Abstract
Background
The oriental fruit moth, Grapholita molesta, is an extremely important oligophagous pest species of stone and pome fruits throughout the world. As a host-switching species, adult moths, especially females, depend on olfactory cues to a large extent in locating host plants, finding mates, and selecting oviposition sites. The identification of olfactory genes can facilitate investigation on mechanisms for chemical communications.
Methodology/Principal Finding
We generated transcriptome of female antennae of G.molesta using the next-generation sequencing technique, and assembled transcripts from RNA-seq reads using Trinity, SOAPdenovo-trans and Abyss-trans assemblers. We identified 124 putative olfactory genes. Among the identified olfactory genes, 118 were novel to this species, including 28 transcripts encoding for odorant binding proteins, 17 chemosensory proteins, 48 odorant receptors, four gustatory receptors, 24 ionotropic receptors, two sensory neuron membrane proteins, and one odor degrading enzyme. The identified genes were further confirmed through semi-quantitative reverse transcription PCR for transcripts coding for 26 OBPs and 17 CSPs. OBP transcripts showed an obvious antenna bias, whereas CSP transcripts were detected in different tissues.
Conclusion
Antennal transcriptome data derived from the oriental fruit moth constituted an abundant molecular resource for the identification of genes potentially involved in the olfaction process of the species. This study provides a foundation for future research on the molecules involved in olfactory recognition of this insect pest, and in particular, the feasibility of using semiochemicals to control this pest.
Introduction
The sensitive olfactory system plays a predominant role in insect behavior, such as seeking host plants, finding mates, selecting oviposition sites, recognizing kins, and escaping from predators and toxic compounds [1]. Antennae are specialized organs of insect for chemical sensing, especially for olfaction. The surface of antennae is covered with different types of sensilla, which are a specialized hair-like, multi-pore structures in which olfactory receptor neurons (ORNs) extend dendrites into the antennal lymph where peripheral olfactory signal transduction occur [2]. ORNs can recognize relevant volatiles and generate an electrical impulse that is transported to the primary olfactory center in the antennal lobe [3]. Within the sensilla-ORN structure, a number of gene families are involved in different steps in signal transduction pathways, such as the genes encoding odorant binding proteins (OBPs), chemosensory proteins (CSPs), odorant receptors (ORs), ionotropic receptors (IRs), sensory neuron membrane proteins (SNMPs), and odorant degrading enzymes (ODEs) [4].
OBPs belong to a group of small water-soluble proteins that are secreted by the accessory cells around the ORNs and impregnated in the sensilla lymph [5]. OBPs are considered to be the first group of proteins that participate in the olfactory signal transduction pathway in insects, which can selectively transport hydrophobic odorant molecules through the sensillum lymph to the surface of ORNs as the odor molecules diffuse through the pores on sensilla [6]. Like OBPs, the CSPs are another class of hydrophilic proteins that are enriched in the sensillum lymph. However, its function in olfactory transduction and non-olfactory procedures remains largely unknown [7]. ORs are embeded in the dendrite membrane of ORNs in the antennae, and play a central role as a bio-transducer in chemosensory signal transduction [8]. In insects, it is generally believed that ORs function as heterodimers, with highly conserved and broadly expressed protein (originally called Or83b but now with the generic name ORCO [9]) serve as a ligand-gate channel with a various partner (OrX) that can distinctly determine ligang-binding specificity [10]. In addition, ORs could recognize odorants and therefore are also involved in odor recognition [11]. Typically, there are three transmembrane domains and a bipartite ligand-binding domain with two lobes in IRs [12]. IRs act in complex of three subunits, which can be composed of individual odor-specific receptors, and one or two of the broadly expressed coreceptors (IR8a, IR25, and IR76b) in one IR-expressing neuron [11].
The development of Next Generation Sequencing (NGS) technologies has greatly improved the efficiency and speed of gene discovery in recent years [13]. De novo assembly of transcripts provides a workable solution to transcriptome analysis. At present, a lot of de novo transcriptome assemblers available are designed for Roche 454, Illuminia Solexa, and SOLID. SOAPdenovo, SOAPdenovo-Trans, Velvet Oases, ABySS, trans-ABySS, and trinity have been successfully applied to de novo transcriptome assembly form short-read RNA-Seq data of model and non-model organisms [3,14–18].
The oriental fruit moth Grapholita molesta Busck is an economically important oligophagous pest species of stone and pome fruits throughout the world, causing substantial losses in fruit yields [19]. Peach (Prunus persica L) is considered the primary host, and pear (Pyrus communis L.) and apple (Malus domestica L.) are secondary hosts [20,21]. In some parts of their geographic range, the adult can migrate from peach orchards to pear or apple orchards by detecting and following changes of volatile components emitted by these host plants [22]. A lot of pests with multiple generations, such as G.molesta, that can annually survive and reproduce on different hosts, are confronted with hight-variability in the volatile blends emitted by different host plant species at specific periods, as well as by the same host plant species across a growing season [23]. One way to adapt these variations in host plant volatile blends is to respond to a specific set of compounds common to all host plants [24]. A three-component mixure of (Z)-3-hexen-1-yl acetate, benzaldehyde and (Z)-3-hexen-1-ol elicited a similar attractant effect on G.molesta as the natural blend from peach shoots [25]. The mixture of (Z)-3-hexenyl acetate: (Z)-β-ocimene: (E)-β-farnesene in the proportion 1:2:2 can attract mated G. molesta males [26]. Small amounts of benzonitrile can convert an unattractive four-compound mixture ((Z)-3-hexen-1-yl acetate, (Z)-3-hexen-1-ol, benzaldehyde, and (E)-2-hexenal, ratio 69.84:14.64:13.26:2.26) to a bioactive five-compound mixture that is attractive to mated G.molesta females as good as natural blends [27]. Volatiles blends from the various attractive stages of peach and pear shared a common set of five aldehydes, suggesting the C6-C10 aliphatic aldehydes play a key role in G.molesta females attraction to host plants [28]. Butyl hexanoate makes up about 10% of the total volatiles emitted from peach shoots and ripe pears. Mated G. molesta females are attracted to butyl hexanoate at intermediate dosages [29]. However, limited information is available on the olfactory recognition mechanism for host plant’s volatiles at molecular levels.
The monitoring of G.molesta mainly lies on pheromone trapping of males. However, the flight performance of this species exhibits remarkable differences between males and females. The proportion of long-flying females was three to six times greater than males, and gravid females can be considered to be the main colonists [30]. In the field, female moths have the capacity to make inter-orchard flights [31], causing a serious threat to pear or apple orchards especially in the vicinity of peach crops. Female moths are more easily to tolerate a modulation of the ratios of volatile compounds with distinct threshold values [32]. Therefore, identification of a wider range of olfactory genes of female moths will enable a better understanding of the mechanisms of olfactory recognition at the molecular level, which could ultimately lead to the development of new environment-friendly control strategies.
To identify genes likely involved in olfaction in species with no sequenced genome available like G. molesta, we sequenced and analyzed an antennal transcriptome of adult females using Illumina Miseq sequencing. We reported here the identification of 28 OBP genes, 17 CSP genes, 48 OR genes, 24 IR genes, four GR genes, two SNMP genes, and one ODE gene in the female antennal transcriptome.
Methods
Insect rearing
G. molesta in all experiments were obtained from a laboratory colony maintained at the College of Plant Protection, Northwest A&F University, Yangling, Shaanxi, China. The colony of G. molesta has been maintained for more than 60 generations in the laboratory. Larvae were reared on artificial diet at 25 ± 1°C, 70 ± 5% RH under a photoperiod of 15:9 (L:D). After pupation, male and female pupae were placed in separate glass tubes and maintained in the conditions described previously. The adults were fed with a cotton swab dipped in 5% honey solution and changed daily. Antennae of 3–4 day-old female moths were dissected after eclosion, immediately frozen in liquid nitrogen, and stored at –80°C until RNA was extracted.
Extraction of total RNA
Frozen antennae were immediately transferred into a 1.5 mL Eppendorf tube immersed in liquid nitrogen and ground with a pestle. Total RNA was extracted using RNAiso Plus Total RNA extraction reagent (TaKaRa, Shiga, Japan) following the manufacturer’s instructions. The residual genomic DNA in total RNA was removed by DNase I (MBI Fermentas, Glen Burnie, MD, USA). Total RNA was dissolved in RNase-free water and RNA integrity was measured using Agilent 2100 bioanalyzer (Quantifluor-ST fluorometer, Promega, E6090). The high quality RNA (RIN number: 9.3) was used for cDNA library construction and Illumina sequencing.
Sequencing
Poly (A) mRNA was isolated from 12 μg of total RNA extracted from approximately 1200 antennae of 3–4 days-old adult female moths using the PolyA+Tract mRNA Isolation System (Illumina, San Diego, CA), and further purified using the RNeasy MinElute Clean up Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. Fragmentation buffer was added to cleave mRNA into short fragments, and then, these fragments were used to synthesize first-strand cDNA using random hexamer primers, which was further transformed into double stranded cDNA with RHase H and DNA polymerase I. A paired-end library was constructed from the cDNA synthesized using the Genomic Sample Prep Kit (Illumina). Fragments larger than 375 bp were purified with QIAquick PCR Extraction Kit (Qiagen), end-repaired, and linked with sequencing adapters. AMPureXP beads were used to remove the unsuitable fragments, and then, the sequencing library was constructed with PCR amplification. After being validated using Pico green staining (Quant-iT PicoGreen dsDNA Assay Kit, Invitrogen, P7589) and fluorospectrophotometry, and quantified using Agilent 2100 (Quantifluor-ST fluorometer, Promega, E6090)), the library was sequenced using Illumina Miseq platform (Shanghai Personal Biotechnology Cp., Ltd. Shanghai, China). For subsequent analysis, 1/2 run data was generated.
Unigene generation
Raw reads were filtered using a stringent process and subsequent de novo assembly. The reads were screened from the 39 to 59 to trim the bases with a quality score of Q<20 using 5 bp windows, and the reads with final length less than 50 bp were removed. In order to accurately discover and reduce false positive olfactory gene detection, we evaluate the performance of de novo transcriptome assembly using SOAPdenovo-Trans, Trans-ABySS, and Trinity form short-read RNA-Seq data of G.molesta antennae. The de novo transcriptome assembly was further analyzed using DETONATE (de novo transcriptome RNA-seq assembly with or without the truth evaluation) [33] and Transrate (http://hibberdlab.com/transrate/index.html). All derived transcript sequences were used to search NCBI non-redundant (NR) database (ftp://ftp.ncbi.nlm.nih.gov/blast/db/) with the BLASTn program (E-value,1E-5), and the top-hit transcripts were selected as unigenes. For the unigenes that failed to be aligned with any sequence in the databases, the software GetORF was used to predict their open reading frames (ORFs) and ascertain their coding orientations with default settings.
Gene identification and functional annotation
The annotation of all derived sequences were executed using the BLASTX program against the NCBI non-redundant database (NR) and SwissProt protein sequences with e-value<1e-5. The BLASTX results were then imported into Blast2GO suite for GO Annotation. Open reading frames (ORFs) of the unigenes were predicted using ORF Finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html) based on the results given by BLASTX. ClustalX version 1.83 and MAFFT version 6.0 (http://mafft.cbrc.jp/alignment/server/) were used to conduct multiple sequence alignments. Signal peptides of the protein sequences were identified using SignalP 4.0 (http://www.cbs.dtu.dk/services/SignalP/) with default parameters. The transmembrane-domains (TMDs) of annotated genes were predicted using TMHMM version 2.0 (http://www.cbs.dtu.dk/services/TMHMM), while the ORFs were translated to amino acid sequences using ExPASy (http://www.expasy.org/translate/).
Phylogenetic analyses
To verify the annotation of the candidate olfaction genes and to search for orthologs, phylogenetic analyses were conducted among G. molesta and other species with close genetic relationships. The species we selected is all belong to Lepidoptera. The genomes of Bombyx mori and Danaua plexippus have been published. The transcriptomes of Agrotis ypsilon, Heliothis virescens, Heliothis armigera, Cydia pomonella and Manduca sexta concentrated on olfactory genes have been well studied, as well as the function of these genes [16,18,34–37]. The OBP data set contained 28 sequences which are identified as candidate GmolOBPs, four sequences from C. pomonella [34], nine sequences from D. plexippus [35], 16 sequences from H. virescens [36], 13 sequences from M. sexta [37], 20 sequences from H. armigera [20], and 13 sequences from A. ipsilon [18]. All together, the OBP data set contained 103 sequences. The CSP data set contained 77 sequences, including 16 sequences from B. mori [7], four sequences from C. fumiferana [38], 17 sequences from H. armigera [39], nine sequences from H. virescens [36], 14 sequences from S. littoralis [15], and 17 sequences which are identified as candidate GmolCSPs. The OR and GR data set contained OR (or GR) sequences identified in Lepidoptera (one from M. sexta [14], 41 from C. pomonella [16], five from S. littoralis [15] and 69 from B. mori [40]). The OR data set contained a total of 168 sequences. In the IR data set, 24 sequences of candidate IRs from G. molesta were added to the number of sequences identified in B. mori [40], M. sext [14], C. pomonella [16], and S. littoralis [15]. Since IRs are more conserved than ORs among insects, IR sequences from non-Lepidoptera species (D. melanogaster [11], A. mellifera [41] and T. castaneum) were also included in the data set, and the final data set contained 175 sequences. Amino acid sequences of proteins used in building the phylogenetic tree are listed in S1 File. Amino acid sequences were aligned using MAFFT version 6.0, while the unrooted trees were constructed by the neighbor-joining method, with observed correction of distances, as implemented in Seaview v.4 software. The node support was assessed using a bootstrap procedure base on 1000 replicates, and the tree was drawn using Adobe Photoshop CS5.
Expression analysis of the candidate OBPs and CSPs by semi-quantitative reverse transcription PCR
To confirm and compare the tissue expression of putative GmolOBPs and GmolCSPs identified from the transcriptome, semi-quantitative reverse transcription PCR was performed using cDNAs template prepared from male antennae, female antennae, and remaining bodies (without antennae) of the moth. Each experiment contained two biological repeats, three technical duplications, and controls were PCR with no template. Total RNA was extracted as described previously, treated with DNAse (RQ1, Promega, Madison, WI, USA), and corresponding cDNAs were synthesized using the First Strand cDNA Synthesis Kit (TaKaRa, Shiga, Japan) following the recommended protocol. Primers were designed using the Primer Premier 5 software and sequences are available in S1 Table. PCR was performed with GeneAmp PCR system 9700 under the general 3-step amplification of 95°C for 5 min, followed by 30 cycles of 95°C for 30 s, 50–60°C for 30 s; 72°C for 30 s, and final extension of 72°C for 10 min. The PCR cycle-numbers were adjusted respectively for each gene. For most chemosensory genes, cycle-numbers were within the range of 30 to 35, but for some genes with high levels of expression, cycle-numbers were reduced to 25. PCR products were analyzed on 1.2% agarose gels electrophoresis and verified by direct DNA sequencing.
Results
Sequencing and de novo assemblies
A total of 5.6 million raw reads (average read length 251 bp) were obtained from the libraries of female antennae. After removing low quality, adaptor, and contaminating sequence reads and reads shorter than 50 bp, about 5.2 million clean-reads comprised the 2.2 gigabases were generated. In total, 114263, 79209 and 71086 transcripts, with the mean length of 630, 510, and 619 bp, were obtained from assembled with Trinity, SOAPdenovo-trans and Trans-Abyss (Table 1). The raw data from IIIumina Miseq sequencing was deposited in the NCBI Short Read Archive (SRA) database with accession number SRR1424578. The gene lengths, reads number of each unigene, and the abundance of the unigenes based on reads were integrated in S2 Table.
Table 1. Assembly summary of G. molesta antenna transcriptome using three different assemblers.
| Assemblers | Total Length (bp) | Transcripts No. | Max Length (bp) | Mean Length (bp) | N50 | >1K reads No. |
|---|---|---|---|---|---|---|
| Trinity | 71953993 | 114263 | 24397 | 630 | 996 | 17955 |
| SOAPdenovo-trans | 40414085 | 79209 | 21711 | 510 | 751 | 8883 |
| Abyss-trans | 43992300 | 71086 | 26749 | 619 | 940 | 10766 |
Quality assessment of de novo transcriptome assemblies
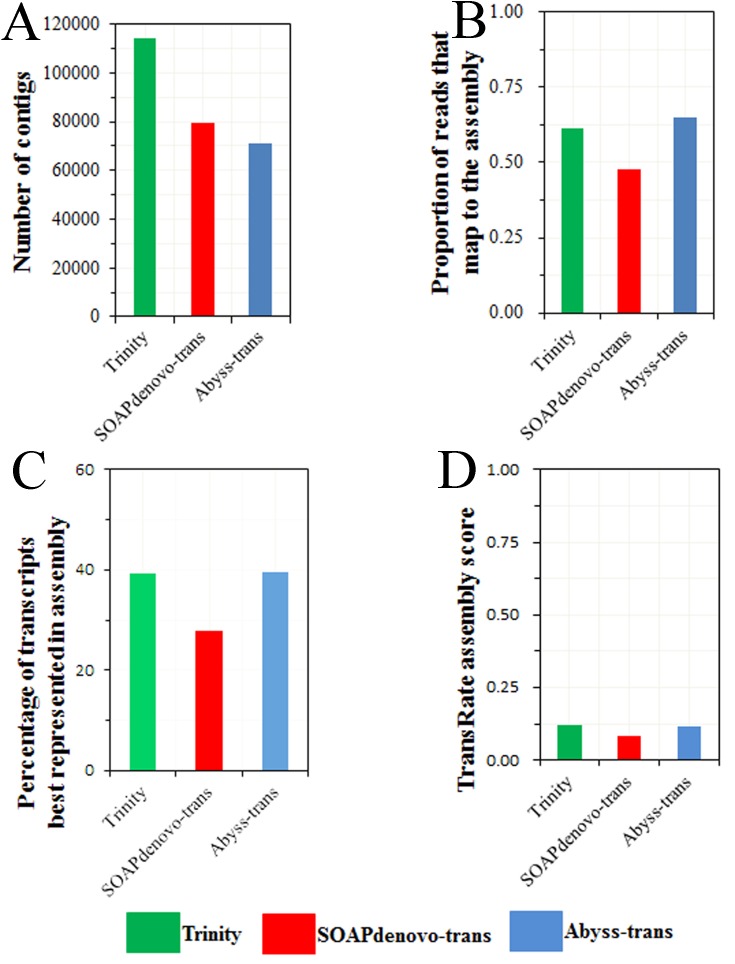
Trinity assembly produced the most transcripts, the longest transcripts in average and the largest N50, followed by Abyss-trans, while SOAPdenovo-trans yielded the worst in very category (Table 1). Among the three different de novo assemblers, we obtained the same number transcripts annotated to putative olfactory genes by the search against the non-redundant protein database. Most of the cover percent transcripts annotated to olfactory genes were greater than 70% among the three assemblers (S3 Table). We evaluated the transcriptome assembly generated using DETONATE and TransRate. The results showed that calculated TransRate assembly scores of Trinity and Abyss-trans had no much difference, but greater than score of SOAPdenovo-trans (Fig 1), revealing the Trinity and Abyss-trans conducted more accurately and completely on individual contigs level than SOAPdenovo-trans. The likelihood score, which the dominant term in the RSEM-EVAL score, was much higher in Trinity and Abyss-trans assembly than SOAPdenovo-Trans (Table 2), indicating that the Trinity and Abyss-trans are more accurate in assembly-level than SOAPdenovo-trans. Overall, Trinity is the most suitable software for de novo RNA-seq assembly for G.molesta without sequenced genomes.
Fig 1. TransRate assembly scores.
(A) Number of contigs for three representative assemblies from RNA-seq data of G.molesta. (B) Proportion of reads that map to each assembly. (C) Percentage of transcripts best represented in the assembly. (D) Final TransRate assembly scores for the three different assemblies.
Table 2. RSEM-EVAL evaluating de novo assemblies from Trinity, SOAPdenovo-Trans and Abyss-trans.
| Assembler | Likehood score | Prior score | BIC score | RSEM-EVAL score |
|---|---|---|---|---|
| Trinity | -2590055043 | -99749414 | -887936 | -2690692395 |
| SOAPdenovo-trans | -2978072059 | -55965628 | -615534 | -3034653222 |
| Abyss-trans | -2618491605 | -60986277 | -552411 | -2680030294 |
RSEM-EVAL was run on each assembly and the likelihood, prior, BIC and total RSEM-EVAL scores were recorded. BIC, Bayesian information criterion.
Gene identification and functional annotation
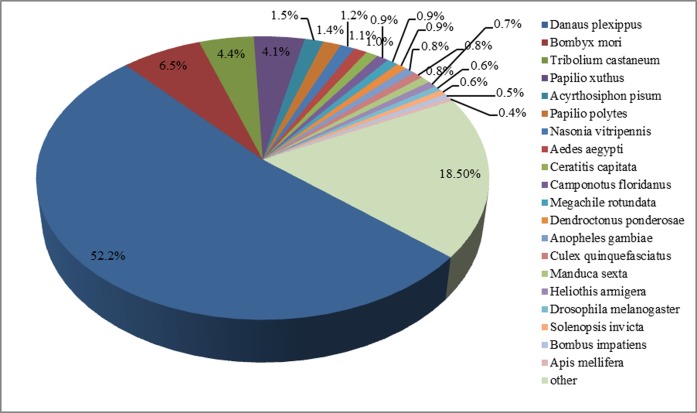
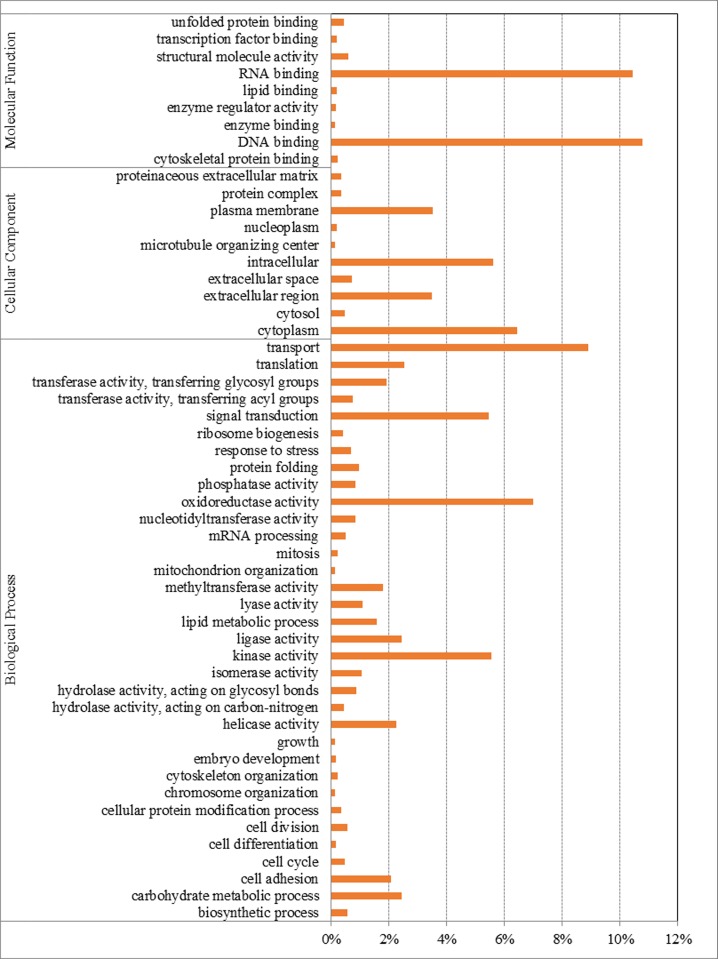
A total of 16,215 unigenes matched to known proteins in Genbank. Among the annotated unigenes, 64.3% had a first hits to Lepidopteran sequences. The top matched species were Danaus plexippus (52.2%), Bombyx mori (6.5%), Tribolium castaneum (4.4%), Papilio xuthus (4.1%), and Acyrthosiphon pisum (1.5%) (Fig 2). Fig 3 illustrated the distribution of the unigenes in GO terms. Among the 16,215 unigenes, 11,569 (71.35%) were assigned to 55,382 GO term annotations, with biological processes 26,297 terms; molecular function 15,062 terms; and cellular component 14,023 terms. In the biological process terms, transport, signal transduction and oxidoreductase activity were the mostly represented, while in the cellular component terms, the cytoplasm and intracellular were the most abundant. In the molecular function category, the genes expressed in the antennae were mostly enriched to DNA binding and RNA binding activity.
Fig 2. Top 20 best hits of the BLASTx results.
All G. molesta antennal unigenes were used in BLASTX search in NR database.
Fig 3. Gene Ontology (GO) analysis of G. molesta antennal transcripts.
GO terms assigned to biological process, cellular component and molecular functions.
Identification of putative odorant-binding proteins
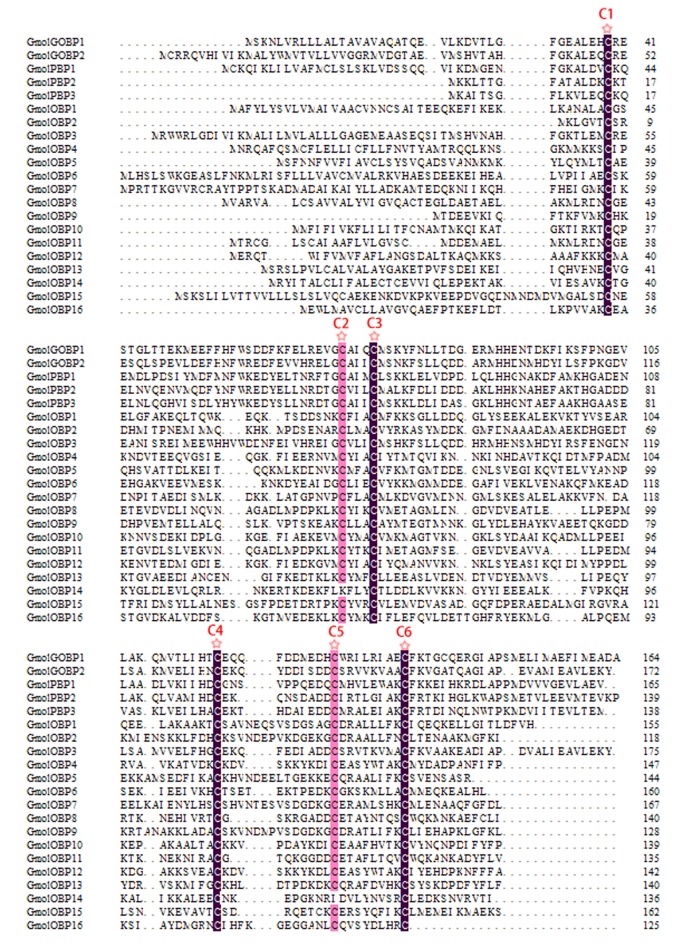
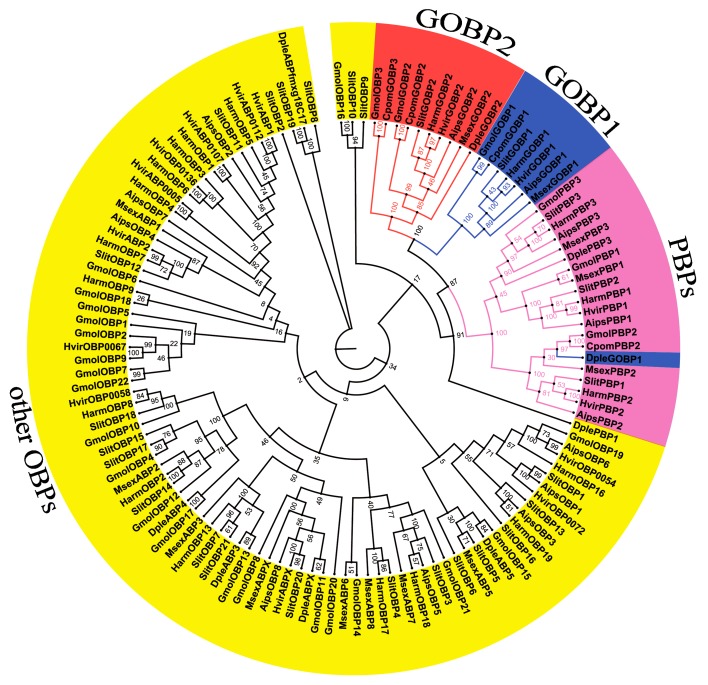
First, we used motif scanning to detect the conserved six cysteine residues pattern (C1-X20-66-C2-X3-C3-X21-43-C4-X8-14-C5-X8-C6 or C1-X15-39-C2-X3-C3-X21-44-C4-X7-12-C5-X8-C6, where X is any amino acid) of the candidate odorant-binding proteins [42]. We then used keyword searching and PSI-Blast. Twenty-eight sequences encoding putative odorant-binding proteins were identified, including two GOBPs and three PBPs. Of the 28 sequences, 21 had full ORFs, four unigenes had full-length ORFs, but without a signal peptide. Sequence alignment showed that almost all the putative OBPs shared the classic six-cysteine motif, except GmolOBP14, which was grouped into the “minus-C” subgroup with the second and fifth cysteine residues missing [43] (Fig 4). In the phylogenetic tree, as expected, the PBP and GOBP sequences were clustered into separate clades away from other OBPs. All the candidate OBP sequences with at least one lepidopteran ortholog were clustered in congruence with the BLAST results (Fig 5). Comparing our candidate OBPs with previously recorded OBPs of G.molesta in NCBI, 23 sequences as new genes, including GmolPBP1, GmolOBP1, GmolOBP2, and GmolOBP4 to GmolOBP23. The information on the OBPs is listed in Table 3. The nucleotide sequences are listed in S2 File.
Fig 4. Sequences alignment of candidate GmolOBPs.
The six conserved cysteine residues were marked with “☆”. As GmolOBP17, 18, 19, 20, 21, 22 and 23 are not intact sequences, those sequences are not included in the multisequence alignment.
Fig 5. Neighbor-joining tree of candidate OBP genes from G. molesta and other Lepidoptera.
The tree was drawn using Adobe Photoshop CS5, based on the unrooted tree constructed using BioNJ algorithm in Seaview v.4. The unrooted tree was constructed based on the sequence alignment obtained using MAFFT version 6. Gmol, Grapholita molesta, Cpom, Cydia pomonella, Dple, Danaus plexippus, Hvir, Heliothis virescens, Msex, Manduca sexta, Harm, Helicoverpa armigera, Aips, Agrotis ipsilon. The clade in blue indicates the GOBP1 gene clade; the clade in red indicates the GOBP2 clade, the clade in fuchsia indicates the PBPs clade, the yellow indicates other OBPs clade.
Table 3. Unigenes of candidate odorant binding proteins.
| Unigene reference | Gene name | Length (nt) | ORF (aa) | BLASTx best hit (Reference/Name/Species) | E-value | Identify | Full length | Signal Peptide |
|---|---|---|---|---|---|---|---|---|
| Pheromone binding protein | ||||||||
| Comp42972_c0_seq8 | GmolPBP1 | 2123 | 166 | gb|AAF06142.1|pheromone binding protein [Synanthedon exitiosa] | 9e-62 | 63% | Yes | Yes |
| Comp43708_c4_seq3 | GmolPBP2 | 2409 | 141 | gb|AFL91693.1|pheromone binding protein 2 [Cydia pomonella] | 1e-96 | 97% | Yes | No |
| comp35908_c0_seq2 | GmolPBP3 | 665 | 139 | gb|AFD34183.1|pheromone binding protein 2 [Argyresthia conjugella] | 1e-100 | 100% | Yes | Yes |
| General odorant binding protein | ||||||||
| Comp43859_c0_seq4 | GmolGOBP1 | 2512 | 165 | gb|AFH02841.1|general odorant binding protein 1 [Grapholita molesta] | 7e-90 | 99% | Yes | Yes |
| comp35716_c0_seq1 | GmolGOBP2 | 1063 | 173 | gb|AFH02842.1|general odorant binding protein 2 [Grapholita molesta] | 5e-111 | 99% | Yes | Yes |
| Other odorant binding protein | ||||||||
| comp34669_c0_seq1 | GmolOBP1 | 980 | 156 | gb|AFD34177.1|odorant binding protein 1 [Argyresthia conjugella] | 4e-26 | 42% | Yes | Yes |
| comp32473_c0_seq5 | GmolOBP2 | 505 | 119 | ref|NP_001140188.1|odorant-binding protein 4 [Bombyx mori] | 2e-39 | 52% | Yes | No |
| comp38984_c0_seq2 | GmolOBP3 | 2608 | 176 | gb|AFP66959.1|general odorant binding protein 3 [Cydia pomonella] | 2e-90 | 99% | Yes | Yes |
| comp21044_c0_seq1 | GmolOBP4 | 798 | 148 | gb|AAL60415.1|AF393490_1antennal binding protein 4 [Manduca sexta] | 7e-66 | 75% | Yes | Yes |
| comp35668_c0_seq1 | GmolOBP5 | 588 | 145 | ref|NP_001140189.1|odorant-binding protein 5 precursor [Bombyx mori] | 3e-28 | 41% | Yes | Yes |
| comp35533_c1_seq1 | GmolOBP6 | 849 | 161 | gb|AGH70102.1|odorant binding protein 6 [Spodoptera exigua] | 9e-20 | 36% | Yes | Yes |
| comp35668_c0_seq2 | GmolOBP7 | 906 | 168 | gb|EHJ67765.1|odorant binding protein [Danaus plexippus] | 7e-55 | 63% | Yes | No |
| comp37376_c1_seq1 | GmolOBP8 | 832 | 141 | gb|AFD34174.1|antennal binding protein X [Argyresthia conjugella] | 3e-49 | 62% | Yes | Yes |
| Comp39973_c0_seq2 | GmolOBP9 | 631 | 129 | gb|AEX07279.1|odorant-binding protein [Helicoverpa armigera] | 9e-54 | 64% | Yes | No |
| comp40035_c5_seq2 | GmolOBP10 | 1799 | 140 | gb|AFG73000.1|odorant-binding protein 2 [Cnaphalocrocis medinalis] | 3e-65 | 72% | Yes | Yes |
| comp41846_c1_seq1 | GmolOBP11 | 788 | 136 | gb|AGK24582.1|antennal-binding protein X [Chilo suppressalis] | 5e-63 | 78% | Yes | Yes |
| Comp43336_c6_seq1 | GmolOBP12 | 862 | 143 | gb|AEB54586.1|OBP2 [Helicoverpa armigera] | 2e-60 | 64% | Yes | Yes |
| comp26714_c0_seq1 | GmolOBP13 | 1387 | 141 | gb|AFD34173.1|odorant binding protein 5 [Argyresthia conjugella] | 9e-74 | 77% | Yes | Yes |
| comp28072_c0_seq1 | GmolOBP14 | 548 | 137 | gb|AGK24577.1|odorant-binding protein 1 [Chilo suppressalis] | 3e-11 | 29% | Yes | Yes |
| comp36383_c0_seq1 | GmolOBP15 | 1331 | 163 | gb|AGI37367.1|pheromone binding protein 3 [Cnaphalocrocis medinalis] | 3e-39 | 51% | Yes | Yes |
| comp39806_c0_seq1 | GmolOBP16 | 1232 | 242 | gb|AGH70107.1|odorant binding protein 11 [Spodoptera exigua] | 8e-63 | 71% | Yes | Yes |
| comp41079_c0_seq11 | GmolOBP17 | 1094 | 96 | gb|AFG72998.1|odorant-binding protein 1 [Cnaphalocrocis medinalis] | 8e-63 | 71% | No | Yes |
| comp61252_c0_seq1 | GmolOBP18 | 250 | 76 | gb|EHJ64212.1|odorant-binding protein 2 [Danaus plexippus] | 2e-34 | 76% | No | Yes |
| comp2648_c0_seq1 | GmolOBP19 | 421 | 94 | gb|AGK24580.1|odorant-binding protein 4 [Chilo suppressalis] | 2e-52 | 80% | No | No |
| comp31482_c0_seq1 | GmolOBP20 | 422 | 124 | gb|AFD34182.1|odorant binding protein 6 [Argyresthia conjugella] | 5e-54 | 66% | No | Yes |
| comp33414_c0_seq1 | GmolOBP21 | 324 | 95 | gb|EHJ66992.1|antennal binding protein [Danaus plexippus] | 1e-27 | 61% | No | Yes |
| comp33722_c0_seq2 | GmolOBP22 | 373 | 123 | ref|NP_001153664.1|odorant binding protein [Bombyx mori] | 2e-22 | 44% | No | No |
| comp41079_c0_seq1 | GmolOBP23 | 1640 | 75 | gb|EHJ65654.1|antennal binding protein 4 [Danaus plexippus] | 9e-12 | 61% | No | No |
Identification of candidate chemosensory proteins
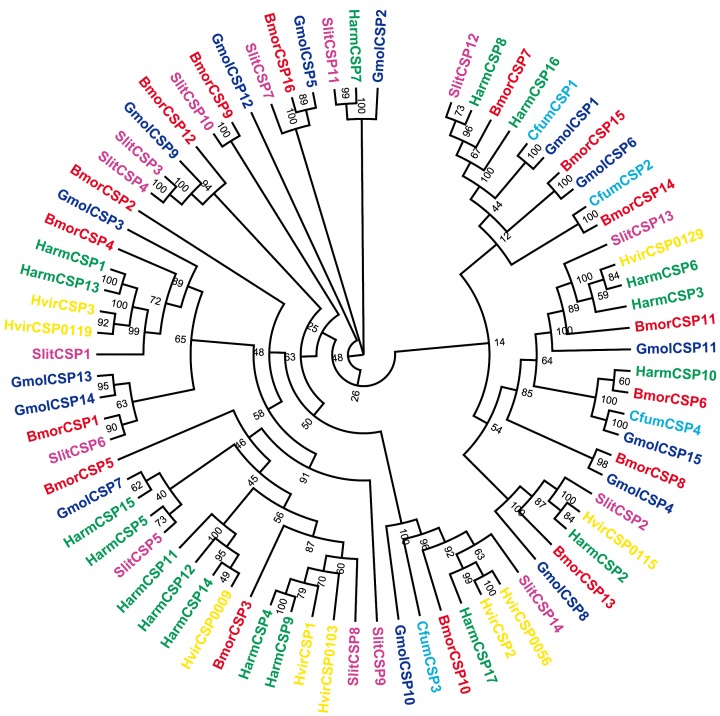
Seventeen different sequences encoding putative chemosensory proteins were identified within the G. molesta antennal transcriptome. Sequence analysis identified 15 unigenes with full-length ORFs and 16 unigenes with predicted signal peptide. One unigene without signal peptide since truncate at the 5'-end. The four conserved cysteine (with a pattern of C1-X6-8-C2-X18-19-C3-X2-C4) were found in all the 17 candidate GmolCSPs [44] (Fig 6). In addition to the conserved cysteine residues, a lysine located between the second and third cysteine was also conserved in all sequences. Neighbor-joining tree analysis showed that all of the 17 sequences were clustered with Lepidopteran orthologous genes (Fig 7). These candidate CSPs were named as “GmolCSP” followed by a numeral. The information on the CSPs is listed in Table 4. The sequences are listed in S2 File.
Fig 6. Sequences alignment of candidate GmolCSPs.
The four conserved cysteine residues were marked with “☆”, a conserved lysine was indicated with a box.
Fig 7. Neighbor-joining tree of candidate CSP genes from G. molesta and other Lepidoptera.
The tree was drawn with Adobe Photoshop CS5, based on the unrooted tree constructed using the BioNJ algorithm in Seaview v.4. The unrooted tree was constructed based on the sequence alignment produced using MAFFT version 6. Gmol, Grapholita molesta, Bmor, Bombyx mori, Cfum, Choristioneure fumiferana, Harm, Helicoverpa armigera, Hvir, Heliothis virescens, Slit, Spodoptera Iittoralis. Color of gene names indicates species.
Table 4. Unigenes of candidate chemosensory proteins.
| Unigene reference | Gene name | Length (nt) | ORF (aa) | BLASTx best hit (Reference/Name/Species) | E-value | Identify | Full length | Signal Peptide |
|---|---|---|---|---|---|---|---|---|
| comp29970_c0_seq1 | GmolCSP1 | 384 | 120 | gb|AAR84077.1|chemosensory protein 1, partial [Choristoneura fumiferana] | 2e-44 | 69% | Yes | Yes |
| comp30965_c0_seq1 | GmolCSP2 | 689 | 116 | gb|AGI37361.1|chemosensory protein 1 [Cnaphalocrocis medinalis] | 1e-06 | 36% | Yes | Yes |
| comp38507_c0_seq3 | GmolCSP3 | 1506 | 130 | gb|AAK53762.1|AF368375_1chemosensory protein [Helicoverpa armigera] | 1e-47 | 70% | Yes | Yes |
| comp31529_c0_seq2 | GmolCSP4 | 520 | 125 | gb|EHJ70186.1|chemosensory protein [Danaus plexippus] | 6e-46 | 70% | Yes | Yes |
| comp33568_c0_seq1 | GmolCSP5 | 427 | 106 | ref|NP_001091782.1|chemosensory protein 16 precursor [Bombyx mori] | 2e-52 | 88% | Yes | Yes |
| comp39117_c0_seq1-1 | GmolCSP6 | 970 | 121 | gb|AAW23971.1|chemosensory protein 4 [Choristoneura fumiferana] | 2e-70 | 88% | Yes | Yes |
| comp32406_c0_seq1 | GmolCSP7 | 527 | 128 | gb|ABM67687.1|chemosensory protein CSP2 [Plutella xylostella] | 4e-54 | 73% | Yes | Yes |
| comp35615_c0_seq1 | GmolCSP8 | 956 | 120 | gb|AEX07265.1|CSP2 [Helicoverpa armigera] | 3e-43 | 60% | Yes | Yes |
| comp37677_c0_seq3 | GmolCSP9 | 1441 | 156 | dbj|BAM18557.1|protein serine/threonine kinase [Papilio xuthus] | 7e-48 | 71% | Yes | Yes |
| comp41217_c1_seq3 | GmolCSP10 | 1262 | 127 | gb|AFQ32775.1|chemosensory protein [Grapholita molesta] | 2e-48 | 60% | Yes | Yes |
| comp41050_c0_seq1 | GmolCSP11 | 681 | 124 | emb|CAJ01505.1|hypothetical protein [Manduca sexta] | 1e-55 | 80% | Yes | Yes |
| comp40028_c0_seq2 | GmolCSP12 | 1396 | 154 | gb|EHJ76400.1|hypothetical protein KGM_11196 [Danaus plexippus] | 6e-44 | 60% | Yes | Yes |
| comp38507_c0_seq4 | GmolCSP13 | 828 | 124 | ref|NP_001037065.1|chemosensory protein 1 [Bombyx mori] | 7e-42 | 63% | Yes | Yes |
| comp38507_c0_seq2 | GmolCSP14 | 830 | 124 | dbj|BAF91711.1|chemosensory protein [Papilio xuthus] | 1e-40 | 61% | Yes | Yes |
| comp39117_c0_seq1-2 | GmolCSP15 | 970 | 125 | gb|AAW23971.1|chemosensory protein 4 [Choristoneura fumiferana] | 2e-70 | 88% | Yes | Yes |
| comp20508_c0_seq1 | GmolCSP16 | 308 | 95 | ref|NP_001037062.1|chemosensory protein 5 precursor [Bombyx mori] | 2e-28 | 53% | No | Yes |
| comp26710_c0_seq1 | GmolCSP17 | 358 | 76 | gb|AAR84078.1|chemosensory protein 2 [Choristoneura fumiferana] | 1e-66 | 91% | No | No |
Identification of candidate odorant receptors
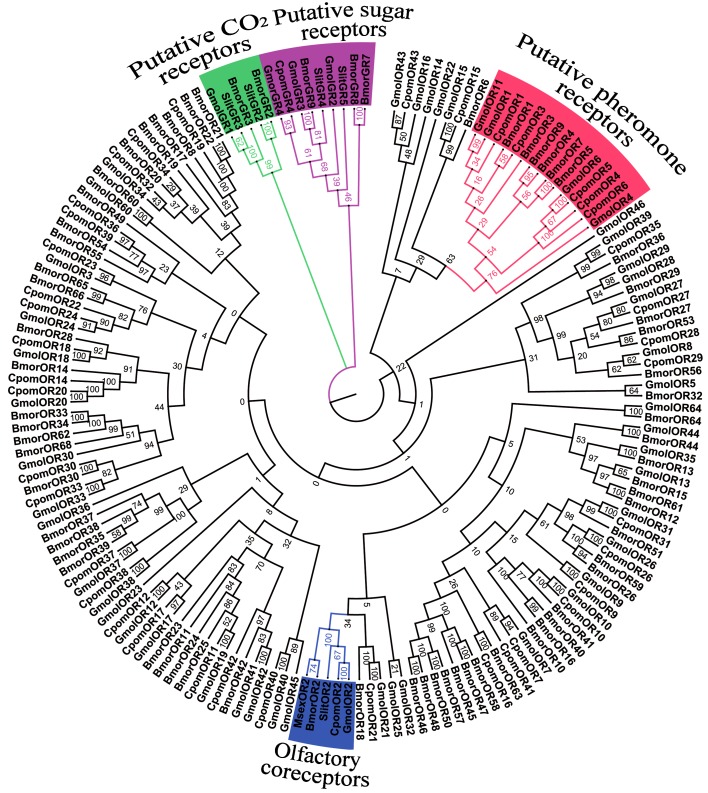
Bioinformatic analysis identified 48 different sequences encoding putative ORs and four sequences encoding putative GRs. GmolORs and GmolGRs were named according to their similarities with previously annotated Lepidoptera ORs and the topology predictions from TMpred as observed from other insect ORs [45]. Twelve of these sequences appeared to contain full lengths genes since they had full length ORFs with 5–8 transmembrane domains (Table 5). The co-receptor of G.molesta showed 95% identity to C. pomonella co-receptor, CpomOR2, one of the most conserved co-receptors in insect species. While similar with other insect ORs, most GmolORs were highly divergent and shared low similarity with other insect ORs, except for closely related species such as Cydia pomonella. Four candidate GmolORs sequences (GmolOR1, GmolOR4, GmolOR6, and GmolOR11) tended to be pheromone receptors (PRs) as they are highly conserved with CpomOR fragments and BmorPRs amino acid sequences from other species [46]. These four sequences were clusterd into one subgroup in the phylogenetic tree (Fig 8). The gustatory receptors that we identified (GmolGR2, GmolGR3, and GmolGR4) were found in a clade with sugar receptors, which included gustatory receptors identified from other moth antennae; these gustatory receptors were also clustered in this clade (Fig 8) [39,47,48]. Another putative GR, named GmolGR1, was clustered with putative CO2 receptors. The information of ORs and GRs are given in Table 5, the nucleotide sequences are listed in S2 File.
Table 5. Unigenes of candidate ordoart receptors and gustatory receptors.
| Unigene reference | Gene name | Length (nt) | ORF (aa) | BLASTx best hit (Reference/Name/Species) | E-value | Identify | Full length | TMD (No) |
|---|---|---|---|---|---|---|---|---|
| Co-receptor | ||||||||
| comp42201_c0_seq1 | GmolOR2 | 2730 | 473 | gb|AFC91712.1| putative odorant receptor OR2 [Cydia pomonella] | 0.0 | 95% | Yes | 7 |
| Pheromone receptors | ||||||||
| comp44030_c0_seq5 | GmolOR1 | 884 | 88 | gb|AFC91711.1| putative odorant receptor OR1, partial [Cydia pomonella] | 3e-29 | 52% | No | 1 |
| comp37031_c0_seq3 | GmolOR4 | 1624 | 430 | gb|AGG91650.1| odorant receptor [Ostrinia furnacalis] | 6e-103 | 40% | No | 5 |
| comp52214_c0_seq1 | GmolOR6 | 377 | 118 | gb|AFC91716.1| putative odorant receptor OR6, partial [Cydia pomonella] | 2e-53 | 70% | No | 0 |
| comp36128_c0_seq1 | GmolOR11 | 536 | 135 | gb|AFK30397.1| odorant receptor 4 [Ostrinia furnacalis] | 4e-36 | 53% | No | 3 |
| Other odorant receptors | ||||||||
| comp39216_c0_seq2 | GmolOR3 | 1400 | 420 | gb|EHJ75140.1| olfactory receptor [Danaus plexippus] | 5e-65 | 70% | Yes | 6 |
| comp40794_c1_seq1 | GmolOR5 | 1980 | 414 | dbj|BAH66328.1| olfactory receptor [Bombyx mori] | 3e-119 | 51% | No | 6 |
| comp42525_c0_seq1 | GmolOR7 | 1213 | 395 | gb|AFC91717.1| putative odorant receptor OR7, partial [Cydia pomonella] | 3e-124 | 86% | Yes | 8 |
| comp37543_c0_seq4 | GmolOR8 | 615 | 199 | ref|NP_001166617.1| olfactory receptor 56 [Bombyx mori] | 2e-82 | 59% | No | 2 |
| comp36812_c0_seq5 | GmolOR9 | 1707 | 395 | gb|AFC91718.1| putative odorant receptor OR9, partial [Cydia pomonella] | 3e-99 | 94% | Yes | 5 |
| comp43031_c0_seq3 | GmolOR10 | 2363 | 397 | gb|AFC91719.1| putative odorant receptor OR10 [Cydia pomonella] | 0.0 | 84% | Yes | 7 |
| comp40338_c0_seq2 | GmolOR12 | 1224 | 399 | gb|AFC91721.1| putative odorant receptor OR12 [Cydia pomonella] | 0.0 | 90% | Yes | 7 |
| comp42372_c0_seq4 | GmolOR13 | 958 | 210 | tpg|DAA05974.1| TPA_exp: odorant receptor 15 [Bombyx mori] | 6e-74 | 53% | No | 4 |
| comp36822_c0_seq2 | GmolOR14 | 439 | 136 | gb|EHJ67735.1| olfactory receptor [Danaus plexippus] | 1e-04 | 28% | No | 2 |
| comp41698_c0_seq2 | GmolOR15 | 946 | 227 | gb|AFC91723.1| putative odorant receptor OR15 [Cydia pomonella] | 5e-159 | 89% | No | 1 |
| comp42516_c0_seq34 | GmolOR16 | 1458 | 177 | gb|AFC91724.1| putative odorant receptor OR16 [Cydia pomonella] | 6e-89 | 77% | No | 0 |
| comp30377_c0_seq2 | GmolOR17 | 953 | 254 | gb|AFC91725.1| putative odorant receptor OR17 [Cydia pomonella] | 7e-174 | 84% | No | 4 |
| comp40766_c0_seq4 | GmolOR18 | 998 | 238 | gb|AFC91726.1| putative odorant receptor OR18 [Cydia pomonella] | 0.0 | 90% | No | 3 |
| comp42164_c0_seq3 | GmolOR19 | 1274 | 398 | emb|CAG38113.1| putative chemosensory receptor 12 [Heliothis virescens] | 3e-12 | 43% | No | 4 |
| comp43037_c0_seq1 | GmolOR20 | 1501 | 427 | gb|AFC91728.1| putative odorant receptor OR20 [Cydia pomonella] | 0.0 | 87% | Yes | 7 |
| comp41733_c0_seq1 | GmolOR21 | 1334 | 376 | gb|AFC91729.1| putative odorant receptor OR21 [Cydia pomonella] | 0.0 | 90% | Yes | 5 |
| comp36128_c0_seq3 | GmolOR22 | 842 | 279 | gi|205361596|dbj|BAG71417.1| olfactory receptor-1 [Diaphania indica] | 3e-40 | 32% | No | 2 |
| comp20621_c0_seq1 | GmolOR23 | 419 | 139 | emb|CAD31850.1| putative chemosensory receptor 1 [Heliothis virescens] | 6e-19 | 45% | No | 0 |
| comp42767_c0_seq45 | GmolOR24 | 1716 | 259 | gb|AFC91732.1| putative odorant receptor OR24 [Cydia pomonella] | 8e-163 | 74% | No | 3 |
| comp33674_c0_seq3 | GmolOR25 | 1054 | 279 | gb|EHJ78030.1| olfactory receptor 29 [Danaus plexippus] | 4e-155 | 75% | No | 0 |
| comp41262_c0_seq4 | GmolOR26 | 1462 | 315 | gb|AFC91734.1| putative odorant receptor OR26 [Cydia pomonella] | 6e-128 | 87% | No | 5 |
| comp39775_c0_seq1 | GmolOR27 | 1281 | 402 | ref|NP_001166893.1| olfactory receptor 27 [Bombyx mori] | 1e-152 | 64% | No | 5 |
| comp39835_c0_seq3 | GmolOR28 | 1686 | 398 | ref|NP_001166894.1| olfactory receptor 29 [Bombyx mori] | 2e-142 | 49% | No | 5 |
| comp41754_c3_seq2 | GmolOR29 | 738 | 238 | tpg|DAA05985.1| TPA_exp: odorant receptor 29 [Bombyx mori] | 1e-95 | 59% | No | 3 |
| comp36822_c0_seq1 | GmolOR30 | 1081 | 181 | gb|AFC91738.1| putative odorant receptor OR30 [Cydia pomonella] | 1e-28 | 40% | No | 0 |
| comp44215_c1_seq7 | GmolOR31 | 1380 | 402 | gb|AFC91739.1| putative odorant receptor OR31 [Cydia pomonella] | 0.0 | 79% | No | 4 |
| comp43530_c1_seq3 | GmolOR32 | 1420 | 444 | ref|NP_001116817.1| olfactory receptor-like [Bombyx mori] | 3e-174 | 58% | Yes | 6 |
| comp41607_c0_seq3 | GmolOR33 | 2364 | 374 | gb|AFC91741.1| putative odorant receptor OR33 [Cydia pomonella] | 4e-141 | 76% | Yes | 6 |
| comp38817_c0_seq3 | GmolOR34 | 1415 | 424 | gb|AFC91742.1| putative odorant receptor OR34 [Cydia pomonella] | 3e-102 | 43% | No | 3 |
| comp41935_c2_seq6 | GmolOR35 | 1198 | 334 | ref|XP_004067596.1| PREDICTED: protein LOC101171734 [Oryzias latipes] | 3e-113 | 55% | Yes | 6 |
| comp40388_c2_seq1 | GmolOR36 | 1277 | 355 | gb|AET06162.1| odorant receptor 3, partial [Planotortrix notophaea] | 0.0 | 82% | No | 6 |
| comp44364_c1_seq2 | GmolOR37 | 1938 | 398 | gb|AFC91745.1| putative odorant receptor OR37 [Cydia pomonella] | 3e-173 | 82% | No | 4 |
| comp44203_c1_seq3 | GmolOR38 | 2158 | 437 | gb|AFC91746.1| putative odorant receptor OR38 [Cydia pomonella] | 0.0 | 65% | No | 5 |
| comp40263_c1_seq4 | GmolOR39 | 1403 | 414 | ref|NP_001166892.1| olfactory receptor 36 [Bombyx mori] | 2e-127 | 48% | No | 4 |
| comp43824_c0_seq2 | GmolOR40 | 1841 | 394 | gb|AFC91748.1| putative odorant receptor OR40 [Cydia pomonella] | 3e-123 | 71% | No | 4 |
| comp38057_c0_seq1 | GmolOR41 | 462 | 108 | ref|NP_001091818.1| olfactory receptor 42 [Bombyx mori] | 7e-39 | 60% | No | 1 |
| comp38057_c0_seq2 | GmolOR42 | 773 | 252 | gb|AFC91750.1| putative odorant receptor OR42 [Cydia pomonella] | 7e-123 | 83% | No | 4 |
| comp33512_c0_seq2 | GmolOR43 | 741 | 90 | gb|AFC91751.1| putative odorant receptor OR43 [Cydia pomonella] | 1e-09 | 57% | No | 0 |
| comp34069_c0_seq3 | GmolOR44 | 1532 | 436 | gb|AGG08877.1| putative olfactory receptor 44 [Spodoptera litura] | 0.0 | 72% | No | 6 |
| comp41837_c0_seq1 | GmolOR45 | 1455 | 401 | emb|CBW30700.1| odorant receptor [Drosophila simulans] | 3e-23 | 26% | No | 5 |
| comp36742_c0_seq1 | GmolOR46 | 1739 | 495 | dbj|BAM19586.1| similar to CG13607, partial [Papilio xuthus] | 0.0 | 80% | No | 5 |
| comp41612_c0_seq4 | GmolOR60 | 1647 | 450 | ref|NP_001155301.1| olfactory receptor 60 [Bombyx mori] | 0.0 | 70% | Yes | 6 |
| comp39091_c0_seq2 | GmolOR64 | 1298 | 416 | ref|NP_001166621.1| olfactory receptor 64 [Bombyx mori] | 9e-97 | 53% | No | 6 |
| Gustatory receptors | ||||||||
| comp29992_c0_seq1 | GmolGR1 | 1047 | 331 | gb|EHJ78216.1|gustatory receptor 24 [Danaus plexippus] | 3e-138 | 77% | No | 5 |
| comp32055_c0_seq3 | GmolGR2 | 407 | 96 | tpg|DAA06387.1|TPA_inf: gustatory receptor 50 [Bombyx mori] | 1e-33 | 66% | No | 2 |
| comp39217_c0_seq1 | GmolGR3 | 1186 | 258 | gb|AGA04648.1|gustatory receptor [Helicoverpa armigera] | 3e-152 | 76% | No | 3 |
| comp25108_c0_seq3 | GmolGR4 | 706 | 136 | gb|AFC91733.1| putative odorant receptor OR25 [Cydia pomonella] | 2e-36 | 46% | No | 2 |
Fig 8. Neighbor-joining tree of candidate odorant receptor (OR) and gustatory receptor (GR) from G. molesta and other Lepidoptera.
The tree was drawn using Adobe Photoshop CS5, based on an unrooted tree constructed using the BioNJ algorithm in Seaview v.4. The unrooted tree was constructed based on the sequence alignment produced using MAFFT version 6. Gmol, Grapholita molesta, Msex, Manduca sexta, Cpom, Cydia pomonella, Slit, Spodoptera Iittoralis, Bmor, Bombyx mori.
Identification of candidate ionotropic receptors
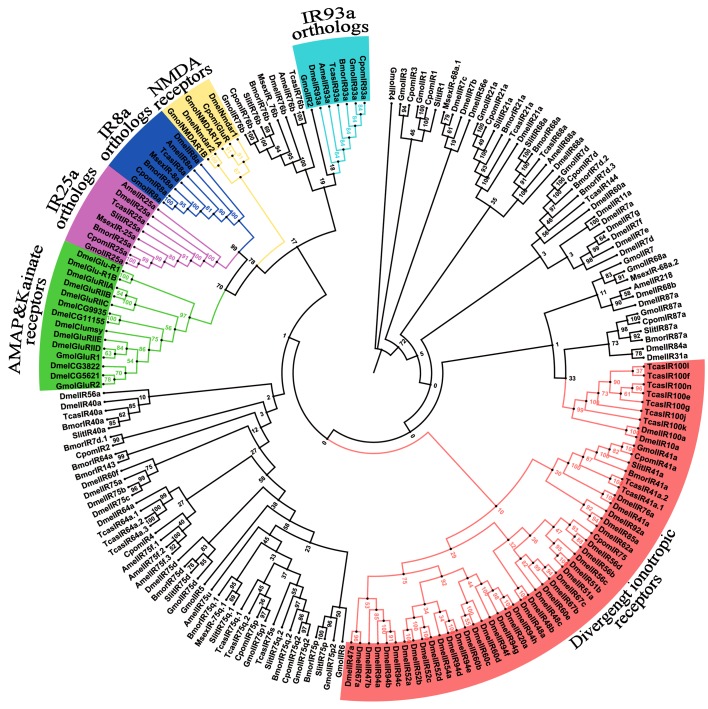
Twenty four sequences encoding putative IRs proteins were also identified in the G. molesta antennal transcriptome. The alignment revealed that all the 24 sequences represent unique genes since they possessed overlapping regions without identity. Ten of the IRs appeared to contain a full length ORFs (GmolIR8a, 25a, 21a, 75p2, 76b, 87a, 93a, 7d, 5 and NMDAR1B), and were longer than 1700 bp in general. In addition, it was predicted that three transmembrane domains existed in all 10 sequences by TMHMM 2.0, the typical characteristic of IRs. The remaining 14 sequences were truncate at either 5' or 3' terminus. Neighbor-joining tree analysis revealed that all putative IRs were found to have orthologoues from B. mori, M. sexta, C. pomonella, S. littoralis, D. melanogaster, A. mellifera, and T. castaneum. According to their positions in the phylogenetic tree and the strong bootstrap support, 15 of 24 putative G. molesta IRs were given names that are consistent with the number and suffix of the Dmel/Bmor/Slit/Cpom/Amel/Tcas IR orthologues in the same clade. Two of the remaining nine IR sequences, comp37980_c0_seq2 and comp40732_c0_seq6, clustered with their ionotropic receptor orthologues into N-methyl-D-aspartic acid receptor (NMDA receptors) clade, and these were named “GmolNMDAR1A” and “GmolNMDAR1B”. The other two sequences, comp5756_c0_seq1 and comp5467_c0_seq1, clustered with their ionotropic receptor orthologues of α-Amino-3-hydroxy-5-methylisoxazole-4-propionic acid (AMAP) and Kainate receptors clade, and these were named “GmolGluIR1” and “GmolGluIR2”, respectively. The remaining five unigenes, comp52557_c0_seq1, comp36336_c0_seq3, comp41705_c0_seq15, comp43295_c0_seq6, and comp43552_c0_seq13, did not show meanful similarity with known IR encoding genes but with conserved structural features, and thus were named as “GmolIR2”, “GmolIR4”, “GmolIR5”, “GmolIR6” and “GmolIR7”, respectively (Fig 9). The information including the unigene reference, length, and first BLASTX hit of all the 24 IRs are given in Table 6. The sequences of all the 24 IRs are listed in S2 File.
Fig 9. Neighbor-joining tree of candidate ionotropic receptor (IR) genes from G. molesta and other insects.
The tree was drawn using Adobe Photoshop CS5, based on an unrooted tree constructed using the BioNJ algorithm in Seaview v.4. The unrooted tree was constructed based on a sequence alignment produced using MAFFT version 6. Gmol, Grapholita molesta, Msex, Manduca sexta, Cpom, Cydia pomonella, Slit, Spodoptera Iittoralis, Bmor, Bombyx mori, Dmel, Drosophila melanogaster, Amel, Apis mellifera, Tcas, Tribolium castaneum.
Table 6. Unigenes of candidate ionotropic receptors.
| Unigene reference | Gene name | Length (nt) | ORF (aa) | BLASTx best hit (Reference/Name/Species) | E-value | Identify | Full length | TMD (No) |
|---|---|---|---|---|---|---|---|---|
| comp44508_c0_seq1 | GmolIR8a | 2971 | 892 | gb|AFC91764.1|putative ionotropic receptor IR8a, partial [Cydia pomonella] | 2e-22 | 54% | Yes | 3 |
| comp43733_c0_seq1 | GmolIR25a | 3490 | 923 | gb|AFC91757.1|putative ionotropic receptor IR25a [Cydia pomonella] | 0.0 | 96% | Yes | 3 |
| comp42937_c0_seq1 | GmolIR21a | 2722 | 853 | gb|AFC91761.1|putative ionotropic receptor IR21a, partial [Cydia pomonella] | 0.0 | 88% | Yes | 3 |
| comp41847_c1_seq3 | GmolIR41a | 1846 | 536 | gb|AFC91758.1|putative ionotropic receptor IR41a [Cydia pomonella] | 0.0 | 91% | No | 2 |
| comp34810_c0_seq9 | GmolIR68a | 1425 | 411 | gb|ADR64682.1|putative chemosensory ionotropic receptor IR68a [Spodoptera littoralis] | 4e-173 | 66% | No | 0 |
| comp43328_c0_seq3 | GmolIR75d | 1225 | 380 | gb|ADR64683.1|putative chemosensory ionotropic receptor IR75d [Spodoptera littoralis] | 6e-105 | 50% | No | 1 |
| comp43295_c0_seq14 | GmolIR75p1 | 523 | 174 | gb|AFC91755.1|putative ionotropic receptor IR75p, partial [Cydia pomonella] | 5e-119 | 95% | No | 0 |
| comp43826_c0_seq19 | GmolIR75p2 | 1958 | 605 | gb|ADR64684.1|putative chemosensory ionotropic receptor IR75p [Spodoptera littoralis] | 2e-143 | 43% | Yes | 3 |
| comp43424_c0_seq31 | GmolIR75q2 | 2528 | 313 | gb|AFC91752.1|putative ionotropic receptor IR75q2 [Cydia pomonella] | 0.0 | 89% | No | 1 |
| comp41154_c0_seq4 | GmolIR76b | 3326 | 440 | gb|AFC91765.1|putative ionotropic receptor IR76b [Cydia pomonella] | 0.o | 84% | Yes | 3 |
| comp44120_c0_seq14 | GmolIR87a | 1734 | 513 | gb|AFC91760.1|putative ionotropic glutamate receptor 87a, partial [Cydia pomonella] | 0.0 | 88% | Yes | 3 |
| comp44415_c0_seq2 | GmolIR93a | 2713 | 870 | gb|AFC91753.1|putative ionotropic receptor IR93a, partial [Cydia pomonella] | 0.0 | 92% | Yes | 4 |
| comp37980_c0_seq2 | GmolNMDAR1A | 1626 | 361 | gb|EHJ78211.1|putative NMDA-type glutamate receptor 1 [Danaus plexippus] | 0.0 | 96% | No | 2 |
| comp40732_c0_seq6 | GmolNMDAR1B | 1780 | 309 | ref|NP_001040129.2|glutamate [NMDA] receptor-associated protein 1 [Bombyx mori] | 5e-108 | 78% | Yes | 3 |
| comp5756_c0_seq1 | GmolGluR1 | 339 | 113 | gb|EGI61384.1|Glutamate receptor, ionotropic kainate 1 [Acromyrmex echinatior] | 1e-64 | 91% | No | 2 |
| comp5467_c0_seq1 | GmolGluIR2 | 358 | 118 | ref|XP_001655460.1|ionotropic glutamate receptor subunit ia [Aedes aegypti] | 2e-60 | 81% | No | 1 |
| comp42373_c0_seq9 | GmolIR1 | 1444 | 458 | gb|AFC91754.1|putative ionotropic receptor IR1, partial [Cydia pomonella] | 2e-154 | 73% | No | 2 |
| comp43818_c0_seq8 | GmolIR3 | 1042 | 138 | gb|AFC91767.1|putative ionotropic receptor IR3, partial [Cydia pomonella] | 2e-41 | 69% | No | 0 |
| comp27491_c0_seq3 | GmolIR7d | 1854 | 549 | gb|AFC91766.1|putative ionotropic receptor IR7d, partial [Cydia pomonella] | 1e-113 | 87% | Yes | 4 |
| comp52557_c0_seq1 | GmolIR2 | 401 | 133 | gb|EHJ63562.1|metabotropic glutamate receptor B [Danaus plexippus] | 7e-87 | 97% | No | 2 |
| comp36336_c0_seq3 | GmolIR4 | 560 | 186 | gb|EHJ74994.1|putative ionotropic glutamate receptor-invertebrate [Danaus plexippus] | 3e-10 | 55% | No | 0 |
| comp41705_c0_seq15 | GmolIR5 | 2529 | 579 | ref|XP_001651553.1|ionotropic glutamate receptor-invertebrate [Aedes aegypti] | 1e-75 | 32% | Yes | 3 |
| comp43295_c0_seq6 | GmolIR6 | 996 | 331 | gb|EHJ72019.1|putative ionotropic glutamate receptor-invertebrate [Danaus plexippus] | 2e-104 | 74% | No | 1 |
| comp43552_c0_seq13 | GmolIR7 | 985 | 239 | gb|ADM88008.1|ionotropic GABA-aminobutyric acid receptor RDL1-3b6a [Bombyx mori] | 2e-142 | 96% | No | 0 |
Identification of candidate sensory neuron membrane proteins
Both SNMP1 and SNMP2 were obtained from G. molesta antennal transcriptome. In comparison, GmolSNMP1 has 68% identity with SNMP1 of Plutella xylostella (GenBank accession number: E2IHA6.1), while GmolSNMP2 has 69% identity with SNMP2 of Ostrinia nubilalis (GenBank accession number: E5EZW9.1). GmolSNMP1 had a full ORF with two transmembrane domains at N-terminus and C-terminus, respectively, while GmolSNMP2 was incomplete due to truncation at the 3' terminus. In addition, we also identified an odor degrading enzyme gene (ODE). ODE is responsible in the signal inactivation step and it rapidly degrades the stimulatory odorant molecules. The information including the unigene reference, length, and best BLASTX hit of SNMPs and ODE was listed in Table 7. The sequences of two SNMPs and an ODE were listed in S2 File.
Table 7. Unigenes of candidate sensory neuron membrane proteins and odor degrading enzyme.
| Unigene reference | Gene name | Length (nt) | ORF (aa) | BLASTx best hit (Reference/Name/Species) | E-value | Identify | Full length | TMD (No) |
|---|---|---|---|---|---|---|---|---|
| Sensory neuron membrane proteins | ||||||||
| comp44414_c0_seq1 | GmolSNMP1 | 1816 | 489 | gnl|BL_ORD_ID|1375793 sensory neuron membrane protein-1 [Plutella xylostella] | 0.0 | 68% | Yes | 2 |
| comp36266_c0_seq1 | GmolSNMP2 | 1868 | 518 | gnl|BL_ORD_ID|1536434 sensory neuron membrane protein 2 [Ostrinia nubilalis] | 0.0 | 69% | No | 1 |
| Odor degrading enzyme | ||||||||
| comp41848_c0_seq1 | GmolODE | 1923 | 540 | gb|AAM14415.1|putative odorant-degrading enzyme [Antheraea polyphemus] | 0.0 | 65% | yes | |
Tissue and sex-specific expression of candidate OBP and CSP genes
The sex and tissue-specific expression of GmolPBP2 and GmolPBP3 had been studied previously [49]. In this work, the expression patterns of the candidate genes encoding 26 OBPs and 17 CSPs in male antennae, female antennae, and the remaining bodies were analyzed by semi-quantitative reverse transcription PCR (Fig 10). The results indicated that these OBP-encoding genes were expressed exclusively in antennae except for GmolOBP1, GmolOBP2, GmolOBP6, and GmolOBP18. Interestingly, GmolOBP2 was expressed in both female antennae and the remaining body; whereas GmolOBP10 and GmolOBP11 showed an antenna-specific expression in females. In addition to the expression in both male and female antennae, GmolOBP1 was also expressed in the female body. The remaining OBPs were expressed with similar levels in the antennae of both sexes. Compared with OBPs, almost all candidate CSPs were expressed in the antennae and body of both sexes, appearing no significant differences between males and females.
Fig 10. Tissue and sex specific expressions of G. molesta OBPs and CSPs.
Fa: female antennae, Ma: male antennae, Fb: female boby (without antennae), Mb: male boby (without antennae), NTC: No template control.
Discussion
We used antennal transcriptomic sequencing to identify patutive olfactory genes and identified genes encoding 28 OBPs, 17 CSPs, 48 ORs, four GRs, 24 IRs, two SNMPs and one ODE in female antennae of G. molesta. The olfactory system genes identified in this work were comparable to recent reports from H. armigera (47 ORs, 12 IRs, 26 OBPs, and 12 CSPs), C. suppressalis (47 ORs, 20 IRs, 26 OBPs, 21 CSPs, and 2 SNMPs), M. sexta (47 ORs, six IRs, 18 OBPs, and 21 CSPs), and C. pomonella. (43 ORs, 15 IRs, and 1 GR) [3,14,16,17]. Our transcriptome data set appears quite comprehensive since all of the previously annotated G. molesta olfactory genes available in NCBI were identified in the present antennal transcriptome.
The alignment of the predicted GmolOBPs showed low sequence identity among OBP family members (Fig 4). The predicted proteins have molecular masses ranged between14 to18 kDa. All putative proteins have a signal peptide sequence in the hydrophobic N-terminus. According to the standard established by Hekmat-Scafe [43], insect OBPs can be classified into classical OBPs and atypical OBPs. The six-cysteine signature is the most typical feature of classical insect OBPs [50,51], including GOBP/PBP, CRLBP, ABPI and ABPII. Atypical OBPs families include Minus-C (missing C2 and C5) and Plus-C (carry additional conserved cysteine located between C1 and C2 and after C6). In our work, the classical GmolOBPs were named PBP, GOBP and other OBPs, since the spacing pattern of conserved cysteine in these typical OBPs is C1-X25–68-C2-X3-C3-X31–46-C4-X8–29-C5-X8-C6 (where X is any amino acid). There is only one Minus-C OBPs (named GmolOBP14) that is missing the second and the fifth cysteine. All of the sequences were clustered into GOBP1, GOBP2, PBPs and other OBPs clades in the phylogenetic tree. PBPs are a subfamily of OBPs and constituted of three members in Lepidopteran, PBP1, PBP2, and PBP3. GmolPBP2 (accession number: KF365878) and GmolPBP3 (accession number: KF365879) had already been logged in NCBI database. Another PBP gene was identified by GO annotation and alignment analysis in our antennal transcriptome. We named the unigene Comp42972_c0_seq8 as GmolPBP1, since the coding region was structurally similar to GmolPBP2 and GmolPBP3, although slightly longer than the latter two PBPs. Recent studies have shown that the PBPs subfamily of proteins mainly bind to sex pheromones. Fluorescence binding assays showed that GmolPBP2 had strong binding affinities with (Z)-8-dodecenyl acetate (Z8-12:Ac) and (E)-8-dodecenyl acetate (E8-12:Ac), and the binding constants were 1.09 and 1.10 μmol/L, respectively. The affinity of GmolPBP3 to both main sex pheromones was very weak, the binding constants was only 19.32 μmol/L for Z8-12:Ac and 22.70 μmol/L for E8-12:Ac [49]. Silkworm BmorPBP1 is capable of enhancing sensitivity and selectively mediating the response to bombykol [52]. Bollworm HarmPBP1 binds strongly with two principal pheromone components (Z)-11-tetradecenal and (Z)-9-hexadecenal [53], but the HarmPBP2 and HarmPBP3 showed only weak affinities with the tested ligand. It seems that HarmPBP1 plays a key role in sex pheromone recognition. In A. pernyi and A. polyphemus, the binding constants value of PBP1 for principal pheromone component E6,Z11-hexadecadienyl acetate was 1.83 and 0.63 μmol/L, respectively [54]. These results illustrated that the insect PBP1 was the most important pheromone binding proteins. Thus, the affinity of GmolPBP1 with sex pheromone was worth studying.
GOBPs are a subfamily of OBPs, consisting of two members, GOBP1 and GOBP2 in most Lepidopterans. But in tortricid moths and the codling moth (Cydia pomonella), which are closely related to G.molesta, three different transcripts were found to encode putative GOBPs. The GOBPs subfamily can be divided into GOBP1, GOBP2 and GOBP3 [34]. The sequence, which was identified in our antennal transcriptome sequencing and homology-based cloning in female antenna, was named GmolOBP3 (accession number:KF395363), sharing 99% identity with CpomGOBP3 and clustered into GOBPs clade in neighbor-joining tree (Fig 5). Phylogenetic analysis of GmolOBP3 protein showed orthology with GOBPs subfamily genes and probably had similar functions to other GOBP members. GOBPs show spatial specificity in expression, and are localized mainly in adult female and male antennae in Lepidopteran [52]. CpomGOBP3 was detected in antennae, late stage pupal heads, mouthparts and female abdomen tips. It has been speculated that CpomGOBP3 might have a role in oviposition and pheromone production or release, in addition to chemosensation. The GmolOBP3 was only highly expressed in male and female antennae. It has also been hypothesized that GmolOBP3 had a potential role in binding pheromones and plant general odor molecules, and these potential specialized functions in G.molesta will need to be addressed in future studies.
The tissue expression patterns of the 26 putative OBPs in G. molesta may help to characterize the function of these OBPs in future research. In this study, semi-quantitative reverse transcription PCR was used to evaluate tissue and sex specific expression levels and abundance of the identified OBPs. Except for GmolPBP2 and GmolPBP3, 22 of the 26 identified OBPs displayed highly antenna-biased expression. The other four genes, HarmOBP1, HarmOBP2, HarmOBP6 and HarmOBP18, were not only highly expressed in antennae, but also expressed equally highly in the remaining bodies. In A. mellifera, only 9 of 21 OBPs are antenna-specific, and the remaining genes are either expressed ubiquitously or are strictly regulated in specialized tissues or during development. Many reports suggest that OBPs are expressed in taste tissues [55,56], and these genes may play an important role in tasting function and gustatory reorganization.
CSPs were highly and almost ubiquitously distributed in olfactory tissues as well as in non-olfactory tissues, suggesting that CSPs in insects may also participate in other functions in addition to chemosensation [57], such as limb regeneration in Periplaneta Americana [58], female survival and reproduction in Spodoptera exigua [59], embryo development in Apis mellifera [60], migratory behavioral in Locusta migratoria [61]. Almost all deduced protein sequences have the characteristic features of CSPs: the presence of a signal peptide and the highly conserved four cysteine profiles (Table 4, Fig 6). Twenty-two putative CSPs have been annotated in B. mori [16], 21 in M. sexta [14], 12 in H. armigera [3], and 21 in C. suppressalis [17], while we identified 17 candidate CSPs is quite reasonable. Interestingly, tissue- and sex-specific expression demonstrated that GmolCSP7, GmolCSP9 and GmolCSP17 are likely antenna-specific; and these genes perhaps have special roles in detection and transduction of host plant odors molecules.
In G. molesta, a previously neuroanatomical and computational study found that 48–49 ordinary glomeruli and one large glomerulus situated at the entrance of the antennal nerve in males, and 49–52 ordinary glomeruli and one large glomerulus in the ventro-medial part of the antennal lobe in females [62]. Considering the one receptor-one glomerulus paradigm [63], by which the number of expected ORs in a given species should correlate with the number of glomeruli in the antennal lobe (meaning that one olfactory receptor type is expressed in OSN type), our OR dataset of 48 sequences indicates that there is at least 48 OSN types. These numbers are comparable to the reported numbers in M. sexta [14], C. pomonella [17] and H. armigera [3]. Phylogenetic analysis of the G. molesta ORs, four of them grouped into a conserved clade containing lepidopteran pheromone receptors (PRs) (Fig 8), and we thus speculate that some or all of them are involved in pheromone recognition. The female G.molesta moths emit four pheromone compounds, (Z)-8-dodecenyl acetate, (E)-8-dodecenyl acetate, (Z)-8-dodecenyl alcohol and dodecanol [64]. However, the OSNs which are involved in detection of these compounds have not been found in this study. Functional analyses of candidate ORs are usually performed by using heterologous expression in Xenopus oocytes and electrophysiology. A distinct receptor of silkworm moth Bombyx mori, BmOR3, is expressed in the second ORN, and binds pheromone compound bombykal can inhibit male behavioural response [65]. Candidate pheromone receptors of tobacco budworm Heliothis virescens (HvORs), HvOR6 was found to be highly tuned to pheromone (Z)-9-tetradecenal, while HvOR13, HvOR14 and HvOR16 showed specificity for (Z)-11-hexadecenal, (Z)-11-hexadecenyl acetate and (Z)-11-hexadecen-1-ol, respectively [66]. A honey bee Apis mellifera ORs, AmOR11 responded specifically to the main “queen substance” component 9-oxo-2-decenoic that maintains the queen’s dominance in the colony and also acts as a long-distance sex pheromone [67]. The Drosophila OR, Or43a, has been found that only four odors molecules (cyclohexanol, cyclohexanone, benzaldehyd, and benzyl alcohol) can activate the receptor at a low micromolar concentration, as demonstrated using two-electrode voltage-clamp recording [68]. Electroantennogram (EAG) had illustrated that G. molesta more sensitive to sex pheromone than females, Z8-12:Ac elicited the strongest antennal response in male [49]. Up to now, the functional of pheromone receptors of G. molesta is lacking.
The gustatory receptors we identified here, including GmolGR2, GmolGR3 and GmolGR4, were found in a clade with putative sugar receptors (Fig 8). This clade includes the newly characterized B. mori fructose receptor (BmorGR9) and inositol receptor (BmorGR8) [39,47]. In addition, other identified gustatory receptors (SlitGR4, SlitGR5 and CpomOR25) in moth antennae were also clustered in this family [15,16]. Sugars and other carbohydrates have been shown to influence host preference and oviposition in codling female moths [69]. An artificial mixture of six metabolites of apple, including the three sugar alcohols sorbitol, quebrachitol, and myo-inositol and three sugars glucose, fructose, and sucrose, did stimulate the laying of eggs of codling female moths. Fructose, sorbitol and myo-inositol are important components of the stimulatory blend. The mated female of G. molesta prefer egg-laying to surfaces of ripe apple or peach fruit or secrete high carbohydrate on immature fruit. The function of these gustatory receptors seemed related to the recognition of these carbohydrates. In addition, one putative GR receptor (GmolGR1) was identified as a putative CO2 receptor, and the protein shares high sequence identity (79%) with the S. Iittoralis CO2 receptor, SlitGR3 [15]. Until this study, moth sensory neurons specific for CO2 have been described only on labial palps [36].
Ionotropic receptors represent a novel member of the chemosensory receptor family, which were first discovered in D. melanogaster by bioinformatics screen genomic data for insect-specific genes with enriched expression in OSNs [11]. These were then found in several other species using genome analyses and antennal transcriptome sequencing. The ionotropic receptor is a variant of the iGluR subfamily. Animal iGluRs have been best characterized for their essential roles in synaptic transmission as receptors for the excitatory neurotransmitter glutamate [70]. In D. melanogaster, 66 IRs were identified, 15 of which proved to be antenna-specific [11]. Twelve IRs were identified in the antennae of S. littoralis [15], 15 IRs in the antennae of C. pomonella [16], 20 IRs in the antennae of C. suppressalis [17], and 12 IRs in the antennae of H. armigera [3]. In our study, we found 24 putative IRs in G. molesta antennae, including two co-receptors, IR8a and IR25a. Compared to ORs, the IR family is relatively conserved in sequence. Among the 24 GmolIRs we discovered, GmolIR8a, GmolIR25a, GmolIR21a, GmolIR41a, GmolIR68a, GmolIR87a, GmolIR93a, GmolIR75d, GmolIR75p1, GmolIR75q.1, GmolIR75q.2, GmolIR76b, GmolIR7d, GmolIR1, and GmolIR3 were clustered in separate clades in neighbor-joining tree with Amel/Bmor/Cpom/Dmel/Msex/Slit/Tcas IRs, respectively. Considering the relatively high sequence conservation and similarities in expression, the functions of GmolIRs are probably conserved as IRs in other Lepidopterans.
Conclusion
The main purpose of this study was to identify the genes involved in the reception, processing, and degradation of volatiles by analyzing the antennal transcriptome sequence from G. molesta. The number of OBPs, CSPs, ORs, IRs, GRs, and SNMPs genes that were identified in this species are close to the complete repertoire of olfactory genes from the antennae identified from other Lepidopteran species. The results demonstrated that Illumina Miseq sequencing was successful in the recovery of low-expressing putative olfactory genes, especially in a non-model pest species without an available genome sequence. Our findings made it possible for future research on the molecular level of olfactory system of G. molesta, and in particular, the discovery of receptor genes will also contribute to the identification of novel volatile host compounds, which would gain new options for controlling insects by mass trapping or disruption.
Supporting Information
(TXT)
(TXT)
(XLSX)
(XLSB)
(XLSX)
Acknowledgments
We thank Prof. Ming-Shun Chen (The University of Kansas, USA) for valuable advice and comments on the manuscript.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by National Natural Science Foundation of China (Grant No. 31272043) and Public Welfare Project from Ministry of Agriculture of the People’s Republic of China (Grant No. 201103024). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Sato K, Touhara K. Insect olfaction: receptors, signal transduction, and behavior. Results and Problems in Cell Differentiation. 2009; 47:203–220 [DOI] [PubMed] [Google Scholar]
- 2. Gao Q, Chess A. Identification of candidate Drosophila olfactory receptors from genomic DNA sequence. Genomics. 1999; 60(1):31–39. [DOI] [PubMed] [Google Scholar]
- 3. Liu Y, Gu S, Zhang Y, Guo Y, Wang G. Candidate olfaction genes identified within the Helicoverpa armigera antennal transcriptome. PLOS One. 2012; 7(10):e48260 10.1371/journal.pone.0048260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bruyne M, Baker TC. Odor detection in insects: volatile codes. J Chem Ecol. 2008; 34(7):882–897. 10.1007/s10886-008-9485-4 [DOI] [PubMed] [Google Scholar]
- 5. Klein U. Sensillum-lymph proteins from antennal olfactory hairs of the moth Antheraea polyphemus (Saturniidae). Insect Biochem. 1987; 17(8):1193–1204. [Google Scholar]
- 6. Laughlin JD, Ha TS, Jones DNM, Smith DP. Activation of pheromone-sensitive neurons is mediated by conformational activation of pheromone binding protein. Cell. 2008; 133(7):1255–1265. 10.1016/j.cell.2008.04.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Forêt S, Wanner KW, Maleszka R. Chemosensory proteins in the honey bee: Insights from the annotated genome, comparative analyses and expressional profiling. Insect Biochem Mol Biol. 2007; 37(1):19–28. [DOI] [PubMed] [Google Scholar]
- 8. Vosshall LB, Amrein H, Morozov P, Rzhetsky A, Axel R. A spatial map of olfactory receptor expression in the Drosophila antenna. Cell. 1999; 96(5):725–736. [DOI] [PubMed] [Google Scholar]
- 9. Wicher D, Schäfer R, Bauernfeind R, Stensmyr MC, Heller R, Heinemann SH, et al. Drosophila odorant receptors are both ligand-gated and cyclic-nucleotide-activated cation channels. Nature. 2008; 452:1007–1011. 10.1038/nature06861 [DOI] [PubMed] [Google Scholar]
- 10. Benton R, Sachse S, Michnick SW, Vosshall LB. Atypical membrane topology and heteromeric function of Drosophila odorant receptors in vivo. PLOS Biology. 2006; 4(2):e20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Benton R, Vannice KS, Gomez-Diaz C, Vosshall LB. Variant ionotropic glutamate receptors as chemosensory receptors in Drosophila . Cell. 2009; 136(1):149–162. 10.1016/j.cell.2008.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Abuin L, Bargeton B, Ulbrich MH, Isacoff EY, Kellenberger S, Benton R. Functional architecture of olfactory ionotropic glutamate receptors. Neuron. 2011; 69:44–60. 10.1016/j.neuron.2010.11.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhao QY, Wang Y, Kong YM, Luo D, Li X, Hao P. Optimizing de novo transcriptome assembly from short-read RNA-Seq data: a comparative study. BMC Bioinformatics. 2011; 12(Suppl 14):S2 10.1186/1471-2105-12-S14-S2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Grosse-Wilde E, Kuebler LS, Bucks S, Vogel H, Wicher D, Hansson BS. Antennal transcriptome of Manduca sexta . P Nati Acad Sci USA. 2011; 108(18):7449–7454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Legeai F, Malpel S, Montagné N, Monsempes C, Cousserans F, Merlin C, et al. An expressed sequence tag collection from the male antennae of the Noctuid moth Spodoptera littoralis: a resource for olfactory and pheromone detection research. BMC genomics. 2011; 12:86 10.1186/1471-2164-12-86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bengtsson JM, Trona F, Montagné N, Anfora G, Ignell R,Witzgall P, et al. Putative chemosensory receptors of the codling moth, Cydia pomonella, identified by antennal transcriptome analysis. PLOS One. 2012; 7(2):e31620 10.1371/journal.pone.0031620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cao DP, Liu Y, Wei JJ, Liao XY, Walker WB, Li JH, et al. Identification of candidate olfactory genes in Chilo suppressalis by antennal transcriptome analysis. Int J Biol Sci. 2014; 10(8):846–860. 10.7150/ijbs.9297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gu SH, Wu KM, Guo YY, Pickett JA, Field LM, Zhou JJ, et al. Identification of genes expressed in the sex pheromone gland of the black cutworm Agrotis ipsilon with putative roles in sex pheromone biosynthesis and transport. BMC Genomics. 2013; 14(1):636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mazzi D, Dorn S. Movement of insect pests in agricultural landscapes. Ann Appl Biol. 2012; 160(2):97–113. [Google Scholar]
- 20. Rice RE, Doyle J, Jones RA. Pear as a host of the oriental fruit moth Lepidoptera-Olethre in California. J Econ Entomol. 1972; 65(4):1212–1213. [Google Scholar]
- 21. Zhao Z, Wang Y, Yan G. A preliminary report on the oriental fruit moth in north Jiangsu. Insect Knowledge. 1989; 26:17–19. [Google Scholar]
- 22. Myers CT, Hull LA, Krawczyk G. Effects of orchard host plants (apple and peach) on development of oriental fruit moth (Lepidoptera: Tortricidae). J Econ Entomol. 2007; 100(2):421–430. [DOI] [PubMed] [Google Scholar]
- 23. Piñero JC, Dorn S. Response of female oriental fruit moth to volatiles from apple and peach trees at three phonological stages. Entomol Exp Appl. 2009; 131(1):67–74. [Google Scholar]
- 24. Rajapakse CNK, Walter GH, Moore CJ, Hull CD, Cribb BW. Host recognition by a polyphagous lepidopteran (Helicoverpa armigera): primary host plants, host produced volatiles and neurosensory stimulation. Physiol Entomol. 2006; 31(3):270–277. [Google Scholar]
- 25. Natale D, Mattiacci L, Hern A, Pasqualini E, Dorn S. Response of female Cydia molesta (Lepidoptera: Tortricidae) to plant derived volatiles. Bull Entomol Res. 2003; 93(4):335–342. [DOI] [PubMed] [Google Scholar]
- 26. Il’ichev AL, Kugimiya S, Williams DG, Takabayashi J. Volatile compounds from young peach shoots attract males of oriental fruit moth in the field. J Plant Interact. 2009; 4(4):289–294. [Google Scholar]
- 27. Piñero JC, Galiziab Galizia C, Dorn S. Synergistic behavioral responses of female oriental fruit moths (Lepidoptera:Tortricidae) to synthetic host plant-derived mixtures are mirrored by odor-evoked calcium activity in their antennal lobes. J Insect Physiol. 2008; 54(2):333–343. [DOI] [PubMed] [Google Scholar]
- 28. Najar-Rodriguez A, Orschel B, Dorn S. Season-long volatile emissions from peach and pear trees in situ, overlapping profiles, and olfactory attraction of an oligophagous fruit moth in the laboratory. J Chem Ecol. 2013; 39(3):418–429. 10.1007/s10886-013-0262-7 [DOI] [PubMed] [Google Scholar]
- 29. Natale D, Mattiacci L, Pasqualini E, Dorn S. Apple and peach fruit volatiles and the apple constituent butyl hexanoate attract female oriental fruit moth, Cydia molesta, in the laboratory. J Appl Entomol. 2004; 128(1):22–27. [Google Scholar]
- 30. Dorn S, Hughes J, Molinari F, Cravedi P. Cydia molesta and Cydia pomonella: comparison of adult behavior. IOBC-WPRS Bulletin. 2001; 24:133–137. [Google Scholar]
- 31. Steiner LF, Yetter WP. Second report on the efficiency of bait traps for the oriental fruit moth as indicated by the release and capture of marked adults. J Econ Entomol. 1993; 26(4):774–788. [Google Scholar]
- 32. Najar-Rodriguez AJ, Galizia CG, Stierle J, Dorn S. Behavioral and neurophysiological responses of female oriental fruit moths to changing ratios of a key component in a bioactive mixture. J Exp Biol. 2011; 214:162. [DOI] [PubMed] [Google Scholar]
- 33. Li B, Fillmore N, Bai YS, Collins M, Thomson JA, Stewart R, et al. Evaluation of de novo transcriptome assemblies from RNA-Seq data. Genome Biol. 2014; 15:553 10.1186/s13059-014-0553-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Garczynski SF, Coates BS, Unruh TR, Schaeffer S, Jiwan D, Koepke T, et al. Application of Cydia pomonella expressed sequence tags: identification and expression of three general odorant binding proteins in codling moth. Insect Sci. 2013; 20(5): 559–574. 10.1111/j.1744-7917.2012.01560.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Zhan S, Merlin C, Boore JL, Reppert SM. The monarch butterfly genome yields insights into long-distance migration. Cell. 2011; 147(5):1171–1185. 10.1016/j.cell.2011.09.052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Picimbon JF, Dietrich K, Krieger J, Breer H. Identity and expression pattern of chemosensory proteins in Heliothis virescens (Lepidoptera, Noctuidae). Insect Biochem Molec Biol. 2011; 31(12): 1173–1181. [DOI] [PubMed] [Google Scholar]
- 37. Wanner KW, Nichols AS, Allen JE, Bunger PL, Garczynski SF, Linn CE, et al. Sex pheromone receptor specificity in the European corn borer moth, Ostrinia nubilalis . PLOS ONE. 2010; 5(1): e8685 10.1371/journal.pone.0008685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gong DP, Zhang HJ, Zhao P, Xia QY, Xiang ZH. The odorant binding protein gene family from the genome of silkworm, Bombyx mori . BMC Genomics. 2009; 10:332 10.1186/1471-2164-10-332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhang HJ, Anderson AR, Trowell SC, Luo AR, Xiang ZH, Xia QY. Topological and functional characterization of an insect gustatory receptor. PLOS One. 2011; 6(9):e24111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tanaka K, Uda Y, Ono Y, Nakagawa T, Suwa M, Yamaoka R, et al. Highly selective tuning of a silkworm olfactory receptor to a key mulberry leaf volatile. Curr Biol. 2009; 19(11):881–890. 10.1016/j.cub.2009.04.035 [DOI] [PubMed] [Google Scholar]
- 41. Croset V, Rytz R, Cummins SF, Budd A, Brawand D, Kaessmann H, et al. Ancient protostome origin of chemosensory ionotropic glutamate receptors and the evolution of insect taste and olfaction. PLOS Genet. 2009; 6(8): e1001064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zhou JJ, He XL, Pickett JA, Field LM. Identification of odorant-binding proteins of the yellow fever mosquito Aedes aegypti: genome annotation and comparative analyses. Insect Mol Biol. 2008; 17(2):147–163. 10.1111/j.1365-2583.2007.00789.x [DOI] [PubMed] [Google Scholar]
- 43. Hekmat-Scafe DS, Scafe CR, Mckinney AJ, Tanouye MA. Genome-wide analysis of the odorant-binding protein gene family in Drosophila melanogaster . Genome Res. 2002; 12(9):1357–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Zhou JJ, Kan YC, Antoniw J, Pickett JA, Field LM. Genome and EST analyses and expression of a gene family with putative functions in insect chemoreception. Chem Senses. 2006; 31(5):453–465. [DOI] [PubMed] [Google Scholar]
- 45. Zhou X, Slone JD, Rokas A, Berger SL, Liebig J, Ray A, et al. Phylogenetic and transcriptomic analysis of chemosensory receptors in a pair of divergent ant species reveals sex-specific signatures of odor coding. PLOS Genet. 2012; 8(8):e1002930 10.1371/journal.pgen.1002930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Garczynski SF, Wanner KW, Unruh TR. Identification and initial characterization of the 3' end of gene transcripts encoding putative members of the pheromone receptor subfamily in Lepidoptera. Insect Sci. 2011; 19(1):64–74. [Google Scholar]
- 47. Sato K, Tanaka K, Touhara K. Sugar-regulated cation channel formed by an insect gustatory receptor. P Nati Acad Sci USA. 2011; 108(28):11680–11685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Krieger J, Raming K, Dewer YME, Bette S, Conzelmann S, Breer H. A divergent gene family encoding candidate olfactory receptors of the moth Heliothis virescens . Eur J Neurosci. 2002; 16(4):619–628. [DOI] [PubMed] [Google Scholar]
- 49. Song YQ, Dong JF, Qiao HL, Wu JX. Molecular characterization, expression patterns and binding properties of two pheromone-binding proteins from the oriental fruit moth, Grapholita molesta (Busck). J Integr Agr. 2014; 13(12):2709–2720. [Google Scholar]
- 50. Leal WS, Nikonova L, Peng GH. Disulfide structure of the pheromone binding protein from the silkworm moth, Bombyx mori . FEBS Lett. 1999; 464(1–2):85–90. [DOI] [PubMed] [Google Scholar]
- 51. Pelosi P, Maida R. Odorant-binding proteins in insects. Comp Biochem Physiol B Biochem Mol Biol. 1995; 111(3):503–514. [DOI] [PubMed] [Google Scholar]
- 52. He XL, Tzotzos G, Woodcock C, Pickett JA, Hooper T, Field LM, et al. Binding of the general odorant binding protein of Bombyx mori BmorGOBP2 to the moth sex pheromone components. J Chem Ecol. 2010; 36(12):1293–1305. 10.1007/s10886-010-9870-7 [DOI] [PubMed] [Google Scholar]
- 53. Zhang TT, Mei XD, Feng JN, Berg BG, Zhang YJ, Guo YY, et al. Characterization of three pheromone-binding proteins (PBPs) of Helicoverpa armigera (Hübner) and their binding properties. J Insect Physiol. 2012; 58(7):941–948. 10.1016/j.jinsphys.2012.04.010 [DOI] [PubMed] [Google Scholar]
- 54. Du GH, Prestwich GD. Protein structure encodes the ligand binding specificity in pheromone binding proteins. Biochemistry. 1995; 34(27):8726–8732. [DOI] [PubMed] [Google Scholar]
- 55. Galindo K, Smith DP. A large family of divergent drosophila odorant-binding proteins expressed in gustatory and olfactory sensilla. Genetics. 2001; 159(3):1059–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shanbhag S, Park SK, Pikielny C, Steinbrecht R. Gustatory organs of Drosophila melanogaster: Fine structure and expression of the putative odorant-binding protein PBPRP2. Cell Tissue Res. 2001; 304(3):423–437. [DOI] [PubMed] [Google Scholar]
- 57. Gu SH, Wang SY, Zhang XY, Ji P, Liu JT, Wang GR, et al. Functional characterizations of chemosensory proteins of the alfalfa plant bug Adelphocoris lineolatus indicate their involvement in host recognition. PLOS One. 2012; 7(8): e42871 10.1371/journal.pone.0042871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nomura A, Kawasaki K, Kubo T, Natori S. Purification and localization of p10, a novel protein that increases in nymphal regenerating legs of Periplaneta americana (American cockroach). Int J Dev Biol. 1992; 36(3):391–398. [PubMed] [Google Scholar]
- 59. Gong L, Luo Q, Rizwan-Ul-Haq M, Hu MY. Cloning and characterization of three chemosensory proteins from Spodoptera exigua and effects of gene silencing on female survival and reproduction. B Entomol Res. 2012; 102(5):1–10. [DOI] [PubMed] [Google Scholar]
- 60. Maleszka J, Forêt S, Saint R, Maleszka R. RNAi-induced phenotypes suggest a novel role for a chemosensory protein CSP5 in the development of embryonic integument in the honeybee (Apis mellifera). Dev Genes Evol. 2007; 217(3):189–196. [DOI] [PubMed] [Google Scholar]
- 61. Guo W, Wang X, Ma Z, Xue L, Han J, Yu D, et al. CSP and takeout genes modulate the switch between attraction and repulsion during behavioral phase change in the migratory locust. PLOS Genet. 2011; 7(2): e1001291 10.1371/journal.pgen.1001291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Varela N, Couton L, Gemeno C, Avilla J, Rospars JP, Anton S. Three-dimensional antennal lobe atlas of the oriental fruit moth, Cydia molesta (Busck) (Lepidoptera: Tortricidae): comparison of male and female glomerular organization. Cell Tissue Res. 2009; 337(3):513–526. 10.1007/s00441-009-0839-1 [DOI] [PubMed] [Google Scholar]
- 63. Vosshall LB, Wong AM, Axel R. An olfactory sensory map in the fly brain. Cell. 2000; 102(2):147–159. [DOI] [PubMed] [Google Scholar]
- 64. Cardé AM, Baker TC, Cardé RT. Identification of a four component sex pheromone of the female oriental fruit moth Grapholitha molesta (Lepidoptera: Tortricidae). J Chem Ecol. 1979; 5(3):423–427. [Google Scholar]
- 65. Nakagawa T, Sakurai T, Nishioka T, Touhara K. Insect sex-pheromone signals mediated by specific combinations of olfactory receptors. Science. 2005; 307(5715):1638–1642. [DOI] [PubMed] [Google Scholar]
- 66. Wang G, Vásquez GM, Schal C, Zwiebel LJ, Gould F. Functional characterization of pheromone receptors in the tobacco budworm Heliothis virescens . Insect Mol Biol. 2011; 20(1):125–133. 10.1111/j.1365-2583.2010.01045.x [DOI] [PubMed] [Google Scholar]
- 67. Wanner KW, Nichols AS, Walden KO, Brockmann A, Luetje CW, Robertson HM. A honey bee odorant receptor for the queen substance 9-oxo-2-decenoic acid. P Nati Acad Sci USA. 2007; 104(36):14383–14388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wetzel CH, Behrendt HJ, Gisselmann G, Störtkuhl KF, Hovemann B, Hatt H, et al. Functional expression and characterization of a Drosophila odorant receptor in a heterologous cell system. P Nati Acad Sci USA. 2001; 98(16):9377–9380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Lombarkia N, Derridj S. Resistance of apple trees to Cydia pomonella egg laying due to leaf surface metabolites. Entomol Exp Appl. 2008; 128(1):57–65. [Google Scholar]
- 70. Madden DR. The structure and function of glutamate receptor ion channels. Nat Rev Neurosci. 2002; 3(2):91–101. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(TXT)
(TXT)
(XLSX)
(XLSB)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.