Abstract
Three half-sandwich iridium and ruthenium organometallic complexes with high cytotoxicity are synthesized, and their anticancer mechanisms are elucidated. The organometallic complexes can interact with DNA through coordination or intercalation, thereby inducing apoptosis and inhibiting proliferation of resistant cancer cells. The organometallic complexes are then incorporated into polymeric micelles through the polymer-metal coordination between poly(ethylene glycol)-b-poly(glutamic acid) [PEG-b-P(Glu)] and organometallic complexes to further enhance their anticancer effects as a result of the enhanced permeability and retention (EPR) effect. The micelles with particle sizes of ~60 nm are more efficiently internalized by cancer cells than the corresponding complexes, and selectively dissociate and release organometallic anticancer agents within late endosomes and lysosomes, thereby enhancing drug delivery to the nuclei of cancer cells and facilitating their interactions with DNA. Thus, the micelles display higher antitumor activity than the organometallic complexes alone with the lack of systemic toxicity in a mouse xenograft model of cisplatin-resistant human ovarian cancer. These results suggest that the polymeric micelles carrying anticancer organometallic complexes provide a promising platform for the treatment of resistant ovarian cancer and other hard-to-treat solid tumors.
Keywords: DNA targeting organometallic complexes, polymeric micelles, coordination, drug delivery, resistant ovarian cancer
1. Introduction
The serendipitous discovery of the antitumor properties of cis-diamminedichloroplatinum(II) (cisplatin, CDDP) by Rosenberg in 1969 led to its adoption for clinical use in 1978.[1] However, the clinical use of platinum-based drugs is limited by the intrinsic and acquired resistance[2–4] and severe side effects, such as acute nephrotoxicity and chronic neurotoxicity,[5–8] which has motivated chemists to develop new metal-based anticancer agents with different mechanisms of action. The diverse structural types and ligand bonding modes of transition metals complexes allow the fine-tuning of metal-based anticancer drugs to broaden the spectrum of treatable cancers, reduce side effects, and overcome drug resistance.[9–12]
In the search for new metal-based anticancer drugs, iridium and ruthenium complexes have attracted much attention due to their high anticancer activities.[13–18] These organometallic complexes are believed to be activated by hydrolysis of the metal-Cl bond prior to binding to DNA via coordination, intercalation, or a combination of both.[19–22] The biological activities of these organometallic complexes can be further fine-tuned by varying the ancillary ligands.[23–29] For example, the introduction of a phenyl or biphenyl substituent to the cyclopentadienyl ring of the half-sandwich [(η5-C5Me4R)IrCl(XY)]n+ (R = H, phenyl, biphenyl; n = 0, 1; XY = N,N- or C,N-chelating ligands) complexes dramatically reduces the IC50 values (the concentration for 50% growth inhibition) toward A2780 human ovarian cancer cells.[24, 25] Replacing the N,N-chelating ligand 2,2’-bipyridine (bpy) in the complex [(η5-C5Me5)Ir(bpy)Cl]+ with the C,N-chelating ligand 2-phenylpyridine (ppy) enhances the cytotoxic activity toward A2780 cells, with the IC50 value decreasing from >100 µM to 10.8 µM.[26] The complexes bearing the large N,N-chelating polypyridyl ligands generally exhibit enhanced anti-proliferative effects.[27–29]
Since DNA is the potential target of organometallic complexes, loading this kind of anticancer agents in nanoparticles that can facilitate their access to DNA will further enhance their anticancer efficacies and facilitate their clinical translation.[30–33] Previous research has demonstrated that polymeric micelles formed by the coordination between the oxaliplatin and poly(ethylene glycol)-b-poly(glutamic acid) [PEG-b-P(Glu)] copolymer selectively dissociated within late endosomes and enhanced drug delivery to the nearby nucleus, thereby exhibiting high antitumor activity.[30] In addition, the PEG-modified long circulating nanoparticles provide an effective strategy to enhance in vivo therapeutic efficacy[34–37] and have been widely studied in preclinical evaluations and clinical trials as promising nanocarriers for tumor-targeted therapy.[38–40] These materials can increase blood circulation time, reduce nonspecific uptake in normal tissues, and enhance tumor accumulation by taking advantage of the enhanced permeability and retention (EPR) effect.[41–43] Clinical studies have demonstrated that polymeric micelles incorporating paclitaxel, SN-38, doxorubicin, or cisplatin can reduce the toxic side effects of the loaded drugs while maintaining appreciable antitumor efficacy.[44, 45]
In this work, we describe the synthesis of three DNA-targeting half-sandwich iridium and ruthenium organometallic complexes and their incorporation into polymeric micelles for achieving enhanced anticancer efficacy. The half-sandwich iridium and ruthenium complexes containing coordinatively stable N,N-chelating ligands, hydrophobic Cp or arene groups, and labile halide components could interact with DNA, induce cell apoptosis and inhibit cell proliferation. After incorporation into polymeric micelles, the organometallic complexes were more efficiently internalized by ovarian cancer cells, and selectively released within late endosomes and lysosomes, leading to higher cytotoxicity than organometallic complexes alone due to their easier access to the DNA. The in vivo efficacy study showed that the micelle exhibited higher antitumor activity than the organometallic complex alone with the lack of systemic toxicity in a mouse xenograft model of cisplatin-resistant human ovarian cancer.
2. Results and Discussion
2.1. Synthesis of Organometallic Complexes
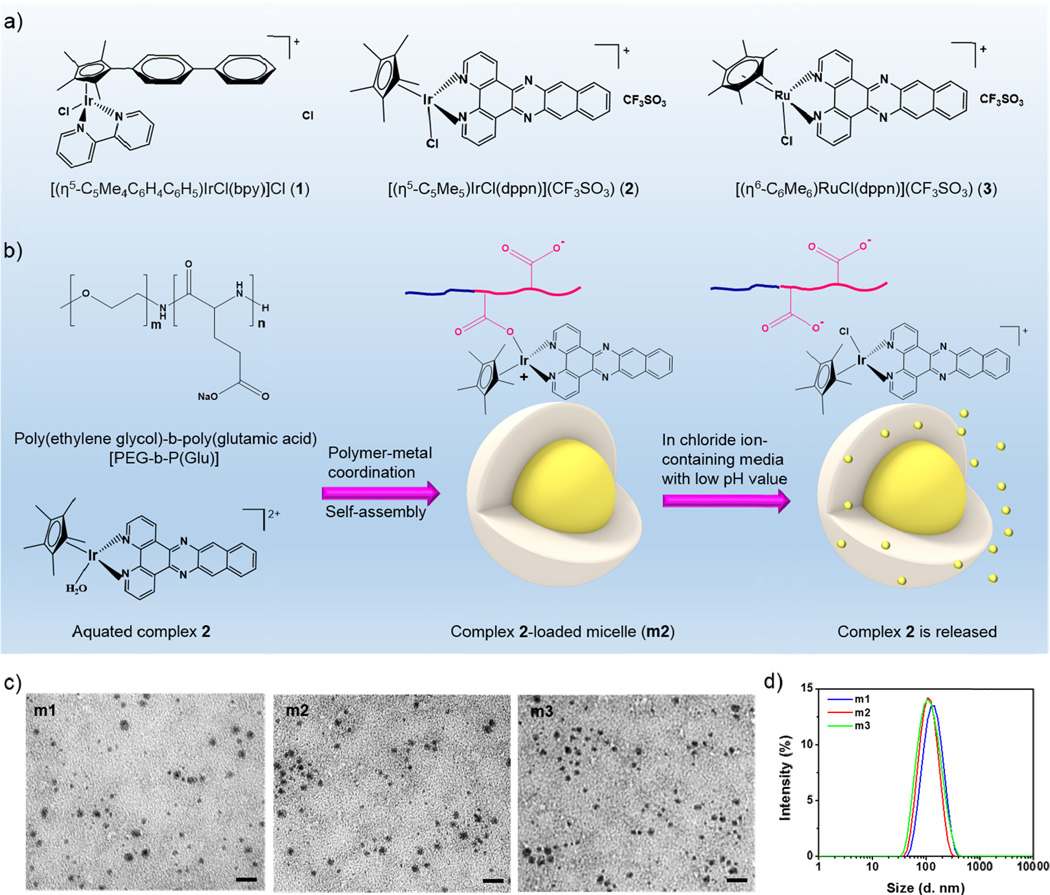
Three half-sandwich iridium and ruthenium complexes [(η5-C5Me4C6H4C6H5)IrCl(bpy)]Cl (1), [(η5-C5Me5)IrCl(dppn)](CF3SO3) (2), and [(η6-C6Me6)RuCl(dppn)](CF3SO3) (3) (Figure 1a) were chosen as anticancer agents based on previous structure-activity studies that established a strong correlation between high cytotoxicity and complexes with coordinatively stable N,N-chelating ligands and hydrophobic Cp or arene groups. The ancillary ligands on the iridium- and ruthenium-centers facilitate the interactions between these complexes and DNA molecules.[23, 28, 29] Complexes 1–3 were synthesized in good yields by reactions of the chelating ligands with the [(η5-C5Me4C6H4C6H5)IrCl2]2, [(η5-C5Me5)IrCl2]2 or [(η6-C6Me6)RuCl2]2 dimer using previously established procedures,[25, 28, 29] and fully characterized by mass spectrometry and 1H NMR.
Figure 1.
a) Chemical structures of complexes 1–3. b) Scheme showing the formation of complex-loaded micelles (m1–m3) and the proposed release of the complex from micelles in chloride ion-containing or/and low pH media. The micelles are spontaneously formed via a ligand exchange reaction of metal from the chloride to the carboxylates in the copolymer, and micelle dissociation is accompanied by the release of complex via an inverse ligand exchange reaction of metal from the carboxylates in the copolymer to the chloride in the surrounding media. c) TEM images of micelles showing the well-defined and spherical morphology. Scale bars: 100 nm. d) Size distribution of micelles as determined by DLS.
2.2. DNA Binding Studies
Knowing that DNA is a potential target for transition metal anticancer complexes,[46–48] we investigated the binding profiles of complexes 1–3 to calf thymus DNA to provide insight into the mechanisms of action.[37] An increase in the absorbance was recorded for complex 1 in the 320–440 nm range, indicating thermodynamically favroable coordinative Ir-N (nucleobase) binding to the DNA (Figure S1, Supporting Information). In contrast, a pronounced decrease in absorbance at about 327, 403 and 425 nm and shifts of these absorption maxima to higher wavelengths were observed after titrating complexes 2 and 3 with calf thymus DNA (Figure S2 and S3, Supporting Information). The spectral changes observed here for 2 and 3 suggested that these complexes may bind to DNA either by intercalation or by surface interaction, involving strong π-π stacking interactions between the complex and the DNA base pairs. We confirmed these results by ethidium bromide (EtBr) displacement experiments (Figure S4, Supporting Information). The fluorescence intensity of EtBr-bound DNA dramatically decreased in the presence of increasing amounts of complexes 2 and 3, as a result of the displacement of dye from the DNA, leading to 51.3% and 58.5% decrease, respectively, when the molar ratio of complex to DNA reached 0.1. In contrast, the fluorescence intensity of EtBr-bound DNA only showed slight decrease after titration with complex 1 (12.8% decrease). These data collectively show that complex 2 and 3 bind to DNA through intercalation while complex 1 mainly interacts with DNA through coordination.
2.3. Preparation and Characterization of Micelles
Iridium and ruthenium complex-loaded micelles m1–m3 were spontaneously formed from the coordination between the metal centers of complexes 1–3 and the carboxylic moieties of the PEG-b-P(Glu) copolymer (Figure 1b). After coordination, the P(Glu) became hydrophobic while the PEG remained hydrophilic, facilitating the self-assembly of the copolymers into micellar structures. The polymeric micelles m1–m3 contained the organometallic complex in the hydrophobic core and the hydrophilic PEG on the surface, which affords long circulation times in the bloodstream.[49–51] Transmission electron microscopy (TEM) images showed that micelles m1–m3 were generally spherical in shape with diameters of 46.8 ± 9.8, 41.6 ± 7.3, 45.3 ± 6.9 nm, respectively (Figure 1c). Dynamic light scattering (DLS) results also showed that m1–m3 were relatively small in size with narrow size distributions (Figure 1d and Table 1). Micelles m1–m3 had number-average diameters of 68.8 ± 6.1, 57.9 ± 3.7, and 60.2 ± 3.9 nm by DLS, respectively. The corresponding polydispersity indexes of m1–m3 were 0.19 ± 0.01, 0.21 ± 0.01, and 0.16 ± 0.01, respectively.
Table 1.
Characterization of m1–m3. Data are expressed as means ± S.D. (n = 3).
| TEM diameter [nm] | Number-Ave diameter [nm] |
PDIa) | ς potential [mV] | Drug loadingb) | [Drug]/[Glu] | CMC [µg/mL]c) | |
|---|---|---|---|---|---|---|---|
| m1 | 46.8 ± 9.8 | 68.8 ± 6.1 | 0.19 ± 0.01 | −9.54 ± 0.59 | 36.0 ± 0.9 wt.% | 0.61 ± 0.02 | 6.3 |
| m2 | 41.6 ± 7.3 | 57.9 ± 3.7 | 0.21 ± 0.01 | −9.55 ± 0.07 | 35.1 ± 0.4 wt.% | 0.49 ± 0.01 | 5.1 |
| m3 | 45.3 ± 6.9 | 60.2 ± 3.9 | 0.16 ± 0.01 | −11.9 ± 0.21 | 34.2 ± 0.8 wt.% | 0.45 ± 0.01 | 7.6 |
Polydispersity index;
Weight ratio of complex to micelle;
Weight concentration of micelles.
The relatively small particle sizes of micelles m1–m3 are potentially beneficial for tumor-targeting delivery. They are large enough to avoid renal filtration and yet still small enough to penetrate through the leaky vasculatures in the tumor region. The PEG coatings on these micelle surfaces can resist protein adsorption to reduce mononuclear phagocytic system (MPS)-mediated clearance.[36, 52, 53] In addition, micelles m1–m3 exhibited negative zeta potentials (~ −10 mV), presumably due to the presence of uncoordinated carboxylic groups at or near the micelle surfaces. We also determined the drug loading as well as the molar ratio of the incorporated complex to the total Glu residues in the micelles ([Drug]/[Glu]) using inductively coupled plasma mass spectrometry (ICP-MS). Micelles m1–m3 displayed high drug loading capacity in the range of 34–36 wt.%, with a [Drug]/[Glu] ratio of 0.45–0.61. Furthermore, the micelles exhibited low critical micelle concentrations (CMCs), which allow them to avoid breakdown and premature drug leakage before reaching the diseased site after injection (Table 1 and Figure S5–S7, Supporting Information).
2.4. Release of Organometallic Complexes from Micelles
The micelles are expected to be exposed to different pH and chloride ion concentrations during subcellular trafficking after internalization by cancer cells,[54, 55] so we investigated the release profiles of the micelles under conditions that mimic early endosomes, late endosomes, and lysosomes (Figure S8–S10, Supporting Information). All three micelles displayed similar release profiles. The releases of complexes from micelles were found to be slow in buffer solution that models the early endosomes (pH 6.9, 10 mM PBS and 20 mM NaCl at 37°C); only a small percentage of drug release (18.2%, 19.8% and 15.3% for 1–3, respectively) was observed after 120 h of incubation. The releases of complexes became faster in a buffer that simulates the conditions in the late endosomes and lysosomes of target cells (pH 5.5, 10 mM PBS and 70 mM NaCl at 37°C); 28.8% of 1, 30.3% of 2 and 27.4% of 3, respectively, were released in a sustained manner after 120 h of incubation. These results indicate that the releases of complexes 1–3 from m1–m3 depend on the pH and chloride concentration in the medium, as a result of ligand substitution of the metals from the carboxylates in the micelle core with the chloride ions in the medium. High chloride concentration and low pH would enhance drug release. The increased dissociation of micelles within late endosomes and lysosomes might facilitate drug delivery close to the nucleus and enhance drug interaction with DNA, thereby exhibiting higher antitumor activity than the corresponding complex alone.[30, 56]
2.5. Cellular Uptake
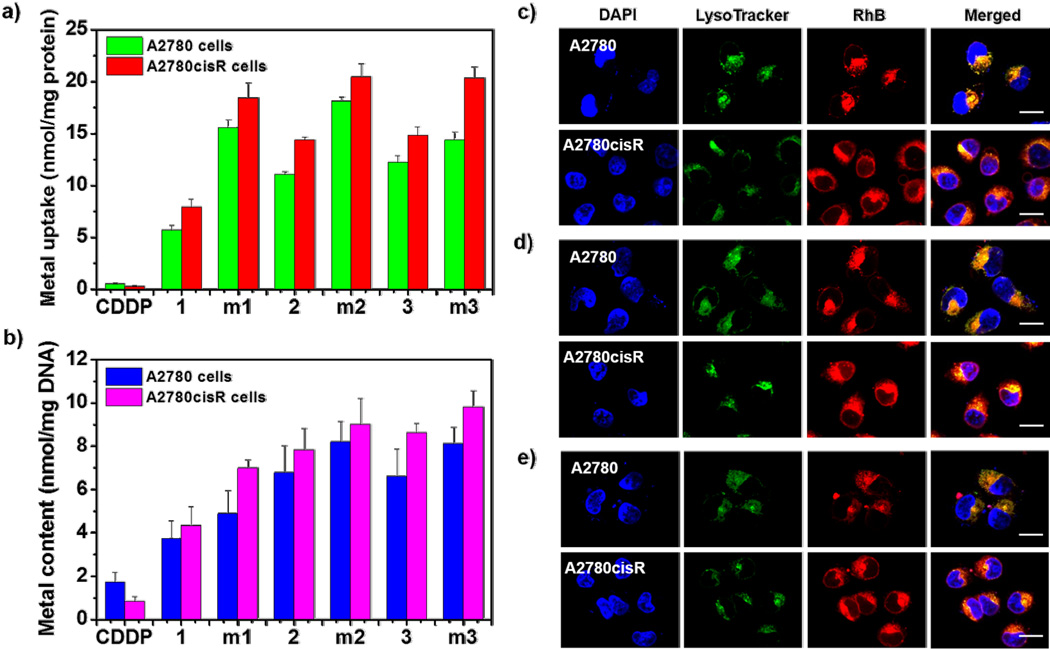
The cellular uptake efficiency of complexes 1–3 and micelles m1–m3 in cisplatin-sensitive A2780 and cisplatin-resistant A2780cisR cells was analyzed by measuring the metal content in the cells by ICP-MS, and compared to free CDDP (Figure 2a). After the cells were incubated with CDDP at a concentration of 20 µM for 4 h, the Pt content was 0.55 ± 0.07 nmol/mg protein in A2780 cells and 0.27 ± 0.07 nmol/mg protein in A2780cisR cells, respectively. The uptake of CDDP in A2780cisR cells was only half of that in A2780 cells, which is in part responsible for cisplatin-resistance in A2780cisR cells. Interestingly, all of the three complexes exhibited significantly higher uptakes than cisplatin in both cell lines after incubation at the same metal concentration for 4 h. The contents of complexes 1–3 were 7.95 ± 0.73, 14.39 ± 0.25, and 14.89 ± 0.78 nmol/mg protein in A2780cisR cells, and 5.73 ± 0.40, 11.11 ± 0.21, and 12.24 ± 0.64 nmol/mg protein in A2780 cells, respectively. The uptakes of complexes 1–3 in A2780 cells were slightly lower than those in A2780cisR cells, but their uptakes in both cell lines were significantly higher than that of CDDP. The enhanced cellular uptake of these complexes is believed to originate from the hydrophobicity and positive charge. Like other small molecules, the uptake of organometallic complexes by tumor cells was reported to be dominated by passive diffusion,[57] some organic and metal transporters may also be involved in the internalization of organometallic complex.[58] The partition coefficients (log P) of complex 1–3 were 0.32 ± 0.01, 0.54 ± 0.04 and 0.54 ± 0.03, respectively, while the log P value of CDDP was 0.23 ± 0.01 (Figure S11, Supporting Information). The higher log P values of the complexes lead to higher cellular uptake, which supports the passive diffusion of complex into the cell. In addition, the complexes were positively charged, which further facilitates the interaction with the negatively charged cell membrane, and enhances cellular uptake. Furthermore, the complexes investigated here may not be cross-resistant to platinum containing compounds,[59, 60] which means that the resistant factors that prevent the cellular uptake of cisplatin may not impact the uptake of these complexes. The results of cellular uptake studies also indicate that the micelles were internalized by the cells at higher efficiency than corresponding complexes. The contents of metal in A2780 cells treated with m1–m3 were 15.59 ± 0.78, 18.12 ± 0.36, and 14.40 ± 0.76 nmol/mg protein, respectively. The uptakes of micelles in A2780cisR cells were slight higher than that in A2780 cells, with the metal contents of 18.43 ± 1.45, 20.46 ± 1.23, and 20.34 ± 1.07 nmol/mg protein for m1–m3, respectively.
Figure 2.
a) In vitro cellular accumulation of metal in A2780 and A2780cisR cells after 4 h incubation with CDDP, complexes 1–3, or m1–m3 measured by ICP-MS. b) Metal content in the DNA collected from the A2780 and A2780cisR cells treated with CDDP, complexes 1–3, or m1–m3 for 4 h. CLSM images of colocalization of RhB (red) from m1 c), m2 d) and m3 e) with a late endosome and lysosome marker, LysoTracker (green), in A2780 and A2780cisR cells after incubation for 2 h. Scale bars: 50 µm.
2.6. Metal-DNA Adduct Formation
We also quantified the levels of bioavailable metals for the internalized complexes and micelles. After incubation with complexes and micelles, the total DNA in the cells was extracted using a DNA purification kit, and the metal content in the extracted DNA was measured by ICP-MS. Metallodrugs are known to cause apoptosis of cancer cells via chelation with the DNA strands to form metal-DNA adducts. Therefore, the metal content in the DNA sample can be a measure of the amount of metal that is responsible for cell death (i.e., therapeutically effective amount of metal). As shown in Figure 2b, the Pt contents in the DNA extracted from A2780 and A2780cisR cells were 1.74 ± 0.44 and 0.86 ± 0.19 nmol/mg DNA, respectively, after incubation for 4 h at a concentration of 20 µM. In contrast, the metal contents in the DNA extracted from cells incubated with complexes 1–3 were 3.73 ± 0.80, 6.79 ± 1.23, and 6.62 ± 1.24 nmol/mg DNA for A2780 cells, and 4.35 ± 0.86, 7.84 ± 0.96, and 8.63 ± 0.42 nmol/mg DNA for A2780cisR cells, respectively. These data indicate that all of the complexes formed more metal-DNA adducts than CDDP did, suggesting they may be more potent inducers of cell apoptosis. In addition, the metal contents in DNA from cells incubated with m1–m3 were a little higher than that incubated with corresponding complexes, 4.89 ± 1.06, 8.21 ± 0.94, and 8.13 ± 0.73 nmol/mg DNA for A2780 cells, and 6.99 ± 0.38, 9.03 ± 1.17, and 9.81 ± 0.73 nmol/mg DNA for A2780cisR cells, respectively. These data demonstrate that polymeric micelles may be able to further enhance the cytotoxicity of complexes through forming more metal-DNA adducts.
2.7. Intracellular Localization
We constructed fluorescently-labeled micelles to visualize the intracellular localization of micelles by confocal laser scanning microscopy (CLSM). To construct the fluorescently-labeled block copolymer, we conjugated the fluorescent dye Rhodamine B (RhB) isothiocynate to the ω-end amine groups of the PEG-b-P(Glu) copolymer, and prepared the RhB-micelles using the same method as described for the non-labeled micelles. The RhB-micelles had similar properties to the non-labeled micelles (Table S1, Supporting Information). After incubation for 2 h, bright red fluorescence from RhB was clearly observed for all three RhB-micelles in both A2780 and A2780cisR cells, indicating that the micelles could be quickly and efficiently internalized by cancer cells. In addition, most of the internalized micelles were colocalized with the acidic organelles labeled with Lysotracker Green (green), suggesting that endocytic pathways are involved in the internalization of micelles (Figure 2c–2e). The involvement of the endocytic pathways would be beneficial to the targeted release of organometallic complexes within the late endosomes and lysosomes, thereby enhancing their perinuclear delivery, facilitating their interaction with DNA, and consequently increasing their anticancer activities.[30, 56]
2.8. Cytotoxicity Assay
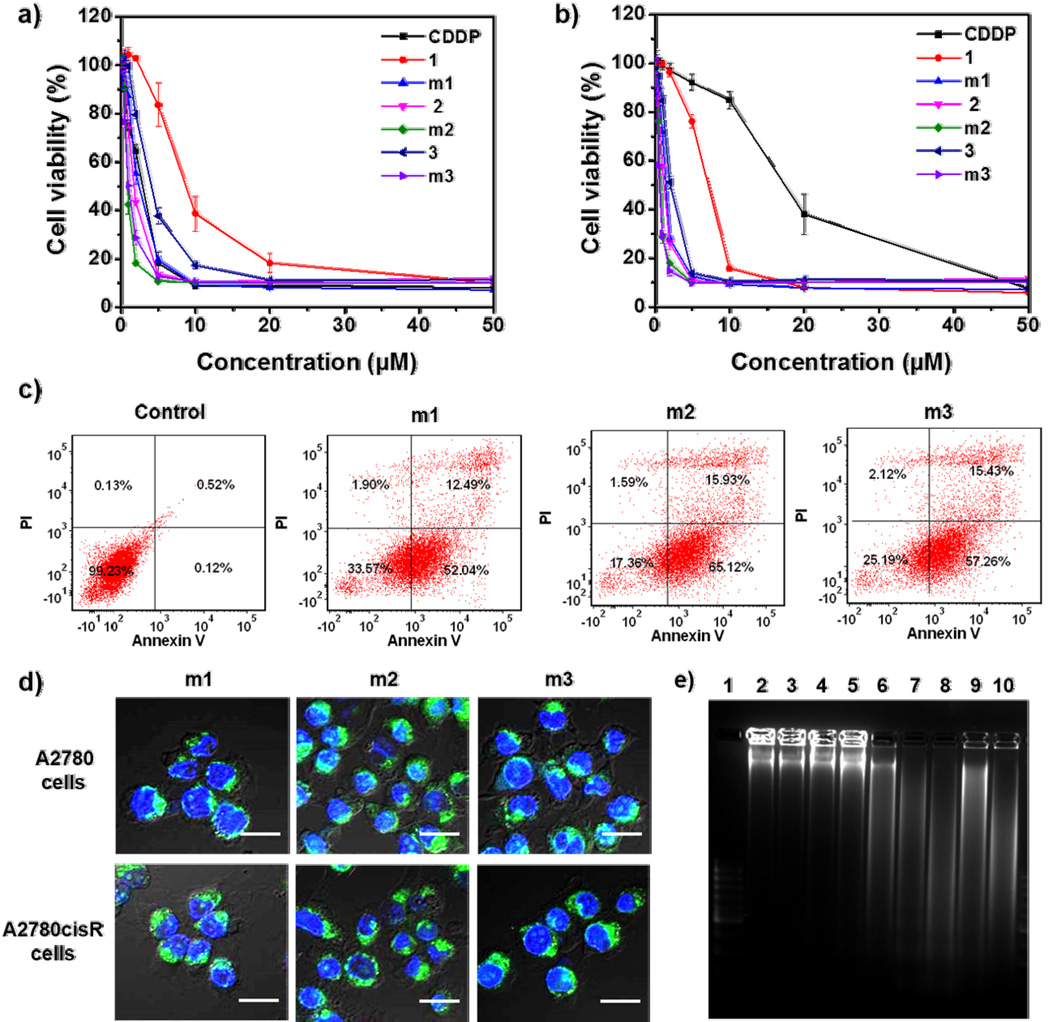
We studied the cytotoxicity of the complexes and micelles in both A2780 and A2780cisR cell lines by (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) (MTS) assay (Figure 3a and 3b, and Table 2). In A2780cisR cells, the IC50 value of free CDDP was as high as 18.6 µM, which was ~6.5-fold more resistant to CDDP in comparison with the parent A2780 cells (IC50 = 2.9 µM). However, the IC50 values of the complexes 1–3 in A2780cisR cells were 7.09 ± 0.11, 1.35 ± 0.08, and 2.03 ± 0.06 µM, respectively, which were 2.2-, 13.8- and 9.2-fold lower than that of CDDP, respectively. These results indicate that the organometallic complexes, especially 2 and 3, have much higher cytotoxicity than CDDP, which may be attributed to their higher cellular uptake and increased metal-DNA adduct formation. The cytotoxicity was increased further after loading the complexes into micelles. The IC50 values of m1–m3 in A2780cisR cells were 1.53 ± 0.02, 0.79 ± 0.01, and 0.72 ± 0.01 µM, respectively, which were 12.2-, 23.5-, and 25.8-fold lower compared to CDDP, respectively. The enhanced cytotoxicity of m1–m3 is probably due to more efficient intercellular accumulation and perinuclear subcellular localization.
Figure 3.
Cytotoxicity of complexes and micelles against A2780 cells a) and A2780cisR cells b). The cells were incubated with free CDDP, complexes 1–3, or m1–m3 for 48 h followed by the MTS assay. c) Annexin V/PI analysis of A2780cisR cells after incubation with m1–m3 for 48 h. The quadrants from lower left to upper left (counter clockwise) represent healthy, early apoptotic, late apoptotic, and necrotic cells, respectively. The percentage of cells in each quadrant was shown on the graphs. d) CLSM images showing cell apoptosis in A2780 and A2780cisR cells after incubation with m1–m3 for 48 h. Cells were stained with Alexa Fluor 488 conjugated Annexin V and the nuclei were stained with DAPI. Scale bars: 50 µm. e) Analysis of DNA ladder on 2% (w/v) agarose gel at 35 V for 3 h after DNA extraction from the A2780cisR cells treated with complexes and micelles. Lanes 1–10: DNA marker, control, PEG-b-p(Glu) copolymer, CDDP, 1, m1, 2, m2, 3, and m3, respectively.
Table 2.
IC50 values (µM) of CDDP, complexes 1–3, and m1–m3 against A2780 and A2780cisR cells after 48 h incubation, as determined by MTS assay. Data are expressed as means ± S.D. (n = 3).
| CDDP | 1 | m1 | 2 | m2 | 3 | m3 | |
|---|---|---|---|---|---|---|---|
| A2780 cells | 2.90 ± 0.30 | 9.21 ± 0.49 | 2.36 ± 0.24 | 1.83 ± 0.04 | 0.97 ± 0.07 | 4.89 ± 0.20 | 1.27 ± 0.06 |
| A2780cisR cells | 18.60 ± 0.23 | 7.09 ± 0.11 | 1.53 ± 0.02 | 1.35 ± 0.08 | 0.79 ± 0.01 | 2.03 ± 0.06 | 0.72 ± 0.01 |
We also evaluated pharmacological activities of the complexes and micelles on other cisplatin-resistant ovarian cancer cell lines, including OVCAR-3 and SKOV-3 cells (Figure S12 and S13, and Table S2, Supporting Information). CDDP displayed very low cytotoxicity on both cell lines, with IC50 values of 15.5 ± 0.14 µM and 37.07 ± 1.32 µM for OVCAR-3 and SKOV-3 cells, respectively. However, the IC50 values of complexes 1–3 were 4.14 ± 0.33, 1.92 ± 0.03, and 5.19 ± 0.21 µM in OVCAR-3 cells, and 16.05 ± 0.75, 5.41 ± 0.22, and 16.33 ± 0.53 µM in SKOV-3 cells, respectively, which demonstrates that the complexes possess much higher cytotoxicity against these two cell lines when compared to CDDP. The IC50 values of m1–m3 were 1.65 ± 0.03, 1.33 ± 0.11, and 2.76 ± 0.36 µM in OVCAR-3 cells, and 4.76 ± 0.14, 3.61 ± 0.08, and 10.61 ± 0.73 µM in SKOV-3 cells, respectively; the incorporation of the complexes into micelles further enhanced their cytotoxicity. All of these results indicate that the complexes and micelles were able to enhance the anticancer efficacy in different types of ovarian cancer cells (A2780 and A2780cisR are from undifferentiated carcinoma, while OVCAR-3 and SKOV-3 are from clear cell adenocarcinoma). The cytotoxicity of copolymer on these cell lines was also evaluated (Figure S14, Supporting Information). No decrease in cell viability was observed at the copolymer concentration as high as 100 µM (equivalent to 1.2 mM 1, 1.0 mM 2 and 0.9 mM 3, respectively), indicating that the enhanced anticancer efficacy of micelles results from the cytotoxicity of corresponding complexes, rather than from the copolymer.
2.9. Cell Apoptosis
The cell apoptosis induced by the complexes and micelles was quantified by flow cytometry. After incubation with the complexes and micelles for 48 h, cells were collected, stained with Annexin V and prodium iodide (PI), and then analyzed by flow cytometry. Annexin V was used to measure early apoptosis by detecting phosphatidylserine expression, and PI was used to detect membrane permeability. As shown in Figure 3c and Figure S15 and S16, Supporting Information, free CDDP induced 41.3% cell apoptosis and 5.8% cell necrosis in A2780 cells, but showed negligible pro-apoptotic effect in A2780cisR cells at a concentration of 0.5 µM. However, complexes 2–3 and m1–m3 all showed high levels of apoptosis in both cell lines at the same concentration. Complexes 2–3 and m1–m3 induced 66.75%, 49.97%, 57.27%, 88.3%, and 72.5% cell apoptosis in A2780 cells, and 66.45%, 62.02%, 64.53%, 80.05%, and 62.69% cell apoptosis in A2780cisR cells, respectively.
The cell apoptosis induced by micelles was confirmed by confocal image (Figure 3d). After incubation for 48 h, cells were stained with Alexa Fluor 488 conjugated Annexin V, and observed using confocal microscopy. The presence of the bright green fluorescence from Annexin V suggests that the micelles successfully induce cancer cell apoptosis.
2.10. DNA Ladder Detection
DNA fragmentation is considered as a hallmark of apoptosis. During apoptosis, activated nucleases degrade the higher order chromatin structure of DNA into fragments, which can be extracted from cells and visualized by gel electrophoresis followed by EtBr staining. As shown in Figure 3e and Figure S17, Supporting Information, the presence of the characteristic DNA ladder indicates that the complexes 2–3 and m1–m3 caused significant cell apoptosis.
2.11. Biodistribution
The biodistribution of m2 in A2780cisR tumor model at different times after intravenous injection was investigated. Figure S18, Supporting Information, showed that m2 could be rapidly distributed to tumor site. As early as 5 min post-injection, about 2.3 ± 0.11 % ID/g tissue of iridium was detected in tumor, and the amount of iridium in tumor tissue increased with time. After 3 h and 8 h post administration, the iridium uptake in tumor tissue increased to 3.2 ± 0.17 and 4.5 ± 0.75 % ID/g tissue, respectively, which indicates the high accumulation and long retention of m2 in tumor tissue. Meanwhile, m2 was also observed in highly perfused organs, such as liver, spleen and lung, which could be due to the combined activity of circulating blood passing through these organs and the unavoidable uptake by the MPS in these organs. The high tumor accumulation and long tumor retention of m2 would enhance their antitumor effect in vivo.
2.12. In Vivo Anticancer Efficacy
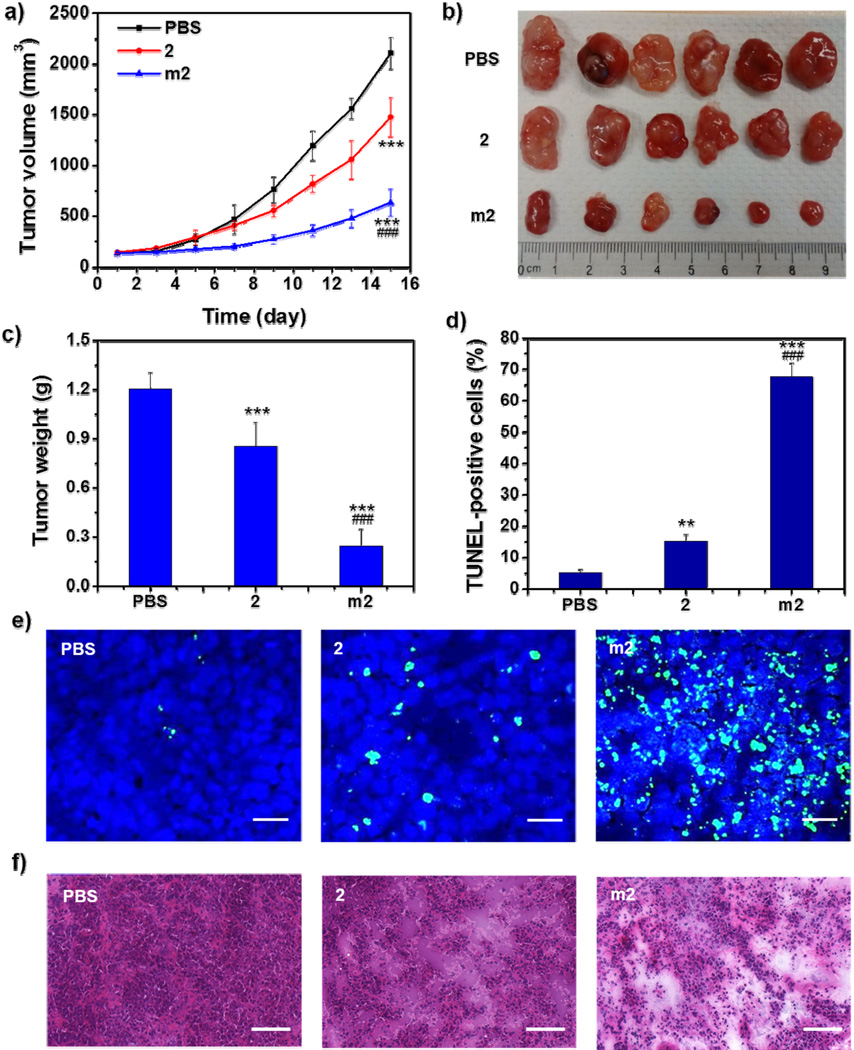
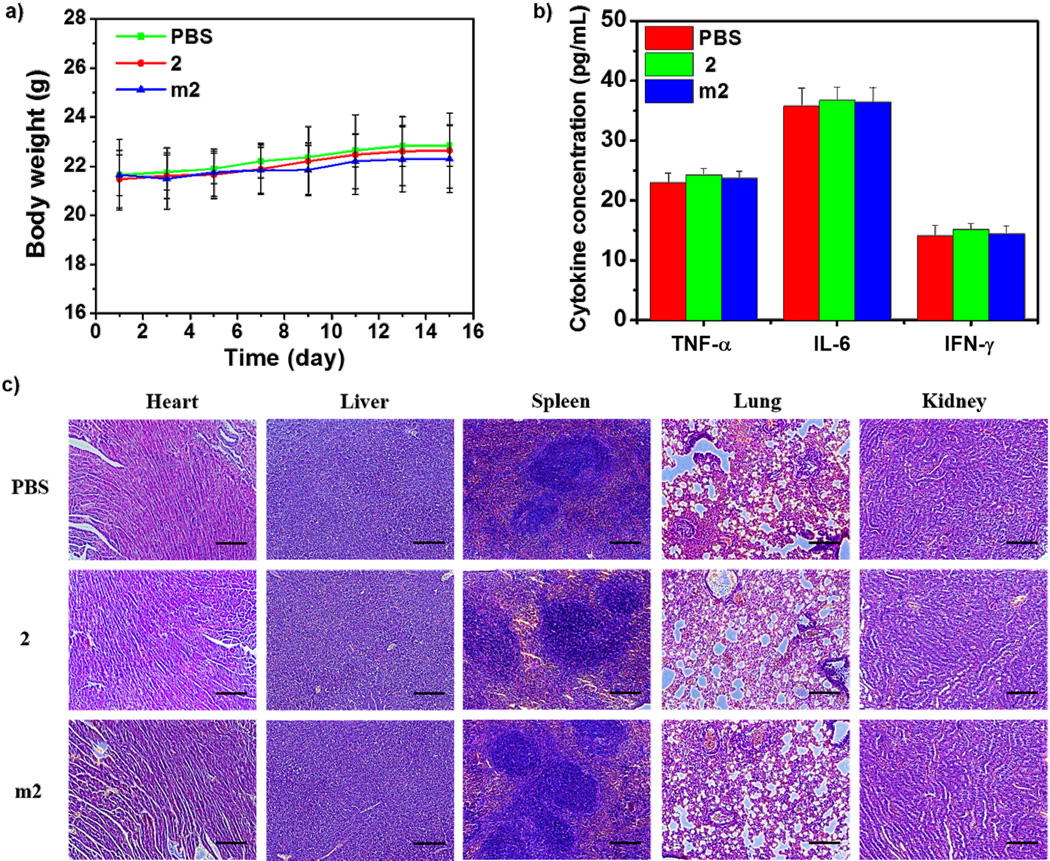
Encouraged by significantly enhanced in vitro anticancer efficacy, we evaluated the in vivo antitumor effect of m2 in cisplatin-resistant A2780cisR subcutaneous xenografts. As depicted in Figure 4a, tumors in PBS-treated mice showed rapid growth, and treatment with free 2 resulted in a slight decrease in tumor growth (29.9% reduction when compared to the PBS-treated group). However, when 2 was loaded into micelles, its effect on the inhibition of tumor growth was significantly improved. At the end of experiment, the average tumor volume in the m2-treated group was reduced by 70.2% and 56.9% compared to the PBS- and 2-treated group (P < 0.001), respectively (Figure 4a and 4b). Likewise, the average tumor weight was also reduced significantly, with 83.5% and 74.3% reduction in the m2-treated group when compared to the PBS- and 2-treated group (P < 0.001), respectively (Figure 4c). The TdT-mediated dUTP nick end labeling (TUNEL) assay showed that the fluorescence intensity of DNA fragmentation and the relative percentage of apoptotic cells in m2-treated group (67.7 ± 4.5%) were higher than those in the other groups (5.2 ± 1.1% and 15.3 ± 2.1% for PBS and 2, respectively), indicating superior anticancer efficacy of m2 (Figure 4d and 4e). The histological analysis of tumor tissues also showed that m2 induced tumor tissue apoptosis and necrosis at the highest level, while 2 only induced slight tumor tissue apoptosis and necrosis, when compared to PBS-treated group (Figure 4f). Furthermore, no obvious body weight loss, immunogenic response, and histological toxicity were observed after repeated treatment with m2, suggesting it is safe when applied in vivo (Figure 5).
Figure 4.
In vivo anticancer effect of m2 on the A2780cisR subcutaneous xenograft. a) Tumor growth curves of the mice receiving intravenous injection of PBS, 2, or m2 (n = 6). b) Photograph of the excised tumors on day 15. c) Weights of the excised tumors on Day 15 (n = 6). d) The percentage of TUNEL-positive cells in tumor tissues (n = 3). e) Representative CLSM images of TUNEL assays of tumor tissues. DNA fragment in apoptotic cells was stained with fluorescein-conjugated deoxynucleotides (green) and the nuclei were stained with DAPI (blue). Scale bars: 50 µm. f) Representative images of histological assays of tumor tissues. Scale bars: 100 µm. Statistical significance: **P < 0.01 and ***P < 0.001 compared to the control group (PBS), ###P < 0.001 compared to the 2-treated group.
Figure 5.
a) Body weight changes of A2780cisR tumor bearing mice after the treatment with 2 and m2 (n = 6). b) TNF-α, IL-6, and IFN-γ levels in the serum of A2780cisR tumor bearing mice receiving intravenous injection of PBS, 2 and m2 determined by ELISA. c) Histological analysis of excised organs from A2780cisR tumor bearing mice after treatment with PBS, 2 and m2. Scale bars: 50 µm.
NC-6004, a polymeric micelle formulation of cisplatin, is currently being tested in clinical trial. Previous studies showed micelles remarkably prolonged blood circulation and effectively accumulated in solid tumors, thereby demonstrating good tolerance and low toxicity. However, the micelles only resulted in comparable antitumor activity to free CDDP.[61–64] In addition, NC-6004 was mainly tested on sensitive solid tumors while the present micelle system showed significant higher antitumor effect than free drug with no obvious systemic toxicity on resistant ovarian cancer.
3. Conclusion
In summary, we successfully prepared polymeric micelles that are loaded with three DNA targeting organometallic complexes with high anticancer activity against ovarian cancer using coordination chemistry. The micelles had suitable sizes and low CMCs for tumor-targeting delivery, and selectively released the loaded complexes within the late endosomes and lysosomes in a sustained fashion. In line with these release profiles, the in vitro cytotoxicity assay against four ovarian cancer cell lines showed appreciably lowered IC50 values compared to that of CDDP. The micelles also displayed a higher propensity for inducing cell apoptosis, as shown by flow cytometry, Annexin V staining, and DNA ladder. More importantly, the micelles showed high antitumor effect in cisplatin-resistant subcutaneous xenografts with the lack of systemic toxicity. These findings indicate that such micelles have the potential to enhance the drug efficacy and overcome drug resistance.
4. Experimental Section
Materials, Cell Lines, and Animals
All of the starting materials were purchased from Sigma-Aldrich and Fisher (USA), unless otherwise noted, and used without further purification.
Cisplatin-sensitive human ovarian cancer cells A2780 and cisplatin-resistant human ovarian cancer cells A2780cisR were obtained from Developmental Therapeutics Core, Northwestern University, and cultured in RPMI 1640 containing 10% FBS (FBS, Gibco, Grand Island, NY), 100 U/mL penicillin G sodium and 100 µg/mL streptomycin sulfate. Human ovarian cancer cells OVCAR-3 and SKOV-3 were obtained from the American Type Culture Collection (ATCC, Rockville, MD), and cultured in RPMI 1640 and McCoy’s 5A, respectively, supplemented with 10% FBS, 100 U/mL penicillin G sodium and 100 µg/mL streptomycin sulfate. All cells were cultured in a humidified atmosphere containing 5% CO2 at 37°C.
Athymic female nude mice (6 weeks, 18–22 g) were provided by Harlan Laboratories, Inc. (USA). The study protocol was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Chicago.
Synthesis of Organometallic Complex and Copolymer
The synthesis of the [(η5-C5Me4C6H4C6H5)IrCl(bpy)]Cl (1), [(η5-C5Me5)IrCl(dppn)](CF3SO3) (2), [(η6-C6Me6)RuCl(dppn)](CF3SO3) (3), and PEG-b-P(Glu) copolymer were described in the Supporting Information.
DNA Titration
Solutions of 10 µM complexes in PBS (pH 7.2) were titrated with calf thymus DNA (R&D Systems, Minneapolis, MN) at 0–120 µM (in terms of nucleotide). The DNA and metal complex were continuously mixed using a motorized stirrer in order to ensure a homogeneous distribution of components. The UV-vis spectra were measured on a Shimadzu UV-2401 spectrophotometer (Shimadzu, Tokyo, Japan) after equilibration, i.e. no further change in the monitored absorbance. During titration, DNA was added to both working and reference cells to ensure that the change of absorbance was due to the interaction of complex with DNA, not because of the higher concentration of DNA.
EtBr Displacement Assay
The solution of DNA and EtBr at molar ratio of 3:1 (saturated binding level[65]) was titrated by complexes; the molar ratio of complex to DNA was from 0 to 0.1. The experiments were conducted at 37°C with continuous stirring in 0.4 M NaCl to avoid secondary binding of EtBr to DNA. After equilibration, fluorescence measurements were performed at an excitation wavelength of 546 nm, and the emission was analyzed at 590 nm using a Shimadzu RF-5301 spectrofluorophotometer (Shimadzu). The extent of complex binding to DNA was determined from the ratio of the observed fluorescence signal upon the addition of the complex, relative to the initial fluorescence of EtBr-bound DNA.
Preparation and Characterization of Complex-Loaded Polymeric Micelles
Organometallic complex (5 mM) was suspended in distilled water and mixed with silver nitrate ([AgNO3]/[Complex] = 1.0) in the dark at 25°C for 24 h to form the aquated complex. The mixture was centrifuged at 13,000 rpm for 10 min to remove the AgCl precipitate, and the supernatant was purified by passing through a 0.22 µm filter. PEG-b-P(Glu) was then added to the aqueous solution ([Complex]/[Glu] = 1.0) and reacted for 72 h to prepare the organometallic complex-loaded micelle. After that, the solution was dialyzed against deionized water (MWCO: 10 kDa) to remove the uncoordinated complex. The RhB-micelles were prepared in the same way as described for the non-labeled micelles except that the copolymer was labeled by RhB through conjugating Rhodamine B isothiocyanate to the amino group at the ω-end of the copolymer. The drug loading of the micelles was determined by ICP-MS. The size distribution and Zeta potential of drug-loaded micelles were measured by DLS using a Zetasizer (Nano-ZS, Malvern, UK) with a scatter angle of 90° at 25°C. The morphologic examination of micelles was performed by TEM (JEM 100CX-II, JOEL, Japan).
CMC Determination
The CMCs of the micelles were determined using pyrene as a fluorescent probe. The concentration of the copolymer was varied from 0.1 to 100 µg/mL, and the concentration of pyrene was fixed at 7 × 10−6 M. The fluorescence spectra were recorded using a spectrofluorophotometer (Shimadzu) with an excitation wavelength of 334 nm. The emission fluorescence at 372 and 382 nm was monitored. The CMC was estimated as the extrapolated cross-point of the intensity ratio I372/I382 at low and high concentration regions.
Release of Organometallic Complexes from Micelles
The release profiles of organometallic complexes from micelles were evaluated in PBS at 37°C. Briefly, a micelle solution of known complex concentration was sealed in a dialysis bag (MWCO: 10 kDa) and immersed in 200 mL PBS under different conditions mimicking the early endosomes (10 mM PBS, pH 6.9, and 20 mM NaCl), and late endosomes and lysosomes (10 mM PBS, pH 5.5, and 70 mM NaCl) at 37°C. Periodically, 1 mL aliquots of the solution were sampled, and equal volume of fresh media was replenished. The concentration of the metal present in the dialysate was determined by ICP-MS.
Cytotoxicity Assay
A2780, A2780cisR, OVCAR-3 and SKOV-3 cells were seeded in 96-well plates at a density of 2 × 103 cells per well. After overnight incubation, the cells were treated with different concentrations of CDDP, complexes, or micelles for 48 h. At the end of the incubation period, cell viability was measured by MTS (Promega, Madison, WI) according to the manufacturer’s instructions. IC50 values were calculated from curves constructed by plotting cell survival (%) versus drug concentration (µM). All experiments were performed in triplicate.
Cellular Uptake
A2780 and A2780cisR cells seeded on 6-well plates (5 × 104 cells/well) were incubated with CDDP, complexes, or micelles (20 µM) for 4 h. Cells were then collected and washed three times with PBS. Half of the cells were dried and digested with concentrated nitric acid for metal analysis by ICP-MS. The other half were lysed with 0.5% (w/v) sodium dodecyl sulfate (SDS, pH 8.0) for analysis of protein content, as quantified by the BCA protein kit (Thermo Pierce, Rockford, IL). Uptake level was expressed as the amount of metal uptake for each mg of cellular protein.
Metal-DNA Adduct Formation
In order to evaluate the metal content in cellular DNA, A2780 and A2780cisR cells seeded on 6-well plates (5 × 104 cells/well) were incubated with CDDP, complexes, or micelles (20 µM) for 4 h. Cells were then collected, and DNA was isolated by using a DNA purification kit (Invitrogen, Grand Island, NY) according to the manufacturer’s protocol. The amount and purity of DNA were determined by measuring absorption at 260 and 280 nm with NanoDrop (ND-1000). The DNA was digested by nitric acid, and the metal content was determined by ICP-MS. The amount of metal-DNA adduct was expressed as nmol of metal per mg of DNA.
log P Determination
Octanol-saturated water (OSW) and water-saturated octanol (WSO) were prepared using analytical grade octanol and 0.2 M NaCl aqueous solution (to suppress hydrolysis of the chloride complexes). Aliquots of complexes in OSW were added to equal volumes of WSO and shaken for 24 h to allow partition at ambient temperature. The aqueous layer was carefully separated from the octanol layer for metal analysis. Metal was quantified from aliquots taken from the octanol-saturated aqueous samples before and after partition. Partition coefficients of complexes were calculated using the equation log P = log ([metal]WSO/[metal]OSW), where [metal]WSO was obtained by subtraction of the metal content of the aqueous layer after partition from the metal content of the aqueous layer before partition.
Intracellular Localization
A2780 and A2780cisR cells were seeded on 10 mm2 glass coverslips placed in 6-well plates and incubated with RhB-micelles (5 µM) for 2 h. Cells were then washed three times with PBS and fixed with 4% paraformaldehyde, followed by staining with LysoTracker Green (100 nM) and DIPA (10 µg/mL) for additional 0.5 h in the dark. The cells were washed twice with PBS, mounted on glass slides, and visualized by CLMS (FluoView TM FV1000, Olympus, Japan).
Apoptosis Analysis
A2780 and A2780cisR cells seeded in 6-well plates (5 × 104 cells/well) were treated with CDDP, complexes, or micelles (0.5 µM) for 48 h. Treated cells were harvested, washed twice with ice-cold PBS, stained with Alexa Fluor 488 conjugated Annexin V and PI for 15 min at room temperature in the dark, and then analyzed by flow cytometry.
Annexin V Staining
A2780 and A2780cisR cells were seeded on 10 mm2 glass coverslips placed in 6-well plates at a density of 5 × 104 cells per well. The cells were incubated at 37°C and 5% CO2 for 24 h prior to treatment. Micelles were incubated with cells at a concentration of 0.5 µM for 48 h. Then, the cells were washed with PBS, fixed with 4% paraformaldehyde, and stained with 10 µg/mL of DAPI and Alexa Fluor 488 conjugated Annexin V. The cells were observed using CLSM at excitation wavelengths of 405 nm and 488 nm to visualize nuclei (blue fluorescence) and cell apoptosis (green fluorescence), respectively.
DNA Ladder Detection
A2780 and A2780cisR cells were seeded in 6-well plates at a density of 5 × 104 cells per well. After 24 h incubation, culture media was replaced by 2 mL of fresh culture media containing 10% FBS. Free CDDP, complexes, or micelles were added to the cells (final concentration: 0.5 µM), respectively. Following incubation for 48 h, total DNA of cancer cells was extracted using a DNA ladder isolation kit (Life Technologies, Grand Island, NY) according to the manufacturer’s instructions and examined for DNA fragmentation on a 2% (w/v) agarose gel electrophoresis at 35 V for 3 h. The DNA ladder induced by the copolymer was also investigated to determine its pro-apoptotic effect.
Biodistribution
Tumor bearing mice were established by subcutaneous inoculation of A2780cisR cell suspension (5 × 106 cells per mouse) into the right flank region of 6-week athymic female nude mice. The tumors were allowed to grow to approximately 100 mm3 before experiment. To assess the tissue distribution, A2780cisR tumor-bearing mice were intravenously injected via tail vein with m2 at a dose of 5 mg Ir/kg. Mice were sacrificed at 5 min, 3 h and 8 h after administration, and the heart, liver, spleen, lung, kidney, bladder and tumor were excised, washed with cold saline and digested with nitrate acid for ICP-MS analysis. The content of Ir in each tissue was expressed as percentage of the injected dose per gram of tissue (% ID/g tissue).
In Vivo Anticancer Efficacy
The A2780cisR tumor-bearing mice were randomly divided into 3 groups (n = 6) and intravenously injected with PBS, complex 2, and m2 at an equivalent dose of 5 mg/kg based on iridium once every three days with a total of three injections. Tumor volumes and body weights were monitored every other day. Tumor volumes were calculated as follows: (width2 × length)/2. Finally, all mice were sacrificed on day 15, and the excised tumors were weighed.
TUNEL Assay
TUNEL analysis was performed on 5-µm frozen tumor sections using DNA Fragmentation Detection Kit (Life Technologies) according to the manufacturer’s instructions and observed under CLSM. DNA fragment in apoptotic cells was stained with fluorescein-conjugated deoxynucleotides (green) and the nuclei were stained with DAPI (10 µg/mL). The percentage of apoptotic cells was determined by the number ratio of TUNEL-positive cells/total cells by Image J.
Histological Analysis of tumor tissue
5-µm frozen tumor sections were stained with hematoxylin and erosin (H&E) and observed with light microscopy.
Histocompatibility Testing
Heart, liver, lung, spleen, and kidney were excised after the mice were sacrificed, and 5-µm tissue sections were stained with H&E and observed for toxicity with light microscopy.
Immunogenic Response
Blood was collected at the endpoint of in vivo antitumor efficacy experiment, and the serum was collected. The serum concentrations of TNF-α, IL-6, and IFN-γ were detected by ELISA (R&D Systems) to evaluate the immunogenic response evoked by 2 and m2.
Supplementary Material
Acknowledgements
We thank NIH (UO1-CA151455) for funding support. We thank C. Poon for the assistance in ICP-MS analysis and Shirley Bond from Integrated Microscopy Core Facility at the University of Chicago for the help with CLSM analysis.
Footnotes
Supporting Information is available from the Wiley Online Library or from the author.
References
- 1.Rosenberg B, Vancamp L, Trosko JE, Mansour VH. Nature. 1969;222:385. doi: 10.1038/222385a0. [DOI] [PubMed] [Google Scholar]
- 2.Giaccone G. Drugs. 2000;59:9. doi: 10.2165/00003495-200059004-00002. [DOI] [PubMed] [Google Scholar]
- 3.Köberle B, Tomicic MT, Usanova S, Kaina B. BBA-Rev. Cancer. 2010;1806:172. doi: 10.1016/j.bbcan.2010.07.004. [DOI] [PubMed] [Google Scholar]
- 4.Marrache S, Pathak RK, Dhar S. Proc. Natl. Acad. Sci. U.S.A. 2014;111:10444. doi: 10.1073/pnas.1405244111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartmann JT, Lipp HP. Pediatr. Hematol. Oncol. 2005;22:441. [Google Scholar]
- 7.Dhar S, Kolishetti N, Lippard SJ, Farokhzad OC. Proc. Natl. Acad. Sci. U.S.A. 2011;108:1850. doi: 10.1073/pnas.1011379108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dhar S, Gu FX, Langer R, Farokhzad OC, Lippard SJ. Proc. Natl. Acad. Sci. U.S.A. 2008;105:17356–17361. doi: 10.1073/pnas.0809154105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bruijnincx PCA, Sadler PJ. Curr. Opin. Chem. Biol. 2008;12:197. doi: 10.1016/j.cbpa.2007.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Muhammad N, Guo Z. Curr. Opin. Chem. Biol. 2014;19:144. doi: 10.1016/j.cbpa.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 11.Garbutcheon-Singh KB, Grant MP, Harper BW, Krause-Heuer AM, Manohar M, Orkey N, Aldrich-Wright JR. Curr. Top. Med. Chem. 2011;11:521. doi: 10.2174/156802611794785226. [DOI] [PubMed] [Google Scholar]
- 12.Ott I, Gust R. Arch. Pharm. 2007;340:117. doi: 10.1002/ardp.200600151. [DOI] [PubMed] [Google Scholar]
- 13.Amouri H, Moussa J, Renfrew AK, Dyson PJ, Rager MN, Chamoreau L-M. Angew. Chem. 2010;122:7692. doi: 10.1002/anie.201002532. [DOI] [PubMed] [Google Scholar]
- 14.Kastl A, Wilbuer A, Merkel AL, Feng L, Fazio PD, Ocker M, Meggers E. Chem. Commun. 2012;48:1863. doi: 10.1039/c1cc15378a. [DOI] [PubMed] [Google Scholar]
- 15.Mangiapia G, D’Errico G, Simeone L, Irace C, Radulescu A, Di Pascale A, Colonna A, Montesarchio D, Paduano L. Biomaterials. 2012;33:3770. doi: 10.1016/j.biomaterials.2012.01.057. [DOI] [PubMed] [Google Scholar]
- 16.Lentz F, Drescher A, Lindauer A, Henke M, Hilger RA, Hartinger CG, Scheulen ME, Dittrich C, Keppler BK, Jaehde U. Anti-Cancer Drug. 2009;20:97. doi: 10.1097/CAD.0b013e328322fbc5. [DOI] [PubMed] [Google Scholar]
- 17.Han Ang W, Dyson PJ. Eur. J. Inorg. Chem. 2006;2006:4003. [Google Scholar]
- 18.Dougan SJ, Sadler PJ. Inter. J. Chem. 2007;61:704. [Google Scholar]
- 19.Aird RE, Cummings J, Ritchie AA, Muir M, Morris RE, Chen H, Sadler PJ, Jodrell DI. Brit. J. Cancer. 2002;86:1652. doi: 10.1038/sj.bjc.6600290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang F, Chen H, Parsons S, Oswald IDH, Davidson JE, Sadler PJ. Chem.-Eur. J. 2003;9:5810. doi: 10.1002/chem.200304724. [DOI] [PubMed] [Google Scholar]
- 21.Bacac M, Hotze ACG, Schilden K, Haasnoot JG, Pacor S, Alessio E, Sava G, Reedijk J. J Inorg Biochem. 2004;98:402–412. doi: 10.1016/j.jinorgbio.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 22.Groessl M, Hartinger CG, Dyson PJ, Keppler BK. J Inorg. Biochem. 2008;102:1060. doi: 10.1016/j.jinorgbio.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 23.Habtemariam A, Melchart M, Fernández R, Parsons S, Oswald IDH, Parkin A, Fabbiani FPA, Davidson JE, Dawson A, Aird RE, Jodrell DI, Sadler PJ. J Med. Chem. 2006;49:6858. doi: 10.1021/jm060596m. [DOI] [PubMed] [Google Scholar]
- 24.Liu Z, Habtemariam A, Pizarro AM, Fletcher SA, Kisova A, Vrana O, Salassa L, Bruijnincx PCA, Clarkson GJ, Brabec V, Sadler PJ. J Med. Chem. 2011;54:3011. doi: 10.1021/jm2000932. [DOI] [PubMed] [Google Scholar]
- 25.Liu Z, Habtemariam A, Pizarro AM, Clarkson GJ, Sadler PJ. Organometallics. 2011;30:4702. [Google Scholar]
- 26.Liu Z, Salassa L, Habtemariam A, Pizarro AM, Clarkson GJ, Sadler PJ. Inorg. Chem. 2011;50:5777. doi: 10.1021/ic200607j. [DOI] [PubMed] [Google Scholar]
- 27.Scharwitz MA, Ott I, Geldmacher Y, Gust R, Sheldrick WS. J Organomet. Chem. 2008;693:2299. [Google Scholar]
- 28.Schäfer S, Sheldrick WS. J Organomet. Chem. 2007;692:1300. [Google Scholar]
- 29.Schäfer S, Ott I, Gust R, Sheldrick WS. Eur. J. Inorg. Chem. 2007;2007:3034. [Google Scholar]
- 30.Murakami M, Cabral H, Matsumoto Y, Wu S, Kano MR, Yamori T, Nishiyama N, Kataoka K. Sci. Transl. Med. 2011;3:64ra2. doi: 10.1126/scitranslmed.3001385. [DOI] [PubMed] [Google Scholar]
- 31.Han S, Li Z, Zhu J, Han K, Zeng Z, Hong W, Li W, Jia H, Liu Y, Zhuo R, Zhang X. Small [Google Scholar]
- 32.Han S, Liu Y, Nie X, Xu Q, Jiao F, Li W, Zhao Y, Wu Y, Chen C. Small. 2012;8:1596. doi: 10.1002/smll.201102280. [DOI] [PubMed] [Google Scholar]
- 33.Yan L, Zhang J, Lee C, Chen X. Small. 2014;10:4487. doi: 10.1002/smll.201401532. [DOI] [PubMed] [Google Scholar]
- 34.Torchilin VP. Nat. Rev. Drug Discov. 2005;4:145. doi: 10.1038/nrd1632. [DOI] [PubMed] [Google Scholar]
- 35.Kataoka K, Harada A, Nagasaki Y. Adv. Drug Deliv. Rev. 2001;47:113. doi: 10.1016/s0169-409x(00)00124-1. [DOI] [PubMed] [Google Scholar]
- 36.Duan X, Li Y. Small. 2013;9:1521. doi: 10.1002/smll.201201390. [DOI] [PubMed] [Google Scholar]
- 37.He Q, Zhang Z, Gao F, Li Y, Shi J. Small. 2011;7:271. doi: 10.1002/smll.201001459. [DOI] [PubMed] [Google Scholar]
- 38.Kim D, Lee ES, Oh KT, Gao ZG, Bae YH. Small. 2008;4:2043. doi: 10.1002/smll.200701275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yokoyama M. Expert Opin. Drug Deliv. 2010;7:145. doi: 10.1517/17425240903436479. [DOI] [PubMed] [Google Scholar]
- 40.Gupta S, Schade B, Kumar S, Böttcher C, Sharma SK, Haag R. Small. 2013;9:894. doi: 10.1002/smll.201201253. [DOI] [PubMed] [Google Scholar]
- 41.Matsumura Y, Maeda H. Cancer Res. 1986;46:6387. [PubMed] [Google Scholar]
- 42.Attia ABE, Oh P, Yang C, Tan JPK, Rao N, Hedrick JL, Yang YY, Ge R. Small. 2014;10:4281. doi: 10.1002/smll.201401295. [DOI] [PubMed] [Google Scholar]
- 43.Zhang Y, Yin Q, Lu H, Xia H, Lin Y, Cheng J. ACS Macro. Lett. 2013;2:809. doi: 10.1021/mz4003672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matsumura Y, Kataoka K. Cancer Sci. 2009;100:572. doi: 10.1111/j.1349-7006.2009.01103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gong J, Chen M, Zheng Y, Wang S, Wang Y. J Control. Release. 2012;159:312. doi: 10.1016/j.jconrel.2011.12.012. [DOI] [PubMed] [Google Scholar]
- 46.Pluim D, van Waardenburg RAM, Beijnen J, Schellens JM. Cancer Chemother. Pharmacol. 2004;54:71. doi: 10.1007/s00280-004-0773-6. [DOI] [PubMed] [Google Scholar]
- 47.Brabec V, Nováková O. Drug Resist. Update. 2006;9:111. doi: 10.1016/j.drup.2006.05.002. [DOI] [PubMed] [Google Scholar]
- 48.Zhang CX, Lippard SJ. Curr. Opin. Chem. Biol. 2003;7:481. doi: 10.1016/s1367-5931(03)00081-4. [DOI] [PubMed] [Google Scholar]
- 49.Schluep T, Cheng J, Khin K, Davis M. Cancer Chemother. Pharmacol. 2006;57:654. doi: 10.1007/s00280-005-0091-7. [DOI] [PubMed] [Google Scholar]
- 50.Schluep T, Hwang J, Cheng J, Heidel JD, Bartlett DW, Hollister B, Davis ME. Clin. Cancer Res. 2006;12:1606. doi: 10.1158/1078-0432.CCR-05-1566. [DOI] [PubMed] [Google Scholar]
- 51.Xiao J, Duan X, Yin Q, Zhang Z, Yu H, Li Y. Biomaterials. 2013;34:9648. doi: 10.1016/j.biomaterials.2013.08.056. [DOI] [PubMed] [Google Scholar]
- 52.Tang L, Yang X, Yin Q, Cai K, Wang H, Chaudhury I, Yao C, Zhou Q, Kwon M, Hartman JA, Dobrucki IT, Dobrucki LW, Borst LB, Lezmi S, Helferich WG, Ferguson AL, Fan TM, Cheng J. Proc. Natl. Acad. Sci. U.S.A. 2014;111:15344. doi: 10.1073/pnas.1411499111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tang L, Gabrielson NP, Uckun FM, Fan TM, Cheng J. Mol. Pharma. 2013;10:883. doi: 10.1021/mp300684a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sakai-Katoa K, Un K, Nanjo K, Nishiyama N, Kusuhara H, Kataoka K, Kawanishi T, Goda Y, Okuda H. Biomaterials. 2014;35:1347. doi: 10.1016/j.biomaterials.2013.11.027. [DOI] [PubMed] [Google Scholar]
- 55.Cui C, Xue YN, Wu M, Zhang Y, Yu P, Liu L, Zhuo RX, Huang SW. Biomaterials. 2013;34:3858. doi: 10.1016/j.biomaterials.2013.01.101. [DOI] [PubMed] [Google Scholar]
- 56.Fan Y, Li C, Cao H, Li F, Chen D. Biomaterials. 2012;33:4220. doi: 10.1016/j.biomaterials.2012.02.038. [DOI] [PubMed] [Google Scholar]
- 57.Puckett CA, Barton JK. Biochemistry. 2008;47:11711. doi: 10.1021/bi800856t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Puckett CA, Ernst RJ, Barton JK. Dalton Trans. 2010;39:1159. doi: 10.1039/b922209j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aird RE, Cummings J, Ritchie AA, Muir M, Morris RE, Chen H, Sadler PJ, Jodrell DI. Brit. J. Cancer. 2002;86:1652. doi: 10.1038/sj.bjc.6600290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Groessl M, Zava O, Dyson PJ. Metallomics. 2011;3:591. doi: 10.1039/c0mt00101e. [DOI] [PubMed] [Google Scholar]
- 61.Endo K, Ueno T, Kondo S, Wakisaka N, Murono S, Ito M, Kataoka K, Kato Y, Yoshizaki T. Cancer Sci. 2013;104:369. doi: 10.1111/cas.12079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nishiyama N, Kato Y, Sugiyama Y, Kataoka K. Pharm. Res. 2001;18:1035. doi: 10.1023/a:1010908916184. [DOI] [PubMed] [Google Scholar]
- 63.Nishiyama N, Okazaki S, Cabral H, Miyamoto M, Kato Y, Sugiyama Y, Nishio K, Matsumura Y, Kataoka K. Cancer Res. 2003;63:8977. [PubMed] [Google Scholar]
- 64.Plummer R, Wilson RH, Calvert H, Boddy AV, Griffin M, Sludden J, Tilby MJ, Eatock M, Pearson DG, Ottley CJ, Matsumura Y, Kataoka K, Nishiya T. Brit. J. Cancer. 2011;104:593. doi: 10.1038/bjc.2011.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Zeng YB, Yang N, Liu WS, Tang N. J Inorg. Biochem. 2003;97:258. doi: 10.1016/s0162-0134(03)00313-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.