Abstract
ATP6AP2, an essential accessory component of the vacuolar H+ ATPase (V-ATPase), has been associated with intellectual disability (ID) and Parkinsonism. ATP6AP2 has been implicated in several signalling pathways; however, little is known regarding its role in the nervous system. To decipher its function in behaviour and cognition, we generated and characterized conditional knockdowns of ATP6AP2 in the nervous system of Drosophila and mouse models. In Drosophila, ATP6AP2 knockdown induced defective phototaxis and vacuolated photoreceptor neurons and pigment cells when depleted in eyes and altered short- and long-term memory when depleted in the mushroom body. In mouse, conditional Atp6ap2 deletion in glutamatergic neurons (Atp6ap2Camk2aCre/0 mice) caused increased spontaneous locomotor activity and altered fear memory. Both Drosophila ATP6AP2 knockdown and Atp6ap2Camk2aCre/0 mice presented with presynaptic transmission defects, and with an abnormal number and morphology of synapses. In addition, Atp6ap2Camk2aCre/0 mice showed autophagy defects that led to axonal and neuronal degeneration in the cortex and hippocampus. Surprisingly, axon myelination was affected in our mutant mice, and axonal transport alterations were observed in Drosophila. In accordance with the identified phenotypes across species, genome-wide transcriptome profiling of Atp6ap2Camk2aCre/0 mouse hippocampi revealed dysregulation of genes involved in myelination, action potential, membrane-bound vesicles and motor behaviour. In summary, ATP6AP2 disruption in mouse and fly leads to cognitive impairment and neurodegeneration, mimicking aspects of the neuropathology associated with ATP6AP2 mutations in humans. Our results identify ATP6AP2 as an essential gene for the nervous system.
Introduction
ATP6AP2 has been associated with cognitive disorders based on a mutation identified in one family with seven affected males presenting with mild-to-moderate intellectual disabilities, delayed motor milestones and speech acquisition, and generalised tonic-clonic seizures (1,2). However, the implication of ATP6AP2 in intellectual disabilities has been questioned after an ATP6AP2 mutation was identified in a single male within a cohort of 6500 sequenced control exomes (3). More recently, another family with Parkinsonian features and variably penetrant spasticity was reported to have an ATP6AP2 mutation (4,5). The mutations identified in these two families are different splicing mutations that give rise to skipping of exon 4 of the ATP6AP2 gene in both cases.
ATP6AP2 was described as a receptor for renin and prorenin ((P)RR). The binding of renin or (P)RR to the ATP6AP2 extracellular domain activates the renin–angiotensin system, leading to the production of angiotensin II and also to activation of other pathways, such as the mitogen-activated protein kinase (MAPK) pathway (6). However, ATP6AP2 is also found in invertebrates such as Drosophila and Caenorhabditis elegans, which do not possess the renin–angiotensin system (7), suggesting alternative functions for ATP6AP2. Indeed, this protein was also isolated as an accessory component of the vacuolar H+ ATPase (V-ATPase) (6,8). V-ATPase is a multisubunit proton pump that consists of a V0 proton-translocation domain, a V1 pump domain and two associated proteins, Ac45 and ATP6AP2. This pump is responsible for the acidification of intracellular and some extracellular compartments and thus is crucial for many fundamental processes that require an intact membrane-trafficking system (9), including endocytosis, protein and signalling molecule processing, protein sorting and trafficking, and lysosomal and autophagosomal enzyme activation (10).
In agreement with its fundamental role, several studies regarding ATP6AP2 in mouse, zebrafish, frog and Drosophila have shown that its full deletion is lethal (reviewed in 11). Moreover, the tissue-specific deletion of Atp6ap2 in mouse cardiomyocytes and podocytes also leads to lethality, as both conditional knockout mice died between 2 and 3 weeks after birth (12–14). Concomitantly, multivesicular bodies and autophagic vacuoles accumulated in the tissues with deleted Atp6ap2 owing to autophagic flux alterations (12,14). Recent work in the Drosophila wing disc epithelium has demonstrated that ATP6AP2 (VhaM8.9/VhaPRR) is required for V-ATPase function in vesicular acidification, endosomal sorting and protein degradation (15). In addition, ATP6AP2 was shown to be essential for both of the Wnt/β-catenin and Wnt/planar cell polarity (PCP) signalling pathways in Xenopus and Drosophila (16–18). Whereas Cruciat and collaborators (17) identified Xenopus ATP6AP2/(P)RR as an adaptor between V-ATPase and the Wnt complex, Hermle and collaborators (18) found that its function in proper trafficking of the planar cell polarity molecular core machinery in the Drosophila wing disc is independent of V-ATPase.
Surprisingly little is known regarding the function of ATP6AP2 in the nervous system, the tissue type most prominently affected in humans with ATP6AP2 mutations. In zebrafish, the absence of ATP6AP2/(P)RR induces necrosis in the central nervous system, eventually resulting in embryonic lethality (19). In Xenopus, ATP6AP2/(P)RR is required for mediating Wnt signalling during early CNS development, including neural patterning (17,20). In mouse, Atp6ap2 regulates adult hippocampal neurogenesis via Wnt/β-catenin and Wnt/PCP signalling pathways (21). Whether ATP6AP2 executes specific roles in postmitotic neurons that can directly underlie impaired cognitive and motor functions in individuals with ATP6AP2 mutations is unclear; however, this question is important in the light of the unclear causative role of this gene in intellectual disabilities (ID) and Parkinsonism with spasticity. To decipher the function of ATP6AP2 in behaviour and cognition, we generated and characterized conditional ATP6AP2 knockouts in Drosophila and mouse nervous system models.
Results
Drosophila ATP6AP2 knockdown in the eye causes defective phototaxis and ultrastructural defects, and knockdown in the mushroom body causes defects in short- and long-term memory
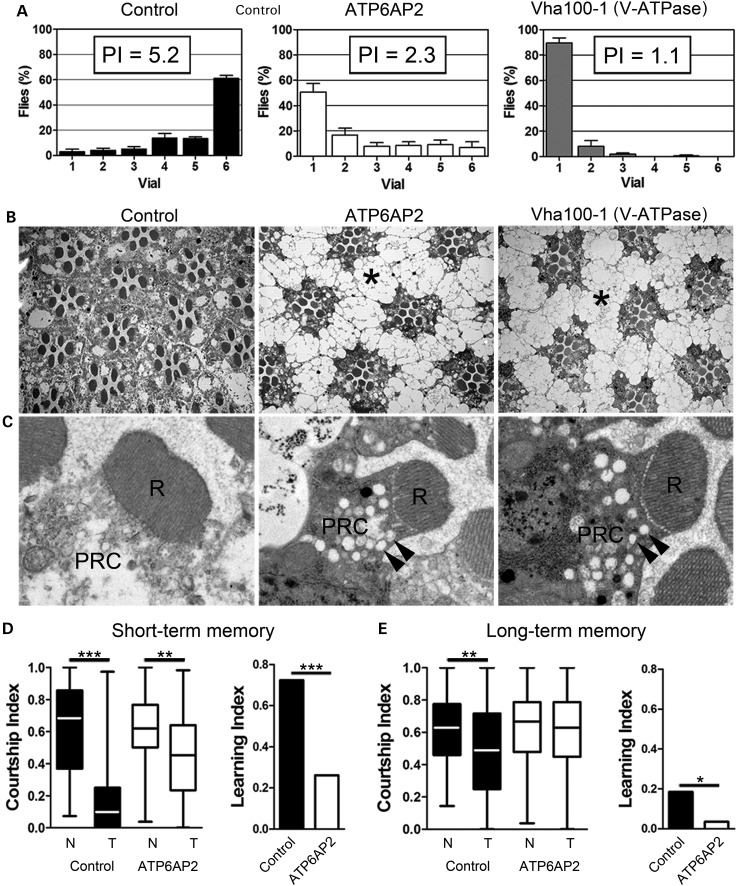
First, we investigated the Drosophila orthologue of the ID candidate gene ATP6AP2, also termed VhaM8.9/VhaPRR and from here on referred to as ATP6AP2, in a combination of Drosophila phototactic behaviour and morphological studies on the fly eye as primary readouts (22). We used the UAS-Gal4 system and two independent conditional RNAi constructs, UAS-ATP6AP2RNAi1 and UAS-ATP6AP2RNAi2, to induce ATP6AP2 knockdown in the developing eye with the GMR-Gal4; UAS-Dicer-2 driver. The potency of both lines to induce knockdown was confirmed by qPCR (see Materials and Methods). Both lines were effective, with UAS-ATP6AP2RNAi1 inducing slightly stronger knockdown compared with UAS-ATP6AP2RNAi2 (Supplementary Material, Fig. S2A). ATP6AP2 eye-specific knockdown did not result in external eye morphology phenotypes (data not shown) but dramatically affected performance in the phototaxis assay. Whereas the genetic background control animals were strongly attracted by light [phototaxis index (PI) = 5.2], the majority of ATP6AP2 eye-specific knockdown animals remained in the first vial, resulting in PIs of merely 2.3 for both RNAi conditions (Fig. 1A and data not shown). This finding suggested potential dysfunction of ATP6AP2-deficient photoreceptor (PR) neurons. To obtain further insights into the cellular basis of these defects, we analysed the structural integrity of PRs by histological semi- and ultrathin sectioning. While ATP6AP2-deficient ommatidia and PRs showed overall intact organization, PR-surrounding pigment cells were highly vacuolated (Fig. 1B, asterisk). Electron microscopy of PRs revealed a large number of accumulated vesicular structures in PR cytoplasm (Fig. 1C, arrowheads). Interestingly, identical phenotypes, low PI, vacuolated pigment cells and large vesicular structures in PRs were observed upon knockdown of the Drosophila V-ATPase subunit Vha100-1 (Fig. 1A–C, Supplementary Material, Fig. S2A, 22), whose homologue, ATP6V0A2, is also implicated in ID (23).
Figure 1.
Knockdown of Drosophila ATP6AP2 disrupted phototaxis and short- and long-term memory. (A) Distribution of adult flies in the phototaxis assay measuring their visually induced activity and resulting phototaxis indexes (PI). Control flies (GMR-Gal4/+;UAS-Dicer-2/+) and flies with either eye-specific knockdown of ATP6AP2 (UAS-ATP6AP2RNAi1/GMR-Gal4;UAS-Dicer-2/+) or Vha100-1 (GMR-Gal4/+;UAS-Dicer-2/Vha100-1RNAi) are shown. ATP6AP2 and Vha100-1 knockdown animals showed dramatically decreased PIs, revealing functional photoreceptor neuron defects. (B and C) Histological and high-magnification electron microscopy images of the same genotypes presented in A. The pigment cells surrounding the photoreceptor neurons were strongly vacuolated (asterisk). Vesicular structures were present in the PR cytoplasm (PRC) (arrowheads). Rhabdomeres (R), the apical photosensitive membrane domains of photoreceptors, were intact in ATP6AP2 and Vha100-1 knockdown eyes. (A–C) Note the strikingly similar phenotypes caused by knockdown of ATP6AP2 and Vha100-1. (D) Short-term and (E) long-term memory in control (w+,UAS-Dicer-2/Y;247-Gal4/+;+/+) and ATP6AP2 mushroom body-specific knockdown flies (w+,UAS-Dicer-2/Y; 247-Gal4/UAS-ATP6AP2RNAi1; +/+) in the courtship conditioning paradigm. Left panels show boxplots of the courtship indexes (CI) of naïve (N) and trained (T) males. Significance of courtship suppression upon training was assessed with ANOVA and post hoc Tukey's multiple comparison test. Panels on the right side represent the learning indexes, which were calculated from CIs as specified in the Material and Methods section. The loss of ATP6AP2 caused a statistically significant reduction in courtship-based short-term memory (STM, A) and long-term memory (LTM, B) compared with controls. Statistical test: bootstrap analysis with 10 000 replicates. *P < 0.05, **P < 0.01, ***P < 0.001.
To address the role of ATP6AP2 in the development and function of the nervous system, we knocked down ATP6AP2 in postmitotic neurons. The induction of UAS-ATP6AP2RNAi1 using the elav-Gal4 promoter in the presence of UAS-Dicer-2 induced lethality at different developmental stages, primarily during larval development. The few adult survivors that were obtained showed strongly reduced spontaneous movements and poor climbing abilities. The induction of the somewhat less potent UAS-ATP6AP2RNAi2 line under the same conditions did not result in lethality, but the progenies showed similar, albeit less affected, locomotion defects, together indicating that ATP6AP2 is required in neurons for survival and normal fly behaviour.
Next, we asked whether ATP6AP2 is required for cognitive processes in Drosophila. We chose the stronger RNAi line UAS-ATP6AP2RNAi1 and the 247-Gal4 promoter to limit ATP6AP2 knockdown to the mushroom bodies (MBs), the major learning and memory centre of the Drosophila brain. Interestingly, we noticed that ATP6AP2 MB-specific knockdown flies showed elevated activity in response to stimulation such as banging or soft-tapping against the vials, with repeated jumping, wing flapping and crashing. Then, we tested memory in the courtship conditioning paradigm (24,25). In this paradigm, the courtship behaviour of male flies is conditioned by exposure to non-receptive females, which suppresses courtship and thus decreases the courtship index (CI, the percentage of time spent on courtship during a 10-min interval) in subsequent encounters when tested after 1 h (short-term memory, Fig. 1D) and 24 h (long-term memory, Fig. 1E). Naïve ATP6AP2RNAi1 males (N) courted at similar level as naïve controls (N). Nevertheless, when tested for short-term memory, they partially suppressed courtship after training (T) but failed to suppress courtship in the long-term memory regime. Calculating the learning indexes for ATP6AP2RNAi1 flies against genetic background control flies revealed significant defects in short- and long-term memory.
Mouse Atp6ap2 deletion in glutamatergic neurons causes increased spontaneous locomotor activity and altered contextual and cued fear memory
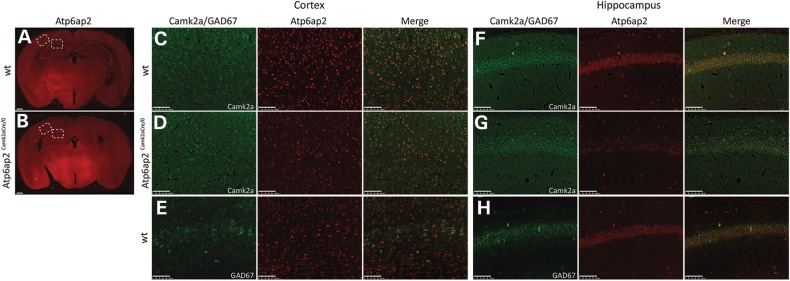
To address whether ATP6AP2 controls similar functions in mammals, we investigated its role in mouse. First, we analysed its expression pattern in adult mouse brain sections by immunohistochemistry. We showed that Atp6ap2 is expressed in most tested brain areas with the highest expression observed in the cortex, the hippocampus, the medial habenular nucleus, the cerebellum, the medulla and the olfactory bulb (Fig. 2A and data not shown). Then, to determine which cell types express Atp6ap2, we performed colocalization studies. In the cortex and the hippocampus, we did not detect Atp6ap2 expression in astrocytes (GFAP marker, data not shown); however, considering the limited sensitivity of immunohistochemistry, we cannot exclude that Atp6ap2 is expressed at low level. We observed Atp6ap2 expression in glutamatergic and GABAergic neurons as revealed by its colocalization with Camk2a-positive cells (Fig. 2C and F) and GAD67-positive cells (Fig. 2E and H), respectively. All Atp6ap2-positive cells were either positive for Camk2a or GAD67, suggesting that Atp6ap2 is expressed primarily in neurons in the cortex and hippocampus.
Figure 2.
Atp6ap2 is expressed in both glutamatergic and GABAergic neurons in the adult mouse brain. (A) Immunohistochemical analysis of adult mouse brain sections showed Atp6ap2 expression in most tested brain areas. Strong expression of Atp6ap2 was detected in the cortex, hippocampus, thalamus and hypothalamus of wt mice (A), and net decreases in Atp6ap2 expression were observed in the cortex and hippocampus of Atp6ap2Camk2aCre/0(B). Scale bars: 500 µm. (C–H) High-magnification images of the cortex and the CA1 area corresponding to the dashed boxes drawn in A and B. Atp6ap2 is expressed in glutamatergic neurons as revealed by its colocalization with Camk2a (a marker of glutamatergic neurons)-positive cells in the cortex and in the CA1 area of wt mice (C and F, respectively). In Atp6ap2Camk2aCre/0 mice, almost no Camk2a-positive cells from the cortex and CA1 expressed Atp6ap2 (D and G, respectively). Atp6ap2 is also expressed in GABAergic cells as revealed by Atp6ap2 colocalization with GAD67 (a marker of GABAergic neurons)-positive cells in the cortex and in the CA1 area (E and H, respectively). Scale bars: 100 µm.
Thus, we decided to inactivate Atp6ap2 gene function specifically in glutamatergic neurons using a conditional approach by crossing a floxed Atp6ap2 transgenic mouse line with a Camk2aCre transgenic mouse line, which expressed the Cre recombinase in glutamatergic neurons (26). Atp6ap2 deletion was first analysed in Atp6ap2Camk2aCre/0 and Atp6ap2Camk2aCreERT2/0 brain tissues (cortex, hippocampus, hypothalamus, striatum and cerebellum) by RT droplet digital PCR. We observed a strong decrease of Atp6ap2 expression in the cortex and the hippocampus, and lower or no decrease in the other brain tissues (Supplementary Material, Fig. S2B). We noticed less Atp6ap2 deletion in all brain tissues analysed in Atp6ap2Camk2aCreERT2/0 mice compared with Atp6ap2Camk2aCre/0 mice (Supplementary Material, Fig. S2B). Next, we confirmed Atp6ap2 deletion by immunohistochemistry. In the cortex and the hippocampus, we showed a net decrease in the number of Atp6ap2-positive cells in Atp6ap2Camk2aCre/0 mice when compared with wt mice (Fig. 2B and A, respectively). Moreover, colocalization analyses showed that Atp6ap2 was only poorly detected in Camk2a-positive cells (Fig. 2D and G) but remained present in GAD67-positive cells (data not shown). In the other brain regions, we did not observe any changes in Atp6ap2 expression, except a decreased expression in the granule cell layer of the olfactory bulb (data not shown). Finally, to determine Atp6ap2 deletion efficiency in Camk2a-positive cells, we quantify the number of Camk2a-positive cells that still expressed Atp6ap2 in the cortex and the hippocampus of Atp6ap2Camk2aCre/0 mice by immunohistochemistry with anti-Camk2a and anti-Atp6ap2 antibodies. We observed that 42.76 ± 1.55% of Camk2a-positive cells still expressed Atp6ap2 in the cortex, and only 16.19 ± 3.93% of Camk2a-positive cells still expressed Atp6ap2 in the hippocampus, confirming a higher deletion efficiency in the hippocampus than in the cortex.
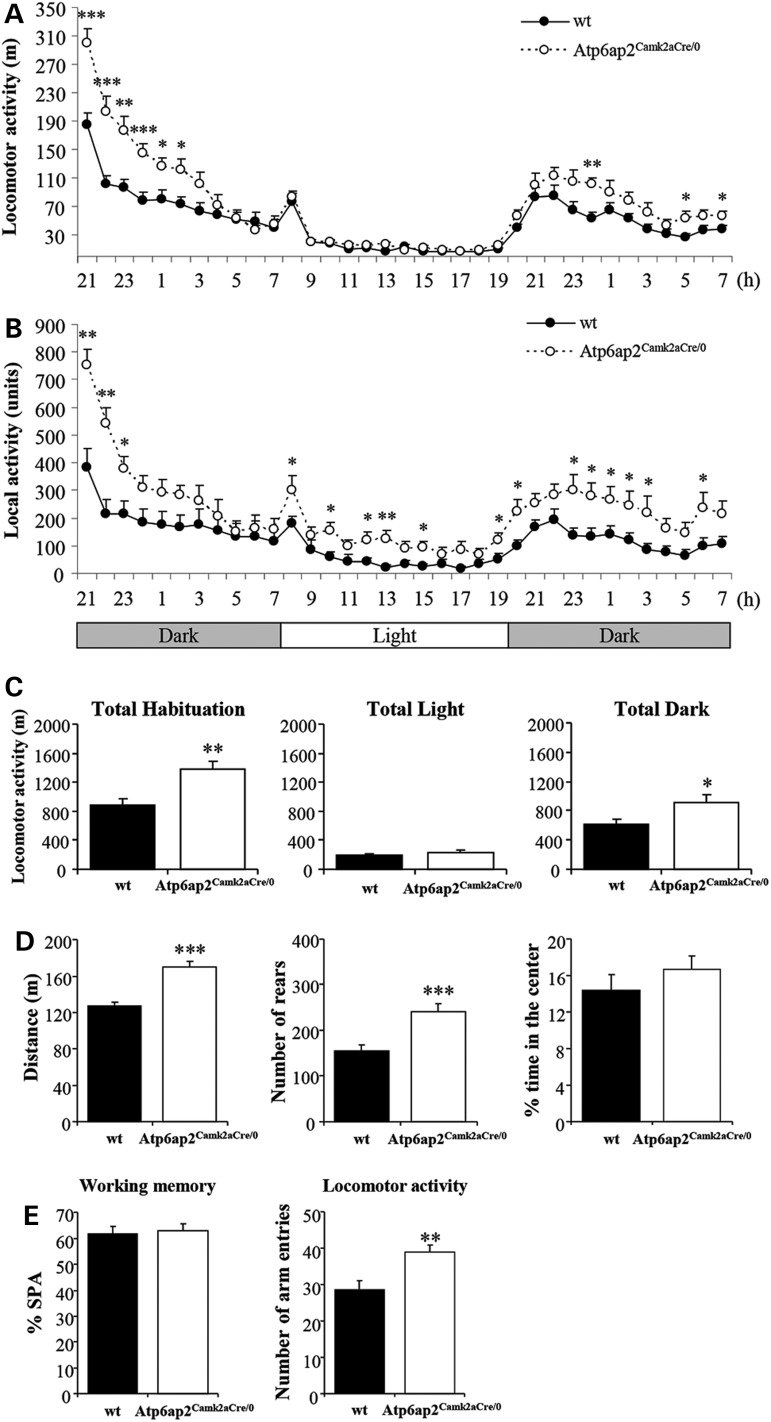
Atp6ap2Camk2aCre/0 males physical appearance and body weights were normal and did not show any obvious sign of altered sensory or vestibular functions. To assess mouse locomotor activity, Atp6ap2Camk2aCre/0 mice were submitted to several tests. When circadian activity was examined, overall locomotor activity (Fig. 3A and C) significantly increased in Atp6ap2Camk2aCre/0 males compared with wt counterparts (t22 ≥ 2.93, P < 0.01); the difference was specifically observed during the dark phases (F1,22 ≥ 6.00, P < 0.05 for locomotion; F1,22 ≥ 4.21, P < 0.05 for rears; Fig. 3C). Atp6ap2Camk2aCre/0 males also showed significantly increased local activity in the front part of the cages throughout the light/dark cycle (t22 = 2.65, P < 0.05; Fig. 3B), which might be interpreted as stereotypic behaviour. When Atp6ap2 was deleted in adults using tamoxifen injection, similar locomotor patterns were observed (data not shown). In the open-field arenas, on one hand, Atp6ap2Camk2aCre/0 males showed a significantly higher level of locomotor activity and rears (t22 ≥ 4.09, P < 0.001; Fig. 3D). On the other hand, the percentage of time spent in the centre of the open-field arena was comparable between genotypes (t22 = 1.02, NS; Fig. 3D), suggesting normal anxiety-related behaviour in Atp6ap2Camk2aCre/0 males. In the Y-maze, the number of visited arms significantly increased in mutants (t22 = 3.26, P < 0.01; Fig. 3E), thus confirming increased locomotor activity following Atp6ap2 deletion in glutamatergic neurons.
Figure 3.
Atp6ap2Camk2aCre/0 mouse line showed increased spontaneous activity in different behavioural experiments. (A–C) In the circadian activity test, Atp6ap2Camk2aCre/0 mice showed significantly increased locomotor activity specifically during the dark phase (A and C) and increased local activity in the front part of the cages throughout the light/dark cycle (B). (D) The distance travelled and the rear number in the open field significantly increased in Atp6ap2Camk2aCre/0 mice compared with wt. (E) In the Y-maze, working memory was comparable between genotypes, as reflected by the percentage of spontaneous alternations (SPAs). The number of visited arms significantly increased in mutants. *P < 0.05, **P < 0.01, ***P < 0.001 versus wt.
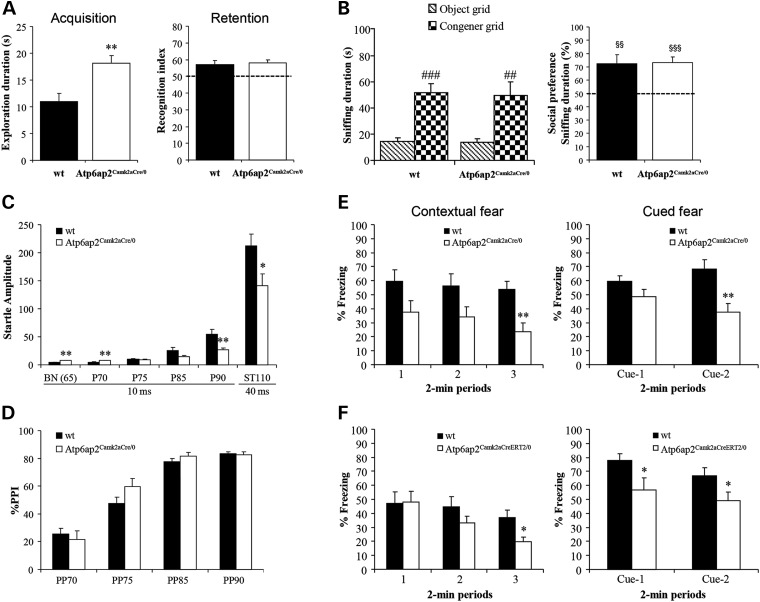
Different assays were performed to test learning and memory abilities of Atp6ap2Camk2aCre/0 mutants. In the Y-maze, the percentage of spontaneous alternations was comparable between genotypes (t22 = 0.22, NS; Fig. 3E), indicating that working memory was not affected in Atp6ap2Camk2aCre/0 mice. In the new object recognition test, Atp6ap2 mutants showed increased object exploration durations compared with wt during the acquisition trial (t22 = 3.30, P < 0.01; Fig. 4A), which could be due to their increased locomotor activity. When the animals were allowed to discriminate between a novel and a familiar object 3 h after acquisition, object recognition performance was comparable between genotypes (t22 = 0.34, NS; Fig. 4A), suggesting that recognition memory is not affected in Atp6ap2 mutants. In the water maze, both wt and Atp6ap2Camk2aCre/0 mice were able to acquire spatial allocentric learning over the training sessions (Supplementary Material, Fig. S3A and B). In contrast, Atp6ap2 mutants showed reduced speed (F1,22 = 17.72, P < 0.001) and higher latencies (F4,88 = 2.50, P < 0.05) to reach the platform position than wt while having the same distance travelled (Supplementary Material, Fig. S3A–C), suggesting altered swimming abilities. When the platform was removed from the pool during the probe trial 1 to 2 h after training, wt and Atp6ap2Camk2aCre/0 mice showed a comparable number of platform crosses and a comparable preference for the target quadrant (Supplementary Material, Fig. S3D and E). These data confirm normal spatial learning in Atp6ap2Camk2aCre/0 mice.
Figure 4.
The Atp6ap2Camk2aCre/0 mouse line showed altered contextual and cued fear memory and increased startle responses. (A) In the object recognition task, the duration of object exploration during acquisition significantly increased in Atp6ap2Camk2aCre/0 mice. Object recognition performance was not affected in mutants. (B) In the social recognition test, both genotypes spent more time exploring the congener than an object and showed comparable preference indexes for the congener. (C) Atp6ap2Camk2aCre/0 mutants showed significantly lower startle reactivity for higher pre-pulses and lower startle responses for high sudden acoustic stimulus. (D) The level of PPI was comparable between genotypes. (E) In the fear conditioning paradigm, when animals were re-exposed in the same context 24 h after conditioning, contextual freezing performance significantly decreased in Atp6ap2Camk2aCre/0 mutants. When the animals were placed in a new context, Atp6ap2Camk2aCre/0 mice also showed lower cued freezing performance than wt. (F) Atp6ap2Camk2aCreERT2/0 mice also showed decreased contextual and cued freezing performances (P < 0.05). *P < 0.05, **P < 0.01 versus wt; ##P < 0.01, ###P < 0.001 versus the object grid; §§P < 0.01, §§§P < 0.001 versus the chance level.
To determine Atp6ap2Camk2aCre/0 males’ social behaviour, the animals were allowed to explore freely a congener and an object placed simultaneously at two opposite goal boxes of a runway. Wt and mutants explored the congener grid significantly more than the object grid (sniffing duration: wt males: t11 = 5.34, P < 0.001; Atp6ap2Camk2aCre/0 males: t11 = 3.96, P < 0.01; paired t-test), reflecting no difference in social recognition performance between both genotypes (% sniffing duration: t ≥ 3.19, P < 0.01, one group t-test; Fig. 4B).
Acoustic startle and prepulse inhibition (PPI) of the startle reflex were also evaluated. Atp6ap2Camk2aCre/0 mice showed significantly lower startle reactivity for higher pre-pulses and lower startle responses for high sudden acoustic stimulus (t22 ≥ 2.45, P < 0.05; Fig. 4C). The level of PPI was comparable between genotypes (F1,22 = 0.44, NS; Fig. 4D), suggesting normal sensorimotor gating in mutants.
In the fear conditioning test, the baseline level of immobility during the habituation to the conditioning cages and the suppression of activity in response to footshocks during conditioning were lower in Atp6ap2Camk2aCre/0 compared with wt mice (F1,22 = 4.32, P < 0.05; t22 = 3.72, P < 0.001). When the animals were re-exposed to the same context 24 h after conditioning, both wt and Atp6ap2Camk2aCre/0 mice showed significant increases in immobility compared with the baseline. However, contextual freezing performance significantly decreased in Atp6ap2 mutants (F1,22 = 6.72, P < 0.05; Fig. 4E). When the animals were placed in a new context, the presentation of conditioned stimuli (CS) alone induced clear fear reaction in both genotypes; nevertheless, Atp6ap2Camk2aCre/0 mice showed lower cued freezing performance than wt (F1,22 = 3.50, P < 0.01; Fig. 4E). Interestingly, the adult deleted Atp6ap2Camk2aCreERT2/0 mice also showed decreased contextual and cued freezing performances (P < 0.05; Fig. 4F). Overall, Atp6ap2 deletion in glutamatergic neurons induced substantial deficits in both cued and contextual fear conditioning, suggesting altered emotional learning and memory.
ATP6AP2 is required for presynaptic transmission in Drosophila and mouse
We used electrophysiological approaches to further investigate and determine the cause of behavioural and cognitive deficits in Drosophila and mouse models. First, we performed electroretinograms (ERGs) (27) on Drosophila control and ATP6AP2 knockdown eyes. ERGs are extracellular field recordings that assess the potential difference between the photoreceptor layer and the remainder of the fly body during light stimulation, revealing photoreceptor transients (de- and repolarization) and synaptic communication (‘on’ and ‘off’ transients). The presynaptic knockdown of ATP6AP2 in photoreceptors using either UAS-ATP6AP2RNAi1 or UAS-ATP6AP2RNAi2 caused synaptic defects characterized by a complete absence of ‘on’ and ‘off’ transients (Fig. 5A and data not shown).
Figure 5.
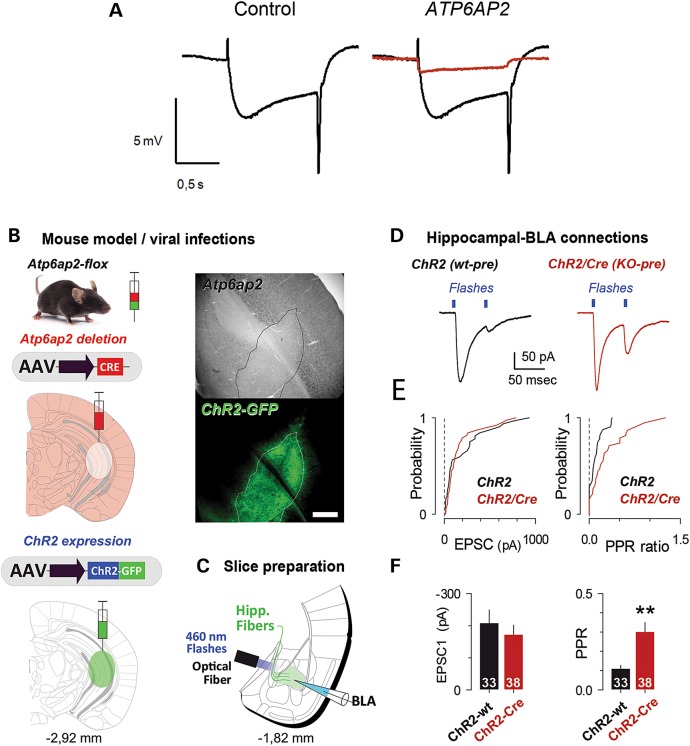
ATP6AP2 deletion induced presynaptic transmission defects in fly and mouse models. (A) Electroretinograms (ERGs) from control flies (GMR-Gal4/+; UAS-Dicer-2/+) in black and from flies with eye-specific knockdown of ATP6AP2 (ATP6AP2 RNAi1/GMR-Gal4; UAS-Dicer-2/+) in red. ATP6AP2 knockdown caused a complete absence of ‘on’ and ‘off’ transients and diminished amplitude photoreceptor de- and repolarization. (B) Specific presynaptic deletion of Atp6ap2 was achieved by the simultaneous viral infection of AAV-Cre and AAV-ChR2-GFP constructs within the caudal hippocampus. An in situ control of Atp6ap2 deletion in the GFP area was tested using immunohistochemistry. Scale bar: 500 µm. (C) Excitatory synaptic transmission at hippocampal to BLA projections was examined using coronal amygdala-containing acute slices. Synaptic transmission was evoked by short flashes of 460-nm light delivered through an optical fibre. (D) Postsynaptic responses to paired light stimulations separated by 50 ms were used to test for release probability at hippocampal terminals. Recordings were performed in Atp6ap2 flx mice infected solely with AAV-ChR2 (ChR2) or AAV-ChR2 and AAV-Cre (ChR2/Cre). (D and E) In the absence of presynaptic Atp6ap2, we observed a change in the EPSC2/EPSC1 ratio (PPR ratio). Statistics includes results from three experimental replicates. The number of recorded cells is indicated.
Then, we analysed synaptic transmission in hippocampal projections to the basolateral amygdala (Hip-BLA) in mice. Indeed, Atp6ap2Camk2aCre/0 mice exhibit pronounced deficits in freezing behaviour when re-exposed to a context in which they received electric footshock previously (Fig. 4E). Recently, we demonstrated that efficacy at hippocampal projections to the basolateral amygdala is of crucial importance for the aversive conditioned response in mice (28). We defined some experimental conditions that allow the functional isolation of Hip-BLA projections using opsin-based strategies combined with genetic deletion of Atp6ap2 in the presynaptic compartment. Specifically, co-injections of AAV-Cre and AAV-ChR2-GFP viruses were performed in the caudal hippocampus, and after a delay allowing both opsin expression and Atp6ap2 removal, electrophysiological recordings were performed in BLA principal neurons (Fig. 5B and C). To verify that the AAV-Cre strategy efficiently depleted endogenous Atp6ap2 proteins, we performed immunodetection of Atp6ap2 at the infection site by visualizing GFP expression associated with AAV-ChR2 infection. As shown in Figure 5B, there is a clear absence of Atp6ap2 labelling in the GFP-expressing area. Next we compared synaptic transmission between groups of mice in which AAV-ChR2-GFP was used solely (wt-pre) or together with AAV-Cre (KO-pre) (Fig. 5D–F). First, to assess for global changes in synaptic efficacy, we applied single, short, 460-nm light flashes of increasing amplitude while recording BLA principal cells at −70 mV in the voltage-clamp mode, thereby building so-called input/output (I/O) curves. We did not observe any difference between the two recording groups (data not shown), suggesting that the presence of Atp6ap2 within the pre-synapse is not essential for basal neurotransmission. Next, to investigate whether Atp6ap2 would affect short-term plasticity, we tested responses to paired stimulations applied at short (50 ms) time intervals (Fig. 5D). A very pronounced paired pulse depression (PPD) was observed in the control group (wt-pre) (wt-pre: Peak 2: 0.11 ± 0.02% of Peak 1, n = 33, Fig. 5D–F). Interestingly, the PPD was less pronounced in KO-pre recordings (KO-pre: Peak 2: 0.30 ± 0.05% of Peak 1, n = 38, P < 0.01 compared with wt-pre, Fig. 5D–F). These results suggested that Atp6ap2 could be essential for maintaining the release probability at excitatory synapses. Interestingly, when stimulations were applied at lower frequencies, no difference was observed (data not shown).
As one of the families with ATP6AP2 mutation presented generalized tonic-clonic seizures, we evaluated Atp6ap2Camk2aCre/0 males for susceptibility to seizures either spontaneously using electroencephalography (EEG) recording with cortical electrodes or following injection of a pro-convulsive compound, the pentylenetetrazol (PTZ). After >24 h of EEG recording, we did not observe any spontaneous seizures both in wt and Atp6ap2Camk2aCre/0 mice (Supplementary Material, Fig. S4A). We also tested the PTZ, a pro-epileptic drug, at two different doses (30 and 40 mg/kg; Supplementary Material, Fig. S4B–G), and we did not find increased susceptibility to seizures. On the contrary, the percentage of animals with seizures was reduced and the latency of seizures was increased in the Atp6ap2Camk2aCre/0 mice compared with wt mice (Supplementary Material, Fig. S4B–G). Altogether, the data suggest that the deletion Atp6ap2 in glutamatergic neurons mice is unlikely to increase susceptibility to seizures.
Atp6ap2 localizes primarily in the endoplasmic reticulum in the soma and in axons and the presynaptic compartment
To better understand the function of Atp6ap2 in neurons, we analysed its subcellular localization in cortical and hippocampal neurons from primary neuronal cultures. First, we performed colocalization studies with markers of different cellular compartments. We showed that Atp6ap2 is predominantly localised in the endoplasmic reticulum (ER) where it colocalized with the ER marker KDEL-ER (Supplementary Material, Fig. S5A) but not in the Golgi because it did not colocalize with the Golgi marker GM130 (data not shown). We performed additional colocalization analyses with markers revealing the different vesicular compartments: Rab5b and EEA1 for early endosomes, Rab7 for late endosomes, lamp1 for lysosomes and LC3B for autophagosomes. Atp6ap2 partially colocalized with all the different vesicular compartments (Supplementary Material, Fig. S5B–E), including lysosomes (Supplementary Material, Fig. S5D) and autophagosomes (Supplementary Material, Fig. S5E). Our results are in agreement with the colocalization quantification studies published by Kinouchi and collaborators (29). Immunogold electron microscopy showed high gold particle concentration in axons and, in particular, in myelinated axons where the gold particles were present in both axons and myelin sheaths (Supplementary Material, Fig. S5J and K). In addition, colocalization studies with pre- and post-synaptic markers revealed that Atp6ap2 colocalized with several presynaptic markers, including synaptophysin (Supplementary Material, Fig. S5G), VGLUT1 (Supplementary Material, Fig. S5H) and VGLUT2 (Supplementary Material, Fig. S5I), and only poorly colocalized with PSD95, a postsynaptic marker (Supplementary Material, Fig. S5F). In conclusion, Atp6ap2 is present in dendrites and axons, with predominance in axons.
Presynaptic knockdown of Drosophila ATP6AP2 disrupts synapse morphology, ultrastructural organization and axonal transport of the active zone component BRP
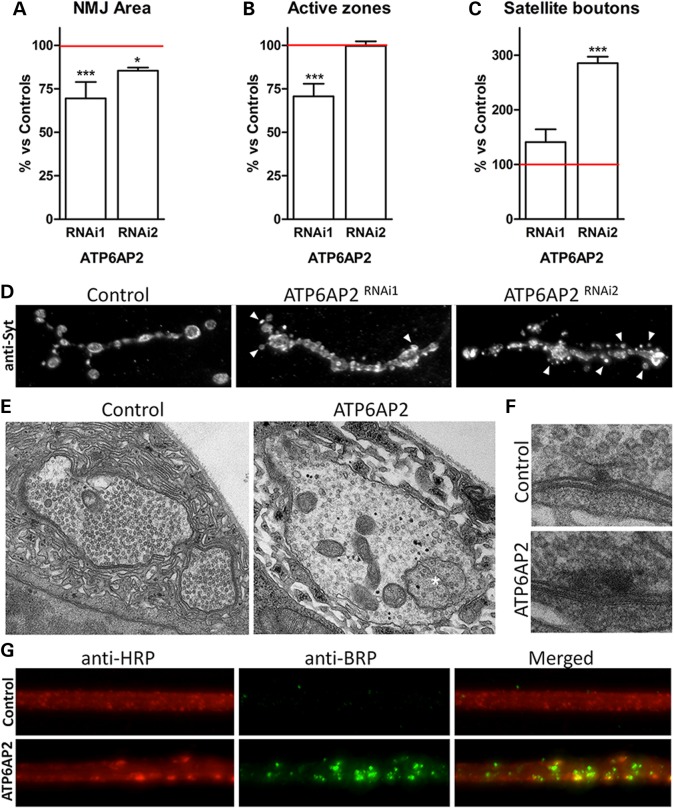
Based on ATP6AP2 electrophysiology in Drosophila and localization in mouse, we examined the consequences of ATP6AP2 loss on synapse development and morphology. The Drosophila neuromuscular junction (NMJ), which is a well-established model system for mammalian central excitatory/glutamatergic synapses, has already provided major insights into synapse biology underlying ID (30). The NMJ synaptic terminal is organized into synaptic boutons that resemble pearls on a string, each containing numerous chemical synapses for vesicle release, so-called active zones (Supplementary Material, Fig. S6). We knocked down ATP6AP2 presynaptically using the pan-neuronal elav-Gal4 driver and examined NMJ morphology. Computer-assisted quantification of immunolabelling with the postsynaptic marker DLG, highlighting the overall morphology of the synaptic terminal, and with the key presynaptic scaffold protein BRP (31), an integral component of active zones, revealed undergrowth of the synaptic terminal in both ATP6AP2RNAi1 and ATP6AP2RNAi2 conditions and a reduced number of active zones in the stronger knockdown condition (ATP6AP2RNAi1) (Fig. 6A and B). Furthermore, we noticed a high number of so-called satellite boutons (32), small immature extra-boutons splitting off from the primary synaptic terminal (Fig. 6C). Because satellite boutons, to our surprise, seemed more frequent in the otherwise weaker ATP6AP2RNAi2-induced knockdown condition, we followed up with a second presynaptic marker, synaptotagmin (anti-Syt), which highlights bouton organization (Fig. 6D). Anti-Syt staining revealed a striking increase in satellite boutons also in the ATP6AP2RNAi1 knockdown synapses. Frequently, satellites remain extremely small in this severe condition (Fig. 6D, arrowheads). Bouton organization of the primary synaptic terminal was also affected consistently in both ATP6AP2 knockdown conditions, with boutons not being properly separated and spaced but fused, with some fused to the degree that individual boutons were no longer quantifiable (Fig. 6D). Ultrastructural analysis of ATP6AP2 knockdown synapses revealed the presence of large multivesicular body-like organelles (Fig. 6E, asterisk). The ultrastructure of T-bars at active zones was also perturbed (Fig. 6F). We conclude that the loss of ATP6AP2 impairs the growth of synaptic terminals, promotes satellite bouton formation and affects bouton, subsynaptic and ultrastructural organization.
Figure 6.
Presynaptic knockdown of Drosophila ATP6AP2 disrupted synapse morphology and ultrastructural organization and affected BRP transport. (A–C) Quantitative assessment of morphological parameters at the NMJ upon presynaptic ATP6AP2 knockdown (ATP6AP2RNAi1: UAS-Dicer-2/+; elav-Gal4/UAS-ATP6AP2RNAi1, ATP6AP2RNAi2:UAS-Dicer-2/+; elav-Gal4/UAS-ATP6AP2RNAi2). The area of synaptic terminals, number of active zones and number of satellite boutons, as quantified based on NMJs immunostained with anti-DLG (postsynaptic) and anti-BRP (active zone) markers (see Supplementary Material, Fig. S6), were normalized against appropriate genetic background controls (UAS-Dicer-2/+; elav-Gal4/+). Bar graphs represent averages, and error bars represent SEM. Significance was determined using a one-way ANOVA and post hoc Tukey's multiple comparison test. *P < 0.5, **P < 0.01, ***P < 0.001. (D) Representative NMJs of control, ATP6AP2RNAi1 and ATP6AP2RNAi2 animals labelled with anti-synaptotagmin (anti-syt). This marker readily revealed evident abnormal bouton morphology and a highly increased number of satellite buttons (arrowheads) in both ATP6AP2 knockdown conditions. (E) Ultrastructural analysis of ATP6AP2RNAi1 versus control synaptic boutons showed the appearance of large multivesicular body-like structures (asterisk) and (F) abnormal T-bar morphology. (E and F) Images were acquired at 15 000× and 30 000× magnification, respectively. (G) Motor axons of control and ATP6AP2RNAi1 animals were immunolabelled with anti-HRP (red) and anti-BRP (nc82 anti-bruchpilot, green). ATP6AP2RNAi1 showed abnormal accumulation of the presynaptic marker BRP, which is indicative of axonal transport defects. All analyses were performed in third instar larvae muscle 4 type 1b NMJs.
While inspecting NMJ morphology in pan-neuronally induced ATP6AP2RNAi1 animals, which are characterized by a decreased amount of BRP-positive active zones, we found that BRP strongly accumulated in motor axons (Fig. 6G). In contrast, BRP, which is required for the structural integrity and function of the synaptic active zone (31,33), is transported to the presynaptic terminal with high efficiency in wild-type animals, indicating that ATP6AP2 is required for BRP axonal transport.
Mouse Atp6ap2 deletion in glutamatergic neurons causes an autophagy defect leading to axonal and neuronal degeneration and to synapse splitting
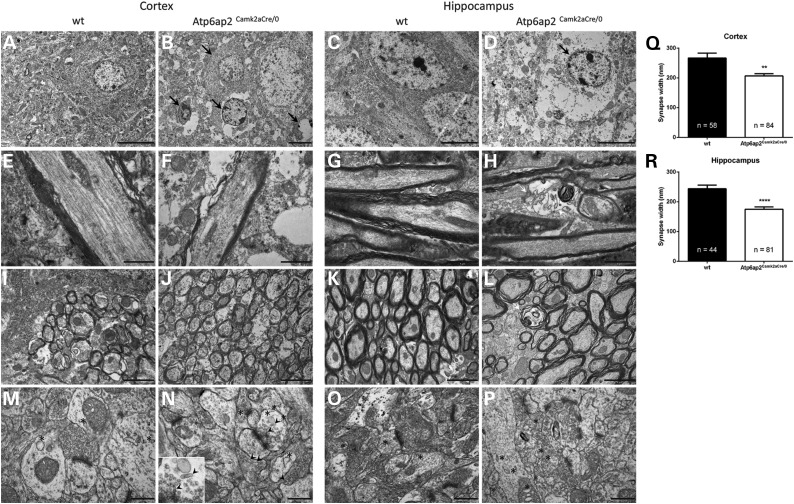
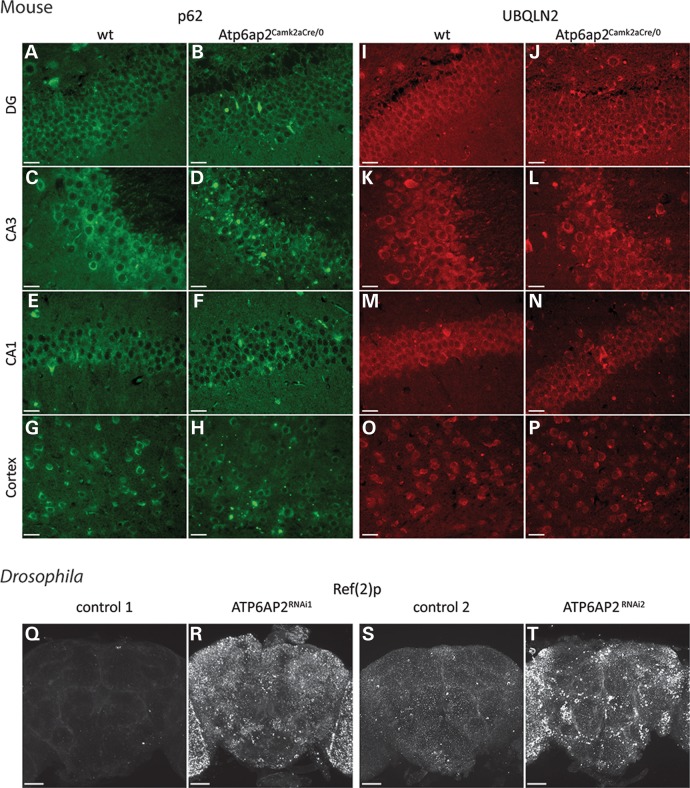
To analyse the consequences of Atp6ap2 deletion in the mouse brain, we performed electron microscopy morphological studies in the cortex and hippocampus, the two regions with the highest amount of Atp6ap2 deletion in our mouse model. Several alterations were observed in the Atp6ap2Camk2aCre/0 mice. First, we revealed the presence of an important number of degenerating neurons in both the cortex and hippocampus of the Atp6ap2Camk2aCre/0 mice but not in the wt mice (Fig. 7A–D); the hippocampus seemed to be more affected than the cortex. The process of neuronal death was not apoptosis, as determined by the lack of cleaved caspase 3 signal in the cortex and the hippocampus of Atp6ap2Camk2aCre/0 mice (data not shown). Rather, cells die from necrosis as suggested in electron microscopy images showing features of necrotic cells: absence of fragmented nuclei, presence of empty cytoplasm and dilatation of mitochondria and of the rough ER (Fig. 7B and D). In addition, we showed that neuronal degeneration is accompanied by the degeneration of axons, particularly myelinated axons (Fig. 7E–L). We also observed reductions in myelin thickness in both degenerating and non-degenerating axons (Fig. 7F, H, J and L). As shown in Figure 7F and 7H, the reduced myelin thickness is due to a decreased number of myelin wraps around axons. Some axons also had disturbed microtubules (Fig. 7F). Finally, we observed the presence of an increased number of vacuoles in the neurons of mutant mice (Fig.7Q–T). Because several publications reported the involvement of the autophagy pathway in the phenotype induced by Atp6ap2 deletion in podocytes and cardiomyocytes (12–14) and because we observed an increased number of vacuoles in neurons in the Atp6ap2Camk2aCre/0 cortex and hippocampus, we tested whether autophagy would be perturbed in our mouse model. We used immunohistochemistry to examine the protein levels of p62 and UBQLN2, two autophagy substrates reflecting autophagy activity, in brain sections from Atp6ap2Camk2aCre/0 and wt mice. We observed increased p62 and UBQLN2 staining in the cortex and hippocampus of mutant mice in comparison with the wt mice (Fig. 8). The most affected area was the CA3 area of the hippocampus, with massive increases in p62 and UBQLN2 staining, which corresponded to an increased number of dying neurons in this region, as shown in Figure 8D and L. The dentate gyrus was also highly affected, and the CA1 area and cortex seemed to be less affected (Fig. 8). To test whether autophagy was also affected in Drosophila, we stained adult brains of pan-neuronally induced ATP6AP2RNAi1 and ATP6AP2RNAi2 flies and their appropriate background controls for the p62 orthologue Ref(2)p. Both ATP6AP2RNAi1 and ATP6AP2RNAi2 conditions showed a highly significantly increase in Ref(2)p levels (Fig. 8Q–T and Supplementary Material, Fig. S7). Thus, loss of ATP6AP2 in mouse and Drosophila impairs autophagy.
Figure 7.
Axonal and neuronal degeneration in the Atp6ap2Camk2aCre/0 mouse cortex and hippocampus. Electron microscopy analysis of adult cortical and hippocampal sections revealed several alterations in the Atp6ap2Camk2aCre/0 mice. (A–D) We observed the presence of degenerating neurons (black arrows) in the cortex and hippocampus of Atp6ap2Camk2aCre/0 mice (B and D, respectively) compared with the morphology of neurons from the cortex and hippocampus of wt mice (A and C, respectively). Image B shows neurons at different stages of degeneration. Scale bar: 5 µm. (E–L) Myelinated axons showed decreased myelin thickness owing to a reduction in the number of myelin sheaths in both the Atp6ap2Camk2aCre/0 cortex and hippocampus (F and H, respectively) compared with the number of myelin sheaths in the cortex and hippocampus of wt mice (E and G, respectively). Some axons also had disturbed microtubules (F). A considerable number of axons were degenerating (white arrows) or already disappeared in Atp6ap2Camk2aCre/0 mice (J and L, respectively). Scale bars: 1 µm (E–H), 2 µm (I–L). (M–P) High-magnification images showed the normal morphology of synapses from the cortex and hippocampus of wt mice (M and O, respectively) and the altered morphology of synapses from the cortex and hippocampus of Atp6ap2Camk2aCre/0 mice with synapse width reduction and synapse splitting (N and P, respectively). Some synapses in mutant mice presented strange elongated vesicles (arrowhead) (N) that were absent in the synapses of wt mice (M). The number of vacuoles (asterisk) increased in the cortex and hippocampus of Atp6ap2Camk2aCre/0 mice (N and P, respectively) compared with the number of vacuoles in the cortex and hippocampus of wt mice (M and O, respectively). Scale bar: 500 nm. Analysis of high-magnification electron microscopy images of synapses from wt and Atp6ap2Camk2aCre/0 mice confirmed the reduction in the synapses width in the cortex (Q) and hippocampus (R) of mutant mice. Bar graphs represent averages, and error bars represent SEM. Significance was determined using unpaired non-parametric t-test. **P < 0.01, ***P < 0.001 versus wt.
Figure 8.
Deletion of ATP6AP2 leads to defective autophagy in neurons. Immunohistochemical analysis of adult mouse brain sections using the autophagy markers p62 and UBQLN2 revealed perturbed autophagy in the Atp6ap2Camk2aCre/0 mouse cortex and hippocampus. (A–H) Immunohistochemical analysis revealed increased p62 staining in Atp6ap2Camk2aCre/0 mice. The region most affected was the CA3 region, with massive increases in p62 staining and cell death (D) that were not observed in the CA3 region of wt mice (C). The DG was also highly affected, with a considerable increase in p62 staining (B). The CA1 (F) and the cortex (H) were less affected, with small increases in p62 staining compared with p62 staining in those regions in wt mice (A, E and G, respectively). (I–P) Immunohistochemistry with the marker UBQLN2 also showed increases in staining, with a massive increase in the CA3 area (L), a strong increase in the DG (J) and a small increase in the CA1 area (N) and cortex (P) compared with UBQLN2 staining in those regions in wt mice (K, I, M and O, respectively). Scale bar (A–P): 100 µm. Immunohistochemical analysis of Drosophila adult brains using the p62 orthologue Ref(2)p as autophagy marker in pan-neuronally induced ATP6AP2 knockdown (ATP6AP2RNAi1: UAS-Dicer-2/+; elav-Gal4/UAS-ATP6AP2RNAi1, ATP6AP2RNAi2: UAS-Dicer-2/+; elav-Gal4/UAS-ATP6AP2RNAi2) and their appropriate genetic background controls. Representative images are shown for the indicated genotypes in (Q–T). In ATP6AP2 knockdown brains, a significant increase in the intensity of Ref(2)p signal is observed, revealing widespread autophagy defects. Scale bar (Q–T): 40 µm. A minimum of 14 images was collected per genotype. Quantitative assessments and statistical analysis of the images are shown in Supplementary Material, Figure S7.
To determine whether synapses were also affected in mouse, we analysed the morphology and size of synapses from the cortex and hippocampus of Atp6ap2Camk2aCre/0 and control mice. We assessed the width of the synapses [active zone/postsynaptic density (PSD)] and the size (diameter) of the synaptic vesicles on electron microscopy images. We found a significant reduction in the mean width of synapses in the cortex [mean width: 266.3 ± 128.2 nm for wt (n = 1 animal, 58 synapses) and 206.2 ± 71.4 nm for Atp6ap2Camk2aCre/0 (n = 2 animals, 84 synapses), unpaired t-test, P = 0.0062, Fig. 7U] and an even stronger reduction in the mean width in the hippocampus [mean width: 243.5 ± 84.3 nm for wt (n = 1 animal, 45 synapses) and 174.7 ± 70.9 nm for Atp6ap2Camk2aCre/0 (n = 2 animals, 81 synapses), unpaired t-test, P < 0.0001, Fig. 7V] of Atp6ap2Camk2aCre/0 mice compared with wt mice. In parallel, we observed an increased number of split synapses, corresponding to the presence of more than one synapse within a single synaptic terminal/spine contact, in the cortex and hippocampus of Atp6ap2Camk2aCre/0 mice, whereas split synapses were only rarely observed in the cortex and hippocampus of wt mice (Fig. 7M–P). In contrast, we did not detect any change in the size of synaptic vesicles (data not shown). However, in synaptic buttons, we noticed the presence of atypically elongated vesicles whose diameters were similar to that of synaptic vesicles (Fig. 7N).
Atp6ap2Camk2aCre/0 transcriptome deregulation correlates with Atp6ap2Camk2aCre/0 mouse behavioural and physiological phenotypes
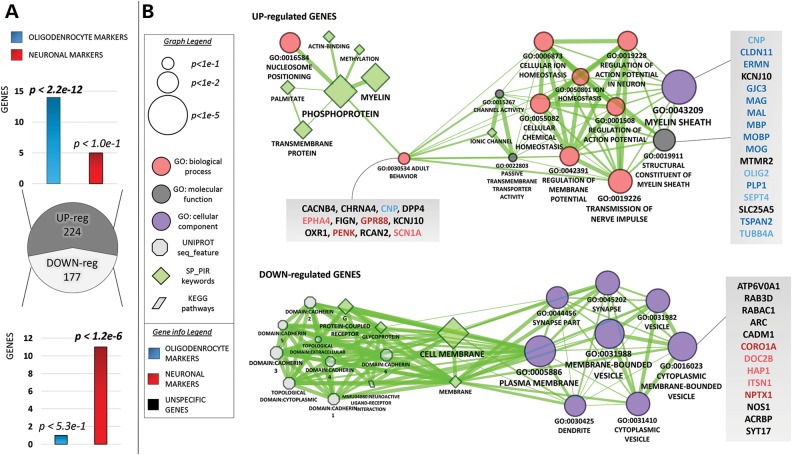
As a global approach to assess ATP6AP2-associated brain pathology and to identify processes dependent on ATP6AP2 unbiasedly, we performed genome-wide transcriptome profiling by mRNA-seq analysis of the Atp6ap2Camk2aCre/0 hippocampus, the brain area most affected in our mouse model, at postnatal day 30. To study the changes in mature mRNA levels, we compared the exon reads of transcripts in wild-type and mutant samples (DEseq algorithm, P < 0.05, Fig. 9) and identified 224 up-regulated and 177 down-regulated genes in mutant hippocampi compared with wt. To determine which brain cell type was affected by those dysregulations directly or indirectly, we performed enrichment tests of either astrocytes, neuronal- or oligodendrocyte-specific markers (34) on the set of up- and down-regulated genes (Fig. 9A). Interestingly, the up-regulated genes were highly enriched in oligodendrocyte-expressed genes (hypergeometric test, P < 2.2E-12) and simultaneously had no enrichment for neuronal markers. In contrast, down-regulated genes were enriched in neuronal markers (P < 1.2E-6) but only slightly enriched in oligodendrocyte markers. Then, we performed GO/KEGG/UniProt enrichment analyses of the lists of up- and down-regulated genes, using a Benjamini cut-off of P < 0.05. The lists of up- and down-regulated genes per GO term are presented in Supplementary Material, Excel File S1 for enrichment calculated in the DAVID website and in Supplementary Material, Excel File S2 for enrichment calculated in the Bingo tools for Cytoscape. A visual representation of the GO enrichment for the up- and down-regulated genes is shown in Figure 9B, together with the lists of individual genes underlying the primary enrichments. The strongest enrichments for the up-regulated genes were related to myelination, regulation of action potential and cellular ion homeostasis. Among those up-regulated genes, 17 genes were specifically linked to the myelin sheath (P < 2.2E-5). Thirteen of these genes were predominantly expressed in oligodendrocytes compared with other cell types (Fig. 9B), suggesting that the physiological equilibrium of oligodendrocytes was altered in Atp6ap2Camk2aCre/0 mice. In total, 18 genes were related to action potential and cellular ion homeostasis, and 2 of them were neuron-specific genes. Among the observed enrichments of the up-regulated genes, 12 genes affecting motor behaviour (P < 9.1E-3) were present (Fig. 9B), including primarily neuron-expressed genes. Down-regulated genes showed a striking enrichment in the cellular components related to vesicles, synapses and dendrites. In particular, we found 13 genes related to membrane-bound vesicles (P < 3.4E-3), including 5 neuronal markers. Interestingly, among the down-regulated genes appeared the ATP6V0A1 gene, which codes for the A1 subunit of the V0 proton-translocation domain of V-ATPase, suggesting that Atp6ap2 deletion in the hippocampal glutamatergic neurons would perturb the expression of its binding partner. In conclusion, the up- and down-regulated genes highlight molecular processes such as myelination, action potential regulation, motor behaviour and membrane-bound vesicles, which are in agreement with the primary phenotypes that we were able to associate with the mouse and fly ATP6AP2 models.
Figure 9.
Transcriptome analysis of the Atp6ap2Camk2aCre/0 mouse hippocampus showed the deregulation of genes related to myelination, behaviour and membrane-bound vesicles. (A) Atp6ap2Camk2aCre/0 hippocampus RNA-seq. The pie chart shows the number of dysregulated genes, with P < 0.05 (DEseq2). The bar charts display the enrichments of up-regulated and down-regulated genes with either neuronal- or oligodendrocyte-specific markers (i.e. genes expressed 16-fold higher in the latter cells compared with other cell types). (B) Visual representation of the GO enrichments for the Atp6ap2Camk2aCre/0 up-regulated and down-regulated genes. The network connectivity is shaped using an overlap score that highlights term similarities; thicker edges represent higher similarity between connected terms/functions. The single genes underlying the primary enrichments are listed in the grey shaded boxes; the gene names are coloured in blue (light blue) when expressed 16-fold (4-fold) more times in oligodendrocytes compared with other cell types. Neuronal markers are coloured in red (light red) using the same principle.
Discussion
Here, we investigated the role of ATP6AP2 in the nervous system of two animal models, mouse and Drosophila. The identified phenotypes are largely consistent in both organisms and suggest an essential role of ATP6AP2 in nervous system development, basal synaptic transmission and cognitive functions.
ATP6AP2 was found to play an evolutionarily conserved role in the regulation of spontaneous motor activity. MB-specific ATP6AP2 knockdown in Drosophila caused elevated activity in response to stimulation, and mice with Atp6ap2 deletion in glutamatergic neurons exhibited increased spontaneous locomotor and vertical activities in several tests. ATP6AP2 is also crucial for short- and long-term memory, as shown in the contexts of Drosophila courtship conditioning and mouse fear conditioning. The fear conditioning defect observed in the mouse model could be attributed to the defective hippocampal-to-basolateral amygdala transmission. Indeed, the efficiency at hippocampal projections to the basolateral amygdala was shown to be of crucial importance for the aversive conditioned response in mice (28).
At the cellular level, ATP6AP2 appears to play a crucial and pleiotropic role at various membrane compartments. In neurons, it localized primarily to the ER and to a lesser extent to lysosomes and autophagosomes. Its deletion in mouse glutamatergic neurons induced an increase in the number vacuoles in cortical and hippocampal neurons. Similarly, its knockdown in Drosophila eye led to vacuolated photoreceptor-surrounding pigment cells and induced ectopic multivesicular body-like membrane compartments in photoreceptor neurons. In addition, we observed strong accumulation of two autophagy substrates, p62 and UBQLN2, in the cortex and hippocampus of Atp6ap2Camk2aCre/0 mice. These results suggest a defect in lysosome-mediated protein degradation and, thus, autophagic activity. Interestingly, this defect corresponds to the situation observed in the Parkinsonism and spasticity (XPDS) patient brains where Korvatska and collaborators (5) observed a massive accumulation of p62/SQSTM1. Autophagy defects have also been observed in cardiomyocyte and podocyte conditional Atp6ap2 mouse models (12–14). In Drosophila, Hermle and collaborators (15) also suggested a role for ATP6AP2/(P)RR in protein degradation. We propose that one primary function of ATP6AP2 in neurons is to regulate autolysosomal material digestion. This hypothesis is supported by the tight physical and functional association of ATP6AP2 with V-ATPase and by the identification of V-ATPase V0 subunit A1 among the down-regulated genes in Atp6ap2Camk2aCre/0 hippocampi. Indeed, the V-ATPase complex is a major element of lysosomes and autolysosomes, where it acidifies the vesicle lumen. Vesicle acidification is required for the activation of lysosomal enzymes. The absence of Atp6ap2 in glutamatergic neurons may induce a perturbation of V-ATPase function in lysosomes, resulting in defective lysosome acidification and, thus, defective activation of lysosomal enzymes and defective digestion of lysosomal substrates. We suggest that this deregulated autophagy induces the axonal and neuronal degeneration observed in the cortex and hippocampus of Atp6ap2Camk2aCre/0 mice. Indeed, neurons, particularly axons, are well known to be highly sensible to autophagy dysregulation, resulting in degeneration (35).
One unexpected finding of this study is the consequence of Atp6ap2 neuronal inactivation on myelin. Indeed, Atp6ap2Camk2aCre/0 mice showed decreased amount of myelin layers and reduction in the total thickness of myelin sheath in the cortex and hippocampus. In addition, we identified the enrichment of dysregulated genes expressed predominantly in oligodendrocytes and implicated in regulating myelination. Nevertheless, the decreased thickness of the myelin sheath in Atp6ap2Camk2aCre/0 mice seems to be a secondary effect of Atp6ap2 loss in glutamatergic neurons, and the up-regulation of oligodendrocyte myelination genes may be a compensatory effect of oligodendrocytes to counteract decreased myelination. Myelin deficits or defective glial/neuronal interactions have already been found in neurodevelopmental disorders including autism spectrum disorder (36) and Rett syndrome (37) and in adult-onset neurodegenerative diseases including Parkinson's disease (38).
At the synapse level, we demonstrated both morphological and functional aberrations in mouse and Drosophila models. In mouse, we observed reduced synapse width in the cortex and hippocampus of Atp6ap2Camk2aCre/0 mice with synapse splitting. Similarly, pan-neuronal ATP6AP2 knockdown in Drosophila led to a reduction in presynaptic terminal size, with abnormal organization of boutons and an increased number of satellite boutons. We also observed defects in subsynaptic organization, including the presence of atypically elongated vesicles in the mouse cortical and hippocampal presynaptic compartments and abnormal active zones in presynaptic terminals of Drosophila NMJs. These converging data suggest a role for ATP6AP2 in synaptic vesicle homeostasis, which is in agreement with a close functional relationship of ATP6AP2 with V-ATPase and its role in synaptic vesicle acidification. V-ATPase is required for synaptic vesicles loading with neurotransmitters (39). In addition, the V0 domain of V-ATPase is part of a complex involved in the fusion of synaptic vesicles with the plasma membrane and thus participates in the regulation of neurotransmitter release (40). Poea-Guyon and collaborators (41) also proposed that V-ATPase, as a sensor of intravesicular pH, informs the exocytosis machinery of the contents of synaptic vesicles to avoid exocytosis of unloaded vesicles. The absence of ATP6AP2 could perturb V-ATPase function in synaptic vesicles and thus disturb synaptic vesicle loading and/or fusion. In addition to morphological alteration, we also provided evidence for defective synaptic transmission. In Drosophila, ATP6AP2 knockdown in photoreceptor neurons caused synaptic transmission defects. In mouse, we showed a decreased release probability of excitatory synapses at the hippocampal to basolateral amygdala connections. However, the reduced presynaptic release probability could also be partly explained by the reduced number of excited fibres owing to axonal degeneration. Altogether, our results confirm an important role for ATP6AP2 in regulating synaptic size, connections and synaptic vesicles, with consequences for presynaptic transmission.
One family with ATP6AP2 mutation was reported to have epilepsy, hence, we tested the mouse model for epilepsy susceptibility. We did not observed spontaneous epilepsy in Atp6ap2Camk2aCre/0 mice and mutant mice even had reduced susceptibility to epilepsy induced by PTZ administration. In our mouse model, Atp6ap2 is deleted only in Camk2a-positive neurons, i.e. excitatory neurons, but not in inhibitory neurons. Thus, considering the important role of inhibitory neurons in preventing epilepsy, we cannot exclude that the absence of epilepsy could be due to the absence of Atp6ap2 deletion in the inhibitory neurons.
Transcriptome analysis of Atp6ap2Camk2aCre/0 hippocampi revealed up-regulated genes related to myelination, motor behaviour and action potential regulation and down-regulated genes implicated in membrane-bound vesicles, showing a striking correlation with the behavioural and physiological phenotypes observed in mouse and fly. This finding suggests that some of the observed phenotypes could be linked to aberrant signalling converging onto transcription. The effects of Atp6ap2 deletion on transcription regulation may be direct or indirect. Atp6ap2 deletion could have indirect effect on gene transcription as suggested for the transcriptional changes observed in genes implicated in myelination mainly expressed in oligodendrocyte; however, this deletion could also affect transcriptional responses directly, e.g. via the Wnt signalling pathway or the MAP kinases ERK1/2 pathway. Indeed, several studies have implicated ATP6AP2 in Wnt signalling (16–18,21). Notably, with its smaller synaptic terminals and many satellite boutons, Drosophila ATP6AP2 knockdown synapses showed striking similarities to mutants defective in Wnt signalling (42–44). In addition, Atp6ap2Camk2aCre/0 mice showed partially overlapping behavioural phenotypes with Wnt/β-catenin mouse models because down- or up-regulation of the Wnt pathway (with injection of the Wnt antagonist Dkkk-1 or Wnt1 peptide) affected contextual and cued fear memory, spatial memory in the Morris water maze and novel object recognition but did not affect baseline locomotor activity and anxiety behaviour (45). ATP6AP2, also known as (pro)renin receptor, is also involved in the renin-angiotensin system (RAS). Activation of ATP6AP2 by (pro)renin binding could trigger the activation of two independent pathways, one resulting in the generation of angiotensin at the cell surface and the activation of its receptors and signalling pathways (including the MAP kinases ERK1/2 pathway) and another resulting in direct activation of the MAP kinases ERK1/2 pathway and the promyelocytic leukaemia zinc finger transcription factor (PLZF) by activated ATP6AP2 (46–49). It has been shown that the RAS plays an important role in brain physiological processes such as learning, memory and cognitive functions (50). Moreover, several members of MAP kinases ERK1/2 pathway have been implicated in ID, including RSK2, which is responsible for the Coffin Lowry syndrome. Further studying the role of ATP6AP2 in transcription regulation and, specifically, to what extent the synaptic and behavioural phenotypes observed in Drosophila and mouse models result from abnormal Wnt and/or MAP kinases ERK1/2 signalling will be extremely interesting.
We demonstrated that ATP6AP2 depletion in Drosophila and mouse induced behavioural, neuronal and synaptic phenotypes that lead to cognitive impairment and neurodegeneration, which greatly recapitulates the phenotypes associated with ATP6AP2 mutations in human. The two ATP6AP2 mutations identified in humans so far are splicing mutations, both leading to the skipping of the exon 4 of the gene (2,5). This results in significant overexpression (∼50%) of a 150-bp minor splice isoform, which produces a protein with an internal deletion of 32 amino acids, and concomitant reduction of the major full size transcripts (2,5). Contrepas and collaborators suggested that ATP6AP2 protein lacking exon 4 (Δ4 ATP6AP2) would act as a dominant negative through dimerization with ATP6AP2 wild-type proteins (51). In XPDS patients, a reduction of the total amount of ATP6AP2 proteins was observed in the brain (5). In contradiction, another ATP6AP2 mutation was identified in a male within a cohort of 6500 sequenced control exomes (3). This mutation is also a splicing mutation affecting the last exon of the gene; however, it has never been validated and the consequences of the mutation remain unclear. As we demonstrated that ATP6AP2 deletion in Drosophila and mouse has drastic consequences on neurons, supporting its involvement as a causative gene for ID and Parkinsonism, we suggest that ATP6AP2 should be considered when screening patients with X-linked intellectual disabilities with epilepsy and/or Parkinsonism with spasticity (2,3,5). Indeed, another member of the V-ATPase complex, ATP6V0A2, has also already been implicated in ID (23).
Materials and Methods
Drosophila stocks and genetics
Conditional RNAi lines targeting ATP6AP2 (stocks 105281 and 5830, referred to as UAS-ATP6AP2RNAi1 and UAS-ATP6AP2RNAi2, respectively) and UAS-Vha100-1RNAi (22924), genetic background controls (60100, 60000) and UAS-Dicer-2 (60008, 60009), were obtained from the Vienna Drosophila RNAi Centre (VDRC, Vienna, Austria). GMR-Gal4 (1104) and elav-Gal4 (8760) strains were obtained from the Bloomington Drosophila Stock Center (Indiana University). Drivers GMR-Gal4; UAS-Dicer-2, act-Gal4/CyO-GFP; UAS-Dicer-2/CyO and UAS-Dicer-2; elav-Gal4 were assembled in-house. The mushroom body-specific driver w+,UAS-Dicer-2; 247-Gal4 was a gift from K. Keleman. Flies were cultured according to standard procedures and raised at 28°C unless indicated otherwise.
Drosophila phototaxis
UAS-ATP6AP2RNAi1, UAS-ATP6AP2RNAi2, UAS-Vha-100-1RNAi lines and their genetic background control lines were crossed with GMR-Gal4;UAS-Dicer-2 virgins and raised at 28°C. In the fast phototaxis assay (52,53), a countercurrent apparatus was used to fractionate genotypes among six tubes according to their visual activity. The phototaxis index (PI) is calculated as ∑i*Ni)/N, where N is the number of flies, i is the tube number and Ni is the number of flies in the ith tube. The average PI and standard deviation were calculated from three independent experiments on different test days. Assays were performed as described previously (22).
Drosophila electroretinograms
UAS-ATP6AP2RNAi1, UAS-ATP6AP2RNAi2 and their control lines were crossed with GMR-Gal4;UAS-Dicer-2 virgins and raised at 28°C. ERG recordings were performed as described previously (27). Flies were tested at day 1 post-eclosion. In total, 8–10 flies were used per genotype and the average of 5 recordings is presented.
Drosophila courtship conditioning
Courtship conditioning (25) was performed as described previously (24) using males of the genotypes w+,UAS-Dicer-2/Y; 247-Gal4/UAS-ATP6AP2RNAi1; +/+ and w+,UAS-Dicer-2/Y; 247-Gal4/+; +/+ (control). Males were collected at eclosion and kept in isolation until 4 days of age. Then, the males were trained by pairing with a single pre-mated female for 1 h to induce short-term memory and for 7 h to induce long-term memory. After training, the males were isolated for 1 h (short-term memory, STM) or 24 h (long-term memory, LTM), transferred to a 1-cm diameter chamber and filmed for 10 min in the presence of a new pre-mated female. The courtship behaviour of male flies towards females was quantified using Actual FlyTrack software (Actual Analytics Ltd). The mean courtship indexes (CI, the percentage of time spent on courtship during a 10-min interval) of trained males and of socially naïve males were used to calculate the learning index (LI), which is defined as the per cent reduction in mean courtship activity in trained males compared with naïve males; LI = (CInaive–CItrained)/CInaive. For all conditions, male–female pairs were analysed on four different test days, and the data were pooled. For non-parametric statistical comparison of the STM and LTM LIs of ATP6AP2 knockdown and control flies, we used a custom R script to perform bootstrapping as described previously (54). Briefly, CI values were randomly sampled with replacement to generate 10 000 hypothetical LIs, which were used to determine the 95% confidence interval of the difference between the index of the control and the index of the knockdown and the probability that this difference is >0.
Mouse lines, ethical statement and genotyping
The Atp6ap2tm1.ICS conditional knockout mouse line (noted Atp6ap2cKO) was generated at the Institut Clinique de la Souris (ICS; llkirch, France) in collaboration with Merck & Co and purchased via Taconic (www.taconic.com). The targeting vector was constructed by the successive cloning of PCR products and contained 4.2-kb 5′ and 3.3-kb 3′ homology arms and a Neomycin selection cassette. Two loxP sequences were located 682 bp before exon 2 and 449 bp after exon 2 (Supplementary Material, Fig. S1). The linearized construct was electroporated into C57BL/6N (B6N) mouse embryonic stem (ES) cells. After selection, targeted clones were identified by PCR using external primers and further confirmed by Southern blot with a Neo internal probe and with a 5′ external probe (data not shown). A positive ES clone was injected into BALBc/J blastocysts, and male chimaeras derived from this procedure gave germline transmission. The resulting line was verified by PCR using external primers and further confirmed by Southern blot with a Neo internal probe. Progeny mice were on the B6N background.
The flipped Neo selection cassette was removed in vivo by breeding the chimaeras directly with a Flp deleter line (57). The Flp transgene was segregated by a further breeding step. We used the transgenic Tg(Camk2aCre)4Gsc mouse line (26), as a glutamatergic neuron-specific Cre driver to inactivate the Atp6ap2 gene spatially and temporally. A Tg(Camk2CreERT2)Ics mouse line was also generated at ICS and used to evaluate the effects of Atp6ap2 excision in adults after tamoxifen (Tam) injection. For the study, we used males that have only one copy of Atp6ap2<cKO/+> gene, i.e. floxed Atp6ap2, and either hemizygote for Tg(Camk2aCre)4Gsc/0 or Tg(Camk2CreERT2)Ics. The corresponding males were noted Atp6ap2Camk2aCre/0 or Atp6ap2Camk2aCreERT2/0 as mutant and compared with wild-type littermate (wt) as controls.
The mice were housed in groups (2–4 per cage) and allowed 2 weeks of acclimation in the phenotyping area with a controlled temperature (21–22°C) under a 12–12 light/dark cycle (lights on at 7:00 a.m.), with food and water available ad libitum. Behavioural experiments started at 13 weeks old and included a pipeline of 12 tests. The behavioural pipeline was performed in agreement with the EC directive 2010/63/UE86/609/CEE and was approved by the local animal care, use and ethic committee of the IGBMC (Com'Eth) under accreditation number (2012–139).
Mutant mice were identified by PCR analysis of tail genomic DNA. Forward (5′ AGCACTCTCTTCCAGGTATGTTGTG 3′) and reverse (5′ CTGGATCCCGGAGCATGGGTAAAGG 3′) primers set 1 generate a 280-bp band from the wild-type allele (Atp6ap2wt) and a 330-bp band from the conditional (Atp6ap2cKO) allele; forward (5′ CAGGTGTGCTGCTATTAATAGG 3′) and reverse (5′ GCCCCTCTCTTACAGTTCTATCAGT 3′) primers set 2 generate a 289-bp band from the Atp6ap2cKO allele and a 179-bp band from the wild-type (wt) allele; forward (5′ AGCACTCTCTTCCAGGTATGTTGTG 3′) and reverse (5′ GCCCCTCTCTTACAGTTCTATCAGT 3′) primers set 3 generate a 1640-bp, a 326-bp and a 1480-bp band from the Atp6ap2 cKO, KO and wt alleles, respectively.
Mouse behavioural analysis
We initially performed a thorough behavioural characterization of Atp6ap2Camk2aCre/0 mice. A cohort of 12 wt and 12 Atp6ap2Camk2aCre/0 mice were submitted to a battery of tests to evaluate a wide range of CNS and PNS functions or their pathologies: circadian activity (13 weeks), object recognition (14 weeks), gross neurological examination grip strength and rotarod (15 weeks), Y maze and prepulse inhibition (16 weeks), fear conditioning (17 weeks), social recognition (23 weeks), Morris water maze (26 weeks) and hot plate (29 weeks). The temporally controlled mutants (Atp6ap2Camk2aCreERT2/0, 11 wt and 10 mutants) were also submitted to selected behavioural tests [circadian activity (31 weeks), fear conditioning and shock threshold (32 weeks), water maze and hot plate (33 weeks)] to evaluate whether the potential phenotypes observed in constitutively mutants are also found when Atp6ap2 deletion is induced in adults after tamoxifen administration. Tamoxifen, which is used to activate the CreERT2 fusion protein, was dissolved in a mixture of ethanol/oil and administered twice a day for five consecutive days. The mice were allowed 2 months to recover after Tam treatment and then transferred for phenotyping and behavioural testing.
Gross neurological examination: the general health and basic sensory motor functions of the mice were evaluated using a modified SHIRPA protocol (EMPRESS, eumorphia.org, 55). This protocol provides an overview of the physical appearance, body weight, body temperature, neurological reflexes and sensory abilities of the mice.
Circadian activity and ingestive behaviours: spontaneous locomotor activity and rears were measured using individual boxes equipped with infrared captors. The quantities of water and food consumed were measured during the test period using an automated pellet feeder and a lickometer (Imetronic, Pessac, France). The mice were tested for 35 h to measure apparatus habituation and nocturnal and diurnal activities. The results are expressed per 1-h period.
The open-field test: mice were tested in automated open fields (Panlab, Barcelona, Spain); each field was divided virtually into central and peripheral regions. The open fields were placed in a room homogeneously illuminated at 70 Lux. Each mouse was placed in the periphery of the open field and allowed to explore the apparatus freely for 30 min with the experimenter out of the animal's sight. The distance travelled, the number of rears and time spent in the central and peripheral regions were recorded over the test session. The number of entries and the percentage of time spent in centre area are used as indexes of emotionality/anxiety.
Social recognition test: this task is used to evaluate the preference of a mouse for a congener compared with an object placed in an opposite compartment. The apparatus is a transparent runway composed of a central starting box and two goal boxes delimited by a sliding grid at each extremity of the runway. Testing was performed for two consecutive days. On the first day, fresh bedding was placed in the goal boxes. The mouse was placed in the start box for 30 s and then allowed to explore the apparatus freely for 10 min to attenuate their emotionality and to evaluate any potential preference between the two compartments. On the second day, a B6N congener of the same gender was placed in one goal box, and an object (dice for example) was placed in the opposite goal box. The mouse was placed in the start box for 30 s and then allowed to explore the apparatus freely for 10 min. The position of the congener and object boxes was counterbalanced to avoid any potential spatial preference. The duration of exploration of each goal box (when the mouse is sniffing the grid delimiting the goal box) was measured, and the percentage of time the mouse took to explore the congener was used as an index of social preference (recognition preference). The number of entries, the time spent in each goal arm and the vertical activity in each arm were also measured. The social recognition index (SR) is defined as [time congener/(time object + time congener)] × 100.
Object recognition task: the object recognition task was performed in automated open fields (see above). The open fields were placed in a room homogeneously illuminated at 70 Lux at the level of each open field. The objects to be discriminated were a glass marble (2.5 cm diameter) and a plastic dice (2 cm). The animals were first habituated to the open field for 30 min. The next day, they were submitted to a 10-min acquisition trial during which they were placed in the open field in the presence of Object A (marble or dice). The time the animal took to explore Object A (when the animal's snout was directed towards the object at a distance of ≤1 cm) was recorded manually. A 10 min retention trial was performed 3 h later. During this trial, Object A and Object B are placed in the open field, and the times tA and tB that the animal takes to explore the two objects are recorded. The recognition index (RI) is defined as [tB/(tA + tB)] × 100.
Auditory startle reflex reactivity and prepulse inhibition: acoustic startle reactivity and prepulse inhibition (PPI) of the startle reflex were assessed in a single session using standard startle chambers (SR-Lab Startle Response System, San Diego Instruments, USA). Ten different trial types were used: acoustic startle pulse alone (110 db); eight different prepulse trials in which either 70, 75, 85 or 90 dB stimuli were presented alone or preceded the pulse and finally one trial (NOSTIM) in which only the background noise (65 dB) was presented to measure the baseline movement in the Plexiglas cylinder. In the startle pulse or prepulse alone trials, the startle reactivity was analysed, and in the prepulse plus startle trials, the amount of PPI was measured and expressed as a percentage of the basal startle response.
Pavlovian fear conditioning: polymodal operant chambers (Coulbourn Instruments, Allentown, PA) were used for this experiment. Each chamber (18.5 × 18 × 21.5 cm) consisted of aluminium side walls and Plexiglas rear and front (the door) walls. A loudspeaker and a bright light constituted the sources of the cues during conditioning and cue testing. The general activity of the animals was recorded through the infrared cell placed at the ceiling of the chambers and was recorded directly on a PC computer using Graphic State (Coulbourn). For conditioning, the mice were allowed to acclimate for 4 min, and then a light/tone (10 kHz) CS was presented for 20 s and co-terminated by a mild (1 s, 0.4 mA) footshock (US). The mice were returned to their home cages 2 min later. Testing was performed 24 h following the conditioning session. Testing for the context was performed in the morning. The mouse was placed back into the same chamber that was used for the conditioning and allowed to explore for 6 min without presentation of the light/auditory CS. Testing for the cue was performed in the afternoon (∼5 h after the context testing). The contextual environment of the chambers was changed (wall colour, odour and floor texture). The mouse was placed in the new chamber, allowed to habituate for 2 min, and then presented with light/auditory cues for 2 min. This sequence was repeated once again. At the end of the testing, the animals were returned to their home cages.
Mouse electrophysiology
Stereotaxic viral infections: adeno-associated viruses (AAV2/9.CAG.ChR2-Venus.W.SV40-p1468, ref Addgene-20071, 5.82E12 vector genomes (vg)/ml) and AAV-Cre (AAV2/1.CMV.HI.eGFP-Cre.WPRE.SV40, PennVector P1415, 1.33E13 vg/ml) were packaged at the University of Pennsylvania Vector Core. Seven-week-old mice (over 20 g) were prepared for stereotaxic injection. Beforehand, the mice were treated with buprenorphine (0.1 mg/kg, i.p.) and positioned in a stereotaxic apparatus (David Kopf Instruments, Tujunga, CA) under continuous anaesthesia with isoflurane. During the surgery, the mice were warmed on a 33–35°C heating pad. The virus was bilaterally pressure-injected through glass pipettes (Hirschmann Laborgerate, ringcaps, tips were pulled from OD 30–50 µm) using a Picospritzer (Parker, Co). The positions of bregma and lambda points were defined and adjusted to the same horizontal level. Injection coordinates for the caudal hippocampus were [AP] −3.1–3.3 mm, [ML] ±3.2–3.4 mm and [DV] −4.0 mm.
Acute slice preparations: AAV-infected mice were used to prepare coronal basolateral amygdala (BLA)-containing acute slices. Owing to animal age (3 months old at the time of dissection), we used the ‘protective recovery method’ described at http://www.brainslicemethods.com. Briefly, mice were anesthetised with a mixture of ketamine/xylazine (100 and 10 mg/kg, respectively) and cardiac-perfused with ice-cold, oxygenated (95% O2, 5% CO2) cutting solution (NMDG) containing 93 mm NMDG, 93 mm HCl, 2.5 mm KCl, 1.2 mm NaH2PO4, 30 mm NaHCO3, 25 mm glucose, 10 mm MgSO4, 0.5 mm CaCl2, 5 mm sodium ascorbate, 3 mm sodium pyruvate, 2 mm thiourea and 12 mm N-acetyl-l-cysteine (pH 7.3–7.4, with osmolarity of 300–310 mOsm). The brains were removed rapidly and placed in the ice-cold and oxygenated NMDG cutting solution described earlier. Coronal slices (300 μm) were prepared using a vibratome (VT1200S, Leica Microsystems, USA) and transferred to a 32°C incubation chamber containing the same NMDG cutting solution. After the slides were incubated, they were maintained at room temperature in oxygenated modified ACSF containing 92 mm NaCl, 2.5 mm KCl, 1.2 mm NaH2PO4, 30 mm NaHCO3, 20 mm HEPES, 25 mm glucose, 2 mm MgSO4, 2 mm CaCl2, 5 mm sodium ascorbate, 3 mm sodium pyruvate, 2 mm thiourea and 12 mm N-acetyl-l-cysteine (pH 7.3–7.4, with osmolarity of 300–310 mOsm) until the recordings were performed.
Recordings: whole-cell recordings from BLA principal neurons were performed at 30–32°C in a superfusing chamber as described previously (28). Neurons were identified visually by infrared videomicroscopy using an upright microscope equipped with a 60× objective. Patch electrodes (3–5 MΩ) were pulled from borosilicate glass tubing and filled with a low-chloride solution containing 140 mm Cs-methylsulfonate, 5 mm QX314-Cl, 10 mm HEPES, 10 mm phosphocreatine, 4 mm Mg-ATP and 0.3 mm Na-GTP (pH adjusted to 7.25 with CsOH, 295 mOsm). Hippocampo-BLA monosynaptic EPSCs were elicited by 1 ms light stimulations delivered by an ultra-high power 460 nm LED (Prizmatix Ltd, Israel) at maximal intensity or increasing intensity when indicated.
Immunohistochemistry
Drosophila NMJ: the following primary antibodies were used: anti-DLG (1:200, mouse, DSHB), anti-HRP (1:500, rabbit, Jackson Immuno Research), anti-BRP (nc82, 1:125, mouse, DSHB) and anti-Syt (syn/DSYT2, 1:100, rabbit, H. Bellen). Type 1b NMJs of muscle 4 were analysed after dissection of wandering L3 larvae and fixation in 3.7% PFA for 30 min. Preparations were rinsed with PBS, blocked and permeabilized with 0.3% Triton X-100 in PBS (PBST) supplemented with 5% normal goat serum (NGS) for 2 h at room temperature. Primary antibodies were diluted in PBST with 1% NGS and incubated with the preparations overnight at 4°C. Alexa Fluor 488 and 568 secondary antibodies (1:1000; Invitrogen Molecular Probes) were incubated with the preparations for 2 h at room temperature. Then, the preparations were extensively washed and mounted in Vectashield (Vector Labs). Co-labelling with anti-DLG and anti-BRP, both monoclonal antibodies, was performed by direct conjugation of the primary antibody to Alexa 568 using a Zenon Kit (Life Technologies) according to the manufacturer's instructions. Fluorescence images were acquired using a ZEISS bright-field multi-colour epifluorescence microscope. At least 10 synaptic terminals were analysed per genotype and quantified in ImageJ.
Drosophila brains: adult brains were dissected in HL3 dissection buffer on ice and immediately fixed in cold 4% PFA for 20 min at room temperature. Samples were then rinsed with PBS containing 1% Tween (PBSTween) and blocked and permeabilized in 0.3% PBSTween with 10% NGS for 2 h at room temperature. For primary antibody treatment, samples were incubated in 0.3% PBTween5% NGS and the anti-Ref(2)p (1:5000, rabbit, G. Juhasz) for 48 h at room temperature. Secondary antibody Cy3 (1:400, Invitrogen Molecular Probes) was incubated for 24 h at room temperature in 0.3% PBTween with 5% NGS. Brains were then mounted in Vectashield® (Vector Labs) for 24 h at −20°C before confocal scanning. Image stacks of specimens were imaged on a Leica TCS SP8X confocal microscope (Leica Microsystems) using a 20× glycerol objective for whole brain imaging. Images were quantified using Fiji software (Fiji.sc/Fiji). Briefly, confocal stacks were merged into a single plane by using the maximum projection function. Subsequently, region of central brain was manually selected (using the free hand tool) and fluorescence intensity (arbitrary units) was measured and normalized to the area of the central brain for each brain. Non-parametric test and means comparison were made using Mann–Whitney test.
Mouse: after the behavioural pipeline, 34-week-old Atp6ap2Camk2aCre/0 and wt males (four of each genotype) were deeply anesthetised with pentobarbital and perfused intracardially with 30 ml of PBS, followed by perfusion with 30 ml of 4% PFA in PBS. The brains were dissected, postfixed for at least 48 h in 4% formalin solution and washed in 70% ethanol. After the brains were embedded in paraffin, they were cut into 5-µm thick frontal sections. Immunohistochemistry was performed using a standard protocol. Briefly, antigen retrieval was performed by heating the slides in citric acid buffer (10 mm citric acid, 0.05% Tween 20, pH 6.0) or Tris/EDTA buffer (10 mm Tris Base, 1 mm EDTA, 0.05% Tween 20, pH 9.0) for 45 min in a 94°C water bath. Then, the sections were blocked with 10% normal horse serum and 0.3% Triton X-100 in 1× PBS for 1 h at room temperature. The sections were incubated overnight at 4°C with the appropriate primary antibody (see Supplementary Material, Table S1). The primary antibodies were detected by incubating the sections with Alexa Fluor 488- or 555-conjugated secondary antibody (1:500; Invitrogen Molecular Probes) for 1 h at room temperature. Then, the sections were mounted with Mowiol mounting medium [0.1 m Tris (pH 8.5), 25% glycerol, 10% w/v Mowiol 4–88 (Citifluor)] containing DAPI (5 μg/ml), and images were acquired using a Hamamatsu NanoZoomer 2.0 (Hamamatsu, Hamamatsu City, Japan) and a Leica DMRXA2 epifluorescence microscope (Leica Microsystems, Heidelberg, Germany).
Electron microscopy
Drosophila eye: UAS-ATP6AP2RNAi1, UAS-ATP6AP2RNAi2 and UAS-Vha100-1RNAi with their corresponding genetic background control lines were crossed with GMR-Gal4 virgins and raised at 25°C. Dissected heads from 3- to 4-day-old females were prefixed in 2% glutaraldehyde (GA) buffered with 0.1 m sodium cacodylate (pH 7.4). After the heads were incubated for 30 min, they were taken out, dissected and then put back into the fixative for 24 h. Subsequently, the dissected heads were postfixed for 1 h in 1% osmium tetroxide in Palade buffer (pH 7.4) with 1% kaliumhexacyanoferrat(III)-trihydrat, dehydrated in ethanol and propylene oxide and embedded in a single drop of Epon. Semi-thin (1 µm thick) transverse and longitudinal sections were stained with 1% toluidine blue. Ultrathin sections were stained with uranyl acetate and lead citrate and then examined using a JEOL 1200 electron microscope.
Drosophila NMJ: UAS-ATP6AP2RNAi1 and its corresponding genetic background control line were crossed with UAS-Dicer-2; elav-Gal4 virgins and raised at 28°C. Open book preparations of third instar larvae were fixed in 4% PFA and 0.5% GA for 10 min, followed by 1 h fixation in 2% GA in 0.1 m sodium cacodylate buffer at room temperature. The larvae were postfixed on ice in the dark with 1% OsO4 in 0.8% KFeCN, washed with NaCac buffer for 1 h and stained with 1% uranyl acetate (UAc). Subsequently, the preparations were dehydrated in ethanol and propylene oxide and embedded in a single drop of Epon. Muscle 4 of abdominal segments 2 to 5 was dissected, and multiple muscles were stacked and embedded in an Epon block, which was heated overnight at 60°C for polymerization. Ultrathin sections (50–70 nm) were prepared using a Reichert Ultracut E ultramicrotome (Leica), mounted on copper electron microscopy grids and counter stained using UAc and lead citrate. The sections were examined at varying nominal magnifications using a 100 kV Philips CM100 microscope equipped with a F114 Fastscan CCD camera (TVIPS) or using a 120 kV Tecnai Spirit microscope equipped with a 2 k Eagle CCD camera (FEI).
Mice: 12- to 14-week-old mice (three of each genotype) were deeply anesthetised with pentobarbital and perfused intracardially with 30 ml of PBS, followed by perfusion with 30 ml of 4% PFA in PBS. The brain was dissected and put in 4% PFA solution in PBS at 4°C. Then, one portion of the cortex and hippocampus was processed for morphological analysis by immersion in 2.5% glutaraldehyde and 2.5% paraformaldehyde in cacodylate buffer (0.1 m, pH 7.4), postfixed in 1% osmium tetroxide in 0.1 m cacodylate buffer for 1 h at 4°C and dehydrated through graded ethanol (50, 70, 90 and 100%) and propylene oxide for 30 min each. The samples were embedded in Epon 812. Semi-thin sections were cut at 2 µm with an ultramicrotome (Leica Ultracut UCT), stained with toluidine blue and analysed histologically by light microscopy. Ultrathin sections were cut at 70 nm, contrast strained with uranyl acetate and lead citrate and examined at 70 kv using a Morgagni 268D electron microscope. Images were captured digitally using a Mega View III camera (Soft Imaging System). The other portion of the cortex and hippocampus was processed for immunogold experiment by fixation for 1 h in 2% PFA—0.2% glutaraldehyde solution in PBS. Then, the brains were blocked in a solution of 0.1% NaBH4 in PBS and in a second solution composed of 5% BSA, 0.1% CWFSkin gelatin and 1% NGS (Aurion). Next, the brains were incubated at room temperature for 1 h with ATP6ap2 antibody (see Supplementary Material, Table S1), which was diluted in 1% NGS in PBS and then incubated with secondary goat anti-rabbit antibody, which was coupled to gold ultra-small particles (Aurion) and diluted in 1% NGS in PBS, for 2 h 30 min at room temperature. After the brains were washed, they were fixed in 2% glutaraldehyde solution in PBS. Silver enhancement of gold ultra-small particles was performed using R-Gent SE-EM (Aurion). Then, the samples were processed as described earlier starting from post-fixation in 1% osmium tetroxide in 0.1 m cacodylate buffer.
RNA sequencing
Total RNA was extracted from two wild-type and two mutant samples (frozen hippocampus) using TRIzol. Then total RNA was treated with DNase (Qiagen, 79254) and purified using an RNeasy MinElute Cleanup Kit (Qiagen, 74204). In total, 2000 ng of total RNA was treated with a Ribo-Zero rRNA Removal Kit (human/mouse/rat; Illumina MRZH11124). The depleted RNA was precipitated for 1 h at −80°C in three volumes of ethanol plus 1 µg of glycogen. Then, the RNA was washed and resuspended in 36 µl of RNase-free water. RNA fragmentation buffer (NEBNext® Magnesium RNA Fragmentation Module, E6150S) was added to the solution, and the RNA was fragmented by incubation at 95°C for 3 min. For reverse transcription, cDNA first-strand synthesis was performed with random hexamer primers. cDNA second-strand synthesis was performed with dNTPs to ensure strand specificity. The RNA-seq library was synthesized using a KAPA Hyper prep kit (KK8504), and a treatment with USER enzyme (NEB, M5505L) was added to digest the non-specific strand. The libraries were pooled (4/lane), sequenced on an Illumina HiSeq and yielded ∼40 million single-end reads per sample. The complete data set and the processed data were submitted to Geo (GSE 67541). The RNA-seq libraries were mapped using GSNAP, the gene expression counts were calculated with a custom C++ script and DEseq2 was used to call the dysregulated genes. The GO enrichments were calculated in the DAVID website (http://david.abcc.ncifcrf.gov/) and in the Bingo tools for Cytoscape (http://apps.cytoscape.org/apps/bingo) and by performing a standard hypergeometric test on the Mus musculus GO annotations (updated to version 22/02/2015). The results of all the enrichments can be found in the Supplementary Material.
Statistical analysis
The data were analysed using unpaired Student's t-test or repeated-measures analysis of variance (ANOVA) with one between factors (genotype) and one within factor (time, quadrant) unless stated otherwise. Qualitative parameters (e.g. clinical observations) were analysed using the χ2 test. The level of significance was set at P < 0.05.
Supplementary Material
Funding
This work was supported by the EU funded project ‘Genetic and Epigenetic Networks in Cognitive Dysfunction’ (GENCODYS) consortium (http://www.gencodys.eu/) (FP7-COLLABORATION PROJECT-2009-2.1.1-1/241995 to A.S. and Y.H.), VIDI and TOP grants from The Netherlands Organization for Scientific Research (NWO) (917-96-346, 912-12-109 to A.S.) and the French state funds through the ‘Agence Nationale de la Recherche’ under the frame programme Investissements d'Avenir labelled (ANR-10-IDEX-0002-02, ANR-10-LABX-0030-INRT, ANR-10-INBS-07 PHENOMIN to Y.H.). The funders had no role in the study design, data collection and analysis, decision to publish or manuscript preparation. Funding to pay the Open Access publication charges for this article was provided by the European Commission.
Supplementary Material
Acknowledgements
We thank Bonnie Nijhof, Judith Besseling and Lilian Eshuis for help with Drosophila NMJ, courtship conditioning and histological analyses; Thomas Rosahl and John Mudget for allowing the use of the Atp6ap2cKO mouse line; Loic Lindner and Guillaume Pavlovic for technical help on the ddPCR experiments; Karim Hnia and Gabor Juhasz for providing the antibodies for autophagy analysis; Josiane Hergueux and Jean-Luc Weickert for technical help with electron microscopy; the Genetic Engineering and Model Validation Department for generating the mouse model; and the employees of the ICS animal facility for their technical assistance with mouse care.
Conflict of Interest statement. None declared.
References
- 1.Hedera P., Alvarado D., Beydoun A., Fink J.K. (2002) Novel mental retardation-epilepsy syndrome linked to Xp21.1-p11.4. Ann. Neurol., 51, 45–50. [DOI] [PubMed] [Google Scholar]
- 2.Ramser J., Abidi F.E., Burckle C.A., Lenski C., Toriello H., Wen G., Lubs H.A., Engert S., Stevenson R.E., Meindl A. et al. (2005) A unique exonic splice enhancer mutation in a family with X-linked mental retardation and epilepsy points to a novel role of the renin receptor. Hum. Mol. Genet., 14, 1019–1027. [DOI] [PubMed] [Google Scholar]
- 3.Piton A., Redin C., Mandel J.L. (2013) XLID-causing mutations and associated genes challenged in light of data from large-scale human exome sequencing. Am. J. Hum. Genet., 93, 368–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Poorkaj P., Raskind W.H., Leverenz J.B., Matsushita M., Zabetian C.P., Samii A., Kim S., Gazi N., Nutt J.G., Wolff J. et al. (2010) A novel X-linked four-repeat tauopathy with Parkinsonism and spasticity. Mov. Disord., 25, 1409–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Korvatska O., Strand N.S., Berndt J.D., Strovas T., Chen D.H., Leverenz J.B., Kiianitsa K., Mata I.F., Karakoc E., Greenup J.L. et al. (2013) Altered splicing of ATP6AP2 causes X-linked parkinsonism with spasticity (XPDS). Hum. Mol. Genet., 22, 3259–3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nguyen G., Delarue F., Burckle C., Bouzhir L., Giller T., Sraer J.D. (2002) Pivotal role of the renin/prorenin receptor in angiotensin II production and cellular responses to renin. J. Clin. Invest., 109, 1417–1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bader M. (2007) The second life of the (pro)renin receptor. J. Renin. Angiotensin Aldosterone Syst., 8, 205–208. [DOI] [PubMed] [Google Scholar]
- 8.Ludwig J., Kerscher S., Brandt U., Pfeiffer K., Getlawi F., Apps D.K., Schagger H. (1998) Identification and characterization of a novel 9.2-kDa membrane sector-associated protein of vacuolar proton-ATPase from chromaffin granules. J. Biol. Chem., 273, 10939–10947. [DOI] [PubMed] [Google Scholar]
- 9.Forgac M. (2007) Vacuolar ATPases: rotary proton pumps in physiology and pathophysiology. Nat. Rev. Mol. Cell. Biol., 8, 917–929. [DOI] [PubMed] [Google Scholar]
- 10.Nishi T., Forgac M. (2002) The vacuolar (H+)-ATPases--nature's most versatile proton pumps. Nat. Rev. Mol. Cell. Biol., 3, 94–103. [DOI] [PubMed] [Google Scholar]
- 11.Sihn G., Rousselle A., Vilianovitch L., Burckle C., Bader M. (2010) Physiology of the (pro)renin receptor: Wnt of change? Kidney Int., 78, 246–256. [DOI] [PubMed] [Google Scholar]
- 12.Kinouchi K., Ichihara A., Sano M., Sun-Wada G.H., Wada Y., Kurauchi-Mito A., Bokuda K., Narita T., Oshima Y., Sakoda M. et al. (2010) The (pro)renin receptor/ATP6AP2 is essential for vacuolar H+-ATPase assembly in murine cardiomyocytes. Circ. Res., 107, 30–34. [DOI] [PubMed] [Google Scholar]
- 13.Oshima Y., Kinouchi K., Ichihara A., Sakoda M., Kurauchi-Mito A., Bokuda K., Narita T., Kurosawa H., Sun-Wada G.H., Wada Y. et al. (2011) Prorenin receptor is essential for normal podocyte structure and function. J. Am. Soc. Nephrol., 22, 2203–2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Riediger F., Quack I., Qadri F., Hartleben B., Park J.K., Potthoff S.A., Sohn D., Sihn G., Rousselle A., Fokuhl V. et al. (2011) Prorenin receptor is essential for podocyte autophagy and survival. J. Am. Soc. Nephrol, 22, 2193–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hermle T., Guida M.C., Beck S., Helmstadter S., Simons M. (2013) Drosophila ATP6AP2/VhaPRR functions both as a novel planar cell polarity core protein and a regulator of endosomal trafficking. Embo J., 32, 245–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Buechling T., Bartscherer K., Ohkawara B., Chaudhary V., Spirohn K., Niehrs C., Boutros M. (2010) Wnt/Frizzled signaling requires dPRR, the Drosophila homolog of the prorenin receptor. Curr. Biol., 20, 1263–1268. [DOI] [PubMed] [Google Scholar]
- 17.Cruciat C.M., Ohkawara B., Acebron S.P., Karaulanov E., Reinhard C., Ingelfinger D., Boutros M., Niehrs C. (2010) Requirement of prorenin receptor and vacuolar H+-ATPase-mediated acidification for Wnt signaling. Science, 327, 459–463. [DOI] [PubMed] [Google Scholar]
- 18.Hermle T., Saltukoglu D., Grunewald J., Walz G., Simons M. (2010) Regulation of Frizzled-dependent planar polarity signaling by a V-ATPase subunit. Curr. Biol., 20, 1269–1276. [DOI] [PubMed] [Google Scholar]
- 19.Amsterdam A., Nissen R.M., Sun Z., Swindell E.C., Farrington S., Hopkins N. (2004) Identification of 315 genes essential for early zebrafish development. Proc. Natl Acad. Sci. USA, 101, 12792–12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dietzl G., Chen D., Schnorrer F., Su K.C., Barinova Y., Fellner M., Gasser B., Kinsey K., Oppel S., Scheiblauer S. et al. (2007) A genome-wide transgenic RNAi library for conditional gene inactivation in Drosophila. Nature, 448, 151–156. [DOI] [PubMed] [Google Scholar]
- 21.Schafer S.T., Han J., Pena M., von Bohlen Und Halbach O., Peters J., Gage F.H. (2015) The Wnt adaptor protein ATP6AP2 regulates multiple stages of adult hippocampal neurogenesis. J. Neurosci., 35, 4983–4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oortveld M.A., Keerthikumar S., Oti M., Nijhof B., Fernandes A.C., Kochinke K., Castells-Nobau A., van Engelen E., Ellenkamp T., Eshuis L. et al. (2013) Human intellectual disability genes form conserved functional modules in Drosophila. PLoS Genet., 9, e1003911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kornak U., Reynders E., Dimopoulou A., van Reeuwijk J., Fischer B., Rajab A., Budde B., Nurnberg P., Foulquier F., Lefeber D. et al. (2008) Impaired glycosylation and cutis laxa caused by mutations in the vesicular H+-ATPase subunit ATP6V0A2. Nat. Genet., 40, 32–34. [DOI] [PubMed] [Google Scholar]
- 24.Keleman K., Kruttner S., Alenius M., Dickson B.J. (2007) Function of the Drosophila CPEB protein Orb2 in long-term courtship memory. Nat. Neurosci., 10, 1587–1593. [DOI] [PubMed] [Google Scholar]
- 25.Hall R.W.S.a.J.C. (1979) Conditioned responses in courtship behavior of normal and mutant Drosophila. Proc. Natl Acad. Sci. USA, 76, 3430–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mantamadiotis T., Lemberger T., Bleckmann S.C., Kern H., Kretz O., Martin Villalba A., Tronche F., Kellendonk C., Gau D., Kapfhammer J. et al. (2002) Disruption of CREB function in brain leads to neurodegeneration. Nat. Genet., 31, 47–54. [DOI] [PubMed] [Google Scholar]
- 27.Heisenberg M. (1971) Separation of receptor and lamina potentials in the electroretinogram of normal and mutant drosophila. J. Exp. Biol., 55, 85–100. [DOI] [PubMed] [Google Scholar]
- 28.Zhang C.L., Houbaert X., Lepleux M., Deshors M., Normand E., Gambino F., Herzog E., Humeau Y. (2014) The hippocampo-amygdala control of contextual fear expression is affected in a model of intellectual disability. Brain Struct. Funct., 220, 3673–3682. [DOI] [PubMed] [Google Scholar]
- 29.Kinouchi K., Ichihara A., Sano M., Sun-Wada G.H., Wada Y., Ochi H., Fukuda T., Bokuda K., Kurosawa H., Yoshida N. et al. (2013) The role of individual domains and the significance of shedding of ATP6AP2/(pro)renin receptor in vacuolar H(+)-ATPase biogenesis. PLoS One, 8, e78603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van der Voet M., Nijhof B., Oortveld M.A., Schenck A. (2014) Drosophila models of early onset cognitive disorders and their clinical applications. Neurosci. Biobehav. Rev., 46(Pt 2), 326–342. [DOI] [PubMed] [Google Scholar]
- 31.Kittel R.J., Wichmann C., Rasse T.M., Fouquet W., Schmidt M., Schmid A., Wagh D.A., Pawlu C., Kellner R.R., Willig K.I. et al. (2006) Bruchpilot promotes active zone assembly, Ca2+ channel clustering, and vesicle release. Science, 312, 1051–1054. [DOI] [PubMed] [Google Scholar]
- 32.Dickman D.K., Lu Z., Meinertzhagen I.A., Schwarz T.L. (2006) Altered synaptic development and active zone spacing in endocytosis mutants. Curr. Biol., 16, 591–598. [DOI] [PubMed] [Google Scholar]
- 33.Wagh D.A., Rasse T.M., Asan E., Hofbauer A., Schwenkert I., Durrbeck H., Buchner S., Dabauvalle M.C., Schmidt M., Qin G. et al. (2006) Bruchpilot, a protein with homology to ELKS/CAST, is required for structural integrity and function of synaptic active zones in Drosophila. Neuron, 49, 833–844. [DOI] [PubMed] [Google Scholar]
- 34.Cahoy J.D., Emery B., Kaushal A., Foo L.C., Zamanian J.L., Christopherson K.S., Xing Y., Lubischer J.L., Krieg P.A., Krupenko S.A. et al. (2008) A transcriptome database for astrocytes, neurons, and oligodendrocytes: a new resource for understanding brain development and function. J. Neurosci., 28, 264–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nikoletopoulou V., Papandreou M.E., Tavernarakis N. (2015) Autophagy in the physiology and pathology of the central nervous system. Cell Death Differ., 22, 398–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Owen J.P., Marco E.J., Desai S., Fourie E., Harris J., Hill S.S., Arnett A.B., Mukherjee P. (2013) Abnormal white matter microstructure in children with sensory processing disorders. Neuroimage Clin., 2, 844–853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mahmood A., Bibat G., Zhan A.L., Izbudak I., Farage L., Horska A., Mori S., Naidu S. (2010) White matter impairment in Rett syndrome: diffusion tensor imaging study with clinical correlations. Am. J. Neuroradiol., 31, 295–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim H.A., Mindos T., Parkinson D.B. (2013) Plastic fantastic: Schwann cells and repair of the peripheral nervous system. Stem Cells Transl. Med., 2, 553–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morel N., Poea-Guyon S. (2015) The membrane domain of vacuolar HATPase: a crucial player in neurotransmitter exocytotic release. Cell. mol. Life Sci, 72, 2561–2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Poea-Guyon S., Ammar M.R., Erard M., Amar M., Moreau A.W., Fossier P., Gleize V., Vitale N., Morel N. (2013) The V-ATPase membrane domain is a sensor of granular pH that controls the exocytotic machinery. J. Cell. Biol., 203, 283–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Poea-Guyon S., Amar M., Fossier P., Morel N. (2006) Alternative splicing controls neuronal expression of v-ATPase subunit a1 and sorting to nerve terminals. J. Biol. Chem., 281, 17164–17172. [DOI] [PubMed] [Google Scholar]
- 42.Ataman B., Ashley J., Gorczyca D., Gorczyca M., Mathew D., Wichmann C., Sigrist S.J., Budnik V. (2006) Nuclear trafficking of Drosophila Frizzled-2 during synapse development requires the PDZ protein dGRIP. Proc. Natl Acad. Sci. USA, 103, 7841–7846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Korkut C., Ataman B., Ramachandran P., Ashley J., Barria R., Gherbesi N., Budnik V. (2009) Trans-synaptic transmission of vesicular Wnt signals through Evi/Wntless. Cell, 139, 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Packard M., Koo E.S., Gorczyca M., Sharpe J., Cumberledge S., Budnik V. (2002) The Drosophila Wnt, wingless, provides an essential signal for pre- and postsynaptic differentiation. Cell, 111, 319–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Maguschak K.A., Ressler K.J. (2012) A role for WNT/beta-catenin signaling in the neural mechanisms of behavior. J. Neuroimmune Pharmacol., 7, 763–773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Feldt S., Maschke U., Dechend R., Luft F.C., Muller D.N. (2008) The putative (pro)renin receptor blocker HRP fails to prevent (pro)renin signaling. J. Am. Soc. Nephrol., 19, 743–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Feldt S., Batenburg W.W., Mazak I., Maschke U., Wellner M., Kvakan H., Dechend R., Fiebeler A., Burckle C., Contrepas A. et al. (2008) Prorenin and renin-induced extracellular signal-regulated kinase 1/2 activation in monocytes is not blocked by aliskiren or the handle-region peptide. Hypertension, 51, 682–688. [DOI] [PubMed] [Google Scholar]
- 48.Huang Y., Noble N.A., Zhang J., Xu C., Border W.A. (2007) Renin-stimulated TGF-beta1 expression is regulated by a mitogen-activated protein kinase in mesangial cells. Kidney Int., 72, 45–52. [DOI] [PubMed] [Google Scholar]
- 49.Nguyen G., Danser A.H. (2008) Prorenin and (pro)renin receptor: a review of available data from in vitro studies and experimental models in rodents. Exp. Physiol., 93, 557–563. [DOI] [PubMed] [Google Scholar]
- 50.McKinley M.J., Albiston A.L., Allen A.M., Mathai M.L., May C.N., McAllen R.M., Oldfield B.J., Mendelsohn F.A., Chai S.Y. (2003) The brain renin-angiotensin system: location and physiological roles. Int. J. Biochem. Cell Biol., 35, 901–918. [DOI] [PubMed] [Google Scholar]
- 51.Contrepas A., Walker J., Koulakoff A., Franek K.J., Qadri F., Giaume C., Corvol P., Schwartz C.E., Nguyen G. (2009) A role of the (pro)renin receptor in neuronal cell differentiation. Am. J. Physiol. Regul. Integr. Comp. Physiol., 297, R250–R257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Benzer S. (1967) Behavioral mutants of Drosophila isolated by countercurrent distribution. Proc. Natl Acad. Sci. USA, 58, 1112–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Galy A., Roux M.J., Sahel J.A., Leveillard T., Giangrande A. (2005) Rhodopsin maturation defects induce photoreceptor death by apoptosis: a fly model for RhodopsinPro23His human retinitis pigmentosa. Hum. Mol. Genet., 14, 2547–2557. [DOI] [PubMed] [Google Scholar]
- 54.Gregor A., Kramer J.M., van der Voet M., Schanze I., Uebe S., Donders R., Reis A., Schenck A., Zweier C. (2014) Altered GPM6A/M6 dosage impairs cognition and causes phenotypes responsive to cholesterol in human and Drosophila. Hum. Mutat., 35, 1495–1505. [DOI] [PubMed] [Google Scholar]
- 55.Brown S.D., Chambon P., de Angelis M.H. (2005) EMPReSS: standardized phenotype screens for functional annotation of the mouse genome. Nat. Genet., 37, 1155. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.