Abstract
Human thymidylate synthase (hTS), a target for antiproliferative drugs, is an obligate homodimer. Single-point mutations to alanine at the monomer–monomer interface may enable the identification of specific residues that delineate sites for drugs aimed at perturbing the protein–protein interactions critical for activity. We computationally identified putative hotspot residues at the interface and designed mutants to perturb the intersubunit interaction. Dimer dissociation constants measured by a FRET-based assay range from 60 nM for wild-type hTS up to about 1 mM for single-point mutants and agree with computational predictions of the effects of these mutations. Mutations that are remote from the active site retain full or partial activity, although the substrate KM values were generally higher and the dimer was less stable. The lower dimer stability of the mutants can facilitate access to the dimer interface by small molecules and thereby aid the design of inhibitors that bind at the dimer interface.
Graphical abstract
INTRODUCTION
Protein–protein interfaces are important but difficult targets for inhibitor design. Mutagenesis provides a means to probe protein–protein interfaces to identify potential sites for interface-perturbing drugs. A challenge is that the interface area is usually much larger than a small molecule inhibitor or a single residue that might be mutated. Obligate homodimeric enzymes in which, by definition, residues of both subunits contribute to and are essential for activity present a special class of protein–protein interfaces where disruption of the interface results in inhibition of the enzyme. In such cases, the subunits of the dimer generally bind each other with high affinity. In a homodimer, a single-point mutation affects both sides of the interface (as opposed to just one in a heterodimer). Moreover, mutation at the interface may affect enzyme activity either directly by perturbing the active site or indirectly by perturbing subunit–subunit interactions. Finally, a single point mutation can influence the affinity for inhibitors designed to bind at the interface to destabilize the homodimer or to stabilize an inactive form of the enzyme. Here, we address these issues by identifying and characterizing interfacial hotspots in the obligate homodimeric enzyme, human thymidylate synthase (hTS), with the goal of providing new sites for drug targeting of an essential function in the cell.
Thymidylate synthase (TS) (E.C. 2.1.1.45) is a highly conserved enzyme, essential for cell survival because of its important role in DNA biosynthesis.1 The methylation reaction that it catalyzes provides the only de novo source of thymidylate (dTMP; 2′-deoxythymidine 5′-monophosphate). hTS is a target for anticancer agents, some of which are clinical drugs. These compounds are inhibitors that bind in the active site and that mostly resemble either the substrate 2′-deoxyuridine 5′-monophosphate (dUMP) or the cofactor 5,10-methylene-5,6,7,8-tetrahydrofolate (mTHF). Treatment with these compounds, however, gives rise to problems of resistance and toxicity. Thus, other strategies to target hTS are being pursued.2,3
In the hTS dimer, two residues from the cognate subunit, R175 and R176, form part of the dUMP binding site in the other subunit.4 The dimeric form is therefore stabilized by substrate binding. The hTS homodimer can adopt active and inactive conformations that differ primarily in the conformation of the active site loop (residues 181–197).5 In the inactive form, the loop twists so that instead of pointing into the active site, the catalytic C195 points toward the dimer interface. Cardinale et al.2 identified peptides that bind at the dimer interface of hTS and thereby stabilize the inactive conformation of the protein. It has been shown that the TS dimer can be denatured into unfolded monomers and refolded to its native state.6 Subunit exchange between TS dimers has been observed and is even enhanced for certain mutants relative to the wild-type (WT).7,8 The fast subunit exchange of some mutants and their greater susceptibility to trypsin digestion may result from shifting the monomer–dimer equilibrium toward the monomeric form.9 Recently, we developed a fluorescence resonance energy transfer (FRET) based assay to quantitatively characterize the monomer–dimer equilibrium of hTS,10 which was used to determine a dissociation constant for WT hTS of 100 nM at room temperature.
In addition to its catalytic activity, hTS functions as a regulatory protein that binds RNA, including its own cognate mRNA.11,12 It has been proposed that inhibitor binding to the active site of hTS reduces the binding of hTS to its mRNA, leading to release of translational arrest, higher hTS protein levels, and thus, cellular drug resistance to hTS inhibitors used in anticancer therapy.11 The exact binding site of the TSmRNA on the hTS protein is not known. However, there is evidence that residues at the hTS dimer interface play an important role in hTS-mRNA recognition, perhaps by controlling a conformational change of the protein that exposes the mRNA binding site.13–15 Thus, it may be possible to design compounds that bind at the dimer interface of hTS and thereby facilitate hTS-mRNA interactions and the regulation of protein expression. Such compounds could overcome drug resistance problems, as suggested by the model proposed by Chu.11
It has been demonstrated that the binding free energy of protein–protein complexes is not evenly distributed over protein–protein interfaces but that interfaces contain “hotspots”, subsets of usually conserved residues that account for most of the binding free energy of the complex.16–18 Experimentally, hotspots can be determined by measuring the change in binding free energy, ΔΔG0d, upon mutating a residue to alanine. A hotspot is usually defined as a residue that contributes at least 4.2 kJ/mol to the binding free energy.19–22 It has been found that hotspot residues at protein–protein interfaces often protrude into favorable sites for ligand binding on the binding partner protein.23 Therefore, the identification of hotspot residues can aid the identification of binding pockets for the design of ligands to perturb protein–protein interfaces. Several computational methods for predicting hotspot residues have been developed.21,24–27 We used computational modeling to identify hotspot residues and to design interface mutants to destabilize the dimer. The WT and mutant forms of hTS were characterized with biochemical and biophysical methods, and the monomer–dimer equilibrium was measured by FRET. The crystal structures of two mutants were determined. Enzyme kinetics gave information on the effect of the mutations on substrate binding and transformation. The structural and functional characterization discussed herein is consistent with other work by some of us in which protein–peptide interactions at the subunit–subunit interface were shown to be reduced by point mutations that truncate residues to alanine.28 The results of the present study provide the first detailed structural and energetic characterization of the determinants of the hTS monomer–dimer equilibrium and show that the observed effects of the hotspot mutations can be a valuable guide for designing anticancer agents that may avoid the onset of drug resistance.
RESULTS
Hotspot Identification and Mutant Design
Most residues are highly conserved among TS species, and almost all the interface residues are highly or at least partly conserved between TS species. The hTS dimer interface contains seven arginines and six aromatic residues from each subunit. These residue types are often enriched at hotspots (see Figure S1 in Supporting Information). Putative hotspot residues were predicted using the computational alanine scanning tools FoldX27,29 and Robetta21,30 (see Materials and Methods) for both the dimeric and monomeric forms of the enzyme. The predictions for the monomer indicate the importance of the residue for the stability of the protein to thermal or chemical unfolding, whereas those for the dimer show the residues important for dimerization, i.e., the hotspots. A particular residue in the dimer was predicted as a hotspot if, for mutation to alanine, computed ΔΔG0d ≥ 4.2 kJ/mol for dimer dissociation and the effect on the protein stability was small (ΔΔG0s, the change in the stability of the hTS monomer upon mutation to alanine, was ≤ΔΔG0d for all cases except W182, which was retained for experimental characterization because it is the sole tryptophan at the interface). The predictions for the putative hotspot residues that were similar for both servers were considered and are given in Table 1 and shown in Figure 1 (see Table S2 for the monomer stability calculations). In addition to these predicted hotspots that were mostly aromatic or hydrophobic residues, R175 was also chosen as a putative hotspot because of its important role at the interface and in substrate binding. FoldX did not identify this residue as a hotspot, whereas Robetta and CUPSAT31 predicted it to be very important for the stability of the dimer. The presence of dUMP, which interacts with R175, was not accounted for in any of these calculations, as the computational methods used cannot treat the energetic contributions of ligands. Although R176 is also directly involved in dUMP binding, it was not selected as a putative hotspot because neither FoldX nor Robetta identified it as a hotspot.
Table 1.
Characterization of hTS Dimer Interface Mutantsa
| mutant | predicted ΔΔG0d(A/B) (kJ/mol)b |
measured ΔΔG0d (kJ/mol) |
Kd (M) | ΔG0d (kJ/mol) |
kcat (s−1) |
KM(mTHF) (µM) |
KM(dUMP) (µM) |
SA (unit/mg) |
|---|---|---|---|---|---|---|---|---|
| WTc | 5 ± 1 × 10−8 | −41.6 | 1.00 ± 0.01 | 6 ± 1 | 11 ± 1 | 1.00 ± 0.3 | ||
| K47A | 7.9/10.0 | 6.1 ± 0.9 | 6 ± 2 × 10−7 | −35.5 | 1.00 ± 0.2 | 18 ± 2 | 22 ± 2.5 | 0.65 ± 0.2 |
| F59A | 15.9/16.3 | 19.3 ± 0.4 | 13 ± 2 × 10−5 | −22.3 | 0.27 ± 0.05 | 15 ± 1.5 | 11 ± 1 | 0.29 ± 0.05 |
| R175A | −5.4/0.8 | >24.4 | >10−3f | <−17.2 | inactive | inactive | inactive | inactive |
| 14.2/21.3d | ||||||||
| 6.2/7.2e | ||||||||
| I178A | 9.2/9.2 | 13.4 ± 0.4 | 12 ± 2 × 10−6 | −28.1 | 0.30 ± 0.05 | 15 ± 2 | 20.8 ± 2 | 0.20 ± 0.05 |
| W182A | 9.6/8.8 | 11.2 ± 0.3 | 47 ± 5 × 10−7 | −30.4 | 0.02 ± 0.01 | ndg | ndg | 0.01 ± 0.005 |
| L198A | 8.8/8.8 | 10.6 ± 0.4 | 37 ± 6 × 10−7 | −31.0 | 0.31 ± 0.07 | 14 ± 1.5 | 21 ± 2 | 0.24 ± 0.06 |
| Y202A | 10.4/13.0 | 7.1 ± 0.3 | 9 ± 1 × 10−7 | −34.5 | 1.00 ± 0.05 | 15 ±1 | 9 ± 1 | 0.96 ± 0.12 |
| Y213A | 13.2/13.0 | ndg | ndg | ndg | 0.04 ± 0.01 | ndg | ndg | 0.03 ± 0.01 |
For each of the predicted hotspot mutants, equilibrium constants, Kd, measured by FRET at 26 ± 1 °C for hTS dimer dissociation, the corresponding free energy ΔG0d, and the measured enzymatic activity parameters are given. Predicted and measured free energies for dimer dissociation relative to WT hTS, ΔΔG0d, are given for each mutant in columns 2 and 3, respectively. See Table S2 for predictions of the change in stability of the hTS monomer fold upon mutation of the putative hotspot residues, ΔΔG0s. The uncertainties in Kd and ΔΔG0d values were estimated from the standard errors on the slopes of the ΦET vs (ΦET/CT)1/2 plot (see Figure S2). kcat, KM, and SA values are expressed as the mean ± SEM; n = 3.
Predicted effect of a single point mutation in the A or in the B subunit of the A:B hTS dimer on the dimer stability. Positive ΔΔG0d values indicate destabilization. All values were computed with FoldX unless indicated otherwise.
Kd and ΔG0d values differ with respect to ref 9 based on several new experimental results.
Robetta prediction.
CUPSAT prediction, denaturant method.
Assuming the protein to be fully dissociated (dissociation degree of >0.9) at the concentration used for the F and T conjugation (40 µM).
nd: not determined.
Figure 1.
Structure of the hTS homodimer (PDB code 1HVY) (left panel) and location of the mutations (red) in one monomer (right panel). Molecular surface representations are shown under the respective cartoon representations. The monomer interface view was created from the dimer view by removing the opposite monomer chain and rotating the structure by 90° to the left. See also Figure S1.
Enzyme Activities
The WT hTS and the mutants listed in Table 1 were expressed. In addition, the catalytic C195 was mutated to Ser and Ala and analyzed to distinguish interfacial and catalytic effects. All alanine hotspot mutants could be purified to more than 90% purity except the Y213A mutant which was difficult to purify. The enzymatic assays show that all the mutants, except for the K47A and Y202A mutants, have a lower kcat value than the WT hTS; see Table 1. R175A is inactive, presumably because the absence of the Arg side chain coordinating the phosphate group of dUMP weakens dUMP binding so much that no activity is measurable. The C195S and C195A mutants are also inactive, as expected for mutation of the catalytic cysteine. For the active mutants K47A, F59A, I178A, L198A, and Y202A, the KM value for folate is up to about 3-fold higher than for WT hTS, while the KM value for dUMP is up to about 2-fold higher. The KM values are similar for all of these mutants, implying that the mutations have little effect on the affinity of the substrates for the active sites. The specific activity (SA) remained high for the mutants with a high kcat value (K47A and Y202A), whereas it was lower for the other mutants.
Monomer–Dimer Equilibrium Constants from Biophysical Experiments
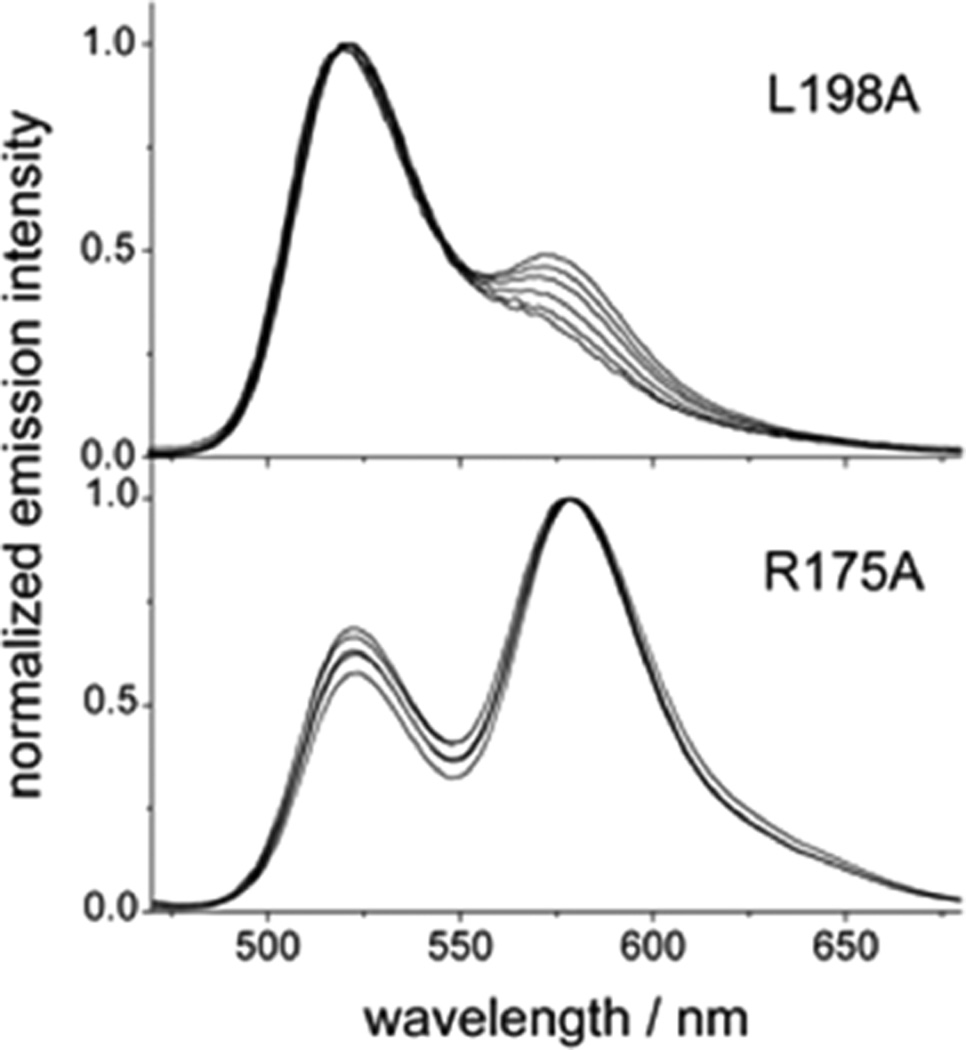
To measure the equilibrium constant for dimer dissociation, we employed a FRET assay in which hTS (WT and mutants) was tagged with fluorescein (F, excitation energy donor) and tetramethylrhodamine (T, acceptor) probes as described by Genovese et al.10 Tagging yielded samples with spectrophotometrically determined F:T:protein dimer concentration ratios near the expected 1:1:1 values for the WT and the K47A, F59A, I178A, W182A, L198A, and Y202A mutants. The only exception among the mutants measured was the R175A mutant, for which a ratio of 1:1:0.3 was obtained, indicating that about three cysteine residues per protein monomer bind a probe molecule (see “Supporting Experimental Procedures” in Supporting Information). This observation suggests that the conditions adopted to conjugate the probes only to C43 of hTS10 are appropriate for all the alanine mutants except R175A. The exceptional nature of the R175A mutant was confirmed by its emission spectra (Figure 2). All other proteins (Figures 2 and S3) display a gradual reduction in the relative emission of T (maximum at 580 nm) with respect to the emission of F (maximum at 520 nm) as the protein is progressively diluted. This observation is in accordance with the law of mass action: the reduction in protein concentration leads to a decrease in the fraction of the dimer and, correspondingly, of the F-to-T FRET efficiency. In contrast, the R175A mutant displays a concentration-independent emission spectrum, with a marked prevalence of T emission. Analysis of these data with a procedure that provided reasonable results with hTS10 yielded meaningless FRET efficiencies around 1.6 for the R175A mutant. On the other hand, these data can be rationalized if we suppose that this mutant is essentially monomeric even at the relatively high concentration (40 µM) at which conjugation with F and T maleimides was performed. Then the peculiarly small F-to-T relative emission of the R175A mutant can be accounted for using simple statistical considerations under the assumption that three Cys residues per monomer, rather than one, are accessible to a fluorescent probe molecule (see “Supporting Experimental Procedures” for details). Indeed, C180, which lies protected at the monomer/monomer interface in the hTS dimer, becomes accessible to the probe maleimides in hTS monomers. Furthermore, because of the loss of integrity of the catalytic pocket, the catalytic C195 is likely to lose protection by active site ligands and thereby become accessible to probe maleimides.
Figure 2.
Emission spectra of hTS mutants at different total protein concentrations. The spectra of the L198A mutant are representative of the spectra of all the alanine mutants except R175A (see Figure S3). L198A shows a decrease in the relative emission of the T probe (maximum at 580 nm) with respect to the emission of the F probe (maximum at 520 nm) as the protein is diluted. Total protein concentrations are the following: for L198A, in order of decreasing emission intensity at 580 nm, 2.7, 1.9, 1.4, 0.9, 0.6, 0.35 µM; for R175A, in order of decreasing emission intensity at 520 nm, 0.5–3 µM. M = moles of dimer per liter. λexc = 450 nm. The spectra are normalized at the maximum intensities for ease of comparison.
Analysis of the dependence of the observed fluorescence intensity ratio, I580/I520, on protein concentration (Figure S2) enabled the tested proteins to be ordered according to decreasing dimer stability: hTS WT > K47A ~ Y202A > W182A ~ L198A ~ I178A > F59A. For the reasons given above, R175A is the least stable dimer and R175 thus makes the greatest contribution to dimer stability. The FRET efficiency, ΦET, which equals the mole fraction of protein dimers, is related to the total protein concentration, CT (in moles of dimers/liter), and the dimer dissociation constant, Kd, through eq 1:10
| (1) |
The Kd values, determined from the slopes of plots of ΦET vs (ΦET /CT)1/2 (Figure S2), are reported in Table 1 along with the corresponding ΔG0d values.
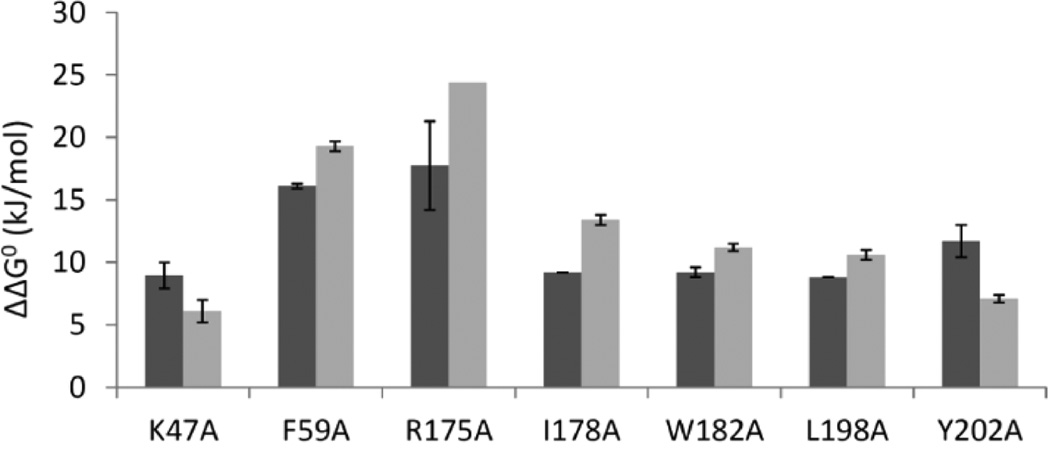
The experimentally determined relative dimer dissociation standard free energies, ΔΔG0d, for the hotspots are in good agreement with the computed trends; see Figure 3. All of the mutants have a less stable dimer than WT hTS, with experimentally measured ΔΔG0d values of >6 kJ/mol. The relative ranking by ΔΔG0d was generally correctly predicted for all mutants except Y202A.
Figure 3.
Predicted (black) and experimental (gray) differences, ΔΔG0d (kJ/mol), in the dimer dissociation energies of the hTS alanine mutants relative to WT hTS; see Table 1. The computed values are shown as an average of the predictions for the A and the B subunits of the hTS homodimer, and the error bars indicate the maximum and minimum predicted values given in Table 1. The computed values thus correspond to the mutation of one residue at the subunit–subunit interface, whereas the experiments are for mutation of two (identical) residues at the interface. All predicted values were calculated with the FoldX Web server except for R175A (predicted with Robetta). The experimental values were determined by FRET. Uncertainties in ΔΔG0d were derived by error propagation from those for Kd; see Table 1.
Structural Characterization
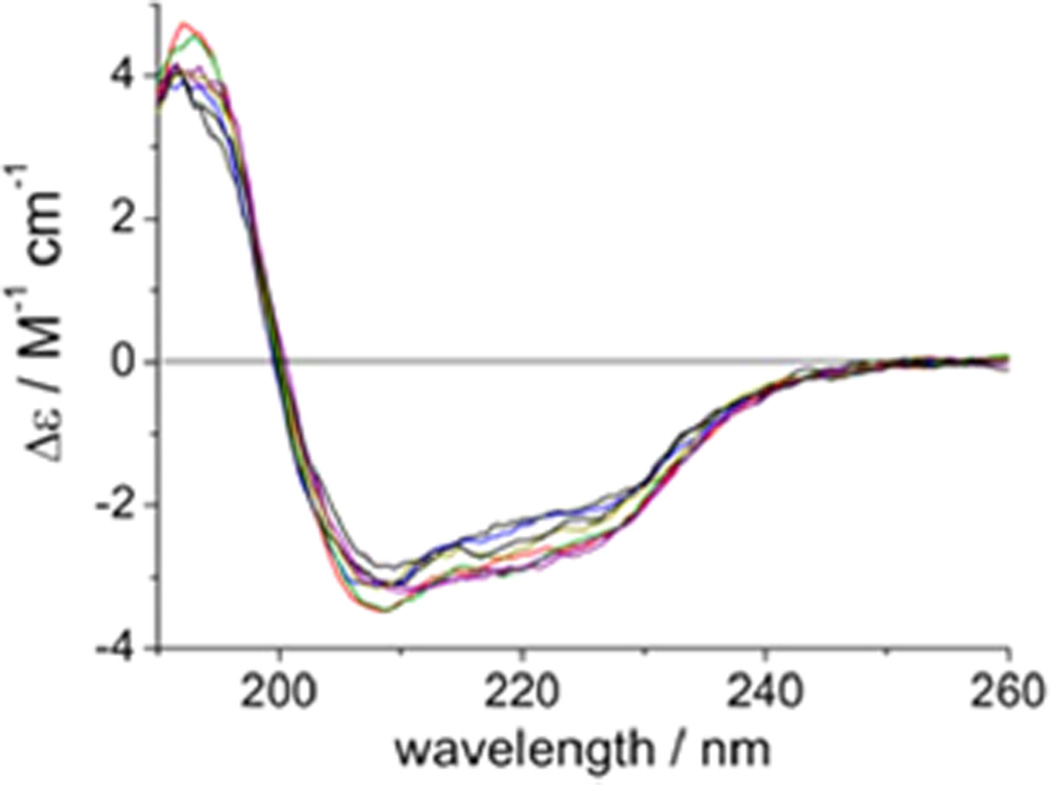
To investigate whether the mutations perturbed the hTS three-dimensional structure, we recorded circular dichroism (CD) spectra. The similarity of the far-UV CD spectra of the WT hTS and seven alanine mutants (Figure 4) indicates that the mutations do not significantly affect the secondary structure of hTS. Because the CD measurements were performed at concentrations around 1 µM at which, based on the dimer dissociation constants given in Table 1, many of the mutants are largely, if not completely, monomeric, we conclude that the monomers of the mutants are folded, homogeneous, and soluble.
Figure 4.
Far-UV CD spectra of hTS (black) and seven Ala mutants (R175A, blue; K47A, green; W182A, gray; L198A, orange; I178A, red; F59A, purple; Y202A, pink).
We determined the crystal structures of the R175A and Y202A mutants; see Table S1. As substrates stabilize the active conformation of the hTS dimer, the enzymes were crystallized in the absence of substrates. Both mutants crystallized at high-salt conditions, yielding crystals isomorphous to those of WT hTS (PDB codes 1HW3, 1HW4, 1YPV, 3NG5)2,32,33 and dimeric structures in the inactive conformation. Neither mutation induced any major change to the overall fold (e.g., the R175A structure has a Cα rmsd of 0.21 Å (with a maximal displacement of 0.87 Å from the WT hTS structure (PDB code 3N5G)2). The Fourier difference maps clearly show the mutations, but there was also no structural change or perturbation around the position of the Y202 mutation and little for the R175 mutation; see Figure 5.
Figure 5.
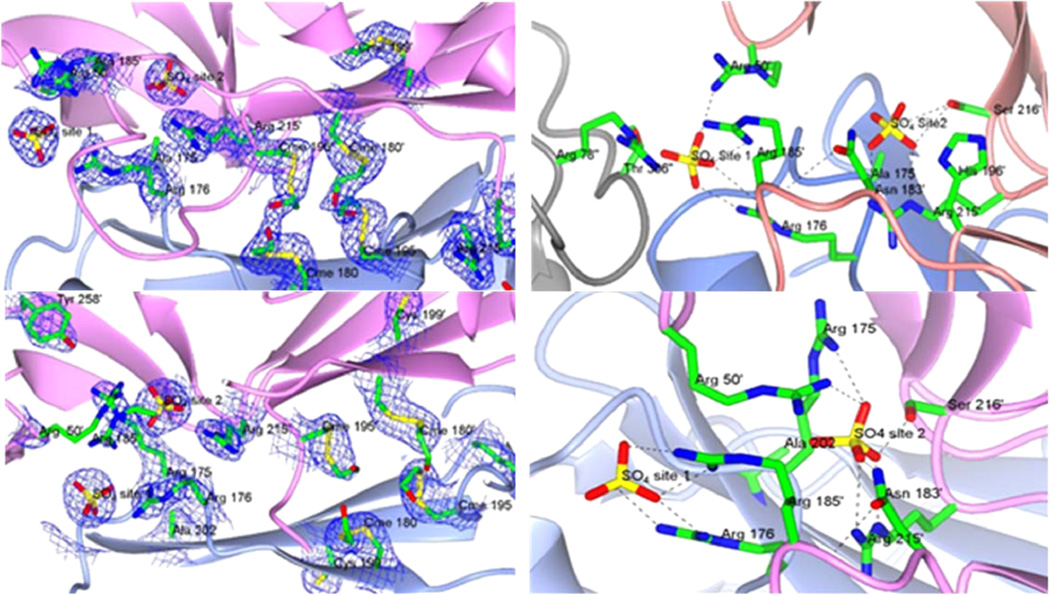
Crystal structures of the (a (top left), b (top right)) R175A and (c (bottom left), d (bottom right)) Y202A mutants of hTS. (a) View of the interface between two symmetry-related subunits where the R175A mutation is located, showing the chemically modified Cys residues, the mutated Ala 175, and the sulfate anions. (c) View of the analogous region of the Y202A mutant. In both (a) and (c), the model of the enzyme is superimposed onto the 2Fo − Fc electron density map contoured at 1.0σ (blue wire); see Figure S4 for stereoimages. (b, d) Detailed views of the salt links and hydrogen bonds (dashed lines) connecting the two sulfate anions to the residues at the interface. The first sulfate ions (site 1) are coordinated by the same five residues in both mutants R176 from one subunit and in R50′, R78′, R185′, T306′ from two different symmetry related subunits. R50′ and R185′ belong to the partner subunit of the hTS dimer, while R78″ and T306″ come from a protein molecule involved in crystal packing (gray ribbon). In the R175A mutant, the second sulfate ion (site 2) is bound to N183, H196, R215, and S216, all from the same subunit. In the Y202A mutant, the second sulfate ion makes hydrogen bonds with R50, R215, S216, N183 of one subunit and with R175 of the opposite subunit. The symmetry-related protein subunits are shown as pink and light blue ribbons. Selected residues are shown in stick representation color-coded by atom type (carbon green; nitrogen blue; oxygen red; sulfur yellow). The primes on the residue labels indicate that the residue is from the symmetry-related subunit. Cme labels in (a) and (c) refer to Cys residues chemically modified to S,S-(2-hydroxyethyl)thiocysteine by reaction with β-mercaptoethanol. Data collection and refinement statistics are given in Table S1.
As previously observed for the WT enzyme when crystallized in the inactive conformation (PDB code 3NG5),2 two sulfate ions are bound at the dimer interface in the mutants, labeled site 1 and site 2 in Figure 5. The positions and coordination of these sulfate ions are not significantly affected by the mutations. In the WT hTS and the Y202A mutant, the anion bound to this second site makes a salt bridge to R175 of the facing subunit, which is obviously missing in the structure of the R175A mutant. The R175A mutation, therefore, does not prevent the binding of the anion. On the other hand, this second binding site for anions is also close (~2.0 Å deeper in the cavity) to the site where the phosphate group of dUMP binds in the active conformation of the WT. Comparison with the dUMP-bound WT protein (PDB codes 1HVY, 1I00, 1JUJ)4,34,35 shows that R175 not only binds the dUMP phosphate with a salt link but also is H-bonded to the Y258 side chain, stabilizing it in a position where it donates a H-bond to the O3′ of the dUMP ribose. It is thus conceivable that the R175A mutant destabilizes the binding of dUMP more than the sulfate (or phosphate) anion alone and thus strongly affects enzymatic activity.
The similarity of the two crystal structures of the alanine mutants, and of the CD spectra of hTS and all tested mutants, supports the rationale for the dimer dissociation energy computations in which it was assumed that the mutations to alanine do not affect the three-dimensional structure of the protein. The active conformation was used for the calculations, whereas the inactive conformation is observed for these mutants. However, it is known that the binding of ligands in the active site favors the active conformation and the dimeric form whereas the crystal structures were determined without ligands other than sulfate in the active site. Furthermore, high concentrations of ammonium sulfate favor the inactive dimeric conformation. Thus, it is likely that, like WT hTS, the mutants can adopt the active conformation as well as the inactive conformation, depending on the presence or absence of ligands and where they bind. We nevertheless tested the dependence of the predictions on conformation by computing ΔΔG0d values for an inactive conformation (PDB code 1YPV) with FoldX. All of the residues predicted to be hotspots using the active conformation were also predicted to be hotspots using the inactive conformation (ΔΔG0d > 4.2 kJ/mol) except for W182, which is not at the interface in the inactive structure studied (see below). The quality of the agreement with the experimental values (see next paragraph) was similar, with the ranking being correct except for the Y202A mutation whose effect was similarly overpredicted.
Correlation of the Effects of Hotspot Mutations on Enzymatic Activity and Dimer Stability
The FRET experiments were aimed at providing an experimental check of the computationally predicted effects of point mutations on the stability toward dissociation of the dimeric protein. They were therefore carried out with no bound substrates. On the other hand, the kinetic experiments reflect the effects of point mutations on substrate binding affinities and transformation rates. The hTS substrates, dUMP and mTHF, are known to significantly stabilize the active dimeric form of hTS,32 and as a consequence, their concentrations are expected to affect the observed kcat and KM values. For these reasons, the relationship between the fraction of dimer present at a given enzyme concentration, calculated from the equilibrium constants derived from the FRET-based assay in the absence of substrates, and the kinetic parameters, measured in their presence, is likely complex and mutant specific. Nonetheless, we attempted to find a correlation between these two sets of results by plotting the catalytic rate constants, obtained as kcat = Vmax/CT from kinetic experiments performed at several different enzyme concentrations, CT, vs the protein dimer fractions calculated at each enzyme concentration from the FRET-derived dimer/monomer equilibrium constants; see Figure 6.
Figure 6.
Relationship between the catalytic rate constant (Vmax = maximum reaction rate, CT = total enzyme concentration in moles of dimers per liter) and the calculated protein dimer fraction for WT hTS (unfilled square) and the five active mutants Y202A (unfilled triangle), K47A (filled circle), F59A (filled triangle), I178A (unfilled circle), and L198A (filled square). The dimer fraction was obtained from the dimer/monomer equilibrium constants determined by FRET (Kd in Table 1) and the total protein concentration at which the kinetic measurements were performed.
As expected, WT hTS and the K47A mutant exhibit a rather steep increase of kcat with increasing dimer fraction but in different protein concentration ranges. On the other hand, the I178A, L198A, and Y202A mutants exhibit a remarkably low sensitivity of kcat to the protein dimer fraction. The effect of substrate binding appears to be particularly pronounced for the Y202A mutant, whose high kinetic activity indicates that the protein is fully dimeric under the conditions of the kinetic measurements, even at concentrations at which the dimer fraction is only of the order of 10% in the absence of the substrates. For the I178A and L198A mutants, the kcat value reaches a saturation value of around 0.3 s−1 at protein concentrations corresponding to dimer fractions calculated from the FRET assay results of about 0.25 and 0.35, respectively. This suggests that the dimeric form of these mutants is also stabilized by the substrates but that the mutations result in decreased catalytic rate constants. For the F59A mutant, the range of concentrations that could be explored in the kinetic experiments is too small to allow for a limiting kcat value to be extrapolated. So while it appears clear that even in the presence of the substrates, an increase in protein concentration, corresponding to an increase of the dimer fraction, correlates with an increase of kinetic activity, it is not possible to conclude whether the kinetic activity of the fully dimeric protein is comparable with those of the WT and the K47A and Y202A mutants, or it remains much lower, as for the I178A and L198A mutants.
Of the interfacial mutants, only R175A is expected to have a direct effect on the hTS active site and the binding of substrate/cofactor. None of the other residues mutated interact directly with the active site, and consistent with this, the KM values for the substrate and the cofactor are not affected much by the mutations. Thus, the effect of these mutations on activity must come primarily from their effect on the protein–protein interface.
DISCUSSION
Mutation to alanine of hot spot residues identified by computational analysis lead to measured changes in dimer stability in general agreement with predicted trends. Some of the mutants have marked decreases in catalytic activity with respect to the WT enzyme, thus supporting their crucial importance for the stability of the catalytically active hTS dimer. In a previous study, we identified seven peptides designed from the dimer interface that inhibit the catalytic activity of the protein by binding at the monomer–monomer interface of the di-inactive form of the protein.2 The interaction of these peptides with four dimer interface mutants, K47A, F59A, L198A, and Y202A, was explored by Tochowicz et al.28 The results of the present hot-spot structural and functional analysis are here analyzed in combination with those of the above peptide inhibitor studies.
Mutants involving the active site loop residues, L198 and W182, have markedly reduced activities and dimer stabilities with respect to WT hTS. L198 is at the end of the active site loop, which undergoes a large conformational change in the transition between active and inactive conformations of hTS. In both conformations, the side chains of the L198 residues on the two subunits come into contact across the interface (Cδ −Cδ distance ~4 Å). Lovelace et al. designed the hTS L198P mutant to stabilize the inactive conformation of the loop and found that it has a kcat value only 2 times lower than that of WT hTS but a KM value for dUMP 6 times higher than that of WT hTS.36 We observed the same trends in kinetic parameters for L198A, suggesting that part of the effect of this mutation may be to shift the equilibrium toward the inactive conformation of the protein. L198 mutations provide additional examples of the connection between hotspot mutation and the binding affinity versus inhibitors. Almost all peptides that inhibit WT hTS showed no activity versus the L198A mutant, possibly because of the loss of hydrophobic interaction between the L198 side chain and the peptides.28 Moreover, mutants L198I/T/F have previously been shown to be drug resistant, whereas mutants with charged residues at this position are inactive,37 thus suggesting that this residue is critical for active site binding inhibitors.
The W182A mutation was predicted to destabilize both the monomer and the dimer. The experiments show that the dimer is destabilized and that activity is almost completely lost. The W182 residues from the two subunits are about 10 Å apart (Cγ–Cγ distance) in the active conformation with the side chains pointing away from each other, and 15 Å apart in the inactive conformation with the side chains pointing toward each other. In the inactive form, there are no residues from the opposite monomer within a 4 Å sphere of W182, while in the active form, I178′ and R175′, for example, are within a 4 Å sphere of W182. In simulations of the active/inactive transition of hTS, we found that the conformational change is hindered by the two W182 side chains which must avoid steric clashes as they reorient.38 The W182A mutation introduces space at the dimer interface region, and this, besides relaxing the steric hindrance to the active/inactive transformation, likely favors the more extended inactive conformation of the active site loop.
The K47A mutant dimer is only slightly destabilized with respect to the WT dimer, retains a good level of activity, with the kcat unaffected by the mutation, and has a KM value for dUMP 2-fold higher than for WT hTS. The crystal structure of the K47A mutant with two peptidic inhibitors bound at the dimer interface displays a symmetric homodimer in the inactive conformation.39 In WT hTS, K47 forms a hydrogen bond across the dimer interface with the backbone carbonyl of Asp173′ in the second monomer. In the K47A mutant, the R175′ residue rearranges in the space formerly occupied by K47 and forms a hydrogen bond with the same backbone carbonyl group of the other monomer. This mutant, while falling into the category of “hotspot residue” as regards the dimer interface, is less perturbative than the other mutants studied here. Consistently, the interfacial peptides showed the same inhibitory effect against the K47A mutant as against WT hTS.28 Moreover, the K47E mutant has previously been shown to be active and not resistant to drugs.40 On the other hand, the double mutant K47Q/D48E exhibits a high level of resistance to 5-FdUR.41
The F59A and I178A mutants have reduced enzyme activity and strongly reduced dimer stability. Consistently, the hTS F59L mutant was void of activity.40 In EcTS, the corresponding F30A/G/P/C/S/Q/H/K/R/E mutants were found to lose their activity, whereas the F30L/Y mutants remained active.1,42 Residues 59 and 178 are located on the first and last strands of the interfacial β;-sheet of hTS, respectively, where their side chains point toward the interface. I178, being located in the proximity of the active-site loop, has different interactions on the interface depending on the conformational state of the loop. It makes contacts with W182′, A197′, and R215′ in the active state of the loop and with P194′ and R215′ in the inactive state. In contrast, F59 maintains its favorable aromatic stacking interaction with Y202 in both the active and the inactive states of the enzyme. Thus, the F59A mutation has a negative impact on the inhibition activities of peptides:28 F59 interacts with Y202 of the opposite monomer; it was expected that the mutation Y202A would be similarly critical. However, the Y202A mutation shows much weaker effects on enzyme activity than F59A, possibly due to its distance from the active site. This result suggests that the structural/functional effects of the F59A mutation are mainly due to a rearrangement induced by the mutation on the same monomer and are only partially explainable with changes in the direct binding pattern of the residue with Y202 and other residues of the opposite monomer.
The R175A mutation inactivates the enzyme, weakens dimerization 1000-fold, and results in identifiable hTS monomers because R175 extends across the dimer interface to coordinate the phosphate group of dUMP and hydrogen-bonds with Y258, which in turn makes a hydrogen-bond with O3′ of dUMP. The absence of this Arg side chain weakens dUMP binding so much that no activity is measurable. So, accounting for the dUMP in making the predictions would be expected to increase the magnitude of the computed binding free energy change associated with this hotspot residue. Studies on the corresponding arginine residues in E. coli and L. casei TS also showed a marked decrease, or a complete loss, of activity upon mutation of this to a number of other residues, due to perturbation of the binding of the dUMP phosphate moiety.1,43–47 hTS is the only TS known to adopt active and inactive conformations. The effect of the R175A mutation in hTS on the equilibrium between these two conformations is not known. However, because it perturbs dUMP binding, it is likely to push the equilibrium toward the inactive conformation, which is seen in the crystal structure. This provides an additional mechanism, not present in EcTS and LcTS, by which the R175 mutation results in enzyme inactivation.
The Y202A mutation somewhat weakens the dimer stability, but it does not affect the enzyme activity. Y202 is situated on the fourth strand of the interfacial β-sheet, and its side chain lies parallel and in contact with that of F59′ from the other subunit in WT hTS. Thus, the mutation to Ala probably does not abolish the subunit contacts and does not affect activity much because Y202 is positioned rather peripherally on the interface and distant from the active site. Previously, an hTS mutant has been reported in which the Y202F mutation was part of a multiple mutant C199L/Y202F/V204L/S206N which caused 5-FdUR resistance, indicating that the residue is crucial for FdUR phosphate binding.48 The inhibition profile of the seven peptides against the Y202A mutant was similar to that of WT hTS.28 Thus, the Y202A mutant may be a useful mutant for drug design, mimicking the WT protein but with lower dimer stability.
CONCLUSIONS
We have designed and experimentally investigated interfacial hotspot mutations in hTS. Here, we provide evidence that hotspot mutations affect the stability of the protein fold, the protein monomer–dimer equilibrium, the active–inactive conformational equilibrium of the dimer, and the binding of substrates in the active site to varying extents. The net result of each mutation on the enzyme activity emerges from the combination of these factors, as well as the concentrations of substrate, ions, and reducing agents present. The agreement between predicted and measured trends in dimer dissociation energies enabled us to rationalize the different effects of the individual mutations studied. The results provide a rather comprehensive mapping of the hTS dimer interface and pinpoint regions of the interface lined by hotspot residues, which can be explored for the design of anticancer agents targeting the hot spot area. Our mutational analysis study has identified mutants that show activity but have a less stable dimer than WT hTS, such as F59A, I178A, and Y202A, that can be useful for the design of compounds perturbing the interface, either to stabilize an inactive conformation of the WT hTS dimer or to destabilize the WT hTS dimer and act as a dissociative inhibitor. The ability to experimentally study the binding of compounds to hTS mutants with a range of homodimer stabilities will facilitate the screening and identification of compounds that target the interfacial region of hTS. We expect that the approach taken in this study of hTS can be applied to other obligate homodimeric enzymes.
MATERIALS AND METHODS
Hotspot Prediction and Mutant Design
The crystal structure of the active, closed conformation of hTS (PDB code 1HVY, A/B chains)4,49 was the basis for predictions. All S,S-(2-hydroxyethyl)-thiocysteines were changed in silico to cysteine using the Sybyl software, version 7.2 (Tripos, St. Louis, MO; http://www.tripos.com). The conservation level of the interface residues was calculated with the ConSurf 3.0 server.50,51
Computational alanine scanning was performed with the FoldX27,52 server, version 2.5.2 (http://foldx.crg.es). In addition to the default options, crystallographic water sites were used in the energy calculations. Calculations were performed separately for the A and B chains (“alascan” option: mutating all residues to Ala) and the hTS dimer (“complex alascan”: mutating only residues at the dimer interface). The Robetta Alanine Scanning server21,30 (http://robetta.bakerlab.org) and the CUPSAT31server (http://cupsat.tu-bs.de) were used for comparison.
Protein Preparation
A synthetic gene construct carrying hTS was excised from a pUC57 derivative and cloned into the expression vector pQE80L with an N-terminal 6xHis-tag construct. The plasmid obtained was named hTS-pQE80L. Mutagenesis was done following the QuikChange mutagenesis kit protocol (Stratagene), using this plasmid as a template. WT and all mutants hTS were expressed in E. coli BL21(DE3) cells and grown in standard Luria broth culture medium. Proteins were purified by His-tag affinity and SEC. For details, see “Supporting Experimental Procedures”.
Enzymatic Activity Assays
TS enzymatic activity was determined spectrophotometrically. The KM values of selected interface mutants were determined for mTHF and dUMP by varying the substrate concentrations. Values of Kcat and the specific activity (SA) were determined by varying the enzyme concentration. Varying concentrations of the substrates were mixed with hTS (which was stored in 20 mM phosphate buffer, pH 6.9, at 25 °C), and aliquots of this mixture were assayed for TS activity under standard conditions: an aliquot of enzyme (0.5–1 µg/mL depending on the mutant SA) was added to 1 mL of assay buffer consisting of 50 mM TES, pH 7.4, containing 25 mM MgCl2, 6.5 mM HCHO, 1 mM EDTA, 75 mM β-mercaptoethanol, and variable dUMP and mTHF concentrations. Following addition of the substrate, the increase in absorbance at 340 nm during the oxidation reaction of mTHF to 7,8-dihydrofolate was monitored for 3 min in a UV–visible spectrophotometer.
FRET Assays
Each mutant was labeled with an excitation-energy donor–acceptor pair, fluorescein (F) and tetramethylrhodamine (T).9 The FRET signal, consisting of acceptor emission upon donor excitation, occurs only from the dimeric protein and was used to monitor the dimer/monomer equilibrium. Quantitative analysis of FRET efficiency as a function of total protein concentration yielded the Kd values and the corresponding ΔGd0 values. For details, see “Supporting Experimental Procedures”.
Structural Analysis
The CD spectra of WT hTS and the mutants were measured at room temperature on a Jasco J-810 spectropolarimeter to investigate whether mutation perturbed the protein secondary structure.
The R175A and Y202A mutants were crystallized using hanging drop vapor diffusion at room temperature.52 The crystal structures were determined by molecular replacement. For details and statistics of data collection and refinement, see “Supporting Experimental Procedures” and Table S1.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. James Holton and Dr. George Meigs for assistance during data collection at the Advanced Light Source beamline 8.3.1, and Dr. Janet Finer-Moore (UCSF) for assistance with structure determination. This work was funded by European Union (LIGHTS—A Framework 6 STREP: LSH-2005-2.2.0-8), the Klaus Tschira Foundation (R.C.W., O.M.H.S-A), AIRC IG 10474 (M.P.C.), and the Alexander von Humboldt Foundation, the Finnish Cultural Foundation, the Academy of Finland, and the University of Eastern Finland (O.M.H.S-A). R.M.S. is supported by NIH Grant GM-24485.
ABBREVIATIONS USED
- WT hTS
wild type human thymidylate synthase
- dUMP
2′-deoxyuridine 5′-monophosphate
- mTHF
methyltetrahydrofolate
- β-ME
β-mercaptoethanol (2-mercaptoethanol)
- DMSO
dimethyl sulfoxide
- DDW
distilled deionized water
- FRET
fluorescence resonance energy transfer
Footnotes
ASSOCIATED CONTENT
Figures S1–S4 of residues, emission spectra, FRET analyis, and stereoviews; Tables S1 and S2 of predictions for change in stability and crystallographic data; additional experimental procedures. This material is available free of charge via the Internet at http://pubs.acs.org.
Accession Codes
PDB codes: 4KPW for hTS R175A mutant; 4JEF for Y202A mutant.
The authors declare no competing financial interest.
REFERENCES
- 1.Carreras CW, Santi DV. The catalytic mechanism and structure of thymidylate synthase. Annu. Rev. Biochem. 1995;64:721–762. doi: 10.1146/annurev.bi.64.070195.003445. [DOI] [PubMed] [Google Scholar]
- 2.Cardinale D, Guaitoli G, Tondi D, Luciani R, Henrich S, Salo-Ahen OM, Ferrari S, Marverti G, Guerrieri D, Ligabue A, Frassineti C, Pozzi C, Mangani S, Fessas D, Guerrini R, Ponterini G, Wade RC, Costi MP. Protein-protein interface-binding peptides inhibit the cancer therapy target human thymidylate synthase. Proc. Natl. Acad. Sci. U.S.A. 2011;108:E542–E549. doi: 10.1073/pnas.1104829108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garg D, Henrich S, Salo-Ahen OMH, Myllykallio H, Costi MP, Wade RC. Novel approaches for targeting thymidylate synthase to overcome the resistnace and toxicity of anticancer drugs. J. Med. Chem. 2010;53:6539–6549. doi: 10.1021/jm901869w. [DOI] [PubMed] [Google Scholar]
- 4.Phan J, Koli S, Minor W, Dunlap RB, Berger SH, Lebioda L. Human thymidylate synthase is in the closed conformation when complexed with dUMP and raltitrexed, an antifolate drug. Biochemistry. 2001;40:1897–1902. doi: 10.1021/bi002413i. [DOI] [PubMed] [Google Scholar]
- 5.Schiffer CA, Clifton IJ, Davisson VJ, Santi DV, Stroud RM. Crystal structure of human thymidylate synthase: a structural mechanism for guiding substrates into the active site. Biochemistry. 1995;34:16279–16287. doi: 10.1021/bi00050a007. [DOI] [PubMed] [Google Scholar]
- 6.Perry KM, Pookanjanatavip M, Zhao J, Santi DV, Stroud RM. reversible dissociation and unfolding of the dimeric protein thymidylate synthase. Protein Sci. 1992;1:796–800. doi: 10.1002/pro.5560010611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Maley F, Pedersen-Lane J, Changchien L. Complete restoration of activity to inactive mutants of Escherichia coli thymidylate synthase: evidence that E. coli thymidylate synthase is a half-the-sites activity enzyme. Biochemistry. 1995;34:1469–1474. doi: 10.1021/bi00005a001. [DOI] [PubMed] [Google Scholar]
- 8.Saxl RL, Changchien LM, Hardy LW, Maley F. Parameters affecting the restoration of activity to inactive mutants of thymidylate synthase via subunit exchange: further evidence that thymidylate synthase is a half-of-the-sites activity enzyme. Biochemistry. 2001;40:5275–5282. doi: 10.1021/bi002925x. [DOI] [PubMed] [Google Scholar]
- 9.Saxl RL, Maley GF, Hauer CR, Maccoll R, Changchien L, Maley F. Significance of mutations on the structural perturbation of thymidylate synthase: implications for their involvement in subunit exchange. Protein Sci. 2007;16:1439–1448. doi: 10.1110/ps.062509807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Genovese F, Ferrari S, Guaitoli G, Caselli M, Costi MP, Ponterini G. Dimer-monomer equilibrium of human thymidylate synthase monitored by fluorescence resonance energy transfer. Protein Sci. 2010;19:1023–1030. doi: 10.1002/pro.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chu E, Koeller DM, Casey JL, Drake JC, Chabner BA, Elwood PC, Zinn S, Allegra CJ. Autoregulation of human thymidylate synthase messenger RNA translation by thymidylate synthase. Proc. Natl. Acad. Sci. U.S.A. 1991;88:8977–8981. doi: 10.1073/pnas.88.20.8977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J, Schmitz JC, Lin X, Tai N, Yan W, Farrell M, Bailly M, Chen T, Chu E. Thymidylate synthase as a translational regulator of cellular gene expression. Biochim. Biophys. Acta. 2002;1587:174–182. doi: 10.1016/s0925-4439(02)00080-7. [DOI] [PubMed] [Google Scholar]
- 13.Berger SH, Berger FG, Lebioda L. Effects of ligand binding and conformational switching on intracellular stability of human thymidylate synthase. Biochim. Biophys. Acta. 2004;1696:15–22. doi: 10.1016/j.bbapap.2003.09.005. [DOI] [PubMed] [Google Scholar]
- 14.Lin X, Liu J, Maley F, Chu E. Role of cysteine amino acid residues on the RNA binding activity of human thymidylate synthase. Nucleic Acids Res. 2003;31:4882–4887. doi: 10.1093/nar/gkg678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Voeller DM, Zajac-Kaye M, Fisher RJ, Allegra CJ. The identification of thymidylate synthase peptide domains located in the interface region that bind thymidylate synthase mRNA. Biochem. Biophys. Res. Commun. 2002;297:24–31. doi: 10.1016/s0006-291x(02)02080-6. [DOI] [PubMed] [Google Scholar]
- 16.Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. J. Mol. Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 17.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 18.Keskin O, Ma B, Nussinov R. Hot regions in protein—protein interactions: the organization and contribution of structurally conserved hot spot residues. J. Mol. Biol. 2005;345:1281–1294. doi: 10.1016/j.jmb.2004.10.077. [DOI] [PubMed] [Google Scholar]
- 19.Gao Y, Wang R, Lai L. Structure-based method for analyzing protein-protein interfaces. J. Mol. Model. 2004;10:44–54. doi: 10.1007/s00894-003-0168-3. [DOI] [PubMed] [Google Scholar]
- 20.Halperin I, Wolfson H, Nussinov R. Protein-protein interactions; coupling of structurally conserved residues and of hot spots across interfaces. Implications for docking. Structure. 2004;12:1027–1038. doi: 10.1016/j.str.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 21.Kortemme T, Baker D. A simple physical model for binding energy hot spots in protein-protein complexes. Proc. Natl. Acad. Sci. U.S.A. 2002;99:14116–14121. doi: 10.1073/pnas.202485799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pons J, Rajpal A, Kirsch JF. Energetic analysis of an antigen/antibody interface: alanine scanning mutagenesis and double mutant cycles on the HyHEL-10/lysozyme interaction. Protein Sci. 1999;8:958–968. doi: 10.1110/ps.8.5.958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zerbe BS, Hall DR, Vajda S, Whitty A, Kozakov D. Relationship between hot spot residues and ligand binding hot spots in protein-protein interfaces. J. Chem. Inf. Model. 2012;52:2236–2244. doi: 10.1021/ci300175u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brandsdal BO, Smalas AO. Evaluation of protein-protein association energies by free energy perturbation calculations. Protein Eng. 2000;13:239–245. doi: 10.1093/protein/13.4.239. [DOI] [PubMed] [Google Scholar]
- 25.Darnell SJ, Page D, Mitchell JC. An automated decision-tree approach to predicting protein interaction hot spots. Proteins. 2007;68:813–823. doi: 10.1002/prot.21474. [DOI] [PubMed] [Google Scholar]
- 26.del Sol A, O’Meara P. Small-world network approach to identify key residues in protein-protein interaction. Proteins. 2005;58:672–682. doi: 10.1002/prot.20348. [DOI] [PubMed] [Google Scholar]
- 27.Guerois R, Nielsen JE, Serrano L. Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J. Mol. Biol. 2002;320:369–387. doi: 10.1016/S0022-2836(02)00442-4. [DOI] [PubMed] [Google Scholar]
- 28.Tochowicz A, Matteo Santucci M, Saxena P, Giambattista G, Trande M, Finer-Moore J, Stroud RM, Costi MP. Alanine mutants of the interface residues of human thymidylate synthase decode key features of the binding mode of allosteric anticancer peptides. J. Med. Chem. 2015;58(2):1012–1018. doi: 10.1021/jm5011176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33:W382–W388. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kortemme T, Kim DE, Baker D. Computational alanine scanning of protein-protein interfaces. Sci. STKE. 2004:pl2. doi: 10.1126/stke.2192004pl2. [DOI] [PubMed] [Google Scholar]
- 31.Parthiban V, Gromiha MM, Schomburg D. CUPSAT: prediction of protein stability upon point mutations. Nucleic Acids Res. 2006;34:W239–W242. doi: 10.1093/nar/gkl190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phan J, Steadman DJ, Koli S, Ding WC, Minor W, Dunlap RB, Berger SH, Lebioda L. Structure of human thymidylate synthase suggests advantages of chemotherapy with noncompetitive inhibitors. J. Biol. Chem. 2001;276:14170–14177. doi: 10.1074/jbc.M009493200. [DOI] [PubMed] [Google Scholar]
- 33.Lovelace LL, Minor W, Lebioda L. Structure of human thymidylate synthase under low-salt conditions. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2005;61:622–627. doi: 10.1107/S0907444905005895. [DOI] [PubMed] [Google Scholar]
- 34.Almog R, Waddling CA, Maley F, Maley GF, Van Roey P. Crystal structure of a deletion mutant of human thymidylate synthase delta (7–29) and its ternary complex with Tomudex and dUMP. Protein Sci. 2001;10:988–996. doi: 10.1110/ps.47601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sayre PH, Finer-Moore JS, Fritz TA, Biermann D, Gates SB, MacKellar WC, Patel VF, Stroud RM. Multi-targeted antifolates aimed at avoiding drug resistance form covalent closed inhibitory complexes with human and Escherichia coli thymidylate synthases. J. Mol. Biol. 2001;313:813–829. doi: 10.1006/jmbi.2001.5074. [DOI] [PubMed] [Google Scholar]
- 36.Lovelace LL, Johnson SR, Gibson LM, Bell BJ, Berger SH, Lebioda L. Variants of human thymidylate synthase with loop 181–197 stabilized in the inactive conformation. Protein Sci. 2009;18:1628–1636. doi: 10.1002/pro.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Landis DM, Gerlach JL, Adman ET, Loeb LA. Tolerance of 5-fluorodeoxyuridine resistant human thymidylate synthases to alterations in active site residues. Nucleic Acids Res. 1999;27:3702–3711. doi: 10.1093/nar/27.18.3702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Salo-Ahen OM, Wade RC. The active-inactive transition of human thymidylate synthase: targeted molecular dynamics simulations. Proteins. 2011;79:2886–2899. doi: 10.1002/prot.23123. [DOI] [PubMed] [Google Scholar]
- 39.Carosati E, Tochowicz A, Marverti G, Guaitoli G, Benedetti P, Ferrari S, Stroud RM, Finer-Moore J, Luciani R, Farina D, Cruciani G, Costi MP. Inhibitor of ovarian cancer cells growth by virtual screening: a new thiazole derivative targeting human thymidylate synthase. J. Med. Chem. 2012;55:10272–10276. doi: 10.1021/jm300850v. [DOI] [PubMed] [Google Scholar]
- 40.Tong Y, Liu-Chen X, Ercikan-Abali EA, Capiaux GM, Zhao SC, Banerjee D, Bertino JR. Isolation and characterization of thymitaq (AG337) and 5-fluoro-2-deoxyuridylate-resistant mutants of human thymidylate synthase from ethyl methanesulfonate-exposed human sarcoma HT1080 cells. J. Biol. Chem. 1998;273:11611–11618. doi: 10.1074/jbc.273.19.11611. [DOI] [PubMed] [Google Scholar]
- 41.Landis DM, Heindel CC, Loeb LA. Creation and characterization of 5-fluorodeoxyuridine-resistant Arg50 loop mutants of human thymidylate synthase. Cancer Res. 2001;1:666–672. [PubMed] [Google Scholar]
- 42.Michaels ML, Kim CW, Matthews DA, Miller JH. Escherichia coli thymidylate synthase: amino acid substitutions by suppression of amber nonsense mutations. Proc. Natl. Acad. Sci. U.S.A. 1990;87:3957–3961. doi: 10.1073/pnas.87.10.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.West DK, Porter DC, Saxl RL, Maley F. A Trojan horse approach for silencing thymidylate synthase. Biochemistry. 2004;43:9177–9184. doi: 10.1021/bi049105v. [DOI] [PubMed] [Google Scholar]
- 44.Strop P, Changchien L, Maley F, Montfort WR. Crystal structures of a marginally active thymidylate synthase mutant, Arg 126–>Glu. Protein Sci. 1997;6:2504–2511. doi: 10.1002/pro.5560061203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Michaels ML, Kim CW, Matthews DA, Miller JH. Escherichia coli thymidylate synthase: amino acid substitutions by suppression of amber nonsense mutations. Proc. Natl. Acad. Sci. U.S.A. 1990;87:3957–3961. doi: 10.1073/pnas.87.10.3957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prasanna V, Gopal B, Murthy MR, Santi DV, Balaram P. Effect of amino acid substitutions at the subunit interface on the stability and aggregation properties of a dimeric protein: role of Arg 178 Arg 218 at the Dimer interface of thymidylate synthase. Proteins. 1999;34:356–368. [PubMed] [Google Scholar]
- 47.Kawase S, Cho SW, Rozelle J, Stroud RM, Finer-Moore J, Santi DV. Replacement set mutagenesis of the four phosphate-binding arginine residues of thymidylate synthase. Protein Eng. 2000;13:557–563. doi: 10.1093/protein/13.8.557. [DOI] [PubMed] [Google Scholar]
- 48.Landis DM, Loeb LA. Random sequence mutagenesis and resistance to 5-fluorouridine in human thymidylate synthases. J. Biol. Chem. 1998;273:25809–25817. doi: 10.1074/jbc.273.40.25809. [DOI] [PubMed] [Google Scholar]
- 49.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Glaser F, Pupko T, Paz I, Bell RE, Bechor-Shental D, Martz E, Ben-Tal N. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19:163–164. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- 51.Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Benvenuti M, Mangani S. Crystallization of soluble proteins in vapor diffusion for X-ray crystallography. Nat. Protoc. 2007;2:1633–1651. doi: 10.1038/nprot.2007.198. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.