Abstract
Successful embryo implantation requires synchronous development and communication between the blastocyst and the endometrium, however the mechanisms of communication in humans are virtually unknown. Recent studies have revealed that microRNAs (miRs) are present in bodily fluids and secreted by cells in culture. We have identified that human blastocysts differentially secrete miRs in a pattern associated with their implantation outcome. miR-661 was the most highly expressed miR in blastocyst culture media (BCM) from blastocysts that failed to implant (non-implanted) compared to blastocysts that implanted (implanted). Our results indicate a possible role for Argonaute 1 in the transport of miR-661 in non-implanted BCM and taken up by primary human endometrial epithelial cells (HEECs). miR-661 uptake by HEEC reduced trophoblast cell line spheroid attachment to HEEC via PVRL1. Our results suggest that human blastocysts alter the endometrial epithelial adhesion, the initiating event of implantation, via the secretion of miR, abnormalities in which result in implantation failure.
Keywords: Embryo, Implantation, microRNA, miR-661, Endometrium
Highlights
-
•
microRNAs are secreted by human blastocysts relative to implantation potential during IVF.
-
•
microRNA-661 is secreted by blastocysts that fail to implant and taken up by endometrial epithelial cells via Argonaute 1.
-
•
microRNA-661 reduces adhesion of trophoblast spheroids to endometrial cells.
Implantation failure is a large problem affecting the success rate of in vitro fertilisation (IVF). There are no effective treatments for implantation failure. Our study demonstrated that human embryos secrete microRNA and their expression is differentially expressed in embryos that achieve a successful pregnancy compared to embryos that fail. The study identified that microRNA 661 secreted by embryos, was taken up by human endometrial epithelial cells via attachment to a protein and inhibited endometrial cell adhesiveness. This suggests that abnormally produced microRNA may prevent attachment of human embryos to the endometrial lining and prevent implantation and pregnancy.
1. Introduction
Embryo–endometrial interactions are critical for implantation and subsequent placental development. During the early stages of implantation, the blastocyst enters the uterine cavity, apposes and then adheres to an adequately prepared or ‘receptive’ endometrial uterine luminal epithelium to initiate implantation. Abnormalities in adhesion during the very early stages of implantation result in implantation failure, which is a major cause of infertility (Dimitriadis et al., 2005, Koot et al., 2012). In humans, very little is known of the blastocyst–endometrial interactions, largely due to the difficulty in studying implantation in humans. The influence of human blastocysts on human endometrial receptivity is largely unknown.
The conceptus enters the uterine cavity up to 72 h prior to implantation (Norwitz et al., 2001) and is thought to act on the endometrium at least in part via soluble factors to facilitate receptivity and implantation (Cuman et al., 2013).We have previously published that human blastocysts release soluble factors that alter primary human endometrial epithelial cell (HEEC) gene expression and adhesion, the initiating event of implantation (Cuman et al., 2013).
miRs are short (~ 20–22 nucleotides), highly conserved sequences that regulate the expression of 50% of genes in the human genome (Bartel, 2004). Mature miRs act by binding to complementary regions of mRNAs, inhibiting translation or by destabilising the gene, resulting in down regulation of their target genes (Bohnsack et al., 2004, Chen and Rajewsky, 2007, Kim, 2005, Lee et al., 2003). miR can be secreted by cells, via a number of mechanisms including exosomes, apoptotic bodies and bound to lipid or RNA binding complex (RBC) proteins, such as Argonaute (Ago) 1 and 2 (Arroyo et al., 2011, Vickers et al., 2011). MiRs are present not only within cells but also in body fluids such as saliva, urine, blood, plasma and cell culture media (Hanke et al., 2010, Mitchell et al., 2008, Park et al., 2009, Zubakov et al., 2010).
Analysis of human endometrium and trophectoderm has identified the expression of a large number of miRs (Dior et al., 2014, Galliano and Pellicer, 2014, Kresowik et al., 2014, Rosenbluth et al., 2013), with more recent studies demonstrating that miRs are secreted by human and bovine embryos in culture (Kropp et al., 2014, Rosenbluth et al., 2014). We hypothesised that miRs are released by human blastocysts and are taken up by endometrial surface epithelial cells to regulated endometrial receptivity and implantation. The aim was to identify miR profiles of spent culture media (BCM) from embryos that successfully implanted compared to those that failed to implant. Furthermore, we aimed to determine miR uptake by human endometrial epithelial cells and the effect on adhesion and therefore identify the possible functional consequences relevant to endometrial receptivity and implantation.
2. Materials and Methods
2.1. Ethical Approval
Human ethical approval was obtained for all the studies in this manuscript as follows:
Endometrium collection
Written informed consent was obtained from each patient, before surgery in the case of women with primary infertility, and protocols were approved by the Southern Health Human Research Ethics Committee, Melbourne, Australia.
Blastocyst media collection
Written informed consent was obtained from each patient and the study was approved by the Monash Surgical Private Hospital Human Research Ethics Committee, Melbourne, Australia.
Trophectoderm collection
Written informed consent was obtained from each patient and the study was approved by Monash Health (#12,101) and the Embryo Research Licencing Committee, National Health and Medical Research Council of Australia (#309722).
2.2. Endometrial Collection
Endometrial biopsies (n = 33) were collected at curettage from women with regular menstrual cycles throughout the proliferative and secretory phases of the menstrual cycle (Cuman et al., 2013, Paiva et al., 2009, Van Sinderen et al., 2013). The women had no steroid treatment for at least 2 months prior to tissue collection. An experienced gynaecological pathologist confirmed biopsies showed no evidence of possible endometrial dysfunction. Biopsies were either placed into DMEM F/12 media for further isolation or fixed in Formalin. See supplemental experimental procedures for further details on endometrial isolation.
2.2.1. Spent Conditioned Media (BCM)
Spent blastocyst conditioned media (BCM) were collected from embryos (fertilised by ICSI only) that had been cultured from days 3 to 5 and stored at − 80 °C. Control culture media (not exposed to an embryo) were also collected. BCM were collected from two groups: 1. Blastocysts that successfully implanted (clinical pregnancy carried to term > 36 weeks) (Implanted) and 2. Blastocysts that did not implant and did not result in pregnancy (no biochemical or clinical indications) (non-implanted).
2.2.2. Trophectoderm Collection
Human embryos consented to medical research (Ethics #12101) were thawed, and allowed to expand with assisted hatching overnight. Using in house technique, the inner cell mass was removed from the embryo and allowed to succumb. The remaining trophectoderm cells were collected directly into lysis buffer for PCR use. MicroRNA was isolated from cells using TaqMan Cell to CT kit (Life Technologies) according to manufactures instructions.
2.2.3. BCM microRNA Real Time PCR Arrays
RNA was isolated from BCM (10 μl) using miRCURY RNA Biofluids isolation Kit (Exiqon, Denmark) according to the manufacturer's instructions. cDNA synthesis and RT qPCR on BCM was performed using the miRCURY LNA™ Universal RT microRNA PCR system (Exiqon, Denmark) according to the manufacturer's instructions. In brief, the RNA was tailed with a poly (A) sequence at their 3′end and then reverse transcribed into cDNA using a universal poly (T) primer with a 3′end degenerate anchor and a 5′end universal tag. The cDNA products were subsequently diluted 125 fold and transferred to the ready-to-use microRNA PCR Human Panels (I + II). The qPCRs were run on a 7900HT thermocycler (ABI) using the thermal-cycling parameters recommended by Exiqon. Raw Ct values were calculated as recommended by Exiqon using the RQ manager software v1.2.1 (ABI) with manual settings for threshold and baseline, i.e. all miRCURY assays were analysed using a ΔRn threshold of 60 and baseline subtraction using cycles 1–14. Analysis was performed using the Gene Ex software.
2.2.4. Primary HEEC Isolation
Endometrial epithelial cells were prepared as previously published (Cuman et al., 2013). Briefly, endometrial tissue was digested with collagenase and the suspension was filtered through 43 and 11 mm nylon mesh to collect endometrial epithelial glands. The cells and epithelial fragments were collected and resuspended in a 1:1 mixture of Dulbecco's modified eagle's medium (DMEM)/Hams F-12 (Gibco) supplemented with 10% foetal calf serum (FCS; Invitrogen), and 1% antibiotic–antimycotic? solution (Gibco, Auckland, NZ) and plated. A purity of 95% was necessary for the cells to be used experimentally.
2.2.5. HTR-8/SVneo Trophoblast Cell Line
The HTR-8/SVneo trophoblast cell line exhibits features of invasive trophoblast cells, such as human leukocyte antigen-G (extravillous trophoblast marker) and cytokeratin-7 expression (Hannan et al., 2010). These cells were cultivated and maintained in RPMI 1640 (Sigma-Aldrich, St. Louis, MO, USA) supplemented with 10% FCS, as previously described (Graham et al., 1993).
2.2.6. RNA Isolation and Quantitative PCR
RNA was extracted from cultured cells and conditioned media (excluding BCM and primary trophectoderm cells) using Tri Reagent (Sigma) according to the manufacturer's instructions. Isolated RNA was reversed transcribed into complimentary DNA with M-MLV RT system (Life Technologies) by using the TaqMan primer sets for miRs (Applied Biosystems) or Oligo primers (sigma) for non-miRs. Real time PCR was performed using the TaqMan Fast Universal PCR Master mix (Applied Biosystems) or Power SYBR Green master mix (Applied Biosystems) by using TaqMan probes or specific primer pairs (MTA1, F- TAACAAGCCAAATCCGAACC R- TCCTGGCCTCTCTCCATCTA; MTA2, F- CGGGTGGGAGATTACGTCTA R- TGGCTGCTTTGATTCCTCTTPVRL1 F- AATCGAGAAAGCCAGCTCAA R- CGGATCTCCTGGTACTCTGC; EPHB2- F- GATGGGGCAGTACAAGGAGA, R- AGGCAGGTGAATGTCAAACC). miR expression levels were normalised against control snU6 probes. Expression of MTA2 and PVRL1was normalised against 18S and beta-actin.
2.2.7. miR Uptake by Primary HEEC
Fluorescein (FLC) tagged mIR-661 (Sigma) was transfected into HTR8s using Lipofectamine RNAiMax at a concentration of 100 nM, (based on the manufacturer's instructions). HTR8s were washed with culture media 12 h post transfection and incubated with fresh culture media for 12 h further. HTR8-CM was collected and used to treat HEECs. A scramble microRNA sequence (Life Technologies) was used as a control. HTR8 cells and 1 ml-conditioned media were collected for confirmation of overexpression of miR-661 by RT qPCR. HEECs were treated for 8 h and uptake confirmed by RTqPCR and immunofluorescence (method adapted from (Zhou et al., 2013)).
2.2.8. Immunofluorescence
Visualisation of FLC-miR-661 was confirmed using immunofluorescence. Briefly, HEECs were plated onto chamber slides and treated with HTR8-CM as described above. Following treatment, media removed, cells were washed and the chamber slide fixed in 70% ethanol overnight. Nuc-Red (to visualise nuclei;Invitrogen) was applied to the slide prior to fixing with fluorescent mounting media (Dako).
2.2.9. Ultracentrifugation
HTR8-CM was collected and centrifuged at 1000 g for 10 min to remove cell debris. The supernatant was transferred to a new tube and spun at 120,000 g for 100 min at 4 °C (Arroyo et al., 2011). The supernatant and pellet were collected and RNA isolated to identify miR-661 expression. HEECs were treated with the collected supernatant, and the pellet re-suspended in 5% FBS DMEM/F12.
2.2.10. Proteinase K Treatment
HTR8-CM was treated with proteinase K (20 μg/ml, Invitrogen) following VESICLE separation by ultracentrifugation, at 55 °C for 15 min to digest proteins in the CM (Arroyo et al., 2011). HEECs were treated with or without the proteinase K treated media for 8 h, followed by RNA extraction and PCR (as described above) to determine the effect on miR-661 expression levels in HEEC.
2.2.11. Co-Immunoprecipitation
Co-immunoprecipitation was performed using 800 μl HTR8-CM or 400 μl pooled BCM 200 μl or 100 μl of lysis buffer respectively and 1 μg of Ago1 antibody (Cell signalling technologies), Ago 2 antibody (Cell signalling technologies) or control IgG (Dako). Following Incubation at 4 °C overnight, the immune complexes were pulled down with protein A/G magnetic beads (Thermo Scientific) and serially washed with 0.5% TBS/Tween, followed by TBS and distilled H2O. 500 μl of TriReagent was added to each sample and RNA extracted as per standard protocol described above. Method adapted from (Arroyo et al., 2011).
2.2.12. In-silico Analysis
For computational analysis, we used miRTarbase release 4.5 (Hsu et al., 2014) and DIANA-TarBase v7.0 (Vlachos et al., 2015). A list of the common targets was composed based on the 2 lists.
2.2.13. Immunohistochemistry
Immunohistochemistry for MTA2 and PVRL1 was performed on endometrial tissue from fertile women across the cycle as previously described (Cuman et al., 2013), using antibodies at the following concentrations: MTA2 (0.5 μg/ml rabbit monocolonal, # sc-28731, Santa Cruz) and PVRL1 (1 μg/ml mouse monocolonal, # sc-21722, Santa Cruz). Negative isotype controls of mouse or rabbit IgG (both DakoCytomation, Denmark) were applied at the same concentration as the primary antibodies.
2.2.14. Western Blotting
HEEC lysates were collected using universal lysis buffer following treatment with HTR8-CM. Western blotting was performed as previously described (Van Sinderen et al., 2013). Membranes were probed with antibodies against MTA2 (1:500 # sc-28731, Santa Cruz), PVRL1 (1:250 # sc-21722, Santa Cruz) and GAPDH (1:5000, #3683 cell signalling). Densitometry analysis was performed using Image Lab (BioRad).
2.2.15. Spheroid Adhesion Assay
To determine the effect of miR-661 on the adhesive properties essential for the attachment of the blastocyst to the endometrium, a co-culture model was established based on previous publication (Krishnan et al., 2013).
HEECs were grown to confluence 96-well plate and transfected according to manufacturers instructions using Lipofectamine RNAimax, with; miR-661 mimic only (3 pmol; Life Technologies); miR-661 mimic + miR-661 inhibitor (3 pmol; Life Technologies);miR-661 mimic + PVRL1 miR script Target Protect (4.5 pmol; Qiagen) or vehicle control for 72 h. Spheroids were formed using HTR8sv/Neo cells (2000 cells per spheroid) in a Cellstar U-shaped 96-well Suspension Culture Plate (Greiner Bio-One) and incubated at 37 °C for 48 h. Spheroids (8–10 per well) were transferred into a 96-well plate containing treated HEEC cells. Spheroid number was determined visually prior to incubation at 37 °C for 2 h. Co-culture wells were washed gently, with 150 μl serum-free DMEM/F12 media and the remaining spheroids counted to determine the number of adhered spheroids; Attachment us expressed as a percentage of the original spheroid number.
2.2.16. Sample Size
All sample sizes detailed have been chosen according to our previous experience using these techniques and power calculations (G*Power).
3. Results
3.1. Human Blastocysts Secrete microRNA Relative to Implantation Potential
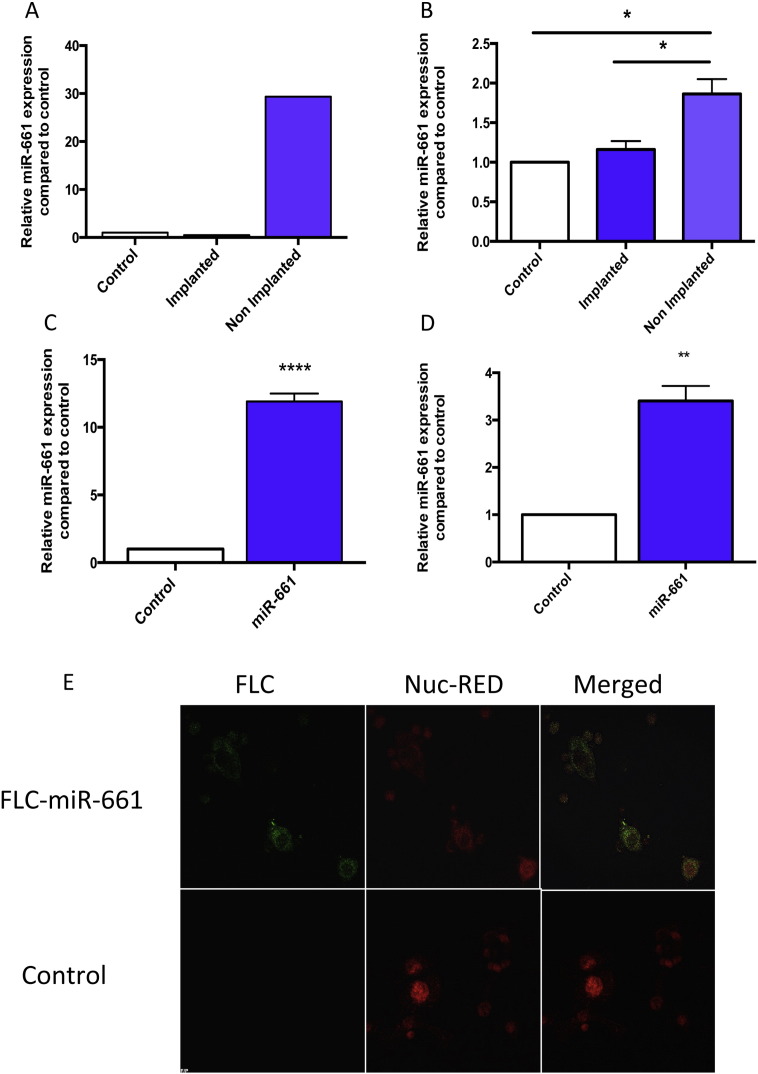
To identify the profile of miRs secreted by blastocysts, real-time PCR miR arrays were used to compare BCM (pooled n = 8, Table S1, patient sample characteristics) collected from blastocysts that successfully implanted (implanted) compared to those which failed to implant (non-implanted). Culture media alone were used as a control. 140 miRs (18% of total 784 on array) were detected across the three media groups. 47 miRs were detected exclusively in the media containing a blastocyst and from these 19 miRs were in the implanted group exclusively, 22 miRs in the non-implanted group exclusively and 6 miRs found in both groups. 22 miRs were solely expressed in the control culture media (Data not shown).miR-661, the highest differentially expressed miR in non-implanted BCM was confirmed by individual real time PCR TaqMan assays of the pooled media sample (Fig. 1A) and its presence in human trophectoderm cells were also confirmed (Table S3). Analysis of additional individual samples (n = 5), showed the presence of miR-661 samples specifically to the non-implanted cohort (Table 1).
Fig. 1.
miR-661 is expressed in non-implanted BCM and HEECs take up secreted miR-661 from conditioned media.
A. RTqPCR validation of miR-661 in implanted and non-implanted BCM vs. control media alone (n = 8, pooled). Data is normalised to endogenous snU6 and presented as mean ± SEM, * p < 0.05, Student's t test).
B. HEEC treated with Pooled BCM (control, implanted and non-implanted BCM) for 24 h increased expression of miR-661 treated with non-implanted compared to implanted and control only media (n = 4).
C. Transfection of HTRsv/neo (HTR8) cell with fluorescein tagged (FLC)-miR-661, increased expression of miR-661 in HTR8 cell conditioned media (CM) compared to control (scrambled miR) CM (n = 3).
D. Uptake of FLC-miR-661 in HEEC when treated with FLC-miR-661 CM for 8 h, compared to treatment with control (scrambled miR) CM (n = 6).
E. Immunofluorescence of FLC-miR-661 in HEEC. FLC-miR-661 (Green), Nuc-Red (Red). Data is presented as mean ± SEM, *p < 0.05, **p < 0.01, ****p < 0.0001, Student's t-test..
Table 1.
Individual BCM miR-661 CT levels.
| Implanted | Non-implanted |
|---|---|
| Undetected CT > 40 | 31.1 |
| Undetected CT > 40 | 27.8 |
| Undetected CT > 40 | 28.6 |
| Undetected CT > 40 | 27.4 |
| Undetected CT > 40 | 26.1 |
3.2. miR-661, Secreted by Human Blastocysts that do not Implant is Taken up by Primary Human Endometrial Epithelial Cells (HEECs)
To determine if HEECs can take up blastocyst secreted miRs, miR-661 uptake was investigated in our in vitro primary human culture models. Cultured HEECs treated with non-implanted BCM (pool of individual BCM samples used in miR arrays) demonstrated a significant increase in intracellular miR-661 mRNA levels compared to treatment with implanted BCM and control media (Fig. 1B). Investigation of endogenous miR-661 expression in HEECs, demonstrated miR-661 expression in the cultured HEECs was detected at very low or undetectable levels by Real time PCR (Data not shown). To further investigate if miRs secreted by blastocysts were taken up by primary HEEC we used fluorescently tagged synthetic miR-661 (FLC-miR-661) which was transfected into the HTR8sv/neo (HTR8) cell line. After transfection, the culture media were refreshed after 12 h to remove free oligonucleotides. The HTR8 CM was collected 12 h after refreshment. Transfection of HTR8 with FLC-miR-661 significantly increased the expression of miR-661 in HTR8 cell CM (Fig. 1C), compared to control (scrambled miR) transfected CM. Primary HEECs were treated with the HTR8 CM which resulted in the expression of miR-661 in HEECs compared to undetectable miR-661 in culture media from control treated cells (Fig. 1D). Fluorescent imaging confirmed the presence of FLC-miR-661 in the cytoplasm of HEECs (Fig. 1E).
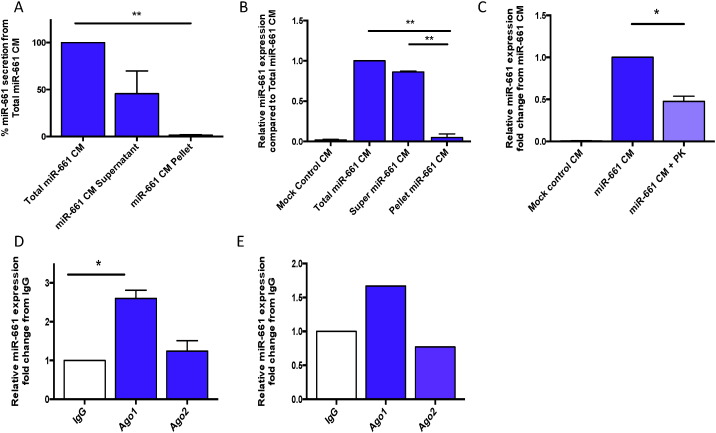
3.3. miR-661 is Secreted and Transported by Human Blastocysts via Argonaute 1 Protein
To determine the possible mechanism by which miR-661 is secreted by blastocysts into culture media, ultracentrifugation was performed on HTR8 FLC-miR-661 CM to separate RBC proteins in the supernatant from micro vesicles (MV) found in the remaining pellet (6). miR-661 expression was significantly higher in the supernatant compared to the remaining MV pellet (Fig. 2A). HEECs were treated with the total (unspun media), supernatant CM or resuspended MV pellet. miR-661 expression significantly increased in the HEEC treated with supernatant CM (Fig. 2B) compared to the resuspended MV pellet. To further prove the hypothesis that miR-661 was transported via RBC proteins, the supernatant CM was digested with proteinase K which significantly decreased miR-661 expression in the culture media (Fig. 2C), thus demonstrating that miR-661 was protected from digestion via its binding to an RBC protein. To determine the RBC protein responsible for the transport of miR-661, co-immunoprecipitation (Co-IP) of the supernatant CM with either Argo 1 or Argo 2 identified Ago 1, but not Ago 2, as the carrier of miR-661 (Fig. 2D). In order to determine if the miR-661 association with Ago 1 was not an artefact of the trophoblast cell line, co-IP on pooled BCM (n = 270), confirmed that miR-661 was bound to Ago 1, thus indicating that human blastocysts transport miR-661 via the RBC protein, Argo-1 (Fig. 2E).
Fig. 2.
miR-661 is bound to Argonaute 1 for extracellular transport. Expression of miR-661 in A. Differentially centrifuged HTR8 CM (n = 3), B. HEEC, treated with differentially centrifuged HTR8 CM (n = 3) (Total miR-661 CM, supernatant only miR-661 CM or re-suspended MV pellet. C. Decreased miR-661 expression in HEEC treated with HTR8-miR-661 CM + Proteinase K (PK) compared to miR-661 CM alone. D Co-immunoprecipitation of FLC-HTR8 CM, miR-661 expression bound to Ago 1 compared to IgG control and Ago2 (n = 3). E. Expression of miR-661 bound to Ago 1 in pooled BCM compared to IgG control (n = 1, pooled 270 samples from non-implanted BCM). Proteinase K (PK), Argonaute (Ago). Data is presented as mean ± SEM*p < 0.05, **p < 0.01, 1-way ANOVA.
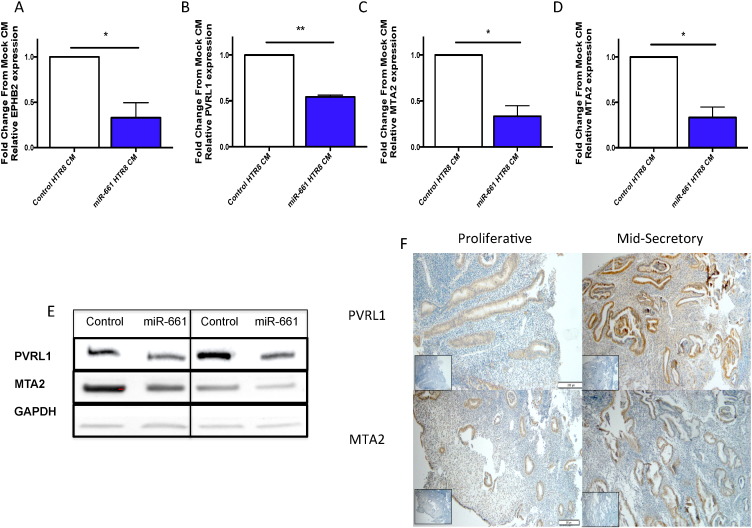
3.4. miR-661 Targets MTA2 and PVRL1 in Human Endometrial Epithelial Cells
In silico bioinformatics analysis of validated miR-661 target genes, identified a number of target genes that have roles in adhesion and invasion (Table S4). Poliovirus receptor-related 1 (PVRL1, also known as Nectin-1), metastasis associated protein (MTA) 1 and 2 and Ephrin type-B receptors 2 (EPHB2) expression were significantly decreased in miR-661 CM treated HEEC compared to control HEEC (Fig. 3A–D). PVRL1 and MTA2 proteins were down regulated (Figs. 3 and S1) and but there was no change in EpBH2 (data not shown). Immunohistochemistry localised MTA2 and PVRL1 to the luminal and glandular epithelium, with no changes in their levels observed across the menstrual cycle in normal fertile endometrial tissue (Fig. 3Fi–iv).
Fig. 3.
miR-661 reduces target expression in HEECs.
HEEC treated with miR-661 CM, significantly decreased expression of A.EBPH2, B. PVRL1, C. MTA1 and D.MTA2 compared to control CM (n = 3). E. Western analysis showed decreased expression of PVRL1 and MTA2 protein from HEEC treated with miR-661 CM compared to control CM. F. Immunohistochemistry (n = 4/phase) localised PVRL1 and MTA2 to glandular (G) and luminal epithelium (LE) in both proliferative and mid-secretory phase of the menstrual cycle. Data is presented as mean ± SEM, * p < 0.05, Students t test.
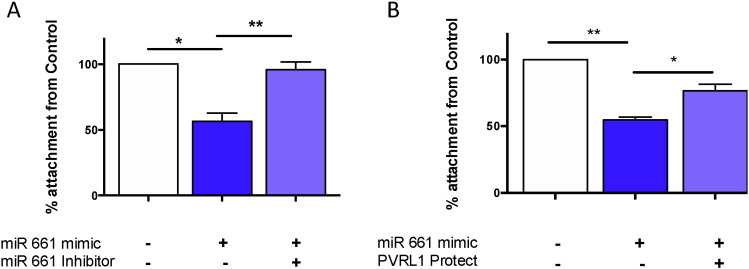
3.5. miR-661 Blocks Adhesion in Primary Human Endometrial Epithelial Cells via PVRL1
To determine the role of miR-661 in implantation, an established trophoblast spheroid-endometrial co-culture adhesion assay (Krishnan et al., 2013) was used to investigate miR-661 ability to inhibit embryo–endometrial adhesion. miR-661 treated HEEC, significantly decreased adhesion of spheroids to HEEC compared to vehicle only HEEC (Fig. 4A). The addition of a miR-661 inhibitor significantly increased the adhesion of the spheroids to HEEC, compared to mimic treatment only HEEC (Fig. 4A). Investigation of PVRL1, previously shown to have a role in cell adhesion (Takai et al., 2003), significantly increased adhesion of spheroids to HEEC treated with PVRL1 target protector, which prevents the binding of miR-661 to the 3′UTR binding site, specifically blocking the down regulation of PVRL1 by miR-661 (Fig. 4B).
Fig. 4.
miR-661 regulates trophoblast spheroid adhesion to HEEC.
A. Decreased adhesion of HTR8 spheroids to HEEC treated with miR-661 mimic compared to control. Addition of miR-661 inhibitor “rescued” adhesion as it was unchanged compared to control (n = 3). B. Increased adhesion of HTR8 spheroids to HEEC treated with miR-661 mimic and PVRL1 target protect, compared to miR-661 mimic only (n = 5). Data is presented as mean ± SEM** p < 0.01, ***p < 0.001, 1-way ANOVA..
4. Discussion
This study has demonstrated that human blastocyst secreted miRs are taken up by primary human endometrial epithelial cells and regulate their adhesive capacity via targeting gene and protein production. We have identified a potential mechanism by which blastocysts communicate with the endometrium, which is likely to facilitate receptivity and implantation in humans. This study has identified a functional role for blastocyst-secreted miRs on endometrial epithelial cell adhesion, the initiating event of implantation.
Our findings demonstrate that blastocysts secrete different miR profiles, in accordance with their implantation outcomes following ART. To date two other papers have examined miR expression in human BCM. Kropp et al., identified only one miR, miR-25, in pooled media from day 5 and day 6 blastocysts, however no correlation was identified in relation to blastocyst quality (Kropp et al., 2014). miR-25 was not identified in our cohort.
One other study used PCR arrays to identify miR expression profiles in BCM (Rosenbluth et al., 2014). They identified two miRs solely expressed in BCM, miR-372 and miR-191 that were not present in control media. miR-372 was expressed in both euploid and aneuploid BCM and was higher in BCM from embryos that failed to implant when correlated with the use of ICSI only embryos. These results are consistent with our finding that miR-372 was detected only in non-implanted BCM samples. miR-191 was not included in the array panel used in our study.
We however identified a large number of miRs that were differentially secreted into BCM from implanted compared to non-implanted BCM. The differences in the miRs detected between our study and the previous studies are likely due to differences in the experimental methods between the studies including differences in RNA extraction, cDNA synthesis, miR array panels used for detecting miRs in BCMand, differences in embryo culture media. Specifically, in our study we cultured embryos for 48 h compared to 24 h, and used different media for culture compared to a previous study (Rosenbluth et al., 2014). This may have affected differences in the miRs detected in BCM between the two studies. In addition, we used a miR array system that required less BCM compared to a previous study suggesting that the array system we used was highly sensitive which may have contributed to differences in detection of specific miRs between the present and other studies (Rosenbluth et al., 2014). In addition, a previous study used embryos that had been frozen at the pronuclear stage, thawed and cultured to blastocyst stage and arrays undertaken on BCM collected at the blastocyst stage (Rosenbluth et al., 2014). By comparison, in our study we did not freeze/thaw the embryos which may have affected the pattern of miR secretion between the present and a previous study (Rosenbluth et al., 2014).
Extracellular miRs are released from cells in membrane bound vesicles (such as exosomes), bound to RBC proteins (Ago1 and Ago2) or attached to high density lipo-proteins. miRs encapsulated in membrane bound vesicles or attached to proteins, protect miRs from RNase activity (Arroyo et al., 2011).Our study demonstrates a mechanism by which human blastocysts secrete miR-661 and transport it for uptake by the primary endometrial epithelial cells. We demonstrated that extracellular miR-661 was bound to the RBC Ago 1 and not Ago 2 or in vesicles, a finding that has not been previously identified in any cell type. Our study however, does not rule out whether primary HEEC take up other miRs or any other factors present in BCM. Our data however does demonstrate that the increased miR-661 expression in the primary HEEC occurs primarily via uptake from the media and not via stimulation of miR-661 expression in HEEC from factors present in the media.
It is unknown if miR transport mechanisms for specific miRs, remain the same in all cell systems or whether a specific cell transports most miRs via one or multiple modes. While this study shows a mechanism of miR transport from human trophectoderm cells in vitro, it remains to be investigated whether this is a generalised phenomenon for most miRs secreted by human blastocysts. Studies investigating the expression of miRs secreted by the endometrium are limited to the capture of exosomes and the miR carried in their cargo (Kresowik et al., 2014, Ng et al., 2013). To date no study, has examined the role Ago proteins play in the communication between the blastocyst and the endometrium.
Studies of the miRs in the endometrium are limited to expression studies, comparing the expression of miRs in receptive with non-receptive phase endometrium (Altmae et al., 2013, Kresowik et al., 2014, Kuokkanen et al., 2010), or in endometrium from fertile, infertile and repeat implantation failure (RIF) women (Dior et al., 2014, Revel et al., 2011, Zhao et al., 2012), with the aim of identifying endometrial receptivity biomarkers. To date only one recent study, has investigated a functional role of miRs in human endometrial cells in vitro, specifically miR-145. miR-145, is a previously identified miR with high expression in endometrium from women with repeat implantation failure (RIF) compared to normal fertile women (Revel et al., 2011). miR-145 overexpression in a human endometrial carcinoma cell line, was shown to inhibit mouse embryo adhesion to the cells (Kang et al., 2015). Our study however, provides evidence of direct uptake of a miR from human BCM by HEEC and demonstrates a functional effect on adhesion.
Mature miRs act by destabilising mRNAs with some degree of complementarity or by repressing protein translation, leading to down regulation of target genes and changes in biological functions (Bohnsack et al., 2004, Chen and Rajewsky, 2007, Kim, 2005, Lee et al., 2003). The expression of miRs is tightly coordinated and each miR has the ability to act on numerous gene targets (Bartel, 2004, Chen and Rajewsky, 2007). miR-661, is predicted to target approximately 1000 target genes (Paraskevopoulou et al., 2013, Reczko et al., 2012, Vlachos et al., 2015), and has been experimentally verified to target 6 genes; MTA1 MTA2, STARD10, VCL and PVRL1 (also known as PVRL1) (Hsu et al., 2014, Reczko et al., 2012, Reddy et al., 2009, Vetter et al., 2010, Vlachos et al., 2015).
The MTA family of proteins, is a central component of the Mi-2Nurd complex, in which their primary role is to regulate gene expression networks, via controlling histone acetylation and by regulating key signalling pathways by acetylation of target networks (Covington and Fuqua, 2014, Sen et al., 2014). MTA1 has been previously shown to be expressed in benign endometrium and in endometrial adenocarcinomas (Balasenthil et al., 2006) and we have identified and localised MTA2 in human endometrial tissue. MTA2 regulates cytoskeletal organisation partly via activation of the Rho signalling pathway (Covington and Fuqua, 2014). Whilst no studies have demonstrated a role of MTA2 or the effects of histone acetylation in human implantation, the Rho signalling pathway has been implicated to have a role in inducing human trophoblast invasion and migration (Saso et al., 2012) The downregulation of MTA2 by miR-661, may therefore inhibit activation of MTA2 target genes, such as Rho, which are required for embryo implantation.
PVRL1 is a membrane bound immunoglobulin-like cell adhesion molecule and modulates cell adhesion (Takai et al., 2003, Yu et al., 2007). It is a validated target of miR-661 breast cancer cells (Vetter et al., 2010). We demonstrated that miR-661 significantly down regulated PVRL1 mRNA and protein in primary endometrial epithelial cells. Nectins regulate the formation of adherens and tight junctions in epithelial cells (Takai and Nakanishi, 2003) and participate in the regulation of cellular activities such as cell polarisation, differentiation and proliferation (Takai et al., 2003, Takai et al., 2008), all of which are know requirements for embryo implantation (Norwitz et al., 2001). This suggests that the repression of PVRL1 by miR-661 may contribute to the disassembly of cell-cell contact and loss of epithelial cell polarity in the endometrial luminal epithelium, thus creating an unstable environment for attachment or loss of the firm adhesion required between the endometrium and trophectoderm for successful implantation. In this regard, we demonstrated that miR-661 blocked HEEC adhesion, at least partly, via PVRL1. Whilst a modest effect on adhesion was noted, there is highly likely to be additional factors regulated by miR-661 that regulate adhesion.
Our findings emphasise the important role that human blastocysts have on regulating the very early stages of implantation, adhesion, abnormalities in which lead to implantation failure and infertility. Our data demonstrate that human blastocysts secrete miRs that likely actively participate in the implantation process. Blastocyst-secreted miR profiles may thus be useful as biomarkers of their implantation potential or as targets to treat implantation failure and infertility, however additional studies are required to explore this further.
The following are the supplementary data related to this article.
miR-661 reduces target expression in HEECSs. Densitometry analysis of MTA-2 and PVRL1 western blots showed decreased expression of PVRL1 and MTA2 protein compared to control, nomalised against GAPDH.
Supplementary tables.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.ebiom.2015.09.003.
Conflict of Interest Statement
Authors declare there are no conflicts of interest.
Author Contributions
CC, MVS, KS and KR performed research. CC, MVS, LR, TO, MG and ED designed the research, CC, ED& MVS analysed research. CC and ED wrote the paper. All authors critically reviewed the final manuscript.
Acknowledgements
We thank the Monash IVF embryology team for their continued assistance in collecting the blastocyst spent media samples. We acknowledge the support of the Victorian State Government Operational Infrastructure Support and Australian Government NHMRC IRIISS. ED and MG were supported by NHMRC Fellowship (#550905) and ARC Fellowship (#140100594) respectively. CC was supported by an Australian Postgraduate Award. This work was supported by project grants awarded by Monash IVF (Grant # P4_FY15).
References
- Altmae S., Martinez-Conejero J.A., Esteban F.J., Ruiz-Alonso M., Stavreus-Evers A., Horcajadas J.A., Salumets A. MicroRNAs miR-30b, miR-30d, and miR-494 regulate human endometrial receptivity. Reprod. Sci. 2013;20:308–317. doi: 10.1177/1933719112453507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arroyo J.D., Chevillet J.R., Kroh E.M., Ruf I.K., Pritchard C.C., Gibson D.F., Mitchell P.S., Bennett C.F., Pogosova-Agadjanyan E.L., Stirewalt D.L. Argonaute2 complexes carry a population of circulating microRNAsIindependent of vesicles in human plasma. Proc. Natl. Acad. Sci. U. S. A. 2011;108:5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasenthil S., Broaddus R.R., Kumar R. Expression of metastasis-associated protein 1 (MTA1) in benign endometrium and endometrial adenocarcinomas. Hum. Pathol. 2006;37:656–661. doi: 10.1016/j.humpath.2006.01.024. [DOI] [PubMed] [Google Scholar]
- Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Bohnsack M.T., Czaplinski K., Gorlich D. Exportin 5 is a RanGTP-dependent dsRNA-binding protein that mediates nuclear export of pre-miRNAs. RNA. 2004;10:185–191. doi: 10.1261/rna.5167604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen K., Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat. Rev. Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- Covington K.R., Fuqua S.A. Role of MTA2 in human cancer. Cancer Metastasis Rev. 2014;33:921–928. doi: 10.1007/s10555-014-9518-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuman C., Menkhorst E.M., Rombauts L.J., Holden S., Webster D., Bilandzic M., Osianlis T., Dimitriadis E. Preimplantation human blastocysts release factors that differentially alter human endometrial epithelial cell adhesion and gene expression relative to IVF success. Hum. Reprod. 2013;28:1161–1171. doi: 10.1093/humrep/det058. [DOI] [PubMed] [Google Scholar]
- Dimitriadis E., White C.A., Jones R.L., Salamonsen L.A. Cytokines, chemokines and growth factors in endometrium related to implantation. Hum. Reprod. Update. 2005;11:613–630. doi: 10.1093/humupd/dmi023. [DOI] [PubMed] [Google Scholar]
- Dior U.P., Kogan L., Chill H.H., Eizenberg N., Simon A., Revel A. Emerging Roles of microRNA in the embryo–endometrium cross talk. Semin. Reprod. Med. 2014;32:402–409. doi: 10.1055/s-0034-1376359. [DOI] [PubMed] [Google Scholar]
- Galliano D., Pellicer A. MicroRNA and implantation. Fertil. Steril. 2014;101:1531–1544. doi: 10.1016/j.fertnstert.2014.04.023. [DOI] [PubMed] [Google Scholar]
- Graham C.H., Hawley T.S., Hawley R.G., MacDougall J.R., Kerbel R.S., Khoo N., Lala P.K. Establishment and characterization of first trimester human trophoblast cells with extended lifespan. Exp. Cell Res. 1993;206:204–211. doi: 10.1006/excr.1993.1139. [DOI] [PubMed] [Google Scholar]
- Hanke M., Hoefig K., Merz H., Feller A.C., Kausch I., Jocham D., Warnecke J.M., Sczakiel G. A robust methodology to study urine microrna as tumor marker: microrna-126 and microrna-182 are related to urinary bladder cancer. urol. oncol. 2010;28:655–661. doi: 10.1016/j.urolonc.2009.01.027. [DOI] [PubMed] [Google Scholar]
- Hannan N.J., Paiva P., Dimitriadis E., Salamonsen L.A. Models for study of human embryo implantation: choice of cell lines? Biol. Reprod. 2010;82:235–245. doi: 10.1095/biolreprod.109.077800. [DOI] [PubMed] [Google Scholar]
- Hsu S.D., Tseng Y.T., Shrestha S., Lin Y.L., Khaleel A., Chou C.H., Chu C.F., Huang H.Y., Lin C.M., Ho S.Y. miRTarBase update 2014: an information resource for experimentally validated miRNA-target interactions. Nucleic Acids Res. 2014;42:D78–D85. doi: 10.1093/nar/gkt1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y.J., Lees M., Matthews L.C., Kimber S.J., Forbes K., Aplin J.D. miR-145 suppresses embryo–epithelial Juxtacrine Communication at Implantation by Modulating Maternal IGF1R. J. Cell Sci. 2015;128:804–814. doi: 10.1242/jcs.164004. [DOI] [PubMed] [Google Scholar]
- Kim V.N. MicroRNA biogenesis: coordinated cropping and dicing. nat. rev. Mol. Cell Biol. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- Koot Y.E., Teklenburg G., Salker M.S., Brosens J.J., Macklon N.S. Molecular aspects of implantation failure. Biochim. Biophys. Acta. 2012;1822:1943–1950. doi: 10.1016/j.bbadis.2012.05.017. [DOI] [PubMed] [Google Scholar]
- Kresowik J.D., Devor E.J., Van Voorhis B.J., Leslie K.K. MicroRNA-31 is significantly elevated in both human endometrium and serum during the window of implantation: apotential biomarker for optimum receptivity. Biol. Reprod. 2014;91:17. doi: 10.1095/biolreprod.113.116590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnan T., Winship A., Sonderegger S., Menkhorst E., Horne A.W., Brown J., Zhang J.G., Nicola N.A., Tong S., Dimitriadis E. The role of leukemia inhibitory factor in tubal ectopic pregnancy. Placenta. 2013;34:1014–1019. doi: 10.1016/j.placenta.2013.09.003. [DOI] [PubMed] [Google Scholar]
- Kropp J., Salih S.M., Khatib H. Expression of microRNAs in bovine and human pre-implantation embryo culture media. Front. Genet. 2014;5:91. doi: 10.3389/fgene.2014.00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuokkanen S., Chen B., Ojalvo L., Benard L., Santoro N., Pollard J.W. Genomic profiling of microRNAs and messenger RNAs reveals hormonal regulation in microRNA expression in human Endometrium. Biol. Reprod. 2010;82:791–801. doi: 10.1095/biolreprod.109.081059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y., Ahn C., Han J., Choi H., Kim J., Yim J., Lee J., Provost P., Radmark O., Kim S. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- Mitchell P.S., Parkin R.K., Kroh E.M., Fritz B.R., Wyman S.K., Pogosova-Agadjanyan E.L., Peterson A., Noteboom J., O'Briant K.C., Allen A. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. U. S. A. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng Y.H., Rome S., Jalabert A., Forterre A., Singh H., Hincks C.L., Salamonsen L.A. Endometrial exosomes/microvesicles in the uterine microenvironment: a new paradigm for embryo-endometrial cross talk at implantation. PLoS One. 2013;8:e58502. doi: 10.1371/journal.pone.0058502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norwitz E.R., Schust D.J., Fisher S.J. Implantation and the survival of early pregnancy. N. Engl. J. Med. 2001;345:1400–1408. doi: 10.1056/NEJMra000763. [DOI] [PubMed] [Google Scholar]
- Paiva P., Menkhorst E., Salamonsen L., Dimitriadis E. Leukemia inhibitory factor and interleukin-11: critical regulators in the establishment of pregnancy. Cytokine Growth Factor Rev. 2009;20:319–328. doi: 10.1016/j.cytogfr.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Paraskevopoulou M.D., Georgakilas G., Kostoulas N., Vlachos I.S., Vergoulis T., Reczko M., Filippidis C., Dalamagas T., Hatzigeorgiou A.G. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013;41:W169–W173. doi: 10.1093/nar/gkt393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park N.J., Zhou H., Elashoff D., Henson B.S., Kastratovic D.A., Abemayor E., Wong D.T. Salivary microRNA: discovery, characterization, and clinical utility for oral cancer detection. Clin. Cancer Res. 2009;15:5473–5477. doi: 10.1158/1078-0432.CCR-09-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reczko M., Maragkakis M., Alexiou P., Grosse I., Hatzigeorgiou A.G. Functional microRNA targets in protein coding sequences. Bioinformatics. 2012;28:771–776. doi: 10.1093/bioinformatics/bts043. [DOI] [PubMed] [Google Scholar]
- Reddy S.D., Pakala S.B., Ohshiro K., Rayala S.K., Kumar R. MicroRNA-661, a c/EBPalpha target, inhibits metastatic tumor antigen 1 and regulates its functions. Cancer Res. 2009;69:5639–5642. doi: 10.1158/0008-5472.CAN-09-0898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revel A., Achache H., Stevens J., Smith Y., Reich R. MicroRNAs are associated with human embryo implantation defects. Hum. Reprod. 2011;26:2830–2840. doi: 10.1093/humrep/der255. [DOI] [PubMed] [Google Scholar]
- Rosenbluth E.M., Shelton D.N., Sparks A.E., Devor E., Christenson L., Van Voorhis B.J. MicroRNA expression in the human blastocyst. Fertil. Steril. 2013;99(855–861):e853. doi: 10.1016/j.fertnstert.2012.11.001. [DOI] [PubMed] [Google Scholar]
- Rosenbluth E.M., Shelton D.N., Wells L.M., Sparks A.E., Van Voorhis B.J. Human embryos secrete microRNAs into culture media—a potential biomarker for implantation. Fertil. Steril. 2014;101:1493–1500. doi: 10.1016/j.fertnstert.2014.01.058. [DOI] [PubMed] [Google Scholar]
- Saso J., Shields S.K., Zuo Y., Chakraborty C. Role of Rho GTPases in human trophoblast migration induced by IGFBP1. Biol. Reprod. 2012;86:1–9. doi: 10.1095/biolreprod.111.094698. [DOI] [PubMed] [Google Scholar]
- Sen N., Gui B., Kumar R. Role of MTA1 in cancer progression and metastasis. Cancer Metastasis Rev. 2014;33:879–889. doi: 10.1007/s10555-014-9515-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai Y., Nakanishi H. Nectin and afadin: novel organizers of intercellular junctions. J. Cell Sci. 2003;116:17–27. doi: 10.1242/jcs.00167. [DOI] [PubMed] [Google Scholar]
- Takai Y., Irie K., Shimizu K., Sakisaka T., Ikeda W. Nectins and nectin-like molecules: roles in cell adhesion, migration, and polarization. Cancer Sci. 2003;94:655–667. doi: 10.1111/j.1349-7006.2003.tb01499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai Y., Miyoshi J., Ikeda W., Ogita H. Nectins and nectin-Like molecules: roles in contact inhibition of cell movement and proliferation. Nat. Rev. Mol. Cell Biol. 2008;9:603–615. doi: 10.1038/nrm2457. [DOI] [PubMed] [Google Scholar]
- Van Sinderen M., Cuman C., Winship A., Menkhorst E., Dimitriadis E. The chondroitin sulfate proteoglycan (CSPG4) regulates human trophoblast function. Placenta. 2013;34:907–912. doi: 10.1016/j.placenta.2013.07.065. [DOI] [PubMed] [Google Scholar]
- Vetter G., Saumet A., Moes M., Vallar L., Le Bechec A., Laurini C., Sabbah M., Arar K., Theillet C., Lecellier C.H. miR-661 expression in SNAI1-induced epithelial to mesenchymal transition contributes to breast cancer cell invasion by targeting PVRL1 and StarD10 messengers. Oncogene. 2010;29:4436–4448. doi: 10.1038/onc.2010.181. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Vickers K.C., Palmisano B.T., Shoucri B.M., Shamburek R.D., Remaley A.T. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat. Cell Biol. 2011;13:423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlachos I.S., Paraskevopoulou M.D., Karagkouni D., Georgakilas G., Vergoulis T., Kanellos I., Anastasopoulos I.L., Maniou S., Karathanou K., Kalfakakou D. DIANA-TarBase v7.0: indexing more than half a million experimentally supported miRNA:mRNA interactions. Nucleic Acids Res. 2015;43:D153–D159. doi: 10.1093/nar/gku1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z., Adusumilli P.S., Eisenberg D.P., Darr E., Ghossein R.A., Li S., Liu S., Singh B., Shah J.P., Fong Y. PVRL1 expression by squamous cell carcinoma is a predictor of herpes oncolytic sensitivity. Mol. Ther. 2007;15:103–113. doi: 10.1038/sj.mt.6300009. [DOI] [PubMed] [Google Scholar]
- Zhao Y., Zacur H., Cheadle C., Ning N., Fan J., Vlahos N.F. Effect of luteal-phase support on endometrial microRNA expression following controlled ovarian stimulation. Reprod. Biol. Endocrinol. 2012;10:72. doi: 10.1186/1477-7827-10-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J., Li Y.S., Nguyen P., Wang K.-C., Weiss A., Kuo Y.-C., Chiu J.-J., Shyy J.Y., Chien S. Regulation of vascular smooth muscle cell turnover by endothelial cell-secreted microRNA-126: role of shear stress. Circ. Res. 2013;113:40–51. doi: 10.1161/CIRCRESAHA.113.280883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zubakov D., Boersma A.W., Choi Y., van Kuijk P.F., Wiemer E.A., Kayser M. MicroRNA markers for forensic body fluid identification obtained from microarray screening and quantitative RT-PCR confirmation. Int. J. Legal Med. 2010;124:217–226. doi: 10.1007/s00414-009-0402-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
miR-661 reduces target expression in HEECSs. Densitometry analysis of MTA-2 and PVRL1 western blots showed decreased expression of PVRL1 and MTA2 protein compared to control, nomalised against GAPDH.
Supplementary tables.