Summary
Mass isotopomer multi-ordinate spectral analysis (MIMOSA) is a step-wise flux analysis platform to measure discrete glycolytic and mitochondrial metabolic rates. Importantly, direct citrate synthesis rates were obtained by deconvolving the mass spectra generated from [U-13C6]-D-glucose labeling for position-specific enrichments of mitochondrial acetyl-CoA, oxaloacetate and citrate. Comprehensive steady-state and dynamic analyses of key metabolic rates (pyruvate dehydrogenase, β-oxidation, pyruvate carboxylase, isocitrate dehydrogenase and PEP/pyruvate cycling) were calculated from the position-specific transfer of 13C from sequential precursors to their products. Important limitations of previous techniques were identified. In INS-1 cells, citrate synthase rates correlated with both insulin secretion and oxygen consumption. Pyruvate carboxylase rates were substantially lower than previously reported but showed the highest fold change in response to glucose stimulation. In conclusion, MIMOSA measures key metabolic rates from the precursor/product position-specific transfer of 13C label between metabolites and has broad applicability to any glucose-oxidizing cell.
Introduction
During his Nobel banquet address in 1953, Sir Hans Krebs modestly understated the importance of citric acid cycle metabolism saying1: “I am convinced than an understanding of the process of energy production will eventually help us in solving some of the practical problems of medicine.” From bacteria to humans, the cores of both oxidative and synthetic metabolism revolve around the sequential enzymatic reactions of the citric acid cycle (tricarboxylic acid, TCA, or Krebs cycle). It is the cantus firmus upon which the rest of organismal metabolism is composed. Each measure of the cycle is described by the ligation of oxaloacetate (OAA) with acetyl-coenzyme A (AcCoA) in order to generate citrate by the mitochondrial matrix enzyme citrate synthase (CS).
Considered in its entirety, the TCA cycle is primarily oxidative. As such, the TCA rate often experimentally correlates to varying degrees with O2 consumption, CO2 production, mitochondrial membrane potential, and ATP synthesis. However, factors such as ROS production, proton leak, ion translocation and citrate lyase activity divert the output of citrate synthesis away from oxidative phosphorylation. Similarly, metabolism flowing through partial segments of the cycle (e.g., into synthetic, signaling, or energy producing pathways) via exchange, entrance (anaplerosis), export (cataplerosis), or futile cycles may have little or no linkage with the more traditional oxidative readouts or even CS. Various isotope-labeling methods have estimated isolated features of the TCA cycle from the interpretation of one or a few metabolite tracers (Bequette et al., 2006). For instance, hyperpolarized NMR has detected positional transfer of label from [1,2-13C2]pyruvate to glutamate via both oxidative and anaplerotic pathways (Chen et al., 2012). While qualitatively informative, these techniques may miss certain aspects of mitochondrial metabolism in part because of unexpected dilutions coming from unlabeled metabolites. A more comprehensive integrated pathway analysis is needed that can account for segmental as well as multi-turn TCA fluxes to allow the quantification of the pleiotropic mitochondrial metabolic pathways, especially in tissues that are not strictly oxidative.
An important early advance in metabolic flux tracking was the use of NMR-based 13C spin-labels for isotopomer analysis (de Graaf et al., 2011; Schryer et al., 2009). Here a major strength is the ability to determine precise position-specific labeling (isotopomeric) patterns of a metabolite. Such data informs the modeling of intracellular metabolic rates from 13C-13C J-coupling patterns that identifies the enrichments of neighboring carbons. Recently there has been an increase in the use of 13C and other mass-labels to chase metabolic fluxes through mass isotopologue (position-independent) analysis (Buescher et al., 2015). Isotopomer analysis, however, need not be limited to NMR, and strategic fragmentation of metabolites can yield position-specific labeling of some metabolites. Key advantages of mass spectroscopy (MS) include simultaneous measurement of both labeled and unlabeled metabolites, improved sensitivity, and the potential to analyze individual isotopomers.
Here, we present a comprehensive and direct platform to track the flow of mass isotope labels through the sequential reactions of glycolysis and the TCA cycle via Mass Isotopomeric Multi-Ordinate Spectral Analysis (MIMOSA). MIMOSA deciphers the glucose-dependent changes in oxidative and non-oxidative metabolic rates in cultured cells.
Results
Isotopomer Analysis
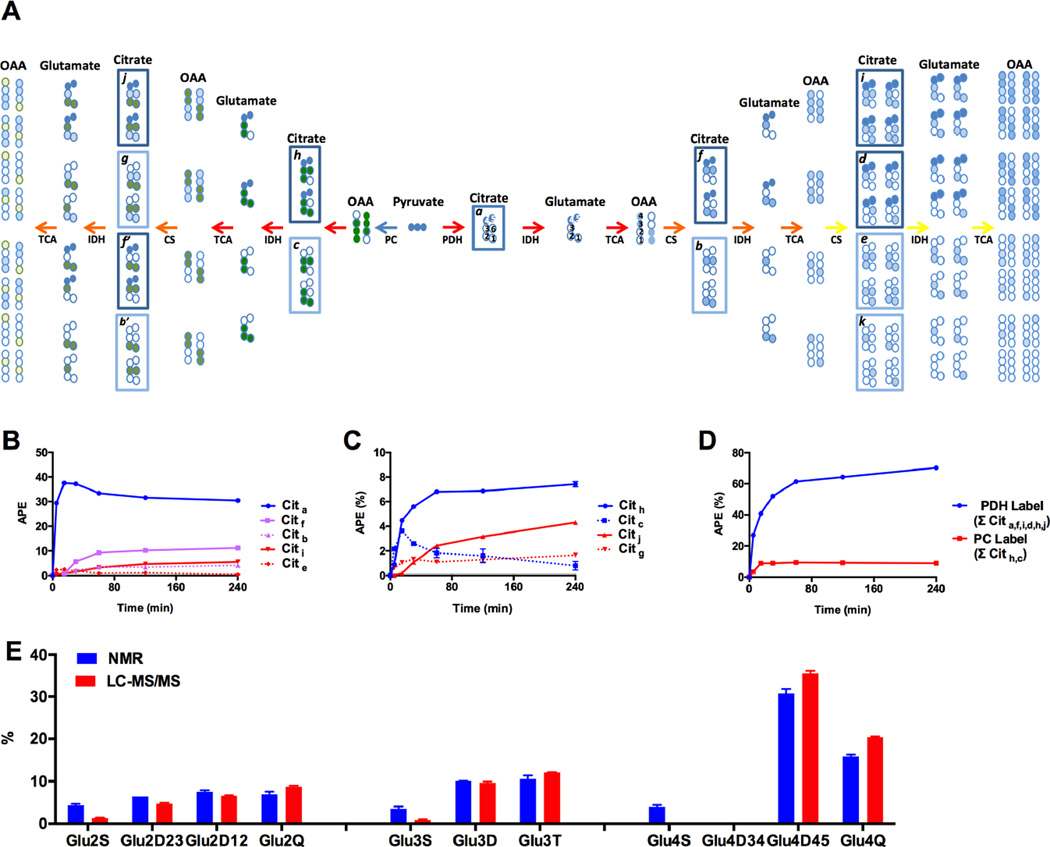
The synthesis of citrate from mitochondrial AcCoA and OAA is the defining reaction of the TCA cycle. The AcCoA is formed via the PDH reaction, β-oxidation, the ligation of free acetate, and/or metabolism of some amino acids. Similarly, the OAA is formed from the oxidation of malate, the deamination of aspartate, and/or the carboxylation of pyruvate. Since pyruvate metabolism generates both AcCoA and OAA, isotope-labeled pyruvate will therefore impact multiple intersecting reactions (Figure 1). Citrate carbon 3 (C3) is stereochemically a prochiral center and C4,5 and C1,2 are biochemically distinguishable. Thus C4,5 arise from AcCoA in the Pro-S arm and those from OAA as the C1,2,3,6 containing the Pro-R carbons. For instance, metabolism of [U-13C6]glucose generates [U-13C3]pyruvate (Figure 1) that enters the TCA cycle via either PDH (dark blue) or PC (dark green). The 13C-label can enter as AcCoA and traverse multiple trips around the TCA cycle (light blue) and recombine to generate the stoichiometric labeling patterns shown to the right. Alternatively, if labeled pyruvate is carboxylated then it generates the labeling patterns in the left branch. Consequently, in the presence of labeled substrates, citrate will be embedded with isotopic information reflecting all of these reactions in proportion to the relative rates they occur. Deconvolving the accumulating isotopomer patterns that ‘mark’ citrate is thus key to understanding multiple crucial mitochondrial reactions.
Figure 1. Citrate isotopomer analysis.
(A) Isotopomers generated during the 1st (red), 2nd (orange), and 3rd (yellow) turn of the TCA cycle. Citrate isotopomers were grouped into families depending on whether the label is originated from PDH flux only (Cita, Citf, Citb, Citi, Citd, Cite and Citk) or from both PDH and PC fluxes (Cith, Citc, Citj, Citg, Citf’, and Citb’). (B) Enrichment time course of the deconvolved citrate isotopomers from PDH flux alone or (C) from both PDH and PC fluxes. (D) Total PDH (∑ Cita, f, i, d, h, j) and PC flux (∑ Cith, c) into citrate. (E) Measured vs. predicted glutamate C2, C3 and C4 NMR multiplets patterns from deconvolved citrate isotopomer data. All data are reported as mean ± S.E.M.
Unlike enzymes, the mass spectrometer does not recognize citrate prochirality during ionization or fragmentation. Nevertheless, because of the stoichiometric relationship between isotopomer family members (Figure 1A, boxed), the unique solution was algebraically decrypted from 13C mass label distribution in citrate (Supplemental Experimental Procedures S2C and Figure S1A). Figure 1B shows the time course of oxidative glucose metabolism generating PDH-derived label. The lag in appearance and sequential dilution of 13C-label with subsequent turns is as expected for repeated cycles (1st cycle (blue) > 2nd cycle (magenta) > 3rd cycle (red)). A similar pattern emerged when following citrate through anaplerotic PC carbon labeling (Figure 1C, 1st cycle (blue) > 2nd cycle (red)). In both cases (Figures 1B and C) the enrichments were higher for those families containing labeled PDH carbons (solid lines) compared to unlabeled (dotted lines) as expected. The summation of all the PDH containing isotopomers and all the PC containing isotopomers are shown (Figure 1D). Since both are derived from the same pyruvate pool it is clear that VPDH is greater than VPC. These data are in direct contrast to the NMR-based measurements where PC was estimated to be even higher than PDH (Cline et al., 2004; Jensen et al., 2006; Lu et al., 2002; Simpson et al., 2006).
In order to understand if the discrepancy between the MIMOSA and NMR-based flux estimates was from differences in positional assignments, the MIMOSA citrate isotopomers were used to predict the 13C-NMR J-coupled multiplet patterns of glutamate and then directly compared to the measured glutamate NMR spectrum (Figure 1E). NMR did have significantly higher singlet enrichments relative to MIMOSA. Unlike NMR, a real strength of the MIMOSA analysis is a more direct natural abundance correction (since MS directly measures both 13C and 12C). Labeling patterns were consistent with expected isotope distributions and all symmetry rules were obeyed. In general the two patterns of glutamate multiplets were concordant and the minor differences observed between the two techniques were not large enough to explain the differences in VPC relative to VPDH.
Mass Isotopomer Distribution Analysis (MIDA) of Citrate and Glutamate
In addition to the citrate isotopomers, the positional enrichments of the precursors to citrate (i.e., the matrix pool of OAA and acetyl-CoA) are essential to fully assess CS metabolism. Unfortunately, direct measurements of these are complicated by their low abundance, fragmentation complexity, and the presence in both mitochondrial and cytosolic pools. To circumvent these difficulties, AcCoA and OAA isotopomers can be calculated directly from MIDA of the enriched products (i.e., citrate or glutamate). MIDA exploits the binomial relationship encoded into the product when two labeled precursors are ligated. This relationship remains discernable even in the setting of dilution of the product by unlabeled metabolites. Consequently, the isotopomers of OAA and AcCoA can be determined within the subcellular compartment where the reaction occurs (Supplemental Experimental Procedures S3, Supplemental Table S1).
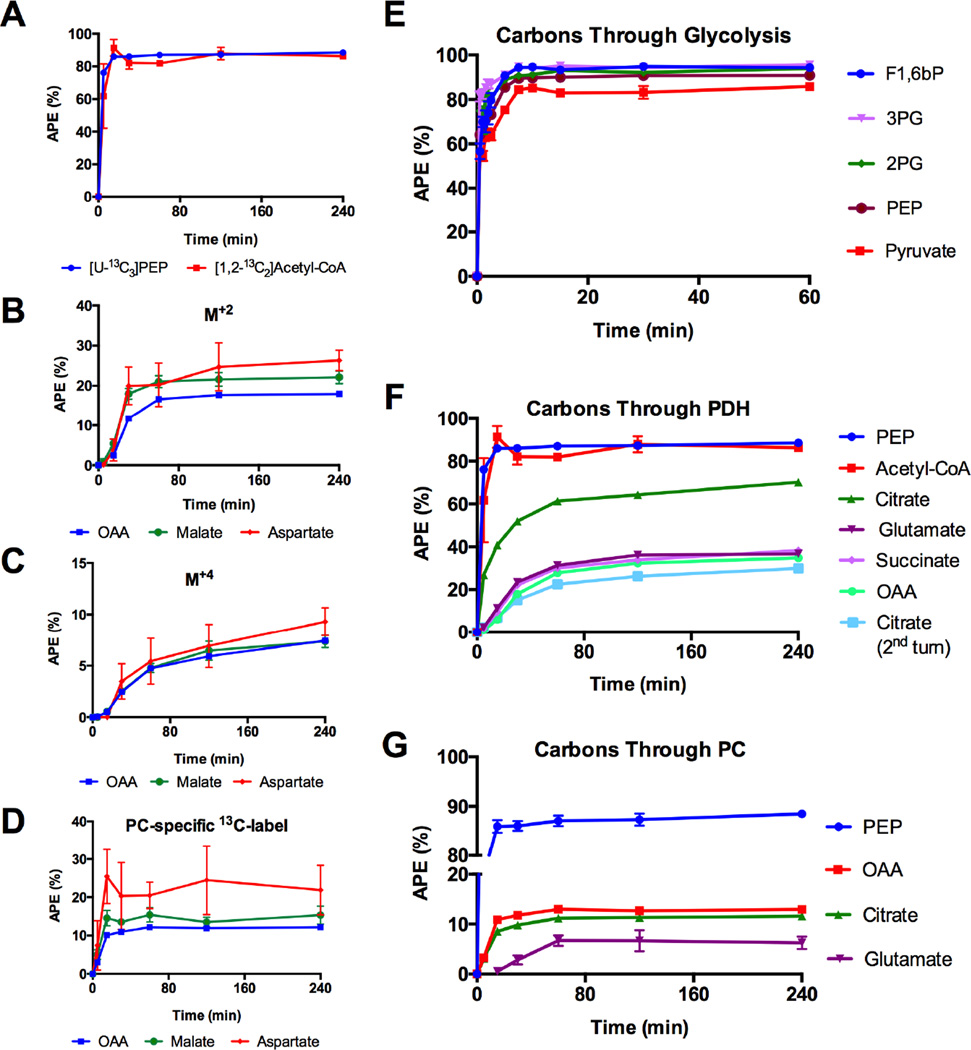
[1,2-13C2]Acetyl-CoA enrichment was determined from the glutamate isotopomer data and compared to the [U-13C3]phosphoenolpyruvate (PEP) enrichments (Figure 2A). PEP was used (rather than pyruvate) to avoid contributions from media pyruvate or lactate at early time points. Notably, the calculated [1,2-13C2]AcCoA enrichments were similar to its precursor PEP at all time points. Identical steady-state [U-13C3]PEP and [1,2-13C2]AcCoA enrichments indicate a virtual absence of lipid oxidation in the presence of 9mM glucose and absence of exogenous fatty acids.
Figure 2. Tracking of 13C-label.
(A) [1,2-13C2]Acetyl-CoA (from glutamate MIDA) compared to its precursor [U-13C3]PEP. (B–D) Mitochondrial M+2, M+4 and [(1,2,3)(2,3,4)-13C3] (from PC flux) isotopomer evolution in OAA, aspartate and malate. (E) Incorporation of [U-13C6]glucose into the glycolytic intermediates fructose-1,6-bis phosphate (F1,6bP, M+6), 3-phosphoglycerate (3PG, M+3), 2-phosphoglycerate (2PG, M+3), PEP(M+3) and pyruvate (M+3). (F) Evolution of 13C-label from glycolysis into the TCA cycle through PDH flux and (G) anaplerotic PC flux. All data are reported as mean ± S.E.M.
Similarly, the mitochondrial OAA isotopomers calculated from citrate MIDA are concordant with the isotopomer kinetics of aspartate and malate (Figures 2B–2D). In contrast to the OAA M+2 and M+4 isotopomers, the OAA coming from PC (denoted as [(1,2,3)(2,3,4)-13C3]OAA) was significantly lower than that for aspartate (Figure 2D). Delayed transmitochondrial shuttle equilibration of aspartate and malate between the cytosol and mitochondrial pools suggest caution should be used if aspartate is surrogated for OAA, particularly in an open system.
Sequential Flux Mapping
Following the addition of labeled glucose, the time-dependent incorporation of 13C-label into the intermediates of glycolysis was measured. The glucose enrichment was a square-wave function that plateaued at ~99 atomic percent enrichment (APE). The glycolytic intermediates approached steady-state within approximately 5 minutes (Figure 2E). Fructose 1,6 bisphosphate (F1,6bP) plateaued at approximately 85% with little or no difference in final enrichment through glycolysis down to pyruvate. Starting with PEP the carbons could be followed through PDH and into Pro-S citrate and around the TCA cycle all the way back to Pro-R citrate after a full turn (denoted as citrate 2nd turn) (Figure 2F). As expected, progressive delays and dilutions were observed in sequential reactions. Similarly, PC-derived carbons flowed from PEP into OAA (via PC) and eventually into pro-R citrate (via CS) followed by C1,2,3 glutamate (Figure 2G). Note that despite originating from the same pyruvate pool, PC-derived OAA had one-eighth the enrichment of AcCoA suggesting a minority contribution of PC flux to the overall TCA cycle.
Steady-State Flux Analysis
The confluences of intersecting metabolic pathways are the sites where label dilutions can occur. Such biochemical nodes may complicate flux determinations though they contain important information about the magnitude of one pathway relative to another. At metabolic and isotopic steady-state, the concentration, flux and enrichment is constant and were confirmed experimentally. If only one metabolite flow contributes to a metabolic pathway, then the ratio of precursor and product positional enrichments should be 1 (Supplemental Experimental Procedures S4). A reduction in Φ between serial metabolites in a pathway/reaction indicates that unlabeled (or differently labeled) carbons from an alternative metabolic source are also contributing to product formation. Such carbon dilutions within the TCA cycle are considered anaplerotic if they are balanced by an equivalent cataplerotic outflow at a different site. In contrast, the label is diluted during an exchange reaction if does not contribute net flux entry and exit of a metabolite at the same site are balanced. For instance, unidirectional entry of glutamate carbons via glutamate dehydrogenase (GDH) is anaplerotic. The reciprocal entry and exit of glutamate carbons via rapid equilibration across transaminase reactions can be considered as an exchange. In the absence of exogenous acetate, ketones, or branched chain amino acids, then PDH and β-oxidation are the only relevant potential suppliers of mitochondrial AcCoA. A nearly 50% APE reduction going from citrate to glutamate is consistent with exchange and/or anaplerosis of similar magnitude to VCS at the level of α-ketoglutarate (αKG) (Figure 2F).
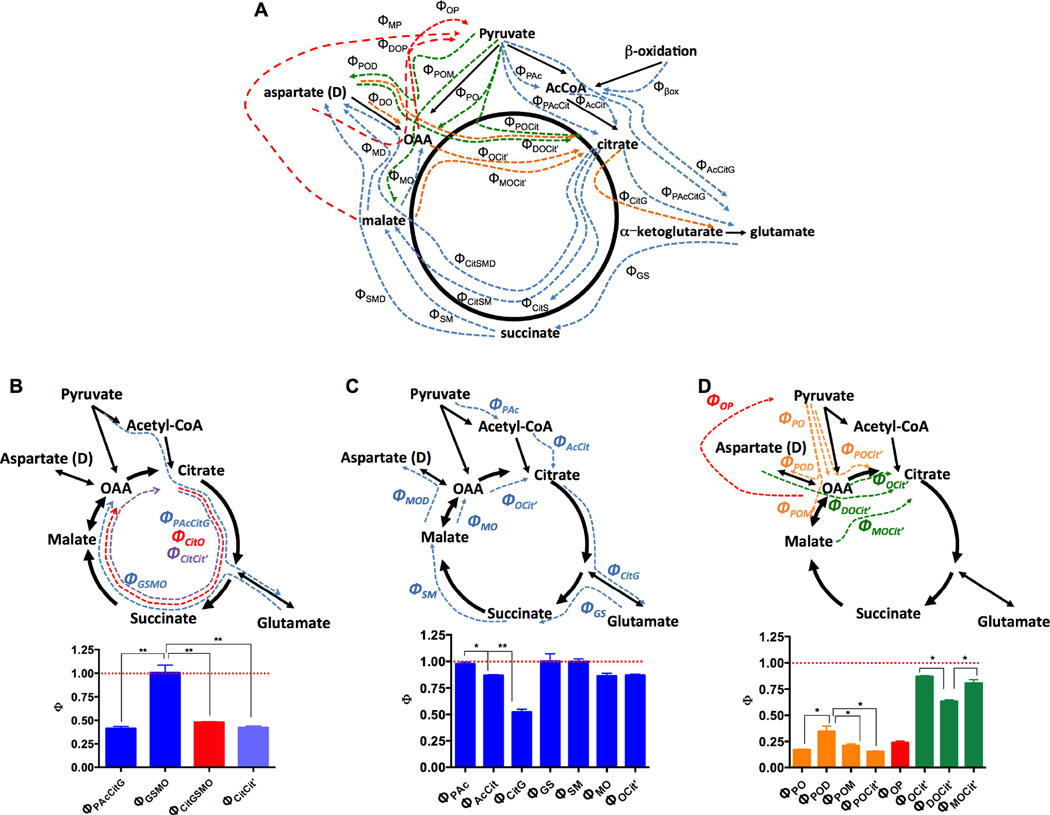
The equations for each of the 52 unique steady state precursor-product relationships (Φ’s) across the TCA cycle were solved for the shown individual and grouped reactions (Figure 3A, Supplemental Table S2, Supplemental Figure S2). This comprehensive map identifies discrete carbon entry points for oxidative reactions (blue), PC-driven anaplerosis (green), PEP/pyruvate cycling (red) and subsequent turns of the TCA cycle (orange) measured by specific steady-state isotopomeric relationships across sequential and serial reactions of the TCA cycle (Fig. 3B–D).
Figure 3. Isotopic steady state analysis.
(A) Graphical scheme of the 52 steady-state precursor-product relationships between the metabolic intermediates contributing to the TCA cycle. (B–D) Map and the ratio of precursor (X) to product (Y) enrichments (ΦX→Y) of (B) segmental (C) serial, and (D) anaplerotic/cataplerotic reactions of the TCA cycle. * P < 0.001, ** P < 0.0001. All data are reported as mean ± S.E.M.
The >50% dilution of isotopomers between [2,3-13C2] PEP and [4,5- 13C2]-L-glutamate labeling (ΦPAcCitG) also indicates a substantial entry of unlabeled carbon somewhere between these two metabolites (Figure 3B). In contrast, the small dilution across the segment from glutamate to OAA (ΦGSMO) indicates very little anaplerosis or exchange after succinyl-CoA. The label dilution from citrate to OAA (ΦCitGSMO), as well as across a full turn of the TCA cycle (ΦCitCit’), was nearly the same as from ΦPAcCitG. Stepwise Φ ratios confirmed and cross-validated the segmental findings (Figures 3B and D). Dilution of αKG can be explained at least in part by a large unlabeled intracellular glutamate pool and exogenous unlabeled glutamine (Supplemental Experimental Procedures S5, Supplemental Figure S3A,B, ). Unfortunately, it is not possible to distinguish anaplerosis from exchange by Φ analysis alone. Glutamate anaplerosis requires a net loss of nitrogen either to ammonia or urea. Consequently, metabolic exchange is the most likely since at 9 mM glucose concentrations aspartate levels are reduced (Supplemental Table S5), transaminases do not generate free ammonia, and GDH is completely inhibited by mitochondrial GTP (Kibbey et al., 2014; Tanizawa et al., 2002). Since there was relatively little dilution in the distal reactions of the TCA cycle (ΦGS, ΦSM and ΦMO; Figures 2F, 3C and Supplemental Figure S2) then glutamate is fully in exchange with αKG at a rate that is very fast relative to the TCA cycle rate.
Both PC and TCA cycle-derived OAA contribute to citrate formation. From independently calculated Φ’s, about 80% of the oxidative TCA flux (green bars) contributes to citrate formation and is balanced by anaplerotic flux of ~20% (orange) (Figure 3D). Interestingly, anaplerotic and oxidative Φ’s involving aspartate (ΦPOD and ΦDOC) were proportionately different suggesting a different carbon flow pattern involving aspartate. At steady state, anaplerosis is balanced by cataplerosis. Since the Φ’s monitoring flow from pyruvate into OAA (orange) were of the same magnitude as those of OAA back to pyruvate (red) suggest that PEPCK-M and ME are the principle cataplerotic pathways. Taken together, the PC contribution to the TCA cycle was confirmed to be approximately 20%. This sets a clear upper limit to anaplerosis contributing to CS and is significantly lower than those suggested previously (Cline et al., 2004; Lu et al., 2002).
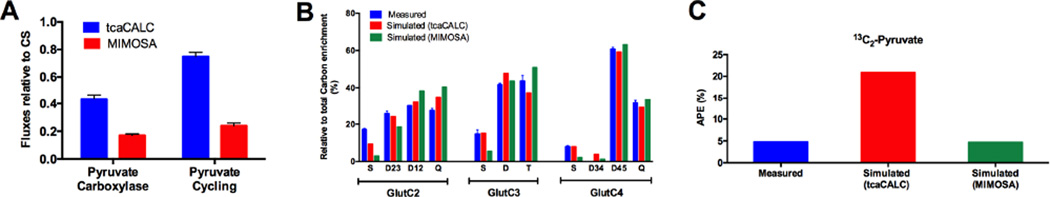
A side-by-side comparison of MIMOSA and NMR methods was performed to better understand the origin of the discrepancy in steady state PC calculations. Because of the inherent low sensitivity of NMR, glutamate isotopomers are often the principle input to calculate fluxes relative to CS. The tcaCALC software was originaly designed to analyze metabolic fluxes based on NMR-derived isotopomer distribution using glutamate as the principle input (Malloy et al., 1988). In order to obtain reasonable fits for insulin secreting cells, tcaCALC required an extra ‘pyruvate cycling’ function (Lu et al., 2002). This ‘pyruvate cycling’ term assumes a metabolic pool of carbon flow from pyruvate to OAA/malate and back to PEP/pyruvate that somehow does not mix with glycolytic pyruvate. While the glutamate isotopomers were similar using either NMR or LC-MS/MS (Figure 1E), the calculated PC and ‘pyruvate cycling’ rates were substantially higher using tcaCALC (Figure 4A). If such high ‘pyruvate cycling’ rates were real (Lu et al., 2002), a predition based on the OAA/malate enrichments would be that pyruvate M+2 enrichments would be high (approaching 22%). Direct pyruvate M+2 is measured at only ~5%, suggesting that tcaCALC overestimates PC.
Figure 4. Comparison of steady state analyses by NMR and MIMOSA.
(A) VPDH, VPC and VPK fluxes relative to VCS calculated from NMR-based tcaCALC simulations (blue) compared to equivalent MIMOSA values (Φ, red). Pyruvate cycling here is denoted as 13C label that cycles out of the TCA cycle into pyruvate via PEPCK and ME. In tcaCALC this is the ‘PK’ term but is distinct from glycolysis. In MIMOSA it is the transfer of oxaloacetate label to the full pyruvate pool, ΦOP. (B) Measured and predicted glutamate NMR multiplet isotopomer distributions from (blue) direct, (red) predicted from tcaSIM using the tcaCALC flux values (PDH=1, PC=0.409, Ys=0.838, PK=0.77) and (green) predicted by tcaSIM using MIMOSA-derived constraints (PDH=1, PC=0.15, Ys=1, PK=0.26). (C) Total enrichments of 13C2-pyruvate isotopomers directly measured by LC-MS/MS (blue), predicted by tcaSIM using tcaCALC fluxes (red), or using steady state MIMOSA values (green). All data are reported as mean ± S.E.M. Predicted values by tcaSIM have no uncertainty levels reported by the software.
We hypothesized that tcaCALC did not have sufficient metabolic information to predict all of the metabolic fluxes from glutamate isotopomers alone. The companion software of tcaCALC, tcaSIM, simulates glutamate isotopomer distributions given different fluxes (Jeffrey et al., 1991). If the fluxes obtained from tcaCALC are used as tcaSIM inputs, it correctly predicts the observed NMR isotopomer distribution (Figure 4B, blue vs. red). However, if the simulations are constrained by the direct MIMOSA Φ values, the measured and predicted glutamate isotopomer distributions are also quite similar (Figure 4B, green). Since there are at least two minima found by the program, there is not a unique solution that can be calculated from the glutamate isotopomers. Using the fluxes from tcaCALC, tcaSIM also predicts total 13C2-pyruvate ~4-fold higher than the measured data (Figure 4C blue vs. red). MIMOSA constrains the predicted 13C2-pyruvate to the observed value (Figure 4C, green). Consequently, without multiple ordinates constraining the flux calculations, the local minimum arrived at by tcaCALC from glutamate isotopomers is substantially different from multiple directly measured values.
Dynamic Metabolic Modeling
A key advantage of MIMOSA flux interrogations is the broad capacity to assess time-dependent positional labeling of multiple ordinates across the TCA cycle. Here, the flow of isotope transfer through mitochondrial reactions was modeled mathematically using the program CWave (Mason et al., 2003). Position-specific (isotopomer) and position-independent (isotopologue) models were used for such comparisons. A final model was built using the mass spectroscopy measured isotopomers through three turns of the TCA cycle (Figure 5A; Supplemental Table S3; Supplemental Figure S4A,B; Supplemental Experimental Procedures S5). Both isotopologue and isotopomeric data obtained at different labeling times were used as separate inputs. The model was then tested for fit quality and proximity to the results obtained from the steady-state analysis.
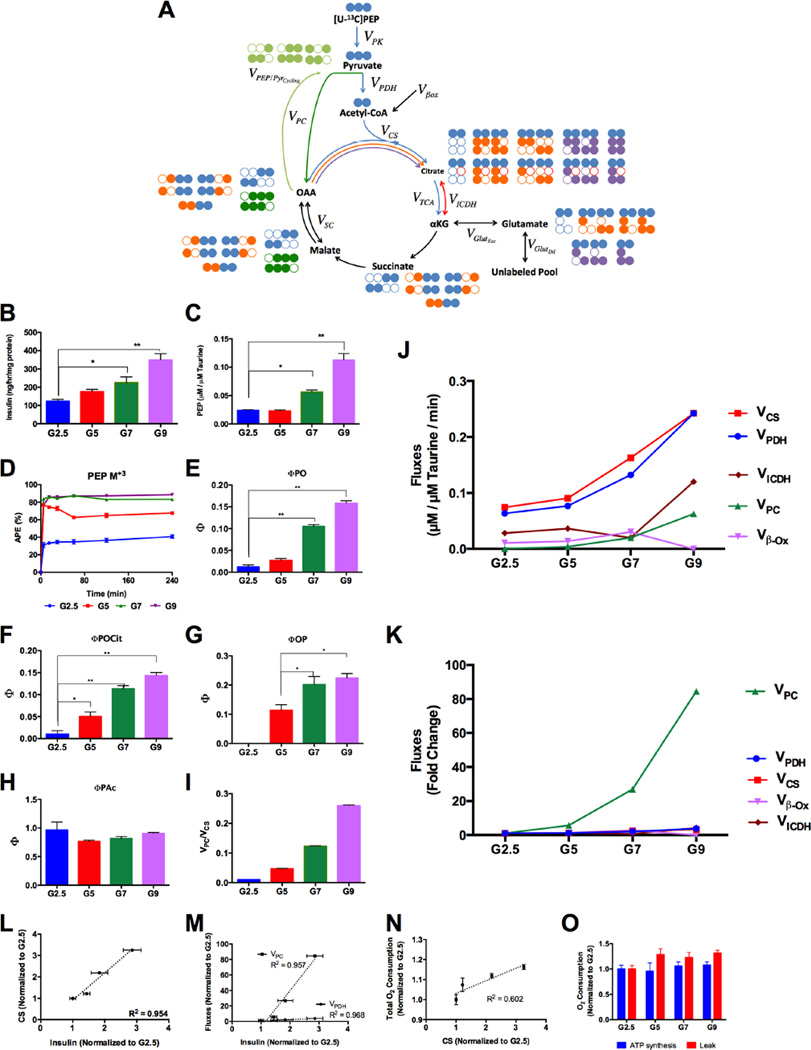
Figure 5. Relationship of MIMOSA modeled mitochondrial rates determined by CWave to insulin secretion.
(A) Overview of the rates and isotopomers considered in the CWave model: Pyruvate kinase (VPK); pyruvate dehydrogenase (VPDH); pyruvate carboxylase (VPC); conversion of oxaloacetate (OAA) into pyruvate (VPEP/PyrCycling); citrate/isocitrate exchange with αKG (VICDH); αKG exchange with glutamate(VGlutExc); dilution of glutamate(VGlutDil); the racemizing exchange between the malate/fumarate and (OAA VSC). The arrows associated with VCS have different colors indicating different turns (blue (1st), orange (2nd), and purple (3rd)). The isotopomers with the greatest impact are displayed next to each pool and their carbons are color-coded to match their associated turn of the TCA cycle. (B) Insulin secretion from cells incubated with 2.5, 5, 7 and 9mM glucose. (C) [PEP] normalized to intracellular [taurine]. (D) PEP M+3 enrichments from glycolysis of glucose M+6. (E–H) precursor/product relationships ΦPO, ΦPOCit, ΦOP and ΦPAc, respectively. (I) VPC/VCS calculated from MIMOSA rates (Table 1) (J) Absolute rates and (K) fold change through CS, PDH, PC, lipid oxidation (β–ox) and ICDH calculated using the model. (L) Correlation between CS and (M) PDH and PC fluxes and insulin secretion. (N) Correlation between O2 consumption and CS. (O) O2 consumption from ATP synthesis and mitochondrial leak. All data are reported as mean ± S.E.M. * P < 0.01, ** P < 0.001.
Using just isotopologue data, the model severely missed some target data. These misses were especially prominent for the anaplerotic containing fragments and in particular citrate M+3 (Supplemental Figure S5A). In the absence of positional data, this poor fit was driven by inappropriately high rates of Vβox (Supplemental Figure S5B) needed to force a dilution of the acetyl-CoA pool. The Vβox rate generated by the model fit is inconsistent with the direct measures of VPDH relative to VCS (VPAc) and overestimates unlabeled acetyl-CoA (Supplemental Figure S5C,D). Subsequently, VCS, VPC and pyruvate cycling are also overestimated (Supplemental Figure S5E,F). By incorporating isotopomeric inputs the overestimation was corrected and lowered these flux values (Figure 5H; Supplemental Experimental Procedures S5, Supplemental Figure S6) to match the direct measures.
Interestingly, ΦAcCit is less than 1 and indicates that citrate is being diluted from an unlabeled source. In the absence of an exogenous anaplerotic citrate pool, the only biologically plausible source of the dilution is from the carboxylation of unlabeled αKG. A consistent 13% reduction in measured relative to predicted citrate for the four isotopomer families affected by C6 carboxylation supports αKG as the diluting source (Supplemental Table S4). Also in agreement, is the time-dependent AcCoA labeling of C4,5 Pro-S citrate that is better described by two processes: a fast component representing VCS and a much smaller and slower component coming from an exchange with αKG (Supplemental Figures S4C–F). The presence of unlabeled glutamate first diluting αKG and then (through reductive/oxidative isocitrate dehydrogenase (ICDH) exchange) diluting citrate was detected in asymmetries of the C6 containing isotopomers of citrate (Supplemental Experimental Procedures S2D). Consequently, a diluting exchange from isocitrate dehydrogenase (ICDH) is needed to interpret the citrate isotopomers fully. Measuring and accounting for these prior to deconvolving the citrate families substantially improved the model fits. Unfortunately, isotopologue-only data sets will have these errors embedded in them without a priori knowledge of the position-specific data. In the final CWave model, fits of each oxidative and anaplerotic ordinate were superior for the isotopomers compared to the isotopologue-only model (Supplemental Figures S5A and S6). Not surprisingly, isotopomeric data was better at predicting the enrichments of other labeled ordinates that were not included as target data (e.g., [13C2]pyruvate in Supplemental Figure S5F). Additionally, the standard deviation of the distribution of values attributed to VPC was also smaller (Supplemental Figure S5G). The overall least square standard deviation (LSSD), was also narrower for the isotopomer data (LSSD 1.39 vs 1.44, respectively). The final model benefitted from 1) isotopomeric citrate, OAA, AcCoA, glutamate, succinate, and aspartate data, 2) inclusion of glutamate, glutamine, and isocitrate exchanges, 3) a balancing anaplerosis and cataplerosis, 4) correction for labeled citrate C6 loss. Importantly, across the physiologic glucose range from 5–9 mM, MIMOSA can detect very small changes in VCS (~8% for P<0.05 and ~12% for P<0.01) and VPC (~37% for P<0.05 and ~60% for P<0.01). These would impart sensitivity to very subtle changes in metabolism given that a 1 mM increase in glucose concentration increases VCS ~40% and VPC >200%. Taken together, the static and dynamic results highlight the importance of using multiple positional inputs while determining metabolic fluxes.
Mitochondrial Metabolism and Insulin Secretion
As a model system, pancreatic β-cells relate glucose-dependent changes in metabolism to the functional output of insulin secretion. The clonal insulinoma cell line INS-1 832/13 has been the subject of many published flux experiments and has a nearly linear release of insulin across physiologic glucose concentrations (2.5 to 9 mM) (Figure 5B, Table 1) (Cline et al., 2004; Jensen et al., 2006; Lorenz et al., 2013; Lu et al., 2002; Pongratz et al., 2007; Stark et al., 2009). Many more metabolomics analyses have endeavored to relate changes in the concentration of metabolite(s) to insulin secretion. As glucose concentration increased, the steady state concentration of all other metabolites increased by varying degrees with the exception of aspartate which declined (Supplemental Table S5). The concentrations remained constant across the time course consistent with metabolic steady state. Differences in concentrations and enrichments of multiple metabolites have been used previously to infer relative differences in metabolic fluxes. However, it is important to point out that incorrect or incomplete conclusions can be drawn from either in isolation. For instance, the concentration in PEP remained unchanged going from 2.5 to 5 mM glucose and nearly doubled going from 7 to 9 mM glucose (Figure 5C, Supplemental Table S5). In contrast, steady-state PEP enrichments nearly doubled over the former, while remaining flat at the latter (Figure 5D).
Table 1. Rates of insulin secretion, oxygen consumption and absolute fluxes, calculated by CWave.
All data are reported as mean ± S.E.M.
| Glucose (mM) |
Insulin (ng/hr/mg protein) |
Oxygen Consumption (pmol/min) |
Fluxes (µM/µM Taurine/min) |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total | ATP Synthesis |
Leak |
VPDH = VPK |
Vβox | VCS |
VPC = VPEP/PyrCycling |
VGlutDil | VICDH | VGlutExc | VSC | ||
| 2.5 | 122.4 ± 11.1 |
0.87 ± 0.03 |
0.57 ± 0.02 | 0.31 ± 0.01 |
0.064 ± 0.002 |
0.011 ± 0.000 |
0.074 ± 0.002 |
0.001 ± 0.000 | 0.331 ± 0.011 |
0.028 ± 0.001 |
7.44 ± 0.23 | 7.44 ± 0.23 |
| 5 | 176.0 ± 12.1 |
0.94 ± 0.06 |
0.54 ± 0.05 | 0.40 ± 0.02 |
0.077 ± 0.000 |
0.014 ± 0.000 |
0.091 ± 0.001 |
0.004 ± 0.000 | 0.108 ± 0.001 |
0.037 ± 0.001 |
7.74 ± 0.46 | 3.83 ± 0.78 |
| 7 | 224.4 ± 32.1 |
0.98 ± 0.04 |
0.60 ± 0.02 | 0.38 ± 0.02 |
0.132 ± 0.001 |
0.031 ± 0.000 |
0.163 ± 0.001 |
0.020 ± 0.000 | 0.167 ± 0.002 |
0.020 ± 0.001 |
12.33 ± 0.91 |
13.13 ± 1.15 |
| 9 | 349.6 ± 33.7 |
1.02 ± 0.03 |
0.61 ± 0.02 | 0.41 ± 0.01 |
0.242 ± 0.002 |
0 | 0.242 ± 0.002 |
0.062 ± 0.001 | 0.273 ± 0.003 |
0.120 ± 0.003 |
18.16 ± 1.69 |
20.27 ± 1.24 |
Steady-state relationships alone can only detect relative changes of one flux versus another but not determine reaction rates. Of the mitochondrial steady-state Φ’s, only those associated with PC flux (ΦPO and ΦPOCit’) and PEP/pyruvate cycling changed proportionately with glucose (Figures 5E–G, Supplemental Table S6) consistent with other steady-state studies (Cline et al., 2004; Lu et al., 2002). PDH flux relative to CS (ΦPAc) did not change with glucose (Figure 5H). In contrast, VCS, VPDH and VPC all increase with glucose (Figure 5J). Even at the highest glucose concentration, VPC approached only ~20% of VCS (Table 1). The relative increase in PC flux exhibited a much higher dynamic range than oxidative metabolism (Figure 5K). Additionally, the ratio between VPC and VCS internally validated the model since it was similar to ΦPO, (Figure 5I). Consequently, all the absolute fluxes (both oxidative and anaplerotic) were strongly correlated with insulin secretion (Figures 5L,M) though VPC had a much steeper slope (Figure 5M). Oxygen consumption modestly increased with glucose (Table 1) and also correlated with VCS (Figure 5N). The oligomycin-sensitive ATP component remained surprisingly flat suggesting that proton leak accounts for the majority of the glucose-dependent change in respiration (Figure 5O).
Metabolic differences are dependent on cell-type, nutrients and hormones
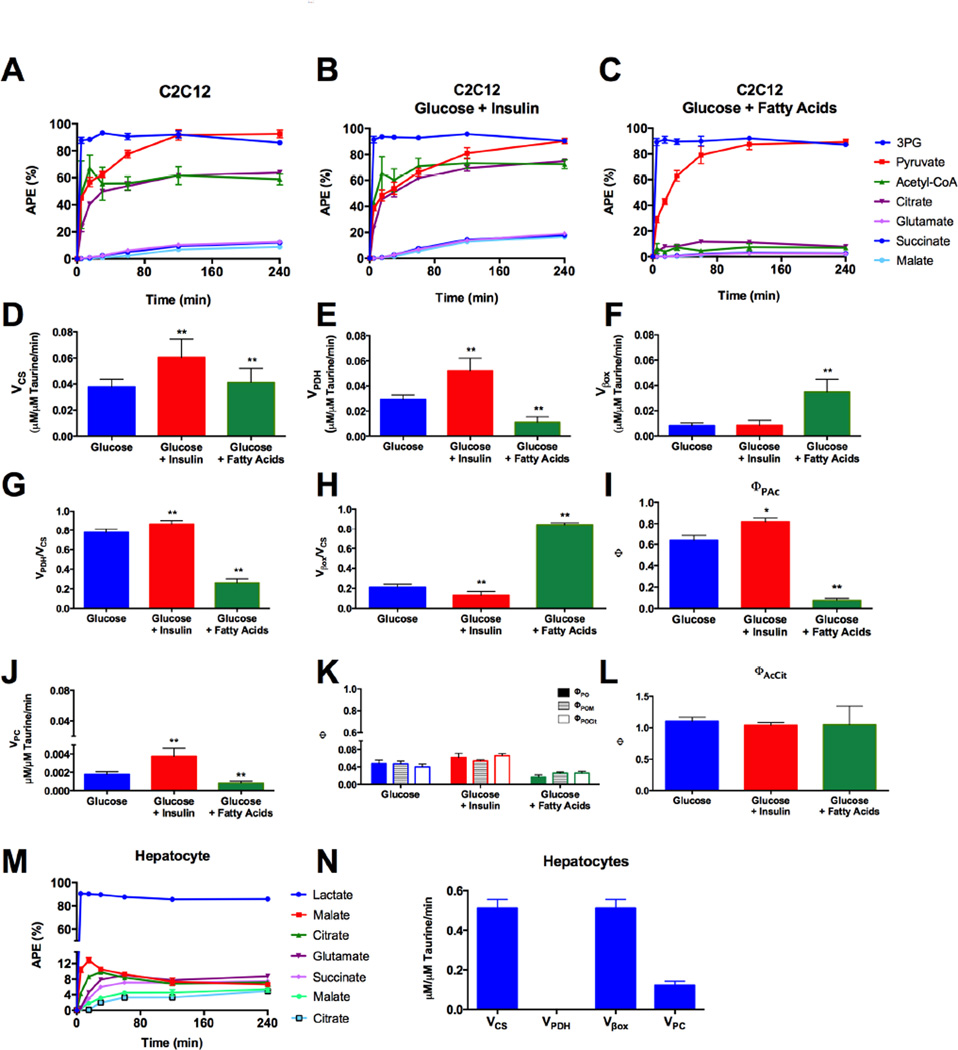
MIMOSA was extended to other cells, nutrient and hormonal conditions (Figure 6). C2C12 myoblast cells were incubated with 5 mM [U-13C6]-D-glucose alone, under the same conditions used for the INS-1 cells (Figure 6A), in the presence of insulin (Figure 6B) or in the presence of fatty acids (Figure 6C). VCS and VPDH rates increased with insulin stimulation as expected (Figure 6D,E). The presence of fatty acids increased Vβox with concomitant inhibition of VPDH (Figure 6E,F). Additionally, the ratios VPDH/VCS and Vβox/VCS (Figures 6G and 6H) obtained from the dynamic analysis were concordant with the ΦPAc calculated from steady state (Figure 6I). VPC was much slower than VPDH, Vβox or VCS but increased with insulin and it was inhibited by the presence of fatty acids (Figure 6J). The same trends were observed in the analysis of ΦPO, ΦPOM and ΦPOCit (Figure 6K). Unlike in the INS-1 cells, ICDH flux was only observed in the direction of αKG in the C2C12 cells (Figure 6L).
Figure 6. Flux analysis in C2C12 myoblast cells and primary rat hepatocytes.
Evolution of 13C-label from glycolysis into the TCA cycle through PDH flux in C2C12 cells incubated with (A) glucose (5mM), (B) glucose and insulin (100nM) and (C) glucose and fatty acids (0.2mM palmitate:oleate (1:2)). Absolute (D) VCS, (E) VPDH and (F) Vβox. Relative contributions of (G) VPDH and (H) Vβox to VCS calculated from by CWave and (I) the equivalent with steady state measurement ΦPAc. (J) Absolute VPC flux and (K) ΦPO calculated using OAA enrichments or malate and citrate as surrogate for OAA. (L) Calculated ΦAcCit to assess the existence of reversed ICDH flux. (M) Evolution of 13C-label from [U-13C3]lactate into the TCA cycle through PC flux in primary rat hepatocytes and (N) respective fluxes calculated by MIMOSA. Absolute flux data is reported as mean ± standard deviation of the flux distribution calculated by CWave. All other data are reported as mean ± S.E.M. * P < 0.01, ** P < 0.001.
In primary rat hepatocytes a similar sequential transfer of label from [U-13C3]-L-lactate to the TCA cycle intermediates was observed (Figures 6M and 6N). The dynamic analysis revealed high rates of VCS (compared to INS-1 and C2D12 cells) and a primary reliance on fatty acid oxidation (Figure 6N). VPC was only a fraction of VCS without hormonal or substrate stimulation under these experimental conditions.
Discussion
MIMOSA is a mass spectroscopic platform to quantitate the essential mitochondrial metabolic fluxes present in most living cells. A key advance is the capacity for unambiguous identification of NMR-validated position-specific enrichments of multiple TCA cycle metabolites. Importantly, the positional AcCoA, OAA and citrate enrichments in the mitochondrial matrix were determined for direct calculation of VCS in the compartment where it actually occurs. In addition, a comprehensive list of steady-state intermolecular sequential and segmental isotopomeric relationships assesses the multiple nodes that occur at biochemical intersections distributed across oxidative, anaplerotic, exchange and cycling reactions. Finally, it incorporates all the positional data into a mathematic model of relevant metabolic reactions to calculate the discrete metabolic flux and exchange rates.
The fully integrated method combines position-specific fragment deconvolution of key intermediates, 52 sequential and segmental steady-state relationships and solves 182 mass and isotope balance differential equations to calculate the fluxes. 13C NMR confirmed isotopomeric assignments and fluxes were cross-validated by correlation of steady-state and dynamic measurements (both isotopologue and fragment deconvolved). Quantitative measurements of glycolysis, pyruvate kinase, PC, PDH, β-oxidation, CS, ICDH, PEP/pyruvate cycling (PEPCK-M + ME), glutamate-αKG and glutamate-glutamine exchanges, ATP synthesis, proton leak and insulin secretion were determined as a function of glucose concentration in a β-cell model and were extended to muscle and liver model systems.
Other models have been created to measure rates from 13C labeling (e.g., Quek et al., 2009; Weitzel et al., 2013). Recently, an improved model was developed which makes use of differential equations to describe the non-steady-state variation of 13C-enrichments (Young, 2014). Unfortunately, these isotopologue-based methods are limited in systems with anaplerotic metabolism especially if there is significant exchange. Similarly, even with position-specific information from a single ordinate such as glutamate, key fluxes were dramatically missed. A significant advantage of MIMOSA is the ability to detect and track position-specific transfer of label across the key mitochondrial reactions. This proved to be essential especially regarding the anaplerotic fluxes (Figure 4 and Supplemental Figure S5). Importantly, in an open in vitro system exchange and/or anaplerosis of unlabeled glutamate can substantially dilute not only subsequent reactions of the TCA cycle but even citrate itself. Consequently, such isotope dilution (or isotope transfer if labeled glutamate is used) through αKG-citrate exchange does not imply net reductive carboxylation. In these studies reductive carboxylation was balanced by oxidative decarboxylation with no net reverse ICDH flux.
Another caveat identified here is the potential problem using one metabolite as surrogate for another. For instance, aspartate enrichments differed from malate and OAA and were excluded from the CWave analysis (Figure 2). Similarly the experimental pyruvate measurements did not behave as the true precursor of AcCoA likely because intra and extracellular pools determined its enrichment. Caution must be used if glutamate is the sole surrogate of AcCoA, particularly in open in vitro systems where exogenous concentrations of glutamine can dilute the pool of glutamate. In the present work, there was no delay or dilution between succinate and glutamate (ΦGS) label transfer indicating that the exchange rate (VGlut_exc) is much greater than VCS, making glutamate an ideal surrogate for αKG here. This situation may differ in some systems such as the perfused heart (Yu et al., 1995).
There have been progressive refinements in the measurement of intracellular metabolic fluxes through examining the metabolism of radioactive, spin and mass labeled substrates. These all have contributed significantly to our understanding of intracellular metabolism and form the theoretical basis upon which MIMOSA is founded. For instance, the importance of anaplerosis in insulin secreting β-cells was first identified using radiolabeled substrates (MacDonald, 1993). This flux was later confirmed by NMR spin-labeling to correlate with insulin secretion (Cline et al., 2004; Lu et al., 2002; Simpson et al., 2006). Importantly, these studies necessitated a re-evaluation of ATP production as the principle mechanisms whereby mitochondrial metabolism regulates rates of insulin secretion. However, these techniques only provided estimates of anaplerosis relative to VCS and not the absolute flux rate. Without a direct measure of VCS or VPDH it is not possible to conclude a stronger relationship of one flux or another with insulin secretion. Here we demonstrate strong correlations between all the anaplerotic and oxidative rates with GSIS making a clear mechanistic linkage with just one more tenuous.
In these same cells, PC rates were previously estimated to exceed the TCA cycle rate. These rates are inconsistent with either the direct steady-state enrichments or the dynamic flux data in the present study (Cline et al., 2004; Jensen et al., 2006; Lu et al., 2002; Simpson et al., 2006). If indeed PC flux were twice that of CS, then M+2 pyruvate should be at least 33%, but in actuality it only approached 4%. MIMOSA uses 1) natural-abundance-corrected singlet glutamate, 2) multiple precursor/product pool enrichments together with 3) other anaplerosis-and exchange-mediated dilutions in order to recalibrate PC and carbon recycling rates. While the overall PC contribution to the TCA cycle is much lower than previously estimated, the large glucose-induced increase in VPC/VCS was confirmed. Of note, since all of these measurements have been made at metabolic steady-state, they can only be mechanistically linked to a PEP/pyruvate cycle involving malic enzyme and/or PEPCK-M (Pongratz et al., 2007; Stark et al., 2009) but not cytosolic ICDH as previously suggested (Jensen et al., 2006; Lu et al., 2002; Ronnebaum et al., 2006; 2008). Since equal amounts of isocitrate exit and return as αKG then ICDH-1 metabolism is not anaplerotic and is simply an extramitochondrial oxidative loop that better correlates with VCS.
MIMOSA has the sensitivity to detect subtle changes in metabolism (as little as 8%) that can be expanded into the analysis of other cellular fluxes or conditions. The platform has broad applicability to flux measurements in any glucose-oxidizing cell (INS-1 832/13, C2C12 and hepatocytes shown here) that contains mitochondria. In comparison to tissue culture, in vivo systems are closed and more complex since label that is metabolized in other tissues can be incorporated into circulating substrates. In addition, disequilibrium of plasma and tissue-specific tracer enrichments, metabolic zonation (especially in tissues such as liver), and tissue-specific as well as substrate-specific times to reach steady state should be considered. It would be a mistake to immediately implement these studies in vivo without a formal validation.
By taking advantage of dynamic accumulation of position-specific enrichment across multiple metabolite ordinates, MIMOSA provides a direct measurement of mitochondrial metabolism. A particular strength is the ability to resolve positional isotopomers of AcCoA, OAA, and citrate in order to capture definitive citrate synthase flux. Recently there has been a surge in the wide spread application of NMR spin labeling and mass isotope labeling strategies to assess different aspects of cellular metabolic flux (Buecher et al. 2015). If interpreted correctly, these have the potential to contribute greatly to our understanding of metabolism. Importantly, conclusions about any steady-state metabolic flux measurement could be substantially flawed without first 1) confirming isotopic and metabolic steady-state, 2) correcting positional natural abundance enrichments, 3) assigning isotopomeric labeling patterns, 4) considering diluting and position-modifying exchange reactions, and 5) correlating precursor and product relationships. Given the complexity of metabolism, the use of orthogonal methods to cross validate should always be considered. When applied to a model of insulin secretion, a significant recalibration of prior flux estimates has identified large changes in the magnitudes of key fluxes most notably pyruvate carboxylase. In addition to studies of β-cell, hepatocyte and muscle metabolism, MIMOSA has broad applicability to potentially characterize the mitochondrial metabolism of any tissue or cell and may be helpful to solve “some of the practical problems of medicine.”1
Experimental Procedures
13C-Isotopomer Isotopomer Labeling Studies
Insulinoma (INS-1) 832/13 cells, primary rat hepatocytes, and C2C12 myocytes were cultured as monolayers in RPMI-1640 complete medium as previously described (Kibbey et al., 2007). 13C-Isotopomer studies were performed in in DMEM medium (D5030, Sigma-Aldrich) supplemented with glucose (2.5, 5, 7 and 9mM), glutamine (4mM), pyruvate (0.05mM) and lactate (0.45mM) with palmitate/oleate, insulin, BSA as described for individual experiments. A metabolic steady state was reached before the addition of [U-13C6]glucose or [U-13C3]lactate (Cambridge Isotope Laboratories) as described (See Supplemental Experimental Procedures, S1).
LC-MS/MS Analysis
Samples were injected onto a Hypercarb column and ionized by electrospray into an SCIEX 5500 QTRAP equipped with a SelexION for differential mobility separation (DMS) and acquired using multiple reaction monitoring (MRM) (Supplemental Experimental Procedures, S2A, Supplemental Table S7). Endogenous taurine, an intracellular osmolyte, was used as internal control for cell density as previously described (Kibbey et al., 2007). 13C incorporation into taurine did not occur under the experimental conditions nor did the absolute concentration of taurine change during the incubations. The atomic percent excess (APE) was calculated and background and natural abundance corrected from an isotopomer matrix accounting for the presence of natural abundance carbons distributed throughout each possible parent/daughter ion combination (Supplemental Experimental Procedures, S2A,B).
NMR Analysis
The isotopomer distribution of glutamate at each carbon position was determined by 13C NMR spectroscopy with an AVANCE 500-MHz spectrometer (Bruker Instruments, Inc., Billerica, MA). Spectra were acquired with TR = 0.5 s, NS = 10,000, 16 K data, and Waltz-16 broadband proton decoupling. The isotopomer pattern obtained from the NMR analysis was used in tcaCALC to calculate metabolic fluxes relative to CS as previously described (Cline et al., 2004; Lu et al., 2002; Simpson et al., 2006).
Insulin Secretion
Media extracts were measured for insulin concentration using the Rat High Range Insulin ELISA kit (ALPCO) per manufacturers instructions. All media extracts were normalized to cell protein concentration using a Micro-BCA Protein Assay Kit (Pierce).
Isotopomer Analysis
Assumptions
The analysis of metabolic fluxes was preceded by the following assumptions:
For the deconvolution of citrate isotopomers, only three turns of the TCA cycle were considered (Figure 1A). This assumption was confirmed by the low percentage of citrate M+1 expected in the third cycle.
In an open system, bicarbonate does not contribute significantly to the labeling patterns.
For calculating citrate and glutamate isotopomers, pyruvate is considered predominantly either M+0 or M+3. M+2 pyruvate from PC-mediated pyruvate recycling will be mostly [2,3-13C2]pyruvate and will label acetyl-CoA the same as M+3. Other M+1 and M+2 labeled pyruvate will make an insignificant contribution (as confirmed by the low enrichments).
Acetyl-CoA only contributes carbons to the Pro-S carbons of citrate.
For the analysis of citrate, aspartate and OAA enrichments, OAA and fumarate are assumed to be in near-equilibrium. Therefore, for any given isotopomer of aspartate, OAA and malate there is an equal percentage of the symmetrical isotopomer. This was confirmed by the modeling fluxes (VSC >> VCS, Table 1, Supplemental Figure S7) and by 13C-NMR analysis of glutamate C3 and C2 (C3/C2 = 0.96 ± 0.06).
Deconvolution of citrate isotopomers
The principles and derivation of mass isotopomeric deconvolution for the indicated metabolites are described in Supplemental Experimental Procedures S2C–H. The enrichments of citrate isotopomers were calculated from the 191/67 fragment according to equation 2 –equation 11. Prior to the calculation of citrate isotopomer families, the citrate spectrum was corrected for C6 loss from reversible exchange across ICDH.
Calculation of acetyl-CoA and OAA enrichments
The fractional enrichments of the mitochondrial matrix pool of acetyl-CoA and OAA were calculated using MIDA (Hellerstein and Neese, 1992). Enrichments were calculated from either glutamate or citrate isotopomers according to the description in Supplemental Experimental Procedures S3.
Steady-State Flux Analysis
Steady-state (Φx) analysis was performed to identify sites of anaplerosis and exchange at intersecting metabolic nodes based on the quotient of product and precursor enrichments (derivations and equations in Supplemental Experimental Procedures S4; Supplemental Table S2).
Metabolic Model for Flux Analysis
The time courses of 13C labeling were fit with a mathematical model of the TCA cycle (described in Supplemental Experimental Procedures S5). The model describes the labeling of the pyruvate pool from a glycolytic precursor and the entry of carbons into the TCA cycle via the PDH and PC reactions using [U-13C3]PEP as a driving function. The label is distributed through all possible isotopomers for citrate, αKG, glutamate, succinate, malate and OAA. The isotopomers were grouped in combination pools based on the number and/or position of labeled carbons and used to fit target data. As target data we used the following time course: [U-13C3]pyruvate, Cita,f,i,d,h,,j, Cita, Citf, Cith+i and Citj, total label in glutamate-C4,5, and 13C2-, 13C3-, 13C4-succinate, malate and OAA. The mass balance and isotope rate equations are shown (Supplemental Experimental Procedures S5). Metabolic modeling was performed using CWave software, version 3.0 (Mason et al., 2003) running in MATLAB (Natick, MA, USA) version R2014b with OS X version 10.9.4. The distributions of uncertainty were calculated using a Monte-Carlo analysis with 20 repetitions (Patel et al., 2010).
Oxygen consumption measurements
Oxygen consumption of INS-1 cells was measured in DMEM media, similar to the described above, and according to the protocol previously described (Wikstrom et al., 2012). Cells were incubated with 2.5 mM glucose for 1 hour before the addition of glucose to a final concentration of 2.5, 5, 7 or 9 mM. Cellular respiration was perturbed at indicated times during the assay by the addition of oligomycin (5µM), FCCP (1µM) and a mixture of rotenone (5µM) and antimycin A (10µM).
Statistical Analysis
All data are reported as mean ± S.E.M. Comparisons of metabolic parameters were made by a one-way analysis of variance corrected for multiple comparisons using Dunnett’s test. Differences were considered to be significant at p<0.05.
Supplementary Material
Highlights.
LC-MS/MS positional 13C-enrichment for steady-state and dynamic flux analysis.
Intersecting metabolic fluxes are disentangled by deciphering citrate isotopomers.
Comprehensive precursor/product positional 13C label transfer analysis.
Quantitative mitochondrial oxidative, anaplerotic, cycling and exchange rates.
Acknowledgments
This work was supported by Public Health Service grants R01 AA-021984 (GM) and R01 DK-092606 and K08 DK-080142 (RGK) and support from the Yale CTSA UL1 RR-0024139, DRC P30 DK-45735, O’Brien Kidney Center P30 DK79310-01A1, Mouse Metabolic Phenotyping Center isotope core U24 DK-059635, Liver Center P30-DK-034989 and Cancer Center IRG-058-012-53. We would like to thank Drs. Gerald Shulman and Douglas Rothman (Yale) for critically reading the manuscript, Anton Bennett (Yale) and Chris Newgard (Duke) for cell lines, and Anthony Romanelli and Keith Goodman (SCIEX) for assistance developing the differential mobility separations.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Hans Krebs- Banquet Speech, Nobelprize.org. Nobel Media AB 2014. Web. 21 Jan 2015. <http://www.nobelprize.org/nobel_prizes/medicine/laureates/1953/krebs-speech.html>
Author Contributions
T.A. and R.G.K. designed experiments, analyzed data, derived relationships and wrote the manuscript. T.A., R.P., X.Z., O.Y. performed experiments. R.P, O.S., and S.S. performed and interpreted O2 consumption experiments. T.A., G.C. and G.M performed/interpreted tcaCALC, tcaSIM and CWave analyses of NMR and LC/MS data and edited the manuscript.
References
- Bequette BJ, Sunny NE, El-Kadi SW, Owens SL. Application of stable isotopes and mass isotopomer distribution analysis to the study of intermediary metabolism of nutrients. J. Anim. Sci. 2006;(84 Suppl):E50–E59. doi: 10.2527/2006.8413_supple50x. [DOI] [PubMed] [Google Scholar]
- Buescher JM, Antoniewicz MR, Boros LG, Burgess SC, Brunengraber H, Clish CB, DeBerardinis RJ, Feron O, Frezza C, Ghesquiere B, et al. A road map for interpreting 13C metabolite labeling patterns from cells. Curr Opin Biotechnol. 2015;34:189–201. doi: 10.1016/j.copbio.2015.02.003. http://dx.doi.org/10.1016/j.copbio.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen AP, Hurd RE, Schroeder MA, Lau AZ, Gu YP, Lam WW, Barry J, Tropp J, Cunningham CH. Simultaneous investigation of cardiac pyruvate dehydrogenase flux, Krebs cycle metabolism and pH, using hyperpolarized [1,2-(13)C2]pyruvate in vivo. NMR in biomedicine. 2012;25:305–311. doi: 10.1002/nbm.1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline GW, Lepine RL, Papas KK, Kibbey RG, Shulman GI. 13C NMR isotopomer analysis of anaplerotic pathways in INS-1 cells. J. Biol. Chem. 2004;279:44370–44375. doi: 10.1074/jbc.M311842200. [DOI] [PubMed] [Google Scholar]
- de Graaf RA, Rothman DL, Behar KL. State of the art direct 13C and indirect 1H-[13C] NMR spectroscopy in vivo. A practical guide. NMR Biomed. 2011;24:958–972. doi: 10.1002/nbm.1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellerstein MK, Neese RA. Mass isotopomer distribution analysis: a technique for measuring biosynthesis and turnover of polymers. Am. J. Physiol. 1992;263:E988–E1001. doi: 10.1152/ajpendo.1992.263.5.E988. [DOI] [PubMed] [Google Scholar]
- Jeffrey FM, Rajagopal A, Malloy CR, Sherry AD. 13C–NMR: a simple yet comprehensive method for analysis of intermediary metabolism. Trends Biochem. Sci. 1991;16:5–10. doi: 10.1016/0968-0004(91)90004-f. [DOI] [PubMed] [Google Scholar]
- Jensen MV, Joseph JW, Ilkayeva O, Burgess S, Lu D, Ronnebaum SM, Odegaard M, Becker TC, Sherry AD, Newgard CB. Compensatory responses to pyruvate carboxylase suppression in islet beta-cells. Preservation of glucose-stimulated insulin secretion. J. Biol. Chem. 2006;281:22342–22351. doi: 10.1074/jbc.M604350200. [DOI] [PubMed] [Google Scholar]
- Kibbey RG, Choi CS, Lee H-Y, Cabrera O, Pongratz RL, Zhao X, Birkenfeld AL, Li C, Berggren P-O, Stanley C, et al. Mitochondrial GTP insensitivity contributes to hypoglycemia in hyperinsulinemia hyperammonemia by inhibiting glucagon release. Diabetes. 2014;63:4218–4229. doi: 10.2337/db14-0783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kibbey RG, Pongratz RL, Romanelli AJ, Wollheim CB, Cline GW, Shulman GI. Mitochondrial GTP regulates glucose-stimulated insulin secretion. Cell Metab. 2007;5:253–264. doi: 10.1016/j.cmet.2007.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz MA, Azzouny El MA, Kennedy RT, Burant CF. Metabolome response to glucose in the β-cell line INS-1 832/13. J. Biol. Chem. 2013;288:10923–10935. doi: 10.1074/jbc.M112.414961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Mulder H, Zhao P, Burgess SC, Jensen MV, Kamzolova S, Newgard CB, Sherry AD. 13C NMR isotopomer analysis reveals a connection between pyruvate cycling and glucose-stimulated insulin secretion (GSIS) Proc. Natl. Acad. Sci. U.S.A. 2002;99:2708–2713. doi: 10.1073/pnas.052005699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald MJ. Metabolism of the insulin secretagogue methyl succinate by pancreatic islets. Arch. Biochem. Biophys. 1993;300:201–205. doi: 10.1006/abbi.1993.1028. [DOI] [PubMed] [Google Scholar]
- Malloy CR, Sherry AD, Jeffrey FM. Evaluation of carbon flux and substrate selection through alternate pathways involving the citric acid cycle of the heart by 13C NMR spectroscopy. J. Biol. Chem. 1988;263:6964–6971. [PubMed] [Google Scholar]
- Mason GF, Falk Petersen K, de Graaf RA, Kanamatsu T, Otsuki T, Shulman GI, Rothman DL. A comparison of (13)C NMR measurements of the rates of glutamine synthesis and the tricarboxylic acid cycle during oral and intravenous administration of [1-(13)C]glucose. Brain Res. Brain Res. Protoc. 2003;10:181–190. doi: 10.1016/s1385-299x(02)00217-9. [DOI] [PubMed] [Google Scholar]
- Patel AB, de Graaf RA, Rothman DL, Behar KL, Mason GF. Evaluation of cerebral acetate transport and metabolic rates in the rat brain in vivo using 1H-[13C]-NMR. J. Cereb. Blood Flow Metab. 2010;30:1200–1213. doi: 10.1038/jcbfm.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pongratz RL, Kibbey RG, Shulman GI, Cline GW. Cytosolic and mitochondrial malic enzyme isoforms differentially control insulin secretion. J. Biol. Chem. 2007;282:200–207. doi: 10.1074/jbc.M602954200. [DOI] [PubMed] [Google Scholar]
- Quek L-E, Wittmann C, Nielsen LK, Krömer JO. OpenFLUX: efficient modelling software for 13C–based metabolic flux analysis. Microb. Cell Fact. 2009;8:25. doi: 10.1186/1475-2859-8-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronnebaum SM, Ilkayeva O, Burgess SC, Joseph JW, Lu D, Stevens RD, Becker TC, Sherry AD, Newgard CB, Jensen MV. A pyruvate cycling pathway involving cytosolic NADP-dependent isocitrate dehydrogenase regulates glucose-stimulated insulin secretion. J. Biol. Chem. 2006;281:30593–30602. doi: 10.1074/jbc.M511908200. [DOI] [PubMed] [Google Scholar]
- Ronnebaum SM, Jensen MV, Hohmeier HE, Burgess SC, Zhou Y-P, Qian S, MacNeil D, Howard A, Thornberry N, Ilkayeva O, et al. Silencing of cytosolic or mitochondrial isoforms of malic enzyme has no effect on glucose-stimulated insulin secretion from rodent islets. J. Biol. Chem. 2008;283:28909–28917. doi: 10.1074/jbc.M804665200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schryer DW, Peterson P, Paalme T, Vendelin M. Bidirectionality and compartmentation of metabolic fluxes are revealed in the dynamics of isotopomer networks. Int J Mol Sci. 2009;10:1697–1718. doi: 10.3390/ijms10041697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson NE, Khokhlova N, Oca-Cossio JA, Constantinidis I. Insights into the role of anaplerosis in insulin secretion: A 13C NMR study. Diabetologia. 2006;49:1338–1348. doi: 10.1007/s00125-006-0216-5. [DOI] [PubMed] [Google Scholar]
- Stark R, Pasquel F, Turcu A, Pongratz RL, Roden M, Cline GW, Shulman GI, Kibbey RG. Phosphoenolpyruvate cycling via mitochondrial phosphoenolpyruvate carboxykinase links anaplerosis and mitochondrial GTP with insulin secretion. J. Biol. Chem. 2009;284:26578–26590. doi: 10.1074/jbc.M109.011775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanizawa Y, Nakai K, Sasaki T, Anno T, Ohta Y, Inoue H, Matsuo K, Koga M, Furukawa S, Oka Y. Unregulated elevation of glutamate dehydrogenase activity induces glutamine-stimulated insulin secretion: identification and characterization of a GLUD1 gene mutation and insulin secretion studies with MIN6 cells overexpressing the mutant glutamate dehydrogenase. Diabetes. 2002;51:712–717. doi: 10.2337/diabetes.51.3.712. [DOI] [PubMed] [Google Scholar]
- Weitzel M, Nöh K, Dalman T, Niedenführ S, Stute B, Wiechert W. 13CFLUX2--high-performance software suite for (13)C-metabolic flux analysis. Bioinformatics. 2013;29:143–145. doi: 10.1093/bioinformatics/bts646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikstrom JD, Sereda SB, Stiles L, Elorza A, Allister EM, Neilson A, Ferrick DA, Wheeler MB, Shirihai OS. A novel high-throughput assay for islet respiration reveals uncoupling of rodent and human islets. PLoS ONE. 2012;7:33023. doi: 10.1371/journal.pone.0033023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young JD. INCA: a computational platform for isotopically non-stationary metabolic flux analysis. Bioinformatics. 2014;30:1333–1335. doi: 10.1093/bioinformatics/btu015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, White LT, Doumen C, Damico LA, LaNoue KF, Alpert NM, Lewandowski ED. Kinetic analysis of dynamic 13C NMR spectra: metabolic flux, regulation, and compartmentation in hearts. Biophys. J. 1995;69:2090–2102. doi: 10.1016/S0006-3495(95)80080-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.