Abstract
Background Recent influenza antiviral resistance studies reveal an alarming increase in both adamantanes and neuraminidase inhibitors (NAIs) resistant viral strains worldwide, particularly in Asia, Europe and the United States.
Objectives In this study, we have evaluated influenza virus resistance in Central and South America.
Methods Influenza viruses, isolated from symptomatic patients throughout Central and South America in 2005–2008 were analyzed for inhibitor resistance. The M2 and NA genes of influenza viruses were sequenced and resistance was inferred by comparison with published sequences and known resistant mutations.
Results Our results indicate that: (i) resistance to adamantanes was seen in the majority (95·5%) of the influenza A/H3N2 isolates but only in one isolate of the influenza A/H1N1 viruses; (ii) resistance to NAIs began to be detected in A/H1N1 isolates from Central America in 2008; and (iii) none of the influenza B viruses analyzed were resistant to NAIs.
Conclusions These findings suggest a limited effectiveness of influenza inhibitors due to the detection of resistance among A/H1 and A/H3 viruses.
Keywords: Adamantanes, amantadine, influenza, neuraminidase Inhibitors, oseltamivir, resistance
Introduction
Influenza is a globally important contagion. Worldwide, each year about 20% of children and 5% of adults develop symptomatic influenza A or B. 1 Of the three types of influenza viruses (influenza A, B, and C), only types A and B typically cause widespread outbreaks. Type A influenza viruses are the major cause of influenza in humans and produce approximately half a million fatalities every year. 2
Influenza viruses have segmented genomes and show great antigenic diversity. Their genome consists of 11 genes encoding for three transcriptases (PB1, PB2, and A), two matrix proteins (M1 and M2), two surface glycoproteins [hemagglutinin (HA) and neuraminidase (NA)], one nucleocapsid protein (NP), and three non‐structural proteins (NS1, NS2, and PB1‐F2). Based upon their antigenic differences in the HA and NA surface glycoproteins, influenza A viruses have been classified into several subtypes; to date, 16 hemagglutinin subtypes (H1–H16) and nine NA subtypes (N1–N9) have been identified. 3 , 4 Only three hemagglutinin subtypes (H1, H2, and H3) and two NA subtypes (N1 and N2) have circulated as stable lineages in human populations.
Although vaccination is the primary method used to prevent influenza infections in human populations, this strategy is not always possible in the developing setting. The use of antiviral agents is an alternative approach that can be utilized to abate infection or reduce severity of illness post‐infection. These agents can be divided into two classes according to the viral protein they target: the M2 blockers or adamantanes (amantadine and rimantadine), and the NA inhibitors (oseltamivir and zanamivir).
Adamantanes inhibit the viral replication by blocking the proton channel formed by the M2 protein of the influenza A virus. Resistance can be achieved by a single substitution of any of the amino acid residues located at positions 26, 27, 30, 31, or 34 of the transmembrane domain of the M2 protein. 5 , 6 Recently, the incidence of adamantane resistance among the influenza A/H3N2 viruses has increased from 0·8% in the early 1990s to approximately 12·3% in 2004, reaching as high as 96% in certain regions of China. 7 , 8 , 9 , 10 Currently, the proportion of adamantane’s resistance among influenza A/H1N1 viruses reaches a global average of only 5·8%. 10 There are three disadvantages in using adamantanes: (i) they have no activity on influenza B viruses because these viruses do not have the M2 protein; (ii) they have adverse side effects; and (iii) drug resistance emerges rapidly during treatment.
Neuraminidase inhibitors (NAIs) inhibit the enzymatic activity of the NA protein preventing the virion’s release form the cell surface and thus, its dissemination and infection of adjacent cells. Resistance to NAIs involves a mutation in the active site of the NA protein at different positions depending on the virus subtype, 11 , 12 , 13 , 14 , 15 altering its sensitivity to inhibition. In contrast to the M2 blockers, the NAIs are licensed for treatment of both influenza A and B, resistance to NAIs is drug specific and they have fewer side effects.
Most influenza antiviral resistance studies have been conducted in East Asia, Australia, Europe and the US and revealed an alarming increase in both M2 and NA inhibitors resistant viral strains. 10 , 16 , 17 , 18 In Central and South America, viral inhibitors are not a commonly used treatment for influenza infection mostly because of their high cost and availability. 19 , 20 However, Deyde et al. 10 found a dramatic increase in M2 resistant A/H3N2 viruses in Central and South America with 7·2% resistance in 2004 and 96% in 2005. In this study we have analyzed the variants of influenza viruses circulating in this region of the world during the 3 year period July 2005–July 2008, focusing on the detection of the most commonly known mutations conferring antiviral resistance (resistance markers).
Material and methods
Specimen collection, isolation and identification of influenza viruses
Influenza A (n = 466) and B (n = 216) viruses were collected from nasopharyngeal and throat swab specimens, at hospitals throughout Central and South America (Nicaragua (n = 29), Honduras (n = 17), El Salvador (n = 6), Venezuela (n = 12), Paraguay (n = 8), Colombia (n = 12), Ecuador (n = 48), Bolivia (n = 12), Peru (n = 520), and Argentina (n = 18)) from patients that were part of the Naval Medical and Research Center Detachment–Lima (NMRCD‐Lima) ‘Influenza Surveillance Network’. These patients presented with a febrile respiratory syndrome, they had a temperature of ≥38°C, cough or sore throat and absence of other diagnosis. Once collected, swabs were placed in viral transport media and stored at −80°C until transported on dry ice to the NMRCD in Lima, Peru.
Virus isolation was carried out by inoculation in Madin‐Darby canine kidney (MDCK) cell line without fetal bovine serum. After 7 days, viral identification was performed by indirect immunofluorescence. Viral isolates obtained from cell culture were used for subtyping the influenza viruses using hemagglutinin type‐specific anti‐sera (D3 ultra DFA respiratory virus screening & ID kit, Diagnostic hybrids).
RNA Extraction and RT‐PCR
Viral RNA extraction was performed in a biosafety level‐3 laboratory. Nucleic acid was extracted with the use of viral RNA kit (QIAamp, Qiagen®, Valencia, CA ) and tested by reverse‐transcriptase‐polymerase‐chain reaction (RT‐PCR). Neuraminidase and matrix protein 2 genes were amplified by RT‐PCR with the following specific primers: bases 617–995 (338 pdb fragment) of the influenza A matrix protein 2, M2‐For3 (5′‐CTAGTCAGGCCAGGCAAATG‐3′) and M2‐Rev (5′‐ACTGTCGTCAGCATCCACAG‐3′); 21 bases 449–1218 (769 pdb fragment) of the influenza A neuraminidase 1, AN1A (5′‐AGGACAGAAGCCCTTATAGG‐3′) and AN1DII (5′‐TTAGCTCAGGATGTTGAACG ‐3′); bases 299–997 (698 pdb fragment) of the influenza A neuraminidase 2, AN2A (5′‐ATTACAGGATTTGCACCTTT‐3′) and H3N2‐NA‐2R (5′‐GGGTGTGTCTCCAACAAGTCTGAGCAC‐3′); bases 352–641 (289 pdb fragment) of the influenza B neuraminidase, NA‐RES‐F (5′‐GCTCTAACCCATTATGCAG‐3′) and NA‐RES‐R (5′‐CTTTCTTGTGTTCTTAGGATG‐3′).
Sequencing and phylogenetic analysis
For direct sequencing of viral nucleic acids from clinical specimens, genes fragments were amplified and sequenced with the use of Big Dye terminator cycle sequencing kit (version 3.1; Applied Biosystems) on a Genetic Analyser system (version 3130xL; Applied Biosystems).
Gene sequences were assembled aligned and edited using Sequencher (version 4.7; Gene Codes Corporation) and BioEdit (version 7.0.0; Isis Pharmaceuticals, Inc.) softwares.
Susceptibility to amantadine
Amantadine sensitivity was determined by plaque assay on MDCK cells. Briefly, 24‐well microplates containing MDCK cell monolayers were inoculated with virus diluted in minimal essential medium (MEM) (Gibco) to give 20–30 plaques per well. Cells were incubated for 1 hour at 37˚C and then overlaid with MEM containing 3% carboxymethyl cellulose, 1 μg/ml L‐1‐(tosylamido‐2‐phenyl)‐ethyl chloromethyl ketone and amantadine (1‐aminoadamantane hydrochloride, Sigma‐Aldrich) at different concentrations (from 0·1 to 1000 μg/ml). After 3 days of incubation at 37˚C, plaques were visualized by staining with naphtol blue solution containing naphtol blue black (1 g/l), sodium acetate (13·6 g/l), and glacial acetic acid (6%). The stain was poured off from the microplates wells and the cell monolayers were gentle washed and allowed to air‐dry and the number of lytic plaques were counted. The percentage of plaque reduction in the amantadine‐treated infected cells relative to the untreated controls was calculated for each drug concentration. The drug concentration resulting in a 50% reduction of plaque number (IC50) was determined.
Results and discussion
Adamantanes resistance among influenza a viruses
There is great controversy about the use of adamantanes for prophylaxis and therapy because their principal attraction is their lower cost and worldwide availability, but drug resistance emerges rapidly during treatment. The latter is of great concern because of the possibility of a highly virulent strain of influenza virus A causing the next influenza pandemic. Being that resistance can be achieved by a single substitution at the transmembrane domain of the M2 protein, we analyzed the M2 protein sequence of 466 influenza A viruses: 167 of which were influenza A/H1N1 subtype and 298 were influenza A/H3N2 subtype.
A total of 166 out of the 167 (99·5%) influenza A/H1N1 viruses analyzed were found to be susceptible to adamantanes. Only one isolate (0·5% of the total A/H1N1 samples) had a substitution in position 26 (L26F) which confers resistance to M2 blockers (Figure 1). In contrast, 285 out of 298 (95·5%) of the A/H3N2 viruses were found to have the specific S31N substitution which is well known to confer resistance to adamantanes. No other mutations conferring resistance were detected on the analyzed M2 gene.
Figure 1.
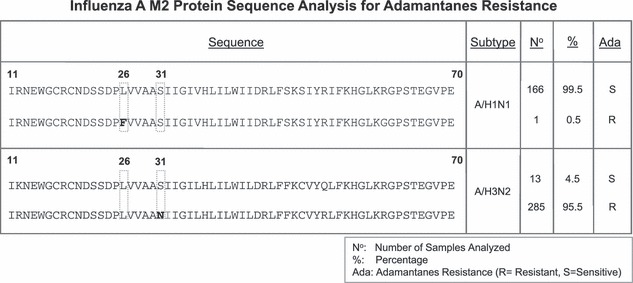
Adamantanes resistance among influenza A viruses. M2 protein sequence analysis for influenza A viruses (A/H1 viruses on upper panel and A/H3 viruses on lower panel). The figure shows the consensus sequence from residues 11 to 70. Positions where a substitution can confer resistance to adamantanes (26 and 31) are boxed and when substituted their position is marked in bold. Number (n) of samples found for each of the strains with their percentage among the population (%), Ada is the susceptibility to adamantanes (R, resistant; S, susceptible).
Resistance to M2 blockers conferred by the S31N or by the L26F substitutions was verified by plaque assay. Ten percent of the samples were tested with increasing amounts of amantadine to determine the IC50. The IC50 for all the sensitive samples tested oscillated between 0·1 and 0·5 μg/ml in agreement to what had been reported by others, 8 , 9 , 22 while the IC50 for the resistant samples (containing the S31N substitution or the influenza B samples (used as negative controls) that are not sensitive to adamantanes) oscillated around 1000 μg/ml or higher (Table 1), thus, showing a decrease in sensitivity of more than 2000 times between the resistant variants and the sensitive variants.
Table 1.
In vitro resistance to amantadine
| Sample | Date | Country | Type/ Subtype | Substitution conferring resistance | Predicted sensitivity | Predicted IC50(μg amantadine/ml) |
|---|---|---|---|---|---|---|
| FLU5148 | 23‐November‐2006 | Ecuador | A/H1N1 | Leu26Phe | R | >1000 |
| FLU4129 | 01‐August‐2006 | Peru | A/H1N1 | – | S | 0·25 |
| FLU5376 | 31‐January‐2007 | Peru | A/ H1N1 | – | S | 0·3 |
| FLU4499 | 13‐October‐2006 | Nicaragua | A/H1N1 | – | S | 0·3 |
| FLU3443 | 18‐April‐2006 | Peru | A/H3N2 | Ser31Asn | R | >1000 |
| FLU3601 | 31‐March‐2006 | Peru | A/H3N2 | Ser31Asn | R | >1000 |
| FLU6219 | 20‐March‐2007 | Venezuela | A/H3/N2 | Ser31Asn | R | >1000 |
| FLU6849 | 09‐May‐2007 | Ecuador | A/H3N2 | Ser31Asn | R | 1000 |
| lQE5463 | 14‐May‐2007 | Peru | A/H3N2 | Ser31Asn | R | >1000 |
| FLU5674 | 06‐March‐2007 | Peru | A/H3N2 | – | S | 0·25 |
| FLU5854 | 08‐March‐2007 | Peru | A/H3N2 | – | S | 0·5 |
| FSC0799 | 30‐July‐2005 | Peru | A/H3N2 | – | S | 0·2 |
| FLU6151 | 17‐April‐2007 | Peru | B | – | R | >1000 |
The table shows the concentration of amantadine giving 50% of plaque assay inhibition (IC50) for different viruses types and subtypes. The table shows the date and country where the samples were collected, the type and subtype determined based on sequence analysis of RT‐PCR amplicons of the corresponding hemagglutinin and neuraminidase genes, the substitution conferring resistance to amantadine, the inferred sensibility to amantadine and finally their sensibility to amantadine found in vitro.
Resistance to neuraminidase inhibitors
Oseltamivir is a licensed NAI for treatment of influenza A and B that has fewer side effects than adamantanes. Influenza A/H1N1 viruses have been demonstrated to be resistant to oseltamivir when the residue Histidine 274 (position 275 of the N1 protein) on the NA gene is replaced by a tyrosine residue, other substitutions of the same residue are also possible. By sequence analysis and comparison with published sequences we found that seven of the 167 (4·2%) analyzed H1N1 isolates presented the H274Y substitution (Figure 2). No other substitutions described in the literature as conferring resistance to oseltamivir were found in these isolates. Interestingly, the seven resistant viruses were detected recently, in 2008, and came from the Central American region (Honduras, Nicaragua and Venezuela).
Figure 2.
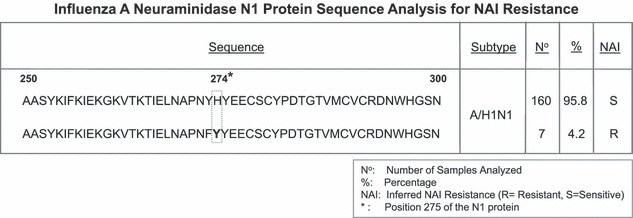
Resistance to NAIs among Influenza A viruses. Neuramidase protein sequence analysis for influenza A/H1N1 viruses. The figure shows the concensus sequence of the neuraminidase N1 gene from residues 250 to 300. Positions where a substitution can confer resistance to neuraminidase inhibitors (NAIs) are boxed and when substituted residues are in bold. Number (n) of isolates found for each of the strains with their percentage among the population (%), Ada is the susceptibility to adamantanes (R, resistant; S, susceptible).
In influenza A/H3N2 viruses, the substitution of many different residues of the NA 2 gene have been reported to confer resistance to oseltamivir. In this study, we have analyzed by sequencing the presence of substitutions on residues R118K, E119Q, D151E, R152K, R224K, E227D, E276D, and R292K and found that none of the 298 A/H3N2 isolates analyzed presented substitutions at any of this positions.
In the same way, we analyzed 216 influenza B viruses for substitution at positions 149, 152, 198, and 203 which can confer resistance to NAIs in influenza B viruses, and found that none of the isolates presented substitutions at these residues.
Our study shows that amantadine resistant strains of influenza have been in circulation in Central and South America during the last 3 years. As most of these resistance strains have the same substitution (Ser31Asn for the M2 gene of A/H3N2 viruses) and antiviral agents are not commonly used in this region, a common origin for these viruses is a strong possibility. 23 In agreement to previously published data, 24 these findings suggest that these resistant variants could have arisen in the absence of any antiviral selective pressure, or if they have a common origin in places where antiviral agents are used, they must have an advantage for their maintenance even in the absence of these agents that contributed to their migration, global distribution and maintenance over a 3 year period.
The situation is slightly different in respect to neuraminidase resistance as oseltamivir resistant viruses were not detected in the first 2 years of influenza surveillance. The detection of resistant variants began only in the period 2007–2008 in Europe 25 , 26 and has remained at a relatively low rate, which is in agreement with the levels of oseltamivir resistance found by others until 2007. 18 , 27 The fact that the seven resistant viruses came from the Central American region strongly suggests that these viruses are arriving from the northern hemisphere. It is important to note that there have been other mutations described in the literature as conferring resistance and is possible that some lesser known mutants may have been missed in this study and, there are likely to be NA mutations that have not yet been described to have an impact on NAI sensitivity. Careful surveillance of influenza viruses in the next months will be needed to further characterize the spreading of this type of viruses among South American countries.
This finding somehow discourage the strategy of stockpiling NAIs, such as is under way in many industrialized countries as part of national influenza pandemic preparedness. 28 Moreover, this NAI is extremely expensive for developing countries as one treatment costs around US$15. 19 In Peru for example, there is an absence of a national sanitary strategy related to the treatment of viral respiratory agents, such as influenza, but even if there was, the high cost of oseltamivir would make it an unsustainable alternative for this low‐income country to use it as common treatment. 14 Furthermore, in Peru, as in many countries in South America, amantadine is the only antiviral agent approved by the government. 20 Therefore, the use of vaccinations in persons at‐risk should be highly encouraged.
Funding Statement
This study was funded by the United States Department of Defense Global Emerging Infections Systems Research Program, work unit number: 847705·82000·25GB.B0016.
Conflict of interest
We declare that we have no conflict of interest.
Disclaimers
The views expressed in this article are those of the authors and do not necessarily reflect the official policy or position of the Department of the Navy, Department of Defense, nor the US Government.
The study protocol was approved by the Naval Medical Research Center Institutional Review Board (Protocols NMRCD.2002·0019) in compliance with all applicable Federal regulations governing the protection of human subjects.
Disclosure
None of the authors has a financial or personal conflict of interest related to this study. The corresponding author had full access to all data in the study and final responsibility for the decision to submit this publication.
Copyright Statement
Authors Tadeusz J. Kochel, Alberto Laguna and Merly Sovero are military service members or employees of the US Government. This work was prepared as part of their official duties. Title 17 U.S.C. § 105 provides that ‘Copyright protection under this title is not available for any work of the US Government’. Title 17 U.S.C. § 101 defines a US Government work as a work prepared by a military service members or employees of the US Government as part of those person’s official duties.
Acknowledgements
We are very grateful to Ms María Ester Gamero for cell culture support in in vitro adamantanes resistance experiments and to Mrs Gloria Chauca for laboratory and administrative support.
References
- 1. Turner D, Wailoo A, Nicholson K, Cooper N, Sutton A, Abrams K. Systematic review and economic decision modelling for the prevention and treatment of influenza A and B. Health Technol Assess 2003; 7:1–170. iii‐iv, xi‐xiii [DOI] [PubMed] [Google Scholar]
- 2. Englund JA. Antiviral therapy of influenza. Semin Pediatr Infect Dis 2002; 13:120–128. [DOI] [PubMed] [Google Scholar]
- 3. Fouchier RA, Munster V, Wallensten A et al. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black‐headed gulls. J Virol 2005; 79:2814–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Nicholson KG, Wood JM, Zambon M. Influenza. Lancet 2003; 362:1733–1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Boivin G, Goyette N, Bernatchez H. Prolonged excretion of amantadine‐resistant influenza a virus quasi species after cessation of antiviral therapy in an immunocompromised patient. Clin Infect Dis 2002; 34:E23–E25. [DOI] [PubMed] [Google Scholar]
- 6. Hay AJ, Zambon MC, Wolstenholme AJ, Skehel JJ, Smith MH. Molecular basis of resistance of influenza A viruses to amantadine. J Antimicrob Chemother 1986; 18(Suppl. B):19–29. [DOI] [PubMed] [Google Scholar]
- 7. Bright RA, Shay DK, Shu B, Cox NJ, Klimov AI. Adamantane resistance among influenza A viruses isolated early during the 2005–2006 influenza season in the United States. JAMA 2006; 295:891–894. [DOI] [PubMed] [Google Scholar]
- 8. Bright RA, Medina MJ, Xu X et al. Incidence of adamantane resistance among influenza A (H3N2) viruses isolated worldwide from 1994 to 2005: a cause for concern. Lancet 2005; 366:1175–1181. [DOI] [PubMed] [Google Scholar]
- 9. Ziegler T, Hemphill ML, Ziegler ML et al. Low incidence of rimantadine resistance in field isolates of influenza A viruses. J Infect Dis 1999; 180:935–939. [DOI] [PubMed] [Google Scholar]
- 10. Deyde VM, Xu X, Bright RA et al. Surveillance of resistance to adamantanes among influenza A(H3N2) and A(H1N1) viruses isolated worldwide. J Infect Dis 2007; 196:249–257. [DOI] [PubMed] [Google Scholar]
- 11. Hayden F, Klimov A, Tashiro M et al. Neuraminidase inhibitor susceptibility network position statement: antiviral resistance in influenza A/H5N1 viruses. Antivir Ther 2005; 10:873–877. [PubMed] [Google Scholar]
- 12. Carr MJ, Sayre N, Duffy M, Connell J, Hall WW. Rapid molecular detection of the H275Y oseltamivir resistance gene mutation in circulating influenza A (H1N1) viruses. J Virol Methods 2008; 153:257–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Aoki FY, Boivin G, Roberts N. Influenza virus susceptibility and resistance to oseltamivir. Antivir Ther 2007; 12:603–616. [PubMed] [Google Scholar]
- 14. Oshitani H, Kamigaki T, Suzuki A. Major issues and chalenges of influenza pandemic preparedness in developing countries. Emerg Infect Dis 2008; 14:875–880. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mishin VP, Hayden FG, Gubareva LV. Susceptibilities of antiviral‐resistant influenza viruses to novel neuraminidase inhibitors. Antimicrob Agents Chemother 2005; 49:4515–4520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Monto AS, McKimm‐Breschkin JL, Macken C et al. Detection of influenza viruses resistant to neuraminidase inhibitors in global surveillance during the first 3 years of their use. Antimicrob Agents Chemother 2006; 50:2395–2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Saito R, Li D, Suzuki Y et al. High prevalence of amantadine‐resistance influenza a (H3N2) in six prefectures, Japan, in the 2005–2006 season. J Med Virol 2007; 79:1569–1576. [DOI] [PubMed] [Google Scholar]
- 18. World Health Organization . WHO/ECDC frequently asked questions for oseltamivir resistance. Available at: (http://www.who.int/csr/disease/influenza/oseltamivir_faqs/en/), [accessed on 04 February 2008].
- 19. Enserink M. Oseltamivir becomes plentiful–but still not cheap. Science 2006; 312:382–383. [DOI] [PubMed] [Google Scholar]
- 20. Ministerio de Salud del Peru . Plan nacional de preparación y respuesta frente a una potencial pandemia de influenza. Perú: Gobierno, 2005. epi.minsal.cl/webInfluenza/enelmundo/Plan_Influenza_Peru.pdf. [Google Scholar]
- 21. Saito R, Oshitani H, Masuda H, Suzuki H. Detection of amantadine‐resistant influenza A virus strains in nursing homes by PCR‐restriction fragment length polymorphism analysis with nasopharyngeal swabs. J Clin Microbiol 2002; 40:84–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ilyushina NA, Govorkova EA, Webster RG. Detection of amantadine‐resistant variants among avian influenza viruses isolated in North America and Asia. Virol 2005; 341:102–106. [DOI] [PubMed] [Google Scholar]
- 23. Barr IG, Komadina N, Durrant C, Sjogren H, Hurt AC, Shaw RP. Circulation and antigenic drift in human influenza B viruses in SE Asia and Oceania since 2000. Commun Dis Intell 2006; 30:350–357. [PubMed] [Google Scholar]
- 24. Simonsen L, Viboud C, Grenfell BT et al. The genesis and spread of reassortment human influenza A/H3N2 viruses conferring adamantane resistance. Mol Biol Evol 2007; 24:1811–1820. [DOI] [PubMed] [Google Scholar]
- 25. Lackenby A, Hungnes O, Dudman SG et al. Emergence of resistance to oseltamivir among influenza A(H1N1) viruses in Europe. Euro Surveill 2008; 13:8026. [DOI] [PubMed] [Google Scholar]
- 26. Rimmelzwaan GF, De Jong JC, Donker GA, Meijer A, Fouchier RA, Osterhaus AD. Influenza season 2007/’08 in the Netherlands: antigenic variation, oseltamivir resistance and vaccine composition for the 2008/’09 season. Ned Tijdschr Geneeskd 2008; 152:2138–2144. [PubMed] [Google Scholar]
- 27. European Center for Disease Prevention and Control . Antivirals and antiviral resistance‐influenza. Resistance to Oseltamivir (tamiflu) found in some European influenza virus samples. Available at: (http://ecdc.europa.eu/Health_topics/influenza/antivirals.html), [accessed on 21 February 2008].
- 28. Mounier‐Jack S, Coker RJ. How prepared is Europe for pandemic influenza? Analysis of national plans Lancet 2006; 367:1405–1411. [DOI] [PubMed] [Google Scholar]


