Abstract
Carnitine is a quaternary amine compound found at high concentration in animal tissues, particularly muscle, and is most well studied for its contribution to fatty acid transport into mitochondria. In bacteria, carnitine is an important osmoprotectant, and can also enhance thermotolerance, cryotolerance and barotolerance. Carnitine can be transported into the cell or acquired from metabolic precursors, where it can serve directly as a compatible solute for stress protection or be metabolized through one of a few distinct pathways as a nutrient source. In this review, we summarize what is known about carnitine physiology and metabolism in bacteria. In particular, recent advances in the aerobic and anaerobic metabolic pathways as well as the use of carnitine as an electron acceptor have addressed some long-standing questions in the field.
Introduction
Carnitine (γ-trimethylamino-β-hydroxybutyric acid) (Fig. 1) is a quaternary amine compound that can be produced by all domains of life, and was discovered in muscle extract in 1905 by Gulewitsch & Krimberg (1905) and Kutscher (1905). It was shown to be essential for larval development of the mealworm Tenebrio molitor and was originally designated vitamin BT based on this requirement. Later, it was discovered that carnitine can be synthesized in mammals and is now considered to be a quasi-nutrient or conditionally essential nutrient (Flanagan et al., 2010), as neonates have reduced biosynthesis and rely on placental transfer of carnitine in utero and exogenous sources after birth (Combs, 2012). Fifty years after the discovery of carnitine, it was demonstrated that assorted Gram-positive and Gram-negative bacteria could use carnitine in either aerobic or anaerobic environments for a variety of cellular functions, including as an electron acceptor, as a compatible solute to survive environmental insults or as a sole carbon, nitrogen and energy source. Bacterial carnitine metabolism was most recently reviewed in 1998 (Bieber, 1988; Bremer, 1983; Kleber, 1997; Rebouche & Seim, 1998) and the field has seen important advances. This review summarizes what we knew at the time of the previous reviews and emphasizes what we have learned since, including: (i) how anaerobic bacteria synthesize and utilize crotonobetaine and carnitine as final electron acceptors, (ii) the impact of carnitine degradation by the intestinal microbiota and the genes responsible for this anaerobic conversion, (iii) the genes involved in aerobic degradation of carnitine, and (iv) how carnitine as a compatible solute impacts survival within and outside of the host.
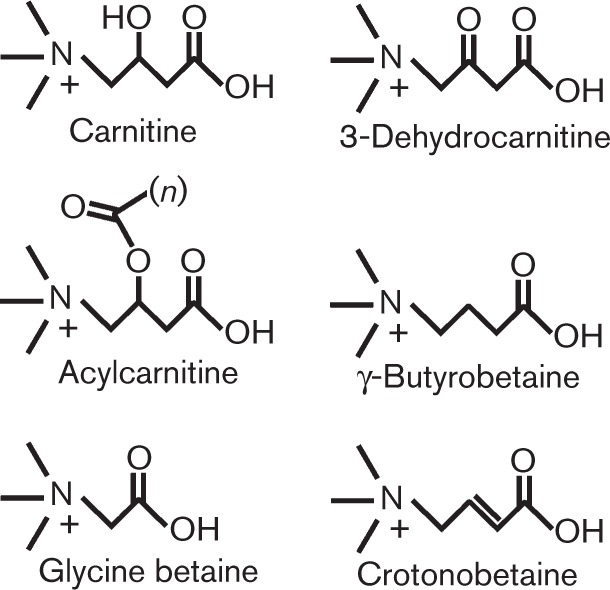
Fig. 1. Structures of carnitine and related compounds discussed in this review.
Carnitine in the environment
Recent work makes it clear that while animals represent the most readily accessible source of carnitine, carnitine is often present and sometimes abundant in soil and natural waters. Quaternary ammonium compounds are abundant in a number of soil ecosystems, including comprising a quarter of the most abundant organic nitrogen compounds in the soil water of a subalpine grassland (Warren, 2013a). In this environment, carnitine was the most abundant quaternary ammonium compound (0.49 μM) and third most abundant soluble nitrogen compound overall, while acetylcarnitine was present at a slightly lower concentration (0.33 μM) (Warren, 2013a). It is apparent that carnitine concentration varies depending on sample location (Warren, 2013b), but there is need for a more thorough quantification of carnitine in other environments. The carnitine levels in soil and water may vary depending on the bacterial flora at the site and whether the bacteria inhabiting those environments are capable of carnitine metabolism. The presence and utility of carnitine in the environment is supported, in part, by the number of bacteria capable of carnitine metabolism, including a few newly identified species such as Burkholderia caribensis (Achouak et al., 1999), Bacillus decisifrondis (Zhang et al., 2007), Pseudomonas kilonensis (Sikorski et al., 2001) from soil, and Shewanella pacifica from sea water (Ivanova et al., 2004).
Importance of carnitine to animals (including humans)
Carnitine is most abundantly associated with animals and its physiology in animals provides an important backdrop to our review of microbial processes. Carnitine is a zwitterion and can exist as either d- or l-enantiomers, but the d stereoisomer is not utilized for normal physiology in animals and can inhibit acylcarnitine transferases, thereby resulting in tissue depletion of l-carnitine (Bieber, 1988). Therefore, unless specifically noted, we are discussing l-carnitine. Animals use the l-carnitine shuttle to transport long- to short-chain fatty acids in and out of the mitochondria by reversibly esterifying the β-carbon hydroxyl group with a fatty acid to form O-acylcarnitine (Fig. 1). β-Oxidation of very-long-chain fatty acids starts in the peroxisome, and once they have been converted to medium- or short-chain fatty acids, carnitine is employed to traffic them out of the peroxisome and into the mitochondria where β-oxidation is completed (Reddy & Hashimoto, 2001; Steiber et al., 2004; Wanders & Waterham, 2006). Acetyl-CoA derived from β-oxidation can be used to generate ATP via the TCA cycle, while β-oxidation allows the cell to maintain the acetyl-CoA: CoA ratio and enables removal of specific harmful acylcarnitines derived from endogenous substances or from xenobiotics (Bieber, 1988).
In humans, ∼95 % of total carnitine is found in skeletal and cardiac muscle, with the remaining 5 % circulating in the plasma (Cave et al., 2008). Approximately 75 % of carnitine is obtained through diet, with foods from animal origin (e.g. meat and dairy products) having the highest carnitine content (Steiber et al., 2004). The remaining 25 % of carnitine is synthesized endogenously from the essential amino acids l-methionine and l-lysine in the liver, kidney, testes, and brain (Bremer, 1983; Flanagan et al., 2010; Rebouche, 2014). The body maintains homeostatic levels of carnitine by balancing carnitine absorption from the small intestine lumen, reabsorption by the kidneys, and modest endogenous synthesis (Rebouche & Seim, 1998). Carnitine and acylcarnitines are primarily absorbed from the lumen of the small intestine, where they are actively transported into enterocytes and diffuse past the serosal membrane into the circulatory system so they can then be transported into all other cells (Marciani et al., 1991; Rebouche, 2004). Dietary carnitine that is not absorbed in the small intestines is metabolized by bacteria in the large intestine – there are no animal enzymes to break down carnitine (Rebouche & Chenard, 1991). Over the past decade, gut microbiome metabolism has become a topic receiving close review, and recently Koeth et al. (2013) associated the degradation of carnitine by intestinal microbiota with cardiovascular disease and the promotion of atherosclerosis. Some intestinal bacteria can convert carnitine to trimethylamine, which is subsequently oxidized in the liver to the proatherogenic species trimethylamine-N-oxide (TMAO) (Koeth et al., 2013) – a metabolic pathway we discuss in more detail below. The role of bacterial metabolism of carnitine directly promoting cardiovascular disease has been shown in multiple studies (Hartiala et al., 2014; Koeth et al., 2013; Kuka et al., 2014), but is still up for discussion in the scientific and medical communities (Johri et al., 2014). However, there is no debate that bacteria, intestinal and otherwise, utilize carnitine in many different ways for their benefit (Ussher et al., 2013).
Physiological benefits of carnitine for bacteria
Animals have been the focus of research on carnitine since its discovery in 1905 (Gulewitsch & Krimberg, 1905) and microbial carnitine metabolism was not described until more than 50 years later (Fraenkel & Friedman, 1957). In the intervening years, microbiologists have described the roles for carnitine in bacteria, where it can be used as a final electron acceptor, a compatible solute, or as a sole carbon, nitrogen, and energy source. Regardless of its eventual role, carnitine is transported into the bacterial cytosol by one of two different mechanisms. The first method is an ATP-dependent ATP-binding cassette (ABC) transport system utilizing the canonical three subunits – a transmembrane domain, an ATPase, and a periplasmic binding protein (also known as a substrate binding protein). The second method of import is by a betaine/choline/carnitine transporter (BCCT) that can be driven by the sodium or proton motive force, but in many cases where carnitine is the substrate often functions as a carnitine: γ-butyrobetaine antiporter (Ziegler et al., 2010). Carnitine import or import of immediate precursors is critical for bacterial acquisition of carnitine, as Verheul et al. (1998) demonstrated that de novo synthesis of carnitine does not occur in Escherichia coli and, to the best of our knowledge, de novo synthesis of carnitine has not been demonstrated in any bacterial species.
Carnitine: an organic compatible solute
Outside of the cosy confines of the laboratory, bacteria are subject to constantly changing environments to which they must respond rapidly to survive and thrive. One way to accommodate stresses caused by changing water content, salt, temperature or pressure is by synthesis or import of compatible solutes. Compatible solutes can be accumulated at high concentrations in the cytoplasm while not interfering with cellular processes, and thus contribute to proper protein function and cellular homeostasis (Brown & Simpson, 1972). Compatible solutes are typically organized into categories: carbohydrates, amino acids, methylamines, methylsulfonium solutes, and specific inorganic ions. Organic solutes that are either non-charged or zwitterions with no net charge at physiological pH are preferred compatible solutes, the intracellular concentrations of which are carefully regulated to maintain protein stability and cell physiology (Fitzsimmons et al., 2012; Hoffmann et al., 2013). Carnitine is an ideal compatible solute that can be imported and/or generated from direct precursors by many bacteria, and its abundance for infectious microbes in the host and presence in many natural environments suggests it is readily accessible (Warren, 2013a, b).
Osmoprotection
Drastic changes in water content in the environment can result in osmotic stress to the cell. Low external solute concentration is hypo-osmotic, with resulting pressure to drive water into the cell causing an increase in cell turgor pressure. Conversely, increased external solute concentration due to added solutes or loss of water is hyper-osmotic, with resulting pressure leading to water efflux from the cell, reducing turgor pressure. In addition to the altered physiology imparted by these turgor changes, both conditions can lead to cell death due to irreversible plasmolysis (loss of water) or cytolysis (too much water). To overcome osmotic stress, bacteria can either acquire or synthesize osmoprotectants – a process that is universal for bacteria. One way bacteria can overcome osmotic stress is by utilizing carnitine as an osmoprotectant and/or osmolyte.
Gram-negative osmoprotection by carnitine
The Gram-negative opportunistic pathogen Pseudomonas aeruginosa uses carnitine as both an osmoprotectant and an osmolyte. Intracellular carnitine can be accumulated directly via transport from extracellular sources through a BCCT (Malek et al., 2011) or ABC transporter (Chen et al., 2010), or indirectly by degradation of O-acylcarnitines (Lucchesi et al., 1995; Meadows & Wargo, 2013). Pseudomonas aeruginosa, as with many environmental proteobacteria, can also metabolize carnitine to the osmolyte glycine betaine (Fig. 2) (Aurich et al., 1967; Bastard et al., 2014; Wargo & Hogan, 2009). E. coli also uses carnitine as an osmolyte, where it functions in aerobic, anaerobic, and high salt conditions (Verheul et al., 1998). Import is primarily mediated through the ProU transport system, a substrate binding protein-dependent ABC transporter, along with modest uptake from the major facilitator superfamily transporter ProP in anaerobic and osmostressed aerobic cells. Although CaiT functions as a carnitine antiporter, its activity is not sufficient to confer osmoprotection (Verheul et al., 1998). Yersinia enterocolitica also employs carnitine as an osmolyte, where unlike most Gram-negative bacteria, NMR observations demonstrated that added carnitine was not metabolized to the more potent osmolyte glycine betaine (Park et al., 1995).
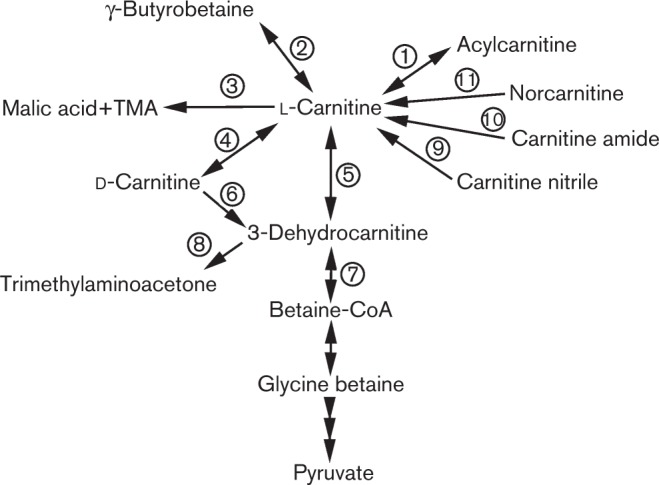
Fig. 2. Diagram of the multiple pathways for carnitine metabolism in bacteria based on Uanschou et al. (2005). Metabolic steps described in this review are noted with numbered circles: (1) acylcarnitine hydrolase, HocS, EC 3.1.1.28; (2) γ-butyrobetaine hydroxylase, EC 1.14.11.1; (3) oxidoreductase CntAB, EC 1.14.13.- (TMA, trimethylamine); (4) carnitine racemase, EC 5.1.-.-; (5) l-carnitine dehydrogenase, EC 1.1.1.108; (6) d-carnitine dehydrogenase, EC 1.1.1.108; (7) β-keto acid cleavage enzyme, EC 3.-.-.-; (8) no ATP, spontaneous decarboxylation; (9) nitrilase, EC 3.5.5.1; (10) carnitine amidase, EC 3.5.1.73; (11) carnitine methylase. Note that not all enzymic steps have full EC descriptors at this time.
Gram-positive osmoprotection by carnitine
The lactic acid bacteria Tetragenococcus halophile and Lactobacillus plantarum can import and use d- and l-carnitine as osmolytes (Kets et al., 1994; Robert et al., 2000). Brevibacterium linens can also use d- and l-carnitine as osmolytes, and while l-carnitine can be metabolized to glycine betaine, it is further metabolized as a sole carbon or nitrogen source (Jebbar et al., 1998). Bacillus subtilis imports d- and l-carnitine, acetylcarnitine, crotonobetaine, γ-butyrobetaine and octanoylcarnitine via the ABC transporter OpuC. Both stereoisomers and the carnitine resulting from acetyl- and octanoylcarnitine degradation function as osmolytes to protect the cell from hyper-osmotic stress (Kappes & Bremer, 1998). The carnitine generated or imported by Bacillus subtilis cannot be metabolized further and therefore is thought to function primarily as an osmoprotectant. Staphylococcus aureus, a common member of the skin and nasal flora that is an important opportunistic pathogen, uses carnitine as a compatible solute in high salt (Vilhelmsson & Miller, 2002).
Carnitine appears to be particularly important for the common foodborne pathogen Listeria monocytogenes. Listeria monocytogenes primarily imports carnitine through the OpuC ABC transporter (Fraser & O'Byrne, 2002; Fraser et al., 2000; Verheul et al., 1995, 1997), where it functions as an osmolyte in high salt conditions (Beumer et al., 1994). Mice infected with Listeria monocytogenes perorally show that OpuC is important for survival and infection in the small intestine, and for the proliferation and dissemination of the bacteria into other organs, including the liver and spleen (Sleator et al., 2001, 2003; Wemekamp-Kamphuis et al., 2002). One of these studies postulated that bacterial destruction of the intestinal epithelial layer allows carnitine release, which then alleviates the effects of the hyper-osmotic environment of the small intestine, where salt concentrations are two times higher than the blood (Sleator et al., 2001). Additional mouse infections comparing mutants in the glycine betaine transport systems BetL and Gbu to mutants of OpuC led to the conclusion that carnitine transport is more important than glycine betaine for Listeria during infection (Sleator et al., 2003; Wemekamp-Kamphuis et al., 2002).
Beyond salt: role of carnitine in cryoprotection, bile tolerance and barotolerance
In addition to osmoprotection, compatible solutes can protect bacteria from additional sources of stress. Listeria and Bacillus have both been well studied in relation to carnitine uptake, and are good examples of bacteria that employ carnitine to protect against multiple stress conditions.
Listeria is a foodborne pathogen that has multiple lifestyles, thriving on decaying plant material and transiently living in the gastrointestinal tract of some animals, including humans. Any bacterium that lives or passes through the small intestine has to cope with bile salts, which aid in digestion of lipids, have antimicrobial activity, disrupt membranes and proteins, and induce oxidative stress and DNA damage (Begley et al., 2005). The OpuC carnitine transport system in Listeria monocytogenes is important for protecting the bacteria against bile stress and its expression is co-regulated with the bile efflux system BilE (Watson et al., 2009). Mice infected perorally with an opuC mutant strain had significantly reduced numbers of bacteria in the faeces and a decrease in systemic infection, measured as bacteria in the liver and spleen (Watson et al., 2009). Carnitine enhances bile tolerance, and is important for dissemination and survival in the small intestine (Gahan & Hill, 2014; Sleator & Hill, 2010; Watson et al., 2009).
Unlike many bacteria, Listeria monocytogenes is capable of growing at refrigeration temperatures (4 °C), making it a common contaminant of dairy, meats, fruits and vegetables (Ryser & Marth, 2007). Cold or chill stress affects protein structure (Jaenicke et al., 1990), protein stability (Privalov & Gill, 1988), and membrane fluidity (Rudolph et al., 1986), and compatible solutes, such as carnitine, can alleviate these negative effects due to cold (Bayles & Wilkinson, 2000). The expression of the carnitine uptake system opuC is increased at low temperatures, allowing the cell to acquire carnitine and maintain growth (Angelidis & Smith, 2003; Angelidis et al., 2002; Sleator et al., 2009; Wemekamp-Kamphuis et al., 2004).
The ability to survive low-temperature stress is not the sole domain of bacteria in our refrigerators, but rather a required feature of bacteria that inhabit the environment, where mean soil surface temperatures range from 0 to 18 °C, mean freshwater ranges from 3 to 25 °C and while the mean ocean surface temperature is ∼17 °C, the bulk of ocean water maintains a temperature of ∼3 °C. Therefore, it is not surprising that successful environmental bacteria have also evolved pathways for cold tolerance that involve carnitine. In Bacillus subtilis, carnitine and its related metabolites crotonobetaine and γ-butyrobetaine have been shown to protect against both cold stress (15 °C) and heat stress (52 °C), via uptake through the OpuC transporter (Hoffmann & Bremer, 2011). It is likely that utilization of carnitine as a cryo- and thermoprotectant is not restricted to Listeria and Bacillus, and the taxonomic breadth and ecological impact of this process require further examination.
Pressure is another assault that Listeria has adapted to, as food processing uses high pressure for food preservation. Listeria exposed to elevated osmolarity and high pressure survive better and demonstrate substantial barotolerance when the compatible solute carnitine is present and imported into the cell (Smiddy et al., 2004).
Carnitine metabolism
Carnitine can be utilized in different metabolic pathways to play a variety of physiological roles. Fig. 2 illustrates multiple described pathways for carnitine metabolism and highlights bacteria in which particular reactions have been demonstrated. For each of the pathways described below, it is important to note that strain-specific utilization of carnitine via particular pathways can be governed by alternative regulation or the presence/absence of metabolic genes. For example, Pseudomonas syringae B728a carries the genes for carnitine conversion to glycine betaine, while Pseudomonas syringae DC3000 has lost the entire carnitine catabolic operon (Chen & Beattie, 2007; Chen et al., 2010).
Carnitine as a nutrient
Carnitine can be catabolized for use as a carbon source via two routes. The first pathway cleaves the backbone carbon–nitrogen bond to yield trimethylamine and malic semialdehyde, in which the latter enters the TCA cycle. The second route begins with the metabolism of carnitine to glycine betaine, which is then subjected to three successive demethylations to yield glycine, which can enter central metabolism. Carnitine can also be used as a sole nitrogen source, most commonly via the glycine betaine route, where glycine conversion to serine is followed by deamination to form pyruvate and ammonia. Formation of trimethylamine from carnitine leaves the nitrogen unusable for most of the organisms covered in this review, but many bacteria can use trimethylamine as a nitrogen source, carbon source, osmoprotectant and, when oxidized to TMAO, as an electron acceptor (as reviewed by Barrett & Kwan, 1985; Strøm et al., 1979; Yancey, 2005).
Carnitine to glycine betaine: a gateway reaction
Carnitine can be metabolized to glycine betaine via a multistep process, the first steps of which are encoded on genes located in the carnitine catabolism operon (Uanschou et al., 2005; Wargo & Hogan, 2009). First, carnitine is converted to 3-dehydrocarnitine by the enzyme carnitine dehydrogenase (CDH, EC 1.1.1.108), an NAD+-dependent oxidoreductase. Pseudomonas aeruginosa (Aurich et al., 1967; Kleber & Aurich, 1967; Kleber et al., 1967), Xanthomonas translucens (Arima et al., 2010; Mori et al., 1988), Enterobacter sp. (Hwang & Bang, 1997), Pseudomonas putida (Kleber et al., 1978), Pseudomonas fluorescens (Hung & Kleber, 1985), Burkholderia cepacia (Dalmastri et al., 2003), Rhizobium sp. (Arima et al., 2010) and Agrobacterium sp. (Hanschmann et al., 1996) all encode CDH enzymes that are specific for l-carnitine. 3-Dehydrocarnitine is relatively unstable, therefore if there is no ATP or CoA present, it will spontaneously decarboxylate to yield trimethylaminoacetone (N,N,N-trimethylaminopropanone) and carbon dioxide (Lindstedt et al., 1967). If sufficient ATP and CoA are present, then 3-dehydrocarnitine is converted to acetylacetone and glycine betaine-CoA by the β-keto acid cleavage enzyme (BKACE). The gene encoding the carnitine-specific BKACE is located directly upstream of the CDH gene(s) in the carnitine catabolism operon (Uanschou et al., 2005) and is designated cdhC in Pseudomonas aeruginosa (Bastard et al., 2014; Wargo & Hogan, 2009). The identification of this gene was part of a massive biochemical screen of various bacterial BKACE members and proved very valuable for the identification of these CoA-dependent cleavage enzyme substrates (Bastard et al., 2014). The glycine betaine-CoA derived from BKACE activity is then converted to glycine betaine and CoA by a CoA transferase, likely the DhcAB enzyme in Pseudomonas aeruginosa (Wargo & Hogan, 2009), which appears to function as a general amino acid CoA transferase (Palmer et al., 2013). After carnitine is metabolized to glycine betaine, it can function as an osmolyte (as described above) or be utilized as a sole carbon, nitrogen, and energy source if the organism encodes the necessary enzymes, as in the case for Pseudomonas aeruginosa (Wargo et al., 2008), Xanthamonas translucens (Arima et al., 2010; Mori et al., 1988), Enterobacter sp. (Hwang & Bang, 1997), Rhizobium sp. (Arima et al., 2010; Goldmann et al., 1991), Sinorhizobium meliloti (Goldmann et al., 1991), Burkholderia cepacia (Dalmastri et al., 2003) and Agrobacterium sp. (Hanschmann et al., 1996; Nobile & Deshusses, 1986). The metabolism and homeostasis of glycine betaine has been reviewed recently (Wargo, 2013).
Cleaving the backbone carbon–nitrogen bond
An alternative pathway to use carnitine as a sole carbon source is to cleave the carbon–nitrogen bond of carnitine to form trimethylamine and malic acid. It has been known since the mid-1960s that Serratia marcescens aerobically splits the carbon–nitrogen bond of both d- and l-carnitine to form trimethylamine and malic acid, and that this reaction does not occur under anaerobic conditions as it requires uptake of oxygen (Unemoto et al., 1966). Through the same reaction, Acinetobacter calcoaceticus can also use both d- and l-carnitine as sole carbon sources (Kleber et al., 1977; Miura-Fraboin et al., 1982), yet the responsible enzymes remained elusive until recently. The genes encoding the enzymes that are essential for the conversion of carnitine to trimethylamine and malic semialdehyde were recently identified as a two-subunit oxidoreductase in Acinetobacter baumannii (Zhu et al., 2014), with CntA being a Rieske-family iron–sulfur cluster oxygenase and CntB being the reductase. The CntAB proteins likely function analogously to GbcAB, which comprise the oxidoreductase that demethylates glycine betaine to dimethylglycine (Wargo et al., 2008). Both sets of proteins cleave one of the carbon–nitrogen bonds in a quaternary amine compound and these enzymes may represent a general evolutionary strategy for quaternary amine metabolism. Orthologues and homologues of the cntAB genes are found in a variety of gut microbiota: Gammaproteobacteria (Klebsiella pneumoniae, E. coli, Citrobacter, Providencia and Shigella), Betaproteobacteria (Achromobacter) and Firmicutes (Sporosarcina) (Zhu et al., 2014). The malic semialdehyde produced during this reaction is converted to malic acid, which enters the TCA cycle. Trimethylamine formed from carnitine by gut bacteria is correlated with human cardiovascular health (Koeth et al., 2013), where it is oxidized to TMAO by hepatic flavin monooxygenases in the liver (Bennett et al., 2013). Therefore, the identification of CntAB provides a target for monitoring gut microbiota capacity for trimethylamine production and is a critical step forward in our understanding of carnitine metabolism by bacteria.
Carnitine as a final electron acceptor
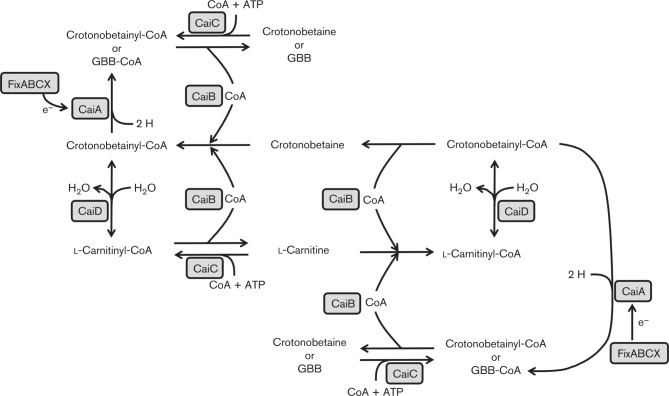
Bacteria that live strictly or transiently in anaerobic environments, where oxygen cannot serve as the final electron acceptor, can use alternate electron acceptors, including sulfates, nitrates, ferric iron, carbon dioxide, and fumarate, amongst others. In cases where the common electron acceptors are not present, some Enterobacteriaceae (E. coli, Salmonella typhimurium and Proteus spp.) can use carnitine and its catabolic product crotonobetaine as final electron acceptors in the absence of oxygen, and in the presence of additional carbon and nitrogen sources (Seim et al., 1982a, b, c). Use of these compounds as electron acceptors is regulated by the transcriptional activator, CaiF, and the regulatory proteins CRP and FNR, which regulate expression of the divergently transcribed operons caiTABCDE and fixABCX, both required for carnitine metabolism in E. coli (Buchet et al., 1998, 1999; Eichler et al., 1995, 1996). For its role as an electron acceptor, carnitine is imported by the substrate: product antiporter CaiT, which exchanges carnitine for its metabolic product γ-butyrobetaine (Jung et al., 1990, 2002; Kalayil et al., 2013). Co-expression of these two operons is induced by carnitine and crotonobetaine, but repressed by oxygen, glucose, γ-butyrobetaine and more desirable final electron acceptors, such as nitrate and fumarate (Jung et al., 1987; Seim et al., 1982a, b). Expression of the carnitine metabolism genes in E. coli and Proteus spp. is also detectable in aerobic environments in the presence of inducing compounds, but to a much lesser extent than under anaerobic conditions (Elssner et al., 1999; Engemann & Kleber, 2001; Obón et al., 1999).
The fix operon is predicted to be involved in transferring electrons to carnitine (Eichler et al., 1995, 1996; Walt & Kahn, 2002). The transformation of carnitine to crotonobetaine is reversible (Jung et al., 1989) and was originally thought to occur via the single carnitine dehydratase CaiB with the co-substrates γ-butyrobetainyl-CoA or crotonobetainyl-CoA (Eichler et al., 1994a; Elssner et al., 2000; Jung et al., 1989). However, Elssner et al. (2001) demonstrated that ‘l-carnitine dehydratase does not exist’ and two enzymes are responsible for the reversible conversion of carnitine to crotonobetaine. CaiB is a CoA transferase that transfers CoA from γ-butyrobetainyl-CoA, crotonobetainyl-CoA or carnitinyl-CoA to form γ-butryobetaine, crotonobetaine or carnitine. CaiD has enoyl-CoA hydratase activity and can dehydrate carnitinyl-CoA to crotonobetainyl-CoA, or can hydrate crotonobetainyl-CoA to carnitinyl-CoA; thus, CaiD has two potential names: crotonobetainyl-CoA hydratase or carnitinyl-CoA dehydratase. These enzymic processes also allow carnitine to be synthesized from crotonobetaine and vice versa (Fig. 3) (Elssner et al., 2001; Engemann et al., 2001, 2005). Crotonobetaine can be reduced to γ-butyrobetaine by two proteins: CaiB, with one of the co-substrates γ-butyrobetainyl-CoA or crotonobetainyl-CoA, and the unidirectional enzyme crotonobetaine reductase CaiA, which reduces crotonobetainyl-CoA to γ-butyrobetainyl-CoA (Engemann et al., 2005; Preusser et al., 1999; Roth et al., 1994). Two other proteins necessary for anaerobic carnitine catabolism are CaiC and CaiE. CaiC is a betaine: CoA ligase with CoA transferase activity in vitro, but is not sufficient in vivo to compensate for a caiB deletion. Therefore, CaiC is likely required for activation of trimethylammonium compounds (Bernal et al., 2008; Eichler et al., 1994b). The function of CaiE is still somewhat mysterious, but it is postulated to be required for activation or synthesis for an unknown cofactor necessary for carnitine metabolism (Eichler et al., 1994b).
Fig. 3. Diagram of the γ-butyrobetaine–crotonobetaine–carnitine cycle primarily based on Elssner et al. (2001), with additions based on Bernal et al. (2008) and Canovas et al. (2003). GBB, γ-butyrobetaine; CaiA, crotonobetaine reductase; CaiB, CoA transferase; CaiC, betaine:CoA ligase; CaiD, enoyl-CoA hydratase; FixABCX, electron transfer flavoproteins. Electron movement is denoted by e− .
The enzymes for converting γ-butyrobetaine into carnitine, best described in the Enterobacteriaceae, are predicted to be present in other bacterial groups. The betaproteobacterium Achromobacter cycloclast and the gammaproteobacterium Acinetobacter calcoaceticus are predicted to have similar enzymes as no γ-butyrobetaine hydroxylase was detected during degradation of γ-butyrobetaine to carnitine. For Acinetobacter calcoaceticus, after γ-butyrobetaine is degraded to carnitine, the carnitine can then be broken down to trimethylamine and malic acid where it uses it as a carbon source (Miura-Fraboin et al., 1982), whereas the fate of the newly synthesized carnitine was not investigated in Acinetobacter cycloclast (Naidu et al., 2001).
Sensing and binding carnitine
For osmotic adaptation to, or metabolism of, carnitine, it is important for bacteria to regulate the expression of genes involved in these processes. To our knowledge, carnitine is sensed to regulate activity at the transcriptional level using so-called ‘one-component’ regulators, i.e. single polypeptides that sense ligand and regulate transcription. However, compared with our understanding of carnitine transport and metabolism, the detection of carnitine for transcriptional regulation is much more poorly understood. We know of only two transcription factors that sense carnitine to regulate transcription: CaiF in E. coli and CdhR in Pseudomonas aeruginosa. Both are transcriptional activators and induce transcription in response to carnitine (Buchet et al., 1999; Wargo & Hogan, 2009), although importantly, direct interaction of either protein with carnitine has not been demonstrated.
In E. coli, the cai and fix operons (described above) are regulated by the global regulator CRP and the carnitine-responsive activator CaiF. The CaiF protein is small and is predicted to contain two AraC-like helix–turn–helix (HTH) domains and likely functions similar to the MarR ‘HTH-only’ activators (Buchet et al., 1999). Interestingly, the lack of a canonical ligand-binding domain suggests that detection of the ligand occurs within what is typically thought of as the DNA-binding domain of the protein or is mediated by an independently encoded sensor protein. Using the Phyre2 protein prediction server (Kelley & Sternberg, 2009), CaiF has high structural homology to GrlA from enterohaemorrhagic E. coli, where the two-component response regulator GrlR has been shown to interact with GrlA, modulating transcriptional regulation (Creasey et al., 2003; Russell et al., 2007). While we have found no obvious GlrR homologues in the fix and cai operons, it remains a possibility that carnitine detection occurs via a GlrR orthologue or a cognate sensor kinase, such as GlrK (Yamamoto et al., 2005).
In Pseudomonas aeruginosa, the transcription factor CdhR functions as the carnitine-responsive activator of the genes encoding the CDH enzymic pathway (Wargo & Hogan, 2009). CdhR is a member of the glutamine amidotransferase-1 subfamily of the AraC transcription factor family and has the canonical AraC family structure consisting of an N-terminal ligand sensing and dimerization domain, and a C-terminal DNA-binding domain composed of two HTH motifs. Orthologues of CdhR are found divergently transcribed from the aerobic CDH genes in a variety of organisms, including many species in the families Pseudomonadaceae, Burkholderiaceae and Rhizobiaceae, as well as in Mesorhizobium loti and Silicibacter sp. (Uanschou et al., 2005). Given the conservation of the gene arrangement and predicted functions, we propose that further understanding of CdhR will yield insights into this regulation beyond Pseudomonas.
In Gram-positive bacteria, the CDH operons contain a TetR family transcription regulator that likely functions as the carnitine sensor for regulated expression of these operons. Compared with the Gram-negative bacteria, presence of the CDH and its cognate TetR-like regulator is much more restricted, occurring in Brevibacterium linens, Staphylococcus epidermidis, Streptomyces coelicolor and Oceanobacillus sp. (Uanschou et al., 2005). For the first two organisms on this list, acquisition of this predicted horizontally acquired operon makes teleological sense given the prevalence of carnitine in milk and in the skin, respectively.
An integral part of transport, metabolism, and transcriptional activation is binding of the target molecule. In the case of carnitine, crystal structures have been solved in complex that include carnitine in complex with the periplasmic binding protein OpuCC (Du et al., 2011), the carnitine:γ-butryobetaine antiporter CaiT (Schulze et al., 2010; Tang et al., 2010) and similar aromatic cage architecture of the quaternary amine-binding site with γ-butyrobetaine has been determined for γ-butyrobetaine hydroxylase (Tars et al., 2014). Therefore, as in the situation for glycine betaine (Schiefner et al., 2004), high-affinity binding of carnitine and its metabolites seems to occur via an aromatic cage enabling the cation–π interaction with the trimethylamine moiety coupled with properly spaced hydrogen-bonding residues to coordinate the carboxylic acid. While this binding arrangement has been demonstrated for transport and metabolic proteins, neither carnitine nor the related compounds choline or glycine betaine have been crystallized with their cognate transcriptional regulators. Given the specificity and affinity provided by the cation–π interaction, it has been hypothesized that glycine betaine and carnitine-sensitive transcription factors utilize a functionally similar binding site (Bremer, 2011).
Multiple paths for carnitine acquisition
De novo carnitine synthesis
While bacteria can synthesize the quaternary amine compounds choline and glycine betaine from single unrelated carbon sources during growth on minimal media, such de novo synthesis of carnitine has not been demonstrated. Rather, as discussed below, carnitine is typically generated by metabolizing appropriate trimethylated precursors. While evidence for true de novo synthesis is lacking, N ε-trimethyllysine is the starting point for mammalian carnitine synthesis (Vaz & Wanders, 2002) and bacteria are known to synthesize N ε-trimethyllysine (Barbier et al., 2013; Klagsbrun & Furano, 1975). Therefore, it remains a formal possibility that bacteria can use this N ε-trimethyllysine to synthesize carnitine in a manner similar to mammals.
d-Carnitine … not a dead end after all
Animals only synthesize and respond physiologically to l-carnitine, thus the presence of the d stereoisomer arises from bacterial processes on carnitine and its derivatives or from ingested food based on abiotic racemization. Despite the relative abundance of l-carnitine, some bacteria are capable of utilizing d-carnitine as a sole carbon and nitrogen source, including Agrobacterium sp., Agrobacterium radiobacter and Enterobacter (Hanschmann & Kleber, 1997; Hwang & Bang, 1997; Klüttermann et al., 2002). Agrobacterium expresses both an l-CDH and a d-CDH, and its utilization of d-carnitine is dependent on the loss of chirality upon conversion of d-carnitine to the achiral 3-dehydrocarnitine (Hanschmann & Kleber, 1997). Enterobacter sp. KC-006 is also able to use d-carnitine as a sole carbon and nitrogen source. Mutations that significantly impair l-CDH activity permitted growth on d-carnitine as well as wild type, suggesting that a carnitine racemase was likely not responsible (Hwang & Bang, 1997). However, the d-carnitine metabolic pathway in Enterobacter has not been described fully. While Enterobacter likely does not have a carnitine racemase, there are bacteria that employ such an enzyme for d-carnitine metabolism. In Pseudomonas sp. AK1, a cytoplasmic carnitine racemase converts d-carnitine to l-carnitine, which is subsequently metabolized to glycine via glycine betaine to supply the cell with carbon and nitrogen (Monnich et al., 1995). E. coli 044 K74 also expresses carnitine racemase activity, which is induced in the presence of l-carnitine or crotonobetaine, and repressed by glucose, oxygen and fumarate (Canovas et al., 2003; Castellar et al., 1998; Jung & Kleber, 1991). CaiD was initially suggested to function as the racemase, as the caiD gene is required for racemase activity (Eichler et al., 1994b; Jung & Kleber, 1991; Jung et al., 1987); however, this has not been tested directly. CaiD is still postulated to be involved in racemization of d-carnitine, as CaiC was shown to activate d-carnitine by adding a CoA group to produce d-carnitinyl-CoA, which is then theorized to be converted to l-carnitinyl-CoA by CaiD (Bernal et al., 2008).
Carnitine synthesis by γ-butyrobetaine hydroxylase
The direct route to the formation of carnitine from γ-butyrobetaine occurs through the enzyme γ-butyrobetaine hydroxylase (EC 1.14.11.1). It has been identified in Pseudomonas sp. AK1 (Lindstedt et al., 1970a, b; Rüetschi et al., 1993) and Pseudomonas sp. L1 (Lu et al., 2012), and both enzymes are homologous to the animal γ-butyrobetaine hydroxylase in their requirement for oxygen and the cofactors iron, ascorbate, and α-ketogluterate (Lindstedt & Lindstedt, 1970; Lindstedt et al., 1968).
Acylcarnitine
A fatty acid moiety can be conjugated to the third carbon of carnitine resulting in O-acylcarnitines (Fig. 1), which can serve as sources of carnitine, but also can alter bacterial physiology directly (Nguyen et al., 2012). Pseudomonas aeruginosa can utilize acylcarnitines with 2–16 fatty acid chain lengths as sole carbon, nitrogen and energy sources, with the exception of octanoylcarnitine, although the reason for this utilization gap is unknown (Meadows & Wargo, 2013). Short-chain acylcarnitines (acetyl- and butyrylcarnitine) are hydrolysed to l-carnitine and a short-chain fatty acid by the esterase HocS (Meadows & Wargo, 2013), while the medium- and long-chain acylcarnitine hydrolase(s) has not yet been identified. Pseudomonas putida can utilize l-acylcarnitines with 10–16 fatty acid chain lengths as sole carbon and nitrogen sources (Kleber et al., 1978). The l-enantiomers of short-chain acylcarnitines acetyl-, propionyl-, butyryl-, and iso-butyrylcarnitine, the medium-chain lauroylcarnitine, and the long-chain palmitoylcarnitine can be hydrolysed to carnitine in Acinetobacter calcoaceticus (Kleber et al., 1977). Hydrolysis of d- and l-octanoylcarnitine has been assessed for a consortium of yeast, bacteria, and fungi, resulting in the findings that Bacillus subtilis ATCC 6633, Bacillus subtilis sp. IMAM and Penicillium notatum IMAM were capable of hydrolysing d- and l-octanoylcarnitine, whilst Pseudomonas fluorescens IMAM, Rhodotorula gracilis IMAM and Fusarium oxysporum sp. lini IMAM had specificity for the l-enantiomer only. However, in this study, the biological function of the resulting carnitine or fatty acid in these strains was not assessed (Aragozzini et al., 1986). An acylcarnitine hydrolase has also been purified from an Alcaligenes species, and was capable of hydrolysing acetyl-, propinoyl-, hexanoyl-, octanoyl-, decanoyl-, lauroyl-, myristoyl-, palmitoyl-, and stearoylcarnitine (Takahashi & Ueda, 1995); however, the gene encoding this enzyme was not determined.
Other carnitine derivatives
Carnitine is used in nutritional supplements, energy drinks, to replace carnitine lost during dialysis, and in treatments for carnitine uptake disorders. As d-carnitine is inhibitory to uptake and metabolism in mammals, these industries have searched for cost-effective methods to synthesize the l-carnitine enantiomer. One strategy for l-carnitine synthesis is to start with a more easily synthesized enantiomeric precursor, such as l-carnitine nitrile or l-carnitine amide, in order to decrease the cost and streamline the production of l-carnitine. A bacterium isolated from the soil, DSM 6230 (no taxonomic classification has been published), metabolizes l-carnitine amide into l-carnitine and ammonia by a novel enzyme l-carnitine amidase (Kula & Joeres, 1993; Joeres & Kula, 1994). The identified carnitine amidase is highly specific for l-carnitine amide, and the d-enantiomer inhibits the enzyme and cannot be used as a substrate (Kula et al., 1996). Another compound that can be metabolized to form carnitine is carnitine nitrile. A nitrilase from Corynebacterium, carnitine nitrile hydrolase, converts d- or l-carnitine nitrile into its corresponding d- or l-carnitine and ammonia (Kakayama et al., 1991). Norcarnitine is a derivative of carnitine that has a diethylamino group instead of trimethylamino group, and can be used as a sole carbon and nitrogen source in Pseudomonas putida (Kleber et al., 1978). The biological importance of these carnitine-related enzymes has not yet been examined, but the findings suggest that there is either enzymic flexibility in some of the carnitine metabolic enzymes or that there are additional, naturally occurring carnitine-like compounds in the environment.
Conclusions and future directions
Bacteria import, synthesize, and metabolize carnitine through various pathways that have different physiological effects. Our understanding of carnitine transport and metabolism is derived from studying extracellular or facultative intracellular bacteria and examining how carnitine is obtained from either the environment or within an animal host. However, there are a number of important questions that remain to be addressed related to carnitine-dependent transcriptional regulation, the ecological roles of carnitine, the role of carnitine in obligate intracellular bacteria and its importance during non-pathogenic interactions, such as symbioses.
In relation to obligate intracellular pathogens, we know surprisingly little. For instance, to date, no spirochaetes have been identified to use carnitine, but the causative agent of syphilis, Treponema pallidum, is predicted to harbour a carnitine transporter (Saier & Paulsen, 2000; Smajs et al., 2005). With Treponema pallidum only having 1000 genes (Fraser et al., 1998), carnitine transport may be important for survival in certain environments and for establishing infection. Furthermore, it is tempting to speculate that intracellular pathogens and symbionts can likely use carnitine based on the role of carnitine:fatty acid transport systems in the mitochondria, particularly in light of the endosymbiotic source of these organelles. As such, one might expect a role for carnitine import in the Rickettsiales.
Beyond pathogens and symbionts, the impact of environmental metabolism of the osmoprotectants glycine betaine and dimethylsulfionylproprionate has been reasonably well studied (reviewed by Curson et al., 2011; Welsh, 2000), but carnitine has not received much attention in relation to its contributions outside of animal infection and likely deserves additional scrutiny. For instance, the source of carnitine detected in the environment (Warren, 2013a, b) is unknown and we know very little about its half-life as a soluble compound in the environment or the flux rate of carnitine in any environment. A priori, one would assume an important role for carnitine metabolism, both aerobic and anaerobic, during animal decomposition on land or in marine environments. In the deep oceans in particular, carnitine utilization from fish and marine mammal carcasses might represent an important pathway to scavenge all available nutrients in this harsh environment.
Finally, we know almost nothing about the mechanisms by which carnitine is bound and detected to mediate transcriptional regulation. Further investigations into CdhR-like, CaiF-like and the predicted carnitine-sensing TetR family regulators in Gram-positives will be needed to understand how ligand detection is accomplished and converted into regulation of gene expression. Of particular import will be understanding the binding pocket of these regulators to determine if they maintain the cation–π binding aromatic cage that typifies the known quaternary amine-binding proteins crystallized to date. From an evolutionary perspective, CdhR likely arose after gene duplication from a GbdR-like ancestor, while the Gram-positive TetR family proteins are reasonably similar to the choline-binding transcription factor BetI. Thus, characterization of any of these quaternary amine-binding transcription factors will provide crucial understanding of ligand binding and, by homology, present likely binding residues in their respective paralogues. Beyond direct ligand-sensing transcription regulators, we do not know of any two-component systems or chemotaxis regulators that sense and respond to carnitine. The GlrA/GlrK system may represent the actual carnitine-sensing input that impinges on CaiF to establish carnitine-sensitive gene induction. Additionally, many bacteria have been shown to chemotax towards the quaternary amines dimethylsulfionylproprionate and glycine betaine (Miller et al., 2004; Seymour et al., 2010; Stocker & Seymour, 2012), and therefore any discovery of chemotaxis towards carnitine will further our understanding of its role in bacterial biology.
Acknowledgements
J. A. M. was supported by a National Institutes of Health Institutional NRSA Fellowship (T32 AI055402). M. J. W. was supported for research related to this review by grants from the National Center for Research Resources (P20 RR021905), the National Institute of General Medical Sciences (P20 GM103496) and the National Institute of Allergy and Infectious Disease (R01 AI103003).
Abbreviations:
- ABC
ATP-binding cassette
- BKACE
β-keto acid cleavage enzyme
- CDH
carnitine dehydrogenase
- HTH
helix–turn–helix
- TMAO
trimethylamine-N-oxide
References
- 1. Achouak W., Christen R., Barakat M., Martel M. H., Heulin T. (1999). Burkholderia caribensis sp. nov., an exopolysaccharide-producing bacterium isolated from vertisol microaggregates in Martinique. Int J Syst Bacteriol 49, 787–794 10.1099/00207713-49-2-787 . [DOI] [PubMed] [Google Scholar]
- 2. Angelidis A. S., Smith G. M. (2003). Role of the glycine betaine and carnitine transporters in adaptation of Listeria monocytogenes to chill stress in defined medium. Appl Environ Microbiol 69, 7492–7498 10.1128/AEM.69.12.7492-7498.2003 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Angelidis A. S., Smith L. T., Hoffman L. M., Smith G. M. (2002). Identification of opuC as a chill-activated and osmotically activated carnitine transporter in Listeria monocytogenes . Appl Environ Microbiol 68, 2644–2650 10.1128/AEM.68.6.2644-2650.2002 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Aragozzini F., Manzoni M., Cavazzoni V., Craveri R. (1986). d,l-Carnitine resolution by Fusarium oxysporum . Biotechnol Lett 8, 95–97 10.1007/BF01048463. [DOI] [Google Scholar]
- 5. Arima J., Uesumi A., Mitsuzumi H., Mori N. (2010). Biochemical characterization of l-carnitine dehydrogenases from Rhizobium sp. and Xanthomonas translucens . Biosci Biotechnol Biochem 74, 1237–1242 10.1271/bbb.100072 . [DOI] [PubMed] [Google Scholar]
- 6. Aurich H., Kleber H. P., Schöpp W. D. (1967). An inducible carnitine dehydrogenase from Pseudomonas aeruginosa . Biochim Biophys Acta 139, 505–507. 10.1016/0005-2744(67)90054-X [DOI] [PubMed] [Google Scholar]
- 7. Barbier M., Owings J. P., Martínez-Ramos I., Damron F. H., Gomila R., Blázquez J., Goldberg J. B., Albertí S. (2013). Lysine trimethylation of EF-Tu mimics platelet-activating factor to initiate Pseudomonas aeruginosa pneumonia. MBio 4, e00207–e00213. 10.1128/mBio.00207-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Barrett E. L., Kwan H. S. (1985). Bacterial reduction of trimethylamine oxide. Annu Rev Microbiol 39, 131–149. 10.1146/annurev.mi.39.100185.001023 [DOI] [PubMed] [Google Scholar]
- 9. Bastard K., Smith A. A., Vergne-Vaxelaire C., Perret A., Zaparucha A., De Melo-Minardi R., Mariage A., Boutard M., Debard A., other authors (2014). Revealing the hidden functional diversity of an enzyme family. Nat Chem Biol 10, 42–49. 10.1038/nchembio.1387 [DOI] [PubMed] [Google Scholar]
- 10. Bayles D. O., Wilkinson B. J. (2000). Osmoprotectants and cryoprotectants for Listeria monocytogenes . Lett Appl Microbiol 30, 23–27. 10.1046/j.1472-765x.2000.00646.x [DOI] [PubMed] [Google Scholar]
- 11. Begley M., Gahan C. G., Hill C. (2005). The interaction between bacteria and bile. FEMS Microbiol Rev 29, 625–651. 10.1016/j.femsre.2004.09.003 [DOI] [PubMed] [Google Scholar]
- 12. Bennett B. J., de Aguiar Vallim T. Q., Wang Z., Shih D. M., Meng Y., Gregory J., Allayee H., Lee R., Graham M., other authors (2013). Trimethylamine-N-oxide, a metabolite associated with atherosclerosis, exhibits complex genetic and dietary regulation. Cell Metab 17, 49–60. 10.1016/j.cmet.2012.12.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bernal V., Arense P., Blatz V., Mandrand-Berthelot M. A., Cánovas M., Iborra J. L. (2008). Role of betaine: CoA ligase (CaiC) in the activation of betaines and the transfer of coenzyme A in Escherichia coli . J Appl Microbiol 105, 42–50. 10.1111/j.1365-2672.2008.03740.x [DOI] [PubMed] [Google Scholar]
- 14. Beumer R. R., Te Giffel M. C., Cox L. J., Rombouts F. M., Abee T. (1994). Effect of exogenous proline, betaine, and carnitine on growth of Listeria monocytogenes in a minimal medium. Appl Environ Microbiol 60, 1359–1363. 10.1146/annurev.bi.57.070188.001401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Bieber L. L. (1988). Carnitine. Annu Rev Biochem 57, 261–283. 10.1111/j.1758-2229.2010.00236.x [DOI] [PubMed] [Google Scholar]
- 16. Bremer J. (1983). Carnitine – metabolism and functions. Physiol Rev 63, 1420–1480. [DOI] [PubMed] [Google Scholar]
- 17. Bremer E. (2011). Crystal ball – 2011. Environ Microbiol Rep 3, 1–26. 10.1099/00221287-72-3-589 [DOI] [PubMed] [Google Scholar]
- 18. Brown A. D., Simpson J. R. (1972). Water relations of sugar-tolerant yeasts: the role of intracellular polyols. J Gen Microbiol 72, 589–591. 10.1046/j.1365-2958.1999.01622.x [DOI] [PubMed] [Google Scholar]
- 19. Buchet A., Eichler K., Mandrand-Berthelot M. A. (1998). Regulation of the carnitine pathway in Escherichia coli: investigation of the cai–fix divergent promoter region. J Bacteriol 180, 2599–2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Buchet A., Nasser W., Eichler K., Mandrand-Berthelot M. A. (1999). Positive co-regulation of the Escherichia coli carnitine pathway cai and fix operons by CRP and the CaiF activator. Mol Microbiol 34, 562–575. 10.1016/S0032-9592(03)00080-3 [DOI] [PubMed] [Google Scholar]
- 21. Canovas D., Castellar M. R., Obon T., Torroglosa C., Olivares J. L., Iborra J. L. (2003). Racemisation of d(+)-carnitine into l( − )-carnitine by Escherichia coli strains. Process Biochem 39, 287–293. 10.1046/j.1365-2672.1998.00601.x [DOI] [Google Scholar]
- 22. Castellar M. R., Cánovas M., Kleber H. P., Iborra J. L. (1998). Biotransformation of d(+)-carnitine into l( − )-carnitine by resting cells of Escherichia coli O44 K74. J Appl Microbiol 85, 883–890. 10.1177/011542650802300116 [DOI] [PubMed] [Google Scholar]
- 23. Cave M. C., Hurt R. T., Frazier T. H., Matheson P. J., Garrison R. N., McClain C. J., McClave S. A. (2008). Obesity, inflammation, and the potential application of pharmaconutrition. Nutr Clin Pract 23, 16–34. 10.1128/JB.00763-07 [DOI] [PubMed] [Google Scholar]
- 24. Chen C., Beattie G. A. (2007). Characterization of the osmoprotectant transporter OpuC from Pseudomonas syringae and demonstration that cystathionine-beta-synthase domains are required for its osmoregulatory function. J Bacteriol 189, 6901–6912. 10.1111/j.1365-2958.2009.06962.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chen C., Malek A. A., Wargo M. J., Hogan D. A., Beattie G. A. (2010). The ATP-binding cassette transporter Cbc (choline/betaine/carnitine) recruits multiple substrate-binding proteins with strong specificity for distinct quaternary ammonium compounds. Mol Microbiol 75, 29–45. 10.1099/mic.0.26355-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Combs G. F., Jr (2012). The Vitamins: Fundamental Aspects in Health and Nutrition, 4th edn London: Academic Press. [Google Scholar]
- 27. Creasey E. A., Delahay R. M., Daniell S. J., Frankel G. (2003). Yeast two-hybrid system survey of interactions between LEE-encoded proteins of enteropathogenic Escherichia coli . Microbiology 149, 2093–2106. 10.1038/nrmicro2653 [DOI] [PubMed] [Google Scholar]
- 28. Curson A. R., Todd J. D., Sullivan M. J., Johnston A. W. (2011). Catabolism of dimethylsulphoniopropionate: microorganisms, enzymes and genes. Nat Rev Microbiol 9, 849–859. 10.1016/S0168-6496(03)00211-3 [DOI] [PubMed] [Google Scholar]
- 29. Dalmastri C., Fiore A., Alisi C., Bevivino A., Tabacchioni S., Giuliano G., Sprocati A. R., Segre L., Mahenthiralingam E., other authors (2003). A rhizospheric Burkholderia cepacia complex population: genotypic and phenotypic diversity of Burkholderia cenocepacia and Burkholderia ambifaria . FEMS Microbiol Ecol 46, 179–187. 10.1042/BJ20102097 [DOI] [PubMed] [Google Scholar]
- 30. Du Y., Shi W. W., He Y. X., Yang Y. H., Zhou C. Z., Chen Y. (2011). Structures of the substrate-binding protein provide insights into the multiple compatible solute binding specificities of the Bacillus subtilis ABC transporter OpuC. Biochem J 436, 283–289. 10.1111/j.1365-2958.1994.tb00470.x [DOI] [PubMed] [Google Scholar]
- 31. Eichler K., Schunck W. H., Kleber H. P., Mandrand-Berthelot M. A. (1994a). Cloning, nucleotide sequence, and expression of the Escherichia coli gene encoding carnitine dehydratase. J Bacteriol 176, 2970–2975. 10.1002/jobm.3620350404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Eichler K., Bourgis F., Buchet A., Kleber H. P., Mandrand-Berthelot M. A. (1994b). Molecular characterization of the cai operon necessary for carnitine metabolism in Escherichia coli . Mol Microbiol 13, 775–786. 10.1016/S0378-1097(99)00151-2 [DOI] [PubMed] [Google Scholar]
- 33. Eichler K., Buchet A., Bourgis F., Kleber H. P., Mandrand-Berthelot M. A. (1995). The fix Escherichia coli region contains four genes related to carnitine metabolism. J Basic Microbiol 35, 217–227. 10.1021/bi000776c [DOI] [PubMed] [Google Scholar]
- 34. Eichler K., Buchet A., Lemke R., Kleber H. P., Mandrand-Berthelot M. A. (1996). Identification and characterization of the caiF gene encoding a potential transcriptional activator of carnitine metabolism in Escherichia coli . J Bacteriol 178, 1248–1257. 10.1021/bi0108812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Elssner T., Preusser A., Wagner U., Kleber H. P. (1999). Metabolism of l( − )-carnitine by Enterobacteriaceae under aerobic conditions. FEMS Microbiol Lett 174, 295–301. 10.1111/j.1574-6968.2001.tb10531.x [DOI] [PubMed] [Google Scholar]
- 36. Elssner T., Hennig L., Frauendorf H., Haferburg D., Kleber H. P. (2000). Isolation, identification, and synthesis of gamma-butyrobetainyl-CoA and crotonobetainyl-CoA, compounds involved in carnitine metabolism of E. coli . Biochemistry 39, 10761–10769. 10.1007/s002030100272 [DOI] [PubMed] [Google Scholar]
- 37. Elssner T., Engemann C., Baumgart K., Kleber H. P. (2001). Involvement of coenzyme A esters and two new enzymes, an enoyl-CoA hydratase and a CoA-transferase, in the hydration of crotonobetaine to l-carnitine by Escherichia coli . Biochemistry 40, 11140–11148. 10.1007/s00203-005-0760-2 [DOI] [PubMed] [Google Scholar]
- 38. Engemann C., Kleber H. P. (2001). Epigenetic regulation of carnitine metabolising enzymes in Proteus sp. under aerobic conditions. FEMS Microbiol Lett 196, 1–6. 10.1128/JB.00596-12 [DOI] [PubMed] [Google Scholar]
- 39. Engemann C., Elssner T., Kleber H. P. (2001). Biotransformation of crotonobetaine to l( − )-carnitine in Proteus sp. Arch Microbiol 175, 353–359. 10.1186/1743-7075-7-30 [DOI] [PubMed] [Google Scholar]
- 40. Engemann C., Elssner T., Pfeifer S., Krumbholz C., Maier T., Kleber H. P. (2005). Identification and functional characterisation of genes and corresponding enzymes involved in carnitine metabolism of Proteus sp. Arch Microbiol 183, 176–189. 10.1016/S0083-6729(08)60508-7 [DOI] [PubMed] [Google Scholar]
- 41. Fitzsimmons L. F., Hampel K. J., Wargo M. J. (2012). Cellular choline and glycine betaine pools impact osmoprotection and phospholipase C production in Pseudomonas aeruginosa . J Bacteriol 194, 4718–4726. 10.1111/j.1574-6968.2002.tb11223.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Flanagan J. L., Simmons P. A., Vehige J., Willcox M. D., Garrett Q. (2010). Role of carnitine in disease. Nutr Metab (Lond) 7, 30. 10.1126/science.281.5375.375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fraenkel G., Friedman S. (1957). Carnitine. Vitam Horm 15, 73–118. 10.1128/AEM.66.11.4696-4704.2000 [DOI] [PubMed] [Google Scholar]
- 44. Fraser K. R., O'Byrne C. P. (2002). Osmoprotection by carnitine in a Listeria monocytogenes mutant lacking the OpuC transporter: evidence for a low affinity carnitine uptake system. FEMS Microbiol Lett 211, 189–194. 10.3389/fcimb.2014.00009 [DOI] [PubMed] [Google Scholar]
- 45. Fraser C. M., Norris S. J., Weinstock G. M., White O., Sutton G. G., Dodson R., Gwinn M., Hickey E. K., Clayton R., other authors (1998). Complete genome sequence of Treponema pallidum, the syphilis spirochete. Science 281, 375–388. 10.1094/MPMI-4-571 [DOI] [PubMed] [Google Scholar]
- 46. Fraser K. R., Harvie D., Coote P. J., O'Byrne C. P. (2000). Identification and characterization of an ATP binding cassette l-carnitine transporter in Listeria monocytogenes . Appl Environ Microbiol 66, 4696–4704. 10.1515/bchm2.1905.45.3-4.326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Gahan C. G., Hill C. (2014). Listeria monocytogenes: survival and adaptation in the gastrointestinal tract. Front Cell Infect Microbiol 4, 9. 10.1016/S0167-4838(96)00161-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Goldmann A., Boivin C., Fleury V., Message B., Lecoeur L., Maille M., Tepfer D. (1991). Betaine use by rhizosphere bacteria: genes essential for trigonelline, stachydrine, and carnitine catabolism in Rhizobium meliloti are located on pSym in the symbiotic region. Mol Plant Microbe Interact 4, 571–578. 10.1016/0304-4165(96)00020-7 [DOI] [PubMed] [Google Scholar]
- 49. Gulewitsch W., Krimberg R. (1905). [On carnitine]. Hoppe Seylers Z Physiol Chem 45, 326–330 (in German) 10.1161/ATVBAHA.114.303252 [DOI] [Google Scholar]
- 50. Hanschmann H., Kleber H. P. (1997). Purification and characterization of d(+)-carnitine dehydrogenase from Agrobacterium sp. – a new enzyme of carnitine metabolism. Biochim Biophys Acta 1337, 133–142. 10.1128/JB.01319-10 [DOI] [PubMed] [Google Scholar]
- 51. Hanschmann H., Ehricht R., Kleber H. P. (1996). Purification and properties of l( − )-carnitine dehydrogenase from Agrobacterium sp. Biochim Biophys Acta 1290, 177–183. 10.1128/JB.01505-12 [DOI] [PubMed] [Google Scholar]
- 52. Hartiala J., Bennett B. J., Tang W. H., Wang Z., Stewart A. F., Roberts R., McPherson R., Lusis A. J., Hazen S. L., Allayee H., CARDIoGRAM Consortium (2014). Comparative genome-wide association studies in mice and humans for trimethylamine N-oxide, a proatherogenic metabolite of choline and l-carnitine. Arterioscler Thromb Vasc Biol 34, 1307–1313. 10.1099/ijs.0.02993-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hoffmann T., Bremer E. (2011). Protection of Bacillus subtilis against cold stress via compatible-solute acquisition. J Bacteriol 193, 1552–1562. 10.1098/rstb.1990.0030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hoffmann T., Wensing A., Brosius M., Steil L., Völker U., Bremer E. (2013). Osmotic control of opuA expression in Bacillus subtilis and its modulation in response to intracellular glycine betaine and proline pools. J Bacteriol 195, 510–522. 10.1016/S0923-2508(98)80081-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Hung K., Kleber H. P. (1985). [Occurrence and regulation of carnitine dehydrogenase of Pseudomonas species]. Wiss Z Karl-Marx-Univ Leipzig Math-Nat R 34, 293–296 (in German). [Google Scholar]
- 56. Hwang K. C., Bang W. (1997). Optimal resolution of l-carnitine from racemic dl-carnitine by Enterobacter sp. assimilating d-carnitine. J Microbiol Biotechnol 7, 318–322. [Google Scholar]
- 57. Ivanova E. P., Gorshkova N. M., Bowman J. P., Lysenko A. M., Zhukova N. V., Sergeev A. F., Mikhailov V. V., Nicolau D. V. (2004). Shewanella pacifica sp. nov., a polyunsaturated fatty acid-producing bacterium isolated from sea water. Int J Syst Evol Microbiol 54, 1083–1087. 10.1007/BF00173315 [DOI] [PubMed] [Google Scholar]
- 58. Jaenicke R., Heber U., Franks F., Chapman D., Griffin M. C. A., Hvidt A., Cowan D. A. (1990). Protein structure and function at low temperatures. Philos Trans R Soc Lond B Biol Sci 326, 535–551. 10.1016/j.numecd.2014.03.007 [DOI] [PubMed] [Google Scholar]
- 59. Jebbar M., Champion C., Blanco C., Bonnassie S. (1998). Carnitine acts as a compatible solute in Brevibacterium linens . Res Microbiol 149, 211–219. 10.1007/BF00172731 [DOI] [PubMed] [Google Scholar]
- 60. Joeres U., Kula M. R. (1994). Purification and characterisation of a microbial l-carnitine amidase. Appl Microbiol Biotechnol 40, 606–610. 10.1002/jobm.3620270303 [DOI] [PubMed] [Google Scholar]
- 61. Johri A. M., Heyland D. K., Hétu M. F., Crawford B., Spence J. D. (2014). Carnitine therapy for the treatment of metabolic syndrome and cardiovascular disease: evidence and controversies. Nutr Metab Cardiovasc Dis 24, 808–814. 10.1016/0005-2760(89)90232-4 [DOI] [PubMed] [Google Scholar]
- 62. Jung H., Kleber H. P. (1991). Metabolism of d(+)carnitine by Escherichia coli . Appl Microbiol Biotechnol 35, 391–395. 10.1002/jobm.3620300711 [DOI] [PubMed] [Google Scholar]
- 63. Jung K., Jung H., Kleber H. P. (1987). Regulation of l-carnitine metabolism in Escherichia coli . J Basic Microbiol 27, 131–137. 10.1074/jbc.M206319200 [DOI] [PubMed] [Google Scholar]
- 64. Jung H., Jung K., Kleber H. P. (1989). Purification and properties of carnitine dehydratase from Escherichia coli – a new enzyme of carnitine metabolization. Biochim Biophys Acta 1003, 270–276. 10.1073/pnas.1309071110 [DOI] [PubMed] [Google Scholar]
- 65. Jung H., Jung K., Kleber H. P. (1990). l-Carnitine uptake by Escherichia coli . J Basic Microbiol 30, 507–514. 10.1099/00221287-144-1-83 [DOI] [PubMed] [Google Scholar]
- 66. Jung H., Buchholz M., Clausen J., Nietschke M., Revermann A., Schmid R., Jung K. (2002). CaiT of Escherichia coli, a new transporter catalyzing l-carnitine/gamma-butyrobetaine exchange. J Biol Chem 277, 39251–39258. 10.1038/nprot.2009.2 [DOI] [PubMed] [Google Scholar]
- 67. Kakayama K., Honda H., Ogawa Y., Ohta T., Ozawa T. (1991). Method of producing carnitine., US Patent 5,041,375. [Google Scholar]
- 68. Kalayil S., Schulze S., Kühlbrandt W. (2013). Arginine oscillation explains Na+ independence in the substrate/product antiporter CaiT. Proc Natl Acad Sci U S A 110, 17296–17301. 10.1007/BF00301845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kappes R. M., Bremer E. (1998). Response of Bacillus subtilis to high osmolarity: uptake of carnitine, crotonobetaine and γ-butyrobetaine via the ABC transport system OpuC. Microbiology 144, 83–90. 10.1016/0003-9861(75)90196-4 [DOI] [PubMed] [Google Scholar]
- 70. Kelley L. A., Sternberg M. J. (2009). Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4, 363–371. 10.1111/j.1574-6968.1997.tb10212.x [DOI] [PubMed] [Google Scholar]
- 71. Kets E. P. W., Galinski E. A., de Bont J. A. M. (1994). Carnitine: a novel compatible solute in Lactobacillus plantarum . Arch Microbiol 162, 243–248. 10.1007/BF00429336 [DOI] [Google Scholar]
- 72. Klagsbrun M., Furano A. V. (1975). Methylated amino acids in the proteins of bacterial and mammalian cells. Arch Biochem Biophys 169, 529–539. 10.1007/BF00406039 [DOI] [PubMed] [Google Scholar]
- 73. Kleber H. P. (1997). Bacterial carnitine metabolism. FEMS Microbiol Lett 147, 1–9. [DOI] [PubMed] [Google Scholar]
- 74. Kleber H. P., Aurich H. (1967). [Damped oscillations in the synthesis of carnitine dehydrogenase by Pseudomonas aeruginosa]. Hoppe Seylers Z Physiol Chem 348, 1727–1729 (in German). [PubMed] [Google Scholar]
- 75. Kleber H. P., Schöpp W., Sorger H., Tauchert H., Aurich H. (1967). [Formation of 3-dehydrocarnitine from l-carnitine through the action of a Pseudomonas aeruginosa enzyme]. Acta Biol Med Ger 19, 659–667 (in German). [PubMed] [Google Scholar]
- 76. Kleber H. P., Seim H., Aurich H., Strack E. (1977). [Utilization of trimethylammonium-compounds by Acinetobacter calcoaceticus (author's transl)]. Arch Microbiol 112, 201–206 (in German). 10.1038/nm.3145 [DOI] [PubMed] [Google Scholar]
- 77. Kleber H. P., Seim H., Aurich H., Strack E. (1978). [Interrelationships between carnitine metabolism and fatty acid assimilation in Pseudomonas putida (author's transl)]. Arch Microbiol 116, 213–220 (in German). 10.1016/j.lfs.2014.09.028 [DOI] [PubMed] [Google Scholar]
- 78. Klüttermann K., Tauchert H., Kleber H. P. (2002). Synthesis of poly-beta-hydroxybutyrate by Agrobacterium radiobacter after growth on d-carnitine. Acta Biotechnol 22, 261–269. 10.1111/j.1749-6632.1996.tb33281.x [DOI] [Google Scholar]
- 79. Koeth R. A., Wang Z., Levison B. S., Buffa J. A., Org E., Sheehy B. T., Britt E. B., Fu X., Wu Y., other authors (2013). Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat Med 19, 576–585. 10.1021/bi00857a006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kuka J., Liepinsh E., Makrecka-Kuka M., Liepins J., Cirule H., Gustina D., Loza E., Zharkova-Malkova O., Grinberga S., other authors (2014). Suppression of intestinal microbiota-dependent production of pro-atherogenic trimethylamine N-oxide by shifting l-carnitine microbial degradation. Life Sci 117, 84–92. 10.1016/0304-4165(68)90317-6 [DOI] [PubMed] [Google Scholar]
- 81. Kula M. R., Joeres U. (1993). l-Carnitine amidase produced by a microorganism., US Patent 5,238,838. [Google Scholar]
- 82. Kula M. R., Joeres U., Stelkes-Ritter U. (1996). New microbial amidases. Ann N Y Acad Sci 799, 725–728. 10.1021/bi00824a014 [DOI] [Google Scholar]
- 83. Kutscher F. (1905). [Zur Kenntnis des Novains]. Hoppe-Seyler's Z Physiol Chem 49, 47–49 (in German). [Google Scholar]
- 84. Lindstedt G., Lindstedt S. (1970). Cofactor requirements of gamma-butyrobetaine hydroxylase from rat liver. J Biol Chem 245, 4178–4186. [PubMed] [Google Scholar]
- 85. Lindstedt G., Lindstedt S., Midtvedt T., Tofft M. (1967). The formation and degradation of carnitine in Pseudomonas . Biochemistry 6, 1262–1270. 10.1007/BF00294525 [DOI] [Google Scholar]
- 86. Lindstedt G., Lindstedt S., Olander B., Tofft M. (1968). Alpha-ketoglutarate and hydroxylation of gamma-butyrobetaine. Biochim Biophys Acta 158, 503–505. [DOI] [PubMed] [Google Scholar]
- 87. Lindstedt G., Lindstedt S., Tofft M. (1970a). Gamma-butyrobetaine hydroxylase from Pseudomonas sp AK 1. Biochemistry 9, 4336–4342. [DOI] [PubMed] [Google Scholar]
- 88. Lindstedt G., Lindstedt S., Midtvedt T., Tofft M. (1970b). Inducible gamma-butyrobetaine-degrading enzymes in Pseudomonas species AK 1. J Bacteriol 101, 1094–1095. 10.1128/JB.00160-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Lu X., Zhang P., Li Q., Liu H., Lin X., Ma X. (2012). [Cloning, expression and characterization of a gamma-butyrobetaine hydroxylase gene bbh from Pseudomonas sp. L-1]. Wei Sheng Wu Xue Bao 52, 602–610 (in Chinese). 10.1016/S1043-6618(05)80117-3 [DOI] [PubMed] [Google Scholar]
- 90. Lucchesi G. I., Lisa T. A., Casale C. H., Domenech C. E. (1995). Carnitine resembles choline in the induction of cholinesterase, acid phosphatase, and phospholipase C and in its action as an osmoprotectant in Pseudomonas aeruginosa . Curr Microbiol 30, 55–60. 10.1128/AEM.03943-12 [DOI] [PubMed] [Google Scholar]
- 91. Malek A. A., Chen C., Wargo M. J., Beattie G. A., Hogan D. A. (2011). Roles of three transporters, CbcXWV, BetT1, and BetT3, in Pseudomonas aeruginosa choline uptake for catabolism. J Bacteriol 193, 3033–3041. 10.1128/AEM.70.8.4692-4701.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Marciani P., Lindi C., Marzo A., Arrigoni Martelli E., Cardace G., Esposito G. (1991). l-Carnitine and carnitine ester transport in the rat small intestine. Pharmacol Res 23, 157–162. 10.1007/BF00415004 [DOI] [PubMed] [Google Scholar]
- 93. Meadows J. A., Wargo M. J. (2013). Characterization of Pseudomonas aeruginosa growth on O-acylcarnitines and identification of a short-chain acylcarnitine hydrolase. Appl Environ Microbiol 79, 3355–3363. 10.1016/0378-1097(95)00286-E [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Miller T. R., Hnilicka K., Dziedzic A., Desplats P., Belas R. (2004). Chemotaxis of Silicibacter sp. strain TM1040 toward dinoflagellate products. Appl Environ Microbiol 70, 4692–4701. 10.1271/bbb1961.52.249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Miura-Fraboin J., Kleber H. P., Englard S. (1982). Assimilation of gamma-butyrobetaine, and d- and l-carnitine by resting cell suspensions of Acinetobacter calcoaceticus and Pseudomonas putida . Arch Microbiol 133, 217–221. 10.1038/sj.jim.7000135 [DOI] [Google Scholar]
- 96. Monnich K., Hanschmann H., Kleber H. P. (1995). Utilization of d-carnitine by Pseudomonas sp. AK 1. FEMS Microbiol Lett 132, 51–55. 10.1128/AEM.07227-11 [DOI] [Google Scholar]
- 97. Mori N., Kasugai T., Kitamoto Y., Ichikawa Y. (1988). Purification and some properties of carnitine dehydrogenase from Xanthomonas translucens . Agric Biol Chem 52, 249–250. 10.1007/s002530051459 [DOI] [Google Scholar]
- 98. Naidu G. S., Lee I. Y., Cho O. K., Park Y. H. (2001). Conversion of gamma-butyrobetaine to l-carnitine by Achromobacter cycloclast . J Ind Microbiol Biotechnol 26, 309–315. 10.1099/mic.0.063065-0 [DOI] [PubMed] [Google Scholar]
- 99. Nguyen U. T., Wenderska I. B., Chong M. A., Koteva K., Wright G. D., Burrows L. L. (2012). Small-molecule modulators of Listeria monocytogenes biofilm development. Appl Environ Microbiol 78, 1454–1465. 10.1016/S0167-4838(99)00032-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Nobile S., Deshusses J. (1986). Transport of gamma-butyrobetaine in an Agrobacterium species isolated from soil. J Bacteriol 168, 780–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Obón J. M., Maiquez J. R., Cánovas M., Kleber H. P., Iborra J. L. (1999). High-density Escherichia coli cultures for continuous l( − )-carnitine production. Appl Microbiol Biotechnol 51, 760–764. 10.1016/S0065-3233(08)60377-0 [DOI] [PubMed] [Google Scholar]
- 102. Palmer G. C., Jorth P. A., Whiteley M. (2013). The role of two Pseudomonas aeruginosa anthranilate synthases in tryptophan and quorum signal production. Microbiology 159, 959–969. 10.1196/annals.1320.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Park S., Smith L. T., Smith G. M. (1995). Role of glycine betaine and related osmolytes in osmotic stress adaptation in Yersinia enterocolitica ATCC 9610. Appl Environ Microbiol 61, 4378–4381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Preusser A., Wagner U., Elssner T., Kleber H. P. (1999). Crotonobetaine reductase from Escherichia coli consists of two proteins. Biochim Biophys Acta 1431, 166–178. 10.1146/annurev.nutr.18.1.39 [DOI] [PubMed] [Google Scholar]
- 105. Privalov P. L., Gill S. J. (1988). Stability of protein structure and hydrophobic interaction. Adv Protein Chem 39, 191–234. 10.1146/annurev.nutr.21.1.193 [DOI] [PubMed] [Google Scholar]
- 106. Rebouche C. J. (2004). Kinetics, pharmacokinetics, and regulation of l-carnitine and acetyl-l-carnitine metabolism. Ann N Y Acad Sci 1033, 30–41. 10.1128/AEM.66.2.509-517.2000 [DOI] [PubMed] [Google Scholar]
- 107. Rebouche C. J. (2014). Modern Nutrition in Health and Disease, 11th edn Baltimore, MD: Lippincott Williams & Wilkins. [Google Scholar]
- 108. Rebouche C. J., Chenard C. A. (1991). Metabolic fate of dietary carnitine in human adults: identification and quantification of urinary and fecal metabolites. J Nutr 121, 539–546. [DOI] [PubMed] [Google Scholar]
- 109. Rebouche C. J., Seim H. (1998). Carnitine metabolism and its regulation in microorganisms and mammals. Annu Rev Nutr 18, 39–61. 10.1007/BF00878280 [DOI] [PubMed] [Google Scholar]
- 110. Reddy J. K., Hashimoto T. (2001). Peroxisomal beta-oxidation and peroxisome proliferator-activated receptor alpha: an adaptive metabolic system. Annu Rev Nutr 21, 193–230. 10.1016/0003-9861(86)90197-9 [DOI] [PubMed] [Google Scholar]
- 111. Robert H., Le Marrec C., Blanco C., Jebbar M. (2000). Glycine betaine, carnitine, and choline enhance salinity tolerance and prevent the accumulation of sodium to a level inhibiting growth of Tetragenococcus halophila . Appl Environ Microbiol 66, 509–517. 10.1111/j.1432-1033.1993.tb17855.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Roth S., Jung K., Jung H., Hommel R. K., Kleber H. P. (1994). Crotonobetaine reductase from Escherichia coli – a new inducible enzyme of anaerobic metabolization of l( − )-carnitine. Antonie van Leeuwenhoek 65, 63–69. 10.1128/JB.00553-07 [DOI] [PubMed] [Google Scholar]
- 113. Rudolph A. S., Crowe J. H., Crowe L. M. (1986). Effects of three stabilizing agents – proline, betaine, and trehalose – on membrane phospholipids. Arch Biochem Biophys 245, 134–143. 10.1074/jbc.M309771200 [DOI] [PubMed] [Google Scholar]
- 114. Rüetschi U., Nordin I., Odelhög B., Jörnvall H., Lindstedt S. (1993). γ-Butyrobetaine hydroxylase. Structural characterization of the Pseudomonas enzyme. Eur J Biochem 213, 1075–1080. 10.1038/nature09310 [DOI] [PubMed] [Google Scholar]
- 115. Russell R. M., Sharp F. C., Rasko D. A., Sperandio V. (2007). QseA and GrlR/GrlA regulation of the locus of enterocyte effacement genes in enterohemorrhagic Escherichia coli . J Bacteriol 189, 5387–5392. 10.1007/BF00690825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Ryser E. T., Marth E. H. (2007). Listeria, Listeriosis, and Food Safety, 3rd edn Boca Raton, FL: CRC Press. [Google Scholar]
- 117. Saier M. H., Jr, Paulsen I. T. (2000). Whole genome analyses of transporters in spirochetes: Borrelia burgdorferi and Treponema pallidum . J Mol Microbiol Biotechnol 2, 393–399. [PubMed] [Google Scholar]
- 118. Schiefner A., Breed J., Bösser L., Kneip S., Gade J., Holtmann G., Diederichs K., Welte W., Bremer E. (2004). Cation–pi interactions as determinants for binding of the compatible solutes glycine betaine and proline betaine by the periplasmic ligand-binding protein ProX from Escherichia coli . J Biol Chem 279, 5588–5596. 10.1126/science.1188418 [DOI] [PubMed] [Google Scholar]
- 119. Schulze S., Köster S., Geldmacher U., Terwisscha van Scheltinga A. C., Kühlbrandt W. (2010). Structural basis of Na+-independent and cooperative substrate/product antiport in CaiT. Nature 467, 233–236. 10.1186/1757-4749-2-20 [DOI] [PubMed] [Google Scholar]
- 120. Seim H., Löster H., Claus R., Kleber H. P., Strack E. (1982a). Stimulation of the anaerobic growth of Salmonella typhimurium by reduction of l-carnitine, carnitine derivatives and structure-related trimethylammonium compounds. Arch Microbiol 132, 91–95. 10.1128/AEM.67.6.2692-2698.2001 [DOI] [PubMed] [Google Scholar]
- 121. Seim H., Loster H., Claus R., Kleber H. P., Strack E. (1982b). Formation of gamma-butryobetaine and trimethylamine from quaternary ammonium compounds structure-related to l-carnitine and choline by Proteus vularis . FEMS Microbiol Lett 13, 201–205. [Google Scholar]
- 122. Seim H., Loster H., Kleber H. P. (1982c). [Reductive metabolism of l-carnitine and structure-related trimethylammonium compounds in Escherichia coli]. Acta Biol Med Ger 41, 1009–1019 (in German). [PubMed] [Google Scholar]
- 123. Seymour J. R., Simó R., Ahmed T., Stocker R. (2010). Chemoattraction to dimethylsulfoniopropionate throughout the marine microbial food web. Science 329, 342–345. 10.1046/j.1365-2672.2003.02056.x [DOI] [PubMed] [Google Scholar]
- 124. Sikorski J., Stackebrandt E., Wackernagel W. (2001). Pseudomonas kilonensis sp. nov., a bacterium isolated from agricultural soil. Int J Syst Evol Microbiol 51, 1549–1555. [DOI] [PubMed] [Google Scholar]
- 125. Sleator R. D., Hill C. (2010). Compatible solutes: the key to Listeria's success as a versatile gastrointestinal pathogen? Gut Pathog 2, 20. 10.1128/JB.187.5.1866-1874.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Sleator R. D., Wouters J., Gahan C. G., Abee T., Hill C. (2001). Analysis of the role of OpuC, an osmolyte transport system, in salt tolerance and virulence potential of Listeria monocytogenes . Appl Environ Microbiol 67, 2692–2698. 10.1128/AEM.70.12.7555-7557.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Sleator R. D., Francis G. A., O'Beirne D., Gahan C. G., Hill C. (2003). Betaine and carnitine uptake systems in Listeria monocytogenes affect growth and survival in foods and during infection. J Appl Microbiol 95, 839–846. 10.1016/j.mam.2004.06.006 [DOI] [PubMed] [Google Scholar]
- 128. Sleator R. D., Banville N., Hill C. (2009). Carnitine enhances the growth of Listeria monocytogenes in infant formula at 7 degrees C. J Food Prot 72, 1293–1295. [DOI] [PubMed] [Google Scholar]
- 129. Smajs D., McKevitt M., Howell J. K., Norris S. J., Cai W. W., Palzkill T., Weinstock G. M. (2005). Transcriptome of Treponema pallidum: gene expression profile during experimental rabbit infection. J Bacteriol 187, 1866–1874. 10.1128/MMBR.00029-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130. Smiddy M., Sleator R. D., Patterson M. F., Hill C., Kelly A. L. (2004). Role for compatible solutes glycine betaine and l-carnitine in listerial barotolerance. Appl Environ Microbiol 70, 7555–7557. 10.1099/00221287-112-2-315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Steiber A., Kerner J., Hoppel C. L. (2004). Carnitine: a nutritional, biosynthetic, and functional perspective. Mol Aspects Med 25, 455–473. 10.1038/nsmb.1788 [DOI] [PubMed] [Google Scholar]
- 132. Stocker R., Seymour J. R. (2012). Ecology and physics of bacterial chemotaxis in the ocean. Microbiol Mol Biol Rev 76, 792–812. 10.1021/jm401603e [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Strøm A. R., Olafsen J. A., Larsen H. (1979). Trimethylamine oxide: a terminal electron acceptor in anaerobic respiration of bacteria. J Gen Microbiol 112, 315–320. 10.1007/s00706-005-0331-x [DOI] [PubMed] [Google Scholar]
- 134. Takahashi M., Ueda S. (1995). Method of assaying for acyl-l-carnitine and short-chain acyl-carnitine., US Patent 5,385,829. [Google Scholar]
- 135. Tang L., Bai L., Wang W. H., Jiang T. (2010). Crystal structure of the carnitine transporter and insights into the antiport mechanism. Nat Struct Mol Biol 17, 492–496. 10.1016/0304-4165(66)90382-5 [DOI] [PubMed] [Google Scholar]
- 136. Tars K., Leitans J., Kazaks A., Zelencova D., Liepinsh E., Kuka J., Makrecka M., Lola D., Andrianovs V., other authors (2014). Targeting carnitine biosynthesis: discovery of new inhibitors against γ-butyrobetaine hydroxylase. J Med Chem 57, 2213–2236. 10.1016/j.atherosclerosis.2013.10.013 [DOI] [PubMed] [Google Scholar]
- 137. Uanschou C., Frieht R., Pittner F. (2005). What to learn from comparative genomic sequence analysis of l-carnitine dehydrogenase. Monatsh Chem 136, 1365–1381. 10.1042/0264-6021:3610417 [DOI] [Google Scholar]
- 138. Unemoto T., Hayashi M., Miyaki K., Hayashi M. (1966). Formation of trimethylamine from dl-carnitine by Serratia marcescens . Biochim Biophys Acta 121, 220–222. 10.1111/j.1365-2672.1998.tb05269.x [DOI] [PubMed] [Google Scholar]
- 139. Ussher J. R., Lopaschuk G. D., Arduini A. (2013). Gut microbiota metabolism of l-carnitine and cardiovascular risk. Atherosclerosis 231, 456–461. 10.1128/JB.184.14.4044-4047.2002 [DOI] [PubMed] [Google Scholar]
- 140. Vaz F. M., Wanders R. J. (2002). Carnitine biosynthesis in mammals. Biochem J 361, 417–429. 10.1146/annurev.biochem.74.082803.133329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Verheul A., Rombouts F. M., Beumer R. R., Abee T. (1995). An ATP-dependent l-carnitine transporter in Listeria monocytogenes Scott A is involved in osmoprotection. J Bacteriol 177, 3205–3212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Verheul A., Glaasker E., Poolman B., Abee T. (1997). Betaine and l-carnitine transport by Listeria monocytogenes Scott A in response to osmotic signals. J Bacteriol 179, 6979–6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Verheul A., Wouters J. A., Rombouts F. M., Abee T. (1998). A possible role of ProP, ProU and CaiT in osmoprotection of Escherichia coli by carnitine. J Appl Microbiol 85, 1036–1046. 10.1128/AEM.03565-12 [DOI] [PubMed] [Google Scholar]
- 144. Vilhelmsson O., Miller K. J. (2002). Humectant permeability influences growth and compatible solute uptake by Staphylococcus aureus subjected to osmotic stress. J Food Prot 65, 1008–1015. [DOI] [PubMed] [Google Scholar]
- 145. Walt A., Kahn M. L. (2002). The fixA and fixB genes are necessary for anaerobic carnitine reduction in Escherichia coli . J Bacteriol 184, 4044–4047. 10.1099/mic.0.028787-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Wanders R. J., Waterham H. R. (2006). Biochemistry of mammalian peroxisomes revisited. Annu Rev Biochem 75, 295–332. 10.1128/JB.01393-07 [DOI] [PubMed] [Google Scholar]
- 147. Wargo M. J. (2013). Homeostasis and catabolism of choline and glycine betaine: lessons from Pseudomonas aeruginosa . Appl Environ Microbiol 79, 2112–2120. 10.1016/j.soilbio.2012.09.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148. Wargo M. J., Hogan D. A. (2009). Identification of genes required for Pseudomonas aeruginosa carnitine catabolism. Microbiology 155, 2411–2419. 10.1111/nph.12171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Wargo M. J., Szwergold B. S., Hogan D. A. (2008). Identification of two gene clusters and a transcriptional regulator required for Pseudomonas aeruginosa glycine betaine catabolism. J Bacteriol 190, 2690–2699. 10.1128/IAI.00153-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Warren C. R. (2013a). High diversity of small organic N observed in soil water. Soil Biol Biochem 57, 444–450. 10.1111/j.1574-6976.2000.tb00542.x [DOI] [Google Scholar]
- 151. Warren C. R. (2013b). Quaternary ammonium compounds can be abundant in some soils and are taken up as intact molecules by plants. New Phytol 198, 476–485. 10.1128/AEM.68.10.4710-4716.2002 [DOI] [PubMed] [Google Scholar]
- 152. Watson D., Sleator R. D., Casey P. G., Hill C., Gahan C. G. (2009). Specific osmolyte transporters mediate bile tolerance in Listeria monocytogenes . Infect Immun 77, 4895–4904. 10.1128/AEM.70.5.2912-2918.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Welsh D. T. (2000). Ecological significance of compatible solute accumulation by micro-organisms: from single cells to global climate. FEMS Microbiol Rev 24, 263–290. 10.1074/jbc.M410104200 [DOI] [PubMed] [Google Scholar]
- 154. Wemekamp-Kamphuis H. H., Wouters J. A., Sleator R. D., Gahan C. G., Hill C., Abee T. (2002). Multiple deletions of the osmolyte transporters BetL, Gbu, and OpuC of Listeria monocytogenes affect virulence and growth at high osmolarity. Appl Environ Microbiol 68, 4710–4716. 10.1242/jeb.01730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Wemekamp-Kamphuis H. H., Sleator R. D., Wouters J. A., Hill C., Abee T. (2004). Molecular and physiological analysis of the role of osmolyte transporters BetL, Gbu, and OpuC in growth of Listeria monocytogenes at low temperatures. Appl Environ Microbiol 70, 2912–2918. 10.1099/ijs.0.64440-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Yamamoto K., Hirao K., Oshima T., Aiba H., Utsumi R., Ishihama A. (2005). Functional characterization in vitro of all two-component signal transduction systems from Escherichia coli . J Biol Chem 280, 1448–1456. 10.1073/pnas.1316569111 [DOI] [PubMed] [Google Scholar]
- 157. Yancey P. H. (2005). Organic osmolytes as compatible, metabolic and counteracting cytoprotectants in high osmolarity and other stresses. J Exp Biol 208, 2819–2830. 10.1242/jeb.01730 [DOI] [PubMed] [Google Scholar]
- 158. Zhang L., Xu Z., Patel B. K. (2007). Bacillus decisifrondis sp. nov., isolated from soil underlying decaying leaf foliage. Int J Syst Evol Microbiol 57, 974–978. 10.1099/ijs.0.64440-0 [DOI] [PubMed] [Google Scholar]
- 159. Zhu Y., Jameson E., Crosatti M., Schäfer H., Rajakumar K., Bugg T. D., Chen Y. (2014). Carnitine metabolism to trimethylamine by an unusual Rieske-type oxygenase from human microbiota. Proc Natl Acad Sci U S A 111, 4268–4273. 10.1073/pnas.1316569111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160. Ziegler C., Bremer E., Krämer R. (2010). The BCCT family of carriers: from physiology to crystal structure. Mol Microbiol 78, 13–34. [DOI] [PubMed] [Google Scholar]