Abstract
Alzheimer's disease (AD) and related tauopathies comprise a large group of neurodegenerative diseases associated with the pathological aggregation of tau protein. While much effort has focused on understanding the function of tau, little is known about the endogenous mechanisms regulating tau metabolism in vivo and how these contribute to disease. Previously, we have shown that the microRNA (miRNA) cluster miR-132/212 is downregulated in tauopathies such as AD. Here, we report that miR-132/212 deficiency in mice leads to increased tau expression, phosphorylation and aggregation. Using reporter assays and cell-based studies, we demonstrate that miR-132 directly targets tau mRNA to regulate its expression. We identified GSK-3β and PP2B as effectors of abnormal tau phosphorylation in vivo. Deletion of miR-132/212 induced tau aggregation in mice expressing endogenous or human mutant tau, an effect associated with autophagy dysfunction. Conversely, treatment of AD mice with miR-132 mimics restored in part memory function and tau metabolism. Finally, miR-132 and miR-212 levels correlated with insoluble tau and cognitive impairment in humans. These findings support a role for miR-132/212 in the regulation of tau pathology in mice and humans and provide new alternatives for therapeutic development.
Introduction
The microtubule-associated protein tau functions to promote microtubule assembly and stabilization in neurons, which is necessary for neurite outgrowth and axonal transport (1–3). The abnormal aggregation of hyperphosphorylated tau is a common pathological feature of neurodegenerative disorders collectively known as tauopathies, including Alzheimer's disease (AD), progressive supranuclear palsy (PSP) and frontotemporal lobe dementia (FTLD-Tau) (3–5). Tau is encoded by an alternatively spliced gene (MAPT) located on chromosome 17. Missense or splice site mutations in MAPT cause frontotemporal dementia and parkinsonism linked to chromosome 17 (FTDP-17) (6,7). An imbalance between tau kinases and phosphatases is also thought to participate in tau dysfunction and disease pathophysiology; however, the molecular mechanisms leading to tau hyperphosphorylation and aggregation remain poorly understood. Interestingly, copy number variations or partial deletion of MAPT has been identified in AD and FTLD cases (8–11), suggesting that changes in tau gene dosage are sufficient to trigger disease. Consistent with this hypothesis, specific MAPT haplotypes are associated with increased expression of tau and risk for AD (12,13). The identification of endogenous mechanisms that participate in tau metabolism (dys)regulation leading to aggregation is therefore of high interest.
The short (∼22 nucleotides) regulatory miRNAs are abundantly expressed in the brain (14,15) and are essential for brain development and maintenance (16,17). MiRNAs function to repress protein output by binding to target mRNA sequences, typically within the 3′ untranslated region (3′ UTR). Each miRNA can regulate multiple genes, thus potentially acting on biological pathways (17,18). Alternatively, some miRNAs can function through specific key target genes or ‘master switches’ (19). MiRNAs are clearly essential for post-mitotic neuronal survival (20), whereas abnormal miRNA expression patterns are observed in neurodegenerative disorders in humans (21). While several miRNAs can participate in the regulation of disease-related genes (22–24), understanding the cause and effect of relationship between miRNA dysregulation and disease development in vivo, particularly in mammals, remains an unresolved issue (25,26).
miR-132 and miR-212 are expressed from a bicistronic locus located on chromosome 17 in humans (chromosome 11 in mice) (27). Both miRNAs have similar mature sequences and share the same seed region, although miR-132 is the major functional species in the brain (28). While the role of miR-132 (and miR-212) in the central nervous system is not fully understood, it is proposed to function in neurite outgrowth (29), dendritic growth and arborization (28), synaptic transmission and plasticity (30,31), learning and memory (32,33), apoptosis (34) and neuroprotection (35). Interestingly, the miR-132/212 cluster is frequently downregulated in AD and other tauopathies (34,36–41). In AD, miR-132 or miR-212 levels correlate with the severity of tau pathology (38,39), whereas miR-132 and tau are co-expressed in the same neuronal populations (39). Previously, we have shown that miR-132 can regulate tau alternative splicing in vitro by targeting polypyrimidine tract-binding protein 2 (PTBP2), whereas miR-132 levels correlate with tau splicing defects in PSP cases (40). Despite these observations, a direct functional link between miR-132 or miR-212 and tau is still lacking.
In the present study, we evaluated the effects of miR-132/212 genetic deletion on tau metabolism in mice. We show that miR-132/212 deficiency leads to disease-related changes in tau expression, phosphorylation and aggregation. Interestingly, tau hyperphosphorylation and aggregation is exacerbated in a combined AD/FTLD mouse model lacking the miR-132/212 cluster. This latter model is also associated with long-term memory deficits, whereas injection of miR-132 mimics ameliorates memory retention in diseased mice. In humans, miR-132 and miR-212 levels correlate with insoluble tau and cognitive decline. Collectively, these results highlight the potential importance of miR-132/212 in tauopathies and open the door to the development of new therapies for neurodegenerative disorders.
Results
Abnormal tau metabolism in miR-132/212 knockout mice
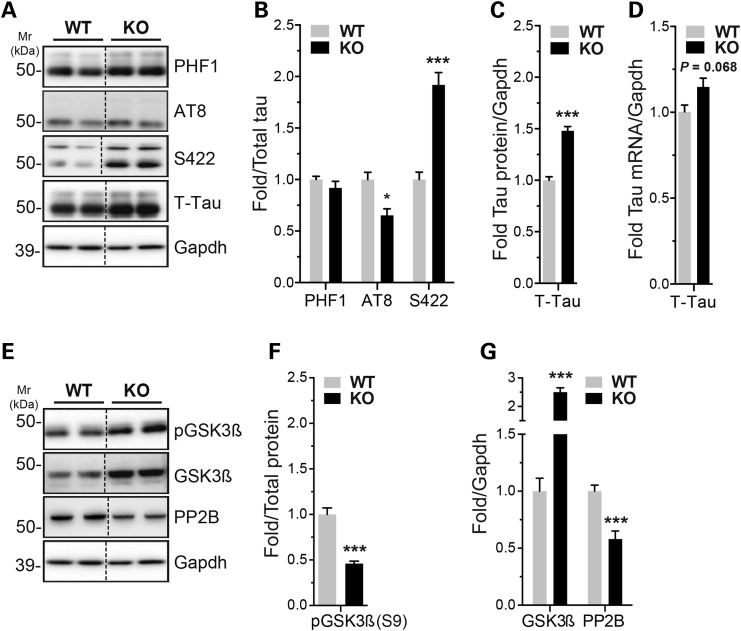
In the present study, we used miR-132/212 knockout (KO) mice (28), in which both miR-132 and miR-212 are absent from the brain (33). By western blot, we observed an increase in total tau protein levels in adult KO mice when compared with controls (WT) (Fig. 1A and C). A non-statistical trend towards increased Mapt (tau) mRNA levels was observed as well (Fig. 1D). Tau phosphorylation at S422 epitope was significantly upregulated in the KO mice. Tau AT8 was slightly downregulated, whereas PHF1 remained unchanged, after normalization to total tau (although both epitopes remain higher compared with wild-type mice) (Fig. 1A and B). Analysis of tau kinases and phosphatases for these epitopes (42) identified glycogen synthase kinase 3 beta (GSK-3β) and calcineurin/PP2B as effectors of tau hyperphosphorylation (Fig. 1E–G). No changes were observed in extracellular signal-regulated kinases 1 and 2 (ERK1/2) (data not shown). Notably, mice lacking miR-132/212 displayed no significant differences in body weight, body temperature or blood glucose levels when compared with littermate controls (Supplementary Material, Fig. S1). Controlling these latter parameters is important, as they are known to influence tau phosphorylation in vivo (43,44).
Figure 1.
miR-132/212 deficiency alters tau expression and phosphorylation in mice. (A, B, C) Representative western blot of endogenous tau and phospho-tau in 6-month-old miR-132/212 KO mice and wild-type littermate controls (n = 8/group, cortex, mixed gender). Total tau or Gapdh was used as normalization control, and quantifications are shown. (D) Real-time quantitative RT-PCR of endogenous tau mRNA in WT and KO mice aged 6 months (n = 5/group, mixed gender). Gapdh mRNA was used as normalization control. (E, F, G) Representative western blot of endogenous GSK-3β, phospho-GSK-3β S9 (inactivated form) and PP2B (n = 8/group, cortex, mixed gender). Gapdh was used as normalization control, and relative quantifications are shown. Statistical significance was determined by Student's unpaired t-test, where *P < 0.05 and ***P < 0.001
Tau is a direct target of miR-132
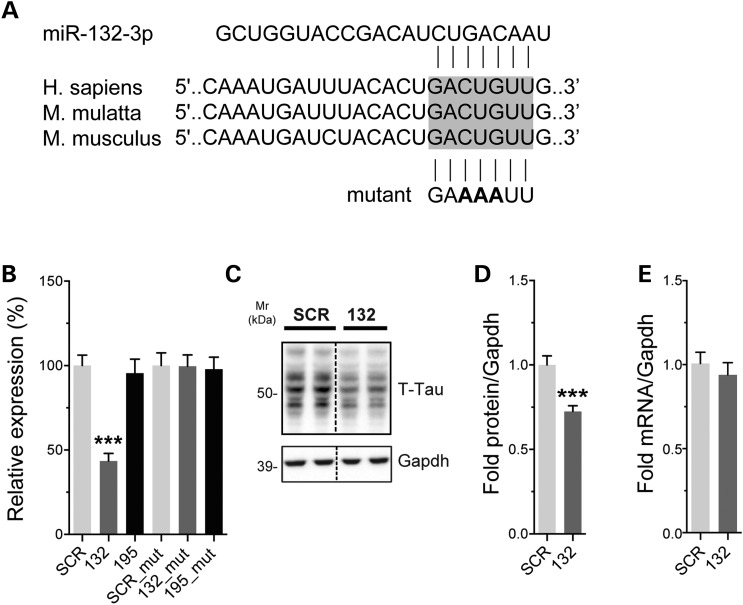
The increase of total tau levels prompted us to search for potential miR-132/212-binding sites in tau mRNA. Bioinformatics analysis using TargetScan (www.targetscan.org) identified a putative miR-132/212 target site located in the 3′ UTR of mapt (Fig. 2A). To confirm these predictions, we used a luciferase reporter gene containing the mouse tau 3′ UTR. Co-transfection of miR-132 mimics with the reporter construct caused a significant downregulation of luciferase activity (tau expression) in Neuro2a cells (Fig. 2B). Here, a scrambled miRNA mimic was used as negative control. An unrelated miRNA, which is not predicted to bind to the 3′ UTR of tau (miR-195), was used as an additional control. Mutating the predicted miRNA:mRNA pairing region abolished the regulatory effect of miR-132 on tau (Fig. 2B). In another set of experiments, we transfected miR-132 mimics into naive Neuro2a cells. Introduction of miR-132 caused a significant reduction in endogenous tau protein levels (Fig. 2C and D), consistent with our previous observations (40). Tau mRNA remained unchanged in these conditions (Fig. 2E). These in vitro assays indicate that miR-132 binds directly to tau mRNA to regulate its expression, providing a mechanism for the observed higher levels of tau in the KO mice.
Figure 2.
Tau is a direct target of miR-132. (A) Sequence of the tau 3′ UTR showing the predicted miR-132/212-binding site. This region is highly conserved (shown here is human, monkey and mouse sequences). The miR-132 site mutation is shown in bold. (B) Left panel: Neuro2a cells were transfected with 50 nm final concentration of miR-132 or miR-195 mimics. Twenty-four hours post-transfection, luciferase signal was measured. Signals were normalized to Renilla luminescence for transfection efficiency, and graph represents the relative luciferase signals compared with a scrambled mimic control (SCR). Right panel: luciferase assays were performed using a mutant 3′ UTR construct for tau. Here, cells were treated with and 10 nm of miR-132 or miR-195 mimics. The tau mutation completely blocked the effects of miR-132. (C, D) Representative western blot of naive Neuro2a cells treated with 50 nm final concentration of miR-132 or SCR control. Cells were lysed 24 h post-transfection. Gapdh was used as normalization control, and quantifications are shown (n = 3 in triplicate). (E) Tau mRNA quantification after miR-132 overexpression (50 nm) in Neuro2a cells compared with SCR control. Gapdh served as normalization control (n = 2 in triplicate). Statistical significance was assessed by one-way ANOVA with Bonferroni multiple comparison test, where ***P < 0.001 and by Student's unpaired t-test, where ***P < 0.001.
miR-132/212 loss accentuates tau hyperphosphorylation in diseased mice
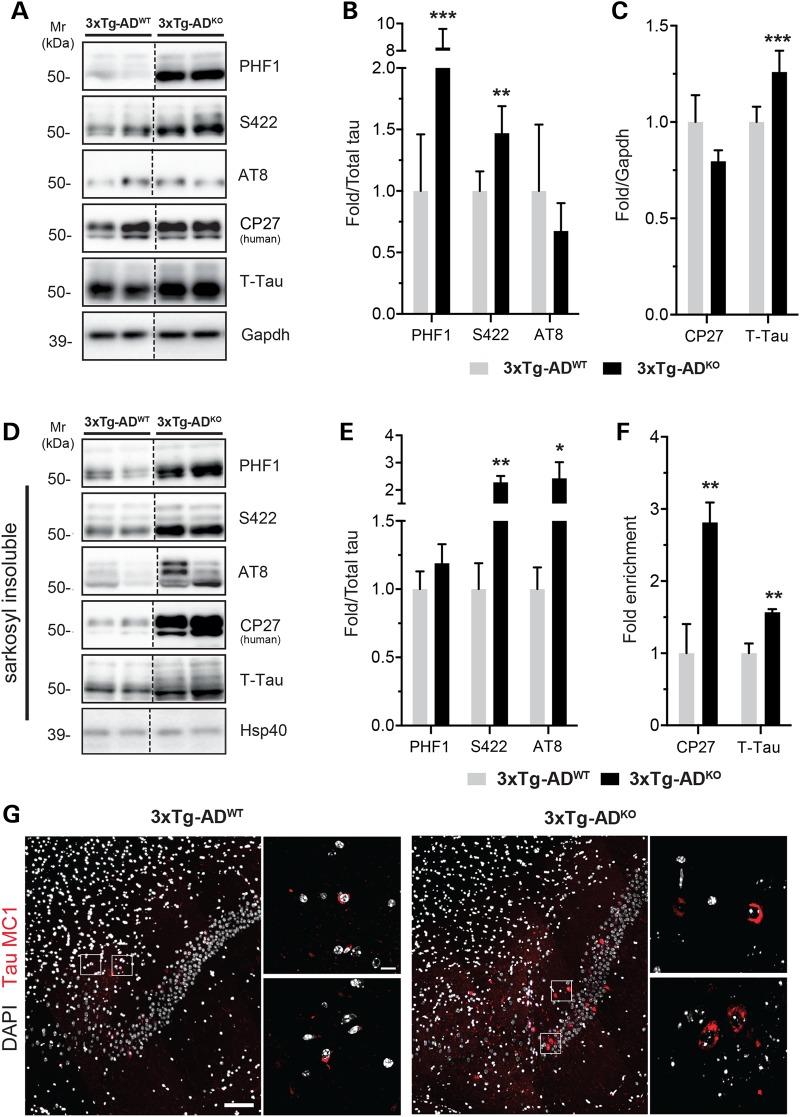
We next studied the effects of miR-132/212 deficiency in disease settings. To this end, we generated triple transgenic AD mice (3xTg-AD) (45) lacking the miR-132/212 cluster. The 3xTg-AD mice harbor both AD (APPSwe/PSENM146V) and FTLD (TauP301L) related genes and thus provide an interesting model to study AD and tauopathies in general by expressing human mutant tau. The 3xTg-AD/miR-132/212 knockout mice (3xTg-ADKO) had increased levels of total tau when compared with controls (3xTg-ADWT) (Fig. 3A and C). This effect was specific for endogenous mouse tau, as human tau remained unchanged in these conditions (as determined using the human-specific CP27 antibody) (46). Interestingly, both S422 and PHF1 tau phospho-epitopes were upregulated in the 3xTg-ADKO (Fig. 3A and B). Tau AT8 phospho-epitope remained unchanged in these conditions. Overall, tau hyperphosphorylation is accentuated in an AD model deficient for the miR-132/212 cluster.
Figure 3.
miR-132/212 deletion promotes tau phosphorylation and aggregation. (A, B, C) Representative western blot of endogenous tau, human tau (CP27) and phospho-tau (PHF1, S422, AT8) in 6-month-old 3xTg-ADKO and 3xTg-ADWT mice (n = 8/group, cortex, mixed gender). Gapdh and total tau were used as normalization control, and quantifications are shown. (D, E, F) Immunoblot analysis of sarkosyl-insoluble total tau, human tau and phospho-tau in the same samples shown in A. Total tau was used as normalization, and quantifications are shown. Heat shock protein of 40 kDa (Hsp40/DnaJ) was used as internal loading control. (F) Quantifications of total tau and human-specific tau. (G) Representative images of tau MC1 immunostainings (red) of 18-month-old 3xTg-ADWT and 3xTg-ADKO mice in the subiculum dorsal brain region (n = 4/genotype, mixed gender). Tau tangles are zoomed. Scale bars are 100 and 10 μm (zoomed panels). Statistical significances were assessed by Student's unpaired t-test, where *P < 0.05, **P < 0.01, and ***P < 0.001.
Deletion of miR-132/212 promotes tau aggregation in mice
We also measured tau aggregation in the different mouse models. Sarkosyl-insoluble mouse tau accumulated in non-transgenic miR-132/212-deficient mice (Supplementary Material, Fig. S2). Interestingly, human tau (CP27 epitope) was strongly enriched in the sarkosyl-insoluble fraction of 3xTg-ADKO mice when compared with 3xTg-ADWT mice (Fig. 3D and F). As control for these experiments, we analyzed soluble and sarkosyl-insoluble tau in non-demented and AD individuals (Supplementary Material, Fig. S2). Notably, hyperphosphorylated tau (PHF1 and AT8 epitopes) accumulated in the sarkosyl-insoluble fraction of 3xTg-ADKO mice when compared with respective controls (Fig. 3D and E). In aged 3xTg-ADKO mice, we observed a strong increase of MC1 (Fig. 3G), a conformation-dependent tau antibody that recognizes an early pathological tau species (47,48). Thus, miR-132/212 loss is associated with disease-relevant changes in tau aggregation and deposition.
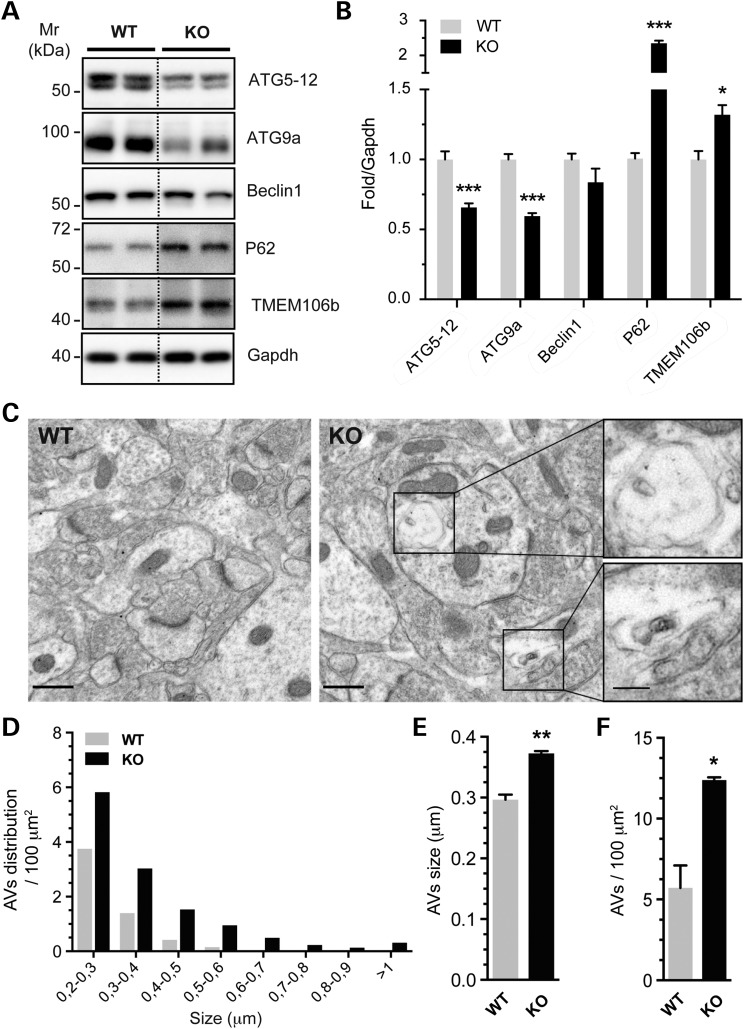
Although increased expression of tau would be sufficient to cause its aggregation, we wanted to explore if other mechanisms could participate in this process. Among known factors involved in tau aggregation, autophagy is of particular interest (49–52). Interestingly, Ucar and colleagues have recently shown that autophagy is defective in miR-132/212 KO cardiomyocytes in mice (53). We therefore sought to analyze autophagy in the brains of miR-132/212-deficient mice. Western blot analysis was performed using a panel of well-characterized autophagy markers, including p62/SQSTM1, Atg9a, Atg5–12, Beclin-1 and Tmem106b. We observed a significant upregulation of p62 and Tmem106b in the KO mice, whereas Atg9a and Atg5-12 were downregulated (Fig. 4A and B). Electron microscopy analysis revealed a net increase in the size and number of autophagic vacuoles (Fig. 4C–F), confirming that the autophagic process is impaired in the absence of miR-132/212. Taken together, these results indicate that loss of miR-132/212 function in the brain leads to pathological-like aggregation of tau, likely via multiple mechanisms including autophagy impairment.
Figure 4.
miR-132/212 knockout is prone to autophagy alteration in the brain. (A, B) Western blot analysis of autophagic component proteins (ATG5–12, ATG9a, Beclin1, P62) and TMEM106b in 6-month-old miR-132/212 KO mice (n = 8/group, cortex, mixed gender). Gapdh served as normalization control, and relative quantifications are shown. (C) (right panel) Representative EM images of 12-month-old miR-132/212 KO CA1 brain region (n = 3/genotype, mixed gender). Autophagic vacuoles (AVs) are zoomed. Scale bar are 500 and 100 nm (zoomed) respectively. EM images of control from the same brain region are shown in left panel. (D) Distribution number organized by AVs size is shown. Size (E) and number in the field (F) are measured from EM images. Statistical significances were assessed by Student's unpaired t-test, where *P < 0.05, **P < 0.01 and ***P < 0.001.
miR-132 treatment improves long-term memory in diseased mice
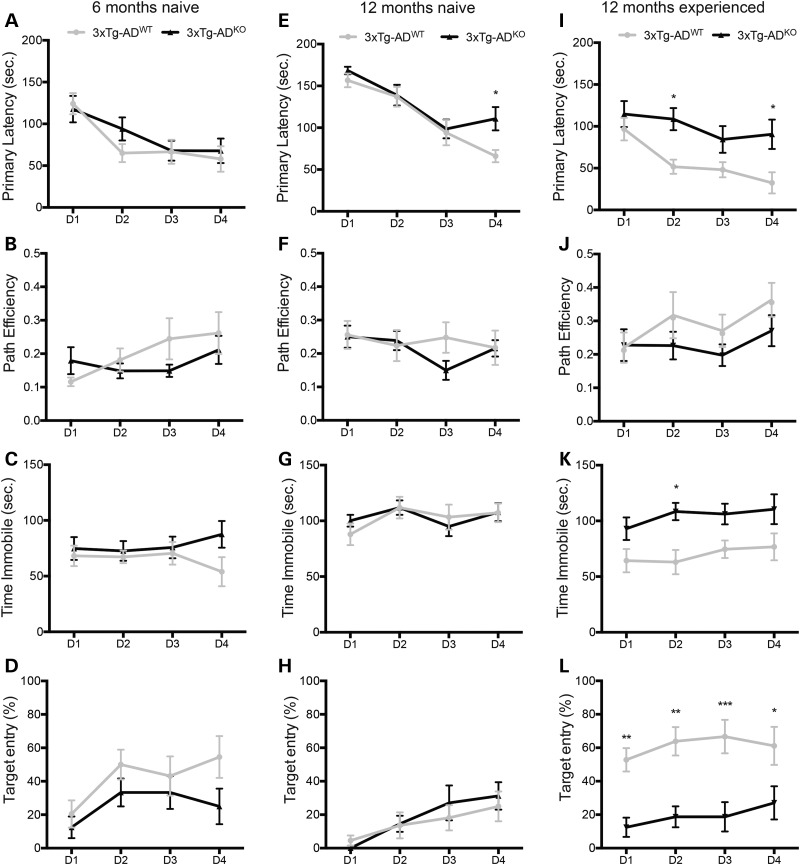
We have previously shown that miR-132/212 knockout mice have impaired memory formation and retention (33). We therefore sought to determine whether miR-132/212 deficiency could exacerbate cognitive deficits in 3xTg-AD mice. We observed little or no differences in (poor) memory performances between 3xTg-ADWT and 3xTg-ADKO mice in the Barnes maze at 6 and 12 months of age (Fig. 5A–H). As positive control, we compared 3xTg-ADWT mice with non-transgenic mice (Supplementary Material, Fig. S3). Interestingly, 12-month-old experienced (pre-conditioned) 3xTg-ADWT mice performed better in various behavioral tasks when compared with age-matched experienced 3xTg-ADKO mice (Fig. 5I–L). No significant changes were observed at the probe trial (Day 5) between naive and experienced 3xTg-ADWT mice and 3xTg-ADKO mice (Supplementary Material, Fig. S4). Taken together, these results indicate that long-term memory retention is affected by miR-132/212 loss in diseased mice.
Figure 5.
Long-term memory is affected in diseased mice lacking miR-132/212. Learning paradigm representations of naive 6-month-old (A–D) and 12-month-old (E–H) 3xTg-ADWT mice and 3xTg-ADKO mice (n = 12/group, mixed gender). (I–L) Learning phase representations of experienced 12-month-old 3xTg-ADWT mice and 3xTg-ADKO mice (n = 12/group, mixed gender). The ‘naive’ mice had no prior experience in the Barnes maze or any other behavioral tasks. The ‘experienced’ mice performed the Barnes maze test (learning phase) 6 months earlier. Statistical significances were assessed by two-way ANOVA (repeated measures) with Bonferroni multiple comparison test, where *P < 0.05, **P < 0.01 and ***P < 0.001. D, day.
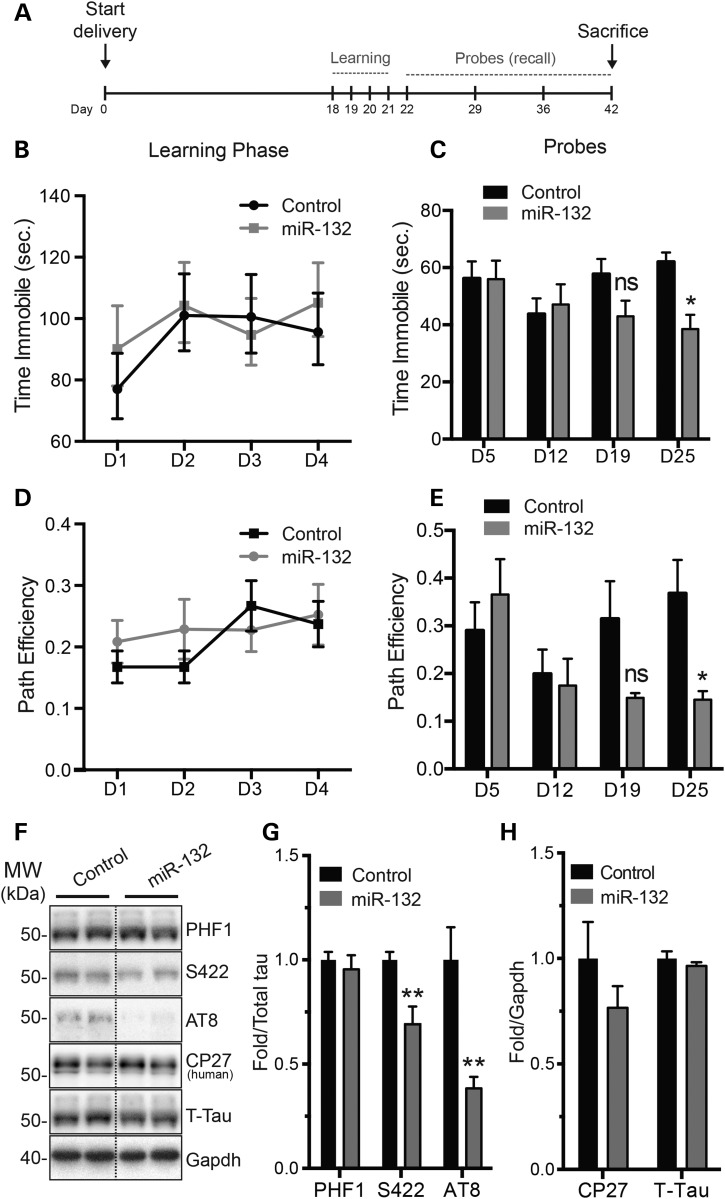
To define whether miR-132 treatment could improve memory deficits in diseased mice, miR-132 mimics were delivered into 3xTg-ADWT mice using osmotic pumps, which allows widespread delivery in the brain [(54) and Supplementary Material, Fig. S5]. No obvious improvement was observed in the Barnes maze during the learning phase (Fig. 6A, B and D). However, when the probe test (recall) was performed 3 weeks later, miR-132-treated mice performed significantly better than controls (Fig. 6C and E). These results are consistent with a role for miR-132 in the regulation of long-term memory. Notably, treatment with miR-132 mimics induced a significant reduction of phosphorylated tau at AT8 and S422 epitopes (Fig. 6F and G). Mouse and human tau protein levels did not change in these conditions (Fig. 6H). Lastly, miR-132 mimic treatment induced various changes in autophagy markers (Supplementary Material, Fig. S6).
Figure 6.
miR-132 brain injections partially rescue memory deficits in 3xTg-AD. (A) Time scale of our experimental paradigms, including the learning and probe phases in the Barnes maze. (B, D) Learning phase of 12-month-old 3xTg-ADWT mice brain treated with miR-132 mimics compared with 3xTg-ADWT controls (n = 11/group, mixed gender). No significant changes were observed during this period. (C, E) Probe trials were done at Day 1, 1 week, 2 weeks and 3 weeks after the learning phase. Here, a significant difference was observed in mobility and path efficiency at 3 weeks. Other behavioral parameters were unchanged (data not shown). (F–H) Western blot analysis of tau expression and phosphorylation after sacrifice. Quantifications are shown, where Gapdh and T-tau were used as normalization controls. Statistical significances were assessed by Student's unpaired t-test, where *P < 0.05, **P < 0.01 and ns = P > 0.05. D, day.
Analysis of miR-132 in Alzheimer's disease
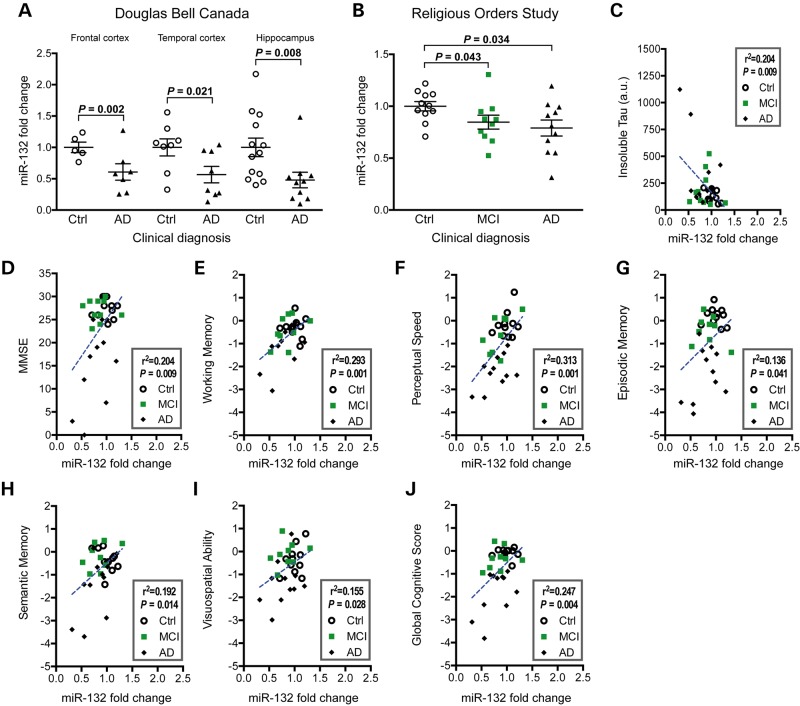
Finally, we asked whether our observations had clinical significance. To validate and extend earlier findings (34,36,37,39), we measured miR-132 levels in various brain regions of AD individuals and non-demented controls in two independent cohorts. In the Douglas Bell Canada Brain Bank samples, we observed lower levels of miR-132 in the temporal cortex (P = 0.021, n = 5–7/group) (Fig. 7A), validating our recent data (36). In the same cohort, miR-132 was lower in the frontal cortex (P = 0.002, n = 8/group) and hippocampus (P = 0.008, n = 10–13/group) (Fig. 6A). In the Religious Orders Study samples, we also observed a downregulation of miR-132 in the temporal cortex (P = 0.034, n = 11–12/group) (Fig. 7B). Interestingly, a similar result was seen in brains from persons with mild cognitive impairment (MCI) (P = 0.043, n = 10–12/group) (Fig. 7B), suggesting that miR-132 is affected early-on in disease course in a subset of patients, which is consistent with earlier reports (39,55,56). No correlation between miR-132 and postmortem delay (PMD) was detected in humans and mice (Supplementary Material, Fig. S7), demonstrating that miR-132 loss is not a consequence of RNA degradation or poor tissue quality. Interestingly, miR-212 was lower in MCI and AD cases as well (Supplementary Material, Fig. S6). These results suggest that lower levels of the miR-132/212 cluster are a rather generalized phenomenon in AD pathogenesis.
Figure 7.
miR-132 correlates with cognitive decline in AD patients. (A) Relative miR-132 expression levels in various brain regions of postmortem tissues from the Douglas Bell Canada Brain Bank (frontal lobe: n = 5 Ctrl, 7 = AD; temporal lobe: n = 8 Ctrl, 8 = AD; hippocampus: n = 13 Ctrl, 10 = AD). The miRNA let-7a was used as normalization control (using the average of controls as 1-fold). (B) Relative miR-132 expression levels in temporal lobe region of non-demented (n = 11), MCI (n = 10) and AD (n = 11) groups from the Religious Orders Study patients. Statistical significances were obtained with a Mann–Whitney test, and exact P-values are shown. The miRNA let-7a was used as normalization control (using the average of controls as 1-fold). Correlation between temporal lobe miR-132 levels and detergent-insoluble tau (C), MMSE (D), working memory (E), perceptual speed (F), episodic memory (G), semantic memory (H) visuospatial ability (I) or global cognitive scores (J) in the Religious Orders Study cohort. Statistical significances were determined using a linear regression analysis, and exact R2 or P-values are given. Ctrl: non-demented controls, AD: Alzheimer disease, MCI: mild cognitive impairment.
Using previous data from the Religious Orders Study cohort [(57,58) and Supplementary Material, Table S1], we noted a significant correlation between miR-132 and insoluble tau in all cases (P = 0.009, n = 32) (Fig. 7C). No correlation was observed with soluble tau (data not shown). Furthermore, miR-132 was correlated with mini-mental state examination (MMSE) (P = 0.009, n = 32), working memory (P = 0.001, n = 32), perceptual speed (P = 0.001, n = 32), episodic memory (P = 0.041, n = 32), semantic memory (P = 0.014, n = 32), visuospatial ability (P = 0.028, n = 32) and global cognitive score (P = 0.004, n = 32), the latter which is based on the average of 19 cognitive function tests scores (Fig. 7D–J) (Supplementary Material, Table S2). Similar observations were obtained with miR-212 (Supplementary Material, Fig. S8). These observations provide further evidence that miR-132/212 expression plays an important role in AD and is closely related to the progression of the disease.
Discussion
The miR-132/212 family is emerging as an important player in neural plasticity, memory formation and neuroprotection. However, little is known about the precise role of this family in vivo and, more importantly, what are the consequences of miR-132/212 deficiency in the brain. In the current study, we focused on tau, which is associated with >20 neurodegenerative disorders. These experiments are a direct follow-up of our previous findings showing that miRNAs can indirectly regulate tau exon 10 alternative splicing and phosphorylation in vitro (20,40). Now, we demonstrate that miR-132 (and possibly miR-212) is directly implicated in endogenous tau expression, phosphorylation and aggregation. While screening for known tau kinases and phosphatases, we identified GSK3β and PP2B that were altered in the absence of miR-132/212 in mice. We also provide substantial evidence that tau is a direct target of miR-132. Of note, we observed more mouse and human tau insolubility in the absence of miR-132/212 in the brain, as seen in AD. Such defects were associated with autophagy dysfunction. Introduction of miR-132 mimics rescued in part long-term memory deficits associated with diseased mice. Finally, we observed a significant correlation between miR-132, insoluble tau and cognitive impairment in humans, providing clinical relevance to our findings. Taken together, these results implicate the miR-132/212 cluster in many aspects of tau metabolism regulation, whereas loss of miR-132/212 function could contribute significantly to tau pathology and likely other important signaling pathways in the brain.
Previous studies have hinted that tau is prone to miR-132 regulation; however, whether this effect was direct or mediated by other factors remained unclear. For instance, Lau P et al. (39) showed that miR-132 could regulate human tau expression in luciferase-based reporter assays. Dickson JR et al. (59) observed similar results with mouse tau, although these authors could not abrogate the effects of miR-132 following mutagenesis of the tau 3′ UTR. In our opinion, these discrepancies are attributable to technical issues (e.g. choice of site mutagenesis, miRNA mimic dosage) and possibly cell-type effects. Using a more physiological approach, Chi SW et al. (60) documented a direct interaction between endogenous miR-132 and tau mRNA in the mouse brain using Argonaute HITS-CLIP assays. Combined with the results herein, we are therefore confident that tau is a bona fide target of miR-132. It remains to be tested whether miR-212 can regulate tau, which is likely based on seed sequence conservation. It should be pointed out that miR-212 is only weakly expressed in neurons (28). The use of double KO mice is nonetheless essential to avoid potential compensatory mechanisms of miR-212. Of course, our results cannot exclude a role for miR-212 in the brain, and perhaps in non-neuronal cell types. In this regard, miR-212 can also correlate with tau pathology in humans [(38) and results herein]. Another unanswered question is the potential developmental consequences related to miR-132/212 deletion. Conditional knockout of the miR-132/212 locus in adulthood will be necessary to address this issue. It's noteworthy that adult miR-132/212 knockout mice do not display overt changes in brain morphology or cell structure (33), suggesting that miR-132 and miR-212 are dispensable for brain development.
Analysis of tau phosphorylation identified epitope-specific changes in the absence of miR-132/212 in mice. Such changes coincided with (epitope) specific tau kinases and phosphatases (42). Interestingly, GSK-3β and PPP3R1/PPP3R2 (two regulatory subunits of PP2B/calcineurin) genes contain a conserved miR-132/212 target site (www.targetscan.org). The effects of miR-132/212 loss on endogenous signaling pathways therefore need to be scrutinized. Previously we showed that certain memory genes of the miR-132/212 pathway are disrupted in adult miR-132/212 knockout mice, including CREB, BDNF and MeCP2 (33). Since not all tau phosphorylation sites tested are affected (even sometimes inversed) in the miR-132/212 deficient mice, other (or additional) mechanisms are likely at play in tauopathies. In the future, it will be interesting to identify all the factors involved in tau phosphorylation regulation in the miR-132/212-deficient mice, and whether these are affected in disease settings (e.g. with Aβ).
The 3xTg-AD model is commonly used to understand the underlying mechanisms of AD (61). In the present study, we investigated tau metabolism mainly prior to AD pathology (i.e. with Aβ plaques and neurofibrillary tangles) to exclude potential downstream or indirect effects. It's noteworthy that miR-132/212 misregulation is associated with early stages of tau pathology (i.e. Braak stages) in humans (38,39). These experiments showed that miR-132/212 deficiency in 3xTg-AD mice results in increased aggregation of phosphorylated tau. The effects of miR-132/212 loss on human tau are likely independent of gene expression regulation, as the 3xTg-AD mice, as with most tauopathy models, do not contain the human tau 3′ UTR. This is consistent with the observation that total (soluble) human tau levels are not changed in the absence of (or overexpression of) miR-132/212 in vivo.
The results presented herein implicate autophagy alterations in miR-132/212-deficient mice. Interestingly, deregulation of autophagy is observed in AD and various other neurodegenerative disorders (62–64), whereas a genetic link between autophagy and neurodegenerative disorders (some of which have lower miR-132/212 levels) is well documented (65). Obviously, we cannot exclude that autophagy defects may be a secondary event (e.g. abnormal protein synthesis), and other indirect factors or pathways might participate in tau aggregation in vivo. For instance, endogenous mouse tau might influence human tau aggregation (66,67). Aβ peptides can influence human tau aggregation via different signaling pathways (68), alone or in combination with miR-132/212 targets. One should keep in mind, however, that mouse tau is also prone to aggregation, independent of Aβ. In any case, our results are consistent with the notion that human mutant tau is ‘naturally’ more prone to aggregation and that miR-132/212 loss affects the brain at various levels. Furthermore, our overexpression studies in mice further implicate miR-132 in the regulation of autophagy via still undetermined mechanisms.
The treatment of 3xTg-AD mice with miR-132 mimics significantly improved long-term memory deficits, which coincided with reduced tau phosphorylation. Higher doses of miR-132 mimics could equally reduce mouse tau protein levels in vitro (herein) and in vivo (Goupil C., unpublished observations). How miR-132 regulates long-term memory in 3xTg-AD mice is uncertain. One possibility is via acetylcholinesterase (AChE), a previously identified miR-132 target (69) implicated in modulating cholinergic signaling and memory in humans and 3xTg-AD mice (70,71). The downregulation of phosphorylated tau can also participate in memory retention by promoting microtubule stability and axonal transport.
In addition to miR-132, other miRNAs are likely involved in the regulation of tau metabolism in normal or disease conditions. As shown by us earlier, members of the miR-15 family can regulate tau phosphorylation by targeting extracellular signal-regulated kinase 1 (ERK1) (20). Wang et al. (72) showed that miR-138 can induce tau phosphorylation in non-neuronal cells by targeting retinoic acid receptor alpha (RAR-α). Introduction of miR-125b in mice induced tau phosphorylation likely via multiple mechanisms (73). Dickson et al. (59) identified a functional miR-34a target site in the 3′ UTR of human tau. Zhao et al. (74) demonstrated that miR-922 could induce tau phosphorylation by targeting ubiquitin carboxy-terminal hydrolase L1 (UCHL1). More recently, it was shown that miR-219 targets tau and promotes tau-induced toxicity in flies (75). It should be noticed, however, that all previous studies have relied on artificial overexpression paradigms and, therefore, the clinical significance of these observations is uncertain. One example is miR-219, which can modulate tau pathology in Drosophila but is mainly expressed in oligodendrocytes in mammals (76). Also, not all reported miRNAs are shown consistently to be misregulated in AD or other tauopathies.
Using a variety of methods (e.g. microarrays, RNA-Seq, qRT-PCR), several groups have now convincingly demonstrated that miR-132 and/or miR-212 is frequently downregulated in AD and various other tauopathies. These findings, combined with our current data, suggest that miR-132/212 dysregulation might occur early in disease course as well as in different brain regions. It should be stressed, however, that MCI does not necessarily reflect prodromal AD. Previous reports have found a direct relationship between tau pathology (i.e. Braak stages) and miR-132/212 expression levels using unbiased screens; in other words, when compared with all other brain miRs. Whether miR-132/212 can be used in diagnostics is an interesting possibility (55,56). The mechanism(s) involved in miR-132/212 downregulation remain(s), however, to be identified. Mild or advanced tau pathology per se is insufficient to cause miR-132/212 downregulation in mice (data not shown). Our results also exclude long PMDs as a causative factor. Transcription factors such as cAMP-response element binding protein (CREB) (29,77), neurotrophins such as brain-derived neurotrophic factor (BDNF) (78,79) and inflammation mediators such as lipopolysaccharide (LPS) (69) have all been associated with miR-132/212 expression regulation. Interestingly, a recent study has shown that miR-132 reduction occurs in neurons predestined to die, possibly downstream of REST/NRSF (35).
Loss of miR-132/212 expression is reported in various other neurological disorders, including Huntington's disease (80), schizophrenia (81) and amyotrophic lateral sclerosis (82). Interestingly, tau pathology is found with these diseases as well (83,84). Of course, we cannot conclude definitively that miR-132/212 deficiency will lead to increased tau expression and/or abnormal tau metabolism in the brain or other tissues, as miRNAs targets may differ in pathological settings. Studies are underway to find additional miR-132/212 targets in the brain, which will help elucidate the role of these intriguing miRNAs in health and disease.
Material and Methods
Study approval
All human and mouse studies were approved by the national ethical committee protocols and in agreement with the Université Laval ethical committee.
Humans
Brain tissue from the first cohort of patients came from the Douglas Bell Canada Brain Bank, Montreal, Canada, and included non-dementia controls and AD cases, based on neuropathological diagnosis. Patient information is available elsewhere (40,58). Brain tissue from a second, independent cohort of persons came from the Religious Orders Study, Chicago, USA, which consisted of non-demented controls, MCI and AD cases, based on detailed clinical diagnosis. Dementia and AD diagnosis required evidence of decline in cognitive function based on the results of 21 cognitive performance tests. MCI refers to participants with cognitive impairment as assessed by the review of the cognitive performance tests. A global measure of cognition was based on 19 cognitive tests, which were used to summarize 5 cognitive ability categories: episodic memory, semantic memory, working memory, perceptual speed and visuospatial ability (57,85,86). Patient-related data and performance tasks deployed are listed in Supplementary Material, Table S1 and S2, respectively. Blocks of tissue from temporal cortex, prefrontal cortex and hippocampus were dissected and snap-frozen in liquid nitrogen until use.
Mice
The generation of full miR-132/212 knockout mice was described previously (28,33). The miR-132/212 knockout mice (a kind gift from Dr R. H. Goodman, Vollum Institute, USA) were bred with homozygous 3xTg-AD mice (JAX Stock No. 004807). First-generation (F1) offspring gave double heterozygous mice, which were crossed with homozygous 3xTg-AD mice. Second-generation (F2) offspring (heterozygous for miR-132/212 and homozygous for 3xTg-AD) were used in all our experimental groups, unless stated otherwise. The mice were group-housed (2–5 per cage) in a temperature-controlled room (22°C) with 12/12 light–dark cycles. All animals of this study had access to food and water ad libitum. Wild-type or 3xTg-AD mice deficient for miR-132/212 were compared with age-matched littermate controls. All mice were killed without anesthesia to avoid tau hyperphosphorylation unless specified otherwise. The brains were removed, dissected on ice and frozen on dry ice. All tissues were stored at −80°C until use.
Cell culture, transfection and mutagenesis
Mouse Neuroblastoma 2a (Neuro2a) were purchased from ATCC (#CCL-131) and cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Cells were tested negative for mycoplasma contamination. Before transfection, 200 000 cells were seeded into 6-well plates. The next day, cells were transfected with 50 nm of miRNA mimics (Pre-miR miRNA precursor molecules, Life technologies) using Lipofectamine® 2000 (Life Technologies). A scrambled miRNA mimic (Life technologies) was used as negative control. Forty-eight hours post-transfection, cells were processed for western blot analysis. For luciferase assays, 50 nm of mimics or scrambled control was co-transfected in Neuro2a cells with 200 ng of the mouse Mapt 3′ UTR luciferase vector encoding 1944bp (Cat# MmiT025143b-MT01 GeneCopoeia™, Rockville, MD). Mutagenesis and sequencing were performed by TOP Gene Technologies, Inc. (Montréal, Canada). The miR-132 seed region was changed from GACTGTT to GAAAATT. Twenty-four hours post-transfection, the measurements were obtained using Dual-luciferase® reporter assay kit (Promega) and read using an Infinite F200 plate reader (Tecan Group Ltd).
Real-time quantitative RT-PCR
Total RNA from mouse and human brain as well as from Neuro2a cells was extracted with Trizol® Reagent (Life technologies) according to the manufacturer's instructions. Relative expression of mRNA targets was determined by quantitative PCR. One microgram of total RNA was reverse-transcribed using the iScript™ cDNA Synthesis Kit (Bio-RAD). cDNA was diluted 1:50 prior to PCR amplification. The following primers were used: mouse Gapdh 5′ CTTTTGTCAAGCTCATTTCCTGG 3′ forward and 5′ TCTTGCTCAGTGTCCTGGC 3′ reverse; mouse tau 5′ CGCTGGGCATGTGACTCAA 3′ forward and 5′ CGAGGTGTGGCGATCTTCG 3′ reverse. Quantitative detection of PCR products was performed on a lightCycler480 (Roche Life Science) using SsoFast™ EvaGreen® Supermix (Bio-RAD) according to the manufacturer's instructions. TaqMan® miRNA assays (Life Technologies) were used for miR-132 (#000457) quantifications following manufacturer's protocol. RNU19 probe (#001003) (for mouse) or let-7a probe (#000377) (for human) was used as normalization control. Relative expression was calculated by the 2-ΔΔCT method.
Western blotting
Neuro2a cells were rinsed with cold PBS and lysed with a Sonic Dismembrator model 500 (Thermo Scientific) in RIPA buffer containing: 50 mm Tris–HCl, pH 7.4, 1 mm ethylenediaminetetraacetic acid (EDTA), 150 mm NaCl, 0.5% sodium deoxycholate, 1% octylphenol ethoxylate (IGEPAL CA630, Bioshop Canada), phosphatase inhibitors (1 mm activated sodium orthovanadate, 1 mm sodium fluoride), 1 mm phenylmethylsulfonyl fluoride and a complete mini EDTA-free protease inhibitor cocktail tablet (Roche life science). Lysates were incubated on ice for 20 min and spun at 20 000×g for 15–20 min at 4°C. Then, the supernatant was kept and 30 µg of protein was mixed to the NuPAGE® LDS sample buffer (Life technologies) with 5% final volume of ß-mercaptoethanol for immunoblot analysis. Frozen tissues from mice or humans were mechanically homogenized in 5× vol./weight of RIPA buffer, put on ice for 20 min and then centrifuged at 20 000×g for 15 min (low speed lysate). For the insoluble fraction, an aliquot of supernatants from low speed lysates were mixed to 1% N-Lauroylsarcosine sodium salt (Sarkosyl, Sigma) and incubated at 37°C for 1 h on a rotarod. Then, the mixes were centrifuged at 100 000×g at 20°C for 1 h using Sorvall® mTX 150 Ultra Centrifuge (ThermoScientific). After centrifugation, the pellet were washed with 1% sarkosyl and dissolved in a sample protein buffer. The Fusion FX5 imaging system (Vilbert Lourmat, France) and Immobilon™ Western Chemiluninescent HRP Substrate (EMD Millipore) were used for immunoblot detection.
Immunochemistry
Brains were fixed with 4% paraformaldehyde for 72 h and embedded in paraffin. Five-micrometer serial sagittal sections were collected with a microtome. Slices were rehydrated, incubated in blocking solution (7.5% NGS; 0.4% Triton; 1% BSA; PBS) for 2 h and in MC1 antibody solution (5% NGS; 0.4% Triton; PBS) overnight. Slices were incubated 2 h 30 min in the secondary antibody solution (AlexaFluor 568, #A11031; Invitrogen) and 5 min in 30 nm DAPI. Slices were observed using a Zeiss AxioImager M2 microscope under 20× and 63× magnification, and images were processed with a computerized image analysis system (ZEN 2012 SP2 Software, Zeiss).
Antibodies
Antibodies directed against tau include: AT8 (Ser202/Thr205, ThermoFisher), PHF-1 [Ser396/Ser404, (87)], phosphoSer422 (#OPA-03151; ThermoScientific), CP27 (specific to human total tau) and total tau (A0024, Dako Cytomation). The MC1, PHF-1 and CP27 antibodies were a generous gift from Dr Peter Davies (Albert Einstein University, New York, NY). Phospho-ERK (#9101), ERK (#4696), GSK3β-S9 (#9336), PP2B (#2614) and SQSTM1/P62 (#5141) were purchased from Cell Signaling. GSK3β antibody (#610202) was purchased from BD Biosciences. Autophagy Antibody Pack #NB910-94877 (ATG5-12, ATG9a, Beclin1; Novus Biologicals) and TMEM106b (#A303-439A, Bethyl Laboratories) were used. Hsp40/Dnaj antibody (#MAB4145) was bought from R&D Systems. Gapdh antibody was purchased from Millipore.
Electronic microscopy
All procedures were conducted as described previously (88). Three mice per experimental group were anesthetized with sodium pentobarbital (80 mg/kg, i.p.) and perfused through the aortic arch with 3.5% acrolein followed by 4% paraformaldehyde. Transverse sections of the brain (50 mm thick) were cut in sodium phosphate buffer (PBS; 50 mm at pH 7.4) using a vibratome and stored at −20°C in cryoprotectant (30% glycerol and 30% ethylene glycol in PBS) until further processing. In preparation for imaging, the sections were washed with PBS, post-fixed flat in 1% osmium tetroxide and dehydrated in ascending concentrations of ethanol. They were treated with propylene oxide, impregnated in Durcupan (Electron Microscopy Sciences) overnight at RT, mounted between ACLAR embedding films (Electron Microscopy Sciences) and cured at 55°C for 72 h. Areas of CA1 stratum radiatum, at a level approximating the transverse planes Bregma −3.27 to −4.03 (89), were excised from the embedding films and re-embedded at the tip of resin blocks. Ultrathin (65–80 nm) sections were cut with an ultramicrotome (Leica Ultracut S), collected on bare square-mesh grids and examined at 80 kV with an FEI Tecnai Spirit G2 transmission electron microscope equipped with an ORCA-HR digital camera (10 MP; Hamamatsu). For analysis, 30 randomly selected electronic microscopy (EM) images per animal were captured at final magnification of 6800×. In each captured field, the number of Autophagic Vacuoles (AVs) was counted by visual inspection, and the size of each AVs was measured using ImageJ software. All vacuoles under 200 µm were excluded to avoid vacuoles from the endosomal pathway.
miRNA delivery studies in vivo
Twelve-month-old 3xTg-ADWT mice (mixed gender) were used in all experiments. The mini-pumps (ALZET® model 2006) and brain infusion kits (cat#8663) were purchased from Durect (Denmark). Pre-operative procedure included 30 µl of Anafen (1 mg/ml), 100 µl Marcaine (5.0 mg/ml) and 500 µl saline (0.9%). ALZET® mini-osmotic pumps were implanted subcutaneously according to the manufacturer's instructions. miR-132 mimics (CONmir® miRNA mimics, Riboxx, Germany) were administrated into the brain (coordinates: ventricle A/P = −0.22 M/l = 0.0 D/v = −3.5) for 42 days at a rate of 1.8 µg/day (n = 10). Five percent final volume of in vivo jetPEI® (Polyplus®, France) was added to the mixture before delivery. Mice treated with vehicle alone (diluent + in vivo jetPEI®) served as negative controls (n = 11). During the post-operative procedure, mice were treated with 50 µl Anafen (1 mg/ml) and 500 µl saline (0.9%). Mice were sacrificed without anesthesia 42 days post-injection.
Memory tests
3xTg-AD mice were subjected to spatial memory tasks in the Barnes maze, as described previously (33). Experienced mice (3xTg-ADWT or 3xTg-ADKO) received a 5-day learning trial 6 months before the second protocol (n = 12/group). For mice treated with miR-132 mimics or vehicle, long-term memory retention was tested using additional probes (recalls) at Days 29, 36 and 42 (i.e. 1, 2 and 3 weeks later).
Statistics
Western blot quantifications were done using ImageJ 1.6 (http://imagej.nih.gov/ij/). The data were analyzed with the two-tailed unpaired Student t-test with Prism 6.0d software (GraphPad Software, San Diego, CA). For multiple group comparisons, the one-way analysis of variance (one-way ANOVA) followed by the Bonferroni post hoc test was used. For Barnes Maze data analysis, the two-way ANOVA (repeated measures) followed by the Bonferroni post hoc test was used. The data are expressed as means ± SEM with P-values of <0.05 considered significant. Correlations were calculated using the Prism 6.0d software.
Supplementary Material
Funding
This work was supported by the Canadian Institutes of Health Research (grant 00892-000), the Alzheimer Society of Canada (grants 1428 and 1236) and the Fonds de Recherche du Québec Santé/INSERM (grants 23635 and 31199). The Religious Orders Study is supported by National Institute on Aging grants P30AG10161 and RF1AG15819.
Supplementary Material
Acknowledgments
Conflict of Interest statement. None declared.
References
- 1.Buee L., Bussiere T., Buee-Scherrer V., Delacourte A., Hof P.R. (2000) Tau protein isoforms, phosphorylation and role in neurodegenerative disorders. Brain Res. Rev., 33, 95–130. [DOI] [PubMed] [Google Scholar]
- 2.Schraen-Maschke S., Sergeant N., Dhaenens C.M., Bombois S., Deramecourt V., Caillet-Boudin M.L., Pasquier F., Maurage C.A., Sablonniere B., Vanmechelen E. et al. (2008) Tau as a biomarker of neurodegenerative diseases. Biomark. Med., 2, 363–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sergeant N., Delacourte A., Buee L. (2005) Tau protein as a differential biomarker of tauopathies. Biochimica et Biophysica Acta, 1739, 179–197. [DOI] [PubMed] [Google Scholar]
- 4.Spillantini M.G., Goedert M. (1998) Tau protein pathology in neurodegenerative diseases. Trends Neurosci., 21, 428–433. [DOI] [PubMed] [Google Scholar]
- 5.Tolnay M., Probst A. (1999) Review: tau protein pathology in Alzheimer's disease and related disorders. Neuropathol. Appl. Neurobiol., 25, 171–187. [DOI] [PubMed] [Google Scholar]
- 6.Gallo J.M., Noble W., Martin T.R. (2007) RNA and protein-dependent mechanisms in tauopathies: consequences for therapeutic strategies. Cell. Mol. Life Sci., 64, 1701–1714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu F., Gong C.X. (2008) Tau exon 10 alternative splicing and tauopathies. Mol. Neurodegener., 3, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hooli B.V., Kovacs-Vajna Z.M., Mullin K., Blumenthal M.A., Mattheisen M., Zhang C., Lange C., Mohapatra G., Bertram L., Tanzi R.E. (2014) Rare autosomal copy number variations in early-onset familial Alzheimer's disease. Mol. Psychiatry, 19, 676–681. [DOI] [PubMed] [Google Scholar]
- 9.Rovelet-Lecrux A., Lecourtois M., Thomas-Anterion C., Le Ber I., Brice A., Frebourg T., Hannequin D., Campion D. (2009) Partial deletion of the MAPT gene: a novel mechanism of FTDP-17. Hum. Mutat., 30, E591–E602. [DOI] [PubMed] [Google Scholar]
- 10.Rovelet-Lecrux A., Campion D. (2012) Copy number variations involving the microtubule-associated protein tau in human diseases. Biochem. Soc. Trans., 40, 672–676. [DOI] [PubMed] [Google Scholar]
- 11.Rovelet-Lecrux A., Hannequin D., Guillin O., Legallic S., Jurici S., Wallon D., Frebourg T., Campion D. (2010) Frontotemporal dementia phenotype associated with MAPT gene duplication. J. Alzheimers Dis., 21, 897–902. [DOI] [PubMed] [Google Scholar]
- 12.Myers A.J., Kaleem M., Marlowe L., Pittman A.M., Lees A.J., Fung H.C., Duckworth J., Leung D., Gibson A., Morris C.M. et al. (2005) The H1c haplotype at the MAPT locus is associated with Alzheimer's disease. Hum. Mol. Genet., 14, 2399–2404. [DOI] [PubMed] [Google Scholar]
- 13.Allen M., Kachadoorian M., Quicksall Z., Zou F., Chai H.S., Younkin C., Crook J.E., Pankratz V.S., Carrasquillo M.M., Krishnan S. et al. (2014) Association of MAPT haplotypes with Alzheimer's disease risk and MAPT brain gene expression levels. Alzheimers Res. Ther., 6, 39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sempere L.F., Freemantle S., Pitha-Rowe I., Moss E., Dmitrovsky E., Ambros V. (2004) Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol., 5, R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ziats M.N., Rennert O.M. (2014) Identification of differentially expressed microRNAs across the developing human brain. Mol. Psychiatry, 19, 848–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Follert P., Cremer H., Beclin C. (2014) MicroRNAs in brain development and function: a matter of flexibility and stability. Front. Mol. Neurosci., 7, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dorval V., Smith P.Y., Delay C., Calvo E., Planel E., Zommer N., Buee L., Hebert S.S. (2012) Gene network and pathway analysis of mice with conditional ablation of Dicer in post-mitotic neurons. PloS one, 7, e44060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thomas M., Lieberman J., Lal A. (2010) Desperately seeking microRNA targets. Nat. Struct. Mol. Biol., 17, 1169–1174. [DOI] [PubMed] [Google Scholar]
- 19.Vidigal J.A., Ventura A. (2015) The biological functions of miRNAs: lessons from in vivo studies. Trends Cell Biol., 25, 137–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hebert S.S., Papadopoulou A.S., Smith P., Galas M.C., Planel E., Silahtaroglu A.N., Sergeant N., Buee L., De Strooper B. (2010) Genetic ablation of Dicer in adult forebrain neurons results in abnormal tau hyperphosphorylation and neurodegeneration. Hum. Mol. Genet., 19, 3959–3969. [DOI] [PubMed] [Google Scholar]
- 21.Abe M., Bonini N.M. (2013) MicroRNAs and neurodegeneration: role and impact. Trends Cell Biol., 23, 30–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Delay C., Dorval V., Fok A., Grenier-Boley B., Lambert J.C., Hsiung G.Y., Hebert S.S. (2014) MicroRNAs targeting Nicastrin regulate Abeta production and are affected by target site polymorphisms. Front. Mol. Neurosci., 7, 67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hebert S.S., Horre K., Nicolai L., Papadopoulou A.S., Mandemakers W., Silahtaroglu A.N., Kauppinen S., Delacourte A., De Strooper B. (2008) Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer's disease correlates with increased BACE1/beta-secretase expression. Proc. Natl Acad. Sci. USA, 105, 6415–6420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hebert S.S., Horre K., Nicolai L., Bergmans B., Papadopoulou A.S., Delacourte A., De Strooper B. (2009) MicroRNA regulation of Alzheimer's Amyloid precursor protein expression. Neurobiol. Dis., 33, 422–428. [DOI] [PubMed] [Google Scholar]
- 25.Hebert S.S., De Strooper B. (2007) Molecular biology. miRNAs in neurodegeneration. Science, 317, 1179–1180. [DOI] [PubMed] [Google Scholar]
- 26.Delay C., Mandemakers W., Hebert S.S. (2012) MicroRNAs in Alzheimer's disease. Neurobiol. Dis., 46, 285–290. [DOI] [PubMed] [Google Scholar]
- 27.Wanet A., Tacheny A., Arnould T., Renard P. (2012) miR-212/132 expression and functions: within and beyond the neuronal compartment. Nucl. Acids Res., 40, 4742–4753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Magill S.T., Cambronne X.A., Luikart B.W., Lioy D.T., Leighton B.H., Westbrook G.L., Mandel G., Goodman R.H. (2010) microRNA-132 regulates dendritic growth and arborization of newborn neurons in the adult hippocampus. Proc. Natl Acad. Sci. USA, 107, 20382–20387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vo N., Klein M.E., Varlamova O., Keller D.M., Yamamoto T., Goodman R.H., Impey S. (2005) A cAMP-response element binding protein-induced microRNA regulates neuronal morphogenesis. Proc. Natl Acad Sci. USA, 102, 16426–16431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Remenyi J., van den Bosch M.W., Palygin O., Mistry R.B., McKenzie C., Macdonald A., Hutvagner G., Arthur J.S., Frenguelli B.G., Pankratov Y. (2013) miR-132/212 knockout mice reveal roles for these miRNAs in regulating cortical synaptic transmission and plasticity. PloS One, 8, e62509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lambert T.J., Storm D.R., Sullivan J.M. (2010) MicroRNA132 modulates short-term synaptic plasticity but not basal release probability in hippocampal neurons. PloS One, 5, e15182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang R.Y., Phang R.Z., Hsu P.H., Wang W.H., Huang H.T., Liu I.Y. (2013) In vivo knockdown of hippocampal miR-132 expression impairs memory acquisition of trace fear conditioning. Hippocampus, 23, 625–633. [DOI] [PubMed] [Google Scholar]
- 33.Hernandez-Rapp J., Smith P.Y., Filali M., Goupil C., Planel E., Magill S.T., Goodman R.H., Hebert S.S. (2015) Memory formation and retention are affected in adult miR-132/212 knockout mice. Behav. Brain Res., 287, 15–26. [DOI] [PubMed] [Google Scholar]
- 34.Wong H.K., Veremeyko T., Patel N., Lemere C.A., Walsh D.M., Esau C., Vanderburg C., Krichevsky A.M. (2013) De-repression of FOXO3a death axis by microRNA-132 and -212 causes neuronal apoptosis in Alzheimer's disease. Hum. Mol. Genet., 22, 3077–3092. [DOI] [PubMed] [Google Scholar]
- 35.Hwang J.Y., Kaneko N., Noh K.M., Pontarelli F., Zukin R.S. (2014) The gene silencing transcription factor REST represses miR-132 expression in hippocampal neurons destined to die. J. Mol. Biol., 426, 3454–3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hebert S.S., Wang W.X., Zhu Q., Nelson P.T. (2013) A study of small RNAs from cerebral neocortex of pathology-verified Alzheimer's disease, dementia with lewy bodies, hippocampal sclerosis, frontotemporal lobar dementia, and non-demented human controls. J. Alzheimers Dis., 35, 335–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cogswell J.P., Ward J., Taylor I.A., Waters M., Shi Y., Cannon B., Kelnar K., Kemppainen J., Brown D., Chen C. et al. (2008) Identification of miRNA changes in Alzheimer's disease brain and CSF yields putative biomarkers and insights into disease pathways. J. Alzheimers Dis., 14, 27–41. [DOI] [PubMed] [Google Scholar]
- 38.Wang W.X., Huang Q., Hu Y., Stromberg A.J., Nelson P.T. (2011) Patterns of microRNA expression in normal and early Alzheimer's disease human temporal cortex: white matter versus gray matter. Acta Neuropathologica, 121, 193–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lau P., Bossers K., Janky R., Salta E., Frigerio C.S., Barbash S., Rothman R., Sierksma A.S., Thathiah A., Greenberg D. et al. (2013) Alteration of the microRNA network during the progression of Alzheimer's disease. EMBO Mol. Med., 5, 1613–1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith P.Y., Delay C., Girard J., Papon M.A., Planel E., Sergeant N., Buee L., Hebert S.S. (2011) MicroRNA-132 loss is associated with tau exon 10 inclusion in progressive supranuclear palsy. Hum. Mol. Genet., 20, 4016–4024. [DOI] [PubMed] [Google Scholar]
- 41.Chen-Plotkin A.S., Unger T.L., Gallagher M.D., Bill E., Kwong L.K., Volpicelli-Daley L., Busch J.I., Akle S., Grossman M., Van Deerlin V. et al. (2012) TMEM106B, the risk gene for frontotemporal dementia, is regulated by the microRNA-132/212 cluster and affects progranulin pathways. J. Neurosci., 32, 11213–11227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Planel E., Xiaoyan S., Takashima A. (2002) Role of GSK-3b in Alzheimer's disease pathology. Drug Develop.Res., 56, 491–510. [Google Scholar]
- 43.Papon M.A., El Khoury N.B., Marcouiller F., Julien C., Morin F., Bretteville A., Petry F.R., Gaudreau S., Amrani A., Mathews P.M. et al. (2013) Deregulation of protein phosphatase 2A and hyperphosphorylation of tau protein following onset of diabetes in NOD mice. Diabetes, 62, 609–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Planel E., Miyasaka T., Launey T., Chui D.H., Tanemura K., Sato S., Murayama O., Ishiguro K., Tatebayashi Y., Takashima A. (2004) Alterations in glucose metabolism induce hypothermia leading to tau hyperphosphorylation through differential inhibition of kinase and phosphatase activities: implications for Alzheimer's disease. J. Neurosci., 24, 2401–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Oddo S., Caccamo A., Shepherd J.D., Murphy M.P., Golde T.E., Kayed R., Metherate R., Mattson M.P., Akbari Y., LaFerla F.M. (2003) Triple-transgenic model of Alzheimer's disease with plaques and tangles: intracellular Abeta and synaptic dysfunction. Neuron, 39, 409–421. [DOI] [PubMed] [Google Scholar]
- 46.Duff K., Knight H., Refolo L.M., Sanders S., Yu X., Picciano M., Malester B., Hutton M., Adamson J., Goedert M. et al. (2000) Characterization of pathology in transgenic mice over-expressing human genomic and cDNA tau transgenes. Neurobiol. Dis., 7, 87–98. [DOI] [PubMed] [Google Scholar]
- 47.Jicha G.A., Bowser R., Kazam I.G., Davies P. (1997) Alz-50 and MC-1, a new monoclonal antibody raised to paired helical filaments, recognize conformational epitopes on recombinant tau. J. Neurosci. Res., 48, 128–132. [DOI] [PubMed] [Google Scholar]
- 48.Weaver C.L., Espinoza M., Kress Y., Davies P. (2000) Conformational change as one of the earliest alterations of tau in Alzheimer's disease. Neurobiol. Aging, 21, 719–727. [DOI] [PubMed] [Google Scholar]
- 49.Ambegaokar S.S., Jackson G.R. (2012) The downward spiral of tau and autolysosomes: a new hypothesis in neurodegeneration. Autophagy, 8, 1144–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Caccamo A., Magri A., Medina D.X., Wisely E.V., Lopez-Aranda M.F., Silva A.J., Oddo S. (2013) mTOR regulates tau phosphorylation and degradation: implications for Alzheimer's disease and other tauopathies. Aging Cell, 12, 370–380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jo C., Gundemir S., Pritchard S., Jin Y.N., Rahman I., Johnson G.V. (2014) Nrf2 reduces levels of phosphorylated tau protein by inducing autophagy adaptor protein NDP52. Nat. Commun., 5, 3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ozcelik S., Fraser G., Castets P., Schaeffer V., Skachokova Z., Breu K., Clavaguera F., Sinnreich M., Kappos L., Goedert M. et al. (2013) Rapamycin attenuates the progression of tau pathology in P301S tau transgenic mice. PloS One, 8, e62459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ucar A., Gupta S.K., Fiedler J., Erikci E., Kardasinski M., Batkai S., Dangwal S., Kumarswamy R., Bang C., Holzmann A. et al. (2012) The miRNA-212/132 family regulates both cardiac hypertrophy and cardiomyocyte autophagy. Nat. Commun., 3, 1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Koval E.D., Shaner C., Zhang P., du Maine X., Fischer K., Tay J., Chau B.N., Wu G.F., Miller T.M. (2013) Method for widespread microRNA-155 inhibition prolongs survival in ALS-model mice. Hum. Mol. Genet., 22, 4127–4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sheinerman K.S., Tsivinsky V.G., Abdullah L., Crawford F., Umansky S.R. (2013) Plasma microRNA biomarkers for detection of mild cognitive impairment: biomarker validation study. Aging, 5, 925–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sheinerman K.S., Tsivinsky V.G., Crawford F., Mullan M.J., Abdullah L., Umansky S.R. (2012) Plasma microRNA biomarkers for detection of mild cognitive impairment. Aging, 4, 590–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tremblay C., Pilote M., Phivilay A., Emond V., Bennett D.A., Calon F. (2007) Biochemical characterization of Abeta and tau pathologies in mild cognitive impairment and Alzheimer's disease. J. Alzheimers Dis., 12, 377–390. [DOI] [PubMed] [Google Scholar]
- 58.Julien C., Tremblay C., Bendjelloul F., Phivilay A., Coulombe M.A., Emond V., Calon F. (2008) Decreased drebrin mRNA expression in Alzheimer disease: correlation with tau pathology. J. Neurosci. Res., 86, 2292–2302. [DOI] [PubMed] [Google Scholar]
- 59.Dickson J.R., Kruse C., Montagna D.R., Finsen B., Wolfe M.S. (2013) Alternative polyadenylation and miR-34 family members regulate tau expression. J. Neurochem., 127, 739–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chi S.W., Zang J.B., Mele A., Darnell R.B. (2009) Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature, 460, 479–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gotz J., Ittner L.M. (2008) Animal models of Alzheimer's disease and frontotemporal dementia. Nat. Rev. Neurosci., 9, 532–544. [DOI] [PubMed] [Google Scholar]
- 62.Peric A., Annaert W. (2015) Early etiology of Alzheimer's disease: tipping the balance toward autophagy or endosomal dysfunction? Acta Neuropathologica, 129, 363–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Perluigi M., Di Domenico F., Butterfield D.A. (2015) mTOR signaling in aging and neurodegeneration: At the crossroad between metabolism dysfunction and impairment of autophagy. Neurobiol. Dis., in press. [DOI] [PubMed] [Google Scholar]
- 64.Nixon R.A. (2013) The role of autophagy in neurodegenerative disease. Nat. Med., 19, 983–997. [DOI] [PubMed] [Google Scholar]
- 65.Hardy J., Rogaeva E. (2014) Motor neuron disease and frontotemporal dementia: sometimes related, sometimes not. Exp. Neurol., 262(Pt B), 75–83. [DOI] [PubMed] [Google Scholar]
- 66.de Calignon A., Polydoro M., Suarez-Calvet M., William C., Adamowicz D.H., Kopeikina K.J., Pitstick R., Sahara N., Ashe K.H., Carlson G.A. et al. (2012) Propagation of tau pathology in a model of early Alzheimer's disease. Neuron, 73, 685–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Baglietto-Vargas D., Kitazawa M., Le E.J., Estrada-Hernandez T., Rodriguez-Ortiz C.J., Medeiros R., Green K.N., LaFerla F.M. (2014) Endogenous murine tau promotes neurofibrillary tangles in 3xTg-AD mice without affecting cognition. Neurobiol. Dis., 62, 407–415. [DOI] [PubMed] [Google Scholar]
- 68.Stancu I.C., Vasconcelos B., Terwel D., Dewachter I. (2014) Models of beta-amyloid induced Tau-pathology: the long and ‘folded’ road to understand the mechanism. Mol. Neurodegenerat., 9, 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Shaked I., Meerson A., Wolf Y., Avni R., Greenberg D., Gilboa-Geffen A., Soreq H. (2009) MicroRNA-132 potentiates cholinergic anti-inflammatory signaling by targeting acetylcholinesterase. Immunity, 31, 965–973. [DOI] [PubMed] [Google Scholar]
- 70.Schliebs R., Arendt T. (2011) The cholinergic system in aging and neuronal degeneration. Behav. Brain Res., 221, 555–563. [DOI] [PubMed] [Google Scholar]
- 71.Girao da Cruz M.T., Jordao J., Dasilva K.A., Ayala-Grosso C.A., Ypsilanti A., Weng Y.Q., Laferla F.M., McLaurin J., Aubert I. (2012) Early increases in soluble amyloid-beta levels coincide with cholinergic degeneration in 3xTg-AD mice. J. Alzheimers Dis., 32, 267–272. [DOI] [PubMed] [Google Scholar]
- 72.Wang X., Tan L., Lu Y., Peng J., Zhu Y., Zhang Y., Sun Z. (2015) MicroRNA-138 promotes tau phosphorylation by targeting retinoic acid receptor alpha. FEBS Lett., 589, 726–729. [DOI] [PubMed] [Google Scholar]
- 73.Banzhaf-Strathmann J., Benito E., May S., Arzberger T., Tahirovic S., Kretzschmar H., Fischer A., Edbauer D. (2014) MicroRNA-125b induces tau hyperphosphorylation and cognitive deficits in Alzheimer's disease. EMBO J., 33, 1667–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhao Z.B., Wu L., Xiong R., Wang L.L., Zhang B., Wang C., Li H., Liang L., Chen S.D. (2014) MicroRNA-922 promotes tau phosphorylation by downregulating ubiquitin carboxy-terminal hydrolase L1 (UCHL1) expression in the pathogenesis of Alzheimer's disease. Neuroscience, 275, 232–237. [DOI] [PubMed] [Google Scholar]
- 75.Santa-Maria I., Alaniz M.E., Renwick N., Cela C., Fulga T.A., Van Vactor D., Tuschl T., Clark L.N., Shelanski M.L., McCabe B.D. et al. (2015) Dysregulation of microRNA-219 promotes neurodegeneration through post-transcriptional regulation of tau. J. Clin. Investig., 125, 681–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dugas J.C., Cuellar T.L., Scholze A., Ason B., Ibrahim A., Emery B., Zamanian J.L., Foo L.C., McManus M.T., Barres B.A. (2010) Dicer1 and miR-219 Are required for normal oligodendrocyte differentiation and myelination. Neuron, 65, 597–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wayman G.A., Davare M., Ando H., Fortin D., Varlamova O., Cheng H.Y., Marks D., Obrietan K., Soderling T.R., Goodman R.H. et al. (2008) An activity-regulated microRNA controls dendritic plasticity by down-regulating p250GAP. Proc. Natl Acad. Sci. USA, 105, 9093–9098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kawashima H., Numakawa T., Kumamaru E., Adachi N., Mizuno H., Ninomiya M., Kunugi H., Hashido K. (2010) Glucocorticoid attenuates brain-derived neurotrophic factor-dependent upregulation of glutamate receptors via the suppression of microRNA-132 expression. Neuroscience, 165, 1301–1311. [DOI] [PubMed] [Google Scholar]
- 79.Remenyi J., Hunter C.J., Cole C., Ando H., Impey S., Monk C.E., Martin K.J., Barton G.J., Hutvagner G., Arthur J.S. (2010) Regulation of the miR-212/132 locus by MSK1 and CREB in response to neurotrophins. Biochem. J., 428, 281–291. [DOI] [PubMed] [Google Scholar]
- 80.Lee S.T., Chu K., Im W.S., Yoon H.J., Im J.Y., Park J.E., Park K.H., Jung K.H., Lee S.K., Kim M. et al. (2011) Altered microRNA regulation in Huntington's disease models. Exp. Neurol., 227, 172–179. [DOI] [PubMed] [Google Scholar]
- 81.Miller B.H., Zeier Z., Xi L., Lanz T.A., Deng S., Strathmann J., Willoughby D., Kenny P.J., Elsworth J.D., Lawrence M.S. et al. (2012) MicroRNA-132 dysregulation in schizophrenia has implications for both neurodevelopment and adult brain function. Proc. Natl Acad. Sci. USA, 109, 3125–3130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Freischmidt A., Muller K., Ludolph A.C., Weishaupt J.H. (2013) Systemic dysregulation of TDP-43 binding microRNAs in amyotrophic lateral sclerosis. Acta Neuropathologica Commun., 1, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Casanova M.F., Stevens J.R., Brown R., Royston C., Bruton C. (2002) Disentangling the pathology of schizophrenia and paraphrenia. Acta Neuropathologica, 103, 313–320. [DOI] [PubMed] [Google Scholar]
- 84.Fernandez-Nogales M., Cabrera J.R., Santos-Galindo M., Hoozemans J.J., Ferrer I., Rozemuller A.J., Hernandez F., Avila J., Lucas J.J. (2014) Huntington's disease is a four-repeat tauopathy with tau nuclear rods. Nat.Med., 20, 881–885. [DOI] [PubMed] [Google Scholar]
- 85.Wilson R.S., Barnes L.L., Mendes de Leon C.F., Aggarwal N.T., Schneider J.S., Bach J., Pilat J., Beckett L.A., Arnold S.E., Evans D.A. et al. (2002) Depressive symptoms, cognitive decline, and risk of AD in older persons. Neurology, 59, 364–370. [DOI] [PubMed] [Google Scholar]
- 86.Ohta Y., Tremblay C., Schneider J.A., Bennett D.A., Calon F., Julien J.P. (2014) Interaction of transactive response DNA binding protein 43 with nuclear factor kappaB in mild cognitive impairment with episodic memory deficits. Acta Neuropathologica Commun., 2, 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Otvos L. Jr, Feiner L., Lang E., Szendrei G.I., Goedert M., Lee V.M. (1994) Monoclonal antibody PHF-1 recognizes tau protein phosphorylated at serine residues 396 and 404. J. Neurosci. Res., 39, 669–673. [DOI] [PubMed] [Google Scholar]
- 88.Tremblay M.E., Riad M., Majewska A. (2010) Preparation of mouse brain tissue for immunoelectron microscopy. J. Visual. Exp., 41, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Paxinos G., Franklin K.B. (2013) The Mouse Brain in Stereotaxic Coordinates. Academic Press, New York. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.