Abstract
Piliation is an important virulence determinant for Neisseria gonorrhoeae. PilE polypeptide is the major protein subunit in the pilus organelle and engages in extensive antigenic variation due to recombination between pilE and a pilS locus. pilS were so-named as they are believed to be transcriptionally silent, in contrast to the pilE locus. In this study, we demonstrate the presence of a small, pil-specific RNA species. Through using a series of pilE deletion mutants, we show by Northern blotting and quantitative reverse transcriptase PCR analysis (qRT-PCR), that these smaller RNA species are not derived from the primary pilE transcript following some processing events, but rather, arose through transcription of the pilS loci. Small transcriptome analysis, in conjunction with analysis of pilS recombinants, identified both sense and anti-sense RNAs originating from most, but not all, of the pilS gene copies. Focusing on the MS11 pilS6 locus, we identified by site-directed mutagenesis a sense promoter located immediately upstream of pilS6 copy 2, as well as an anti-sense promoter immediately downstream of pilS6 copy 1. Whole transcriptome analysis also revealed the presence of pil-specific sRNA in both gonococci and meningococci. Overall, this study reveals an added layer of complexity to the pilE/pilS recombination scheme by demonstrating pil-specific transcription within genes that were previously thought to be transcriptionally silent.
Introduction
Neisseria gonorrhoeae causes the sexually transmitted disease gonorrhoea. Uncomplicated gonorrhoea is a mucosal infection of either the urethra in males, or of the cervix in women. Piliation facilitates the infectious process by providing attachment of the bacterium to the mucosal epithelium (Meyer & Hill, 2003). The pilus organelle is a complicated structure, with PilE polypeptide being the major component. PilE polypeptide changes chemically due to recombination between the pilE locus and one of several silent pil genes (pilS) located elsewhere on the chromosome (Hill & Davies, 2009). The pilS genes are different from pilE in that no apparent promoter element is present, and that the pilS gene copies only comprise the 3′ two-thirds (or the variable region) of pilE. Consequently, pilS genes are believed to be transcriptionally silent.
There are multiple pilS loci in the chromosome, most of which consist of several pil gene copies in a tandem array (Haas & Meyer, 1986; Haas et al., 1992). Interspersed within the pil gene copies are several different repeat elements (e.g. RS1, RS2, RS4) (Haas & Meyer, 1986; Haas et al., 1992). The exact location of the repeat elements within a pilS locus appears to be random, with little conformity being seen across the different strains that have been sequenced. Currently, the precise role of these repeat elements is unknown, however, it is believed they may play a role in the pilE/pilS recombination process.
Transcription within the pil system is believed to be confined to the pilE locus (Meyer et al., 1984; Haas & Meyer, 1986; Haas et al., 1992). Despite three fully functional promoters being present upstream of the pilE gene, only a single promoter, P1, is apparently used in the gonococcus (Fyfe et al., 1995; Carrick et al., 1997). No regulator has been identified to moderate expression (Fyfe et al., 1995; Carrick et al., 1997; Laskos et al., 1998). However, binding of integration host factor (IHF) upstream of the P1 promoter potentiates pilE transcription by facilitating interaction of UP-elements with RNA polymerase (Hill et al., 1997; Fyfe & Davies, 1998).
The basis of this study was the identification of novel pil-specific RNAs when Northern blots of total RNA samples were probed sequentially with oligonucleotide probes. These RNAs were noticeably smaller than the full-length pilE message. Through a combination of mutational analysis, Northern blotting, in vitro transcription, quantitative reverse transcriptase PCR analysis (qRT-PCR) and transcriptome analysis, we show that these small RNAs are comprised of both sense and anti-sense small RNAs that originate from the pilS loci. Consequently, these observations add a further degree of complexity to not only pilE transcriptional regulation, but also to the pilE/pilS gene variation paradigm.
Methods
Strains and growth conditions.
N. gonorrhoeae strain MS11 strain 7/30 : 2 was used in this study (Bergström et al., 1986). Gonococci were passaged daily on gonococcal typing medium (Swanson, 1982) at 37 °C in 5 % CO2. When grown in liquid culture, the agar was omitted and the medium was supplemented with NaHCO3 at a final concentration of 420 ng ml−1. In studies using antibiotics, rifampicin was added at a final concentration of 200 µg ml−1; erythromycin was added at a final concentration of 5 µg ml−1.
For studying transcription within a recombinant setting, Escherichia coli DH5α was used. Plasmids were maintained by the presence of antibiotics within the medium; ampicillin was added at a final concentration of 100 µg ml−1; erythromycin was added at a final concentration of 200 µg ml−1. All genetic manipulations utilized standard molecular biological procedures.
Construction of gonococcal pilE deletion mutants.
The pilE deletion mutants were all similarly generated; defined PCR fragments were derived using plasmid DNA that carried the pilE gene cloned from MS11 strain 7/30 : 2 as template (primers described in Table S1, available in the online Supplementary Material). Two PCR fragments were ligated such that a small fragment of the pilE gene was deleted. The ligated fragments were then cloned into the pCRII (Invitrogen) vector. Successful transformants were then linearized using SmaI; an opaE : : ermC fragment derived from pNG3005 (Wainwright et al., 1994) was then inserted. The correct orientation of the construct was confirmed by PCR analysis. The plasmids carrying the ΔpilE opaE : : ermC constructs were then used to transform gonococci to erythromycin resistance using standard Neisseria protocols; gonococcal transformants were then tested for the incorporation of the deletion into the pilE chromosomal locus by Southern analysis using defined oligonucleotide probes (Table S2) .
Cloning and site-directed mutagenesis.
All pilS6 constructs were derived from the pilS6 locus previously cloned from a N. gonorrhoeae strain MS11 genome library into the pBR322 vector (S.A. Hill, unpublished observations). PCR amplicons were ligated into the pSMART (Lucigen) vector to create five pilS6 subclones which progressively removed potential promoters by shortening the 5′ end of the locus (pilS6 : 1, pilS6 : 2, pilS6 : 3, pilS6 : 4, pilS6 : 6 and pilS6:SM1) (primers described in Table S3). Successful transformants were used to introduce point mutations into putative promoter sequences with QuikChangeII or QuikChange Lightning Mutagenesis kits (Agilent) (primers described in Table S4). Potential promoter sequences were identified with the BPROM bacterial recognition program available through the SoftBerry tool package (Solovyev & Salamov, 2011).
RNA analysis.
RNA was prepared from gonococci either by using TRIzol (Gibco), an Ambion total RNA kit or an Ambion MirVana Mira RNA kit (Applied Biosystems) according to the manufacturer’s instructions. Purified RNA preparations were then treated with RNase-free DNase I (10 units) for 30 min at 37 °C, extracted with phenol/chloroform, precipitated with ethanol, air-dried and resuspended in DEPC-treated water at a concentration of 10–20 mg ml−1. The total RNA preparations were fractionated on 1.0 % agarose gels containing 1.1 M formaldehyde using 1× MOPS buffer containing 1.1 M formaldehyde. Prior to blotting, the gels were washed in DEPC-treated water for 5 min, followed by 45 min in 0.5 M NaOH/1.5 M NaCl, followed by 45 min in 0.1M Tris pH 7.4 and a final washing for 1 h in 20× SSC. All solutions were prepared with DEPC-treated water. The RNAs were transferred to nitrocellulose membranes by capillary action using 20× SSC as buffer. Strand-specific oligonucleotides (Table S2) were end-labelled with [32P]-γ-ATP using T4 polynucleotide kinase (New England Biolabs). Following hybridization with the labelled primers (Table S3), blots were washed under high stringency conditions (0.1 % SSC at 40 °C). Primer extension analysis was performed as previously described (Bergström et al., 1986; Hill et al., 1997) using primer 5 as the extension primer for reverse transcriptase using RNA prepared from strain MS11 7/30 : 2 that had been grown in liquid culture for 12 h. The DNA sequencing ladder template utilized pVD203, which contains the pilE gene cloned from MS11 7/30 : 2 chromosomal DNA (Bergström et al., 1986).
qRT-PCR analysis.
Total RNAs were prepared from cells grown on solid medium for 12 h. The amount of RNA was estimated through comparison with an O’Gene Ruler ladder (Fermentas, Thermo Fisher Scientific). One microgram of RNA was then treated with RQ1 DNase (Promega), and 1 mM dithiothreitol (Acros Organics) for supernatant RNA, at 37 °C for 1 h to remove any contaminating chromosomal DNA. RQ1 DNase Stop Solution (Promega) was then added with further incubation at 65 °C for 15 min. Following DNase treatment, the RNA was ethanol precipitated, washed twice with 70 % (DEPC-treated) ethanol, dried in a speedvac and resuspended in 12 µl aliquots of DEPC-treated water. Complementary DNA (cDNA) synthesis was performed under the following conditions: 8 µl of DNase-treated RNA, 1 µg of an oligonucleotide random decamer, 2 µl of DNase reaction, 1 µl RNA3 preparation (1000× dilution of in vitro transcribed RNA; French & Ahlquist, 1987) and 2.5 µl of DEPC-treated water. The mixture was heated at 70 °C for 5 min and placed on ice. Following heat denaturation, 10.5 µl of common components (0.5 µl rRNasin [Promega], 5 µl 5× M-MLV RT Buffer [Promega] and 5 µl 2.5 mM dNTPs [dATP, dGTP, dCTP, dTTP]) were added to the reaction tube along with 1 µl of M-MLV RT (Promega) followed by a 1 h incubation at 42 °C followed by 5 min at 95 °C. The resulting single-stranded cDNA was diluted 10× in ddH2O (25 µl cDNA in 225 µl ddH2O) for real-time quantitative PCR (qRT-PCR).
A standard qRT-PCR involved adding 3 µl of cDNA (10× dilution), 9.5 µl of primer mix (1.25 µl of 10 ng µl−1 forward primer, 1.25 µl of 10 ng µl−1 reverse primer, 0.25 µl of 2× Internal Reference Dye R4526 included in a SYBR Green JumpStar ReadyMix kit (Sigma) and 6.75 µl of ddH2O) and 12.5 µl of SYBR Green JumpStart Taq ReadyMix S9939 (20 mM Tris/HCl, pH 8.3, 100 mM KCl, 7 mM MgCl2, 0.4 mM each dNTP [dATP, dCTP, dGTP, TTP], stabilizers, 0.05 unit µl−1 Taq DNA Polymerase, JumpStart Taq antibody and SYBR Green I) from a SYBR Green JumpStar Taq ReadyMix kit (Sigma). Primers for all qRT-PCR are presented in Table S1. A typical qRT-PCR entailed 40 cycles of a 94 °C denaturation step for 30 s, a 60 °C annealing step for 30 s, and a 30 s extension step at 72 °C. All qRT-PCRs were carried out using a Realtime PCR System Mx3000 P (Stratagene).
Prior to using the experimental primers, control experiments were performed against the RNA3 external control to determine the quality of the cDNA synthesis reaction. All newly designed primers and all 16S rRNA primers underwent positive and negative control qRT-PCRs with plasmid DNA and water to ensure appropriate primer-template interactions and product size. Prior to performing cDNA reactions with primers designed to amplify specific regions of the target DNA, a control qRT-PCR was performed with cDNA (10× dilution) and DNase-treated RNA (31.25× dilution) using primers designed to amplify regions of 16S rRNA to ensure all the contaminating DNA had been removed during DNase treatment prior to synthesis of the cDNA. The fold difference was computed assuming 100 % PCR amplification efficiency which leads to a doubling of template per cycle (manufacturers guide; Stratagene). The oligonucleotide primers that were used are described in Table S2.
5′ RACE analysis.
To identify the 5′-end point(s) of the small RNA species, total RNA was extracted from WT GC following 18 h of growth on solid medium. A commercially available 45 base RNA adaptor oligonucleotide 5′-GCUGAUGGCGAUGAAUGAACACUGCGUUUGCUGG CUUUGAUGAAA-3′ was ligated to all mRNA transcripts using T4 RNA ligase (Ambion) at 37 °C for 1 h. Adaptor-linked mRNAs were reverse transcribed at 42 °C for 1 h using random decamers as primers (provided by the manufacturer). The pilE cDNA was then amplified through nested PCR using the following primers: forward primers were complementary to the adaptor RNA (provided by the manufacturer) and reverse primers were complementary to 3′ end pilE sequence (primer 246 and primer Sma/Cla III). The cDNA products were ligated into pCRIITM cloning vector (Invitrogen), with the inserts being sequenced using the M13 universal primers.
RNA circularization.
RNA circularization experiments were used to determine the 5′ and 3′ end points of single-stranded RNA molecules. Total RNA was prepared as described above. Following DNase treatment, 25 pmol of RNA was treated with 10 units of tobacco acid pyrophosphatase (Epicenter Biotechnologies) for 1–2 h at 37 °C to remove the 5′ pyrophosphate group. The 5′-monophosphorylated RNA molecule was then treated with T4 RNA ligase (Epicenter Biotechnologies) at 37 °C for 1 h which creates a circular single-stranded RNA molecule; the RNA ligase enzyme was then heat inactivated by incubating at 65 °C for 10 min. The self-ligated RNA was then converted into cDNA by reverse transcription using a RNA-specific primer and M-MLV reverse transcriptase (Promega). The resulting RNA–cDNA molecule is then incubated with RNase H (New England Biolabs) at 37 °C in 20 min to degrade the RNA strand. A total of 2μl of the reaction mix was used as template for a conventional PCR. The PCR product was then column purified and cloned into pCRII–TOPO vector (Invitrogen). Plasmids carrying the appropriately sized inserts were then sequenced using the M13 forward and reverse primers to locate the transcription start point of the RNA molecule.
RNA preparation, library construction and sequencing.
Following small RNA extraction of N. gonorrhoeae MS11 (Wachter & Hill, 2015) with a MirVana Mira kit; AM 1560 (Life Technologies), transcripts larger than 250–300 bp were excluded from analysis by size fractionation. Small RNA libraries were constructed and sequenced on an Illumina HiSeq2000 with chemistry version 4 and analysed with pipeline 1.8 (Wachter & Hill, in press). The error rate of the runs was <0.3 %. For the WT sample, the small RNA library coverage was 24 592 832 reads. For analysis, the adaptor sequences were subtracted from the read and only sequenced RNA transcripts that mapped to N. gonorrhoeae strain MS11 chromosome assembly GCA_000156855.2 were kept. Additionally, sequencing data for N. gonorrhoeae NCCP11945 was obtained from NCBI Gene Expression Omnibus series GSE58650 on October 6, 2014. The transcriptomes of N. gonorrhoeae FA1090 and Neisseria meningitidis FAM18 were generously made available by John Davies at Monash University through the Victorian Bioinformatics Consortium (http://www.vicbioinformatics.com). All RNA sequence analysis focused initially on unambiguous reads (i.e. transcript reads needed to map to regions of the chromosome with no lapse in read depth). Small transcriptomes are available at the GEO accession number GSE62926.
Mapping and visualization of reads.
All reference genomes used for transcriptome assembly were obtained from the NCBI Genome database. The N. gonorrhoeae strain MS11 small transcriptome sequencing data were mapped to the reference genome assembly GCA_00156855.2; the FA1090 whole transcriptome sequencing data were mapped to the reference genome assembly GCA_0000068451; the NCCP11945 whole transcriptome sequencing data were mapped to the reference genome assembly GCA_000020105.1; and, the N. meningitidis FAM18 whole trancriptome sequencing data were mapped to the reference genome assembly GCA_000009465.1. All transcriptome maps were created using the Nesoni data analysis toolset available through the Victorian Bioinformatics Consortium (http://www.vicbioinformatics.com/software.nesoni.shtml) which uses the SHRiMP read aligner and produces files that may be supplied to Artemis for graphic visualization (Rumble et al., 2009; Carver et al., 2012). Visualization of genomic locations of transcripts and histograms of transcript read depth were generated using the Circos software package version 0.66 (Krzywinski et al., 2009).
Transcript quantification.
Genomic coordinates were obtained from NCBI GenBank CP003909.1, (MS11), AE004969.1 (FA1090), CP001050.1 (NCCP11945) and AM421808.1 (FAM18). The amount of transcripts within genomic regions was determined, and transcripts per million (TPM) values were calculated in order to compare expression levels among the transcripts (Wagner et al., 2012).
Promoter analysis.
To determine potential promoter sequences that gave rise to pil-derived sRNAs, sequences spanning 100 base pairs upstream of these sRNA were analysed. Therefore, a total of 177 sequences were analysed for potential promoter regions that were responsible for the 177 ambiguous sRNAs detected within the pil genes. Tools available from meme (Multiple EM for Motif Elicitation) Suite were used to determine significant enriched sequences (motifs) present within the putative pil promoter regions (Bailey et al., 2009). A local installation of meme version 4.10.0 was used for primary analysis of potential motifs (Bailey et al., 2009, Bailey & Elkan, 1994). For this analysis, a zero or one occurrence model was used to detect motifs spanning between 5 and 20 bases with an E-value threshold of 0.001. Additionally, sequence composition was examined by determining the frequency of bases within the putative promoter regions. These sequences were also analysed to detect any regions bearing sequence similarity to the Pribnow box, as this sequence was found to be highly conserved within promoter elements of N. gonorrhoeae (Remmele et al., 2014). A multiple sequence alignment of these regions was performed with mafft and the sequence alignment was visualized with the WebLogo server (Katoh et al., 2002; Crooks et al., 2004).
Results
pilE transcription – identification of novel pil-specific RNAs
In order to determine whether pilE transcription showed any growth phase effects, N. gonorrhoeae strain MS11 was grown in liquid culture and cells were harvested after 12 h and after 24 h. Total RNA was extracted from each set of cells and initially analysed by Northern blotting using the 5′ pilE-specific probe 245 (Fig. 1a) where a strong signal at approximately 650 bp was apparent in the RNA prepared from 12 h grown cells and was absent in the RNA prepared from 24 h grown cells. In data not shown, control blots probed with a 16S RNA probe confirmed the presence of RNA in the 24 h sample. When cells that were grown for 24 h were subcultured into fresh medium, and grown for an additional 12 h, pilE transcription re-established itself (Fig. 1a). When the same blot was reprobed with an oligonucleotide that targets the 3′-end of the pilE transcript (probe 246), additional RNA species of varying sizes can be seen, with a prominent signal(s) migrating around 200–450 bp (Fig. 1b). Furthermore, additional, larger sized transcripts (approximately 2 kb) are also evident within the blots. As pilE is believed to be the only pil locus that engages in transcription, the appearance of the smaller RNA species was initially assumed to have arisen from the pilE transcript through some processing event. However, when pilE mRNA turnover was artificially induced (by treating exponentially growing cultures with rifampicin (200 µg ml−1) the novel smaller RNA species did not accumulate as would be expected under these conditions, indicating that their appearance were not a consequence of pilE mRNA turnover (data not shown).
Fig. 1. pilE transcription over time. Total RNAs were prepared at the indicated time points and were assessed by Northern blotting. (a) A 5′-pilE-specifc probe (245) was used; this probe detects full-length pilE transcript. (b) A 3′-pil-specific probe (246) was used; this probe detects all pil sense transcripts within the cell. Arrows indicate location of RNA signals. Curved arrows indicate passage of bacteria.
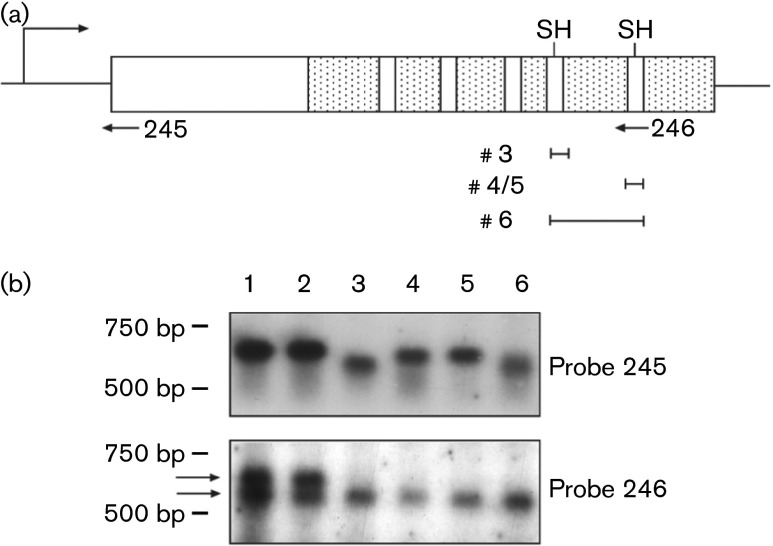
To further investigate the origin of this smaller pil-specific RNA species, we constructed a series of pilE deletion mutants (Fig. 2a). Four pilE deletion mutants were constructed that either deleted the pilE DNA that encodes the more upstream cysteine residue within the PilE polypeptide (mutant #3), that encodes the more downstream cysteine residue within the PilE polypeptide (mutants #4/5) or that encodes the hypervariable region of PilE polypeptide (mutant #6); these small deletions were then crossed into the gonococcal chromosome using a linked erythromycin gene as a selection marker. The pilE DNA that was deleted in mutants 4, 5 and 6 removes the DNA sequence that is complementary to probe 246; DNA sequencing of PCR products confirmed the deletions in pilE following transformation of gonococci to erythromycin resistance. When total RNA was prepared from each mutant plus a WT control (lane 1) as well as from an appropriate erythromycin-resistant control (lane 2), full-length message (or near full-length message for each mutant) was detected in a Northern blot when probed with the 5′ pilE-specific probe 245 (Fig. 2b). The aberrant migration, and, apparent transcript instability, of mutants 3 and 6 is believed to be due to the deletion of an embedded promoter element described below. However, when the same RNA samples were probed with the 3′ pil-specific probe 246, the RNA obtained from WT and the erythromycin-resistant control presented two signals, full-length message as well as the smaller RNA species, yet in contrast to these observations, deletion mutants 4, 5 and 6 only displayed a single signal which corresponded to the smaller RNA species (Fig. 2b). As the complementary DNA to probe 246 was deleted from the pilE gene in these mutants, these data indicate that the smaller RNA species are not derived from the full-length pilE message.
Fig. 2. Presence or absence of pil-specific RNA in pilE deletion mutants. (a) Schematic of the pilE gene indicating the location of the specific pilE deletions (#3, #4/5 and #6). Note deletions 4/5 and 6 remove the complementary DNA to oligonucleotide probe 246. (b) Northern analysis of the various mutant RNAs using the indicated probes. Lane 1, WT; lane 2, erythromycin-resistant control; lane 3, deletion 3; lanes 4/5, deletions 4/5; lane 6, deletion 6. The arrows indicate full-length pilE transcript and the smaller pilS derived transcripts. SH indicates the relative position of the cysteine residues in the PilE polypeptide.
Given the data presented in Fig. 2(b), we further investigated deletion mutants 4, 5 and 6 using qRT-PCR (Fig. 3). The pil-specific RNAs from these mutants were assayed using a series of pil primer pairs (Fig. 3a) with the cycle threshold (Ct) values being compared to the Ct value obtained for recA within the same sample (Fig. 3b, c). When primer 1 was utilized in conjunction with primers 2, 3, 4 and 5, such qRT-PCR analysis determined the relative amounts of full-length (or, approximately full-length depending upon the deletion) message. These data are shown in Fig. 3(b). As expected for deletion mutant 4, a strong signal relative to recA was obtained for primer pairs 1/2, 1/3 and 1/4, with a poor signal being observed for primer pair 1/5, as the sequence that is complementary to primer 5 was deleted from the pilE gene. In fact, there was approximately a six orders of magnitude difference for primer pair 1/5 when compared to primer pair 1/2. Likewise for deletion mutant 6, strong signals were obtained for primer pairs 1/2 and 1/3, with poor signals obtained for primer pairs 1/4 and 1/5. In this mutant there was in the order of a three orders of magnitude difference for primer pair 1/4 compared to primer pair 1/2, and a four orders of magnitude difference for primer pair 1/5 compared to primer pair 1/2. Again, the poor signals reflect that the complementary DNA to primers 4 and 5 was deleted from the pilE gene in mutant 6. In contrast to these observations, when primer pairs 6/5, 7/5 and 8/5 were used, a strong signal is observed for primer pair 8/5 in both sets of RNA, with the signal strength weakening depending upon the location of the upstream primer (primers 6 and 7) (Fig. 3c). Presumably, the signals detected in Fig. 3(c) reflect pil-specific RNA that originated elsewhere on the chromosome rather than from the pilE locus. For comparison, the qRT-PCR profiles using total RNA from WT cells are shown in Fig. S1. Therefore, collectively, these data support the Northern analysis presented in Fig. 2(b), and further indicate that the small RNA species are not derived from the full-length pilE message.
Fig. 3. qRT-PCR analysis of RNAs derived from the pilE deletion mutants. (a) Schematic showing the relative location of the primer pairs used in the qRT-PCR experiments. Tsp indicates the transcription start point; Tep indicates the transcription end point. The locations of the deletions are also indicated. (b) qRT-PCR analysis of RNA preparations extracted from deletion mutant 4 (the brick-like shading) and deletion mutant 6 (the diagonal shading). The forward primer in each case was primer 1 which assesses the relative amount of full-length (or near full-length transcript depending upon the deletion) in the sample. The data are presented as a difference in the pilE Ct scores verses the recA Ct score. The recA Ct scores were in the order of 24 cycles. A negative value indicates a lower Ct score for the pilE primer pair than that obtained for recA which equates to more RNA being present within the sample.
The novel RNA species is derived from multiple pilS loci
We next explored the possibility that the small pil RNA arose from one of the several pilS loci distributed around the chromosome. Our initial experiments exploited a property associated with the RNA derived from the pilE deletion mutants described above (mutants 4/5) where the complementary sequence to primer 5 is deleted from pilE which allowed us to perform 5′ RACE analysis using primer 5 as an amplifying primer. In addition, RNA circularization studies were also employed to define the 5′ and 3′ ends of the small RNA molecules. Several products were obtained using these strategies with products mapping to several different pilS. For example, one product mapped to the intergenic region between copy 3 and copy 2 located in pilS1 (the actual site within pilS1 being within the RS2 element, approximately 6 nt downstream of a potential −10 sequence 5′-CAAAAT-3′; data not shown). Subsequent qRT-PCR assays were then performed on E. coli recombinants carrying two different pilS present in strain MS11 (pilS2 and pilS6 cloned into plasmid pBR322 such that the resident tet promoter was inactivated as well as being in the opposite orientation to the β-lactamase gene) with each pilS locus showing active transcription when queried by qRT-PCR analysis using the primer pairs shown in Fig. 3(c) (data not shown). RNA sequencing (RNA Seq) was also used to extract pil-specific RNAs from whole transcriptomes that were available in the database; sense and anti-sense RNAs mapped to various pilS loci as well as pilE in the gonococcal strains FA1090 and NCCP 11945 and the meningococcal strain FAM18 that expresses class II pili (data not shown). Collectively, these observations strongly indicated that the small pil-specific RNAs arose from transcription within the pilS loci.
Molecular characterization of transcription from the MS11 pilS6 locus
N. gonorrhoeae strain MS11 contains five different pilS loci within the chromosome (Haas & Meyer, 1986; Haas et al., 1992). Consequently, investigating individual pilS promoter usage in gonococci initially proved to be problematic. However, a recombinant approach alleviated such problems. The MS11 pilS6 locus was chosen for a more in-depth analysis of its transcription profile. pilS6 contains three pil gene copies (Haas et al., 1992). Variously sized fragments of the pilS6 locus were cloned into a pSMART vector where overspill transcription from other genes located on the plasmid is prevented by the presence of transcriptional terminators that flank the multiple cloning site. An initial qRT-PCR analysis of these various subclones provided a crude assessment as to the location of the promoter elements. Two subclones were subsequently selected for further study (pilS6 : 4 and pilS6 : 6; for schematic, see Fig. 4a). A promoter was identified in plasmid pilS6 : 4 within the copy 2 pil segment. This putative promoter yielded RNA when assessed by qRT-PCR analysis with a greatly reduced signal in the pilS6 : 6 construct where the promoter was absent (Fig. 4b). Site-directed mutagenesis was employed to mutate the putative promoter, which eliminated the majority of RNA that arose from the construct (Fig. 4c, d).
Fig. 4. In-depth investigation of the pilS6 locus of strain N. gonorrhoeae MS11. (a) Schematic representation of the pilS6 : 4 and pilS6 : 6 constructs cloned from N. gonorrhoeae MS11 pilS locus. The vertical lines designating pilS6 : 4 and pilS6 : 6 indicate the ends of the cloned DNA fragments. The location of the primer pairs utilized in qRT-PCR are indicated. The white boxes represent constant regions, striped boxes represent the semivariable regions and double-striped boxes represent the hypervariable regions. The numbers beneath the figure indicate the relative location within the entire pilS6 locus as previously described (Haas et al., 1992). Tsp indicates the transcription start point. (b) qRT-PCR analysis of the recombinant clones pilS6 : 4 and pilS6 : 6 using the indicated primer pairs. The number of biological replicates is three and the number of technical replicates is ten. (c) Schematic representation of the sequence changes within the putative promoter in the pilS6 : 4 construct following site-directed mutagenesis. (d) qRT-PCR analysis following site-directed mutation of the putative promoter using the defined primer pairs. Bars represent means+SD.
However, pil-specific RNA was still detectable in pilS6 : 6 (Fig. 4b). Northern analysis was then performed on total RNA extracts from pilS6 : 4 and pilS6 : 6 cultures using strand-specific oligonucleotide probes (Fig. 5a). pilS6 : 4 yielded a sense transcript, whereas an anti-sense transcript was observed from pilS6 : 6 (Fig. 5a); production of anti-sense RNA apparently arising from a hitherto silent promoter which was present in the pilS6 : 4 construct. Two closely linked anti-sense promoters were identified immediately downstream of pilS6 copy 1 (Fig. 5b, c). Site-directed mutagenesis of each putative promoter eliminated anti-sense RNA production (Fig. 5d). Therefore, from this analysis we conclude for the MS11 pilS6 locus that, (i) both sense and anti-sense RNA is produced; (ii) the transcriptional profile varies depending upon the presence or absence of promoter elements; and, (iii) a hierarchy appears to exist with regard to promoter usage when all promoters are present.
Fig. 5. Analysis of anti-sense promoter located at the 3′ end of pilS6 gene copy 1. (a) Northern blot analysis of the pilS6 : 4 (lane 1) and pilS6 : 6 (lane 2) constructs. Total RNAs were prepared and probed with a sense and anti-sense oligonucleotide probes. (b) Schematic representation of the constant regions (underlined in red) containing the verified sense promoter in pilS6 copy 2 and the putative anti-sense promoter in pilS6 copy 1. All mutational constructs are italicized and bolded. All minicassettes are shown in grey. The Sma/Cla repeat is shown in light yellow. (c) qRT-PCR analysis following site-directed mutagenesis of the putative anti-sense promoters using the indicated primers. The number of biological replicates is three and the number of technical replicates is ten. Bars represent means+SD.
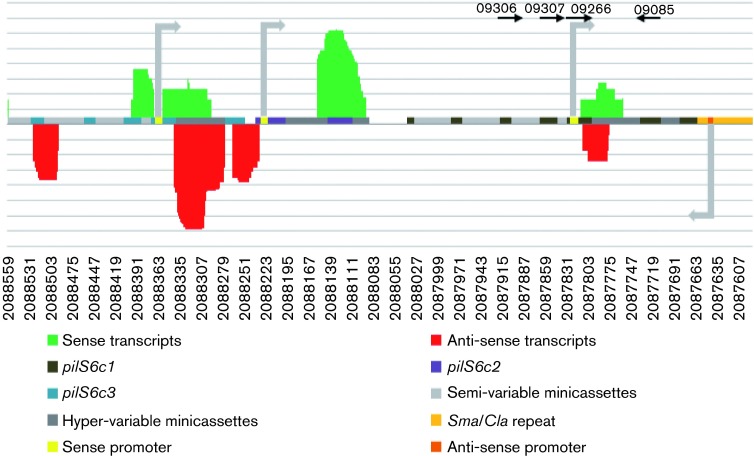
Confirmation of pilS6 transcription within a recombinant setting utilized small transcriptomic analysis. A small RNA transcriptome from strain MS11 WT cells underwent high-throughput sequencing (Wachter & Hill, 2015). The pilS6-specific sRNAs were extracted from the sequences and are displayed using RNA Seq where multiple sense and anti-sense RNAs were observed (Fig. 6), and correspond to the sRNAs identified within a recombinant context. However, the sensitivity of the transcriptomic analysis allowed other pilS6-derived RNAs to be also identified as well. Primer extension analysis was also performed utilizing primer 246 as the extension primer (Fig. 7a). Two end points could be identified (Fig. 7b); one that terminated within the so-called hypervariable region of pilE (Hagblom et al., 1985), with the end point being adjacent to the −10 promoter motif that was identified in plasmid pilS6 : 4 (Fig. 4c), a less prominent end point being observed further upstream within a semi-variable region (Fig. 7a). Therefore, these data confirm the previous recombinant analysis and demonstrate that in WT gonococci, the pilS6 locus actively engages in transcription and transcripts can be identified that may have arisen from a previously identified promoter.
Fig. 6. Small unambiguous RNAs mapping to the pilS6 locus in N. gonorrhoeae MS11. Small RNAs were isolated from N. gonorrhoeae strain MS11. RNA Seq analysis was employed to retrieve unambiguous transcripts that mapped to the pilS6 locus. Gonococcal transcripts that mapped sense to the pilS6 locus are shown as green, while anti-sense transcripts are shown as red. The pilS6 copies are shown in light blue (pilS6c3), purple (pilS6c2) and brown (pilS6c1), while the location of the minicassettes are shown in light grey (semi-variable) and dark grey (hyper-variable). The location of the experimentally verified pilS6c2 promoter is shown in yellow with the direction of ensuing transcripts shown with the faint grey arrow. Conserved regions in the pilS6 locus that share sequence identity to the pilS6c2 promoter are also shown in yellow with grey arrows indicating the direction of transcription. The experimentally verified anti-sense promoter is shown in orange with a grey arrow indicating the direction of transcription. The primer pairs utilized for qRT-PCR transcript detection from the pilS6c2 promoter are shown above.
Fig. 7. Primer extension analysis of pil sRNA. (a) Schematic of the pilE gene indicating the relative position of the oligonucleotide probes used in this study. Also indicated are the relative locations of the two identified primer extension products. (b) Total RNA was extracted from WT cells grown in liquid culture for 12 h. Primer extension analysis utilized oligonucleotide 246 as extension primer to determine the 5′ end(s) of pil sRNA. The horizontal arrows adjacent to the DNA sequence indicate the 5′ ends of two primer extension products. The shaded box next to the sequence indicates the upstream constant region that flanks the pilE hypervariable region. This constant region corresponds to the location of oligonucleotide 248 in panel A. PE, primer extension.
The WT small RNA transcriptome from strain MS11 was further interrogated to extract other pil-specific RNAs (Table 1). Analysis of 13 pilS gene copies revealed multiple pil-specific sRNAs within the cell. Transcript amounts varied depending upon the pilS gene copy, with sense sRNA apparently being more abundant overall than anti-sense sRNAs. Interestingly, sense and anti-sense sRNA was also observed at the pilE locus. Promoter searches were performed for the various pilS sRNAs and focused on regions spanning upwards of 100 base pairs upstream of pil-derived sRNAs, as previous small transcriptome promoter analysis had revealed a high incidence of non-canonical promoter usage (Wachter & Hill, in press). Primary motif analysis revealed that the experimentally verified pilS6 promoter sequence was present within one of the most abundant motifs (n = 19) within the dataset. Further analysis utilizing the Fisher’s exact test identified short regular expression motifs within the primary motif dataset, where two prevalent motifs were identified that may act as promoter elements (Fig. S2). The first motif, 5′–GTAAAAT–3′ (n = 34, P-value = 2.3×10−10) shares sequence similarity to the identified pilS6 promoter elements, while the second motif 5′–ATATT–3′ (n = 34, P-value = 1.3×10−10) bears slight homology to the Pribnow box. However, the majority of these putative promoter regions lacked a canonical Pribnow box sequence, which may indicate that these intragenic pil-specific promoters differ from the majority of protein coding genes.
Table 1. Combined length and quantity of ambiguous N. gonorrhoeae MS11 pil-specific sRNAs.
| Transcripts sense to pil genes | Transcripts anti-sense to pil genes | |||
| pil | Combined length of all sRNAs* | TPM | Combined length of all sRNAs | TPM |
| pilS7 | 309 | 9 | 423 | 1 |
| pilS6c3 | 280 | 531 | 297 | 1 |
| pilS6c2 | 128 | 626 | 93 | 2 |
| pilS6c1 | 411 | 3 | 393 | 3 |
| pilS5 | 368 | 666 | 496 | 1 |
| pilS2c2 | 61 | 51 | 369 | 66 |
| pilS2c1 | 419 | 1 | 339 | 1 |
| pilS1c6 | 173 | 113 | 33 | 1 |
| pilS1c5 | 329 | 2 | 268 | 28 |
| pilS1c4 | 329 | 7 | 312 | 24 |
| pilS1c3 | 340 | 114 | 307 | 12 |
| pilS1c2 | 388 | 42 | 280 | 22 |
| pilS1c1 | 479 | 23 | 421 | 1 |
| pilE | 573 | 50 | 587 | 97 |
Combined length includes unambiguous reads coupled to ambiguous reads, which primarily consist of the constant gene segments located within pil genes.
TPM, Number of transcripts per million mapped reads.
Discussion
Despite the fact that the pilE gene possesses three functional promoters, albeit only one of which is utilized in the gonococcus, plus the presence of an IHF binding site, which, when absent causes an approximate tenfold reduction in pilE transcript levels, very little regulation apparently has been defined. An alternative regulatory scheme could be at the level of transcript stability, which was initially explored with the growth phase experiment presented in Fig. 1. Clearly, the pilE transcript turned-over with transcription being reinstated when cells were subcultured into fresh medium. What was unexpected was that multiple pil-specific RNAs became evident when the 3′ end of the pilE transcript was examined, with turnover of the full-length message apparently not being involved in their formation. Currently, why the pilE message disappears so abruptly is unknown, but our initial studies indicate that it may involve sense and anti-sense RNAs pairing, which would yield substrates for RNA degradation (data not shown). Nonetheless, our subsequent experiments indicated that these novel small RNAs arose due to active transcription from the pilS loci, yielding novel small RNA species that consist of both sense and anti-sense pil-specific RNAs. Consequently, the paradigm that was established for the pilin antigenic variation scheme, where pilE was believed to be the only transcribed pil gene within the chromosome, now requires re-evaluation (Meyer et al., 1984; Haas & Meyer, 1986).
From the small RNA transcriptome analysis, multiple pil gene copies were found to produce sense and anti-sense sRNAs (Table 1). Initial attempts at analysing the transcripts in total gonococcal RNA extracts using RNA ligation, as well as 5′ and 3′ RACE analysis proved to be problematic due to the number of pilS-specific transcripts that are found in the cell and the relative high degree of similarity between the pilS gene copies. Consequently, a recombinant approach initially allowed specific promoters to be molecularly characterized within an individual pilS locus. The identification of the pilS6 : 4 sense promoter proved to be illuminating as the −10 region (–TAAAAT–) is present in all pilS gene copies as well as in all pilE genes as this stretch of nucleotides is located within a constant region within the variable gene segments (it is the segment of DNA that specifies the 5′ cysteine residue in the PilE polypeptide). Similarly, the identification of the downstream promoters in pilS6 : 6 are also conserved between the pilE locus and all the pilS loci, as a Sma/Cla repeat element is found downstream of each pil locus. Consequently, there appears to be conservation of these three characterized promoter elements. Whether these potential promoters are actually utilized in a constitutive manner in the gonococcus is currently unknown, especially as promoter silencing appeared to occur when pilS6 : 4 and pilS6 : 6 transcription was compared; transcription from the downstream anti-sense promoters in pilS6 : 4 was not observed when the pilS6 : 4 sense promoter was active (Fig. 5a). However, subsequent RNA Seq analysis confirmed the presence of these pilS6-derived sRNAs in WT gonococci (Fig. 6) and further identified multiple other pil-specific sRNAs within the gonococcus (Table 1).
In a previous small RNA transcriptome study, promoter usage generating sRNAs did not appear to conform to conventional established rules (Wachter & Hill, 2015). Promoter elements often did not reside immediately adjacent to the identified sRNA transcripts. Furthermore, exact spacing between −35 and −10 elements, if a −35 element was present, was rarely observed as is the case with the experimentally identified promoter in pilS6 : 4. Consequently, in light of these previous observations, promoter analysis of the gonococcal small RNA transcriptome focused on regions spanning 100 base pairs upstream of pil-derived sRNAs. Primary motif analysis revealed that the experimentally verified pilS6 : 4 promoter sequence was present within one of the most abundant motifs (n = 19) within the dataset, as would be expected as this promoter motif is located within one of the conserved segments of pil genes. Further analysis to identify short regular expression motifs within the primary motif dataset utilizing the Fisher’s exact test revealed two significantly prevalent motifs that may act as promoter elements. The first motif, 5′–GTAAAAT–3′ (n = 34, P-value = 2.3×10−10) shares sequence similarity to the identified pilS6 promoter elements, while the second motif 5′–ATATT–3′ (n = 34, P-value = 1.3×10−10) bears slight homology to the Pribnow box. However, as previously observed, the majority of these putative promoter regions lacked a conserved Pribnow box sequence.
Unambiguous pilS-specific transcripts could also be identified in the whole transcriptomes of gonococcal strains FA1090 and NCCP 11945, as well as in the class II meningococcal strain FAM18 (data not shown). The demand for absolute identity between the sRNAs and the corresponding pilS gene copy reduced the number of identified transcripts, as transcripts were found to map to additional pilS gene copies when the stringency of RNA Seq analysis was relaxed (data not shown). Besides pilS-specific RNAs, small sense and anti-sense RNAs were also observed at the pilE locus in each of the Neisseria strains. Currently, the role, if any, for pilE anti-sense RNA is unknown. However, what became apparent was that no conformity appears to be applied regarding pilS transcription between the different Neisseria strains and species, indicating that even though –TAAAAT– is present in each pilS gene copy, it is not universally utilized as a promoter element, which may reflect differences in the upstream DNA harbouring the −35 element as this stretch of DNA is located within the variable segments of the pil genes.
The demonstration of pilS transcripts, both sense and anti-sense, is a prime example of a new phenomenon known as pervasive transcription. Pervasive transcription has emerged as a major theme in transcriptomics following the advent of deep RNA sequencing technologies (Sorek & Cossart, 2010). As many of the newly identified RNAs map outside of conventional gene boundaries, pervasive transcription was initially thought to be an artefact of the sequencing process. However, with the availability of more transcriptomes for analysis, across multiple genera of bacteria, the identification of transcription start sites in non-conventional locations within a gene has become a common feature (Wade & Grainger, 2014). Currently, no consensus has been reached on the role of pervasive transcription (Sorek & Cossart, 2010; Wade & Grainger, 2014). However, given that its occurrence has not been selected against in prokaryotes, and that many of the small RNAs arise from intragenic promoters yielding anti-sense RNAs that theoretically can pair with the sense transcript, it is suggested that these novel small RNA molecules may play a role in RNA regulation (Lasa & Villanueva, 2014; Lybecker et al., 2014). If duplex RNAs are created between the sense transcript and these small anti-sense RNAs, these molecules would then provide substrates for RNase III degradation. With respect to the pil system, regulation of pilE expression does not appear to involve a classic regulator protein. Therefore, as the presence of multiple anti-sense RNA species within the cell could bind to the pilE sense transcript in vivo, these transcripts may act in a regulatory manner to maintain optimal levels of pilE sense RNA. Furthermore, duplex RNA formation may also regulate expression of PilE polypeptide by impeding efficient translation of the sense transcript through occlusion of the ribosome-binding site on the message. Consequently, there exists the potential for multiple, overlapping layers of pilE regulation due to pervasive transcription occurring within the pilS loci.
It has also been suggested that pervasive transcription may allow genomes a certain degree of plasticity (Wade & Grainger, 2014). As shown in Fig. 5(a), the loss of the sense promoter in plasmid pilS6 : 6 allowed a hitherto silent anti-sense promoter to be utilized. Such on/off promoter switching could provide a multi-layered effect with respect to gene expression. Consequently, pervasive transcription may allow for new gene functions to emerge that may prove beneficial to the cell through the use of unorthodox promoters located within a gene. Alternatively, because transcription-coupled DNA repair occurs in most species, then promiscuous transcription may also be a mechanism that reduces the mutation load within a cell, as the repair machinery is actively being recruited to otherwise transcriptionally silent regions of the chromosome (Wade & Grainger, 2014). A recent study using high-throughput DNA sequencing has provided the most detailed analysis of pilE gene variants that arise through pilE/pilS recombination (Davies et al., 2014). This led us to explore the possibility that pilS transcription may facilitate a pilE/pilS recombination event by providing denatured pilS duplexes with which pilE could engage in the recombination process. However, transcriptome analysis found no correlation between the extent of pilS transcription and the frequency with which pilE recombined with any particular pilS locus (data not shown).
Acknowledgements
The authors would like to thank John Davies for sharing unpublished data and allowing access to the FA1090 and FAM18 whole transcriptome data, as well as Scott Grayburn for technical help. The work was supported by NIH grant 1R15 AI072720-01A1 to S. A. H. and Northern Illinois University's graduate student grant program.
Footnotes
Edited by: L. Ruiting
References
- Bailey, T. L. & Elkan, C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. In Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology, pp. 28–36, AAAI Press, Menlo Park, California. [PubMed] [Google Scholar]
- Bailey T. L., Boden M., Buske F. A., Frith M., Grant C. E., Clementi L., Ren J., Li W. W., Noble W. S. ( 2009. ). meme SUITE: tools for motif discovery and searching. Nucleic Acids Res 37 (Web Server issue), W202–W208. 10.1093/nar/gkp335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergström S., Robbins K., Koomey J. M., Swanson J. ( 1986. ). Piliation control mechanisms in Neisseria gonorrhoeae. Proc Natl Acad Sci U S A 83, 3890–3894. 10.1073/pnas.83.11.3890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrick C. S., Fyfe J. A. M., Davies J. K. ( 1997. ). The normally silent σ54 promoters upstream of the pilE genes of both Neisseria gonorrhoeae and Neisseria meningitidis are functional when transferred to Pseudomonas aeruginosa. Gene 198, 89–97. 10.1016/S0378-1119(97)00297-7 [DOI] [PubMed] [Google Scholar]
- Carver T., Harris S. R., Berriman M., Parkhill J., McQuillan J. A. ( 2012. ). Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics 28, 464–469. 10.1093/bioinformatics/btr703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. ( 2004. ). WebLogo: a sequence logo generator. Genome Res 14, 1188–1190. 10.1101/gr.849004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J. K., Harrison P. F., Lin Y.-H., Bartley S., Khoo C. A., Seemann T., Ryan C. S., Kahler C. M., Hill S. A. ( 2014. ). The use of high-throughput DNA sequencing in the investigation of antigenic variation: application to Neisseria species. PLoS ONE 9, e86704. 10.1371/journal.pone.0086704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- French R., Ahlquist P. ( 1987. ). Intercistronic as well as terminal sequences are required for efficient amplification of brome mosaic virus RNA3. J Virol 61, 1457–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyfe J. A. M., Davies J. K. ( 1998. ). An AT-rich tract containing an integration host factor-binding domain and two UP-like elements enhances transcription from the pilEp1 promoter of Neisseria gonorrhoeae. J Bacteriol 180, 2152–2159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fyfe J. A. M., Carrick C. S., Davies J. K. ( 1995. ). The pilE gene of Neisseria gonorrhoeae MS11 is transcribed from a σ 70 promoter during growth in vitro. J Bacteriol 177, 3781–3787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas R., Meyer T. F. ( 1986. ). The repertoire of silent pilus genes in Neisseria gonorrhoeae: evidence for gene conversion. Cell 44, 107–115. 10.1016/0092-8674(86)90489-7 [DOI] [PubMed] [Google Scholar]
- Haas R., Veit S., Meyer T. F. ( 1992. ). Silent pilin genes of Neisseria gonorrhoeae MS11 and the occurrence of related hypervariant sequences among other gonococcal isolates. Mol Microbiol 6, 197–208. 10.1111/j.1365-2958.1992.tb02001.x [DOI] [PubMed] [Google Scholar]
- Hagblom P., Segal E., Billyard E., So M. ( 1985. ). Intragenic recombination leads to pilus antigenic variation in Neisseria gonorrhoeae. Nature 315, 156–158. 10.1038/315156a0 [DOI] [PubMed] [Google Scholar]
- Hill S. A., Davies J. K. ( 2009. ). Pilin gene variation in Neisseria gonorrhoeae: reassessing the old paradigms. FEMS Microbiol Rev 33, 521–530. 10.1111/j.1574-6976.2009.00171.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill S. A., Samuels D. S., Carlson J. H., Wilson J., Hogan D., Lubke L., Belland R. J. ( 1997. ). Integration host factor is a transcriptional cofactor of pilE in Neisseria gonorrhoeae. Mol Microbiol 23, 649–656. 10.1046/j.1365-2958.1997.2321612.x [DOI] [PubMed] [Google Scholar]
- Katoh K., Misawa K., Kuma K., Miyata T. ( 2002. ). mafft: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res 30, 3059–3066. 10.1093/nar/gkf436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M., Schein J. E., Birol I., Connors J., Gascoyne R., Horsman D., Jones S. J., Marra M. A. ( 2009. ). Circos: an information aesthetic for comparative genomics. Genome Res 19, 1639–1645. 10.1101/gr.092759.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasa I., Villanueva M. ( 2014. ). Overlapping transcription and bacterial RNA removal. Proc Natl Acad Sci U S A 111, 2868–2869. 10.1073/pnas.1324236111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskos L., Dillard J. P., Seifert H. S., Fyfe J. A. M., Davies J. K. ( 1998. ). The pathogenic Neisseriae contain an inactive rpoN gene and do not utilize the pilE σ54 promoter. Gene 208, 95–102. 10.1016/S0378-1119(97)00664-1 [DOI] [PubMed] [Google Scholar]
- Lybecker M., Zimmermann B., Bilusic I., Tukhtubaeva N., Schroeder R. ( 2014. ). The double-stranded transcriptome of Escherichia coli. Proc Natl Acad Sci U S A 111, 3134–3139. 10.1073/pnas.1315974111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer T. F., Hill S. A. ( 2003. ). Genetic variation in the Pathogenic Neisseria species. In Antigenic Variation, pp. 142–164. Edited by Craig A., Scherf A. San Diego: Academic Press; 10.1016/B978-012194851-1/50033-0 [DOI] [Google Scholar]
- Meyer T. F., Billyard E., Haas R., Storzbach S., So M. ( 1984. ). Pilus genes of Neisseria gonorrheae: chromosomal organization and DNA sequence. Proc Natl Acad Sci U S A 81, 6110–6114. 10.1073/pnas.81.19.6110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmele C. W., Xian Y., Albrecht M., Faulstich M., Fraunholz M., Heinrichs E., Dittrich M. T., Müller T., Reinhardt R., Rudel T. ( 2014. ). Transcriptional landscape and essential genes of Neisseria gonorrhoeae. Nucleic Acids Res 42, 10579–10595. 10.1093/nar/gku762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumble S. M., Lacroute P., Dalca A. V., Fiume M., Sidow A., Brudno M. ( 2009. ). SHRiMP: accurate mapping of short color-space reads. PLOS Comput Biol 5, e1000386. 10.1371/journal.pcbi.1000386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovyev V., Salamov A. ( 2011. ). Automatic annotation of microbial genomes and metagenomic sequences. In Metagenomics and its Applications in Agriculture, Biomedicine and Environmental Studies, pp. 61–78. Edited by Li R. W. Hauppauge, NY: Nova Science Publishers. [Google Scholar]
- Sorek R., Cossart P. ( 2010. ). Prokaryotic transcriptomics: a new view on regulation, physiology and pathogenicity. Nat Rev Genet 11, 9–16. 10.1038/nrg2695 [DOI] [PubMed] [Google Scholar]
- Swanson J. ( 1982. ). Colony opacity and protein II compositions of gonococci. Infect Immun 37, 359–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wachter, J. & Hill, S. A. (2015). Small transcriptome analysis indicates that the enzyme RppH influences both the quality and quantity of sRNAs in Neisseria gonorrhoeae FEMS Microbiol Letts 362, 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade J. T., Grainger D. C. ( 2014. ). Pervasive transcription: illuminating the dark matter of bacterial transcriptomes. Nat Rev Microbiol 12, 647–653. 10.1038/nrmicro3316 [DOI] [PubMed] [Google Scholar]
- Wagner, G. P., Kin, K. & Lynch, V. J. (2012). Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theor Biosci 131, 281–285. [DOI] [PubMed] [Google Scholar]
- Wainwright L. A., Pritchard K. H., Seifert H. S. ( 1994. ). A conserved DNA sequence is required for efficient gonococcal pilin antigenic variation. Mol Microbiol 13, 75–87. 10.1111/j.1365-2958.1994.tb00403.x [DOI] [PubMed] [Google Scholar]