Abstract
Glioblastoma (GBM) is the most frequent and malignant form of primary brain tumor. GBM is essentially incurable and its resistance to therapy is attributed to a subpopulation of cells called glioma stem cells (GSCs). To meet the present shortage of relevant GBM cell (GC) lines we developed a library of annotated and validated cell lines derived from surgical samples of GBM patients, maintained under conditions to preserve GSC characteristics. This collection, which we call the Human Glioblastoma Cell Culture (HGCC) resource, consists of a biobank of 48 GC lines and an associated database containing high-resolution molecular data. We demonstrate that the HGCC lines are tumorigenic, harbor genomic lesions characteristic of GBMs, and represent all four transcriptional subtypes. The HGCC panel provides an open resource for in vitro and in vivo modeling of a large part of GBM diversity useful to both basic and translational GBM research.
Keywords: Glioblastoma, Cell culture, Stem cell culture condition, Molecular subtype, Xenograft models
Highlights
-
•
The HGCC resource contains 48 annotated human GBM cell lines and an interactive database
-
•
The GBM cell lines are propagated in stem cell conditions and display GSC characteristics
-
•
The HGCC resource provides cell lines of all molecular (TCGA) subtypes
-
•
All data connected with the HGCC cell lines can be accessed at hgcc.se
1. Introduction
The prognosis for glioblastoma (GBM), the commonest primary malignant brain tumor in adults (Dolecek et al., 2012) is poor with a 1-year survival rate of 36.5% (Ostrom et al., 2014). Standard treatment involves surgery to remove as much of the tumor as possible, followed by combined radiation and chemotherapy with temozolomide (Stupp et al., 2005). GBM is characterized by pronounced invasiveness, as well as extensive intra- and inter-tumor heterogeneity (Patel et al., 2014, Sottoriva et al., 2013, Verhaak et al., 2010).
Transcript profiling in combination with analysis of genomic aberrations have revealed distinct molecular subtypes of GBM (Brennan et al., 2013, Phillips et al., 2006, Verhaak et al., 2010). The most commonly used classification is that presented by The Cancer Genome Atlas Research Network (TCGA), which describes four subtypes (Verhaak et al., 2010), i.e. Proneural, Classical, Mesenchymal and Neural GBM. These are defined primarily on the basis of their particular transcriptional signatures but can be associated, at least statistically, with distinctive genetic aberrations. Proneural tumors exhibit a higher frequency of PDGFRA or IDH1 mutations, as well as a G-CIMP+ (glioma-CpG island methylator phenotype) subgroup that displays global hypermethylation, which overlaps with IDH1 mutations. Patients with G-CIMP+ tumors are younger at the time of diagnosis and have a survival advantage (Brennan et al., 2013, Noushmehr et al., 2010). Classical tumors demonstrate high rates of EGFR amplification and homozygous deletions of CDKN2A; mesenchymal samples often harbor hemizygous deletions of NF1; whereas no distinctive mutations have yet been found in neural GBMs. Subtyping is complicated by the pronounced intratumor heterogeneity and different regions of one and the same tumor can be classified differently (Sottoriva et al., 2013). In addition, single-cell sequencing of cells from five patients has revealed a mixture of subtypes even at the cellular level in each individual patient (Patel et al., 2014).
There is a high demand for readily available and relevant cell models of GBM. For 30 years, several GC lines, including U87 (> 1900 citations in PubMed), U251 (> 1100 citations) and T98G (> 900 citations), have been employed extensively in this context, providing valuable knowledge about this type of tumor. However, these models are imperfect for several reasons. First, the serum-containing medium in which these standard cell lines are cultured alters both their genomes and transcriptomes and causes depletion of stem cell-like tumor cells (Lee et al., 2006). Secondly, tumors formed by injection of such cell lines into the brains of mice fail to develop the defining morphological features of GBM, such as diffuse infiltration into surrounding healthy tissue and microvascular proliferations (Lee et al., 2006, Mahesparan et al., 2003, Pontén and Macintyre, 1968, Westermark et al., 1973). And third, the lack of systematic clinical characterization of the tumors from which the current GC lines derive makes it impossible to correlate findings with these models to patient parameters.
There is ample evidence that a minor subset of GBM cells (GCs), denoted glioblastoma stem cells (GSCs) are likely responsible for relapse because they possess a unique capacity for growth and progression (Beier et al., 2007, Galli et al., 2004, Lathia et al., 2010, Ogden et al., 2008, Singh et al., 2004, Son et al., 2009) and are particularly resistant to therapy (Bao et al., 2006, Bleau et al., 2009, Chen et al., 2012). Research designed to improve GBM culture conditions has shown that GSCs can be readily cultured as spheres, utilizing the same conditions as for normal neural stem cells (Hemmati et al., 2003, Ignatova et al., 2002, Singh et al., 2003). Moreover, orthotopical transplantation of such spheres into mice generated secondary tumors that retained the features of the primary tumor (Galli et al., 2004), which was also the case after injection of adherent cultures of GSC (Fael Al-mayhani et al., 2009, Pollard et al., 2009). Recent extensive work characterizing orthotopic xenograft models showed that acutely transplanted patient-derived GCs mimicked well the histopathology, genomics and phenotypic properties of the corresponding patient's primary tumor (Joo et al., 2013). This study provides an important platform for accurate in vivo modeling of GBM but cannot fully meet the need for cell-based models.
Since GSCs cultured under stem cell conditions more accurately mirror GBM biology and because such models are increasingly in demand, we have created a novel library of well-characterized GC cultures that we make publicly available here. We describe the establishment and characterization of 48 sustainable GC lines, derived from Swedish patients during the period of 2009–2012, and including all four molecular subtypes, a biobank we refer to as the Human Glioblastoma Cell Culture (HGCC) resource. This information, along with clinical variables, is also available online (www.hgcc.se). The utility of this database is reflected in the fact that several of these cell lines have already been shared and used to discover a novel potent candidate drug for treatment of GBM (Kitambi et al., 2014), as well as in a number of other studies (Wee et al., 2014, Babateen et al., 2015, Schmidt et al., 2013, Savary et al., 2013, Yu et al., 2013).
2. Materials & Methods
2.1. GBM Patients and Glioma Cell Cultures
Surgical specimens and clinical records for 102 adult patients with glioma were obtained from Uppsala University Hospital in accordance with protocols approved by the regional ethical review board and after obtaining written consent by all of the patients. Most of the tumor specimens were obtained directly from the operating theater, but in some cases from Clinical Pathology. Following World Health Organization (WHO) guidelines (Louis et al., 2007) neuropathologists classified the tumors as grades II–IV. The surgical samples were rendered anonymous and coded. A piece of each was stored at − 70 °C for later RNA extraction and another piece fixed with formalin and embedded in paraffin for histological analysis. The remainder of the specimen was explanted as described in detail in the Extended Experimental Procedures.
2.2. Analysis of Global Gene Expression and Classification of the Molecular Subtype of the GC Lines
Total RNA extracted from 48 GC lines using the RNeasy Mini kit (Qiagen) was labeled and hybridized on Affymetrix GeneChip Human Exon 1.0 ST arrays. Expression levels were RMA-normalized employing the Affymetrix Expression Console software. The GC lines were classified with the k-nearest neighbor approach, and bootstrap aggregation in which the classification was repeated 1000 times, each time using a subsampled version of the TCGA dataset (529 randomly selected cases, sampled with replacement from the original dataset). Isomap analysis was applied to visualize the GC lines and TCGA samples in two dimensions. The details of data analysis are provided in the Extended Experimental Procedures. Expression data is made available via NCBI Gene Expression Omnibus (GSE72217) and hgcc.se.
2.3. Analysis of Gene Expression by NanoString Technology and Assignment of a Subtype to the Surgical Samples and GC Lines
To determine molecular subtypes, RNA extracted from 22 specimens of fresh frozen human glioma using TRIzol and from the corresponding GC lines in the same manner as described above, was used in a custom-made assay by NanoString Technology. For further details, see the Extended Experimental Procedures.
2.4. Proliferation Assay
The proliferation of 13 GC lines was assessed by the AlamarBlue assay (Invitrogen) and that of 18 other lines by Trypan blue exclusion on Countess Cell Counting Chamber Slides (Invitrogen). See the Extended Experimental Procedures for additional details.
2.5. Analysis of the In Vivo Tumorigenicity of the GC Lines
All animal experiments were performed in accordance with the rules and regulations of Uppsala University and approved by the local animal ethics committee. Neonatal non-obese diabetic-severe combined immunodeficiency (NOD-SCID) mice (P1–3) were injected intracerebrally with 1.0 × 105 human GCs, as summarized in Table S6. The mice were sacrificed when they showed symptoms or otherwise 20 weeks after injection and their brains were analyzed for xenograft tumors. See the Extended Experimental Procedures for additional details.
2.6. Analysis of Aberrations in DNA Copy Number
DNA isolated from 48 GC lines using the DNeasy blood and tissue kit (Qiagen) was profiled on Affymetrix Cytoscan arrays at the Uppsala Academic Hospital Array and Analysis facility, in accordance with the manufacturer's instructions. Identification of segments carrying altered numbers of copies was achieved with the Patchwork R package (Mayrhofer et al., 2013), which quantifies the log-relative change in DNA content for each chromosomal region. CNA data is made available via NCBI Gene Expression Omnibus (GSE72209) and hgcc.se. See the Extended Experimental Procedures for additional details.
2.7. Subtype Stability In Vitro and In Vivo
RNA was prepared from cells and tumor tissue and analyzed on the Affymetrix HTA 2.0 platform. See the Extended Experimental Procedures for additional details. Expression data is made available via NCBI Gene Expression Omnibus (GSE72218) and hgcc.se.
2.8. Statistical Analyses
The unpaired t-test was used for comparison of two groups, a one-way ANOVA for comparison of more than two groups; and the log-rank test for survival curves. Statistical significance was defined as p ≤ 0.05. Cox regression was performed using the package rms in R 3.1.2. All statistical analyses were carried out with the GraphPad Prism6.0 software.
3. Results
3.1. The Cohort of Patients and In Vitro Cell Cultures
From 101 patients (including 13 duplicate samples from the same individuals and 9 relapse samples) who underwent surgery for suspected high-grade glioma, 114 surgical specimens were explanted (Table S1). Following dissociation, the tumor cells were first cultured as spheres in defined serum-free media and after 5–7 days transferred to laminin-coated dishes for further propagation as adherent monolayer cultures. In connection with both of these steps, the pattern of growth of cells from different patients varied extensively. Most of the primary cell cultures did not readily form spheres, but instead formed smaller aggregates of cells, many of which could still be effectively expanded on laminin. Most cultures that ceased to grow did so early in the process, usually not even producing an adherent culture that could be subjected to the first passage. After a culture had undergone 8–10 passages and continued to proliferate, it was considered a sustainable GC line. Many have now undergone 20 passages or more. From 94 GBM surgical specimens (82 patients) 53 GC lines were established. Two parallel lines were produced from each of two GBM patients (U3084MG and U3086MG; U3117MG and U3118MG), and one (U3054MG) was established from a GBM that recurred after the patient had undergone surgery and radiotherapy (50.4 Gy in total). We also established one cell line from a grade III tumor but none from grade II tumors.
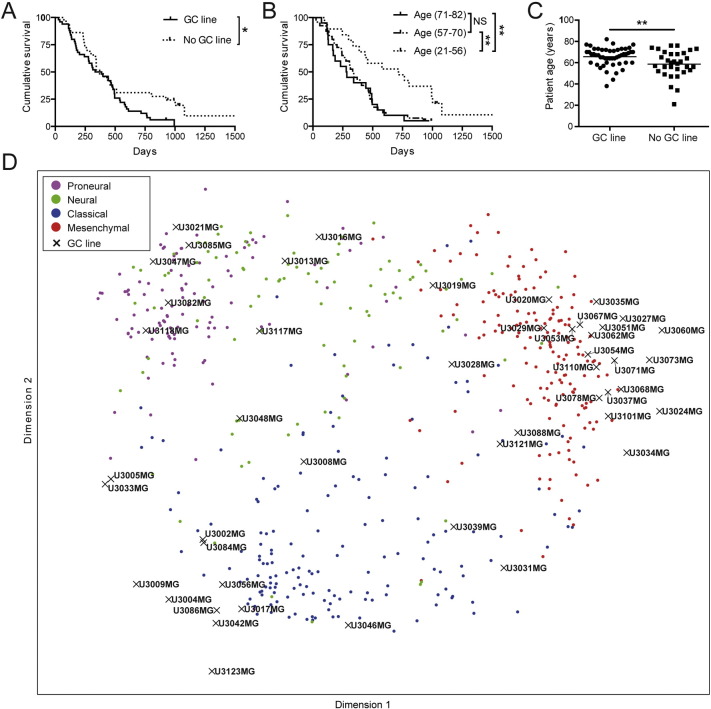
Here, we describe 48 of the GBM-derived GC lines obtained (Table S2). Initially, for identification and future authentication, multiplex short tandem repeat (STR) profiling was performed on most of these (Table S2, Table S3). The ability of GCs to form renewable neurospheres has been correlated to shorter GBM patient survival (Laks et al., 2009). To evaluate if also the ability of GCs to be maintained as adherent cultures could be a predictor of survival we compared patients from which one or more GC lines could be established to those from which no cell line was produced. We found a significant correlation between successful establishment of a GC line from a patient's tumor and a shorter patient survival (Fig. 1A). It is well established that age at the time of diagnosis is a major determinant of GBM patient survival (Brennan et al., 2013) and to investigate if this also holds true for our patient cohort all GBM patients were divided by quartiles based on age, and survival was compared between the youngest patients below the first quartile, the intermediate patients between the first and third quartiles, and the oldest patients above the third quartile. As expected, our younger GBM patients survived significantly longer (Fig. 1B). The age of patients from whose tumors a GC line could be established was significantly higher (Fig. 1C), and, accordingly, a Cox multivariate analysis of both these parameters (ability to be propagated in culture and age) showed that only age was a significant predictor of survival. This is an interesting observation and could indicate that shorter survival in the older population is due to inherently more aggressive tumor cells rather than factors such as differences in the extent of resection or type of treatment, although the influence by changes in the microenvironment due to aging cannot be excluded.
Fig. 1.
The GBM patient cohort: Molecular diversity and correlations between establishment of GC lines, survival and age. (A) Kaplan–Meier comparison of the survival of patients with GBM from whom sustainable cell lines could (solid line) and could not (dotted line) be established. Log-rank test, *p < 0.05. (B) Kaplan–Meier comparison of survival versus age. The oldest patients (71–82 years old, average 74) are above the third quartile, the intermediate ones (57–70, average 64) between first and third quartiles, and the youngest (21–56 years old, average 48) below the first quartile. Log-rank test, **p < 0.01. (C) Age distribution of the patients from whom a sustainable GC line could (average age 66 years) and could not (average age 59 years) be established. Student's t-test, **p < 0.01. (D) Isomap analysis of 48 GC lines and 529 TCGA tissue samples of known molecular subtypes.
3.2. Subtyping of the GC Lines and Tumor Samples From Which They Were Derived
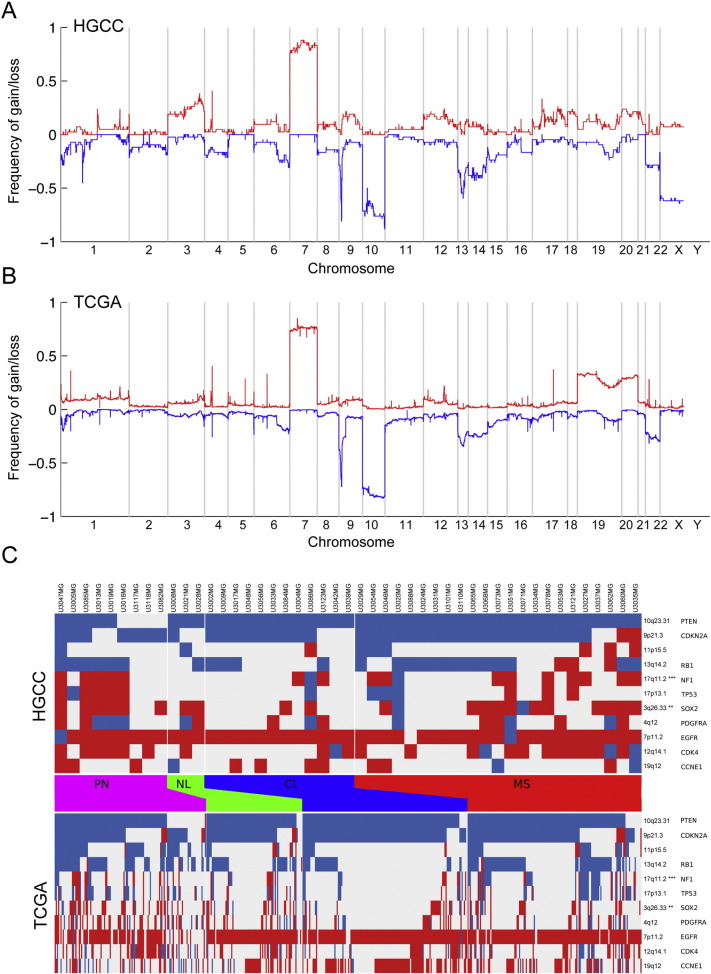
Global gene expression (employing Affymetrix GeneChip Human Exon 1.0ST Arrays) followed by subtype classification was performed on 48 of the GC lines. In a first unsupervised analysis we combined z-scores for mRNA expression levels for 765 subtype classification genes (Verhaak et al., 2010) across 529 TCGA tissue samples and our 48 GC lines, and mapped them into a 2-dimensional space using Isomap, a dimensionality reduction technique applicable to grouping of cancer samples (Nilsson et al., 2004, Tenenbaum et al., 2000). The resulting Isomap (Fig. 1D) confirmed a clear separation of the Classical, Mesenchymal and Proneural TCGA subtypes and showed that our GC lines with similar transcriptional profiles were interspersed among them, suggesting that these three subtypes are well represented in our collection.
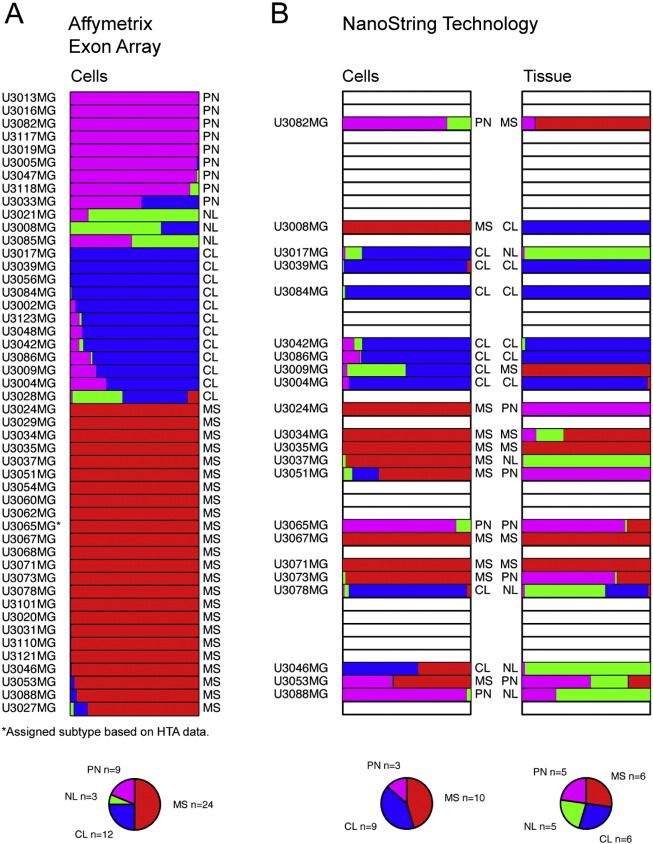
We then classified each individual GC line as Proneural, Classical, Mesenchymal or Neural using the TCGA cohort of GBM (n = 529) as a reference. For this purpose, we applied a supervised classification method, the k-nearest neighbor algorithm (KNN) to assign each line to a class (Fig. 2A). To assess the stability of these assignments we utilized statistical bootstrapping (James et al., 2013); applied in this context, bootstrapping means that we repeated the classification 1000 times for each cell line, each time using a subsampled version of the TCGA data set (Materials and Methods). With this simulation, 38 of the cell lines were assigned to the same classification at least 90% of the time (i.e. same subtype in at least 900 out of 1000 bootstrap simulations), whereas the classification of the remaining 10 varied. For instance, U3033MG was classified as intermediate between Classical and Proneural (Fig. 2A). Overall, the distribution of the cell lines between different subtypes was similar to the TCGA analysis of tumor tissue (Verhaak et al., 2010) with the most being Mesenchymal, followed by Classical, Proneural, and Neural in that order. The cell lines derived from two separate samples from the same patient (U3084MG and U3086MG; U3117MG and U3118MG) were of the same subtype. The cell line derived from a recurrent GBM (U3054MG) was classified as Mesenchymal.
Fig. 2.
Molecular subtyping of GC lines and their corresponding tumor samples. (A) Forty-eight GC lines were analyzed by Affymetrix GeneChip Human Exon 1.0ST Arrays, followed by subtyping (indicated to the right of each bar) by applying a k-nearest neighbor classification in combination with bootstrapping. The variation in the fraction of times each cell line classified as Mesenchymal (MS, red bars), Neural (NL, green bars), Proneural (PN, magenta bars) or Classical (CL, blue bars) reflects the uncertainty of the assignment. The subtype assignment of each GC line is indicated to the right of each bar. * = sample that failed to be analyzed on the Exon Array, the assigned subtype is based on gene expression analyzed on Human Transcriptome Array (see Fig. 7B). (B) Comparison of the predicted subtypes of 22 GC lines and the corresponding tumor tissue based on gene expression analysis by NanoString Technology. The fraction of times each cell line or tissue sample was assigned to the Mesenchymal (MS, red bars), Neural (NL, green bars), Proneural (PN, purple bars) or Classical (CL, blue bars) subtype is depicted and the final assignment indicated to the right or left of the bars, respectively. See also Table S2.
For one subset of samples (n = 22) we analyzed the molecular subtype of both the primary tumor and the corresponding GC line (Fig. 2B). Since the tissue RNA was too degraded (due to long handling times in connection with surgery) for microarray hybridization, we assessed gene expression employing NanoString Technology and a custom set of probes (Table S4) designed by Drs. Cameron Brennan and Jason Huse at the Brain Tumor Center, Memorial Sloan-Kettering Cancer Center (Kastenhuber et al., 2014). In 10 cases the tissue and corresponding GC line were of the same subtype. The differences in the remaining 12 cases could have many causes, including intratumoral heterogeneity (Patel et al., 2014, Sottoriva et al., 2013) and clonal selection during culturing (Meyer et al., 2015). The slight discrepancy of 23% in classification of GC lines when comparing the results from the Affymetrix and NanoString Technology approaches probably reflects the different gene signatures used. This difference is modest and as a benchmark we also classified the TCGA GBM samples for which there were data from two different platforms (Affymetrix and Agilent), which showed a discrepancy of 29% between the two platforms although the same gene signature was used in this case (Fig. S1). Altogether, these analyses demonstrate that much of the inter-tumoral GBM heterogeneity can be modeled by using neural stem cell culture conditions. The subtype that we finally assigned each GC line and use hereafter is based on the Affymetrix array (Fig. 2A).
Molecular subtyping of GBM tissue has proven to be of only limited value in predicting patient survival, with the most striking difference being a less favorable survival 15 months after diagnosis for patients with G-CIMP+ and IDH1 mutated Proneural GBM in comparison to all other GBM patients (Brennan et al., 2013). Analysis of our patients revealed a trend towards worse survival among patients from whose tumors GCs of the Proneural subtype were obtained (Fig. 3A). This observation suggests that most of our Proneural GC lines were derived from GBMs carrying wildtype IDH1, in line with a previous report that showed that G-CIMP+ GCs survive poorly under defined culture conditions (Balvers et al., 2013). Indeed, exome sequencing of our cell lines did not detect any mutations at codon R132, the most frequently mutated site in the IDH1 (Weller et al., 2011) (Table S1, Table S5), indicating that all of our lines are G-CIMP− (Noushmehr et al., 2010).
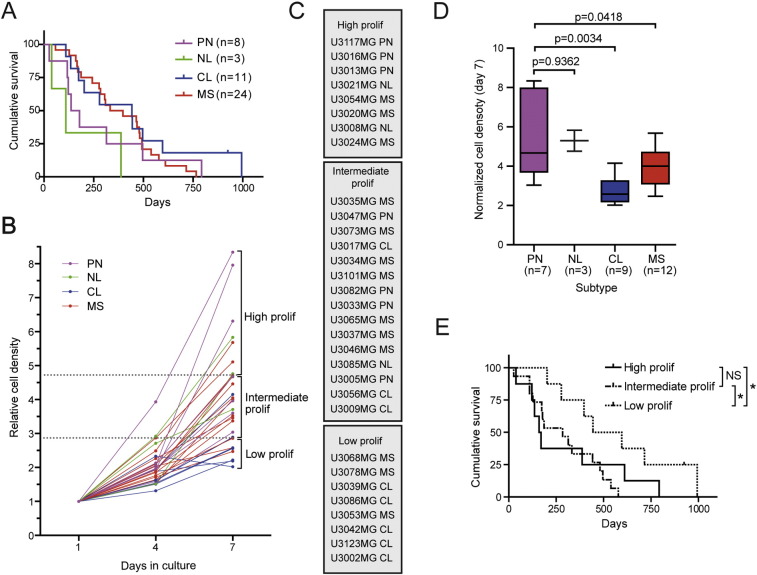
Fig. 3.
Proliferation capacity of GC lines in relation to subtype and survival. (A) Kaplan–Meier comparing the survival of patients in relationship to the TCGA molecular subtype of the GC lines established from their tumors. (B) Growth curves for 31 GC lines of differing TCGA molecular subtypes. Based on their proliferative capacity on day 7, the cell lines below the first quartile were designated as “low proliferation”, between the first and third quartiles as “intermediate proliferation”, and above the third quartile as “high proliferation”. (C) The GC lines categorized were listed in the order of highest to lowest proliferative capacity. (D) Analysis (Student's t-test) of the proliferative capacity of GC lines in relationship to their TCGA molecular subtype. (E) Kaplan–Meier plot comparing the survival of patients with GBM in relationship to the proliferative capacity (high, intermediate, and low) of the corresponding GC lines. Log-rank test, *p < 0.05.
3.3. A Low Rate of Proliferation Predicts Better Survival
We evaluated the growth rates of 31 GC lines by measuring cell density and normalized the results from days 4 and 7 to that on day 1 (Fig. 3B). On the basis of their proliferative capacity on day 7, cell lines below the first quartile were designated “low proliferation”, between the first and third quartiles “intermediate proliferation”, and above the third quartile “high proliferation” (Fig. 3B), and the cell lines in these groups are listed from the highest to the lowest proliferative capacity in Fig. 3C. The Proneural GC lines grew more rapidly than the other three subtypes, while the Classical lines grew most slowly (Fig. 3D). Moreover, we found that patients whose tumors gave rise to GC lines designated “low proliferation” exhibited significantly better survival than the “high” and “intermediate proliferation” groups (Fig. 3E), which were similar in this respect. There was no significant difference in age between these three groups, but when both age and proliferative capacity were included in a multivariate Cox-regression, only the effect of age remained significant, indicating that this factor is the most important predictor of survival.
3.4. All of the GC Lines Expressed SOX2 and NESTIN
Expression by 27 GC lines of the commonly used cell lineage markers SOX2 (sex determining region Y (SRY)-box 2), NESTIN, GFAP (glial fibrillary acidic protein), OLIG2 (oligodendrocyte transcription factor 2), S100B (S100 calcium binding protein B) and TUBB3 (beta 3 class III tubulin) was examined by immunostaining and Fig. 4A depicts representative staining patterns for one cell line of each subtype. SOX2, a marker of neural stem and progenitor cells (Graham et al., 2003), was expressed by almost all of the cells in all lines stained (Fig. 4B), indicating that an immature stem cell-like population of GCs had been propagated. NESTIN, a stem- and progenitor cell marker (Dahlstrand et al., 1992, Lendahl et al., 1990), was also expressed by most of the cells in all lines (Fig. 4C). While the expression of SOX2 and NESTIN was highly uniform, expression of GFAP (Fig. 4D) and OLIG2 (Fig. 4E) varied widely. Further, a large proportion of Mesenchymal lines show a lower expression of Olig2 (Fig. S2A–B). The neuronal lineage marker TUBB3 and the astrocytic marker S100B (Fig. 4A) were expressed to varying extents within and between the GC lines, being co-expressed in the same cells in approximately 80% of the cultures and thereby displaying aberrant lineage commitment, a feature characteristic of cancer stem cells (Galli et al., 2004).
Fig. 4.
Expression of neural and glial protein markers by the GC lines. (A) Representative double immunofluorescence staining of cultured cells for SOX2 and GFAP, OLIG2 and NESTIN, and S100B and TUBB3. Scale bar = 50 μm. (B–E). The percentage of cells in 27 GC lines staining positively for (B) SOX2, (C) NESTIN, (D) GFAP, and (E) OLIG2.
3.5. Tumorigenicity of the GC Lines
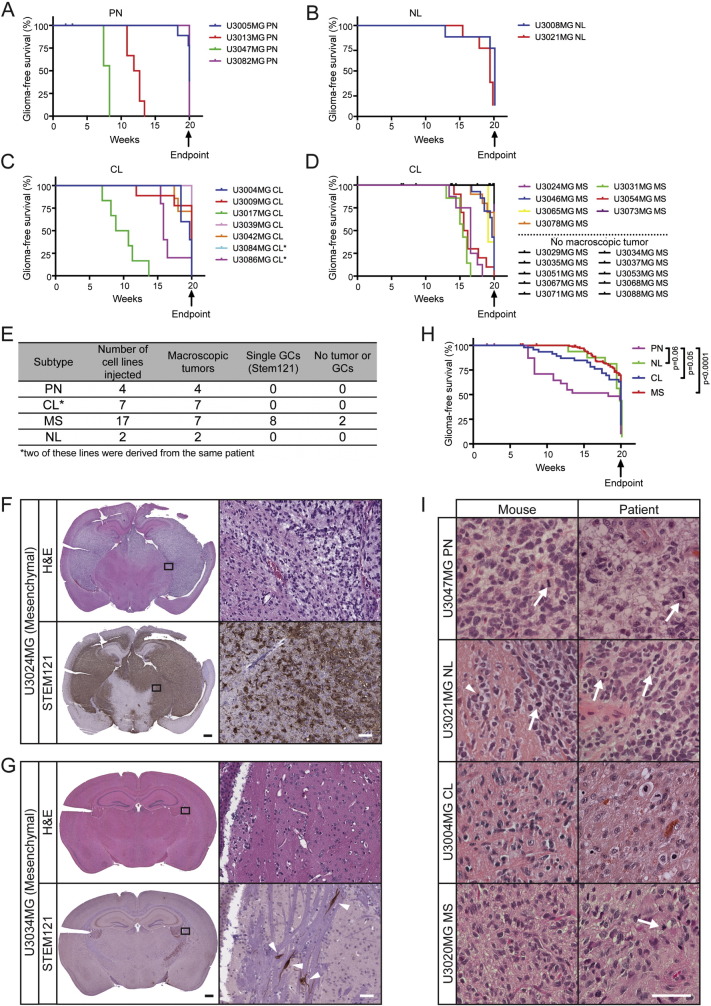
To test the capacity of our GC lines to produce secondary tumors, 1 × 105 cells from 30 different GC lines (of which two, U3084MG and U3086MG were derived from the same patient) were injected intracranially into NOD-SCID mice (Table S6). These animals were sacrificed when they showed symptoms, or, otherwise, at 20 weeks after the injection (Fig. 5A–D). Interestingly, all of the Proneural, Neural and Classical cell lines gave rise to macroscopic tumors that could be visualized by hematoxylin and eosin (H&E) staining, whereas only 7 out of the 17 Mesenchymal cell lines generated such tumors (Fig. 5D–E). To distinguish xenografted human tumor cells from murine cells we stained tissue sections with the human-specific STEM121 antibody (Fig. 5F–G and S3A–C, lower panels), and found that this staining overlapped well with the H&E staining (Figs. 5F, S3A–C). At the same time we could detect single human GCs in the brains of the mice injected with 8 of the cell lines that did not generate macroscopic tumors (Fig. 5G, lower right panel).
Fig. 5.
The tumorigenicity of the GC lines. (A–D) Kaplan–Meier graphs illustrating the survival of NOD-SCID mice injected intracranially with GC lines of the (A) Proneural (PN), (B) Neural (NL), (C) Classical (CL) and (D) Mesenchymal (MS) subtypes. (E) Summary of tumor formation and staining for STEM121 (human-specific). (F–G) H&E staining and immunostaining for STEM121 in the brains of mice injected with the U3024MG (F) or U3034MG (G) Mesenchymal GC lines. Black scale bar = 500 μm, white scale bar = 50 μm. (H) Kaplan–Meier graph comparing survival of NOD-SCID mice injected with GC lines of different subtypes. (I) H&E stainings of secondary tumors in NOD-SCID mice (left column) and the human tumor from which the cell line was derived (right column). Scale bar = 50 μm. See also Fig. S3.
Mice with tumors caused by Proneural GC lines displayed the shortest survival compared to mice injected with cells of the other subtypes (Fig. 5H), with the most significant difference being to brain tumors caused by Mesenchymal GC lines. This goes well in line with the higher proliferative capacity of Proneural lines (Fig. 3D) and the reduced survival of non-G-CIMP Proneural patients (Brennan et al., 2013), suggesting a more aggressive GC phenotype in non-G-CIMP Proneural GBM.
Twenty of the 30 GC lines gave rise to tumors that were often large, cell-dense, diffuse and invasive within 20 weeks (Figs. 5F and S3A–C). Their histopathology was similar to that of gliomas grades III–IV with characteristic features such as cellular pleomorphism (Fig. S3D–G), atypical nuclei (Fig. S3D–G), diffuse infiltration (Figs. S3A, S3C, S3D), mitotic figures (Fig. S3E), increased vascularity (Fig. S3F) and in some cases, pseudopalisading necroses (Fig. S3G). Some had spread diffusely throughout the brain parenchyma (Fig. S3A), while others grew expansively in nodular patterns, clearly demarcated from the normal parenchyma (Fig. S3B). Although there were such differences between GC lines, individual tumors derived from the same cell line exhibited highly similar histopathology. These differences in tumorigenicity and histopathology most likely reflect the extensive heterogeneity of GBMs and indicate that we can maintain their distinct properties both in vitro and in vivo.
The histopathology of a subset of these secondary tumors in NOD-SCID mice was compared to that of the corresponding primary patient tumor (Fig. 5I). Mitotic tumor cells (white arrows), and apoptotic cells with condensed nuclei (white arrowheads) were numerous in both types of tumors. With certain GC lines (e.g. U3047MG, U3021MG and U3020MG) we observed striking similarities between the human GBM and corresponding mouse tumor while others (e.g. U3004MG) were less similar (Fig. 5I). To summarize, the xenografted tumors in mice had the hallmarks of high-grade gliomas but did not always display the same histopathology as the primary tumor from which the GC line was isolated.
3.6. Genomic Similarity of the GC Lines to the TCGA GBM Cohort
Analysis of copy number aberration (CNA) in all of our 48 GC lines employing Affymetrix Cytoscan HD arrays containing more than 2.6 million probes revealed changes characteristic of glioblastoma, such as chromosome 7 gains and chromosome 10 losses, present in more than 70% of the lines (Fig. 6A). This pattern was strikingly similar to that of the tumors in the TCGA cohort (Fig. 6B, Pearson correlation coefficient 0.72 across the genome) indicating that, in terms of genomic aberrations, our GC lines constitute a diverse and representative sampling of human GBM. Focusing on regions known to be recurrently altered in GBM, we analyzed the amplification and deletion frequency in the significantly altered regions reported by Brennan et al. (Brennan et al., 2013). Regions containing the CDKN2A, EGFR and PDGFRA loci, showed no significant difference in aberration frequency (Fig. 6C). Loci with a significant difference in alteration frequency included 3q26.33 (containing SOX2) which was more frequently amplified in GC lines than in the TCGA material (p < 0.01) and 17q11.2 (containing NF1) which was also more frequently amplified (p < 0.001) (Fig. 6C). This discrepancy could reflect differences between the sample populations, or an enrichment of tumor cells in HGCC cultures. The HGCC and TCGA CNA patterns were largely similar at the level of individual subtypes (Table S7), with significant differences only in 17q11.2 (p < 0.01), 3q13.31 (p < 0.05) and 3p21.31 (p < 0.05) which were more frequently amplified in HGCC Mesenchymal samples when compared to TCGA Mesenchymal samples (Table S8). The fact that most of the Mesenchymal GC lines did not form intracranial tumors in the NOD-SCID mice raised the concern that many of these lines might have originated from normal (non-cancerous) cells. To examine this possibility, we compared the CNA data for the GC lines that could (Fig. S4A) or could not (Fig. S4B) form macroscopic tumors and found highly similar genomic aberrations between the groups, demonstrating that the GC lines that did not form tumors were nonetheless also cancer cells.
Fig. 6.
Genomic similarity of the GC lines to the TCGA GBM tissue samples. Genomic copy number variation analysis in (A) the 48 GC lines, and (B) 509 GBM samples from the TCGA tumor tissue cohort. (C) CNA variation in the GC lines of different subtypes shows similar trends as the TCGA data; the heat map shows amplifications (red) and deletions (blue) for a selected set of regions (rows) previously reported (Brennan et al., 2013). Significant difference (Chi-squared p-value, Holm correction) in alteration frequency was found for 3q26.33 (containing the SOX2 locus, p < 0.01) and 17q11.2 (containing NF1, p < 0.001) which was amplified at a higher frequency in our GC lines compared to TCGA.
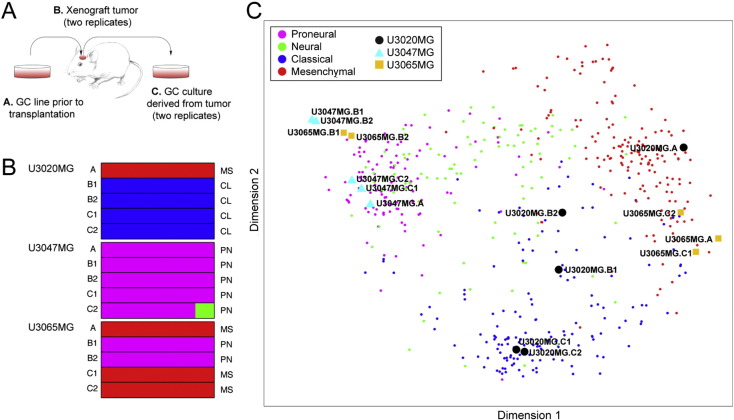
3.7. Subtype Stability of the GC Lines In Vitro and In Vivo
To explore the degree to which the GC lines remained transcriptionally stable under diverse experimental conditions, we transplanted three different lines (U3020MG, U3047MG and U3065MG) intracranially to NOD-SCID mice; explanted the resulting tumors and cultured the cells for two passages, and then isolated RNA from the cell line prior to transplantation (U3020MG-p10, U3047MG-p7, U3065MG-p10), from the xenograft tumor and from the explanted cells (Fig. 7A). Transcript profiling followed by molecular subtype analysis (Fig. 7B) and Isomap representation (Fig. 7C) revealed that the Proneural U3047MG line remained Proneural throughout the entire experiment (Fig. 7B–C). The Mesenchymal U3020MG line changed to the Classical subtype in the xenograft tumor and remained Classical in the explanted cells (Fig. 7B–C). The other Mesenchymal line, U3065MG, changed to the Proneural phenotype in the xenograft tumor, but reverted to Mesenchymal the explanted GCs (Fig. 7B–C).
Fig. 7.
Stability of the molecular subtypes in GC cell line culture and in vivo. (A) Schematic overview of the procedure. (B) Molecular subtype based on gene expression as determined with the Affymetrix Human Transcriptome Array 2.0. The proportion of times each cell line or tissue was assigned to the Mesenchymal (MS, red bars), Neural (NL, green bars), Proneural (PN, magenta bars) or Classical (CL, blue bars) subtype is depicted by the bars, and the final assignment is denoted to the right of the bars. (C) Isomap of samples from the analysis of subtype stability with 529 TCGA GBM tissue samples as reference.
Altogether, these findings confirm to some extent the data by Bhat et al. where Proneural GCs were found to be more transcriptionally stable than Mesenchymal GCs when transferred from culture to xenograft (Bhat et al., 2013). The drift in subtype could reflect a clonal selection similar to the mechanism of developing drug resistance (Bhang et al., 2015), but could also be due to a differential sensitivity of the cells to respond to a new microenvironment, in particular in the case of U3065MG, since GCs were only cultured for two passages after the xenograft tumor was explanted. Systematic analysis of the plasticity and genetic diversity of our GC lines will be the subject of future work.
4. Discussion
The development and pre-clinical validation of novel anti-cancer drugs require scalable, representative, and reproducible experimental models, typically established cell lines. Current large-scale efforts in this connection include the collection of NCI-60 human tumor cell lines (containing 6 GBM lines) (Scherf et al., 2000), the translational Cancer Genomics program (Garnett et al., 2012), and the Broad-Novartis Cancer Cell Line Encyclopedia (CCLE) (Barretina et al., 2012) which includes 62 GBM samples. All these initiatives involve cultures grown in serum, which do not demonstrate the phenotypic properties of the tumor of origin. Furthermore, the origin of several of the cell lines has been questioned; in several instances STR profiling has revealed cross contamination or misidentification of established cell lines, including several GBM lines (Capes-Davis et al., 2010). Finally, for most of the publically available cell lines associated clinical data are lacking, which further reduces their value.
Since the initial reports on culturing human GC lines in serum-free neural stem cell medium (Lee et al., 2006), several groups have demonstrated the usefulness of this approach, where the phenotype of the cultured cells resembles that of the tumor of origin (Balvers et al., 2013, Fael Al-mayhani et al., 2009, Meyer et al., 2015, Pollard et al., 2009). However, to date these cultures have not been established with the intention to be made publically available to researchers in general. Here, we have established a biobank of 48 cell lines derived from GBM tumors (the HGCC resource, Table S2), employing the neurosphere assay and the adherent monolayer protocol, two validated techniques for culturing GSCs. A large part of the HGCC panel is linked to the U-CAN biobank initiative (www.u-can.uu.se), which, in addition to providing samples of frozen tumor tissue and patient blood, makes annotated clinical data for each patient available.
As a first step, sphere formation to remove tissue debris and red blood cells, as well as to enrich for GSCs was utilized (Fael Al-Mayhani et al., 2009). Although this neurosphere technique has been successful in establishing cultures with stem cell characteristics from malignant brain tumors (Ignatova et al., 2002), it has certain limitations. If not closely monitored neurospheres grow too large and begin to differentiate (Suslov, 2002). Moreover, inadvertent fusion of spheres (Singec et al., 2006) can contribute to an undesirable heterogeneity in sphere size. Adherent cultures (Conti et al., 2005) allow the cells to be uniformly exposed to nutrients, oxygen and growth factors, in addition to monitoring of individual cells. Therefore, we subsequently plated the spheres onto laminin-coated dishes, thereby generating 53 expandable cell lines from 94 surgical GBM samples (Table S1), of which 48 were characterized in greater details (Table S2). The observation that patients whose tumors produced sustainable cultures exhibited poorer survival (Fig. 1A) shows that our culture conditions are appropriate for cells from the more aggressive tumors, although we also have cell lines from the tumors of long-term survivors (> 2 years). A future challenge will be to improve the development of sustainable cultures from younger patients in order to extend the coverage of GBM heterogeneity in our biobank. Achieving this goal probably require extensive modification of the culture medium and solid substrate.
With few exceptions, our GC lines could be reliably classified with respect to molecular subtype, with all four subtypes being represented at about the same proportions as in the TCGA database. Because of the extensive intra-tumor heterogeneity of GBM tumors, with different samples and even individual cells from the same tumor being of different subtypes (Sottoriva et al., 2013), we anticipated that our GC lines might differ phenotypically from the tumor of origin. Indeed, gene expression profiling revealed that only 45% of these lines displayed the same subtype as the tumor of origin, probably not only because of sample heterogeneity but also due to the presence of non-neoplastic cells (inflammatory cells, reactive astrocytes, vascular cells) in the tissue samples.
When subtype stability was tested in a small subset of GC lines (Fig. 7), two out of three were found to change their subtype following orthotopic xenotransplantation to NOD-SCID mice, although one of these reverted to the original subtype after explantation. Thus, in addition to local heterogeneity of cells of a particular subtype at any given timepoint, the profile of RNA expression within a given cell population can also alter. It is possible that in addition to tumor-stromal interactions, the tumor microenvironment may not only facilitate progression of GBM (Charles et al., 2011) but also influence classifier gene expression that leads to a shift in subtype.
All of the cells in our GC lines express the stem cell markers NESTIN and SOX2, indicating enrichment of GSCs in the cultures. This conclusion gains further support for the finding that most of the cell lines tested gave rise to large tumors that infiltrated the brain tissue, including the contralateral hemisphere. Since the invasive character of GBM is such a key factor in its dismal outcome, human cell lines that mimic this behavior in the mouse brain are potentially highly useful tools. Interestingly, most of the Mesenchymal GC lines injected did not give rise to macroscopic tumors after 20 weeks, even though these were confirmed to be tumor cells by CNA (Fig. S4). Instead, we detected viable human GCs, often diffusely spread, in most of the brains from these mice (Fig. 5G). The observation that all the cell lines that failed to produce a tumor were Mesenchymal suggests that this subtype may contain phenotypically distinct subgroups that should be explored. Other challenges will be to determine whether, given enough time, these scattered human GCs can give rise to GBM in mice, and whether they represent a distinct subset of the parental cell line.
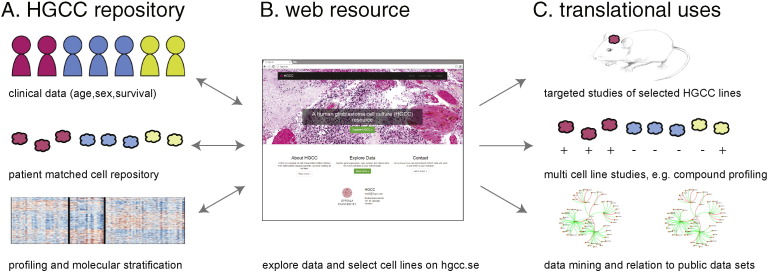
In summary, we have created a highly relevant biobank of human GC lines that retain the most prominent characteristics of their original tumors. Our goal is to make this an open-source repository that will enable stratified studies of disease mechanisms and facilitate the development of novel treatment strategies. All ethical, legal and practical matters regarding sharing the cell lines are in place. A number of the HGCC cell lines have already been used in different types of investigations ranging from basic understanding of GBM biology (Savary et al., 2013, Babateen et al., 2015, Wee et al., 2014), drug screening (Schmidt et al., 2013), drug discovery (Kitambi et al., 2014) and development of vectors for gene therapy (Yu et al., 2013). The HGCC resource enables accurate cell-based modeling of GBM and is connected with a database (www.hgcc.se), which provides current molecular, phenotypic and clinical data on each cell line in a searchable fashion (Fig. 8A–B). This website will be continuously updated with links to future datasets. Specific ways in which the cell repository can be used as a resource for translational studies include i) selection of individual lines for in vivo modeling of GBM, ii) the use of cell line panels for comparative testing of compounds or candidate genes; or, iii) integrated analysis of HGCC lines in conjunction with public datasets, for instance to relate variability in tumor cells to that obtained in tumor tissue repositories such as TCGA (Fig. 8C).
Fig. 8.
Overview of how the HGCC biobank will support translational brain tumor research. We provide a large-scale, open access repository of patient-derived GBM cell cultures matched with clinical data (A) that enables accurate cell-based modeling of GBM diversity. Coupled to the cell bank we make available a user-friendly data repository (B) to support users in their selection of HGCC lines with particular properties, molecular subtype or marker expression. We foresee a multitude of applications (C), such as single/oligo cell line studies of e.g. candidate genes and mechanisms in vitro and in vivo (xenograft modeling), multi-cell line studies, e.g. screening for inhibitory compounds or siRNAs, and data mining in relation to other publically available data sets.
GBM is only one of several types of cancer for which current therapy is inadequate and we hope that our present initiative will encourage other investigators to emulate and improve upon our model.
Author Contributions
Contributions are only specified for each first, second and last author. L.U., B.W., K.F.N., and S.N. conceived the project and designed experiments. Y.X., T.B., and Y.J. designed and performed in vitro and in vivo experiments and analyzed data. P.J., V.D.M., and S.N. performed molecular profiling, genomic characterization and data analyses. P.J. established the web resource. All first and last authors wrote the manuscript.
Acknowledgments
This study was supported by grants from the Swedish Cancer Society (#140787, #140743, #140678, #140385), the Swedish Research Council (#K2015-99X-20774-08-4, #K2014-61X-15274-07-4, #K2013-62X-21473-04-3), and the Knut and the Alice Wallenberg Foundation (#KAW2013.0280). We thank Drs. Cameron Brennan and Jason Huse, Memorial Sloan-Kettering Cancer Center, for sharing the NanoString probe set with us. Array measurements, normalization and copy number identification were performed at the SciLifeLab Uppsala Array and Analysis Facility. Sequencing of the IDH1 locus was performed at the National Genomics Infrastructure and SciLifeLab Uppsala Genome Center. The tissue slides were scanned at the SciLifeLab Tissue Profiling Facility in Uppsala.
Footnotes
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2015.08.026.
Contributor Information
Sven Nelander, Email: sven.nelander@igp.uu.se.
Lene Uhrbom, Email: lene.uhrbom@igp.uu.se.
Appendix A. Supplementary Data
Supplementary material.
References
- Babateen O., Jin Z., Bhandage A., Korol S.V., Westermark B., Forsberg Nilsson K., Uhrbom L., Smits A., Birnir B. Etomidate, propofol and diazepam potentiate GABA-evoked GABAA currents in a cell line derived from human glioblastoma. Eur. J. Pharmacol. 2015;748:101–107. doi: 10.1016/j.ejphar.2014.12.001. [DOI] [PubMed] [Google Scholar]
- Balvers R.K., Kleijn A., Kloezeman J.J., French P.J., Kremer A., Van Den Bent M.J., Dirven C.M., Leenstra S., Lamfers M.L. Serum-free culture success of glial tumors is related to specific molecular profiles and expression of extracellular matrix-associated gene modules. Neuro Oncol. 2013;15:1684–1695. doi: 10.1093/neuonc/not116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S., Wu Q., Mclendon R.E., Hao Y., Shi Q., Hjelmeland A.B., Dewhirst M.W., Bigner D.D., Rich J.N. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- Barretina J., Caponigro G., Stransky N., Venkatesan K., Margolin A.A., Kim S., Wilson C.J., Lehar J., Kryukov G.V., Sonkin D., Reddy A., Liu M., Murray L., Berger M.F., Monahan J.E., Morais P., Meltzer J., Korejwa A., Jane-Valbuena J., Mapa F.A., Thibault J., Bric-Furlong E., Raman P., Shipway A., Engels I.H., Cheng J., Yu G.K., Yu J., Aspesi P., Jr., De Silva M., Jagtap K., Jones M.D., Wang L., Hatton C., Palescandolo E., Gupta S., Mahan S., Sougnez C., Onofrio R.C., Liefeld T., Macconaill L., Winckler W., Reich M., Li N., Mesirov J.P., Gabriel S.B., Getz G., Ardlie K., Chan V., Myer V.E., Weber B.L., Porter J., Warmuth M., Finan P., Harris J.L., Meyerson M., Golub T.R., Morrissey M.P., Sellers W.R., Schlegel R., Garraway L.A. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier D., Hau P., Proescholdt M., Lohmeier A., Wischhusen J., Oefner P.J., Aigner L., Brawanski A., Bogdahn U., Beier C.P. CD133(+) and CD133(−) glioblastoma-derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 2007;67:4010–4015. doi: 10.1158/0008-5472.CAN-06-4180. [DOI] [PubMed] [Google Scholar]
- Bhang H.E.C., Ruddy D.A., Radhakrishna V.K., Caushi J.X., Zhao R., Hims M.M., Singh A.P., Kao I., Rakiec D., Shaw P., Balak M., Raza A., Ackley E., Keen N., Schlabach M.R., Palmer M., Leary R.J., Chiang D.Y., Sellers W.R., Michor F., Cooke V.G., Korn J.M., Stegmeier F. Studying clonal dynamics in response to cancer therapy using high-complexity barcoding. Nat. Med. 2015;21:440-448. doi: 10.1038/nm.3841. [DOI] [PubMed] [Google Scholar]
- Bhat K.P., Balasubramaniyan V., Vaillant B., Ezhilarasan R., Hummelink K., Hollingsworth F., Wani K., Heathcock L., James J.D., Goodman L.D., Conroy S., Long L., Lelic N., Wang S., Gumin J., Raj D., Kodama Y., Raghunathan A., Olar A., Joshi K., Pelloski C.E., Heimberger A., Kim S.H., Cahill D.P., Rao G., Den Dunnen W.F., Boddeke H.W., Phillips H.S., Nakano I., Lang F.F., Colman H., Sulman E.P., Aldape K. Mesenchymal differentiation mediated by NF-kappaB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24:331–346. doi: 10.1016/j.ccr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleau A.M., Hambardzumyan D., Ozawa T., Fomchenko E.I., Huse J.T., Brennan C.W., Holland E.C. PTEN/PI3K/Akt pathway regulates the side population phenotype and ABCG2 activity in glioma tumor stem-like cells. Cell Stem Cell. 2009;4:226–235. doi: 10.1016/j.stem.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan C.W., Verhaak R.G.W., McKenna A., Campos B., Noushmehr H., Salama S.R., Zheng S.Y., Chakravarty D., Sanborn J.Z., Berman S.H., Beroukhim R., Bernard B., WU C.J., Genovese G., Shmulevich I., Barnholtz-Sloan J., Zou L.H., Vegesna R., Shukla S.A., Ciriello G., Yung W.K., Zhang W., Sougnez C., Mikkelsen T., Aldape K., Bigner D.D., Van Meir E.G., Prados M., Sloan A., Black K.L., Eschbacher J., Finocchiaro G., Friedman W., Andrews D.W., Guha A., Iacocca M., O'neill B.P., Foltz G., Myers J., Weisenberger D.J., Penny R., Kucherlapati R., Perou C.M., Hayes D.N., Gibbs R., Marra M., Mills G.B., Lander E., Spellman P., Wilson R., Sander C., Weinstein J., Meyerson M., Gabriel S., Laird P.W., Haussler D., Getz G., Chin L., Network T.R. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capes-Davis A., Theodosopoulos G., Atkin I., Drexler H.G., Kohara A., Macleod R.A., Masters J.R., Nakamura Y., Reid Y.A., Reddel R.R., Freshney R.I. Check your cultures! A list of cross-contaminated or misidentified cell lines. Int. J. Cancer. 2010;127:1–8. doi: 10.1002/ijc.25242. [DOI] [PubMed] [Google Scholar]
- Charles N.A., Holland E.C., Gilbertson R., Glass R., Kettenmann H. The brain tumor microenvironment. Glia. 2011;59:1169–1180. doi: 10.1002/glia.21136. [DOI] [PubMed] [Google Scholar]
- Chen J., Li Y., Yu T.S., Mckay R.M., Burns D.K., Kernie S.G., Parada L.F. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488:522–526. doi: 10.1038/nature11287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conti L., Pollard S.M., Gorba T., Reitano E., Toselli M., Biella G., Sun Y., Sanzone S., Ying Q.-L., Cattaneo E., Smith A. Niche-independent symmetrical self-renewal of a mammalian tissue stem cell. PLoS Biol. 2005;3:e283. doi: 10.1371/journal.pbio.0030283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlstrand J., Collins V.P., Lendahl U. Expression of the class VI intermediate filament nestin in human central nervous system tumors. Cancer Res. 1992;52:5334–5341. [PubMed] [Google Scholar]
- Dolecek T.A., Propp J.M., Stroup N.E., Kruchko C. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2005–2009. Neuro-Oncology. 2012;14(Suppl. 5):v1–v49. doi: 10.1093/neuonc/nos218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fael Al-mayhani T.M., Ball S.L.R., Zhao J.-W., Fawcett J., Ichimura K., Collins P.V., Watts C. An efficient method for derivation and propagation of glioblastoma cell lines that conserves the molecular profile of their original tumours. J. Neurosci. Methods. 2009;176:192–199. doi: 10.1016/j.jneumeth.2008.07.022. [DOI] [PubMed] [Google Scholar]
- Galli R., Binda E., Orfanelli U., Cipelletti B., Gritti A., De Vitis S., Fiocco R., Foroni C., Dimeco F., Vescovi A. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 2004;64:7011–7021. doi: 10.1158/0008-5472.CAN-04-1364. [DOI] [PubMed] [Google Scholar]
- Garnett M.J., Edelman E.J., Heidorn S.J., Greenman C.D., Dastur A., Lau K.W., Greninger P., Thompson I.R., Luo X., Soares J., Liu Q., Iorio F., Surdez D., Chen L., Milano R.J., Bignell G.R., Tam A.T., Davies H., Stevenson J.A., Barthorpe S., Lutz S.R., Kogera F., Lawrence K., McLaren-Douglas A., Mitropoulos X., Mironenko T., Thi H., Richardson L., Zhou W., Jewitt F., Zhang T., O'Brien P., Boisvert J.L., Price S., Hur W., Yang W., Deng X., Butler A., Choi H.G., Chang J.W., Baselga J., Stamenkovic I., Engelman J.A., Sharma S.V., Delattre O., Saez-Rodriguez J., Gray N.S., Settleman J., Futreal P.A., Haber D.A., Stratton M.R., Ramaswamy S., McDermott U., Benes C.H. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham V., Khudyakov J., Ellis P., Pevny L. SOX2 functions to maintain neural progenitor identity. Neuron. 2003;39:749–765. doi: 10.1016/s0896-6273(03)00497-5. [DOI] [PubMed] [Google Scholar]
- Hemmati H.D., Nakano I., Lazareff J.A., Masterman-Smith M., Geschwind D.H., Bronner-Fraser M., Kornblum H.I. Cancerous stem cells can arise from pediatric brain tumors. Proc. Natl. Acad. Sci. U. S. A. 2003;100:15178–15183. doi: 10.1073/pnas.2036535100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ignatova T.N., Kukekov V.G., Laywell E.D., Suslov O.N., Vrionis F.D., Steindler D.A. Human cortical glial tumors contain neural stem-like cells expressing astroglial and neuronal markers in vitro. Glia. 2002;39:193–206. doi: 10.1002/glia.10094. [DOI] [PubMed] [Google Scholar]
- James G., Witten D., Hastie T., Tibshirani R. Springer; New York: 2013. An Introduction to Statistical Learning: With Applications in R. [Google Scholar]
- Joo K.M., Kim J., Jin J., Kim M., Seol H.J., Muradov J., Yang H., Choi Y.L., Park W.Y., Kong D.S., Lee J.I., Ko Y.H., Woo H.G., Lee J., Kim S., Nam D.H. Patient-specific orthotopic glioblastoma xenograft models recapitulate the histopathology and biology of human glioblastomas in situ. Cell Rep. 2013;3:260–273. doi: 10.1016/j.celrep.2012.12.013. [DOI] [PubMed] [Google Scholar]
- Kastenhuber E.R., Huse J.T., Berman S.H., Pedraza A., Zhang J., Suehara Y., Viale A., Cavatore M., Heguy A., Szerlip N., Ladanyi M., Brennan C.W. Quantitative assessment of intragenic receptor tyrosine kinase deletions in primary glioblastomas: their prevalence and molecular correlates. Acta Neuropathol. 2014;127:747–759. doi: 10.1007/s00401-013-1217-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitambi S.S., Toledo E.M., Usoskin D., Wee S., Harisankar A., Svensson R., Sigmundsson K., Kalderen C., Niklasson M., Kundu S., Aranda S., Westermark B., Uhrbom L., Andang M., Damberg P., Nelander S., Arenas E., Artursson P., Walfridsson J., Forsberg Nilsson K., Hammarstrom L.G., Ernfors P. Vulnerability of glioblastoma cells to catastrophic vacuolization and death induced by a small molecule. Cell. 2014;157:313–328. doi: 10.1016/j.cell.2014.02.021. [DOI] [PubMed] [Google Scholar]
- Laks D.R., Masterman-Smith M., Visnyei K., Angenieux B., Orozco N.M., Foran I., Yong W.H., Vinters H.V., Liau L.M., Lazareff J.A., Mischel P.S., Cloughesy T.F., Horvath S., Kornblum H.I. Neurosphere formation is an independent predictor of clinical outcome in malignant glioma. Stem Cells. 2009;27:980–987. doi: 10.1002/stem.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lathia J.D., Gallagher J., Heddleston J.M., Wang J., Eyler C.E., Macswords J., Wu Q., Vasanji A., Mclendon R.E., Hjelmeland A.B., Rich J.N. Integrin alpha 6 regulates glioblastoma stem cells. Cell Stem Cell. 2010;6:421–432. doi: 10.1016/j.stem.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Kotliarova S., Kotliarov Y., Li A., Su Q., Donin N.M., Pastorino S., Purow B.W., Christopher N., Zhang W., Park J.K., Fine H.A. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006;9:391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- Lendahl U., Zimmerman L.B., McKay R.D. CNS stem cells express a new class of intermediate filament protein. Cell. 1990;60:585–595. doi: 10.1016/0092-8674(90)90662-x. [DOI] [PubMed] [Google Scholar]
- Louis D.N., Ohgaki H., Wiestler O.D., Cavenee W.K., Burger P.C., Jouvet A., Scheithauer B.W., Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahesparan R., Read T.-A., Lund-Johansen M., Skaftnesmo K., Bjerkvig R., Engebraaten O. Expression of extracellular matrix components in a highly infiltrative in vivo glioma model. Acta Neuropathol. 2003;105:49–57. doi: 10.1007/s00401-002-0610-0. [DOI] [PubMed] [Google Scholar]
- Mayrhofer M., Dilorenzo S., Isaksson A. Patchwork: allele-specific copy number analysis of whole-genome sequenced tumor tissue. Genome Biol. 2013;14:R24. doi: 10.1186/gb-2013-14-3-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M., Reimand J., Lan X., Head R., Zhu X., Kushida M., Bayani J., Pressey J.C., Lionel A.C., Clarke I.D., Cusimano M., Squire J.A., Scherer S.W., Bernstein M., Woodin M.A., Bader G.D., Dirks P.B. Single cell-derived clonal analysis of human glioblastoma links functional and genomic heterogeneity. Proc. Natl. Acad. Sci. U. S. A. 2015;112:851–856. doi: 10.1073/pnas.1320611111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson J., Fioretos T., Hoglund M., Fontes M. Approximate geodesic distances reveal biologically relevant structures in microarray data. Bioinformatics. 2004;20:874–880. doi: 10.1093/bioinformatics/btg496. [DOI] [PubMed] [Google Scholar]
- Noushmehr H., Weisenberger D.J., Diefes K., Phillips H.S., Pujara K., Berman B.P., Pan F., Pelloski C.E., Sulman E.P., Bhat K.P., Verhaak R.G.W., Hoadley K.A., Hayes D.N., Perou C.M., Schmidt H.K., Ding L., Wilson R.K., Van Den Berg D., Shen H., Bengtsson H., Neuvial P., Cope L.M., Buckley J., Herman J.G., Baylin S.B., Laird P.W., Aldape K., Network C.G.A.R. Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17:510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogden A.T., Waziri A.E., Lochhead R.A., Fusco D., Lopez K., Ellis J.A., Kang J., Assanah M., Mckhann G.M., Sisti M.B., McCormick P.C., Canoll P., Bruce J.N. Identification of AB25 + CD133-tumor-initiating cells in adult human gliomas. Neurosurgery. 2008;62:505–515. doi: 10.1227/01.neu.0000316019.28421.95. [DOI] [PubMed] [Google Scholar]
- Ostrom Q.T., Gittleman H., Liao P., Rouse C., Chen Y., Dowling J., Wolinsky Y., Kruchko C., Barnholtz-Sloan J. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2007–2011. Neuro Oncol. 2014;Suppl. 4:iv1–iv63. doi: 10.1093/neuonc/nou223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A.P., Tirosh I., Trombetta J.J., Shalek A.K., Gillespie S.M., Wakimoto H., Cahill D.P., Nahed B.V., Curry W.T., Martuza R.L., Louis D.N., Rozenblatt-Rosen O., Suva M.L., Regev A., Bernstein B.E. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344:1396–1401. doi: 10.1126/science.1254257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips H.S., Kharbanda S., Chen R., Forrest W.F., Soriano R.H., Wu T.D., Misra A., Nigro J.M., Colman H., Soroceanu L., Williams P.M., Modrusan Z., Feuerstein B.G., Aldape K. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- Pollard S.M., Yoshikawa K., Clarke I.D., Danovi D., Stricker S., Russell R., Bayani J., Head R., Lee M., Bernstein M., Squire J.A., Smith A., Dirks P. Glioma stem cell lines expanded in adherent culture have tumor-specific phenotypes and are suitable for chemical and genetic screens. Cell Stem Cell. 2009;4:568–580. doi: 10.1016/j.stem.2009.03.014. [DOI] [PubMed] [Google Scholar]
- Pontén J., Macintyre E.H. Long term culture of normal and neoplastic human glia. Acta Pathol. Microbiol. Scand. 1968;74:465–486. doi: 10.1111/j.1699-0463.1968.tb03502.x. [DOI] [PubMed] [Google Scholar]
- Savary K., Caglayan D., Caja L., Tzavlaki K., Bin Nayeem S., Bergstrom T., Jiang Y., Uhrbom L., Forsberg-Nilsson K., Westermark B., Heldin C.H., Ferletta M., Moustakas A. Snail depletes the tumorigenic potential of glioblastoma. Oncogene. 2013;32:5409–5420. doi: 10.1038/onc.2013.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherf U., Ross D.T., Waltham M., Smith L.H., Lee J.K., Tanabe L., Kohn K.W., Reinhold W.C., Myers T.G., Andrews D.T., Scudiero D.A., Eisen M.B., Sausville E.A., Pommier Y., Botstein D., Brown P.O., Weinstein J.N. A gene expression database for the molecular pharmacology of cancer. Nat. Genet. 2000;24:236–244. doi: 10.1038/73439. [DOI] [PubMed] [Google Scholar]
- Schmidt L., Kling T., Monsefi N., Olsson M., Hansson C., Baskaran S., Lundgren B., Martens U., Haggblad M., Westermark B., Forsberg Nilsson K., Uhrbom L., Karlsson-Lindahl L., Gerlee P., Nelander S. Comparative drug pair screening across multiple glioblastoma cell lines reveals novel drug–drug interactions. Neuro Oncol. 2013;15:1469–1478. doi: 10.1093/neuonc/not111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singec I., Knoth R., Meyer R.P., Maciaczyk J., Volk B., Nikkhah G., Frotscher M., Snyder E.Y. Defining the actual sensitivity and specificity of the neurosphere assay in stem cell biology. Nat. Methods. 2006;3:801–806. doi: 10.1038/nmeth926. [DOI] [PubMed] [Google Scholar]
- Singh S.K., Clarke I.D., Terasaki M., Bonn V.E., Hawkins C., Squire J., Dirks P.B. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- Singh S.K., Hawkins C., Clarke I.D., Squire J.A., Bayani J., Hide T., Henkelman R.M., Cusimano M.D., Dirks P.B. Identification of human brain tumour initiating cells. Nature. 2004;432:396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- Son M.J., Woolard K., Nam D.-H., Lee J., Fine H.A. SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell. 2009;4:440–452. doi: 10.1016/j.stem.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sottoriva A., Spiteri I., Piccirillo S.G., Touloumis A., Collins V.P., Marioni J.C., Curtis C., Watts C., Tavare S. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc. Natl. Acad. Sci. U. S. A. 2013;110:4009–4014. doi: 10.1073/pnas.1219747110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupp R., Mason W.P., Van Den Bent M.J., Weller M., Fisher B., Taphoorn M.J.B., Belanger K., Brandes A.A., Marosi C., Bogdahn U., Curschmann J., Janzer R.C., Ludwin S.K., Gorlia T., Allgeier A., Lacombe D., Cairncross J.G., Eisenhauer E., Mirimanoff R.O., European Organisation For Research And Treatment Of Cancer Brain Tumor And Radiotherapy Groups & National Cancer Institute Of Canada Clinical Trials Group Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- Suslov O.N. Neural stem cell heterogeneity demonstrated by molecular phenotyping of clonal neurospheres. Proc. Natl. Acad. Sci. 2002;99:14506–14511. doi: 10.1073/pnas.212525299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenenbaum J.B., De Silva V., Langford J.C. A global geometric framework for nonlinear dimensionality reduction. Science. 2000;290:2319–2323. doi: 10.1126/science.290.5500.2319. [DOI] [PubMed] [Google Scholar]
- Verhaak R.G.W., Hoadley K.A., Purdom E., Wang V., Qi Y., Wilkerson M.D., Miller C.R., Ding L., Golub T., Mesirov J.P., Alexe G., Lawrence M., O'kelly M., Tamayo P., Weir B.A., Gabriel S., Winckler W., Gupta S., Jakkula L., Feiler H.S., Hodgson J.G., James C.D., Sarkaria J.N., Brennan C., Kahn A., Spellman P.T., Wilson R.K., Speed T.P., Gray J.W., Meyerson M., Getz G., Perou C.M., Hayes D.N. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee S., Niklasson M., Marinescu V.D., Segerman A., Schmidt L., Hermansson A., Dirks P., Forsberg-Nilsson K., Westermark B., Uhrbom L., Linnarsson S., Nelander S., Andang M. Selective calcium sensitivity in immature glioma cancer stem cells. PLoS One. 2014;9:e115698. doi: 10.1371/journal.pone.0115698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller M., Wick W., Von Deimling A. Isocitrate dehydrogenase mutations: a challenge to traditional views on the genesis and malignant progression of gliomas. Glia. 2011;59:1200–1204. doi: 10.1002/glia.21130. [DOI] [PubMed] [Google Scholar]
- Westermark B., Ponten J., Hugosson R. Determinants for the establishment of permanent tissue culture lines from human gliomas. Acta Pathol. Microbiol. Scand. A. 1973;81:791–805. doi: 10.1111/j.1699-0463.1973.tb03573.x. [DOI] [PubMed] [Google Scholar]
- Yu D., Jin C., Ramachandran M., Xu J., Nilsson B., Korsgren O., Le Blanc K., Uhrbom L., Forsberg-Nilsson K., Westermark B., Adamson R., Maitland N., Fan X.L., Essand M. Adenovirus serotype 5 vectors with Tat-PTD modified hexon and serotype 35 fiber show greatly enhanced transduction capacity of primary cell cultures. PLoS One. 2013;8 doi: 10.1371/journal.pone.0054952. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material.