Abstract
The myelin proteolipid protein gene (Plp1) encodes the most abundant protein found in CNS myelin, accounting for nearly one-half of the total protein. Its expression in oligodendrocytes is developmentally regulated – peaking during the active myelination period of CNS development. Previously we have identified a novel enhancer (designated ASE) in intron 1 DNA that appears to be important in mediating the surge of Plp1 gene activity during the active myelination period. Evidence suggests that the ASE participates in the formation of a specialized multi-protein/DNA complex called an enhanceosome. The current study describes an optimized, five-step, DNA affinity chromatography purification procedure to purify nuclear proteins from mouse brain that bind to the 85-bp ASE sequence, specifically. EMSA analysis demonstrated that specific DNA binding activity was retained throughout the purification procedure, resulting in concomitant enrichment of nucleoprotein complexes. Identification of the purported regulatory factors was achieved through mass spectrometry analysis and included over twenty sequence-specific DNA-binding proteins. Supplementary Western blot analyses to determine which of these sequence-specific factors are present in oligodendrocytes, and their developmental and regional expression in whole brain, suggest that Purα and Purβ rank highest among the candidate factors as constituents of the multi-protein complex formed on the ASE.
Keywords: DNA affinity chromatography, DNA-binding proteins, enhanceosome, intronic enhancer, mass spectrometry, multi-protein complex, myelin proteolipid protein gene, proteomics
INTRODUCTION
Myelin is a specialized multilayer membranous structure emanating from glia that provides the physiological basis for rapid unidirectional conduction of action potentials along axons. In the CNS, oligodendrocytes are responsible for the synthesis of myelin, with the peak rate of myelin deposition occurring around three weeks postnatal in rodents (Norton and Poduslo 1973). Since oligodendrocytes can myelinate multiple axons, large quantities of myelin components such as lipids and proteins must be synthesized during this stage. Many of the “myelin-specific” genes are coordinately regulated at the transcriptional level to ensure for sufficient and synchronous expression during the active myelination period of CNS development (Macklin et al. 1991; Shiota et al. 1991). Elucidation of the pertinent mechanisms regulating their transcription should aid in rational design of therapeutic strategies that promote remyelination in adults faced with demyelinating conditions such as multiple sclerosis (Zhao et al. 2005) and may reveal the etiological basis for some dysmyelinating disorders when the process goes awry.
Work from our laboratory has focused on the myelin proteolipid protein gene (Plp1) as a model system to study myelin-specific gene regulation. However it is particularly important to understand the mechanisms controlling Plp1 gene expression since 10% of the total mRNA present in oligodendrocytes during the active myelination period emanate from the gene (Baumann and Pham-Dinh 2001). In mice, the level of Plp1 gene transcription in brain is highest during the third postnatal week (Macklin et al. 1991; Shiota et al. 1991). To determine how the gene is regulated in oligodendrocytes, expression of Plp1-lacZ fusion genes have been examined in transgenic mice, as well as by deletion-transfection analysis. When mouse Plp1 genomic sequences spanning the proximal 2.4 kb of 5’-flanking DNA, downstream to the first 37 bp of exon 2 were used to drive lacZ reporter gene expression in transgenic mice, the developmental profile of lacZ expression in brain recapitulated that of the endogenous Plp1 gene (Wight et al. 1993, 2007). Thus the transgene, designated PLP(+)Z because it contains all of Plp1 intron 1 DNA, must contain regulatory element(s) important for governing its temporal regulation. Conversely, expression of the PLP(+)Z-derived transgene PLP(−)Z, which is missing the intron, remained relatively low and stagnant throughout the active myelination period of CNS development (Li et al. 2002). Because both transgenes demonstrated similar levels of lacZ expression in brain prior to the onset of the active myelination period, but only the PLP(+)Z transgene showed a significant increase in expression during the active myelination period, element(s) important in regulating the temporal expression of Plp1 must reside somewhere within the first intron. The intron contains a single positive regulatory element as identified by deletion-transfection analysis in N20.1 oligodendroglial cells (Verity et al. 1993) using a battery of PLP(+)Z-derived constructs containing partial deletion of intron 1 sequence (Dobretsova and Wight 1999; Dobretsova et al. 2000). The regulatory element was minimally mapped to Plp1 intron 1 positions 1,093–1,177 (Dobretsova et al. 2004), based on numbering the intron from positions 1 to 8,140 (Wight and Dobretsova 1997). We have dubbed the element, ASE, for antisilencer/enhancer due to its functional properties in N20.1 cells. The ASE contains a plethora of potential target sites for DNA-binding proteins as determined in silico (Dobretsova et al. 2000), including binding sites for transcription factors as well as architectural proteins.
Evidence suggests that the ASE participates in formation of an enhanceosome. An enhanceosome is defined as a multi-protein/DNA complex, possessing a specific three-dimensional structure that ultimately is responsible for enhancing the level of transcription from an inducible gene in cis (reviewed in Carey 1998; Merika and Thanos 2001; Alvarez et al. 2003). Assembly of the complex is highly cooperative and the ensuing transcriptional synergy is dependent upon the precise arrangement of factors within the nucleoprotein complex. All of the factors must be present for the enhanceosome to be functional, in vivo. DNase I footprinting studies (Dobretsova et al. 2004) show that the vast majority of the ASE can be protected by nuclear proteins from N20.1 cells. However mutating an AP-1-like binding site in the middle of the ASE negates any protection from DNase I in footprinting studies. Functionally, this mutation obliterates the ability of the ASE to augment Plp1-lacZ gene transcription in N20.1 cells (Dobretsova et al. 2004). Typically, enhanceosomes contain architectural proteins which bend DNA (enhancer sequence) so that synergistic effects can occur between some of the factors within the complex. Similarly, the ASE contains putative target sites for architectural proteins such as Sox proteins, which have been shown to be important in activating myelin-specific gene expression, including Plp1 (Stolt et al. 2004). Moreover, the ASE contains three non-overlapping sub-regions (27 bp, each) which bind nuclear proteins in a sequence-specific manner. Electrophoretic mobility shift assays (EMSA) demonstrate that when one of the sub-regions is used as a probe, the other two can compete for specific DNA binding of nuclear proteins (Dobretsova et al. 2000), suggesting that the factors together form a single unit (i.e., a multi-protein complex) on the ASE. Transfection analysis suggests that the functional activity of the ASE is strongly dependent upon the precise arrangement of bound factors, since removal of the sequences (7 bp, each) separating the sub-regions caused a significant diminishment in activity (Dobretsova et al. 2000). Overall, these results suggest that the ASE functions as an enhanceosome.
In order to determine the composite of factors from mouse brain that bind the ASE and ultimately enhance Plp1 gene transcription during development, DNA affinity chromatography was optimized to permit formation of a multi-protein complex on the 85-bp ASE sequence. Identification of the bound nuclear proteins was achieved by mass spectrometry analysis. We took this unbiased approach for several reasons. First, it permits detection of the exact subunits/factors involved which is important when a target site can be recognized by a transcription factor composed of various subunit combinations, such as AP-1, or altogether by different transcription factors. Second, the unbiased nature is essential for identifying transcription factors whose target sites have not yet been characterized and, as such, would be overlooked when the ASE sequence was scrutinized for putative transcription factor binding sites. Third, the procedure approximates the physiological salt concentration present in the nucleus, which is important for purification of salt-sensitive protein complexes. Here we report the results of the optimized procedure for DNA affinity purification under conditions of low stringency to support formation of a multi-protein complex on the ASE. Mass spectrometry analysis was performed to identify nuclear proteins that selectively bind the ASE, followed by Western blot analysis of a select subset of these sequence-specific transcription factors to ascertain their potential importance for regulation of Plp1 gene activity in oligodendrocytes, first by determining whether they are present in nuclei from oligodendrocytes, and then if so, their developmental and regional expression profiles in nuclei isolated from mouse brain.
MATERIALS AND METHODS
Animals
Breeding colonies of mice were established onsite. C57BL/6 mice were used for preparation of all nuclear extracts used for DNA affinity purification. The CD-1 mouse strain was used in the Western blot studies. All procedures involving the use of mice were approved by the Institutional Animal Care and Use Committee at the University of Arkansas for Medical Sciences. Anesthetized pups were sacrificed by decapitation and brains removed quickly. Brains were either used in their entirety (whole brain) or dissected on ice into the following regions: hindbrain (medulla oblongata and pons), cerebellum, midbrain (thalamus, subthalamus, midbrain, hypothalamus), cortex (anterior and posterior cortex, hippocampus), forebrain (striatum-enriched preparation), and olfactory bulb, similar to that as described by Glowinski and Iversen (1966).
Preparation of nuclear extracts
Nuclear extracts were prepared from whole brains (or brain regions) of mice between postnatal days 17–25 of age according to the methods of Gorski et al. (1986) except that nuclei were purified by a single round of centrifugation through a 2 M sucrose cushion, instead of two rounds. Nuclear extracts were snap-frozen in liquid nitrogen and stored at −70°C for no longer than six months. Multiple preparations of nuclear extracts were pooled together for subsequent use in DNA affinity purification. Typically, 100 µg of nuclear proteins were obtained from 100 mg of brain tissue. Thus, roughly 400 mice were sacrificed in order to obtain enough nuclear extract (40 mg of protein) necessary for DNA affinity purification/mass spectrometry analysis.
Nuclei from immunosorted cells (O1-enriched and O1-depleted populations) were isolated as previously described (Macklin et al. 1991), with minor adjustments. Cells (2–4 × 106) were collected by centrifugation and resuspended in 3 ml of PIPES Buffer Solution (10 mM PIPES, pH 7.0, 10 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 3 mM MgCl2, 1 mM DTT, 1 mM spermidine, 0.1 mM PMSF, 0.32 M sucrose). After incubation on ice for 20 min, cells were disrupted by 20 strokes in a Dounce homogenizer (pestle A). The mixture was then transferred to a centrifuge tube and Nonidet P40 added to a final concentration of 0.5% while vortexing. Nuclei were pelleted at 3,000 × g for 20 min at 4°C. Nuclear proteins were separated from genomic DNA as described by Gorski et al. (1986) and dialyzed overnight in mini Slide-A-Lyzer cassette, MWCO 10,000 Da (Pierce Biotechnology, Rockford, IL). Protein concentration was determined using Coomassie Plus Protein Assay Reagent (Pierce Biotechnology) per manufacturer’s directions. Nuclear extracts were stored in liquid nitrogen until further use. Typically, 10–25 µg of protein were obtained per preparation.
ASE Column Preparation
Synthetic oligonucleotides (The Midland Certified Reagent Company, Inc., Midland, TX) identical to mouse Plp1 intron 1 DNA positions 1,093–1,177 (Dobretsova et al. 2004) were used to generate the double-stranded ASE (dsASE) column. The 3’-end of the sense strand was extended with three polyethylenglycol spacers and a terminal amino-C7 linker. The 5’-end of the antisense strand was radiolabeled using γ-32P-ATP and T4 polynucleotide kinase. The oligonucleotides (20 nmols each) were annealed in a total volume of 540 µl of STE buffer (10 mM Tris, pH 8.0, 1 mM EDTA, 50 mM NaCl) by heating to 95°C for 5 min followed by slow cool down, and then concentrated using a Microcon YM-10 concentrator (Millipore, Bedford, MA) (170 µl final volume). Annealed DNA (dsASE) was purified by size exclusion chromatography with Superdex 75 (Amersham Biosciences, Piscataway, NJ) equilibrated in 100 mM sodium phosphate buffer, pH 7.2, 150 mM NaCl. Chromatography was performed using a 0.5-cm diameter column (Bio-Rad, Hercules, CA) with an 18-ml bed volume, precisely per the manufacturer’s directions. Forty-eight 0.5-ml fractions were collected and 10-µl aliquots from each fraction measured for radioactivity in a liquid scintillation counter. Peak radioactivity was present in fractions 10–15, consistent with elution of dsASE in the void volume. Only fractions 10–13, which accounted for 90% of the total input radioactivity, were combined to eliminate possible contamination with unannealed single-stranded DNA. The resulting yield of dsASE was approximately 17 nmols. The amino group at the 3’-terminus was reacted with the bifunctional cross-linker succinimidyl 4-hydrazinonicotinate acetone hydrazone (SANH) to produce a hydrazine moiety by treatment with 2% SANH at room temperature for 90 min, followed by 4% SANH at room temperature for 90 min, and then dialysis against DNA Binding Buffer (20 mM sodium acetate buffer, pH 4.9, 100 mM NaCl).
Two grams of cellulose (Fibrous cellulose CF11; Whatmann, Florham Park, NJ) per 50 ml conical centrifuge tube was swelled overnight in water, in the cold. The cellulose was cleared from fine particles by 5 washes in water (cellulose was mixed and allowed to settle for 10 min after each wash prior to aspiration of the upper layer). The cellulose was then activated by incubation in 20 mM sodium (meta)periodate (Fluka, Allentown, PA) overnight in the dark with constant agitation. Activated cellulose (4 ml packed volume) was washed 8 times in 10 mM sodium acetate buffer, pH 4.9, 100 mM NaCl and then incubated with dsASE DNA in DNA Binding Buffer (or buffer alone) for 4 h at room temperature with constant agitation. Subsequently, the buffer was replaced with Gel Shift Buffer (50 mM potassium phosphate buffer, pH 7.4, 20 mM KCl, 1 mM EDTA) containing 0.425 mM of sodium cyanoborohydride and incubation continued, overnight. Unreacted aldehyde groups from the matrix were then neutralized by the addition of sodium borohydride to a final concentration of 100 mM, and the conjugated matrix (dsASE-cellulose) or matrix alone (cellulose) incubated at 4°C, overnight, in an open tube. Matrices were washed twice in 25 ml of Gel Shift Buffer and a small amount of the dsASE matrix measured for radioactivity. It was estimated that approximately 232 pmols of dsASE bound per gram of wet cellulose.
DNA Affinity Chromatography
Pooled nuclear extract containing a total of 40 mg of protein was diluted to a final concentration of 8 mg/ml in Protein Binding Buffer (25 mM potassium phosphate buffer, pH 7.6, 50 mM KCl, 1 mM EDTA, 5 % glycerol, 0.5 mM DTT, 0.01% Triton X-100) and equally distributed between four columns (1-ml bed volume) of double-stranded calf thymus DNA-cellulose (dsDNA) (Sigma, St. Louis, MO), run in parallel. Each column was washed with 25 ml of Protein Binding Buffer and bound proteins eluted in 12 ml of Elution Buffer [20 mM sodium phosphate buffer, pH 7.2, 1.7 M KCl, 1 mM EDTA, 0.5 mM DTT, 0.5 mM PMSF, 7% glycerol, protease inhibitors (1 µg/ml leupeptin, 2 µg/ml antipain, 10 µg/ml benzamidine, 10 µg/ml antipain, 1 µg/ml chymostatin, 1 µg/ml pepstatin)]. Eluted proteins were concentrated immediately using an Amicon Ultra-15 (10,000 MWCO) concentrator (Millipore) and dialyzed overnight at 4°C against Protein Binding Buffer (without Triton X-100) containing one-tenth the amount of the protease inhibitor cocktail present in Elution Buffer. Nearly 11 mg of protein were eluted from the dsDNA columns. Eluted proteins were diluted to a final volume of 40 ml with Protein Binding Buffer and passed over a column (0.5-ml bed volume) of single-stranded calf thymus DNA-cellulose (ssDNA) (Sigma) to deplete the mixture of DNA/RNA single-stranded binding proteins, abundant within the mixture. The column was washed with 15 ml of Protein Binding Buffer and the bound proteins eluted in 5 ml of Elution Buffer containing additional KCl (2M final concentration). The ssDNA eluate was dialyzed overnight against Protein Binding Buffer (without Triton X-100). Unbound proteins (3.58 mg) present in the flow-through and wash from the ssDNA column were concentrated to a final volume of 40 ml and “pre-cleared” by mixing with matrix alone (unconjugated cellulose) for 15 min at room temperature on a rotator. The mixture was then poured in a column (3-ml bed volume) and washed with 20 bed volumes of Protein Binding Buffer. Proteins bound to the unconjugated matrix were eluted in 5 bed volumes of Elution Buffer, and concentrated and dialyzed as described earlier. Unbound proteins (2.45 mg) present in the flow-through and wash of the unconjugated matrix were concentrated to a final volume of 20 ml and mixed with dsASE-cellulose for 1 h at room temperature using a rotator. The material was then poured into a column (1-ml bed volume) and the bound and unbound protein fractions processed similarly to those with the unconjugated matrix. Protein concentrations were determined in all fractions after concentration and dialysis. A total of 4.8 µg of protein eluted from the unconjugated matrix and 36 µg of protein eluted from the dsASE column (ASE-1). Thirty micrograms of protein from the ASE-1 eluate were subjected to a second round of DNA affinity purification using a fresh dsASE column, resulting in a total of 11.74 µg of protein in the second (ASE-2) eluate.
Mass spectrometry analysis of affinity chromatography fractions
Purified proteins were resolved by SDS-PAGE (4% stacking, 11% resolving gel) and stained with Coomassie blue. Gels were placed on a clean plastic plate and cut into 3-mm bands along the entire lane with a razor blade. Each band was then cut in half and washed until colorless in 50 mM ammonium bicarbonate, 50% acetonitrile and then dried completely in 100% acetonitrile. Gel bands were digested robotically using a ProGest instrument (Genomic Solutions, Ann Arbor, MI) and the resultant peptides analyzed by nano LC-MS/MS as previously described (Gambus et al. 2006). Briefly, 45 µl was loaded from an autosampler onto a 0.5 mm × 0.75 mm vented-column at 15 µl/min and peptides separated at 200 nl/min on a 150 mm × 0.75 mm column with a 45 min gradient into an LCQ Deca XP Plus ion trap mass spectrometer (Thermo, San Jose, CA).
The mass spectrometry data was processed as described previously (Gambus et al. 2006). Briefly, the product ion data were searched against the IPI Mouse protein database v3.13 (containing concatenated forward and reversed sequences) using a locally stored copy of the Mascot search engine v2.0.04 (Matrix Science, London, UK) via Mascot Daemon v2.0.0. Mascot LCQ_DTA executable generates both doubly and triply charged versions of each ion selected in the DDA experiment, unless no ions are observed above the parent m/z, in which case it is assigned as singly charged. Search parameters were: precursor mass tolerance 2.5 Da, product ion mass tolerance 0.6 Da, 2 missed cleavages allowed, fully tryptic peptides only, fixed modification of carbamidomethyl cysteine, variable modifications of oxidized methionine, N-terminal acetylation and pyro-glutamic acid on N-terminal glutamine. Mascot search result flat files (.DAT) were parsed to an Oracle database using in-house software called ProteinTrack; all peptides with a Mascot score of 23 or greater were saved. ProteinTrack allows multi-function querying of Mascot data sets including the generation of protein summary lists based on user-defined scoring criteria, comparison of multiple experiments, and output of peptide sequence lists (including charge state, Mascot score). The criteria for accepting a protein match were determined by calculating the false discovery rate (FDR) through searching data against the concatenated forward and reversed IPI mouse protein database. The FDR was calculated by the following formula: (# reverse hits *2)/(# total hits) (Peng et al. 2003). The following cut-off values were used in this analysis: if there was more than one peptide for a particular protein then the total score (sum of the peptide scores) had to be at least 57 and each peptide had to score 23 or greater; if only one peptide was identified, it had to score 45 or greater. Peptides had to have a mass of at least 600 Da. In each Mascot search, a protein needed at least one bold red peptide in each DAT if it was to be considered (bold red peptides represent a unique highest ranking assignment in Mascot search results). These criteria resulted in FDR of less than 0.1% for this data set. Relative quantization was based on spectral counting (Liu et al. 2004).
Electrophoretic Mobility Shift Assays (EMSA)
EMSA was performed as previously described (Dobretsova et al. 2004) to monitor DNA binding activity present within the various chromatographic steps of protein purification. PCR-generated ASE DNA (intron 1 positions 1,093–1,177) was end-labeled with (γ−32P)ATP (PerkinElmer, Waltham, MA) by T4 polynucleotide kinase (New England BioLabs) and used as a probe. DNA-protein binding reactions (20 µl) were performed at room temperature in siliconized microcentrifuge tubes (MidWest Scientific, St. Louis, MO) and consisted of Protein Binding Buffer containing an aliquot of protein from a given chromatography fraction (final concentration adjusted to 2 µg with BSA), 1 µg poly(dI-dC), and 2 × 104 cpm of probe. DNA-protein complexes were allowed to form for 20 min at room temperature and then resolved on a 4.5% polyacrylamide, non-denaturing gel. The gel was dried and subjected to autoradiography.
Isolation of O1-Positive Cells from Mouse Brain
O1-positive cells were isolated as described earlier for oligodendrocytes (Berti-Mattera et al. 1984; Sato-Bigbee et al. 1999) and microglia (Carson et al. 1998), with several modifications. Mice between P16–P21 of age were anesthetized and perfused intracardially with approximately 20 ml of PBS injected directly into the left ventricle. The following steps were performed on ice unless noted otherwise. Brains were removed immediately after perfusion and finely minced on ice in a glass Petri dish filled with HBSS buffer pH 7.4 (without calcium, magnesium or phenol red; Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum (HyClone, Logan, UT). Minced tissue (from up to 20 brains) was sequentially forced through 140-µm and 70-µm Nitex nylon meshes (Sefar, Kansas City, MO) with the bottom of a 5 ml glass beaker and collected in a 300 ml glass beaker on ice. Sufficient amounts of HBSS containing 10% FBS were used to keep the tissue wet at all times. Tissue particles were pelleted by centrifugation in 50 ml plastic conical tubes in a table-top centrifuge with swinging bucket rotor (Eppendorf, Westbury, NY; Models 5810R and A-4-62) at 1,000 rpm for 10 min at 4°C. The pellet was washed once in serum-free HBSS and digested in a solution of 28 U/ml of DNase I, 0.2 mg/ml of collagenase in serum-free HBSS (10 ml/brain) for 30 min at 37°C with constant agitation. Digestion was stopped by a 2-fold dilution with HBSS containing 20% FBS. The digested material often appeared as clumps due to the absence of divalent cations, which could be disrupted by several strokes in a 25 ml plastic pipet. Cellular material was collected by centrifugation at 1,000 rpm for 10 min at 4°C, washed once in HBSS with 10% FBS and once in serum-free HBSS, and the pellet resuspended in 6 ml/brain of HBSS supplemented with 2 mM EDTA (pH 8.0). Percoll solution (1.122 g/ml Percoll in HBSS supplemented with 2 mM EDTA) was added (3 ml/brain) to the cell suspension and mixed by inverting the tube several times. Live cells were separated from myelin debris and other material by centrifugation at 4,000 rpm for 20 min at 4°C in the swinging bucket rotor and collected in the bottom 7 ml of solution. Percoll was removed by diluting the cell suspension six-fold with HBSS containing 10% FBS and 2 mM EDTA (HBSS/FBS/EDTA), and centrifugation at 1,000 rpm for 10 min at 4°C, followed by an additional wash in HBSS/FBS/EDTA. Cells were resuspended in HBSS/FBS/EDTA and mixed with the monoclonal anti-oligodendrocyte marker O1 antibody (R&D Systems, Minneapolis, MN) per the manufacturer’s specifications for flow cytometry (0.5 µg of primary antibody per 2.5 × 105 cells in a total reaction volume up to 200 µl) on a rotary shaker for 15 min at 4°C. Unbound antibodies were removed by two 10 ml washes in HBSS/FBS/EDTA followed by centrifugation. Magnetic microbeads containing rat anti-mouse IgM secondary antibody (Miltenyi Biotec, Auburn, CA) were washed three times with 0.5 ml of HBSS/FBS/EDTA to remove sodium azide and collected in at least 0.3 ml of HBSS/FBS/EDTA. It was empirically determined that twice the amount of magnetic beads was needed (40 µl microbeads per 1 × 107 cells) as recommended by the manufacturer, probably due to a partial loss of microbeads during the wash procedure. The microbeads were incubated with cells pre-treated with O1 antibody per the manufacturer’s recommendations and washed twice in HBSS/FBS/EDTA. Prior to sorting, cells were resuspended (1 ml per 8 × 107 cells) in HBSS containing 5% FBS, then diluted two-fold with serum-free HBSS and filtered through a 50-µm mesh. The resulting suspension was loaded onto a magnetic column in 500 µl increments and unbound cells were collected in six washes (0.5 ml) of HBSS containing 2.5% FBS (O1-depleted fraction). The magnetic field was then removed and O1+ cells collected in 1.0 ml of HBSS containing 2.5% FBS (O1-enriched fraction). Cells in both fractions were washed in HBSS/FBS, collected by centrifugation, and live cells counted using a solution of 0.4% Trypan Blue (Invitrogen) in PBS. The procedure described above resulted in recovery of 86% of the initial Percoll-isolated cells in both fractions, combined. From 20 brains of mice at P21 of age, approximately 9 × 107 total cells were obtained after the Percoll gradient, and approximately 4 × 106 cells in the O1-enriched fraction. The efficiency of cell sorting was analyzed by immunocytochemistry.
Western Blot Analysis
Nuclear extracts prepared from the brains of mice at specific ages were snap-frozen in liquid nitrogen and stored at −70°C. Protein concentrations were determined simultaneously as described earlier to minimize variation between the extracts. Nuclear proteins were denatured by heat (98°C for 5 min) in 31.25 mM Tris pH 6.8, 1% SDS, 5% glycerol, 0.05 mg/ml bromophenol blue, and 0.785% β-mercaptoethanol (Gel Loading Buffer). Nuclear proteins (equal amounts) isolated from immunosorted cells or specific brain regions were precipitated with trichloroacetic acid as described by Bensadoun and Weinstein (1976), washed in 100% acetone pre-chilled at −20°C, and resuspended in Gel Loading Buffer to a final concentration of 0.5 µg/µl. Proteins were denatured by heating, fractionated on an SDS-PAGE gel (10% polyacrylamide, Bio-Rad) and subsequently transferred to a nitrocellulose membrane (Optitran BA-S 85, Schleicher & Schuell, Florham Park, NJ) per manufacturer’s recommendations. Proteins were visualized by staining with a 0.1% (w/v) solution of Ponceau S (Sigma) in 5% acetic acid. Membranes were blocked with 10% non-fat dry milk or with 10% heat-inactivated goat serum and 1% BSA in TBST (20 mM Tris, pH 7.6, 150 mM NaCl, 0.1% Tween-20) for 1 h at room temperature, washed three times (15 min each) in TBST and subsequently incubated with primary antibody diluted in TBST plus 5% BSA overnight at 4°C with constant agitation. Membranes were washed three times (5 min each) in TBST at room temperature and then incubated with secondary antibodies (Jackson Immunoresearch) for 40 min. Secondary antibodies were diluted in TBST containing 5% (anti-mouse antibody) or 10% (anti-rabbit antibody) non-fat dry milk or 10% heat-inactivated goat serum plus 1% BSA (anti-goat antibody or immune serum) and used at the dilution as recommended by the supplier. Blots were washed three times (15 min each) in TBST and immunoreactive bands were visualized using Western Lightning Chemiluminescence Reagent Plus detection reagents (PerkinElmer). Primary antibodies included rabbit anti-c-Jun, anti-JunD, anti-ATF2, anti-CREB-1, and anti-NF-1, mouse anti-ATF-1, goat anti-MeCP2 and anti-hnRNP U (Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-ZONAB (MSY4; Invitrogen) and anti-PARP1 (Upstate, Charlottesville, VA). Peptide antibodies against mouse Purα (residues 291–313), Purβ (residues 302–324), and MSY1 (residues 242–267 or 276–302) (Kelm et al. 1999) were a kind gift from Dr. Robert J. Kelm, Jr. (University of Vermont College of Medicine, Colchester). Other primary antibodies were generously provided by the following researchers: anti-Foxk1/MNF (Bassel-Duby et al. 1994) from Dr. Rhonda Bassel-Duby (University of Texas Southwestern Medical Center, Dallas), anti-n-PAC/NP60 (Fu et al. 2006) from Dr. Jun Gu (Peking University, Beijing), anti-Pax6 (Tang et al. 1997) from Dr. Yi-Hong Zhou (University of California, Irvine), and anti-SNEV1/Pso4 (Mahajan and Mitchell 2003) from Dr. Kiran Mahajan (University of North Carolina, Chapel Hill).
RESULTS
DNA affinity purification of proteins that form a complex on the ASE
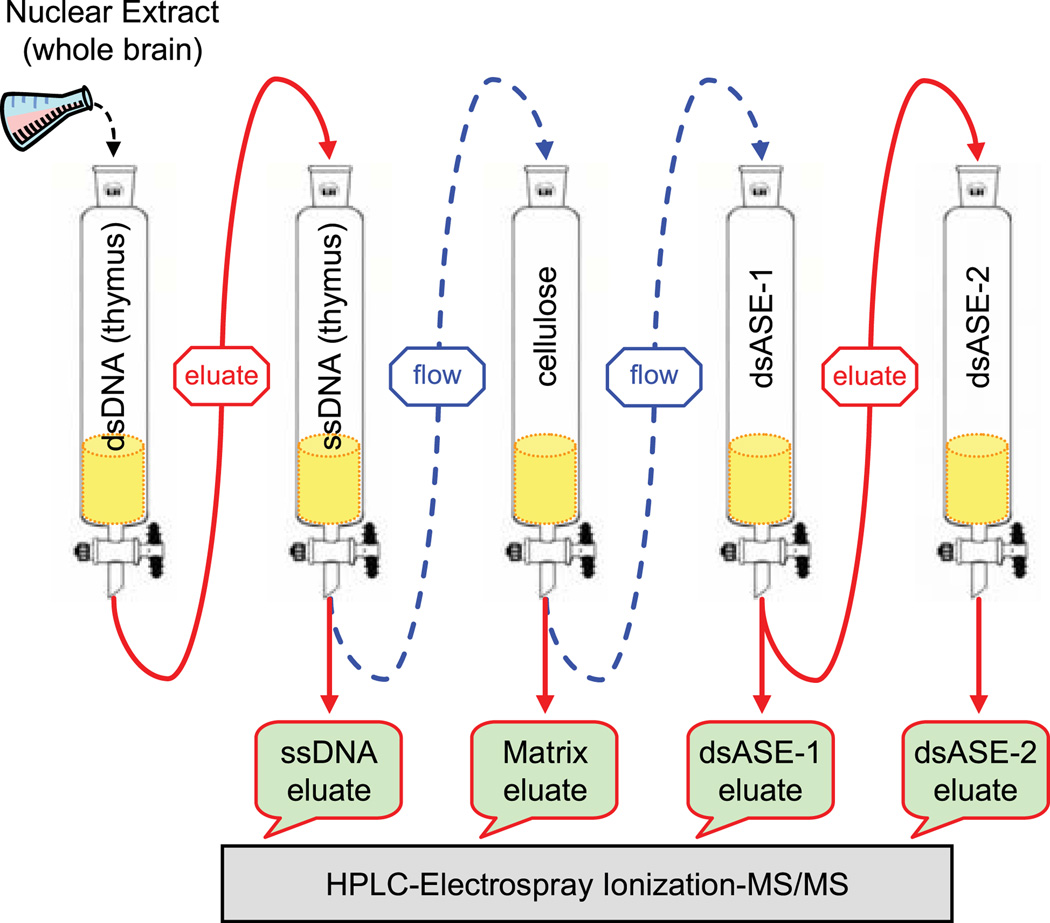
Analysis of proteins within higher-order complexes has been greatly aided by the advent of proteomics. In order to identify the proteins that form a complex on the ASE, the multi-protein complex was first enriched through a series of DNA affinity chromatographic steps. The purification scheme is depicted in Fig. 1. The initial starting material consisted of nuclear extracts prepared from the brains of mice around postnatal day 21 (P21) of age, which corresponds to the active myelination period of CNS development. [Roughly 400 mice were sacrificed in order to obtain enough starting material (40 mg of nuclear proteins) to ensure that sufficient quantities of proteins remained at the end of the purification process for subsequent mass spectrometry analysis.] At the outset, nuclear extracts were applied to a broad-spectrum, double-stranded DNA (dsDNA) column containing calf thymus DNA to enrich for DNA-binding proteins. Bound proteins were subsequently eluted and passed over a broad-spectrum, single-stranded (ssDNA) column containing calf thymus DNA to selectively reduce the amount of RNA- and single-stranded DNA-binding proteins in the fraction. The ensuing flow-through material and associated washes were combined and then applied to a column containing unconjugated cellulose (matrix alone) to remove proteins that inherently stick to the cellulose matrix, which were used later on to quantify the relative amounts of proteins bound between the matrix alone column and those containing the ASE sequence. The resulting flow-through material was then loaded onto a column of cellulose conjugated to the double-stranded ASE sequence (dsASE-1). Bound proteins were eluted and identified directly by mass spectrometry analysis or subjected to a second round of DNA affinity purification using a new column (dsASE-2).
Fig. 1.
DNA affinity purification scheme. Nuclear proteins that bind the ASE sequence specifically were purified through a series of chromatographic steps. Initially, nuclear extracts prepared from the brains of mice at P17–P25 of age were loaded onto a broad-spectrum, double-stranded DNA cellulose column (dsDNA from calf thymus) to enrich for DNA-binding proteins. Bound proteins were eluted and passed over a broad-spectrum, single-stranded (calf thymus) DNA cellulose column (ssDNA) to retain RNA/DNA single-strand binding proteins. The flow-through, enriched for dsDNA-binding proteins, was then passed over unconjugated cellulose to capture proteins that stuck to the matrix itself. The flow-through was subsequently loaded onto a cellulose column conjugated to the double-stranded ASE sequence (dsASE-1). Proteins that bound to the dsASE-1 column were eluted and a portion of the eluate subjected to a second round of purification using a fresh dsASE column (dsASE-2). Proteins eluted from the ssDNA, matrix alone (cellulose), dsASE-1, and dsASE-2 columns were fractionated by SDS-PAGE and subsequently identified by mass spectrometry analysis (high-performance liquid chromatography electrospray ionization mass spectrometry (HPLC-ESI-MS/MS). Arrows with dashed lines indicate flow-through material, while arrows with solid lines represent eluates.
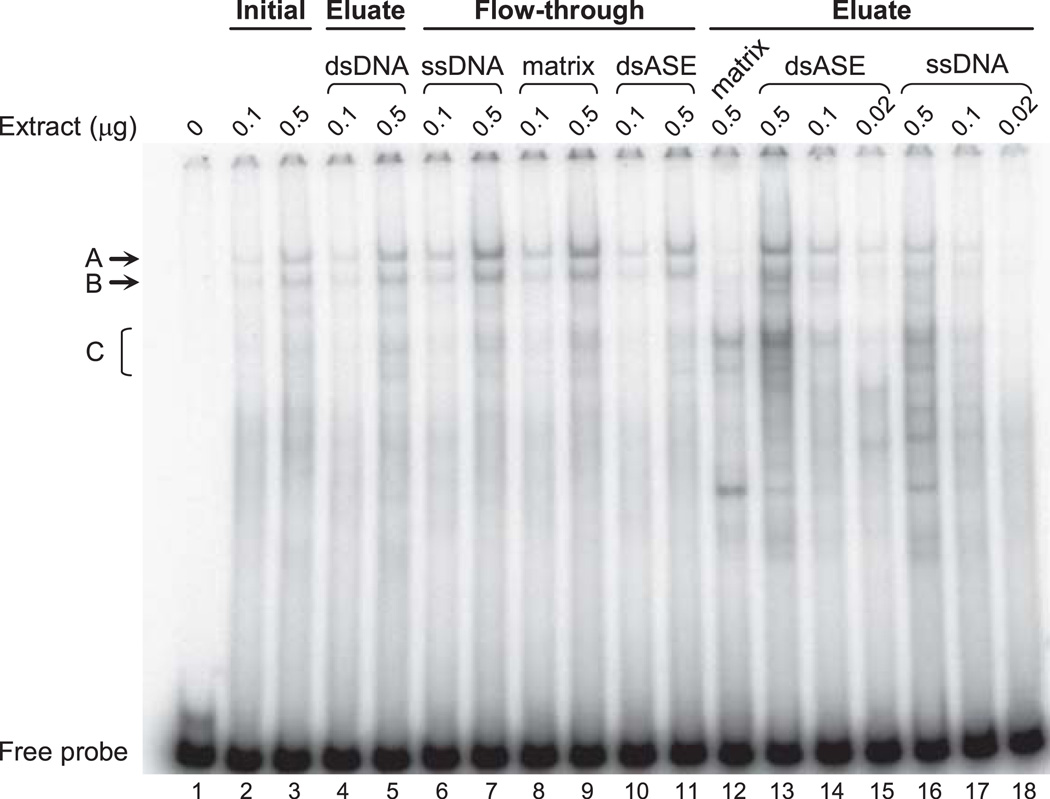
The steps along the DNA affinity purification process were monitored by EMSA for integrity of nucleoprotein complexes formed using the 85-bp ASE sequence (intron 1 positions 1,093–1,177) as a probe. Previously we have identified EMSA complexes which are formed due to sequence-specific interactions with nuclear proteins from mouse brain (Dobretsova et al. 2004). The sequence-specific complexes are indicated by the letters A, B and C in Fig. 2. EMSA analysis showed that nuclear proteins specifically bound to the ASE sequence, retain their DNA-binding activity throughout the purification process (Fig. 2). Moreover, there was enrichment in these factors during the purification process; compare intensities for complexes A, B and C from the dsASE eluate (lanes 13–15) with those from the initial nuclear extract (lanes 2–3). After the first purification step (to enrich for DNA-binding proteins with the dsDNA column), total protein content was only 27% compared with the initial starting material, without any loss in specific DNA-binding activity (compare lanes 2–3 with lanes 4–5). Subsequent passage of the dsDNA eluate over a ssDNA column resulted in further depletion of RNA- and single-stranded DNA-binding proteins from the mixture and a concomitant increase in specific DNA-binding activity [notice the relative increase in Complexes A and B in the ssDNA flow-through (lanes 6–7) compared to the dsDNA eluate (lanes 4–5), with preferential retention of faster mobility, nonspecific, DNA-protein complexes in the ssDNA eluate (lanes 16–18)]. Whereas proteins that bind the ASE sequence specifically did not demonstrate an inherent stickiness to the cellulose matrix itself [compare the low levels of Complex A and B using the matrix eluate (lane 12) to the high levels observed with the matrix flow-through (lanes 8–9)]. Thus the purification procedure, which has been optimized to support formation of a multi-protein complex, still retains proteins that bind to the ASE, specifically. Remarkable enrichment in sequence-specific binding was noted with the dsASE-1 column compared to the matrix alone (compare lanes 13–15 to lane 12). Significant levels of specific binding were still present in the flow-through from the dsASE-1 column (lanes 10–11) suggesting that the “ligand” (nuclear proteins forming the ASE complex) was present in excess relative to the “receptor” (ASE sequence), thereby reducing the chance for non-specific binding by virtue of titrating out proteins that bind specifically. The majority of eluate obtained from the dsASE-1 column was passed over a fresh dsASE column (dsASE-2) to further enrich for proteins that selectively bind the ASE. The resulting dsASE-2 flow-through showed a significant reduction in DNA binding activity relative to that from the dsASE-1 eluate (data not shown). The eluate from the dsASE-2 column was not analyzed by EMSA due to the limited amount of protein in this fraction.
Fig. 2.
EMSA analysis of fractions obtained during the DNA affinity purification process. Nuclear extracts prepared from the brains of mice (P17 to P25 of age) were subjected to DNA affinity purification using the scheme depicted in Fig. 1. Proteins from the initial nuclear extract and specified chromatographic fractions (eluate and flow-through) were analyzed for formation of nucleoprotein complexes using the 85-bp ASE sequence (intron 1 positions 1,093–1,177) as a probe. The amount (µg) of nuclear extract (protein) used in the reactions is indicated above each lane. DNA-protein complexes marked by the letters A, B and C have previously been shown to be due to sequence-specific interactions (Dobretsova et al. 2004). Fractions denoted dsASE were obtained with the dsASE-1 column, while ‘matrix’ fractions were obtained from the unconjugated cellulose column.
Identification of DNA affinity purified proteins by mass spectrometry analysis
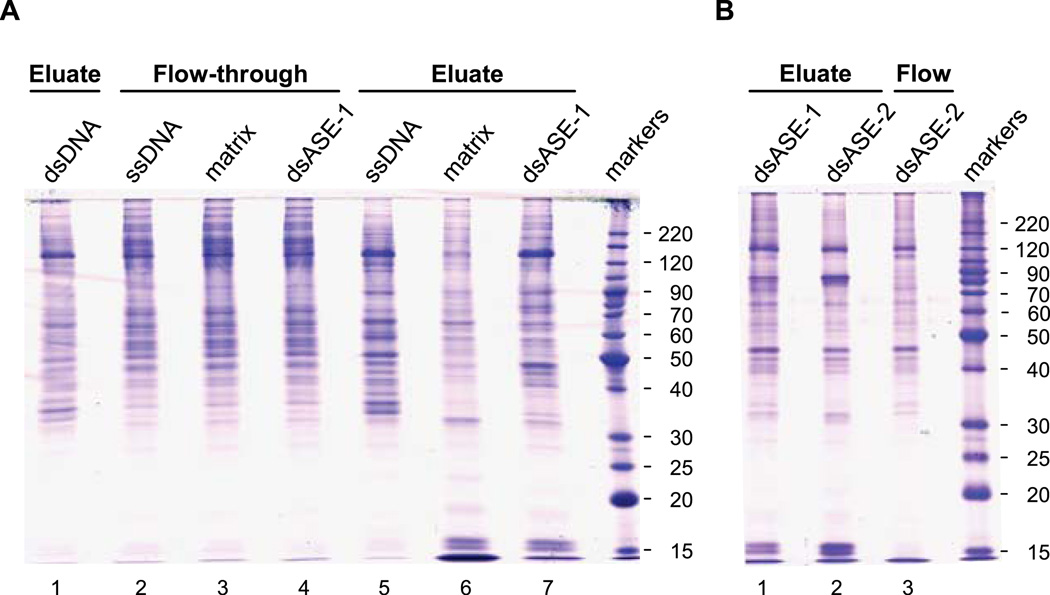
Proteins from the different fractions of DNA affinity purification were separated on SDS-PAGE gels and stained with Coomassie Blue. Equal amounts of total protein were loaded between the lanes of a given gel (5 and 3 µg/lane in Fig. 3A and B, respectively). A multifaceted staining pattern was observed, with perceptible differences between the fractions (Fig. 3). Several bands appeared enriched in eluates from the dsASE columns. However to obtain a complete list of eluted proteins, an entire lane was cut lengthwise into 3-mm gel slices and the proteins digested (in-gel) with trypsin. The resulting peptides were eluted from each gel slice and analyzed by mass spectrometry. This approach allows for high resolution peptide analysis of the eluates and the ability to correlate protein identification (i.e., peptide “hits”) with a particular gel slice, and hence the apparent molecular weight as determined by SDS-PAGE.
Fig. 3.
SDS-PAGE gels of DNA affinity purification fractions. Proteins from the indicated chromatographic fractions were resolved on an 11% SDS-PAGE gel and visualized by staining with Coomassie Blue. The gel in panel A contained a total of 5 µg of nuclear proteins per lane, while the gel in panel B contained only 3 µg/lane. The gels were placed on a clean plastic plate, and individual lanes cut sequentially into (3-mm) gel slices across their entire length. Proteins within the gel slices were digested (in-gel) with trypsin and the eluted peptides analyzed by mass spectrometry.
Proteins eluted from the various columns were identified by high-performance liquid chromatography electrospray ionization mass spectrometry (HPLC-ESI-MS/MS) analysis. Analysis 1 consisted of a crosswise comparison of the proteins eluted from the matrix alone (Fig. 3A, lane 6) and the dsASE-1 column (Fig. 3A, lane 7). Analysis 2 consisted of a crosswise comparison of proteins eluted from the matrix alone (Fig. 3A, lane 6) and those eluted from dsASE-1 and dsASE-2 columns (Fig. 3B, lanes 1 and 2, respectively). Due to the limited amount of protein eluted from the unconjugated matrix, there was not enough material to run this fraction on the gel depicted in Fig. 3B. [This further exemplifies the usefulness of cellulose as a matrix due to the low level of background binding.] Analysis 3 consisted of a crosswise comparison of the proteins eluted from the ssDNA and dsASE-1 columns (Fig. 3A, lanes 5 and 7, respectively). Table 1 shows a summary of the results of the three trials. Included in the table is the strategy for sorting the proteins “hits” deemed to be most relevant; i.e., those proteins that are enriched in the dsASE eluate fractions and retained fairly well on the dsASE-2 column.
Table 1.
Number of unique proteins identified in DNA affinity eluates by mass spectrometry*
| Analysis 1 | Analysis 2 | Analysis 3 | Combined | |||||
|---|---|---|---|---|---|---|---|---|
| Eluate | Matrix Fig. 3A, lane 6 |
dsASE-1 Fig. 3A, lane 7 |
Matrix Fig. 3A, lane 6 |
dsASE-1 Fig. 3B, lane 1 |
dsASE-2 Fig. 3B, lane 2 |
ssDNA Fig. 3A, lane 5 |
dsASE-1 Fig. 3A, lane 7 |
|
| Total number of proteins identified | 347 | 390 | 565 | 468 | 264 | 404 | 422 | 916 |
| Proteins in dsASE-1 & dsASE-2 combined | 589 | |||||||
| Proteins enriched in dsASE over matrix | dsASE-1 : matrix ≥ 1.5 | dsASE-1 : matrix ≥ 0.9** | 464 | |||||
| Proteins present in dsASE-2 | 207 | |||||||
| Proteins appreciably retained in dsASE-2 | dsASE-2 : dsASE-1 ≥0.5 | 143 | ||||||
The mass spectrometry analyses resulted in a total of 916 proteins being identified (Table 1). Proteins detected solely in the eluate from the ssDNA column (105) or unconjugated matrix (135), or only in both of these eluates (39), were not considered further thereby dropping the number of proteins of interest to 637. In addition, protein identifications made on the basis of a single peptide from the dsASE-1 eluate in only one of the three mass spectrometry trials were discarded from further analysis. All other proteins identified were considered further. This refinement reduced the total number down to 589 proteins of interest (i.e., proteins present in dsASE-1 and/or dsASE-2 eluates). The 589 proteins were analyzed for the total number of peptides per protein (peptide hits) as a rough estimate of their relative abundance within a given fraction. Proteins were considered potentially enriched if the ratio of number of peptides for dsASE-1 to matrix was ≥ 1.5 in Analysis 1 and ≥ 0.9 in Analysis 2 due to differences in the quantity of protein loaded between the gels, or simply greater than matrix in the case of dsASE-2 (see Table 2). By these criteria a total of 464 proteins appeared to be enriched in the dsASE eluates, of which, 207 were detected in the dsASE-2 eluate. The 207 remaining proteins were further sorted based on their relative “retention” (abundance) in the dsASE-2 eluate (number of peptide hits per protein ≥ 0.5 for dsASE-2 compared to dsASE-1 eluates). A total of 143 proteins met this criterion (Table 1). Since the ASE functions in a sequence-specific manner, the 143 remaining proteins were searched for reports of sequence-specific dsDNA binding using the International Protein Index (IPI) database housed on the European Bioinformatics Institute (EBI) server (http://www.ebi.ac.uk/IPI/IPIhelp.html) as well as by searching the scientific literature. Twenty-one sequence-specific DNA-binding factors were identified out of the 143 proteins (Table 2). In some cases, isoform-specific peptides generated by splice variants were detected (supplemental Table 1). The most abundant class of transcription factors detected were those that contain a basic leucine zipper (bZIP) domain, but other classes containing winged helix, homeobox, zinc finger, basic region-helix-loop-helix-leucine zipper (b/HLH/LZ) and HMG-I/(Y)/AT-hook domains were also identified (supplemental Table 1).
Table 2.
Number of peptide hits of sequence-specific DNA-binding proteins enriched in dsASE eluates*
| IPI ID # | Protein Name | Analysis 1 | Analysis 2 | Analysis 3 | ||||
|---|---|---|---|---|---|---|---|---|
| Matrix | dsASE-1 | Matrix | dsASE-1 | dsASE-2 | ssDNA | ASE-1 | ||
| IPI00131063 | Methyl CpG binding protein 2 (MeCP2) | 131 | 155 | 212 | 222 | 270 | 103 | 173 |
| IPI00458583 | hnRNP U | 58 | 204 | 164 | 264 | 259 | 283 | 235 |
| IPI00329976 | PREDICTED: Forkhead box K2 (Foxk2) | 4 | 19 | 13 | 34 | 32 | 9 | 33 |
| IPI00119924 | Isoform 1 of CREB | 6 | 27 | 8 | 34 | 31 | 5 | 14 |
| IPI00112473 | Poly [ADP-ribose] polymerase 1 (PARP1) | 4 | 16 | 9 | 14 | 35 | 31 | 20 |
| IPI00118447 | Transcriptional activator protein Purα | 0 | 7 | 9 | 17 | 22 | 16 | 12 |
| IPI00656285 | Forkhead box K1 (Foxk1) | 0 | 13 | 4 | 18 | 21 | 0 | 13 |
| IPI00230233 | Activating transcription factor 2 (ATF2) | 0 | 9 | 4 | 11 | 15 | 4 | 11 |
| IPI00111821 | Cytokine-like nuclear factor n-PAC homolog | 1 | 3 | 2 | 17 | 15 | 0 | 8 |
| IPI00128867 | Transcriptional activator protein Purβ | 1 | 11 | 1 | 8 | 12 | 16 | 17 |
| IPI00117877 | Transcription factor MafG | 2 | 2 | 6 | 8 | 9 | 0 | 6 |
| IPI00120886 | DNA-binding protein B (MSY1) | 0 | 5 | 3 | 8 | 8 | 0 | 3 |
| IPI00130597 | Bromodomain adjacent to zinc finger domain, 1B | 7 | 25 | 10 | 7 | 7 | 3 | 34 |
| IPI00126223 | Transcription factor JunD | 0 | 6 | 5 | 11 | 7 | 4 | 7 |
| IPI00274739 | DNA-binding protein A (MSY4) | 0 | 0 | 0 | 0 | 6 | 0 | 0 |
| IPI00137503 | Isoform 1 of Nuclear factor 1 X-type (NF1/X1) | 0 | 0 | 0 | 8 | 5 | 13 | 13 |
| IPI00118638 | Isoform 1 of Paired box protein 6 (Pax6) | 0 | 0 | 0 | 3 | 3 | 0 | 4 |
| IPI00310962 | Myc-associated protein X (Max) | 0 | 4 | 2 | 4 | 3 | 0 | 4 |
| IPI00117062 | Upstream stimulatory factor 1 (USF1) | 0 | 3 | 1 | 6 | 3 | 3 | 2 |
| IPI00121829 | Transcription factor AP-1 (c-Jun) | 0 | 0 | 0 | 2 | 2 | 0 | 0 |
| IPI00319017 | Transcription factor Sox2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
Proteins enriched in dsASE eluates compared to the unconjugated matrix eluate
As a means to evaluate, in silico, the effectiveness of the DNA affinity purification procedure for its capacity to preserve multi-protein complexes, the transcription factors listed in Table 2 were searched for possible interactions with other proteins enriched in the dsASE eluates (Table 1) using the IntAct database at EBI (http://ebi.ac.uk/intact/site). While not all of the factors listed in Table 2 were present in the database, a number of possible interacting proteins (from 9 to 64 potential partners) could be identified for MSY4, PARP1, Purα, Purβ, MSY1 and hnRNP U in the dsASE eluates (supplemental Table 2). Additionally, a search of iHOP (http://www.ihop-net.org; Hoffmann and Valencia 2004) and the scientific literature indicate that: 1) Sox2 and Pax6 are interacting partners (Kamachi et al. 2001); 2) ATF2 forms heterodimers with CREB and c-Jun; and 3) CREB is able to interact with c-Jun (Abdel-Hafiz et al. 1993). Which of these interactions are relevant in regulating Plp1 gene expression remains to be determined. However, the data strongly suggest that the DNA affinity purification procedure is permissive for the formation of multi-protein complex(es).
Acutely isolated oligodendrocytes from brain at the active myelination stage
Since purification of factors that selectively bind the ASE was performed using nuclear extracts prepared from whole brain, the possibility remains that some of the proteins identified in the mass spectrometry analyses are not relevant (i.e., they are expressed in non-oligodendroglia and are capable of binding to the dsASE sequence, in vitro). To address this issue, oligodendrocytes from the brains of mice at P16–21 of age were isolated via Percoll gradient centrifugation followed by indirect immunosorting using magnetic beads. The procedure was optimized for maximal yield of live cells with the O1 antigen being targeted for cell sorting. This oligodendrocyte marker (O1) reacts with galactosylcerebroside which is not expressed in late progenitor cells (Miller 2002), only in oligodendrocytes when they enter terminal differentiation (Bansal et al. 1989). In order to evaluate the effectiveness of cell sorting, fractions of freshly isolated cells were stained with an antibody specific for myelin basic protein (MBP), a marker for pre- and myelinating oligodendrocytes (see Supplementary Methods for details). Representative results obtained with mice at P17 of age are shown in supplemental Fig. 1. In the absence of primary antibody, very little background staining was observed: 0% in the initial (presorted) mixture (0/2083) and O1-depleted fraction (0/1948), and 2.5% (49/1970) in the O1-enriched fraction. Staining for reactivity with the MBP antibody showed that approximately 14% (350/2552) of the cells were MBP+ in the initial mixture, before sorting. [In general, glial cells are preferentially enriched in the initial mixture since neurons do not appear to survive the Percoll gradient isolation procedure very well, which is consistent with an earlier report in which neurons did not survive tissue dissociation (McCarthy and de Vellis 1980).] After sorting, 71% of cells in the O1-enriched pool were MBP+ (1564/2197), while only 0.8% (23/2978) of cells in the O1-depleted fraction stained with the MBP antibody. Consequently, there was a 5-fold increase in oligodendrocytes in the O1 enriched pool of cells compared to the initial mixture of cells isolated from brain, which was comprised mainly of glia.
Analysis of transcription factor expression in oligodendrocytes
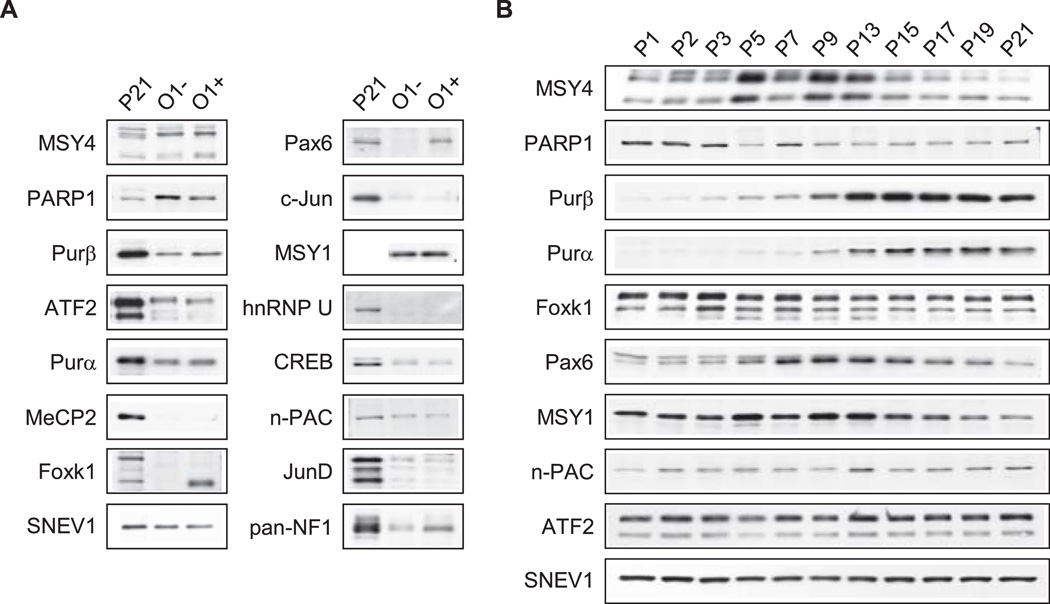
Nuclear extracts were prepared from O1-enriched and O1-depleted pools of cells obtained from brains of mice at P16–21 of age and assessed by Western blot analysis for expression of the transcription factors listed in Table 2, provided that suitable antibodies were readily available. Nuclear extracts were also prepared directly from whole brain of P21 mice (without any prior isolation of cells through a Percoll gradient) for comparison. This extract would contain nuclear proteins from all of the cells in the brain, including neurons, which are preferentially lost in the O1-enriched and O1-depleted fractions as indicated earlier. Hence, the nuclear proteins isolated from whole brain represent the pool of proteins used in the DNA affinity purification procedure. However, certain proteins may be absent in both the O1-enriched and O1-depleted fractions due to selective loss of some cell types. Results from the Western blot analysis are shown in Fig. 4. Due to interference in determination of nuclear protein concentrations with the O1-sorted fractions, approximately 2-fold less protein was loaded compared to whole brain as determined by Ponceau S staining of the membranes following protein transfer (data not shown), and supported by Western blot analysis for SNEV1, which appears to be constitutively expressed in nuclei. The interference is likely due to the presence of small amounts of antibody-bound magnetic beads that co-precipitated with nuclei during extraction. Even so, differences in several transcription factors were evident between the various extracts (Fig. 4A). Pax6 and Foxk1 were detected in nuclear extracts prepared from whole brain (P21) and the O1-positive pool, but not from the O1-depleted pool, suggesting that they are mainly expressed in pre- and myelinating oligodendrocytes. Levels of Purβ and Purα were higher in the O1-enriched pool compared to the O1-depleted pool, while MSY1, MSY4, and n-PAC demonstrated relatively equal distribution between the sorted pools. PARP1 levels were highest in nuclear extracts from O1-depleted cells, although the level from the O1-enriched pool was more abundant than for whole brain. Very low levels of c-Jun, CREB, JunD and ATF2 were detected in the sorted pools compared to those from whole brain. MeCP2 and hnRNP U were not detected in nuclear extracts prepared from either of the O1-sorted pools, although both were present in nuclear extracts from whole brain indicating that the proteins are expressed in other (non-oligodendroglial) cell types. We were not able to find a specific antibody directed against NF1/X1, while data with a pan-specific NF1 antibody were inconclusive.
Fig. 4.
Western blot analysis of transcription factors in brain. Western blot analysis was performed with some of the transcription factor “candidates” listed in Table 2. A: Cells were isolated from the brains of mice between P16 and P21 of age and sorted into O1-depleted (O1−) and O1-enriched (O1+) pools. Nuclear extracts were prepared from the sorted pools, and directly from whole brain (P21), and analyzed for expression of the indicated transcription factors. Despite every attempt to load equal amounts of protein, the immunosorted fractions contained roughly 2-fold less protein compared to nuclear extracts from whole brain as determined by Ponceau S staining of the membranes following protein transfer (data not shown), and supported by Western blot analysis for SNEV1, which appears to be constitutively expressed in nuclei. New blots were used for all analyses except SNEV1, which was first incubated with an anti-Purβ antibody, then stripped and incubated with an antibody against SNEV1. [Nuclear extract from P21 brain was not included in the analysis of MSY1]. B: Nuclear extracts were prepared from the brains of mice at the indicated ages (postnatal days) and analyzed by Western blot for expression of transcription factors. New blots were used for all analyses except SNEV1, which was first incubated with an anti-Purα antibody, then stripped and incubated with an antibody against SNEV1.
Developmental profile of transcription factor expression in brain
Whole brain nuclear extracts were prepared from mice between P1 and P21 of age to determine the developmental profile of transcription factor candidates that bind to the ASE. Western blot analysis was performed to assess the relative levels of transcription factors during the pre- and active myelination period of CNS development (Fig. 4B); in mice, Plp1 gene transcription in brain rises significantly two weeks after birth and peaks during the third postnatal week (Macklin et al. 1991). Factors important in regulating Plp1 gene activity may also be regulated similarly, although constitutive expression of a candidate factor would not necessarily rule it out as a potential regulator. Purα and Purβ demonstrated developmental profiles consistent with that for PLP. Several proteins exhibited transient elevation in their levels: MSY1 and MSY4 (elevated between P5–13), and one isoform of Pax6 (elevated between P5–19). The Foxk1 isoform present in nuclei from O1-enriched cells also exhibited a transient increase between P3–7; another Foxk1 isoform (faint lower band in Fig. 4B), which is not present in the O1-enriched pool, showed a transient increase between P3–P13. PARP1 levels were higher in pre-myelinating brain (P1–3), while n-PAC levels were slightly higher during the later time points. Levels for ATF2 and SNEV1 remained constant throughout the study, strongly suggesting that the factors are constitutively expressed in brain during this period.
Regional distribution of transcription factors in brain
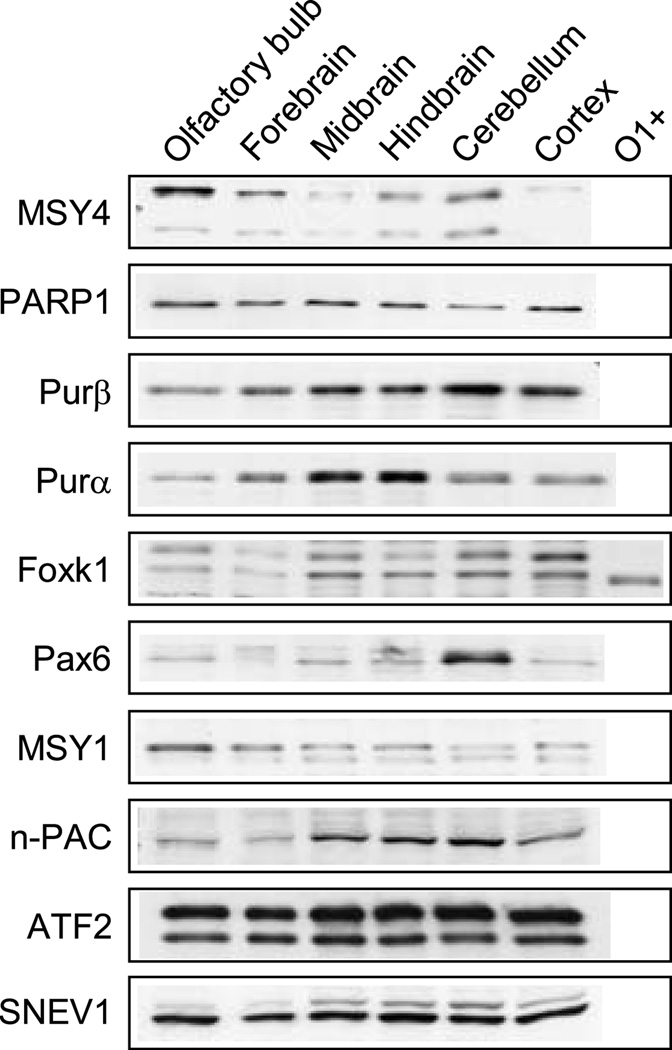
Since oligodendrocytes are differentially distributed in brain, brains from mice at P19–21 of age were broadly dissected into several regions according to the methods of Glowinski and Iverse (1966). Nuclear extracts were prepared from the regions (olfactory bulb, forebrain, midbrain, hindbrain, cerebellum, and cortex) and individually assessed for transcription factor expression by Western blot analysis. Only factors which demonstrated enhanced or sustained levels of expression in the O1-enriched fraction in Fig. 4A were examined. As shown in Fig. 5, all of the factors exhibited regional differences in their levels except for PARP1, ATF2 and SNEV1. Levels of MSY4 and MSY1 were highest in olfactory bulb, while the cortex demonstrated extremely low levels of MSY4 expression. In contrast, Pax6 expression was significantly elevated in cerebellum compared with the other regions. Levels of the Foxk1 isoform present in oligodendrocytes appeared highest in midbrain, hindbrain, cerebellum, and cortex. Purβ expression also showed a similar expression pattern, with the highest level present in cerebellum. Relative levels of Purα were highest in midbrain and hindbrain, while those for n-PAC were highest in midbrain, hindbrain and cerebellum. Regional distribution of individual transcription factors in brain have previously been noted for Foxk1 (Wijchers 2006) and Sox2 (Komitova and Eriksson 2004), as well as globally, in microarray studies (Sandberg et al. 2000).
Fig. 5.
Western blot analysis of transcription factors in brain regions. Nuclear extracts were prepared from the indicated brain regions obtained from mice at P19–21 of age and analyzed for expression of the specified transcription factors. Nuclear extract prepared from O1-enriched cells (O1+) was also included in the analysis of Foxk1 expression (see Fig. 4 for further details). New blots were used for all analyses except in the case of SNEV1, where the blot was first incubated with an anti-n-PAC antibody. The SNEV1-reactive band is the lower, darker band, while the upper band signifies residual staining against n-PAC.
DISCUSSION
A number of genes, including the Plp1 gene, contain intronic enhancers which are important cis-regulatory elements required to achieve maximal levels of expression, usually in a time- or tissue-specific manner (reviewed in Fedorova and Fedorov 2003). The intronic enhancer located within intron 1 of the Plp1 gene, which we refer to as ASE, is an important mediator of Plp1 gene activity in oligodendrocytes. Specifically, we believe the enhancer helps govern the surge in Plp1 gene activity during the active myelination period of CNS development. DNA affinity chromatography was optimized to enrich for factors that bind the 85-bp ASE sequence, specifically. Nuclear proteins extracted from whole brain were used as the starting material since it is not feasible to obtain enough protein for DNA affinity purification from myelinating oligodendrocytes. Brains were harvested from mice at ages consistent with the active myelination period. Salt concentrations used during the chromatographic DNA binding reactions approximated that in the nucleus to sustain formation of a multi-protein complex on the ASE. EMSA analysis, used to monitor DNA binding activity during the purification process, confirmed that specific nucleoprotein complexes were still present in eluates from the dsASE columns (Fig. 2). DNA affinity purification protocols often use salt concentrations that exceed physiological conditions and contain detergent concentrations that supersede micelle formation. These conditions effectively reduce non-specific protein binding since they prevent weak interactions. However this approach is generally counterproductive for the purification of large protein complexes, where weak interactions are often involved. Therefore the procedure used here focused on optimizing a purification scheme which uses conditions of low stringency for binding to allow formation of large protein complexes on the ASE sequence. DNA binding activity, as monitored by EMSA, demonstrated continuous enrichment in specific binding throughout the purification procedure (Fig. 2). It is important to note that proteins were eluted from columns with high salt solutions (up to 2 M), which inevitably causes irreversible denaturation of a proportion of proteins present in the eluate. Despite this shortcoming we were able to observe enrichment in specific binding activity, but precise quantization by EMSA is not feasible.
We conclude that the DNA affinity purification procedure was successful based on the following criteria: 1) the total yield of protein eluted from the dsASE-1 column was 7.5 times higher than that from the unconjugated matrix column (see Experimental Methods) indicating a higher level of binding to DNA compared to the matrix, which exhibits low levels of background binding; 2) consistently, 73% of the proteins detected (464/637) were enriched in the dsASE eluates compared to matrix alone; 3) 45% of these proteins (207/464) were detected in the dsASE-2 eluate, which represents 39% of the total amount loaded on the dsASE-2 column and demonstrates consistent binding to the same DNA sequence; 4) EMSA analysis demonstrated noticeable enrichment of specific binding activity in the dsASE-1 eluate, concomitant with a decrease in specific binding activity in the dsASE flow-through fractions; 5) proteins known to interact with some of the sequence-specific DNA-binding proteins enriched in the dsASE-2 eluate were also detected in the purified pool of proteins, suggesting that the procedure supports in vitro formation of stable multi-protein complexes on the dsASE sequence (supplemental Table 2). To the best of our knowledge this is one of the first examples of DNA affinity chromatography using such a long unconcatamerized regulatory sequence as bait.
From the 143 proteins consistently identified in the dsASE-2 eluate, 24 were sequence-specific DNA-binding proteins. Three of them showed a significant drop in the number of peptide hits compared with the dsASE-1 eluate (indicative of inconsistent retention) and thus were excluded from further analysis. [The total number of peptides detected by mass spectrometry for a given protein provides a rough estimate of the protein’s relative abundance (Gao et al. 2005) and was used merely to rank order the candidate transcription factors worthy of further study, based on the assumption that all proteins had an equal chance of being identified with the purification procedure described here.] Fifteen out the 21 remaining sequence-specific DNA-binding proteins were evaluated by Western blot analysis based on their relatively high abundance in the dsASE-2 eluate and the availability of specific antibodies. Initially, the factors were screened for presence in oligodendrocytes using nuclear extracts prepared from O1-enriched cells obtained from mouse brain. The procedure used to isolate cells from brain is very harsh on neurons. Thus the O1-depleted pool of cells does not represent the entire population of non-oligodendroglial cells from brain. For instance MeCP2, which is highly expressed in neurons (Johnston et al. 2005), was neither detected in the O1-enriched or O1-depleted pools by Western blot analysis, although it was present in nuclear extracts prepared directly from whole brain, as expected (Fig. 4A). Hence, MeCP2 was excluded as a potential regulator of Plp1 gene activity in oligodendrocytes.
Nine of the topmost transcription factors detected in nuclei from the O1-enriched pool of cells were evaluated to assess their developmental profile and regional distribution in brain. We consider transcription factors which demonstrate developmental dynamics consistent with Plp1 gene expression and are present at higher levels in heavily myelinated brain regions to be among the top candidates as regulators of Plp1 gene expression. As such, Purα and Purβ showed an increase in expression consistent with the active myelination period of CNS development (Fig. 4B), with Purα (and Purβ to a lesser extent) being preferentially expressed in heavily myelinated brain regions (hindbrain and midbrain; Fig. 5). These factors, along with MSY1 and MSY4 (which were also detected in the dsASE eluates), are known to form homo- and heterodimers through DNA-dependent and independent means that have varying effects on transcription dependent upon the particular subunit pairing (IntAct database). Binding to DNA is through purine/pyrimidine-rich elements. Consistently, the ASE contains a couple of purine/pyrimidine-rich stretches (positions 1,101–1,106 and 1,160–1,164) within two sub-regions which: 1) are necessary for orientation-independent function of the ASE; 2) demonstrate similar patterns of protein binding by EMSA; 3) are effective cross-competitors for specific binding to nuclear proteins by EMSA (Dobretsova and Wight 1999; Dobretsova et al. 2000). Purα, Purβ and MSY1 have been reported to activate expression of a number of genes in the CNS, including Mbp (reviewed in Gallia et al. 2000). These proteins were also identified as part of a repressor complex for the vascular smooth muscle α-actin gene enhancer (Kelm et al. 1999), and their human counterparts have been shown to repress fas promoter activity (Lasham et al. 2000). Interestingly, when expression constructs of the relevant transcription factors were co-transfected along with a fas promoter-reporter construct, considerably lower levels of repression were obtained with expression constructs derived from different species (human and rat). This is somewhat akin to the ASE, where substitution of the human orthologous sequence in lieu of the mouse ASE sequence in a Plp1-lacZ fusion gene failed to enhance reporter gene expression in transfected N20.1 cells, whilst the more conserved orthologous sequence from rat, did so (Meng et al. 2005). Presumably a multi-protein complex would be far more sensitive to differences between species compared to the binary interaction between a single factor and its cognate regulatory element.
Other similarities also exist between the fas promoter and the ASE. In addition to Pur and cold-shock domain proteins, the human fas promoter is also regulated by the bZIP proteins c-Jun and c-Fos (Lasham et al. 2000). A functional AP-1 element is located in very close proximity to where Pur/cold-shock proteins bind, and as a result, affect fas promoter activity. Likewise, five bZIP family members were selectively retained in the dsASE eluates: ATF2, c-Jun, CREB, JunD and MafG (Table 2). Previous results with site-directed mutagenesis to disrupt AP-l-like motifs within the ASE suggest that these sites are critical for the enhancer’s ability to function (Dobretsova et al. 2004).
Purα deficient mice die prematurely, around 3 weeks after birth, and exhibit a number of CNS abnormalities including reduced numbers of myelinated tracts and MBP-positive cells in the subcortical white matter (Khalili et al. 2003). Our results indicate that the purine/pyrimidine track binding factors, Purα and Purβ, and associated cold-shock domain factors, MSY1 and MSY4, are not uniformly distributed in brain around 3 weeks postnatal (Fig. 5). The functional significance of these intriguing regional differences remains to be elucidated. This interesting group of multi-potential proteins can shuttle between the nucleus and cytoplasm and have an effect on a wide variety of processes including transcription, DNA replication, RNA transport and translation (Khalili et al. 2003).
The transcription factors Foxk1 and Pax6 were also selectively retained in the dsASE eluates (Table 2) and demonstrated much higher levels in nuclei from oligodendrocytes (O1-enriched cells) compared to the O1-depleted pool of cells from brain (Fig. 4A). Foxk2, as well, was selectively retained in the dsASE eluates, but was not evaluated by Western blot analysis due to lack of a specific antibody immunoreactive against the mouse protein. Foxk2 is the mouse ortholog of human interleukin enhancer binding factor 1 (ILF1), demonstrating 93% identity at the amino acid level. ILF1 binds to purine-rich sequences in the interleukin-2 (IL-2) promoter and to several similar motifs in the HIV long terminal repeat (LTR), and affects the expression of genes in cis (Li et al. 1991). In yeast, the Foxk2 homolog (Fkh2p) has been shown to control transcription elongation of target genes (Morillon et al. 2003).
The closest relative to Foxk2 in the forkhead/winged-helix (Fox) transcription factor family (Katoh and Katoh 2004) is Foxk1/MNF (myocyte nuclear factor). However, Foxk1 binds DNA through an entirely different motif (Bassel-Duby et al. 1994). Two alternative splice isoforms (α and β) can be generated from the Foxk1 primary transcript encoding proteins that may exert opposing effects on expression of target genes (Garry et al. 2000; Yang et al. 2000). It is intriguing that both isoforms were detected in the dsASE-2 eluate (supplemental Table 1). It is possible that different cell types in brain express different isoforms or that there is an interchange between isoform expression during development for a given cell type. Interestingly, only a single isoform was detected in nuclei from oligodendroglial (O1-enriched) cells (Fig. 4). However, the exact identity of the Foxk1 isoform expressed in the O1-enriched pool awaits further study.
Cytokine-like nuclear factor n-PAC or NP60 was also selectively retained in dsASE eluates (Table 2). The nuclear protein was only recently identified (Fu et al. 2006) and contains a unique collection of motifs (Conserved Domain Database; Marchler-Bauer et al. 2007) including two nuclear localization sequences, a PWWP domain (PF00855) and an AT-hook (PF02178) at the N-terminus, and an NAD-binding domain (PF03446) similar to that in 6-phosphogluconate dehydrogenase at its C-terminus. The protein specifically interacts with p38α, leading to phosphorylation of p38α, which subsequently results in the phosphorylation and activation of ATF2, although n-PAC/NP60 itself does not possess kinase activity (Fu et al. 2006). The initial report by Fu et al. (Fu et al. 2006) only described n-PAC/NP60 expression in various cells lines, primarily those derived from the immune system. Here we demonstrate that the protein is expressed in nuclei from brain, with the highest levels occurring in heavily myelinated regions (midbrain, hindbrain and cerebellum) from mice at ages consistent with the active myelination period of development (Fig. 5).
PARP1 was also selectively retained in dsASE eluates (Table 2). The protein, which contains an NAD-binding domain, is an abundant nuclear factor that exhibits both sequence-specific and non-specific DNA binding. It is involved in a wide variety of processes including DNA repair, cell death, mitotic apparatus function, chromatin regulation, and transcription regulation (reviewed in Kim et al. 2005). In the process of optimizing conditions for DNA chromatography, PARP1 was also selectively retained when a single-stranded oligo(dT) linker was used to attach the dsASE sequence to the cellulose matrix (Dobretsova et al. 2005), instead of the polyethylene glycol linker used in the current study. Initially we thought that PARP1 might have been selectively retained on the dsASE column containing the oligo(dT) linker not due to sequence-specific recognition of the ASE sequence itself, but because attachment of the single stranded oligo(dT) linker to the dsASE sequence mimics a strand break that might attract PARP1. However a single strand break would not be present with the current arrangement using a polyethylene glycol linker to attach the dsASE sequence to the matrix. Thus, selective retention of PARP1 in the current studies suggests that PARP1 may actually play a role in regulating Plp1 gene expression. This notion is further supported by selective retention of partners that interact with PARP1 after two successive rounds of purification with the dsASE column (supplemental Table 2). However in addition to single strand breaks, PARP1 can also recognize DNA double strand breaks (Vidaković et al. 2005). Therefore PARP1 may have been retained on the dsASE columns simply by virtue of its binding to the free terminus of the ASE sequence. Chromatin immunoprecipitation studies are currently underway in our laboratory to help resolve this issue.
In conclusion, the current studies have resulted in the identification of multiple transcription factor candidates that bind to the ASE and as a result may augment the level of Plp1 gene transcription in oligodendrocytes during development. Importantly, these factors were identified from nuclei directly isolated from brain during the active myelination period of development. Results presented here are an important first step in the identification of factors that regulate myelin gene activity during this developmental phase. Future studies will reveal which of these factors modulate Plp1 gene activity, in vivo. However in the current report, we have demonstrated that nuclear expression of several of the factors is developmentally regulated in whole brain, and that most of the factors are not uniformly distributed throughout the brain. Moreover we have established which of these factors are present in nuclei from oligodendrocytes (O1-enriched pool) freshly isolated from whole brain of mice around P21 of age. To the best of our knowledge, this is the first report to characterize transcription factors present in nuclei of oligodendrocytes, which were acutely isolated from brains of post-neonatal mice. These results, undoubtedly, will be applicable in understanding the regulation of a much broader array of genes expressed in brain. As mentioned earlier, this study is also one of the few to use such as long, unique sequence as bait to capture sequence-specific DNA-binding proteins. Wang and colleagues (2007) recently used a 151-bp biotinylated probe to purify proteins that interact with the promoter region of the tr1 gene from A. phagocytophilum by DNA affinity chromatography. However in that study, only two proteins from the two most intensely stained, low molecular weight bands could be identified by mass spectrometry analysis. Hence, the methods for DNA affinity purification/mass spectrometry used here should be of general interest to researchers engaged in the purification and subsequent identification of DNA-binding proteins, especially with regards to those proteins involved in multi-protein complexes.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Kelly McCastlain and G. Bruno Pereira for technical assistance and to Drs. Michael Jennings, Vladimir Lupashin and Kevin Raney for helpful advice regarding protein purification. We are also indebted to Drs. Robert J. Kelm, Jr., Rhonda Bassel-Duby, Jun Gu, Yi-Hong Zhou, and Kiran Mahajan for gifts of antibodies. This work was supported by grants from the National Institutes of Health (NS037821 and NS047546) and a grant from the National Multiple Sclerosis Society (RG 2705).
Abbreviations used
- AP1
activator protein 1
- β-gal
β-galactosidase
- b/HLH/LZ
basic region-helix-loop-helix-leucine
- bZIP
basic leucine zipper
- EMSA
electrophoretic mobility shift assay(s)
- ESI
electrospray ionization
- FBS
fetal bovine serum
- FDR
false discovery rate
- HMG
high mobility group
- HPLC
high-performance liquid chromatography
- MBP
myelin basic protein
- MS
mass spectrometry
- P
postnatal day
- PLP
myelin proteolipid protein
- Plp1
myelin proteolipid protein gene
REFERENCES
- Abdel-Hafiz HA-M, Chen C-y, Marcell T, Kroll DJ, Hoeffler JP. Structural determinants outside of the leucine zipper influence the interactions of CREB and ATF2: interaction of CREB with ATF2 blocks E1a–ATF-2 complex formation. Oncogene. 1993;8:1161–1174. [PubMed] [Google Scholar]
- Alvarez M, Rhodes SJ, Bidwell JP. Context-dependent transcription: all politics is local. Gene. 2003;313:43–57. doi: 10.1016/s0378-1119(03)00627-9. [DOI] [PubMed] [Google Scholar]
- Bansal R, Warrington AE, Gard AL, Ranscht B, Pfeiffer SE. Multiple and novel specificities of monoclonal antibodies O1, O4, and R-mAb used in the analysis of oligodendrocyte development. J. Neurosci. Res. 1989;24:548–557. doi: 10.1002/jnr.490240413. [DOI] [PubMed] [Google Scholar]
- Bassel-Duby R, Hernandez MD, Yang Q, Rochelle JM, Seldin MF, Williams RS. Myocyte nuclear factor, a novel winged-helix transcription factor under both developmental and neural regulation in striated myocytes. Mol. Cell. Biol. 1994;14:4596–4605. doi: 10.1128/mcb.14.7.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumann N, Pham-Dinh D. Biology of oligodendrocyte and myelin in the mammalian central nervous system. Physiol. Rev. 2001;81:871–927. doi: 10.1152/physrev.2001.81.2.871. [DOI] [PubMed] [Google Scholar]
- Bensadoun A, Weinstein D. Assay of proteins in the presence of interfering materials. Anal. Biochem. 1976;70:241–250. doi: 10.1016/s0003-2697(76)80064-4. [DOI] [PubMed] [Google Scholar]
- Berti-Mattera LN, Larocca JN, Pellegrino de Iraldi A, Pasquini JM, Soto EF. Isolation of oligodendroglial cells from young and adult whole rat brains using an in situ generated Percoll density gradient. Neurochem. Int. 1984;6:41–50. doi: 10.1016/0197-0186(84)90024-x. [DOI] [PubMed] [Google Scholar]
- Carey M. The enhanceosome and transcriptional synergy. Cell. 1998;92:5–8. doi: 10.1016/s0092-8674(00)80893-4. [DOI] [PubMed] [Google Scholar]
- Carson MJ, Reilly CR, Sutcliffe JG, Lo D. Mature microglia resemble immature antigen-presenting cells. Glia. 1998;22:72–85. doi: 10.1002/(sici)1098-1136(199801)22:1<72::aid-glia7>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- Dobretsova A, Kokorina NA, Wight PA. Functional characterization of a cis-acting DNA antisilencer region that modulates myelin proteolipid protein gene expression. J. Neurochem. 2000;75:1368–1376. doi: 10.1046/j.1471-4159.2000.0751368.x. [DOI] [PubMed] [Google Scholar]
- Dobretsova A, Kokorina NA, Wight PA. Potentiation of myelin proteolipid protein (Plp) gene expression is mediated through AP-1-like binding sites. J. Neurochem. 2004;90:1500–1510. doi: 10.1111/j.1471-4159.2004.02683.x. [DOI] [PubMed] [Google Scholar]
- Dobretsova A, Lichti CF, Wight PA. Characterization of the enhanceosome formed on the myelin proteolipid protein gene by DNA affinity chromatography and mass spectrometry. J. Neurochem. 2005;94(Suppl. 1):35. [Google Scholar]
- Dobretsova A, Wight PA. Antisilencing: myelin proteolipid protein gene expression in oligodendrocytes is regulated via derepression. J. Neurochem. 1999;72:2227–2237. doi: 10.1046/j.1471-4159.1999.0722227.x. [DOI] [PubMed] [Google Scholar]
- Fedorova L, Fedorov A. Introns in gene evolution. Genetica. 2003;118:123–131. [PubMed] [Google Scholar]
- Fu J, Yang Z, Wei J, Han J, Gu J. Nuclear protein NP60 regulates p38 MAPK activity. J. Cell Sci. 2006;119:115–123. doi: 10.1242/jcs.02699. [DOI] [PubMed] [Google Scholar]
- Gallia GL, Johnson EM, Khalili K. Purα: a multifunctional single-stranded DNA- and RNA-binding protein. Nuc. Acids Res. 2000;28:3197–3205. doi: 10.1093/nar/28.17.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gambus A, Jones RC, Sanchez-Diaz A, Kanemaki M, van Deursen F, Edmondson RD, Labib K. GINS maintains association of CDC45 with MCM in replisome progression complexes at eukaryotic DNA replication forks. Nat. Cell Biol. 2006;8:358–366. doi: 10.1038/ncb1382. [DOI] [PubMed] [Google Scholar]
- Gao J, Friedrichs MS, Dongre AR, Opiteck GJ. Guidelines for the routine application of the peptide hits technique. J. Am. Soc. Mass Spectrom. 2005;16:1231–1238. doi: 10.1016/j.jasms.2004.12.002. [DOI] [PubMed] [Google Scholar]
- Garry DJ, Meeson A, Elterman J, Zhao Y, Yang P, Bassel-Duby R, Williams RS. Myogenic stem cell function is impaired in mice lacking the forkhead/winged helix protein MNF. Proc. Natl. Acad. Sci. USA. 2000;97:5416–5421. doi: 10.1073/pnas.100501197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glowinski J, Iversen LL. Regional studies of catecholamines in the rat brain–I. The disposition of [3H]norepinephrine, [3H]dopamine and [3H]DOPA in various regions of the brain. J. Neurochem. 1966;13:655–669. doi: 10.1111/j.1471-4159.1966.tb09873.x. [DOI] [PubMed] [Google Scholar]
- Gorski K, Carneiro M, Schibler U. Tissue-specific in vitro transcription from the mouse albumin promoter. Cell. 1986;47:767–776. doi: 10.1016/0092-8674(86)90519-2. [DOI] [PubMed] [Google Scholar]
- Hoffmann R, Valencia A. A gene network for navigating the literature. Nat. Genet. 2004;36:664. doi: 10.1038/ng0704-664. [DOI] [PubMed] [Google Scholar]
- Johnston MV, Blue ME, Naidu S. Rett syndrome and neuronal development. J. Child Neurol. 2005;20:759–763. doi: 10.1177/08830738050200091101. [DOI] [PubMed] [Google Scholar]
- Kamachi Y, Uchikawa M, Tanouchi A, Sekido R, Kondoh H. Pax6 and SOX2 form a co-DNA-binding partner complex that regulates initiation of lens development. Genes Dev. 2001;15:1272–1286. doi: 10.1101/gad.887101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh M, Katoh M. Human FOX gene family (Review) Int. J. Oncol. 2004;25:1495–1500. [PubMed] [Google Scholar]
- Kelm RJ, Jr, Cogan JG, Elder PK, Strauch AR, Getz MJ. Molecular interactions between single-stranded DNA-binding proteins associated with an essential MCAT element in the mouse smooth muscle α-actin promoter. J. Biol. Chem. 1999;274:14238–14245. doi: 10.1074/jbc.274.20.14238. [DOI] [PubMed] [Google Scholar]
- Khalili K, Del Valle L, Muralidharan V, Gault WJ, Darbinian N, Otte J, Meier E, Johnson EM, Daniel DC, Kinoshita Y, Amini S, Gordon J. Purα is essential for postnatal brain development and developmentally coupled cellular proliferation as revealed by genetic inactivation in the mouse. Mol. Cell. Biol. 2003;23:6857–6875. doi: 10.1128/MCB.23.19.6857-6875.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MY, Zhang T, Kraus WL. Poly(ADP-ribosyl)ation by PARP-1: ‘PAR-laying’ NAD+ into a nuclear signal. Genes Dev. 2005;19:1951–1967. doi: 10.1101/gad.1331805. [DOI] [PubMed] [Google Scholar]
- Komitova M, Eriksson PS. Sox-2 is expressed by neural progenitors and astroglia in the adult rat brain. Neurosci. Lett. 2004;369:24–27. doi: 10.1016/j.neulet.2004.07.035. [DOI] [PubMed] [Google Scholar]
- Lasham A, Lindridge E, Rudert F, Onrust R, Watson J. Regulation of the human fas promoter by YB-1, Purα, and AP-1 transcription factors. Gene. 2000;252:1–13. doi: 10.1016/s0378-1119(00)00220-1. [DOI] [PubMed] [Google Scholar]
- Li C, Lai C, Sigman DS, Gaynor RB. Cloning of a cellular factor, interleukin binding factor, that binds to NFAT-like motifs in the human immunodeficiency virus long terminal repeat. Proc. Natl. Acad. Sci. USA. 1991;88:7739–7743. doi: 10.1073/pnas.88.17.7739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Moore CL, Dobretsova A, Wight PA. Myelin proteolipid protein (Plp) intron 1 DNA is required to temporally regulate Plp gene expression in the brain. J. Neurochem. 2002;83:193–201. doi: 10.1046/j.1471-4159.2002.01142.x. [DOI] [PubMed] [Google Scholar]
- Liu H, Sadygov RG, Yates JR., 3rd A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- Macklin WB, Gardinier MV, Obeso ZO, King KD, Wight PA. Mutations in the myelin proteolipid protein gene alter oligodendrocyte gene expression in jimpy and jimpymsd mice. J. Neurochem. 1991;56:163–171. doi: 10.1111/j.1471-4159.1991.tb02576.x. [DOI] [PubMed] [Google Scholar]
- Mahajan KN, Mitchell BS. Role of human Pso4 in mammalian DNA repair and association with terminal deoxynucleotidyl transferase. Proc. Natl. Acad. Sci. USA. 2003;100:10746–10751. doi: 10.1073/pnas.1631060100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, Hao L, He S, Hurwitz DI, Jackson JD, Ke Z, Krylov D, Lanczycki CJ, Liebert CA, Liu C, Lu F, Lu S, Marchler GH, Mullokandov M, Song JS, Thanki N, Yamashita RA, Yin JJ, Zhang D, Bryant SH. CDD: a conserved domain database for interactive domain family analysis. Nucleic Acids Res. 2007;35:D237–D240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy KD, de Vellis J. Preparation of separate astroglial and oligodendroglial cell cultures from rat cerebral tissue. J. Cell Biol. 1980;85:890–902. doi: 10.1083/jcb.85.3.890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F, Kokorina NA, Dobretsova A, Wight PA. Characterization of an intronic enhancer that regulates myelin proteolipid protein (Plp) gene expression in oligodendrocytes. J. Neurosci. Res. 2005;82:346–356. doi: 10.1002/jnr.20640. [DOI] [PubMed] [Google Scholar]
- Merika M, Thanos D. Enhanceosomes. Curr. Opin. Genet. Dev. 2001;11:205–208. doi: 10.1016/s0959-437x(00)00180-5. [DOI] [PubMed] [Google Scholar]
- Miller RH. Regulation of oligodendrocyte development in the vertebrate CNS. Prog. Neurobiol. 2002;67:451–467. doi: 10.1016/s0301-0082(02)00058-8. [DOI] [PubMed] [Google Scholar]
- Morillon A, O’Sullivan J, Azad A, Proudfoot N, Mellor J. Regulation of elongating RNA polymerase II by forkhead transcription factors in yeast. Science. 2003;300:492–495. doi: 10.1126/science.1081379. [DOI] [PubMed] [Google Scholar]
- Norton WT, Poduslo SE. Myelination in rat brain: changes in myelin composition during brain maturation. J. Neurochem. 1973;21:759–773. doi: 10.1111/j.1471-4159.1973.tb07520.x. [DOI] [PubMed] [Google Scholar]
- Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- Sandberg R, Yasuda R, Pankratz DG, Carter TA, Del Rio JA, Wodicka L, Mayford M, Lockhart DJ, Barlow C. Regional and strain-specific gene expression mapping in the adult mouse brain. Proc. Natl. Acad. Sci. USA. 2000;97:11038–11043. doi: 10.1073/pnas.97.20.11038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato-Bigbee C, Pal S, Chu AK. Different neuroligands and signal transduction pathways stimulate CREB phosphorylation at specific developmental stages along oligodendrocyte differentiation. J. Neurochem. 1999;72:139–147. doi: 10.1046/j.1471-4159.1999.0720139.x. [DOI] [PubMed] [Google Scholar]
- Shiota C, Ikenaka K, Mikoshiba K. Developmental expression of myelin protein genes in dysmyelinating mutant mice: analysis by nuclear run-off transcription assay, in situ hybridization, and immunohistochemistry. J. Neurochem. 1991;56:818–826. doi: 10.1111/j.1471-4159.1991.tb01997.x. [DOI] [PubMed] [Google Scholar]
- Stolt CC, Lommes P, Friedrich RP, Wegner M. Transcription factors Sox8 and Sox10 perform non-equivalent roles during oligodendrocyte development despite functional redundancy. Development. 2004;131:2349–2358. doi: 10.1242/dev.01114. [DOI] [PubMed] [Google Scholar]
- Tang HK, Chao L-Y, Saunders GF. Functional analysis of paired box missense mutations in the PAX6 gene. Hum. Mol. Genet. 1997;6:381–386. doi: 10.1093/hmg/6.3.381. [DOI] [PubMed] [Google Scholar]
- Verity AN, Bredesen D, Vonderscher C, Handley VW, Campagnoni AT. Expression of myelin protein genes and other myelin components in an oligodendrocyte cell line conditionally immortalized with a temperature-sensitive retrovirus. J. Neurochem. 1993;60:577–587. doi: 10.1111/j.1471-4159.1993.tb03188.x. [DOI] [PubMed] [Google Scholar]
- Vidaković M, Poznanović G, Bode J. DNA break repair: refined rules of an already complicated game. Biochem. Cell Biol. 2005;83:365–373. doi: 10.1139/o05-044. [DOI] [PubMed] [Google Scholar]
- Wang X, Kikuchi T, Rikihisa Y. Proteomic identification of a novel Anaplasma phagocytophilum DNA binding protein that regulates a putative transcription factor. J. Bacteriol. 2007;189:4880–4886. doi: 10.1128/JB.00318-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wight PA, Dobretsova A. The first intron of the myelin proteolipid protein gene confers cell type-specific expression by a transcriptional repression mechanism in non-expressing cell types. Gene. 1997;201:111–117. doi: 10.1016/s0378-1119(97)00435-6. [DOI] [PubMed] [Google Scholar]
- Wight PA, Duchala CS, Readhead C, Macklin WB. A myelin proteolipid protein-lacZ fusion protein is developmentally regulated and targeted to the myelin membrane in transgenic mice. J. Cell Biol. 1993;123:443–454. doi: 10.1083/jcb.123.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wight PA, Duchala CS, Shick HE, Gudz TI, Macklin WB. Expression of a myelin proteolipid protein (Plp)-lacZ transgene is reduced in both the CNS and PNS of Plpjp mice. Neurochem. Res. 2007;32:343–351. doi: 10.1007/s11064-006-9202-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijchers PJEC, Hoekman MFM, Burbach JPH, Smidt MP. Identification of forkhead transcription factors in cortical and dopaminergic areas of the adult murine brain. Brain Res. 2006;1068:23–33. doi: 10.1016/j.brainres.2005.11.022. [DOI] [PubMed] [Google Scholar]
- Yang Q, Kong Y, Rothermel B, Garry DJ, Bassel-Duby R, Williams RS. The winged-helix/forkhead protein myocyte nuclear factor β (MNF-β) forms a co-repressor complex with mammalian Sin3B. Biochem. J. 2000;345:335–343. [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Fancy SPJ, Kotter MR, Li W-W, Franklin RJM. Mechanisms of CNS remyelination–the key to therapeutic advances. J. Neurol. Sci. 2005;233:87–91. doi: 10.1016/j.jns.2005.03.008. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.