Abstract
Congenital sodium diarrhea (CSD) refers to an intractable diarrhea of intrauterine onset with high fecal sodium loss. CSD is clinically and genetically heterogeneous. Syndromic CSD is caused by SPINT2 mutations. While we recently described four cases of the non-syndromic form of CSD that were caused by dominant activating mutations in intestinal receptor guanylate cyclase C (GC-C), the genetic cause for the majority of CSD is still unknown. Therefore, we aimed to determine the genetic cause for non-GC-C non-syndromic CSD in 18 patients from 16 unrelated families applying whole-exome sequencing and/or chromosomal microarray analyses and/or direct Sanger sequencing. SLC9A3 missense, splicing and truncation mutations, including an instance of uniparental disomy, and whole-gene deletion were identified in nine patients from eight families with CSD. Two of these nine patients developed inflammatory bowel disease (IBD) at 4 and 16 years of age. SLC9A3 encodes Na+/H+ antiporter 3 (NHE3), which is the major intestinal brush-border Na+/H+ exchanger. All mutations were in the NHE3 N-terminal transport domain, and all missense mutations were in the putative membrane-spanning domains. Identified SLC9A3 missense mutations were functionally characterized in plasma membrane NHE null fibroblasts. SLC9A3 missense mutations compromised NHE3 activity by reducing basal surface expression and/or loss of basal transport function of NHE3 molecules, whereas acute regulation was normal. This study identifies recessive mutations in NHE3, a downstream target of GC-C, as a cause of CSD and implies primary basal NHE3 malfunction as a predisposition for IBD in a subset of patients.
Introduction
Congenital intractable diarrheas with known etiology are monogenic disorders. Clinical and histological criteria as well as molecular testing for mutations in known disease genes are used to classify these disorders (1). The rare diarrheal disorder, congenital sodium diarrhea (CSD; MIM #270420), was first described in two sporadic patients in 1985 (2,3), and subsequently <40 cases have been reported in the literature (4). Jejunal brush-border (BB) membrane studies suggested a defect in sodium/proton exchange activity in three sporadic patients diagnosed with CSD (2,5,6). In 2000, we identified five patients with CSD from a large consanguineous Austrian kindred, demonstrating autosomal recessive inheritance in that family. Linkage analyses in that family excluded all known sodium/proton exchanger genes including SLC9A3 (solute carrier family 9, subfamily A, member 3; MIM #182307), the gene encoding sodium/proton antiporter 3 (NHE3) (4). Instead, the positional candidate approach identified a homozygous splice-site mutation in SPINT2 (serine peptidase inhibitor, Kunitz type, 2; MIM #605124) in this family (7). Pathogenic biallelic SPINT2 mutations were identified in 19 additional families (7–9). Patients with SPINT2 mutations were clinically characterized as a form of ‘syndromic’ CSD, in which intractable diarrhea was associated with superficial punctate keratitis, choanal atresia, intestinal atresia, as well as other sporadic abnormalities. This form of CSD is also referred to as a syndromic form of congenital tufting enteropathy (intestinal epithelial dysplasia; MIM #613217), as it often shows clustered enterocytes that form ‘tufts’ with branching crypts on histology (8,10).
Classical CSD (also called the non-syndromic phenotype) resembles congenital chloride diarrhea (MIM #214700) (11), but is distinguished by having high fecal loss of Na+, and is excluded from enterocyte differentiation and polarization disorders such as microvillus inclusion disease (MIM #251850) (12,13) and non-syndromic tufting enteropathy (10) by histopathology. Very recently, dominant activating mutations in receptor guanylate cyclase C (GC-C; MIM #601330) were found to cause a spectrum of secretory diarrheas including non-syndromic CSD in four patients (14,15). These mutations were associated with elevated intracellular cyclic guanosine monophosphate (cGMP) levels (14,15), an effect of which is the inhibition of NHE3 via its phosphorylation by cGMP kinase II (MIM #601591) as shown in PS120 fibroblasts and Caco-2/Bbe cells (16,17), Here, we identified deletion, truncating and missense mutations in SLC9A3, the gene encoding NHE3, in nine patients including one sib-pair with CSD. Reduced Na+/H+ exchange activity was demonstrated by the in vitro expression of identified SLC9A3 missense mutants. We thus show that recessive mutations in SLC9A3 are a cause of CSD. Interestingly, inflammatory bowel disease (IBD) developed in a number of patients with dominant GC-C mutations, and we also report IBD in two of nine patients with recessive SLC9A3 mutations, implicating NHE3 in the pathogenesis of IBD in a subset of patients.
Results
Clinical summary
In our cohort of 18 patients from 16 unrelated families with CSD, we identified SLC9A3 mutations in 9 patients from 8 unrelated families. One family had two affected children; this sib-pair is denoted as Patients 4 and 5 in Table 1, and both sibs harbored the same homozygous SLC9A3 mutation. Detailed clinical findings in patients with SLC9A3 mutations are shown in Table 1, including the two patients (Patients 2 and 3) originally described with this disorder (2,3). There was a uniform history of maternal polyhydramnios, and all CSD patients had watery secretory diarrhea and prominent abdominal distension after birth owing to dilated fluid-filled loops of intestine, indicating that secretory diarrhea begins prenatally.
Table 1.
Clinical findings in nine CSD patients with SLC9A3 mutations
| Parameter | Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5a | Patient 6 | Patient 7 | Patient 8 | Patient 9 |
|---|---|---|---|---|---|---|---|---|---|
| Consanguinity | No | No | No | Yes | Yes | Yes | Yes | No | No |
| Nationality/ethnicity | Canadian Caucasian | German/Finnish Caucasian | British Caucasian | Turkish | Turkish | Turkish Kurd Caucasian | German Caucasian | Serbian Caucasian | Canadian Caucasian |
| Current age (years) | 2 ½ | 37 | 33 | 1 ½ | Neonate | 6 ½ | 4 | 19 | 1 ½ |
| Sex | F | F | F | F | M | M | M | M | M |
| Gestational age (weeks) | 37 | 33 | 36 | 33 | 32 | 33 | 34 | 34 | 34 |
| Birth weight (g) | 2285 | 2530 | 2800 | 2560 | 2160 | 3210 | 2615 | 2125 | 2700 |
| Weight (kg, centile) | 12, 25th | n.i. | n.i. | 7.34, 90th | 2.84, 75th | 32.7, >97th | 16, 25th | 72, 50th | 9.02, 10th |
| Height (cm, centile) | 83.2, <10th | n.i. | n.i. | 63, 50th–75th | 48, 25th–50th | 119, 50th | 102, 10th | 175, 50th | 80, 25th |
| Polyhydramnios | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes | Yes |
| Plasma [Na+] (132–155 mmol/l) | 138 | 140 | 138 | 135–8 | 135 | 132 | 140 | 140 | 138 |
| Plasma pH | 7.28 | 7.34 | 7.30 | 7.33 | 7.30 | 7.37 | 7, 25 | n.i. | 7.33 |
| Plasma [HCO3−] (mmol/l) | 11.8 | n.i. | 16.0 | 21.1 | n.i. | 24 | 20,5 | n.i. | 23 |
| Fecal [Na+] (20–50 mmol/l) | 110 | 145 | 123 | 30 | 144 | 147 | 68 | 64 | 151 |
| Fecal [Cl−] (5–25 mmol/l) | <50 | 75 | 42 | 31 | 90 | 63 | 33 | 51 | 101 |
| Fecal pH (6–7) | 8 | 7 | 9 | 6 | n.i. | n.i. | 8 | 7 | n.i. |
| Fecal osmolality (320–370 mosmol/kg) | 271 | n.i. | n.i. | 353 | n.i. | 290 | 304 | n.i. | 315 |
| Urinary [Na+] (2–28 mmol/l) | <10 | 28 | n.i. | 2 | n.i. | 19 | 8 | n.i. | <10 |
| Histology | Normal | Not done | Nonspecific focal inflammation | Focal lymphatic hyperplasia | n.d | Partial villous atrophy ileal ulcerations, inflammatory infiltration with increase of eosinophils | Mild nonspecific inflammation | Ulceration in the rectum and sigmoid at 16 years, nodular lymphoid hyperplasia with ileal granulomas colonic ulcers at age 17 years | Normal |
| Parenteral fluids till (weeks) | Intermittent | None | 5 | 3 | 2 | 237 | 20 | None | Current |
| Current treatment | Oral | Oral | Oral | Oral | Oral | Oral + i.v. | Oral | Oral | i.v. |
| Current Na supplement (mmol/kg/d) | None | 4 | n.i. | n.i. | None | 6.5 | None | None | 13 |
| Outcome | Growth retardation | Mild watery diarrhea, otherwise normal life | Mild watery diarrhea, otherwise normal life | Watery diarrhea, hyperaldosteronism | Normal, fully breast-fed | Watery diarrhea, partial PN; ileal ulcerations since the age of 4 years | Mild watery diarrhea, otherwise normal life | Mild watery diarrhea; budesonide treatment; hyperaldosteronism | Total parenteral nutrition, growth retardation |
n.d.: not determined; n.i.: no information.
aPatients 4 and 5 are sibs.
The patients studied here did not have clinical features associated with the ‘syndromic form’ of CSD. While serum Na+ was normal in all patients, fecal Na+ concentrations were high in all patients except one, and urinary Na+ concentrations, as a marker of body Na+ depletion, were low. Patient 6 developed IBD at 4 years of age leading to ileo-caecal resection and temporary ileostomy because of recurrent episodes of small bowel obstruction. Patient 8 was diagnosed with IBD at 16 years of age, when he presented with bloody diarrhea and ulceration in the rectum and sigmoid. One year later he had nodular lymphoid hyperplasia with ileal granulomas and colonic ulcers. At present, he passes three to four watery stools per day without bleeding. Intestinal anatomy, histology and transit functions were normal in the other patients.
Mutation identification
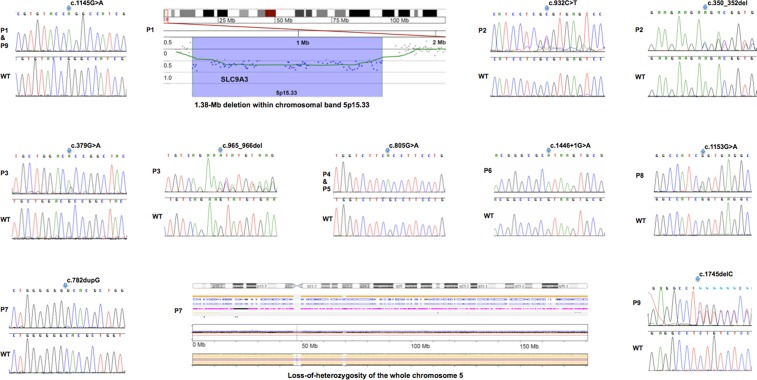
To identify the molecular basis of CSD in patients without GC-C mutations, we performed whole-exome sequencing in Patient 9 and in Patient 2 who was originally reported as having CSD (3). Studies of purified BB membrane vesicles of the proximal intestine from three reported CSD patients had pointed to impaired Na+/H+ exchanger activity previously (2,5,6), but genetic defects in NHE isoforms were not identified. Filtering exome data for private or rare variants in genes encoding Na+/H+ exchangers identified compound-heterozygous SLC9A3 mutations in Patients 2 and 9. Independently, chromosomal microarray analysis revealed a heterozygous, paternally inherited 1.383-Mb deletion on chromosome 5p15.33 encompassing SLC9A3 in Patient 1; Sanger sequencing of the remaining copy of SLC9A3 identified a maternally inherited missense mutation.
We subsequently identified another five SLC9A3 mutations in five unrelated CSD patients by Sanger-sequencing the coding region and flanking intronic regions of SLC9A3 in our patient cohort. A total of eight different SLC9A3 point mutations, a 3-bp deletion and a whole-gene deletion were identified in eight unrelated CSD families (Table 2, Fig. 1). One mutation, p.Arg382Gln, was observed in two seemingly unrelated patients of Canadian origin (Patients 1 and 9). Both affected siblings (Patients 4 and 5) of a consanguineous family were homozygous for the missense mutation p.Ala269Thr. Nine patients from 8 families of our cohort of 16 unrelated CSD families did not harbor either SLC9A3 or GUCY2C mutations indicating that other genes are responsible for the disease in these patients. Sequencing of DNA samples from family members confirmed the bi-allelic inheritance of the NHE3 mutations in six families and demonstrated autosomal recessive inheritance of CSD. A rare disease mechanism of a recessive disorder was revealed in Patient 7, who was homozygous for a SLC9A3 frame-shift mutation that was heterozygous in the mother but absent in the father. SNP array analysis in this family resolved this discrepancy by revealing maternal isodisomy of the entire chromosome 5 in this patient resulting in the maternal SLC9A3 mutation acting as a homozygous mutation in the patient. Monosomy rescue causes isodisomy, in which one chromosome is present in duplicate. In monosomy rescue, a nullisomic gamete is fertilized with a haploid gamete. The single chromosome from the other parent is duplicated and produces isodisomy of the chromosome.
Table 2.
SLC9A3 mutations in nine CSD patients
| Patient 1 | Patient 2 | Patient 3 | Patient 4 | Patient 5a | Patient 6 | Patient 7b | Patient 8 | Patient 9 | |
|---|---|---|---|---|---|---|---|---|---|
| Allele 1 (exon) | c.1145G>A (6) | c.932C>T (5) | c.[379G>A; 963_964delGT] (2; 6) | c.805G>A (5) | c.805G>A (5) | c.1446+1G>A (8) | c.782dupG (5) | c.1153G>A (6) | c.1145G>A (6) |
| Protein alteration/consequence | p.Arg382Gln | p.Ala311Val, splicing?c | p.[Ala127Thr; Tyr322Argfs*83] | p.Ala269Thr | p.Ala269Thr | Splicing, truncation | p.Thr262Hisfs*1 44 | p.Gly385Ser, splicing?c | p.Arg382Gln |
| Polyphen2 | Probably damaging | Probably damaging | p.Ala127Thr: possibly damaging | Probably damaging | Probably damaging | n.a. | n.a. | Probably damaging | Probably damaging |
| SIFT | Not tolerated | Not tolerated | Tolerated | Not tolerated | Not tolerated | n.a. | n.a. | Tolerated | Not tolerated |
| CADD PHRED | 29.9 | 29.7 | 24.3 | 34.0 | 34.0 | 23.3 | 27.3 | 29.9 | |
| Allele 2 (exon) | Gene deletion | c.350_352del (2) | n.d. | c.805G>A (5) | c.805G>A (5) | c.1446+1 G>A (8) | c.782dupG (5) | n.d. | c.1745delC |
| Protein alteration | No protein | p.Phe117del | n.d. | p.Ala269Thr | p.Ala269Thr | Splicing, truncation | p.Thr262Hisfs*144 | n.d. | p.Ser582Leufs*6 |
aPatients 4 and 5 are sibs.
bhomozygous mutation owing to maternal isodisomy of the entire chromosome 5.
cDNA mutation affects the most 3′ nucleotide in the exon and might interfere with proper splicing in addition to creating an amino acid change.
n.d., not determined; n.a., not applicable.
Figure 1.
Detection of SLC9A3 mutations and that of a whole SLC9A3 gene deletion in CSD patients. Identified compound-heterozygous and homozygous mutations in SLC9A3 identified by Sanger sequencing in eight unrelated patients, and a heterozygous whole-gene deletion identified in Patient 1 (Table 1) by array comparative genomic hybridization, and loss-of-heterozygosity for the whole chromosome 5 owing to uniparental isodisomy identified in Patient 7.
Patients 3 and 8 are heterozygous for one mutation with no second allelic mutation identified after sequencing the complete coding region and multiplex ligation-dependent probe amplification (MLPA) analysis of SLC9A3 exons. Most likely, we failed to detect the two allelic mutations with the current methodology, which does not cover deep intronic or promoter sequences, and which might be identified by whole genome sequencing. With the exception of the observation of mutation p.Arg382Gln in two seemingly unrelated patients, the identified SLC9A3 mutations were all private and not previously identified in several public databases [dbSNP, the 1000 Genomes Project, the National Heart, Lung, and Blood Institute (NHLBI) Exome Sequencing Project (ESP), the exome aggregation consortium server (EXAC)]. The description of SLC9A3 sequence variants is based on NCBI reference sequence NM_004174.2.
Functional studies of NHE3
The apical membrane Na+/H+ exchanger NHE3 carries out the majority of intestinal Na+ absorption. It is regulated by changes in its continual trafficking to and from the brush border, which results in changes in its surface expression (18). When the intestinal mucosa is stimulated by neurotransmitters as part of normal digestion, it is acutely stimulated or inhibited, and when exposed to enterotoxins or drugs that increase intracellular cGMP, Ca2+ or cAMP and mimic the pathophysiology of diarrhea, electroneutral NaCl absorption and the NHE3 contribution are inhibited (18). NHE3 consists of 12 N-terminal transmembrane domains that carry out Na+/H+ exchange, followed by a long cytoplasmic domain that interacts with the cytoskeleton and confers acute regulation of NHE3. This NHE3 regulation is critical for systemic volume and acid–base homeostasis (18,19).
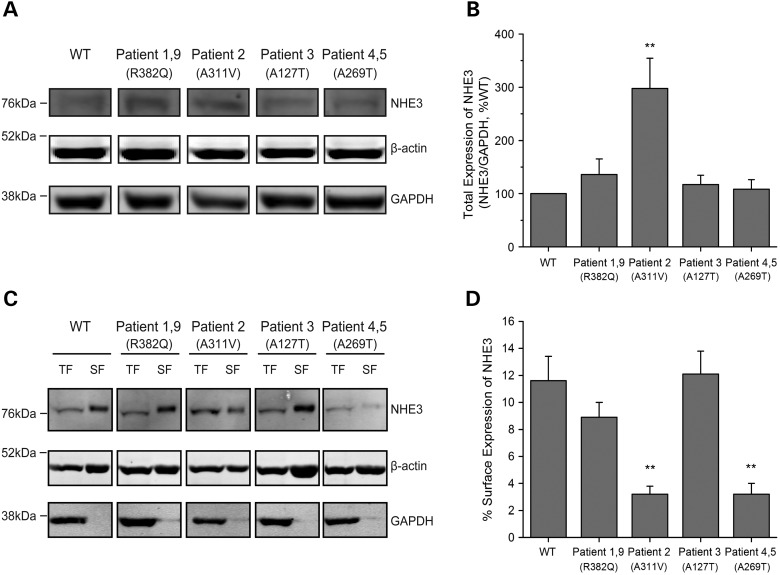
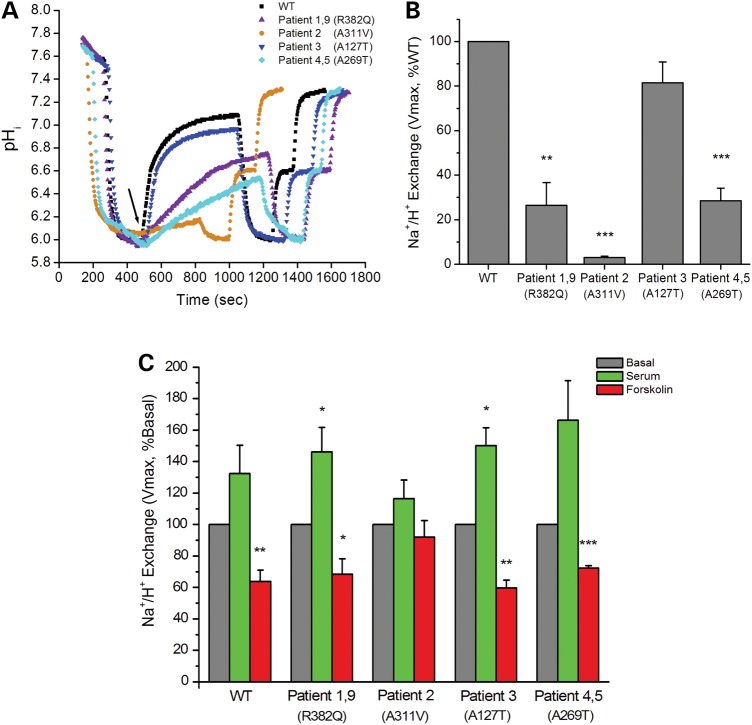
SLC9A3 mutations identified included one whole-gene deletion, one splicing and two frame-shift mutations, all of which would be expected to abolish protein production from these alleles. We further investigated the effect of four identified missense mutations or variants, p.Arg382Gln, p.Ala311Val, p.Ala269Thr, p.Ala127Thr on NHE3 activity. Wild-type and variant forms of NHE3 were stably expressed in PS120/Flag-NHERF2 fibroblasts that lack all endogenous plasma membrane NHEs (20). NHERF2 was also expressed because much of NHE3 regulation is NHERF2 dependent (18). NHE3 total protein expression was comparable in all cell lines, except for the p.Ala311Val cell line, which had increased protein expression (Fig. 2, Table 3). The percentage of surface expression of NHE3, determined by cell surface biotinylation, was reduced in cells expressing p.Ala311Val and p.Ala269Thr and was normal in the other two mutant cell lines studied (Fig. 2, Table 3). Basal Na+/H+ exchange activity was significantly decreased in three of the four cell lines studied (Fig. 3, Table 3). NHE3 activity was normal in the cell line expressing p.Ala127Thr. Note that p.Ala127Thr was present in-cis with a p.Tyr322Argfs*83 frame-shift mutation indicating that the latter compromised NHE3 activity and p.Ala127Thr represents a benign variant.
Figure 2.
Total and surface expression of NHE3 in PS120/FLAG-NHERF2 cells stably transfected with human NHE3-WT and mutations. Total expression (above). (A) A representative immunoblot with total cell lysates. Proteins were separated by SDS–PAGE and transferred onto nitrocellulose membranes. Protein expression was detected with primary antibodies to NHE3 (1:500, AB 6347) and β-actin (1:5000) and GAPDH (1:5000) as the loading controls. (B) Quantitative analysis of total expression of NHE3. The expression of NHE3/GAPDH was calculated, and NHE3-WT was set as 100% for each experiment. Results are mean ± SEM of nine independent experiments. Of the four NHE3 missense variants, one (Patient 2) had higher expression whereas the other three had comparable expression to NHE3-WT. Analysis of the same data normalized to β-actin (1:5000) gave similar results and is shown in Supplementary Material (see Supplementary Material, Fig. S3). **P < 0.01 compared with NHE3-WT (unpaired Student's t test). Surface expression (below). (C) A representative immunoblot with biotinylated cell lysates. Cells were biotinylated with NHS-SS-biotin and washed with quenching buffer to remove unbound NHS-SS-biotin. Cells were then lysed and divided into the total fractions and surface fractions. The surface fraction was incubated with avidin-agarose beads, and biotinylated proteins were eluted into SDS buffer. Total and surface fractions of each cell line were separated by SDS–PAGE, transferred onto nitrocellulose membranes, and probed with primary antibodies to NHE3, β-actin and GAPDH as mentioned above. TF, total fraction; SF, surface fraction. (D) Quantitative analysis of surface expression of NHE3. The percentage of surface expression of NHE3 was determined as in Supplementary Material. Results are mean ± SEM of five independent experiments. Two mutant forms of NHE3 (Patient 2 and Patients 4 and 5) showed significantly lower plasma membrane expression than NHE3-WT. **P < 0.01 compared with NHE3-WT (unpaired Student's t test).
Table 3.
Comparison of transport characteristics in NHE3 wild type and mutations
| Point mutation | Amino acid alteration | Basal activity |
Total NHE3 expression (%WT) | % Surface NHE3 expression | Basal activity/surface NHE3 expression (%WT) | ||||
|---|---|---|---|---|---|---|---|---|---|
| Vmax (µm/s) | Vmax (%WT) | K′ (H+)i (µm) | K′(H+)i (%WT) | ||||||
| WT | – | – | 474.9 ± 40.7 | 100 | 0.43 ± 0.05 | 100 | 100 | 11.6 ± 1.8 | 100 |
| Patients 1 and 9 | c.1145G>A | p.Arg382Gln | 123.6 ± 47.4 | 26.5 ± 10.2 | 0.76 ± 0.18 | 192.7 ± 71.9 | 135.8 ± 29.3 | 8.9 ± 1.1 | 25.4 |
| Patient 2 | c.932C>T | p.Ala311Val | 13.8 ± 1.9 | 3.0 ± 0.6 | 0.84 ± 0.10 | 208.7 ± 53.7 | 297.7 ± 56.6 | 3.2 ± 0.6 | 3.7 |
| Patient 3 | c.379G>pA | p.Ala127Thr | 393.8 ± 78.9 | 81.5 ± 9.4 | 0.5 ± 0.06 | 115.2 ± 4.1 | 117.3 ± 17.5 | 12.1 ± 1.7 | 66.6 |
| Patients 4 and 5 | c.805G>A | p.Ala269Thr | 131.1 ± 18.3 | 28.5 ± 5.6 | 0.77 ± 0.11 | 177.3 ± 6.3 | 108.3 ± 18.1 | 3.2 ± 0.8 | 95.4 |
Figure 3.
Functional study of NHE3-WT and mutations. (A) The basal activity of NHE3 was studied fluorometrically using the pH-sensitive dye BCECF. A representative experiment is shown demonstrating that the mutations detected in five patients (Patients 1, 2, 4, 5, 9) had reduced Na+/H+ exchange activities compared with NHE3-WT. In contrast, the NHE3 mutation identified in Patient 3 had similar activity with NHE3-WT. The black arrow indicates the start of Na+-dependent intracellular alkalinization. (B) Quantitative analysis of Na+/H+ exchange activities. Basal NHE3 activity (Vmax, μm/s) of each cell line was determined, and NHE3-WT was set as 100% for each experiment. Results are mean ± SEM of three independent experiments. **P < 0.01, ***P < 0.001 compared with NHE-WT (unpaired Student's t test). (C) PS120/FLAG-NHERF2 cells stably transfected with human NHE3-WT and mutations were treated with dialyzed serum (10%) or forskolin (10 μm) for 30 min following serum starvation for 3 h. NHE3 activity was determined and compared with the basal activity (Vmax, μm/s) of each cell line which was set as 100%. All cell lines responded normally by up-regulating activity following serum stimulation and down-regulating activity following forskolin inhibition except for the mutation identified in Patient 2. Results are mean ± SEM of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001 compared with basal activity (unpaired Student's t test).
For normalization of the basal activity with the amount of surface-expressed NHE3, the relative number of plasma membrane NHE3 molecules (compared with wild-type NHE3) was calculated as total expression of NHE mutant times the percentage of total NHE3 on the plasma membrane. p.Arg382Gln and p.Ala311Val had reduced, and p.Ala127Thr and p.Ala269Thr had normal Na+/H+ exchange function per surface molecule expressed compared with wild-type function (Table 3). Acute stimulation by dialyzed serum and inhibition by forskolin occurred normally on all point mutations except p.Ala311Val in which the Na+/H+ exchange rate was so low that accurate changes could not be determined (Fig. 3).
Discussion
This study shows that absent or mutated NHE3 protein, owing to recessive SLC9A3 mutations, is a cause of CSD. The loss or reduced NHE3 function acts similarly to the NHE3 inhibition observed in the post-prandial state or in diarrhea. Similar to these NHE3-deficient patients, Nhe3−/− mice exhibit diarrhea, metabolic acidosis, low blood pressure and reduced body fat and die rapidly when placed on a low Na+ diet (21,22). Interestingly, activating GC-C mutations can result in a CSD phenotype indistinguishable from patients with recessive NHE3 mutations (15). Owing to the small number of CSD patients with GUCY2C or SLC9A3 mutations known, a genotype–phenotype correlation cannot be provided. As activating GC-C mutations were shown to cause elevated cGMP levels (14,15), they are expected to result in inhibition of NHE3 at the apical enterocyte membrane (16), thereby providing an explanation for the secretory diarrhea by abrogated Na+ absorption (23).
There were two patterns to the abnormal transport function attributed to the NHE3 missense mutations, reduced transport function of individual NHE3 molecules present on the plasma membrane (abnormal turnover number) (Patients 1, 2 and 9) and reduced surface expression of NHE3 (Patients 2, 4 and 5). The explanation for the reduced surface expression could be due to either abnormal trafficking to the membrane (reduced exocytosis or increased endocytosis) or reduced BB stability. The functionally abnormal missense mutations that compromised NHE3 function all are present in one of the twelve membrane-spanning domains (MSD) of NHE3, based on homology modeling with bacterial Na+/H+ antiporters, which have had their structures solved (see Supplementary Material). The mutations are in putative MSD II (Patients 2 and 3), VI (Patients 4 and 5), VIII (Patient 2) and X (Patients 1, 8 and 9). How these mutations compromise NHE3 function at an atomic level has not been determined. However, MSD II and VIII form the funnel through which Na+/H+ exchange occurs, MSD II on the cytoplasmic and MSD VIII on the extracellular surfaces (24–26).
An additional important component of the phenotype associated with the NHE3 mutations was early and adolescent IBD in two of nine patients indicating a role of NHE3 deficiency as a predisposition to IBD. Interestingly, large-scale IBD genome-wide association studies have shown a strong replicated association between ulcerative colitis and the SLC9A3 locus (27,28). This association between compromised NHE3 function and IBD is further supported by a number of rodent and human studies. Nhe3−/− mice develop spontaneous distal chronic colitis and have increased susceptibility to dextran sulfate (DSS)-induced mucosal injury (29). An abnormal microbiome contributes to the Nhe3−/− colitis with no colitis reported in a very clean facility and an altered microbiome documented in mice in a facility in which colitis occurred (29–31). Consistent with the animal studies, NHE3 activity is reduced in both ulcerative colitis and Crohn's disease with activity reduced in areas of inactive colitis as well as in the areas of active inflammation (32,33). The mechanism is somewhat controversial with reports of both reduced message and protein (32) and reduced NHE3 activity with normal message, amount of protein and surface expression (33). In addition, 6 out of 36 patients with chronic diarrhea owing to activating mutations of GCC including 1 of 4 patients with CSD had IBD (14,15) further supporting the role of reduced sodium absorption in the pathogenesis of IBD possibly following changes in the intestinal microbial composition owing to alterations in electrolyte transport and in mucosal pH as demonstrated in Nhe3−/− mice (30).
In conclusion, we demonstrate genetic locus heterogeneity for CSD; CSD can either be caused by recessive mutations in NHE3 or by dominant mutations in GC-C, thereby stimulating further search for mutations in these genes as well as future research on drugs and peptides to restore or enhance Na+ absorption of diarrheal disorders.
Materials and Methods
Ethics statement
The study was approved by the ethics committees of the Medical University of Innsbruck, the Johns Hopkins University School of Medicine and the Hospital for Sick Children.
Patients
CSD was diagnosed in 18 patients from 16 unrelated families by gastroenterologists on characteristic clinical findings, which are described in the results section.
Genetic screening and verification
Exome capture was performed with the Illumina Truseq Exome Enrichment Kit and HiSeq2000 sequencer. Chromosomal microarray analysis (Genome-wide copy-number variant detection) and loss-of-heterozygosity analysis was performed with CytoSNP-12v2 BeadChip SNP arrays (Illumina, Little Chesterford, UK). Primers and conditions for SLC9A3 Sanger sequencing and MLPA analysis are provided in Supplementary Material, Tables S1 and S2.
Molecular characterization
PS120 cells stably expressing triple FLAG-tagged human NHERF2, as described, were stably transfected with human wild-type and four NHE3 mutants (20,34). Trafficking of NHE3 has been characterized in PS120 cells and shown to be similar to that in Caco-2 and OK cells as related to regulation of NHE3 exocytosis and endocytosis by elevated Ca2+, cAMP and cGMP (16,35–37). Na+/H+ exchange activity was determined fluorometrically using the intracellular pH-sensitive dye BCECF-AM, as described (38). Immunoblotting was performed with primary antibodies to NHE3 (rabbit polyclonal 6347, 1:500) (35) GAPDH (mouse-monoclonal Abcam, Cambridge, MA; 1:5000) and β−actin (rabbit polyclonal, Sigma).
Experimental procedures are described in detail in Supplementary Material, Methods.
Supplementary Material
Web Resources
dbSNP: http://www.ncbi.nlm.nih.gov/projects/SNP/, Ensembl: http://www.ensembl.org/, Exome Aggregation Consortium (ExAC) Browser: http://exac.broadinstitute.org/, NHLBI Exome Sequencing Project (ESP) Exome Variant Server: http://snp.gs.washington.edu/EVS, Combined Annotation Dependent Depletion (CADD) score: http://cadd.gs.washington.edu/score.
Funding
This work was supported by grants from Jubiläumsfonds der Österreichischen Nationalbank (grants no. 14496 and no. 15627), the Tiroler Wissenschaftsfonds (grant no. UNI-0404/1286), Else Kröner-Fresenius-Stiftung Nr. 2013_A230. This work was also supported in part from NIH/NIDDK grants RO1DK 26523, RO1 DK 61765, P01DK 72084, KO8DK 088950, R03DK 099566, T32 DK2007632 and Conte GI Core Center grant P30DK 89502, The Hopkins Digestive Diseases Basic and Translational Research Core Center. A.M.M. is funded by a CIHR – Operating Grant (MOP119457) and by the Leona M. and Harry B. Helmsley Charitable Trust to study VEOIBD.
Supplementary Material
Acknowledgments
Conflict of Interest statement. None declared.
References
- 1.Berni Canani R., Terrin G., Cardillo G., Tomaiuolo R., Castaldo G. (2010) Congenital diarrheal disorders: improved understanding of gene defects is leading to advances in intestinal physiology and clinical management. J. Pediatr. Gastroenterol. Nutr., 50, 360–366. [DOI] [PubMed] [Google Scholar]
- 2.Booth I.W., Stange G., Murer H., Fenton T.R., Milla P.J. (1985) Defective jejunal brush-border Na+/H+ exchange: a cause of congenital secretory diarrhoea. Lancet, 1, 1066–1069. [DOI] [PubMed] [Google Scholar]
- 3.Holmberg C., Perheentupa J. (1985) Congenital Na+ diarrhea: a new type of secretory diarrhea. J. Pediatr., 106, 56–61. [DOI] [PubMed] [Google Scholar]
- 4.Muller T., Wijmenga C., Phillips A.D., Janecke A., Houwen R.H., Fischer H., Ellemunter H., Fruhwirth M., Offner F., Hofer S. et al. (2000) Congenital sodium diarrhea is an autosomal recessive disorder of sodium/proton exchange but unrelated to known candidate genes. Gastroenterology, 119, 1506–1513. [DOI] [PubMed] [Google Scholar]
- 5.Keller K.M., Wirth S., Baumann W., Sule D., Booth I.W. (1990) Defective jejunal brush border membrane sodium/proton exchange in association with lethal familial protracted diarrhoea. Gut, 31, 1156–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fell J.M., Miller M.P., Finkel Y., Booth I.W. (1992) Congenital sodium diarrhea with a partial defect in jejunal brush border membrane sodium transport, normal rectal transport, and resolving diarrhea. J. Pediatr. Gastroenterol. Nutr., 15, 112–116. [DOI] [PubMed] [Google Scholar]
- 7.Heinz-Erian P., Muller T., Krabichler B., Schranz M., Becker C., Ruschendorf F., Nurnberg P., Rossier B., Vujic M., Booth I.W. et al. (2009) Mutations in SPINT2 cause a syndromic form of congenital sodium diarrhea. Am. J. Hum. Genet., 84, 188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salomon J., Goulet O., Canioni D., Brousse N., Lemale J., Tounian P., Coulomb A., Marinier E., Hugot J.P., Ruemmele F. et al. (2013) Genetic characterization of congenital tufting enteropathy: epcam associated phenotype and involvement of SPINT2 in the syndromic form. Hum. Genet., 133, 299–310. [DOI] [PubMed] [Google Scholar]
- 9.Sivagnanam M., Janecke A.R., Muller T., Heinz-Erian P., Taylor S., Bird L.M. (2010) Case of syndromic tufting enteropathy harbors SPINT2 mutation seen in congenital sodium diarrhea. Clin. Dysmorphol., 19, 48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sivagnanam M., Mueller J.L., Lee H., Chen Z., Nelson S.F., Turner D., Zlotkin S.H., Pencharz P.B., Ngan B.Y., Libiger O. et al. (2008) Identification of EpCAM as the gene for congenital tufting enteropathy. Gastroenterology, 135, 429–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoglund P., Haila S., Socha J., Tomaszewski L., Saarialho-Kere U., Karjalainen-Lindsberg M.L., Airola K., Holmberg C., de la Chapelle A., Kere J. (1996) Mutations of the down-regulated in adenoma (DRA) gene cause congenital chloride diarrhoea. Nat. Genet., 14, 316–319. [DOI] [PubMed] [Google Scholar]
- 12.Muller T., Hess M.W., Schiefermeier N., Pfaller K., Ebner H.L., Heinz-Erian P., Ponstingl H., Partsch J., Rollinghoff B., Kohler H. et al. (2008) MYO5B mutations cause microvillus inclusion disease and disrupt epithelial cell polarity. Nat. Genet., 40, 1163–1165. [DOI] [PubMed] [Google Scholar]
- 13.Wiegerinck C.L., Janecke A.R., Schneeberger K., Vogel G.F., van Haaften-Visser D.Y., Escher J.C., Adam R., Thoni C.E., Pfaller K., Jordan A.J. et al. (2014) Loss of syntaxin 3 causes variant microvillus inclusion disease. Gastroenterology, 147, 65–68 e10. [DOI] [PubMed] [Google Scholar]
- 14.Fiskerstrand T., Arshad N., Haukanes B.I., Tronstad R.R., Pham K.D., Johansson S., Havik B., Tonder S.L., Levy S.E., Brackman D. et al. (2012) Familial diarrhea syndrome caused by an activating GUCY2C mutation. N. Engl. J. Med., 366, 1586–1595. [DOI] [PubMed] [Google Scholar]
- 15.Müller T., Rasool I., Heinz-Erian P., Mildenberger E., Hülstrunk C., Müller A., Michaud L., Koot B.G.P., Ballauff A., Vodopiutz J. et al. (2015) Congenital Secretory Diarrhea caused by activating germline mutations in GUCY2C. Gut, doi:10.1136/gutjnl-2015-309441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cha B., Kim J.H., Hut H., Hogema B.M., Nadarja J., Zizak M., Cavet M., Lee-Kwon W., Lohmann S.M., Smolenski A. et al. (2005) cGMP inhibition of Na+/H+ antiporter 3 (NHE3) requires PDZ domain adapter NHERF2, a broad specificity protein kinase G-anchoring protein. J. Biol. Chem., 280, 16642–16650. [DOI] [PubMed] [Google Scholar]
- 17.Chen T., Kocinsky H.S., Cha B., Murtazina R., Yang J., Tse C.M., Singh V., Cole R., Aronson P.S., de Jonge H. et al. (2015) Cyclic GMP Kinase II (cGKII) inhibits NHE3 by altering its trafficking and phosphorylating NHE3 at three required sites: identification of a multifunctional phosphorylation site. J. Biol. Chem., 290, 1952–1965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Donowitz M., Mohan S., Zhu C.X., Chen T.E., Lin R., Cha B., Zachos N.C., Murtazina R., Sarker R., Li X. (2009) NHE3 regulatory complexes. J. Exp. Biol., 212, 1638–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cabado A.G., Yu F.H., Kapus A., Lukacs G., Grinstein S., Orlowski J. (1996) Distinct structural domains confer cAMP sensitivity and ATP dependence to the Na+/H+ exchanger NHE3 isoform. J. Biol. Chem., 271, 3590–3599. [DOI] [PubMed] [Google Scholar]
- 20.Murtazina R., Kovbasnjuk O., Donowitz M., Li X. (2006) Na+/H+ exchanger NHE3 activity and trafficking are lipid Raft-dependent. J. Biol. Chem., 281, 17845–17855. [DOI] [PubMed] [Google Scholar]
- 21.Schultheis P.J., Clarke L.L., Meneton P., Miller M.L., Soleimani M., Gawenis L.R., Riddle T.M., Duffy J.J., Doetschman T., Wang T. et al. (1998) Renal and intestinal absorptive defects in mice lacking the NHE3 Na+/H+ exchanger. Nat. Genet., 19, 282–285. [DOI] [PubMed] [Google Scholar]
- 22.Noonan W.T., Woo A.L., Nieman M.L., Prasad V., Schultheis P.J., Shull G.E., Lorenz J.N. (2005) Blood pressure maintenance in NHE3-deficient mice with transgenic expression of NHE3 in small intestine. Am. J. Physiol. Regul. Integr. Comp. Physiol., 288, R685–R691. [DOI] [PubMed] [Google Scholar]
- 23.Arshad N., Visweswariah S.S. (2012) The multiple and enigmatic roles of guanylyl cyclase C in intestinal homeostasis. FEBS Lett., 586, 2835–2840. [DOI] [PubMed] [Google Scholar]
- 24.Padan E. (2014) Functional and structural dynamics of NhaA, a prototype for Na(+) and H(+) antiporters, which are responsible for Na(+) and H(+) homeostasis in cells. Biochim. Biophys. Acta, 1837, 1047–1062. [DOI] [PubMed] [Google Scholar]
- 25.Landau M., Herz K., Padan E., Ben-Tal N. (2007) Model structure of the Na+/H+ exchanger 1 (NHE1): functional and clinical implications. J. Biol. Chem., 282, 37854–37863. [DOI] [PubMed] [Google Scholar]
- 26.Lee C., Yashiro S., Dotson D.L., Uzdavinys P., Iwata S., Sansom M.S., von Ballmoos C., Beckstein O., Drew D., Cameron A.D. (2014) Crystal structure of the sodium-proton antiporter NhaA dimer and new mechanistic insights. J. Gen. Physiol., 144, 529–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Anderson C.A., Boucher G., Lees C.W., Franke A., D'Amato M., Taylor K.D., Lee J.C., Goyette P., Imielinski M., Latiano A. et al. (2011) Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat. Genet., 43, 246–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jostins L., Ripke S., Weersma R.K., Duerr R.H., McGovern D.P., Hui K.Y., Lee J.C., Schumm L.P., Sharma Y., Anderson C.A. et al. (2012) Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature, 491, 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Laubitz D., Larmonier C.B., Bai A., Midura-Kiela M.T., Lipko M.A., Thurston R.D., Kiela P.R., Ghishan F.K. (2008) Colonic gene expression profile in NHE3-deficient mice: evidence for spontaneous distal colitis. Am. J. Physiol. Gastrointest. Liver Physiol., 295, G63–G77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Larmonier C.B., Laubitz D., Hill F.M., Shehab K.W., Lipinski L., Midura-Kiela M.T., McFadden R.M., Ramalingam R., Hassan K.A., Golebiewski M. et al. (2013) Reduced colonic microbial diversity is associated with colitis in NHE3-deficient mice. Am. J. Physiol. Gastrointest. Liver Physiol., 305, G667–G677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kiela P.R., Laubitz D., Larmonier C.B., Midura-Kiela M.T., Lipko M.A., Janikashvili N., Bai A., Thurston R., Ghishan F.K. (2009) Changes in mucosal homeostasis predispose NHE3 knockout mice to increased susceptibility to DSS-induced epithelial injury. Gastroenterology, 137, 965–975, 975 e961–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sullivan S., Alex P., Dassopoulos T., Zachos N.C., Iacobuzio-Donahue C., Donowitz M., Brant S.R., Cuffari C., Harris M.L., Datta L.W. et al. (2009) Downregulation of sodium transporters and NHERF proteins in IBD patients and mouse colitis models: potential contributors to IBD-associated diarrhea. Inflamm. Bowel Dis., 15, 261–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yeruva S., Farkas K., Hubricht J., Rode K., Riederer B., Bachmann O., Cinar A., Rakonczay Z., Molnar T., Nagy F. et al. (2010) Preserved Na(+)/H(+) exchanger isoform 3 expression and localization, but decreased NHE3 function indicate regulatory sodium transport defect in ulcerative colitis. Inflamm. Bowel Dis., 16, 1149–1161. [DOI] [PubMed] [Google Scholar]
- 34.Zhu X.C., Sarker R., Horton J.R., Chakraborty M., Chen T.E., Tse C.M., Cha B., Donowitz M. (2015) Nonsynonymous single nucleotide polymorphisms of NHE3 differentially decrease NHE3 transporter activity. Am. J. Physiol. Cell Physiol., 308, C758–C766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sarker R., Valkhoff V.E., Zachos N.C., Lin R., Cha B., Chen T.E., Guggino S., Zizak M., de Jonge H., Hogema B. et al. (2011) NHERF1 and NHERF2 are necessary for multiple but usually separate aspects of basal and acute regulation of NHE3 activity. Am. J. Physiol. Cell Physiol., 300, C771–C782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zachos N.C., van Rossum D.B., Li X., Caraveo G., Sarker R., Cha B., Mohan S., Desiderio S., Patterson R.L., Donowitz M. (2009) Phospholipase C-gamma binds directly to the Na+/H+ exchanger 3 and is required for calcium regulation of exchange activity. J. Biol. Chem., 284, 19437–19444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhu X., Cha B., Zachos N.C., Sarker R., Chakraborty M., Chen T.E., Kovbasnjuk O., Donowitz M. (2011) Elevated calcium acutely regulates dynamic interactions of NHERF2 and NHE3 proteins in opossum kidney (OK) cell microvilli. J. Biol. Chem., 286, 34486–34496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Levine S.A., Montrose M.H., Tse C.M., Donowitz M. (1993) Kinetics and regulation of three cloned mammalian Na+/H+ exchangers stably expressed in a fibroblast cell line. J. Biol. Chem., 268, 25527–25535. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.