Abstract
Behavior genetics is the study of the relationship between genetic variation and psychological traits. Turkheimer (2000) proposed “Three Laws of Behavior Genetics” based on empirical regularities observed in studies of twins and other kinships. On the basis of molecular studies that have measured DNA variation directly, we propose a Fourth Law of Behavior Genetics: “A typical human behavioral trait is associated with very many genetic variants, each of which accounts for a very small percentage of the behavioral variability.” This law explains several consistent patterns in the results of gene discovery studies, including the failure of candidate gene studies to robustly replicate, the need for genome-wide association studies (and why such studies have a much stronger replication record), and the crucial importance of extremely large samples in these endeavors. We review the evidence in favor of the Fourth Law and discuss its implications for the design and interpretation of gene-behavior research.
Keywords: behavior genetics, genome-wide association studies, polygenic architecture, individual differences, molecular genetics
Behavior genetics is the study of the manner in which genetic variation affects psychological phenotypes (traits), including cognitive abilities, personality, mental illness, and social attitudes. In a seminal article published in this journal, Turkheimer (2000) noted three robust empirical regularities that had by then emerged from the literature on behavior genetics. He dubbed these regularities the “Three Laws of Behavior Genetics.” They are:
-
1
All human behavioral traits are heritable. [That is, they are affected to some degree by genetic variation.]
-
2
The effect of being raised in the same family is smaller than the effect of genes.
-
3
A substantial portion of the variation in complex human behavioral traits is not accounted for by the effects of genes or families.
These observations surprised many outsiders to the field of behavior genetics at the time, yet they remain an accurate broad-brush summary of the empirical evidence fourteen years later. Indeed, they have attained the status of “null hypotheses”—the most reasonable a priori expectations to hold in the absence of contrary evidence (Turkheimer, Pettersson, & Horn, 2014).
The original Three Laws summarized results from biometrical studies of twins, adoptees, and other kinships. These research designs have many valuable uses, but they cannot discover particular genomic regions or specific variants that are causally responsible for downstream phenotypic variation. Since the completion of the Human Genome Project, numerous studies of behavioral traits have directly measured DNA variation among individuals in an attempt to take this logical next step. While there are many types of genetic variants, most studies have assayed single-nucleotide polymorphisms (SNPs), which are sites in the genome where single DNA base pairs carried by distinct individuals may differ.2 Virtually all SNPs have two different possible base pairs, called alleles. The less frequent allele in the population is called the minor allele of the SNP. If the frequency of a SNP’s minor allele in the population exceeds 1%, the SNP is called a common variant. Among individuals of European descent, there are approximately 8 million common variants in the human genome.
The results of studies searching for SNP-behavior associations have disappointed any hope that a small number of these common variants are responsible for a large percentage of cross-sectional trait variability. Instead, the evidence to date is consistent with what we propose as the Fourth Law of Behavior Genetics:
-
4
A typical human behavioral trait is associated with very many genetic variants, each of which accounts for a very small percentage of the behavioral variability.3
For purposes of the law, a “typical human behavioral trait” is one that is (a) customarily measured by psychometric methods, (b) a serious psychiatric disease, or (c) a social outcome, such as educational attainment, that is plausibly related to a person’s behavioral dispositions. As is customary in behavioral science, we use the word law to describe what we consider to be a very robust empirical regularity (not a universal, mechanistic truth). In the remainder of this article we will summarize the mounting empirical evidence in favor of the Fourth Law, consider what gene-behavior research strategies are likely to be profitable in light of the Fourth Law, and briefly consider possible explanations for the Fourth Law.
Empirical Evidence for the Fourth Law
Each person inherits two copies of any DNA segment, one from each parent, and therefore may carry 0, 1, or 2 copies of the minor allele at a particular SNP. The number of minor alleles can be taken to characterize the individual’s genotype at that SNP. The straight line that best fits the overall genotype-phenotype relationship is called the average effect of gene substitution (Fisher, 1941; Lee & Chow, 2013). The true genotype-phenotype relationship will certainly not be precisely linear, but the slope of the best-fitting straight line is equal to a weighted average of the phenotypic changes following from the possible gene substitutions. In principle, it is also possible to estimate the nonlinear effects of genotype, as well as gene-gene and gene-environment interactions. In practice, however, given the staggering combinatorial explosion of possible hypotheses, it will normally be a useful first step to estimate the average effects in order to identify a subset of SNPs that should be studied in greater detail (e.g., Rietveld, Esko, et al., 2014).
For example, a SNP designated “rs9320913” is located on chromosome 6 and has two alleles: C (cytosine) and A (adenine). It was identified in a recent genome-wide association study (GWAS) of educational attainment (Rietveld et al., 2013). A GWAS is a hypothesis-free analysis of the predictive power of each of approximately one million (or more) individual SNPs spread across the genome. The goal of a GWAS is to discover which SNPs are associated with a trait of interest (e.g., educational attainment, intelligence, extraversion, or schizophrenia). Because so many statistical tests are performed in a GWAS, the significance threshold is typically set at a stringent 5×10−8 (“p < .00000005”) rather than the 5×10−2 (“p < .05”) that is standard for behavioral studies assumed to be testing a single hypothesis; this practice is analogous to a Bonferroni correction. The association between rs9320913 and education reached the GWAS significance threshold and was also replicated at a conventional level in two follow-up studies of separate samples (Rietveld et al., 2013; Rietveld, Conley, et al., 2014).
Strikingly, each additional copy of A (the education-increasing allele) is associated with only one additional month of schooling. Note that a combined sample size of 126,599 participants from over 50 cohorts in 15 countries was used to discover and initially replicate this gene-education association; an additional sample of 34,428 participants was used for a second replication. The SNP rs9320913 is estimated to account for only 0.02% of the overall variability in educational attainment, but biometrical studies show that the total percentage of variability owed to genetic differences is three orders of magnitude larger (Heath et al., 1985; Rietveld et al., 2013). Since the SNPs with the largest effects are the easiest to find, these results suggest that educational attainment is a phenotype affected by thousands of undiscovered genetic variants, each responsible for a minuscule fraction of individual differences.
The story is similar for the better-studied phenotype of schizophrenia. Studies of DNA from over 36,000 diagnosed cases and 113,000 controls have so far identified 108 SNPs associated with schizophrenia at a strict evidentiary threshold, yet these 108 SNPs jointly account for only 3.4% of the variance of the trait (measured on a liability scale; Ripke et al., 2014). Each of these 108 “hits” has a small effect size: typically, less than a 1.1-fold increase in the odds of a schizophrenia diagnosis with each additional risk-conferring allele.
As GWAS sample sizes get larger, will more variants (presumably with even smaller effect sizes) be identified? The answer is surely yes, since the fraction of trait variance captured by currently known SNPs falls well below the traits’ heritabilities. This quantity—the total fraction of variance in a trait attributable to genetic factors—is traditionally estimated using behavioral genetics methods. In recent years, however, a novel technique has been developed that uses GWAS data from nominally unrelated individuals to estimate an even more relevant benchmark: the total fraction of variance in a trait that is attributable to the average effects of SNPs (Yang et al., 2010; Lee, Wray, Goddard, & Visscher, 2011; Speed, Hemani, Johnson, & Balding, 2012; Lee & Chow, 2014). This technique is called genomic-relatedness-matrix restricted maximum likelihood (GREML).4 In essence, GREML determines whether the randomly-arising genetic similarity between pairs of unrelated individuals predicts the phenotypic similarity between those individuals; a stronger relationship between genetic and phenotypic similarity implies a greater influence of the measured SNPs on the trait of interest. Importantly, the magnitude of this influence can be accurately estimated even if the sample size is too small to identify all (or any) of the associated variants at a high level of confidence. GREML is perhaps the most important innovation in quantitative genetics to have been introduced in the last dozen years, and it has provided convincing evidence that the heritabilities of many psychological traits are more or less accurately estimated in traditional family studies (e.g., Lee et al., 2011; Davies et al., 2011; Chabris et al., 2012; Rietveld et al., 2013; Cross-Disorder Group of the Psychiatric Genomics Consortium, 2013).
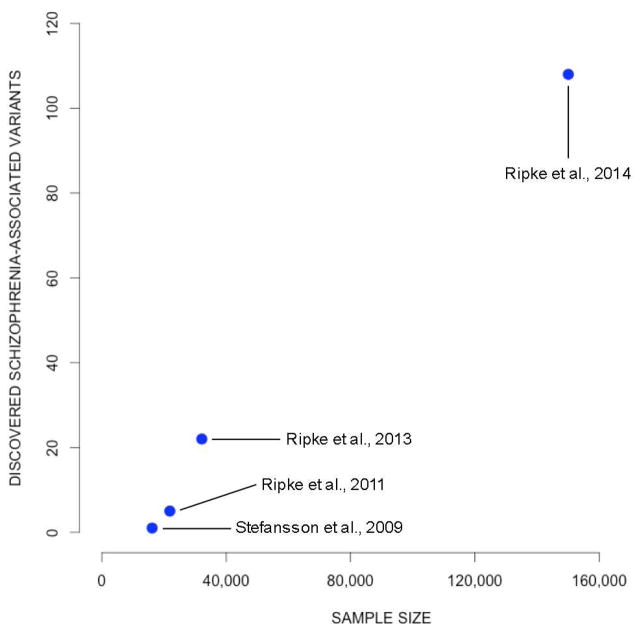
It is also noteworthy that even if few individual SNPs contributing to the GREML-estimated heritability meet the evidentiary threshold of a hit, it is possible to use summary statistics from GWAS results to estimate coarse-grained quantities such as the total number of SNPs that will ultimately be found to be associated with the trait (Stahl et al., 2012). Applying this method to a subset of the schizophrenia data produced an estimate of 8,300 trait-associated SNPs (Ripke et al., 2013). Figure 1 demonstrates that the number of schizophrenia-associated SNPs discovered has increased substantially as GWAS sample sizes have increased. The results of Ripke et al. (2013) and the GREML studies cited above tell us is that there is every reason to expect the trend in Figure 1 to continue.
Figure 1.
Number of schizophrenia-associated SNPs clearing the strict GWAS significance threshold (p < 5×10−8) as a function of discovery-stage sample size, which has increased over time. Although the four studies presented are not methodologically identical (because of different ratios of cases to controls), the increasing number of “hits” with sample size is nevertheless informative.
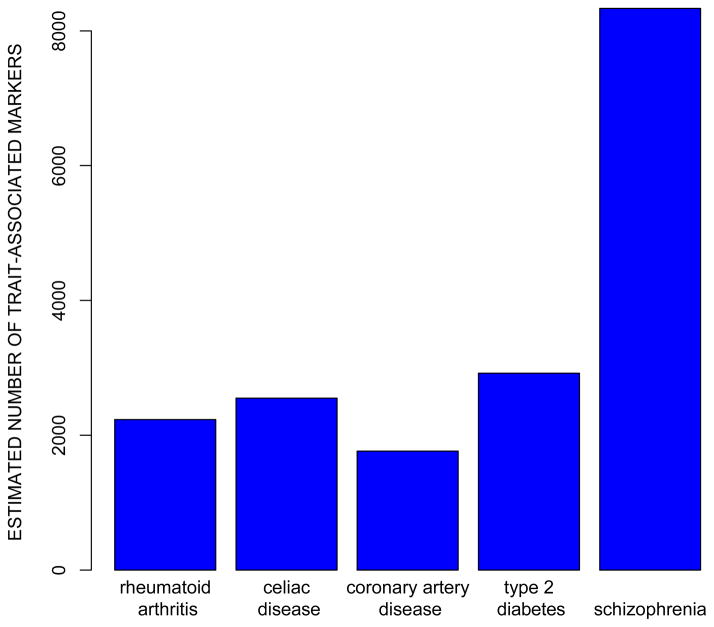
The genetic architectures of many human phenotypes tend to be highly polygenic (influenced by many different variants; Gottesman & Shields, 1967; Visscher, Brown, McCarthy, & Yang, 2012), and what the Fourth Law states is that this is also true for behavioral phenotypes. Since the number of common variants is finite (albeit large), the Fourth Law suggests in turn that the genetic architectures of human phenotypes are also pleiotropic (each variant affects many different traits). Moreover, evidence to date, implies that behavioral phenotypes are even more polygenic than typical physical and medical traits. For example, the SNPs that have the largest effects on education (Rietveld et al., 2013) account for roughly one tenth as much variability (0.02%) as those with the largest effects on physical traits such as height and body mass index (roughly 0.3%). Moreover, as shown in Figure 2, a comparison of schizophrenia with a number of physical diseases suggests that its genetic variability is distributed among a greater number of variants with smaller effects (Ripke et al., 2013).
Figure 2.
Estimated total number of SNPs associated with five disease phenotypes based on results of genome-wide association studies (based on a figure in Ripke et al., 2013).
Besides the small effect sizes of positive results in well-powered (large-sample) studies, null results (at the 5×10−8 level) in less powerful studies provide converging evidence that variants with large effects are unlikely to exist. For example, null results have been obtained in GWAS of personality (de Moor et al., 2012), suggesting that these traits are not exceptions to the pattern described here.
It is possible that rare SNPs (with <1% frequencies of the minor allele) make an important contribution to the heritabilities of behavioral traits; such SNPs are not well-represented in the GWAS we have cited. However, recent studies comprehensively assaying both common and rare variants in certain regions of the genome have failed to find any variants accounting for large portions of phenotypic variability (e.g., Purcell et al., 2014). Thus it seems that the inclusion of more variants in whole-genome sequencing studies will not alter the conclusion that individual genetic polymorphisms with effects on cognition, personality, education, or psychiatric disease accounting for even 1% of the variability are unlikely to exist. At this point, claims to the contrary should be considered extraordinary, and require corresponding amounts of evidence.
The Fourth Law also explains why the results of “candidate gene” studies, which focus on a handful of genetic variants, usually fail to replicate in independent samples. The main problem is that such studies tend to have insufficient statistical power. If well-powered studies that search the entire genome for associations find only tiny effects, then large effects found in studies with sample sizes in the dozens to hundreds (e.g., Kogan et al., 2011; Skafidas et al., 2012) are likely to be false positives. This was shown empirically for the trait of general intelligence (g) by Chabris et al. (2012), who, using a sample of about 10,000 participants, failed to replicate published associations between g and 12 genetic variants. Accordingly, results of small-sample genetic studies should be regarded with great caution, especially in studies claiming to have identified interactions (Duncan & Keller, 2012).
However, candidate gene studies can succeed when the sample is large and the candidate variants to be investigated have high prior probabilities of being associated with the trait—for example, when they consist of hits from a previous GWAS of a “proxy phenotype” that is itself strongly associated with the trait of interest. For example, Rietveld, Esko, et al. (2014) began with 69 SNPs that were associated with educational attainment (in a subset of the data from Rietveld et al., 2013) and tested them for association with g (which is correlated with educational attainment) in a separate sample of 24,189 individuals. Three of those SNPs were significant hits after adjustments for multiple hypothesis testing, and represent the first robust discovery of common genetic variants associated with normal-range, non-age-related variation in general cognitive ability.
There are caveats to the use of the causality-implying term “effect” when discussing GWAS. One is that a SNP-phenotype association may reflect the causal effect of a correlated but unmeasured variant in the same genomic region. For example, if rs9320913 turns out to be a mere proxy for the causal variant, then the average effect of the true causal SNP on education may be somewhat larger than estimated by Rietveld et al. (2013). It is natural to wonder whether the empirical findings summarized by the Fourth Law might be artifacts attributable to the attenuation of larger effect sizes at unmeasured causal variants, but careful investigation has shown that a multitude of causal variants with small effects must still be invoked to explain GWAS results (Wray, Purcell, & Visscher, 2011).
The Relevance of GWAS in Light of the Fourth Law
It has been argued that the empirical regularity summarized by the Fourth Law strengthens the case for deemphasizing gene-mapping studies (Turkheimer, 2012). We believe that the appropriate response to the Fourth Law is instead to pursue research strategies suited to the reality that most genetic effects on behavioral traits are very small.
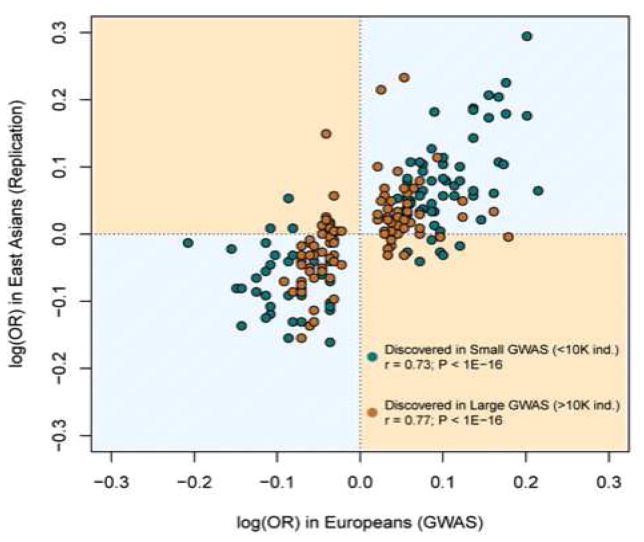
Critics of GWAS findings have argued that they have a poor record of replication (McClellan & King, 2010; Turkheimer, 2012). It is true that the small SNP-trait association signals described by the Fourth Law are impossible to distinguish from noise in poorly-powered studies. In our view, however, the Fourth Law makes clear that one solution is larger samples—much larger than most psychologists contemplate in the normal course of their research. Indeed, when GWAS are conducted with adequate sample sizes and strict evidentiary standards, the degree of quantitative replication is extremely strong (e.g., Rietveld, Conley, et al., 2014).5 In a GWAS of height, the correlation among strictly significant hits between initial-sample and replication-sample estimates of SNP effects was nearly .97 (Lango Allen et al., 2010)—almost perfect agreement. Such precision holds even when comparing groups of different geographic origin (see Figure 3). The correlation between effects measured in European and East Asian samples is only about .75 because the disease studies represented in Figure 3 employed smaller sample sizes than the height study. Nevertheless it is evident that the best-fitting straight line in Figure 3 would approximate an intercept of zero and a slope of one, as would be expected if European effect sizes were estimated with no error and were equal to East Asian effect sizes (up to sampling noise). This concordance is particularly remarkable because East Asians differ from Europeans in genetic background and environmental exposures. Evidence from studies of schizophrenia (de Candia et al., 2013; Ripke et al., 2014) suggests that this pattern will generalize to behavioral traits.
Figure 3.
The strong agreement between genetic effects on 28 diseases estimated in Europeans and East Asians (reproduced from Marigorta & Navarro, 2013). The x-axis corresponds to the increment in the odds (on a logarithmic scale) of suffering from the disease for each additional copy of the reference allele, as estimated in Europeans. The y-axis corresponds to the same quantity estimated in East Asians. The increment in log(odds) is the equivalent of the “additive effect” for dichotomously scored traits such as disease status (affected versus unaffected). The gap in the data on the x-axis results from the fact that non-significant genetic effects would have odds ratios near 1 (and thus logarithms of 0) and therefore would not be included in the results of European samples.
Skeptics also caution that the problem of distinguishing causation from correlation in genetic association studies may be intractable (Charney, 2012; Turkheimer, 2012). Since observational studies can usually only reveal correlation, what is the justification for inferring that a SNP-phenotype association reflects an effect on the trait? Here we consider the issue of whether we can be confident that an association signal from some genomic region reflects the causal effect of a gene in that region rather than confounding with trait-affecting environmental conditions (Lee, 2012).
One major potential confound of this type is population stratification: If a SNP is more common in individuals of a certain ancestry or region, then it may falsely appear that the SNP is associated with a trait when the trait is actually associated with a pattern of ancestry or region of origin. Modern GWAS of unrelated individuals seek to account for this possibility by controlling for several principal components of the massive correlation matrix of all the assayed SNPs. These principal components typically identify geographic ancestry of individuals in the sample (Price et al., 2006). There is a more elegant study design, however, that can provide powerful evidence in followup studies against confounding as an explanation for genetic associations.
When an organism becomes a parent, it passes on a randomly chosen allele from each of its pairs to a given offspring. Because the offspring’s genotype is randomly assigned conditional on the parental genotypes, a significant association between the presence of a particular allele and the focal phenotype within families is strong evidence for the presence of a nearby causal variant (Fisher, 1952; Ewens, Li, & Spielman, 2008; Lee & Chow, 2013). And indeed, family-based genetic studies have so far affirmed the results of GWAS that use unrelated individuals. For example, alleles identified as increasing liability to schizophrenia in standard GWAS were more likely, in a separate sample of families, to be transmitted from parents to affected children (Ripke et al., 2014; see also Benyamin, Visscher, & McRae, 2009; Lango Allen et al., 2010; Turchin et al., 2012; Wood et al., 2014). There is also evidence that the top SNPs identified by Rietveld et al. (2013) are jointly predictive of sibling differences in educational attainment (Rietveld, Conley, et al., 2014).
It has been argued that the effect sizes discovered by GWAS are so small that they are scientifically inert (e.g., Turkheimer, 2012). However, a small effect size from variation across individuals in a gene does not rule out a major qualitative role of the gene product itself in the relevant biological pathway. The principle is that making a small change to one step in a process may cause only a small change in the final output of the process, but making a large change, or omitting the step entirely, can halt the entire process. For example, by using molecular biological techniques to silence zebrafish genes whose orthologs were discovered in a GWAS to contain variants with small effects on human platelet count, researchers were able to abolish platelet production in this model organism (Gieger et al., 2011). Whether such cases will be numerous or rare for behavioral traits is a question that future GWAS-based research may soon answer.
Moreover, even though individual genetic variants have small effects, GWAS results can be used to construct a polygenic score, a variable that exploits the joint predictive power of many variants and therefore can explain substantial variance in the trait or outcome. The simplest polygenic score for a trait is constructed by adding up the individual effects measured for all of the SNPs in a GWAS (Dudbridge, 2013). Rietveld et al. (2013) found that such a polygenic score for educational attainment explains 2–3% of the variation across people—one hundred times more than was explained by the most predictive individual SNP. The power of polygenic scores increases with the size of the GWAS samples they are derived from; e.g., 15% of the variance in educational attainment could be explained if the scores came from a sample of 1 million individuals (Rietveld et al., 2013). Polygenic scores could have value for predicting disease or disability risk (Ripke et al., 2014), for identifying individuals who might benefit from early treatment or intervention, for studying gene-environment interactions, and for modeling genetic differences among individuals in epidemiological and experimental studies of behavioral and biological treatments (Rietveld et al., 2013; Rietveld, Conley, et al., 2014).
When the early height GWAS were published, the confirmed SNP associations had a combined explanatory power of only 2–3%. This low predictive power was thought by some to illustrate fundamental limitations of GWAS methodology, such as its inability to estimate interactions and nonlinear effects that were hypothesized to account for the apparently “missing heritability.” This may turn out to be true for some phenotypes, but GREML studies have now confirmed that that for many phenotypes, including height, intelligence and schizophrenia, the combined additive effects of common SNPs do account for a large fraction of the variance. The most recent height GWAS (Wood et al., 2014) brings this fraction to about 17% (if the criterion is out-of-sample predictive accuracy) and 29% (if the criterion is GREML-estimated heritability attributable to 10,000 SNPs with the strongest evidence for association). Thus, much of the heritability of height was not missing, but merely hiding in the form of small but additive effect sizes. This can be seen as yet another illustration of the Fourth Law in action.
The Importance of the Fourth Law
We have recently highlighted two possible explanations for the Fourth Law (Chabris et al., 2013). First, causal chains from DNA variation to behavioral phenotypes are likely very long (longer than with physical traits, such as eye color), so the effect of any one variant on any one such trait is likely to be small. For example, a SNP may have a substantial effect on the concentration of an enzyme in cortical synapses, but that proximal biochemical phenotype is only one of many factors that explain why some people score higher than others on paper-and-pencil IQ tests, so the SNP has only a tiny effect on the distal phenotype of cognitive function.
Second, when a population is already well-adapted to its environment, mutations with large effects on a focal trait are likely to have deleterious side effects (Fisher, 1930). If the effect of a genetic variant is small enough, however, then its population frequency has some chance of drifting upward to a detectable level (Kimura, 1983).6 These forces could conspire to keep variants of large effect on a trait at negligible frequency, while allowing some variants of small effect to become relatively common. Some support for this hypothesis comes from two observations: (1) SNPs discovered in GWAS that have larger additive effects tend to have lower frequencies of the minor allele (Park et al., 2011), and (2) variants with very large phenotypic effects, such as those causing mental retardation, are always very rare and thus contribute little to overall population variability, or they have their effects later in life (e.g, the well-documented relationship between variants of the APOE gene and cognitive decline). These examples suggest that valuable evolutionary insights might flow from the detailed analysis of GWAS data (Turchin et al., 2012).
In conclusion, we shall place the Fourth Law in the context of what has long been well-understood about the relationship between genes and human behavior, namely that it is mistaken to believe that there might be a gene “for” one complex trait or another (for an eloquent statement of this basic point, see Dawkins, 1979, p. 189). What the Fourth Law adds to this understanding is that most genetic variability in behavior between individuals is attributable to genetic differences that are each responsible for very small behavioral differences.
The law we have proposed here provides a unified conceptual explanation for several consistent patterns in the results of the past two decades of gene discovery studies, including the failure of candidate gene studies to replicate, the need for genome-wide association studies (and why they actually do replicate), and the crucial importance of extremely large samples in these endeavors. We believe that compelling motives for pursuing gene-mapping studies of behavioral traits can be found in the promise of learning more about the evolutionary trajectory of the human species, the formulation of new biological hypotheses regarding cognition and neural function, and the value of polygenic scores in the social and medical sciences. The Fourth Law of Behavior Genetics provides fundamental guidance for how research in all of these areas can most efficiently progress.
Acknowledgments
This work was supported by the Pershing Square Fund for Research on the Foundations of Human Behavior, Ragnar Söderberg Foundation Grant E9/11, Swedish Research Council Grant 412-2013-1061, and National Institute on Aging Grants P01AG005842, P01AG005842-20S2, P30AG012810, R01AG021650, and T32AG000186-23.
Recommended Reading
Chabris, C. F., Hebert, B. M., Benjamin, D. J., Beauchamp, J., Cesarini, D., van der Loos, M., … Laibson, D. (2012). (See References). An empirical demonstration that genetic studies of general cognitive ability employing small sample sizes cannot be trusted to produce replicable results.
Cross-Disorder Group of the Psychiatric Genomics Consortium (2013). (See References). An elegant application of the GREML/GCTA method to understanding the genetic architecture of behavioral traits.
Lee, J. J. (2012). (See References). A target article on the notion of causality in the study of individual differences, accompanied by commentaries and author response.
Rietveld, C. A., Medland, S. E., Derringer, J., Yang, J., Esko, T., Martin, N. W., … Koellinger, P. D. (2013). (See References). A study of over 125,000 individuals reporting three single-nucleotide polymorphisms (SNPs) associated with educational attainment. The supplemental online material contains much valuable additional information.
Ripke, S., Neale, B. M., Corvin, A., Walters, J. R., Farh, K.-H., Holmans, P. A., … O’Donovan, M. C. (2014). (See References). A study reporting 108 SNPs associated with schizophrenia and implicating neuronal calcium signaling and acquired immunity as important biological processes in the etiology of this disorder.
Visscher, P. M., Brown, M. A., McCarthy, M. I., & Yang, J. (2012). (See References). An overview of what genome-wide association studies (GWAS) have discovered, and a response to criticisms of this methodology.
Footnotes
There are other kinds of genetic variants besides SNPs, including insertions, deletions, and variable-length repeats of one or more base pairs or short sequences, but SNPs are the most abundant and readily assayed kind of variant.
A recent review of genetic research on intelligence has formulated a similar generalization: “A third law has emerged from has emerged from molecular genetic research that attempts to identify specific genes responsible for widespread heritability, especially genome-wide association (GWA) studies of the past few years: The heritability of traits is caused by many genes of small effect” (Plomin & Deary, in press). Note that the parallel is between Plomin and Deary’s “third law” and what we are calling the “Fourth Law of Behavior Genetics.”
This technique is also sometimes called GCTA, for Genomewide Complex Trait Analysis (e.g., Plomin and Deary, in press).
Additionally, when the results of GWAS and candidate gene studies are compared directly, GWAS hits are usually more likely to replicate (e.g., in a study of SNPs reportedly associated with glioma, Walsh et al., 2013).
This explanation was first put forth by Lande (1983) to explain, for instance, why pesticide resistance is polygenic in some insect populations and dependent on variants of large effect in other populations that have been disturbed by humans.
Author Contributions: C. F. Chabris and J. J. Lee contributed equally to this work.
The content of this publication is solely the responsibility of the authors and does not necessarily represent the views of any of these funding organizations.
Contributor Information
Christopher F. Chabris, Union College.
James J. Lee, University of Minnesota Twin Cities
David Cesarini, New York University.
Daniel J. Benjamin, Cornell University and University of Southern California
David I. Laibson, Harvard University
References
- Benyamin B, Visscher PM, McRae AF. Family-based genome-wide association studies. Pharmacogenomics. 2009;10:181–190. doi: 10.2217/14622416.10.2.181. [DOI] [PubMed] [Google Scholar]
- Chabris CF, Hebert BM, Benjamin DJ, Beauchamp JP, Cesarini D, van der Loos MJHM, Laibson DI. Most reported genetic associations with general intelligence are probably false positives. Psychological Science. 2012;23:1314–1323. doi: 10.1177/0956797611435528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabris CF, Lee JJ, Benjamin DJ, Beauchamp JP, Glaeser EL, Borst G, Pinker S, Laibson DI. Why it is hard to find genes that are associated with social science traits: Theoretical and empirical considerations. American Journal of Public Health. 2013;103:S152–S166. doi: 10.2105/AJPH.2013.301327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charney E. Behavior genetics and postgenomics (with discussion) Behavioral and Brain Sciences. 2012;35:331–410. doi: 10.1017/S0140525X11002226. [DOI] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics Consortium. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nature Genetics. 2013;45:984–995. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawkins R. Twelve misunderstandings of kin selection. Zeitschrift für Tierpsychologie. 1979;51:184–200. [Google Scholar]
- Davies G, Tenesa A, Payton A, Yang J, Harris SE, Goddard ME, Deary IJ. Genome-wide association studies establish that human intelligence is highly heritable and polygenic. Molecular Psychiatry. 2011;16:996–1005. doi: 10.1038/mp.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Candia TR, Lee SH, Yang J, Browning BL, Gejman PV, Levinson DF, Keller MC. Additive genetic variation in schizophrenia risk is shared by populations of African and European descent. American Journal of Human Genetics. 2013;93:463–470. doi: 10.1016/j.ajhg.2013.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Moor MHM, Costa PT, Terracciano A, Krueger RF, de Geus EJC, Toshiko T, Boomsma DI. Meta-analysis of genome-wide association studies for personality. Molecular Psychiatry. 2012;17:337–349. doi: 10.1038/mp.2010.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudbridge F. Power and predictive accuracy of polygenic risk scores. PLoS Genetics. 2013;9:e1003348. doi: 10.1371/journal.pgen.1003348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan LE, Keller MC. A critical review of the first 10 years of candidate gene-by-environment interaction research in psychiatry. American Journal of Psychiatry. 2011;168:1041–1049. doi: 10.1176/appi.ajp.2011.11020191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewens WJ, Li M, Spielman RS. A review of family-based tests for linkage disequilibrium between a quantitative trait and a genetic marker. PLoS Genetics. 2008;4:e1000180. doi: 10.1371/journal.pgen.1000180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher RA. The genetical theory of natural selection. Oxford, UK: Oxford University Press; 1930. [Google Scholar]
- Fisher RA. Average excess and average effect of a gene substitution. Annals of Eugenics. 1941;11:53–63. [Google Scholar]
- Fisher RA. Statistical methods in genetics. Heredity. 1952;6:1–12. [Google Scholar]
- Gieger C, Radhakrishnan A, Cvejic A, Tang W, Porcu E, Pistis G, Soranzo N. New gene functions in megakaryopoiesis and platelet formation. Nature. 2011;480:201–208. doi: 10.1038/nature10659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman II, Shields J. A polygenic theory of schizophrenia. Proceedings of the National Academy of Sciences USA. 1967;58:199–205. doi: 10.1073/pnas.58.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath AC, Berg K, Eaves LJ, Solaas MH, Corey LA, Sundet JM, Nance WE. Education policy and the heritability of educational attainment. Nature. 1985;314:734–736. doi: 10.1038/314734a0. [DOI] [PubMed] [Google Scholar]
- Kimura M. The neutral theory of molecular evolution. New York: Cambridge University Press; 1983. [Google Scholar]
- Kogan A, Saslow LR, Impett EA, Oveis C, Keltner D, Rodrigues Saturn S. Thin-slicing study of the oxytocin receptor (OXTR) gene and the evaluation and expression of the prosocial disposition. Proceedings of the National Academy of Sciences. 2011;108:19189–19192. doi: 10.1073/pnas.1112658108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lande R. The response to selection on major and minor mutations affecting a metrical trait. Heredity. 1983;50:47–65. [Google Scholar]
- Lango Allen H, Estrada K, Lettre G, Berndt SI, Weedon MN, Rivadeneira F, Hirschhorn JN. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JJ. Correlation and causation in the study of personality (with discussion) European Journal of Personality. 2012;26:372–412. [Google Scholar]
- Lee JJ, Chow CC. The causal meaning of Fisher’s average effect. Genetics Research. 2013;95:89–109. doi: 10.1017/S0016672313000074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JJ, Chow CC. Conditions for the validity of SNP-based heritability estimation. Human Genetics. 2014;133:1011–1022. doi: 10.1007/s00439-014-1441-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. American Journal of Human Genetics. 2011;88:294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marigorta UM, Navarro A. High trans-ethnic replicability of GWAS results implies common causal variants. PLoS Genetics. 2013;9:e1003566. doi: 10.1371/journal.pgen.1003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClellan J, King MC. Genetic heterogeneity in human disease. Cell. 2010;141:210–217. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]
- Park JH, Gail MH, Weinberg CR, Carroll RJ, Chung CC, Wang Z, Chatterjee N. Distribution of allele frequencies and effect sizes and their interrelationships for common genetic susceptibility variants. Proceedings of the National Academy of Sciences USA. 2011;108:18026–18031. doi: 10.1073/pnas.1114759108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plomin R, Deary IJ. Genetics and intelligence: Five special findings. Molecular Psychiatry. doi: 10.1038/mp.2014.105. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, Sklar P. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietveld CA, Medland SE, Derringer J, Yang J, Esko T, Martin NW, Koellinger PD. GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science. 2013;340:1467–1471. doi: 10.1126/science.1235488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietveld CA, Conley D, Eriksson E, Esko T, Medland SE, Vinkhuyzen AAE Social Science Genetic Association Consortium. Replicability and robustness of GWAS for behavioral traits. Psychological Science. 2014;25:1975–1986. doi: 10.1177/0956797614545132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietveld CA, Esko T, Davies G, Pers TH, Benyamin B, Chabris CF, Koellinger PD. Proxy-phenotype method identifies common genetic variants associated with cognitive performance. Proceedings of the National Academy of Sciences. 2014;111:13790–13794. doi: 10.1073/pnas.1404623111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, Sanders AR, Kendler KS, Levinson DF, Sklar P, Holmans PA, Gejman PV. Genome-wide association study identifies five new schizophrenia loci. Nature Genetics. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, O’Dushlaine C, Chambert K, Moran JL, Kähler AK, Akterin S, Sullivan PF. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nature Genetics. 2013;45:1150–1159. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, Neale BM, Corvin A, Walters JR, Farh KH, Holmans PA, O’Donovan MC. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skafidas E, Testa R, Zantomio D, Chana G, Everall IP, Panelis C. Predicting the diagnosis of autism spectrum disorder using gene pathway analysis. Molecular Psychiatry. 2012;19:504–510. doi: 10.1038/mp.2012.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speed D, Hemani G, Johnson MR, Balding DJ. Improved heritability estimation from genome-wide SNPs. American Journal of Human Genetics. 2012;91:1011–1021. doi: 10.1016/j.ajhg.2012.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl EA, Wegmann D, Trynka G, Gutierrez-Achury J, Do R, Voight BF, Plenge RM. Bayesian inference analyses of the polygenic architecture of rheumatoid arthritis. Nature Genetics. 2012;44:483–489. doi: 10.1038/ng.2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D, Collier DA. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turchin MC, Chiang CWK, Palmer CD, Sankararaman S, Reich D, Hirschhorn JN Genetic Investigation of Anthropometric Traits Consortium. Evidence of widespread selection on standing variation in Europe at height-associated SNPs. Nature Genetics. 2012;44:1015–1019. doi: 10.1038/ng.2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkheimer E. Three laws of behavior genetics and what they mean. Current Directions in Psychological Science. 2000;9:160–164. [Google Scholar]
- Turkheimer E. Genome wide association studies of behavior are social science. In: Plaisance KS, Reydon TAC, editors. Philosophy of behavioral biology. Dordrecht, The Netherlands: Springer; 2012. pp. 43–64. [Google Scholar]
- Turkheimer E, Pettersson E, Horn EE. A phenotypic null hypothesis for the genetics of personality. Annual Review of Psychology. 2014;65:515–540. doi: 10.1146/annurev-psych-113011-143752. [DOI] [PubMed] [Google Scholar]
- Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. American Journal of Human Genetics. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh KM, Anderson E, Hansen HM, Decker PA, Kosel ML, Kollmeyer T, Wrensch MR. Analysis of 60 reported glioma risk SNPs replicates published GWAS findings but fails to replicate associations from published candidate-gene studies. Genetic Epidemiology. 2013;37:222–228. doi: 10.1002/gepi.21707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood AR, Esko T, Yang J, Vedantam S, Pers TH, Frayling TM. Defining the role of common variation in the genomic and biological architecture of adult human height. Nature Genetics. 2014;46:1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray NR, Purcell SM, Visscher PM. Synthetic associations created by rare variants do not explain most GWAS results. PLoS Biology. 2011;9:e1000579. doi: 10.1371/journal.pbio.1000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Benyamin B, McEvoy B, Gordon S, Henders AK, Nyholt DR, Visscher PM. Common SNPs explain a large proportion of the heritability for human height. Nature Genetics. 2010;42:565–569. doi: 10.1038/ng.608. [DOI] [PMC free article] [PubMed] [Google Scholar]