Springer Link (http://link.springer.com) databases using the keywords “breast cancer”, “C677T”, “methylenetetrahydrofolate reductase” and “MTHFR” published up to March 31, 2015. In addition references of reviews and meta-analyses were examined to identify potential additional studies.
The inclusion criteria for the present meta-analysis were: (a) studies should investigated associations between MTHFR C677T polymorphism and BC; (b) studies should provide complete data on genotype number and frequencies of cases and controls for calculation of odd ratios (ORs) with 95% confidence intervals (CIs); (c) studies should be case–control studies. Exclusion criteria were as follows: (a) study design other than case–control (e.g., case reports, cohort study design without control group); (b) main outcome other than the risk of BC among genotypes (e.g., pharmacogenetic studies); and (c) reports were further excluded if they evaluated the role of MTHFR variants in other cancers. For duplicate publications, study with small sample size was excluded.
2.2. Extraction of data
The characteristics of the included studies were independently extracted by two investigators (UY and VR) through a standardized protocol. They independently extracted the following data from each publication: author name; country of origin; selection and characteristics of cases and controls; source of control, demographic information; racial descent of the study population; numbers of eligible and genotyped cases and controls; and numbers of cases and controls for each MTHFR genotype. Number and frequency of genotypes and alleles in both case and control groups were extracted or calculated from published data to re-calculate crude ORs and their 95% confidence intervals (95% CIs). Results were compared and minor disagreements were resolved by discussion. If essential information was missing from the article, the authors of the respective papers were contacted and asked to provide additional data.
2.3. Statistical analysis
The strength of association between the MTHFR C677T polymorphism and BC was estimated using odds ratios (OR), with the corresponding 95% confidence intervals (95% CI). We estimated the risk of C677T polymorphism using all genetic models viz. allele contrast/additive model (T vs. C), homozygote model (TT vs. CC), co-dominant/heterozygote model (CT vs. CC), dominant model (TT + CT vs. CC) and recessive model (TT vs. CT + CC). We tested heterogeneity between studies using Cochran's chi-square-based Q-statistic and estimated the degree of heterogeneity with I2. I2 ranges from 0% to 100 (Huedo-Medina et al., 2006, Higgins and Thompson, 2002). When low heterogeneity (I2 < 50%) was observed, then overall OR was estimated under the fixed-effects model (Mantel and Haenszel, 1959), otherwise (I2°≥°50%) under the random-effects model (DerSimonian and Laird, 1986).
Two methods were used to detect possible publication bias in meta-analysis: graphical and statistical. The funnel plot is a commonly used graphical test and Egger's (Egger et al., 1997) and Begg and Mazumdar (Begg and Mazumdar, 1994) are statistical methods. Pearson's x2 test was used to determine whether genotype of control population were in Hardy–Weinberg equilibrium (HWE) or not (P > 0.05). Sensitivity analyses were performed by excluding studies with a small number of cases (n < 100) and studies with control population violating HWE. Subgroup analyses based on ethnicity were also performed to investigate the cause of heterogeneity.
Meta-analysis was performed using Open Meta Analyst (Wallace et al., 2013) and publication bias analysis was performed using Mix version 1.7 (Bax et al., 2006). All P values are two-tailed with a significance level at 0.05.
2.4. Quality score assessment
Method of Guo et al. (2012) was adopted for quality score assessment. The quality scores ranged from 0 to 10 and studies with score < 5 was defined as low quality, and studies with score ≥ 7 was defined as high quality.
3. Results
3.1. Characteristics of included studies
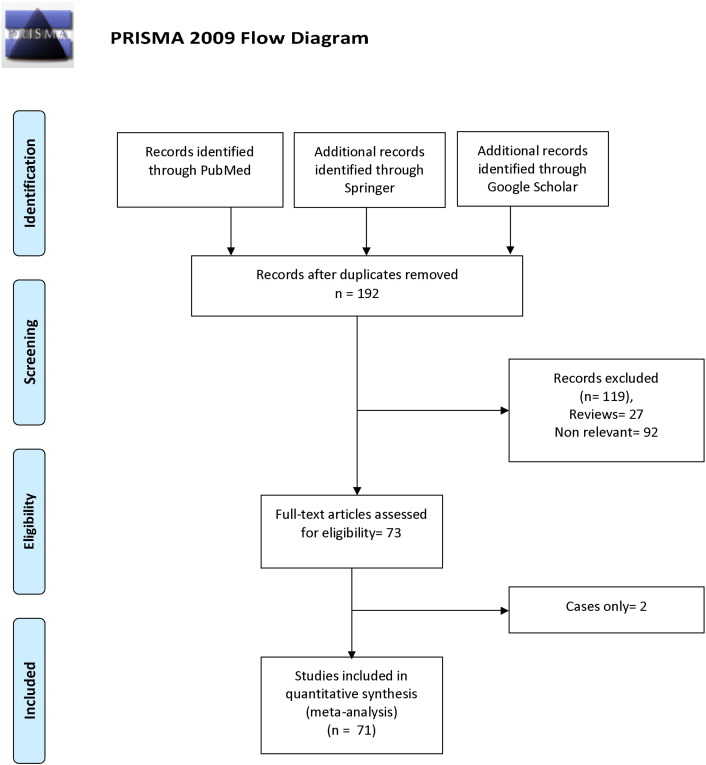
A flow chart summarizing the process of study selection is shown in Fig. 1. Initially, the highly sensitive search strategy of Pubmed, Google Scholar, and Springer Link databases, 192 articles were retrieved. After screening the titles and abstracts of all retrieved articles, 119 articles were excluded. Then full texts were reviewed and 2 articles (only cases) were further excluded. Based on the inclusion and exclusion criteria, finally, seventy one studies were included in the present met-analysis (Kakkoura et al., 2015, Lin et al., 2015, López-Cortés et al., 2015, Lu et al., 2015, Singh et al., 2015, He et al., 2014, Huang et al., 2014, Jiang-Hua et al., 2014, Wang et al., 2014, Weiwei et al., 2014, Liu et al., 2013, Ozen et al., 2013, Akram et al., 2012, Barbosa Rde et al., 2012, Diakite et al., 2012, Jakubowska et al., 2012, Lajin et al., 2012, Wu et al., 2012, Batschauer et al., 2011, Cerne et al., 2011, Hosseini et al., 2011, Hua et al., 2011, Naushad et al., 2011, Prasad and Wilkhoo, 2011, Alshatwi, 2010, Bentley et al., 2010, Sangrajrang et al., 2010, Vainer et al., 2010, Wu et al., 2010, Cam et al., 2009, Ericson et al., 2009, Gao et al., 2009, Hennquez-Hernandez et al., 2009, Jin et al., 2009, Li and Chen, 2009, Ma et al., 2009a, Ma et al., 2009b, Maruti et al., 2009, Platek et al., 2009, Yuan et al., 2009, Cheng et al., 2008, Inoue et al., 2008, Kotsopoulos et al., 2008, Langsenlehner et al., 2008, Mir et al., 2008, Suzuki et al., 2008, Hekim et al., 2007, Kan et al., 2007, Lissowska et al., 2007, Macis et al., 2007, Reljic et al., 2007, Stevens et al., 2007, Xu et al., 2007, Yu et al., 2007, Chou et al., 2006, Kalyankumar and Jamil, 2006, Chen et al., 2005, Deligezer et al., 2005, Justenhoven et al., 2005, Kalemi et al., 2005, Forsti et al., 2004, Grieu et al., 2004, Lee et al., 2004, Le Marchand et al., 2004, Lin et al., 2004, Qi et al., 2004, Shrubsole et al., 2004, Ergul et al., 2003, Langsenlehner et al., 2003, Semenza et al., 2003, Campbell et al., 2002, Sharp et al., 2002). One author (eLe Marchand et al., 2004) investigated fiv different population. We included each population as separate article so total seventy five article were included in the present meta-analysis (Table 1).
Fig. 1.
Flow diagram of study search and selection process.
Table 1.
Characteristics of the eligible studies considered in the meta-analysis.
| Study ID | Country | Ethnicity | Case/control | Control source | Genotyping method | HWE | Study quality |
|---|---|---|---|---|---|---|---|
| Sharp et al. (2002) | UK | Caucasian | 54/57 | PB | PCR-RFLP | 0.10 | 4 |
| Campbell et al. (2002) | Australia | Caucasian | 335/233 | HB | PCR-RFLP | 0.41 | 6.5 |
| Semenza et al. (2003) | USA | Caucasian | 105/247 | HB | PCR-RFLP | 0.64 | 6 |
| Langsenlehner et al. (2003) | Austria | Caucasian | 494/495 | PB | PCR-RFLP | 0.33 | 7 |
| Ergul et al. (2003) | Turkey | Caucasian | 118/193 | HB | PCR-RFLP | 0.16 | 6.5 |
| Shrubsole et al. (2004) | China | Asian | 1112/1160 | PB | PCR-RFLP | 0.44 | 8.5 |
| Forsti et al. (2004) | Poland | Caucasian | 223/298 | NR⁎ | PCR-RFLP | 0.68 | 7 |
| Lee et al. (2004) | Australia | Caucasian | 186/147 | HB | PCR-RFLP | 0.07 | 7.5 |
| Grieu et al. (2004) | Korea | Asian | 334/551 | PB | PCR-RFLP | 0.10 | 7 |
| Lin et al. (2004) | Taiwan | Asian | 88/342 | PB | PCR-RFLP | 0.38 | 7 |
| Le Marchand et al. (2004) | Hawaiian | Caucasian | 1189/2414 | PB | TaqMan | 0.75 | 8.5 |
| Qi et al. (2004) | China | Asian | 217/218 | PB | PCR-RFLP | 0.59 | 4.5 |
| Chen et al. (2005) | USA | Caucasian | 1063/1104 | PB | PCR-RFLP | 0.68 | 9.5 |
| Kalemi et al. (2005) | Greece | Caucasian | 42/51 | NR⁎ | PCR-RFLP | 0.31 | 5 |
| Deligezer et al. (2005) | Turkey | Caucasian | 189/223 | NR⁎ | PCR-RFLP | 0.75 | 7 |
| Justenhoven et al. (2005) | Germany | Caucasian | 557/633 | PB | MALDI-TOF | 0.19 | 8 |
| Chou et al. (2006) | China | Asian | 142/285 | HB | PCR-RFLP | 0.47 | 7 |
| Kalyankumar and Jamil (2006) | India | Asian | 88/95 | HB | PCR-RFLP | 0.69 | 6.5 |
| Xu et al. (2007) | USA | Caucasian | 1063/1104 | PB | PCR-RFLP | 0.68 | 8.5 |
| Hekim et al. (2007) | Turkey | Caucasian | 40/68 | NR⁎ | PCR-RFLP | 0.87 | 6 |
| Kan et al. (2007) | China | Asian | 125/103 | PB | PCR-RFLP | 0.04 | 7 |
| Lissowska et al. (2007) | Poland | Caucasian | 1974/2282 | PB | TaqMan | 0.01 | 8 |
| Macis et al. (2007) | Italy | Caucasian | 46/80 | PB | TaqMan | 0.51 | 4 |
| Reljic et al. (2007) | Croatia | Caucasian | 93/65 | PB | PCR-RFLP | 0.11 | 6 |
| Stevens et al. (2007) | USA | Others | 494/494 | PB | TaqMan | 0.01 | 7 |
| Yu et al. (2007) | Taiwan | Asian | 119/420 | PB | PCR-RFLP | 0.33 | 7.5 |
| Inoue et al. (2008) | Singapore | Asian | 380/662 | PB | TaqMan | 0.17 | 9 |
| Kotsopoulos et al. (2008) | Canada | Caucasian | 944/680 | HB | Mass-array system | 0.08 | 7.5 |
| Suzuki et al. (2008) | Japan | Asian | 454/909 | HB | TaqMan | 0.52 | 9.5 |
| Cheng et al. (2008) | Taiwan | Asian | 349/530 | HB | PCR-RFLP | 0.62 | 6.5 |
| Langsenlehner et al. (2008) | Austria | Caucasian | 105/105 | NR⁎ | PCR-RFLP | 0.68 | 6 |
| Mir et al. (2008) | India | Asian | 35/33 | HB | PCR-RFLP | 0.95 | 4 |
| Ericson et al. (2009) | Sweden | Caucasian | 540/1074 | PB | MALDI-TOF | 0.70 | 6 |
| Gao et al. (2009) | China | Asian | 624/624 | PB | PCR-RFLP | 0.59 | 9 |
| Ma et al. (2009) | Japan | Asian | 388/387 | HB | TaqMan | 0.66 | 6.5 |
| Platek et al. (2009) | USA | Caucasian | 994/1802 | PB | TaqMan | 0.39 | 9 |
| Hennquez-Hernandez et al. (2009) | Spain | Caucasian | 135/292 | PB | PCR-RFLP | 0.82 | 7 |
| Cam et al. (2009) | Turkey | Caucasian | 110/95 | NR⁎ | PCR-RFLP | 0.39 | 4.5 |
| Maruti et al. (2009) | USA | Caucasian | 318/647 | PB | ASPE | 0.67 | 8.5 |
| Ma et al. (2009) | Brazil | Others | 458/458 | HB | NR⁎ | 0.30 | 8.5 |
| Li et al. (2009) | China | Asian | 65/143 | PB | PCR-RFLP | 0.18 | 7 |
| Yuan et al. (2009) | China | Asian | 80/80 | HB | PCR-RFLP | 0.51 | 6.5 |
| Jin et al. (2009) | China | Asian | 41/100 | NR⁎ | PCR-RFLP | 0.74 | 7.5 |
| Bentley et al. (2010) | USA | Caucasian | 939/1163 | HB | PCR-RFLP | 0.05 | 8 |
| Alshatwi (2010) | Arab | Asian | 100/100 | HB | TaqMan | 0.80 | 6.5 |
| Sangrajrang et al. (2010) | Indian | Asian | 563/487 | HB | TaqMan | 0.42 | 9 |
| Weiner et al. (2010) | Russia | Asian | 837/778 | PB | PCR-RFLP | 0.80 | 8 |
| Wu et al. (2010) | China | Asian | 80/80 | HB | PCR-RFLP | 0.51 | 5.5 |
| Batschauer et al. (2011) | Brazil | Others | 68/85 | PB | PCR-RFLP | 0.59 | 4.5 |
| Cerne et al. (2011) | Caucasian | Caucasian | 522/269 | PB | Sequencing | 0.88 | 6 |
| Hosseini et al. (2011) | Iran | Asian | 294/300 | HB | PCR-RFLP | < 0.0001 | 3 |
| Hua et al. (2011) | China | Asian | 95/90 | PB | PCR-RFLP | 0.02 | 6.5 |
| Nausad et al. (2011) | India | Asian | 244/244 | HB | PCR-RFLP | 0.17 | 7 |
| Prasad et al. (2011) | India | Asian | 130/125 | PB | PCR-RFLP | 0.06 | 6 |
| Akram et al. (2012) | Pakistan | Asian | 110/110 | HB | PCR-RFLP | 0.85 | 5 |
| Barbosa et al. (2012) | Mixed, Caucasian | Caucasian | 176/176 | HB | PCR-RFLP | 0.38 | 5.5 |
| Diakite et al. (2012) | Morocco | Others | 96/117 | HB | PCR-RFLP | 0.78 | 7 |
| Jakubowska et al. (2012) | Mixed, Caucasian | Caucasian | 4778/3350 | PB | TaqMan | 0.15 | 7 |
| Lajin et al. (2012) | Syria | Asian | 119/126 | HB | PCR-RFLP | 0.35 | 6.5 |
| Wu et al. (2012) | China | Asian | 32/37 | NR⁎ | PCR-RFLP | 0.03 | 6 |
| Liu et al. (2013) | China | Asian | 435/435 | HB | PCR-RFLP | 0.57 | 6 |
| Ozen et al. (2013) | Turkey | Caucasian | 51/106 | NR⁎ | Strip-assay | 0.08 | 4 |
| He et al. (2014) | China | Asian | 310/381 | HB | Sequenom | < 0.0001 | 6 |
| Huang et al. (2014) | Taiwan | Asian | 1232/1232 | HB | PCR-RFLP | 0.01 | 8 |
| Jiang-hua et al. (2014) | China | Asian | 535/673 | HB | PCR-RFLP | < 0.0001 | 7 |
| Wang et al. (2014) | China | Asian | 435/435 | HB | Sequenom | 0.22 | 6 |
| Weiwei et al. (2014) | China | Asian | 297/306 | HB | Sequenom | 0.00 | 8 |
| Kakkoura et al. (2015) | Cyprus | Caucasian | 1065/1157 | PB | TaqMan | 0.09 | 9.5 |
| Lopez-Cortes et al. (2015) | Ecuador | Others | 114/195 | HB | PCR-RFLP | 0.00 | 7 |
| Lu et al. (2015) | China | Asian | 560/560 | HB | TaqMan | 0.27 | 9 |
| Singh et al. (2015) | India | Asian | 588/508 | HB | PCR-RFLP | 0.37 | 5.5 |
NR = not reported.
In seventy five studies included in the present meta-analysis, the smallest case sample size was 32 (Wu et al., 2012) and highest sample size was 4778 (Jakubowska et al., 2012). In included studies, total cases were 31,315 with CC (13,960), CT (13,328) and TT (4027), and controls were 35,608 with CC (16,527), CT (14,868), and TT (4213). In controls genotype percentage of CC, CT and TT were 46.41%, 41.75% and 11.83% respectively. In cases genotype percentage of CC, CT and TT were 44.58%, 42.56% and 12.86% respectively. Frequencies of CC genotype and C allele were highest in both cases and controls.
Out of 75 studies, only twenty studies reported OR above one and significant association between C677T polymorphism and BC (López-Cortés et al., 2015, Lu et al., 2015, He et al., 2014, Jiang-Hua et al., 2014, Weiwei et al., 2014, Liu et al., 2013, Ozen et al., 2013, Lajin et al., 2012, Naushad et al., 2011, Wu et al., 2010, Gao et al., 2009, Maruti et al., 2009, Li and Chen, 2009, Yuan et al., 2009, Xu et al., 2007, Chen et al., 2005, Deligezer et al., 2005, Qi et al., 2004). Control population of eleven studies (López-Cortés et al., 2015, He et al., 2014, Jiang-Hua et al., 2014, Wang et al., 2014, Weiwei et al., 2014, Wu et al., 2012, Hosseini et al., 2011, Hua et al., 2011, Lissowska et al., 2007, Stevens et al., 2007) was not in Hardy–Weinberg equilibrium (Table 1).
3.2. Meta-analysis
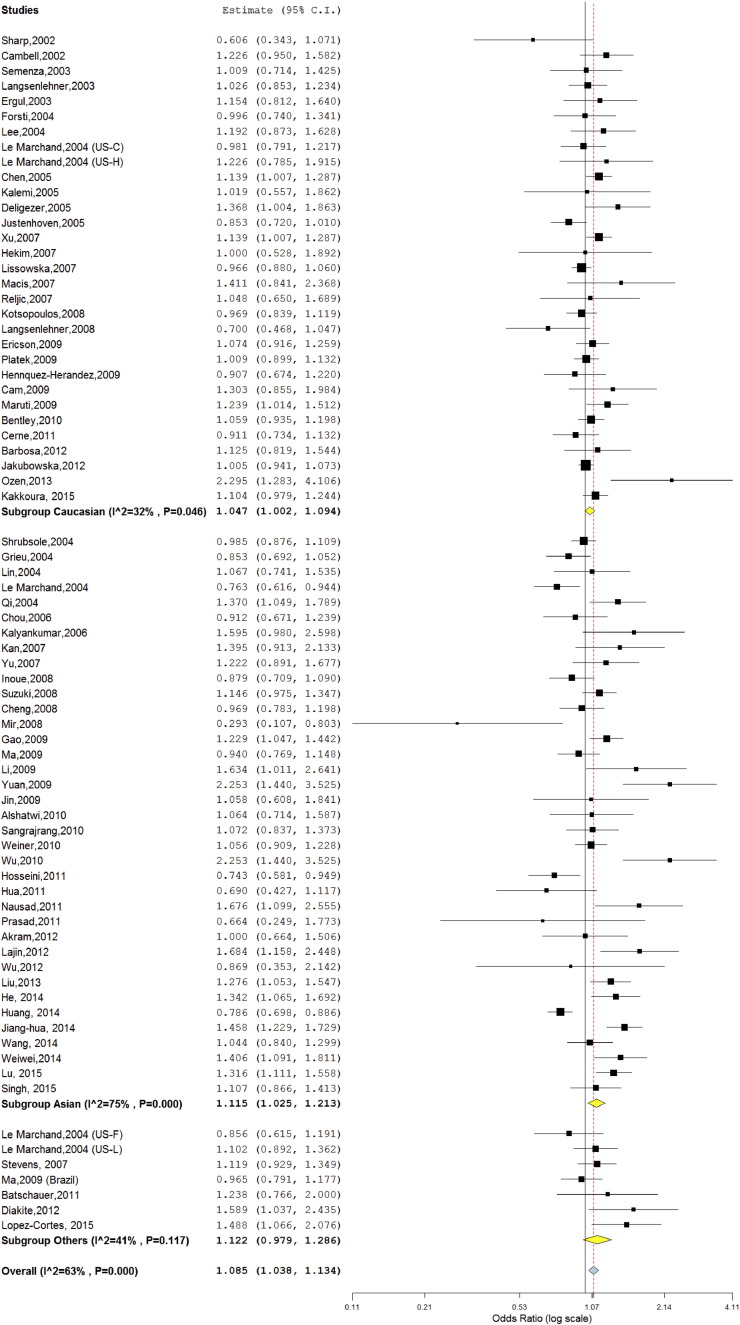
The meta-analysis was carried out using all five genetic models- allele contrast (T vs. C), co-dominant (CT vs. CC), homozygote (TT vs. CC), dominant (TT + CT vs. CC), and recessive (TT vs. CT + CC) models. Meta-analysis with allele contrast (T vs. C) showed moderate significant association with both fixed effect (OR = 1.05; 95%CI = 1.02–1.07; p = < 0.001) and random effect model (OR = 1.08; 95% CI = 1.03–1.13; p = < 0.001). Subjects with T allele showed a slightly increased risk of BC (Table 2; Fig. 2).
Table 2.
Summary estimates for the odds ratio (OR) of MTHFR C677T in various allele/genotype contrasts, the significance level (p value) of heterogeneity test (Q test), and the I2 metric: overall analysis, and subgroup analyses.
| Genetic contrast | Fixed effect OR (95% CI), p |
Random effect OR (95% CI), p |
Heterogeneity p-value (Q test) | I2 (%) | |
|---|---|---|---|---|---|
| All (75 studies) |
Allele contrast (T vs. C) | 1.05 (1.02–1.07), < 0.001 | 1.08 (1.03–1.13), < 0.001 | < 0.001 | 63 |
| Dominant (TT + CT vs. CC) | 1.06 (1.02–1.09), < 0.001 | 1.08 (1.03–1.14), 0.002 | < 0.001 | 48 | |
| Homozygote (TT vs. CC) | 1.10 (1.04–1.16), < 0.001 | 1.17 (1.06–1.28), 0.001 | < 0.001 | 60 | |
| Co-dominant (CT vs. CC) | 1.05 (1.01–1.08), 0.005 | 1.05 (1.01–1.10), 0.01 | 0.01 | 29 | |
| Recessive (CC + CT vs. TT) | 1.07 (1.02–1.13), 0.002 | 1.12 (1.03–1.22), 0.005 | < 0.001 | 55 | |
| Ethnicity | |||||
| Asian (37 studies) |
Allele contrast (T vs. C) | 1.06 (1.02–1.11), < 0.001 | 1.11 (1.02–1.21), 0.01 | < 0.001 | 75 |
| Dominant (TT + CT vs. CC) | 1.07 (1.01–1.12), 0.009 | 1.10 (1.00–1.20), 0.04 | < 0.001 | 64 | |
| Homozygote (TT vs. CC) | 1.15 (1.06–1.25), < 0.001 | 1.26 (1.06–1.51), 0.009 | < 0.001 | 71 | |
| Co-dominant (CT vs. CC) | 1.04 (0.99–1.10), 0.08 | 1.05 (0.97–1.14), 0.19 | 0.003 | 43 | |
| Recessive (CC + CT vs. TT) | 1.12 (1.04–1.21), 0.003 | 1.21 (1.04–1.40), 0.01 | < 0.001 | 65 | |
| Caucasian (31 studies) |
Allele contrast (T vs. C) | 1.03 (1.00–1.06), 0.02 | 1.04 (1.00–1.09), 0.04 | 0.04 | 32 |
| Dominant (TT + CT vs. CC) | 1.04 (1.00–1.08), 0.05 | 1.04 (1.00–1.08), 0.05 | 0.56 | 0 | |
| Homozygote (TT vs. CC) | 1.06 (0.99–1.14), 0.05 | 1.08 (0.97–1.21), 0.12 | 0.007 | 43 | |
| Co-dominant (CT vs. CC) | 1.03 (0.99–1.08), 0.12 | 1.03 (0.99–1.08), 0.12 | 0.79 | 0 | |
| Recessive (CC + CT vs. TT) | 1.05 (0.99–1.12), 0.09 | 1.07 (0.96–1.19), 0.18 | 0.002 | 47 | |
| Others (7 studies) |
Allele contrast (T vs. C) | 1.10 (0.99–1.21), 0.05 | 1.12 (0.97–1.28), 0.09 | 0.11 | 41 |
| Dominant (TT + CT vs. CC) | 1.17 (1.02–1.33), 0.01 | 1.23 (0.99–1.53), 0.05 | 0.03 | 57 | |
| Homozygote (TT vs. CC) | 1.14 (0.91–1.42), 0.23 | 1.14 (0.91–1.44), 0.22 | 0.40 | 2 | |
| Co-dominant (CT vs. CC) | 1.19 (1.03–1.36), 0.01 | 1.24 (1.00–1.55), 0.04 | 0.03 | 55 | |
| Recessive (CC + CT vs. TT) | 1.03 (0.84–1.26), 0.74 | 1.03 (0.84–1.27), 0.73 | 0.77 | 0 | |
| Study design | |||||
| Hospital based (34 studies) |
Allele contrast (T vs. C) | 1.07 (1.03–1.12), < 0.001 | 1.14 (1.05–1.23), < 0.001 | < 0.001 | 73 |
| Dominant (TT + CT vs. CC) | 1.08 (1.03–1.14), 0.001 | 1.14 (1.04–1.26), 0.004 | < 0.001 | 65 | |
| Homozygote (TT vs. CC) | 1.15 (1.06–1.25), < 0.001 | 1.27 (1.08–1.50), 0.003 | < 0.001 | 69 | |
| Co-dominant (CT vs. CC) | 1.06 (1.01–1.12), 0.02 | 1.10 (1.01–1.20), 0.02 | < 0.001 | 54 | |
| Recessive (CC + CT vs. TT) | 1.12 (1.04–1.21), 0.002 | 1.20 (1.05–1.39), 0.008 | < 0.001 | 63 | |
| Population based (32 studies) |
Allele contrast (T vs. C) | 1.03 (1.00–1.06), 0.04 | 1.03 (0.98–1.09), 0.15 | 0.001 | 48 |
| Dominant (TT + CT vs. CC) | 1.04 (0.99–1.08), 0.05 | 1.04 (0.98–1.09), 0.12 | 0.13 | 22 | |
| Homozygote (TT vs. CC) | 1.06 (0.99–1.13), 0.07 | 1.08 (0.96–1.21), 0.16 | < 0.001 | 50 | |
| Co-dominant (CT vs. CC) | 1.03 (0.99–1.08), 0.10 | 1.03 (0.99–1.08), 0.10 | 0.61 | 0 | |
| Recessive (CC + CT vs. TT) | 1.04 (0.97–1.10), 0.20 | 1.05 (0.95–1.16), 0.27 | 0.003 | 45 | |
| Menopausal status | |||||
| Pre-menopausal (9 studies) |
Allele contrast (T vs. C) | 1.01 (0.90–1.12), 0.84 | 1.01 (0.98–1.12), 0.84 | 0.68 | 0 |
| Dominant (TT + CT vs. CC) | 1.00 (0.87–1.17), 0.90 | 1.00 (0.86–1.16), 0.92 | 0.45 | 0 | |
| Homozygote (TT vs. CC) | 1.01 (0.80–1.28), 0.89 | 1.01 (0.80–1.28), 0.89 | 0.63 | 0 | |
| Co-dominant (CT vs. CC) | 1.00 (0.85–1.17), 0.99 | 1.01 (0.84–1.21), 0.88 | 0.25 | 20 | |
| Recessive (CC + CT vs. TT) | 1.02 (0.82–1.27), 0.83 | 1.02 (0.81–1.27), 0.84 | 0.50 | 0 | |
| Post-menopausal (9 studies) |
Allele contrast (T vs. C) | 1.03 (0.95–1.12), 0.40 | 1.05 (0.92–1.20), 0.39 | 0.03 | 51 |
| Dominant (TT + CT vs. CC) | 1.08 (0.96–1.20), 0.16 | 1.09 (0.95–1.25), 0.20 | 0.22 | 24 | |
| Homozygote (TT vs. CC) | 1.01 (0.85–1.20), 0.87 | 1.06 (0.77–1.45), 0.71 | 0.01 | 57 | |
| Co-dominant (CT vs. CC) | 1.10 (0.98–1.23), 0.10 | 1.10 (0.98–1.23), 0.10 | 0.62 | 0 | |
| Recessive (CC + CT vs. TT) | 0.96 (0.82–1.13), 0.68 | 0.99 (0.75–1.30), 0.96 | 0.04 | 48 | |
Fig. 2.
Random effect Forest plot of allele contrast model (T vs. C) of MTHFR C677T polymorphism.
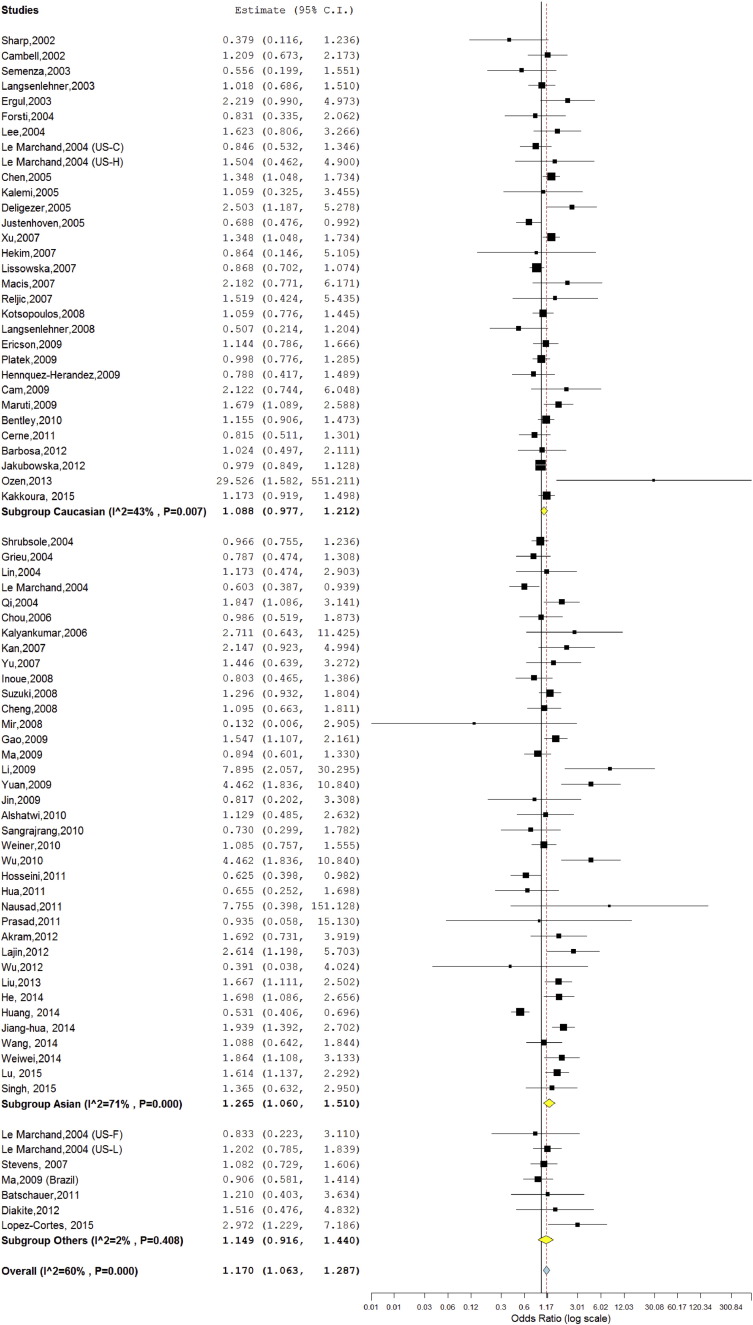
An increased significant association was found between BC and mutant genotype (TTvs.CC; homozygote model) with both fixed (OR = 1.10; 95%CI = 1.04–1.16; p = < 0.001) and random (OR = 1.17; 95%CI = 1.06–1.28; p = 0.001) effect models (Table 2, Fig. 3). Association of mutant heterozygous genotype (CT vs.CC; co-dominant model) was observed significant with BC using fixed (OR = 1.05; 95%CI = 1.01–1.08; p = 0.005) and random (OR = 1.05; 95%CI = 1.01–1.10; p = 0.01) effect models (Table 2). Combined mutant genotypes (TT + CT vs. CC; dominant model) showed positive association with BC using both fixed (OR = 1.06; 95%CI = 1.02–1.09; p = < 0.001) and random (OR°=°1.08; 95%CI = 1.03–1.14; p = < 0.001) effect models. Similarly the recessive genotypes model (TT vs. CT + CC) also showed positive association fixed (OR = 1.07; 95%CI = 1.02–1.13; p = 0.002) and random (OR°=°1.12; 95%CI = 1.03–1.22; p = 0.005) effect models (Table 2). In allele contrast cumulative meta-analysis, after addition of Bentley et al. (2010) study, the pooled turned statistically significant and remained significant after addition of subsequent studies (details not given).
Fig. 3.
Random effect Forest plot of homozygote model (TT vs. CC) of MTHFR C677T polymorphism.
A true heterogeneity existed between studies for allele contrast (Pheterogeneity = < 0.001, Q = 203.99, I2 = 63%, t2 = 0.019, z = 3.73), homozygote (Pheterogeneity = < 0.001, Q = 186.33, I2 = 60%, t2 = 0.079, z = 3.24), dominant (Pheterogeneity = < 0.001, Q = 147.7, I2 = 48%, t2 = 0.019, z = 3.29) and recessive (Pheterogeneity = < 0.001, Q = 163.7, I2 = 55%, t2 = 0.054, z = 2.83) comparisons.
3.3. Sensitivity analysis
Sensitivity analysis was performed by eliminating studies with small sample size (< 100) and control population deviating from HWE. Control population of eleven studies was not in HWE (López-Cortés et al., 2015, He et al., 2014, Huang et al., 2014, Jiang-Hua et al., 2014, Weiwei et al., 2014, Wu et al., 2012, Hosseini et al., 2011, Hua et al., 2011, Kan et al., 2007, Lissowska et al., 2007, Stevens et al., 2007) and heterogeneity was decreased after exclusion of these studies (I2 = 52%; p = < 0.001). Sample size of seventeen studies was less than 100 (Ozen et al., 2013, Diakite et al., 2012, Wu et al., 2012, Batschauer et al., 2011, Hua et al., 2011, Wu et al., 2010, Jin et al., 2009, Li and Chen, 2009, Yuan et al., 2009, Mir et al., 2008, Hekim et al., 2007, Macis et al., 2007, Reljic et al., 2007, Kalyankumar and Jamil, 2006, Kalemi et al., 2005, Le Marchand et al., 2004, Sharp et al., 2002) and after exclusion of these studies heterogeneity was slightly decreased (I2 = 61%; p = 0.002).
3.4. Subgroup analysis
Out of 75 studies included in the present meta-analysis, 37 studies were carried out on Asian population, and 31 studies were carried out on Caucasian population and other studies were carried on other ethnic group and we grouped those studies in mixed population subgroup (7 studies). The subgroup analysis by ethnicity revealed significant association between MTHFR C677T polymorphism and BC in Asian population (T vs. C: OR = 1.11; 95% CI = 1.02–1.21; p = 0.01; I2 = 75%; Pheterogeneity = < 0.001; CT vs. CC: OR = 1.04; 95% CI = 0.99–1.10; p = 0.08; I2 = 43%; Pheterogeneity = 0.003; TT vs. CC: OR = 1.26; 95% CI = 1.06–1.51; p = 0.009; I2 = 71%; Pheterogeneity = < 0.001; TT + CT vs. CC: OR = 1.10; 95% CI = 1.00–1.20; p = 0.04; I2 = 64; Pheterogeneity = < 0.001; TT vs. CT + CC: OR = 1.21; 95% CI = 1.04–1.40; p = 0.01; I2 = 65%; Pheterogeneity = < 0.001) (Table 2). In Caucasian subgroup analysis, heterogeneity was low and except allele contrast model, significant association was not found between C677T polymorphism and BC risk (T vs. C: OR = 1.03; 95% CI = 1.00–1.06; p = 0.02; I2 = 32%; Pheterogeneity = 0.04; CT vs. CC: OR = 1.03; 95% CI = 0.99–1.12; p = 0.09; I2 = 0%; Pheterogeneity = 0.79; TT vs. CC: OR = 1.06; 95% CI = 0.99–1.14; p = 0.05; I2 = 43%; Pheterogeneity = 0.007; TT + CT vs. CC: OR = 1.04; 95% CI = 1.00–1.08; p = 0.05; I2 = 0; Pheterogeneity = 0.56; TT vs. CT + CC: OR = 1.05; 95% CI = 0.99–1.12; p = 0.09; I2 = 47%; Pheterogeneity = 0.002). In mixed subgroup analysis, significant association was found in allele contrast, co-dominant and dominant models (T vs. C: OR = 1.10; 95% CI = 0.99–1.21; p = 0.05; I2 = 41%; Pheterogeneity = 0.11; CT vs. CC: OR = 1.24; 95% CI = 1.0–1.55; p = 0.04; I2 = 55%; Pheterogeneity = 0.03; TT vs. CC: OR = 1.14; 95% CI = 0.91–1.42; p = 0.23; I2 = 2%; Pheterogeneity = 0.40; TT + CT vs. CC: OR = 1.23; 95% CI = 0.99–1.53; p = 0.05; I2 = 57%; Pheterogeneity = 0.03; TT vs. CT + CC: OR = 1.03; 95% CI = 0.84–1.26; p = 0.74; I2 = 0%; Pheterogeneity = 0.77) (Table 2; Fig. 2, Fig. 3).
Sub-group analysis based on menstrual status i.e. premenopausal and postmenopausal was performed. Out of 75 included studies, in 9 studies BC cases was from premenopausal group and in other 9 studies BC cases was from postmenopausal group. In remaining 57 studies menstrual status was not mentioned. In both the group, pre and post-menopausal groups no significant association was observed using all five genetic models.
Sub-group analysis based on source of control population i.e. hospital based or population based was also performed. Out of 75 included studies, control population in 34 studies was hospital based and in 32 studies control population was from population and in 9 studies source of controls was not mentioned. In hospital based control group studies, (number of studies = 34; 12,515/13,560 cases/controls), allele contrast meta-analysis showed significant association (ORTvsC = 1.14; 95%CI = 1.05–1.23; p < 0.001). In population based control group studies, (number of studies = 32; 2916/4300 cases/controls), allele contrast meta-analysis did not show significant association (ORTvsC = 1.03; 95%CI = 0.98–1.09; p = 0.15).
3.5. Publication bias
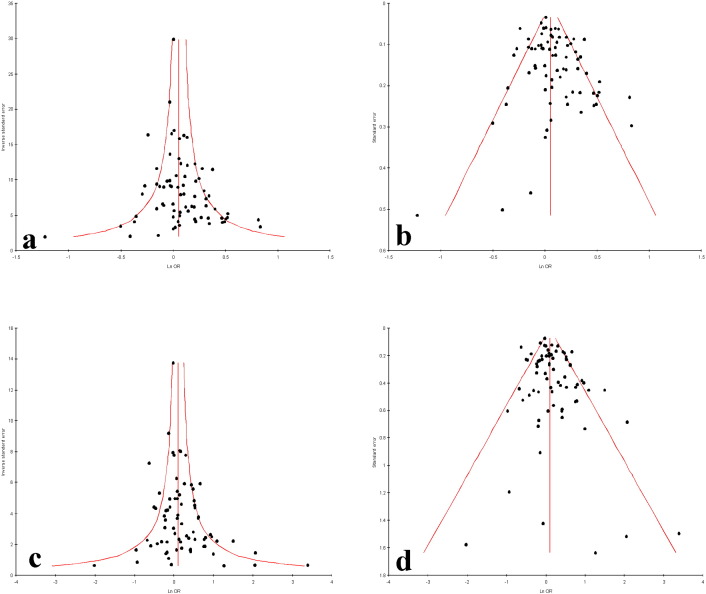
Funnel plots and Egger's test were performed to estimate the risk of publication bias. Except allele contrast and homozygote model, publication bias was absent (T vs. C: PBegg's test = 0.03, PEgger's test = 0.03; CT vs. CC: PBegg's test = 0.41, PEgger's test = 0.29; TT vs. CC: PBegg's test = 0.10, PEgger's test = 0.03; Dominant model TT + CT vs. CC:, PBegg's test = 0.27, PEgger's test = 0.06; Recessive model TT vs. CT + CC: PBegg's test = 0.18, PEgger's test = 0.05) (Fig. 4).
Fig. 4.
Funnel plots of a. precision by OR; b. standard error by OR of MTHFR C677T allele contrast model (T vs. C); c. precision by OR; d. standard error by OR of MTHFR C677T homozygote model (TT vs. CC).
4. Discussion
Present meta-analysis investigated association of the MTHFR C677T polymorphism with BC risk (31,315 patients and 35,608 controls from 75 case–control studies). Results of meta-analysis suggested moderate significant genetic association between the MTHFR C677T polymorphism and BC. This result is in line with that of eight other previously published meta-analyses that had included fewer case control studies of the MTHFR C677T polymorphism and BC (Li et al., 2014, Liang et al., 2014, Rai, 2014, Yu and Chen, 2012, Qi et al., 2011, Zhang et al., 2010, Macis et al., 2007, Zintzaras, 2006). This is the largest meta-analysis carried out so far to investigate the association between MTHFR and BC.
In subgroup analysis based of ethnicity, we find significant association between C677T polymorphism and BC risk in Asian population but did not find such association in Caucasian population. These discrepancies in the results could be arise because of the multitude of the factors such as the differences in the allele frequencies due to ethnic variations, nutritional status especially folate intake and sample size studied etc. Frequency of MTHFR C677T polymorphism varies in different ethnic populations. Recently, Yadav et al. (2014) reported that T allele and TT genotype frequencies in Asian population (37.2% and 16.9%) are higher in comparison to Caucasian populations (33.6% of T allele and 12.1% of TT genotype).
MTHFR enzyme function may influence cancer risk in two ways. The substrate of MTHFR enzyme, 5,10- methylenetetrahydrofolate, is involved in the conversion of deoxyuridylate monophosphate to deoxythymidylate monophosphate, and low levels of 5,10-methylenetetrahydrofolate would lead to an increased deoxyuridylate monophosphate/deoxythymidylate monophosphate ratio. In this situation, increased incorporation of uracil into DNA in place of thymine may follow, resulting in an increased chance of point mutations and DNA/chromosome breakage (Sohn et al., 2009, Boccia et al., 2008, Blount et al., 1997). The second way by which dysfunctional MTHFR increases risk of cancer is determined by the level of SAM, which is necessary for maintenance of the methylation patterns in DNA. Altered methylation pattern may modify DNA conformation and gene expression. A less active form of MTHFR leads to lower SAM levels and consequently to hypomethylation and increase the risk of cancers (Boccia et al., 2008, Stern et al., 2000, Duthie, 1999).
The role of folate in breast cancer has been investigated in several dietary studies and most have shown folate consumption to be inversely related to breast cancer risk (Zhang et al., 1999, Rohan et al., 2000, Goodman et al., 2001, Xu et al., 2007) and adequate folate intake has been associated with a substantially decreased risk of cancer. Cancer risk modification conferred by C677T polymorphism is further modified by the status of folate and nutrients involved in one-carbon and folate metabolism (Ueland et al., 2001, Robien and Ulrich, 2003, Sharp and Little, 2004). We did not done sub group analysis on the basis of folate concentrations. In total 75 included studies, folate intake information was reported only in 12 studies, out of which few authors reported folate uptake dose and others reported blood level of folate. With increased folic acid fortification in the Caucasian population, the general intake of folate may be higher than that from the Asian population, whose folate intake is primarily obtained from unfortified diets. Further, in Asian population malnutrition, low folate intake and impaired folate absorption due to infectious diseases were already reported (Rosenberg et al., 2002, Wilcken et al., 2003). Folate supplementation would outweigh the negative effects of C677T polymorphism. Hence the effect of MTHFR on breast cancer risk in a particular population may depend on the intake level of folate food in that population.
Meta-analysis is a powerful tool for analyzing cumulative data of studies where the individual sample sizes are small and the statistical power low (Yadav et al., 2015, Rai et al., 2014). Several meta-analyses were published to assess the role of MTHFR polymorphism in cancer development like: lung cancer (Boccia et al., 2009), pancreatic cancer (Tu et al., 2012), prostate cancer (Zhang et al., 2012), esophageal cancer (Wen et al., 2013), ovarian cancer (Ding et al., 2012) and cervical cancer (Mei et al., 2012).
We identified ten meta-analyses (Singh et al., 2015, Liang et al., 2014, Li et al., 2014, Rai, 2014, Yu and Chen, 2012, Liang et al., 2014; Zhang et al., 2010, Lissowska et al., 2007, Macis et al., 2007, Zintzaras, 2006) identified concerning similar topic as we did during the literature search. A comparative details of all the meta-analysis published so far (including present) were presented in Table 3. Zintzaras (2006) carried out first meta-analysis of MTHFR C677T genotype of 18 studies and reported significant heterogeneity (p = 0.08, I2 = 34%) and non-significant association (OR = 1.02; 95% confidence interval (0.95–1.10) in allele contrast model. Lissowska et al. (2007) carried out meta-analysis of 22 studies and showed no association between TT (mutant homozygote) vs. CC genotypes and breast cancer risk (OR = 0.99; 95% CI = 0.86–1.15), based on 8330 cases and 10,825 controls. Macis et al. (2007) performed a meta-analysis of 18 studies examining the association between polymorphisms C677T and BC risk and found positive association between the TT genotype BC risk. A meta-analysis of 41 retrospective studies (16,480 cases and 22,388 controls) was carried out by Qi et al. (2011) and reported significant elevated breast cancer risk using all five genetic model (TT vs. CC: OR = 1.13, 95% CI = 1.01–1.25). Zhang et al. (2010) reported significant association between 677T polymorphism with BC (TT vs. CC: OR = 1.11, 95% CI = 1.01–1.23 and suggested MTHFR T allele as a low-penetrant risk factor for developing breast cancer. Yu and Chen (2012) carried out meta-analysis of 51 studies including 20,907 cases and 23,905 controls and reported significant associations between MTHFR C677T polymorphism and BC risk. Liang et al., 2014, Li et al., 2014, Rai, 2014 and Singh et al. (2015) conducted meta-analyses on 37 studies (15,260 cases and 20,411 controls), 57 studies (25,877 breast cancer cases and 29,781 controls), 36 studies (8040 cases and 10,008 controls) and 41 studies (16,480 cases and 22,388 controls), and 61 studies (28,031 Cases and 31,880 Controls), respectively, and except Singh et al. (2015), all were reported significant association between C677T polymorphism and BC risk. Compared with present meta-analysis, most of these meta-analyses included less number of studies and smaller total sample was analyzed.
Table 3.
A comparative analysis of details of odds ratio, 95% CI, genetic models reported in total 11 (including present) meta-analysis published so far analyzing case–control studies of MTHR C677T polymorphism and breast cancer.
| SN | Author | No. of studies | Sample size |
OR | 95% confidence interval | Model | I2 | ||
|---|---|---|---|---|---|---|---|---|---|
| Case | Control | Total | |||||||
| 1 | Zintzaras (2006) | 18 | 5467 | 7336 | 12,803 | 1.03 | 0.97–1.08 | T vs. C | 34 |
| 1.07 | 0.95–1.20 | TT vs. CC | 36 | ||||||
| 1.06 | 0.95–1.19 | TT vs. CT + CC | 33 | ||||||
| 1.02 | 0.95–1.10 | TT + CT vs. CC | 14 | ||||||
| 2 | Lissowska et al. (2007) | 22 | 8330 | 10,825 | 19,155 | 1.01 | 0.95–1.08 | CT vs. CC | NA |
| 0.99 | 0.86–1.15 | TT vs. CC | NA | ||||||
| 3 | Macis et al. (2007) | 18 | 1.01 | 0.87–1.18 | TT vs. CT + CC | NA | |||
| 1.04 | 0.97–1.11 | TT + CT vs. CC | NA | ||||||
| 4 | Qi et al. (2011) | 41 | 16,480 | 22,388 | 38,868 | 1.04 | 1.00–1.07 | T vs. C | NA |
| 1.13 | 1.01–1.25 | TT vs. CC | NA | ||||||
| 1.03 | 0.99–1.07 | TT + CT vs. CC | NA | ||||||
| 1.11 | 1.01–1.23 | TT vs. CT + CC | NA | ||||||
| 5 | Zhang et al. (2010) | 37 | 15,260 | 20,411 | 35,671 | 1.04 | 0.99–1.08 | CT vs. CC | NA |
| 1.11 | 1.01–1.23 | TT vs. CC | NA | ||||||
| 1.04 | 1.00–1.09 | TT + CT vs. CC | NA | ||||||
| 1.09 | 0.99–1.20 | TT vs. CT + CC | NA | ||||||
| 6 | Yu and Chen (2012) | 51 | 20,907 | 23,905 | 44,812 | 0.93 | 0.88–0.98 | T vs. C | NA |
| 0.96 | 0.92–1.01 | CT vs. CC | NA | ||||||
| 0.87 | 0.78–0.95 | TT vs. CC | NA | ||||||
| 0.89 | 0.82–0.97 | TT vs. CT | NA | ||||||
| 0.88 | 0.80–0.96 | TT + CT vs. CC | NA | ||||||
| 0.94 | 0.89–0.99 | TT vs. CT + CC | NA | ||||||
| 7 | Liang et al. (2014) | 13 | 3273 | 4419 | 7692 | 1.12 | 1.02–1.23 | T vs. C | NA |
| 1.35 | 1.10–1.67 | TT vs. CC | NA | ||||||
| 1.37 | 1.11–1.70 | TT vs. CT + CC | NA | ||||||
| 8 | Li et al. (2014) | 57 | 25,877 | 29,781 | 55,658 | 0.94 | 0.89–0.98 | T vs. C | NA |
| 0.98 | 0.96–1.00 | CT vs. CC | NA | ||||||
| 0.98 | 0.96–0.99 | TT vs. CC | NA | ||||||
| 0.98 | 0.96–1.00 | TT vs. CT | NA | ||||||
| 0.95 | 0.92–0.99 | TT + CT vs. CC | NA | ||||||
| 0.99 | 0.98–0.99 | TT vs. CT + CC | NA | ||||||
| 9 | Rai (2014) | 36 | 8040 | 10,008 | 18,048 | 1.23 | 1.13–1.37 | T vs. C | 77.3 |
| 1.03 | 0.97–1.10 | CT vs. CC | 33.7 | ||||||
| 1.38 | 1.16–1.63 | TT vs. CC | 58.2 | ||||||
| 1.12 | 1.01–1.23 | TT + CT vs. CC | 51.5 | ||||||
| 1.33 | 1.15–1.43 | TT vs. CT + CC | 50.3 | ||||||
| 10 | Singh et al. (2015) | 61 | 28,031 | 31,880 | 59,911 | 0.97 | 0.93–1.00 | TT + CT vs. CC | 29.5 |
| 1.05 | TT vs. CT + CC | 29.5 | |||||||
| 11 | Present study, 2015 | 75 | 31,315 | 35,608 | 66,923 | 1.08 | 1.03–1.13 | T vs. C | 63 |
| 1.05 | 1.01–1.08 | CT vs. CC | 29 | ||||||
| 1.17 | 1.06–1.28 | TT vs. CC | 60 | ||||||
| 1.06 | 1.02–1.09 | TT + CT vs. CC | 48 | ||||||
| 1.12 | 1.03–1.22 | TT vs. CT + CC | 55 | ||||||
NA = not given in paper.
Presence of higher heterogeneity showed that there were significant differences between individual studies. Hence, sensitivity and subgroup analyses were performed to explore the causes of heterogeneity. Sensitivity analysis showed that even after excluding studies with a small number of cases (n < 100), or having controls violating the HWE, the heterogeneity decreased slightly. However, the larger sample size does not mean the study is without limitations. The current meta-analysis has few limitations also like - (i) only published studies were included, thus possibility of publication bias cannot be excluded, (ii) single gene polymorphism of folate metabolic pathway was considered, and (iii) finally, due to lack of data, gene–gene and gene–environment interactions could not be included.
We hope that this meta-analysis of the most comprehensive literature addressing the association is yielded convincing evidence to determine the role of MTHFR C677T polymorphism in BC risk. In summary, results of present meta-analysis showed modest association between MTHFR C677T polymorphism with breast cancer in total studies. However, sub-group analysis results based on ethnicity showed strong significant association between TT genotype and breast cancer (TT vs. CC; OR°=°1.26; 95% CI: 1.06–1.51; p = 0.009) in Asian population but in Caucasian population such association was not observed (TT vs. CC; OR°=°1.08; 95% CI: 0.99–1.14; p = 0.05). However, presence of publication bias and higher between study heterogeneity suggested that results should be interpreted cautiously and also indicated that the observed association may differ in strength between populations, or may not exist at all in some populations.
Conflict of interest
None.
Acknowledgments
The authors are highly grateful to Leon Bax (Chief Scientific Officer at BiostatXL, UMC Utrecht) for his valuable suggestions, which help us in statistical analysis. We also thank all authors of the included studies for their cooperation.
References
- Akram M., Malik F.A., Kayani M.A. Mutational analysis of the MTHFR gene in breast cancer patients of Pakistani population. Asian Pac. J. Cancer Prev. 2012;13:1599–1603. doi: 10.7314/apjcp.2012.13.4.1599. [DOI] [PubMed] [Google Scholar]
- Alshatwi A.A. Breast cancer risk, dietary intake, and methylenetetrahydrofolate reductase (MTHFR) single nucleotide polymorphisms. Food Chem. Toxicol. 2010;48:1881–1885. doi: 10.1016/j.fct.2010.04.028. [DOI] [PubMed] [Google Scholar]
- Barbosa Rde C., Menezes D.C., Freire T.F., Sales D.C., Alencar V.H. Associations of polymorphisms of folate cycle enzymes and risk of breast cancer in a Brazilian population are age dependent. Mol. Biol. Rep. 2012;39:4899–4907. doi: 10.1007/s11033-011-1285-1. [DOI] [PubMed] [Google Scholar]
- Batschauer A.P., Cruz N.G., Oliveira V.C., Coelho F.F., Santos I.R. HFE, MTHFR, and FGFR4 genes polymorphisms and breast cancer in Brazilian women. Mol. Cell. Biochem. 2011;357:247–253. doi: 10.1007/s11010-011-0895-1. [DOI] [PubMed] [Google Scholar]
- Bax L., Yu L.M., Ikeda N., Tsuruta H., Moons K.G. Development and validation of MIX: comprehensive free software for meta-analysis of causal research data. BMC Med. Res. Methodol. 2006;6:50. doi: 10.1186/1471-2288-6-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg C.B., Mazumdar M. Operating characteristics of a rank correlation test for publication bias. Biometrics. 1994;50:1088–1101. [PubMed] [Google Scholar]
- Bentley A.R., Raiszadeh F., Stover P.J., Hunter D.J., Hankinson S.E. No association between cSHMT genotypes and the risk of breast cancer in the Nurses' Health Study. Eur. J. Clin. Nutr. 2010;64:108–110. doi: 10.1038/ejcn.2009.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount B.C., Mack M.M., Wehr C.M., MacGregor J.T., Hiatt R.A., Wang G. Folate deficiency causes uracil misincorporation into human DNA and chromosome breakage: implications for cancer and neuronal damage. Proc. Natl. Acad. Sci. U. S. A. 1997;94:3290–3295. doi: 10.1073/pnas.94.7.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccia S., Boffetta P., Brennan P., Ricciardi G., Gianfagna F., Matsuo K. Meta-analyses of the methylenetetrahydrofolate reductase C677T and A1298C polymorphisms and risk of head and neck and lung cancer. Cancer Lett. 2009;273:55–61. doi: 10.1016/j.canlet.2008.07.026. [DOI] [PubMed] [Google Scholar]
- Boccia S., Hung R., Ricciardi G., Gianfagn F., Ebert M.P.A., Fang J.Y. Meta- and pooled analyses of the methylenetetrahydrofolate reductase C677T and A1298C polymorphisms and gastric cancer risk: a huge-GSEC review. Am. J. Epidemiol. 2008;167(5):505–516. doi: 10.1093/aje/kwm344. [DOI] [PubMed] [Google Scholar]
- Cam R., Eroglu A., Egin Y., Akar N. Dihydrofolate reductase (DHRF) 19-bp intron-1 deletion and methylenetetrahydrofolate reductase (MTHFR) C677T polymorphisms in breast cancer. Breast Cancer Res. Treat. 2009;115:431–432. doi: 10.1007/s10549-008-0054-x. [DOI] [PubMed] [Google Scholar]
- Campbell I.G., Baxter S.W., Eccles D.M., Choong D.Y. Methylenetetrahydrofolate reductase polymorphism and susceptibility to breast cancer. Breast Cancer Res. 2002;4:R14. doi: 10.1186/bcr457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerne J.Z., Pohar-Perme M., Cerkovnik P., Gersak K., Novakovic S. Age at menarche and menopause is not associated with two common genetic variants in the methylenetetrahydrofolate reductase (MTHFR) gene. Eur. J. Contracept. Reprod. Health Care. 2011;16(4):241–247. doi: 10.3109/13625187.2011.575481. [DOI] [PubMed] [Google Scholar]
- Chen J., Gammon M.D., Chan W., Palomeque C., Wetmur J.G., Kabat G.C. One-carbon metabolism, MTHFR polymorphisms, and risk of breast cancer. Cancer Res. 2005;65:1606–1614. doi: 10.1158/0008-5472.CAN-04-2630. [DOI] [PubMed] [Google Scholar]
- Cheng C.W., Yu J.C., Huang C.S., Shieh J.C., Fu Y.P., Wang H.W. Polymorphism of cytosolic serine hydroxymethyltransferase, estrogen and breast cancer risk among Chinese women in Taiwan. Breast Cancer Res. Treat. 2008;111:145–155. doi: 10.1007/s10549-007-9754-x. [DOI] [PubMed] [Google Scholar]
- Chou Y.C., Wu M.H., Yu J.C., Lee M.S., Yang T., Shih H.L. Genetic polymorphisms of the methylenetetrahydrofolate reductase gene, plasma folate levels, and breast cancer susceptibility: a case–control study in Taiwan. Carcinogenesis. 2006;27:2295–2300. doi: 10.1093/carcin/bgl108. [DOI] [PubMed] [Google Scholar]
- Collaborative Group on Hormonal Factors in Breast Cancer Breast cancer and hormone replacement therapy: collaborative reanalysis of data from 51 epidemiological studies of 52 705 women with breast cancer and 108 411 women without breast cancer. Lancet. 1997;350:1047–1059. [PubMed] [Google Scholar]
- Deligezer U., Akisik E.E., Dalay N. Homozygosity at the C677T of the MTHFR gene is associated with increased breast cancer risk in the Turkish population. In Vivo. 2005;19:889–893. [PubMed] [Google Scholar]
- DerSimonian R., Laird N. Meta-analysis in clinical trials. Control. Clin. Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- Diakite B., Tazzite A., Hamzi K., Jouhadi H., Nadifi S. Methylenetetrahydrofolate reductase C677T polymorphism and breast cancer risk in Moroccan women. Afr. Health Sci. 2012;12(2):204–209. doi: 10.4314/ahs.v12i2.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X.P., Feng L., Ma L. MTHFR C677T polymorphism and ovarian cancer risk: a meta-analysis. Asian Pac. J. Cancer Prev. 2012;13:3937–3942. doi: 10.7314/apjcp.2012.13.8.3937. [DOI] [PubMed] [Google Scholar]
- Duthie S.J. Folic acid deficiency and cancer: mechanisms of DNA instability. Br. Med. Bull. 1999;55:578–592. doi: 10.1258/0007142991902646. [DOI] [PubMed] [Google Scholar]
- Egger M., Davey S., Schneider G., Minder M.C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ergul E., Sazci A., Utkan Z., Canturk N.Z. Polymorphisms in the MTHFR gene are associated with breast cancer. Tumor Biol. 2003;24:286–290. doi: 10.1159/000076460. [DOI] [PubMed] [Google Scholar]
- Ericson U., Sonestedt E., Ivarsson M.I., Gullberg B., Carlson J., Olsson H. Folate intake, methylenetetrahydrofolate reductase polymorphisms, and breast cancer risk in women from the Malmö Diet and Cancer cohort. Cancer Epidemiol. Biomark. Prev. 2009;18:1101–1110. doi: 10.1158/1055-9965.EPI-08-0401. [DOI] [PubMed] [Google Scholar]
- Ferlay J., Bray F., Pisani P., Parkin D.M. IARC Cancer Base No. 5 [CD-ROM]. Version 1.1. IARC Press; Lyon: 2000. Cancer incidence, mortality and prevalence worldwide. [Google Scholar]
- Forsti A., Angelini S., Festa F., Sanyal S., Zhang Z., Grzybowska E. Single nucleotide polymorphisms in breast cancer. Oncol. Rep. 2004;11:917–922. [PubMed] [Google Scholar]
- Frankenburg F.R. The role of one-carbon metabolism in schizophrenia and depression. Harv. Rev. Psychiatry. 2007;15(4):146–160. doi: 10.1080/10673220701551136. [DOI] [PubMed] [Google Scholar]
- Frosst P., Bloom H.J., Milos R., Goyette P., Sheppard C.A., Matthews R.G. A candidate genetic risk factor for vascular disease: a common mutation in methylenetetrahydrofolate reductase. Nat. Genet. 1995;10:111–113. doi: 10.1038/ng0595-111. [DOI] [PubMed] [Google Scholar]
- Gao C.M., Tang J.H., Cao H.X., Ding J.H., Wu J.Z., Wang J. MTHFR polymorphisms, dietary folate intake and breast cancer risk in Chinese women. J. Hum. Genet. 2009;5:414–418. doi: 10.1038/jhg.2009.57. [DOI] [PubMed] [Google Scholar]
- Gilbody S., Lewis S., Lightfoot T. Methylenetetrahydrofolate reductase (MTHFR) genetic polymorphisms and psychiatric disorders: a HuGE review. Am. J. Epidemiol. 2007;165(1):1–13. doi: 10.1093/aje/kwj347. [DOI] [PubMed] [Google Scholar]
- Goodman J.E., Lavigne J.A., Wu K., Helzlsouer K.J., Strickland P.T., Selhub J. COMT genotype, micronutrients in the folate metabolic pathway and breast cancer risk. Carcinogenesis. 2001;22(10):1661–1665. doi: 10.1093/carcin/22.10.1661. [DOI] [PubMed] [Google Scholar]
- Grieu F., Powell B., Beilby J., Iacopetta B. Methylenetetrahydrofolate reductase and thymidylate synthase polymorphisms are not associated with breast cancer risk or phenotype. Anticancer Res. 2004;24:3215–3219. [PubMed] [Google Scholar]
- Guo J., Jin M., Zhang M., Chen L.A. Genetic variant in miR-196a2 increased digestive system cancer risks: a meat-analysis of 15 case–control studies. PLoS ONE. 2012;7(1):e30585. doi: 10.1371/journal.pone.0030585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J.M., Pu Y.D., Wu Y.J., Qin R., Zhang Q.J., Sun Y.S. Association between dietary intake of folate and MTHFR and MTR genotype with risk of breast cancer. Genet. Mol. Res. 2014;13(4):8925–8931. doi: 10.4238/2014.October.31.7. [DOI] [PubMed] [Google Scholar]
- Hekim N., Ergen A., Yaylim I., Yilmaz H., Zeybek U., Oztürk O. No association between methylenetetrahydrofolate reductase C677T polymorphism and breast cancer. Cell Biochem. Funct. 2007;25:115–117. doi: 10.1002/cbf.1274. [DOI] [PubMed] [Google Scholar]
- Hennquez-Hernandez L.A., Murias-Rosales A., Hernandez Gon-zalez A., Cabrera De Leon A., Dıaz-Chico B.N., Mori De Santiago M. Gene polymorphisms in TYMS, MTHFR, p53 and MDR1 as risk factors for breast cancer: a case–control study. Oncol. Rep. 2009;22:1425–1433. doi: 10.3892/or_00000584. [DOI] [PubMed] [Google Scholar]
- Higgins J.P., Thompson S.G. Quantifying heterogeneity in a meta-analysis. Stat. Med. 2002;21:1539–1558. doi: 10.1002/sim.1186. [DOI] [PubMed] [Google Scholar]
- Holmes M.V., Newcombe P., Hubacek J.A., Sofat R., Ricketts S.L., Cooper J. Effect modification by population dietary folate on the association between MTHFR genotype, homocysteine, and stroke risk: a meta-analysis of genetic studies and randomized trials. Lancet. 2011;378:584–594. doi: 10.1016/S0140-6736(11)60872-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosseini M., Houshmand M., Ebrahimi A. MTHFR polymorphisms and breast cancer risk. Arch. Med. Sci. 2011;7:134–137. doi: 10.5114/aoms.2011.20618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z., Wang Y., Ni J., Ge F., Zou T. Serum folate, vitamin B12 concentration and MTHFR, MS gene polymorphism associated with risk of breast cancer research. Mod. Oncol. 2011;19:L428–L431. [Google Scholar]
- Huang C.Y., Chang W.S., Shui H.A., Hsieh Y.H., Loh C.H., Wang H.C. Evaluation of the contribution of methylenetetrahydrofolate reductase genotypes to Taiwan breast cancer. Anticancer Res. 2014;34:4109–4115. [PubMed] [Google Scholar]
- Huedo-Medina T.B., Sanchez-Meca J., Marin-Martinez F., Botella J. Assessing heterogeneity in meta-analysis: Q statistic or I2 index? Psychol. Methods. 2006;11:193–206. doi: 10.1037/1082-989X.11.2.193. [DOI] [PubMed] [Google Scholar]
- Hulka B.S., Stark A.T. Breast cancer: cause and prevention. Lancet. 1995;346:883–887. doi: 10.1016/s0140-6736(95)92713-1. [DOI] [PubMed] [Google Scholar]
- Inoue M., Robien K., Wang R., Van Den Berg D.J., Koh W.P., Yu M.C. Green tea intake, MTHFR/TYMS genotype and breast cancer risk: the Singapore Chinese health study. Carcinogenesis. 2008;29:1967–1972. doi: 10.1093/carcin/bgn177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowska A., Rozkrut D., Antoniou A., Hamann U., Scott R.J., McGuffog L. Association of PHB 1630 C > T and MTHFR 677 C > T polymorphisms with breast and ovarian cancer risk in BRCA1/2 mutation carriers: results from a multicenter study. Br. J. Cancer. 2012;106:2016–2024. doi: 10.1038/bjc.2012.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang-Hua Q., De-Chuang J., Zhen-Duo L., Shu-de C., Zhenzhen L. Association of methylenetetrahydrofolate reductase and methionine synthase polymorphisms with breast cancer risk and interaction with folate, vitamin B6, and vitamin B 12 intakes. Tumor Biol. 2014;35(12):11895–11901. doi: 10.1007/s13277-014-2456-1. [DOI] [PubMed] [Google Scholar]
- Jin Z.Z., Lu Q., Ge D.H., Zong M., Zhu Q.H. Effect of the methylenetetrahydrofolate reductase gene C677T polymorphism on C-erbB-2 methylation status and its association with cancer. Mol. Med. Rep. 2009;2:283–289. doi: 10.3892/mmr_00000097. [DOI] [PubMed] [Google Scholar]
- Justenhoven C., Hamann U., Pierl C.B., Rabstein S., Pesch B. One-carbon metabolism and breast cancer risk: no association of MTHFR, MTR, and TYMS polymorphisms in the GENICA study from Germany. Cancer Epidemiol. Biomark. Prev. 2005;14:3015–3018. doi: 10.1158/1055-9965.EPI-05-0592. [DOI] [PubMed] [Google Scholar]
- Kakkoura M.G., Demetriou C.A., Loizidou M.A., Loucaides G., Neophytou I., Marcou Y., Hadjisavvas A. Single-nucleotide polymorphisms in one-carbon metabolism genes, Mediterranean diet and breast cancer risk: a case–control study in the Greek-Cypriot female population. Genes Nutr. 2015;10(2):453. doi: 10.1007/s12263-015-0453-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalemi T.G., Lambropoulos A.F., Gueorguiev M., Chrisafi S., Papazisis K.T. The association of p53 mutations and p53 codon 72, Her 2 codon 655 and MTHFR C677T polymorphisms with breast cancer in Northern Greece. Cancer Lett. 2005;222:57–65. doi: 10.1016/j.canlet.2004.11.025. [DOI] [PubMed] [Google Scholar]
- Kalyankumar C., Jamil K. Methylene tetrahydofolate reductase (MTHFR) C677T and A1298C polymorphisms and breast cancer in South Indian population. Int. J. Cancer Res. 2006;2:143–151. [Google Scholar]
- Kan X., Zou T., Wu X., Wang X. Yunnan methylenetetrahydrofolate reductase gene polymorphism associated with breast cancer susceptibility. Cancer Res. 2007;34:716–718. [Google Scholar]
- Kang S.S., Zhou J., Wong P., Kowalisyn J., Strokosch G. Intermediate homocysteinemia: a thermolabile variant of methylenetetrahydrofolate reductase. Am. J. Hum. Genet. 1988;43:414. [PMC free article] [PubMed] [Google Scholar]
- Kelsey J.L. Breast cancer epidemiology: summary and future directions. Epidemiol. Rev. 1993;15:256–263. doi: 10.1093/oxfordjournals.epirev.a036112. [DOI] [PubMed] [Google Scholar]
- Kim Y.I. Folate and carcinogenesis: evidence, mechanisms, and implications. J. Nutr. Biochem. 1999;10:66–88. doi: 10.1016/s0955-2863(98)00074-6. [DOI] [PubMed] [Google Scholar]
- Kotsopoulos J., Zhang W.W., Zhang S., McCready D., Trudeau M., Zhang P. Polymorphisms in folate metabolizing enzymes and transport proteins and the risk of breast cancer. Breast Cancer Res. Treat. 2008;8:9895–9896. doi: 10.1007/s10549-008-9895-6. [DOI] [PubMed] [Google Scholar]
- Laanpere M., Altmäe S., Stavreus-Evers A., Nilsson T.K., Yngve A., Salumets A. Folate-mediated one-carbon metabolism and its effect on female fertility and pregnancy viability. Nutr. Rev. 2010;68(2):99–113. doi: 10.1111/j.1753-4887.2009.00266.x. [DOI] [PubMed] [Google Scholar]
- Lajin B., Sakur A.A., Ghabreau L., Alachkar A. Association of polymorphisms in one-carbon metabolizing genes with breast cancer risk in Syrian women. Tumor Biol. 2012;33:1133–1139. doi: 10.1007/s13277-012-0354-y. [DOI] [PubMed] [Google Scholar]
- Langsenlehner U., Krippl P., Renner W., Yazdani-Biuki B., Wolf G., Wascher T.C. The common 677C > T gene polymorphism of methylenetetrahydrofolate reductase gene is not associated with breast cancer risk. Breast Cancer Res. Treat. 2003;81:169–172. doi: 10.1023/A:1025752420309. [DOI] [PubMed] [Google Scholar]
- Langsenlehner T., Renner W., Yazdani-Biuki B., Langsenlehner U. Methylenetetrahydrofolatereductase (MTHFR) and breast cancer risk: a nested-case–control study and a pooled meta-analysis. Breast Cancer Res. Treat. 2008;107:459–460. doi: 10.1007/s10549-007-9564-1. [DOI] [PubMed] [Google Scholar]
- Le Marchand L., Haiman C.A., Wilkens L.R., Kolonel L.N., Henderson B.E. MTHFR polymorphisms, diet, HRT, and breast cancer risk: the multiethnic cohort study. Cancer Epidemiol. Biomark. Prev. 2004;13:2071–2077. [PubMed] [Google Scholar]
- Lee S.A., Kang D., Nishio H., Lee M.J., Kim D.H., Han W. Methylenetetrahydrofolate reductase polymorphism, diet, and breast cancer in Korean women. Exp. Mol. Med. 2004;36:116–121. doi: 10.1038/emm.2004.17. [DOI] [PubMed] [Google Scholar]
- Li W.D., Chen S.Q. Association of methylenetetrahydrofolate reductase C677T polymorphism and breast cancer risk. J. Prac. Med. 2009;25:2031–2033. [Google Scholar]
- Li K., Li W., Dong X. Association of 677 C > T (rs1801133) and 1298A > C (rs1801131) polymorphisms in the MTHFR gene and breast cancer susceptibility: a meta-analysis based on 57 individual studies. PLoS ONE. 2014;9:e71290. doi: 10.1371/journal.pone.0071290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang H., Yan Y., Li T., Li R., Li M., Li S. Methylenetetrahydrofolate reductase polymorphisms and breast cancer risk in Chinese population: a meta-analysis of 22 case–control studies. Tumor Biol. 2014;35:1695–1701. doi: 10.1007/s13277-013-1234-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W., Cen Y.L., Lin Y., Su F.X., Wu B.H., Tang L.Y. Joint effects between urinary selenium and polymorphisms in methylation related genes on breast cancer risk. Neoplasma. 2015 doi: 10.4149/neo_2015_059. (Epub ahead of print: DOI:10.4149/neo_2015_059) [DOI] [PubMed] [Google Scholar]
- Lin W.Y., Chou Y.C., Wu M.H., Huang H.B., Jeng Y.L., Wu C.C. The MTHFR C677T polymorphism, estrogen exposure and breast cancer risk: a nested case–control study in Taiwan. Anticancer Res. 2004;24:3863–3868. [PubMed] [Google Scholar]
- Lissowska J., Gaudet M.M., Brinton L.A., Chanock S.J., Peplonska B. Genetic polymorphisms in the one carbon metabolism pathway and breast cancer risk: a population-based case–control study and metaanalyses. Int. J. Cancer. 2007;120:2696–2703. doi: 10.1002/ijc.22604. [DOI] [PubMed] [Google Scholar]
- Liu Y., Zhou L.S., Xu X.M., Deng L.Q., Xiao Q.K. Association of dietary intake of folate, vitamin B6 and B12 and MTHFR genotype with breast cancer risk. Asian Pac. J. Cancer Prev. 2013;14:5189–5192. doi: 10.7314/apjcp.2013.14.9.5189. [DOI] [PubMed] [Google Scholar]
- Lohmueller K.E., Pearce C.L., Pike M., Lander E.S., Hirschhorn J.N. Meta-analysis of genetic association studies supports a contribution of common variants to susceptibility to common disease. Nat. Genet. 2003;33(2):177–182. doi: 10.1038/ng1071. [DOI] [PubMed] [Google Scholar]
- López-Cortés A., Echeverría C., Oña-Cisneros F., Sánchez M.E., Herrera C., Cabrera-Andrade A. Breast cancer risk associated with gene expression and genotype polymorphisms of the folate-metabolizing MTHFR gene: a case–control study in a high altitude Ecuadorian mestizo population. Tumor Biol. 2015 doi: 10.1007/s13277-015-3335-0. (Epub ahead of print: DOI 10.1007/s13277-015-3335-0) [DOI] [PubMed] [Google Scholar]
- Lu Q., Jiang K., Li Q., Ji Y.J., Chen W.L., Xue X.H. Polymorphisms in the MTHFR gene are associated with breast cancer risk and prognosis in a Chinese population. Tumor Biol. 2015 doi: 10.1007/s13277-014-3016-4. (Epub ahead of print: DOI 10.1007/s13277-014-3016-4) [DOI] [PubMed] [Google Scholar]
- Ma E., Iwasaki M., Junko I., Hamada G.S., Nishimoto I.N. Dietary intake of folate, vitamin B6, and vitamin B12, genetic polymorphism of related enzymes, and risk of breast cancer: a case–control study in Brazilian women. BMC Cancer. 2009;9:122. doi: 10.1186/1471-2407-9-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma E., Iwasaki M., Kobayashi M., Kasuga Y., Yokoyama S., Onuma H. Dietary intake of folate, vitamin B2, vitamin B6, vitamin B12, genetic polymorphism of related enzymes, and risk of breast cancer: a case control study in Japan. Nutr. Cancer. 2009;61:447–456. doi: 10.1080/01635580802610123. [DOI] [PubMed] [Google Scholar]
- Macis D., Maisonneuve P., Johansson H., Bonanni B., Botteri E., Iodice S. Methylenetetrahydrofolate reductase (MTHFR) and breast cancer risk: a nested-case–control study and a pooled meta-analysis. Breast Cancer Res. Treat. 2007;106:263–271. doi: 10.1007/s10549-006-9491-6. [DOI] [PubMed] [Google Scholar]
- Mantel N., Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J. Natl. Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- Maruti S.S., Ulrich C.M., Jupe E.R., White E. MTHFR C677T and postmenopausal breast cancer risk by intakes of one-carbon metabolism nutrients: a nested case–control study. Breast Cancer Res. 2009;11:R9. doi: 10.1186/bcr2462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei Q., Zhou D., Gao J., Shen S., Wu J., Guo L. The association between MTHFR 677C > T polymorphism and cervical cancer: evidence from a meta-analysis. BMC Cancer. 2012;12:467–476. doi: 10.1186/1471-2407-12-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mir M.M., Dar J.A., Dar N.A., Dar M.S., Salam I., Lone M.M. Combined impact of polymorphism of folate metabolism genes; glutamate carboxypeptidase, methylene tetrahydrofolate reductase and methionine synthase reductase on breast cancer susceptibility in Kashmiri women. In. J. Health Sciences. 2008;2:3–14. [PMC free article] [PubMed] [Google Scholar]
- Naushad S.M., Reddy C.A., Rupasree Y., Pavani A., Digumarti R.R., Gottumukkala S.R. Cross-talk between one-carbon metabolism and xenobiotic metabolism: implications on oxidative DNA damage and susceptibility to breast cancer. Cell Biochem. Biophys. 2011;61:715–723. doi: 10.1007/s12013-011-9245-x. [DOI] [PubMed] [Google Scholar]
- Ozen F., Erdis E., Sik E., Silan F., Uludag A., Ozdemir O. Germ-line MTHFR C677T, FV H1299R and PAI-1 5G/4G variations in breast carcinoma. Asian Pac. J. Cancer Prev. 2013;14:2903–2908. doi: 10.7314/apjcp.2013.14.5.2903. [DOI] [PubMed] [Google Scholar]
- Platek M.E., Shields P.G., Marian C., McCann S.E., Bonner M.R., Nie J. Alcohol consumption and genetic variation in methylenetetrahydrofolate reductase and 5-methyltetrahydrofolate-homocysteine methyltransferase in relation to breast cancer risk. Cancer Epidemiol. Biomarkers Prev. 2009;18(2459):2453–2459. doi: 10.1158/1055-9965.EPI-09-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad V.V., Wilkhoo H. Association of the functional polymorphism C677T in the methylenetetrahydrofolate reductase gene with colorectal, thyroid, breast, ovarian, and cervical cancers. Onkologie. 2011;34:422–426. doi: 10.1159/000331131. [DOI] [PubMed] [Google Scholar]
- van der Put N., van Straaten M., Trijbels H.W., Blom F.J., H.J. Folate, homocysteine and neural tube defects: an overview. Exp. Biol. Med. (Maywood) 2001;226(4):243–270. doi: 10.1177/153537020122600402. [DOI] [PubMed] [Google Scholar]
- Qi X., Ma X., Yang X., Fan L., Zhang Y. Methylenetetrahydrofolate reductase polymorphisms and breast cancer risk: a meta-analysis from 41 studies with 16,480 cases and 22,388 controls. Breast Cancer Res. Treat. 2011;123:499–506. doi: 10.1007/s10549-010-0773-7. [DOI] [PubMed] [Google Scholar]
- Qi J., Miao X.P., Tan W., Yu C.Y., Liang G., Lü W.F. Association between genetic polymorphisms in methylenetetrahydrofolate reductase and risk of breast cancer. Zhonghua Zhong Liu Za Zhi. 2004;26:287–289. [PubMed] [Google Scholar]
- Rai V. Evaluation of methylenetetrahydrofolate reductase gene variant (C677T) as risk factor for bipolar disorder. Cell. Mol. Biol. 2011 (Noisy-le-grand). 57, Suppl:OL1558-OL1566. [PubMed] [Google Scholar]
- Rai V. The methylenetetrahydrofolate reductase C677T polymorphism and breast cancer risk in Asian populations. Asian Pac. J. Cancer Prev. 2014;15:5853–5860. doi: 10.7314/apjcp.2014.15.14.5853. [DOI] [PubMed] [Google Scholar]
- Rai V., Yadav U., Kumar P., Yadav S.K., Mishra O.P. Maternal methylenetetrahydrofolate reductase C677T polymorphism and down syndrome risk: a meta-analysis from 34 studies. PLoS ONE. 2014;9:e108552. doi: 10.1371/journal.pone.0108552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reljic A., Simundic A.M., Topic E., Nikolac N., Justinic D. The methylenetetrahydrofolate reductase (MTHFR) C677T polymorphism and cancer risk: the Croatian case–control study. Clin. Biochem. 2007;40:981–985. doi: 10.1016/j.clinbiochem.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Robien K., Ulrich C.M. 5,10-Methylenetetrahydrofolate reductase polymorphisms and leukemia risk: a HuGE minireview. Am. J. Epidemiol. 2003;157(7):571–582. doi: 10.1093/aje/kwg024. [DOI] [PubMed] [Google Scholar]
- Rohan T.E., Jain M.G., Howe G.R., Miller A.B. Dietary folate consumption and breast cancer risk. J. Natl. Cancer Inst. 2000;92:266–269. doi: 10.1093/jnci/92.3.266. [DOI] [PubMed] [Google Scholar]
- Rosenberg N., Murata M., Ikeda Y., Opare-Sem O., Zivelin A., Geffen E. The frequent 5,10-methylenetetrahydrofolate reductase C677T polymorphism is associated with a common haplotype in whites, Japanese, and Africans. Am. J. Hum. Genet. 2002;70:758–762. doi: 10.1086/338932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sangrajrang S., Sato Y., Sakamoto H., Ohnami S., Khuhaprema T., Yoshida T. Genetic polymorphisms in folate and alcohol metabolism and breast cancer risk: a case–control study in Thai women. Breast Cancer Res. Treat. 2010;123:885–893. doi: 10.1007/s10549-010-0804-4. [DOI] [PubMed] [Google Scholar]
- Semenza J.C., Delfino R.J., Ziogas A., Anton-Culver H. Breast cancer risk and methylenetetrahydrofolate reductase polymorphism. Breast Cancer Res. Treat. 2003;77:217–223. doi: 10.1023/a:1021843019755. [DOI] [PubMed] [Google Scholar]
- Sen S., Duman R., Sanacora G. Serum BDNF, depression and anti-depressant medications: meta-analyses and implications. Biol. Psychiatry. 2008;64(6):527–532. doi: 10.1016/j.biopsych.2008.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharp L., Little J. Polymorphisms in genes involved in folate metabolism and colorectal neoplasia: a HuGE review. Am. J. Epidemiol. 2004;159(5):423–443. doi: 10.1093/aje/kwh066. [DOI] [PubMed] [Google Scholar]
- Sharp L., Little J., Schofield A.C., Pavlidou E., Cotton S.C., Meidzybrodzka Z. Folate and breast cancer: the role of polymorphisms in methylenetetrahydrofolate reductase (MTHFR) Cancer Lett. 2002;181:65–71. doi: 10.1016/s0304-3835(02)00030-7. [DOI] [PubMed] [Google Scholar]
- Shrubsole M.J., Gao Y.T., Cai Q., Shu X.O., Dai Q., Hébert J.R. MTHFR polymorphisms, dietary folate intake, and breast cancer risk: results from the Shanghai Breast Cancer Study. Cancer Epidemiol. Biomark. Prev. 2004;13:190–196. doi: 10.1158/1055-9965.epi-03-0273. [DOI] [PubMed] [Google Scholar]
- Singh P., Carlus J., Sekhar D., Francis A., Gupta N., Konwar R. MTHFR 677C > T polymorphism and the risk of breast cancer: evidence from an original study and pooled data for 28031 cases and 31880 controls. PLoS ONE. 2015;10(3):e0120654. doi: 10.1371/journal.pone.0120654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn K.J., Jang H., Campan M., Weisenberger D.J., Dickhout J., Wang Y.C. The methylenetetrahydrofolate reductase C677T mutation induces cell-specific changes in genomic DNA methylation and uracil misincorporation: a possible molecular basis for the site-specific cancer risk modification. Int. J. Cancer. 2009;124:1999–2005. doi: 10.1002/ijc.24003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern L.L., Mason J.B., Selhub J., Choi S.W. Genomic DNA hypomethylation, a characteristic of most cancers, is present in peripheral leukocytes of individuals who are homozygous for the C677T polymorphism in the methylenetetrahydrofolate reductase gene. Cancer Epidemiol. Biomark. Prev. 2000;9:849–853. [PubMed] [Google Scholar]
- Stevens V.L., McCullough M.L., Pavluck A.L., Talbot J.T., Feigelson H.S., Thun M.J. Association of polymorphisms in one-carbon metabolism genes and postmenopausal breast cancer incidence. Cancer Epidemiol. Biomark. Prev. 2007;16:1140–1147. doi: 10.1158/1055-9965.EPI-06-1037. [DOI] [PubMed] [Google Scholar]
- Sturgeon S.R., Schairer C., Grauman D., El Ghormli L., Devesa S. Trends in breast cancer mortality rates by region of the United States, 1950–1999. Cancer Causes Control. 2004;15:987–995. doi: 10.1007/s10552-004-1092-2. [DOI] [PubMed] [Google Scholar]
- Suzuki T., Matsuo K., Hirose K., Hiraki A., Kawase T., Watanabe M. One-carbon metabolism-related gene polymorphisms and risk of breast cancer. Carcinogenesis. 2008;2:356–362. doi: 10.1093/carcin/bgm295. [DOI] [PubMed] [Google Scholar]
- Tu Y.L., Wang S.B., Tan X.L. MTHFR gene polymorphisms are not involved in pancreatic cancer risk: a meta-analysis. Asian Pac. J. Cancer Prev. 2012;13:4627–4630. doi: 10.7314/apjcp.2012.13.9.4627. [DOI] [PubMed] [Google Scholar]
- Ueland P.M., Hustad S., Schneede J., Refsum H., Vollset S.E. Biological and clinical implications of the MTHFR C677T polymorphism. Trends Pharmacol. Sci. 2001;22:195–201. doi: 10.1016/s0165-6147(00)01675-8. [DOI] [PubMed] [Google Scholar]
- Vainer A.S., Boiarskikh U.A., Voronina E.N., Selezneva I.A., Sinkina T.V. Polymorphic variants of folate metabolizing genes (C677T and A1298C MTHFR, C1420T SHMT1 and G1958A MTHFD) are not associated with the risk of breast cancer in West Siberian Region of Russia. Mol. Biol. (Mosk) 2010;44:816–823. [PubMed] [Google Scholar]
- Wallace B.C., Dahabreh I.J., Trikalinos T.A., Lau J., Trow P., Schmid C.H. Closing the gap between methodologists and end-users: R as a computational back-end. J. Stat. Softw. 2013;49:1–15. [Google Scholar]
- Wang Z.G., Cui W., Yang L.F., Zhu Y.Q., Wei W.H. Association of dietary intake of folate and MTHFR genotype with breast cancer risk. Genet. Mol. Res. 2014;13:5446–5451. doi: 10.4238/2014.July.24.24. [DOI] [PubMed] [Google Scholar]
- Weiwei Z., Liping C., Dequan L. Association between dietary intake of folate, vitamin B6, B12 & MTHFR, MTR genotype and breast cancer risk. Pak. J. Med. Sci. 2014;30:106–110. doi: 10.12669/pjms.301.4189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen Y.Y., Yang S.J., Zhang J.X., Chen X.Y. Methylenetetrahydrofolate reductase genetic polymorphisms and esophageal squamous cell carcinoma susceptibility: a meta-analysis of case–control studies. Asian Pac. J. Cancer Prev. 2013;14:21–25. doi: 10.7314/apjcp.2013.14.1.21. [DOI] [PubMed] [Google Scholar]
- Wilcken B., Bamforth F., Li Z., Zhu H., Ritvanen A., Renlund M. Geographical and ethnic variation of the 677C > T allele of 5,10 methylenetetrahydrofolate reductase (MTHFR): findings from over 7000 newborns from 16 areas worldwide. J. Med. Genet. 2003;40(8):619–625. doi: 10.1136/jmg.40.8.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X.Y., Ni J., Xu W.J., Zhou T., Wang X. Interactions between MTHFR C677T-A1298C variants and folic acid deficiency affect breast cancer risk in a Chinese population. Asian Pac. J. Cancer Prev. 2012;13:2199–2206. doi: 10.7314/apjcp.2012.13.5.2199. [DOI] [PubMed] [Google Scholar]
- Wu Y., Yuan X., Zheng H. Northeast methylenetetrahydrofolate reductase gene c677t single nucleotide polymorphisms and susceptibility to breast cancer research. Mod. Oncol. 2010;18:2375–2378. [Google Scholar]
- Xu X., Gammon M.D., Zhang H., Wetmur J.G., Rao M. Polymorphisms of one-carbon-metabolizing genes and risk of breast cancer in a population-based study. Carcinogenesis. 2007;28:1504–1509. doi: 10.1093/carcin/bgm061. [DOI] [PubMed] [Google Scholar]
- Yadav U., Kumar P., Rai V. Global prevalence of MTHFR C677T gene polymorphism: a meta-analysis of population based studies. Indian J. Clin. Biochem. 2014;29(1):123–124. [Google Scholar]
- Yadav U., Kumar P., Yadav S.K., Mishra O.P., Rai V. Polymorphisms in folate metabolism genes as maternal risk factor for neural tube defects: an updated meta-analysis. Metab. Brain Dis. 2015;30:7–14. doi: 10.1007/s11011-014-9575-7. [DOI] [PubMed] [Google Scholar]
- Yang X., Lippman M.E. BRCA1 and BRCA2 in breast cancer. Breast Cancer Res. Treat. 1999;54:1–10. doi: 10.1023/a:1006189906896. [DOI] [PubMed] [Google Scholar]
- Yu L., Chen J. Association of MHTFR Ala222Val (rs1801133) polymorphism and breast cancer susceptibility: an update meta-analysis based on 51 research studies. Diagn. Pathol. 2012;7:171. doi: 10.1186/1746-1596-7-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C.P., Wu M.H., Chou Y.C., Yang T., You S.L., Chen C.J. Breast cancer risk associated with multigenotypic polymorphisms in folate metabolizing genes: a nested case–control study in Taiwan. Anticancer Res. 2007;27:1727–1732. [PubMed] [Google Scholar]
- Yuan H., Xu X.Y., Wang Z.L. The relation between polymorphisms of methylenetetrahydrofolate reductase C677T and the risk of breast cancer. J. MuDan Jiang Med. Univ. 2009;30:2–4. [Google Scholar]
- Zhang S., Hunter D.J., Hankinson S.E., Giovannucci E.L., Rosner B.A. A prospective study of folate intake and the risk of breast cancer. J. Am. Med. Assoc. 1999;281:1632–1637. doi: 10.1001/jama.281.17.1632. [DOI] [PubMed] [Google Scholar]
- Zhang T., Lou J., Zhong R., Wu J., Zou L., Sun Y. Genetic variants in the folate pathway and the risk of neural tube defects: a meta-analysis of the published literature. PLoS ONE. 2013;8:e59570. doi: 10.1371/journal.pone.0059570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Qiu L.X., Wang Z.H., Wu X.H., Liu X.J. MTHFR C677T polymorphism associated with breast cancer susceptibility: a meta-analysis involving 15,260 cases and 20,411 controls. Breast Cancer Res. Treat. 2010;123:549–555. doi: 10.1007/s10549-010-0783-5. [DOI] [PubMed] [Google Scholar]
- Zhang W.B., Zhang J.H., Pan Z.Q., Yang Q.S., Liu B. The MTHFR C677T polymorphism and prostate cancer risk: new findings from a meta-analysis of 7306 cases and 8062 controls. Asian Pac. J. Cancer Prev. 2012;13:2597–2604. doi: 10.7314/apjcp.2012.13.6.2597. [DOI] [PubMed] [Google Scholar]
- Zintzaras E. Methylenetetrahydrofolate reductase gene and susceptibility to breast cancer: a meta-analysis. Clin. Genet. 2006;69:327–336. doi: 10.1111/j.1399-0004.2006.00605.x. [DOI] [PubMed] [Google Scholar]