Abstract
Background Minimal influenza surveillance has been carried out in sub‐Saharan Africa to provide information on circulating influenza subtypes for the purpose of vaccine production and monitoring trends in virus spread and mutations.
Objective The aim of this study was to investigate a surveillance program in Kenya to isolate and characterize influenza viruses.
Results In the 2006–2007 influenza season, nine influenza A viruses were isolated. All were of H3N2 subtype with key amino acid (aa) changes indicating that they were more closely related to recent World Health Organization recommended vaccine strains than to older vaccine strains, and mirroring the evolution of circulating influenza A globally. Hemagglutination inhibition data showed that the 2006 Kenya isolates had titers identical to the 2005–2006 H3N2 vaccine strain but two‐ to threefold lower titers to the 2006–2007 vaccine strain, suggesting that the isolates were antigenic variants of the 2006–2007 vaccine strains. Analysis of aa substitutions of hemagglutinin‐1 (HA1) protein of the 2006 Kenyan viruses revealed unique genetic variations with several aa substitutions located at immunodominant epitopes of the HA1 protein. These mutations included the V112I change at site E, the K 173 E substitution at site D and N 278 K change at site C, mutations that may result in conformational change on the HA molecule to expose novel epitopes thus abrogating binding of pre‐existing antibodies at these sites.
Conclusion Characterization of these important genetic variations in influenza A viruses isolated from Kenya highlights the importance of continuing surveillance and characterization of emerging influenza drift variants in sub‐Saharan Africa.
Keywords: Genetic variant, H3N2, influenza A, Kenya
Background
Identification and characterization of circulating serotypes and genotypes of influenza viruses can assist detection of viral antigenic drifts and shifts – phenomena that underlie epidemics and pandemics. 1 Influenza surveillance also allows for the identification and selection of appropriate virus components for inclusion in the annual formulation of influenza vaccines. 2 , 3 The recent emergence of novel, highly virulent H5N1 influenza A subtype has heightened global concern about the onset of an influenza pandemic. 4 , 5 , 6 In response, influenza monitoring and pandemic preparedness are being implemented globally. 7 , 8 , 9
Despite strong anecdotal evidence of frequent acute upper respiratory infections, sub‐Saharan Africa has not conducted influenza surveillance. To begin to address this, a partnership between the US Army Medical Research Unit‐Kenya, Kenya Medical Research Institute (KEMRI), Kenya Ministry of Health and the Centers for Disease Control and Prevention‐Kenya was established to undertake influenza surveillance activities in Kenya, under the World Health Organization’s (WHO) global influenza surveillance program. 2 , 3 In this report, we present genetic analysis data of the initial nine influenza A isolates, all belonging to the H3N2 subtype.
Materials and methods
Collection of clinical samples and influenza virus isolations
The following hospitals comprised the active surveillance field sites: Mbagathi, New Nyanza, Malindi, Isiolo, Mombasa Port Reitz, and Kericho (Figure 1). Study sites were selected based on geographic regions and population demographics.
Figure 1.
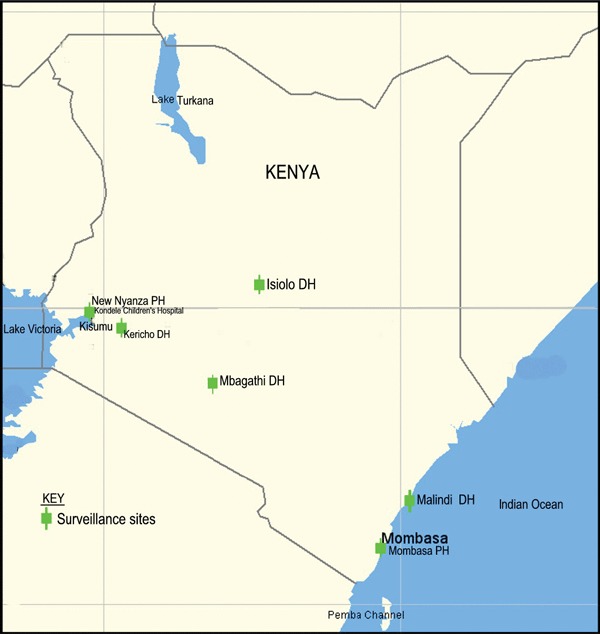
Site locations for the influenza surveillance in Kenya. The sites cover regions of the country where majority of the populace live.
Patients over 2 months of age and presenting at hospital outpatient clinics with fever (≥38°C) and either cough or sore throat were recruited by dedicated project Clinical Officers. Subjects with exudative pharyngitis or tonsillitis or symptom onset >72 hours were excluded from enrollment. None of the patients sampled had previously been vaccinated against influenza. Informed consent and assent approved by the KEMRI and the Walter Reed Army Institute of Research institutional review boards was obtained from all participating patients. Patients’ age, sex, occupation, residence, workplace, history of influenza‐like illness within household, travel history, and animal contact were ascertained and a clinical examination completed.
Duplicate nasopharyngeal samples were collected using a cotton swab and placed in a 1 ml cryovial containing virus transport medium, kept at 4°C, and stored in a liquid nitrogen dry shipper within 8 hours of collection. All samples were transported from the surveillance sites to the National Influenza Center (NIC) laboratory within one week. The cold chain was maintained throughout. Specimens were received weekly and tested for both influenza A and B.
Virus isolation
Influenza virus isolations were performed at the NIC located at KEMRI Nairobi Kenya, using Madin‐Darby canine kidney (MDCK) cells. Serotyping was performed by hemagglutination inhibition assay (HAI) using guinea pig red blood cells and reference antiserum in accordance with Centers for Disease Control and Prevention (CDC) protocols. 10 Briefly, inoculated cells were incubated at 37°C with 5% CO2 for 10 days to allow development of cytopathic effects (CPE). Supernatant fluid was collected from MDCK cultures showing CPE and the hemagglutination titer determined. 10 Subsequent HAI testing was only performed on specimens showing high titers of virus. Isolates with low hemagglutination titers were passaged further in MDCK cells. The guinea pigs used to provide blood for HA and HAI assays were reared under animal care in accordance with KEMRI guidelines.
Polymerase chain reaction
Isolates were sent to the Air Force Institute of Operational Health (AFIOH), San Antonio, TX where they were genotyped. 11 , 12 Isolates confirmed at AFIOH were forwarded to the WHO Influenza Collaborating Centre at CDC, Atlanta, USA.
RNA was extracted from 48‐hour shell vial cultures at AFIOH using the M48 BioRobot and Mag Attract Virus Mini M48 Kit (Qiagen, Inc., Valencia, CA, USA) according to the manufacturers’ protocols. For RT‐PCR, the hemagglutinin‐1 (HA1) gene forward primer designated H3‐F7 (5′‐ACTATCATTGCTTTGAGC‐3′) and the reverse primer H3R‐1184 (5′‐ATGGCTGCTTGAGTGCTT3′) were used. The reaction consisted of 5 μl RNA, 400 nmol/l of each of the primers and the master mixture containing buffer and enzymes following the Superscript III One‐Step RT‐PCR protocol from the manufacturer (Invitrogen, Carlsbad, CA, USA). The thermocycling conditions included 40 cycles of 94°C for 15 seconds, 52°C for 30 seconds, and 68°C for 75 seconds, with a final extension cycle at 68°C for 5 minutes. All PCR products were purified and sequenced using the Big Dye Terminator v3·1 Kit (Applied Biosystems, Foster City, CA, USA). The sequencing primers included the H3‐F7 and H3R‐1184 primers and two internal primers designated as H3R‐466 (5′‐GGTGCAACCAATTCAATC‐3′) and H3F‐282 (5′‐CAGCAACTGTTACCC‐3′).
HA1 nucleotide sequencing and genetic analyses
Nucleotide sequencing was performed and analyzed using an ABI 3130xl Genetic Analyzer (Applied Biosystems) according to the manufacturers’ specifications. Nucleotide contigs were assembled using the Contig Assembly program of Bioedit. 13 Protein translations were performed using DS Gene version 1.5 (Accelrys Inc., Cambridge, UK). Multiple sequence alignments were performed with muscle version 3.6. 14 Phylogenetic analyses were performed with the mega version 3.1. 15 Three‐dimensional HA protein structures were generated using Swiss Model 16 , 17 and visualized and annotated by the rasmol version 2.7.3. 18 , 19 The predicted amino acid (aa) sequences for the 2006–2007 Kenya isolates were submitted to GenBank under accession numbers ABM98421.1, ABM98422.1, ABW34445.1, ABW34446.1, ABW34450, ABS00300, ABS00301, ABS00302, and ABS00303.
Results
A total of 1014 nasopharyngeal specimens were obtained from patients with influenza like illness at outpatient clinics of the six hospitals during the first 9 months of the surveillance program. Although the ages of the subjects ranged from 2 months to 36 years, most (82·8%) were below 5 years of age and 519 (51%) were male. None were previously vaccinated against influenza. Nine specimens (0·9%) tested positive for influenza A (H3N2) by isolation. The first four influenza A isolates designated as A/Nairobi/2041/2006 (H3N2), A/Nairobi/5842/2006 (H3N2), A/Kenya/AF5626/2006 (H3N2), and A/Kenya/AF5627/2006 (H3N2) were obtained between July and September 2006. Five other isolates designated as A/Kenya/AF1055/2007 (H3N2), A/Kenya/AF1056/2007 (H3N2), A/Kenya/AF1057/2007 (H3N2), A/Kenya/AF1058/2007 (H3N2), and A/Kenya/AF5618/2007 (H3N2) were isolated during the first quarter of 2007. All isolates obtained in 2006 were obtained from Nairobi. In contrast, the 2007 isolates came from across the country, namely: two from western Kenya (New Nyanza Provincial hospital), one from Rift Valley (Kericho district hospital) and two from Nairobi (Mbagathi district hospitals) (Figure 1).
The HAI assays were performed using post‐infection ferret antisera provided by WHO with reference antigens that included the 2005–2006 southern and northern hemisphere H3N2 vaccine strains [A/California/7/2004 (H3N2) and A/New York/55/2004 (H3N2)], which are antigenically similar. All the 2007 isolates were antigenically equivalent to A/Wisconsin/67/2005, the vaccine strain of 2007 (data not shown). Antigenically, all the four 2006 Kenyan isolates were identical to A/California/07/2004 and A/New York/55/2004 – variants of the 2006 vaccine component, but showed a two fold reduction in HAI titers when compared with A/Wisconsin/67/2005, which was the vaccine strain for 2007 (Table 1). Compared to A/Hiroshima/52/2005, A/Nairobi/2041/2006 and A/Kenya/AF5626/2006 displayed a three fold reduction in HAI titers while A/Nairobi/5842/2006 and A/Kenya/AF5627/2006 had twofold lower HAI titers (Table 1). These results indicate that as expected, the 2006 Kenyan isolates were antigenically identical to the 2006 vaccine strains.
Table 1.
Hemagglutination inhibition reactions of the 2006 Kenyan influenza A (H3N2) viruses whereas A/Nairobi/2041/2006 is antigenically identical to A/Kenya/AF5626/2006, A/Nairobi/5842/2006 is identical to A/Kenya/AF5627/2006
| Reference ferret antisera | ||||
|---|---|---|---|---|
| A/California/ 07/2004 | A/New York/ 55/2004 | A/Wisconsin/ 67/2005 | A/Hiroshima/ 52/2005 | |
| Reference antigens | ||||
| A/California/07/2004 | 1280 | 320 | 320 | 320 |
| A/New York/55/2004 | 640 | 320 | 320 | 320 |
| A/Wisconsin/67/2005 | 320 | 160 | 1280 | 640 |
| A/Hiroshima/52/2005 | 160 | 160 | 640 | 1280 |
| Test antigens | ||||
| A/Nairobi/5842/2006 | 1280 | 320 | 320 | 320 |
| A/Nairobi/2041/2006 | 1280 | 320 | 320 | 160 |
| A/Kenya/AF5626/2006 | 1280 | 320 | 320 | 160 |
| A/Kenya/AF5627/2006 | 1280 | 320 | 320 | 320 |
Molecular subtyping confirmed the H3N2 subtyping for all the nine Kenya isolates. The viruses were further characterized by comparing the nucleotide (nt) sequences of the HA1 region of the HA gene. The A/Nairobi/2041/2006 had 100% nt sequence identity with A/Kenya/AF5626/2006 whereas A/Nairobi/5842/2006 was also identical (100%) to A/Kenya/AF5627/2006. Minimal (0·1%) nt sequence variations were observed between A/Nairobi/5842/2006 and A/Nairobi/2041/2006, and between A/Kenya/AF5626/2006 and A/Kenya/AF5627/2006. All the 2006 Kenyan isolates had 100% aa sequence identity with each other. At the aa level, the five 2007 isolates were identical except for A/Kenya/AF1056/2007 (H3N2) and A/Kenya/AF1057/2007 (H3N2) which had a G50E change. This aa change is present in the A/Brisbane/10/2007 (H3N2) strain which is the recommended 2008 southern hemisphere vaccine strain (Figure 2).
Figure 2.

The major amino acid changes within region 1 of the HA proteins of influenza A (H3N2) Kenyan isolates (2006–2007), South African isolates (2004–2006), and southern hemisphere vaccine strains (1999–2008). All isolate names bear corresponding gi database accession numbers. Amino acid positions are shown vertically.
To determine the phylogenetic relationship of the Kenyan isolates with other circulating viruses globally, the predicted aa sequences for HA1 proteins of the four 2006 isolates were compared with the HA1 protein of the five 2007 Kenya isolates, South African isolates of the past 4 years, and the northern and southern hemisphere vaccine strains from 1999 to 2007 (Figure 3). As shown in Figure 3, the 2006 Kenya isolates clustered together with the 2006 South African isolates, forming a genetic variant distinct from the H3N2 component of the northern and southern hemisphere vaccine strains for 2005–2008 (A/Wellington/1/2004, A/California/7/2004, A/Wisconsin/67/2005, and A/Brisbane/10/2007).
Figure 3.
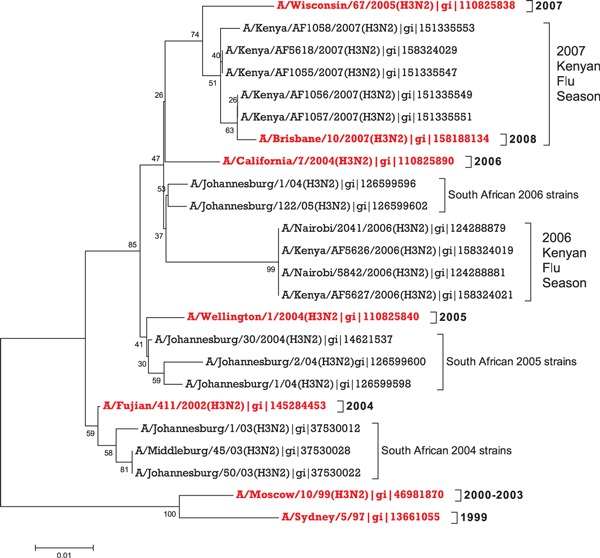
The un‐rooted consensus phylogenetic tree expressing the relationship between the HA1 amino acid sequences of the Kenyan 2006–2007 isolates to southern hemisphere vaccine strains used from 1999 to 2008 and South African isolates (2004–2006). The tree was produced using Neighbor Joining (NJ) method generated by mega 3.1 software. 15 The phylogeny test was done by Bootstrap based on 500 replications. The accession numbers corresponding to the protein sequences are given by the gi numbers. The year for each southern hemisphere vaccine strain is indicated on the branches.
Compared with vaccine strains from 1999 to 2007, 30 aa substitutions were identified in the HA1 region of the Kenya 2006 isolates. Six of the 30 aa substitutions; K2E, I25V, V112I, K173E, S199P, and N278K were unique, never having been detected in any vaccine strains of the past eight years (2, 4). More importantly, four of the six aa substitutions (except K2E and I25V) were also found in 2006–2007 H3N2 isolates from Eastern Europe (internal WHO‐share document). The other 22 aa substitution occurred more randomly among isolates evaluated. In contrast, the aa sequence of the 2007 Kenya isolates were identical to A/Brisbane/10/2007 (H3N2) – the 2008 southern hemisphere recommended vaccine strain – except for a single L194P change. This aa change is present at the immunodominant site B.
Figure 4.
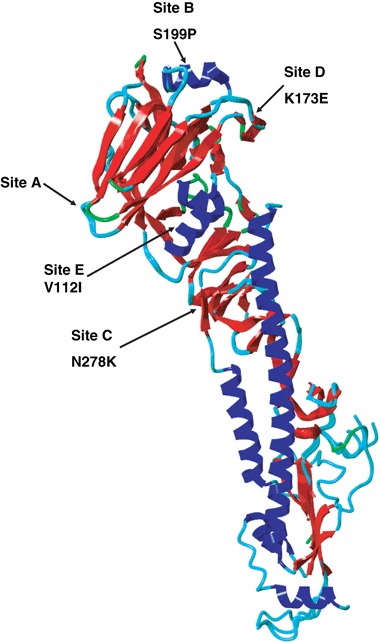
Three dimensional model (ribbon view) showing the aa changes in the HA1 molecule observed in the 2006 Kenyan isolates: V112I, K173E, S199P, and N278K. These occupy the immunodominant sites B (Pro 199), C (Lys 278), D (Pro199), and E (Glu 173).25 Figure generated and visualized using YASARA (http://www.yasara.org) and PovRay (http://www.povray.org).
To better understand the immunologic and epidemiologic relevance of the unique aa substitutions observed in the 2006 Kenya isolates, we used the predicted aa sequence of one of the 2006 isolates to develop a 3D model of the HA protein, based on the X‐ray crystal structural coordinates of protein precursor; 1HA0. 20 This 3D model revealed that four of the six novel aa substitutions present in the 2006 Kenya isolates occupied immunodominant sites of the HA protein (Figure 4). 21 Thus, the V112I substitution was located at site E, K173E at site D, S199P immediately adjacent to site B, and N278K adjacent to site C of the HA protein (Figure 4).
Discussion
Kenya and most of Africa have not provided the much needed information on circulating influenza A subtypes and their variants in the region for the purpose of vaccine production, and monitoring trends in virus spread and mutations. In this report, the first characterization of human influenza A viruses from East Africa, we demonstrate prevalence of the H3N2 subtypes, in agreement with data from the rest of the world. In addition, the findings show key aa changes exhibited by the 2006 Kenyan isolates indicating that these viruses were more closely related to recent WHO vaccine strains than to older strains, mirroring the evolution of circulating influenza A globally. Amino acid changes by recent vaccine strains away from earlier vaccine strains have been localized at key immunodominant sites, including mutations at positions 75, 83, 144, and 172 of antigenic determinant site A, mutations at positions 131, 155, 156, 159, 189, 192, 196, and 202 of site B, and at positions 222 and 227 of site D. The HAI data showing that the Kenya isolates had titers identical to the H3N2 isolates of the 2005–2006 vaccine strain but two‐ to threefold lower titers to the 2006–2007 vaccine strain suggest that the Kenyan isolates were antigenic variants of the 2006–2007 vaccine strains.
Six of the 30 aa substitutions identified in the Kenyan viruses were unique and located within known antibody‐binding sites. These changes, including V112I and K173E substitutions at site D, S199P at site B, and N278K at site C have also been observed in viruses from Ukraine (internal WHO‐share document) and represent a novel genetic variant different from the current vaccine strains. Whereas mutations at dominant sites are believed to be the evolutional outcome of constant immunological and environmental pressures on the viruses, they also represent a public health threat of possible emergence of more virulent isolates that are not covered by available vaccines.
More specifically, the V112I change at site E may result in a conformational change on the HA molecule to expose novel epitopes, the substitution at position 173 is within site D that converts a basic R group (K) to an acidic (E) one. This substitution can alter electrostatic properties of the HA molecule to abrogate the binding of antibodies that previously recognized the epitope at site D. Similarly, the asparagine to lysine change at position 278, at site C which is the hinge separating the globular receptor head from the stem of the HA1 protein likely results in an electrostatic changes that alter antigenicity. In contrast, the 3D modeling suggests that the S199P substitution at position 199 of site B did no result in notable conformational change of the HA protein. Such findings emphasize the importance of continuous molecular surveillance and characterization of emerging influenza drift variants in sub‐Saharan Africa.
Although the 2006 Kenyan isolates had 100% aa sequence identity at HA1 protein, they had disparate antigenic profiles by hemagglutination inhibition using the A/Hiroshima/52/2005 (H3N2) reference antisera. This antigenic difference may have been induced by genetic and antigenic variation in the other surface proteins such as the neuraminidase, HA2 and M2 proteins known to contribute to antigenic/immunologic properties of influenza viruses. 22 , 23 , 24 Work to characterize these other surface glycoproteins is ongoing.
The phylogenetic analysis indicates that the Kenyan isolates are the result of point mutational drift from the A/California/7/04 and A/Wisconsin/67/05 vaccine strains. Although genetically distinct, the 2006 Kenyan isolates were antigenically similar to the 2005–2006 vaccine strains, implying that vaccination with either the northern or southern hemisphere influenza vaccines were protective for local residents and travelers to the region against seasonal influenza viruses.
Author Contributions
W. D. Bulimo was responsible for the supervision and performance of laboratory evaluations, data analysis, and manuscript preparation. J. L. Garner was responsible for molecular genotyping of viruses, data analysis, and manuscript preparation. D. C. Schnabel and S. A. Bedno was responsible for study design, data analysis, and manuscript preparation. M. K. Njenga and W. O. Ochieng was responsible for data analysis and manuscript preparation. E. Amukoye was responsible for laboratory evaluation, data analysis, and manuscript preparation. J. M. Magana and J. M. Simwa was responsible for subtyping of influenza specimens and data analysis. V. O. Ofula, S. M. Lifumo was responsible for specimen analysis and data analysis. J. Wangui was responsible for QA/QC functions and critical data analysis. R. F. Breiman was responsible for critical revision of the intellectual content. S. K. Martin was responsible for study design, data analysis, and critical revision of the intellectual content.
Acknowledgements
Funds for this study were provided by US Department of Defense through the Global Emerging Infections Surveillance and Response System (DoD‐GEIS). The views expressed are those of the authors and should not be construed to represent the position of the United States Department of the Army or Department of Defense. The Division of Epidemiology, Air Force Institute of Operational Health, Brooks City Base, San Antonio Texas carried out quality control and quality assurance and sequenced the HA1 gene segment of the viruses described in this publication. This manuscript has been published with permission of Director, KEMRI, Kenya.
References
- 1. Hilleman MR. Realities and enigmas of human viral influenza: pathogenesis, epidemiology and control. Vaccine 2002; 20:3068–3087. [DOI] [PubMed] [Google Scholar]
- 2. Stohr K. Overview of the WHO Global Influenza Programme. Dev Biol (Basel) 2003; 115:3–8. [PubMed] [Google Scholar]
- 3. Stohr K. The WHO Global Influenza Program and its animal influenza network. Avian Dis 2003; 47:934–938. [DOI] [PubMed] [Google Scholar]
- 4. CDC . Isolation of avian influenza A (H5N1) viruses from humans – Hong Kong, May–December. MMWR Morb Mortal Wkly Rep 1997; 46:1204–1207. [PubMed] [Google Scholar]
- 5. Subbarao K, Klimov A, Katz J et al. Characterization of an avian influenza A (H5N1) virus isolated from a child with a fatal respiratory illness. Science 1998; 279:393–396. [DOI] [PubMed] [Google Scholar]
- 6. De Jong JC, Claas EC, Osterhaus AD. Influenza A (H5N1) in Hong Kong: forerunner of a pandemic or just a scientifically interesting phenomenon and a useful exercise in pandemiology? Ned Tijdschr Geneeskd 1998; 142:1252–1256. [PubMed] [Google Scholar]
- 7. Claas EC, Osterhaus AD, Van Beek R et al. Human influenza A H5N1 virus related to a highly pathogenic avian influenza virus. Lancet 1998; 351:472–477. [DOI] [PubMed] [Google Scholar]
- 8. Stohr K. The global agenda on influenza surveillance and control. Vaccine 2003; 21:1744–1748. [DOI] [PubMed] [Google Scholar]
- 9. Fauci AS. Pandemic influenza threat and preparedness. Emerg Infect Dis 2006; 12:73–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kendal AP, Pereira MS, Skehel JJ. Concepts and Procedures for Laboratory‐based Influenza Surveillance. Atlanta, GA: Centers for Disease Control and Prevention, 1982. [Google Scholar]
- 11. Yamada A, Imanishi J, Nakajima E, Nakajima K, Nakajima S. Detection of influenza viruses in throat swab by using polymerase chain reaction. Microbiol Immunol 1991; 35:259–265. [DOI] [PubMed] [Google Scholar]
- 12. Zhang WD, Evans DH. Detection and identification of human influenza viruses by the polymerase chain reaction. J Virol Methods 1991; 33:165–189. [DOI] [PubMed] [Google Scholar]
- 13. Hall TA. BioEdit: a user‐friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 1999; 41:95–98. [Google Scholar]
- 14. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 2004; 32:1792–1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 2004; 5:150–163. [DOI] [PubMed] [Google Scholar]
- 16. Kopp J, Schwede T. The SWISS‐MODEL repository of annotated three‐dimensional protein structure homology models. Nucleic Acids Res 2004; 32:D230–D234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schwede T, Kopp J, Guex N, Peitsch MC. SWISS‐MODEL: an automated protein homology‐modeling server. Nucleic Acids Res 2003; 31:3381–3385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bernstein HJ. Recent changes to RasMol, recombining the variants. Trends Biochem Sci 2000; 25:453–455. [DOI] [PubMed] [Google Scholar]
- 19. Sayle RA, Milner‐White EJ. RASMOL: biomolecular graphics for all. Trends Biochem Sci 1995; 20:374. [DOI] [PubMed] [Google Scholar]
- 20. Chen J, Lee KH, Steinhauer DA, Stevens DJ, Skehel JJ, Wiley DC. Structure of the hemagglutinin precursor cleavage site, a determinant of influenza pathogenicity and the origin of the labile conformation. Cell 1998; 95:409–417. [DOI] [PubMed] [Google Scholar]
- 21. Wiley DC, Skehel JJ. The structure and function of the hemagglutinin membrane glycoprotein of influenza virus. Annu Rev Biochem 1987; 56:365–394. [DOI] [PubMed] [Google Scholar]
- 22. Hughey PG, Roberts PC, Holsinger LJ, Zebedee SL, Lamb RA, Compans RW. Effects of antibody to the influenza A virus M2 protein on M2 surface expression and virus assembly. Virology 1995; 212:411–421. [DOI] [PubMed] [Google Scholar]
- 23. Webster RG, Brown LE, Laver WG. Antigenic and biological characterization of influenza virus neuraminidase (N2) with monoclonal antibodies. Virology 1984; 135:30–42. [DOI] [PubMed] [Google Scholar]
- 24. Zebedee SL, Lamb RA. Influenza A virus M2 protein: monoclonal antibody restriction of virus growth and detection of M2 in virions. J Virol 1988; 62:2762–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]


