Abstract
Patients with 46,XY gonadal dysgenesis (GD) exhibit genital anomalies, which range from hypospadias to complete male-to-female sex reversal. However, a molecular diagnosis is made in only 30% of cases. Heterozygous mutations in the human FGFR2 gene cause various craniosynostosis syndromes including Crouzon and Pfeiffer, but testicular defects were not reported. Here, we describe a patient whose features we would suggest represent a new FGFR2-related syndrome, craniosynostosis with XY male-to-female sex reversal or CSR. The craniosynostosis patient was chromosomally XY, but presented as a phenotypic female due to complete GD. DNA sequencing identified the FGFR2c heterozygous missense mutation, c.1025G>C (p.Cys342Ser). Substitution of Cys342 by Ser or other amino acids (Arg/Phe/Try/Tyr) has been previously reported in Crouzon and Pfeiffer syndrome. We show that the ‘knock-in’ Crouzon mouse model Fgfr2cC342Y/C342Y carrying a Cys342Tyr substitution displays XY gonadal sex reversal with variable expressivity. We also show that despite FGFR2c-Cys342Tyr being widely considered a gain-of-function mutation, Cys342Tyr substitution in the gonad leads to loss of function, as demonstrated by sex reversal in Fgfr2cC342Y/− mice carrying the knock-in allele on a null background. The rarity of our patient suggests the influence of modifier genes which exacerbated the testicular phenotype. Indeed, patient whole exome analysis revealed several potential modifiers expressed in Sertoli cells at the time of testis determination in mice. In summary, this study identifies the first FGFR2 mutation in a 46,XY GD patient. We conclude that, in certain rare genetic contexts, maintaining normal levels of FGFR2 signaling is important for human testis determination.
Introduction
Disorders of sex development (DSD) are congenital conditions in which development of chromosomal, gonadal or anatomical sex is atypical (1–3). Perturbations in testis development can lead to 46,XY DSD with gonadal dysgenesis (46,XY GD) frequently associated with infertility and gonadal cancer (1–4). In the partial form of 46,XY GD, low levels of androgens produced by the dysgenic testes can result in babies born with ambiguous genitalia. DSD conditions are difficult to manage often requiring multiple surgery, and ongoing endocrine and psychosocial care. In the complete form of 46,XY GD, individuals have completely undeveloped ‘streak’ gonads and are phenotypically female. 46,XY GD is caused by mutations in genes belonging to the testis-determining network. The major breakthrough in elucidating this network came with the discovery of the human testis-determining gene SRY on the Y chromosome, mutation or deletion of which accounts for ∼15% of all 46,XY GD cases (5–8). Since the discovery of SRY, causative mutations in several other testis genes have been identified, including SOX9, NR5A1 and MAP3K1 (3,7,9). Despite this, a molecular diagnosis is so far possible for only ∼30% of XY GD patients.
The testis-determining network is best characterized in mice. During mammalian embryogenesis, the developing gonads are bipotential and develop into either testes (XY) or ovaries (XX). In the XY mouse gonad at embryonic day 11.5 (E11.5) (in humans at 7 weeks gestation), the SRY protein is expressed in the bipotential supporting cell lineage, which triggers Sertoli cell fate by directly upregulating the transcription of the pivotal testis-determining gene Sox9 (10–14). Extensive Sertoli cell proliferation ensues, with the development of testis cords, the future sperm-producing seminiferous tubules (15). Within fetal testis cords, Sertoli cells surround the germ cells, which become mitotically arrested. Sertoli cells produce anti-Müllerian hormone (AMH), which regresses the female reproductive tract, while outside the testis cords, Leydig cells begin to differentiate and produce androgens leading to the differentiation of the male internal and external genitalia (16). In the female mouse XX gonad, two major independent pathways are activated at E11.5, RSPO1-WNT4 and FOXL2 (17,18). As a consequence, the bipotential supporting cells differentiate into granulosa cells resulting in the development of the ovarian follicles after birth. The fetal ovary is unstructured and germ cells enter prophase of meiosis before arrest (16).
Downstream of SRY and SOX9, the Fibroblast Growth Factor 9 (FGF9)/FGF receptor 2 (FGFR2) signaling pathway represses Wnt4 to promote testis development in the mouse (19,20). Both Fgf9 and Fgfr2 knockout mice show XY gonadal male-to-female sex reversal, highlighting the importance of FGF signaling in mouse testis determination (21–24). However, the role of FGF9 and FGFR2 in human sex development and 46,XY GD remains elusive. No FGF9 mutations were identified in a large cohort of 46,XY GD patients (25). Clinically, human FGFR2 mutations lead to loss- or gain of function. In lacrimoauriculodentodigital (LADD) syndrome (OMIM #149730) characterized by tear tract, ear, teeth and digit abnormalities, FGFR2 substitutions occur in the tyrosine kinase (TK) domain, resulting in reduced activity (26,27). In craniosynostosis syndromes (premature fusion of the skull bones), including Crouzon, Pfeiffer, Apert and Antley-Bixler syndromes (OMIM #123500, #101600, #101200 and #207410), FGFR2 substitutions occur predominantly in or near the immunoglobulin-like domain 3 (D3) (28–32), leading to FGFR2 activation and increased osteogenesis (33–35). However, gonadal defects have not been described in LADD or craniosynostosis syndromes. Here, we describe a patient with FGFR2-related craniosynostosis and XY sex reversal. We would suggest this novel syndrome be designated CSR.
Results
FGFR2c mutation in a female patient with Crouzon-like features and 46,XY complete GD
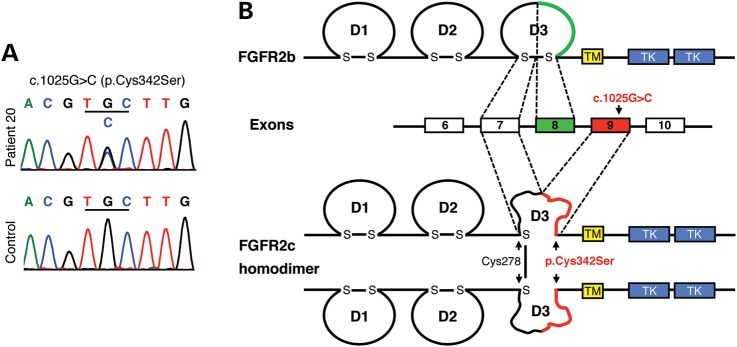
In a cohort of 44 patients with 46,XY GD of unknown etiology (36), we noted a case with complete 46,XY GD associated with craniosynostosis (Patient 20, Fig. 1). The Crouzon-like features in this patient combined with the established role of FGF signaling in mouse testis determination suggested a mutation in FGFR2. DNA sequencing of the entire FGFR2 gene identified the heterozygous missense mutation, c.1025G>C (p.Cys342Ser) (Fig. 2A), affecting the FGFR2c isoform (Fig. 2B). Cysteine 342 substitutions for serine or other amino acids (Arg/Phe/Trp/Tyr; or R/F/W/Y) occur frequently in FGFR2-related craniosynostosis syndromes, including Crouzon and Pfeiffer syndromes (28–30). Biochemically, these substitutions are functionally identical and lead to loss of the essential Cys278–Cys342 disulfide bond of the third immunoglobulin-like domain, D3 (Fig. 2B). These mutant receptors can no longer be activated by ligands. Instead, the unpaired Cys278 forms an intermolecular disulfide bond to a second mutant receptor, resulting in constitutively activated stable FGFR2c homodimers (33,34).
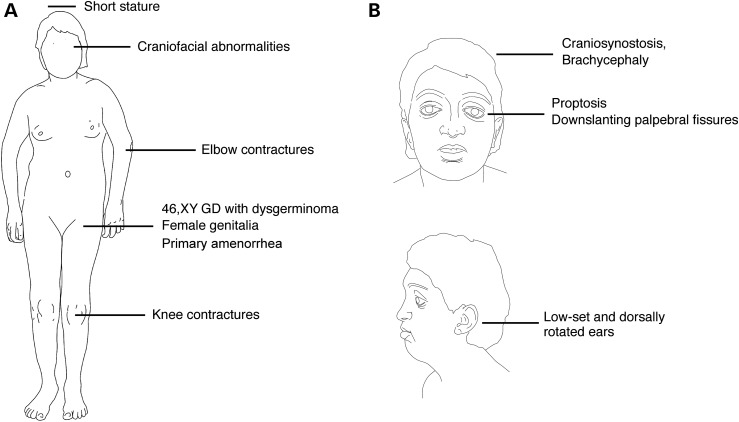
Figure 1.
(A) Schematic representation of abnormalities of the patient. (B) Schematic diagram of the facial characteristics of the patient.
Figure 2.
DNA sequencing identifies a heterozygous c.1025G>C (p.Cys342Ser) missense mutation in the FGFR2c isoform. (A) Electropherograms showing a doublet G/C peak at c.1025 in Patient 20 and a single G peak in the control. (B) Location and biochemical consequences of the FGFR2c-Cys342Ser substitution: FGFR2 contains an extracellular ligand-binding region composed of three immunoglobulin-like domains (D1–D3), a transmembrane domain (TM) and a split cytoplasmic TK domain. FGFR2 exists in two isoforms, FGFR2b and FGFR2c, which are generated by alternative splicing of the C-terminal half of the D3. The c.1025G>C mutation leads to a cysteine-to-serine substitution at position 342 (p.Cys342Ser) in the FGFR2c isoform. Cys342Ser results in loss of the D3 intrachain disulfide bond in FGFR2c and thus to major disruption of the D3. However, the unpaired Cys278 residues form intermolecular disulfide bonds between the mutant receptors resulting in constitutively activated stable FGFR2c homodimers.
Heterozygous XY Fgfr2cC342Y/+ mice develop normal testes
To investigate whether a Cys342 substitution in FGFR2c could disrupt testis development, we analyzed a well-characterized knock-in mouse model carrying the most commonly observed substitution in Crouzon syndrome patients, Cys342Tyr (or C342Y) (35,37,38). Cys342Tyr and Cys342Ser substitutions have similar phenotypic spectra (Crouzon and Pfeiffer syndromes) though they preferentially lead to Crouzon and Pfeiffer phenotypes, respectively (30,39,40). Heterozygous Fgfr2cC342Y/+ mice (CD1) faithfully recapitulate most of the craniofacial features of Crouzon syndrome patients including craniosynostosis, shallow orbits and ocular proptosis.
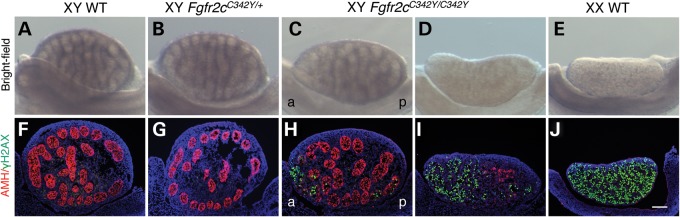
At E15.5, XY testes are larger than XX ovaries and characterized by the presence of testis cords, whereas XX ovaries are unstructured (Fig. 3A and E). Within testis cords, Sertoli cells surround the germ cells that enter mitotic arrest. While in ovaries, due to the absence of testis cords, the germ cells enter prophase of meiosis (16). Upon gross examination, heterozygous XY Fgfr2cC342Y/+ testes (n = 40) appeared indistinguishable from XY wild-type (WT) testes, with testis cords evident throughout the gonad (Fig. 3A and B). In support, double immunofluorescence analyses for the Sertoli cell marker AMH and the germ cell meiotic prophase marker γH2AX revealed no differences between heterozygous XY Fgfr2cC342Y/+ (n = 11) and WT XY testes. Both XY Fgfr2cC342Y/+ and XY WT testes express AMH throughout the gonad (Fig. 3F and G), and WT XX ovaries express γH2AX uniformly (Fig. 3J).
Figure 3.
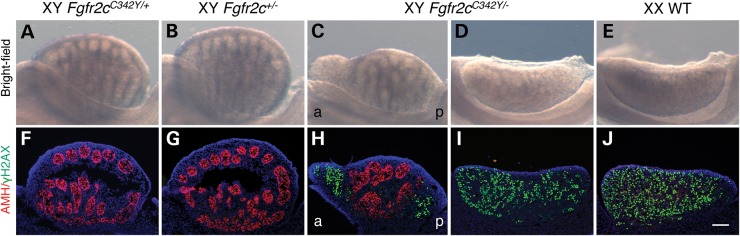
Homozygous Fgfr2cC342Y/C342Y mice show variable XY gonadal sex reversal at E15.5. (A–E) Bright-field images of XY WT, XY Fgfr2cC342Y/+, XY Fgfr2cC342Y/C342Y and XX WT gonads at E15.5. In the vast majority of XY Fgfr2cC342Y/C342Y embryos (15 out of 16), testis cords were less defined or absent at the anterior pole when compared with XY WT and Fgfr2cC342Y/+ testes. In one XY Fgfr2cC342Y/C342Y embryo, testis cords could not be identified and the gonads resembled ovaries. (F–J) Double immunofluorescence staining for the Sertoli cell marker AMH (red, cytoplasmic) and γ-H2AX (green, nuclear), a marker of female meiotic germ cells, in E15.5 XY WT, XY Fgfr2cC342Y/+, XY Fgfr2cC342Y/C342Y and XX WT gonads. In the vast majority of XY Fgfr2cC342Y/C342Y embryos (15 out of 16), AMH expression was reduced or absent at the anterior pole and to a lesser extent at the posterior pole, which expressed γH2AX. In one XY Fgfr2cC342Y/C342Y embryo, only a few AMH-expressing cells could be detected, whereas widespread γH2AX expression was observed. Sections were stained with DAPI (blue, nuclear) to visualize the cell nuclei. a, anterior; p, posterior. Scale bar is 100 µm.
Homozygous XY Fgfr2cC342Y/C342Y mice display variable disruption of testicular development
The absence of an abnormal testicular phenotype in heterozygous XY Fgfr2cC342Y/+ embryos could be a dosage effect. Indeed, features of the phenotypic spectrum of Crouzon syndrome including cleft lip, cleft palate and maxillary hypoplasia are only revealed in homozygous Fgfr2cC342Y/C342Y mice (CD1) (35). In 1 of 16 XY Fgfr2cC342Y/C342Y embryos, testis cords could not be identified and the gonads resembled ovaries (Fig. 3D and E). Double immunofluorescence analyses for AMH and γH2AX revealed only a few Sertoli cells/testis cords, while widespread γH2AX expression was observed (Fig. 3I), suggesting near-complete failure of testis differentiation. The majority of Fgfr2cC342Y/C342Y embryos (15 out of 16) presented with mild testicular defects. In these gonads, testis cords were poorly defined or absent at the anterior pole (Fig. 3C). In agreement, AMH was reduced or absent at the anterior pole, and to a lesser extent at the posterior pole (Fig. 3H), indicating a failure of Sertoli cell differentiation. Also at the poles, germ cells appear to have entered meiosis as shown by ectopic γH2AX staining (Fig. 3H).
XY Fgfr2cC342Y/C342Y gonads show sex reversal of the supporting cell lineage and reduced numbers of androgen-producing Leydig cells
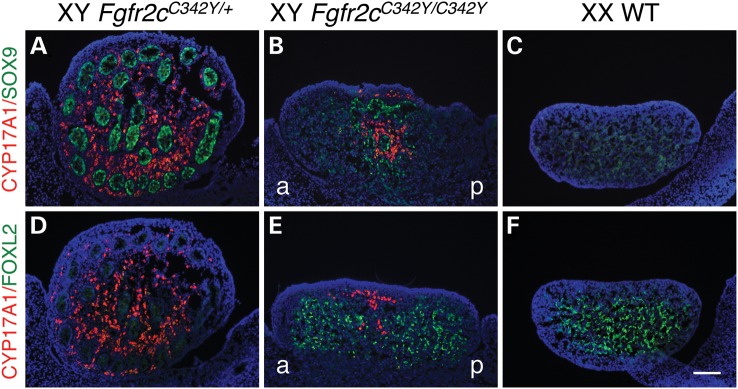
A critical step during testis determination is the differentiation of the bipotential supporting cell lineage into Sertoli cells, driven by the expression of SOX9 (11). In the XX female gonad, in the absence of SOX9 expression, female-specific proteins such as FOXL2 are expressed in the supporting cells, which differentiate into granulosa cells (13,17,41). Sertoli cells also secrete factors to trigger the differentiation of Leydig cells (42). Leydig cells produce steroidogenic enzymes such as CYP17A1 required for the biosynthesis of androgens to masculinize the male internal and external genitalia (16). Because FGF9/FGFR2 signaling is important for SOX9 expression in the XY gonad (22–24,43), we speculated that loss of SOX9 expression could be responsible for the partial failure of Sertoli cell differentiation in E15.5 XY Fgfr2cC342Y/C342Y gonads. To test this and to examine the consequence of reduced numbers of Sertoli cells in these gonads upon Leydig cell differentiation, we performed immunofluorescence analyses for CYP17A1 in combination with either SOX9 or FOXL2 on gonad sections. E15.5 heterozygous XY Fgfr2cC342Y/+ testes were indistinguishable from E15.5 XY WT testes and used as controls. They express SOX9 in the Sertoli cells of the testis cords and CYP17A1 in Leydig cells surrounding the cords (Fig. 4A and D), whereas E15.5 XX WT ovaries express FOXL2 in the granulosa cells throughout (Fig. 4F). In E15.5 XY Fgfr2cC342Y/C342Y gonads, SOX9 is expressed in areas with testis cords, but not in areas devoid of cords (Fig. 4B), which instead expressed FOXL2 (Fig. 4E), indicating that the supporting cell lineage had differentiated into granulosa cells. In addition, although CYP17A1-expressing cells were found in the SOX9-expressing testicular regions of the gonads (Fig. 4B), they were absent in the FOXL2-positive ovarian regions of the gonads (Fig. 4E). Thus, XY Fgfr2cC342Y/C342Y gonads show an overall reduction in the number of Leydig cells when compared with heterozygous XY Fgfr2cC342Y/+ testes.
Figure 4.
Homozygous Fgfr2cC342Y/C342Y gonads show sex reversal of the supporting cell lineage and reduced numbers of androgen-producing Leydig cells. (A–C) Double immunofluorescence staining for the critical Sertoli cell protein SOX9 (green, nuclear) and the Leydig cell marker CYP17A1 (red, cytoplasmic) in E15.5 XY Fgfr2cC342Y/+, XY Fgfr2cC342Y/C342Y and XX WT gonads. In E15.5 XY Fgfr2cC342Y/C342Y mice, SOX9 was expressed in testis cords, but was not detectable in the gonadal areas devoid of cords, which instead expressed FOXL2 (see E). In addition, CYP17A1-expressing cells were restricted to the SOX9-expressing testicular regions of the gonads (D–F): Double immunofluorescence staining for the Granulosa cell marker FOXL2 (green, nuclear) and the Leydig cell marker CYP17A1 (red, cytoplasmic) in E15.5 XY Fgfr2cC342Y/+, XY Fgfr2cC342Y/C342Y and XX WT gonads. In XY Fgfr2cC342Y/C342Y gonads, CYP17A1-expressing cells are absent in the FOXL2-positive ovarian regions of the gonads. Sections were stained with DAPI (blue, nuclear) to visualize the cell nuclei. a, anterior; p, posterior. Scale bar is 100 µm.
Taken together, XY Fgfr2cC342Y/C342Y gonads display partial male-to-female gonadal sex reversal and thus develop as ovotestes containing both testicular and ovarian tissue. The failure of testicular development specifically at the gonad poles is typical for ovotestis formation in mice, because SRY and SOX9 are expressed from center to pole, rendering the poles more sensitive to XY sex reversal (23,44,45).
WT FGFR2c can rescue sex reversal in XY Fgfr2cC342Y/− gonads
Despite FGFR2c-C342Y being widely considered a gain-of-function mutation, the gonadal abnormalities in XY Fgfr2cC342Y/C342Y mice phenocopy those observed in both Fgf9 and Fgfr2 knockout mice (21,23,24,43). The loss-of-function phenotype could be explained by either the detrimental effects of FGFR2c overactivation or by FGFR2c loss of function. To distinguish between these possibilities, we applied a classical genetic approach and crossed heterozygous Fgfr2cC342Y/+ knock-in mice (CD1) with heterozygous Fgfr2c knockout mice (46) (Fgfr2c+/−, maintained on a C57BL/6 genetic background) to generate compound heterozygous Fgfr2cC342Y/− embryos. E15.5 gonads were analyzed by gross examination and by double immunofluorescence for AMH and γH2AX. As expected, heterozygous XY Fgfr2cC342Y/+ knock-in (n = 6) (Fig. 5A and F) and heterozygous XY Fgfr2c+/− knockout (n = 5) embryos (Fig. 5B and G) developed normal testes. In contrast, XY compound heterozygous Fgfr2cC342Y/− embryos displayed either ovotestes (n = 6 embryos) or completely sex-reversed ovaries (n = 2 embryos). In XY Fgfr2cC342Y/− ovotestes, testis cords were less defined or absent at the poles (Fig. 5C) with AMH expression reduced or absent, and γH2AX ectopically expressed (Fig. 5H). In completely sex-reversed XY Fgfr2cC342Y/− ovaries, testis cords were not apparent (Fig. 5D); AMH expression was completely absent and γH2AX expression was expressed throughout the gonad (Fig. 5I), similar to XX WT ovaries (Fig. 5J). These data demonstrate that sex reversal in XY Fgfr2cC342Y/− mice is rescued by WT FGFR2c (Fgfr2cC342Y/+ mice) suggesting that C342Y leads to FGFR2c loss-of-function activity in the developing XY gonads. It is noteworthy that the XY sex reversal phenotype in Fgfr2cC342Y/− mice was generally more severe than in Fgfr2cC342Y/C342Y mice, which is likely attributable to the 50% contribution of C57BL/6, a mouse strain exquisitely sensitive to XY gonadal sex reversal (47).
Figure 5.
XY compound heterozygous Fgfr2cC342Y/− gonads show sex reversal. (A–E) Bright-field images of XY Fgfr2cC342Y/+, XY Fgfr2c+/−, XY Fgfr2cC342Y/− and XX WT gonads at E15.5. In XY compound heterozygous Fgfr2cC342Y/− embryos, testis cords were less defined or absent at the poles (n = 6), or testis cords could not be identified and the gonads resembled ovaries (n = 2). (F–J) Double immunofluorescence staining for the Sertoli cell marker AMH (red, cytoplasmic) and γ-H2AX (green, nuclear), a marker of female meiotic germ cells, in E15.5 XY Fgfr2cC342Y/+, XY Fgfr2c+/−, XY Fgfr2cC342Y/− and XX WT gonads. In XY compound heterozygous Fgfr2cC342Y/− ovotestes, AMH expression was reduced or absent at the poles, which expressed γH2AX. In XY compound heterozygous Fgfr2cC342Y/− ovaries, AMH expression was absent and γH2AX expression was expressed throughout the gonad. Sections were stained with DAPI (blue, nuclear) to visualize the cell nuclei. a, anterior; p, posterior. Scale bar is 100 µm.
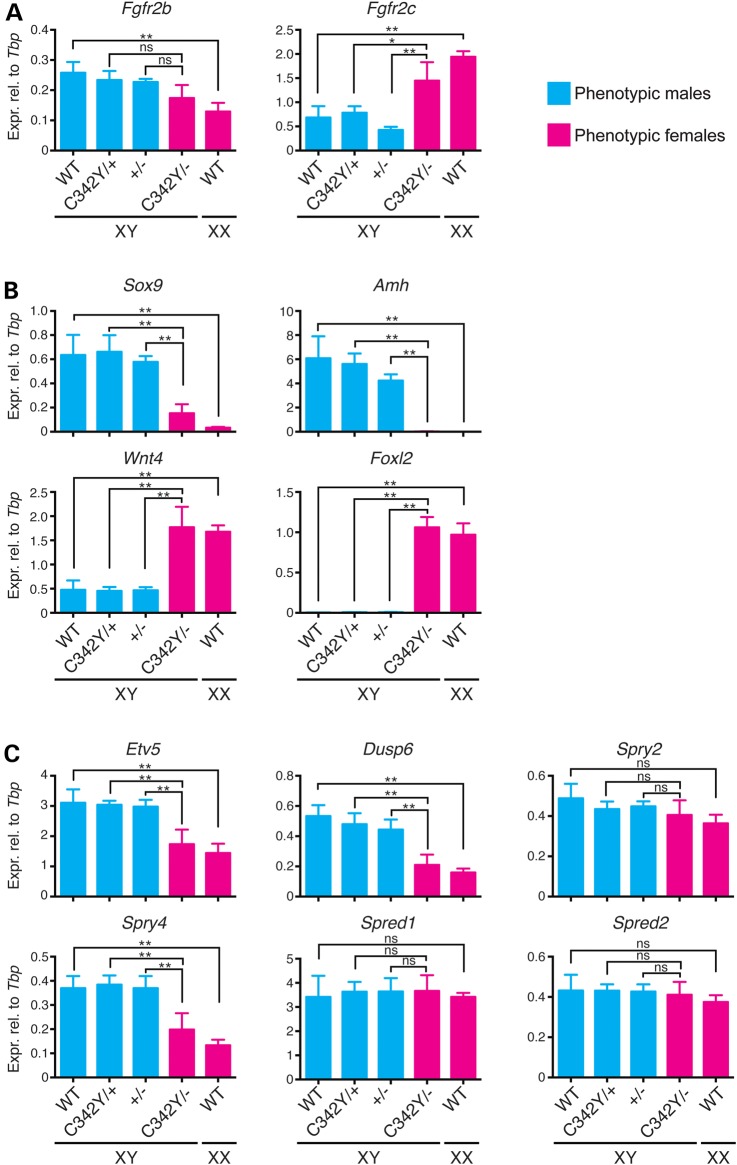
To examine the sex reversal of XY Fgfr2cC342Y/− ovaries in more detail, the expression levels of specific, potentially relevant genes were measured by quantitative real-time polymerase chain reaction (qRT-PCR). As controls, data were compared with WT XY testes, heterozygous XY Fgfr2cC342Y/+ knock-in testes and heterozygous XY Fgfr2c+/− knockout testes (Fig. 6, in blue) and also with XX WT ovaries (Fig. 6, in pink). Statistical significance for all 10 genotype comparisons was determined by one-way analysis of variance (ANOVA), of which the three most critical comparisons are indicated in Figure 6. Similar results were obtained for all three XY testis genotypes (Fig. 6) whose expression profiles did not significantly differ from each other. We, therefore, refer to them as XY control testes hereafter. Comparison of XY control testes versus XX WT ovaries identifies nine genes with sexually dimorphic expression—Fgfr2b, Fgfr2c, Sox9, Amh, Wnt4, Foxl2, Etv5, Dusp6 and Spry4 (Fig. 6).
Figure 6.
Gene expression analyses in XY Fgfr2cC342Y/− ovaries by qRT-PCR. mRNA expression levels in E13.5 XY Fgfr2cC342Y/− ovaries were measured for the two Fgfr2 isoforms, Fgfr2b and Fgfr2c (A), for the Sertoli cell markers Sox9 and Amh and the granulosa cell markers Wnt4 and FoxL2 (B) and for the FGF-responsive genes Etv5, Dusp6, Spry2, Spry4, Spred1 and Spred2 (C). Pink bars represent phenotypic females and blue bars phenotypic males. Mean ± SEM, n = 3 (except n = 2 for XY WT). *P < 0.05, **P < 0.01 (one-way ANOVA with Tukey's range test); ns, not significant.
In the mouse, cerebral cortex deletion of the Fgfr2c isoform causes a splicing switch and consequent increase in Fgfr2b isoform expression (48). We examined this possibility in XY Fgfr2cC342Y/− gonads (Fig. 6A). Both isoforms show sexually dimorphic expression, with Fgfr2b expression higher in XY control testes and Fgfr2c expression higher in XX WT ovaries. Compared with XY control testes, Fgfr2b expression in XY Fgfr2cC342Y/− ovaries was similar, whereas Fgfr2c expression was higher (Fig. 6A). This suggests that an Fgfr2c/Fgfr2b isoform switch in the gonads of the Crouzon model is unlikely to explain the sex reversal.
Next, we analyzed gene expression of the Sertoli cell identity marker Sox9 and its direct target gene Amh and also of the granulosa cell differentiation factors Wnt4 and FoxL2 representing the two independent ovarian pathways (17). Sox9 and Amh showed higher expression in XY control testes, whereas Wnt4 and FoxL2 showed higher expression in XX WT ovaries, as expected (Fig. 6B). In XY Fgfr2cC342Y/− ovaries, Sox9 expression was lower than in XY control testes, and Amh expression was virtually absent while Wnt4 and Foxl2 expression was increased to levels similar to those of XX WT ovaries (Fig. 6B). This indicates that these XY Fgfr2cC342Y/− gonads are completely sex reversed at a molecular level.
Lastly, we investigated changes in expression of FGF receptor signaling components in the supporting cell lineage, specifically Etv5, Dusp6, Spry2, Spry4, Spred1 and Spred2. All these genes are reported to be expressed higher in Sertoli than in granulosa cells (49), except for Spry2. Also, FGF9 potently induces the expression of Dusp6 in XX ex vivo gonad cultures (50). XY Fgfr2cC342Y/− ovaries showed reduced expression of Etv5, Dusp6, and Spry4 when compared with XY control testes (Fig. 6C). This suggests that loss of FGF-signaling components contributes to sex reversal.
In summary, XY Fgfr2cC342Y/− ovaries show reduced expression of Sertoli cell markers and FGF-responsive genes and increased expression of granulosa cell markers when compared with heterozygous XY Fgfr2c+/− knockout testes. In addition, these expression changes in XY Fgfr2cC342Y/− ovaries are restored to XY WT levels by the addition of the WT allele found in the heterozygous Crouzon model (XY Fgfr2cC342Y/+), supporting the C342Y mutant receptor showing loss-of-function activity in the gonads.
Whole exome sequencing identifies potential modifier genes expressed in pre-Sertoli cells
Although our Crouzon mouse models show that Cysteine 342 substitution in FGFR2c can disrupt testicular development, the rarity of our patient suggests the influence of modifier genes, which exacerbated the testicular phenotype. To identify potential variants, we performed whole exome sequencing. We identified novel single nucleotide variants (SNVs) or INDELs in 193 genes (see Supplementary Material, Table S1). None of these SNVs/INDELs are located in 63 DSD genes recently defined by Baxter et al. (51), which include all known 46,XY GD genes (SRY, MAP3K1, NR5A1, SOX9, WT1, DHH, GATA4 and CBX2). The patient carries novel non-synonymous SNVs or INDELs in 35 genes expressed in pre-Sertoli cells at the time of sex determination in mice (see Supplementary Material, Table S1). Of these 35 genes, 5 show enriched expression in mouse pre-Sertoli cells (WIPF3, ERCC6, TPD52, HDHD2 and PRKRA) (49) and 3 have known (CITED2 and WIPF3) or postulated (EP300) functions in Sertoli cells (see Supplementary Material, Table S1).
Discussion
We hypothesized that human FGFR2 is a good candidate gene to examine for mutations in unsolved XY DSD cases given that both Fgf9 and Fgfr2 knockout mice show XY gonadal male-to-female sex reversal, either partial or complete (21–24). Evidence for the involvement of FGF9 and FGFR2 in human testis development remained elusive. No FGF9 mutations were identified in a large cohort of 46,XY GD patients (n = 33) (25). Clinically, FGFR2 mutations can lead to loss- or gain of function. LADD syndrome is caused by loss-of-function mutations (26,27), whereas craniosynostosis syndromes including Crouzon and Pfeiffer are caused by gain-of-function mutations (28–30). Gonadal defects had not been described in any of these syndromes.
In this study, we report a unique case of 46,XY GD with craniosynostosis. Due to the highly unusual features of the patient, a definitive assignment to a specific craniosynostosis syndrome was not achieved. Based on craniosynostosis, brachycephaly, proptosis, downslanting palpebral fissures and low-set dorsally rotated ears, the patient was initially diagnosed with FGFR2-related craniosynostosis syndrome (52). Although the absence of hand and feet anomalies is characteristic of Crouzon syndrome (52), reduced extension at the elbows is a hallmark feature of the exceptionally rare Antley-Bixler syndrome (31,32) and is common in Apert and Pfeiffer syndromes (53). Elbow anomalies also occur in Crouzon syndrome, but are less frequent and severe. A detailed radiographic study of 22 Crouzon patients revealed that 8 patients had elbow anomalies, including 1 patient who had elbow synostosis (53). Short stature is extremely rare in Crouzon syndrome and has been reported in only two cases. In both patients, short stature is most likely caused by growth hormone deficiency (54,55). Due to the additional presence of 46,XY GD, we would suggest that our patient has a novel syndrome—craniosynostosis with XY male-to-female sex reversal (CSR).
DNA sequencing revealed a cysteine-to-serine substitution at position 342 in the FGFR2c isoform (Cys342Ser). Cys342 substitutions by Ser or other amino acids (Arg/Phe/Trp/Tyr) occur frequently in the craniosynostosis syndromes Crouzon and Pfeiffer (28–30). Using the ‘knock-in’ Crouzon mouse models Fgfr2cC342Y/− and Fgfr2cC342Y/C342Y, we show that Cys342 substitution in FGFR2c leads to disruption of testicular development. The testicular defects were most likely caused by a loss of the pivotal sex determining protein SOX9, resulting in a reduction in the number of Sertoli cells and testis cords with concomitant loss of the androgen-producing Leydig cells. The mutation in the 2c isoform is in agreement with knockout data showing that FGFR2c is the critical isoform during sex determination in the mouse (S. Bagheri-Fam et al., unpublished). Taken together, these data suggest that the FGFR2c c.1025G>C (p.Cys342Ser) mutation contributed to 46,XY GD in the sex-reversed patient.
FGFR2c-Cys342 substitutions are widely considered gain-of-function mutations leading to increased osteogenesis (33–35). It was, therefore, surprising that the testicular abnormalities and molecular changes in XY Fgfr2cC342Y/− and Fgfr2cC342Y/C342Y mice phenocopy those observed in knockout mice with FGFR2 loss of function (23). The downstream mediators of FGF signaling in the developing mouse testis remain largely unknown. However, it is well established that the main function of FGF9/FGFR2 in the XY gonad is to repress expression of the granulosa cell marker Wnt4 (19,20,23,24). The fact that in XY Fgfr2cC342Y/− ovaries, Wnt4 expression was increased to levels similar to WT XX ovaries supports the notion that the C342Y substitution in FGFR2c leads to loss-of-function rather than gain-of-function activity in the gonads. We also observed reduced expression of the FGF-responsive genes Etv5, Dusp6 and Spry4 in XY Fgfr2cC342Y/− ovaries when compared with XY control testes. However, interpretation of these data is complicated by the fact that all three FGF-responsive genes are expressed in multiple gonadal cell types (49), potentially masking specific effects in supporting cells. Moreover, Etv5 (in Sertoli, interstitial and germ cells), Dusp6 (in Sertoli and germ cells) and Spry4 (in Sertoli and endothelial cells) show marked male-specific overexpression in gonadal cell types (49). Thus, downregulation of these genes in XY Fgfr2cC342Y/− ovaries to nearly WT female levels (Fig. 6C) might be a consequence of sex reversal (e.g. switch to a female transcriptional profile). Snyder-Warwick et al. (38) observed that homozygous Fgfr2cC342Y/C342Y mice exhibited high incidence of cleft palate, like Fgfr1/Fgfr2 double knockout mice, also implying loss-of-function activity. The authors proposed a model whereby during palate development, C342Y acts as a gain-of-function mutation FGFR2c signaling increasing initially, but decreasing after reaching a threshold, through feedback loops (38). Our data, however, do not support such a ‘threshold model’ to explain loss-of-function receptor activity in the XY gonad. Sex reversal in XY Fgfr2cC342Y/− mice was actually rescued by the addition of the WT Fgfr2 allele (Fgfr2cC342Y/+), supporting C342Y being a loss-of-function mutation in the developing gonads. This suggests that craniosynostosis and 46,XY GD in our patient are caused by distinct mechanisms. Although craniosynostosis is reportedly the result of the mutant FGFR2c protein acting in a dominant manner, 46,XY GD is likely caused by haploinsufficiency. In contrast to the patient, XY gonadal sex reversal in the heterozygous Crouzon mouse model (Fgfr2cC342Y/+ mice) was not observed, which is consistent with previous reports showing that testis development in humans is more susceptible to dosage effects than in mice. For example, loss-of-function mutations in SOX9 cause XY sex reversal when heterozygous in humans, but only when homozygous in mice (12,13,56,57).
The question remains as to how a mutation in FGFR2c that leads to gain of function in the sutures can lead to loss of function in the gonads. One possible explanation could be the fact that Cys342 substitutions are initially loss-of-function mutations. They lead to loss of the Cys278–Cys342 disulfide bond and thus to major disruption of the ligand-binding D3 (Fig. 2). In the sutures, it is believed that the unpaired Cys278 forms an intermolecular disulfide bond to a second mutant receptor, resulting in constitutively activated stable FGFR2c homodimers (33,34). However, in the gonads, such FGFR2c homodimer formation might not occur due to the presence of tissue-specific factors such that the mutant receptor is unable to signal. Another possibility for the loss-of-function gonadal phenotype in Fgfr2cC342Y/− and Fgfr2cC342Y/C342Y mice is that either no or limited amounts of mutant FGFR2c protein are actually present in the gonad. This could be due to more rapid internalization of the mutant receptor by endocytosis followed by degradation inside lysosomes (58,59). Reduced stability of the Fgfr2c transcript or a switch from the Fgfr2c to the Fgfr2b isoform, which occurs in an Apert/Pfeiffer mouse model (48), is less likely. When compared with XY Fgfr2c+/− control gonads, in XY Fgfr2cC342Y/− ovaries, Fgfr2b transcript levels were unchanged, whereas Fgfr2c transcript levels were increased. This increase is most likely due to higher Fgfr2c expression levels in ovaries than testes.
Human XY individuals with heterozygous Cys342 substitutions are reportedly male (28–30). Based on our mouse models, this can be explained by their single WT FGFR2 allele, which appears to be sufficient to maintain FGFR2 signaling above a critical threshold required for normal testis function. However, testis/male development in these patients will be more susceptible to modifiers than in normal individuals. The patient presented here is the first case where FGFR2-related craniosynostosis is associated with XY male-to-female sex reversal. The rarity was reflected in homozygous Fgfr2cC342Y/C342Y mice where only one in 16 XY embryos displayed severe disruption of testicular development (near-complete sex reversal). In the majority of Fgfr2cC342Y/C342Y embryos, XY gonadal sex reversal was mild. Fgfr2cC342Y/C342Y mice die shortly after birth (35), but would likely develop as normal males with normal or slightly smaller testes, because the ovarian portions of ovotestes in fetal mice tend to regress over time due to apoptosis (60,61). Therefore, XY patients with FGFR2-related craniosynostosis may develop testicular defects in utero that might be transient or mild and thus go undiagnosed. Our patient might harbor a unique set of modifier genes, which exacerbate this testicular phenotype. In support of this, in Fgfr2 knockout mice, expressivity of the XY gonadal sex reversal phenotype was greatly dependent on the genetic background (23). In search for potential modifiers, we undertook whole exome analysis and identified novel non-synonymous SNVs or INDELs in 35 genes expressed in mouse pre-Sertoli cells, a feature shared by all known 46,XY GD genes. Five of these genes (WIPF3, ERCC6, TPD52, HDHD2 and PRKRA) show enriched expression in this cell lineage, and three have known (CITED2 and WIPF3) or postulated (EP300) functions in Sertoli cells. Mutations in CITED2 can lead to congenital heart disease (OMIM #614433 and #614431) (62–64), and Cited2 knockout embryos die with heart malformations (65). In the absence of one gene copy of either Nr5a1 or Wt1, known 46,XY GD genes in humans (4) Cited2 knockout embryos show partial XY sex reversal, identifying Cited2 as a modifier during mouse sex determination (66). Although the CITED2 c.364C>G (p.Gln122Glu) missense variant was predicted damaging by all three in silico algorithms, the clinical significance remains unknown due to the lack of heart defects detected in our patient. EP300 interacts and cooperates with CITED2 during heart development (67,68). Mutations in EP300 lead to Rubinstein–Taybi syndrome 2 (OMIM #613684), which is characterized by congenital heart defects, skeletal abnormalities, cryptorchidism in males (undescended testes) and intellectual disability (69–71). Ep300 knockout mice die before sex determination with defects in heart development, neurulation and cell proliferation (72). The EP300 acetyl-transferase interacts with SRY to increase SRY DNA-binding activity and acetylates SRY to promote SRY nuclear import in cell culture experiments (73). However, because the EP300 c.457A>G (p.Met153Val) missense variant was predicted non-damaging by all three in silico algorithms and due to lack of classical Rubinstein–Taybi features in our patient, we would classify this variant as likely benign. In humans, the WIPF3 protein is expressed in Sertoli cell–spermatogenic cell junctions. Although no human mutations have been reported for WIPF3, its expression is reduced in testes of men with idiopathic azoospermia (74). In the mouse, Wipf3 expression is enriched in pre-Sertoli cells at the time of testis determination (E11.5–E13.5) (49). Wipf3 knockout mice have no apparent defects during development, but adult mice display male-specific sterility due to Sertoli cell dysfunction (75). The WIPF3 c.178C>T (p.Arg60Cys) missense variant was predicted damaging by two out of the three in silico algorithms (SIFT and PolyPhen2) and thus is classified as a variant of unknown clinical significance.
In summary, we report the first case of FGFR2-related craniosynostosis with XY sex reversal. Similarly, a corresponding Fgfr2c knock-in craniosynostosis mouse model also shows XY sex reversal. This strongly suggests that the heterozygous FGFR2c mutation contributed to XY sex reversal in the craniosynostosis patient. Given the rarity of our patient, additional mutations (such as the variants identified in CITED2 and EP300) are likely to contribute along with the FGFR2c mutation to explain the sex reversal.
Conclusion
We identified a novel syndrome: craniosynostosis with XY sex reversal, arising from an FGFR2c mutation (c.1025G>C (p.Cys342Ser)). The heterozygous mutation leads to gain of function in the skull, but to loss of function in the developing gonads. This study identifies the first FGFR2 mutation in a 46,XY GD patient and suggests that, in certain rare genetic contexts, maintaining normal levels of FGFR2 signaling is important for human testis determination. Our results exemplify the need to include FGF-signaling components in diagnostic protocols for 46,XY DSD with partial or complete GD.
Materials and Methods
Case report
The 15-year-old female (Patient 20) was presented with delayed puberty, primary amenorrhea, female external genitalia and Müllerian structures (Fig. 1). She underwent gonadectomy due to the presence of bilateral ovarian tumors. Subsequent chromosome analysis revealed the karyotype 46,XY suspecting XY GD. Histological analyses of the dysgenic gonads revealed bilateral dysgerminoma, which apparently developed from preexisting gonadoblastoma. The gonads lacked seminiferous tubules and only a few Sertoli- and Leydig-like cells were detected, confirming 46,XY complete GD. Previously overlooked, the patient was also diagnosed with Crouzon-like syndrome based on craniosynostosis requiring surgery, brachycephaly, proptosis, downslanting palpebral fissures, low-set and dorsally rotated ears, and the absence of hand and feet anomalies. Additional features included limited movement of elbows and knees, and short stature (148 cm, −2.46 SD). Informed consent was obtained from the parents (legal guardians) of the proband aged 15 years. The study was approved by the Monash Health Human Research Ethics Committee (Australia) and by the Ethical Commission of the University of Freiburg (Germany).
Patient DNA sequence analysis
Genomic DNA of Patient 20 was extracted from peripheral blood by standard methods. Genomic DNA from the parents was not available for analysis. The entire FGFR2 gene was screened for mutations by DNA sequence analysis using the Sanger Method. Primer sequences and conditions for genomic amplification of FGFR2 (21 exons) were as described (30). PCR products were sequenced using an Applied Biosystems 3130xl Genetic Analyzer.
Whole exome sequencing and analysis
Whole exome capture was performed using the 64 Mb Nimblegen SeqCap EZ Exome Library V3.0 (Nimblegen SeqCap EZ Library SR Protocol Version 4.0, January 2013). The exome was sequenced on the Illumina HiSeq1500 as 100-bp paired-end runs. Initial quality screening of the sequence reads using FastQC (Andrews, S., http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc/) led to the identification of a few base pairs with highly variable base quality scores. Upon further investigation, this artifact was linked to a tile-specific error. Therefore, for the purpose of this study, reads overlapping these tiles were filtered out. The alignment to the human reference genome (hg19, GRCh37) was performed using BWA (v0.5.9) (76).
SNVs and INDELs were identified using a set of tools included in Picard (http://broadinstitute.github.io/picard) and the Genome Analysis Toolkit (GATK) (77,78). Variant call was performed using the UnifiedGenotyper walker from GATK with a minimum base quality of 20 (Phred scale), a call confidence threshold of 50 (Phred-scaled) and an emission confidence threshold of 10 (Phred-scaled). SNVs and INDELs were separately filtered using GATK's VariantFiltration walker [SNVs: SNP cluster size = 10; depth of coverage: ≥5; Qual: ≥50; strand bias: Fisher's exact test, ≥60, quality by depth, >2, u-based z-approximation from the Mann–Whitney Rank Sum Test for the distance from the end of the read for reads with the alternate allele (ReadPosRankSum) < −8.0, u-based z-approximation from the Mann–Whitney Rank Sum Test for mapping qualities (MQRankSum) < −12.5; indels: quality by depth >2.0, ReadPosRankSum < −20 and Strand bias: Fisher's exact test >200]. Functional annotation was performed using ANNOVAR (79) based on NCBI's reference sequence database (http://www.ncbi.nlm.nih.gov/refseq), the database of single nucleotide polymorphisms (dbSNP) (dbSNP 137; http://www.ncbi.nlm.nih.gov/SNP) and dbNSFP (80,81) for SIFT (82), Polyphen-2 (83) and Mutation Taster (84) scores. Visualization was performed using the Integrative Genomics Viewer (IGV, Broad Institute; http://www.broadinstitute.org/igv/) (85,86).
A total of 18 596 SNVs/INDELs in exons including the FGFR2c mutation were identified of which 231 variants in 193 genes were novel (see Supplementary Material, Table S1). To determine the best candidate modifiers in the patient, we have ranked these 193 genes including all of their variants into four main groups based on their function and expression in Sertoli cells (see Supplementary Material, Table S1). This strategy was chosen because all known 46,XY GD genes (SRY, MAP3K1, NR5A1, SOX9, WT1, DHH, GATA4 and CBX2) and FGFR2 are expressed in this cell lineage. Genes expressed in Sertoli cells were identified using the transcriptional expression profile of purified E11.5–E13.5 mouse Sertoli cells (49). Genes were considered to be expressed in Sertoli cells that have a log base 2 normalized expression value >7 at least at one of the three developmental stages (Groups 1–3) (n = 50 genes). All remaining genes were placed into Group 4 (n = 143 genes). The first group contains genes (n = 3 genes) (see Supplementary Material, Table S1), which have known or postulated functions in Sertoli cells. These genes were identified by searching PubMed (http://www.ncbi.nlm.nih.gov/pubmed/) and the online database OMIM (http://www.ncbi.nlm.nih.gov/omim/) with the key words ‘sex’, ‘testis’, ‘Sertoli’ or ‘SRY’. The second group harbors genes (n = 6 genes) with enriched expression in Sertoli cells compared with all other gonadal cell types, and the third group contains all remaining genes expressed in this cell lineage (n = 41 genes) (see Supplementary Material, Table S1). Within each group, variants were ranked based on their possible impact on protein structure and function in the order (i) deletions/insertions/stopgain/splicing, (ii) non-synonymous substitutions and (iii) synonymous substitutions; non-synonymous substitutions were ranked based on their functional impact scores as determined by the in silico algorithms SIFT (82), PolyPhen-2 (83) and Mutation Taster (84). Following American College of Medical Genetics guidelines (87), all 231 novel exonic variants were assessed against four main categories (pathogenic, likely pathogenic, variants of uncertain clinical significance (VUS) and likely benign). The categories pathogenic or likely pathogenic were not assigned because none of the variants were located in known 46,XY GD genes. Synonymous substitutions and all non-synonymous substitutions predicted to be non-disease causing by all three in silico algorithms are classified likely benign, whereas all other variants are classified as VUS. Genes known to be associated with a human disease (see Supplementary Material, Table S1) were identified by searching the online database OMIM (http://www.ncbi.nlm.nih.gov/omim/). All 231 novel variants were submitted to dbSNP (http://www.ncbi.nlm.nih.gov/SNP).
Generation and genotyping of mouse embryos
Heterozygous Fgfr2cC342Y/+ mice (35) were maintained on a CD1 genetic background and intercrossed to generate E15.5 WT, heterozygous Fgfr2cC342Y/+ and homozygous Fgfr2cC342Y/C342Y embryos. Heterozygous Fgfr2cC342Y/+ (CD1) mice were crossed with heterozygous Fgfr2c+/− mice (46) maintained on a C57BL/6 genetic background to generate E15.5 WT, heterozygous Fgfr2cC342Y/+, heterozygous Fgfr2c+/− and compound heterozygous Fgfr2cC342Y/− embryos. For genotyping, genomic DNA was isolated from the yolk sac. For sexing, embryos were genotyped for the presence of the Sry gene as previously described (88). Primers and PCR conditions for the Fgfr2cC342Y and Fgfr2c knockout alleles were used as previously described (35,46). Embryos were harvested at 15.5 days gestation with noon of the day on which plug was identified designated as 0.5 day. Institutional Animal Care and Use Committee of Yale University approved all procedures involving mice.
Gross examination of gonads
E15.5 embryos were collected in cold phosphate-buffered saline (PBS). The gonads were dissected out of the embryos and documented using a Zeiss Stemi SV11 stereomicroscope. At least three independent gonads were examined per genotype.
Immunofluorescence
Testes were fixed, processed and sectioned as described previously (23). For indirect immunofluorescence staining, frozen sections were incubated overnight at room temperature with the following primary antibodies. Anti-AMH goat polyclonal (1:200; Santa Cruz, sc-6886), anti-CYP17A1 goat polyclonal (1:800; Santa Cruz, sc-46081), anti γ-H2AX rabbit monoclonal (1:200; Cell signaling, #9718), anti-SOX9 rabbit polyclonal (1:400; kindly provided by Francis Poulat) and anti-FOXL2 rabbit polyclonal (1:200; kindly provided by Dagmar Wilhelm). After washing for 30 min in PBS, primary antibodies were detected by incubating sections for 1 h at room temperature with Alexa dye-linked secondary antibodies (1000; Molecular Probes). Cell nuclei were visualized with DAPI (4′,6-Diamidino-2-phenylindole; Invitrogen) at a concentration of 0.6 µg/ml in Dako Fluorescent Mounting Medium. Images were captured using fluorescence microscopy (Olympus Corp., New York). Immunofluorescence staining was performed on at least three independent gonad samples per genotype.
Quantitative RT-PCR
mRNA expression analysis was performed as described (89). Briefly, total RNA was extracted from a pair of fetal gonads (with mesonephros removed) at E13.5 using RNeasy Micro kit (Qiagen). cDNA was synthesized using a high-capacity cDNA kit (Life Tech) and quantitative PCR was conducted with SYBR Green mix (Life Tech) on a ViiA7 machine (Life Tech). All primer sequences are shown in Supplementary Material, Table S2. Relative expression was calculated using ΔCt method with Tbp as the normalizing control. Statistical significance was determined with one-way ANOVA with Tukey's range test for multiple comparisons performed using GraphPad Prism 6.
Supplementary Material
Funding
This work was supported by the National Health and Medical Research Council (NHMRC, Australia) Program Grant 1074258 to V.R.H. and P.K., NHMRC, Project Grant 1058548 to V.R.H. and S.B.-F., the Victorian Government's Operational Infrastructure Support Program and the National Institute of Dental and Craniofacial Research (NIDCR) Grant DE020823 to J.V.P.E. V.R.H. is the recipient of the NHMRC (Australia) Research Fellowship 441102.
Supplementary Material
Acknowledgements
We thank Dagmar Wilhelm and Francis Poulat for antibodies, Sue Panckridge for graphic artwork, and Vivien Vasic and David Powell for whole exome sequence analysis.
Conflict of Interest statement. None declared.
References
- 1.Hughes I.A., Houk C., Ahmed S.F., Lee P.A. (2006) Consensus statement on management of intersex disorders. J. Pediatr. Urol., 2, 148–162. [DOI] [PubMed] [Google Scholar]
- 2.Vilain E., Achermann J.C., Eugster E.A., Harley V.R., Morel Y., Wilson J.D., Hiort O. (2007) We used to call them hermaphrodites. Genet. Med., 9, 65–66. [DOI] [PubMed] [Google Scholar]
- 3.Ostrer H. (2014) Disorders of sex development (DSDs): an update. J. Clin. Endocrinol. Metab., 99, 1503–1509. [DOI] [PubMed] [Google Scholar]
- 4.Ono M., Harley V.R. (2013) Disorders of sex development: new genes, new concepts. Nat. Rev. Endocrinol., 9, 79–91. [DOI] [PubMed] [Google Scholar]
- 5.Berta P., Hawkins J.R., Sinclair A.H., Taylor A., Griffiths B.L., Goodfellow P.N., Fellous M. (1990) Genetic evidence equating SRY and the testis-determining factor. Nature, 348, 448–450. [DOI] [PubMed] [Google Scholar]
- 6.Sinclair A.H., Berta P., Palmer M.S., Hawkins J.R., Griffiths B.L., Smith M.J., Foster J.W., Frischauf A.M., Lovell-Badge R., Goodfellow P.N. (1990) A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature, 346, 240–244. [DOI] [PubMed] [Google Scholar]
- 7.Baxter R.M., Vilain E. (2013) Translational genetics for diagnosis of human disorders of sex development. Annu. Rev. Genomics Hum. Genet., 14, 371–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jager R.J., Anvret M., Hall K., Scherer G. (1990) A human XY female with a frame shift mutation in the candidate testis-determining gene SRY. Nature, 348, 452–454. [DOI] [PubMed] [Google Scholar]
- 9.Pearlman A., Loke J., Le Caignec C., White S., Chin L., Friedman A., Warr N., Willan J., Brauer D., Farmer C. et al. (2010) Mutations in MAP3K1 cause 46,XY disorders of sex development and implicate a common signal transduction pathway in human testis determination. Am. J. Hum. Genet., 87, 898–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koopman P., Gubbay J., Vivian N., Goodfellow P., Lovell-Badge R. (1991) Male development of chromosomally female mice transgenic for Sry. Nature, 351, 117–121. [DOI] [PubMed] [Google Scholar]
- 11.Vidal V.P., Chaboissier M.C., de Rooij D.G., Schedl A. (2001) Sox9 induces testis development in XX transgenic mice. Nat. Genet., 28, 216–217. [DOI] [PubMed] [Google Scholar]
- 12.Chaboissier M.C., Kobayashi A., Vidal V.I., Lutzkendorf S., van de Kant H.J., Wegner M., de Rooij D.G., Behringer R.R., Schedl A. (2004) Functional analysis of Sox8 and Sox9 during sex determination in the mouse. Development, 131, 1891–1901. [DOI] [PubMed] [Google Scholar]
- 13.Barrionuevo F., Bagheri-Fam S., Klattig J., Kist R., Taketo M.M., Englert C., Scherer G. (2006) Homozygous inactivation of Sox9 causes complete XY sex reversal in mice. Biol. Reprod., 74, 195–201. [DOI] [PubMed] [Google Scholar]
- 14.Sekido R., Lovell-Badge R. (2008) Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature, 453, 930–934. [DOI] [PubMed] [Google Scholar]
- 15.Cool J., DeFalco T., Capel B. (2012) Testis formation in the fetal mouse: dynamic and complex de novo tubulogenesis. Wiley Interdiscip. Rev. Dev. Biol., 1, 847–859. [DOI] [PubMed] [Google Scholar]
- 16.Wilhelm D., Palmer S., Koopman P. (2007) Sex determination and gonadal development in mammals. Physiol. Rev., 87, 1–28. [DOI] [PubMed] [Google Scholar]
- 17.Ottolenghi C., Pelosi E., Tran J., Colombino M., Douglass E., Nedorezov T., Cao A., Forabosco A., Schlessinger D. (2007) Loss of Wnt4 and Foxl2 leads to female-to-male sex reversal extending to germ cells. Hum. Mol. Genet., 16, 2795–2804. [DOI] [PubMed] [Google Scholar]
- 18.Chassot A.A., Ranc F., Gregoire E.P., Roepers-Gajadien H.L., Taketo M.M., Camerino G., de Rooij D.G., Schedl A., Chaboissier M.C. (2008) Activation of beta-catenin signaling by Rspo1 controls differentiation of the mammalian ovary. Hum. Mol. Genet., 17, 1264–1277. [DOI] [PubMed] [Google Scholar]
- 19.Kim Y., Kobayashi A., Sekido R., DiNapoli L., Brennan J., Chaboissier M.C., Poulat F., Behringer R.R., Lovell-Badge R., Capel B. (2006) Fgf9 and Wnt4 act as antagonistic signals to regulate mammalian sex determination. PLoS Biol., 4, e187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jameson S.A., Lin Y.T., Capel B. (2012) Testis development requires the repression of Wnt4 by Fgf signaling. Dev. Biol., 370, 24–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colvin J.S., Green R.P., Schmahl J., Capel B., Ornitz D.M. (2001) Male-to-female sex reversal in mice lacking fibroblast growth factor 9. Cell, 104, 875–889. [DOI] [PubMed] [Google Scholar]
- 22.Schmahl J., Kim Y., Colvin J.S., Ornitz D.M., Capel B. (2004) Fgf9 induces proliferation and nuclear localization of FGFR2 in Sertoli precursors during male sex determination. Development, 131, 3627–3636. [DOI] [PubMed] [Google Scholar]
- 23.Bagheri-Fam S., Sim H., Bernard P., Jayakody I., Taketo M.M., Scherer G., Harley V.R. (2008) Loss of Fgfr2 leads to partial XY sex reversal. Dev. Biol., 314, 71–83. [DOI] [PubMed] [Google Scholar]
- 24.Kim Y., Bingham N., Sekido R., Parker K.L., Lovell-Badge R., Capel B. (2007) Fibroblast growth factor receptor 2 regulates proliferation and Sertoli differentiation during male sex determination. Proc. Natl Acad. Sci. USA, 104, 16558–16563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Machado A.Z., da Silva T.E., Frade Costa E.M., Dos Santos M.G., Nishi M.Y., Brito V.N., Mendonca B.B., Domenice S. (2012) Absence of inactivating mutations and deletions in the DMRT1 and FGF9 genes in a large cohort of 46,XY patients with gonadal dysgenesis. Eur. J. Med. Genet., 55, 690–694. [DOI] [PubMed] [Google Scholar]
- 26.Rohmann E., Brunner H.G., Kayserili H., Uyguner O., Nurnberg G., Lew E.D., Dobbie A., Eswarakumar V.P., Uzumcu A., Ulubil-Emeroglu M. et al. (2006) Mutations in different components of FGF signaling in LADD syndrome. Nat. Genet., 38, 414–417. [DOI] [PubMed] [Google Scholar]
- 27.Shams I., Rohmann E., Eswarakumar V.P., Lew E.D., Yuzawa S., Wollnik B., Schlessinger J., Lax I. (2007) Lacrimo-auriculo-dento-digital syndrome is caused by reduced activity of the fibroblast growth factor 10 (FGF10)-FGF receptor 2 signaling pathway. Mol. Cell. Biol., 27, 6903–6912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reardon W., Winter R.M., Rutland P., Pulleyn L.J., Jones B.M., Malcolm S. (1994) Mutations in the fibroblast growth factor receptor 2 gene cause Crouzon syndrome. Nat. Genet., 8, 98–103. [DOI] [PubMed] [Google Scholar]
- 29.Rutland P., Pulleyn L.J., Reardon W., Baraitser M., Hayward R., Jones B., Malcolm S., Winter R.M., Oldridge M., Slaney S.F. et al. (1995) Identical mutations in the FGFR2 gene cause both Pfeiffer and Crouzon syndrome phenotypes. Nat. Genet., 9, 173–176. [DOI] [PubMed] [Google Scholar]
- 30.Kan S.H., Elanko N., Johnson D., Cornejo-Roldan L., Cook J., Reich E.W., Tomkins S., Verloes A., Twigg S.R., Rannan-Eliya S. et al. (2002) Genomic screening of fibroblast growth-factor receptor 2 reveals a wide spectrum of mutations in patients with syndromic craniosynostosis. Am. J. Hum. Genet., 70, 472–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee H.J., Cho D.Y., Tsai F.J., Shen W.C. (2001) Antley-Bixler syndrome, description of two new cases and review of the literature. Pediatr. Neurosurg., 34, 33–39. [DOI] [PubMed] [Google Scholar]
- 32.McGlaughlin K.L., Witherow H., Dunaway D.J., David D.J., Anderson P.J. (2010) Spectrum of Antley-Bixler syndrome. J. Craniofac. Surg., 21, 1560–1564. [DOI] [PubMed] [Google Scholar]
- 33.Mangasarian K., Li Y., Mansukhani A., Basilico C. (1997) Mutation associated with Crouzon syndrome causes ligand-independent dimerization and activation of FGF receptor-2. J. Cell. Physiol., 172, 117–125. [DOI] [PubMed] [Google Scholar]
- 34.Galvin B.D., Hart K.C., Meyer A.N., Webster M.K., Donoghue D.J. (1996) Constitutive receptor activation by Crouzon syndrome mutations in fibroblast growth factor receptor (FGFR)2 and FGFR2/Neu chimeras. Proc. Natl Acad. Sci. USA, 93, 7894–7899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Eswarakumar V.P., Horowitz M.C., Locklin R., Morriss-Kay G.M., Lonai P. (2004) A gain-of-function mutation of Fgfr2c demonstrates the roles of this receptor variant in osteogenesis. Proc. Natl Acad. Sci. USA, 101, 12555–12560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Georg I., Bagheri-Fam S., Knower K.C., Wieacker P., Scherer G., Harley V.R. (2010) Mutations of the SRY-responsive enhancer of SOX9 are uncommon in XY gonadal dysgenesis. Sex. Dev., 4, 321–325. [DOI] [PubMed] [Google Scholar]
- 37.Perlyn C.A., DeLeon V.B., Babbs C., Govier D., Burell L., Darvann T., Kreiborg S., Morriss-Kay G. (2006) The craniofacial phenotype of the Crouzon mouse: analysis of a model for syndromic craniosynostosis using three-dimensional MicroCT. Cleft Palate Craniofac. J., 43, 740–748. [DOI] [PubMed] [Google Scholar]
- 38.Snyder-Warwick A.K., Perlyn C.A., Pan J., Yu K., Zhang L., Ornitz D.M. (2010) Analysis of a gain-of-function FGFR2 Crouzon mutation provides evidence of loss of function activity in the etiology of cleft palate. Proc. Natl Acad. Sci. USA, 107, 2515–2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Roscioli T., Elakis G., Cox T.C., Moon D.J., Venselaar H., Turner A.M., Le T., Hackett E., Haan E., Colley A. et al. (2013) Genotype and clinical care correlations in craniosynostosis: findings from a cohort of 630 Australian and New Zealand patients. Am. J. Med. Genet. C Semin. Med. Genet., 163C, 259–270. [DOI] [PubMed] [Google Scholar]
- 40.Lajeunie E., Heuertz S., El Ghouzzi V., Martinovic J., Renier D., Le Merrer M., Bonaventure J. (2006) Mutation screening in patients with syndromic craniosynostoses indicates that a limited number of recurrent FGFR2 mutations accounts for severe forms of Pfeiffer syndrome. Eur. J. Hum. Genet., 14, 289–298. [DOI] [PubMed] [Google Scholar]
- 41.Lavery R., Lardenois A., Ranc-Jianmotamedi F., Pauper E., Gregoire E.P., Vigier C., Moreilhon C., Primig M., Chaboissier M.C. (2011) XY Sox9 embryonic loss-of-function mouse mutants show complete sex reversal and produce partially fertile XY oocytes. Dev. Biol., 354, 111–122. [DOI] [PubMed] [Google Scholar]
- 42.Svingen T., Koopman P. (2013) Building the mammalian testis: origins, differentiation, and assembly of the component cell populations. Genes Dev., 27, 2409–2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Siggers P., Carre G.A., Bogani D., Warr N., Wells S., Hilton H., Esapa C., Hajihosseini M.K., Greenfield A. (2014) A novel mouse Fgfr2 mutant, hobbyhorse (hob), exhibits complete XY gonadal sex reversal. PLoS One, 9, e100447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bullejos M., Koopman P. (2001) Spatially dynamic expression of Sry in mouse genital ridges. Dev. Dyn., 221, 201–205. [DOI] [PubMed] [Google Scholar]
- 45.Wilhelm D., Martinson F., Bradford S., Wilson M.J., Combes A.N., Beverdam A., Bowles J., Mizusaki H., Koopman P. (2005) Sertoli cell differentiation is induced both cell-autonomously and through prostaglandin signaling during mammalian sex determination. Dev. Biol., 287, 111–124. [DOI] [PubMed] [Google Scholar]
- 46.Eswarakumar V.P., Monsonego-Ornan E., Pines M., Antonopoulou I., Morriss-Kay G.M., Lonai P. (2002) The IIIc alternative of Fgfr2 is a positive regulator of bone formation. Development, 129, 3783–3793. [DOI] [PubMed] [Google Scholar]
- 47.Munger S.C., Aylor D.L., Syed H.A., Magwene P.M., Threadgill D.W., Capel B. (2009) Elucidation of the transcription network governing mammalian sex determination by exploiting strain-specific susceptibility to sex reversal. Genes Dev., 23, 2521–2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hajihosseini M.K., Wilson S., De Moerlooze L., Dickson C. (2001) A splicing switch and gain-of-function mutation in FgfR2-IIIc hemizygotes causes Apert/Pfeiffer-syndrome-like phenotypes. Proc. Natl Acad. Sci. USA, 98, 3855–3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jameson S.A., Natarajan A., Cool J., DeFalco T., Maatouk D.M., Mork L., Munger S.C., Capel B. (2012) Temporal transcriptional profiling of somatic and germ cells reveals biased lineage priming of sexual fate in the fetal mouse gonad. PLoS Genet., 8, e1002575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hiramatsu R., Harikae K., Tsunekawa N., Kurohmaru M., Matsuo I., Kanai Y. (2010) FGF signaling directs a center-to-pole expansion of tubulogenesis in mouse testis differentiation. Development, 137, 303–312. [DOI] [PubMed] [Google Scholar]
- 51.Baxter R.M., Arboleda V.A., Lee H., Barseghyan H., Adam M.P., Fechner P.Y., Bargman R., Keegan C., Travers S., Schelley S. et al. (2015) Exome sequencing for the diagnosis of 46,XY disorders of sex development. J. Clin. Endocrinol. Metab., 100, E333–E344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Robin N.H., Falk M.J., Haldeman-Englert C.R. (2011) FGFR-related craniosynostosis syndromes. Gene Rev., [Internet]. [Google Scholar]
- 53.Anderson P.J., Hall C.M., Evans R.D., Hayward R.D., Jones B.M. (1998) The elbow in syndromic craniosynostosis. J. Craniofac. Surg., 9, 201–206. [DOI] [PubMed] [Google Scholar]
- 54.Feild C.R., Leiber A., Toniges C. (1991) Case report: Crouzon syndrome with short stature. Am. J. Med. Sci., 302, 101–102. [DOI] [PubMed] [Google Scholar]
- 55.Wen M.H., Hsiao H.P., Chao M.C., Tsai F.J. (2010) Growth hormone deficiency in a case of Crouzon syndrome with hydrocephalus. Int. J. Pediatr. Endocrinol., 2010, 876514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wagner T., Wirth J., Meyer J., Zabel B., Held M., Zimmer J., Pasantes J., Bricarelli F.D., Keutel J., Hustert E. et al. (1994) Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell, 79, 1111–1120. [DOI] [PubMed] [Google Scholar]
- 57.Foster J.W., Dominguez-Steglich M.A., Guioli S., Kwok C., Weller P.A., Stevanovic M., Weissenbach J., Mansour S., Young I.D., Goodfellow P.N. et al. (1994) Campomelic dysplasia and autosomal sex reversal caused by mutations in an SRY-related gene. Nature, 372, 525–530. [DOI] [PubMed] [Google Scholar]
- 58.Bache K.G., Slagsvold T., Stenmark H. (2004) Defective downregulation of receptor tyrosine kinases in cancer. EMBO J., 23, 2707–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haugsten E.M., Sorensen V., Brech A., Olsnes S., Wesche J. (2005) Different intracellular trafficking of FGF1 endocytosed by the four homologous FGF receptors. J. Cell Sci., 118, 3869–3881. [DOI] [PubMed] [Google Scholar]
- 60.Nagamine C.M., Capehart J., Carlisle C., Chang D. (1998) Ovotestes in B6-XXSxr sex-reversed mice. Dev. Biol., 196, 24–32. [DOI] [PubMed] [Google Scholar]
- 61.Gregoire E.P., Lavery R., Chassot A.A., Akiyama H., Treier M., Behringer R.R., Chaboissier M.C. (2011) Transient development of ovotestes in XX Sox9 transgenic mice. Dev. Biol., 349, 65–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sperling S., Grimm C.H., Dunkel I., Mebus S., Sperling H.P., Ebner A., Galli R., Lehrach H., Fusch C., Berger F. et al. (2005) Identification and functional analysis of CITED2 mutations in patients with congenital heart defects. Hum. Mutat., 26, 575–582. [DOI] [PubMed] [Google Scholar]
- 63.Liu Y., Wang F., Wu Y., Tan S., Wen Q., Wang J., Zhu X., Wang X., Li C., Ma X. et al. (2014) Variations of CITED2 are associated with congenital heart disease (CHD) in Chinese population. PLoS One, 9, e98157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen C.M., Bentham J., Cosgrove C., Braganca J., Cuenda A., Bamforth S.D., Schneider J.E., Watkins H., Keavney B., Davies B. et al. (2012) Functional significance of SRJ domain mutations in CITED2. PLoS One, 7, e46256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bamforth S.D., Braganca J., Eloranta J.J., Murdoch J.N., Marques F.I., Kranc K.R., Farza H., Henderson D.J., Hurst H.C., Bhattacharya S. (2001) Cardiac malformations, adrenal agenesis, neural crest defects and exencephaly in mice lacking Cited2, a new Tfap2 co-activator. Nat. Genet., 29, 469–474. [DOI] [PubMed] [Google Scholar]
- 66.Buaas F.W., Val P., Swain A. (2009) The transcription co-factor CITED2 functions during sex determination and early gonad development. Hum. Mol. Genet., 18, 2989–3001. [DOI] [PubMed] [Google Scholar]
- 67.Bhattacharya S., Michels C.L., Leung M.K., Arany Z.P., Kung A.L., Livingston D.M. (1999) Functional role of p35srj, a novel p300/CBP binding protein, during transactivation by HIF-1. Genes Dev., 13, 64–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Su D., Li Q., Guan L., Gao X., Zhang H., Dandan E., Zhang L., Ma X. (2013) Down-regulation of EBAF in the heart with ventricular septal defects and its regulation by histone acetyltransferase p300 and transcription factors smad2 and cited2. Biochim. Biophys. Acta, 1832, 2145–2152. [DOI] [PubMed] [Google Scholar]
- 69.Roelfsema J.H., White S.J., Ariyurek Y., Bartholdi D., Niedrist D., Papadia F., Bacino C.A., den Dunnen J.T., van Ommen G.J., Breuning M.H. et al. (2005) Genetic heterogeneity in Rubinstein-Taybi syndrome: mutations in both the CBP and EP300 genes cause disease. Am. J. Hum. Genet., 76, 572–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Negri G., Milani D., Colapietro P., Forzano F., Della Monica M., Rusconi D., Consonni L., Caffi L.G., Finelli P., Scarano G. et al. (2015) Clinical and molecular characterization of Rubinstein-Taybi syndrome patients carrying distinct novel mutations of the EP300 gene. Clin. Genet., 87, 148–154. [DOI] [PubMed] [Google Scholar]
- 71.Zimmermann N., Acosta A.M., Kohlhase J., Bartsch O. (2007) Confirmation of EP300 gene mutations as a rare cause of Rubinstein-Taybi syndrome. Eur. J. Hum. Genet., 15, 837–842. [DOI] [PubMed] [Google Scholar]
- 72.Yao T.P., Oh S.P., Fuchs M., Zhou N.D., Ch'ng L.E., Newsome D., Bronson R.T., Li E., Livingston D.M., Eckner R. (1998) Gene dosage-dependent embryonic development and proliferation defects in mice lacking the transcriptional integrator p300. Cell, 93, 361–372. [DOI] [PubMed] [Google Scholar]
- 73.Thevenet L., Mejean C., Moniot B., Bonneaud N., Galeotti N., Aldrian-Herrada G., Poulat F., Berta P., Benkirane M., Boizet-Bonhoure B. (2004) Regulation of human SRY subcellular distribution by its acetylation/deacetylation. EMBO J., 23, 3336–3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xiang W., Wen Z., Pang W., Hu L., Xiong C., Zhang Y. (2011) CR16 forms a complex with N-WASP in human testes. Cell Tissue Res., 344, 519–526. [DOI] [PubMed] [Google Scholar]
- 75.Suetsugu S., Banzai Y., Kato M., Fukami K., Kataoka Y., Takai Y., Yoshida N., Takenawa T. (2007) Male-specific sterility caused by the loss of CR16. Genes Cells, 12, 721–733. [DOI] [PubMed] [Google Scholar]
- 76.Li H., Durbin R. (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics, 25, 1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., Philippakis A.A., del Angel G., Rivas M.A., Hanna M. et al. (2011) A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet., 43, 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M. et al. (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res., 20, 1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang K., Li M., Hakonarson H. (2010) ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res., 38, e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu X., Jian X., Boerwinkle E. (2013) dbNSFP v2.0: a database of human non-synonymous SNVs and their functional predictions and annotations. Hum. Mutat., 34, E2393–E2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu X., Jian X., Boerwinkle E. (2011) dbNSFP: a lightweight database of human nonsynonymous SNPs and their functional predictions. Hum. Mutat., 32, 894–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kumar P., Henikoff S., Ng P.C. (2009) Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc., 4, 1073–1081. [DOI] [PubMed] [Google Scholar]
- 83.Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. (2010) A method and server for predicting damaging missense mutations. Nat. Methods, 7, 248–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schwarz J.M., Cooper D.N., Schuelke M., Seelow D. (2014) MutationTaster2: mutation prediction for the deep-sequencing age. Nat. Methods, 11, 361–362. [DOI] [PubMed] [Google Scholar]
- 85.Thorvaldsdottir H., Robinson J.T., Mesirov J.P. (2013) Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform., 14, 178–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. (2011) Integrative genomics viewer. Nat. Biotechnol., 29, 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Richards C.S., Bale S., Bellissimo D.B., Das S., Grody W.W., Hegde M.R., Lyon E., Ward B.E. (2008) ACMG recommendations for standards for interpretation and reporting of sequence variations: revisions 2007. Genet. Med., 10, 294–300. [DOI] [PubMed] [Google Scholar]
- 88.Lavrovsky Y., Song C.S., Chatterjee B., Roy A.K. (1998) A rapid and reliable PCR-based assay for gene transmission and sex determination in newborn transgenic mice. Transgenic Res., 7, 319–320. [DOI] [PubMed] [Google Scholar]
- 89.Zhao L., Ng E.T., Davidson T.-L., Longmuss E., Urschitz J., Elston M., Moisyadi S., Bowles J., Koopman P. (2014) Structure–function analysis of mouse Sry reveals dual essential roles of the C-terminal polyglutamine tract in sex determination. Proc. Natl Acad. Sci. USA, 111, 11768–11773. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.