Abstract
Two-component signal transduction involves phosphoryl transfer between a histidine kinase sensor and a response regulator effector. The nitrate-responsive two-component signal transduction systems in Escherichia coli represent a paradigm for a cross-regulation network, in which the paralogous sensor–response regulator pairs, NarX–NarL and NarQ–NarP, exhibit both cognate (e.g. NarX–NarL) and non-cognate (e.g. NarQ–NarL) interactions to control output. Here, we describe results from bacterial adenylate cyclase two-hybrid (BACTH) analysis to examine sensor dimerization as well as interaction between sensor–response regulator cognate and non-cognate pairs. Although results from BACTH analysis indicated that the NarX and NarQ sensors interact with each other, results from intragenic complementation tests demonstrate that they do not form functional heterodimers. Additionally, intragenic complementation shows that both NarX and NarQ undergo intermolecular autophosphorylation, deviating from the previously reported correlation between DHp (dimerization and histidyl phosphotransfer) domain loop handedness and autophosphorylation mode. Results from BACTH analysis revealed robust interactions for the NarX–NarL, NarQ–NarL and NarQ–NarP pairs but a much weaker interaction for the NarX–NarP pair. This demonstrates that asymmetrical cross-regulation results from differential binding affinities between different sensor–regulator pairs. Finally, results indicate that the NarL effector (DNA-binding) domain inhibits NarX–NarL interaction. Missense substitutions at receiver domain residue Ser-80 enhanced NarX–NarL interaction, apparently by destabilizing the NarL receiver–effector domain interface.
Introduction
Anaerobic metabolism in Escherichia coli is regulated in part by dual nitrate- and nitrite-responsive two-component signal transduction systems, comprised of the sensor–response regulator pairs NarX–NarL and NarQ–NarP. Signal transmission within each cognate pair proceeds via histidyl-aspartyl phosphotransfer from the sensor transmitter module to the response regulator receiver domain, resulting in target operon activation or repression. The sensor and regulator components share substantial similarities in structure and function, and interact closely in a global cross-regulation network that differentially controls target operon expression in response to nitrate and nitrite (Stewart & Rabin, 1995).
The paralogous response regulators NarL and NarP share co-linear amino acid sequences of about 215 residues, in which the amino-terminal receiver domain (120 residues) is connected via a linker to the carboxyl-terminal GerE family helix–turn–helix DNA-binding domain (50 residues). The sequences for both domains share 48 % identity, whereas the linker sequences are divergent. Receiver phosphorylation by the sensors results in effector domain dimerization, enhancing affinity for DNA binding (Lin & Stewart, 2010; Maris et al., 2002). The two proteins recognize dissimilar DNA-binding site architectures (Darwin et al., 1997), contributing to differential target operon control.
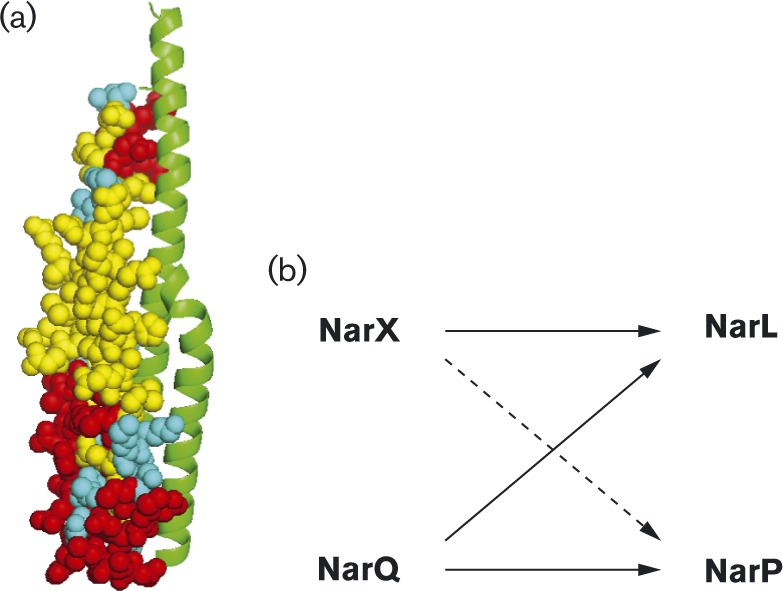
The paralogous sensors NarX (598 residues) and NarQ (566 residues) likewise share identical architecture and about 30 % overall sequence identity (Stewart, 2003). Each contains two transmembrane helices that delimit a periplasmic dimer of symmetrical four-helix bundles, with the nitrate-binding site at the dimer interface (Cheung & Hendrickson, 2009). The cytoplasmic portions include a dimeric four-helix bundle HAMP domain (histidine kinases, adenylyl cyclases, methyl-accepting cyclases and certain phosphatases) that converts input signal to output response (Parkinson, 2010); a GAF-like domain (cGMP-specific phosphodiesterases, certain adenylate cyclases and F hlA) of unknown function (Heikaus et al., 2009); and a transmitter module comprising an amino-terminal DHp (dimerization and histidyl phosphotransfer) domain connected to a carboxyl-terminal CA (catalytic and ATP-binding) domain. The DHp domain is a homodimeric bundle of two antiparallel helix–turn–helix subunits (Gao & Stock, 2009) (Fig. 1a), in which the α1 helices contain most of the interaction and specificity determinants for binding the cognate response regulator receiver domain (Podgornaia & Laub, 2013). The NarX and NarQ sensors both are classified in the HPK7 transmitter subfamily (Grebe & Stock, 1999), which corresponds to the Pfam database HisKA_3 DHp domain subfamily (Punta et al., 2012).
Fig. 1. (a) Model of the NarX DHp domain homodimer. One monomer is shown as a ribbon diagram, the other in spheres. Residues identical and similar between NarX and NarQ are shown in yellow and cyan, respectively. Dissimilar residues, shown in red, provide specificity determinants both for homodimer formation and for response regulator interaction. (b) Interactions in the NarX–NarL and NarQ–NarP cross regulation network. The dashed arrow represents relatively low phosphoryl-transfer rates between NarX and NarP. Adapted from Noriega et al. (2010).
Due to their multiple interactions with functional consequences on gene expression, the NarX–NarL and NarQ–NarP systems are an excellent model for studying cross-regulation in two-component signalling (Laub & Goulian, 2007). We set out to characterize intra- and intermolecular interactions between these cognate pairs, in order to gain insights into specificity and cross-regulatory determinants. For this purpose, we employed the BACTH method (bacterial adenylate cyclase two-hybrid) (Karimova et al., 1998), a widely used genetic approach for monitoring protein–protein interactions (Stynen et al., 2012). This assay reconstitutes Bordetella pertussis CyaA activity from trypsin-defined amino-terminal and carboxyl-terminal subdomains of 25 and 18 kDa, respectively (Guo et al., 2005). Interaction between tbl25 and tbl18 hybrid proteins generates cAMP, which activates lacZYA operon expression in an E. coli Δcya host strain lacking endogenous adenylate cyclase activity (Brickman et al., 1973). The BACTH method is particularly well-suited for analysing cytoplasmic membrane proteins (Karimova et al., 2005), because the two-hybrid interaction does not have to take place near the transcription complex as is required for most other interaction assays, including the yeast two-hybrid method (Stynen et al., 2012).
The magnitude of IPTG-induced lacZYA operon expression depends upon cAMP concentration over the nonsaturating range (Wanner et al., 1978; You et al., 2013). Thus, LacZ activity in the BACTH assay can be taken as a measure of protein interaction strength (Karimova et al., 2005). Indeed, BACTH output levels correlate with results from surface plasmon resonance analysis of interaction between VirB components (Sivanesan et al., 2010). On the other hand, BACTH measurements are conducted with stationary phase cultures and thus reflect long-term cAMP accumulation, report interactions between hybrid proteins expressed from multicopy plasmids and depend on protein stability and conformation.
Although the NarX and NarQ sensors independently detect nitrate and phosphorylate the NarL and NarP response regulators, they exhibit different specificity for their signal ligands and target proteins. NarX is specialized, responding preferentially to nitrate and displaying kinetic preference for NarL, whereas NarQ is generalized, responding to both nitrate and nitrite and phosphorylating both response regulators with similar kinetics (Noriega et al., 2010; Williams & Stewart, 1997b) (Fig. 1b).
Here, we describe experimental results demonstrating that NarX and NarQ do not form functional heterodimers, that relative affinity provides the basis for observed sensor–response regulator kinetic preferences and that the NarL DNA-binding domain modulates the cognate interaction between NarX and NarL.
Methods
Molecular cloning
Full-length and truncated nar alleles were amplified by PCR with oligonucleotide primers containing restriction endonuclease sites for subsequent cloning into plasmid vectors (Stewart & Chen, 2010). Oligonucleotide-directed site-specific mutagenesis also was performed by PCR as described previously (Stewart & Chen, 2010). After cloning, amplified regions were subjected to chain-termination DNA sequence analysis, performed by the UC Davis College of Biological Sciences DNA sequencing facility, to ensure that no spurious alterations were introduced.
Functionality of narX and narQ constructs was tested by complementation of the narX narQ double null alleles in the Φ(narG-lacZ) host strain VJS11210, whereas functionality of narL and narP constructs was tested by complementation of the narL narP double null alleles in the Φ(napF Hi -lacZ) host strain VJS9284 (Table 1), as described previously (Lin & Stewart, 2010; Williams & Stewart, 1997a).
Table 1. Strains, plasmids and oligonucleotide primers.
| Element | Genotype or sequence | Reference |
| Escherichia coli K-12 strains | ||
| VJS632 | F – λ– prototroph; isolate of strain W1485 F – | Stewart & Parales (1988) |
| VJS676 | As VJS 632; Δ(argF–lac)U169 | Stewart & Parales (1988) |
| Derivatives of VJS632 | ||
| VJS711 | Δcya-854 | Stewart et al. (2009) |
| VJS10304 | Δcya-854 ΔnarX263 ΔnarQ264 | Huynh et al. (2013) |
| VJS11768 | Δcya-854 ΔnarL261 ΔnarP262 ΔnarX263 ΔnarQ264 | This study |
| Derivatives of VJS676 | ||
| VJS3041 | λΦ(narG–lacZ) ΔnarX242 narQ251::Tn10d(Tc) | Rabin & Stewart (1993) |
| VJS9284 | λΦ(napF Hi –lacZ) ΔnarL261 ΔnarP262 | Lin & Stewart (2010) |
| VJS11210 | λΦ(narG–lacZ) ΔnarX263 narQ251::Tn10d(Tc) pcnB1 zad-981::Tn10d(Km) | This study |
| Plasmid cloning vectors: | ||
| With ori pMB1 | ||
| pHG165 | Apr; lacZα pUC8 polylinker | Stewart et al. (1986) |
| pHG329 | Apr; lacZα pUC19 polylinker | Stewart et al. (1986) |
| pUT18 | Apr; CyaA codons 225–399 for N-terminal tbl18 fusion | Karimova et al. (1998) |
| pUT18C | Apr; CyaA codons 225–399 for C-terminal tbl18 fusion | Karimova et al. (1998) |
| pVJS5317 | As pHG329; CyaA codons 225–399 between HindIII and AatII sites for C-terminal tbl18 fusion | This study |
| With ori p15A | ||
| pSU18 | Cmr; lacZα pUC18 polylinker | Bartolomé et al. (1991) |
| pKNT25 | Kmr; CyaA codons 1–224 for N-terminal tbl25 fusion | Karimova et al. (1998) |
| pKT25 | Kmr; CyaA codons 1–224 for C-terminal tbl25 fusion | Karimova et al. (1998) |
| Plasmid constructs for BACTH analysis: | ||
| With ori pMB1 | ||
| pUT18C-zip | As pUT18C; Zip–T18 | Karimova et al. (1998) |
| pVJS5319 | As pVJS5317; NarX–T18 (NarX codons 1–598) | This study |
| pVJS5364 | As pVJS5317; NarQ–T18 (NarQ codons 1–566) | This study |
| pVJS5432 | As pVJS5317; NarL–T18 (NarL codons 1–216) | This study |
| pVJS5751 | As pUT18; NarLREC–T18 (NarL codons 1–142) | This study |
| pVJS5752 | As pUT18C; tbl18–NarL (NarL codons 1–216) | This study |
| pVJS5753 | As pUT18C; tbl18–NarLREC (NarL codons 1–142) | This study |
| pVJS5769 | As pUT18C; tbl18–NarP (NarP codons 1–215) | This study |
| pVJS5770 | As pUT18C; tbl18–NarPREC (NarP codons 1–142) | This study |
| With ori p15A | ||
| pKT25-zip | As pKT25; tbl25-Zip | Karimova et al. (1998) |
| pVJS5321 | As pKNT25; NarX–T25 (NarX codons 1–598) | This study |
| pVJS5365 | As pKNT25; NarQ–T25 (NarQ codons 1–566) | This study |
| pVJS5433 | As pKNT25; NarL–T25 (NarL codons 1–216) | This study |
| Plasmid constructs for intragenic complementation: | ||
| With ori pMB1 | ||
| pVJS2474 | As pHG165; narX + between EcoRI and HindIII sites | Stewart et al. (2003) |
| pVJS5501 | As pHG165; narX (H399Q) DHp– | This study |
| pVJS5502 | As pHG165; narX (H513Q) CA– | This study |
| pVJS5511 | As pHG165; narQ + between HindIII and EcoRI sites | This study |
| pVJS5512 | As pHG165; narQ (H370Q) DHp– | This study |
| pVJS5513 | As pHG165; narQ (H484Q) CA– | This study |
| With ori p15A | ||
| pVJS5503 | As pSU18; narX + between EcoRI and HindIII sites | This study |
| pVJS5504 | As pSU18; narX (H399Q) DHp– | This study |
| pVJS5505 | As pSU18; narX (H513Q) CA– | This study |
| pVJS5508 | As pSU18; narQ + between HindIII and EcoRI sites | This study |
| pVJS5509 | As pSU18; narQ (H370Q) DHp– | This study |
| pVJS5510 | As pSU18; narQ (H484Q) CA– | This study |
| Oligonucleotide primers* | ||
| LLC2907 | 5′-gggaagcttgATGCTTAAACGTTGTCTCTCTC | This study |
| LLC2908 | 5′-ggaattcgACTCATGGGTATCTCCTTGGACGTCTGTGA | This study |
| LLC2914 | 5′-gggaagcttgGTGATTGTTAAACGACCCGTCTCGGCCAGTCTG | This study |
| LLC2915 | 5′-ggaattcgACATTAACTGACTTTCCTCACCCTC | This study |
| LLC2963 | 5′-gggaagcttgATGAGTAATCAGGAACCGGCTAC | This study |
| LLC2965 | 5′-ggaattcgAGAAAATGCGCTCCTGATGCACCCATAC | This study |
| TNH3469 | 5′-gggtctagagATGAGTAATCAGGAACCGGCTAC | This study |
| TNH3470 | 5′-ggaattcgaCAAGCTGGCGGCCAGAACAGGCGTTAAT | This study |
| TNH3471 | 5′-accgagctcgaattcaATGAGTAATCAGGAACCGGCTACTATC | This study |
| TNH3472 | 5′-atatcgatgaattcgaGAAAATGCGCTCCTGATGCACC | This study |
| TNH3475 | 5′-atatcgatgaattcgaCAAGCTGGCGGCCAGAACAGGCGTTAAT | This study |
| TNH3485 | 5′-accgagctcgaattcaATGCCTGAAGCAACACCTTTTCAGGTG | This study |
| TNH3486 | 5′-atatcgatgaattcgaTTGTGCCCCGCGTTGTTGCAGG | This study |
| TNH3487 | 5′-atatcgatgaattcgaTTCACGTAAGTACTGATTGACGCGTTCG | This study |
* Lower-case, added sequence; upper-case, native sequence.
BACTH assay: strains and plasmids
Host strains for BACTH assays (Table 1) carry the Δcya-854 allele (Brickman et al., 1973), and are derivatives of strain VJS623, a W1485 F – isolate (Stewart & Parales, 1988). Strains were constructed by bacteriophage P1-mediated generalized transduction (Miller, 1972), and carry our laboratory's standard null alleles for nar regulatory genes (Lin & Stewart, 2010).
The BACTH assay employs moderate copy number p15A replicon plasmids (Chang & Cohen, 1978) for making tbl25 hybrids (plasmids pKNT25 and pKT25), and high copy number pMB9 replicon plasmids (Lin-Chao et al., 1992) for making tbl18 hybrids (plasmids pUT18 and pUT18C) (Table 1). The two replicons, both of the ColE1 type, are fully compatible (Chang & Cohen, 1978). These plasmids were purchased from Euromedex. In order to avoid potential artefacts resulting from high-level expression of NarX–T18 and NarQ–T18 hybrid proteins (Gueguen et al., 2011), we cloned the tbl18-encoding fragment into an intermediate copy number pMB9 replicon vector (Stewart et al., 1986), creating plasmid pVJS5317.
To construct NarX–CyaA and NarQ–CyaA hybrids, the narX and narQ genes were amplified by PCR and cloned as HindIII–EcoRI fragments at the 5′-ends of the tbl18- and tbl25-encoding fragments in plasmids pVJS5317 and pKNT25, respectively. Oligonucleotide primers for narX were LLC2907 (HindIII) and LLC2908 (EcoRI), and those for narQ were LLC2914 (HindIII) and LLC2915 (EcoRI) (Table 1). The upstream (HindIII) primers were designed to place the narX and narQ initiation codons in-frame in the cyaA polylinker sequence, which lies just downstream of the native lacZ gene transcription and translation initiation signals. Accordingly, the nar-cyaA hybrid genes encode an amino-terminal octapeptide (MTMITPSL) at the amino-termini of the native NarX and NarQ sequences. The downstream (EcoRI) primers fuse the last codon of the narX and narQ sequences in-frame to the polylinker sequence upstream of the cyaA initiation codon.
An analogous strategy was used to construct the NarL–T18 and NarL–T25 hybrids, employing oligonucleotide primers LLC2963 (HindIII) and LLC 2965 (EcoRI) (Table 1). These encode the same amino-terminal octapeptide sequence as described above.
The NarLREC–T18 hybrid was constructed by cloning PCR-amplified narL sequence spanning codons 1–142 as an XbaI–EcoRI fragment into plasmid pUT18. Oligonucleotide primers were TNH3469 (XbaI) and TNH3470 (EcoRI) (Table 1). The resulting narL REC–cyaA hybrid gene encodes an amino-terminal hexadecapeptide (MTMITPSLHACRSTLE) derived from the polylinker sequence.
To construct CyaA–NarL and CyaA–NarP hybrids, the narL and narP genes were amplified by PCR, and cloned as SacI–ClaI fragments at the 3′-end of the tbl18-encoding fragment in the high copy number plasmid pUT18C in order to mimic the relatively higher cellular concentration of response regulators relative to sensors (Ishihama et al., 2014; Wayne et al., 2010). Oligonucleotide primers were designed to place the nar sequence in-frame with the cyaA and polylinker sequence, and to utilize the ochre (TAA) codon just downstream from the ClaI site in plasmid pUT18C. Hence, the resulting nar–cyaA hybrid genes encode a carboxyl-terminal pentapeptide (SNSSI). The upstream (SacI) primers were TNH3471 and TNH3485 for narL–cyaA and narP–cyaA hybrids, respectively (Table 1). The downstream (ClaI) primers were TNH3472 and TNH3475 for narL–cyaA and narL REC–cyaA hybrids, respectively, and TNH3486 and TNH3487 for narP–cyaA and narP REC–cyaA hybrids, respectively (Table 1).
Intragenic complementation assay: strains and plasmids
The host strain VJS11210 (Table 1) carries the nitrate-inducible Φ(narG–lacZ) gene fusion in monocopy at the bacteriophage λ attachment site, null alleles for both narX and narQ, and the pcnB1 mutation that reduces ColE1-type plasmid copy number to approximately one per cell (Williams & Stewart, 1997b). WT and mutant narX and narQ genes are cloned into the compatible medium copy number vectors pHG165 (Stewart et al., 1986) and pSU18 (Bartolomé et al., 1991).
Culture media and conditions
Defined medium to grow cultures for enzyme assays was buffered with MOPS as described previously (Stewart & Parales, 1988). Glucose was added at 80 mM. NaNO3 (40 mM) was added as indicated. IPTG was included at 0.5 mM in cultures grown for BACTH assays. TY broth contained tryptone (8 g L–1), yeast extract (5 g L–1) and NaCl (5 g L–1).
Plasmid-containing strains were cultured in medium prepared by mixing equal volumes of defined (MOPS) and complex (TY) media. This medium produces a favourable combination of rapid growth and robust response to nitrate (10). Antibiotics were added as appropriate to maintain selection for plasmid-bearing cells: ampicillin (Ap; 100 μg ml–1), chloramphenicol (Cm; 25 μg ml–1) and kanamycin (Km; 50 μg ml–1).
Culture densities were monitored with a Klett–Summerson photoelectric colorimeter (Klett Manufacturing Co.) equipped with a number 66 (red) filter. Anaerobic cultures were grown in filled screw-cap tubes as described previously (Stewart & Parales, 1988). Cultures to monitor Φ(narG–lacZ) and Φ(napF Hi–lacZ) expression were grown at 37 °C to the early exponential phase, about 25–35 Klett units. In comparing different growth phases and temperatures, we obtained the most reproducible BACTH results from cultures grown overnight (at least 16 h) at 30 °C, as recommended (Karimova et al., 1998).
Enzyme assay
β-Galactosidase activities were measured in CHCl3-SDS-permeabilized cells (Miller, 1972). Assays were performed at room temperature, approximately 21 °C. Reported values for BACTH assays are mean values of at least three independent cultures grown on different days, whereas those for Φ(narG–lacZ) and Φ(napF Hi–lacZ) assays are mean values of at least two independent cultures grown on different days. Each culture was assayed in duplicate, with one assay reaction containing twice as much cell extract as the other.
Specific activities are calculated in arbitrary (Miller) units (Miller, 1972). Graphical data are normalized relative to the value obtained in the same experiment for the tbl25-Zip and Zip-T18 control hybrids, typically about 3500 Miller units.
DHp domain model
The NarX DHp domain model (Fig. 1a) was constructed in swiss-model (Bordoli et al., 2009), and is based on the X-ray structure of the homologous DesK sensor (Albanesi et al., 2009), PDB accession number 3EHJ.
Results and Discussion
NarX and NarQ homomeric interaction
We first evaluated the properties of NarX–CyaA and NarQ–CyaA hybrid proteins. The sensor amino-terminus is close to the cytoplasmic membrane and the dimer interface, so we fused the CyaA tbl25 and tbl18 subdomains to the carboxyl-terminus. Hybrid proteins were expressed from the native lacZ gene transcription and translation initiation signals, and were encoded on medium copy number plasmids for balanced gene dosage. Note that other transmembrane sensors may require different fusion points. For example, DcuS–CyaA hybrids conferred toxicity to the host strain, whereas CyaA–DcuS hybrids were stable and functional (Scheu et al., 2012).
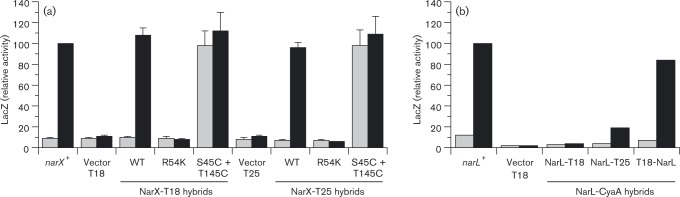
In order to determine if the fusion proteins retain normal sensor activity, we measured Φ(narG–lacZ) induction in response to nitrate. Results for both the NarX–T25 and NarX–T18 were indistinguishable from those for native NarX (Fig. 2a). Analogous results were obtained for the corresponding NarQ–CyaA hybrids (not shown). We additionally examined two different mutational alterations, both within the transmembrane signalling module, that affect NarX nitrate response. The R54K substitution at the nitrate-binding site results in the uninducible phenotype (Williams & Stewart, 1997b), whereas the dual S43C plus tbl145C substitutions result in the constitutive phenotype (T. N. Huynh and V. Stewart, unpublished). Overall, these tests confirm that the hybrid proteins were stable and functional.
Fig. 2. Functionality of NarX–CyaA and NarL–CyaA hybrid proteins. (a) Sensor function of NarX–CyaA hybrids. NarX+, vector, NarX–T18 and NarX–T25 plasmids were introduced into strain VJS3041 [Φ(narG–lacZ) ΔnarX ΔnarQ] to monitor target operon activation. Cultures were grown in the absence or presence of nitrate (grey bars and black bars, respectively). Relative LacZ activity of 100 corresponds to 1890 Miller units. Controls: narX +, WT narX gene on plasmid pVJS2474; tbl18 vector, plasmid pVJS5317; tbl25 vector, plasmid pKNT25.Error bars show sd of mean values determined from four independent assays. (b) Transcription activation function of NarL–CyaA hybrids. NarL+, vector, NarL–T18, NarL–T25 and tbl18–NarL plasmids were introduced into strain VJS9284 [Φ(napF Hi–lacZ)260 ΔnarL ΔnarP]. Cultures were grown in the absence or presence of nitrate (grey bars and black bars, respectively). Relative LacZ activity of 100 corresponds to 970 Miller units. Controls: narL +, WT narL gene on plasmid pVJS4781; vector, plasmid pUT18. Results were mean values from two independent experiments, so sd values were not calculated.
BACTH assay functionality is documented by the tbl25–Zip and Zip–T18 hybrids, in which leucine zipper elements form dimeric coiled-coils to mediate strong CyaA subdomain interaction (Karimova et al., 1998). The NarX–T25 plus NarX–T18 hybrids generated a similarly high level of BACTH output, as did the NarQ–T25 plus NarQ–T18 hybrids (Fig. 3). Thus, both NarX and NarQ displayed robust homodimeric interaction, as anticipated.
Fig. 3. BACTH output for NarX–CyaA and NarQ–CyaA hybrid pairs. NarX and NarQ sequences carry the tbl25 and tbl18 domains fused to the carboxyl teminus. Tests include self interactions between NarX fusions (grey bars) and NarQ fusions (black bars), and mixed interactions between NarX and NarQ fusions (open hatched bars). Cultures were grown in the absence of nitrate. Interactions were tested in strain VJS10304 (Δcya ΔnarX ΔnarQ). Controls (open bars): vectors, empty tbl25 and tbl18 vectors; Zip, tbl25-Zip and Zip-T18. Error bars show the sd of mean values determined from three independent assays.
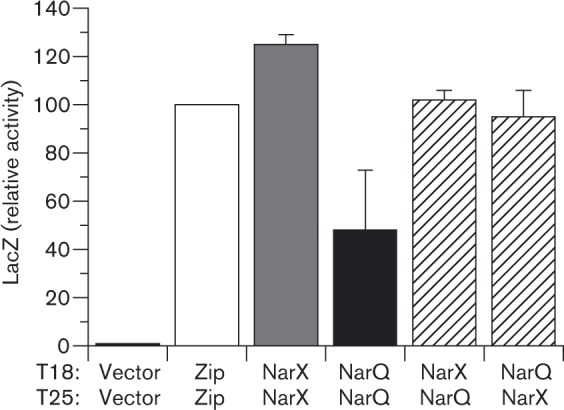
NarX and NarQ heteromeric interaction
We observed high-level BACTH output in cultures co-expressing either NarX–T25 plus NarQ–T18 or NarX–T18 plus NarQ–T25 hybrid proteins (Fig. 3). This result is consistent with two distinct hypotheses (Goodman et al., 2009): (A) sensor monomers associate to form heterodimers and (B) sensor homodimers associate to form hetero-oligomers.
We conducted intragenic complementation experiments in order to distinguish these hypotheses. Intragenic complementation can occur for multidomain gene products when two alleles inactivate different domains (Yanofsky et al., 1971). Yang & Inouye (1991) applied this logic to the sensor transmitter domain, demonstrating intragenic complementation between alleles that inactivate either the DHp or the CA domain. For the narX and narQ genes, we used missense substitutions that remove the DHp domain phospho-accepting His residue (residues His-399 and His-370 in NarX and NarQ, respectively) together with substitutions that abolish CA domain function (residues His-513 and His-484, respectively) (Huynh et al., 2010). Allele pairs were expressed from the compatible plasmids pHG165 and pSU18 (Table 1), and sensor function was examined in a Φ(narG–lacZ) ΔnarX ΔnarQ double null strain background.
Pairs that result in homodimer formation (narX–narX or narQ–narQ) yielded the expected results, as substitutions in the different domains complemented each other (Table 2; lines 11–12 and 22–23). Contrariwise, for pairs that would result in heterodimer formation (narX–narQ), the different substitutions failed to complement (Table 3; lines 15–18). Although complemented expression did not reach the WT level in some instances, the large Φ(narG–lacZ) induction ratio allows even these cases to be clearly distinguished from non-complementation (e.g. Table 2 lines 9–12).
Table 2. Intragenic complementation test for NarX and NarQ homodimers.
| Allele on plasmid* | LacZ activity† | |||
| Line | pSU18 | pHG165 | − Nitrate | + Nitrate |
| 1 | Vector only | Vector only | 12 | 13 |
| 2 | narX+ | Vector only | 55 | 2270 |
| 3 | Vector only | narX+ | 34 | 2900 |
| 4 | narX+ | narX+ | 30 | 3120 |
| 5 | narX+ | narX DHp– | 30 | 2890 |
| 6 | narX DHp– | narX+ | 30 | 2780 |
| 7 | narX+ | narX CA– | 22 | 2610 |
| narX CA– | narX+ | 19 | 1990 | |
| 9 | narX CA– | narX CA– | 18 | 23 |
| 10 | narX DHp– | narX DHp– | 20 | 23 |
| 11 | narX CA– | narX DHp– | 20 | 820 |
| 12 | narX DHp– | narX CA– | 24 | 1290 |
| 13 | narQ+ | Vector only | 180 | 2080 |
| 14 | Vector only | narQ+ | 140 | 1670 |
| 15 | narQ+ | narQ+ | 150 | 1590 |
| 16 | narQ+ | narQ DHp– | 180 | 1680 |
| 17 | narQ DHp– | narQ+ | 160 | 1660 |
| 18 | narQ+ | narQ CA– | 130 | 1450 |
| 19 | narQ CA– | narQ+ | 83 | 1400 |
| 20 | narQ CA– | narQ CA– | 14 | 14 |
| 21 | narQ DHp– | narQ DHp– | 13 | 14 |
| 22 | narQ CA– | narQ DHp– | 40 | 1490 |
| 23 | narQ DHp– | narQ CA– | 100 | 1300 |
* Indicates allele-plasmid combination in strain VJS11210 {Φ(narG–lacZ) ΔnarX263 narQ251::Tn10 narL + pcnB1}. Missense substitutions corresponding to the DHp– and CA– phenotypes are shown in Table 1.
† Activities in Miller units. Strains cultured anaerobically to the mid-exponential phase in enriched medium (1 : 1 mixture of MOPS-glucose plus TY) containing Ap and Cm. Nitrate added as indicated at 40 mM. Values are mean values of two independent experiments.
Table 3. Intragenic complementation test for NarX–NarQ heterodimers.
| Allele on plasmid* | LacZ activity† | |||
| Line | pSU18 | pHG165 | − Nitrate | + Nitrate |
| 1 | narX+ | narQ+ | 20 | 690 |
| 2 | narQ+ | narX+ | 29 | 1270 |
| 3 | narX+ | narQ DHp– | 16 | 400 |
| 4 | narQ+ | narX DHp– | 70 | 320 |
| 5 | narX DHp– | narQ+ | 37 | 1220 |
| 6 | narQ DHp– | narX+ | 16 | 450 |
| 7 | narX+ | narQ CA– | 14 | 720 |
| 8 | narQ+ | narX CA– | 17 | 750 |
| 9 | narX CA– | narQ+ | 16 | 310 |
| 10 | narQ CA– | narX+ | 13 | 1590 |
| 11 | narX DHp– | narQ DHp– | 13 | 12 |
| 12 | narQ DHp– | narX DHp– | 12 | 13 |
| 13 | narX CA– | narQ CA– | 11 | 11 |
| 14 | narQ CA– | narX CA– | 11 | 12 |
| 15 | narX DHp– | narQ CA– | 11 | 8 |
| 16 | narQ CA– | narX DHp– | 11 | 8 |
| 17 | narX CA– | narQ DHp– | 12 | 10 |
| 18 | narQ DHp– | narX CA– | 10 | 9 |
* Indicated allele–plasmid combination in strain VJS11210 {Φ(narG–lacZ) ΔnarX263 narQ251::Tn10 narL + pcnB1}. Missense substitutions corresponding to the DHp– and CA– phenotypes are shown in Table 1.
† Activities in Miller units. Strains cultured anaerobically to the mid-exponential phase in enriched medium (1 : 1 mixture of MOPS-glucose plus TY) containing Ap and Cm. Nitrate added as indicated at 40 mM. Values are mean values of two independent experiments.
These results provide clear evidence that, despite their sequence similarity, NarX and NarQ do not form functional heterodimers. Prior studies with the paralogous EnvZ and RstB revealed an exquisite specificity of these sensors to form homodimers, and showed that many homodimer specificity determinants reside in the membrane-distal base of the DHp domain (Laub & Goulian, 2007). Indeed, this region displays the highest number of dissimilar residues between the NarX and NarQ DHp sequences (Fig. 1a). Moreover, there likely are other specificity determinants to inhibit formation of NarX–NarQ heterodimers, including the dimeric HAMP domain. Finally, as discussed previously (Williams & Stewart, 1997a), there are no existing data to suggest a functional role for NarX–NarQ heterodimers.
Therefore, the two-hybrid interaction between NarX and NarQ (Fig. 3) results from close proximity of NarX2 and NarQ2 homodimers. It is possible that they form heterologous clusters, as shown for the paralogous CitA and DcuS sensors (Scheu et al., 2008). Heteromeric chemoreceptor clusters have documented roles in signalling (Hazelbauer et al., 2008). However, the physiological function for heteromeric sensor clusters remains obscure (Scheu et al., 2012). Perhaps the association revealed by BACTH analysis is instead an artefact resulting from artificially high expression levels.
NarX and NarQ sensors undergo intermolecular autophosphorylation
The transmitter autokinase reaction initially was thought to occur exclusively by transfer of γ-phosphate from the ATP-bound CA domain of one subunit to the DHp domain of the opposite subunit in a dimer. This mechanism is demonstrable in vitro for NtrB (Ninfa et al., 1993), CheA (Swanson et al., 1993) and EnvZ (Cai & Inouye, 2003) among others. However, intramolecular phosphate transfer within a subunit was later observed for PhoR, HK853 (Casino et al., 2009) and ArcB (Peña-Sandoval & Georgellis, 2010). Subunit exchange analysis of EnvZ and PhoR orthologues suggests that the autophosphorylation mode is determined by the handedness of the loop connecting DHp helices α1 and α2 (Ashenberg et al., 2013). In particular, sensors with right- and left-handed loops are predicted to undergo intermolecular and intramolecular autophosphorylation, respectively.
The Bacillus subtilis thermosensor DesK appears to deviate from this rule. Structural analysis and cross-linking experiments document intermolecular sensor autophosphorylation (Albanesi et al., 2009; Trajtenberg et al., 2010), even though its DHp domain loop is left-handed (Ashenberg et al., 2013). However, unlike the HisKA (HPK1-4) family DHp domains described above, the DesK DHp domain is in the distinct HisKA_3 (HPK7) family together with NarX and NarQ (Grebe & Stock, 1999). Indeed, the three DHp domain sequences, including the loop regions, are co-linear.
Intragenic complementation results described above for both NarX and NarQ homodimers are compatible only with the intermolecular autophosphorylation mechanism (Yang & Inouye, 1991), in accordance with the structural and biochemical data for DesK (Albanesi et al., 2009; Trajtenberg et al., 2010). This highlights the substantial differences in detail for reaction mechanisms employed by the two transmitter sequence families (Huynh et al., 2010).
Sensor–response regulator interactions
Satisfied that we understood the behaviour and properties of sensor-CyaA hybrids, we next used BACTH analysis to characterize sensor–response regulator interactions. We made CyaA fusions with both full-length response regulators and their liberated receiver domains. The response regulator carboxyl-terminal NarL–T25 and NarL–T18 fusions both were inactive for Φ(napF–lacZ) induction (Fig. 2b) and for interaction with the corresponding NarX–CyaA hybrids (not shown), whereas the amino-terminal tbl18–NarL fusion was active in both assays (Figs. 2b and 4). Analogous results were obtained with the analogous NarP fusions (not shown). Thus, fusion design is important for monitoring sensor–response regulator BACTH interaction output, as noted also for other systems, such as VraS–VraR (McCallum et al., 2011) and GraS–GraR (Falord et al., 2012).
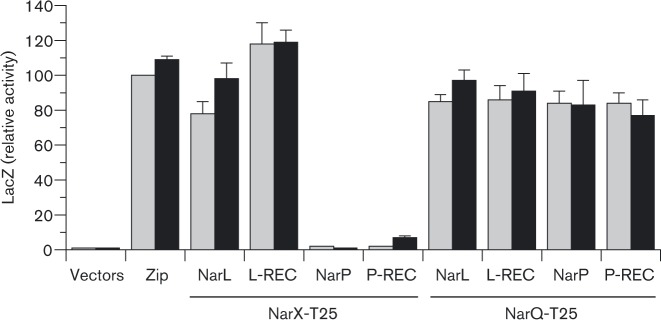
By contrast, both amino-terminal and carboxyl-terminal tbl18 subdomain fusions to the NarL receiver domain (T18–NarLREC and NarLREC–T18, respectively) were active for BACTH interaction output with the NarX–T25 hybrid (Fig. 4 and not shown). Indeed, the tbl18–NarLREC hybrid yielded the highest output values. Based on these results, we chose the tbl18–NarLREC plus NarX–T25 pair for our previously reported study examining interactions with NarX DHp domain missense mutants (Huynh et al., 2013).
Fig. 4. BACTH output for cognate and non-cognate sensor–response regulator pairs. WT NarL and NarP response regulator sequences are full-length (NarL and NarP) or receiver domain only (L-REC and P-REC), and carry the tbl18 domain fused to the amino-terminus. The full-length WT NarX and NarQ sequences carry the T-25 domain fused to the carboxyl teminus. Cultures were grown in the absence or presence of nitrate (grey bars and black bars, respectively). Interactions were tested in strain VJS11768 (Δcya ΔnarL ΔnarP ΔnarX ΔnarQ). Controls: vectors, empty tbl18 and tbl25 vectors; Zip, tbl25-Zip and Zip-T18. Error bars show the sd of mean values determined from three independent assays.
Interactions in the Nar cross-regulation network
Prior studies revealed that the Nar regulatory proteins engage in both cognate (e.g. NarX–NarL) and non-cognate (e.g. NarQ–NarL) interactions to control target operon expression (Noriega et al., 2010; Rabin & Stewart, 1993). This provides a good example of cross-regulation (Laub & Goulian, 2007). However, the interactions are asymmetrical: NarX and NarQ are equivalent substrates for NarL autophosphorylation and similar catalysts for phospho-NarL dephosphorylation. For NarP, by contrast, NarQ is more effective than NarX in both reactions (Noriega et al., 2010; Rabin & Stewart, 1993) (Fig. 1b).
However, these experiments did not resolve the mechanism for differential regulation. The intuitive explanation is that regulatory asymmetry results from differences in direct binding (affinity) between sensor transmitter and response regulator receiver domains. Alternatively, the different pairs might simply exhibit different reaction rates (catalytic efficiency). Distinguishing these hypotheses is important for determining the mechanisms through which sensor–regulator specificity determinants act.
Results from BACTH analysis support the former hypothesis (Fig. 4). The NarQ–T25 hybrid generated high-level BACTH output in combination with either the tbl18–NarL or tbl18–NarP hybrids. By contrast, the NarX–T25 hybrid generated high-level BACTH output only in combination with the tbl18–NarL hybrid. These patterns held for both the full-length and receiver-only versions of the response regulator hybrid proteins. Thus, asymmetrical cross-regulation likely results from differential sensor–regulator interaction affinities.
Note that BACTH output for the sensor–response regulator pairs was not influenced by nitrate (Fig. 4), counter to our initial expectations (Huynh et al., 2013). Upon reflection, however, one recognizes that sensor–regulator docking must occur not only for positive control (phosphotransfer) in the presence of signal, but also for negative control (dephosphorylation) in the absence of signal (Huynh & Stewart, 2011). Both types of interaction are likely to be equally transient.
Differential interactions between NarX and full-length NarL versus the NarL receiver domain
BACTH output for the combination of NarX–T25 plus tbl18–NarL consistently was significantly lower (P < 0.01; Student's t-test) than that for the combination of NarX–T25 plus tbl18–NarLREC (Fig. 4). Since this difference was not observed for combinations with NarQ–T25 (Fig. 4), it is not due to structural interference by the tbl25 or tbl18 subdomains. Apparently, the BACTH output differential between the NarL full-length and receiver hybrid constructs results from inhibition exerted by the DNA-binding effector domain. One idea, elaborated below, is that effector domain binding influences receiver domain conformation to (slightly) decrease its affinity for the cognate DHp domain.
In the X-ray structure, unphosphorylated NarL displays an extensive interaction between the receiver and effector domains, in which the α6 linker helix packs against the effector domain to block access to target DNA (Baikalov et al., 1996). The domain interface comprises residues in the linker helix α6, the α3-β4, α4-β5 and α5-α6 loops in the receiver domain, and the α7–α8 loop in the effector domain. Interdomain contacts are further enhanced by parts of the dimerization helix α10 and the recognition helix α9 (Eldridge et al., 2002). During growth with nitrate, NarL receiver phosphorylation at the invariant residue Asp-59 releases the α6 helix, allowing the effector domain to dimerize along the α10 helix and reorient its recognition helix α9 into the DNA-binding groove (Eldridge et al., 2002; Maris et al., 2002; Zhang et al., 2003). This dimerization is required for high-affinity DNA binding by NarL.
Among several residues involved in NarL interdomain interaction, Ser-80, located in the receiver domain α3-β4 loop, appears to have a key role (Eldridge et al., 2002). This residue likely forms a hydrogen bond with Arg-203 in the dimerization helix α10, and potentially interacts with Lys-192 in the DNA binding helix α9. Tellingly, a previous selection for randomly generated narL constitutive mutants yielded two substitutions for Ser-80, S80L and S80P (Egan & Stewart, 1991). Thus, alterations at Ser-80 may confer the constitutive phenotype by disrupting this inhibitory interdomain interaction.
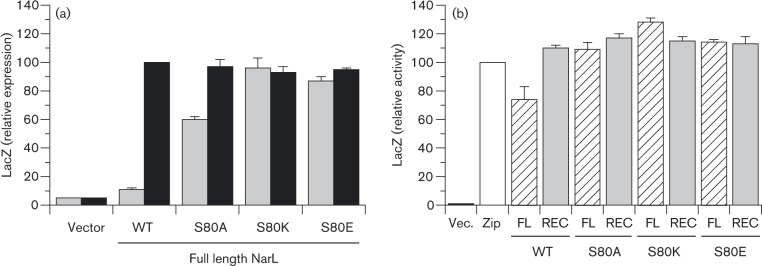
In order to probe the relationship between receiver-effector domain binding and relative BACTH output for NarX interaction, we made three substitutions for NarL Ser-80: Ala, to remove side-chain residues beyond the α carbon; Lys, to substitute positive charge; and Glu, to substitute negative charge. All three substitutions conferred strong constitutive phenotypes, as revealed by elevated levels of uninduced Φ(napF Hi–lacZ) expression (Fig. 5a). This supports the hypothesis, that the S80A, S80K and S80E substitutions converted NarL to a more open conformation competent for transcription activation even in the absence of phosphorylation.
Fig. 5. Effects of NarL Ser-80 substitutions on functionality of tbl18–NarL hybrids and on BACTH output for tbl18–NarL plus NarX–T25 hybrid pairs. (a) tbl18–NarL plasmids were introduced into strain VJS9284 [Φ(napF Hi -lacZ)260 ΔnarL ΔnarP]. Cultures were grown in the absence or presence of nitrate (grey bars and black bars, respectively). Relative LacZ activity of 100 corresponds to 1890 Miller units. Relative LacZ activity of 100 corresponds to 560 Miller units. Controls: narL +, WT narL gene (plasmid pVJS4781); vector, plasmid pHG329. (b) BACTH interaction between tbl18–NarL and NarX–T25 fusion proteins. NarL sequences are full-length (FL; open hatched bars) or receiver domain only (REC; grey bars), and carry the tbl18 domain fused to the amino-terminus. The full-length WT NarX sequence carries the T-25 domain fused to the carboxyl teminus. Cultures were grown in the absence of nitrate. Interactions were tested in strain VJS11768 (Δcya ΔnarX ΔnarQ ΔnarL ΔnarP). Controls (open bars): vectors, empty tbl18 and tbl25 vectors; Zip, tbl25-Zip and Zip-T18. For both panels, error bars show the sd of mean values determined from three independent assays.
Next, we used the BACTH system to test for interactions between the NarX–T25 hybrid and WT and Ser-80 substituted tbl18–NarL hybrids (Fig. 5b). Each of the full-length mutant hybrids generated the higher BACTH output levels equivalent to that of the WT receiver domain hybrid. Thus, missense substitutions designed to abrogate inhibitory interdomain contacts within the response regulator also enhance interaction with the cognate sensor, supporting the overall hypothesis.
Receiver domain inhibition by the effector domain stabilizes the inactive state for diverse response regulators that function as transcriptional factors (Barbieri et al., 2010). This functions to inhibit phosphorylation by metabolites such as acetyl phosphate, without influencing sensor-mediated phosphorylation (Barbieri et al., 2010; Da Re et al., 1999). Indeed, NarX robustly phosphorylates NarL both in vivo and in vitro (Noriega et al., 2010), so the relatively small effect on NarX–NarL interaction implied by the BACTH analysis, if genuine, would not meaningfully influence phosphoryl transfer between this cognate pair.
By contrast to the NarX–NarL pair, BACTH output for the NarQ–NarL pair was not influenced by the presence or absence of the response regulator effector domain (Fig. 4). Unlike NarX, NarQ does not discriminate strongly between the two response regulators (Fig. 1b), implying that the interaction surface is less stringent. If so, then perhaps the NarQ–NarL interaction is less sensitive to the (subtle) conformational changes resulting from receiver–effector domain interaction.
BACTH output for the NarQ–NarP pair likewise was not influenced by the presence or absence of the response regulator effector domain (Fig. 4). This implies that NarP makes different interdomain contacts than NarL, a plausible supposition considering their substantial differences in primary sequence (less than 50 % identity). Indeed, available structures for NarL-type response regulators, those containing a GerE effector domain, reveal nonconserved interdomain packing interactions (Leonard et al., 2013).
Acknowledgements
We thank our colleagues, Drs Alice Lin, Hsia-Yin Lin and Chris Noriega, for helpful advice and support.
This study was supported by the National Institute of General Medical Sciences (Public Health Service grant R01GM036877).
Abbreviations:
- BACTH
bacterial adenylate cyclase two-hybrid
- DHp
dimerization and histidyl phosphotransfer
Footnotes
Edited by: G. Unden
References
- Albanesi D., Martín M., Trajtenberg F., Mansilla M.C., Haouz A., Alzari P.M., de Mendoza D., Buschiazzo A. (2009). Structural plasticity and catalysis regulation of a thermosensor histidine kinase Proc Natl Acad Sci U S A 106 16185–16190 10.1073/pnas.0906699106 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashenberg O., Keating A.E., Laub M.T. (2013). Helix bundle loops determine whether histidine kinases autophosphorylate in cis or in trans J Mol Biol 425 1198–1209 10.1016/j.jmb.2013.01.011 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baikalov I., Schröder I., Kaczor-Grzeskowiak M., Grzeskowiak K., Gunsalus R.P., Dickerson R.E. (1996). Structure of the Escherichia coli response regulator NarL Biochemistry 35 11053–11061 10.1021/bi960919o . [DOI] [PubMed] [Google Scholar]
- Barbieri C.M., Mack T.R., Robinson V.L., Miller M.T., Stock A.M. (2010). Regulation of response regulator autophosphorylation through interdomain contacts J Biol Chem 285 32325–32335 10.1074/jbc.M110.157164 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartolomé B., Jubete Y., Martínez E., de la Cruz F. (1991). Construction and properties of a family of pACYC184-derived cloning vectors compatible with pBR322 and its derivatives Gene 102 75–78 10.1016/0378-1119(91)90541-I . [DOI] [PubMed] [Google Scholar]
- Bordoli L., Kiefer F., Arnold K., Benkert P., Battey J., Schwede T. (2009). Protein structure homology modeling using swiss-model workspace Nat Protoc 4 1–13 10.1038/nprot.2008.197 . [DOI] [PubMed] [Google Scholar]
- Brickman E., Soll L., Beckwith J. (1973). Genetic characterization of mutations which affect catabolite-sensitive operons in Escherichia coli, including deletions of the gene for adenyl cyclase J Bacteriol 116 582–587 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S.J., Inouye M. (2003). Spontaneous subunit exchange and biochemical evidence for trans-autophosphorylation in a dimer of Escherichia coli histidine kinase (EnvZ) J Mol Biol 329 495–503 10.1016/S0022-2836(03)00446-7 . [DOI] [PubMed] [Google Scholar]
- Casino P., Rubio V., Marina A. (2009). Structural insight into partner specificity and phosphoryl transfer in two-component signal transduction Cell 139 325–336 10.1016/j.cell.2009.08.032 . [DOI] [PubMed] [Google Scholar]
- Chang A.C.Y., Cohen S.N. (1978). Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid J Bacteriol 134 1141–1156 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung J., Hendrickson W.A. (2009). Structural analysis of ligand stimulation of the histidine kinase NarX Structure 17 190–201 10.1016/j.str.2008.12.013 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Da Re S., Schumacher J., Rousseau P., Fourment J., Ebel C., Kahn D. (1999). Phosphorylation-induced dimerization of the FixJ receiver domain Mol Microbiol 34 504–511 10.1046/j.1365-2958.1999.01614.x . [DOI] [PubMed] [Google Scholar]
- Darwin A.J., Tyson K.L., Busby S.J., Stewart V. (1997). Differential regulation by the homologous response regulators NarL and NarP of Escherichia coli K-12 depends on DNA binding site arrangement Mol Microbiol 25 583–595 10.1046/j.1365-2958.1997.4971855.x . [DOI] [PubMed] [Google Scholar]
- Egan S.M., Stewart V. (1991). Mutational analysis of nitrate regulatory gene narL in Escherichia coli K-12 J Bacteriol 173 4424–4432 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldridge A.M., Kang H.S., Johnson E., Gunsalus R., Dahlquist F.W. (2002). Effect of phosphorylation on the interdomain interaction of the response regulator, NarL Biochemistry 41 15173–15180 10.1021/bi026254+ . [DOI] [PubMed] [Google Scholar]
- Falord M., Karimova G., Hiron A., Msadek T. (2012). GraXSR proteins interact with the VraFG ABC transporter to form a five-component system required for cationic antimicrobial peptide sensing and resistance in Staphylococcus aureus Antimicrob Agents Chemother 56 1047–1058 10.1128/AAC.05054-11 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao R., Stock A.M. (2009). Biological insights from structures of two-component proteins Annu Rev Microbiol 63 133–154 10.1146/annurev.micro.091208.073214 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman A.L., Merighi M., Hyodo M., Ventre I., Filloux A., Lory S. (2009). Direct interaction between sensor kinase proteins mediates acute and chronic disease phenotypes in a bacterial pathogen Genes Dev 23 249–259 10.1101/gad.1739009 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebe T.W., Stock J.B. (1999). The histidine protein kinase superfamily Adv Microb Physiol 41 139–227 10.1016/S0065-2911(08)60167-8 . [DOI] [PubMed] [Google Scholar]
- Gueguen E., Flores-Kim J., Darwin A.J. (2011). The Yersinia enterocolitica phage shock proteins B and C can form homodimers and heterodimers in vivo with the possibility of close association between multiple domains J Bacteriol 193 5747–5758 10.1128/JB.05080-11 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q., Shen Y., Lee Y.-S., Gibbs C.S., Mrksich M., Tang W.-J. (2005). Structural basis for the interaction of Bordetella pertussis adenylyl cyclase toxin with calmodulin EMBO J 24 3190–3201 10.1038/sj.emboj.7600800 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelbauer G.L., Falke J.J., Parkinson J.S. (2008). Bacterial chemoreceptors: high-performance signaling in networked arrays Trends Biochem Sci 33 9–19 10.1016/j.tibs.2007.09.014 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heikaus C.C., Pandit J., Klevit R.E. (2009). Cyclic nucleotide binding GAF domains from phosphodiesterases: structural and mechanistic insights Structure 17 1551–1557 10.1016/j.str.2009.07.019 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh T.N., Stewart V. (2011). Negative control in two-component signal transduction by transmitter phosphatase activity Mol Microbiol 82 275–286 10.1111/j.1365-2958.2011.07829.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh T.N., Noriega C.E., Stewart V. (2010). Conserved mechanism for sensor phosphatase control of two-component signaling revealed in the nitrate sensor NarX Proc Natl Acad Sci U S A 107 21140–21145 10.1073/pnas.1013081107 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh T.N., Noriega C.E., Stewart V. (2013). Missense substitutions reflecting regulatory control of transmitter phosphatase activity in two-component signalling Mol Microbiol 88 459–472 10.1111/mmi.12195 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihama A., Kori A., Koshio E., Yamada K., Maeda H., Shimada T., Makinoshima H., Iwata A., Fujita N. (2014). Intracellular concentrations of 65 species of transcription factors with known regulatory functions in Escherichia coli J Bacteriol 196 2718–2727 10.1128/JB.01579-14 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova G., Pidoux J., Ullmann A., Ladant D. (1998). A bacterial two-hybrid system based on a reconstituted signal transduction pathway Proc Natl Acad Sci U S A 95 5752–5756 10.1073/pnas.95.10.5752 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova G., Dautin N., Ladant D. (2005). Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis J Bacteriol 187 2233–2243 10.1128/JB.187.7.2233-2243.2005 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laub M.T., Goulian M. (2007). Specificity in two-component signal transduction pathways Annu Rev Genet 41 121–145 10.1146/annurev.genet.41.042007.170548 . [DOI] [PubMed] [Google Scholar]
- Leonard P.G., Golemi-Kotra D., Stock A.M. (2013). Phosphorylation-dependent conformational changes and domain rearrangements in Staphylococcus aureus VraR activation Proc Natl Acad Sci U S A 110 8525–8530 10.1073/pnas.1302819110 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin A.V., Stewart V. (2010). Functional roles for the GerE-family carboxyl-terminal domains of nitrate response regulators NarL and NarP of Escherichia coli K-12 Microbiology 156 2933–2943 10.1099/mic.0.040469-0 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin-Chao S., Chen W.-T., Wong T.-T. (1992). High copy number of the pUC plasmid results from a Rom/Rop-suppressible point mutation in RNA II Mol Microbiol 6 3385–3393 10.1111/j.1365-2958.1992.tb02206.x . [DOI] [PubMed] [Google Scholar]
- Maris A.E., Sawaya M.R., Kaczor-Grzeskowiak M., Jarvis M.R., Bearson S.M., Kopka M.L., Schröder I., Gunsalus R.P., Dickerson R.E. (2002). Dimerization allows DNA target site recognition by the NarL response regulator Nat Struct Biol 9 771–778 10.1038/nsb845 . [DOI] [PubMed] [Google Scholar]
- McCallum N., Meier P.S., Heusser R., Berger-Bächi B. (2011). Mutational analyses of open reading frames within the vraSR operon and their roles in the cell wall stress response of Staphylococcus aureus Antimicrob Agents Chemother 55 1391–1402 10.1128/AAC.01213-10 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.H. (1972). Experiments in Molecular Genetics., Cold Spring Harbor, NY: Cold Spring Harbor Laboratory. [Google Scholar]
- Ninfa E.G., Atkinson M.R., Kamberov E.S., Ninfa A.J. (1993). Mechanism of autophosphorylation of Escherichia coli nitrogen regulator II (NRII or NtrB): trans-phosphorylation between subunits J Bacteriol 175 7024–7032 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noriega C.E., Lin H.-Y., Chen L.-L., Williams S.B., Stewart V. (2010). Asymmetric cross-regulation between the nitrate-responsive NarX-NarL and NarQ-NarP two-component regulatory systems from Escherichia coli K-12 Mol Microbiol 75 394–412 10.1111/j.1365-2958.2009.06987.x . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkinson J.S. (2010). Signaling mechanisms of HAMP domains in chemoreceptors and sensor kinases Annu Rev Microbiol 64 101–122 10.1146/annurev.micro.112408.134215 . [DOI] [PubMed] [Google Scholar]
- Peña-Sandoval G.R., Georgellis D. (2010). The ArcB sensor kinase of Escherichia coli autophosphorylates by an intramolecular reaction J Bacteriol 192 1735–1739 10.1128/JB.01401-09 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podgornaia A.I., Laub M.T. (2013). Determinants of specificity in two-component signal transduction Curr Opin Microbiol 16 156–162 10.1016/j.mib.2013.01.004 . [DOI] [PubMed] [Google Scholar]
- Punta M., Coggill P.C., Eberhardt R.Y., Mistry J., Tate J., Boursnell C., Pang N., Forslund K., Ceric G., other authors (2012). The Pfam protein families database Nucleic Acids Res 40 (D1), D290–D301 10.1093/nar/gkr1065 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabin R.S., Stewart V. (1993). Dual response regulators (NarL and NarP) interact with dual sensors (NarX and NarQ) to control nitrate- and nitrite-regulated gene expression in Escherichia coli K-12 J Bacteriol 175 3259–3268 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheu P., Sdorra S., Liao Y.F., Wegner M., Basché T., Unden G., Erker W. (2008). Polar accumulation of the metabolic sensory histidine kinases DcuS and CitA in Escherichia coli Microbiology 154 2463–2472 10.1099/mic.0.2008/018614-0 . [DOI] [PubMed] [Google Scholar]
- Scheu P.D., Witan J., Rauschmeier M., Graf S., Liao Y.F., Ebert-Jung A., Basché T., Erker W., Unden G. (2012). CitA/CitB two-component system regulating citrate fermentation in Escherichia coli and its relation to the DcuS/DcuR system in vivo J Bacteriol 194 636–645 10.1128/JB.06345-11 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivanesan D., Hancock M.A., Villamil Giraldo A.M., Baron C. (2010). Quantitative analysis of VirB8-VirB9-VirB10 interactions provides a dynamic model of type IV secretion system core complex assembly Biochemistry 49 4483–4493 10.1021/bi902201y . [DOI] [PubMed] [Google Scholar]
- Stewart V. (2003). Biochemical Society Special Lecture. Nitrate- and nitrite-responsive sensors NarX and NarQ of proteobacteria Biochem Soc Trans 31 1–10 10.1042/BST0310001 . [DOI] [PubMed] [Google Scholar]
- Stewart V., Chen L.L. (2010). The shelix mediates signal transmission as a HAMP domain coiled-coil extension in the NarX nitrate sensor from Escherichia coli K-12 J Bacteriol 192 734–745 10.1128/JB.00172-09 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V., Parales J., Jr (1988). Identification and expression of genes narL narX of the nar (nitrate reductase) locus in Escherichia coli K-12 J Bacteriol 170 1589–1597 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart V., Rabin R.S. (1995). Dual sensors and dual response regulators interact to control nitrate- and nitrite-responsive gene expression in Escherichia coli . In Two-component Signal Transduction, pp. 233–252. Edited by Hoch J. A., Silhavy T. J. Washington, DC: American Society for Microbiology. [Google Scholar]
- Stewart G.S., Lubinsky-Mink S., Jackson C.G., Cassel A., Kuhn J. (1986). pHG165: a pBR322 copy number derivative of pUC8 for cloning and expression Plasmid 15 172–181 10.1016/0147-619X(86)90035-1 . [DOI] [PubMed] [Google Scholar]
- Stewart V., Chen L.-L., Wu H.C. (2003). Response to culture aeration mediated by the nitrate and nitrite sensor NarQ of Escherichia coli K-12 Mol Microbiol 50 1391–1399 10.1046/j.1365-2958.2003.03776.x . [DOI] [PubMed] [Google Scholar]
- Stewart V., Bledsoe P.J., Chen L.L., Cai A. (2009). Catabolite repression control of napF (periplasmic nitrate reductase) operon expression in Escherichia coli K-12 J Bacteriol 191 996–1005 10.1128/JB.00873-08 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stynen B., Tournu H., Tavernier J., Van Dijck P. (2012). Diversity in genetic in vivo methods for protein-protein interaction studies: from the yeast two-hybrid system to the mammalian split-luciferase system Microbiol Mol Biol Rev 76 331–382 10.1128/MMBR.05021-11 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson R.V., Bourret R.B., Simon M.I. (1993). Intermolecular complementation of the kinase activity of CheA Mol Microbiol 8 435–441 10.1111/j.1365-2958.1993.tb01588.x . [DOI] [PubMed] [Google Scholar]
- Trajtenberg F., Graña M., Ruétalo N., Botti H., Buschiazzo A. (2010). Structural and enzymatic insights into the ATP binding and autophosphorylation mechanism of a sensor histidine kinase J Biol Chem 285 24892–24903 10.1074/jbc.M110.147843 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wanner B.L., Kodaira R., Neidhardt F.C. (1978). Regulation of lac operon expression: reappraisal of the theory of catabolite repression J Bacteriol 136 947–954 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wayne K.J., Sham L.T., Tsui H.C., Gutu A.D., Barendt S.M., Keen S.K., Winkler M.E. (2010). Localization and cellular amounts of the WalRKJ (VicRKX) two-component regulatory system proteins in serotype 2 Streptococcus pneumoniae J Bacteriol 192 4388–4394 10.1128/JB.00578-10 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S.B., Stewart V. (1997a). Nitrate- and nitrite-sensing protein NarX of Escherichia coli K-12: mutational analysis of the amino-terminal tail and first transmembrane segment J Bacteriol 179 721–729 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams S.B., Stewart V. (1997b). Discrimination between structurally related ligands nitrate and nitrite controls autokinase activity of the NarX transmembrane signal transducer of Escherichia coli K-12 Mol Microbiol 26 911–925 10.1046/j.1365-2958.1997.6262002.x . [DOI] [PubMed] [Google Scholar]
- Yang Y., Inouye M. (1991). Intermolecular complementation between two defective mutant signal-transducing receptors of Escherichia coli Proc Natl Acad Sci U S A 88 11057–11061 10.1073/pnas.88.24.11057 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanofsky C., Horn V., Bonner M., Stasiowski S. (1971). Polarity and enzyme functions in mutants of the first three genes of the tryptophan operon of Escherichia coli Genetics 69 409–433 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- You C., Okano H., Hui S., Zhang Z., Kim M., Gunderson C.W., Wang Y.P., Lenz P., Yan D., Hwa T. (2013). Coordination of bacterial proteome with metabolism by cyclic AMP signalling Nature 500 301–306 10.1038/nature12446 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J.H., Xiao G., Gunsalus R.P., Hubbell W.L. (2003). Phosphorylation triggers domain separation in the DNA binding response regulator NarL Biochemistry 42 2552–2559 10.1021/bi0272205 . [DOI] [PubMed] [Google Scholar]