Abstract
Dysregulation of amyloid-β (Aβ) metabolism is critical for Alzheimer's disease (AD) pathogenesis. Mounting evidence suggests that apolipoprotein E (ApoE) is involved in Aβ metabolism. ATP-binding cassette transporter A1 (ABCA1) is a key regulator of ApoE lipidation, which affects Aβ levels. Therefore, identifying regulatory mechanisms of ABCA1 expression in the brain may provide new therapeutic targets for AD. Here, we demonstrate that microRNA-33 (miR-33) regulates ABCA1 and Aβ levels in the brain. Overexpression of miR-33 impaired cellular cholesterol efflux and dramatically increased extracellular Aβ levels by promoting Aβ secretion and impairing Aβ clearance in neural cells. In contrast, genetic deletion of mir-33 in mice dramatically increased ABCA1 levels and ApoE lipidation, but it decreased endogenous Aβ levels in cortex. Most importantly, pharmacological inhibition of miR-33 via antisense oligonucleotide specifically in the brain markedly decreased Aβ levels in cortex of APP/PS1 mice, representing a potential therapeutic strategy for AD.
SIGNIFICANCE STATEMENT Brain lipid metabolism, in particular Apolipoprotein E (ApoE) lipidation, is critical to Aβ metabolism and Alzheimer's disease (AD). Brain lipid metabolism is largely separated from the periphery due to blood–brain barrier and different repertoire of lipoproteins. Therefore, identifying the novel regulatory mechanism of brain lipid metabolism may provide a new therapeutic strategy for AD. Although there have been studies on brain lipid metabolism, its regulation, in particular by microRNAs, is relatively unknown. Here, we demonstrate that inhibition of microRNA-33 increases lipidation of brain ApoE and reduces Aβ levels by inducing ABCA1. We provide a unique approach for AD therapeutics to increase ApoE lipidation and reduce Aβ levels via pharmacological inhibition of microRNA in vivo.
Keywords: ABCA1, abeta, Alzheimer's disease, ApoE, miR-33
Introduction
Dysregulation of Aβ peptide metabolism is hypothesized to initiate pathogenic cascades that lead to Alzheimer's disease (AD) (Haass and Selkoe, 2007). Therefore, strategies to regulate Aβ production and clearance are actively being pursued as AD therapies. Apolipoprotein E (APOE) genotype is the strongest genetic risk factor for AD, with the ε4 allele acting as an AD risk factor, whereas the ε2 allele is protective (Kim et al., 2009a). Previous studies have suggested that the main effect of APOE isoforms on AD risk is via their effect on Aβ metabolism (Kim et al., 2009a). In addition to APOE isoforms, alterations in ApoE protein levels and lipidation status have been shown to strongly influence Aβ metabolism (Kim et al., 2009a,b; Bien-Ly et al., 2012).
ApoE is a major cholesterol carrier in the brain (Kim et al., 2009a). Mounting evidence has demonstrated that dysregulation of cholesterol metabolism is implicated in AD pathogenesis (Kim et al., 2009a; Di Paolo and Kim, 2011). ATP-binding cassette transporter A1 (ABCA1) is a major cholesterol transporter that transfers cellular cholesterol onto lipid-poor apolipoproteins and regulates ApoE lipidation in the brain (Koldamova et al., 2014). Several genetic association studies have suggested possible association between polymorphisms in ABCA1 and AD risk (Koldamova et al., 2010). Moreover, animal studies have demonstrated that ABCA1 levels regulate Aβ deposition in vivo (Hirsch-Reinshagen et al., 2004; Wahrle et al., 2004, 2005, 2008; Koldamova et al., 2005). Furthermore, several lines of in vitro evidence support the critical role of ABCA1 in Aβ production, although how ABCA1 regulates Aβ production remains to be clarified (Koldamova et al., 2003; Sun et al., 2003; Kim et al., 2007). Given its effects on ApoE and Aβ metabolism, ABCA1 is considered to be a promising therapeutic target for AD. Therefore, identifying the regulatory mechanisms of ABCA1 expression in the brain may provide new therapeutic strategies for AD. In this study, we address this critical gap in our understanding of cerebral ABCA1 regulation.
Recently, microRNAs (miRNAs) have emerged as a new class of therapeutic targets for various diseases, including neurodegenerative disorders (Nelson and Keller, 2007; Hébert et al., 2009). Small noncoding miRNAs bind to their target mRNAs and repress the expression of target genes through translational repression and/or mRNA decay. Previously, we demonstrated that ABCA1 expression is regulated by miR-33 in the periphery (Horie et al., 2010; Rayner et al., 2010, 2011a,b). Pharmacological inhibition of miR-33 increased plasma high-density lipoprotein cholesterol levels in mice (Horie et al., 2010; Najafi-Shoushtari et al., 2010; Rayner et al., 2011a) and primates (Rayner et al., 2011b; Rottiers et al., 2013), leading to a marked reduction of atherosclerosis phenotypes. However, the physiological function of miR-33 in the CNS and whether miR-33 has any pathological role in AD have never been investigated.
Importantly, miRNAs quite often show cell-type, tissue, and species-specific regulation of their target genes due to the differences in their expression levels and the complexity of interaction with their targets in different cellular contexts (Landgraf et al., 2007; Jovičić et al., 2013). Therefore, it is critical to study miRNA function in appropriate cell types and tissues. Given the species-specific phenotypes, it is critical to study the roles of miRNA in human cells particularly for therapeutic approaches. Here, we demonstrated that miR-33 suppresses ABCA1 expression in mouse and human neural cells. We also provide evidence that overexpression of miR-33 increases extracellular Aβ levels by promoting Aβ secretion in neuronal cells and impairing Aβ clearance in both neuronal and glial cells. In contrast, genetic deletion of mir-33 in mice dramatically increases ABCA1 levels and ApoE lipidation while decreasing endogenous Aβ levels in cortex. Most importantly, pharmacological inhibition of miR-33 in the brain dramatically increases ABCA1 levels and effectively reduces Aβ levels in a transgenic mouse model of Aβ amyloidosis.
Materials and Methods
Animals and anti-miR-33 treatment.
All procedures with animals followed the guidelines of the institutional animal care and use committee at Kyoto University and Washington University. We previously reported the generation of the mir-33 knock-out mice (Horie et al., 2010). miR-33 (−/−) mice were killed at 16 weeks of age for analyses. Regulus Therapeutics provided 2′-fluoro/methoxyethyl-modified, phosphorothioate-backbone-modified anti-miR-33 (TGCAATGCAACTACAATGCAC) and the mismatch control (TCCAATCCAACTTCAATCATC). Two-month-old APPsw/PSEN1Δ9 (APP/PS1) transgenic mice overexpressing Swedish mutant APP695 and human PSEN1 with an exon 9 deletion (Jankowsky et al., 2004) were treated with 30 μg/d of anti-miR-33 or mismatch control for 4 weeks via intracerebroventricular infusion using an osmotic pump (Alzet) implanted in the subcutaneous space of the back. Cerebral cortical tissues were dissected out and frozen immediately. Female mice were used for all experiments.
qRT-PCR.
Total RNAs were extracted from mouse cortical tissues using TRIzol Reagent (Invitrogen). For mRNA quantification, the extracted RNAs were reverse transcribed using the High Capacity cDNA Reverse Transcription kit (Applied Biosystems). Quantitative PCR was performed with Power SYBR Green PCR Master Mix (Applied Biosystems) on an ABI 7500 (Applied Biosystems) using the default thermal cycling program. For miR-33 quantification, the total RNAs were reverse transcribed with miScript II RT kit using HiSpec buffer (QIAGEN). Quantitative PCR was performed with miScript SYBR Green PCR kit (QIAGEN) on an ABI 7500 (Applied Biosystems). Relative mRNA and miRNA levels were calculated by comparative Ct method using GeneEx 5.3.2 (multiday analyses). GAPDH and U6 were used as normalization controls for SREBF2 and miR-33, respectively. miR-33 levels in the body were normalized with the total RNA amounts due to the high variation in U6 levels among different tissues.
Luciferase assay.
Luciferase assays were performed as previously described (Kim et al., 2012). Briefly, Neuro2a mouse neuronal (N2a) cells were plated at a density of 4 × 104 cells per well in a 96-well plate 1 d before transfection; 0.12 μg of psiCHECK-2-human ABCA1 3′UTR luciferase reporter vector containing the entire 3′UTR of ABCA1 mRNA downstream of Renilla luciferase was transfected to the cells. After 8 h, media were changed to fresh DMEM/10% FBS, and the cells were allowed to recover overnight. At 24 h after the first transfection, the cells were transfected again with miR-33 or negative control at a final concentration of 75 nm for 24 h. Luciferase activity was measured using the Dual-Glo Luciferase Assay System (Promega). Renilla luciferase activity was normalized with the corresponding firefly luciferase activity. Each transfection condition was performed in triplicate, and the experiments were repeated twice.
Aβ secretion assay.
Aβ secretion assays were performed as previously described (Kim et al., 2012). Briefly, N2a-APPsw and H4-APPsw cells were transfected with miR-33 or scrambled control (Insight Genomics) together with or without mAbca1 open reading frame (ORF) plasmid as indicated using Lipofectamine 2000 (Invitrogen). At 48 h after transfection, media were changed. At 6 h after media change, the cells and media were collected for analyses. All experiments were performed in duplicate or triplicate and repeated independently 3 times.
Aβ clearance assay.
Primary astrocytes were prepared as described previously (Kim et al., 2009b). Primary astrocyte cells were transfected with miR-33 or scrambled control together with or without mAbca1 ORF using Lipofectamine LTX (Invitrogen). At 48 h after transfection, cells were incubated in fresh DMEM/F-12/N2/1 mm sodium pyruvate medium containing 200 nm Aβ40 for 24 h. Aβ clearance assays with N2a and H4 cells were performed with 20 nm Aβ40 as previously described (Kim et al., 2012). Media were applied for Western blot analysis of Aβ. Aβ levels were normalized with total protein levels. All experiments were performed in duplicate or triplicate and repeated independently 3 times. Aβ40 monomers were prepared as previously described (Kim et al., 2012).
Aβ degradation assay.
Astrocytes were transfected with miR-33 or scrambled control. At 48 h after transfection, the cells were incubated in fresh serum-free media. After 24 h incubation, conditioned media were collected and were incubated with 200 nm of synthetic Aβ40. After 24 h of incubation, the remaining Aβ40 levels in the media were analyzed by Western blots. For extracellular Aβ degradation with PBS-soluble fractions from mir-33 knock-out or wild-type (WT) mice, 20 μg of total proteins from each fraction was incubated with 200 nm of synthetic Aβ40 in 200 μl of DMEM/F-12 for the indicated times, and then the remaining Aβ40 levels were analyzed by Western blots.
Cholesterol efflux assay.
Cholesterol efflux assays were performed as described previously (Kim et al., 2012). N2a cells and astrocytes were plated in 12-well plates at a density of 1 × 106 and 4 × 105 cells/well, respectively, and transfected with miR-33 or scrambled control for 24 h. Cells were then loaded with 0.5 μCi/ml 3H-cholesterol (PerkinElmer) for an additional 24 h. Liver X receptor agonist TO901317 (Cayman Chemical) was applied to the cells at 1 μm for 12 h. Cells were then washed twice with PBS and incubated with 2% fatty-acid free BSA (Sigma) in media in the presence of acetyl-coenzyme A acetyltransferase inhibitor (2 μm) for 2 h before the addition of 50 μg/ml human ApoE (Leinco Technologies) in fatty-acid free BSA media. Supernatants were collected after 6 h, and radioactivity was counted and expressed as a percentage of total cell 3H-cholesterol content (total effluxed 3H-cholesterol + cell-associated 3H-cholesterol). Experiments were repeated 3 times.
Western blot analysis.
Western blots were performed as previously described (Kim et al., 2009b, 2012). Cells were lysed in RIPA buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 0.25% deoxycholic acid, 1% NP-40, 0.1% SDS, and 1 mm EDTA) with protease inhibitor mixture (Roche). Brain tissues were sequentially extracted with PBS and RIPA buffer with protease inhibitor mixture. For SDS-PAGE, equal amounts of total protein for each lysate were separated on 4%–20% TGX (Tris-Glycine eXtended) gels (Bio-Rad). For native PAGE, proteins adjusted to equal amounts of ApoE per lane were separated by 4%–20% TGX gels for 11 h at 4°C under nondenaturing condition. After transferring proteins to 0.22 μm pore size nitrocellulose membranes (Bio-Rad), blots were probed with mouse anti-ABCA1 antibody (HJ1, D.M.H. laboratory), rabbit anti-APP antibody (Invitrogen), goat-ApoE antibody (Meridian Life Science), mouse anti-Aβ antibody (82E1, IBL International), mouse anti-actin antibody (AC-15, Sigma), or mouse anti-tubulin antibody (Abcam) at room temperature for 2 h. CTFβ was also detected with 82E1 antibody. 82E1 antibody was raised against Aβ1–16 but does not react with noncleaved APP (IBL International). After secondary antibody incubation, membranes were developed using Lumigen TMA-6 ECL detection kit (Lumigen).
Sandwich ELISA for Aβ.
ELISA for Aβ was performed as previously described (Kim et al., 2009b). Cortical tissues were lysed in 0.2% diethylamine/50 mm NaCl lysis buffer with EDTA-free protease inhibitor mixture (Roche) by sonication, as previously described (Kim et al., 2008). The lysates were centrifuged at 18,000 × g for 30 min, and the supernatants were collected. The level of Aβ was measured by sandwich ELISA. HJ2 (anti-Aβ35-40) and HJ7.4 (anti-Aβ37-42) were used as capture antibodies, and HJ5.1-biotin (anti-Ab13-28) was used as the detection antibody. All antibodies were generated by the D.M.H. laboratory. The specificity of Aβ ELISA was validated with the brain tissue lysates from APP knock-out mice. The levels of Aβ were normalized with the levels of total protein.
Statistical analysis.
To determine the statistical significance, we first tested whether our datasets passed the equal variance test (Levene Median test) and normality test (Kolmogorov–Smirnov test) using SigmaStat 3.5 software. After confirmation that the data did not violate the assumptions of parametric testing, the data were analyzed with a two-tailed Student's t test (GraphPad Prism 5).
Results
miR-33 is highly enriched in the brain
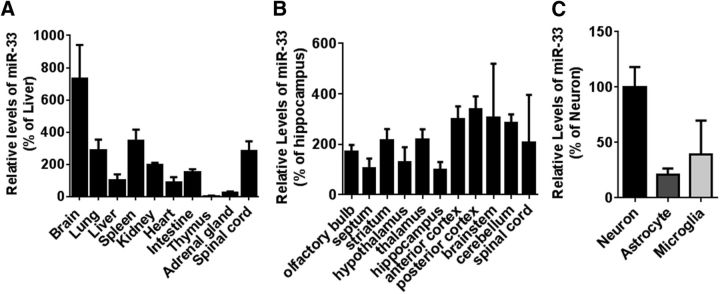
Given the tissue-specific expression of miRNAs and their diverse functions in different tissues (Landgraf et al., 2007), it is important to determine the spatial expression pattern of a miRNA and investigate its function in physiologically relevant cell types. We first measured miR-33 expression among several tissues. Interestingly, miR-33 is highly enriched in the brain (Fig. 1A) and is widely expressed across multiple brain regions (Fig. 1B).
Figure 1.
miR-33 is enriched in the brain. A, Relative expression levels of miR-33 in different mouse tissues. Endogenous levels of miR-33 were analyzed by qRT-PCR in different mouse tissues from 3.5-month-old C57B6 mice (n = 3). Each level was represented as a percentage of liver. B, Relative expression of miR-33 in the mouse brain. Mature miR-33 levels were analyzed by qRT-PCR and were normalized with corresponding U6 levels (n = 2). Data are shown as a percentage of hippocampus. C, Relative expression levels of miR-33 in primary cells isolated from mice brains. miR-33 levels were analyzed in mouse primary neurons, astrocytes, and microglia by qRT-PCR. Data are shown as a percentage of neurons (n = 3). Values are mean ± SEM.
Many miRNAs show unique cell-type-specific expression patterns in different neural cell types, such as neurons, astrocytes, and microglia (Jovičić et al., 2013). To study miR-33 function in relevant cell types, we next measured miR-33 expression in mouse primary neural cells. miR-33 was expressed in all neural cell types with relatively higher expression in neurons, compared with glial cells (Fig. 1C). Therefore, we hypothesized that miR-33 may have a critical role in ABCA1 regulation and Aβ metabolism in the brain.
miR-33 suppresses ABCA1 expression in the brain
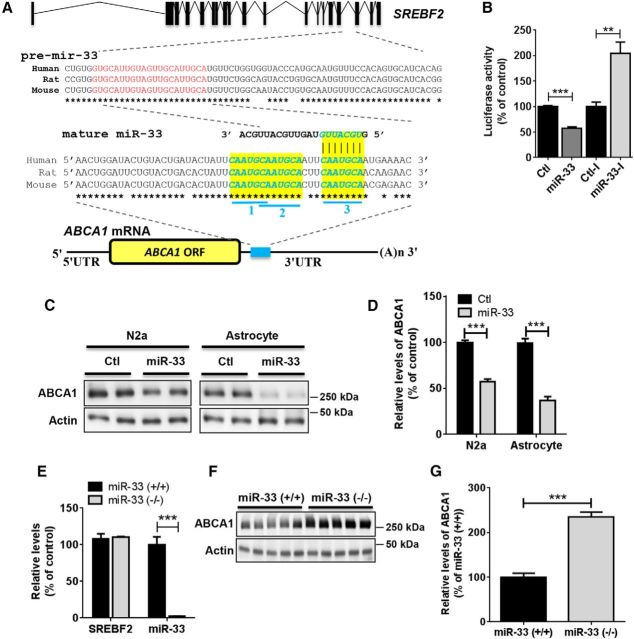
To determine whether miR-33 directly binds to the 3′UTR region of ABCA1 mRNA in neural cells, we performed a luciferase assay with the reporter construct containing the entire 3′UTR of human ABCA1 mRNA downstream of luciferase (Fig. 2A). miR-33 suppressed the luciferase activity, whereas LNA-based miR-33 inhibitor (referred to miR-33-I) increased luciferase expression in N2a cells (Fig. 2B). To determine whether miR-33 regulates ABCA1 expression in neural cells, we transfected synthetic miR-33 or scrambled negative control to N2a and astrocytes. miR-33 overexpression significantly suppressed ABCA1 expression in both cell types (Fig. 2C,D). These results demonstrated that miR-33 directly suppresses ABCA1 expression by targeting its 3′UTR region.
Figure 2.
miR-33 suppresses ABCA1 expression in the brain. A, Schematic diagram of the conserved target sites of miR-33 in the 3′UTR of ABCA1 mRNA. The mir-33 gene is located within the intron of Sterol regulatory element binding transcription factor 2 (SREBF2) gene. The genomic sequences around mir-33 are highly conserved in mammals. The mature sequence of miR-33 is shown in red within pre-mir-33. miR-33 has three potential targeting sites in the ABCA1. Blue underlines indicate 3′UTR. *Conserved nucleotides. B, miR-33 directly targets the 3′UTR of ABCA1 mRNA. (n = 6 per group). Luciferase reporter assays were performed with the reporter construct containing the full-length 3′UTR of hABCA1 mRNA downstream of Renilla luciferase. Renilla luciferase activity was normalized with the corresponding firefly luciferase activity. C, D, miR-33 decreased ABCA1 levels in N2a cells (n = 6) and mouse primary astrocytes (n = 5). Cells were transfected with miR-33 or scrambled negative control (Ctl). At 48 h after transfection, cells were harvested for Western blot analyses. ABCA1 protein levels were normalized by actin levels and quantified as a percentage of control. E, miR-33 is selectively deleted in cortex of miR-33 (−/−) mice without altering the expression of its host gene, SREBF2, in cortex. GAPDH levels were used for normalization. Data are shown as a percentage of WT. F, G, ABCA1 levels were increased in cortex of mir-33 knock-out mice (miR-33 (−/−), n = 5), compared with WT mice (miR-33 (+/+), n = 5). ABCA1 levels were analyzed by Western blot at 4 months of age. Actin levels were used for normalization. Data are shown as a percentage of control, and values are mean ± SEM. **p < 0.01 (t test). ***p < 0.001 (t test).
Previously, we generated mir-33-deficient mice (Horie et al., 2010) in which the mir-33 gene is ablated without altering the expression of the mir-33 host gene, SREBF2 (Fig. 2E). In many cases, miRNA knock-out mice showed no gross phenotype under normal physiological conditions, possibly due to the functional redundancy of multiple miRNAs for the same target gene (Ebert and Sharp, 2012; Mendell and Olson, 2012). Importantly, ABCA1 protein levels were strongly increased in cortex of mir-33-deficient mice (miR-33 (−/−)), compared with WT mice (miR-33 (+/+)) (Fig. 2F,G), suggesting that miR-33 plays a dominant role in regulating cerebral ABCA1 expression even under normal physiological conditions.
Deletion of mir-33 gene increases ApoE lipidation in the brain
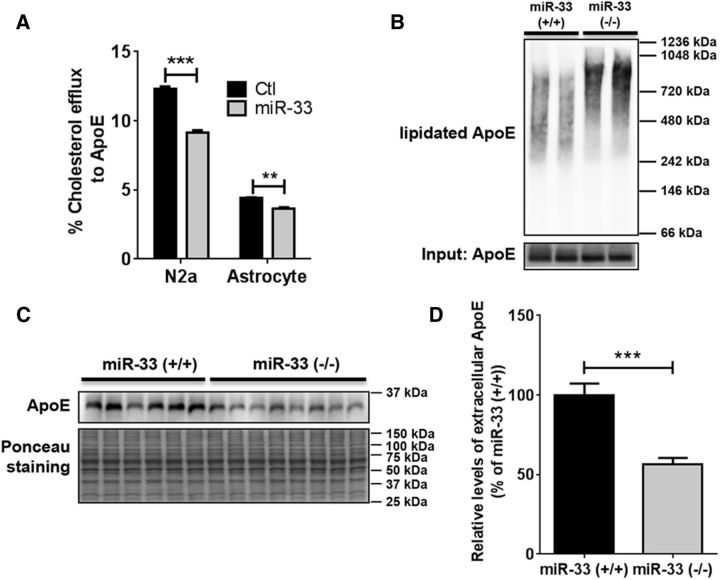
Because ABCA1 transports cellular cholesterol onto apolipoproteins, we assessed whether miR-33 regulates lipid transport in neural cells. We performed cholesterol efflux assays in N2a cells and astrocytes using ApoE as a cholesterol acceptor. miR-33 significantly decreased ApoE-mediated cholesterol efflux in both N2a cells and astrocytes (Fig. 3A).
Figure 3.
miR-33 regulates lipidation and level of ApoE in the brain. A, miR-33 impaired cellular cholesterol efflux in N2a and astrocytes. N2a and astrocytes were transfected with miR-33 or scrambled negative control for 24 h and then incubated the cells with radioactively labeled 3H-cholesterol. After 24 h, cells were incubated with human ApoE3 as a cholesterol acceptor. Data are shown as a percentage of total cellular 3H-cholesterol content (total effluxed 3H-cholesterol + cell-associated 3H-cholesterol). Values are mean ± SEM. **p < 0.01 (t test). ***p < 0.001 (t test). n = 3. B, mir-33 deletion increased highly lipidated ApoE-containing particles. PBS-soluble fractions were analyzed by native PAGE and ApoE-specific Western blot. Total ApoE levels applied for each lane were shown below. C, D, PBS-soluble apoE levels were decreased in mir-33-deficient mice, compared with WT. Extracellular protein-enriched PBS-soluble fraction from cortex of mir-33-deficient (n = 6) and WT mice (n = 8) were analyzed by Western blots. Total protein levels (Ponceau staining) were used for normalization. Data are shown as a percentage of control, and values are mean ± SEM. ***p < 0.001 (t test).
To determine whether mir-33 deletion increases ApoE lipidation in the brain, we performed the native PAGE followed by Western blot for ApoE with extracellular protein-enriched PBS-soluble fractions. ApoE-containing lipoprotein particles from the mir-33-deficient mice tended to be larger in size than the particles from WT mice, indicating that inhibition of miR-33 increases ApoE lipidation in the brain (Fig. 3B). Moreover, mir-33-deficient mice had significantly less PBS-soluble ApoE levels in cortex, compared with WT mice (Fig. 3C,D). Similar phenotypes were observed in our previous study using ABCA1 transgenic mice (Wahrle et al., 2008). Collectively, these data suggest that miR-33 regulates ApoE lipidation as well as extracellular ApoE levels in the brain.
miR-33 regulates Aβ secretion and Aβ clearance in neural cells
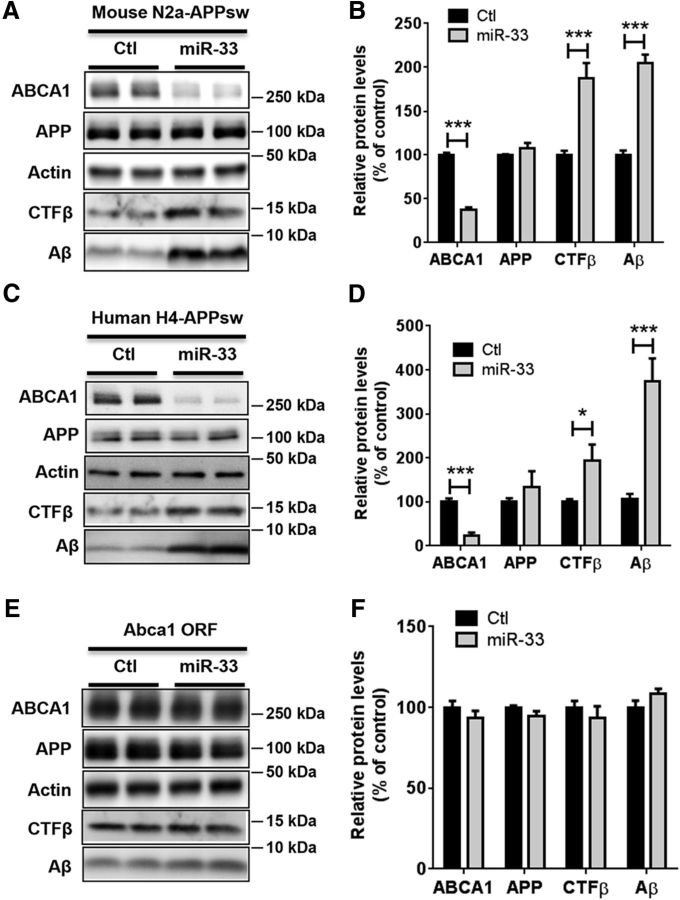
We and others previously demonstrated that ABCA1 decreases Aβ secretion by inhibiting BACE1-mediated APP cleavage without affecting BACE1 expression in neuronal cells (Sun et al., 2003; Kim et al., 2012). To determine whether miR-33 modulates Aβ secretion in neuronal cells, we transfected N2a cells expressing Swedish mutant amyloid precursor protein (APPsw) with miR-33 or scrambled control and then measured the secreted Aβ levels in the media. miR-33 markedly decreased ABCA1 levels and increased the levels of intracellular CTFβ, the cleavage product of APP by BACE1, and the secreted Aβ (Fig. 4A,B). As expected from previous ABCA1 studies (Sun et al., 2003; Kim et al., 2012), miR-33 did not affect the levels of BACE1 (Ctl 100.0 ± 1.774 vs miR-33 101.0 ± 3.652, n = 5, p = 0.8061). Similar results were observed in human H4 neuroglioma cells expressing APPsw transfected with miR-33 (Fig. 4C,D). To determine whether the increase of Aβ levels by miR-33 was mediated by repressing ABCA1, we tested whether the effect of miR-33 on Aβ levels could be rescued by the expression of miR-33-resistant ABCA1. In the presence of miR-33 resistant Abca1 without 3′UTR (Abca1 ORF), miR-33 did not affect ABCA1 levels nor Aβ levels in N2a cells expressing APPsw, suggesting that miR-33 regulates Aβ secretion via modulating ABCA1, rather than other genes (Fig. 4E,F).
Figure 4.
Overexpression of miR-33 increases Aβ secretion by suppressing ABCA1 in neural cells. Mouse N2a (A, B) and human H4 cells (C, D) expressing APPsw were transfected with miR-33 or scrambled control. At 48 h after transfection, media were changed to fresh serum-free media. At 6 h after media change, cells and media were collected for analyses. Intracellular ABCA1, APP, and CTFβ levels and the secreted Aβ levels in the media were measured by Western blots (n = 6 per group). Actin levels were used for normalization. E, F, Overexpression of Abca1 ORF restored miR-33-mediated alteration of secreted Aβ levels. Abca1 ORF plasmids were contransfected with miR-33 or scrambled control to N2a-expressing APPsw, and the secreted Aβ levels in the media were measured by Western blots (n = 5 per group). Actin levels were used for normalization. Data are shown as a percentage of control, and values are mean ± SEM. *p < 0.05 (t test). ***p < 0.001 (t test).
In addition to its role in Aβ secretion, ABCA1 has been also shown to decrease Aβ levels by promoting extracellular Aβ clearance in both neurons and glial cells (Jiang et al., 2008; Kim et al., 2012). Therefore, we examined whether miR-33 regulates Aβ clearance. Cells were first transfected with miR-33 or scrambled control and then incubated with synthetic Aβ. After 24 h of incubation, levels of Aβ remaining in the media were measured by Western blot. We found that miR-33 dramatically impaired Aβ clearance in N2a, H4, and mouse primary astrocytes (Fig. 5A,B). Moreover, expression of miR-33-resistant Abca1 ORF completely restored the miR-33-mediated reduction of Aβ clearance in mouse primary astrocytes (Fig. 5C,D). These data suggest that impairment of Aβ clearance by miR-33 was mediated by ABCA1.
Figure 5.
Overexpression of miR-33 impairs Aβ clearance in neural cells. A, B, Overexpression of miR-33 decreased Aβ clearance in neural cells. N2a, H4, and mouse primary astrocytes were transfected with miR-33 or scrambled control. At 48 h after transfection, cells were incubated with synthetic Aβ40 monomers for 24 h. The remaining Aβ40 levels in the media were analyzed by Western blots (n = 5 per group). C, D, Overexpression of Abca1 ORF restored the reduction of Aβ clearance. Abca1 ORF plasmids were contransfected with miR-33 or scrambled control to mouse primary astrocytes. At 48 h after transfection, cells were incubated with synthetic Aβ40 monomers for 24 h. The remaining Aβ40 levels in the media were analyzed by Western blots (n = 4 per group). Data are shown as a percentage of control, and values are mean ± SEM. **p < 0.01 (t test). ***p < 0.001 (t test).
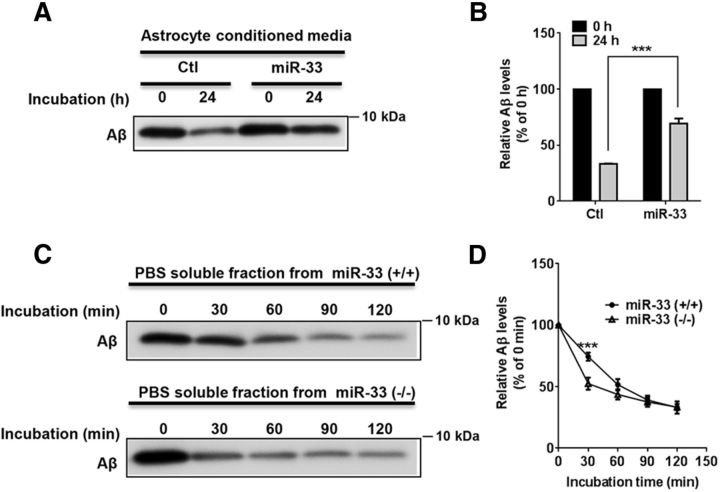
A previous report demonstrated that extracellular Aβ degradation by proteases, such as insulin-degrading enzyme, is influenced by the lipidation status of ApoE (Jiang et al., 2008). Because miR-33 regulates ApoE lipidation (Fig. 3A,B), we next examined whether miR-33 could regulate extracellular Aβ degradation. After transfecting miR-33 or scrambled control miR to astrocytes, we collected conditioned media for in vitro Aβ degradation assay. After incubating synthetic Aβ40 in the conditioned media for 24 h, we measured the levels of Aβ remaining in the media using Western blot. Overexpression of miR-33 significantly impaired Aβ degradation in the astrocyte conditioned media (Fig. 6A,B). To determine whether mir-33 deletion could affect extracellular Aβ degradation, we performed ex vivo Aβ degradation assay using cerebral cortical tissue from WT and mir-33-deficient mice. After incubating synthetic Aβ40 in the extracellular protein-enriched PBS-soluble cortical tissue homogenates, we measured the remaining Aβ levels at different time points. We found that mir-33 deletion significantly facilitated extracellular Aβ degradation, compared with WT (Fig. 6C,D). Together, our data suggest that miR-33 regulates extracellular Aβ levels by modulating Aβ secretion as well as Aβ degradation. Our data also imply that the role of miR-33 in regulating ABCA1 and Aβ levels is conserved in mouse and human neural cells.
Figure 6.
miR-33 regulates extracellular Aβ degradation. A, B, Overexpression of miR-33 impaired extracellular Aβ degradation in the conditioned media from astrocytes. Primary astrocytes were transfected with miR-33 or scrambled control. At 48 h after transfection, conditioned media were collected and were incubated with 200 nm of synthetic Aβ40. After 24 h of incubation, the levels of Aβ40 remaining in the media were analyzed by Western blots (n = 3 per group). C, D, Deletion of miR-33 facilitated extracellular Aβ degradation in PBS-soluble fractions of cortices. PBS-soluble fraction of each mouse cortex was adjusted to the same amount of total proteins and then incubated with 200 nm of synthetic Aβ40 for the indicated time (n = 6 per group). Data are shown as a percentage of control, and values are mean ± SEM. ***p < 0.001 (two-way ANOVA).
Inhibition of miR-33 effectively decreases Aβ levels in the brain
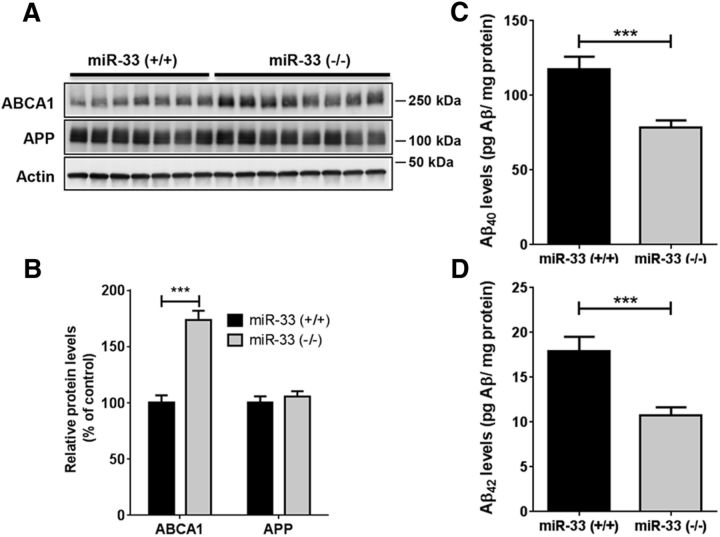
Given strong in vitro evidence that miR-33 regulates Aβ levels in neural cells, we next investigated whether inhibition of miR-33 decreases Aβ levels in vivo. We first assessed whether mir-33 deletion decreases mouse endogenous Aβ levels in the brain. mir-33 gene deletion markedly increased ABCA1 levels (Fig. 7A,B) and decreased endogenous Aβ40 levels by 33% and Aβ42 levels by 40% in cortex (Fig. 7C,D).
Figure 7.
mir-33 deletion decreases mouse endogenous Aβ levels in cortex. Compared with WT mice (n = 8), ABCA1 levels were dramatically increased in cortex of mir-33 knock-out mice (n = 7) at 4 months of age, without altering APP levels (A, B). The levels of Aβ40 (C) and Aβ42 (D) are significantly decreased in cortex of mir-33 knock-out mice (n = 7), compared with WT (n = 9). Actin levels were used for normalization of Western blots. Aβ levels were measured by Aβ-specific ELISA and normalized by total protein levels. Data are shown as a percentage of control, and values are mean ± SEM. ***p < 0.001 (t test).
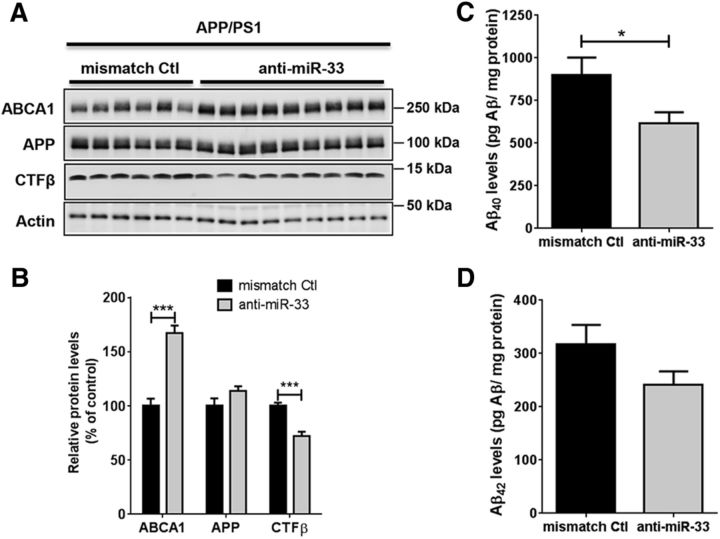
Next, we determined whether inhibition of miR-33 specifically in the brain can decrease human Aβ levels in a mouse model that produces human Aβ in the brain (APP/PS1 mice). Using 2′-fluoro/methoxyethyl-modified- and phosphorothioate-backbone-modified antisense miR-33 (denoted anti-miR-33) (Rayner et al., 2011a,b), we treated 2-month-old APP/PS1 mice with anti-miR-33 or mismatch negative control for 4 weeks via intracerebroventricular infusion using osmotic pumps. Anti-miR-33 significantly increased ABCA1 protein levels and decreased CTFβ levels in cortex, compared with the mismatch control (Fig. 8A,B). Importantly, anti-miR-33 therapy significantly decreased Aβ40 levels in cortex (Fig. 8C). There was also a trend showing that anti-miR-33 decreased Aβ42 levels (24% difference between means) (Fig. 8D). Collectively, these results demonstrated that inhibition of cerebral miR-33 effectively reduces Aβ levels in vivo.
Figure 8.
Brain-specific miR-33 inhibition decreases Aβ levels in cortex of APP/PS1 mice. Two-month-old APP/PS1 mice were treated with anti-miR-33 or the mismatch negative control for 4 weeks via intracerebroventricular infusion. Anti-miR-33 markedly increased ABCA1 levels in cortex of APP/PS1 mice (n = 9), compared with the mismatch control (n = 6) (A, B). In the anti-miR-33 treatment group (n = 9), Aβ40 levels were significantly decreased (p = 0.0286) (C) and Aβ42 levels showed a trend toward decrease (p = 0.0954) (D), compared with the mismatch control (n = 6). Actin levels were used for normalization of Western blots. Aβ levels were measured by Aβ-specific ELISA and normalized by total protein levels. Data are shown as a percentage of control, and values are mean ± SEM. *p < 0.05 (t test). ***p < 0.001 (t test).
Discussion
Despite the critical role of Aβ metabolism in AD pathogenesis, our understanding of how endogenous Aβ levels are regulated is still limited. miRNAs have been identified as a new class of regulatory molecules that regulate many biological functions, including pathogenesis of neurological diseases. Although several in vitro studies have implicated miRNAs in AD pathogenesis, whether pharmacological inhibition of a miRNA can be a practical strategy to regulate brain Aβ levels in vivo has not yet been demonstrated. For the first time, we demonstrate here that genetic deletion and pharmacological inhibition, using antisense oligonucleotide, of miR-33 decreased Aβ levels in the brain. In addition, our study provides the first evidence that miR-33 is an endogenous regulator of ABCA1 in the brain and brain-specific miR-33 antagonism may be an effective strategy to modulate Aβ levels.
miRNAs often show strong cell type- and tissue-specific regulation of their targets (Landgraf et al., 2007; Jovičić et al., 2013). Therefore, ABCA1 regulation by miR-33 in the periphery does not necessarily mean that miR-33 will regulate ABCA1 in the brain. In our current study, we demonstrated that miR-33 effectively suppresses ABCA1 in the brain. miRNA overexpression over a certain physiological level may lead to an artificial effect by surpassing biological buffering capacity of the cells. Therefore, it is crucial to assess the function of miRNA by modulating endogenous miRNA levels. Somewhat unexpectedly, the vast majority of miRNA knock-out mice examined thus far lack any strong phenotypes under normal physiological conditions (Ebert and Sharp, 2012; Mendell and Olson, 2012). This may be due to the functional redundancy of many miRNAs in regulating the same target. In addition, low expression levels of miRNAs in certain cell types and tissues may partially account for the lack of observable effects on their targets (Jovičić et al., 2013). Here, we clearly demonstrated that endogenous miR-33 suppresses ABCA1 expression under basal condition by using mir-33-deficient mice as well as inhibiting endogenous miR-33 specifically in the brain. Therefore, our study suggests that miR-33 is a potent regulator of ABCA1 in the brain under normal physiological condition.
Although how ABCA1 regulates Aβ levels still remains unclear, several in vitro studies have suggested that ABCA1 inhibits Aβ secretion in neuronal cells by modulating APP proteolysis and membrane lipid homeostasis (Di Paolo and Kim, 2011). Moreover, we and others have shown that ABCA1 is required for the effective clearance of Aβ by neuronal and glial cells, and ApoE lipidation by ABCA1 is critical for Aβ clearance (Jiang et al., 2008; Kim et al., 2012). In line with these findings, we observed that overexpression of miR-33 increased Aβ secretion in neuronal cells by increasing β-cleavage of APP. Moreover, overexpression of miR-33 impaired ApoE lipidation and decreased Aβ clearance by neuronal and glial cells. In contrast, inhibition of miR-33 in the brain significantly decreased β-cleavage of APP in APP/PS1 mice. Moreover, deletion of miR-33 increased ApoE lipidation and facilitated extracellular Aβ degradation, leading to significant reduction of brain Aβ levels, compared with WT. We also demonstrated that the roles of miR-33 in Aβ metabolism are conserved in mouse and human neural cells. These data suggest that miR-33 may represent a potential therapeutic target for AD by regulating metabolism of ApoE and Aβ.
Several miRNAs and antisense oligonucleotides (ASOs) are currently under investigation in clinical trials for various diseases (Kanasty et al., 2013; van Rooij and Kauppinen, 2014; Gaudet et al., 2015). Pharmacological modulations of disease-associated miRNAs appeared to be well tolerated with promising outcomes in animal models and human patients (van Rooij and Kauppinen, 2014). In the current study, we demonstrated that ASO-based pharmacological inhibition of miR-33 reduces Aβ levels in the brain after intracerebroventricular delivery of anti-miR-33 ASO. In clinical application for brain diseases, blood–brain barrier is a major obstacle in the delivery of small molecules into the brain. So far, no human clinical trial demonstrated that an ASO can reduce the levels of a target RNA or protein in the brain. However, it is noteworthy that intrathecal ASO injection to nonhuman primates led to accumulation of ASO in the cortical neuronal and glial cells (Rigo et al., 2014). It will be interesting to determine whether intrathecal ASO injection to human can also reproduce the similar data. Recently, intrathecal ASO injection to human was successfully completed in the Phase I and II clinical trials (Faravelli et al., 2015). Both trials demonstrated that intrathecal ASO injection is safe and effective. In addition, there have been substantial advances in the small RNA delivery technology into the brain. For example, conjugation with neurotrophic virus peptide and exosome- or liposome-based nanoparticles are promising avenues of future research (Kumar et al., 2007; Pulford et al., 2010; Alvarez-Erviti et al., 2011; Roshan et al., 2014). In addition to ASOs, there have been several successful cases that have identified small-molecule modulators of miRNAs (Velagapudi et al., 2015), further expanding the roles of miRNAs from diagnostic tools to therapeutic targets of diseases. Therefore, further therapeutic developments are warranted for the clinical application of miR-33 antagonism for AD.
Footnotes
This work was supported by Washington University McDonnell Center for Cellular and Molecular Neurobiology to Jungsu Kim, GHR Foundation to Jungsu Kim, Mayo Clinic Center for Individualized Medicine to Jungsu Kim, Alzheimer's Association to Jungsu Kim, National Institutes of Health Grants NS069329, AG016574, and AG005681 to Jungsu Kim, Grant AG042513 to J.R.C., Grant HL107953 to C.F.-H., Grant AG039708 to H.-S.H., Grant AG13956 to D.M.H., Mayo Graduate School to H.Y., BrightFocus Foundation A2010613 to P.B.V., and Grant A2012421 to Jungsu Kim. Anti-miR-33 reagent was provided by Regulus Therapeutics. We thank Dr. Timothy Miller (Washington University) for technical advice on anti-miR-33 infusion and Kelly Viola (Mayo Clinic) for editing this manuscript.
The authors declare no competing financial interests.
References
- Alvarez-Erviti L, Seow Y, Yin H, Betts C, Lakhal S, Wood MJ. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat Biotechnol. 2011;29:341–345. doi: 10.1038/nbt.1807. [DOI] [PubMed] [Google Scholar]
- Bien-Ly N, Gillespie AK, Walker D, Yoon SY, Huang Y. Reducing human apolipoprotein E levels attenuates age-dependent Abeta accumulation in mutant human amyloid precursor protein transgenic mice. J Neurosci. 2012;32:4803–4811. doi: 10.1523/JNEUROSCI.0033-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Paolo G, Kim TW. Linking lipids to Alzheimer's disease: cholesterol and beyond. Nat Rev Neurosci. 2011;12:284–296. doi: 10.1038/nrn3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebert MS, Sharp PA. Roles for microRNAs in conferring robustness to biological processes. Cell. 2012;149:515–524. doi: 10.1016/j.cell.2012.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faravelli I, Nizzardo M, Comi GP, Corti S. Spinal muscular atrophy: recent therapeutic advances for an old challenge. Nat Rev Neurol. 2015;11:351–359. doi: 10.1038/nrneurol.2015.77. [DOI] [PubMed] [Google Scholar]
- Gaudet D, Alexander VJ, Baker BF, Brisson D, Tremblay K, Singleton W, Geary RS, Hughes SG, Viney NJ, Graham MJ, Crooke RM, Witztum JL, Brunzell JD, Kastelein JJ. Antisense inhibition of apolipoprotein C-III in patients with hypertriglyceridemia. N Engl J Med. 2015;373:438–447. doi: 10.1056/NEJMoa1400283. [DOI] [PubMed] [Google Scholar]
- Haass C, Selkoe DJ. Soluble protein oligomers in neurodegeneration: lessons from the Alzheimer's amyloid beta-peptide. Nat Rev Mol Cell Biol. 2007;8:101–112. doi: 10.1038/nrm2101. [DOI] [PubMed] [Google Scholar]
- Hébert SS, Horré K, Nicolaï L, Bergmans B, Papadopoulou AS, Delacourte A, De Strooper B. MicroRNA regulation of Alzheimer's amyloid precursor protein expression. Neurobiol Dis. 2009;33:422–428. doi: 10.1016/j.nbd.2008.11.009. [DOI] [PubMed] [Google Scholar]
- Hirsch-Reinshagen V, Zhou S, Burgess BL, Bernier L, McIsaac SA, Chan JY, Tansley GH, Cohn JS, Hayden MR, Wellington CL. Deficiency of ABCA1 impairs apolipoprotein E metabolism in brain. J Biol Chem. 2004;279:41197–41207. doi: 10.1074/jbc.M407962200. [DOI] [PubMed] [Google Scholar]
- Horie T, Ono K, Horiguchi M, Nishi H, Nakamura T, Nagao K, Kinoshita M, Kuwabara Y, Marusawa H, Iwanaga Y, Hasegawa K, Yokode M, Kimura T, Kita T. MicroRNA-33 encoded by an intron of sterol regulatory element-binding protein 2 (Srebp2) regulates HDL in vivo. Proc Natl Acad Sci U S A. 2010;107:17321–17326. doi: 10.1073/pnas.1008499107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jankowsky JL, Fadale DJ, Anderson J, Xu GM, Gonzales V, Jenkins NA, Copeland NG, Lee MK, Younkin LH, Wagner SL, Younkin SG, Borchelt DR. Mutant presenilins specifically elevate the levels of the 42 residue beta-amyloid peptide in vivo: evidence for augmentation of a 42-specific gamma secretase. Hum Mol Genet. 2004;13:159–170. doi: 10.1093/hmg/ddh019. [DOI] [PubMed] [Google Scholar]
- Jiang Q, Lee CY, Mandrekar S, Wilkinson B, Cramer P, Zelcer N, Mann K, Lamb B, Willson TM, Collins JL, Richardson JC, Smith JD, Comery TA, Riddell D, Holtzman DM, Tontonoz P, Landreth GE. ApoE promotes the proteolytic degradation of Abeta. Neuron. 2008;58:681–693. doi: 10.1016/j.neuron.2008.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovičić A, Roshan R, Moisoi N, Pradervand S, Moser R, Pillai B, Luthi-Carter R. Comprehensive expression analyses of neural cell-type-specific miRNAs identify new determinants of the specification and maintenance of neuronal phenotypes. J Neurosci. 2013;33:5127–5137. doi: 10.1523/JNEUROSCI.0600-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanasty R, Dorkin JR, Vegas A, Anderson D. Delivery materials for siRNA therapeutics. Nat Mater. 2013;12:967–977. doi: 10.1038/nmat3765. [DOI] [PubMed] [Google Scholar]
- Kim J, Miller VM, Levites Y, West KJ, Zwizinski CW, Moore BD, Troendle FJ, Bann M, Verbeeck C, Price RW, Smithson L, Sonoda L, Wagg K, Rangachari V, Zou F, Younkin SG, Graff-Radford N, Dickson D, Rosenberry T, Golde TE. BRI2 (ITM2b) inhibits Abeta deposition in vivo. J Neurosci. 2008;28:6030–6036. doi: 10.1523/JNEUROSCI.0891-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Basak JM, Holtzman DM. The role of apolipoprotein E in Alzheimer's disease. Neuron. 2009a;63:287–303. doi: 10.1016/j.neuron.2009.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Castellano JM, Jiang H, Basak JM, Parsadanian M, Pham V, Mason SM, Paul SM, Holtzman DM. Overexpression of low-density lipoprotein receptor in the brain markedly inhibits amyloid deposition and increases extracellular A beta clearance. Neuron. 2009b;64:632–644. doi: 10.1016/j.neuron.2009.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Yoon H, Ramírez CM, Lee SM, Hoe HS, Fernández-Hernando C, Kim J. MiR-106b impairs cholesterol efflux and increases Abeta levels by repressing ABCA1 expression. Exp Neurol. 2012;235:476–483. doi: 10.1016/j.expneurol.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WS, Rahmanto AS, Kamili A, Rye KA, Guillemin GJ, Gelissen IC, Jessup W, Hill AF, Garner B. Role of ABCG1 and ABCA1 in regulation of neuronal cholesterol efflux to apolipoprotein E discs and suppression of amyloid-beta peptide generation. J Biol Chem. 2007;282:2851–2861. doi: 10.1074/jbc.M607831200. [DOI] [PubMed] [Google Scholar]
- Koldamova RP, Lefterov IM, Ikonomovic MD, Skoko J, Lefterov PI, Isanski BA, DeKosky ST, Lazo JS. 22R-hydroxycholesterol and 9-cis-retinoic acid induce ATP-binding cassette transporter A1 expression and cholesterol efflux in brain cells and decrease amyloid beta secretion. J Biol Chem. 2003;278:13244–13256. doi: 10.1074/jbc.M300044200. [DOI] [PubMed] [Google Scholar]
- Koldamova R, Staufenbiel M, Lefterov I. Lack of ABCA1 considerably decreases brain ApoE level and increases amyloid deposition in APP23 mice. J Biol Chem. 2005;280:43224–43235. doi: 10.1074/jbc.M504513200. [DOI] [PubMed] [Google Scholar]
- Koldamova R, Fitz NF, Lefterov I. The role of ATP-binding cassette transporter A1 in Alzheimer's disease and neurodegeneration. Biochim Biophys Acta. 2010;1801:824–830. doi: 10.1016/j.bbalip.2010.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koldamova R, Fitz NF, Lefterov I. ATP-binding cassette transporter A1: from metabolism to neurodegeneration. Neurobiol Dis. 2014;72:13–21. doi: 10.1016/j.nbd.2014.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar P, Wu H, McBride JL, Jung KE, Kim MH, Davidson BL, Lee SK, Shankar P, Manjunath N. Transvascular delivery of small interfering RNA to the central nervous system. Nature. 2007;448:39–43. doi: 10.1038/nature05901. [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, Lin C, Socci ND, Hermida L, Fulci V, Chiaretti S, Foà R, Schliwka J, Fuchs U, Novosel A, Müller RU, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendell JT, Olson EN. MicroRNAs in stress signaling and human disease. Cell. 2012;148:1172–1187. doi: 10.1016/j.cell.2012.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najafi-Shoushtari SH, Kristo F, Li Y, Shioda T, Cohen DE, Gerszten RE, Näär AM. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science. 2010;328:1566–1569. doi: 10.1126/science.1189123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson PT, Keller JN. RNA in brain disease: no longer just “the messenger in the middle.”. J Neuropathol Exp Neurol. 2007;66:461–468. doi: 10.1097/01.jnen.0000240474.27791.f3. [DOI] [PubMed] [Google Scholar]
- Pulford B, Reim N, Bell A, Veatch J, Forster G, Bender H, Meyerett C, Hafeman S, Michel B, Johnson T, Wyckoff AC, Miele G, Julius C, Kranich J, Schenkel A, Dow S, Zabel MD. Liposome-siRNA-peptide complexes cross the blood–brain barrier and significantly decrease PrP on neuronal cells and PrP in infected cell cultures. PLoS One. 2010;5:e11085. doi: 10.1371/journal.pone.0011085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner KJ, Suárez Y, Dávalos A, Parathath S, Fitzgerald ML, Tamehiro N, Fisher EA, Moore KJ, Fernández-Hernando C. MiR-33 contributes to the regulation of cholesterol homeostasis. Science. 2010;328:1570–1573. doi: 10.1126/science.1189862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner KJ, Sheedy FJ, Esau CC, Hussain FN, Temel RE, Parathath S, van Gils JM, Rayner AJ, Chang AN, Suarez Y, Fernandez-Hernando C, Fisher EA, Moore KJ. Antagonism of miR-33 in mice promotes reverse cholesterol transport and regression of atherosclerosis. J Clin Invest. 2011a;121:2921–2931. doi: 10.1172/JCI57275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner KJ, Esau CC, Hussain FN, McDaniel AL, Marshall SM, van Gils JM, Ray TD, Sheedy FJ, Goedeke L, Liu X, Khatsenko OG, Kaimal V, Lees CJ, Fernandez-Hernando C, Fisher EA, Temel RE, Moore KJ. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nature. 2011b;478:404–407. doi: 10.1038/nature10486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigo F, Chun SJ, Norris DA, Hung G, Lee S, Matson J, Fey RA, Gaus H, Hua Y, Grundy JS, Krainer AR, Henry SP, Bennett CF. Pharmacology of a central nervous system delivered 2′-O-methoxyethyl-modified survival of motor neuron splicing oligonucleotide in mice and nonhuman primates. J Pharmacol Exp Ther. 2014;350:46–55. doi: 10.1124/jpet.113.212407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roshan R, Shridhar S, Sarangdhar MA, Banik A, Chawla M, Garg M, Singh VP, Pillai B. Brain-specific knockdown of miR-29 results in neuronal cell death and ataxia in mice. RNA. 2014;20:1287–1297. doi: 10.1261/rna.044008.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottiers V, Obad S, Petri A, McGarrah R, Lindholm MW, Black JC, Sinha S, Goody RJ, Lawrence MS, deLemos AS, Hansen HF, Whittaker S, Henry S, Brookes R, Najafi-Shoushtari SH, Chung RT, Whetstine JR, Gerszten RE, Kauppinen S, Näär AM. Pharmacological inhibition of a microRNA family in nonhuman primates by a seed-targeting 8-mer antimiR. Sci Transl Med. 2013;5:212ra162. doi: 10.1126/scitranslmed.3006840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Yao J, Kim TW, Tall AR. Expression of liver X receptor target genes decreases cellular amyloid beta peptide secretion. J Biol Chem. 2003;278:27688–27694. doi: 10.1074/jbc.M300760200. [DOI] [PubMed] [Google Scholar]
- van Rooij E, Kauppinen S. Development of microRNA therapeutics is coming of age. EMBO Mol Med. 2014;6:851–864. doi: 10.15252/emmm.201100899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velagapudi SP, Vummidi BR, Disney MD. Small molecule chemical probes of microRNA function. Curr Opin Chem Biol. 2015;24:97–103. doi: 10.1016/j.cbpa.2014.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahrle SE, Jiang H, Parsadanian M, Legleiter J, Han X, Fryer JD, Kowalewski T, Holtzman DM. ABCA1 is required for normal central nervous system ApoE levels and for lipidation of astrocyte-secreted apoE. J Biol Chem. 2004;279:40987–40993. doi: 10.1074/jbc.M407963200. [DOI] [PubMed] [Google Scholar]
- Wahrle SE, Jiang H, Parsadanian M, Hartman RE, Bales KR, Paul SM, Holtzman DM. Deletion of Abca1 increases Abeta deposition in the PDAPP transgenic mouse model of Alzheimer disease. J Biol Chem. 2005;280:43236–43242. doi: 10.1074/jbc.M508780200. [DOI] [PubMed] [Google Scholar]
- Wahrle SE, Jiang H, Parsadanian M, Kim J, Li A, Knoten A, Jain S, Hirsch-Reinshagen V, Wellington CL, Bales KR, Paul SM, Holtzman DM. Overexpression of ABCA1 reduces amyloid deposition in the PDAPP mouse model of Alzheimer disease. J Clin Invest. 2008;118:671–682. doi: 10.1172/JCI33622. [DOI] [PMC free article] [PubMed] [Google Scholar]