Abstract
Stargardt disease (STGD1) due to mutations in the large ABCA4 gene is the most common inherited macular degeneration in humans. We have shown that dual adeno-associated viral (AAV) vectors effectively transfer ABCA4 to the retina of Abca4−/− mice. However, they express both lower levels of transgene compared with a single AAV and truncated proteins. To increase productive dual AAV concatemerization, which would overcome these limitations, we have explored the use of either various regions of homology or heterologous inverted terminal repeats (ITR). In addition, we tested the ability of various degradation signals to decrease the expression of truncated proteins. We found the highest levels of transgene expression using regions of homology based on either alkaline phosphatase or the F1 phage (AK). The use of heterologous ITR does not decrease the levels of truncated proteins relative to full-length ABCA4 and impairs AAV vector production. Conversely, the inclusion of the CL1 degradation signal results in the selective degradation of truncated proteins from the 5′-half without affecting full-length protein production. Therefore, we developed dual AAV hybrid ABCA4 vectors including homologous ITR2, the photoreceptor-specific G protein-coupled receptor kinase 1 promoter, the AK region of homology and the CL1 degradation signal. We show that upon subretinal administration these vectors are both safe in pigs and effective in Abca4−/− mice. Our data support the use of improved dual AAV vectors for gene therapy of STGD1.
Introduction
Stargardt disease (STGD1; MIM#248200), the most common form of inherited macular degeneration in humans, is caused by mutations in ABCA4 [coding sequence (CDS): 6822 bp], which encodes the photoreceptor (PR)-specific all-trans-retinal transporter (1,2). In the absence of a functional ABCA4 protein, vitamin A aldehyde forms bisretinoid adducts that are deposited in retinal pigment epithelial (RPE) cells during the process of disc shedding and phagocytosis. Consequently, abnormally high levels of lipofuscin pigments, such as A2E and all-trans-retinal dimer-phosphatidylethanolamine, accumulate in the RPE, triggering RPE-cell death and causing secondary PR degeneration (2). Abca4−/− mice (3), the only STGD1 animal model, recapitulate some of the features of the human retinal disease including abnormal lipofuscin accumulation in the RPE.
Sight-restoring therapy for many inherited retinal degenerations (IRDs) including STGD1, is still a major unmet medical need. Gene therapy with adeno-associated viral (AAV) vectors represents, to date, the most promising approach for treatment of many IRDs (4–9). However, one of the main obstacles for the use of AAV is their packaging capacity limited to ∼5 kb. This has become a limiting factor for the development of gene replacement therapy for common IRDs due to mutations in genes with a CDS larger than 5 kb, which include, among others, STGD1. Dual AAV vectors, based on the ability of AAV genomes to concatamerize via intermolecular recombination, have been successfully exploited to overcome this limitation (10–12). Others and we have recently shown the potential of dual AAV vectors in the retina (10,13–15). In particular we have reported that dual AAV hybrid AK vectors which rely on both ITR-mediated concatemerization and homologous recombination mediated by the AK sequence of the F1 phage for the reconstitution of the full-length expression cassette, transduce efficiently mouse and pig PR and rescue the Abca4−/− mouse retinal phenotype (10,15). However, critical issues need to be addressed before considering further clinical translation of this strategy. First, the levels of PR transduction achieved with dual AAV vectors are lower than those achieved with single normal size AAV vectors (10,15). A second major issue associated with the use of dual AAV vectors is the production of truncated proteins from the 5′-half vector that contains the promoter sequence and/or from the 3′-half vector due to the low promoter activity of the inverted terminal repeats (ITR) (10,13,16,17). Both increased transduction efficiency and reduction of truncated protein production may be achieved by boosting the directional formation of productive tail-to-head concatemers, which reduces the number of non-concatemerized single AAV vector halves from which the truncated protein products derive. Since directional reconstitution of dual AAV hybrid vectors relies on recombination mediated by regions of homology included in the two vectors, we sought to evaluate side-by-side the various recombinogenic regions used in the context of dual AAV hybrid vectors which include those derived from either the human placental alkaline phosphatase cDNA (12,18) or the AK sequence from the F1 phage (10).
Another option we have explored to increase the generation of productive tail-to-head AAV genome concatemers is the use of vectors with heterologous ITR (i.e. vectors with ITR from different AAV serotypes at the opposite ends of the viral genome). Indeed, vectors with heterologous ITR from serotypes 2 and 5 (ITR2 and ITR5, respectively), which are highly divergent [58% of homology (19)], show both reduced ability to form circular monomers and increased directional tail-to-head concatamerization than vectors with homologous ITR (20), and reconstitute transgene expression more efficiently than dual AAV vectors with homologous ITR (20,21).
Although highly recombinogenic sequences and/or heterologous ITR can be explored to achieve higher rates of productive dual AAV concatemer formation, the production of truncated protein products might still remain an issue. MicroRNA (miR) target sequences, artificial stop codons or protein ubiquitination signals can be exploited to mediate the degradation of truncated protein products. Among the miR expressed in the retina, miR-let7b or -26a are expressed at high levels (22–25) while miR-124 and -204 have been shown to restrict AAV-mediated transgene expression to either RPE or PR, respectively (26). Signals that mediate the degradation of proteins include: (i) the short degron CL1, a C-terminal destabilizing peptide that shares structural similarities with misfolded proteins and is thus recognized by the ubiquitination system (27–29), (ii) ubiquitin, whose fusion at the N-terminal of a donor protein mediates both direct protein degradation or degradation via the N-end rule pathway (30,31) and (iii) the N-terminal PB29 degron which is a 9 aminoacid-long peptide which, similarly to the CL1 degron, is predicted to fold in structures that are recognized by enzymes of the ubiquitination pathway (32). Additionally, artificial stop codons can be inserted to cause the early termination of an mRNA. We evaluated the ability of these strategies to silence the expression of truncated proteins from dual AAV vectors. Finally, in view of a possible clinical translation of dual AAV vectors for retinal gene therapy, we evaluated the efficacy and the safety of dual AAV hybrid ABCA4 vectors including these optimized features in Abca4−/− mice and in wild-type mice and pigs, respectively.
Results
Dual AAV hybrid ABCA4 vectors which include the AK or AP1 recombinogenic regions show efficient mouse photoreceptor transduction
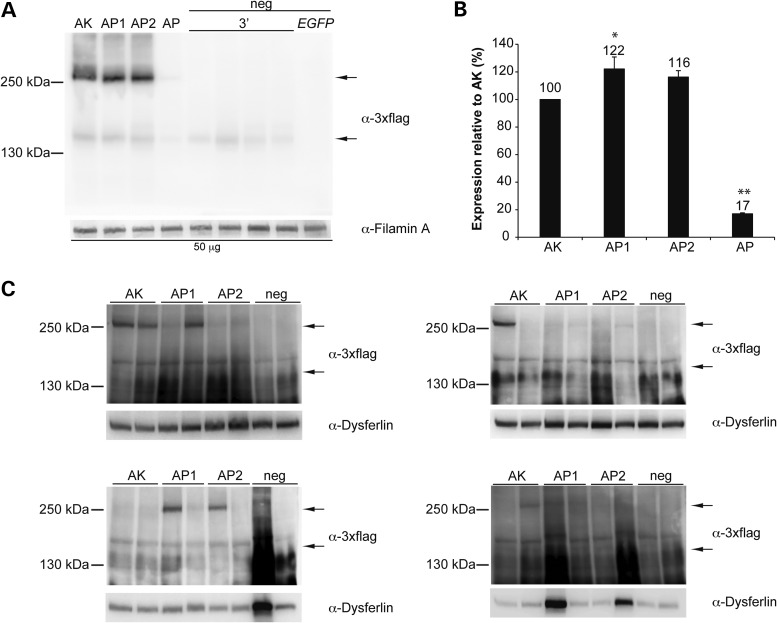
We evaluated in parallel the transduction efficacy of dual AAV hybrid ABCA4 vectors with different regions of homology. The most used derive from either the head or tail (18) of the middle one-third of the human placental alkaline phosphatase cDNA (12). We have previously shown that dual AAV hybrid vectors including the AK sequence from the F1 phage outperform those including the sense placental alkaline phosphatase head region sequence [AP (10), Supplementary Material, Fig. S1], which we generated based on the description provided in Ghosh et al. (18). Recently, the exact sequence of the head and tail placental alkaline phosphatase sequences (AP1 and AP2) have been published by Lostal et al. (16). These correspond to the reverse complementary (antisense) sequences in the middle one-third of the AP cDNA (Supplementary Material, Fig. S1). Thus, we compared the efficacy of these AP1 and AP2 sequences to that of both the AK sequence and of the AP sequence we have previously tested in Trapani et al. (10). For this purpose we generated dual AAV2/2 hybrid vectors that include the ABCA4-3×flag CDS, under the control of the ubiquitous CMV promoter, and either the AK (10), AP (10), AP1 or AP2 (16) regions of homology (Fig. 1 and Supplementary Material, Fig. S1). We used these vectors to infect HEK293 cells [multiplicity of infection, m.o.i.: 5 × 104 genome copies (GC)/cell of each vector]. Cell lysates were analysed by Western blot with anti-3×flag antibodies to detect ABCA4-3×flag (Fig. 2A). Each of the dual AAV hybrid vectors sets resulted in expression of full-length proteins of the expected size that were not detected in the lanes loaded with negative controls (Fig. 2A). Quantification of ABCA4 expression (Fig. 2B) showed that infection with dual AAV hybrid AP1 and AP2 vectors resulted in slightly higher levels of transgene expression than with dual AAV hybrid AK vectors and all significantly outperformed dual AAV hybrid AP vectors. We have previously found that the efficiency of dual AAV vectors which rely on homologous recombination is lower in terminally-differentiated cells as PR than in cell culture (10). We therefore evaluated PR-specific transduction levels in C57BL/6 mice following subretinal administration of dual AAV2/8 AK, AP1 and AP2 vectors which include the PR-specific human G protein-coupled receptor kinase 1 (GRK1) promoter (dose of each vector/eye: 1.9 × 109 GC; Fig. 2C). One month after vector administration we detected efficient ABCA4 protein expression more frequently in retinas treated with dual AAV hybrid AK than AP1 or AP2 vectors (Fig. 2C). However, the average ABCA4 expression levels from all treated eyes were not significantly different between eyes injected with dual AAV hybrid AK, AP1 and AP2 vectors [mean ABCA4 levels: AK: 3.7 ± 0.9 arbitrary units (a.u.); AP1: 3.7 ± 0.9 a.u.; AP2: 2.5 ± 0.5 a.u. No significant differences were found using one-way ANOVA. More details on the quantification and specific statistical values can be found in the Material and Methods section].
Figure 1.
Schematic representation of the dual AAV vectors tested. ITR: inverted terminal repeats from AAV2 or AAV5; Prom: promoter; CDS: coding sequence; SD: splicing donor signal; HR: homology region, AK or from human placental alkaline phosphatase (AP1, AP2 and AP); Deg Sig: degradation signal (see Table 2); SA: splicing acceptor signal; pA: polyadenylation signal.
Figure 2.
Efficient ABCA4 protein expression using the AK, AP1 and AP2 regions of homology. Representative Western blot analysis of (A) HEK293 cells (50 micrograms/lane) infected with dual AAV2/2 hybrid CMV-ABCA4 vectors or (C) C57BL/6 retinas (whole retinal lysates) 1-month post-injection with dual AAV2/8 hybrid GRK1-ABCA4 vectors (dose of each vector/eye: 1.9 × 109 GC). The upper arrow indicates full-length proteins, the lower arrow indicates truncated proteins from the 3′-half vector. Since no truncated proteins can be detected in the mouse retina (10), the lower arrow in (C) points to the level of truncated proteins present in a lysate of infected cells which has been loaded on the same gel as positive control. The molecular weight ladder is depicted on the left. (B) Quantification of ABCA4 protein bands from Western blot analysis in (A). The intensity of the ABCA4 bands in (A) was divided by the intensity of the Filamin A bands. The histograms show the expression of proteins as a percentage relative to dual AAV hybrid AK vectors, the mean value is depicted above the corresponding bar. Values are represented as: mean ± s.e.m. *pANOVA ≤ 0.05, the asterisk indicates significant differences with AK. **pANOVA < 0.001, the asterisks indicate significant differences with AK, AP1 and AP2. More details on the statistical analysis including specific statistical values can be found in the Statistical analysis paragraph of the Materials and Methods section. The Western blot images are representative of: (A) n = 3 independent experiments; (B) n = 8 eyes injected with dual AAV hybrid AK, n = 8 eyes injected with dual AAV hybrid AP1, n = 8 eyes injected with dual AAV hybrid AP2, n = 8 eyes injected with negative control vectors. The quantifications (B) are from n = 3 independent experiments. (A–C) AK: cells infected or eyes injected with dual AAV hybrid AK vectors; AP1: cells infected or eyes injected with dual AAV hybrid AP1 vectors; AP2: cells infected or eyes injected with dual AAV hybrid AP2 vectors; AP: cells infected with dual AAV hybrid AP vectors; neg: negative controls, in (A) cells infected with either the 3′-half vectors (3′) or EGFP-expressing vectors (EGFP), in (B) eyes injected with the 3′-half vectors; α-3×flag: Western blot with anti-3×flag antibodies; α-Filamin A, Western blot with anti-Filamin A antibodies, used as loading control; α-Dysferlin, Western blot with anti-Dysferlin antibodies, used as loading control.
Inclusion of heterologous ITR in dual AAV hybrid ABCA4 vectors affects their production yields and does not reduce levels of truncated proteins
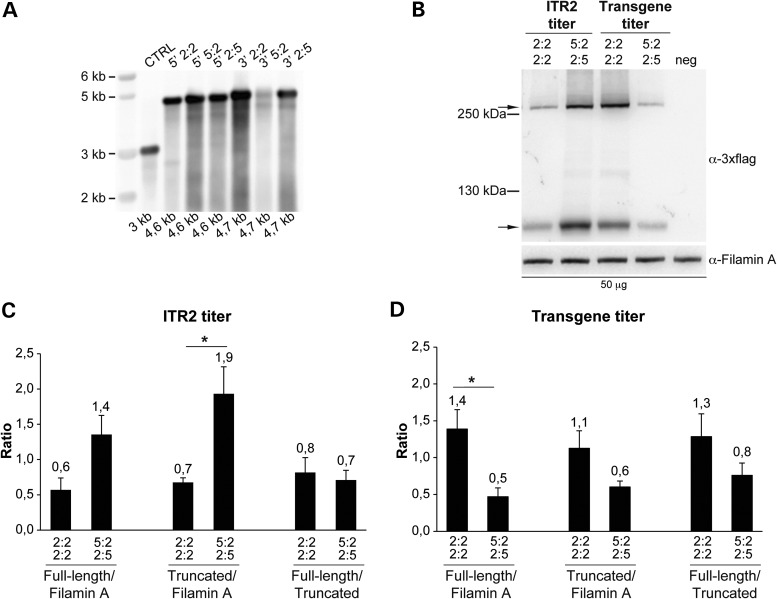
To test if the use of heterologous ITR improves the productive directional concatemerization of dual AAV hybrid ABCA4 vectors, we generated dual AAV2/2 hybrid ABCA4 vectors that included the ABCA4-3×flag CDS with heterologous ITR2 and ITR5 in either the 5:2 (left ITR from AAV5 and right ITR from AAV2) or the 2:5 (left ITR from AAV2 and right ITR from AAV5) configuration. The production of dual AAV vectors bearing heterologous ITR2 and ITR5 requires the simultaneous expression of the Rep proteins from AAV serotypes 2 and 5 which cannot cross-complement virus replication (19). Indeed, it has been shown that Rep2 and Rep5 can bind interchangeably to ITR2 or ITR5, although less efficiently than to homologous ITR, however they cannot cleave the terminal resolution sites of the ITR from the other serotype (33). Therefore, before generating dual AAV hybrid ABCA4 vectors with heterologous ITR2 and ITR5, we assessed the potential competition of (i) Rep5 with Rep2 in the production of AAV2/2-CMV-EGFP vectors (i.e. vectors with homologous ITR2) and (ii) Rep2 with Rep5 in the production of AAV5/2-CMV-EGFP vectors (i.e. vectors with homologous ITR5), using the same amount of the Rep5Cap2 and Rep2Cap2 packaging constructs (ratio1:1). Indeed, when the Rep5Cap2 packaging construct is provided in addition to Rep2Cap2, the total yields of AAV2/2-CMV-EGFP vectors are reduced to 42% of those of control preparations obtained when only Rep2Cap2 is provided as packaging construct (average of four independent preps of each type, p Student's t-test: 0.014). Conversely, no significant differences were found in the total yields of AAV5/2-CMV-EGFP preps obtained when Rep2Cap2 was added to Rep5Cap2, which were 83% of those obtained when Rep5Cap2 was the only packaging construct transfected (average of four independent preps of each type, no significant differences were found using Student's t-test). Given the competition of Rep5 with Rep2 in the production of vectors with ITR2, we tested three different ratios between Rep5 and the Rep2Cap2 packaging constructs in the production of dual AAV hybrid ABCA4 vectors with heterologous ITR2 and ITR5 (Protocol A with 1:1, Protocol B with 1:3 and Protocol C with 1:10 Rep5:Rep2Cap2 ratio). As shown in Supplementary Material, Table S1, viral titres determined by PCR quantification using a probe annealing to ITR2 progressively increased when the amount of Rep5 was decreased, with the best titre obtained with Protocol C. These results confirmed the competition of Rep5 with Rep2 during the production of vectors with ITR2 and led us to follow Protocol C for the production of AAV vectors with heterologous ITR2 and ITR5. However, several AAV preparations obtained with this strategy revealed: (i) up to 6-fold lower titres determined on ITR2 than titres determined on a transgenic sequence in between the ITR (Table 1) which could suggest that the integrity of ITR2 is compromised and (ii) a mean reduction of about 6-fold in the total yields of AAV vectors with heterologous ITR2 and ITR5 compared with those containing homologous ITR2 (p Student's t-test: 0.04; Table 1). However, Southern blot analysis of AAV preparations with heterologous ITR revealed no evident alterations of genome integrity (Fig. 3A). To test if the inclusion of heterologous ITR in dual AAV hybrid ABCA4 vectors enhanced the formation of tail-to-head productive concatemers and full-length protein transduction while reducing the production of truncated proteins, we infected HEK293 cells with dual AAV hybrid ABCA4 vectors with either heterologous ITR2 and ITR5 (in the 5:2/2:5 configuration) or homologous ITR2 (Fig. 3B). Given the difference between the ITR2 and transgene titres for vectors with heterologous but not homologous ITR (Table 1), we infected cells with 104 GC/cell of each vector based on either ITR2 or transgene titres. Western blot analysis of HEK293 cells infected with dual AAV vectors based on ITR2 titres showed that the inclusion of heterologous ITR2 and ITR5 resulted in higher levels of both full-length and truncated protein than the inclusion of homologous ITR2 (Fig. 3B and C). However this was not observed when HEK293 cells were infected with the same dual AAV vector preps based on the transgene titre (Fig. 3B and D). In conclusion, the ratio between full-length and truncated protein expression was similar regardless of the ITR included in the vectors and of the vector titre used to dose cells (Fig. 3C and D).
Table 1.
Low yields and differences between ITR2 and transgene titres of AAV2 with heterologous ITR2 and ITR5
| ID | ITR configuration | ITR2 titre (GC/ml) | Transgene titre (GC/ml) | Yields (GC/ml×3.5 ml) |
|---|---|---|---|---|
| 2101 | 5:2 | 2.0E+12 | 2.5E+12 | 7.9E+12 |
| 2136 | 5:2 | 2.4E+11 | 6.0E+11 | 1.5E+12 |
| 2137 | 5:2 | 4.4E+11 | 2.5E+12 | 5.1E+12 |
| 2140 | 5:2 | 5.2E+10 | 1.5E+11 | 3.5E+11 |
| 2102 | 2:5 | 4.6E+11 | 1.2E+12 | 2.9E+12 |
| 2135 | 2:5 | 1.5E+12 | 2.5E+12 | 7.0E+12 |
| 2138 | 2:5 | 6.8E+11 | 1.2E+12 | 3.3E+12 |
| 2139 | 2:5 | 4.8E+11 | 2.5E+12 | 5.2E+12 |
| AAV2/2 (n = 8) | 2:2 | (8.5 ± 3.7)E+12a | (5.9 ± 2) E+12a | (2.5 ± 0.9)E+13a |
ID: identification number of AAV vectors; GC: genome copies.
aValues represent mean ± s.e.m.
Figure 3.
Genome and transduction efficiency of vectors with heterologous ITR2 and ITR5. (A) Alkaline Southern blot analysis of DNA extracted from 3 × 1010 GC of both 5′- and 3′-ABCA4-half vectors with either homologous (2:2) or heterologous (5:2 or 2:5) ITR, and of a control AAV preparation with homologous ITR2 (CTRL). The expected size of each genome is depicted below each lane. The molecular weight marker (kb) is depicted on the left 5′: 5′-half vector; 3′: 3′-half vector. (B–D) Representative Western blot analysis and quantification of HEK293 cells infected with dual AAV2/2 hybrid ABCA4 vectors with either heterologous ITR2 and ITR5 or homologous ITR2 at m.o.i. based on either the ITR2 (B and C) or the transgene (B and D) titre. The Western blot images (B) are representative of n = 3 independent experiments; the quantifications (C and D) are from n = 3 independent experiments. (B) The upper arrow indicates full-length ABCA4 protein, the lower arrow indicates truncated proteins; the molecular weight ladder is depicted on the left. The micrograms of proteins loaded are depicted below the image. α-3×flag: Western blot with anti-3×flag antibodies; α-Filamin A: Western blot with anti-Filamin A antibodies, used as loading control. (C and D) Quantification of full-length and truncated ABCA4 protein bands from Western blot analysis of cells infected with a dose of vector based on either the ITR2 (C) or the transgene (D) titre. The histograms show either the intensity of the full-length and truncated protein bands divided by that of the Filamin A bands or the intensity of the full-length protein bands divided by that of the truncated protein bands in the corresponding lane. The mean value is depicted above the corresponding bar. Values are represented as: mean ± s.e.m. *p Student's t-test ≤ 0.05. More details on the statistical analysis including specific statistical values can be found in the Statistical analysis paragraph of the Materials and Methods section. 2:2 2:2: cells infected with dual AAV hybrid ABCA4 vectors with homologous ITR from AAV2; 5:2 2:5: cells infected with dual AAV hybrid ABCA4 vectors with heterologous ITR from AAV2 and AAV5; neg: cells infected with EGFP-expressing vectors, as negative controls.
Inclusion of the CL1 degron in the 5′-half vector decreases the production of truncated proteins
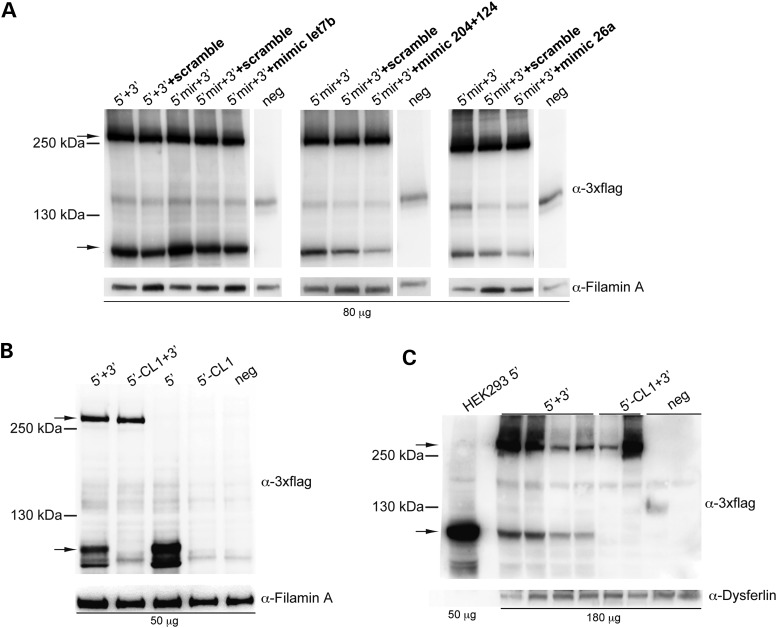
To selectively reduce the levels of truncated ABCA4 proteins produced by each 5′- and 3′-half of dual AAV hybrid ABCA4 vectors (10), we placed putative degradation sequences in the 5′-half vector after the splicing donor signal between AK and the right ITR, and in the 3′-half vector between AK and the splicing acceptor signal (Fig. 1). Thus, the degradation signal will be included in the truncated but not in the full-length protein which results from a spliced mRNA. As degradation signals in the 5′-half vectors we have included: (i) the CL1 degron (CL1), (ii) four copies of the miR-let7b target site (4×let7b), (iii) four copies of the miR-26a target site (4×26a) or (iv) the combination of three copies each of miR-204 and miR-124 target sites (3×204 + 3×124) (Table 2). As degradation signals in the 3′-half vectors we have included: (i) three stop codons (STOP), (ii) PB29 either in a single (PB29) or in three tandem copies (3×PB29) or (iii) ubiquitin (Table 2). We generated dual AAV2/2 hybrid ABCA4 vectors including the various degradation signals and evaluated their efficacy after infection of HEK293 cells [m.o.i.: 5 × 104 GC/cell of each vector]. Since miR-let7b, miR-26a, miR-204 and miR-124 are poorly expressed or completely absent in HEK293 cells [Ambion miRNA Research Guide and (34)], to test silencing of the constructs which contain their corresponding target sites, we transfected cells with miR mimics [i.e. small, chemically modified double-stranded RNAs that mimic endogenous miR (35)], prior to infection with the AAV2/2 vectors containing the corresponding target sites. To define the concentration of miR mimics required to achieve silencing of a gene containing the corresponding miR target sites, we used a plasmid encoding for the reporter EGFP protein and containing the miR target sites before the polyadenylation signal (data not shown). The same experimental settings were used for further evaluation of the miR target sites in the context of dual AAV hybrid ABCA4 vectors. We found that inclusion of miR-204 + 124 and 26a target sequences in the 5′-half of dual AAV hybrid ABCA4 vectors reduced albeit did not abolish the expression of the truncated protein products (Fig. 4A and Supplementary Material, Table S2).
Table 2.
Degradation signals tested in this study
| Degradation signal | Nucleotide sequence | Size (bp) | References | |
|---|---|---|---|---|
| 5′-half vectors | CL1 | gcctgcaagaactggttcagcagcctgagccacttcgtgatccacctg | 48 | (27–29) |
| 3×204 + 3×124 | aggcataggatgacaaagggaacgataggcataggatgacaaagggaa aagcttaggcataggatgacaaagggaaggtaccagatct ggcattcaccgcgtgccttacgat ggcattcaccgcgtgccttaaagcttggcattcaccgcgtgcctta | 158 | (22) | |
| 4×let7b | aaccacacaacctactacctcacgataaccacacaacctactacctca aagcttaaccacacaacctactacctcatcacaaccacacaacctactacctca | 102 | (22–24) | |
| 4×26a | agcctatcctggattacttgaacgat agcctatcctggattacttgaaaagcttagcctatcctggattacttgaatcac agcctatcctggattacttgaa | 102 | (24,25) | |
| 3′-half vectors | 3×STOP | Tga-a-tga-a-tga | 11 | |
| PB29 | atgcacagctggaacttcaagctgtacgtcatgggcagcggc | 42 | (32) | |
| 3×PB29 | atgcacagctggaacttcaagctgtacgtcatgggcagcggcggggtaccatgcac agctggaacttcaagctgtacgtcatgggcagcggcggatgcac agctggaacttcaagctgtacgtcatgggcagcggc | 136 | ||
| ubiquitin | atgcagatcttcgtgaagactctgactggtaagaccatcaccctcgaggtggag cccagtgacaccatcgagaatgtcaaggcaaagatccaagataa ggaaggcattcctcctgatcagcagaggttgatctttgccggaaaacagc tggaagatggtcgtaccctgtctgactacaacatccagaaagagtccaccttgc acctggtactccgtctcagaggtggg | 228 | (30,31) |
The sequences underlined correspond to the degradation signals while those not underlined have been included for cloning purposes.
Figure 4.
Inclusion of the CL1 degradation signal but not of miR target sites in the 5′-half vectors results in significant reduction of truncated proteins. (A) Representative Western blot analysis of HEK293 cells infected with dual AAV2/2 hybrid vectors encoding for ABCA4, containing miR target sites for either miR-let7b (4×, left panel), miR-204 + 124 (3×, central panel) or miR-26a (4×, right panel). 5′ + 3′: cells co-infected with 5′-half vectors without miR target sites and 3′-half vectors; 5′mir + 3′: cells co-infected with 5′-half vectors containing miR target sites and 3′-half vectors; neg: control cells infected with the 3′-half vectors; +scramble: cells infected in the presence of scramble miR mimics; +mimic let7b: cells infected in the presence of miR-let7b mimics; +mimic 204+124: cells infected in the presence of miR-204 and -124 mimics; +mimic 26a: cells infected in the presence of miR-26a mimics. (B and C) Representative Western blot analysis of either HEK293 cells infected with dual AAV2/2 hybrid vectors (B) or pig eyes (c; RPE+retina) 1-month post-injection of dual AAV2/8 hybrid vectors encoding for ABCA4 and containing or not the CL1 degradation signal. 5′ + 3′: cells co-infected or eyes co-injected with 5′-half vectors without CL1 and 3′-half vectors; 5′-CL1 + 3′: cells co-infected or eyes co-injected with 5′-half vectors containing CL1 and 3′-half vectors; 5′: cells infected with 5′-half vectors without CL1; 5′-CL1: cells infected with 5′-half vectors containing CL1; neg: control cells infected or control eyes injected with either the 3′-half vectors or EGFP-expressing vectors, as negative controls. (A–C) The upper arrows indicate full-length ABCA4 proteins, the lower arrows indicate truncated proteins; the molecular weight ladder is depicted on the left. The micrograms of proteins loaded are depicted below the image. α-3×flag: Western blot with anti-3×flag antibodies; α-Filamin A: Western blot with anti-Filamin A antibodies, used as loading control; α-Dysferlin: Western blot with anti-Dysferlin antibodies, used as loading control. (A and B) The Western blot images are representative of n = 3 independent experiments. (C) The Western blot image is representative of n = 5 eyes injected with 5′ + 3′ vectors, n = 2 eyes injected with 5′-CL1 + 3′ vectors and n = 5 of eyes injected with either the 3′-half vectors or EGFP-expressing vectors as negative controls.
Notably, as shown in Figure 4B, we found that the inclusion of the CL1 degradation signal in the 5′-half vector reduced truncated protein expression to undetectable levels. Since differences in the tissue-specific expression of enzymes of the ubiquitination pathway that mediate CL1 degradation (27) may account for changes in CL1 efficacy, we further evaluated the efficacy of the CL1 degron in the pig retina, which has a size and structure similar to human (15,26,36,37) and is therefore an excellent pre-clinical large animal model to evaluate vector safety and efficiency. To this aim, we injected subretinally in Large White pigs AAV2/8 dual AAV hybrid ABCA4 vectors (of which the 5′-half vector included or not the CL1 sequence, dose of each vector/eye: 1 × 1011 GC). Notably, we found that the inclusion of the CL1 degradation signal in the 5′-half vector resulted in reduction of truncated protein expression to levels that are below the detection limit of the Western blot analysis (Fig. 4C). Among the degradation signals tested in the 3′-half vector we identified three (PB29, 3×PB29 and ubiquitin) that reduced both the levels of truncated protein products and of full-length proteins (Supplementary Material, Fig. S2 and Table S3).
Subretinal administration of improved dual AAV vectors reduces lipofuscin accumulation in the Abca4−/− retina
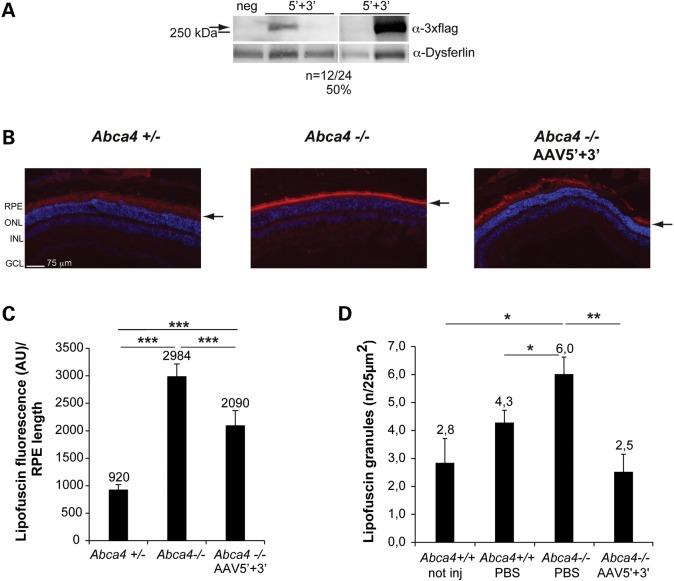
Based on our findings, improved dual AAV hybrid ABCA4 vectors should include homologous ITR2, the AK region of homology and CL1. As ABCA4 is expressed in both rod and cone photoreceptors in humans (38), we identified a suitable promoter for ABCA4 delivery by comparing the PR transduction properties of single AAV2/8 vectors encoding EGFP from either the human GRK1 or IRBP (interphotoreceptor retinoid binding protein) promoters, which have been both described to drive high levels of combined rod and cone PR transduction in various species (39–41). Taking advantage of the pig retinal architecture which includes a streak-like region with a cone:rod = 1:3 (42) similar to the human macula, we injected subretinally 1 × 1011 GC/eye of either AAV2/8-GRK1- or IRBP-EGFP vectors in 3 month-old Large White pigs. Four weeks after the injection, we analysed the corresponding retinal cryosections under a fluorescence microscope. EGFP fluorescence quantification in the PR cell layer (Supplementary Material, Fig. S3a and b) showed that both promoters give comparable levels of PR transduction (predominantly rods in this region). However, when we counted the number of cones labelled with an antibody raised against cone arrestin (CAR) (43) that were also EGFP-positive, we found higher although not statistically significant levels of cone PR transduction with the GRK1 promoter (Supplementary Material, Fig. S3C and D). Based on this, we included the GRK1 promoter in our improved dual AAV hybrid ABCA4 vectors, and investigated their ability to both express ABCA4 and decrease the abnormal content of autofluorescent lipofuscin material in the RPE of Abca4−/− mice. We initially injected subretinally 1 month-old C57/BL6 mice with improved dual AAV vectors (dose of each vector/eye: 2 × 109 GC) and found that 12 out of 24 (50%) injected eyes had detectable albeit variable levels of full-length ABCA4 protein by Western blot [Figure 5A; ABCA4 protein levels in the ABCA4-positive eyes: 2.8 ± 0.7 a.u. (mean ± standard error of the mean)]. This is similar to our previous finding that a different version of the dual AAV platform resulted in 50% ABCA4-expressing eyes (10). We then injected 5.5 month-old pigmented Abca4−/− mice subretinally in the temporal region of the eye with the improved dual AAV vectors (dose of each vector/eye: 1.8 × 109 GC). Three months later we harvested the eyes and measured the levels of lipofuscin fluorescence (excitation: 560 ± 40 nm; emission: 645 ± 75) on retinal cryosections [in either the RPE alone or in RPE + outer segments (OS)] in the temporal region of the eye (Fig. 5B and C and Supplementary Material, Fig. S4). We found that lipofuscin fluorescence intensity in this region of the eye was significantly higher in untreated Abca4−/− than in both Abca4+/− and −/− mice injected with the therapeutic dual AAV hybrid ABCA4 vectors (Fig. 5B and C and Supplementary Material, Fig. S4). Then, using transmission electron microscopy we counted the number of RPE lipofuscin granules. These were increased in 5.5- to 6-month old albino Abca4−/− mice injected with PBS compared with age-matched Abca4+/+ controls (Fig. 5D), at levels similar to those we have independently measured in Abca4−/− mice either uninjected or injected with a control AAV vector (data not shown). The number of lipofuscin granules in Abca4−/− RPE was normalized 3 months post subretinal injection of improved dual AAV hybrid ABCA4 vectors (dose of each vector/eye: 1 × 109 GC, Fig. 5D).
Figure 5.
Subretinal delivery of improved dual AAV vectors results in ABCA4 expression in mouse photoreceptors and significant reduction of lipofuscin accumulation in the Abca4−/− mouse retina. (A) Representative Western blot analysis of C57BL/6 retinas (whole retinal lysates) either injected with dual AAV2/8 hybrid ABCA4 vectors (5′ + 3′) or with negative controls (neg). The arrow indicates full-length proteins, the molecular weight ladder is depicted on the left. α-3×flag: Western blot with anti-3×flag antibodies; α-Dysferlin: Western blot with anti-Dysferlin antibodies, used as loading control. (B and C) Representative pictures (B) and quantification (C) of lipofuscin autofluorescence (red signal) in the retinas (RPE or RPE + OS) of either pigmented Abca4+/− mice not injected or injected with AAV as control (Abca4+/−) or pigmented Abca4−/− mice either not injected (Abca4−/−) or injected with dual AAV hybrid ABCA4 vectors (Abca4−/− AAV5′+3′). (B) The scale bar (75 μm) is depicted in the picture. RPE: retinal pigment epithelium; ONL: outer nuclear layer; INL: inner nuclear layer; GCL: ganglion cell layer. The arrows indicate lipofuscin signal. (C) Mean lipofuscin autofluorescence in the temporal side of three sections for each sample. Mean autofluorescence in each section was normalized for the length of the underlying RPE. The mean value is depicted above the corresponding bar. Values are represented as mean ± s.e.m. ***p ANOVA < 0.0001. n = 4 eyes for each group. (D) Mean number of RPE lipofuscin granules counted in at least 40 fields (25 µm2)/retina of albino Abca4+/+ mice either not injected (Abca4+/+ not inj) or injected with PBS (Abca4+/+ PBS), and albino Abca4−/− mice injected with either PBS (Abca4−/− PBS) or dual AAV hybrid ABCA4 vectors (Abca4−/− AAV5′+3′). The mean value is depicted above the corresponding bar. Values are represented as mean ± s.e.m. *pANOVA ≤ 0.05; **pANOVA ≤0.01. n = 4 eyes from Abca4+/+ not inj; n = 4 eyes from Abca4+/+ PBS; n = 3 eyes from Abca4−/− PBS; n = 3 eyes from Abca4−/− AAV5′ + 3′. More details on the statistical analysis including specific statistical values can be found in the Statistical analysis paragraph of the Materials and Methods section.
Improved dual AAV vectors are safe upon subretinal administration to the mouse and pig retina
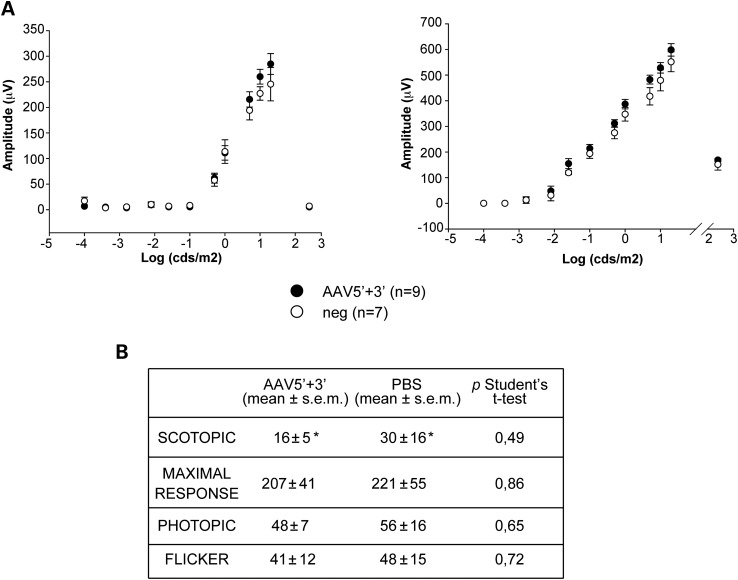
To investigate the safety of improved dual AAV2/8 hybrid ABCA4 vectors, we injected them subretinally in both wild-type C57BL/6 mice and Large White pigs (dose of each vector/eye: 3 × 109 and 1 × 1011 GC, respectively). One month post-injection we measured retinal electrical activity by Ganzfeld electroretinogram (ERG) and found that both the a- and b-wave amplitudes were not significantly different between mouse eyes that were injected with dual AAV hybrid ABCA4 vectors and eyes injected with either negative control AAV vectors or PBS (Fig. 6A and Supplementary Material, Fig. S5A). Similarly, the b-wave amplitude in both scotopic, photopic, maximum response and flicker ERG tests was comparable in pig eyes that were injected with dual AAV hybrid ABCA4 vectors to those of control eyes injected with PBS (Fig. 6B and Supplementary Material, Fig. S5B).
Figure 6.
Similar electrical activity between either negative control or improved dual AAV-treated eyes of mice and pigs. (A) Mean a- (left panel) and b- (right panel) wave amplitudes of C57BL/6 mice 1-month post-injection of either dual AAV hybrid ABCA4 vectors (AAV5′ + 3′) or negative controls (i.e. negative control AAV vectors or PBS; neg). Data are presented as mean ± s.e.m.; n indicates the number of eyes analysed. More details on the statistical analysis can be found in the Statistical analysis paragraph of the Materials and Methods section. (B) Mean b-wave amplitudes (μV) in scotopic, maximal response, photopic and flicker ERG tests in pigs 1-month post-injection of either dual AAV hybrid ABCA4 vectors (AAV5′ + 3′) or PBS. n = 5 eyes injected with dual AAV hybrid ABCA4 vectors; n = 4 injected with PBS; *: n = 2.
Discussion
AAV restricted packaging capacity represents one of the main obstacles to the widespread application of AAV for gene therapy of IRDs including STGD1. However, recently, several groups, including ours, have independently reported effective delivery of large genes to both the mouse and pig retina by dual AAV vectors (10,13,15,44), thus expanding their applicability to diseases like STGD1 due to mutations in genes with a CDS which exceeds the canonical AAV cargo capacity. Here we set-up to overcome some limitations associated with the use of dual AAV vectors, namely their relatively low efficiency when compared with a single vector, and the production of truncated proteins which may raise safety concerns. Strategies aiming at increasing dual AAV genome tail-to-head concatemerization should in theory increase the levels of full-length and reduce those of truncated proteins from free single half-vectors. We set to improve tail-to-head dual AAV hybrid genome concatemerization by including either optimal regions of homology or heterologous ITR. In a side-by-side evaluation of previously described regions of homology, we have found that the AP1 and AP2 sequences recently published by Lostal et al. (16) and the AK sequence from the F1 phage (10) drive overall similar levels of protein expression in vitro, with dual AAV hybrid AK vectors driving efficient ABCA4 expression in the mouse retina more frequently than those with AP1 and AP2 sequences. Independently, the availability of different regions of homology will still be useful to direct proper concatemerization of triple AAV vectors which are being exploited to further expand AAV cargo capacity (16,45). Heterologous ITR2 and ITR5 have been successfully included in dual (20,21) and triple (45) AAV vectors. We found that, although the genome of AAV vectors with heterologous ITR2 and ITR5 is homogeneous in size, the yields of AAV vectors with heterologous ITR2 and ITR5 are lower than those of vectors with homologous ITR2. We also detected less vector genomes with heterologous ITR when we probe their ITR2 than when we probe a different region of their genome. As we show that Rep5 interferes with production of vectors with ITR2, this suggests anomalies at the level of ITR2 included in AAV vectors with heterologous ITR, which are produced in the presence of Rep5, but not in AAV vectors with homologous ITR2, which are produced only in the presence of Rep2 and that showed similar titres whether we probed ITR2 or a different region of the genome. In addition, regardless of the titre used to perform the infections, inclusion of heterologous ITR2 and ITR5 in dual AAV hybrid vectors did not result in significant increase in the rates of full-length to truncated protein production. These results partly differ from those previously reported where dual AAV vectors with heterologous ITR2 and ITR5 had higher transduction efficiency than vectors with homologous ITRs and apparently no production issues (20,21). Besides the different packaging constructs and production protocols, in this study we used dual AAV hybrid vectors which included regions of homology between the two half-vectors as opposed to the trans-splicing (TS) system used in the previous reports which simply relies on the ITR for concatemerization (20,21). As in dual AAV hybrid vectors the reconstitution of the full-length gene is mainly mediated by the region of homology included in the vectors (12) which directs concatemer formation, this may account for the lack in the increase in transgene expression we observed with vectors with heterologous ITR compared with the previous studies that used TS vectors (20,21).
The production of truncated protein products from single half-vectors of dual AAV might raise safety concerns and we aimed at overcoming this by including various degradation signals in our dual AAV vectors. The inclusion of miR target sites in the transcript of a gene has been shown to be an effective strategy to knock-down and restrict transgene expression in various tissues, including the retina (26). However, we achieved only a partial reduction of truncated protein production with miR target sites. Although we cannot rule out that miR target sites lead to a more effective silencing in the mouse retina than in vitro, our in vitro experiments suggest that features of the mRNA external to the miR target sites rather than miRNA levels might affect the efficiency of the silencing. Indeed, while miR mimics provide robust silencing of a canonical EGFP expression cassette containing the miR target sites, this silencing was less robust when we tested the same mimics with dual AAV ABCA4 vectors (Fig. 4A). Along this line, since the truncated protein products that derive from the 5′-half are produced from a vector that is not endowed with a canonical polyadenilation signal, it may be possible that the resulting mRNA cannot undergo an efficient miR-mediated silencing (46,47).
Importantly, we achieved complete degradation of the truncated protein product from the 5′-half vector by inclusion of the CL1 degron. We showed that this signal is effective both in vitro and in the pig retina, indicating that the enzymes of the degradative pathway required for CL1 activity are expressed in various cell types. On the other hand, we found that none of the degradation signals tested in the 3′-half vectors significantly reduced truncated protein expression without affecting full-length protein expression. In addition, we found a negative correlation between the length of the degradation signal included in the 3′-half vector and the levels of protein expression, possibly indicating that increasing the size of the sequences between the ITR and the splicing acceptor signal interferes with mRNA processing. However, we could never abolish truncated protein formation from the 3′-half vectors even using different lengths of degradation sequences. Since 3′-half vectors do not have a canonical promoter with a defined transcription start site, it is possible that start codons downstream of the region of the vector where we included the degradation signals are recognized more efficiently than the ATG of the degradation signals. This would result in exclusion of the degradation signal from the truncated proteins. As the truncated protein product from the 3′-half vector is less abundant than that produced by the 5′-half vector (Fig. 4A and Supplementary Material, Fig. S2), its presence should raise less safety concerns. Indeed, although further formal toxicity studies are required to finally address this issue, data presented here in the mouse and pig retina support the safety of improved dual AAV vectors.
Notably, we found that subretinal administration of improved dual AAV vectors, under the control of the GRK1 promoter, which provides high levels of combined rod and cone transduction, results in effective ABCA4 delivery in mice, although at variable levels. This could be due to both the inherent variability of the subretinal injection in the small murine eye and the overall lower efficacy of the dual AAV system compared with a single AAV vector (10). Despite this variability, we found that dual AAV mediated ABCA4 delivery results in significant lipofuscin reduction in the Abca4−/− retina suggesting that a wide range of transgene expression levels can similarly contribute to therapeutic efficacy. This was observed using two independent techniques, however, more pronounced improvement of the phenotype was observed when we dissected and analysed the AAV transduced area of the retina that indeed showed normalization of the number of lipofuscin granules. HPLC measurement of A2E, the major lipofuscin granules component, has been used to assess the efficacy of therapies in Abca4−/− mice (48–50). However, we found these measurements to be inconsistent in our hands, even between affected and normal retinas (data not shown), thus we were not able to use this technique in our rescue experiments.
In conclusion, our study characterized dual AAV vectors with improved features in view of their potential future clinical application to the therapy of STGD1. In addition, our findings can be used to improve dual AAV vectors for gene therapy of other conditions due to mutations in genes with a CDS which exceeds AAV cargo capacity.
Materials and Methods
Generation of plasmids
The plasmids used for AAV vector production were all derived from dual AAV hybrid AK vector plasmids encoding for the human ABCA4 gene containing the ITR of AAV serotype 2 (10). For cloning purposes, the 77 bp-long AK sequence (10) was followed by a 32 bp-long region (sequence: attaacgtttataatttcaggtggcatctttc) in each of the dual AAV hybrid AK vectors. The 77-bp AK recombinogenic sequence and the following 32-bp-long region were replaced with three different recombinogenic sequences derived from the human placental alkaline phosphatase gene: AP [NM_001632_4, bp 927–1204, (10)]; AP1 [NM_001632_4, bp 1802–1516 (16)]; AP2 [NM_001632_4, bp 1224–938 (16)].
Dual AAV vector plasmids bearing heterologous ITR from AAV serotype 2 (ITR2) and ITR from AAV serotype 5 (ITR5) in the 5:2-2:5 configuration were generated by replacing the left ITR2 in the 5′-half vector plasmid and the right ITR2 in the 3′-half vector plasmids, respectively, with ITR5 (NC_006152.1, bp 1–175). Dual AAV vector plasmids bearing heterologous ITR2 and ITR5 in the 2:5-5:2 configurations were generated by replacing either the right or the left ITR2 with the ITR5, respectively. The pAAV5/2 packaging plasmid containing the Rep5 and AAV2 Cap genes (Rep5Cap2), was obtained from the pAAV2/2 packaging plasmid, containing the Rep and Cap genes from AAV2 (Rep2Cap2), by replacing Rep2 with the Rep5 open reading frame from AAV5 (NC_006152.1, bp 171–2206).
The pZac5:5-CMV-EGFP plasmid containing the EGFP expression cassette with ITR5 was generated from the pAAV2.1-CMV-EGFP plasmid, containing the ITR2 flanking the EGFP expression cassette (51).
Degradation signals were cloned in dual AAV hybrid ABCA4 vectors as follows: in the 5′-half vector plasmids between the AK sequence and the right ITR2; in the 3′-half vector plasmids between the AK sequence and the splice acceptor signal. Details on degradation signal sequences can be found in Table 2.
The ABCA4 protein expressed from dual AAV vectors is tagged with 3×flag at both N- (amino acidic position 590) and C-termini for the experiments shown in Figures 3 and 4 and Supplementary Material, Figure S2, and at the C-terminus alone for the experiments in Figures 2 and 5a.
Dual AAV hybrid ABCA4 vectors included either the ubiquitous CMV (52) or the GRK1 (41) promoters. Single AAV vectors encoding EGFP included either the GRK1 or the IRBP (39) promoters.
AAV vector production and characterization
AAV2/2 and AAV2/8 vector large preparations were produced by the TIGEM AAV Vector Core by triple transfection of HEK293 cells followed by two rounds of CsCl2 purification. AAV vectors bearing homologous ITR2 were obtained as previously described (54).
To obtain AAV vectors bearing heterologous ITR2 and ITR5 a suspension of 1.1 × 109 low-passage HEK293 cells was quadruple-transfected by calcium phosphate with 500 µg of pDeltaF6 helper plasmid which contains the Ad helper genes (55), 260 µg of pAAV cis-plasmid and different amounts of Rep2Cap2 and Rep5 packaging constructs. The amount of Rep2Cap2 and Rep5 packaging constructs was as follows:
PROTOCOL A: 130 µg of each Rep5 and Rep2Cap2 (ratio 1:1)
PROTOCOL B: 90 µg of Rep5 and 260 µg of Rep2Cap2 (ratio 1:3)
PROTOCOL C: 26 µg of Rep5 and 260 µg of Rep2Cap2 (ratio 1:10)
Each AAV preparation was then purified according to the published protocol (54).
The protocols described below were used for the Rep competition experiments:
to assess Rep5 competition with Rep2 for production of AAV vectors with ITR2, HEK293 cells were either quadruple-transfected by calcium phosphate with pDeltaF6, pAAV2.1-CMV-EGFP cis, the Rep2Cap2 and Rep5Cap2 constructs at a weight ratio of 2:1:1.5:1.5 or, as a control, quadruple-transfected with the pDeltaF6, pAAV2.1-CMV-EGFP, the Rep2Cap2 packaging construct and a control irrelevant plasmid at a weight ratio of 2:1:1.5:1.5;
to assess Rep2 competition with Rep5 for production of AAV vectors with ITR5, HEK293 cells were either quadruple-transfected by calcium phosphate with pDeltaF6, pZac5:5-CMV-EGFP, the Rep5Cap2 and Rep2Cap2 constructs at a weight ratio of 2:1:1.5:1.5 or, as a control, quadruple-transfected with pDeltaF6, pZac5:5-CMV-EGFP, the Rep5 construct and a control irrelevant plasmid at a weight ratio of 2:1:1.5:1.5.
For the large-scale AAV vector preparations physical titres [(GC)/mL] were determined by averaging the titre achieved by PCR quantification using TaqMan (Applied Biosystems, Carlsbad, CA, USA) (54) with a probe annealing on ITR2 and that obtained by dot-blot analysis (56) with a probe annealing within 1 kb from ITR2. For the large-scale AAV5:2 vector preparations produced with different Rep5:Rep2Cap2 weight ratio using the 5′-half plasmid of dual AAV vector, physical titres were determined by PCR quantification using TaqMan with a probe annealing on ITR2. For the AAV vector preparations used in the competition experiments physical titres were determined by PCR quantification using TaqMan with a probe annealing on the bovine growth hormone (BGH) polyadenilation signal, included in the EGFP-expressing cassette packaged in the AAV vectors.
AAV infection of HEK293 cells
AAV infection of HEK293 cells was performed as previously described (10). AAV2 vectors bearing heterologous ITR2 and ITR5 and produced according to Protocol C were used to infect HEK293 cells with a multiplicity of infection (m.o.i) of 1 × 104 GC/cell of each vector (i.e. 2 × 104 total GC/cell when we used dual AAV vectors at a 1:1 ratio) calculated considering either the ITR2 or the transgene titre. Infections with AAV2/2 bearing recombinogenic regions and degradation signals were carried out with a m.o.i of 5 × 104 GC/cell of each vector (i.e. 1 × 105 total GC/cell in the case of dual AAV vectors at 1:1 ratio) calculated considering the average titre between TaqMan and dot-blot.
For the experiments using 5′-half vectors containing miR target sites, cells were transfected with the corresponding miR mimics (50 nM; miRIDIAN microRNA mimic hsa-let-7b-5p, hsa-miR-204-5p, hsa-miR-124-3p and hsa-miR-26a-5p; Dharmacon, Lafayette, CO, USA) using calcium phosphate 4 h prior to infection.
Subretinal injection of AAV vectors in mice and pigs
Mice were housed at the Institute of Genetics and Biophysics animal house (Naples, Italy), maintained under a 12-h light/dark cycle (10–50 lux exposure during the light phase). C57BL/6 mice were purchased from Harlan Italy SRL (Udine, Italy). Pigmented Abca4−/− mice were generated through four successive crosses of albino Abca4−/− mice (10) with Sv129 mice and then maintained inbred; breeding was performed crossing Abca4+/− with −/− mice. Albino Abca4−/− mice were generated through successive crosses and backcrossed with BALB/c mice (homozygous for Rpe65 Leu450) and maintained inbred; breeding was performed crossing Abca4+/− mice with −/− mice. C57BL/6 (5 week-old), pigmented Abca4−/− (5.5 month-old) and albino Abca4−/− (2.5- to 3-month old) mice were anaesthetized as previously described (57), then 1 µl of either PBS or AAV2/8 vectors was delivered subretinally to the temporal side of the retina via a trans-scleral trans-choroidal approach as described by Liang et al. (58). AAV2/5-VMD2-human Tyrosinase (59) (dose: 2×10e8 GC/eye) was added to the AAV2/8 vector solution that was subretinally delivered to albino Abca4−/− mice (Fig. 5D). This allowed us to mark the RPE within the transduced part of the eyecup, which was subsequently dissected and analyzed.
The Large White Female pigs used in this study were registered as purebred in the LW Herd Book of the Italian National Pig Breeders’ Association. Pigs were housed at the Cardarelli hospital animal house (Naples, Italy) and maintained under 12-h light/dark cycle (10–50 lux exposure during the light phase). This study was carried out in accordance with the Association for Research in Vision and Ophthalmology Statement for the Use of Animals in Ophthalmic and Vision Research and with the Italian Ministry of Health regulation for animal procedures. All procedures were submitted to the Italian Ministry of Health; Department of Public Health, Animal Health, Nutrition and Food Safety. Surgery was performed under anaesthesia and all efforts were made to minimize suffering. Animals were sacrificed as previously described (36). Subretinal delivery of AAV vectors to 3 month-old pigs was performed as previously described (36). All eyes were treated with 100 µl of either PBS or AAV2/8 vector solution. The AAV2/8 dose was 1 × 1011 GC of each vector/eye therefore co-injection of dual AAV vectors at a 1:1 ratio resulted in a total dose of 2 × 1011 GC/eye.
For the animal studies included in Figures 2C, 4C, 5 and 6 and Supplementary Material, Figures S3–S5, right and left eyes were assigned randomly to the various experimental groups and the researchers conducting and quantifying the experiments were blind to the treatment received by the animals.
Western blot analysis
HEK293 cells, mouse and pig retinas were lysed in RIPA buffer (50 mm Tris–HCl pH 8.0, 150 mm NaCl, 1% NP40, 0.5% Na-Deoxycholate, 1 mm EDTA pH 8.0, 0.1% SDS). Lysis buffers were supplemented with protease inhibitors (Complete Protease inhibitor cocktail tablets; Roche, Milan, Italy) and 1 mm phenylmethylsulfonyl. After lysis, samples of cells containing ABCA4 were denatured at 37°C for 15 min in 1× Laemli sample buffer supplemented with 4 M urea. Lysates were separated by 6% or 8% (the latter used for the WB in Fig. 4C) SDS–polyacrylamide gel electrophoresis. The antibodies used for immuno-blotting are as follows: anti-3×flag (1:1000, A8592; Sigma-Aldrich); anti-Filamin A (1:1000, catalogue #4762; Cell Signaling Technology, Danvers, MA, USA); anti-Dysferlin (1:500, Dysferlin, clone Ham1/7B6, MONX10795; Tebu-bio, Le Perray-en-Yveline, France). Filamin A and Dysferlin were used as loading controls for the in vitro and in vivo experiments, respectively.
The quantification of ABCA4 bands detected by Western blot was performed using ImageJ software (free download available at http://rsbweb.nih.gov/ij/). Quantification of the Western blot experiments has been performed as follows:
– Figure 2A and B: the intensity of the ABCA4 band was normalized to that of Filamin A band in the corresponding lane. Normalized ABCA4 expression was then expressed as percentage relative to dual AAV hybrid AK vectors;
– Figure 2C: the intensity of the ABCA4 band (a.u.) was calculated as fold of increase relative to the mean intensity measured at the same level in the negative control lanes of each gel (the measurement of the negative control sample in lane 7 of the lower left panel was excluded from the analysis given the exceptionally high background signal). Values for each group are represented as mean ± standard error of the mean (s.e.m.);
– Figure 3B–D: the full-length ABCA4 and truncated protein band intensities were divided by those of the Filamin A bands or the intensity of the full-length ABCA4 protein bands was divided by that of the truncated protein bands in the corresponding lane. Values are represented as: mean ± s.e.m;
– Supplementary Material, Table S2: full-length ABCA4 and truncated protein band intensities were measured in cells co-infected with 5′- and 3′-half vectors. The ratio between the intensity of full-length ABCA4 and truncated protein bands in the presence of either the corresponding mimic or a scramble mimic was calculated. Values represent mean ± s.e.m. of the ratios from three independent experiments;
– Supplementary Material, Table S3: full-length ABCA4 and truncated protein band intensities were measured in cells co-infected with 5′- and 3′-half vectors. The ratio between the intensity of the full-length ABCA4 and truncated bands from vectors either with or without the degradation signals was calculated. Values represent mean ± s.e.m. of the ratios from three independent experiments.
– Figure 5A: the intensity of the ABCA4 band (a.u.) was calculated as fold of increase relative to the mean background intensity measured in the negative control lanes of the corresponding gel. Values are expressed as mean ± s.e.m.
Southern blot analysis
Three ×1010 GC of viral DNA were extracted from AAV particles. To digest unpackaged genomes, the vector solution was resuspended in 240 µl of PBS pH 7.4 19 (GIBCO; Invitrogen S.R.L., Milan, Italy) and then incubated with 1 U/µl of DNase I (Roche) in a total volume of 300 µl containing 40 mm TRIS–HCl, 10 mm NaCl, 6 mm MgCl2, 1 mm CaCl2 pH 7.9 for 2 h at 37°C. The DNase I was then inactivated with 50 mm EDTA, followed by incubation with proteinase K and 2.5% N-lauroyl-sarcosil solution at 50°C for 45 min to lyse the capsids. The DNA was extracted twice with phenol-chloroform and precipitated with two volumes of ethanol 100 and 10% sodium acetate (3 M, pH 7). Alkaline agarose gel electrophoresis and blotting were performed as previously described (60). Ten microlitres of the 1 kb DNA ladder (N3232L; New England Biolabs, Ipswich, MA, USA) were loaded as molecular weight marker. Two different double strand DNA fragments were labelled with digoxigenin-dUTP using the DIG high prime DNA labelling and detection starter kit (Roche), and used as probes. The 5′ probe (768 bp) was generated by double digestion of the pZac2.1-CMV-ABCA4_5′ plasmid with SpeI and NotI; the 3′ probe (974 bp) was generated by double digestion of the pZac2.1-ABCA4_3′_3×flag_SV40 plasmid with ClaI and MfeI. Prehybridization and hybridization were performed at 65°C in Church buffer (60) for 1 h and overnight, respectively. Then, the membrane (Whatman Nytran N, charged nylon membrane; Sigma-Aldrich, Milan, Italy) was first washed for 30 min in SSC 29–0.1% SDS, then for 30 min in SSC 0.59–0.1% SDS at 65°C and then for 30 min in SSC 0.19–0.1% SDS at 37°C. The membrane was then analyzed by chemiluminescence detection by enzyme immunoassay using the DIG DNA Labelling and Detection Kit (Roche).
Histological analysis
Mice were euthanized, and their eyeballs were then harvested and fixed overnight by immersion in 4% paraformaldehyde (PFA). Before harvesting the eyeballs, the temporal aspect of the sclerae was marked by cauterization, in order to orient the eyes with respect to the injection site at the moment of the inclusion. The eyeballs were cut so that the lens and vitreous could be removed while leaving the eyecup intact. Mice eyecups were infiltrated with 30% sucrose for cryopreservation and embedded in tissue-freezing medium (O.C.T. matrix; Kaltek, Padua, Italy). For each eye, 150–200 serial sections (10 µm thick) were cut along the horizontal plane and the sections were progressively distributed on 10 slides so that each slide contained 15–20 sections, each representative of the entire eye at different levels. The sections were stained with 4′,6′-diamidino-2-phenylindole (Vectashield; Vector Lab, Peterborough, UK) and were monitored with a Zeiss Axiocam (Carl Zeiss, Oberkochen, Germany) at different magnifications.
Pigs were sacrificed, and their eyeballs were harvested and fixed overnight by immersion in 4% PFA. The eyeballs were cut so that the lens and vitreous could be removed, leaving the eyecups in place. The eyecups were gradually dehydrated by progressively infiltrating them with 10, 20 and 30% sucrose. Tissue-freezing medium (O.C.T. matrix; Kaltek) embedding was performed. Before embedding, the swine eyecups were analyzed with a fluorescence stereomicroscope (Leica Microsystems GmbH, Wetzlar, Germany) in order to localize the transduced region whenever an EGFP-encoding vector was administered. For each eye, 200–300 serial sections (12 µm thick) were cut along the horizontal meridian and the sections were progressively distributed on glass slides so that each slide contained 6–10 sections. Section staining and image acquisition were performed as described for mice.
Cone immunofluorescence staining
Frozen retinal sections were washed once with PBS and then permeabilized for 1 h in PBS containing 0.1% Triton X-100. A blocking solution containing 10% normal goat serum (Sigma–Aldrich) was applied for 1 h. A primary antibody [anti-human CAR (61,62), which also recognizes the porcine CAR (‘Luminaire founders’—hCAR, 1:10.000; kindly provided by Dr Cheryl M. Craft, Doheny Eye Institute, Los Angeles, CA, USA)] was diluted in PBS and incubated overnight at 4°C. The secondary antibody (Alexa Fluor 594, anti-rabbit, 1:1000; Molecular Probes, Invitrogen, Carlsbad, CA, USA) was incubated for 45 min. Sections stained with the anti-CAR antibodies were analyzed at 63× magnification using a Leica Laser Confocal Microscope System (Leica Microsystems GmbH), as previously described (63). Briefly, for each eye six different z-stacks from six different transduced regions were taken. For each z-stack, images from single plans were used to count CAR+/EGFP + cells. In doing this, we carefully moved along the Z-axis to distinguish one cell from another and thus to avoid to count twice the same cell. For each retina we counted the CAR-positive (CAR+)/EGFP-positive (EGFP+) cells on total CAR+ cells. We then calculated the average number of CAR+/EGFP + cells of the three eyes of each experimental group.
EGFP quantification
Fluorescence intensity in PR was rigorously and reproducibly quantified in an unbiased manner as previously described (63). Individual colour channel images were taken using a Leica microscope (Leica Microsystems GmbH). TIFF images were gray-scaled with image analysis software (LAS AF lite; Leica Microsystems GmbH). Six images of each eye were analyzed at 20× magnification by a masked observer. The PR (outer nuclear layer + OS) were selectively outlined in every image, and the total fluorescence for the enclosed area was calculated in an unbiased manner using the image analysis software. The fluorescence in PR was then averaged from six images collected from separate retinal sections from each eye. We then calculated the average fluorescence of the three eyes of each experimental group.
Quantification of lipofuscin autofluorescence
For lipofuscin fluorescence analysis, eyes were harvested from pigmented Abca4+/− and Abca4−/− mice at 3 months after AAV injection. Mice were dark-adapted over-night and sacrificed under dim red-light. For each eye, four overlapping pictures from the temporal side of three sections at different levels of the eye were taken using a Leica DM5000B microscope equipped with a TX2 filter (excitation: 560 ± 40 nm; emission: 645 ± 75) (64–68) and under a 20× objective. The four images for each section were then combined in a single montage used for further fluorescence analysis. Intensity of lipofuscin fluorescence (red signal) in each section was automatically calculated using the ImageJ software and was then normalized for the length of the RPE underlying the area of fluorescence.
Transmission electron microscopy
For electron microscopy analyses eyes were harvested from albino Abca4−/− and Abca4+/+ mice at 3 months after AAV injection. Eyes were fixed in 0.2% glutaraldehyde-2% PFA in 0.1 M PHEM buffer pH 6.9 (240 mm PIPES, 100 mm HEPES, 8 mm MgCl2, 40 mm EGTA) overnight and then rinsed in 0.1 M PHEM buffer. Eyes were then dissected under light microscope to select the tyrosinase-positive portions of the eyecups. The transduced portion of the eyecups were subsequently embedded in 12% gelatin, infused with 2.3 M sucrose and frozen in liquid nitrogen. Cryosections (50 nm) were cut using a Leica Ultramicrotome EM FC7 (Leica Microsystems) and extreme care was taken to align PR connecting cilia longitudinally. To avoid bias in the attribution of morphological data to the various experimental groups, counts of lipofuscin granules were performed by a masked operator (Dr Roman Polishchuk) using the iTEM software (Olympus SYS, Hamburg, Germany). The ‘Touch count’ module of the iTEM software was used to count the number of lipofuscin granules in 25 µm2 areas (at least 40) distributed randomly across the RPE layer. The granule density was expressed as number of granules per 25 µm2.
Electroretinogram recordings
Electrophysiological recordings in mice and pigs were performed as detailed in (69) and in (70), respectively.
Statistical analysis
p-values ≤0.05 were considered statistically significant. One-way ANOVA with post-hoc Multiple Comparison Procedure was used to compare data depicted in Figure 2B (pANOVA = 1.2 × 10−6), Figure 2C (pANOVA = 0.326), Figure 5C (pANOVA = 1.5 × 10−10), Figure 5D (pANOVA = 0.034) and Figure 6A (pANOVA a-wave: 0.5; pANOVA b-wave: 0.8) and Supplementary Material, Table S3 (pANOVA = 0.0135). As the counts of lipofuscin granules (Fig. 5D) are expressed as discrete numbers, these were analyzed by deviance from a Negative Binomial generalized linear models (71). The statistically significant differences between groups determined with the post-hoc Multiple Comparison Procedure are the following: Figure 2B: AP versus AK: 1.08 × 10−5; AP1 versus AK: 0.05; AP2 versus AK: 0.17; AP1 versus AP: 1.8 × 10−6; AP2 versus AP: 2.8 × 10−6; AP2 versus AP1: 0.82. Figure 5C: Abca4+/− versus Abca4−/−: 0.00; Abca4−/− versus Abca4−/− AAV5′+3′: 9.3 × 10−5; Abca4+/− versus Abca4−/− AAV5′ + 3′: 4 × 10−6. Figure 5D: Abca4−/− PBS versus Abca4−/− AAV5′ + 3′: 0.01; Abca4+/+ PBS versus Abca4−/− AAV5′ + 3′: 0.37; Abca4+/+ not inj versus Abca4−/− AAV5′ + 3′: 0.53; Abca4+/+ PBS versus Abca4−/− PBS: 0.05; Abca4+/+ not inj versus Abca4−/− PBS: 0.03; Abca4+/+ not inj versus Abca4+/+ PBS: 0.76. Supplementary Material, Table S3: 3×STOP versus no degradation signal: 0.97; 3×STOP versus PB29: 1.0; 3×STOP versus 3×PB29: 0.15; 3×STOP versus ubiquitin: 0.10; PB29 versus no degradation signal: 1.0; PB29 versus 3×PB29: 0.1; PB29 versus ubiquitin: 0.07; 3×PB29 versus no degradation signal: 0.06; 3×PB29 versus ubiquitin: 1.0; ubiquitin versus no degradation signal: 0.04.
The Student's t-test was used to compare data depicted in Table 1, Figures 3C, D and 6B and in Supplementary Material, Figure S3B–D and Table S2.
Supplementary Material
Funding
The following funding support is gratefully acknowledged: the European Research Council/ERC Grant agreement number: 282085 ‘RetGeneTx’; the Italian Telethon Foundation (grant TGM11MT1) and a grant from Shire. Funding to pay the Open Access publication charges for this article was provided by FONDAZIONE TELETHON.
Supplementary Material
Acknowledgements
We thank Annamaria Carissimo (Bioinformatics Core, TIGEM, Naples, Italy) for the statistical analyses; Monica Doria, Antonella Ferrara and Viola Alba (AAV Vector Core, TIGEM, Naples, Italy) for AAV vector production; Mary D Allen (Laboratory for Vision Research) and Dr Cheryl MCraft (University of Southern California, Los Angeles, CA) for providing the anti-human CAR antibody; Graciana Diez-Roux (Scientific Office, TIGEM, Naples, Italy) for the critical reading of this manuscript.
Conflict of Interest statement. Ivana Trapani, Pasqualina Colella and Alberto Auricchio are co-inventors on patent applications on the dual AAV vector platform.
References
- 1.Allikmets R., Singh N., Sun H., Shroyer N.F., Hutchinson A., Chidambaram A., Gerrard B., Baird L., Stauffer D., Peiffer A. et al. (1997) A photoreceptor cell-specific ATP-binding transporter gene (ABCR) is mutated in recessive Stargardt macular dystrophy. Nat. Genet., 15, 236–246. [DOI] [PubMed] [Google Scholar]
- 2.Molday R.S., Zhang K. (2010) Defective lipid transport and biosynthesis in recessive and dominant Stargardt macular degeneration. Prog. Lipid Res., 49, 476–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weng J., Mata N.L., Azarian S.M., Tzekov R.T., Birch D.G., Travis G.H. (1999) Insights into the function of Rim protein in photoreceptors and etiology of Stargardt's disease from the phenotype in abcr knockout mice. Cell, 98, 13–23. [DOI] [PubMed] [Google Scholar]
- 4.Bainbridge J.W., Smith A.J., Barker S.S., Robbie S., Henderson R., Balaggan K., Viswanathan A., Holder G.E., Stockman A., Tyler N. et al. (2008) Effect of gene therapy on visual function in Leber's congenital amaurosis. N. Engl. J. Med., 358, 2231–2239. [DOI] [PubMed] [Google Scholar]
- 5.Maguire A.M., High K.A., Auricchio A., Wright J.F., Pierce E.A., Testa F., Mingozzi F., Bennicelli J.L., Ying G.S., Rossi S. et al. (2009) Age-dependent effects of RPE65 gene therapy for Leber's congenital amaurosis: a phase 1 dose-escalation trial. Lancet, 374, 1597–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maguire A.M., Simonelli F., Pierce E.A., Pugh E.N. Jr, Mingozzi F., Bennicelli J., Banfi S., Marshall K.A., Testa F., Surace E.M. et al. (2008) Safety and efficacy of gene transfer for Leber's congenital amaurosis. N. Engl. J. Med., 358, 2240–2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cideciyan A.V., Hauswirth W.W., Aleman T.S., Kaushal S., Schwartz S.B., Boye S.L., Windsor E.A., Conlon T.J., Sumaroka A., Pang J.J. et al. (2009) Human RPE65 gene therapy for Leber congenital amaurosis: persistence of early visual improvements and safety at 1 year. Hum. Gene Ther., 20, 999–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simonelli F., Maguire A.M., Testa F., Pierce E.A., Mingozzi F., Bennicelli J.L., Rossi S., Marshall K., Banfi S., Surace E.M. et al. (2010) Gene therapy for Leber's congenital amaurosis is safe and effective through 1.5 years after vector administration. Mol. Ther., 18, 643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trapani I., Puppo A., Auricchio A. (2014) Vector platforms for gene therapy of inherited retinopathies. Prog. Retin. Eye Res., 43, 108–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trapani I., Colella P., Sommella A., Iodice C., Cesi G., de Simone S., Marrocco E., Rossi S., Giunti M., Palfi A. et al. (2014) Effective delivery of large genes to the retina by dual AAV vectors. EMBO Mol. Med., 6, 194–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duan D., Yue Y., Engelhardt J.F. (2001) Expanding AAV packaging capacity with trans-splicing or overlapping vectors: a quantitative comparison. Mol. Ther., 4, 383–391. [DOI] [PubMed] [Google Scholar]
- 12.Ghosh A., Yue Y., Lai Y., Duan D. (2008) A hybrid vector system expands adeno-associated viral vector packaging capacity in a transgene-independent manner. Mol. Ther., 16, 124–130. [DOI] [PubMed] [Google Scholar]
- 13.Dyka F.M., Boye S.L., Chiodo V.A., Hauswirth W.W., Boye S.E. (2014) Dual adeno-associated virus vectors result in efficient in vitro and in vivo expression of an oversized gene, MYO7A. Hum Gene Ther Methods, 25, 166–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lopes V.S., Boye S.E., Louie C.M., Boye S., Dyka F., Chiodo V., Fofo H., Hauswirth W.W., Williams D.S. (2013) Retinal gene therapy with a large MYO7A cDNA using adeno-associated virus. Gene Ther., 20, 824–833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Colella P., Trapani I., Cesi G., Sommella A., Manfredi A., Puppo A., Iodice C., Rossi S., Simonelli F., Giunti M., Bacci M.L., Auricchio A. (2014) Efficient gene delivery to the cone-enriched pig retina by dual AAV vectors. Gene Ther., 21, 450–456. [DOI] [PubMed] [Google Scholar]
- 16.Lostal W., Kodippili K., Yue Y., Duan D. (2014) Full-length dystrophin reconstitution with adeno-associated viral vectors. Hum. Gene Ther., 25, 552–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Flotte T.R., Afione S.A., Solow R., Drumm M.L., Markakis D., Guggino W.B., Zeitlin P.L., Carter B.J. (1993) Expression of the cystic fibrosis transmembrane conductance regulator from a novel adeno-associated virus promoter. J. Biol. Chem., 268, 3781–3790. [PubMed] [Google Scholar]
- 18.Ghosh A., Yue Y., Duan D. (2011) Efficient transgene reconstitution with hybrid dual AAV vectors carrying the minimized bridging sequences. Hum. Gene Ther., 22, 77–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chiorini J.A., Kim F., Yang L., Kotin R.M. (1999) Cloning and characterization of adeno-associated virus type 5. J. Virol., 73, 1309–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yan Z., Zak R., Zhang Y., Engelhardt J.F. (2005) Inverted terminal repeat sequences are important for intermolecular recombination and circularization of adeno-associated virus genomes. J. Virol., 79, 364–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan Z., Lei-Butters D.C., Zhang Y., Zak R., Engelhardt J.F. (2007) Hybrid adeno-associated virus bearing nonhomologous inverted terminal repeats enhances dual-vector reconstruction of minigenes in vivo. Hum. Gene Ther., 18, 81–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karali M., Peluso I., Gennarino V.A., Bilio M., Verde R., Lago G., Dolle P., Banfi S. (2010) miRNeye: a microRNA expression atlas of the mouse eye. BMC Genomics, 11, 715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kutty R.K., Samuel W., Jaworski C., Duncan T., Nagineni C.N., Raghavachari N., Wiggert B., Redmond T.M. (2010) MicroRNA expression in human retinal pigment epithelial (ARPE-19) cells: increased expression of microRNA-9 by N-(4-hydroxyphenyl)retinamide. Mol. Vis., 16, 1475–1486. [PMC free article] [PubMed] [Google Scholar]
- 24.Ragusa M., Caltabiano R., Russo A., Puzzo L., Avitabile T., Longo A., Toro M.D., Di Pietro C., Purrello M., Reibaldi M. (2013) MicroRNAs in vitreus humor from patients with ocular diseases. Mol. Vis., 19, 430–440. [PMC free article] [PubMed] [Google Scholar]
- 25.Sundermeier T.R., Palczewski K. (2012) The physiological impact of microRNA gene regulation in the retina. Cell. Mol. Life Sci., 69, 2739–2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karali M., Manfredi A., Puppo A., Marrocco E., Gargiulo A., Allocca M., Corte M.D., Rossi S., Giunti M., Bacci M.L. et al. (2011) MicroRNA-restricted transgene expression in the retina. PLoS One, 6, e22166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gilon T., Chomsky O., Kulka R.G. (1998) Degradation signals for ubiquitin system proteolysis in Saccharomyces cerevisiae. EMBO J., 17, 2759–2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bence N.F., Sampat R.M., Kopito R.R. (2001) Impairment of the ubiquitin-proteasome system by protein aggregation. Science, 292, 1552–1555. [DOI] [PubMed] [Google Scholar]
- 29.Wang X., Robbins J. (2006) Heart failure and protein quality control. Circ. Res., 99, 1315–1328. [DOI] [PubMed] [Google Scholar]
- 30.Bachmair A., Finley D., Varshavsky A. (1986) In vivo half-life of a protein is a function of its amino-terminal residue. Science, 234, 179–186. [DOI] [PubMed] [Google Scholar]
- 31.Johnson E.S., Bartel B., Seufert W., Varshavsky A. (1992) Ubiquitin as a degradation signal. EMBO J., 11, 497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sadis S., Atienza C. Jr, Finley D. (1995) Synthetic signals for ubiquitin-dependent proteolysis. Mol. Cell Biol., 15, 4086–4094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chiorini J.A., Afione S., Kotin R.M. (1999) Adeno-associated virus (AAV) type 5 Rep protein cleaves a unique terminal resolution site compared with other AAV serotypes. J. Virol., 73, 4293–4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tian W., Dong X., Liu X., Wang G., Dong Z., Shen W., Zheng G., Lu J., Chen J., Wang Y., Wu Z., Wu X. (2012) High-throughput functional microRNAs profiling by recombinant AAV-based microRNA sensor arrays. PLoS One, 7, e29551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Z. (2011) The guideline of the design and validation of MiRNA mimics. Methods Mol. Biol., 676, 211–223. [DOI] [PubMed] [Google Scholar]
- 36.Mussolino C., della Corte M., Rossi S., Viola F., Di Vicino U., Marrocco E., Neglia S., Doria M., Testa F., Giovannoni R. et al. (2011) AAV-mediated photoreceptor transduction of the pig cone-enriched retina. Gene Ther., 18, 637–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hendrickson A., Hicks D. (2002) Distribution and density of medium- and short-wavelength selective cones in the domestic pig retina. Exp. Eye Res., 74, 435–444. [DOI] [PubMed] [Google Scholar]
- 38.Molday L.L., Rabin A.R., Molday R.S. (2000) ABCR expression in foveal cone photoreceptors and its role in Stargardt macular dystrophy. Nat. Genet., 25, 257–258. [DOI] [PubMed] [Google Scholar]
- 39.Beltran W.A., Cideciyan A.V., Lewin A.S., Iwabe S., Khanna H., Sumaroka A., Chiodo V.A., Fajardo D.S., Roman A.J., Deng W.T. et al. (2012) Gene therapy rescues photoreceptor blindness in dogs and paves the way for treating human X-linked retinitis pigmentosa. Proc. Natl. Acad. Sci. USA, 109, 2132–2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Boye S.E., Alexander J.J., Boye S.L., Witherspoon C.D., Sandefer K.J., Conlon T.J., Erger K., Sun J., Ryals R., Chiodo V.A. et al. (2012) The human rhodopsin kinase promoter in an AAV5 vector confers rod- and cone-specific expression in the primate retina. Hum. Gene Ther., 23, 1101–1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khani S.C., Pawlyk B.S., Bulgakov O.V., Kasperek E., Young J.E., Adamian M., Sun X., Smith A.J., Ali R.R., Li T. (2007) AAV-mediated expression targeting of rod and cone photoreceptors with a human rhodopsin kinase promoter. Invest. Ophthalmol. Vis. Sci., 48, 3954–3961. [DOI] [PubMed] [Google Scholar]
- 42.Chandler M.J., Smith P.J., Samuelson D.A., MacKay E.O. (1999) Photoreceptor density of the domestic pig retina. Vet. Ophthalmol., 2, 179–184. [DOI] [PubMed] [Google Scholar]
- 43.Li A., Zhu X., Craft C.M. (2002) Retinoic acid upregulates cone arrestin expression in retinoblastoma cells through a Cis element in the distal promoter region. Invest. Ophthalmol. Vis. Sci., 43, 1375–1383. [PubMed] [Google Scholar]
- 44.Reich S.J., Auricchio A., Hildinger M., Glover E., Maguire A.M., Wilson J.M., Bennett J. (2003) Efficient trans-splicing in the retina expands the utility of adeno-associated virus as a vector for gene therapy. Hum. Gene Ther., 14, 37–44. [DOI] [PubMed] [Google Scholar]
- 45.Koo T., Popplewell L., Athanasopoulos T., Dickson G. (2014) Triple trans-splicing adeno-associated virus vectors capable of transferring the coding sequence for full-length dystrophin protein into dystrophic mice. Hum. Gene Ther., 25, 98–108. [DOI] [PubMed] [Google Scholar]
- 46.Walters R.W., Bradrick S.S., Gromeier M. (2010) Poly(A)-binding protein modulates mRNA susceptibility to cap-dependent miRNA-mediated repression. RNA, 16, 239–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ricci E.P., Limousin T., Soto-Rifo R., Allison R., Poyry T., Decimo D., Jackson R.J., Ohlmann T. (2011) Activation of a microRNA response in trans reveals a new role for poly(A) in translational repression. Nucleic Acids Res., 39, 5215–5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Allocca M., Doria M., Petrillo M., Colella P., Garcia-Hoyos M., Gibbs D., Kim S.R., Maguire A., Rex T.S., Di Vicino U. et al. (2008) Serotype-dependent packaging of large genes in adeno-associated viral vectors results in effective gene delivery in mice. J. Clin. Invest., 118, 1955–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parish C.A., Hashimoto M., Nakanishi K., Dillon J., Sparrow J. (1998) Isolation and one-step preparation of A2E and iso-A2E, fluorophores from human retinal pigment epithelium. Proc. Natl. Acad. Sci. USA, 95, 14609–14613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ben-Shabat S., Parish C.A., Vollmer H.R., Itagaki Y., Fishkin N., Nakanishi K., Sparrow J.R. (2002) Biosynthetic studies of A2E, a major fluorophore of retinal pigment epithelial lipofuscin. J. Biol. Chem., 277, 7183–7190. [DOI] [PubMed] [Google Scholar]
- 51.Auricchio A., Kobinger G., Anand V., Hildinger M., O'Connor E., Maguire A.M., Wilson J.M., Bennett J. (2001) Exchange of surface proteins impacts on viral vector cellular specificity and transduction characteristics: the retina as a model. Hum. Mol. Genet., 10, 3075–3081. [DOI] [PubMed] [Google Scholar]
- 52.Gao G., Qu G., Burnham M.S., Huang J., Chirmule N., Joshi B., Yu Q.C., Marsh J.A., Conceicao C.M., Wilson J.M. (2000) Purification of recombinant adeno-associated virus vectors by column chromatography and its performance in vivo. Hum. Gene Ther., 11, 2079–2091. [DOI] [PubMed] [Google Scholar]
- 53.Young J.E., Vogt T., Gross K.W., Khani S.C. (2003) A short, highly active photoreceptor-specific enhancer/promoter region upstream of the human rhodopsin kinase gene. Invest. Ophthalmol. Vis. Sci., 44, 4076–4085. [DOI] [PubMed] [Google Scholar]
- 54.Doria M., Ferrara A., Auricchio A. (2013) AAV2/8 vectors purified from culture medium with a simple and rapid protocol transduce murine liver, muscle, and retina efficiently. Hum. Gene Ther. Methods, 24, 392–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang Y., Chirmule N., Gao G., Wilson J. (2000) CD40 ligand-dependent activation of cytotoxic T lymphocytes by adeno-associated virus vectors in vivo: role of immature dendritic cells. J. Virol., 74, 8003–8010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Drittanti L., Rivet C., Manceau P., Danos O., Vega M. (2000) High throughput production, screening and analysis of adeno-associated viral vectors. Gene Ther., 7, 924–929. [DOI] [PubMed] [Google Scholar]
- 57.Gargiulo S., Greco A., Gramanzini M., Esposito S., Affuso A., Brunetti A., Vesce G. (2012) Mice anesthesia, analgesia, and care, Part II: anesthetic considerations in preclinical imaging studies. ILAR J., 53, E70–E81. [DOI] [PubMed] [Google Scholar]
- 58.Liang F.Q., Anand V., Maguire A.M., Bennett J. (2001) Intraocular delivery of recombinant virus. Methods Mol. Med., 47, 125–139. [DOI] [PubMed] [Google Scholar]
- 59.Gargiulo A., Bonetti C., Montefusco S., Neglia S., Di Vicino U., Marrocco E., Corte M.D., Domenici L., Auricchio A., Surace E.M. (2009) AAV-mediated tyrosinase gene transfer restores melanogenesis and retinal function in a model of oculo-cutaneous albinism type I (OCA1). Mol. Ther., 17, 1347–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sambrook J., Russell D.W. (2001) Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, New York, USA. [Google Scholar]
- 61.Li A., Zhu X., Brown B., Craft C.M. (2003) Melatonin enhances retinoic acid induction of cone arrestin gene expression in retinoblastoma cells. Adv. Exp. Med. Biol., 533, 361–368. [DOI] [PubMed] [Google Scholar]
- 62.Li A., Zhu X., Brown B., Craft C.M. (2003) Gene expression networks underlying retinoic acid-induced differentiation of human retinoblastoma cells. Invest. Ophthalmol. Vis. Sci., 44, 996–1007. [DOI] [PubMed] [Google Scholar]
- 63.Manfredi A., Marrocco E., Puppo A., Cesi G., Sommella A., Della Corte M., Rossi S., Giunti M., Craft C.M., Bacci M.L. et al. (2013) Combined rod and cone transduction by adeno-associated virus 2/8. Hum. Gene Ther., 24, 982–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sparrow J.R., Wu Y., Nagasaki T., Yoon K.D., Yamamoto K., Zhou J. (2010) Fundus autofluorescence and the bisretinoids of retina. Photochem. Photobiol. Sci., 9, 1480–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sparrow J.R., Duncker T. (2014) Fundus autofluorescence and RPE lipofuscin in age-related macular degeneration. J. Clin. Med., 3, 1302–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Finnemann S.C., Leung L.W., Rodriguez-Boulan E. (2002) The lipofuscin component A2E selectively inhibits phagolysosomal degradation of photoreceptor phospholipid by the retinal pigment epithelium. Proc. Natl. Acad. Sci. USA, 99, 3842–3847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Secondi R., Kong J., Blonska A.M., Staurenghi G., Sparrow J.R. (2012) Fundus autofluorescence findings in a mouse model of retinal detachment. Invest. Ophthalmol. Vis. Sci., 53, 5190–5197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Delori F.C., Dorey C.K., Staurenghi G., Arend O., Goger D.G., Weiter J.J. (1995) In vivo fluorescence of the ocular fundus exhibits retinal pigment epithelium lipofuscin characteristics. Invest. Ophthalmol. Vis. Sci., 36, 718–729. [PubMed] [Google Scholar]
- 69.Allocca M., Manfredi A., Iodice C., Di Vicino U., Auricchio A. (2011) AAV-mediated gene replacement, either alone or in combination with physical and pharmacological agents, results in partial and transient protection from photoreceptor degeneration associated with betaPDE deficiency. Invest. Ophthalmol. Vis. Sci., 52, 5713–5719. [DOI] [PubMed] [Google Scholar]
- 70.Testa F., Surace E.M., Rossi S., Marrocco E., Gargiulo A., Di Iorio V., Ziviello C., Nesti A., Fecarotta S., Bacci M.L. et al. (2011) Evaluation of Italian patients with leber congenital amaurosis due to AIPL1 mutations highlights the potential applicability of gene therapy. Invest. Ophthalmol. Vis. Sci., 52, 5618–5624. [DOI] [PubMed] [Google Scholar]
- 71.Venables V.N., Ripley B.D. (2002) Modern Applied Statistics with S. Springer Science+Business Media, New York, USA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.