Abstract
Cytochromes P450 3A4, 2D6, and 2C9 metabolize a large fraction of drugs. Knowing where these enzymes will preferentially oxidize a molecule, the regioselectivity, allows medicinal chemists to plan how best to block its metabolism. We present QSAR-based regioselectivity models for these enzymes calibrated against compiled literature data of drugs and drug-like compounds. These models are purely empirical and use only the structures of the substrates, in contrast to those models that simulate a specific mechanism like hydrogen radical abstraction and/or use explicit models of active sites. Our most predictive models use three substructure descriptors and two physical property descriptors. Descriptor importance from the random forest QSAR method show that other factors than the immediate chemical environment and the accessibility of the hydrogen affects regioselectivity in all three isoforms. The cross-validated predictions of the models are compared to predictions from our earlier mechanistic model
INTRODUCTION
Oral bioavailability of drugs depends largely on their ability to withstand degradation by intestinal and hepatic enzymes during “first-pass” metabolism. One important class of enzymes is the cytochromes P450 (CYPs). These are heme-containing enzymes that catalyze a number of chemical changes: oxidation, dealkylation, desaturation, etc. 1–4. All probably involve the transfer of the oxygen radical from the heme iron of the enzyme to the molecule as one of the steps. Knowing where a molecule would be preferentially oxidized, i.e. the regioselectivity, by a particular CYP would give medicinal chemists insight on where to block the metabolism and make their drug candidates more stable in vivo. Normally the regioselectivity of CYP-mediated biotransformation is determined experimentally by metabolite identification techniques (for instance reference 2). However, all such experimental techniques are time and labor intensive, and a computational model for regioselectivity could allow chemists to make rational decisions more quickly.
A number of models for predicting regioselectivity by CYPs have been proposed 5–10 and a few commercial systems for doing so have been released. It should be noted that these models usually do not predict whether a molecule will be a substrate for a particular CYP, only where the oxidation will likely occur assuming it is a substrate. Of the CYP isoforms, CYPs 3A4, 2D6, and 2C9 are probably the most important in metabolizing drugs and drug-like molecules 1,3. In some CYPs like 2D6, it has been proposed that there is a “pharmacophore” (i.e. a cation in molecules oxidized by 2D6) that controls regioselectivity by orienting the molecule in the active site such that certain atoms are closer to the heme oxygen (reviewed by Ekins et al. 11). In contrast, others like 3A4 do not have an obvious pharmacophore 4.
Previous work from this laboratory (Singh et al. 7) addressed metabolism by CYP 3A4. That model assumed, based on the observation that 3A4 lacks substrate specificity, that orientation effects from the 3A4 active site are negligible, and that regioselectivity depends primarily on the energy necessary to remove a hydrogen radical (the dehydration energy) from a particular atom, with the stipulation that only those hydrogens with sufficient solvent accessible surface area (≥ 8 Å2) could be attacked. We used AM1 molecular orbital calculations to calculate the dehydrogenation energy. Since even semi-empirical calculations like AM1 would take too long to make a rapid prediction system, we estimated the AM1 dehydrogenation energy with a QSAR model based on the local chemical environment of the atoms.
The Singh et al. model is modestly predictive of 3A4 regioselectivity, but it was clear from the outset that this model has some serious limitations. First and most importantly, given that it depended only on dehydrogenation energy, it could address oxidations only where removal of a hydrogen radical is the proposed mechanism, i.e. sp3 carbons with at least one attached hydrogen, and could not at all address oxidations at aromatic carbons, sulfurs, etc. Second, it was clear that dehydrogenation energies sometimes gave systematically wrong answers for some sp3 carbons. For instance in N-methylpiperidines, the observed action of CYP3A4 is to almost always oxidize the methyl, resulting in an N-dealkylation, while the AM1 dehydrogenation energy always suggested that the piperidine ring carbons adjacent to the N would be slightly more susceptible. However, due to our small sample size at the time (only about 50 examples), we could not confidently add any correction factors to our model. Third, the use of a sharp cutoff on the solvent accessible surface area makes the results sensitive to the starting conformation; sometimes a particular carbon would be marked as a site of oxidation, sometimes not, depending on whether the area of the hydrogen was just above or below the cutoff. Finally, we were not comfortable with completely ignoring orientation effects after crystal structures of CYP 3A4s became available 12,13 and it became clear that the active site is not so large or open as to permit free tumbling of a substrate, or at least rapid exchange of bound with free substrate, as would be required for orientation effects to be neglected.
Almost all models to date of CYP regioselectivity are mechanism-based. That is, one tries to simulate the chemical steps involved with oxidation, or at least the rate-limiting step (e.g. removing the H radical). Mechanism-based models are appealing because they appear to be general and require the least knowledge beforehand. For instance, molecular orbital calculations of dehydrogenation energy ought to give valid predictions for any molecule. However, in practice things are never so simple. For instance, oxidation at sp3 carbons probably involves the removal of a hydrogen radical as the product-determining step, but oxidation at aromatic rings probably occurs by a different mechanism, one proposal being the addition of a hydroxy radical 5. One must find a way to scale the relative importance of the two (or more) mechanisms for a prediction. Also, to match experiment one almost always has to add other effects that are not local to the atom.
Previous efforts developing CYP 3A4 models by one of us (KRK, unpublished work) suggested other factors, e.g. relative position to polar functionalities, whether the atom was part of a piperidine or piperazine ring, etc., were required to make the predicted and observed regioselectivity for 3A4 agree. When empirically-calibrated corrections are added on top of the original mechanism-based parameters, it is not clear that such an approach will result in a better model than that obtained by fitting parameters directly to experimental data and ignoring mechanistic considerations altogether.
In this paper we present QSAR-based regioselectivity models for human CYPs 3A4, 2D6, and 2C9 based on data in the literature plus some proprietary in-house data (for 3A4). The intention is to cover most commonly observed potential sites of oxidation (sp3 carbons, sp2 carbons, sulfurs, etc.). We use descriptors intrinsic only to the candidate substrates and include no information about the active sites of the CYPs. We are able to show that the QSAR models make cross-validated predictions better than the predictions from Singh et al., and at least as good as the predictions from MetaSite, a more mechanism-based method of predicting regioselectivity. We also apply the models to a small set of compounds not in the original set.
METHODS
Datasets
Even more than for the Singh et al. model, we depend here on regioselectivity data in the literature. Gathering the citations was greatly aided by two licensed databases, the Metabolite Database from Molecular Design Limited (www.mdli.com) and the Human Drug Metabolizing Enzyme Database from Fujitsu (www.fqs.fujitsu.com/ccs/ASP_service_eng/ASP_ADMEdatabase/ASP_ADMEdatabase _eng.html). For each molecule, the specific mechanism for oxidation had to be established as native human CYP3A4, 2D6, or 2C9 and the exact site(s) of oxidation of the molecule had to be known. The list of citations is provided as Supplementary Material. The final “calibration” sets consisted of 316 molecules for 3A4 (305 from the literature plus 11 proprietary molecules), 124 molecules for 2D6, and 92 molecules for 2C9. The structures of the molecules (minus the proprietary ones for 3A4) are also in Supplementary Material. One concern is that the molecules in the training set be diverse. Elucidation of metabolic products is difficult and time-consuming, so we are likely to see literature data on a limited number of molecules, drugs or drug candidates far along in their development, which tend to occur in a limited number of families. However, as will be shown, no family dominates any of the calibration sets.
Some months after we generated our original models, during the review process for the first submitted version of this manuscript, we rechecked the literature and found a total of 25 additional compounds: 19 for 3A4, 10 for 2D6, and 9 for 2C9, with some overlap. We will call these the “external” sets. The structures of these are also in Supplementary Material.
Sites of metabolism
In an ideal world, as a QSAR “response” we would like to have a rate of oxidation for every atom in every molecule measured under uniform conditions. However, what can be found in the literature is the elucidation of at most a few major sites of metabolism per molecule. Sometimes the relative amounts of the metabolic products are noted in the citation. In the molecular structure in Supplementary Material we have marked atoms as “1” (primary site), “2” (secondary site), etc. Topologically equivalent atoms are marked identically. However, most of the citations do not contain such detailed information and to maximize the number of molecules our models, we felt it best to treat atoms as having a binary response: “1” if it was noted as a major site of metabolism in the citation and “0” if it was not. We counted N- and O-dealkylations as occurring on the carbon of the leaving group adjacent to the N or O. There are some rarer reactions, such as ring openings, replacements of =S with =O, etc. In those cases the atom was marked which, in the opinion of the authors of the citation, is most likely to receive an oxygen radical from the CYP. Table 1 shows the frequencies of the atoms and observed oxidation sites in the calibration sets divided into major types
Table 1.
Frequency of oxidations in the calibration sets.
| 3A4 | 2D6 | 2C9 | ||||
|---|---|---|---|---|---|---|
| Atom type | Total nonhydrogen atoms | Marked as oxidation Sites | Total nonhydrogen atoms | Marked as oxidation sites | Total nonhydrogen atoms | Marked as oxidation sites |
| sp3C with H (hydroxylation or N,O-dealkylation) | 2792 | 432 | 856 | 119 | 456 | 84 |
| sp2C with H (hydroxylation) | 1817 | 87 | 713 | 55 | 579 | 56 |
| -S- , -S(=O)-(to –S(=O)- , -SO2-) | 44 | 31 | 14 | 8 | 15 | 8 |
| N in 6-membered aromatic ring (e.g. pyridine N to N->O) | 71 | 10 | 19 | 0 | 20 | 1 |
| sp3N (basic) (N to N->O) | 119 | 6 | 69 | 0 | 10 | 0 |
| Other | 3655 | 12 | 1009 | 11 | 956 | 3 |
| Total | 8498 | 566 | 2680 | 193 | 2036 | 152 |
Data of this type has several issues. Foremost is the usual concern whether data from the literature can be sensibly combined. In this case, oxidation products of different molecules are measured in different labs with different techniques. Also, it is not clear that the primary oxidation product is given in some citations because the authors may be trying to identify the CYP responsible for a particular oxidation product without necessarily establishing it as the primary product. Errors in the structures of oxidation products and incorrect assignment of oxidation products to a particular CYP are also possible.
Another major concern is that the data shows relative susceptibilities to oxidation of atoms within each individual molecule, but the meaning of “1” may not be the same between molecules and it is not clear that atoms from different molecules can be pooled in a single training set. For instance, consider a purely aliphatic compound A with an N-methylpiperidine and a methoxy group. The N-methyl is established as a site of oxidation (1). The methoxy methyl is not (0). On the other hand, consider a completely aromatic compound B. One of the aromatic carbons is marked as the site of oxidation (1), although on an absolute basis the methoxy methyl group in A is probably much more susceptible than the aromatic carbon in B. Despite the potential difficulties, however, we have produced a reasonably self-consistent and predictive model.
Descriptors
We have examined several descriptors that describe the local environment around each nonhydrogen atom i in each molecule. The first types (SS, SS-A, SS-B) can be called “detailed substructure descriptors” (SS for substructure). Solvent accessible surface area of hydrogens is obviously important and is represented by the descriptors HYDROGENAREA and NONHYDROGENAREA. The PE (physiochemical environment) and HYDROPHOBICMOMENT descriptors describe the long-range environment around atom i. The final descriptor SPAN has to do with where atom i is placed in a molecule. Only the HYDROPHOBICMOMENT, HYDROGENAREA, and NONHYDROGEN area require the 3D structure of a molecule; all the others use the connection table only. Details follow:
1. SS
This is the nonhydrogen-centric version of the substructure descriptors introduced in Singh et al.7 to describe local chemical environments. They have the form ATi, ATi-ATj, ATi-ATj-ATk, ATi-ATj-ATk-ATm where ATi is the type of atom i, ATj is the atom one bond away, ATk the atom two bonds away, etc. Atom i is always the candidate atom for oxidation. Atom type includes the element, the number of nonhydrogen neighbors and the hybridization of the atom. It might also include some special properties: what kind of ring the atom i is in: a5, a6 for five- and six-membered aromatic rings, and A5, A6 for five and six-membered aliphatic rings. br5, br6 mark bridgehead atoms in 5- and 6-membered rings. The PATTY 14 notation for defining these types is in Supplementary Material. It is established, since one can fit and predict AM1 dehydrogenation energies very well with this type of descriptor 7, that they contain implicit information about dehydrogenation energy.
2. SS-A, SS-B
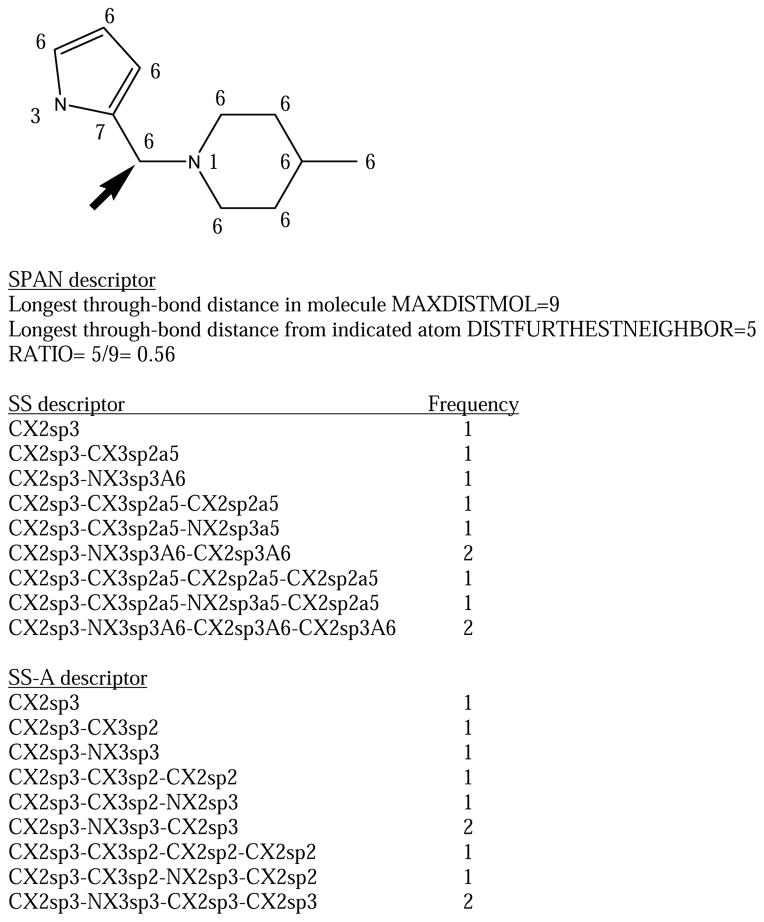
The information in SS can be split into two separate descriptors, one SS-A including the element/neighbors/hybridization information, and one SS-B including the special properties. In the case of SS-B, an atom without any special properties is labelled “n” (for “none”). Figure 1 shows the substructure descriptors for a candidate site in a small example molecule.
Figure 1.
The topological substructure descriptors (SS, SS-A, SS-B) and physiochemical environment (PE) descriptors for an atom (indicated by arrow) in an example molecule. The number near each atom is it's physiochemical type (1=cation, 3=H-bond donor, 6=hydrophobe, 7=other). Also indicated is the ratio for the SPAN descriptor that determines whether an atom is at the end or middle of a molecule based on its topology.
3. HYDROGENAREA
As with Singh et al. we use the areas of attached hydrogens. Not all atoms i have hydrogens, but for the ones that do we note the total area of all the hydrogens attached to atom i (SUM), the mean area (MEAN), and the minimum (MIN) and maximum (MAX) areas. Again, as in Singh et al., the area of any given hydrogen is an average over 25 conformations generated by our Flexibase procedure 15 starting from the CORINA16 conformation. A variation of this is to use the area of a single CORINA conformation.
4. NONHYDROGENAREA
This is the same as the above, except that one looks at the areas of nonhydrogens once the hydrogens are removed from the structure.
5. PE
These are of the form AT_d, where d is the through-bond distance to atom i and AT may be one of the following: 1=cation; 2=anion, 3=neutral H-bond donor, 4=neutral H-bond acceptor, 5=donor/acceptor, 6=hydrophobe, 7=none of the above. Definitions of these types are in reference 14. Figure 1 shows the PE descriptors for a candidate site in the example molecule.
6. HYDROPHOBICMOMENT
Given a CORINA conformation, one calculates the hydrophobic moment vector of a molecule (analogous with dipole moment with atom type replacing charge; type “6” above is hydrophobic). We note the length of the hydrophobic moment, and the projection of atom i on the hydrophobic moment and the projection normalized by the length. This is averaged over 25 conformations. The idea of these descriptors is to distinguish atoms at the hydrophobic ends of molecules vs. those at the hydrophilic end, under the hypothesis that the hydrophobic ends are preferentially oxidized.
7. SPAN
This is a measure of whether the candidate oxidation site is at the end or the middle of a molecule in a topological sense. One notes the longest through-bond distance in the molecule, MAXDISTMOL. One then notes the longest distance from atom i to any other atom in the molecule, DISTFURTHESTNEIGHBOR. The descriptor RATIO= DISTFURTHESTNEIGHBOR/MAXDISTMOL is 0.5 if atom i is exactly at the middle of the molecule and 1.0 at the end. Figure 1 illustrates this for one candidate site.
Descriptors were generated using our in-house modeling infrastructure MIX.
Random forest QSAR method
Random forest 17 is an ensemble recursive partitioning method that constructs predictions by averaging over multiple “trees”. Each tree in the forest is constructed from a different bagged subset of the training set and at each branch point of the tree the method chooses from a random subset of the descriptors. We used the R implementation of random forest (http://cran.r-project.org/src/contrib/Descriptions/randomForest.html). In our experience random forest gives the best cross-validated predictions compared to other major QSAR methods (partial-least squares, k-nearest neighbors, neural networks, etc.), and this is true for our particular dataset as well. Recursive partitioning methods like random forest have the advantage that not all the cases have to be fit by one model (e.g. sp3 carbons and sp2 carbons can have their own set of rules), coupling between descriptors is naturally handled, and it is not assumed that the activity is a linear function of the descriptors. Also, recursive partitioning methods are not affected by having large numbers of irrelevant descriptors, so descriptor elimination is not necessary. The importance of a descriptor for a random forest model may be gotten from the “out-of-bag” predictions during model building (on the average bagging leaves out about one-third of the cases). Each descriptor is in turn randomly reassigned to the wrong case and the accuracy of the prediction (over multiple trees) is monitored. The out-of-bag prediction accuracy will become much worse when an important descriptor is permuted, but will change little when an unimportant one is.
For our QSAR models, each atom was treated as a separate entity with its own descriptors and binary response. Similarly each atom was predicted as a separate entity. We generated the models using 100 trees; having more trees generally does not improve the predictions 14. Predictions were returned as probabilities that a given atom would be a site of metabolism, a number between 0 and 1.
Generally, when we speak of predictions in this paper, it will refer to cross-validated predictions on the calibration set. Half the molecules in the calibration set for a particular CYP were randomly selected, and a QSAR model was constructed from the atoms in those molecules. Then, the responses for atoms in the remaining molecules were predicted. This was repeated 20 times. The predicted response for any particular atom is the mean over the number of predictions for that atom, on the average 10 predictions for the 20 trials. At no time is a molecule being predicted represented in the QSAR model doing the prediction. “Leave-half-out” is usually considered a very conservative method of cross-validation, certainly less likely to overestimate the goodness of prediction than leave-one-out cross-validation.
For the external sets, a QSAR model was made from all the compounds in the calibration set for a particular CYP and atoms in the molecules in the external set for that CYP were predicted against it.
Measures of goodness
One may quantitatively measure the “goodness” of a model in a number of ways. Cross-validated R2 of the cross-validated predictions vs. the observed responses is a standard for QSAR, but this is not appropriate when the activities are binary and we do not expect a particularly linear response. Therefore we use the following methods: 1. One may construct Receiver Operating Characteristic (ROC) curves 18 by ordering the atoms in decreasing predicted probability of being an oxidation site, and monitoring how many true and false positives are found as atoms are checked in that order. Here we will use “molecule-scaled” predictions. The maximum prediction of all atoms in a particular molecule is set to 1.0, the lowest to 0.0, with the other atoms linearly scaled between. For regioselectivity, where we are trying to find the relative probability of oxidation for atoms within a single molecule, this is more appropriate than using the raw predictions. The ROC curve for the case where the predictions are perfect would be the left and top sides of a square (area under the curve=1.0), and the curve for the case where the predictions are no better than random would be a diagonal line (area=0.5). Since the ROC curve pools all atoms regardless of what molecule they are in, not quite what is needed for regioselectivity, we need other measures in addition: 2. In a “Molecule-scaled prediction plot,” atoms are plotted with their molecule-scaled prediction on the y-axis and the molecule they are from on the x-axis. Thus all the atoms in a given molecule are in a single column. Usually the molecules are arranged left to right in order of decreasing Z-score. Z-score=(M1-M0)/S0, where M1 is the mean prediction for the atoms that are oxidation sites in the molecule, and M0 and S0 are the mean and standard deviation prediction for the atoms that are non-sites. Since the Z-score is characteristic of a single molecule, it does not matter if one uses the raw or molecule-scaled prediction. The more positive the Z-score, the better the discrimination of the sites from the non-sites. A prediction that did not discriminate sites at all would have Z-score=0. One can use the mean Z-score over all the molecules as a measure of goodness. 3. Often regioselectivity models are measured by of the percent of the molecules for which at least one of the k atoms in a molecule with the highest predictions is an observed oxidation site. Typically k=2.
Comparison to other methods
We compare our current method against the earlier model of Singh et al. 7 and MetaSite8, which at the time of writing is the only widely distributed package for predicting regioselectivity. For the purposes of generating a ROC curves for Singh et al. predictions, we ordered the nonhydrogen atoms in order of increasing AM1 dehydrogenation energy. In accordance with the model, if the maximum solvent accessible area of all attached hydrogens was < 8 Å2, or if there were no attached hydrogens, the atom was given a dehydrogenation energy of 99 kcal/mol, an arbitrarily high number near the maximum dehydrogenation energy, which serves to put such hydrogens at the end of the sorted list.
A license for MetaSite was obtained from Molecular Discovery (http://www.moldiscovery.com). Here we show results from version 2.7.5. MetaSite can handle sp3 carbons, sp2 carbons, sulfurs, and aromatic nitrogens. We followed the default protocol: “reactivity correction on” and a maximum of 20 conformations. MetaSite produces a prediction for each nonhydrogen atom in each conformation, but there are two types of score averaged over the conformations: Averaged Similarities and Averaged Ranking. It is the latter that is recommended by the vendor to get the best predictivity.
RESULTS
Which descriptors are important?
Building a QSAR model involves relating the activity of interest (here the probability of being an oxidation site) to the structure (here the attributes of nonhydrogen atoms) which is represented as descriptors. In this section we explain which of the descriptors presented in the Methods section are important for activity. Having tried a number of descriptor combinations and checking the cross-validated predictions with ROC curves, we settled on SS-A, SS-B (substructure descriptors), HYDROGENAREA (exposure of hydrogens), PE (physiochemical environment), and SPAN (end-vs-middle) as a reasonable minimum combination of descriptors that gives the best cross-validated predictions. SS-A plus SS-B is slightly superior to SS because making the atom types in the substructure descriptors too specific hurts the ability of the model to extrapolate, especially for the two smaller datasets (2D6 and 2C9). We stay with HYDROGENAREA, the solvent accessible surface area of each hydrogen averaged over multiple conformations, to be consistent with our previous work. However, using only a single conformation also gives reasonably predictive models, as does using the area of the atoms of a molecule from which the hydrogens have been deleted.
One way of appreciating the relative contributions of the descriptors is to look at the descriptor importances in Table 2. Descriptors that are on the average negatively correlated with being an oxidation site (independently of the QSAR model) are marked with *. Since random forest is not a linear method, one cannot always interpret a high descriptor importance for a descriptor as suggesting that higher values of that descriptor means a higher probability of being an oxidation site (or a lower probability for the descriptors marked with *). In some cases it may be that intermediate values of the descriptor give the highest probability.
Table 2.
Twenty most important descriptors for QSAR models.
| Descriptor | Importance |
|---|---|
| 3A4 all nonhydrogen atoms | |
| CX1sp3-NX3sp3 | 19.304 |
| HYDROGENAREA_SUMAREA | 18.269 |
| 1_1 | 15.726 |
| CX1sp3-NX3sp3-CX1sp3 | 14.709 |
| HYDROGENAREA_MAXAREA | 14.368 |
| HYDROGENAREA_MEANAREA | 13.125 |
| HYDROGENAREA_MINAREA | 12.349 |
| CX1sp3-NX3sp3-CX2sp3-CX2sp3 | 11.419 |
| SPAN_RATIO | 10.523 |
| CX1sp3-NX3sp3-CX2sp3 | 10.337 |
| HYDROGENAREA_NHYD | 9.420 |
| SPAN_MAXDISTMOL | 7.294 * |
| CX2sp3-NX3sp3-CX2sp3 | 7.020 |
| SPAN_DISTFURTHESTNEIGHBOR | 6.810 * |
| 6_1 | 6.406 * |
| 6_3 | 5.893 * |
| 6_5 | 5.812 * |
| 6_4 | 5.015 * |
| 6_6 | 4.959 * |
| SX2sp3 | 4.881 |
| 3A4 sp3 carbon with H | |
| 1_1 | 16.368 |
| CX1sp3-NX3sp3-CX2sp3 | 11.224 |
| CX1sp3-NX3sp3 | 10.804 |
| HYDROGENAREA_MEANAREA | 9.568 |
| HYDROGENAREA_SUMAREA | 9.549 |
| 6_1 | 9.214 * |
| CX1sp3-NX3sp3-CX1sp3 | 8.329 |
| SPAN_RATIO | 8.132 |
| HYDROGENAREA_MAXAREA | 7.620 |
| HYDROGENAREA_MINAREA | 7.438 |
| SPAN_MAXDISTMOL | 6.862 * |
| 6_5 | 6.313 * |
| SPAN_DISTFURTHESTNEIGHBOR | 5.189 |
| CX1sp3-NX3sp3-CX2sp3-CX2sp3 | 4.998 |
| CX2sp3-NX3sp3-CX2sp3-CX1sp3 | 4.803 |
| 6_4 | 4.563 * |
| 6_3 | 4.353 * |
| 6_2 | 4.320 * |
| CX2sp3-NX3sp3-CX2sp3 | 4.310 |
| 6_8 | 4.168 |
| 3A4 sp2 carbon with H | |
| REGIO8_MAXDISTMOL | 2.278 * |
| 6_3 | 2.126 * |
| HYDROGENAREA_MEANAREA | 2.071 |
| HYDROGENAREA_MINAREA | 1.964 |
| HYDROGENAREA_MAXAREA | 1.963 |
| SPAN_RATIO | 1.887 |
| SPAN_DISTFURTHESTNEIGHBOR | 1.805 * |
| HYDROGENAREA_SUMAREA | 1.696 |
| CX2sp2-CX3sp2-OX1sp3 | 1.476 |
| 6_6 | 1.053 * |
| 5_2 | 0.990 |
| 6_4 | 0.944 * |
| 7_3 | 0.902 |
| CX2sp2-CX3sp2-ClX1sp3 | 0.878 |
| a6-a6-a6-a6 | 0.844 |
| 6_7 | 0.840 * |
| 6_8 | 0.821 * |
| 6_5 | 0.766 * |
| 3_4 | 0.755 |
| CX2sp2-CX2sp2-CX2sp2 | 0.713 |
| 2D6 all nonhydrogen atoms | |
| HYDROGENAREA_SUMAREA | 10.569 |
| HYDROGENAREA_MEANAREA | 7.888 |
| HYDROGENAREA_MAXAREA | 7.559 |
| CX1sp3-OX2sp3-CX3sp2-CX2sp2 | 6.896 |
| SPAN_RATIO | 6.710 |
| HYDROGENAREA_MINAREA | 6.453 |
| CX1sp3-OX2sp3-CX3sp2 | 5.507 |
| HYDROGENAREA_NHYD | 5.284 |
| CX1sp3 | 4.804 |
| CX1sp3-OX2sp3 | 4.627 |
| CX2sp2-CX2sp2-CX2sp2-CX3sp2 | 3.131 |
| 6_1 | 3.055 * |
| SPAN_MAXDISTMOL | 2.816 * |
| 6_3 | 2.720 * |
| CX1sp3-CX3sp2-CX2sp2 | 2.538 |
| SPAN_DISTFURTHESTNEIGHBOR | 2.531 |
| 6_4 | 2.441 * |
| n-n-a6 | 2.376 |
| 4_1 | 2.337 |
| 6_2 | 2.216 * |
| 2C9 all nonhydrogen atoms | |
| HYDROGENAREA_SUMAREA | 7.104 |
| HYDROGENAREA_MAXAREA | 6.227 |
| HYDROGENAREA_MEANAREA | 5.680 |
| HYDROGENAREA_MINAREA | 5.574 |
| SPAN_RATIO | 5.189 |
| CX1sp3 | 3.729 |
| HYDROGENAREA_NHYD | 3.428 |
| SPAN_MAXDISTMOL | 2.914 * |
| CX1sp3-OX2sp3-CX3sp2-CX2sp2 | 2.777 |
| 6_3 | 2.361 * |
| SPAN_DISTFURTHESTNEIGHBOR | 2.313 |
| CX1sp3-CX3sp2-CX2sp2 | 2.164 |
| CX2sp2-CX2sp2-CX2sp2-CX3sp2 | 1.988 |
| CX1sp3-OX2sp3-CX3sp2 | 1.975 |
| 6_2 | 1.790 * |
| 6_8 | 1.690 |
| 6_1 | 1.689 * |
| 6_4 | 1.642 * |
| CX1sp3-OX2sp3 | 1.614 |
| a6-a6-a6-a6 | 1.603 * |
For 3A4, the most important descriptor is CX1sp3-NX3sp3, which indicates that methyl groups adjacent to sp3 nitrogens with 3 neighbors are the most likely sites of oxidation. This is especially true if the nitrogen is a cation as shown by the PE descriptor 1_1 (one bond away from a cation). This is not surprising given that N-demethylation is a widely observed reaction of CYP3A4. HYDROGENAREA descriptors are important as expected, the more exposure the better. SPAN_RATIO indicates that atoms at the ends of molecules are more likely to be oxidized than atoms in the middle. The importance of SPAN descriptors is evidence that at least some orientation issues are important for 3A4, contrary to the assumptions in Singh et al. The oxidation of –S- (SX2sp3) is among the top 20 descriptors.
There are enough atoms in the 3A4 dataset that one can further dissect the descriptor importances by generating a QSAR model for only a subset of the atoms, here the sp3 carbons with hydrogens and the sp2 carbons with hydrogens. The descriptor importances for sp3 carbons with hydrogens resemble that for the full set (not surprisingly because they account for the majority of the total atoms and 80% of the oxidation sites in 3A4) except that the relative importance of the HYDROGENAREA terms becomes less. Some of the descriptors below the top 20 are interesting relative to some of the systematically wrong predictions of Singh et al.. Descriptors number 21, 25, and 26 are n-n-n, n-n-n-n, and n-n. They indicate that, among sp3 carbons, oxidation is favored on non-ring atoms. We believe this reflects the fact that carbons in piperidines and piperazines (among the most common aliphatic rings in drugs) are rarely oxidation sites for 3A4 despite being adjacent to nitrogens.
For sp2 carbons with hydrogens, HYDROGENAREA terms are still important for 3A4. The site adjacent to a phenolic oxygen is especially favorable as indicated by CX2sp2-CX3sp2-OX1sp3 and 5_2 (two bonds from a donor/acceptor). SPAN parameters are still important, although SPAN_RATIO is no longer clearly the most important among them. The fact that nearly all oxidation sites at sp2 carbons for 3A4 are on 6-membered aromatic rings is reflected by the descriptor a6-a6-a6-a6.
For 2D6, HYDROGENAREA and SPAN descriptors are clearly important. The most important substructure descriptors (CX1sp3-OX2sp3-CX3sp2, CX1sp3-OX3sp3, n-n-a6, 6_4 etc.) are all consistent with the very common O-dealkylation of aromatic methoxy by 2D6. There is evidence for oxidation at the para-position of aromatic rings (CX2sp2-CX2sp2-CX2sp2-CX3sp2) and at aromatic methyl groups (CX1sp3-CX3sp2-CX2sp2) as well. The putative 3D “pharmacophore” for 2D6 oxidation, where oxidation sites are expected to be 5 to 7 Å from a cation 11 is not discernible as such, but the closest is the 22nd descriptor 1_8 (8 bonds from a cation). A through-bond distance of 8 corresponds to a through-space distance of 7.8 + 0.9 Å in CORINA-built structures, somewhat longer than expected. If one builds a QSAR model for 2D6 using only cationic substrates, the 1_8 descriptor is 13th in importance.
The set of top 20 descriptor importances for 2C9 qualitatively resemble those for 2D6. We can discern no descriptor that corresponds to a pharmacophore for 2C9.
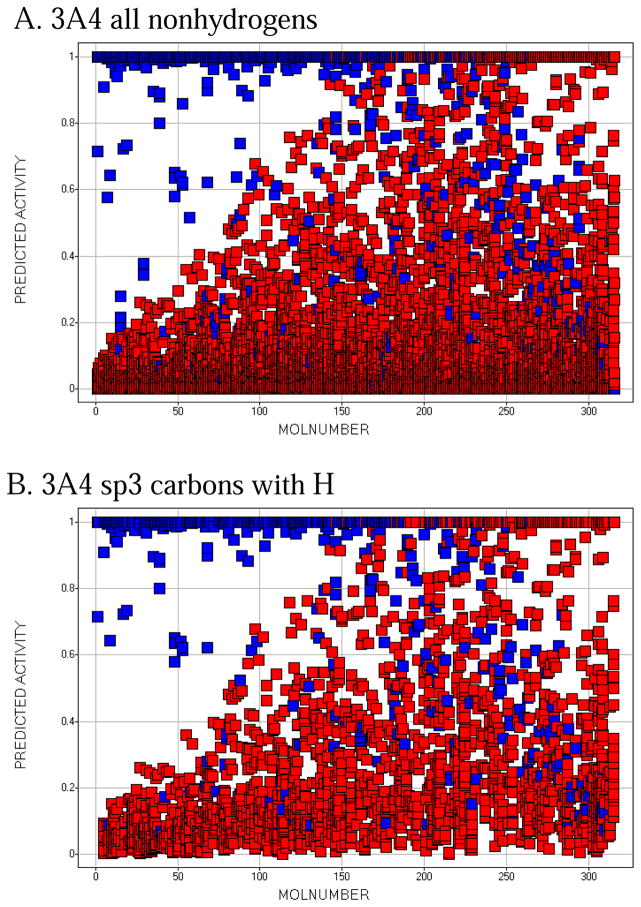
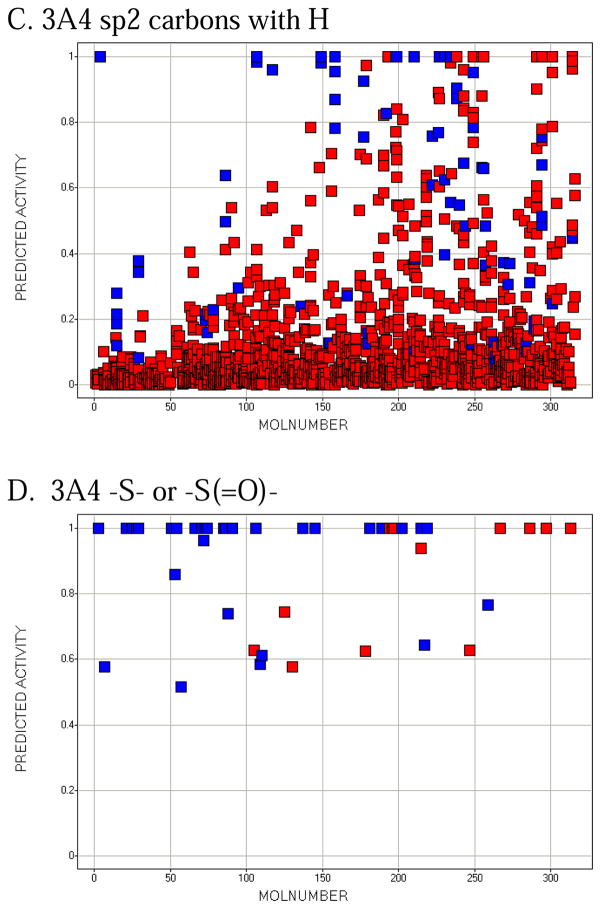
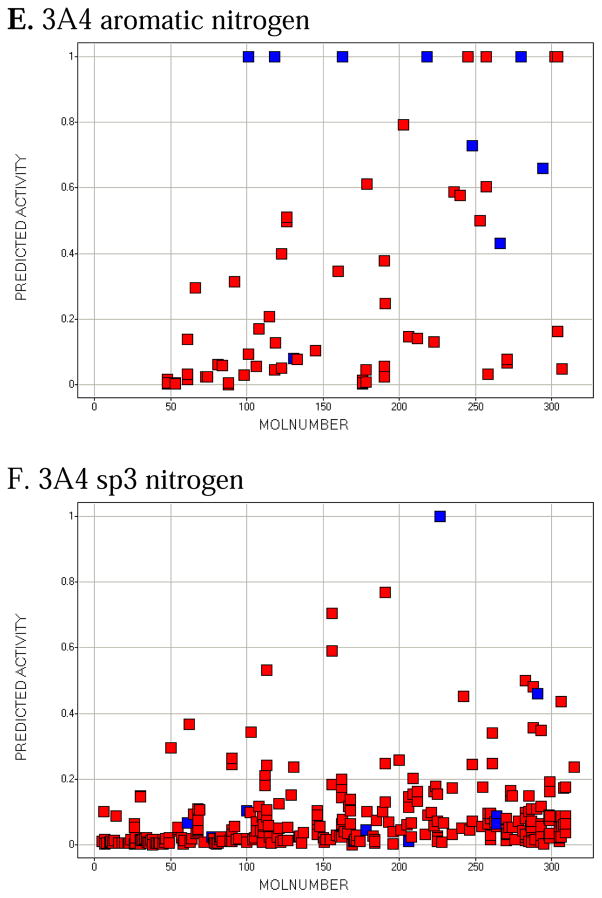
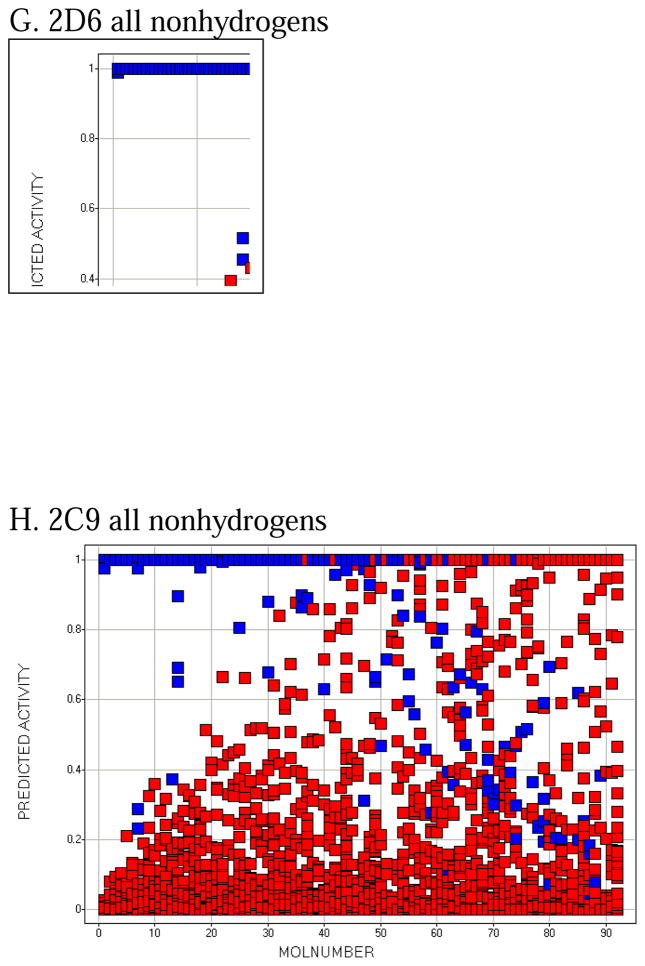
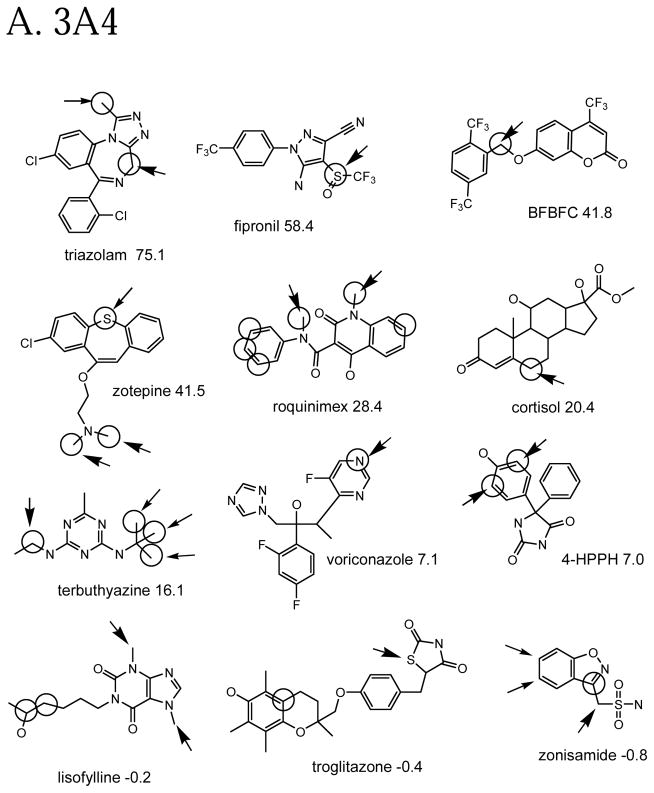
Molecule-scaled cross-validated prediction plots are shown in Figure 2 for the calibration set. If the models were perfectly predictive, all the blue squares would be above all the red squares. Clearly, prediction is far from perfect, but reasonable. For about two-thirds of the 3A4 molecules (Figure 2A), there is a blue square at the top, indicating that the atom with the highest molecule-scaled cross-validated prediction is indeed an oxidation site. However, after that the predictions tend to degrade and at the right side the blue squares are toward the bottom, indicating particularly bad predictions. Again for 3A4, we can further dissect the plot by looking at subclasses of potential sites. Figure 2B shows the plot for sp3 carbons with hydrogens. It resembles the full plot, again not surprisingly, because sp3 carbons account for a large majority of the oxidation sites. Figure 2C shows the plot for sp2 carbons with hydrogens. The model has more trouble predicting these than sp3 carbons, that is, fewer blue squares are near the top. Figure 2D shows the plot for –S- and –S(=O)-. The model does well here. This not surprising since whenever a potentially oxidizable S appears in a molecule, it is likely to be an oxidation site for 3A4 (i.e. there are more blue than red squares), and any statistical model is sure to incorporate that information. The model correctly predicts 5 out of 10 aromatic nitrogen sites in Figure 2E near the top of the plot. However, sp3 nitrogens are poorly predicted, with most of the blue squares at the bottom. Again, not surprising since very few sp3 nitrogens are oxidation sites.
Figure 2.
Molecule-scaled prediction plots for the cross-validated predictions. The y-axis is the molecule-scaled prediction from 0 to 1. Atoms in one molecule fall in a single column. Molecules are ordered from left to right based on the Z-score, how much higher the observed oxidation sites (blue squares) are predicted relative to the other atoms in a molecule (red squares).
The molecule-scaled prediction plot for 2D6 (Figure 2G) also looks very good, with about two-thirds of the compounds having a blue square at the top of the plot. The plot for 2C9 (Figure 2H) looks slightly less good, with only about half of the molecules having a blue square at the top. Given that the 2C9 dataset is the smallest, it is not be surprising that the cross-validated predictions would be poorest. The dissections of 2D6 and 2C9 (not shown) are qualitatively similar to those for 3A4, except that there are few or no examples of aromatic nitrogens or sp3 nitrogens being oxidation sites.
Examples of well-predicted and poorly-predicted molecules by cross-validation
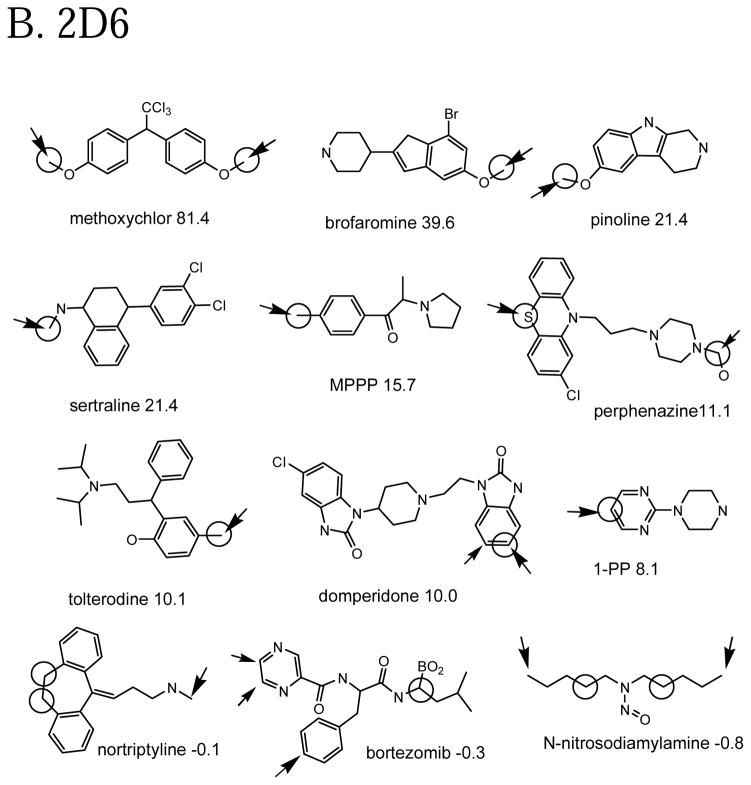
Some example 3A4 molecules from the left and right sides of the plot in Figure 2A are shown in Figure 3A. We try here to show a variety of oxidation sites. Not surprisingly, the molecules with the highest Z-scores ones have commonly oxidized groups (N-alkyls, sulfur, etc.) and/or a few aliphatic carbons among many aromatic ones. However, there are other not so common cases (aromatic oxidations, aromatic nitrogen oxidations) where the Z-score is reasonably high. It is perhaps more interesting to look at the molecules where the prediction fails. For instance in lisofylline, the model would not expect an aliphatic carbon to be oxidized when much more attractive N-methyl groups are present. The model expects –S- to be easily oxidized in troglidazone, while the observed reaction is a rare ring opening. Similarly, the model expects an sp3 carbon near unsaturated carbons in zonisamide to be oxidized, not a ring opening. Troglitazone is an example where, after our citations had been compiled, a different citation with an additional metabolite was brought to our attention. Reddy et al. 19 proposed that an S oxidation, the site predicted by the model, leads to opening of the thiazolidinedione ring and formation of a glutathione conjugate.
Figure 3.
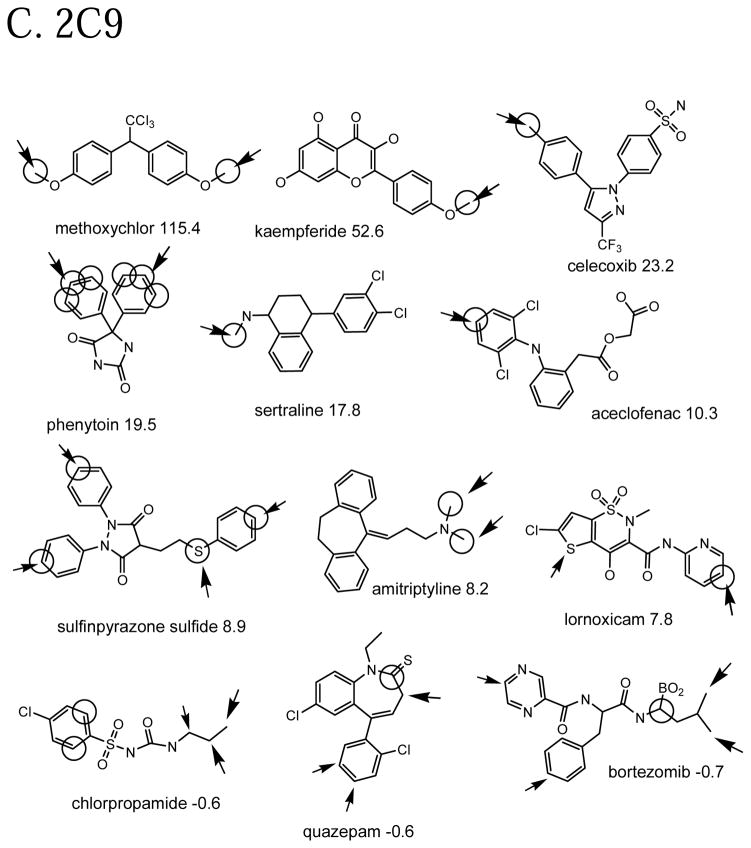
Example molecules in the calibration set that are well-predicted and poorly predicted in cross-validation by the QSAR models. Observed oxidation sites are circled. The large arrow points to the atom with the highest cross-validated prediction in the molecule (molecule-scaled prediction=1). The smaller arrows are for molecule-scaled predictions > 0.5. The number after the molecule name is the Z-score for that molecule. Molecule nomenclature is from the original citations.
Figure 3B shows the same for 2D6. Again, not surprisingly molecules with aromatic methoxy and aromatic methyl have the largest Z-scores, but sulfur oxidations and N-demethylations are also observed. The model expects an N-demethylation instead of a tryciclic ring oxidation in nortriptyline. Bortezomib is unique in having a boron atom, so cross-validated prediction from the other molecules is unlikely to predict it correctly. The model expects atoms at the end of molecules to be oxidized, all else being equal, hence the misprediction for N-nitrosodiamylamine.
Figure 3C shows the same for 2C9. Again we see high Z-scores for aromatic methoxy and aromatic methyl, with sulfur oxidations and N-dealkylation. Again we have a problem with bortezemib. Quazepam is another example of an almost unique reaction that the model does not account for. Chlorpropamide has an end-vs-middle misprediction.
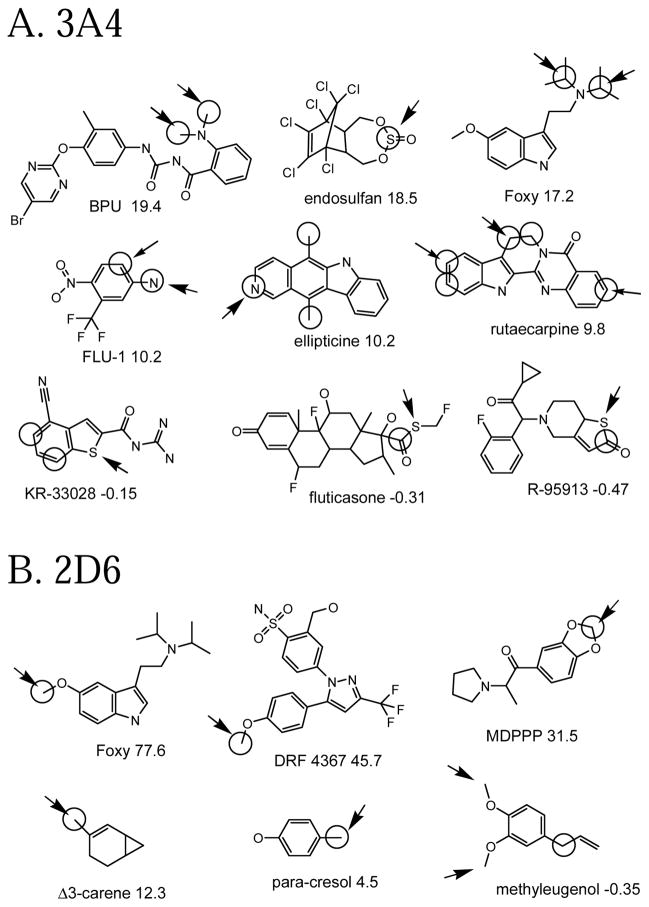
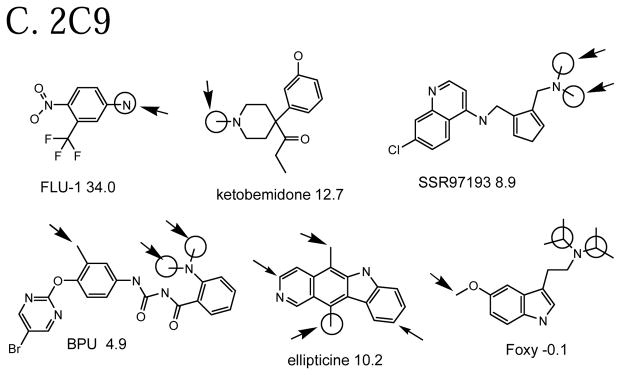
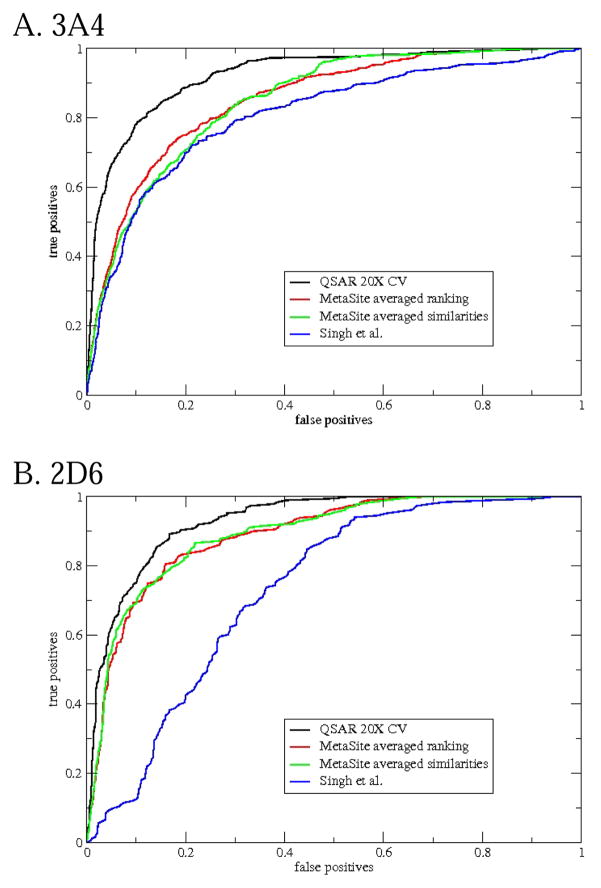
External set compounds
Having external sets gives us a chance to predict compounds that had no participation in any QSAR model. Some well-predicted and poorly-predicted compounds are shown in Figure 4. There seems to be a similar mix as with the cross-validated predictions: a few “easy” examples with high Z-scores (e.g. BPU for 3A4, Foxy for 2D6), some with moderate Z-scores (e.g. FLU-1 for 3A4 ) and a few poorly predicted ones with negative Z-scores (e.g. methyleugenol for 2D6). As with the cross-validated predictions, some of the observed oxidation sites are hard to explain. For instance, given the high propensity of 2D6 for O-demethylation, one expects the methoxy groups of methyleugenol to be the sites of metabolism, but they are not noted as such.
Figure 4.
Example molecules in the external set that are well-predicted and poorly predicted by full QSAR models. The same conventions are used for as for Figure 3.
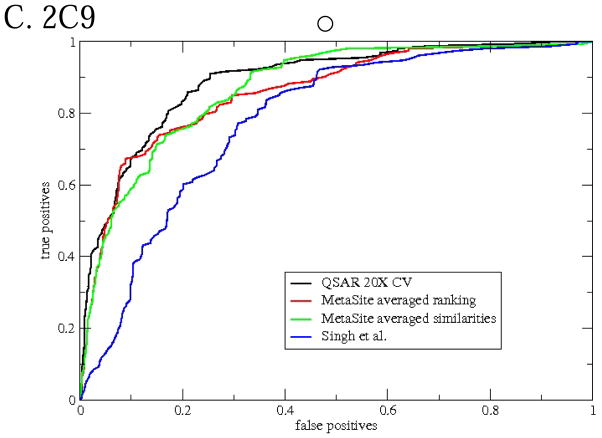
Comparison with other methods for the calibration set
ROC curves for the methods are shown in Figure 5 and measures of goodness in Table 3. The appearance of the ROC curves seems generally consistent with the table, the better the ROC curve the higher the other measures of goodness. The Singh et al. model is strictly speaking applicable to 3A4 only, but we include it for the other CYPs as well. For any of the CYPs the Singh et al. model has the worst ROC curve among the three methods. This is not at all surprising since the model is expected to work only on sp3 carbons with hydrogens. The Singh et al. model seems to do much worse for 2D6 and 2C9 than for 3A4, probably because other factors than the lability and exposure of hydrogens are more important for those CYPs.
Figure 5.
ROC curves (using molecule-scaled predictions) comparing the cross-validated predictions of the QSAR model with predictions from other models of CYP regioselectivity.
Table 3.
Measures of goodness for regioselectivity models for the calibration sets.
| CYP | Method of prediction | Area under ROC curve | Mean Z-score | % of molecules where top 2 atoms contain oxidation site |
|---|---|---|---|---|
| 3A4 N=316 | QSAR 20X cross-validated | 0.924 (0.916)a | 7.83 (5.85) | 77 (74) |
| Singh et al. | 0.803 (0.796) | 1.08 (1.05) | 51 (50) | |
| MetaSite Averaged Ranking | 0.853 (0.854) | 2.72 (2.79) | 62 (61) | |
| 2D6 N=124 | QSAR 20X cross-validated | 0.931 (0.927) | 9.32 (8.05) | 72 (70) |
| Singh et al. | 0.735 (0.746) | 0.86 (0.87) | 24 (25) | |
| MetaSite Averaged Ranking | 0.891 (0.886) | 3.47 (3.39) | 65 (64) | |
| 2C9 N=92 | QSAR 20X cross-validated | 0.894 (0.855) | 6.74 (5.06) | 73 (68) |
| Singh et al. | 0.783 (0.785) | 1.10 (1.11) | 31 (33) | |
| MetaSite Averaged Ranking | 0.862 (0.862) | 3.26 (3.06) | 69 (66) |
Number in parenthesis is for the corresponding diverse dataset.
Of the MetaSite predictions Averaged Ranking seems the better, consistent with the recommendations of the vendor. The cross-validated predictions of the QSAR model for 3A4 are clearly better than MetaSite for this set of compounds. For 2D6 and 2C9, the QSAR predictions are only slightly better than MetaSite. The MetaSite authors claim that in a diverse set of molecules they assembled, the oxidation site is in the top 2 atoms in 78, 86, and 86% of the molecules for 3A4, 2D6, and 2C9, respectively 8. Since the authors did not release the identities of the molecules in their set, we cannot verify this or try our own method on their set. Using our own dataset, the results for MetaSite in Table 3 are 62, 72, and 73%, respectively.
Detailed comparison of the predictions of the QSAR models against MetaSite for any given molecule for any given CYP shows that the predictions are very different. The correlation of molecule-scaled predictions between the QSAR cross-validated predictions and MetaSite predictions is only ~0.5 for any of the CYPs. This is also true for all subclasses of oxidation sites (sp3 carbons, sp2 carbons, etc.) as well. Perhaps this is not surprising given the methods use very different approaches.
A big concern is whether the QSAR method may have an unfair advantage over the other methods, because there are similar molecules in the training set and the test set. One way to evaluate this is to determine the relative contributions of close analogs to the datasets. For instance, if we cluster (using the method of Butina 20) the 3A4 set at 0.7 similarity using the atom pair descriptor 21, we get 243 clusters, of which 198 are singletons. We see that the largest cluster (tricyclics) has 7 members, the next largest (benzodiazepines) has 6 members, the next largest (steroids) has 4 members, etc. Similarly the 2D6 set generates 105 clusters, with the largest cluster containing 5 morphine analogs. The 2C9 set generates 81 clusters, with the largest containing 4 kaempferide analogs. These clusters are small compared to the entire set. Also, for the most part, each molecule in a cluster comes from a different citation, so they are truly independent determinations. A stronger argument comes from redoing the QSAR cross-validation on “diverse” datasets that contain only one molecule from each cluster, so that there is no possibility of a close analog of the compound being predicted being included in the QSAR model. The measures of goodness for the diverse datasets are in parentheses in Table 2. There is a detectable decrease in the measures, but the decrease is not substantial, and the predictions from the QSAR model for 3A4 and 2D6 remain better than those from MetaSite.
We can also look into the question of why the QSAR predictions for 3A4 seem to be significantly better than MetaSite while those for 2D6 and 2C9 are not. One possible explanation is that since the 3A4 dataset is much larger than the other sets, the models built during cross-validation contain more information, and thus the cross-validated predictions are likely to be better. One way to address this is to generate smaller 3A4 sets by randomly selecting 124 and 92 compounds (the sizes of the other sets) and to repeat the 20-fold cross-validation. When we do this, the ROC curve (not shown) for the cross-validated QSAR predictions for 3A4 looks only slightly better than the curve for MetaSite on the same reduced set of compounds, much like the situation with 2D6 and 2C9. Thus, the size of the dataset is at least part of the explanation.
Goodness measures for the external set
Measures of goodness for the external sets are in Table 4. Since the number of external compounds is small, one should not overinterpret the results. However, the goodness measures for QSAR predictions in this table seem about as good as the corresponding measures in Table 3. We see that the QSAR predictions in Table 4 are on a par with MetaSite predictions or slightly better, with the Singh et al. methods doing more poorly (at least in ROC area and Z-score). This is in general agreement with the results in Table 3. Thus, there is no evidence that cross-validated predictions and “real” predictions are fundamentally different for these models.
Table 4.
Measures of goodness for the external sets.
| CYP | Method of prediction | Area under ROC curve | Mean Z-score | % of molecules where top 2 atoms contain oxidation site |
|---|---|---|---|---|
| 3A4 N=19 | QSAR full model | 0.901 | 7.61 | 84 |
| Singh et al. | 0.797 | 1.03 | 58 | |
| MetaSite Averaged Ranking | 0.853 | 1.94 | 58 | |
| 2D6 N=10 | QSAR full model | 0.934 | 17.90 | 70 |
| Singh et al. | 0.842 | 1.38 | 50 | |
| MetaSite Averaged Ranking | 0.949 | 3.38 | 70 | |
| 2C9 N=9 | QSAR full model | 0.937 | 7.43 | 67 |
| Singh et al. | 0.866 | 1.50 | 67 | |
| MetaSite Averaged Ranking | 0.920 | 3.06 | 67 |
DISCUSSION
We have created a purely empirical model for CYP 3A4, 2D6, and 2C9 regioselectivity based on data from the literature. In essence we are asking “What would the literature predict would be oxidation sites in this molecule?” It should be reemphasized that our model can predict only where a molecule might be oxidized assuming it is a substrate, and cannot predict whether a molecule will actually be a substrate of a CYP, or determine which CYP might be more important for the metabolism for a given molecule. Models of whether particular molecules will be substrates or inhibitors of CYPs are under development in this and other laboratories (reviewed by Lewis 22).
Given the potential difficulties mentioned in the Introduction about combining the data of separate molecules, the QSAR approach works remarkably well, making molecule-scaled predictions at least as good as those from MetaSite, which is much more mechanistically-based (more below). This is true even for the very conservative method of cross-validation we use here. Using a less conservative cross-validation, for instance leaving only 20% of the molecules out instead of 50% makes the results appear even better. A reasonable speculation as to why the models work is that, if enough molecules are included, the relative frequencies of an atom environment appearing as the major oxidation sites of different molecules will eventually reflect the relative probabilities of those environments being oxidation sites if they occurred in the same molecule. This is reasonable because most drug molecules contain a variety of environments, for instance, they contain aliphatic and aromatic portions.
The QSAR-based approach is one of a number of valid approaches to the regioselectivity problem, and we know of at least one example where a regioselectivity model was derived from literature databases of oxidation 23. One advantage of the QSAR-based approach is that, as long as the relevant information is implicit in the descriptors used to build the model, it does not require knowledge about which mechanisms are important for regioselectivity or require that we undertake computationally expensive simulations of each mechanism. The downside of this, of course, is that our models contain no mechanistic explanation, only a statistical summary of what is already known. In our particular case, one strong motivation for attacking the problem empirically was to avoid expensive molecular orbital calculations altogether. On the other hand, any kind of QSAR approach depends on having a large body of data from which to calibrate a model, and it is never clear whether the data is sufficiently unbiased or complete enough to allow extrapolation far beyond the types of molecules and atom environments the model was built on. (A specific corollary to this is that models have trouble predicting the more rare reactions, e.g. ring openings in 3A4.) Noise in the data is always an issue unless there are a sufficient number of cases such that the noise can be averaged out. For all these reasons QSAR models are limited to those CYPs for which a great deal of regioselectivity data is already available, currently only 3A4, 2D6, and 2C9, and 2C9 is probably a borderline case.
While tried to be inclusive of all the data in the literature, it is possible to argue that the data sets we have assembled here, while diverse overall, may not be representative of drugs in general. This is to some extent unavoidable because by definition the datasets contain only known substrates for each CYP and are thereby enriched in specific chemical groups. For example the 3A4 set contains more N-alkyl amines and the 2D6 set contains more aromatic methoxy groups and cations than expected in a randomly selected set of drug molecules of the same size. We do not feel that the bias is hurting the applicability of the model, especially since the assumption of the model is that any molecule to be predicted already is a substrate. In any case, as more data is generated in the literature, the datasets will likely become more inclusive and less biased.
MetaSite 8 provides an interesting contrast to our empirical approach in that it does not depend at all on having preexisting data, but is based on first-principle arguments. It makes its predictions based on the lability of hydrogens, plus orientation effects based on the 3D structure of a CYP active site. Specifically this is done by matching the intramolecular environment of a candidate atom in a molecule (encoded by atom types and distances) to the active site environment around the heme oxygen in a CYP active site, atoms that more closely match the heme environment presumably being the ones most likely to be oxidized. Currently, MetaSite can handle 3A4, 2D6, 2C9, 1A2, 2C9, and 2C19, and can be extended to any CYP for which a homology model can be generated. Clearly, MetaSite has the advantage for 1A2 and 2C19, where there is not currently enough data in the literature to generate a QSAR model. There are other methods, which we did not examine, that predict regioselectivity by explicitly docking potential substrates into the active sites of CYPs. Zhou et al. 9 have discussed the GLUE method for predicting 3A4 regioselectivity, and de Graaf et al. 10 discuss a docking method for 2D6.
The influences on regioselectivity are usually thought to have two components, the local reactivity of the atom to be oxidized, and “orientation” effects that make broad regions of the molecule more or less likely to be attacked. We concur with the MetaSite authors’ conclusion that orientation issues are important in all CYPs, though perhaps not as much for 3A4. In the case of MetaSite and the docking-based methods the orientation information is provided by an active site model. One can expect that the results of such models will depend on which specific active site structure is used, and one can also argue that having a single explicit structure for the active site of a CYP would not necessarily provide all the needed orientation information since CYPs, oxidizing a very wide variety of substrates, are likely to have very flexible active sites that can change shape to adapt to specific molecules. Ekroos and Sjorgen 13 have recently confirmed this for 3A4 by x-ray crystallography. The fact that our QSAR models do at least as well as MetaSite suggests that, given a large amount of regioselectivity information to calibrate against, it is not necessary to use explicit CYP active site structure to get reasonable predictions.
Our QSAR model looks only at aspects of the substrate molecules, but we did try to relate the important “environment” descriptors in our QSAR models to the active site structures of the CYPs. Not surprisingly in retrospect, we were not able to do so except in a very broad sense. The active sites of the CYPs examined here are of limited size, and some longer molecules cannot fit into them, so it is not a surprise to see for all CYPs that molecules are more likely to be oxidized at the ends than in the middle. The only specific active site feature we can discern is the presence of an anionic residue in the active site of 2D6 influencing the regioselectivity of cationic substrates.
It should be noted that since neither our model nor MetaSite can explain more than about 70% of the regioselectivity data in the literature (assuming the literature data is for the most part correct), some critical information is likely missing in current modeling efforts, and more work is needed. Certainly, at present, it makes sense for a chemist to look at predictions from all available methods.
Supplementary Material
Acknowledgments
Vladimir Maiorov maintains our random forest scripts. Rhonda Torres thanks the NIH (Grant ES09122) for support. Dan McMasters provided useful discussions.
Footnotes
- A comma-separated table containing the literature references:
- For the large calibration sets.
- For the external sets.
- Mol2 files containing the structures of the molecules with activities
- For the three large calibration sets.
- For three external sets.
- Definitions for atom properties for calculating the descriptors.
References
- 1.Ortiz de Montellano PR. Cytochrome P450: Structure, Mechanism, and Biochemistry. 2. Plenum; New York: 1995. [Google Scholar]
- 2.Triolo A, Altamura M, Dimoulas T, Guidi A, Lecci A, Tramontana M. In vivo metabolite detection and identification in drug discovery via LC-MS/MS with data-dependent scanning and postacquisition data mining. J Mass Spectrom. 2005;40:1572–1582. doi: 10.1002/jms.934. [DOI] [PubMed] [Google Scholar]
- 3.Bertz RJ, Granneman GR. Use of in vitro and in vivo data to estimate the likelihood of metabolic pharmacokinetic interactions. Clin Pharmacokin. 1997;32:210–258. doi: 10.2165/00003088-199732030-00004. [DOI] [PubMed] [Google Scholar]
- 4.Wrighton SA, Schuetz EG, Thummel KE, Shen DD, Korzekwa KR, Watkins PB. The human CYP3A subfamily: practical considerations. Drug Metab Rev. 2000;32:339–361. doi: 10.1081/dmr-100102338. [DOI] [PubMed] [Google Scholar]
- 5.Jones JP, Mysinger M, Korzekwa KR. Computational models for cytochrome P450: a predictive electronic model for aromatic oxidation and hydrogen atom abstraction. Drug Metab Dispos. 2002;30:7–12. doi: 10.1124/dmd.30.1.7. [DOI] [PubMed] [Google Scholar]
- 6.Jones JP, Shou M, Korzekwa KR. Predicting the regioselectivity and stereoselectivity of cytochrome P450-mediated reactions: structural models for bioactivation reactions. Adv Exp Med Biol. 1996;387:355–360. doi: 10.1007/978-1-4757-9480-9_43. [DOI] [PubMed] [Google Scholar]
- 7.Singh SB, Shen LQ, Walker MJ, Sheridan RP. A model for predicting likely sites of CYP3A4-mediated metabolism on drug-like molecules. J Med Chem. 2003;46:1330–1336. doi: 10.1021/jm020400s. [DOI] [PubMed] [Google Scholar]
- 8.Cruciani G, Carosati E, DeBoeck B, Ethirajulu K, Mackie C. MetaSite: Understanding metabolism in human cytochromes from the perspective of the chemist. J Med Chem. 2005;48:6970–6979. doi: 10.1021/jm050529c. [DOI] [PubMed] [Google Scholar]
- 9.Zhou D, Afzelius L, Grimm SW, Andersson TB, Zauhar RJ, Zamora I. Comparison of methods for the prediction of the metabolic sites for CYP3A4-mediated metabolic reactions. Drug Metab Disp. 2006;34:976–983. doi: 10.1124/dmd.105.008631. [DOI] [PubMed] [Google Scholar]
- 10.de Graaf C, Oostenbrink C, Keizers PHJ, van der Wijst T, Jongejan A, Vermeulen NPE. Catatlystic site prediction and virtual screening of cytochrome P450 2D6 substrates by consideration of water and rescoring in automated docking. J Med Chem. 2006;49:2417–2430. doi: 10.1021/jm0508538. [DOI] [PubMed] [Google Scholar]
- 11.Ekins S, De Groot MJ, Jones JP. Pharmacophore and three-dimensional QSAR methods for modeling cytochrome P450 active sites. Drug Metab Disp. 2001:936–944. [PubMed] [Google Scholar]
- 12.Yano JK, Wester MR, Schoch GA, Griffin KJ, Stout CD, Johnson EF. The structure of human microsomal cytochrome P450 3A4 determined by x-ray crystallography to 2.5-A resolution. J Biol Chem. 2004;279:38091–38094. doi: 10.1074/jbc.C400293200. [DOI] [PubMed] [Google Scholar]
- 13.Ekroos M, Sjogren T. Structural basis for ligand promiscuity in cytochrome P450 3A4. Proc Natl Acad Sci USA. 2006;103:13682–13687. doi: 10.1073/pnas.0603236103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bush BL, Sheridan RP. PATTY: a programmable atom typer and language for automatic classification of atoms in molecular databases. J Chem Inf Comput Sci. 1993;33:756–762. [Google Scholar]
- 15.Kearsley SK, Underwood DJ, Sheridan RP, Miller MD. Flexibases: a way to enhance the use of molecular docking methods. J Comput -Aided Molec Design. 1994;8:565–582. doi: 10.1007/BF00123666. [DOI] [PubMed] [Google Scholar]
- 16.Gasteiger J, Rudolf C, Sadowski J. Automatic generation of 3D-atomic coordinates for organic molecules. Tetrahedron Comp Method. 1990;3:537–547. CORINA currently distributed by Molecular Networks ( http://www.mol-net.de/software/corina/) [Google Scholar]
- 17.Svetnik V, Liaw A, Tong C, Culberson JC, Sheridan RP, Feuston BP. Random forest: a classification and regression tool for compound classification and QSAR modeling. J Chem Inf Comput Sci. 2003;43:1947–1958. doi: 10.1021/ci034160g. [DOI] [PubMed] [Google Scholar]
- 18.Triballeau N, Archer F, Brabet I, Pin JP, Bertrand HO. Virtual screening workflow development guided by the ‘receiver operating characteristic’ curve approach. Application to high-throughput docking on metabotropic glutamate receptor subtype 4. J Med Chem. 2005;48:2534–2547. doi: 10.1021/jm049092j. [DOI] [PubMed] [Google Scholar]
- 19.Reddy VBG, Karanam BV, Gruber WL, Wallace MA, Vincent SH, Franklin RB, Baille TA. Mechanistic studies of the metabolic scission of thiozolidinenone derivatives to acyclic thiols. Chem Res Toxicol. 2005;18:880–888. doi: 10.1021/tx0500373. [DOI] [PubMed] [Google Scholar]
- 20.Butina D. Unsupervised database clustering based on Daylight’s fingerprint and Tanimoto dissimilarity: a fast and automated way to cluster small and large data sets. J Chem Inf Comput Sci. 1999;39:747–750. [Google Scholar]
- 21.Carhart RE, Smith DH, Venkataraghavan R. Atom pairs as molecular features in structure-activities studies: definition and application. J Chem Inf Comput Sci. 1985;25:64–74. [Google Scholar]
- 22.Lewis DFV, Dickins M. Substrate SAR in human P450s. Drug Discovery Today. 2002;17:918–925. doi: 10.1016/s1359-6446(02)02412-1. [DOI] [PubMed] [Google Scholar]
- 23.Borodina Y, Rudik A, Filimonov D, Kharchevnikova N, Dmeitriev A, Blinova V, Proikov V. A new statistical approach to predicting aromatic hydroxylation sites. Comparison with model-based approaches. J Chem Inf Comput Sci. 2004;44:1998–2009. doi: 10.1021/ci049834h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.