Abstract
An accurate in vivo measure of myelin content is essential to deepen our insight into the mechanisms underlying demyelinating and dysmyelinating neurological disorders, and to evaluate the effects of emerging remyelinating treatments. Recently [11C]PIB, a positron emission tomography (PET) tracer originally conceived as a beta-amyloid marker, has been shown to be sensitive to myelin changes in preclinical models and humans. In this work, we propose a reference-region methodology for the voxelwise quantification of brain white-matter (WM) binding for [11C]PIB. This methodology consists of a supervised procedure for the automatic extraction of a reference region and the application of the Logan graphical method to generate distribution volume ratio (DVR) maps. This approach was assessed on a test–retest group of 10 healthy volunteers using a high-resolution PET tomograph. The [11C]PIB PET tracer binding was shown to be up to 23% higher in WM compared with gray matter, depending on the image reconstruction. The DVR estimates were characterized by high reliability (outliers <1%) and reproducibility (intraclass correlation coefficient (ICC) >0.95). [11C]PIB parametric maps were also found to be significantly correlated (R2>0.50) to mRNA expressions of the most represented proteins in the myelin sheath. On the contrary, no correlation was found between [11C]PIB imaging and nonmyelin-associated proteins.
Keywords: myelin, parametric imaging, PET, PIB, quantification
Introduction
Myelin is a layered tissue wrapping the axons of neurons. The function of myelin is crucial for a proper transmission of the electric impulses in the central (CNS) and peripheral nervous system. Myelin acts as an axonal electrical insulator, enhancing the neuronal impulse speed as well as balancing the system energy.1 Chemically, myelin is defined as a lipid–protein complex. In human CNS, myelin is composed by a high proportion of lipids (70% to 85%) and a low amount of proteins (15% to 30%), whose composition is simpler than that of other brain membranes.2 The major lipids of myelin are glycolipids, phospholipids, and cholesterol3 while the proteins are the myelin basic protein (MBP) and proteolipid protein (PLP1, PLP2) making up 60% to 80% of the total myelin proteins (TMPs) in most species. An important role is also played by 2′,3′-Cyclic-nucleotide 3'-phosphodiesterase (CNP, 4% of TMPs), myelin-associated glycoprotein (MAG, <1% of TMPs), myelin oligodendrocyte glycoprotein (MOG, <0.05% of TMPs), myelin-associated oligodendrocyte basic protein (MOBP, ~7% of TMPs), and myelin protein zero (MPZ).4 Myelin protein zero is mainly present at the membrane of peripheral nervous system neurons (50% to 70% of TMPs) but negligible in CNS.5
The presence of myelin is fundamental for enabling a correct functioning of the CNS,6 brain maturation,7 as well as nervous tissue plasticity.8 Alteration and damage of the myelin sheaths are linked to several brain disorders such as multiple sclerosis and leukodystrophies.9 A reliable measure of the tissue myelin content would allow us to significantly improve our understanding of the physiopathological mechanisms underlying these disorders and would offer the opportunity to evaluate the effects of neuroprotective or pro-myelinating therapies.10, 11, 12
Positron emission tomography (PET) represents a well-established tool for imaging the CNS at the molecular level,13 and therefore it has the potential for detecting ongoing demyelination and remyelination processes in vivo in human. For this purpose, PET tracers such as [11C]BMB,14 a Congo red derivative, [11C]PIB,15 a Thioflavin derivative, [11C]CIC,16 and [11C]MeDAS,17 two stilbene derivatives, have been explored as myelin imaging biomarkers. Note that Congo-red and Thioflavins have been originally used as histochemical dyes to bind amyloid fibrils rich in β-sheet structures, hallmark of Alzheimer's disease,18, 19, 20, 21, 22 but have also shown significant binding to intact myelin sheaths. This suggests that [11C]PIB, in analogy to its interaction to amyloid fibrils, might have multiple interactions with the myelin structure: on one side, it might get trapped into β-sheet structures of myelin proteins such as MBP,23, 24, 25 and on the other side be highly soluble in the myelin-associated lipid bilayer.26
Myelin-binding properties and uptake of [11C]BMB, [11C]CIC, [11C]MeDAS, and [11C]PIB have been studied in preclinical models of demyelinated diseases,14, 27 but only [11C]PIB has been validated to image myelin and demyelinated lesions in humans.15 The PIB staining of CNS myelin brain sections, both in mice and in baboon postmortem sections, has been shown to be proportional to the tissue myelin content.15 In preclinical models, [11C]PIB tissue uptake reflected the ability to separate normal myelinated tracts, demyelinated lesions, and remyelinated regions15, 27 across the whole brain. In humans, examination of demyelinated plaques in two relapsing-remitting multiple sclerosis patients showed a significant decrease for [11C]PIB uptake compared with the normal-appearing white matter (NAWM) tissues and a different behavior between active and nonactive lesions.15
Analysis of [11C]PIB PET images is well described in the literature and a number of quantitative approaches have been presented. Compartmental modelling and graphical kinetic methods have been employed for the quantification of the tracer binding with arterial input functions.19 Subsequently, simplified kinetic approaches using reference regions (e.g., the cerebellum or gray-matter (GM) voxels selected via statistical clustering of tissue kinetics) have been suggested to avoid arterial cannulation and blood analysis during the PET acquisition.28, 29 Both plasma and reference region approaches provide reliable parametric maps that can be used with high sensitivity to discriminate between the healthy and the pathological state of the tissues. To date, amyloid imaging is the unique context in which the use of [11C]PIB PET methods have been validated.
This work proposes a specifically-designed protocol of data acquisition and analysis aimed at the voxelwise quantification of [11C]PIB PET employed as a specific tracer for myelin in brain tissues.
Two of the major requirements that need to be fulfilled by this methodology are (1) general applicability to a wide range of dysmyelinating pathologies, a condition that is satisfied by avoiding the use of blood sampling during the PET examination; and (2) the generation of reliable and reproducible high-resolution parametric maps to allow application in both cross-sectional and longitudinal studies.
In this work, we have used a high-resolution PET scanner and paid particular attention to the selection of appropriate reconstruction settings to find the best compromise between spatial resolution and data variability. The relationship between resolution (lesion size) and sensitivity of [11C]PIB myelin imaging as well as the partial volume effect on the [11C]PIB uptake were also assessed. Finally, we used the normal [11C]PIB data to construct a myelin density brain template in standardized coordinates and tested its fidelity to the patterns predicted by an atlas of mRNA expression of myelin-associated proteins in the same anatomic space.
Materials and methods
Data Set
Ten healthy control subjects (3 male, 7 female, age: 29.5±6.2 years) were enrolled in this study. Ethical approval was granted by the ethics committee of the Pitie-Salpetriere Hospital (Approval No. P080503) and informed written consent was obtained from all participants. This study was conducted according to the Declaration of Helsinki. Inclusion criteria consisted in an age between 18 and 55 years and the exclusion of any known neurological or psychiatric condition.
Each volunteer involved in the study was scanned twice, with the retest scan performed within approximately 1 month from the baseline.
Positron Emission Tomography and Magnetic Resonance Imaging Data Acquisitions
All PET exams were performed on the high resolution research tomograph (HRRT) (CPS Innovations, Knoxville, TN, USA). This brain dedicated PET scanner achieves an intraslice spatial resolution of ~2.5 mm full width at half maximum, with 25 cm and 31.2 cm of axial and transaxial field of view, respectively.30
The 90-minute emission scan was initiated coincident with a 1-minute intravenous bolus injection of [11C]PIB (mean 358±34 MBq). Images were reconstructed using the 3D ordinary Poisson ordered subset expectation maximization algorithm (POSEM).31 Different number of POSEM iterations (1, 2, 3, 4, 6, and 10) were implemented to find an appropriate trade-off between image resolution, quality of the data, and reliability of results. An additional smoothing filter implementing the point spread function (PSF) was applied to the 10-iteration reconstructed image. This range of reconstruction setting was selected among the common approaches used for HRRT PET scanner in dynamic brain PET imaging.32
All the resulting dynamic PET images consisted of 25 frames of data (6 × 1, 6 × 2, 4 × 3, 6 × 5, 3 × 10 minutes) with a voxel size of 1.22 mm × 1.22 mm × 1.22 mm. Interframe subject motion correction was applied by realigning each PET frame to a common reference space through a procedure similar to those reported by Montgomery et al.33 The first six frames of each scan corresponding to the first 6 minutes of acquisition were moved together because of the lack of spatial information necessary for the image coregistration. The procedure was applied to each PET study and all the reconstructed images but no frame or subject were discarded for exceed of motion. Data were also corrected for carbon-11 decay.
In addition to the PET scans, all subjects underwent a 3D anatomic brain magnetic resonance imaging (MRI) using a 3T Siemens system (Siemens, Erlangen, Germany; TRIO 32-channel TIM system): T1-weighted MPRAGE (echo time/inversion time/repetition time=2.9/900/2,300, flip angle=8°, pixel resolution=1 × 1 × 1.1 mm). Gray matter, white matter (WM), and basal ganglia (BG) were automatically extracted from MRI image by combining outputs from VBM8 toolbox (http://dbm.neuro.uni-jena.de/vbm/download/) and FIRST (http://fsl.fmrib.ox.ac.uk/fsl/) using a probability threshold of 90%. Segmented MRI images were hence coregistered to PET using rigid transformation estimated using Flirt (http://fsl.fmrib.ox.ac.uk/fsl/) maintaining PET data in its original space and avoiding further loss of resolution due to interpolation.
Voxelwise [11C]PIB Positron Emission Tomography Quantification: Analysis Pipeline
To make the procedure of general applicability, hence avoid blood sampling, we focused our methodology development toward reference region approaches.19, 28 In general, a reference region method consists of a two-step procedure that requires: (1) the selection of a brain tissue region containing no or negligible amount of specific binding and (2) the use of a quantification model in which the plasma tracer information can be replaced with the tracer activity in the reference region.
In this work, for the extraction of the reference time-activity curve (TAC), we exploited the supervised clustering method developed by Turkheimer et al34 and already validated for [11C]PIB PET tracer by Ikoma et al.29 This method consists of the multiple regression of all PET image voxel TACs on a predefined set of kinetic classes. After regression, all the voxels associated to the reference class with probability >90% are combined together and the average of their TACs defines the reference input function. Because the approach is implemented as a functional rather than an anatomic segmentation, its use can be easily extended to those pathological situations in which the tissue structure might be altered as in demyelinating diseases.
For this particular study, three classes were defined: normal GM, high-specific binding GM (e.g., amyloid bound fraction), and blood pool. The reference class of choice was normal GM. Details on the definition of the kinetic classes and on the implementation and validation of the procedure may be found in the original methodological work.29 Since the original method was defined on different scanners using filtered back-projection reconstructions we also evaluated the fidelity of the kinetic classes used as a standard to normal GM in this novel context.
Once the reference TAC was extracted, [11C]PIB binding was calculated by application of a kinetic operator to the measured dynamic PET activity. Several quantification approaches based on the reference input are available in the literature: the simplified reference tissue model,35 the rank-shaping spectral analysis,36 and the Logan graphical methods.37 All these methodologies can be applied at the voxel level and can return the distribution volume ratio (DVR), defined as the ratio of the total distribution volume between the target and the reference region. Distribution volume ratio is the parameter of interest because it is directly linked to tracer-target affinity as well as the target density.35 In the instance here, in which [11C]PIB is used as a myelin marker, the DVR becomes a proxy of myelin density.
For the Logan analysis, we chose to apply both the standard Logan with reference (Logan) and the Logan with reference corrected for k2 value (Logan_k2). The latter has showed to recover part of the bias that affects Logan approaches when applied at the voxel level.37 We followed the literature to define the settings19, 29 (k2=0.149 1/min), while for both Logan and Logan_k2 we used the last 60 minutes of the experiment for the DVR calculation.
In parallel to the quantitative PET analysis, maps of standardized uptake value (SUV), defined as the measured activity (from 30 to 70 minutes) normalized by the injected activity and subject weight, were also computed. This implementation was optimized to maximize the contrast between WM and GM.15 A comparison between SUV and DVR parametric maps was performed to understand whether static [11C]PIB PET scans could be used as a surrogate of dynamic [11C]PIB PET imaging.
Statistical Analysis
Quantification methods were evaluated in terms of reliability and reproducibility of the estimates. The first was assessed looking at the failure rate (i.e., fraction of voxels where methods did not converge to solution) and the outlier rate (i.e., fraction of voxels where the methods did not provide a biologically meaningful solution). Voxels with negative estimates for SUV analysis and DVR estimates with coefficient of variation >100% were considered as outliers. Reproducibility was assessed looking at the test–retest repeatability of the results and was measured by the intraclass correlation coefficient (ICC) and the mean relative differences. Both repeatability and reproducibility analyses were performed with varying number of POSEM iterations to assess the impact of reconstruction setting on quantification metrics. To indicate the level of noise produced by each reconstruction setting, the mean of tracer activity over its standard deviation measured across the image voxels was used as proxy of the signal-to-noise ratio (SNR).
Analysis of Local Contrast and Reproducibility
The processing of PET-derived myelin density maps potentially allows the analysis of WM density changes at the local level from large regions to areas of few millimeters size, up to the voxel level. However the technique, in order to be applicable, needs to be sensitive and reproducible at these high resolution levels. Thus, we reanalyzed test–retest reproducibility as well as GM/WM contrast in small regions with volumes equivalent to spheres of 2.5, 5, 7.5, and 10 mm diameter. The procedure was the following:
Given a particular subject, two regions (with volume comparable to a sphere of a given diameter), one in WM and one in GM, were randomly drawn on the individual structural T1-weighted MR image.
These GM and WM regions were moved to the parametric PET space, both in test and in retest images, by coregistering the individual MR on the test and retest PET spaces.
The mean of voxel estimates within each region (respectively GMtest, GMretest, WMtest, and WMretest) was computed. From these results, mean local contrast (mlc%) and mean local reproducibility (mlr%) were defined as:
 |
 |
Similarly, mlr% was computed also for GM simulated regions (using GMtest and GMretest, respectively).
The procedure was repeated 500 times for each subject considering all the tested reconstruction settings and all the preselected region diameters for both SUV and DVR maps. Mean and variability of mlc% and mlr% distributions were compared to determine the effect of region size and reconstruction on analysis.
To combine both contrast and reproducibility in a unique measure, a performance score index (p-index) was defined by normalizing the mean contrast to the test–retest mean and variability as:
 |
where mean and s.d. refer to the mean and the standard deviation of the 500 realizations of mlr and mlr, respectively. Both WM mlr and GM mlr were used as normalization factors in equation (3). Notably, the p-score was defined as an easy metric for the [11C]PIB PET applicability in small regions of interest (ROIs): a value of ⩽1 indicated that the WM–GM method contrast is comparable/inferior to its reproducibility (not adequate for clinical investigation); a value of >1 indicated a contrast power superior to the method variability (necessary condition for clinical investigation).
The spatial variability of DVR voxel estimates across the brain was investigated to assess the impact that regions with low activity concentration (like ventricles or brain sinus) might have on the tracer quantification for the highly myelinated adjacent tissues. Specifically, the mean DVR voxel estimates for GM and WM were analyzed as function to the cerebral spinal fluid (CSF) distance, to estimate the partial volume effect of this tissue on the rest of the brain.
[11C]PIB Positron Emission Tomography Binding: Correlation with Myelin-Associated Protein Gene Expressions
To complement the analysis above and enable comparison of the methodologies used with an independent predictor of myelin density, we tested the spatial brain profile of the myelin maps produced versus the one predicted by the brain mRNA expression maps of a set of myelin-associated proteins contained in the Allen Human Brain Atlas.38 We have previously shown that these maps are highly predictive of protein levels in vivo measured with PET for all those transcripts that do not undergo significant posttranscriptional modifications (R2>90%).39 Indeed there is good evidence of a tight relationship between mRNA transcription and myelin protein synthesis, supporting the use of Allen Brain Atlas as a reference for myelin protein brain distributions.40, 41, 42 Hence, we assumed to find a significant correlation between [11C]PIB imaging and myelin-associated protein gene expressions.
For the purpose, we created from the 10 subjects a [11C]PIB DVR template in stereotaxic MNI (Montreal Neurological Institute) space. We then tested the [11C]PIB binding spatial profile obtained from this standardized map, versus the mRNA protein expression profiles of PLP1, PLP2, MBP, CNP, MAG, MOG, MOBP, and MPZ. These proteins were selected based on their prevalence in the myelin sheath.4 The comparison was performed by correlation analysis at the regional level according to the anatomic segmentation of the Hammersmith anatomic atlas as described in our original reference.39
Results
Impact of Poisson Ordered Subset Expectation Maximization Algorithm Reconstruction Setting on Data Quality
Figure 1 provides an example of the effect of different settings used for POSEM reconstruction on the raw PET images. The images refer to the 60-minute frame of a transaxial brain slice at the cortical level in a representative subject of the data set. As expected, increasing the number of iterations improves the spatial resolution at the cost of higher noise. In the range of POSEM iterations that we considered, the image evolved from a smooth appearance (POSEM 1 iteration, SNR1-iteration=43 unitless) to the characteristic salt and pepper distribution (SNR10-iterations=26 unitless). The introduction of the PSF smoothing filter to the 10-iteration reconstructed image reduced the noise levels with a final result that is comparable in both term of resolution and SNR level to the 4-iteration reconstructed image (SNR relative difference <2%).
Figure 1.
Impact of the reconstruction setting on positron emission tomography (PET) image quality. The figure shows an example of PET images obtained from the same acquisition by modifying the reconstruction setting: Poisson ordered subset expectation maximization algorithm (POSEM) 1, 2, 3, 4, 5, 6, 10 iterations and POSEM with 10 iterations combined with point spread function (PSF) smoothing. Maps refer to the same transaxial slice, 60 minutes after tracer injection, obtained from one representative subject of the study. Trend of SNR (SNR=mean/s.d. of the voxel activity) levels calculated from each image is reported in the bar graph as function of reconstruction setting. SNR, signal-to-noise ratio.
The differences in the noise content of the reconstructed image directly impact on the ROI TAC definitions (Figure 2). Moving from the 1-iteration to the 10-iteration scenario, it can be noticed how GM, BG, and reference regions present a sharpened peak (range 15% to 25%) and a lower tail (range 7% to 15%), while for the WM TACs the opposite is true. The POSEM iterations impact also on the supervised clustering results in term of number and the position of the voxels associated with the reference class (Figure 2). Notably, the higher the number of iterations, the better the agreement of the normalized reference TAC with the reference GM class (Supplementary Material—Figure 1). This consistency between extracted references and GM class supports the use of the supervised clustering even with data acquired with HRRT PET scanner.
Figure 2.
Impact of reconstruction setting on reference region extraction. (A) Spatial distribution of the voxels selected via supervised clustering as a reference region with different Poisson ordered subset expectation maximization algorithm (POSEM) reconstruction settings (1 and 10 iterations, respectively) is shown. (B and C) The time courses of the regional activity obtained from different reconstructed positron emission tomography (PET) images, with POSEM 1-iteration and POSEM 10-iterations, respectively, are shown. Notably, the two reconstruction settings are associated with the highest and lowest image signal-to-noise ratio (SNR) (Figure 1). (D) The fraction of the gray matter (GM) voxels selected as a reference region as function of the reconstruction settings is reported. (E) The normalized TACs of the three kinetic classes used by the supervised clustering in comparison with the normalized reference region time-activity curves (TACs) obtained with POSEM 1-iteration (red line) and POSEM 10-iterations (blue line), respectively, are shown.
[11C]PIB Positron Emission Tomography Data Quantification
The use of different POSEM reconstruction settings led to a plurality of parametric maps for both Logan and SUV. Depending on the number of iterations used for POSEM, it can be observed that the spatial and intensity pattern of the voxel estimates tends to follow the noise in the raw dynamic PET data (Supplementary material—Figures 2 and 3). This effect is more remarkable in SUV maps compared with those obtained from the Logan as the latter methodology exploits the entire kinetic of measured TACs to provide a DVR value.
Analysis of failures and outliers showed that both Logan and SUV were characterized by a negligible rate of not reliable voxel estimates (<0.1% in all the subjects and with all the reconstruction settings). In Logan, this fraction corresponded entirely to not precise DVR estimates, whose coefficients of variation calculated a posteriori were higher than 100%. In the SUV analysis instead, the discarded voxels were those in which the calculated values were negative. Interesting, the application of rank-shaping spectral analysis and simplified reference tissue model (solved with basis function approach) methods for [11C]PIB PET image quantification returned a high number of voxel outliers (up to 80% with POSEM 10 iterations), suggesting the inapplicability of these two methods for the particular data set (Supplementary Material—Figure 4). The result was quite expected as the image noise for some of the reconstruction settings was remarkably higher that what is commonly reported by standard dynamic [11C]PIB PET acquisitions. In light of these results, Logan graphical approach was selected as the method of choice for DVR quantification, due to its superior robustness to measurement noise.
Despite the methodological differences, both Logan and SUV approaches provide a consistent description of PIB uptake across the different subjects (Figures 3A and 3B, ). In Logan analysis, DVR ranged from 1.10 to 1.13 in GM, from 1.22 to 1.33 in BG, and from 1.14 to 1.37 in WM. Standardized uptake value, instead, ranged from 3.65 to 4.00 (g/mL) in GM, from 4.73 to 5.11 (g/mL) in BG, and from 5.59 to 6.15 (g/mL) in WM. Between-subject variability of Logan DVR estimates was ⩽6% for all the ROIs and all the reconstruction settings while for SUV it was consistently around 12%. A summary of the results is reported in Supplementary Table 1 in the Supplementary Material.
Figure 3.
Quantification results. (A and B) Mean and variability across subjects of LOGAN distribution volume ratio (DVR) and standardized uptake value (SUV) estimates as a function of reconstruction setting are reported. Gray bars indicate gray matter (GM), white bars indicate white matter (WM), and black bars indicate basal ganglia (BG). The mean and variability of WM/GM relative differences are also shown. An example of parametric maps computed with both SUV and LOGAN at different axial levels for a representative subject is reported in (C).
Comparing [11C]PIB binding in the GM and in the WM, we found a variable contrast depending on the quantification method and reconstruction setting. In Logan analysis, mean voxel DVR differences ranged from 23±4% (POSEM 1 iteration) to 8±5% (POSEM 10 iterations); with SUV, WM–GM relative differences ranged from 40±8% (POSEM 1 iteration) to 59±12% (POSEM 10 iterations). As for the absolute quantification, the WM–GM contrast showed higher variability in SUV maps compared with the Logan ones. It is noteworthy that, while for Logan analysis the relative differences between WM and GM DVR tended to decrease with increasing number of POSEM iterations, for SUV an opposite effect was observed, with the tissue contrast increasing at higher image resolutions. In both instances, the tracer binding measured in GM was significantly lower than that measured in WM (P value<0.05). A visualization of these differences can be seen in a comparative example reported in Figure 3C. Distribution volume ratio and SUV values were found to be moderately correlated (R2=0.58±0.14, mean±s.d.), with the highest correlation found for POSEM 1-iteration reconstructed PET images (R2=0.79) and the lowest correlation for POSEM 10-iteration reconstructed PET images (R2=0.45). Notably, no effect of reconstruction setting was noticed in term of stability of Logan equilibrating time (t*). Mean relative difference of DVR estimates was <3% with t*=45 minutes and <4% t*=60 min, respect to the setting used for the analysis (t*=30 minutes).
The Logan approach with k2 correction produced comparable estimates with the standard Logan approach with the reference region: the means of voxel estimates within ROIs were almost identical (DVRLogan-k2= 0.98·DVRLogan+0.01, R2>0.99) with the absolute mean relative difference equal to 3±1%. The WM–GM contrast measured by the Logan modified approach was identical (mean relative difference <1%) to the one measured with the standard approach both in amplitude and in variability. Given the reported strong similarity between the two measures, we omitted Logan with k2 correction from subsequent analyses.
Out of the 10 subjects analyzed, only 2 subjects presented a pattern that was remarkably different from the others (Supplementary material—Figure 5). In particular, both DVR and SUV measured in WM were lower to those measured in the rest of dataset (−12±4% and −39±5% for DVR and SUV, respectively) but comparable to the tracer uptake measured in GM (4±2% DVR and −7±5% SUV relative difference, respectively). Notably, these results were consistent for both test and retest scans. The experimental protocol for these two subjects was the same as the one conducted in the rest of the group and [11C]PIB injected dose and tracer-specific activity were in the overall range. These subjects were considered outliers and therefore excluded from the statistical analysis. It is important to note that none of the 20 scans analyzed presented a [11C]PIB uptake consistent with the presence of amyloid plaques in the brain.
Reproducibility
In term of reproducibility, Logan outperformed SUV independently of the reconstruction setting used. For DVR estimates, ICC was always >0.90 while test/retest absolute mean of relative differences was equal to 2±1%. For SUV results instead, ICC ranged from 0.65 up to 0.76, depending on the number of POSEM iterations, with a test/retest mean relative difference of 14±10%. For both methods, both WM and GM showed comparable test/retest repeatability to those found a whole brain scale. For Logan analysis test–retest, mean DVR differences were 1±1% in GM and 2±2% in WM, while for SUV 14±11% in GM and 12±9% in WM.
Regional Analysis
Local contrast analysis confirmed the results obtained at whole brain level: (1) SUV reported the highest contrast (mlc% from 41±32% for 2.5 mm region diameter up to 44±17% for 10 mm region diameter) compared with Logan (mlc% from 13±24% for 2.5 mm region diameter up to 13±15% for 10 mm region diameter); (2) higher number of reconstruction iterations impacted in opposite ways the mlc% of the two methods, ameliorating SUV performance but penalizing the Logan. A summary of these results is reported in Figures 4A and 4B. Notably, for both methods, across all subjects, all region dimensions, and all reconstruction settings, no significant difference in mlc% was found between test and retest data. The major impact of increasing the region size (from 2.5 to 10 mm diameter) was on reducing the mlc% variability: from 27% to 15% with DVR and from 41% to 16% with SUV.
Figure 4.
Local analysis: contrast and reproducibility. The mean and s.d. of white matter (WM)/gray matter (GM) contrast values for Logan distribution volume ratio (DVR) and standardized uptake value (SUV) analysis are reported as function of the positron emission tomography (PET) reconstruction setting and simulated region dimensions (A and B, respectively). Similarly, the mean local test/retest reproducibility is also shown for both GM and WM as obtained with Logan (C and E) and SUV (D and F).
Regional test/retest reproducibility maintained the patterns found for the whole brain analysis: with Logan, mlr% was 3±12% for GM and 4±13% for WM; with SUV, mlr% was 15±18% for GM and 13±15% for WM. For both methods, these results referred to the mean±s.d. across region dimensions and reconstruction settings, while a more exhaustive description of local reproducibility analysis is reported in Figures 4C–4F.
Figure 5 shows the p-index results as a function of POSEM iterations and simulated region dimensions: while the first represents an important variable for Logan performance (for SUV p-indexes remain stable), the second significantly modulates the contrast power in both quantification methods. Consistently with the rest of results, p-index highlights the better WM/GM contrast power of SUV with respect to Logan.
Figure 5.
Local analysis: performance index. The mean and s.d. of the p-index values for Logan and standardized uptake value (SUV) analysis are reported as function of the positron emission tomography (PET) reconstruction setting and simulated region dimensions. Results refer to white-matter normalization for Logan (A) and SUV (B), respectively. Equivalent results are obtained with gray-matter normalization (data not shown).
Analysis of Partial Volume Effect
Analysis of mean DVR values in GM and WM as function of the CSF distance shows the presence of a strong partial volume effect reducing the value of the DVR in the voxels within 2 to 3 mm from the CSF (Figure 6A). Notably, this distance is consistent with the scanner resolution. With regard to the mean DVR values reported within 6 to 8 mm from CSF, the underestimation for these voxel is −38±30% for WM and −20±9% for GM. Moving away from CSF (distance >10 mm), DVR estimates in GM remain stable while the WM DVR significantly increases (+20% mean relative difference respect to 6 to 8 mm WM DVR correspondent to 1.54±0.05 DVR absolute value). These particular highly myelinated brain regions appear to be spatially localized within the spinal-cortical brain tracts (Figure 6B).
Figure 6.
[11C]PIB uptake and partial volume effect. (A) The mean distribution volume ratio (DVR) values measured (open circles) and modelled (dashed lines) for WM and GM, respectively, as function of the distance from the cerebral spinal fluid (CSF) are shown. Data refer to Poisson ordered subset expectation maximization algorithm (POSEM) 10 iterations with point spread function (PSF) smoothing reconstruction setting. (B) The correspondent [11C]PIB uptake map based on the distance model from CSF is shown. Hyperintense regions refer to those voxels with the highest DVR estimates.
[11C]PIB Positron Emission Tomography Binding: Correlation with Myelin-Associated Protein Gene Expressions
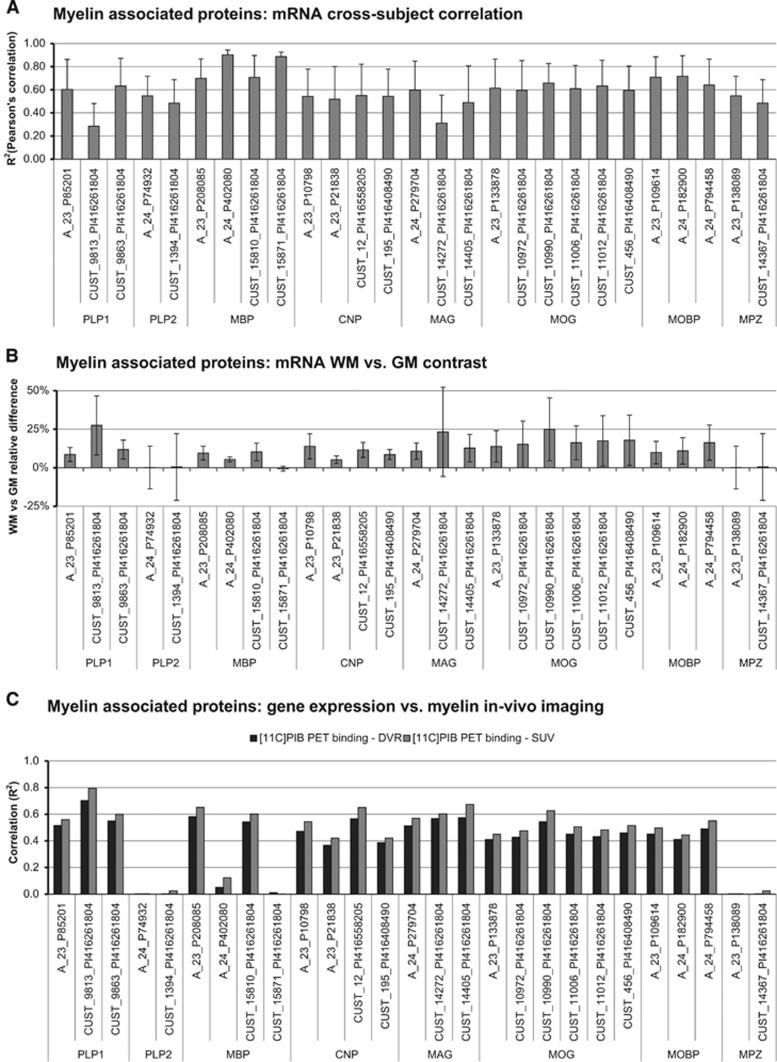
Analysis of mRNA myelin-associated protein expression shows cross-subject consistency for most of the probes analyzed, with Pearson's correlation R2 mean±s.d. across probes equal to 0.60±0.13 (Figure 7A). The maximum R2 was reported for probe A_24_P402080 (MBP, R2=0.90) while the minimum was found for probe CUST_9813_PI416261804 (PLP1, R2=0.28).
Figure 7.
Comparison between [11C]PIB imaging and myelin-associated protein gene expressions. (A) The myelin-associated protein mRNA auto-correlations are shown. Probe selection and their spatial expression patterns are used as reported by the Allen Brain Atlas database (http://human.brain-map.org/). (B) Reports for the same probes the relative differences of mRNA expressions between white matter (WM) and gray matter (GM). (C) mRNA cross-correlation with myelin-associated templates obtained by using Poisson ordered subset expectation maximization algorithm (POSEM) 10 iterations with point spread function (PSF) smoothing reconstruction setting is shown. Black and gray bars refer to correlation with [11C]PIB myelin maps using distribution volume ratio (DVR) and standardized uptake value (SUV), respectively.
The mRNA expressions and spatial distributions of these probes were highly cross-correlated (R2=0.60±0.30, mean±s.d.), with the only exception of A_24_P402080 and CUST_15871_PI416261804, whose spatial expression profiles were inconsistent with the others. Notably these two probes target Golli-MBP isoform 2 (http://www.ncbi.nlm.nih.gov/gene/4155), a transcript variant containing just one exon of the classic MBP, and known to be expressed in cells distinct from myelinating oligodendrocytes.
Analysis of mRNA expression differences between WM and GM shows, which probes better reflect myelin density contrast between these two tissues (Figure 7B). The WM and GM mRNA samples were localized by using two anatomic masks for WM and GM in MNI space (90% of probability threshold). The WM versus GM mRNA expression contrast ranged from −1% to 27% (mean±s.d.: 11±7%). Among the myelin-associated proteins, those with the lowest WM–GM differences were PLP2 and MPZ, while the highest was reported for MOG (18±3%), MAG (16±6%), and MOBP (12±3%). These results reflect the a priori knowledge of the gene activities: while MOG, MAG, and MOBP are specific to myelin synthesis and deposition, PLP2 and MPZ are mainly expressed out of brain tissues.4
This classification was maintained when myelin-associated protein expressions were compared with in vivo [11C]PIB binding estimates (Figure 7C). Myelin oligodendrocyte glycoprotein, MAG, and MOBP were the protein that best correlated with [11C]PIB DVR (R2: 0.48±0.06 for DVR versus mRNA; R2: 0.53±0.07 for SUV versus mRNA), while PLP2 and MPZ registered the lowest correlation (Figure 7C). 2′,3′-Cyclic-nucleotide 3'-phosphodiesterase, PLP1, and MBP correlation with myelin maps was in average relevant (R2>0.30) but highly variable depending on the probes considered.
As a negative control study, we investigated the correlation between [11C]PIB bindings maps and the mRNA expressions of some well-known nonmyelin-associated proteins. For the purpose, we considered two monoaminergic receptor systems (dopamine D1, D2, and D3 receptors and serotonin 5HT1A, 5HT1B, 5HT1D, 5HT1E, and 5HT1F receptors), two astrocyte markers (Aq4L and Aldh1L1) and one oligodendrocyte precursor protein (PDGF receptor alpha, PDGFRA).43 The first two groups were chosen because of their different spatial distributions: while dopamine receptors are mainly concentrated in the striatum, the serotonin ones present a more diffuse distribution across brain tissues. Aq4L, Aldh1L1, and PDGFRA were tested because they are known to be expressed also in the WM, although they are not directly linked to myelin.43
Results confirmed the independency of the systems: notwithstanding the high probe autocorrelations that testified to the quality of mRNA, cross-correlations between [11C]PIB-DVR estimates and all the pre-selected genes were very low (R2: 0.16±0.16 for dopamine; R2: 0.15±0.14 for serotonin; R2: 0.23±0.23 for Aq4L; R2: 0.10±0.5 for Aldh1L1; R2: 0.18±0.11 for PDGFRA). Notably, none of the correlations was significant. A summary of these results is reported in the (Supplementary material—Figure 6).
Discussion
In the current work, we presented a methodological analysis of the quantification of [11C]PIB PET data as a myelin imaging tool. The combination of a supervised clustering approach with the Logan graphical method is proposed as a noninvasive solution for the voxelwise quantification of such data. The performance of the method was assessed in comparison with the use of SUV, a semiquantitative static index whose use is standard in clinical PET routine.
Both SUV and Logan methods showed to be reliable, with the SUV providing a superior contrast between WM and GM tracer uptake, at both global and regional levels, but at the same time presenting a higher sensitivity to the measurement noise. In terms of reproducibility, Logan overperformed SUV presenting a significantly lower test–retest variability. Notably, the two methods were not perfectly correlated. This is justified by the different specificity offered by these measures: while SUV reflects the overall radioactivity within a given volume of interest that integrates both tracer perfusion and binding, the DVR estimate computed by the Logan analysis directly returns the measure of [11C]PIB binding to the target. Dynamic PET imaging and full quantification should be preferred when applicable.
The mismatch between DVR and SUV is already known in PIB data analysis for the characterization of amyloid density44 and may also justify the inconsistent distribution of binding across different WM regions discovered for [11C]PIB in preclinical data when assessed with SUV.27 To overcome the problem, SUV normalized by reference region has been proposed as a more accurate quantification method for [11C]PIB static imaging.28 This was not applicable in our case since the reference region is extracted through tissue kinetic clustering thus requiring dynamic imaging.
Of particular interest is the relationship between SUV and Logan performance and the PET reconstruction settings. A larger number of iterations was shown to improve PET spatial resolution, but led to an expected increase in the image noise. Interestingly, this impacted on SUV and Logan methods with opposite effects. In the case of the SUV, the higher the number of iterations, the better the WM/GM contrast, while for the Logan it was the opposite for the analysis at both global and local levels. The effects of the noise introduced in the reconstructed PET data by the POSEM setting mainly impacted by increasing the DVR voxel estimate variability as the number of iterations increased, with the final results of making the discrimination between different levels of tracer uptake more challenging. In agreement with this, we found that Logan returns the best parametric maps in terms of both quality of voxel estimates and power of differentiate between tracer uptake when the dynamic PET data are reconstructed with the best possible SNR. On the other hand, this configuration led to very smooth and blurred images, making the choice of using a high-resolution scanner like HRRT completely redundant. Among the tested reconstruction settings, we believe that the best compromise between image resolution, image noise, and estimate consistency was the use of 10 iterations with PSF smoothing. It is also worth to underline that HRRT does not represent the standard for PET acquisition but a high-resolution resource to which few centers have access. We are currently running additional studies with [11C]PIB analogs to establish the feasibility of the method with more conventional PET/CT tomographs.
The local analysis, i.e., the investigation of method performance in small size regions, showed that [11C]PIB PET imaging has the potential to be applied for investigations of small lesions down to the pixel level. Nevertheless, the capacity to describe tissue alterations depends not only on the lesion size, but also on its brain localization as well as on the magnitude of signal variations. The first issue is related to the partial volume effect and the impact that CSF has on the quantification of [11C]PIB binding in the adjacent regions. Any cross-sectional comparison between healthy and pathological tissues should not be performed without taking this aspect into account. The second issue is related to the amount of [11C]PIB binding reduction occurring in demyelinated tissues. This cannot be established with a data set of healthy controls, and further studies in pathological conditions are needed to fully characterize the power of the method at the lesion level.
Supervised clustering confirmed to be a robust approach for reference region extraction. Although the different levels of noise introduced by modifying the POSEM settings had an impact on the number of voxel-TACs associated with the reference region (from 15% of brain analyzed voxels with POSEM 1-iteration, to 8% of brain analyzed voxels with POSEM 10-iterations), in all the tested scenarios the extracted reference input functions were consistent with the expected profile. This finding is encouraging for the extension of the method to pathological conditions. Importantly, this procedure also allows an optimal investigation of the cerebellar region which cannot be performed when cerebellum is used as a reference in standard practice. Note that the use of GM as a reference region limits the method applicability to WM only, since the amount of myelin in normal GM represents the lowest boundary of detection of the method. Only quantification methods with arterial input functions can overcome this limitation and be applied to investigate GM myelin density. It is important to remark that the myelin content of GM (although in much smaller amount compared with WM) does not impede its use as a reference: a reference region can contain some specific binding, but this has to remain stable across clinical conditions.45 The use of supervised clustering rather than the anatomic segmentation warrants this assumption and the consistent selection of reference tissue with pre-defined kinetic properties across studies and subjects.
The comparison of [11C]PIB PET binding maps with myelin-associated gene expressions highlights a strong association with the most representative myelin brain proteins and the absence of any significant correlation with myelin-independent proteins, including proteins expressed by astrocytes and oligodendrocyte precursors. Despite the limits of these comparisons (e.g., two different groups of subjects, PET referring to in vivo data, mRNA to post-mortem analysis) this finding supports the specificity of [11C]PIB PET binding to myelin: as demonstrated by Wang et al,46 a sensitive and specific myelin probe cannot be determined only on the basis of its lipophilicity, but it has to be able to interact directly with the protein and the lipidic components of the myelin sheaths. Thus, the fact that [11C]PIB PET binding estimates present a spatial brain pattern consistent with the level of expression of myelin-associated protein is consistent with the previously demonstrated specificity.15
One possible criticism to the method here presented concerns the role that [11C]PIB tracer metabolites might have in [11C]PIB tissue binding. Several lines of evidence convincingly exclude any such contribution: (1) [11C]PIB plasma tracer metabolites are very polar and therefore very unlikely to pass through the blood–brain barrier;18 (2) the kinetic modelling of [11C]PIB tissue tracer uptake with an arterial input function does not require any additional metabolite input in both healthy and pathological conditions;18, 19 (3) the quantification of [11C]PIB uptake with a reference input function is consistent with that returned by metabolite-corrected arterial input function in both healthy and pathological conditions, which attests the good reliability of noninvasive estimation methods even when beta amyloid in absent.19, 29 This last aspect was confirmed by a pilot study (Supplementary Material—Figure 7), performed to verify the reliability and robustness of the implemented PET experimental procedure.
The use of Logan graphical analysis might also appear as another limit of the study. Although we could not fully evaluate the use of more conventional [11C]PIB quantification methods (e.g., RPM or RPM2), it is worth to mention that in the presence of low [11C]PIB uptake, as it is the distribution of the tracer in healthy volunteers when beta-amyloid is absent, Logan represents a reliable and almost unbiased method for [11C]PIB parametric imaging.47
Conclusions
In this work, we showed that [11C]PIB PET binding to WM is reliable and reproducible. Results are dependent on the use of a proper data quantification approach, but [11C]PIB parametric maps can be obtained noninvasively via supervised clustering and Logan graphical analysis. The method is generally applicable as long as beta amyloid plaques are absent within the investigated tissues and the analysis limited to the WM.
We also showed that there is a close relation between the major proteins associated with myelin sheath and [11C]PIB imaging. Future work will elucidate [11C]PIB sensitivity and specificity to myelin dynamics in WM disorders.
Acknowledgments
The authors thank Adrien Clavairoly for the useful discussion on mRNA myelin-associated markers; Carlos Parras, from the ICM, Paris, for helpful suggestions; and the Assistance Publique des Hopitaux de Paris (APHP) for sponsoring the research.
Dr Stankoff reports receiving consulting and lecture fees from Biogen Idec, Novartis, Merck Serono, Sanofi-Aventis, Teva-Pharma and research support from Genzyme and Merck-Serono. The other authors state no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Journal of Cerebral Blood Flow & Metabolism website (http://www.nature.com/jcbfm)
The project described was supported by the following grants: Programme Grant ‘Quantitative methodologies for Positron Emission Tomography' (UK Medical Research Council No. G1100809/1); ELA (European Leukodystrophy Association, grant 2007-0481); INSERM-DHOS (grant 2008-recherche clinique et translationnelle); ANR MNP2008-007125; ECTRIMS post-doctoral research fellowship. The research leading to these results has also received funding from the program ‘Investissements d'avenir' ANR-10-IAIHU-06, ARSEP travel grant, JNLF and FRM.
Supplementary Material
References
- 1Hartline DK. What is myelin? Neuron Glia Biol 2008; 4: 153–163. [DOI] [PubMed] [Google Scholar]
- 2Morell P, Quarles R, Norton W. Myelin formation, structure, and biochemistry. Basic Neurochem 1994; 1994: 117–136. [Google Scholar]
- 3Evans MJ, Finean J. The lipid composition of myelin from brain and peripheral nerve. J Neurochem 1965; 12: 729–734. [DOI] [PubMed] [Google Scholar]
- 4Jahn O, Tenzer S, Werner HB. Myelin proteomics: molecular anatomy of an insulating sheath. Mol Neurobiol 2009; 40: 55–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5Sakamoto Y, Kitamura K, Yoshimura K, Nishijima T, Uyemura K. Complete amino acid sequence of PO protein in bovine peripheral nerve myelin. J Biol Chem 1987; 262: 4208–4214. [PubMed] [Google Scholar]
- 6Sherman DL, Brophy PJ. Mechanisms of axon ensheathment and myelin growth. Nat Rev Neurosci 2005; 6: 683–690. [DOI] [PubMed] [Google Scholar]
- 7Paus T, Collins D, Evans A, Leonard G, Pike B, Zijdenbos A. Maturation of white matter in the human brain: a review of magnetic resonance studies. Brain Res Bull 2001; 54: 255–266. [DOI] [PubMed] [Google Scholar]
- 8Wang S, Young K. White matter plasticity in adulthood. Neuroscience 2013; 276: 148–160. [DOI] [PubMed] [Google Scholar]
- 9Love S. Demyelinating diseases. J Clin Pathol 2006; 59: 1151–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10Dubois-Dalcq M, Williams A, Stadelmann C, Stankoff B, Zalc B, Lubetzki C. From fish to man: understanding endogenous remyelination in central nervous system demyelinating diseases. Brain 2008; 131: 1686–1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11Lubetzki C, Williams A, Stankoff B. Promoting repair in multiple sclerosis: problems and prospects. Curr Opin Neurol 2005; 18: 237–244. [DOI] [PubMed] [Google Scholar]
- 12Zawadzka M, Franklin RJ. Myelin regeneration in demyelinating disorders: new developments in biology and clinical pathology. Curr Opin Neurol 2007; 20: 294–298. [DOI] [PubMed] [Google Scholar]
- 13Bertoldo A, Gaia R, Mattia V. Deriving physiological information from PET images: from SUV to compartmental modelling. Clin Transl Imaging 2014; 2: 239–251. [Google Scholar]
- 14Stankoff B, Wang Y, Bottlaender M, Aigrot M-S, Dolle F, Wu C et al. Imaging of CNS myelin by positron-emission tomography. Proc Natl Acad Sci 2006; 103: 9304–9309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15Stankoff B, Freeman L, Aigrot MS, Chardain A, Dolle F, Williams A et al. Imaging central nervous system myelin by positron emission tomography in multiple sclerosis using [methyl-(1)(1)C]-2-(4'-methylaminophenyl)- 6-hydroxybenzothiazole. Ann Neurol 2011; 69: 673–680. [DOI] [PubMed] [Google Scholar]
- 16Wang Y, Wu C, Caprariello AV, Somoza E, Zhu W, Wang C et al. In vivo quantification of myelin changes in the vertebrate nervous system. J Neurosci 2009; 29: 14663–14669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17Wu C, Wang C, Popescu DC, Zhu W, Somoza EA, Zhu J et al. A novel PET marker for in vivo quantification of myelination. Bioorg Med Chem 2010; 18: 8592–8599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18Klunk WE, Engler H, Nordberg A, Wang Y, Blomqvist G, Holt DP et al. Imaging brain amyloid in Alzheimer's disease with Pittsburgh Compound-B. Ann Neurol 2004; 55: 306–319. [DOI] [PubMed] [Google Scholar]
- 19Price JC, Klunk WE, Lopresti BJ, Lu X, Hoge JA, Ziolko SK et al. Kinetic modeling of amyloid binding in humans using PET imaging and Pittsburgh Compound-B. J Cereb Blood Flow Metab 2005; 25: 1528–1547. [DOI] [PubMed] [Google Scholar]
- 20Flaherty DP, Kiyota T, Dong Y, Ikezu T, Vennerstrom JL. Phenolic bis-styrylbenzenes as β-amyloid binding ligands and free radical scavengers. J Med Chem 2010; 53: 7992–7999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21Puchtler H, Sweat F, Levine M. On the binding of Congo red by amyloid. J Histochem Cytochem 1962; 10: 355–364. [Google Scholar]
- 22Reinke AA, Gestwicki JE. Insight into amyloid structure using chemical probes. Chem Biol Drug Design 2011; 77: 399–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23Beniac DR, Luckevich MD, Czarnota GJ, Tompkins TA, Ridsdale RA, Ottensmeyer FP et al. Three-dimensional structure of myelin basic protein I. Reconstruction via angular reconstitution of randomly oriented single particles. J Biol Chem 1997; 272: 4261–4268. [DOI] [PubMed] [Google Scholar]
- 24Ridsdale RA, Beniac DR, Tompkins TA, Moscarello MA, Harauz G. Three-dimensional structure of myelin basic protein II. Molecular modeling and considerations of predicted structures in multiple sclerosis. J Biol Chem 1997; 272: 4269–4275. [DOI] [PubMed] [Google Scholar]
- 25Stoner GL. Predicted folding of beta-structure in myelin basic protein. J Neurochem 1984; 43: 433–447. [DOI] [PubMed] [Google Scholar]
- 26Biancalana M, Koide S. Molecular mechanism of Thioflavin-T binding to amyloid fibrils. Biochim Biophys Acta 2010; 1804: 1405–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27de Paula Faria D, Copray S, Sijbesma JW, Willemsen AT, Buchpiguel CA, Dierckx RA et al. PET imaging of focal demyelination and remyelination in a rat model of multiple sclerosis: comparison of [(11)C]MeDAS, [ (11)C]CIC and [ (11)C]PIB. Eur J Nucl Med Mol Imaging 2014; 41: 995–1003. [DOI] [PubMed] [Google Scholar]
- 28Lopresti BJ, Klunk WE, Mathis CA, Hoge JA, Ziolko SK, Lu X et al. Simplified quantification of Pittsburgh Compound B amyloid imaging PET studies: a comparative analysis. J Nucl Med 2005; 46: 1959–1972. [PubMed] [Google Scholar]
- 29Ikoma Y, Edison P, Ramlackhansingh A, Brooks DJ, Turkheimer FE. Reference region automatic extraction in dynamic [(11)C]PIB. J Cereb Blood Flow Metab 2013; 33: 1725–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30Wienhard K, Schmand M, Casey M, Baker K, Bao J, Eriksson L et al. The ECAT HRRT: performance and first clinical application of the new high resolution research tomograph. IEEE Trans Nucl Sci 2002; 49: 104–110. [Google Scholar]
- 31Carson R, Barker W, Liow J-S, Johnson C. Design of a motion-compensation OSEM list-mode algorithm for resolution-recovery reconstruction for the HRRT. IEEE Nucl Sci Symp Conf Rec 2003; 5: 3281–3285. [Google Scholar]
- 32OSEM-3D reconstruction strategies for the ECAT HRRTNuclear Science Symposium Conference Record, 2004 IEEE. IEEE: Rome, Italy. 2004. [Google Scholar]
- 33Montgomery AJ, Thielemans K, Mehta MA, Turkheimer F, Mustafovic S, Grasby PM. Correction of head movement on PET studies: comparison of methods. J Nucl Med 2006; 47: 1936–1944. [PubMed] [Google Scholar]
- 34Turkheimer FE, Edison P, Pavese N, Roncaroli F, Anderson AN, Hammers A et al. Reference and target region modeling of [11C]-(R)-PK11195 brain studies. J Nucl Med 2007; 48: 158–167. [PubMed] [Google Scholar]
- 35Lammertsma AA, Hume SP. Simplified reference tissue model for PET receptor studies. NeuroImage 1996; 4: 153–158. [DOI] [PubMed] [Google Scholar]
- 36Turkheimer FE, Hinz R, Gunn RN, Aston JA, Gunn SR, Cunningham VJ. Rank-shaping regularization of exponential spectral analysis for application to functional parametric mapping. Phys Med Biol 2003; 48: 3819–3841. [DOI] [PubMed] [Google Scholar]
- 37Logan J, Fowler JS, Volkow ND, Wang GJ, Ding YS, Alexoff DL. Distribution volume ratios without blood sampling from graphical analysis of PET data. J Cereb Blood Flow Metab 1996; 16: 834–840. [DOI] [PubMed] [Google Scholar]
- 38Hawrylycz MJ, Lein S, Guillozet-Bongaarts AL, Shen EH, Ng L, Miller JA et al. An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 2012; 489: 391–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39Rizzo G, Veronese M, Heckemann RA, Selvaraj S, Howes OD, Hammers A et al. The predictive power of brain mRNA mappings for in vivo protein density: a positron emission tomography correlation study. J Cereb Blood Flow Metab 2014; 34: 827–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40Trapp BD, Hauer P, Lemke G. Axonal regulation of myelin protein mRNA levels in actively myelinating Schwann cells. J Neurosci 1988; 8: 3515–3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41Dracheva S, Davis KL, Chin B, Woo DA, Schmeidler J, Haroutunian V. Myelin-associated mRNA and protein expression deficits in the anterior cingulate cortex and hippocampus in elderly schizophrenia patients. Neurobiol Dis 2006; 21: 531–540. [DOI] [PubMed] [Google Scholar]
- 42Boggs J. Myelin basic protein: a multifunctional protein. Cell Mol Life Sci 2006; 63: 1945–1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43Zeisel A, Muñoz-Manchado AB, Codeluppi S, Lönnerberg P, La Manno G, Juréus A et al. Cell types in the mouse cortex and hippocampus revealed by single-cell RNA-seq. Science 2015; 347: 1138–1142. [DOI] [PubMed] [Google Scholar]
- 44van Berckel BN, Ossenkoppele R, Tolboom N, Yaqub M, Foster-Dingley JC, Windhorst AD et al. Longitudinal amyloid imaging using 11C-PiB: methodologic considerations. J Nucl Med 2013; 54: 1570–1576. [DOI] [PubMed] [Google Scholar]
- 45Turkheimer FE, Selvaraj S, Hinz R, Murthy V, Bhagwagar Z, Grasby P et al. Quantification of ligand PET studies using a reference region with a displaceable fraction: application to occupancy studies with [ 11C]-DASB as an example. J Cereb Blood Flow Metab 2011; 32: 70–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46Wang C, Wu C, Zhu J, Miller RH, Wang Y. Design, synthesis, and evaluation of coumarin-based molecular probes for imaging of myelination. J Med Chem 2011; 54: 2331–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47Yaqub M, van Berckel BN, Schuitemaker A, Hinz R, Turkheimer FE, Tomasi G et al. Optimization of supervised cluster analysis for extracting reference tissue input curves in (R)-[(11)C]PK11195 brain PET studies. J Cereb Blood Flow Metab 2012; 32: 1600–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.