Abstract
The viral tegument is a layer of proteins between the herpesvirus capsid and its outer envelope. According to phylogenetic studies, only a third of these proteins are conserved amongst the three subfamilies (Alpha-, Beta- and Gammaherpesvirinae) of the family Herpesviridae. Although some of these tegument proteins have been studied in more detail, the structure and function of the majority of them are still poorly characterized. VP22 from Herpes simplex virus 1 (subfamily Alphaherpesvirinae) is a highly interacting tegument protein that has been associated with tegument assembly. We have determined the crystal structure of the conserved core domain of VP22, which reveals an elongated dimer with several potential protein–protein interaction regions and a peptide-binding site. The structure provides us with the structural basics to understand the numerous functional mutagenesis studies of VP22 found in the literature. It also establishes an unexpected structural homology to the tegument protein ORF52 from Murid herpesvirus 68 (subfamily Gammaherpesvirinae). Homologues for both VP22 and ORF52 have been identified in their respective subfamilies. Although there is no obvious sequence overlap in the two subfamilies, this structural conservation provides compelling structural evidence for shared ancestry and functional conservation.
Introduction
Human herpesviruses are known to cause vastly different diseases/illnesses that range from mild oral-facial blisters and chicken pox to fatal conditions such as Burkitt’s lymphoma and Kaposi’s sarcoma (Antman & Chang, 2000; Davison, 2007; Whitley & Roizman, 2001). Herpesviruses are large DNA viruses that share an overall common virion structure. The virion consists of a dsDNA encapsidated within an icosahedral capsid (Davison, 2007). Between the capsid and the outer membrane lies a layer of proteins, collectively known as the tegument (Guo et al., 2010). Twenty-four different tegument proteins have been identified in Herpes simplex virus 1 (HSV-1; Human herpesvirus 1) but, judging from sequence alignments, only a third of them are conserved across all the subfamilies (Alpha-, Beta- and Gammaherpesvirinae) of the family Herpesviridae (Kelly et al., 2009). Tegument proteins can occur in several thousands of copies inside the virion, whilst others are less abundant (Elliott & Meredith, 1992). Some tegument proteins have been found to contribute greatly to viral entry, assembly and egress, whilst others play vital roles in viral immune evasion and regulation of viral gene expressions (Kalejta, 2008; Kelly et al., 2009; Sathish et al., 2012).
VP22 is a highly abundant tegument protein in HSV-1 that has been suggested, based on sequence analysis, to be unique to the alphaherpesviruses. VP22 has been suggested to be important for the secondary tegumentation of the virion and the accurate localization of several important herpesviral proteins, including the transcription activating protein VP16, the outer capsid protein VP26, the interesting E3 ubiquitin ligase ICP0, the major transcriptional regulatory protein ICP4 and the essential multifunctional ICP27 (Brignati et al., 2003; Elliott & Meredith, 1992; Farnsworth et al., 2007; Maringer & Elliott, 2010; Potel & Elliott, 2005; Tanaka et al., 2012; Yu et al., 2010). Recently, VP22 has emerged as a key node in the HSV-1 tegument–glycoprotein network, where it makes multiple protein–protein interactions and plays a selective role in the tegument acquisition of viral glycoproteins gE, gD and gM (Chi et al., 2005; Elliott et al., 1995; Farnsworth et al., 2007; Hafezi et al., 2005; Maringer et al., 2012; O’Regan et al., 2007a, 2010; Potel & Elliott, 2005; Stylianou et al., 2009). VP22 also binds directly to cellular proteins like chromatin remodelling protein (TAF-1) and non-muscle myosin II (NMII) (van Leeuwen et al., 2002, 2003). It has also been associated with interactions with cellular membranes, microtubules and nucleic acids (Brignati et al., 2003; Elliott & O’Hare, 1998; Martin et al., 2002; Sciortino et al., 2002). Interestingly VP22 exhibits transfection potential and has been used successfully in several studies to target therapeutic DNA to specific cells of interest, such as stem cells (Bennett et al., 2002; Elliott & O’Hare, 1999; Jin et al., 2013; Lai et al., 2000).
Sequence analysis and secondary structure predictions reveal that VP22 consists of a non-conserved N-terminal domain and a conserved C-terminal domain with the clear presence of secondary structures (O’Regan et al., 2007a). Deletion and functional studies have shown that the conserved C-terminal domain in VP22 is important for binding to VP16 and gE (O’Regan et al., 2007a, b). To generate insight into the VP22 structure and function, we crystallized and solved the structure of the conserved C-terminal domain of this protein, hereafter referred to as VP22core, to a resolution of 1.9 Å. VP22core exists as a dimer with a highly conserved dimerization site. Although sequence homology of VP22 has only been established within the alphaherpesviruses, the crystal structure reveals that it shares extensive structural similarity with ORF52 from Murid herpesvirus 68 (MHV-68) (subfamily Gammaherpesvirinae). ORF52MHV-68 has been found to be essential for replication in MHV-68 in vitro (Song et al., 2005). Similar to VP22core, ORF52MHV-68 is also a highly expressed tegument protein that exists as a dimer made up of two identical monomers (Benach et al., 2007; Bortz et al., 2007). It is well conserved within the gammaherpesviruses, and has been implicated to be important for tegument association and interactions (Bortz et al., 2007; Fossum et al., 2009; Rozen et al., 2008; Uetz et al., 2006). These are coincidentally similar to some of the proposed functions of VP22 (Brignati et al., 2003; Farnsworth et al., 2007). With the VP22core structure in hand, we have been able to compare the two protein structures, revisit the outcome of reported mutational studies as well as identify completely conserved residues that might be important for function.
Results and Discussion
Structure of VP22core
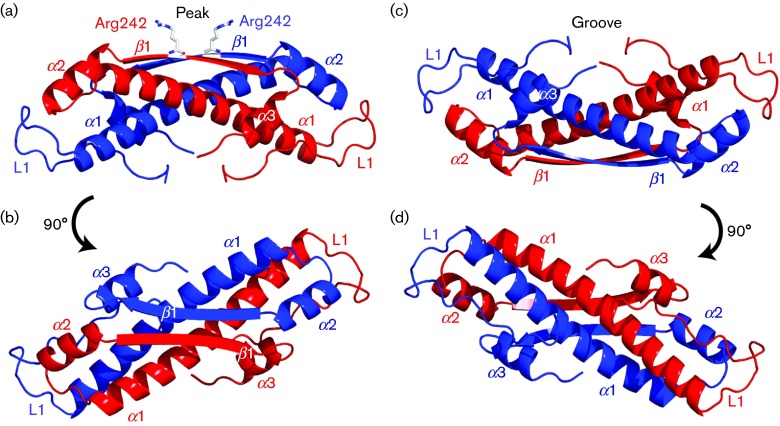
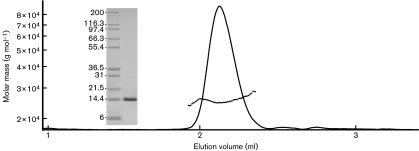
VP22core crystallized in the space group P6122 and the crystal structure [Protein Data Bank (PDB) ID: 4XAL] was determined at a resolution of 1.87 Å using single isomorphous replacement with anomalous scattering (SIRAS). Each asymmetrical unit consists of a molecule of VP22core, with visual electron density for residues 174–260, together with three amino acids from the N-terminal purification tag. The crystallographic data statistics are summarized in Table 1. The structure of VP22core is constituted by a long central α-helix (α1) flanked by a long random coil (L1) at the N terminus, two shorter α-helices (α2 and α3) and one β-strand (β1) at the C terminus (Fig. 1). Two VP22core monomers are related by the crystallographic twofold axis and are slightly twisted around each other, creating an elongated dimer (Fig. 1) where the α1 helices and the β1 interact in an anti-parallel fashion. The dimeric state of VP22core has been proposed previously (Mouzakitis et al., 2005) and our light-scattering results show that VP22core is mono-dispersed with a mean molar mass of ~26 500 g mol−1. This is roughly twice the theoretical molecular mass of the monomeric VP22core including the purification tag and the tobacco etch virus (TEV) protease site (14 551 g mol−1), further confirming that VP22core is a dimer in solution (Fig. 2).
Table 1. Summary of data collection, phasing and refinement statistics.
| Parameter | Native (PDB ID: 4XAL) | Soaked with PbCl2 |
| X-ray source | NSRRC 13C1 | NSRRC 13C1 |
| Wavelength (Å) | 0.9762 | 0.9762 |
| Space group | P6122 | P6122 |
| Unit cell parameters | a = 65.0, b = 65.0, c = 107.9 | a = 65.0, b = 65.0, c = 107.6 |
| α = 90, β = 90, γ = 120 | α = 90, β = 90, γ = 120 | |
| Resolution range (Å) | 30.00–1.87 (1.94–1.87)* | 27.80–1.87 (1.94–1.87)* |
| I/σ(I) | 24.3 (3.2)* | 43.8 (5.81)* |
| Completeness (%) | 99.0 (99.6)* | 100 (97.6) |
| Redundancy | 2.9 (2.8)* | 10.2 (10.2) |
| R sym† | 0.052 (0.289) | 0.047 (0.188) |
| Total reflections | 182 439 | 194 305 |
| Unique reflections | 11 745 | 11 789 |
| AutoSol | ||
| No. of sites | 1 | |
| Initial figure of merit | 0.21 | |
| Figure of merit after density modification | 0.63 | |
| Refinement | ||
| R factor‡/R free§ (%) | 20.8/25.1 | |
| Atoms | 828 | |
| Protein residues | 96 | |
| Solvent molecules | 74 | |
| RMSD bonds (Å) | 0.027 | |
| RMSD angles (°) | 0.77 | |
| Ramachandran quality plot | ||
| In preferred region (%) | 99 | |
| In allowed region (%) | 1 | |
| Outliers (%) | 0 |
NSRRC, National Synchrotron Radiation Research Center (Taiwan, ROC).
Values within parentheses represent the highest resolution shell (1.939–1.872 Å).
R sym = 100×∑(|Ij−[I]|)/∑(|I|), where the sum is calculated over all observations of a measured reflection (Ij) and [I] is the mean intensity of all the measured observations (Ij).
R factor = 100×∑(|F o|−|F c|)/∑(|F o|), where F o and F c are the observed and calculated structure factors, respectively.
R free is equivalent to R factor, but where 5 % of the measured reflections have been excluded from refinement and set aside for cross-validation.
Fig. 1. Different views of the crystal structure of the VP22core dimer. (a) Each monomer (coloured red and blue) consists of three α-helices (α1–α3) and one β-strand (β1). The monomers of VP22core coil around each other. The flat β1 from both monomers create a plateau with a conserved arginine (Arg242) sticking up like a peak. We refer to this face of the protein as the ‘peak side’. (b) A 90° rotation of the dimer gives the top view of the peak side. (c) The opposite side reveals a groove that is created by L1 and α1 from both monomers. (d) A 90° rotation of the dimer gives the top view of the groove side.
Fig. 2. Light-scattering curve of VP22core in solution as a function of its elution volume. The monomeric molar mass of VP22core is 14551 g mol−1 and the light-scattering results show that VP22core is mono-dispersed with an estimated mean molar mass of 26 500±5000 g mol−1. This shows that VP22core is dimeric in solution. The SDS-PAGE gel of the injected VP22core sample and the protein ladder (Mark12 Unstained Standard; kDa) is displayed on the left of the elution peak.
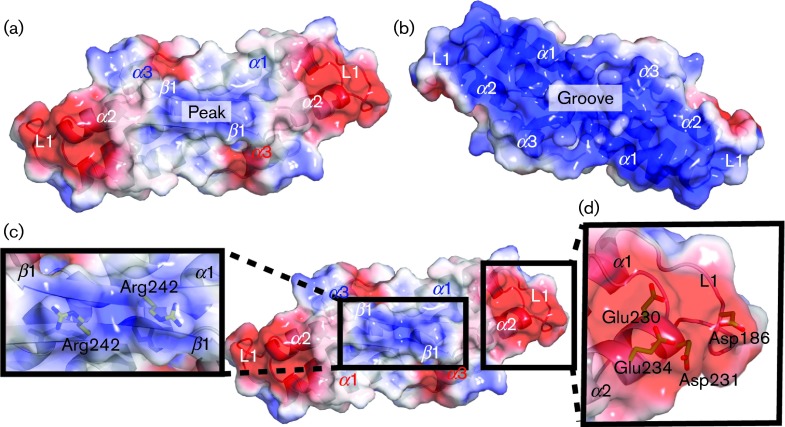
To be able to orientate ourselves in the structure, we have dubbed one side of the dimer the ‘peak’ (Figs 1a, b and 3a) and the other side the ‘groove’ (Figs 1c, d and 3b). On the peak side, the dimerization of β1 creates a flat plateau where two conserved arginines (Arg242) create a positively charged peak in the middle of a less charged area (Figs 1a and 3a, c). Flanking the sides of this peak are two identical negatively charged patches. The residues that contributed to these two patches are Asp186 from L1 of one VP22core monomer and a cluster of negatively charged residues, Glu230, Asp231 and Glu234, from α2 of the other monomer (Figs 1c and 3a, d). The electrostatic potential surface map of the groove, which is created by L1 and α1 from both monomers, shows two large and positively charged patches. In general, distinctly charged patches on a protein surface might indicate potential sites for protein–protein interactions and any of these described areas in VP22core could serve this purpose.
Fig. 3. Electrostatic potential surface maps of the VP22core dimer. (a) The positive, negative and uncharged regions of the surface map are coloured blue, red and white, respectively. The protein is shown in the same orientation as in Fig. 1(b). It reveals a patch of positive charges in the middle of a relatively uncharged surface. Flanking their sides are areas of negatively charged patches. (b) At the groove side, there are two large positively charged patches. The charges on this surface are contributed by the α1 amino acids lining the groove. The protein is shown in the same orientation as in Fig. 1(d). (c) The positively charged patch at the peak side is created by Arg242, whilst (d) the negatively charged patch is created by Asp186 from L1 of one monomer and a cluster of negatively charged residues, Glu230, Asp231 and Glu234, from α2 of the second monomer. These distinctively charged patches on VP22core might be potential molecular interaction sites.
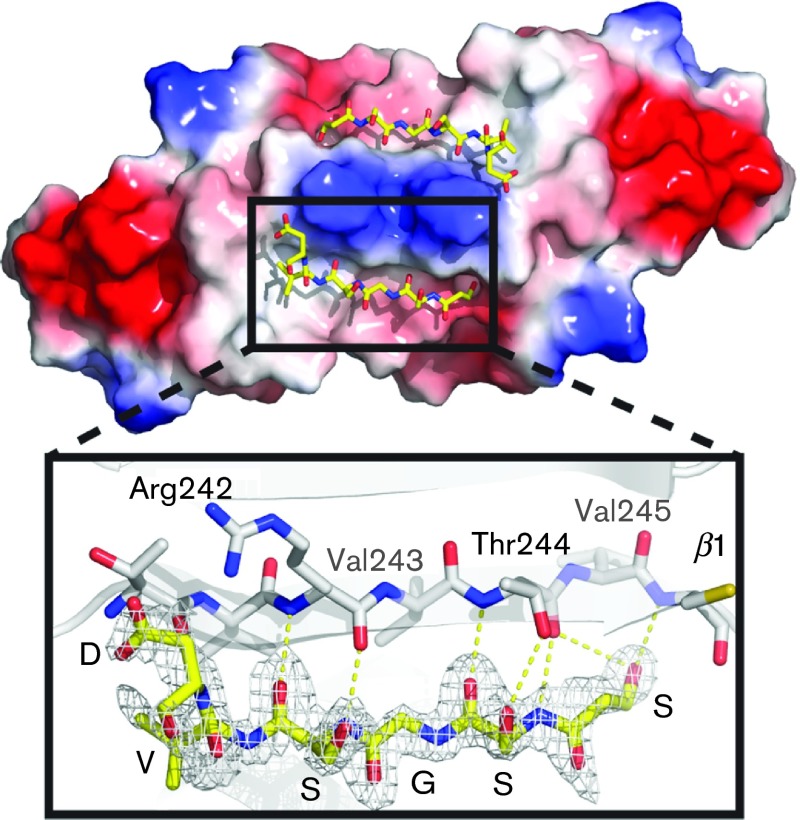
Interestingly, we observed a stretch of unaccounted electron density next to β1 (Fig. 4). The β1 β-sheet forms a tiny β-sheet through interactions with β1 from the other monomer and contributes to the overall dimerization of VP22core. We managed to model a six-amino-acid peptide into this density. This peptide forms a perfect β-strand, expanding the β1 β-sheet to four stands. It corresponds to the sequence SSGSVD, which is a part of the linker region between the N-terminal His6-tag and the TEV protease cleavage site. This peptide is most likely contributed in trans from a neighbouring subunit in the crystal lattice, which is not part of the crystallographic dimer. The peptide is held tightly into place by backbone interactions and the coordination of the hydroxyl group on the N-terminal serine. Although this particular peptide sequence is most likely not of biological relevance, it indicates directly that this peptide-binding cavity could constitute a real site for protein interaction with VP22. A motif similar to the peptide was not identified at the N terminus of VP22, but it is plausible that some as-yet unidentified part of the N terminus could form a β-strand and bind in this location.
Fig. 4. Peptide-binding site of VP22core with the electrostatic potential surface map of the peak side. A peptide consisting of six amino acids was traced from the stretch of unmodelled electron density next to β1. The interaction between the peptide (yellow) and β1 (white) is magnified and displayed below. The peptide fits well into the electron density and the sequence was traced to be SSGSVD. Hydrogen bonds hold the peptide to β1 and these interactions are illustrated by yellow dotted lines.
Conserved residues in VP22core contribute to its fold, oligomerization and interactions
VP22 has many proposed interaction partners. In order to evaluate and differentiate between these interactions, various deletion mutants have been created, described and discussed (Brignati et al., 2003; Elliott et al., 2005; Hafezi et al., 2005; Martin et al., 2002; O’Regan et al., 2007a, b, 2010; Stylianou et al., 2009) (summarized in Table S1, available in the online Supplementary Material). Whilst these studies have laid a foundation for the VP22 protein interaction network, the crystal structure of VP22core can now aid in understanding these interactions at the atomic level. We mapped several of the published mutations onto the VP22core structure to gain more insights into their structure–function relationship.
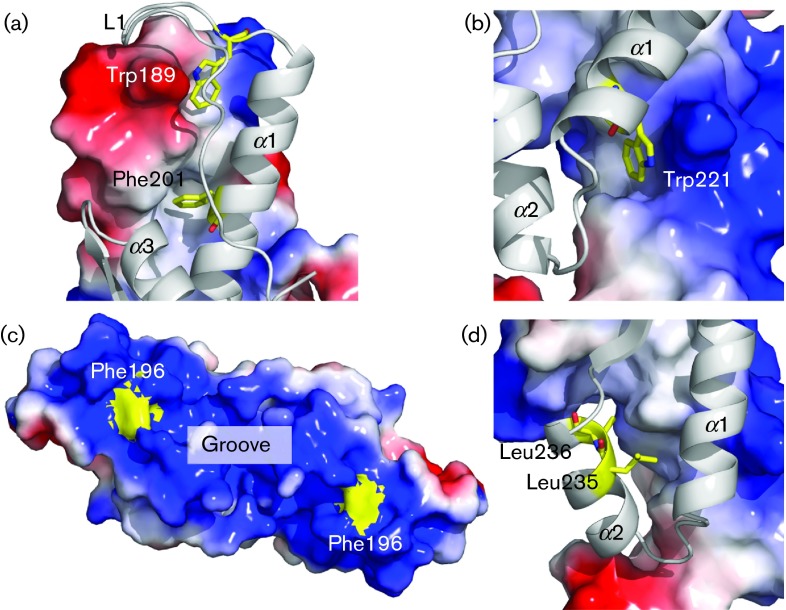
Upon mapping these deletions and truncations, we can now see that most of the mutated residues that yielded in a loss of protein function are located in L1, α1 or α2 (Table S1). In most cases, the reported deletions would have removed parts of the long central helix α1 – a key secondary structure along the dimerization interface. Most of the described point mutations that seem to have an effect on VP22 interactions are also focused on this helix (O’Regan et al., 2007b, 2010; Tanaka et al., 2012). In particular, Trp189, Phe201 and Trp221, which have been found to disrupt the binding between VP22 and gE/VP16, are located along the dimerization interface of α1 (Fig. 5a, b). It is possible that most effects observed in these studies are the result of the distortion of VP22’s dimerization, rather than specific functional effects.
Fig. 5. Point mutations mapped in VP22core. The mapped residues (a) Trp189/Phe201, (b) Trp221, (c) Phe196 and (d) Leu245/Leu246 were found to be important for VP22 protein interactions by O’Regan et al. (2010); (a, b, d) show one VP22core monomer displayed as a cartoon and the other monomer displayed as the electrostatic potential surface map. (a) Trp189/Phe201 and (b) Trp221 are buried in the hydrophobic dimerization interface, rendering them unlikely to participate in any specific protein–protein interactions. Instead, they seem very important for dimerization. However, the surface electrostatic potential map shows that the conserved Phe196 (c) is found on the surface of the VP22core and is likely to participate in protein–protein interactions. However, Leu245/Leu246 (d) are buried in the hydrophobic interface, indicating that the leucine pair is not likely to participate in specific protein–protein interactions.
A residue of particular interest is the conserved and solvent-exposed Phe196. The electrostatic surface potential of VP22core reveals that this hydrophobic Phe196 is located in the middle of the two large and highly positively charged patches at the groove side (Fig. 5c). With a single point mutation of this amino acid, O’Regan et al. (2010) were able to remove the binding between gE and VP22, but not between VP16 and VP22. Moreover, conserved aromatic residues on the surface of a protein have often been shown to be important for protein interactions (Albiston et al., 2010; Cao et al., 2008; Chouljenko et al., 2012; Ferrandon et al., 2003). The functional evidence from O’Regan et al. (2010), in combination with the high degree of conservation and strategic location/orientation of Phe196, suggest its importance in protein interactions. Given that the point mutation on Phe196 only removed the binding between gE and VP22, Phe196 and the surrounding amino acid residues may also play a key role in discriminating between the different interacting proteins of VP22 (O’Regan et al., 2010).
Similarly, the binding between VP22 and VP16 was disrupted when a pair of conserved leucines along α2VP22 (Leu235 and Leu236) was mutated into alanines (O’Regan et al., 2007b). These mutations also altered the localization sites of several HSV-1 proteins, including ICP0, gE, gD, VP16 and vhs, in the host cell (Tanaka et al., 2012). However, these leucines are exposed to the hydrophobic core and do not appear to be able to participate in any direct protein–protein interactions (Fig. 5d). Thus, the loss of protein function may likely have arisen due to either the collapse of the global VP22core structure or local distortions of α-helical stability/positions. If the observed effects are indeed a result of local structural distortions, these mutations highlight the significance of the entire α2 for making interactions with its binding partners.
VP22core is structurally homologous to ORF52 from MHV-68
VP22 consists of two domains where only the C-terminal domain is highly conserved in the alphaherpesviruses. The N-terminal domain is more variable and this domain is completely absent in some alphaherpesviruses (O’Regan et al., 2007a). A structural homology search with the structure of VP22core on the Dali server identified another herpesvirus protein, ORF52 from MHV-68 (PDB ID: 2R3H and 2OA5), with a mean Z score of 5.6 (Holm & Rosenström, 2010). ORF52 from MHV-68 (ORF52MHV-68) is a small viral protein of 21 kDa, making it substantially smaller than the full-length VP22 (35 kDa). As with VP22core, ORF52MHV-68 is also a highly expressed tegument protein that exists as a dimer made up of two identical monomers (Benach et al., 2007; Bortz et al., 2007). Both VP22 and ORF52MHV-68 are well conserved within the alpha- and gammaherpesviruses, respectively, and both proteins seem to share similar functions, such as tegument association and interactions (Bortz et al., 2007; Brignati et al., 2003; Fossum et al., 2009; Rozen et al., 2008; Uetz et al., 2006). For clarity, we use the subscripts ‘VP22’ and ‘ORF52’ to differentiate between the secondary structural elements in the respective proteins.
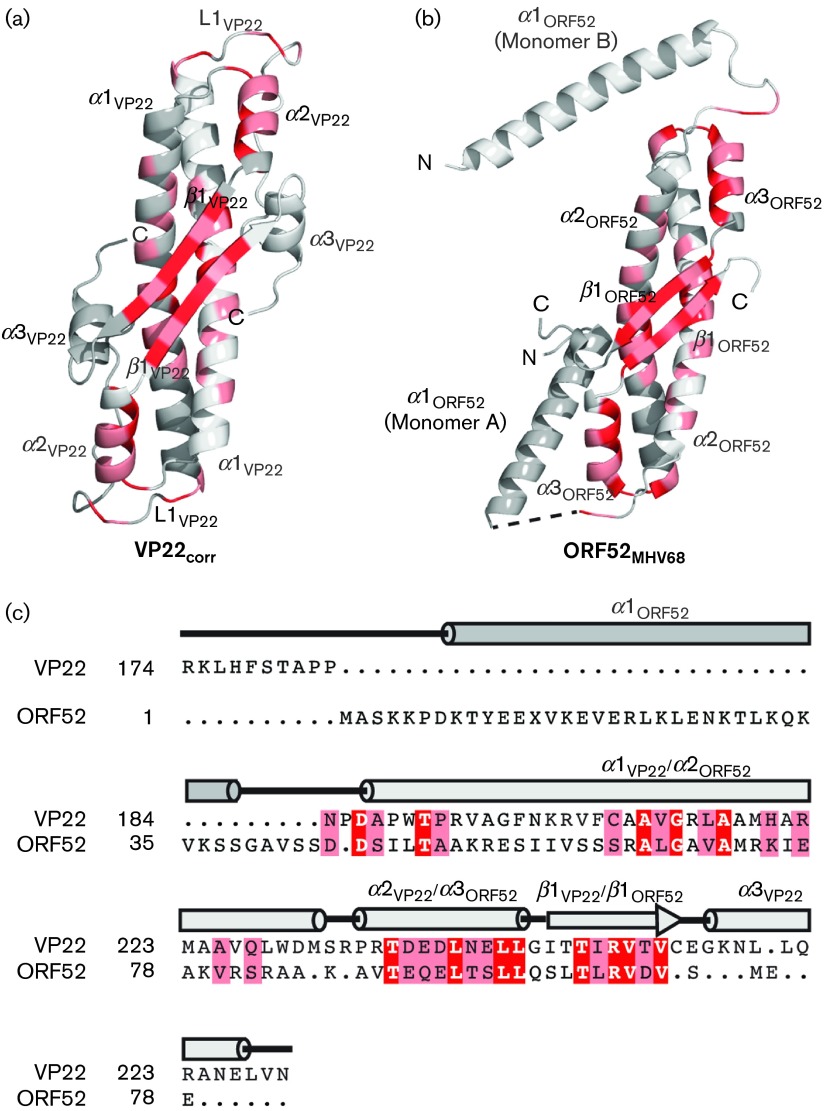
To analyse the structural similarities in detail, the VP22core structure was compared with the published dimer of ORF52MHV-68 (PDB ID: 2OA5) using Coot (Emsley et al., 2010). The α carbons of each VP22core monomer and the individual ORF52MHV-68 monomer align well with a mean root-mean-square deviation (RMSD) of 2.1 Å (Fig. 6a, b). Both VP22core and ORF52MHV-68 have long central α-helices (α1VP22 and α2ORF52) that constitute the core of the dimer interactions. The anti-parallel β-strands (β1VP22 and β1ORF52) also contribute to this dimerization. The helices α2VP22 and α3ORF52 located on the surface of the proteins align well with each other. There is a slight difference at the C terminus of this superposition where we notice that whilst ORF52MHV-68 has an extended loop, HSV-1 VP22core has an α-helix denoted α3VP22. However, the major differences between VP22core and ORF52MHV-68 lie at the N terminus (Fig. 6a, b). At the N terminus, ORF52MHV-68 has an additional helix (α1ORF52), whilst VP22core has a long extended loop (L1VP22). In ORF52MHV-68, this particular helix displays two different conformations by extending in different directions in the dimer structure, suggestive of a flexible N terminus in ORF52MHV-68. L1VP22 stretches in the same direction as α1ORF52 in chain A and in the opposite direction from α1ORF52 of ORF52MHV-68 chain B. VP22core has an additional N-terminal domain, not present in our structure, and secondary structure predictions also indicate a low α-helical propensity along L1VP22 (not shown) (Cole et al., 2008). Thus, there is a possibility that L1VP22 exists as a part of a long and flexible connection between VP22core and its N-terminal domain.
Fig. 6. Structural and sequence alignment of VP22core and ORF52MHV-68. The dimer structures of (a) VP22core and (b) ORF52MHV-68 are shown as cartoons in the same orientation. (c) The structural alignment of VP22core and ORF52MHV-68 is reproduced in a sequence alignment. The completely conserved amino acids are highlighted in red, whilst the other conserved residues are highlighted in pink. The conserved amino acids are mainly concentrated along the hydrophobic dimerization interface at α1VP22, α2VP22 and β1VP22.
Based on the structural similarity, we generated a structure-based sequence alignment between the monomeric VP22core and ORF52MHV-68 yielding a sequence identity of 13 % (Fig. 6c) (Pettersen et al., 2004). As with the conserved residues within the VP22 homologues, most of these residues are clustered throughout the dimerization interface and the hydrophobic core. The structure-based sequence alignment prompted us to try and identify a possible homologue in the betaherpesviruses. However, no homologue could be identified.
To further understand the sequence conservation between the alpha- and gammaherpesviruses, we generated an additional alignment with most homologues from the alpha- and gammaherpesviruses (Fig. S1). Although this sequence alignment displays very low sequence similarity, it does reveal four amino acids that are particularly conserved in both subfamilies. In particular, along α2VP22 and α3MHV-68, a leucine (Leu236VP22/Leu89ORF52) is conserved in both the alpha- and gammaherpesviruses. As in VP22, this conserved leucine in ORF52MHV-68 (coloured red at α3MHV-68 in Fig. 6b) is also exposed to the hydrophobic core of the protein, supporting the importance of oligomerization of this protein for proper function.
The remaining three amino acids that are conserved in both the alpha- and gammaherpesviruses are Arg242VP22/Arg95ORF52, Val243VP22/Val96ORF52 and Val245VP22/Val98ORF52 (Fig. S1). These amino acids are located along β1, where the side chain of the valines stretches into the core of the structure, whilst the side chain of the arginine is solvent-exposed (Figs 4 and 6). The two conserved valines seem to contribute to the fold, but the highly conserved arginine (Arg242VP22/Arg95ORF52) along β1 appears to be important for protein binding. This conserved residue is found next to our proposed peptide-binding site and is what creates the distinct peak of VP22core (Figs 3a, c and 4). To underline the importance of this completely conserved arginine is the fact that Wang et al. (2012) could disrupt the binding between ORF52MHV-68 and ORF42MHV-68 with a single amino acid substitution (Arg→Ala) in this position. Hence, although there is no determined homologue to ORF42MHV-68 in HSV-1, it is likely that the corresponding mutation in VP22 could also disrupt the interaction to one or several of its (un)known binding partners. It would be interesting to see how a mutation of this conserved Arg242VP22 would affect this protein in vivo.
The described conserved structural features and functions of VP22 and ORF52MHV-68 suggest that both proteins could act as protein adaptors in which different proteins are bound. Moreover, being a major tegument protein in HSV-1, VP22 has been associated with multiple protein–protein interactions, several HSV-1 protein localizations as well as protein transportation along the microtubules (Chi et al., 2005; Elliott et al., 1995, 2005; Elliott & O’Hare, 1998; Farnsworth et al., 2007; Hafezi et al., 2005; Kotsakis et al., 2001; Maringer & Elliott, 2010; Maringer et al., 2012; Martin et al., 2002; O’Regan et al., 2007a, 2010; Potel & Elliott, 2005; Stylianou et al., 2009; Tanaka et al., 2012; Yedowitz et al., 2005). It is likely that VP22 and the structural homologue ORF52MHV-68 could be involved in assembling a protein scaffold consisting of other tegument proteins, thereby creating a protein bridge between the capsid and the lipid envelope. This assembly may be important for the intracellular transportation of proteins along the microtubules.
In conclusion, with a three-dimensional structure of a well-studied protein like VP22, we can now start connecting functional data with structural information. We hope that the data presented in this paper might help to spur new and directed efforts to elucidate this protein’s function. The unexpected structural similarities between VP22 and ORF52MHV-68 may contribute to further functional studies of other herpesviral proteins and pose intriguing questions about the evolutionary relationship of the different herpesvirus subfamilies.
Methods
Cloning and protein expression.
The VP22core (residues 174–281) (GenBank accession number BAE87004.1) was cloned into the plasmid pNIC28-Bsa4 (GenBank accession number EF198106) containing a His6-tag and a TEV protease site at the N terminus. Cloning and protein expression of the VP22core were performed as described previously (Hew et al., 2013). The purification of VP22core was performed as follows. The bacterial cells expressing VP22core were pelleted and resuspended in lysis buffer (100 mM HEPES, pH 8.0, 500 mM NaCl, 10 mM imidazole pH 8.0, 10 % glycerol, 0.5 mM TCEP [Tris(2-carboxyethyl)phosphine], 0.1 mg lysozyme ml−1, 1 ml protease inhibitor ml−1 and 25 U Benzonase). Lysates were clarified and loaded on a 1 ml HisTrap HP column (GE Healthcare). The column was washed with 20 ml wash buffer (20 mM HEPES, pH 7.5, 500 mM NaCl, 10 mM imidazole, pH 7.5, 10 % glycerol and 0.5 mM TCEP) and 10 ml second wash buffer (20 mM HEPES, pH 7.5, 500 mM NaCl, 25 mM imidazole, pH 7.5, 10 % glycerol and 0.5 mM TCEP) before eluting with 5 ml elution buffer (20 mM HEPES, pH 7.5, 500 mM NaCl, 500 mM imidazole, pH 7.5, 10 % glycerol and 0.5 mM TCEP). The eluted sample was loaded on a pre-equilibrated (20 mM HEPES, pH 7.5, 300 mM NaCl, 10 % glycerol and 0.5 mM TCEP) size-exclusion column (HiLoad 16/60 Superdex 75; GE Healthcare) and eluted in 2 ml fractions. The fractions were accessed for purity on a SDS-PAGE gel and only pure fractions containing our target protein were pooled. TCEP (2 mM) was added to the pooled sample and the protein was further concentrated to 12 mg ml−1 before storing at −80 °C.
Crystallization and data collection.
Native crystals of VP22core were obtained from a sitting drop experiment with drops containing 1.5 µl purified VP22core protein (12 mg VP22core ml−1) and 1.5 µl reservoir solution (40 % PEG 300 and 0.1 M phosphate citrate, pH 5) was incubated with 300 µl reservoir solution in a 24-wells sitting drop Intelli-plate (Art Robbins) at 20 °C. Native crystals were transferred to a fresh drop of reservoir solution containing 1 mM PbCl2 for 45 min to obtain derivative crystals. No additional cryoprotectant was added to the native and the derivative crystals before flash freezing them in liquid nitrogen.
Diffraction datasets were collected at beamline BL13C1 at the National Synchrotron Radiation Research Center (Taiwan, ROC) with the detector ADSC Quantum-315r CCD. Datasets were collected at 0.97 Å, and integrated and scaled with hkl-2000 (Otwinowski & Minor, 1997).
Structural determination.
The initial crystallographic model of VP22core was obtained with SIRAS using AutoSol wizard and AutoBuild from the phenix suite (Adams et al., 2010). The final structure was obtained after many cycles of automatic and manual structural refinement with refmac (Murshudov et al., 2011) and Coot (Emsley et al., 2010). The structure refinement was validated with sfcheck (Vaguine et al., 1999) and the geometry of the final structure was analysed with rampage (Lovell et al., 2003).
The figures of the final VP22core structure were created and displayed with PyMOL (http://www.PyMOL.org/). The electrostatic potential of the solvent accessible surfaces of the protein were calculated using pdb2pqr (Dolinsky et al., 2004) and the apbs plugin (Baker et al., 2001) in PyMOL. The electrostatic potential contour levels were set at ±3 kT/e and the surface maps were displayed with PyMOL.
Structure-based sequence alignment.
The sequence alignment between VP22core and ORF52MHV-68 was generated with a pair-wise structure-based alignment between the monomers using Chimera (Pettersen et al., 2004). Sequences of the VP22core and ORF52MHV-68 homologues from the alpha- and gammaherpesviruses were aligned by adding their amino acid sequences to the structure-based alignment. The amino acid conservation was mapped and displayed with PyMOL.
Multi-angle light scattering.
Light-scattering data were obtained with analytical size-exclusion chromatography (Superdex 200 5/150 GL; GE Healthcare) coupled with a multi-angle light-scattering detector (MiniDAWN TREOS; Wyatt Technology) and a refractive index detector (Optilab rEX; Wyatt Technology) on an ÄKTAmicro (GE Healthcare). An aliquot of 20 µl VP22core (6 mg VP22core ml−1) was injected onto the pre-equilibrated column (20 mM HEPES, pH 7.5, 300 mM NaCl, 10 % glycerol and 2 mM TCEP) at a flow rate of 0.3 ml min−1. astra 6 (Wyatt Technology) was used to determine the experimental protein molecular mass from the light-scattering data.
Acknowledgements
We would like to thank Jurgen Haas (University of Edinburgh, UK) for providing the full-length clone of VP22, and the Protein Production Platform (Nanyang Technological University, Singapore) for the initial cloning and small-scale expression screening. Portions of this research were carried out at the National Synchrotron Radiation Research Center, a national user facility supported by the National Science Council of Taiwan, ROC. The Synchrotron Radiation Protein Crystallography Facility is supported by the National Core Facility Program for Biotechnology. This research was also undertaken on the MX1 and MX2 beamlines at the Australian Synchrotron, Victoria, Australia.
Footnotes
One supplementary table and one supplementary figure are available with the online Supplementary Material.
References
- Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L.-W., Kapral G. J., et al. ( 2010. ). phenix: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr 66, 213–221. 10.1107/S0907444909052925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albiston A. L., Pham V., Ye S., Ng L., Lew R. A., Thompson P. E., Holien J. K., Morton C. J., Parker M. W., Chai S. Y. ( 2010. ). Phenylalanine-544 plays a key role in substrate and inhibitor binding by providing a hydrophobic packing point at the active site of insulin-regulated aminopeptidase. Mol Pharmacol 78, 600–607. 10.1124/mol.110.065458 [DOI] [PubMed] [Google Scholar]
- Antman K., Chang Y. ( 2000. ). Kaposi’s sarcoma. N Engl J Med 342, 1027–1038. 10.1056/NEJM200004063421407 [DOI] [PubMed] [Google Scholar]
- Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. ( 2001. ). Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A 98, 10037–10041. 10.1073/pnas.181342398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benach J., Wang L., Chen Y., Ho C. K., Lee S., Seetharaman J., Xiao R., Acton T. B., Montelione G. T., et al. ( 2007. ). Structural and functional studies of the abundant tegument protein ORF52 from murine gammaherpesvirus 68. J Biol Chem 282, 31534–31541. 10.1074/jbc.M705637200 [DOI] [PubMed] [Google Scholar]
- Bennett R. P., Dalby B., Guy P. M. ( 2002. ). Protein delivery using VP22. Nat Biotechnol 20, 20. 10.1038/nbt0102-20 [DOI] [PubMed] [Google Scholar]
- Bortz E., Wang L., Jia Q., Wu T. T., Whitelegge J. P., Deng H., Zhou Z. H., Sun R. ( 2007. ). Murine gammaherpesvirus 68 ORF52 encodes a tegument protein required for virion morphogenesis in the cytoplasm. J Virol 81, 10137–10150. 10.1128/JVI.01233-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brignati M. J., Loomis J. S., Wills J. W., Courtney R. J. ( 2003. ). Membrane association of VP22, a herpes simplex virus type 1 tegument protein. J Virol 77, 4888–4898. 10.1128/JVI.77.8.4888-4898.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao L., Bandelac G., Volgina A., Korostoff J., DiRienzo J. M. ( 2008. ). Role of aromatic amino acids in receptor binding activity and subunit assembly of the cytolethal distending toxin of Aggregatibacter actinomycetemcomitans . Infect Immun 76, 2812–2821. 10.1128/IAI.00126-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi J. H., Harley C. A., Mukhopadhyay A., Wilson D. W. ( 2005. ). The cytoplasmic tail of herpes simplex virus envelope glycoprotein D binds to the tegument protein VP22 and to capsids. J Gen Virol 86, 253–261. 10.1099/vir.0.80444-0 [DOI] [PubMed] [Google Scholar]
- Chouljenko D. V., Kim I. J., Chouljenko V. N., Subramanian R., Walker J. D., Kousoulas K. G. ( 2012. ). Functional hierarchy of herpes simplex virus 1 viral glycoproteins in cytoplasmic virion envelopment and egress. J Virol 86, 4262–4270. 10.1128/JVI.06766-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole C., Barber J. D., Barton G. J. ( 2008. ). The Jpred 3 secondary structure prediction server. Nucleic Acids Res 36 (Web Server issue), W197–W201. 10.1093/nar/gkn238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davison A. J. ( 2007. ). Overview of classification. In Human Herpesviruses: Biology, Therapy and Immunoprophylaxis, pp. 3–9. Edited by Arvin A., Campadelli-Fiume G., Mocarski E., Moore P. S., Roziman B., Whitley R., Yamanishi K. Cambridge: Cambridge University Press. [PubMed] [Google Scholar]
- Dolinsky T. J., Nielsen J. E., McCammon J. A., Baker N. A. ( 2004. ). pdb2pqr: an automated pipeline for the setup of Poisson–Boltzmann electrostatics calculations. Nucleic Acids Res 32 (Web Server issue), W665–W667. 10.1093/nar/gkh381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott G. D., Meredith D. M. ( 1992. ). The herpes simplex virus type 1 tegument protein VP22 is encoded by gene UL49. J Gen Virol 73, 723–726. 10.1099/0022-1317-73-3-723 [DOI] [PubMed] [Google Scholar]
- Elliott G., O’Hare P. ( 1998. ). Herpes simplex virus type 1 tegument protein VP22 induces the stabilization and hyperacetylation of microtubules. J Virol 72, 6448–6455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott G., O’Hare P. ( 1999. ). Intercellular trafficking of VP22-GFP fusion proteins. Gene Ther 6, 149–151. 10.1038/sj.gt.3300850 [DOI] [PubMed] [Google Scholar]
- Elliott G., Mouzakitis G., O’Hare P. ( 1995. ). VP16 interacts via its activation domain with VP22, a tegument protein of herpes simplex virus, and is relocated to a novel macromolecular assembly in coexpressing cells. J Virol 69, 7932–7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott G., Hafezi W., Whiteley A., Bernard E. ( 2005. ). Deletion of the herpes simplex virus VP22-encoding gene (UL49) alters the expression, localization, and virion incorporation of ICP0. J Virol 79, 9735–9745. 10.1128/JVI.79.15.9735-9745.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P., Lohkamp B., Scott W. G., Cowtan K. ( 2010. ). Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66, 486–501. 10.1107/S0907444910007493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnsworth A., Wisner T. W., Johnson D. C. ( 2007. ). Cytoplasmic residues of herpes simplex virus glycoprotein gE required for secondary envelopment and binding of tegument proteins VP22 and UL11 to gE and gD. J Virol 81, 319–331. 10.1128/JVI.01842-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandon S., Sterzenbach T., Mersha F. B., Xu M.-Q. ( 2003. ). A single surface tryptophan in the chitin-binding domain from Bacillus circulans chitinase A1 plays a pivotal role in binding chitin and can be modified to create an elutable affinity tag. Biochim Biophys Acta 1621, 31–40. 10.1016/S0304-4165(03)00029-1 [DOI] [PubMed] [Google Scholar]
- Fossum E., Friedel C. C., Rajagopala S. V., Titz B., Baiker A., Schmidt T., Kraus T., Stellberger T., Rutenberg C., et al. ( 2009. ). Evolutionarily conserved herpesviral protein interaction networks. PLoS Pathog 5, e1000570. 10.1371/journal.ppat.1000570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H., Shen S., Wang L., Deng H. ( 2010. ). Role of tegument proteins in herpesvirus assembly and egress. Protein Cell 1, 987–998. 10.1007/s13238-010-0120-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafezi W., Bernard E., Cook R., Elliott G. ( 2005. ). Herpes simplex virus tegument protein VP22 contains an internal VP16 interaction domain and a C-terminal domain that are both required for VP22 assembly into the virus particle. J Virol 79, 13082–13093. 10.1128/JVI.79.20.13082-13093.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hew K., Dahlroth S. L., Venkatachalam R., Nasertorabi F., Lim B. T., Cornvik T., Nordlund P. ( 2013. ). The crystal structure of the DNA-binding domain of vIRF-1 from the oncogenic KSHV reveals a conserved fold for DNA binding and reinforces its role as a transcription factor. Nucleic Acids Res 41, 4295–4306. 10.1093/nar/gkt082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L., Rosenström P. ( 2010. ). Dali server: conservation mapping in 3D. Nucleic Acids Res 38 (Web Server issue), W545–549. 10.1093/nar/gkq366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin G., Zhou Y., Chai Q., Zhu G., Xu F., Liu F. ( 2013. ). VP22 and cytosine deaminase fusion gene modified tissue-engineered neural stem cells for glioma therapy. J Cancer Res Clin Oncol 139, 475–483. 10.1007/s00432-012-1347-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalejta R. F. ( 2008. ). Tegument proteins of human cytomegalovirus. Microbiol Mol Biol Rev 72, 249–265. 10.1128/MMBR.00040-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly B. J., Fraefel C., Cunningham A. L., Diefenbach R. J. ( 2009. ). Functional roles of the tegument proteins of herpes simplex virus type 1. Virus Res 145, 173–186. 10.1016/j.virusres.2009.07.007 [DOI] [PubMed] [Google Scholar]
- Kotsakis A., Pomeranz L. E., Blouin A., Blaho J. A. ( 2001. ). Microtubule reorganization during herpes simplex virus type 1 infection facilitates the nuclear localization of VP22, a major virion tegument protein. J Virol 75, 8697–8711. 10.1128/JVI.75.18.8697-8711.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai Z., Han I., Zirzow G., Brady R. O., Reiser J. ( 2000. ). Intercellular delivery of a herpes simplex virus VP22 fusion protein from cells infected with lentiviral vectors. Proc Natl Acad Sci U S A 97, 11297–11302. 10.1073/pnas.97.21.11297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell S. C., Davis I. W., Arendall W. B., III, de Bakker P. I. W., Word J. M., Prisant M. G., Richardson J. S., Richardson D. C. ( 2003. ). Structure validation by Calpha geometry: ϕ,ψ and Cbeta deviation. Proteins 50, 437–450. 10.1002/prot.10286 [DOI] [PubMed] [Google Scholar]
- Maringer K., Elliott G. ( 2010. ). Recruitment of herpes simplex virus type 1 immediate-early protein ICP0 to the virus particle. J Virol 84, 4682–4696. 10.1128/JVI.00126-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maringer K., Stylianou J., Elliott G. ( 2012. ). A network of protein interactions around the herpes simplex virus tegument protein VP22. J Virol 86, 12971–12982. 10.1128/JVI.01913-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin A., O’Hare P., McLauchlan J., Elliott G. ( 2002. ). Herpes simplex virus tegument protein VP22 contains overlapping domains for cytoplasmic localization, microtubule interaction, and chromatin binding. J Virol 76, 4961–4970. 10.1128/JVI.76.10.4961-4970.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mouzakitis G., McLauchlan J., Barreca C., Kueltzo L., O’Hare P. ( 2005. ). Characterization of VP22 in herpes simplex virus-infected cells. J Virol 79, 12185–12198. 10.1128/JVI.79.19.12185-12198.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G. N., Skubák P., Lebedev A. A., Pannu N. S., Steiner R. A., Nicholls R. A., Winn M. D., Long F., Vagin A. A. ( 2011. ). refmac5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 67, 355–367. 10.1107/S0907444911001314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Regan K. J., Bucks M. A., Murphy M. A., Wills J. W., Courtney R. J. ( 2007. a). A conserved region of the herpes simplex virus type 1 tegument protein VP22 facilitates interaction with the cytoplasmic tail of glycoprotein E (gE). Virology 358, 192–200. 10.1016/j.virol.2006.08.024 [DOI] [PubMed] [Google Scholar]
- O’Regan K. J., Murphy M. A., Bucks M. A., Wills J. W., Courtney R. J. ( 2007. b). Incorporation of the herpes simplex virus type 1 tegument protein VP22 into the virus particle is independent of interaction with VP16. Virology 369, 263–280. 10.1016/j.virol.2007.07.020 [DOI] [PubMed] [Google Scholar]
- O’Regan K. J., Brignati M. J., Murphy M. A., Bucks M. A., Courtney R. J. ( 2010. ). Virion incorporation of the herpes simplex virus type 1 tegument protein VP22 is facilitated by trans-Golgi network localization and is independent of interaction with glycoprotein E. Virology 405, 176–192. 10.1016/j.virol.2010.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z., Minor W. ( 1997. ). Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276, 307–326. 10.1016/S0076-6879(97)76066-X [DOI] [PubMed] [Google Scholar]
- Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. ( 2004. ). UCSF Chimera – a visualization system for exploratory research and analysis. J Comput Chem 25, 1605–1612. 10.1002/jcc.20084 [DOI] [PubMed] [Google Scholar]
- Potel C., Elliott G. ( 2005. ). Phosphorylation of the herpes simplex virus tegument protein VP22 has no effect on incorporation of VP22 into the virus but is involved in optimal expression and virion packaging of ICP0. J Virol 79, 14057–14068. 10.1128/JVI.79.22.14057-14068.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen R., Sathish N., Li Y., Yuan Y. ( 2008. ). Virion-wide protein interactions of Kaposi’s sarcoma-associated herpesvirus. J Virol 82, 4742–4750. 10.1128/JVI.02745-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sathish N., Wang X., Yuan Y. ( 2012. ). Tegument proteins of Kaposi’s sarcoma-associated herpesvirus and related gamma-herpesviruses. Front Microbiol 3, 98. 10.3389/fmicb.2012.00098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sciortino M. T., Taddeo B., Poon A. P., Mastino A., Roizman B. ( 2002. ). Of the three tegument proteins that package mRNA in herpes simplex virions, one (VP22) transports the mRNA to uninfected cells for expression prior to viral infection. Proc Natl Acad Sci U S A 99, 8318–8323. 10.1073/pnas.122231699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M. J., Hwang S., Wong W. H., Wu T.-T., Lee S., Liao H.-I., Sun R. ( 2005. ). Identification of viral genes essential for replication of murine gamma-herpesvirus 68 using signature-tagged mutagenesis. Proc Natl Acad Sci U S A 102, 3805–3810. 10.1073/pnas.0404521102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stylianou J., Maringer K., Cook R., Bernard E., Elliott G. ( 2009. ). Virion incorporation of the herpes simplex virus type 1 tegument protein VP22 occurs via glycoprotein E-specific recruitment to the late secretory pathway. J Virol 83, 5204–5218. 10.1128/JVI.00069-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka M., Kato A., Satoh Y., Ide T., Sagou K., Kimura K., Hasegawa H., Kawaguchi Y. ( 2012. ). Herpes simplex virus 1 VP22 regulates translocation of multiple viral and cellular proteins and promotes neurovirulence. J Virol 86, 5264–5277. 10.1128/JVI.06913-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetz P., Dong Y. A., Zeretzke C., Atzler C., Baiker A., Berger B., Rajagopala S. V., Roupelieva M., Rose D., et al. ( 2006. ). Herpesviral protein networks and their interaction with the human proteome. Science 311, 239–242. 10.1126/science.1116804 [DOI] [PubMed] [Google Scholar]
- Vaguine A. A., Richelle J., Wodak S. J. ( 1999. ). sfcheck: a unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr D Biol Crystallogr 55, 191–205. 10.1107/S0907444998006684 [DOI] [PubMed] [Google Scholar]
- van Leeuwen H., Elliott G., O’Hare P. ( 2002. ). Evidence of a role for nonmuscle myosin II in herpes simplex virus type 1 egress. J Virol 76, 3471–3481. 10.1128/JVI.76.7.3471-3481.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwen H., Okuwaki M., Hong R., Chakravarti D., Nagata K., O’Hare P. ( 2003. ). Herpes simplex virus type 1 tegument protein VP22 interacts with TAF-I proteins and inhibits nucleosome assembly but not regulation of histone acetylation by INHAT. J Gen Virol 84, 2501–2510. 10.1099/vir.0.19326-0 [DOI] [PubMed] [Google Scholar]
- Wang L., Guo H., Reyes N., Lee S., Bortz E., Guo F., Sun R., Tong L., Deng H. ( 2012. ). Distinct domains in ORF52 tegument protein mediate essential functions in murine gammaherpesvirus 68 virion tegumentation and secondary envelopment. J Virol 86, 1348–1357. 10.1128/JVI.05497-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitley R. J., Roizman B. ( 2001. ). Herpes simplex virus infections. Lancet 357, 1513–1518. 10.1016/S0140-6736(00)04638-9 [DOI] [PubMed] [Google Scholar]
- Yedowitz J. C., Kotsakis A., Schlegel E. F. M., Blaho J. A. ( 2005. ). Nuclear localizations of the herpes simplex virus type 1 tegument proteins VP13/14, vhs, and VP16 precede VP22-dependent microtubule reorganization and VP22 nuclear import. J Virol 79, 4730–4743. 10.1128/JVI.79.8.4730-4743.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Liu L., Wu L., Wang L., Dong C., Li W., Li Q. ( 2010. ). Herpes simplex virus type 1 tegument protein VP22 is capable of modulating the transcription of viral TK and gC genes via interaction with viral ICP0. Biochimie 92, 1024–1030. 10.1016/j.biochi.2010.04.025 [DOI] [PubMed] [Google Scholar]