Abstract
The open reading frame 45 (ORF45) of the Kaposi's sarcoma-associated herpesvirus (KSHV) is an immediate-early phosphorylated tegument protein critical for viral escape from host immune surveillance. Its expression is upregulated by the viral replication and transcription activator (RTA), a key protein that controls the switch from latency to lytic replication. We report here that ORF45 expression was not only upregulated by RTA, but ORF45 could also be degraded by RTA in a proteasome-dependent manner. The ORF45 was activated by RTA via activation of the ORF45 promoter, and the promoter region from nt 69 271 to nt 69 026 was involved. In chronic KSHV infected TRE-BCBL-1 RTA cells, the endogenous ORF45 protein increased dramatically after the induction of RTA expression, but then decreased rapidly after 8 h post-induction. Our study suggests that RTA might control the kinetics of viral replication through fine-tuning of the level of ORF45 and other viral/host proteins.
Kaposi's sarcoma-associated herpesvirus (KSHV), also known as human herpesvirus 8, is the aetiological agent of Kaposi's sarcoma, the most common neoplasm in AIDS patients (Chang et al., 1994). The open reading frame 45 of KSHV is an immediate–early viral gene, which encodes a multifunctional phosphorylated tegument protein (ORF45) (Kuang et al., 2008; Zhu et al., 1999; Zhu et al., 2005; Zhu & Yuan, 2003). It functions as a modulator to promote viral escape from immune surveillance by interacting with the inhibitory domain of the cellular interferon-regulatory factor 7 (IRF-7) (Lacoste et al., 2004; Sathish et al., 2011) and inhibits virus-induced type I interferon production by blocking IRF-7 phosphorylation and nuclear translocation (Zhu et al., 2002; Zhu et al., 2010). ORF45 can also interact with other viral proteins such as viral tegument proteins (Rozen et al., 2008). It can also interact with cellular proteins such as the p90 ribosomal S6 kinase, to mediate the phosphorylation of eukaryotic translation initiation factor 4B to facilitate protein translation (Kuang et al., 2011). ORF45 can interact with motor protein KIF3A to play a role in viral maturation and egress (Sathish et al., 2009), and is also required at the early stage of primary infection since ORF45-null virus can neither express any viral gene nor establish latency (Zhu et al., 2006). Recently, KSHV ORF45 was also found to help recruit RNA polymerase II to the HIV-1 LTR to enhance HIV-1 transcription (Karijolich et al., 2014).
The ORF45 expression and function are modulated by both cellular and viral proteins. Its transcription is regulated by cellular chromatin-organizing factor cohesins (Chen et al., 2012), and ORF45 can be ubiquitinated by a cellular ubiquitin E3 ligase known as the seven in absentia homologue to lead to its degradation via the proteasome pathway (Abada et al., 2008). Interestingly, ORF45 is also activated by the KSHV replication and transcription activator (RTA) (Chang et al., 2013). Since RTA controls the switch of KSHV from latency to lytic replication (Gradoville et al., 2000; Lukac et al., 1998; Sun et al., 1998), it is important to determine the interaction between ORF45 and RTA during early viral replication.
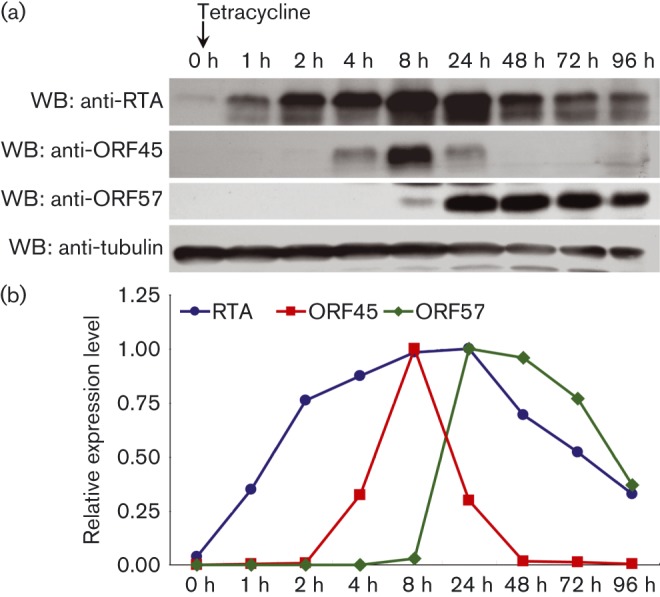
We first detected the endogenous RTA and ORF45 protein levels in TRE × BCBL-1 RTA cells upon induction of viral replication. The RTA gene is integrated into the genome of TRE × BCBL-1 RTA cells and its expression is tightly regulated by tetracycline (Nakamura et al., 2003; Wang et al., 2005). The TRE × BCBL-1 RTA cells were collected at 0, 1, 2, 4, 8, 24, 48, 72 or 96 h post tetracycline treatment, and the expression of RTA and ORF45 were monitored by Western blots (Fig. 1a). The expression levels of RTA and ORF45 were quantified using ImageJ software and normalized to their peak levels, respectively (Fig. 1b). At 1 h post tetracycline treatment, enhanced RTA expression (35 %) was detected as expected. The accumulation of endogenous RTA could be observed (76 %, 87 % and 99 % at 2 h, 4 h and 8 h, respectively), with the highest expression level detected at 24 h (100 %), then followed by a gradual decrease (70 %, 52 % and 33 % at 48 h, 72 h and 96 h, respectively). For ORF45, it was hardly detectable before 4 h, but increased dramatically in the presence of RTA (33 % at 4 h), and peaked at 8 h (100 %) post tetracycline treatment. This increase supports previous reports that suggested that RTA could activate the ORF45 promoter. Interestingly, ORF45 was found to decrease rapidly after peaking at 8 h (30 % at 24 h), and this decrease correlated with the accumulation of RTA expression. The ORF45 was completely undetectable at 48 h and beyond post-induction. These results demonstrated that the expression of ORF45 correlated with RTA levels; the expression of RTA was followed by an increase in ORF45, and supports the hypothesis that RTA can activate ORF45 expression in vivo. However, once ORF45 reached a threshold level it then decreased rapidly, whether this decrease was due to the accumulation of RTA needs to be further analysed. The expression of another lytic protein, ORF57, was also detected. The endogenous ORF57 started to express and peaked at 24 h (100 %), and decreased gradually afterward (96 %, 77 % and 37 % at 48, 72 and 96 h, respectively) (Fig. 1).
Fig. 1. ORF45 expression in the TRE × BCBL-1 RTA cells. (a) Detection of endogenous RTA, ORF45 and ORF57 in the TRE × BCBL-1 RTA cells. TRE × BCBL-1 RTA cells were collected at 0, 1, 2, 4, 8, 24, 48, 72 and 96 h post treatment with tetracycline (5 μg ml− 1), and lysates were analysed by Western blot to detect the endogenous RTA, ORF45, ORF57 and β-tubulin. (b) Quantification of the data from (a). The expressions of each protein were quantified using ImageJ software and were normalized to their peak levels.
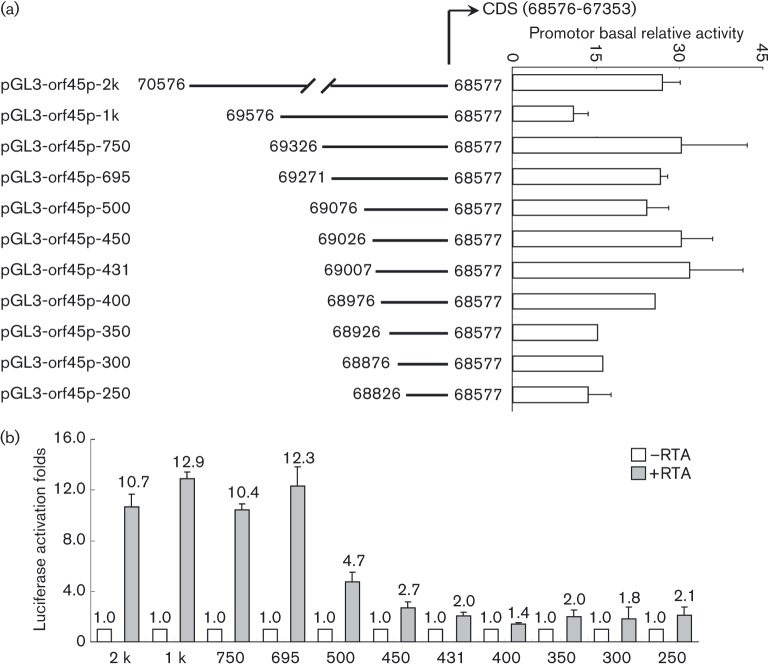
As the initial ORF45 expression appeared to be dependent on the presence of RTA, which was shown to activate ORF45 expression, we then investigated how RTA could activate ORF45 molecularly by further characterizing the activation of the ORF45 promoter by RTA. The ORF45 promoter fragments were amplified using primers listed in Table S1 (available in the online Supplementary Material), and each fragment was inserted into the pGL3-basic vector. The reporter construct, which contains a region upstream of the ORF45 coding region spanning from nt 70 576 to nt 68 577, was constructed and designated pGL3-orf45p-2k (intact fragment). A series of deletion promoter reporter constructs were also made and designated pGL3-orf45p-1k, -750, -695, -500, -450, -431, -400, -350, -300 and -250 bp (Fig. 2a). The 293T cells were transfected with each of the promoter-derived luciferase reporter construct (100 ng), and the relative basal activity was measured for each construct (Fig. 2a, right panel). The 293T cells were co-transfected with 1 μg of the RTA expression plasmid and the luciferase activities were measured and normalized to the basal activities of each reporter construct. As expected, RTA activated pGL3-orf45p-2k by 10.7-fold, and for reporters pGL3-orf45p-1k, pGL3-orf45p-750 and pGL3-orf45p-695, the activations were 12.9-, 10.4-, and 12.3-fold, respectively. However, RTA transactivation decreased to 4.7-fold with construct pGL3-orf45p-500, with − 500 bp of the promoter sequence. Further deletion of the ORF45 promoter to − 450 bp almost completely abolished RTA transactivation activities. These results demonstrated that the ORF45 promoter region from − 450 to − 695, or from nt 69 271 to nt 69 026, is necessary for its responsiveness to RTA transactivation (Fig. 2b).
Fig. 2. RTA activates ORF45 promoter. (a) Schematic representation of ORF45 promoter constructs used in transient transfection analyses. The basal relative activity of each promoter construct is shown on the right. (b) Responsiveness of each reporter to RTA activation. The 293T cells were transfected with each reporter plasmid, with or without RTA expression plasmid. Luciferase activities were measured at 48 h post-transfection, and transfection efficiency was normalized by using the pCMV-β expression plasmid as an internal control. The activation fold was normalized to control in the absence of RTA (white columns). The levels of activation when the reporters were co-transfected with RTA expression plasmid are shown (shaded columns). Results are averages of three independent experiments, and the standard deviations are shown.
A predicted consensus binding sequence of RTA has been found in the ORF45 promoter from nt 69 231 to nt 69 238 (Liu et al., 2008). In addition, the RTA direct binding site on the ORF45 promoter was mapped to between nt 69 000 and nt 69 480 using a ChIP-on-chip approach (Chen et al., 2009). We have now mapped the RTA-responsive element located in the region between nt 69 271 and nt 69 026, which covered the predicted RTA binding consensus sequence proposed by Liu et al. (2008) and is within the previously mapped RTA direct binding site reported by Chen et al. (2009). Recently Chang et al. reported that the ORF45 promoter could be activated by both RTA-dependent and -independent mechanisms (Chang et al., 2013). They showed that two RBP-Jκ binding sites (nt 69 018 to nt 69 012 and nt 69 510 to nt 69 504) in the ORF45 promoter confer RTA-dependent responsiveness via RBP-Jκ, whereas the NF-Y and Sp1-binding sites mediate RTA-independent response to sodium butyrate. Interestingly, the RTA-responsive element we mapped to the ORF45 promoter in this study does not cover these RBP-Jκ binding sites, suggesting that our promoter construct was activated by an RBP-Jκ independent mechanism.
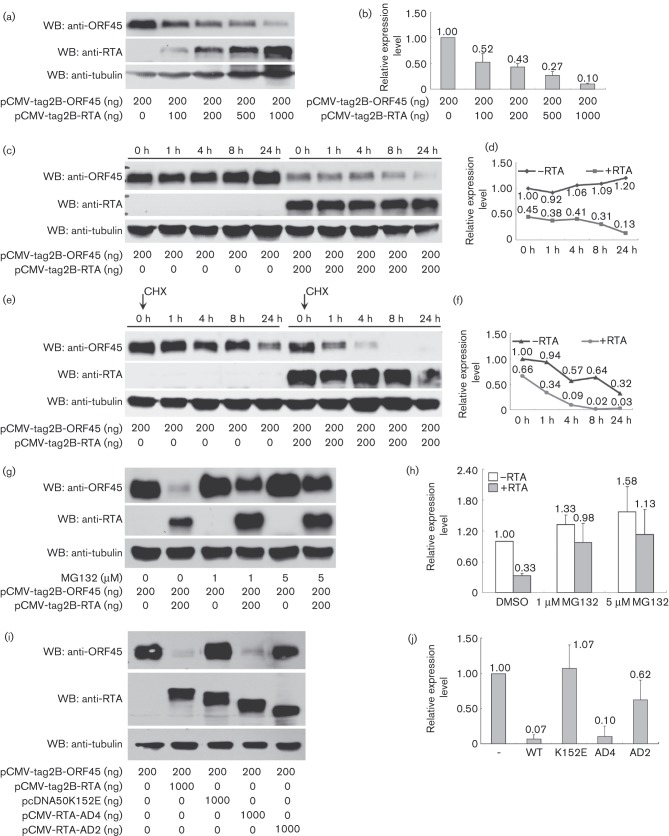
To further analyse what contributes to the rapid decrease of ORF45 after reaching its threshold level at 8 h upon RTA activation and whether RTA plays a role in the decrease, we then studied the effects of RTA on the ORF45 protein level. It has been shown that RTA possesses E3 ubiquitin ligase activities and can also recruit other cellular ubiquitin E3 ligase for the proteasome-dependent degradation of a number of cellular and viral proteins to regulate viral replication and host antiviral responses (Ehrlich et al., 2014; Izumiya et al., 2013; Yang et al., 2008; Yu & Hayward, 2010; Yu et al., 2005). It is possible that ORF45 levels may also be regulated by RTA via a similar mechanism. Therefore, Western blot analyses were conducted using 293T cells co-transfected with the ORF45 expression plasmid (200 ng) and an increasing amount of the RTA expression plasmid (from 0 to 1000 ng). Indeed, the ORF45 protein level decreased when increasing amounts of RTA were expressed (Fig. 3a). This experiment was repeated three times and the ORF45 protein levels were quantified using ImageJ software, by first normalizing the ORF45 to the β-tubulin levels and then to the ORF45 levels without RTA (Fig. 3b). In the presence of 100, 200, 500 or 1000 ng RTA expression plasmid, the ORF45 protein levels decreased to 52 %, 43 %, 27 % and 10 %, respectively, when compared to control in the absence of RTA. Our results thus suggest that RTA downregulates ORF45 protein level in a dose-dependent manner.
Fig. 3. RTA targets ORF45 for degradation. (a) RTA downregulated ORF45 protein level in a dose-dependent manner. The 293T cells were co-transfected with ORF45 expression plasmid (200 ng) and an increasing amount of RTA expression plasmid (0, 100, 200, 500 and 1000 ng). The expressions of ORF45, RTA and β-tubulin were detected with Western blots. (b) Quantification of the data from (a). The ORF45 protein levels were quantified using ImageJ software and normalized first to the levels of β-tubulin and then to the ORF45 level in the absence of RTA. Results are averages of three independent experiments, and the standard deviations are shown. (c) The time-course effects of RTA on the ORF45 protein level. The 293T cells were transfected with 200 ng ORF45 expression plasmid, with or without 200 ng RTA expression plasmid. The 24 h post-transfection was regarded as time point 0, and proteins were collected 0, 1, 4, 8 or 24 h later. The expressions of ORF45, RTA and β-tubulin were detected with Western blots. (d) Quantification of the data from (c). (e) The downregulation of ORF45 protein level by RTA is a result of ORF45 degradation. CHX (100 μg ml− 1) was added to block the de novo protein synthesis. The expressions of ORF45, RTA and β-tubulin were detected. (f) Quantification of the data from (e). (g) The degradation of ORF45 protein level by RTA is dependent on the 26S proteasome pathway. MG132 dissolved in DMSO was added 24 h post-transfection and cells were cultured for another 24 h before being collected for Western blots. MG132 concentrations were 0 μM (lanes 1 and 2), 1 μM (lanes 3 and 4) and 5 μM (lanes 5 and 6). (h) Quantification of the data from (g). (i) Detection of mutant or deletions of RTA in degrading ORF45. RTA ubiquitination mutant K152E (lane 3) and deletions AD4 (lane 4) and AD2 (lane 5) were utilized to co-transfect 293T cells with ORF45 expression plasmid. (j) Quantification of the data from (i).
The effect of RTA on the kinetics of ORF45 protein levels was then investigated. In the absence of RTA expression, the ORF45 protein levels remained constant at 0, 1, 4, and 8 h, but increased slightly at 24 h post-transfection. However, when 200 ng of RTA expression plasmid was co-transfected with the ORF45 plasmid, the ORF45 protein levels decreased with time. Moreover, ORF45 was expressed much lower at each time point in the presence of RTA as compared to those without RTA (Fig. 3c). When quantified, the ORF45 protein levels in the presence of RTA were 45 % (0 h), 38 % (1 h), 41 % (4 h), 31 % (8 h) and 13 % (24 h) of the 0 h ORF45 level without RTA (Fig. 3d).
When protein synthesis inhibitor cycloheximide (CHX) was added to block de novo protein synthesis, ORF45 protein levels decreased with time both in the presence and absence of RTA. However, in the absence of RTA, ORF45 protein levels decreased to 32 % at 24 h post-transfection, whereas in the presence of RTA expression the ORF45 levels decreased much more rapidly, from 66 % (0 h) to undetectable levels at 8 and 24 h post-transfection (Fig. 3e, f). These results suggested that the downregulation of ORF45 protein level by RTA was not attributed to the decrease of protein synthesis, but was due to enhanced ORF45 degradation.
To investigate whether the enhanced degradation of ORF45 protein in the presence of RTA was mediated by proteasomal degradation, an inhibitor MG132, which blocks the 26S proteasome complex proteolytic activity was utilized. In the absence of RTA, the ORF45 protein level increased to 133 % and 158 % of untreated control, when treated with 1 μM or 5 μM of MG132, respectively. In the presence of RTA, the ORF45 levels also increased from 33 % to 98 % and 113 %, respectively, when 1 μM or 5 μM MG132 was added (Fig. 3g, h). RTA ubiquitination mutant K152E, which completely lost its ability to induce protein degradation (Yang et al., 2008), and deletion mutants AD4 and AD2, which have various deletions in their activation domain (Zhang et al., 2005), were tested for their ability to degrade ORF45 (Fig. 3i, j). At 48 h post-transfection, the ORF45 level was decreased to 7 % by the wild-type RTA. The deletion mutant AD4 was capable of degrading ORF45 as effectively as the wild-type (10 %), whereas AD2 could only partially degrade ORF45 (62 %). When the RTA ubiquitination mutant K152E was tested, ORF45 was no longer degraded (107 %), demonstrating that a single amino acid mutation at K152 abolished the ability of RTA to degrade ORF45. These results suggest that the degradation of ORF45 protein is mediated by RTA via the 26S proteasome pathway.
Our results so far have demonstrated that ORF45 expression can be stimulated by RTA (Fig. 2b) but ORF45 protein can be targeted for degradation by RTA (Fig. 3). We have shown that in TRE × BCBL-1 RTA cells, the endogenous ORF45 protein level increased dramatically from 2 to 8 h and then decreased rapidly (Fig. 1). This increase coincided with the accumulation of RTA and suggests that RTA might regulate ORF45 differentially at different stages of infection. At the early phase of viral lytic replication, RTA activates ORF45 promoter to evade the host immune surveillance. However, after the initial infection and lytic replication, KSHV needs to establish latency and persistent infection; viral proteins that are involved in initial lytic replication such as ORF45 and even RTA need to be down modulated. It is possible that the degradation of accumulated ORF45 could also be mediated by RTA, as has been shown with a number of viral and cellular proteins (Yang et al., 2008; Yu & Hayward, 2010; Yu et al., 2005).
As the lytic switch protein, RTA regulates a variety of viral and host proteins. It activates viral gene expressions through direct binding or indirect interaction with viral promoters (Chen et al., 2009; Ziegelbauer et al., 2006). It also regulates a number of viral and host proteins post-translationally (Ehrlich et al., 2014; Izumiya et al., 2013;Yang et al., 2008; Yu & Hayward, 2010; Yu et al., 2005). Here we demonstrate for the first time, to our knowledge, that RTA modulates ORF45 level by activating ORF45 expression initially and then degrading the accumulated ORF45. This could be a common mechanism that RTA has adopted to subtly modulate viral or host gene expression to regulate different stages of viral infection, from lytic replication to latency and subsequent reactivation. In fact, besides modulating ORF45, RTA can modulate itself by auto-activation of its own promoter to increase expression (Deng et al., 2000) and auto-ubiquitination to enhance proteasome-mediated degradation (Yu et al., 2005). Our study will help elucidate the mechanism of how the ORF45 level is subtly modulated during KSHV replication and how RTA controls the different phases of viral infection.
Acknowledgements
This work was supported by NIH PHS grants RO1 TW007294, Natural Science Foundation of China 30570083, 30870129 to J. W.; 30970140, 81470095 and Natural Science Foundation of Tianjin City No. 12JCYBJC15200 to Y. W.; PHS grants CA75903, GM103509 and the Fogarty AIDS International Training and Research Program D43TW01492 from the NIH to C. W.
References
- Abada R., Dreyfuss-Grossman T., Herman-Bachinsky Y., Geva H., Masa S.R., Sarid R. (2008). SIAH-1 interacts with the Kaposi's sarcoma-associated herpesvirus-encoded ORF45 protein and promotes its ubiquitylation and proteasomal degradation J Virol 82 2230–2240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Y., Cesarman E., Pessin M.S., Lee F., Culpepper J., Knowles D.M., Moore P.S. (1994). Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma Science 266 1865–1869. [DOI] [PubMed] [Google Scholar]
- Chang P.J., Wang S.S., Chen L.Y., Hung C.H., Huang H.Y., Shih Y.J., Yen J.B., Liou J.Y., Chen L.W. (2013). ORF50-dependent and ORF50-independent activation of the ORF45 gene of Kaposi's sarcoma-associated herpesvirus Virology 442 38–50. [DOI] [PubMed] [Google Scholar]
- Chen J.G., Ye F.C., Xie J.P., Kuhne K., Gao S.J. (2009). Genome-wide identification of binding sites for Kaposi's sarcoma-associated herpesvirus lytic switch protein, RTA Virology 386 290–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H.S., Wikramasinghe P., Showe L., Lieberman P.M. (2012). Cohesins repress Kaposi's sarcoma-associated herpesvirus immediate early gene transcription during latency J Virol 86 9454–9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng H., Young A., Sun R. (2000). Auto-activation of the rta gene of human herpesvirus-8/Kaposi's sarcoma-associated herpesvirus J Gen Virol 81 3043–3048. [DOI] [PubMed] [Google Scholar]
- Ehrlich E.S., Chmura J.C., Smith J.C., Kalu N.N., Hayward G.S. (2014). KSHV RTA abolishes NFκB responsive gene expression during lytic reactivation by targeting vFLIP for degradation via the proteasome PLoS ONE 9 e91359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gradoville L., Gerlach J., Grogan E., Shedd D., Nikiforow S., Metroka C., Miller G. (2000). Kaposi's sarcoma-associated herpesvirus open reading frame 50/Rta protein activates the entire viral lytic cycle in the HH-B2 primary effusion lymphoma cell line J Virol 74 6207–6212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumiya Y., Kobayashi K., Kim K.Y., Pochampalli M., Izumiya C., Shevchenko B., Wang D.H., Huerta S.B., Martinez A., other authors (2013). Kaposi's sarcoma-associated herpesvirus K-Rta exhibits SUMO-targeting ubiquitin ligase (STUbL) like activity and is essential for viral reactivation PLoS Pathog 9 e1003506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karijolich J., Zhao Y., Peterson B., Zhou Q., Glaunsinger B. (2014). Kaposi's sarcoma-associated herpesvirus ORF45 mediates transcriptional activation of the HIV-1 long terminal repeat via RSK2 J Virol 88 7024–7035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang E., Tang Q.Y., Maul G.G., Zhu F.X. (2008). Activation of p90 ribosomal S6 kinase by ORF45 of Kaposi's sarcoma-associated herpesvirus and its role in viral lytic replication J Virol 82 1838–1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuang E.S., Fu B.S., Liang Q.M., Myoung J., Zhu F.X. (2011). Phosphorylation of eukaryotic translation initiation factor 4B (EIF4B) by open reading frame 45/p90 ribosomal S6 kinase (ORF45/RSK) signaling axis facilitates protein translation during Kaposi sarcoma-associated herpesvirus (KSHV) lytic replication J Biol Chem 286 41171–41182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacoste V., de la Fuente C., Kashanchi F., Pumfery A. (2004). Kaposi's sarcoma-associated herpesvirus immediate early gene activity Front Biosci 9 2245–2272. [DOI] [PubMed] [Google Scholar]
- Liu Y., Cao Y., Liang D., Gao Y., Xia T., Robertson E.S., Lan K. (2008). Kaposi's sarcoma-associated herpesvirus RTA activates the processivity factor ORF59 through interaction with RBP-Jkappa and a cis-acting RTA responsive element Virology 380 264–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukac D.M., Renne R., Kirshner J.R., Ganem D. (1998). Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein Virology 252 304–312. [DOI] [PubMed] [Google Scholar]
- Nakamura H., Lu M., Gwack Y., Souvlis J., Zeichner S.L., Jung J.U. (2003). Global changes in Kaposi's sarcoma-associated virus gene expression patterns following expression of a tetracycline-inducible Rta transactivator J Virol 77 4205–4220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen R., Sathish N., Li Y., Yuan Y. (2008). Virion-wide protein interactions of Kaposi's sarcoma-associated herpesvirus J Virol 82 4742–4750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathish N., Zhu F.X., Yuan Y. (2009). Kaposi's sarcoma-associated herpesvirus ORF45 interacts with kinesin-2 transporting viral capsid-tegument complexes along microtubules PLoS Pathog 5 e1000332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathish N., Zhu F.X., Golub E.E., Liang Q.M., Yuan Y. (2011). Mechanisms of autoinhibition of IRF-7 and a probable model for inactivation of IRF-7 by Kaposi's sarcoma-associated herpesvirus protein ORF45 J Biol Chem 286 746–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun R., Lin S.F., Gradoville L., Yuan Y., Zhu F.X., Miller G. (1998). A viral gene that activates lytic cycle expression of Kaposi's sarcoma-associated herpesvirus Proc Natl Acad Sci U S A 95 10866–10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J.Z., Zhang J., Zhang L.W., Harrington W., Jr, West J.T., Wood C. (2005). Modulation of human herpesvirus 8/Kaposi's sarcoma-associated herpesvirus replication and transcription activator transactivation by interferon regulatory factor 7 J Virol 2420–2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.L., Yan Z.C., Wood C. (2008). Kaposi's sarcoma-associated herpesvirus transactivator RTA promotes degradation of the repressors to regulate viral lytic replication J Virol 82 3590–3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Hayward G.S. (2010). The ubiquitin E3 ligase RAUL negatively regulates type i interferon through ubiquitination of the transcription factors IRF7 and IRF3 Immunity 33 863–877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y.X., Wang S.E., Hayward G.S. (2005). The KSHV immediate-early transcription factor RTA encodes ubiquitin E3 ligase activity that targets IRF7 for proteosome-mediated degradation Immunity 22 59–70. [DOI] [PubMed] [Google Scholar]
- Zhang J., Wang J.Z., Wood C., Xu D.S., Zhang L.W. (2005). Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8 replication and transcription activator regulates viral and cellular genes via interferon-stimulated response elements J Virol 79 5640–5652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F.X., Yuan Y. (2003). The ORF45 protein of Kaposi's sarcoma-associated herpesvirus is associated with purified virions J Virol 77 4221–4230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F.X., Cusano T., Yuan Y. (1999). Identification of the immediate-early transcripts of Kaposi's sarcoma-associated herpesvirus J Virol 73 5556–5567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F.X., King S.M., Smith E.J., Levy D.E., Yuan Y. (2002). A Kaposi's sarcoma-associated herpesviral protein inhibits virus-mediated induction of type I interferon by blocking IRF-7 phosphorylation and nuclear accumulation Proc Natl Acad Sci U S A 99 5573–5578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F.X., Chong J.M., Wu L.J., Yuan Y. (2005). Virion proteins of Kaposi's sarcoma-associated herpesvirus J Virol 79 800–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F.X., Li X.J., Zhou F.C., Gao S.J., Yuan Y. (2006). Functional characterization of Kaposi's sarcoma-associated herpesvirus ORF45 by bacterial artificial chromosome-based mutagenesis J Virol 80 12187–12196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu F.X., Sathish N., Yuan Y. (2010). Antagonism of host antiviral responses by Kaposi's sarcoma-associated herpesvirus tegument protein ORF45 PLoS ONE 5 e10573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegelbauer J., Grundhoff A., Ganem D. (2006). Exploring the DNA binding interactions of the Kaposi's sarcoma-associated herpesvirus lytic switch protein by selective amplification of bound sequences in vitro J Virol 80 2958–2967. [DOI] [PMC free article] [PubMed] [Google Scholar]