Abstract
Please cite this paper as: Yamanaka et al. (2010) Infectivity and pathogenicity of canine H3N8 influenza A virus in horses. Influenza and Other Respiratory Viruses 4(6), 345–351.
Background Equine H3N8 influenza A viruses (EIVs) cause respiratory disease in horses and circulate among horses worldwide. In 2004, an outbreak of canine H3N8 influenza A virus (CIV) occurred among dogs in Florida and has spread among dogs in the United States (US). Genetic analyses revealed that this CIV is closely related to the recent EIVs. Although CIV‐infected dogs could be the source of H3N8 influenza A virus for horses, it remains unclear whether the CIV circulating in the United States still maintains its infectivity and/or pathogenicity in horses. To address this, we investigated the infectivity and pathogenicity of CIV in horses and the receptor binding specificity of CIV.
Materials and methods Three horses were inoculated with A/canine/Colorado/30604/2006 (CO06, H3N8). Clinical signs and nasal swabs were recorded or collected every day. We also evaluated the virus binding to α2‐3‐linked 5‐N‐acetylneuraminic acid (NeuAcα2‐3Gal) and 5‐N‐glycolylneuraminic acid (NeuGcα2‐3Gal) receptor analogues.
Results Although all the three horses inoculated with CO06 seroconverted, they showed only mild clinical signs and two of them showed no virus shedding. CO06 had reduced binding to NeuGcα2‐3Gal.
Discussion Our results demonstrated that CO06 had reduced proliferation ability and pathogenicity in horses. As the recognition of NeuGcα2‐3Gal by EIV is known to be essential for binding to the equine respiratory system, the decreased binding of CO06 to NeuGcα2‐3Gal may be one of the important factors that reduces the proliferation ability and pathogenicity of CO06 in horses.
Keywords: Canine, equine, H3N8, influenza A virus
Introduction
Equine influenza is one of the most important respiratory diseases of horses because of its rapid transmission and results from infection with H7H7 or H3N8 subtype influenza A viruses. 1 , 2 While equine influenza virus (EIV) of the H7N7 subtype may be extinct among horses at the present time, the other subtype, H3N8 that was first isolated from an epidemic in the United States (US) in 1963 continues to circulate among horses worldwide. 2 , 3 Genetic studies revealed that since around the latter half of 1980s, EIVs of the H3N8 subtype have diverged into two evolutionary lineages, designated ‘American’ and ‘Eurasian’ on the basis of the geographic origin of strains comprising the different lineages. 4 Strains within the American lineage further segregate into three sub‐lineages, referred to as ‘Florida’, ‘Kentucky’ and ‘Argentina’. 5 Since around 2003, many EI outbreaks of the Florida sub‐lineage strains have been reported in horses throughout the world. 6 , 7 , 8 , 9 , 10 , 11 , 12 , 13
Canine influenza is also a contagious respiratory disease of dogs resulting from infection with influenza A viruses. 14 , 15 Canine influenza A subtype H3N8 virus (CIV) was first recognized in the United States in 2004 and spread among dogs throughout the United States. 14 , 15 , 16 , 17 At the present time, canine influenza has likely become enzootic among dogs, at least in the United States. 15 The genetic analyses of the CIV strain isolated during the first outbreak in 2004 indicated that all 8 genes were most similar to those from H3N8 EIV. 14 Moreover, phylogenetic analysis showed that the hemagglutinin (HA) gene of several CIV isolates were closely related to the HA genes of EIV belonging to the Florida sub‐lineage. 14 , 17 Therefore, it is likely that CIV emerged recently as the result of a direct inter‐species transmission of contemporary EIV into dogs without genetic reassortment. 14 , 17
The HAs of influenza A viruses from different hosts differ in their ability to bind sialic acid (SA)‐galactose (Gal) linkages (α2‐3 or α2‐6) and SA species such as N‐acetylneuraminic acid (NeuAc) or N‐glycolylneuraminic acid (NeuGc). 18 Accordingly, the HA of influenza A virus is a critical factor as regards the host species specificity of influenza A virus. 18 , 19 , 20 Thus far, the difference between the binding specificities of EIV and CIV has only been deduced by sequence analysis, and direct measurement of this has not been reported.
As CIV‐infected dogs could be the source of infection of H3N8 to horse populations, it is of great interest to see whether CIV circulating in dogs maintains infectivity and/or pathogenicity for horses. To address this, we have investigated the infectivity and pathogenicity of CIV in horses, and also the receptor binding specificity of CIV and EIV.
Materials and methods
Virus preparation
A/canine/Colorado/30604/2006 (CO06, H3N8) was used in this study. CO06 was isolated from a diseased dog in Colorado in the United States, and kindly provided by Dr. Edward J. Dubovi (Cornell University, New York, NY, USA). The stock virus was propagated in 10‐day‐old embryonated hen’s eggs. The stock virus was aliquoted and stored at −80°C before use, following low‐speed centrifugation (1500 × g for 10 minutes) to remove cell debris. The passage number was limited to 4 to avoid adaptation of the virus to eggs. Inoculation of dogs with the stock virus caused clinical signs (pyrexia, cough, nasal and ocular discharges) similar to those reported for natural infections. 14 , 15 , 16 The stock virus was used for experimental inoculation, and also as an antigen in a hemagglutination inhibition (HI) test. HI tests were performed as previously described. 21 The HI titers were expressed as the reciprocals of the highest dilutions of the sera showing HI.
Experimental inoculation of horses with CIV
Three thoroughbred horses (17–20 months old) were used in this study (Horses 1, 2 and 3). All the horses showed no serological evidence of prior H3N8 virus infection or vaccination in HI tests for antibodies to CO06 and A/equine/Ibaraki/1/2007 (IBK07, H3N8) (HI titers <10). The methods of inoculation were conducted similarly to our recent study, where we inoculated three similar aged thoroughbred horses (Horses 4, 5 and 6) with IBK07. 22 Briefly, the horses were inoculated by inhalation of CO06 (108·3 50% egg infectious dose [EID50]/20 ml) using an ultrasonic nebulizer (SONICLIZER305; ATOM, Tokyo, Japan) for 20 minutes. The duration of the experiment was 14 days following inoculation. Rectal temperatures (RTs) were measured each morning throughout the study. We also examined each horse physically on a daily basis with a particular focus on nasal discharge and cough. These findings were scored as follows. Nasal discharge: (−) Nil, (+) serous discharge, (++) mild mucopurulent discharge, (+++) severe mucopurulent discharge. Cough: (−) Nil, (+) 2–5 times per 10 minutes, (++) more than 6 times per 10 minutes. Sera were collected from the horses on days 0 and 14 for the HI test and were stored at −20°C prior to use. The experimental procedures were approved by the Animal Care Committee of Equine Research Institute of the Japan Racing Association.
Virus shedding
To detect virus shedding from the nostrils, nasal samples were collected from the horses using 1·0 × 1·5 cm absorbent cotton swabs (JMS menbou; Japan Medical Supply, Tokyo, Japan) from days 0 to 14 as previously described. 22 Briefly, the swabs collected from horses were immediately immersed in 2·5 ml of transport medium [phosphate‐buffered saline (PBS, pH:7·4) supplemented with 0·6% (w/v) tryptose phosphate broth, 500 unit/ml penicillin, 500 μg/ml streptomycin and 1·25 μg/ml amphotericin B]. The swab samples in transport medium were vortexed for 10 seconds and centrifuged at 1500 × g for 10 minutes for precipitate debris. The supernatant of each sample was aliquoted and stored at −80°C prior to use. Two hundred microliters of the supernatants that had been diluted 1:10 (v/v) in transport medium was inoculated into the allantoic cavities of 10‐day‐old embryonated hen’s eggs (four eggs per sample). Allantoic fluid was harvested after 3 days of incubation at 34°C and examined for the presence of influenza A virus in a hemagglutination test using 0·5% hen’s red blood cells. The virus titers (log10 EID50/200 μl) were determined for nasal swabs samples that were hemagglutination positive. 23
Sialylglycolipids
Sialylglycolipids used in this study were chemically synthesized. 24 They commonly have Galβ1‐4GlcNAcβ1‐3Galβ1‐4Glc as a core structure, a SA residue (NeuAcα2‐3 or NeuGcα2‐3) linked to a non‐reducing terminal galactose, and the ceramide portion substituted by a branched hydrocarbon chain containing 30 carbons. These synthetic sialylglycolipids have been evaluated by influenza A virus binding assays as described previously. 25
Solid‐phase binding assay
Virus binding to sialylglycolipids was determined according to a method described previously. 26 Synthetic sialylglycolipids were serially twofold diluted in 100% ethanol from 0·625 to 10 pmol/μl. Ten microlitres of each diluted sialylglycolipid was placed into wells of polystyrene Universal‐BIND™ microplates (flat‐bottom, Product# 2504; Corning, Tokyo, Japan) and incubated for approximately 1 hour at 37°C until the ethanol had completely evaporated. Sialylglycolipids were covalently immobilized on the surface of plates by exposure for 1 minute under ultraviolet irradiation (254 nm) according to the manufacture’s instruction. Each well was blocked for 1 hour at room temperature with PBS containing 2% bovine serum albumin. After three washes with PBS, the plates were incubated in solutions containing viruses in PBS (32 HA unit/50 μl/well) overnight at 4°C. After seven washes with ice‐cold PBS, the plates were incubated in a substrate solution containing 40 μm 2′‐(4‐methylumbelliferyl)‐α‐D‐N‐acetylneuraminic acid in PBS (50 μl/well) at 37°C for 60 minutes to detect virus neuraminidase (NA) activity associated with bound viruses. The reactions were terminated by addition of 500 mm carbonate buffer (50 μl/well, pH 10·2). Fluorescence intensity of the 4‐methylumbelliferone released by viral NAs was measured at 355 nm (excitation) and 460 nm (emission) with a microplate fluorometer (Fluoroskan Ascent FL; Labysystems, Helsinki, Finland). Duplicate measurements were performed in each assay. The direct virus‐binding activity was calculated as follows: Virus‐binding score = [(average of NA activity of duplicate sialylglycolipid‐immobilized wells) – (average of NA activity of duplicate ethanol‐treated wells)]/[NA activity of applied virus (32 HA units/50 μl)]. The virus‐binding score was expressed as mean score ± SD for three independent experiments.
Phylogenetic analysis of the HA gene
A phylogenetic tree was constructed from the partial HA1 sequences of CIVs and EIVs (947 nucleotides) by a neighbor‐joining method using the MEGA 4·1 program. 27 , 28 Five hundred bootstrap replicates were conducted to assess the statistical support for the tree topology. All the sequences for the phylogenetic analysis were obtained from GenBank. All the accession numbers were shown in parentheses in Figure 3.
Figure 3.
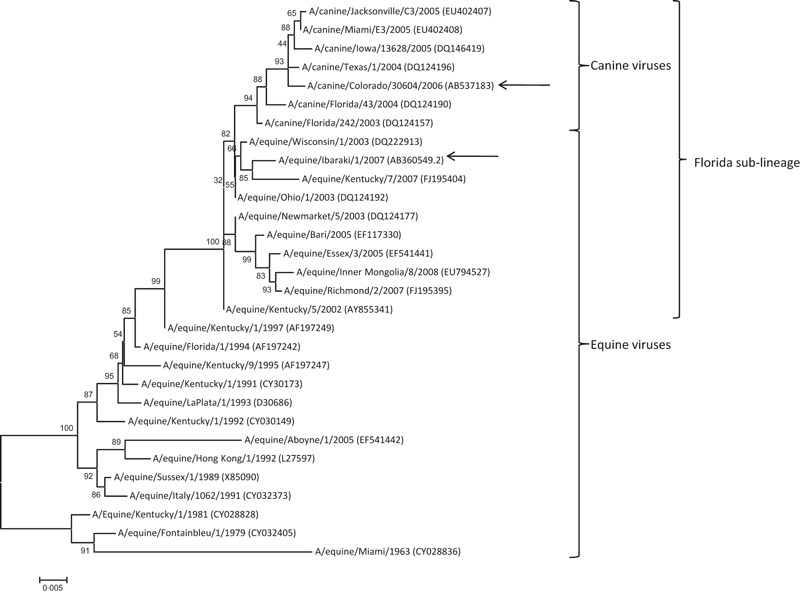
Phylogenetic tree of the partial HA1 sequences of the H3N8 strains from equine and canine hosts (947 nucleotides). The GenBank accession numbers were given in parentheses. Bootstrap values (n = 500) are shown at the branch points. Scale bar indicates nucleotide substitutions per site. Arrows indicate the viruses used in this study.
Statistical analyses
In this study (n = 3), we inoculated the horses with the same titer (108·3 EID50) of CO06 by the same method as in our previous study (n = 3). 22 Also, we have scored the clinical signs (nasal discharge and cough) of the horses inoculated with IBK07, 22 using the same criteria mentioned above as the horses inoculated with CO06 in this study. Therefore, we compared the data of experimental inoculation of horses in this study with the data obtained from our previous report. 22 The mean RTs of animals inoculated with CO06 and IBK07 were compared by two‐way repeated measures analysis of variance followed by Tukey–Kramer multiple comparisons test. The mean durations of nasal discharge and cough between the inoculated viruses, and the differences of virus‐binding scores between NeuAcα2‐3 and NeuGcα2‐3 in each virus were compared by Student’s t‐test. All statistical analyses were performed using SigmaStat 3·5 (Systat Software, Chicago, IL, USA). The level of significance (P) was set at 0·05 in this study.
Results
Figure 1 shows the daily change of mean RT of the horses inoculated with CIV strain CO06 and is compared to those infected with EIV strain IBK07 from our previous study. 22 As we discussed previously, the horses inoculated with EIV strain IBK07 showed a clear bi‐phasic temperature pattern that is often seen with equine cases of equine influenza. The horses inoculated with CIV strain CO06 had a sharp elevation in RT on Day 2, but the magnitude of second phase of elevated RT on Days 6 and 8–10 was significantly (P < 0·05) smaller than that for horses inoculated with EIV strain IBK07. Daily scores of nasal discharge and cough are shown in Table 1. All horses inoculated with IBK07 began showing nasal discharge on Day 2 or 3 which continued until Day 12 or 14. All horses inoculated with IBK07 began coughing on Day 3 and continued to cough until Day 13 or 14. In addition, all the horses inoculated with IBK07 showed mucopurulent nasal discharges (++ or +++) from Days 4 to 13 during the experiment. Horses 4 and 5 coughed more than 6 times per 10 min (++) on Days 6 and 8. On the other hand, no horse inoculated with CIV strain CO06 showed mucopurulent nasal discharge or coughed more than 6 times per 10 min throughout the experiment. The mean durations where the horses inoculated with CIV strain CO06 showed nasal discharges and coughs [2·7 ± 1·5 (mean ± SD) days and 1·7 ± 2·9 days, respectively] were significantly (P < 0·05) shorter than those of the horses inoculated with EIV strain IBK07 (11·0 ± 1·0 days and 10·3 ± 0·6 days, respectively).
Figure 1.
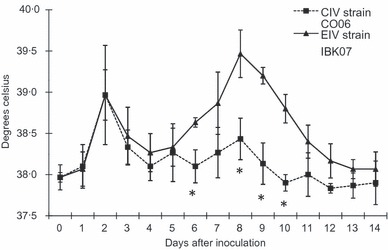
Daily mean rectal temperatures (±SDs) of the horses inoculated with CIV strain CO06 compared to those with EIV strain IBK07. 22 *P < 0·05.
Table 1.
Daily scores of clinical symptoms signs in each horse inoculated with CIV strain CO06, as compared to those with EIV strain IBK07
| Clinical sign* | Inoculated strain | Horse | Days after inoculation | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | |||
| Nasal discharge | CIV strain CO06 | 1 | − | + | + | + | − | − | − | − | − | − | − | − | − | − | − |
| 2 | − | + | + | + | + | − | − | − | − | − | − | − | − | − | − | ||
| 3 | − | + | − | − | − | − | − | − | − | − | − | − | − | − | − | ||
| EIV strain IBK07** | 4 | − | − | − | + | + | + | +++ | ++ | +++ | +++ | ++ | ++ | ++ | ++ | + | |
| 5 | − | − | + | + | + | ++ | + | ++ | +++ | ++ | − | ++ | − | ++ | + | ||
| 6 | − | − | + | + | +++ | +++ | ++ | ++ | ++ | ++ | ++ | − | + | − | − | ||
| Cough | CIV strain CO06 | 1 | − | − | − | − | + | + | + | + | + | − | − | − | − | − | − |
| 2 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | ||
| 3 | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | ||
| EIV strain IBK07** | 4 | − | − | − | + | − | + | ++ | + | ++ | + | + | + | + | + | + | |
| 5 | − | − | − | + | + | + | ++ | − | ++ | + | + | + | + | − | + | ||
| 6 | − | − | − | + | + | + | + | + | − | + | + | + | + | + | − | ||
*See Materials and methods for the scoring systems.
**Data were cited from our previous study 22 and have been scored for this study.
Table 2 shows the results of virus shedding from each horse inoculated with CO06 when compared to the horses inoculated with EIV strain IBK07. Although one of the three horses inoculated with CIV strain CO06 (Horse 1) had detectable virus shedding on Days 1, 2, 3, 5 and 6, those titers were <102·0 EID50/200 μl. The other two horses (Horses 2 and 3) had no detectable virus shedding throughout the experiment. On the other hand, all the horses inoculated with EIV strain IBK07 (Horses 4, 5 and 6) began shedding virus on Day 2 (103·5–104·3 EID50/200 μl) and continued to shed for 5 or 6 consecutive days. During the experiment, the horses inoculated with EIV strain IBK07 demonstrated virus shedding of more than 102·0 EID50/200 μl for 3–5 days.
Table 2.
Virus detection by egg culture and titers (log10 EID50/200 μl) of nasal swabs collected from each horse infected with CIV strain CO06 in comparison with that with EIV strain IBK07
| Inoculated strain | Horse | Days after inoculation | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | ||
| CIV strain CO06 | 1 | –* | ≤0·7 | 1·5 | 1·3 | – | 1·7 | 1·3 | – | – | – | – | – | – | – | – |
| 2 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | |
| 3 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | |
| EIV strain IBK07** | 4 | – | – | 3·7 | 4·5 | 3·4 | 3·5 | 2·5 | – | – | – | – | – | – | – | – |
| 5 | – | – | 4·3 | 3·7 | 2·7 | ≤1·4 | 3·0 | 1·5 | – | – | – | – | – | – | – | |
| 6 | – | – | 3·5 | 1·5 | ≤1·4 | 2·7 | 2·4 | – | – | – | – | – | – | – | – | |
*<0·7 (no virus was isolated from four eggs inoculated with nasal swabs specimen diluted of 1:10).
**Data were cited from our recent report 22.
Table 3 shows the HI titers to CIV strain CO06 of the paired sera collected from horses inoculated with CIV strain CO06 on Days 0 and 14, compared to the data cited from our previous report. 22 Although all the horses inoculated with CIV strain CO06 seroconverted, the HI titers of Horses 2 and 3 on Day 14 were more than eightfold lower than those of the horses inoculated with EIV strain IBK07.
Table 3.
Hemagglutination inhibition (HI) titers* of the paired‐sera collected from each horse inoculated with CIV strain CO06 or EIV strain IBK07
| Inoculated strain | Horse | HA antigen | Days after inoculation | |
|---|---|---|---|---|
| 0 | 14 | |||
| CIV strain CO06 | 1 | CIV strain CO06 | <10 | 640 |
| 2 | <10 | 80 | ||
| 3 | <10 | 20 | ||
| EIV strain IBK07** | 4 | EIV strain IBK07 | <10 | 640 |
| 5 | <10 | 640 | ||
| 6 | <10 | 640 | ||
*HI titers were expressed as the reciprocals of the highest dilutions of the sera showing HI.
**Data were cited from our previous report 22.
The results of solid‐phase binding assays are shown in Figure 2. EIV strain IBK07 binding to NeuGcα2‐3Gal was significantly greater (P < 0·05) than to NeuAcα2‐3Gal. There was no difference in the binding of CO06 to NeuAcα2‐3Gal and NeuGcα2‐3Gal.
Figure 2.
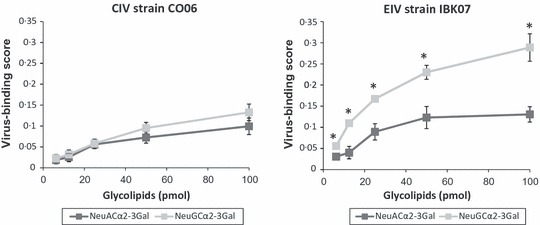
Binding of CO06 and IBK07 to the synthetic sialylglycolipids carrying NeuAcα2‐3Gal and NeuGcα2‐3Gal in solid‐phase binding assays. Virus‐binding activity was determined as described in Materials and methods. The values of y‐axis are expressed as virus‐binding scores (mean ± SDs) for three independent experiments. *P < 0·05.
The phylogenetic tree of the partial HA1 sequences of CIVs and EIVs is shown in Figure 3. All CIVs including CO06 were divergent from the EIVs and formed a cluster within the Florida sub‐lineage. EIV strain IBK07 was in the Florida sub‐lineage outside of the CIV cluster.
Discussion
The horses inoculated with CO06 showed milder and shorter clinical signs compared to the horses inoculated with IBK07. In addition, the horses inoculated with CO06 shed little or no virus from the upper respiratory tract. Therefore, even though the horses were infected with CO06 as supported by seroconversion, the level of infection was variable, and it is evident that the canine virus did not replicate well enough to cause severe disease or virus shedding.
Some researchers previously reported that large amounts of SAα2,3Gal exist on the surfaces of respiratory epithelial cells of dogs by histological analysis with lectin staining. 29 , 30 Furthermore, Song et al. 30 reported that SAα2,6Gal was not detected in the respiratory epithelial cells of dogs. Therefore, the binding to SAα2,3Gal of CIV could be an important factor for the infection of the canine respiratory system. Suzuki et al. 20 reported that the surface of respiratory epithelial cells of horses has an abundant NeuGcα2‐3Gal, and the recognition of NeuGcα2‐3Gal by EIV is essential to bind to the equine respiratory system. Thus, we evaluated the binding of CO06 and IBK07 to sialylglycolipids with two kinds of SAα2,3Gal species (NeuGcα2‐3Gal and NeuAcα2‐3Gal). Our results indicated that CO06 had reduced binding to NeuGcα2‐3Gal when compared to IBK07, while the binding of CO06 to NeuAcα2‐3Gal is almost similar to that of IBK07. Therefore, it is suggested that CO06 has reduced pathogenicity and replication capacity in horses because of its lower binding to NeuGcα2‐3Gal.
To date, the genetic analysis of HA genes of the six CIVs isolated in the field in the United States have been reported. 17 The phylogenetic analysis in this study clearly showed that CO06 was clustered together with the other six CIVs (Bootstrap value = 94). IBK07 was clustered into the Florida sub‐lineage, which is predominantly circulating among horses worldwide, 13 is outside the CIVs cluster. It has been known that five amino acid substitutions (N54K, N83S, W222L, I328T and N483T) distinguish the six CIVs isolated in the United States from the contemporary EIVs. 14 , 17 CO06 also had all these five substitutions (data not shown). In terms of the binding ability to the SA moieties, because position 222 of the HA is located near the receptor binding site, 14 , 17 the mutation (W222L) may lead to the reduced binding of CO06 to NeuGcα2‐3Gal. However, whether the reduction of binding of CIV to NeuGcα2‐3Gal contributes to the adaptation to canine respiratory systems remains unclear because the receptor profile is as yet unknown. Further studies on the receptor binding characteristics of CIV are needed to draw definitive conclusions of the adaptation mechanism.
In summary, it is suggested that CIV circulating in the United States has reduced replication capacity and pathogenicity in horses when compared to the contemporary EIVs. Decreased binding of CIV strain CO06 to NeuGcα2‐3Gal may be one of the important factors that reduces the proliferation ability and pathogenicity of CIV in horses. However, as CIV is still infectious to horses, close‐contact between dogs and horses should be limited to minimize interspecies transmission during an epidemic of canine influenza.
Acknowledgements
The authors thank Dr. Edward J. Dubovi (Cornell University, Ithaca, NY, USA) for kindly providing CO06. We also thank Dr. Shigeo Sugita (Equine Research Institute, Utsunomiya, Japan) and Mr. Manabu Nemoto (Epizootic Research Center, Shimotsuke, Japan) for their assistance in the experiments. We are also grateful to Drs. Alan Hay and Stephen Wharton (National Institute for Medical Research, London, UK) for helping us prepare for this manuscript.
References
- 1. Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev 1992; 56(1):152–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Van Maanen C, Cullinane A. Equine influenza virus infections: an update. Vet Q 2002; 24(2):79–94. [DOI] [PubMed] [Google Scholar]
- 3. Webster RG. Are equine 1 influenza viruses still present in horses? Equine Vet J 1993; 25(6):537–538. [DOI] [PubMed] [Google Scholar]
- 4. Daly JM, Lai AC, Binns MM, Chambers TM, Barrandeguy M, Mumford JA. Antigenic and genetic evolution of equine H3N8 influenza A viruses. J Gen Virol 1996; 77(Pt 4):661–671. [DOI] [PubMed] [Google Scholar]
- 5. Lai AC, Rogers KM, Glaser A, Tudor L, Chambers T. Alternate circulation of recent equine‐2 influenza viruses (H3N8) from two distinct lineages in the United States. Virus Res 2004; 100(2):159–164. [DOI] [PubMed] [Google Scholar]
- 6. Barbic L, Madic J, Turk N, Daly J. Vaccine failure caused an outbreak of equine influenza in Croatia. Vet Microbiol 2009; 133(1–2):164–171. [DOI] [PubMed] [Google Scholar]
- 7. Damiani AM, Scicluna MT, Ciabatti I et al. Genetic characterization of equine influenza viruses isolated in Italy between 1999 and 2005. Virus Res 2008; 131(1):100–105. [DOI] [PubMed] [Google Scholar]
- 8. Ito M, Nagai M, Hayakawa Y et al. Genetic Analyses of an H3N8 Influenza Virus Isolate, Causative Strain of the Outbreak of Equine Influenza at the Kanazawa Racecourse in Japan in 2007. J Vet Med Sci 2008; 70(9):899–906. [DOI] [PubMed] [Google Scholar]
- 9. Martella V, Elia G, Decaro N et al. An outbreak of equine influenza virus in vaccinated horses in Italy is due to an H3N8 strain closely related to recent North American representatives of the Florida sub‐lineage. Vet Microbiol 2007; 121(1–2):56–63. [DOI] [PubMed] [Google Scholar]
- 10. Newton JR, Daly JM, Spencer L, Mumford JA. Description of the outbreak of equine influenza (H3N8) in the United Kingdom in 2003, during which recently vaccinated horses in Newmarket developed respiratory disease. Vet Rec 2006; 158(6):185–192. [DOI] [PubMed] [Google Scholar]
- 11. Yamanaka T, Niwa H, Tsujimura K, Kondo T, Matsumura T. Epidemic of equine influenza among vaccinated racehorses in Japan in 2007. J Vet Med Sci 2008; 70(6):623–625. [DOI] [PubMed] [Google Scholar]
- 12. Cowled B, Ward MP, Hamilton S, Garner G. The equine influenza epidemic in Australia: spatial and temporal descriptive analyses of a large propagating epidemic. Prev Vet Med 2009; 92(1–2):60–70. [DOI] [PubMed] [Google Scholar]
- 13. Bryant NA, Rash AS, Russell CA et al. Antigenic and genetic variations in European and North American equine influenza virus strains (H3N8) isolated from 2006 to 2007. Vet Microbiol 2009; 138(1–2):41–52. [DOI] [PubMed] [Google Scholar]
- 14. Crawford PC, Dubovi EJ, Castleman WL et al. Transmission of equine influenza virus to dogs. Science 2005; 310(5747):482–485. [DOI] [PubMed] [Google Scholar]
- 15. Dubovi EJ, Njaa BL. Canine influenza. Vet Clin North Am Small Anim Pract 2008; 38(4):827–835, viii. [DOI] [PubMed] [Google Scholar]
- 16. Yoon KJ, Cooper VL, Schwartz KJ et al. Influenza virus infection in racing greyhounds. Emerg Infect Dis 2005; 11(12):1974–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Payungporn S, Crawford PC, Kouo TS et al. Influenza A virus (H3N8) in dogs with respiratory disease, Florida. Emerg Infect Dis 2008; 14(6):902–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rogers GN, Pritchett TJ, Lane JL, Paulson JC. Differential sensitivity of human, avian, and equine influenza A viruses to a glycoprotein inhibitor of infection: selection of receptor specific variants. Virology 1983; 131(2):394–408. [DOI] [PubMed] [Google Scholar]
- 19. Ito T, Kawaoka Y. Host‐range barrier of influenza A viruses. Vet Microbiol 2000; 74(1–2):71–75. [DOI] [PubMed] [Google Scholar]
- 20. Suzuki Y, Ito T, Suzuki T, et al. Sialic acid species as a determinant of the host range of influenza A viruses. J Virol 2000; 74(24):11825–11831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Imagawa H, Fukunaga Y, Kamada M, Nanbu M, Kitamura M. Distribution of HI antibody against 3 vaccine strains of equine influenza in light‐breed horses in Japan. J Equine Sci 1993; 4:31–38. [Google Scholar]
- 22. Yamanaka T, Nemoto M, Tsujimura K, Kondo T, Matsumura T. Interspecies transmission of equine influenza virus (H3N8) to dogs by close contact with experimentally infected horses. Vet Microbiol 2009; 139(3–4):351–355. [DOI] [PubMed] [Google Scholar]
- 23. Reed LJ, Müench H. A simple method of estimating fifty per cent of endpoint. AM J Hyg 1938; 27:493. [Google Scholar]
- 24. Tanahashi E, Fukunaga K, Ozawa Y et al. Synthesis of Sialyl‐α‐(2→3)‐Neolactotetraose Derivatives Containing Different Sialic Acids: Molecular Probes for Elucidation of Substrate Specificity of Human α1,3‐Fucosyltransferases. J Carbohydr Chem 2000; 19:747–768. [Google Scholar]
- 25. Masuda H, Suzuki T, Sugiyama Y et al. Substitution of amino acid residue in influenza A virus hemagglutinin affects recognition of sialyl‐oligosaccharides containing N‐glycolylneuraminic acid. FEBS Lett 1999; 464(1–2):71–74. [DOI] [PubMed] [Google Scholar]
- 26. Hidari KI, Murata T, Yoshida K et al. Chemoenzymatic synthesis, characterization, and application of glycopolymers carrying lactosamine repeats as entry inhibitors against influenza virus infection. Glycobiology 2008; 18(10):779–788. [DOI] [PubMed] [Google Scholar]
- 27. Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 2007; 24(8):1596–1599. [DOI] [PubMed] [Google Scholar]
- 28. Saitou N, Nei M. The neighbor‐joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 1987; 4(4):406–425. [DOI] [PubMed] [Google Scholar]
- 29. Daly JM, Blunden AS, Macrae S et al. Transmission of equine influenza virus to English foxhounds. Emerg Infect Dis 2008; 14(3):461–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Song D, Kang B, Lee C et al. Transmission of avian influenza virus (H3N2) to dogs. Emerg Infect Dis 2008; 14(5):741–746. [DOI] [PMC free article] [PubMed] [Google Scholar]


