Summary
MGG_03307 is a lectin isolated from Magnaporte oryzae, a fungus that causes devastating rice blast disease. Its function is associated with protecting M. oryzae from the host immune response in plants. To provide the structural basis of how MGG_03307 protects the fungus, crystal structures of its CVNH-LysM module were determined in the absence and presence of GlcNAc-containing cell wall chitin constituents, which can act as pathogen-associated molecular patterns. Our structures revealed that glycan binding is accompanied by a notable conformational change in the LysM domain and that GlcNAc3 and GlcNAc4 are accommodated similarly. GlcNAc5 and GlcNAc6 interact with the LysM domain in multiple conformations, as evidenced by solution nuclear magnetic resonance studies. No dimerization of MoCVNH3 via its LysM domain was observed upon binding to GlcNAc6, unlike in multiple LysM domain-containing proteins. Importantly, we define a specific consensus binding mode for the recognition of GlcNAc oligomers by single LysM domains.
Introduction
During pathogen invasion, the plant host initiates various self-defense mechanisms, one of which is the expression of various chitinases and their buildup at the site of invasion (Boller and Felix, 2009; Medzhitov and Janeway, 1997a, 1997b). The major targets of these chitin-degrading enzymes are the exoskeleton of insects and the cell walls of fungi, yeast, and algae. The fungal cell wall is primarily composed of chitin, an unbranched β-1,4-linked polymer of N-acetylglucosamine (N-GlcNAc or NAG) (Buist et al., 2008), and chitinases have the ability to degrade chitin directly to low molecular weight chitin oligosaccharides, which, in turn, act as pathogen-associated molecular patterns (PAMPs) that trigger a host immune response in plants (Felix et al., 1993; Kombrink et al., 2011; Shibuya et al., 1993, 1996). To suppress PAMP-triggered host immunity, fungi counteractively secrete effector proteins to aid in establishing productive infection (de Jonge et al., 2011; de Jonge and Thomma, 2009; Tanaka et al., 2013). For example, Ecp6 is a fungal effector protein that shields the biotrophic tomato leaf mold fungus Cladosporium fulvum from carbohydrate-degrading enzymes that are released by the invaded plant. Ecp6 does this by sequestering the released chitin fragments that would otherwise trigger plant immunity (Bolton et al., 2008; de Jonge et al., 2010).
Fungal effector proteins, including Ecp6, frequently contain repeating LysM (lysin motif) domains. The LysM domain is a widely distributed protein domain found in all kingdoms of life (Akcavinar et al., 2015; Buist et al., 2008). To date, more than 4,000 proteins have been identified in both prokaryotes and eukaryotes that contain at least one LysM domain. LysM domains are peptidoglycan- and chitin-binding structures that are found in both effector proteins (de Jonge and Thomma, 2009; Kombrink and Thomma, 2013) and enzymes, such as chitinases and other hydrolases (Bielnicki et al., 2006; Gruber et al., 2011; Iizasa et al., 2010; Ohnuma et al., 2008; Onaga and Taira, 2008; Visweswaran et al., 2013; Wan et al., 2008; Wong et al., 2014). Intriguingly, a LysM domain is also present in modular type III CVNHs (a subset of the Cyanovirin-N homology [CVNH] protein family) as an additional carbohydrate-binding module (Percudani et al., 2005). Among all the CVNH family proteins, only type III members possess a LysM domain.
The LysM domain is a relatively small folded protein unit, comprising only 44–65 amino acids. As of March 2015, only ten different LysM structures have been deposited in the PDB (Berman et al., 2000). All ten structures possess the same architecture comprising a βααβ topology in which the two α helices are packed onto the same side of an antiparallel β sheet (Bateman and Bycroft, 2000; Koharudin et al., 2011; Leo et al., 2015; Liu et al., 2012; Mesnage et al., 2014; Sanchez-Vallet et al., 2013). The loop region that connects the second α helix and the second β strand exhibits some variability, and the differences in this region are most likely caused by amino acid sequence variations (Koharudin et al., 2011).
LysM domains are carbohydrate-interaction domains that are known to bind GlcNAc oligomers (GlcNAcn), oligosaccharide units of chitin, a major component of the fungal cell wall (Koharudin et al., 2011; Ohnuma et al., 2008; Sanchez-Vallet et al., 2013; Wong et al., 2014, 2015), and GlcNAc-MurNAc units, major components of the bacterial peptidoglycan cell wall (Mesnage et al., 2014). A single LysM domain can recognize different lengths of GlcNAc oligomers, with increasing apparent binding affinity for increasing oligomer lengths; the optimal ligand for LysM appears to be a GlcNAc pentasaccharide (GlcNAc5) (Koharudin et al., 2011). Using both solution nuclear magnetic resonance (NMR) spectroscopy and X-ray crystallography, the interacting region on LysM for chitin oligosaccharides has been identified (Koharudin et al., 2011; Liu et al., 2012; Sanchez-Vallet et al., 2013; Wong et al., 2015): the binding site primarily comprises two loop regions, one that connects the first β strand to the first α helix and another that connects the second α helix to the second β strand. Residues in these two loops are positioned such that a shallow groove is formed, which can accommodate the carbohydrate. Indeed, crystallographic studies have revealed details of the interaction between GlcNAc4 and the chitin elicitor receptor kinase 1 ectodomain from Arabidopsis thaliana, AtCERK1-ECD (PDB: 4EBZ) (Liu et al., 2012) and with the extracellular protein 6 of Passalora fulva, PfEcp6 (PDB: 4B8V) (Sanchez-Vallet et al., 2013). Both of these proteins possess three LysM domains in a row. Interestingly, in the AtCERK1-ECD structure, only the second LysM domain (residues D97–C155) interacts with GlcNAc4 (Liu et al., 2012), while in the case of PfEcp6, a single GlcNAc4 molecule is simultaneously bound by the first (residues S9–C61) and third LysM (residues E140–D190) domains, contributing to the observed high (pM) binding affinity (Sanchez-Vallet et al., 2013).
Here, we describe the structural basis of fungal cell wall recognition by a type III CVNH protein, using its single LysM domain. Single LysM domains are exclusively observed in this class of modular proteins (Percudani et al., 2005). We describe crystal structures of the CVNH-LysM module from Magnaporte oryzae appressoria-associated protein MGG_03307, also known as MoCVNH-LysM or MoCVNH3, in the absence or presence of N-acetyl chitotriose (GlcNAc3) or N-acetyl chitotetraose (GlcNAc4). The structures of a crystallization positive construct of MoCVNH3, named Mo0v throughout this article, reveal that binding to chitin oligosaccharides introduces a major conformational change in the LysM domain, especially the loop region connecting helix α2 and strand β2. Structural comparison between the GlcNAc3- and GlcNAc4-bound structures shows that both glycans are accommodated similarly: three GlcNAc moieties interact directly and intimately with the protein while GlcNAc4, the fourth sugar moiety, at the non-reducing end of GlcNAc3, is not directly contacting the protein and is exposed to solvent. Solution NMR studies that probed for binding between the protein and GlcNAc5 and GlcNAc6, revealed that MoCVNH3 interacts with these glycans, and that different conformations of the protein are present in the complexes. Our structures demonstrate that the protein can accommodate GlcNAc5 or GlcNAc6 via different binding modes. Furthermore, unlike previous findings for proteins with multiple LysM domains, MoCVNH3 binding to GlcNAc6 is not accompanied by dimerization via its LysM domain. In conjunction with other available data on LysM-GlcNAcn recognition, our results allow us to propose a specific binding consensus motif for the interaction between any single LysM domain and a GlcNAc oligomer. In addition, our structural findings add direct evidence for MGG_03307 functioning as a fungal effector protein.
Results and Discussion
Design and Characterization of an MoCVNH3 Construct for Crystallization
MoCVNH3 is a member of the type III CVNH lectin family, and comprises a single CVNH domain into which a single LysM domain is inserted between the tandem amino acid sequence repeats that make up all CVNH proteins (Figures 1A and 1B). We initially attempted to crystallize wild-type MoCVNH3 protein in complex with varying oligomers of GlcNAc. Unfortunately, however, all our repeated crystallization efforts over the last five years failed, most likely due to the presence of two highly flexible, seven-residue, Gly-rich linkers that connect the CVNH and LysM domains (Koharudin et al., 2011). We therefore generated a series of MoCVNH3 variants, each with linker lengths shorter than the one found in the wild-type protein (Figure 1B). All MoCVNH3 constructs that contained linkers comprising three Gly residues (Mo3G), one Gly residue (Mo1G), or zero Gly (Mo0G) also failed to crystallize, even after exhaustive screening. Of all our various protein constructs, only one, in which we removed the last two residues (TK) at the C-terminal end of the LysM domain and changed the proline to a glycine, resulted in crystals. This protein was named Mo0v (Figure 1B).
Figure 1. Design, Characterization, and Structure of the MoCVNH3 Variant Amenable to Crystallization.
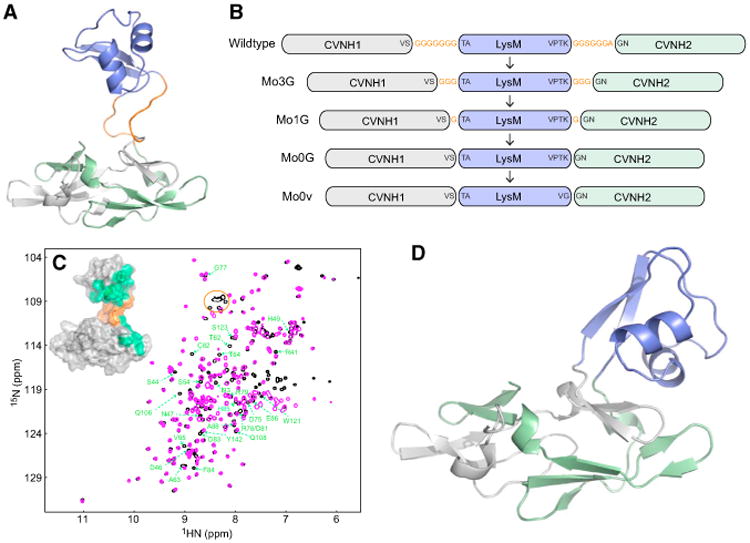
(A)Solution structure of MoCVNH3, with the LysM domain colored blue, the first and second halves of the CVNH tandem repeat amino acid sequence colored gray and green, respectively, and the two glycine-rich, flexible linkers that connect the LysM and CVNH domains colored orange.
(B) Several constructs that were generated in our attempts to crystallize MoCVNH3. Glycine residues were removed from the two flexible linkers in a stepwise manner, and proteins were prepared and set up for crystallization. The construct in the series that yielded crystals was named Mo0v.
(C) Superposition of the 2D 1H-15N HSQC spectra of MoCVNH3 (black) and Mo0v (magenta). Resonances corresponding to the glycine residues in the two flexible linkers of MoCVNH3 are enclosed in an orange circle and are not present in Mo0v. Those residues whose resonances exhibit chemical-shift differences after removal of the two flexible linkers are labeled in green and are mapped onto the NMR structure of MoCVNH3 (inset).
(D) The crystal structure of Mo0v at ∼2.1 Å resolution. The color code is the same as in (A).
Mo0v is soluble and well folded, as indicated by the excellent chemical-shift dispersion and line widths exhibited in its 2D 1H-15N heteronuclear single-quantum coherence (HSQC) spectrum (Figure 1C). Gratifyingly, superposition of the parental MoCVNH3 1H-15N HSQC spectrum with that of Mo0v revealed very similar amide chemical shifts for almost all resonances. The most notable differences in the Mo0v spectrum, compared with the MoCVNH3 spectrum, were the absence of resonances corresponding to the residues in the two Gly-rich linkers of MoCVNH3 (indicated by an orange oval in Figure 1C) and, as expected, changes in the resonances of residues directly adjacent to the missing linkers. Intriguingly, some additional chemical-shift differences were also noted for resonances of several residues from the LysM domain and from subdomain B of the CVNH domain (labeled in green in Figure 1C). They are located relatively far from the linker regions, as shown when mapped onto the structure of the parental MoCVNH3 (inset, Figure 1C). Such additional chemical-shift perturbation, albeit minor, may indicate that removal of the two Gly-rich linkers has altered whether and how the CVNH and the LysM domains communicate; in other words, while no interaction or communication is present in the parental MoCVNH3 (Koharudin et al., 2011), some very transient communication might take place in Mo0v. Indeed, our current crystal structure of Mo0v, determined at 2.09 Å resolution, reveals several close contacts between the LysM domain and subdomain B of the CVNH domain, supporting the possibility that the two domains could also temporarily engage in some contacts in solution (Figure 1D).
The individual domains of Mo0v closely resemble those of the parental MoCVNH3. Superposition of the CVNH domain of the crystal structure of Mo0v with that of the NMR solution structure of MoCVNH3 (residues 7–52 and 104–150 in Mo0v, corresponding to residues 7–52 and 120–166 in the parental protein) resulted in root-mean-square deviation (rmsd) values of ∼1.05 Å and ∼1.38 Å for backbone and heavy atoms, respectively (Figure 2A). Likewise, structural superposition of the LysM domains of the current crystal structure (residues 56–100 in Mo0v) and our previous NMR structure of MoCVNH3 (residues 63–107 in the parental protein) yielded rmsd values of ∼0.75 Å and ∼0.98 Å for backbone and heavy atoms, respectively (Figure 2B). Interestingly, despite the close similarity in the individual domain structures, the relative orientation of the two domains in Mo0v compared with that of the parental MoCVNH3 is distinctly different: as previously noted, the two domains in the parental protein behave as two completely independent units, rather than as a single entity, evidenced by heteronuclear Overhauser effect and relaxation data (Koharudin et al., 2011). For Mo0v, however, this does not appear to be true. Inspection of the crystal structure reveals numerous hydrophobic and polar interactions between the LysM domain and subdomain B of the CVNH domain (Figure 2C). Polar contacts involve hydrogen bonds between the side chain of R83, in the LysM domain, and the backbone carbonyl oxygen O of G101 and the side chain OD1 of N103 of the CVNH domain. In addition, water-mediated contacts between the side chains of D76 of the LysM domain and the backbone amide nitrogen N of N45 of CVNH, between the side chain of E79 of LysM and the backbone amide nitrogen N of N45 or the backbone carbonyl oxygen O of G102 or the backbone carbonyl oxygen O of N103 or the side-chain OG of S107 of CVNH, and between the side chain of R82 of LysM and the backbone carbonyl oxygen O of N103 or the side-chain OG of S107 of CVNH are present (Figure 2C). These data, along with the chemical-shift differences observed for these residues in solution (Figure 1C), support our conclusion that the LysM domain is in contact with the CVNH domain in the Mo0v construct.
Figure 2. Structural Comparison and Carbohydrate Binding.
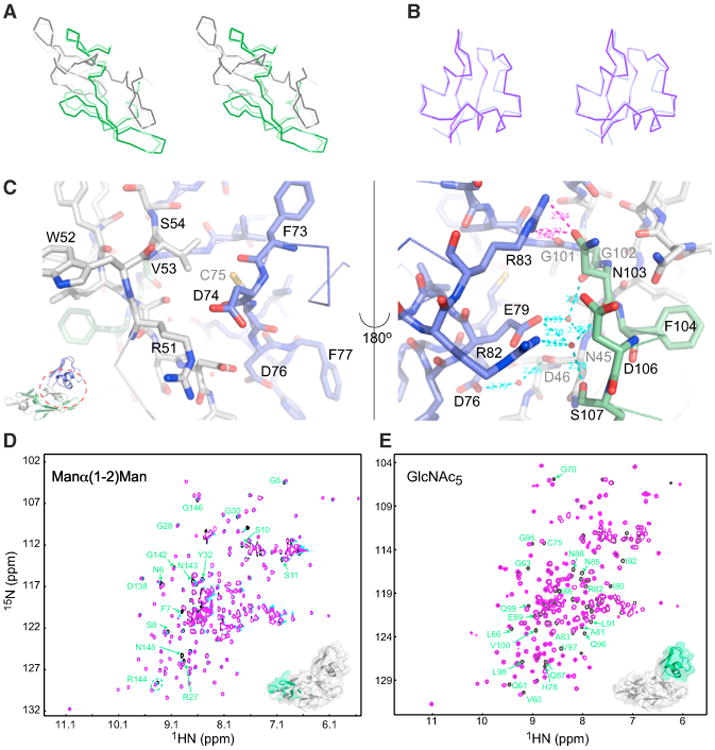
(A and B) Stereo views of best-fit Cα superpositions of the CVNH and LysM domains of MoCVNH3 and Mo0v. The tandem sequence repeats 1 and 2 of the CVNH domain of MoCVNH3 are colored in dark gray and dark green, and those of Mo0v are colored in light gray and light green, respectively (A). The LysM domain of MoCVNH3 and Mo0v is colored in dark and light purple, respectively (B).
(C) Intramolecular direct and water-mediated hydrogen bonds between the LysM and CVNH domains of Mo0v (not present in MoCVNH3).
(D and E) Superpositions of the 2D 1H-15N HSQC spectra of Mo0v in the absence (black, D and E) and presence of Man(α1-2)Man (magenta, D) or GlcNAc5 (magenta, E). Resonances affected by carbohydrate binding are labeled by residue name and number, colored in green and mapped onto the crystal structure of Mo0v (insets in D and E).
To determine whether these structural differences in Mo0v compared with MoCVNH3 affect carbohydrate binding by the two domains, Mo0v was titrated with Man(α1-2)Man and GlcNAc6, and binding was assessed by 2D 1H-15N HSQC spectroscopy. In the parental protein, Man(α1-2)Man addition affects resonances of residues in both CVNH subdomains A and B, while GlcNAc6 only perturbs resonances associated with residues in the LysM domain (Koharudin et al., 2011). Similarly, Mo0v also experiences chemical-shift perturbations upon addition of both Man(α1-2)Man and GlcNAc6 (Figures 2D and 2E). Surprisingly, however, only resonances of residues in subdomain A of the Mo0v CVNH are perturbed by Man(α1-2)Man (Figure 2D), suggesting that CVNH subdomain B is not available for Man(α1-2)Man interaction. The most likely cause for the loss of the binding site for Man(α1-2)Man on subdomain B is due to the fact that this CVNH region of Mo0v is interacting with residues from the LysM domain, similar to what is observed in the crystallographic apo structure (Figure 2C). Despite the observed differences between the parental MoCVNH3 and Mo0v in terms of relative disposition between the CVNH and LysM domains and of the Man(α1-2)Man binding specificity in the CVNH domain, it is important to note that the specificity of our Mo0v construct toward the chitin oligosaccharides is intact and identical to that of the parental MoCVNH3.
Structural Basis for GlcNAc Oligomer Specificity
Given that MoCVNH3 strongly and specifically binds to various GlcNAc oligomers of chitin in solution (Koharudin et al., 2011), we aimed to delineate its recognition specificity at the atomic level by determining the crystal structures of Mo0v in complex with GlcNAc3, GlcNAc4, GlcNAc5, and GlcNAc6. Co-crystallization was carried out using the same crystallization conditions as for the apo structure, with a protein/sugar molar ratio of ∼1:2.5 and a protein concentration ∼10 mg/ml. Co-crystallization with GlcNAc5 and GlcNAc6 appeared to disturb the homogeneity and packing of the crystal (see further discussion below), while GlcNAc3 and GlcNAc4 did not have this effect and yielded well-diffracting crystals with Mo0v.
The structures of GlcNAc3- and GlcNAc4-bound Mo0v revealed one sugar-binding site in the LysM domain of the protein, confirming our previous studies in solution (Koharudin et al., 2011) (Figure 3). High-quality electron density permitted fitting the atomic structures of GlcNAc3 (Figure 3A) and GlcNAc4 (Figure 3B) easily into the map. While the conformation of the CVNH domain in the apo and sugar-bound Mo0v structures is very similar, with rmsd values of ∼0.25 and ∼0.30 Å (between the apo and GlcNAc3-bound chain) and of ∼0.23 and ∼0.35 Å (between the apo and GlcNAc4-bound chain) for backbone and heavy atoms, respectively, the conformation of the LysM domain exhibited some small, but discernible differences. The LysM domain of the apo and GlcNAc3-and GlcNAc4-bound protein exhibited rmsd values of ∼0.53 and ∼0.87 Å and ∼0.75 and ∼1.02 Å for backbone and heavy atoms, respectively. Very little change, however, was seen for the CVNH and LysM domains of the GlcNAc3- and GlcNA4-bound structures, as evidenced by rmsd values of ∼0.26 and ∼0.31 Å and of ∼0.29 and ∼0.45 Å for backbone and heavy atoms, respectively.
Figure 3. Crystal Structures of GlcNAc3 and GlcNAc4 Bound to Mo0v.
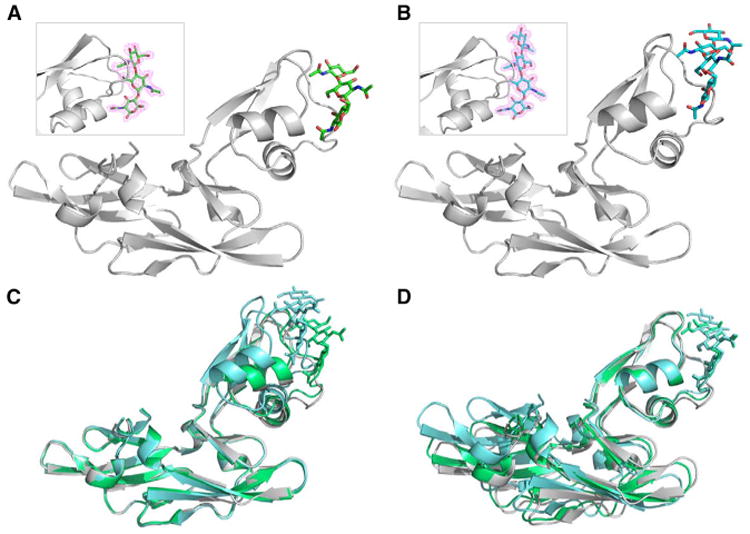
(A and B) Ribbon representation of the protein backbone structure and stick representation of GlcNAc3 (A) and GlcNAc4 (B) in the sugar-bound Mo0v complexes, determined at 2.2 and 1.9 Å resolution, respectively. Electron density maps, contoured at 1.0σ, enclosing the molecular models of GlcNAc3 and GlcNAc4 in stick representation are shown as insets.
(C and D) Superposition of sugar-free (gray), GlcNAc3-bound (green), and GlcNAc4-bound (cyan) structures CVNH (C) and LysM (D) domains.
While the relative orientation between the two domains is very similar in the apo and GlcNAc3-bound structures, a slight reorientation of the CVNH domain relative to the LysM domain is seen in the complex with GlcNAc4 and may be necessary to accommodate the additional GlcNAc moiety in GlcNAc4 (Figures 3C and 3D). This suggests that although the two domains of Mo0v appear to be less flexible in their relative orientation than in the parental MoCVNH3, they are not rigidly fixed, and some degree of plasticity is retained in the short linker in Mo0v, especially for residues V53S54T55 and G101G102N103 (Figure 1B).
The most noticeable conformational change upon ligand binding is seen in the loop region between the second α helix and the second β strand of the LysM domain, comprising residues N85 to I92 (Figure 4A). The associated structural rearrangement includes the loss of two hydrogen bonds between the side chains of R67 and E89, concomitant with side-chain flips of D90 and L91 (Figures 4B–4D). This causes the loop region to “open up” and accommodate the ligand. Loss of the interaction between R67 and E89 permits the accommodation of the acetyl group of the reducing end of GlcNAc3 or GlcNAc4 (labeled as NAG1 in Figures 4E–4H) that is inserted into the binding cleft. Between the reducing end of GlcNAc3/4 and the protein, new hydrogen bonds are formed (discussed below). The flip of the D90 side chain is critical for accommodating the ligand: the side-chain oxygen OD1 hydrogen bonds to the acetyl nitrogen N2 of NAG1 of GlcNAc3/4 (Figures 4E–4H). This is aided by the reorientation of the L91 side chain, which alleviates any steric clash with the ligand.
Figure 4. Details of the Interaction Networks in the GlcNAc3- and GlcNAc4-Bound LysM Domains in the Glycan-Complexed Mo0v Structures.
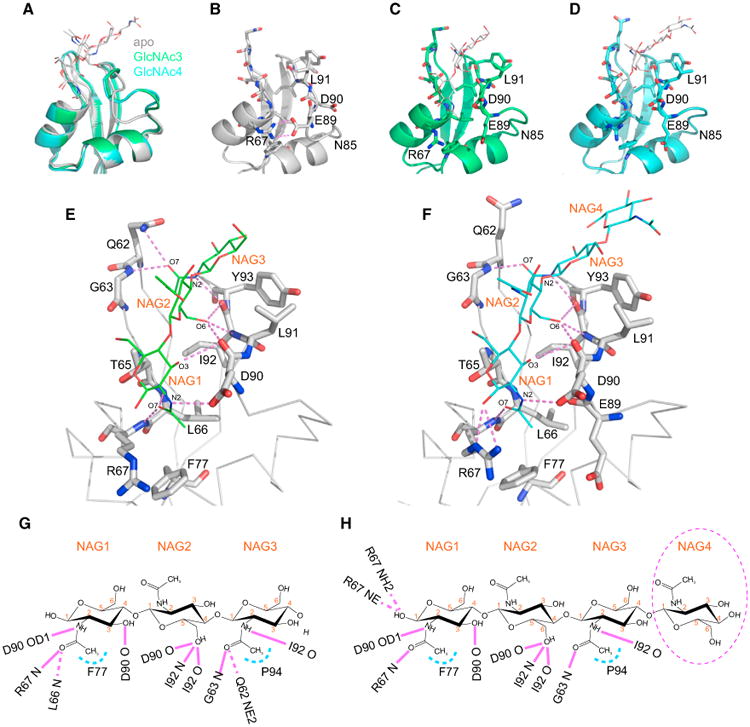
(A) Best-fit superpositions of free (gray), GlcNAc3-bound (green), and GlcNAc4-bound (cyan) structures. The LysM domain and the carbohydrate molecules are shown in ribbon and stick representations, respectively.
(B–D) Free (B), GlcNAc3-bound (C), and GlcNAc4-bound (D) LysM domain structures, illustrating the conformational change in loop α2-β2, comprising residues N85–L91.
(E and F) Intermolecular interactions between the LysM domain and GlcNAc3 (E) and GlcNAc4 (F). LysM residues involved in hydrogen bond formation (indicated by magenta lines) are shown in stick representation and labeled by single-letter code and number. The GlcNAc moiety is labeled as NAG and depicted in line representation.
(G and H)Schematic illustration of the intermolecular atomic contacts between GlcNAc3 (G) or GlcNAc4 (H) and amino acids of the LysM domain. Hydrogen bonds and hydrophobic contacts are indicated by magenta and light blue lines, respectively.
In the GlcNAc3- and GlcNAc4-bound structures, specific contacts primarily involve the main-chain amide groups of the protein. As illustrated in Figures 4E–4H, these include hydrogen bonds between the backbone amide of R67 and the carbonyl O7 of the NAG1 acetyl group, between the backbone carbonyl of D90 and the C3 hydroxyl group of NAG1, between the backbone carbonyl of D90 and the C6 hydroxyl group of NAG2, between the backbone amide of I92 and the C6 hydroxyl group of NAG2, between the backbone carbonyl of I92 and the C6 hydroxyl group of NAG2, between the backbone amide of G63 and the carbonyl O7 of the NAG3 acetyl group, and between the backbone carbonyl of I92 and the acetyl nitrogen N2 of NAG3. These eight hydrogen bonds are conserved in the GlcNAc3-and GlcNAc4-bound structures. In addition to these conserved hydrogen bonds, hydrophobic contacts also appear to play some role in the interaction. For example, the side chains of F77 and P94 are close to the methyl groups of the NAG1 and NAG3 acetyl groups, respectively, and both are present in the GlcNAc3- and GlcNAc4-bound structures.
There are also some hydrogen bonds that are specific to either the GlcNAc3- or GlcNAc4-bound structures. In the GlcNAc3-bound structure, hydrogen bonds between the backbone amide of L66 and the carbonyl O7 of the NAG1 acetyl group (distance of 3.25 Å) and between the side-chain NE2 of Q62 and the carbonyl O7 of the NAG3 acetyl group (distance of 3.30 Å) are seen. In the GlcNAc4-bound structure, the corresponding distances are 3.59 and 5.30 Å, respectively, longer than compatible with hydrogen bonding. Instead, in the GlcNAc4-bound structure, unique hydrogen bonds occur between the side-chain NH2 of R67 and the C1 hydroxyl group of NAG1 (distance of 3.28 Å) and between the side-chain Nε of R67 and the C1 hydroxyl group of NAG1 (distance of 3.10 Å). Thus, equal numbers of hydrogen bonds occur between the protein and the sugars in the GlcNAc3- and GlcNAc4-bound structures, although the identity of the interaction varies to some degree. Interestingly, the additional GlcNAc moiety in GlcNAc4 at the non-reducing end (labeled as NAG4) in the complex structure does not appear to make any additional contacts with the protein and is exposed to solvent (Figures 4F and 4H).
Comparison of Chitin-Binding Specificity of the Mo0v LysM with Other LysM Domains
Understanding chitin and peptidoglycan recognition by LysM domains with regard to specificity, affinity, and binding-site location is of considerable biological and biotechnological interest (de Jonge et al., 2010; Koharudin et al., 2011; Liu et al., 2012; Mesnage et al., 2014; Sanchez-Vallet et al., 2013; Wong et al., 2014, 2015). However, at present only two crystal structures of LysM-containing proteins in complex with GlcNAc oligomers are available in the PDB: the ectodomain of chitin elicitor receptor kinase 1 from A. thaliana, AtCERK1-ECD (PDB: 4EBZ) (Liu et al., 2012) and the extracellular protein 6 of P. fulva, PfEcp6, both in complex with GlcNAc4 (PDB: 4B8V) (Sanchez-Vallet et al., 2013). Both proteins contain three LysM domains, but only one molecule of GlcNAc4 is bound to the protein. In the AtCERK1-ECD structure, GlcNAc4 interacts with the second LysM domain (AtCERK1-ECD LysM2; residues D97–C155) (Figure 5A), and in the PfEcp6 structure, GlcNAc4 is simultaneously bound by the first (PfEcp6 LysM1; residues S9–C61) and third (PfEcp6 LysM3; residues E140–D190) LysM domains (Figure 5B).
Figure 5. Structural and Sequence Alignment of the Chitin Recognition Domains of Mo0v, AtCERK1-ECD, and PfEcp6.
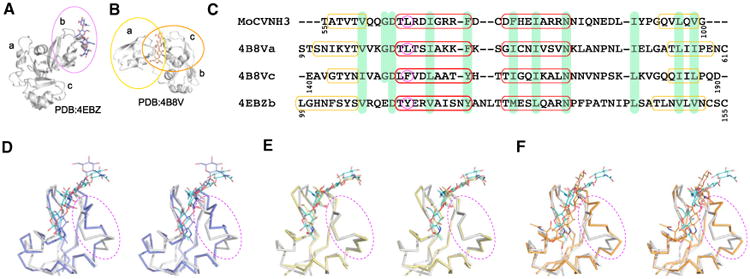
(A and B) Ribbon and stick representations of the AtCERK1-ECD-GlcNAc4 (A) and PfEcp6-GlcNAc4 (B) complex structures. Only a single GlcNAc4 molecule is observed in the complex structures of the three LysM-containing AtCERK1-ECD (PDB: 4EBZ) and the three LysM-containing PfEcp6 (PDB: 4B8V) proteins. GlcNAc4 is bound by the second LysM domain of AtCERK1-ECD (labeled b and enclosed in the purple oval in (A) and simultaneously bound by the first and the third LysM domains of PfEcp6 (labeled a and c and enclosed in the yellow and orange ovals, respectively, in (B).
(C) Sequence alignment of the GlcNAc-binding LysM domains of MoCVNH3 (and Mo0v), the first and the third LysM domains of PfEcp6, and the second LysM domain of AtCERK1-ECD. Residues comprising secondary structural elements in each structure are indicated by yellow and red boxes for β strands and α helices, respectively. Identical residues are highlighted in light green. The pivotal difference in amino acids at the beginning of helix α1 is circled in magenta.
(D–F) Stereo views of best-fit superpositions of GlcNAc4-bound Mo0v (light gray) and the second LysM domain of AtCERK1-ECD (4EBZ, in light purple) (D), the first LysM domain of PfEcp6 (in yellow) (E), and the third LysM domain of PfEcp6 (in orange) (F), respectively. The protein and sugar molecules are displayed as Cα trace and line representations, respectively. The regions of largest conformational change are encircled by dashed purple ovals.
Amino acid sequence alignment of the AtCERK1-ECD LysM2, the PfEcp6 LysM1, and the PfEcp6 LysM3 domains with respect to the Mo0v LysM domain revealed 33%, 23%, and 8% sequence identity, respectively (Figure 5C), although the overall architecture of these LysM domains is well conserved. All structures comprise two α helices and two β strands in a β1-α1-α2-β2 topology. Small differences are seen in the length of β1 and β2, and the number of residues in the loop that connects the α1 and α2 helices is different. Structural superposition yields backbone rmsd values of 0.49, 1.07, and 0.85 Å between the Mo0v LysM domain and the PfEcp6 LysM1 domain, between the Mo0v LysM domain and PfEcp6 LysM3 domain, and between the Mo0v LysM domain and the AtCERK1-ECD LysM2 domain, respectively (Figures 5D–5F). The larger structural difference between the Mo0v LysM domain and the PfEcp6 LysM3 domain or between the Mo0v LysM domain and the AtCERK1-ECD LysM2 domain, compared with the one between the Mo0v LysM domain and the PfEcp6 LysM1 domain in the loop region connecting the α2 helix and β2 strand (Figures 5D–5F), may be related to the identity of the amino acid at position 66 (Figure 5C). In Mo0v, this residue, L66, located at the start of helix α1, makes hydrophobic contacts with residues I86 and I92, both of which are located in the α2-β2 loop. The corresponding three interacting residues are L23, L43, and I49 in the PfEcp6 LysM1 domain, F153, V174, and L179 in the PfEcp6 LysM3 domain, and Y113, F136, and I141 in the AtCERK1-ECD LysM2 domain. Thus, it seems likely that replacement of Leu, present in the Mo0v LysM and the PfEcp6 LysM1 domain, by the larger Phe or Tyr aromatic side chains in the PfEcp6 LysM3 or the AtCERK1-ECD LysM2 domain affects the conformation of the loop region between helix α2 and the β2 strand.
Detailed analysis of the interactions between GlcNAc4 and the AtCERK1-ECD LysM2 domain (Figure 6A), the PfEcp6 LysM1 domain (Figure 6B), and the PfEcp6 LysM3 domain (Figure 6C), respectively, revealed conserved hydrogen bonds that are also present in the current complex of GlcNAc4 with the LysM domain of Mo0v (Figures 4E–4H). Likewise, hydrophobic contacts appear also to be conserved among the various ligand-bound structures. These interactions are detailed below.
Figure 6. Hydrogen Bonding Networks in the GlcNAc4-Bound LysM Structures of AtCERK1-ECD and PfEcp6.
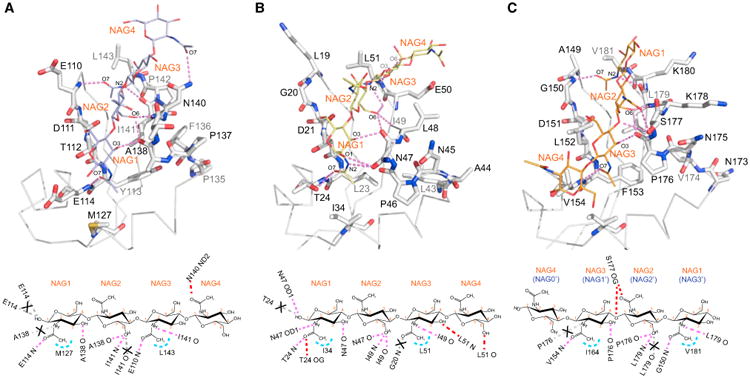
(A–C) Intermolecular interactions between GlcNAc4 and the second LysM domain of AtCERK1-ECD (PDB: 4EBZ) (A), between GlcNAc4 and the first LysM domain of PfEcp6 (PDB: 4B8V) (B), and between GlcNAc4 and the third LysM domain of PfEcp6 (4B8V) (C). The LysM residues involved in hydrogen bond formation (indicated by magenta dashed lines) are shown in stick representation and labeled by single-letter code and number. The GlcNAc moiety is labeled as NAG and depicted in line representation. Below each panel is a schematic representation of the intermolecular contacts formed between GlcNAc4 and the respective LysM domain. Hydrogen bonds and hydrophobic contacts are depicted as magenta and light blue dashed lines, respectively. The hydrogen bonds that are missing when compared with the Mo0v complex structure are shown in gray dashed lines and marked with a cross.
In the GlcNAc4-AtCERK1-ECD LysM2 complex (PDB: 4EBZ), hydrogen bonds are seen between the backbone amide of E114 and the carbonyl O7 of the NAG1 acetyl group, between the backbone carbonyl of A138 and the C3 hydroxyl group of NAG1, between the backbone carbonyl of A138 and the C6 hydroxyl group of NAG2, between the backbone amide of I141 and the C6 hydroxyl group of NAG2, between the backbone amide of E110 and the carbonyl O7 of the NAG3 acetyl group, between the backbone carbonyl of I141 and the acetyl nitrogen N2 of NAG3, and between the side-chain ND2 of N140 and the carbonyl O7 of the NAG4 acetyl group (Figure 6A). In addition, the hydrophobic side chains of M127 and L143 are close to the methyls of the NAG1 and NAG3 acetyl groups (Figure 6A).
In the GlcNAc4-PfEcp6 LysM1 (PDB: 4B8V), hydrogen bonds are present between the side-chain OD1 of N47 and the C1 hydroxyl group of NAG1, between the backbone amide of T24 and the carbonyl O7 of the NAG1 acetyl group, between the sidechain OG of T24 and the carbonyl O7 of the NAG1 acetyl group, between the side-chain OD1 of N47 and the acetyl nitrogen N2 of NAG1, between the backbone carbonyl of N47 and the C3 hydroxyl group of NAG1, between the backbone carbonyl of N47 and the C6 hydroxyl group of NAG2, between the backbone amide of I49 and the C6 hydroxyl group of NAG2, between the backbone carbonyl of I49 and the C6 hydroxyl group of NAG2, between the backbone carbonyl of I49 and the acetyl nitrogen N2 of NAG3, between the backbone amide of L51 and the C3 hydroxyl of NAG3, and between the backbone carbonyl O of L51 and the C6 hydroxyl of NAG4 (Figure 6B). In addition, the hydrophobic side chains of I34 and L51 are in close proximity to the methyls of the NAG1 and NAG3 acetyl groups (Figure 6B).
The interactions between the third LysM domain of PfEcp6 and GlcNAc4 are rather unique, since the GlcNAc4 chain is in reverse orientation when compared with the GlcNAc4-Mo0v, GlcNAc4-AtCERK1-ECD LysM2, or GlcNAc4-PfEcp6 LysM1 complex. For reasons of comparison, we therefore changed the labeling from NAG1, NAG2, NAG3, and NAG4 (used in GlcNAc4-Mo0v, GlcNAc4-AtCERK1-ECD LysM2, and GlcNAc4-PfEcp6 LysM1) to NAG0', NAG1', NAG2', and NAG3', respectively (Figure 6C). NAG0' does not make any contacts with any residues of the third LysM domain of PfEcp6. NAG1', NAG2', and NAG3' engage in interactions similar to those observed in the GlcNAc3-Mo0v complex (Figures 4E and 4G) or in the complexes with GlcNAc4 (Figures 4F, 4H, 6A, and 6B). These involve hydrogen bonds between the backbone amide of N154 and the carbonyl O7 of the NAG1' acetyl group, between the backbone carbonyl of P176 and the C3 hydroxyl group of NAG1', between the backbone carbonyl of P138 and the C6 hydroxyl group of NAG2', between the backbone amide of L179 and the C6 hydroxyl group of NAG2', between the backbone amide of G150 and the carbonyl O7 of the NAG3' acetyl group, and between the backbone carbonyl of L179 and the acetyl nitrogen N2 of NAG3' (Figure 6C). As in other complexes, the hydrophobic side chains of I164 and V181 are close to the methyls of the NAG1' and NAG3' acetyl groups (Figure 6C). In addition, unique hydrogen bonds that are only present in this complex involve the side-chain OG of S177 and the carbonyl O7 of the NAG2' acetyl group, and the side-chain OG of S177 and the C4 hydroxyl of the NAG1' (Figure 6C).
Structural Models of GlcNAc5- and GlcNAc6-Bound Mo0v Based on NMR Spectroscopy Data
All efforts to obtain crystals of Mo0v bound to GlcNAc5 or GlcNAc6 were unsuccessful. However, guided by NMR titration data with GlcNAc4, GlcNAc5, and GlcNAc6, we were able to derive reliable models for the Mo0v-GlcNAc5 and Mo0v-GlcNAc6 complexes, using the structure of the Mo0v-GlcNAc4 complex as a starting point. Titrating Mo0v with GlcNAc4, the affected LysM resonances appear as single bound resonances in the spectrum (Figure 7A), similar to our observations adding GlcNAc3 to Mo0v. On the other hand, addition of GlcNAc5 or GlcNAc6 to Mo0v revealed two sets of resonances for several residues, such as L66 and G70, indicating that these amino acids experience two different chemical environments (Figure 7). Interestingly, the relative amounts of the two states are different in the GlcNAc5-bound protein and the GlcNAc6-bound protein, as judged from relative resonance intensities, with the population shifting from a majority conformation that is similar to the one in the GlcNAc4-bound state toward a predominant alternative conformation present in the GlcNAc6-bound state. Compared with the GlcNAc4 complex, the additional GlcNAc moiety in GlcNAc5 can be considered as occupying either the non-reducing or the reducing end of GlcNAc4. Since the major form of GlcNAc5-bound Mo0v exhibits resonances similar to those seen in GlcNAc4-bound Mo0v, we propose that the additional GlcNAc moiety of GlcNAc5 is located at the non-reducing end of the GlcNAc oligomer (left panel below Figure 7B). This would explain why essentially no frequency differences were observed between the GlcNAc4- and GlcNAc5-bound Mo0v.
Figure 7. Interaction of Mo0v with GlcNAc5 and GlcNAc6 and Structural Models for the Complexes.
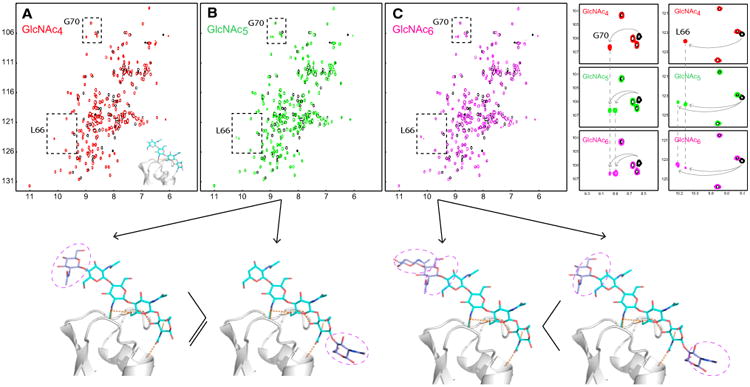
(A–C) Superpositions of 2D 1H-15N HSQC spectra of free (black) and GlcNAc4-bound (red) (A), free (black) and GlcNAc5-bound (green) (B), and free (black) and GlcNAc6-bound (magenta) (C) Mo0v. Doubling of resonances is observed for several resonances of Mo0v (or MoCVNH3) upon interaction with GlcNAc5 (green) and GlcNAc6 (magenta), compared with GlcNAc4 (red). The top right-hand panels illustrate the doubling of resonances for the amide resonances of residues G70 and L66. The relative amount of the alternative conformations (proportional to the resonance intensities) changes from GlcNAc5 (green) to GlcNAc6 (magenta). Based on the NMR titration data, two possible structural models for the Mo0v-GlcNAc5 and Mo0v-GlcNAc6 complexes are indicated below the spectra.
For the GlcNAc6-bound protein, we suggest that the additional two GlcNAc moieties are each positioned on both the reducing and non-reducing ends of the centrally bound GlcNAc4 unit (right panel below Figure 7C). This conclusion is supported by the fact that the majority of amide resonances of GlcNAc6-bound Mo0v residues are exhibiting different chemical shifts. Residues associated with these new resonances are located at the beginning of the first α helix, close to where the NAG1 moiety of GlcNAc4 is positioned in the GlcNAc4-bound Mo0v complex crystal structure (left panel of Figure 7C). Alternatively, a third model of the Mo0v-GlcNAc6 complex could be considered. In this model, the two additional GlcNAc moieties would be positioned at the reducing end of the GlcNAc4 oligomer. If this were the case, the last GlcNAc moiety at the reducing end would have to be exposed to the solvent and not interact with the protein in order to explain our NMR titration data, which did not reveal a third chemical environment for any of the residues. It should also be pointed out that no significant difference in the apparent binding affinity (KD) was observed for GlcNAc5 (28 ± 3 μM) and GlcNAc6 (21 ± 4 μM) binding to MoCVNH3 (Koharudin et al., 2011), indicating that optimal binding to a single LysM domain is reached by a pentasaccharide (GlcNAc5). We therefore suggest that our model for the GlcNAc5 or GlcNAc6 interaction with Mo0v based on the GlcNAc4-Mo0v complex is reasonable and compatible with all the experimental data.
Mo0v Does Not Dimerize on GlcNAc6
It has previously been suggested that dimerization of LysM-containing proteins occurs upon binding to GlcNAc oligomers (Liu et al., 2012). To probe whether this holds true for Mo0v, we first incubated the protein with GlcNAc6 at a 1:1 molar ratio and then added a further equivalent of protein into the initial 1:1 mixture. This 1:2 mixture was analyzed by size-exclusion chromatography coupled with multi-angle light scattering, and 2D 1H-15N HSQC spectroscopy. No larger molecular mass species were detected using light scattering (data not shown), and no linewidth changes were observed for the bound Mo0v. Therefore, it is safe to conclude that a single GlcNAc6 molecule is bound by a single LysM domain, similar to our findings for the parental MoCVNH3. Hence, unlike some multi-LysM-containing proteins, our data show that no GlcNAc6-mediated dimerization of the single LysM-containing Mo0v occurs. This does not, however, discount the possibility that more than one MoCVNH3 can simultaneously bind to long stretches of GlcNAc oligomers (>GlcNAc6). Indeed, our previous studies showed that MoCVNH3 precipitated when interacted with chitin (Koharudin et al., 2011).
Conclusion
To elucidate the structural basis of fungal cell wall protection from carbohydrate-degrading enzymes released by the invaded plant, we solved the crystal structure of a variant of MoCVNH3, named Mo0v, in which the linker regions between the LysM domain and the CVNH domain are shortened. The shorter linker lengths permitted crystallization, which could not be achieved with the parent wild-type protein. Crystal structures of Mo0v were determined for the glycan-free, GlcNAc3-bound, and GlcNAc4-bound protein at atomic resolutions of 2.1, 2.2, and 1.9 Å, respectively. Between free and ligand-bound structures a different conformation of the protein is seen, especially for residues in the loop region connecting helix α2 and strand β2.
Detailed structural comparison of our current GlcNAc4-bound LysM domain with two other GlcNAc4-bound LysM structures (Liu et al., 2012; Sanchez-Vallet et al., 2013) revealed conserved interactions between the proteins and the glycan, despite notable amino acid sequence differences among these proteins. As depicted in Figures 5 and 6, only three GlcNAc moieties, labeled NAG1, NAG2, and NAG3 in this article, are critical and make contact with the protein. This finding allowed us to propose rules that govern LysM-GlcNAc interactions. (1) NAG1 or NAG1' (the GlcNAc moiety at the reducing end of GlcNAc3 or GlcNAc4 in this article) contributes at least two hydrogen bonds via the carbonyl O7 of the acetyl group and the C3 hydroxyl group to the backbone amide (N) of the third amino acid in helix α1 and the backbone carbonyl (O) of a residue in the loop region that connects helix α2 and strand β2, respectively. (2) NAG2 contributes a single hydrogen bond from the C6 hydroxyl group to the backbone carbonyl (O) of the loop residue that is also hydrogen bonded to the C3 hydroxyl group of NAG1. Note that the sugar ring of NAG2 is positioned upside-down with respect to that of NAG1 and, hence, the acetyl group of NAG2 is exposed to the solvent. (3) NAG3 adopts the same conformation as NAG1 and contributes at least one hydrogen bond via the nitrogen atom (N2) of the acetyl group to a backbone carbonyl (O) of another residue in the α2-β2 loop region. (4) NAG4 (or the GlcNAc moiety at the non-reducing end of GlcNAc4) is in the same conformation as NAG2 and makes no direct contacts with any amino acid in the protein. Overall, the acetyl groups of NAG1 and NAG3 and the C6 hydroxyl group of NAG2 are inserted into the binding cleft, which is formed between two loop regions. The first loop connects the β1 strand with helix α1, and the second loop connects helix α2 and strand β2. Hydrogen bonds are formed exclusively from these acetyl and hydroxyl groups to backbone amide or carbonyl groups of the α2-β2 loop residues.
In addition, our NMR solution studies revealed two conformers for the GlcNAc5- and GlcNAc6-bound LysM domain. Since only three GlcNAc moieties make direct contact with the LysM domain, the longer GlcNAc5 or GlcNAc6 oligosaccharides can interact with the LysM domain in two different fashions, with respect to the position of the three GlcNAc units (Figure 7). Interestingly, a recently solved crystal structure of the N-terminal LysM domains from the putative NlpC/P60 D,L endopeptidase from Thermus thermophilus bound to GlcNAc6 (PDB: 4UZ3) also found GlcNAc6 in several alternative conformations (Wong et al., 2015). In this case too, despite the different binding modes, our interaction rules described above are obeyed. Therefore, our proposed structural models for the GlcNAc5- and GlcNAc6-bound complexes, based on our NMR solution data, are clearly valid.
The fact that the LysM domain of MoCVNH3 can contiguously interact with several GlcNAc units, with three units as the central recognition core, suggests that any short GlcNAc oligomers that are created from fungal cell walls, following digestion by host chitinases, will not be able to act as PAMPs and trigger a host immune response. In essence, this permits the fungus to cover up the signal that triggers the host's anti-pathogen response and enhances the probability of a successful fungal infection.
Experimental Procedures
Protein Expression, Purification, and Crystallization
To enable crystallization of MoCVNH3, several constructs were devised by removing the flexible linkers between the LysM and CVNH domains, which previously were shown to interfere with crystallization. The Mo0v construct (see details below) was expressed and purified as described previously for wild-type MoCVNH3 (Koharudin et al., 2011). In brief, Escherichia coli Rosetta2 (DE3) cells (Novagen) were transformed with the pET-15b(+)-Mo0v vector for protein expression. Cells were initially grown at 37°C, induced with 1 mM isopropyl β-D-1-thiogalactopyranoside at an OD600 of ∼0.9, and further grown for ∼18 hrat 16°C for protein expression. Cells were harvested by centrifugation, resuspended in lysis buffer (20 mM Tris-HCl [pH 8.0], 150 mM NaCl, and 1 mM NaN3) and lysed by sonication. The cell lysate was centrifuged to remove cell debris and, after centrifugation, the supernatant was loaded onto an Ni2+-derivatized His Trap column (GE Healthcare), pre-equilibrated with loading buffer (20 mM Tris-HCl [pH 8.0], 150 mM NaCl, and 1 mM NaN3). The protein was eluted using a linear (20–500 mM) imidazole gradient in the same loading buffer and protein-containing fractions were collected for thrombin digestion in 20 mM Tris-HCl buffer (pH 8.0), 100 mM NaCl, and 1 mM NaN3 to remove the N-terminal His tag (13 of the 20 tag residues were removed). Final purification of the cleaved protein was carried out by gel filtration on a Superdex75 column (GE Healthcare) in 20 mM sodium acetate buffer (pH 5.0), 100 mM NaCl, and 1 mM NaN3. Fractions containing pure protein were collected, buffer exchanged into 20 mM sodium acetate (pH 5.0), 20 mM NaCl, 1 mM NaN3, 90%:10% H2O/D2O, and concentrated up to 10 mg/ml using Centriprep devices (Millipore). For 15N isotopic labeling, the bacterial cell culture was grown in modified minimal medium, containing 15NH4Cl as the sole nitrogen source.
Crystallization trials of apo Mo0v were carried out at room temperature by the sitting-drop vapor diffusion method, using 2 μl of protein in 2 μl of reservoir solution, with an initial protein concentration of ∼10 mg/ml. Well-diffracting crystals were obtained in ∼8%–10% (w/v) polyethylene glycol 3350, 0.1 M sodium phosphate-citrate buffer (pH 4.2), and 0.2 M NaCl after approximately 1 week. Crystals of GlcNAc3- and GlcNAc4-bound Mo0v were obtained in the presence of 2.5 M excess of glycan by co-crystallization using the same crystallization conditions as described above for the apo protein.
Diffraction Data Collection and Structure Determination
X-Ray diffraction data for apo and ligand-bound crystals of Mo0v were collected up to 2.09, 2.20, and 1.90 Å resolution, respectively, on flash-cooled (−180°C) crystals, using a Rigaku FR-E generator with a Saturn 944 CCD detector at a wavelength corresponding to the copper edge (1.54 Å). All diffraction data were processed, integrated, and scaled using d*TREK software (Pflugrath, 1999), and eventually converted to mtz format using the CCP4 package (Collaborative Computational Project Number 4, 1994). A summary of unit-cell dimensions for all crystals is provided in Table 1. The apo Mo0v crystal and both glycan-bound crystals contained one polypeptide molecule per asymmetric unit with very similar cell dimensions (Table 1).
Table 1. Data Collection, Refinement, and Ramachandran Statistics for Free, GlcNAc3-bound, and GlcNAc4-bound Mo0v.
| Free Mo0va,b,c | GlcNAc3-Mo0va,c | GlcNAc4-Mo0va,c | |
|---|---|---|---|
| Data Collection | |||
| Space group | P21 | P21 | P21 |
| Cell dimensions | |||
| a, b, c (Å) | 37.41, 51.08, 43.24 | 36.75, 50.60, 43.61 | 36.72, 50.28, 43.13 |
| α, β, γ (°) | 90, 110.62, 90 | 90, 109.86, 90 | 90, 103.53, 90 |
| ASU content | 1 | 1 | 1 |
| Wavelength (Å) | 1.54 | 1.54 | 1.54 |
| Resolution (Å) | 40.48–2.09 (2.16–2.09)d | 41.02–2.20 (2.28–2.20)d | 41.94–1.90 (1.97–1.90)d |
| Rmerge | 0.073 (0.179)d | 0.073 (0.234)d | 0.098 (0.257)d |
| Rmeas | 0.088 (0.252)d | 0.082 (0.276)d | 0.110 (0.320)d |
| <I/σI> | 10.5 (2.1)d | 13.0 (3.5)d | 9.0 (2.2)d |
| Completeness (%) | 99.2 (96.7)d | 98.7 (99.9)d | 98.7 (97.0)d |
| Redundancy | 2.66 (1.68)d | 4.69 (3.50)d | 4.03 (2.51)d |
| Refinement | |||
| Resolution (Å) | 40.48–2.09 (2.14–2.09)d | 41.02–2.20 (2.26–2.20)d | 41.94–1.90 (1.95–1.90)d |
| No. of unique reflections | 9,051 | 7,630 | 11,980 |
| No. of test reflectionse | 906 | 743 | 1,180 |
| Rwork/Rfree | 0.220/0.281 (0.267/0.355)d | 0.232/0.280 (0.318/0.366)d | 0.224/0.256 (0.242/0.326)d |
| No. of atoms | |||
| Protein | 1,227 | 1,201 | 1,201 |
| Ligand/ion | – | 43 | 57 |
| Water | 102 | 50 | 94 |
| Average B factors (Å2) | |||
| Protein | 34.25 | 36.83 | 31.51 |
| Ligand/ion | – | 42.50 | 29.49 |
| Water | 39.54 | 42.34 | 42.01 |
| Rmsd | |||
| Bond lengths (Å) | 0.007 | 0.008 | 0.012 |
| Bond angles (°) | 1.034 | 1.196 | 1.409 |
| MolProbity Statisticsf | |||
| All-atom clashscore | 1.7 | 7.6 | 4.2 |
| Rotamer outliers (%) | 0.8 | 2.4 | 1.6 |
| Cβ deviation | 0 | 0 | 0 |
| MolProbity score | 0.93 (100 percentile) | 1.84 (92 percentile) | 1.57 (93 percentile) |
| Ramachandranf | |||
| Favored region (%) | 99.3 | 97.3 | 96.6 |
| Allowed region (%) | 0.7 | 2.7 | 3.4 |
| Outliers (%) | 0 | 0 | 0 |
| PDB ID | 5C8O | 5C8P | 5C8Q |
Data were obtained from the best diffracting crystal according to crystallization condition (see text for details) and collected at −180°C.
Solved by molecular replacement using the crystal structures of the pseudomonomer of CV-N (PDB: 1L5B) and of the LysM domain (PDB: 1Y7M).
Refinement was carried out using Refmac5.
Values in parentheses are for highest-resolution shell.
Random selection.
Values are obtained from MolProbity.
The crystallographic phase for apo Mo0v crystals was determined by molecular replacement using the coordinates of a pseudomonomeric structure derived from the domain-swapped P51G CV-N structure (PDB: 1L5B) (Barrientos et al., 2002) and those of the Bacillus subtilis YkuD LysM domain structure (residues L2-I45; PDB: 1Y7M) (Bielnicki et al., 2006) as structural probes in Phaser (McCoy, 2007). After generation of the initial model, the chain was rebuilt using the program Coot (Emsley and Cowtan, 2004). Iterative refinement was carried out by alternating between manual rebuilding in Coot (Emsley and Cowtan, 2004) and automated refinement in REFMAC (Murshudov et al., 1997). The structures of the glycan-bound protein were solved similarly, in these cases using the apo Mo0v structure as search probe for molecular replacement.
All final models exhibited clear electron density for all residues of Mo0v (G1-C151). The three additional N-terminal amino acids derived from the His tag exhibited no density. The final apo Mo0v structure is well defined to 2.09 Å resolution with an R factor of 22.0% and a free R of 28.1%. All residues are located in both favored and allowed regions of the Ramachandran plot, respectively, as evaluated by MolProbity (Davis et al., 2007). Similarly, the final ligand-bound protein structures are well defined to 2.20 and 1.90 Å resolution, respectively, with R values of 23.2% and 22.4% and free R values of 28.0% and 25.6% for the GlcNAc3- and GlcNAc4-bound structures, respectively. For these structures too, no residues are located in the disallowed region the Ramachandran plot, as evaluated by MolProbity (Davis et al., 2007). Data obtained for the glycan complexes contained additional density not accounted for by the polypeptide chain, allowing placement of GlcNAc3 or GlcNAc4 into the binding site of the LysM domain of Mo0v. A summary of all data collection parameters as well as pertinent structural statistics for all the structures is provided in Table 1. The atomic coordinates and diffraction data for apo, GlcNAc3-, and GlcNAc4-bound Mo0v structures have been deposited in the Research Collaboratory for Structural Bioinformatics PDB under accession codes PDB: 5C8O, 5C8P, and 5C8Q, respectively. Structural figures were generated using the program PyMOL (DeLano, 2002).
Oligosaccharides
Man(α1-2)Man, GlcNAc3 (N,N′,N″-triacetylchitotriose or β-D-GlcNAc-(1–4)-β-D-GlcNAc-(1–4)-D-GlcNAc), GlcNAc4 (N,N′,N″,N′″-tetraacetylchitotetraose or β-D-GlcNAc-(1–4)-β-D-GlcNAc-(1–4)-D-GlcNAc-(1–4)-D-GlcNAc), GlcNAc5 (N,N′,N″,N′″,N″″-pentaacetylchitopentaose or β-D-GlcNAc-(1–4)-β-D-GlcNAc-(1–4)-β-D-GlcNAc-(1–4)-D-GlcNAc-(1–4)-D-GlcNAc), and GlcNAc6 (N,N′,N″, N′″,-N″″,N′″″-hexaacetylchitohexaose or β-D-GlcNAc-(1–4)-β-D-GlcNAc-(1–4)-β-D-GlcNAc-(1–4)-β-D-GlcNAc-(1–4)-D-GlcNAc-(1–4)-D-GlcNAc) were all purchased from V-lab and dissolved in NMR buffer (20 mM sodium acetate, 20 mM NaCl, 1 mM NaN3, 90%:10% H2O/D2O, pH 5.0). Note that GlcNAc is also interchangeable with NAG (which stands for N-acetylglucosamine) throughout this manuscript, and that the reducing end of GlcNAc oligomers is arbitrarily assigned as NAG1.
Carbohydrate Binding Studies by NMR Spectroscopy
3D CBCANH and CBCA(CO)NH spectra (Bax and Grzesiek, 1993) were recorded at 25°C on a 13C/15N-labeled sample in 20 mM sodium acetate, 20 mM NaCl, 1 mM NaN3, 90%:10% H2O/D2O (pH 5.0) at a protein concentration of ∼0.5 mM and analyzed to yield complete backbone chemical-shift assignment of apo Mo0v, using a Bruker AVANCE 900 spectrometer, equipped with a 5-mm triple-resonance, z-axis gradient cryoprobe.
Binding of Man(α1-2)Man, GlcNAc4, GlcNAc5, and GlcNAc6 to the protein was probed at 25°C by 2D 1H-15N HSQC spectroscopy, using 0.050 mM 15N-labeled protein in 20 mM sodium acetate (pH 5.0), 20 mM NaCl, 1 mM NaN3, and 90%:10% H2O/D2O. All 2D 1H-15N HSQC spectra were recorded on a Bruker AVANCE 700 spectrometer, equipped with a 5-mm triple-resonance, z-axis gradient cryoprobe. For titrations with Man(α1-2)Man, spectra were recorded in the absence and presence of the carbohydrate with molar ratios of 1:30 and 1:60. Similarly, for titrations with GlcNAc oligomers, spectra were recorded in the absence or presence of 5-fold molar excess of GlcNAc4 and 2.5-fold molar excess of GlcNAc5 or GlcNAc6. All spectra were processed with NMRPipe (Delaglio et al., 1995) and analyzed using NMRView (Johnson and Blevins, 1994).
Highlights.
A single LysM domain can efficiently interact with chitin constituents
Crystal structures of apo, GlcNAc3-, and GlcNAc4-bound MoCVNH3 were determined
Mannose and GlcNAc oligomers bind to their respective recognition domains selectively
Consensus binding mode between GlcNAc oligomers and a single LysM domain is defined
Acknowledgments
The authors thank Dr. William Furey for helpful discussion, Doowon Jee and Mike Delk for X-ray and NMR technical support, Dr. Teresa Brosenitsch for critical reading of the manuscript, and Dr. Simone Ottonello for providing the original plasmid of MoCVNH3. K.T.D. was a recipient of a NIH T32 predoctoral traineeship (GM088119). This work was supported by a NIH grant to A.M.G. (GM080642).
References
- Akcavinar GB, Kappel L, Sezerman OU, Seidl-Seiboth V. Molecular diversity of LysM carbohydrate-binding motifs in fungi. Curr Genet. 2015;61:103–113. doi: 10.1007/s00294-014-0471-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrientos LG, Louis JM, Botos I, Mori T, Han Z, O'Keefe BR, Boyd MR, Wlodawer A, Gronenborn AM. The domain-swapped dimer of cyanovirin-N is in a metastable folded state: reconciliation of X-ray and NMR structures. Structure. 2002;10:673–686. doi: 10.1016/s0969-2126(02)00758-x. [DOI] [PubMed] [Google Scholar]
- Bateman A, Bycroft M. The structure of a LysM domain from E. coli membrane-bound lytic murein transglycosylase D (MltD) J Mol Biol. 2000;299:1113–1119. doi: 10.1006/jmbi.2000.3778. [DOI] [PubMed] [Google Scholar]
- Bax A, Grzesiek S. Methodological advances in protein NMR. Acc Chem Res. 1993;26:131–138. [Google Scholar]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielnicki J, Devedjiev Y, Derewenda U, Dauter Z, Joachimiak A, Derewenda ZS. B. subtilis ykuD protein at 2.0 A resolution: insights into the structure and function of a novel, ubiquitous family of bacterial enzymes. Proteins. 2006;62:144–151. doi: 10.1002/prot.20702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller T, Felix G. A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- Bolton MD, van Esse HP, Vossen JH, de Jonge R, Stergiopoulos I, Stulemeijer IJ, van den Berg GC, Borras-Hidalgo O, Dekker HL, de Koster CG, et al. The novel Cladosporium fulvum lysin motif effector Ecp6 is a virulence factor with orthologues in other fungal species. Mol Microbiol. 2008;69:119–136. doi: 10.1111/j.1365-2958.2008.06270.x. [DOI] [PubMed] [Google Scholar]
- Buist G, Steen A, Kok J, Kuipers OP. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol Microbiol. 2008;68:838–847. doi: 10.1111/j.1365-2958.2008.06211.x. [DOI] [PubMed] [Google Scholar]
- Collaborative Computational Project Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, Richardson DC. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–W383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jonge R, Thomma BP. Fungal LysM effectors: extinguishers of host immunity? Trends Microbiol. 2009;17:151–157. doi: 10.1016/j.tim.2009.01.002. [DOI] [PubMed] [Google Scholar]
- de Jonge R, van Esse HP, Kombrink A, Shinya T, Desaki Y, Bours R, van der Krol S, Shibuya N, Joosten MH, Thomma BP. Conserved fungal LysM effector Ecp6 prevents chitin-triggered immunity in plants. Science. 2010;329:953–955. doi: 10.1126/science.1190859. [DOI] [PubMed] [Google Scholar]
- de Jonge R, Bolton MD, Thomma BP. How filamentous pathogens co-opt plants: the ins and outs of fungal effectors. Curr Opin Plant Biol. 2011;14:1–7. doi: 10.1016/j.pbi.2011.03.005. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Nmrpipe—a multidimensional spectral processing system based on Unix Pipes. J Biomol NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL Molecular Graphics System. Version MacPyMol (Schrödinger, LLC) 2002 [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Felix G, Regenass M, Boller T. Specific perception of subnanomolar concentrations of chitin fragments by tomato cells: induction of extracellular alkalinization, changes in protein phosphorylation, and establishment of a refractory state. Plant J. 1993;4:407–416. [Google Scholar]
- Gruber S, Vaaje-Kolstad G, Matarese F, Lopez-Mondejar R, Kubicek CP, Seidl-Seiboth V. Analysis of subgroup C of fungal chitinases containing chitin-binding and LysM modules in the mycoparasite Trichoderma atroviride. Glycobiology. 2011;21:122–133. doi: 10.1093/glycob/cwq142. [DOI] [PubMed] [Google Scholar]
- Iizasa E, Mitsutomi M, Nagano Y. Direct binding of a plant LysM receptor-like kinase, LysM RLK1/CERK1, to chitin in vitro. J Biol Chem. 2010;285:2996–3004. doi: 10.1074/jbc.M109.027540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson BA, Blevins RA. NMRView—a computer-program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- Koharudin LM, Viscomi AR, Montanini B, Kershaw MJ, Talbot NJ, Ottonello S, Gronenborn AM. Structure-function analysis of a CVNH-LysM lectin expressed during plant infection by the rice blast fungus Magnaporthe oryzae. Structure. 2011;19:662–674. doi: 10.1016/j.str.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kombrink A, Thomma BP. LysM effectors: secreted proteins supporting fungal life. PLoS Pathog. 2013;9:e1003769. doi: 10.1371/journal.ppat.1003769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kombrink A, Sanchez-Vallet A, Thomma BP. The role of chitin detection in plant-pathogen interactions. Microbes Infect. 2011;13:1168–1176. doi: 10.1016/j.micinf.2011.07.010. [DOI] [PubMed] [Google Scholar]
- Leo JC, Oberhettinger P, Chaubey M, Schutz M, Kuhner D, Bertsche U, Schwarz H, Gotz F, Autenrieth IB, Coles M, Linke D. The Intimin periplasmic domain mediates dimerisation and binding to peptidoglycan. Mol Microbiol. 2015;95:80–100. doi: 10.1111/mmi.12840. [DOI] [PubMed] [Google Scholar]
- Liu T, Liu Z, Song C, Hu Y, Han Z, She J, Fan F, Wang J, Jin C, Chang J, et al. Chitin-induced dimerization activates a plant immune receptor. Science. 2012;336:1160–1164. doi: 10.1126/science.1218867. [DOI] [PubMed] [Google Scholar]
- McCoy AJ. Solving structures of protein complexes by molecular replacement with phaser. Acta Crystallogr D Biol Crystallogr. 2007;63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R, Janeway CA., Jr Innate immunity: impact on the adaptive immune response. Curr Opin Immunol. 1997a;9:4–9. doi: 10.1016/s0952-7915(97)80152-5. [DOI] [PubMed] [Google Scholar]
- Medzhitov R, Janeway CA., Jr Innate immunity: the virtues of a nonclonal system of recognition. Cell. 1997b;91:295–298. doi: 10.1016/s0092-8674(00)80412-2. [DOI] [PubMed] [Google Scholar]
- Mesnage S, Dellarole M, Baxter NJ, Rouget JB, Dimitrov JD, Wang N, Fujimoto Y, Hounslow AM, Lacroix-Desmazes S, Fukase K, et al. Molecular basis for bacterial peptidoglycan recognition by LysM domains. Nat Commun. 2014;5:4269. doi: 10.1038/ncomms5269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- Ohnuma T, Onaga S, Murata K, Taira T, Katoh E. LysM domains from Pteris ryukyuensis chitinase-A: a stability study and characterization of the chitin-binding site. J Biol Chem. 2008;283:5178–5187. doi: 10.1074/jbc.M707156200. [DOI] [PubMed] [Google Scholar]
- Onaga S, Taira T. A new type of plant chitinase containing LysM domains from a fern (Pteris ryukyuensis): roles of LysM domains in chitin binding and antifungal activity. Glycobiology. 2008;18:414–423. doi: 10.1093/glycob/cwn018. [DOI] [PubMed] [Google Scholar]
- Percudani R, Montanini B, Ottonello S. The anti-HIV cyanovirin-N domain is evolutionarily conserved and occurs as a protein module in eukaryotes. Proteins. 2005;60:670–678. doi: 10.1002/prot.20543. [DOI] [PubMed] [Google Scholar]
- Pflugrath JW. The finer things in X-ray diffraction data collection. Acta Crystallogr D Biol Crystallogr. 1999;55:1718–1725. doi: 10.1107/s090744499900935x. [DOI] [PubMed] [Google Scholar]
- Sanchez-Vallet A, Saleem-Batcha R, Kombrink A, Hansen G, Valkenburg DJ, Thomma BP, Mesters JR. Fungal effector Ecp6 outcompetes host immune receptor for chitin binding through intrachain LysM dimerization. Elife. 2013;2:e00790. doi: 10.7554/eLife.00790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibuya N, Kaku H, Kuchitsu K, Maliarik MJ. Identification of a novel high-affinity binding site for N-acetylchitooligosaccharide elicitor in the membrane fraction from suspension-cultured rice cells. FEBS Lett. 1993;329:75–78. doi: 10.1016/0014-5793(93)80197-3. [DOI] [PubMed] [Google Scholar]
- Shibuya N, Ebisu N, Kamada Y, Kaku H, Conn J, Ito Y. Localization and binding characteristics of a high-affinity binding site for N-acetylchitooligosaccharide elicitor in the plasma membrane from suspension-cultured rice cells suggest a role as a receptor for the elicitor signal at the cell surface. Plant Cell Physiol. 1996;37:894–898. [Google Scholar]
- Tanaka K, Nguyen CT, Liang Y, Cao Y, Stacey G. Role of LysM receptors in chitin-triggered plant innate immunity. Plant Signal Behav. 2013;8:e22598. doi: 10.4161/psb.22598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visweswaran GR, Steen A, Leenhouts K, Szeliga M, Ruban B, Hesseling-Meinders A, Dijkstra BW, Kuipers OP, Kok J, Buist G. AcmD, a homolog of the major autolysin AcmA of Lactococcus lactis, binds to the cell wall and contributes to cell separation and autolysis. PLoS One. 2013;8:e72167. doi: 10.1371/journal.pone.0072167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan J, Zhang XC, Neece D, Ramonell KM, Clough S, Kim SY, Stacey MG, Stacey G. A LysM receptor-like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell. 2008;20:471–481. doi: 10.1105/tpc.107.056754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong JEMM, Alsarraf HM, Kaspersen JD, Pedersen JS, Stougaard J, Thirup S, Blaise M. Cooperative binding of LysM domains determines the carbohydrate affinity of a bacterial endopeptidase protein. FEBS J. 2014;281:1196–1208. doi: 10.1111/febs.12698. [DOI] [PubMed] [Google Scholar]
- Wong JEMM, Midtgaard SR, Gysel K, Thygesen MB, Sorensen KK, Jensen KJ, Stougaard J, Thirup S, Blaise M. An intermolecular binding mechanism involving multiple lysM domains mediates carbohydrate recognition by an endopeptidase. Acta Crystallogr. 2015;D71:592–605. doi: 10.1107/S139900471402793X. [DOI] [PMC free article] [PubMed] [Google Scholar]


