Abstract
Peptidoglycan associated lipoprotein (Pal) of Escherichia coli (E. coli) is a characteristic bacterial lipoprotein, with an N-terminal lipid moiety anchoring it to the outer membrane. Since its discovery over three decades ago, Pal has been well studied for its participation in the Tol–Pal complex which spans the periplasm and has been proposed to play important roles in bacterial survival, pathogenesis and virulence. Previous studies of Pal place the lipoprotein in the periplasm of E. coli, allowing it to interact with Tol proteins and the peptidoglycan layer. Here, we describe for the first time, a subpopulation of Pal which is present on the cell surface of E. coli. Flow cytometry and confocal microscopy detect anti-Pal antibodies on the surface of intact E. coli cells. Interestingly, Pal is surface exposed in an ‘all or nothing’ manner, such that most of the cells contain only internal Pal, with fewer cells ( < 20 %) exhibiting surface Pal.
Introduction
Like all Gram-negative bacteria, Escherichia coli (E. coli) contains two distinct membranes, an outer membrane and an inner membrane, separated by an aqueous layer commonly referred to as the periplasm. The periplasm contains peptidoglycan (also known as murein), a mesh-like layer of amino acids and sugars which provide the cell with structural integrity (Vollmer & Bertsche, 2008) and a unique opportunity to ‘connect’ the two membranes. For example, in the Tol–Pal complex, a combination of lipoproteins, integral membrane proteins and periplasmic proteins interact with each other and the peptidoglycan, forming a web of covalent and noncovalent contacts between the outer and inner membranes (Yeh et al., 2010). The outer membrane peptidoglycan associated lipoprotein (Pal; tethered to the outer membrane via its N-terminal lipid moiety) is one of the key components in the Tol–Pal complex (Godlewska et al., 2009; Gerding et al., 2007) interacting noncovalently with the peptidoglycan layer (Leduc et al., 1992; Bouveret et al., 1999), outer membrane proteins OmpA (Clavel et al., 1998) and Lpp (Cascales et al., 2002), periplasmic protein TolB (Bouveret et al., 1995, 1999) and the inner membrane protein TolA (Cascales et al., 2000; Deprez et al., 2005). Although the exact function of Pal is unknown, Pal deletion mutants in E. coli exhibit cell envelope defects and greater susceptibility to the antibiotic vancomycin (Bernadac et al., 1998; Cascales & Lloubès, 2004). Pal has also been shown to dimerize in the presence of Lpp and OmpA (Cascales et al., 2002), although the physiological significance of dimerization is unknown.
While its periplasmic interactions have been well established, we considered the possibility that Pal may have a second orientation allowing surface exposure. We have recently shown that a homologue protein of Pal called P6 in nontypable Haemophilus influenzae (NTHi) has a dual orientation, existing as a surface-exposed protein and a periplasmic protein (Michel et al., 2013). Cowles et al. (2011) were the first to describe a bacterial lipoprotein existing in two distinct orientations. They studied Lpp in E. coli. It had been known for some time that Lpp existed in two distinct forms – one covalently bound to the peptidoglycan, and a ‘free’ population that did not interact with peptidoglycan (Braun & Rehn, 1969; Braun & Bosch, 1972). The novel observation made by Cowles et al. (2011) was the discovery that the ‘free’ population of Lpp spans the outer membrane, with one end of its trimeric structure exposed to the extracellular space.
In light of these recent Lpp and P6 dual orientation studies, we sought to determine if Pal might be another E. coli protein with a dual orientation. Here we describe experiments that show Pal is a third example of a dual oriented lipoprotein in Gram-negative bacteria, with a smaller population of Pal being surface exposed compared to its periplasmic population. We also show that the transmembrane protein OmpA does not play an important role in surface exposure of Pal. Pal has been shown to be released from E. coli in experimental models of sepsis (Hellman et al., 2000, 2002). We discuss the possibility that the surface form of Pal may allow for its release from the outer membrane during sepsis, thus potentially implicating dual orientation in an important pathological mechanism.
Methods
Bacterial strains and cell culture conditions
All E. coli cultures were grown on Luria Broth (LB) media. The following E. coli strains were utilized for experiments: XL1-Blue Supercompetent cells (Agilent); K1 RS218 cells and OmpA deletion cells (an OmpA-deficient derivative of K1 RS218), which were gifts from Dr Kwang Sik Kim, Johns Hopkins Children's Center (Wang & Kim, 2002); K12 JC1292 and JC7752 (a Pal-deficient derivative of JC1292) cells, which were a gift from Dr Jean-Claude Lazzaroni, University of Lyon (Hellman et al., 2000); Top10 competent cells (Life Technologies); and JM109 competent cells (Agilent or Sigma Aldrich). Cells were cultured on LB under aerobic conditions, shaking (200 r.p.m.) at 37 °C for 2–3 h until the OD600 reached 0.8 (exponential phase). Cells were pelleted gently (5000 g) and washed before further sample preparation.
SDS-PAGE and immunoblot assay
Samples for the 10 % SDS-PAGE experiments were prepared in non-reducing sample buffer (2 × recipe: 0.12 M Tris/HCl pH 6.8, 4 % SDS, 20 % glycerol, 0.01 % bromophenol blue) and boiled for 10 min. Proteins were transferred to a nitrocellulose membrane (Pierce) and blocked with 5 % milk in Tris buffered saline (TBS). The membrane was incubated with anti-Pal sera (Valentine et al., 2006) at a 1 : 4000 dilution, anti-alkaline phophatase (Millipore) at a 1 : 1000 dilution or anti-RNA polymerase B (Santa Cruz Biotechnology) at a 1:200 dilution in 1 % milk and TBS. The membrane was then incubated with HRP-conjugated goat anti-mouse IgG (Bethyl Laboratories) at a 1:12 000 dilution in 1 % milk and TBST (TBS with 0.05 % Tween-20). The membrane was washed with TBS or TBST between antibody incubations. The blot was visualized using the Lumiglo Reserve HRP chemiluminescent substrate kit (KPL) according to the manufacturer's instructions.
Flow cytometry
Bacterial samples were cultured as described above and used for surface expression analysis by flow cytometry as described in the literature (Michel et al., 2013). Briefly, approximately 250 μl of cells (OD600 of approximately 0.8) were washed twice with PBS/1 % BSA pH 7.2, and then incubated with 250 μl of primary antibody (anti-Pal) in serum at a 1:200 dilution for 1 h at room temperature. The cells were washed twice and then incubated with either 250 μl of either Alexa Fluor 488-labelled (Invitrogen) or Alexa Fluor 647-labelled (Life Technologies) goat anti-mouse IgG at 1:167 or 1:350 dilutions, respectively. After two additional washes, the cells were fixed using 250 μl of 4 % paraformaldehyde fixative reagent (BioLegend) for 30 min at room temperature. For the vancomycin experiments, cells were incubated with 17 μg ml− 1 of BODIPY FL vancomycin (Life Technologies) for 30 min prior to fixation. Permeabilized cells were treated with 0.1 % Triton X-100 (US Biologicals) in PBS (final concentration of approximately 1.7 mM Triton X-100) for 30 min at room temperature prior to fixation. All samples were resuspended in 200–250 μl of PBS and read on a LSR II flow cytometer (BD Biosciences).
Confocal microscopy
Bacterial samples were cultured and prepared as described above for flow cytometry. The confocal images were collected on a Leica TCS SP5 II AOBS Filter-free Tunable Spectral Confocal Research Microscope with Resonant Scanner and Hybrid Detectors (Leica Microsystems) attached to a Leica DMI6000 Fully Automated Microscope using Leica LAS system software and a 60 × oil immersion objective.
Biotinylation experiment: Sulfo-NHS-SS-biotin
A Cell Surface Protein Isolation kit (Pierce) was used according to the manufacturer's instructions, with some modifications. E. coli bacteria were cultured as described above. Ten millilitres of cells was pelleted (5000 g), washed in PBS and incubated with 10 ml of freshly prepared Sulfo-NHS-SS-biotin (0.6 mg ml− 1) for 30 min. Quenching solution (1.5 ml) was added to the cells which were then washed with TBS. Cells were resuspended in 10 ml TBS and lysed via sonication (15 s on, 45 s off, for 20 cycles) in the presence of 30 μl of Triton X-100. The cells were pelleted (15 000 g), and the supernatant was further concentrated to 500 μl using an Amicon Ultra-15 Centrifugal Filter unit (10 kDa MWCO). The supernatant was then added to a NeutrAvidin Agarose column (500 μl of beads, prepared as described in the manufacturer's instructions). After a 1 h incubation, the nonbiotinylated proteins were eluted off the column, added to 500 μl of 2 × SDS-PAGE sample buffer and stored at 4 °C. The biotinylated proteins were eluted off the column after a 1 h incubation with 500 μl of 1 M DTT and 2 × SDS-PAGE sample buffer. Before running the nonbiotinylated and biotinylated samples on a 10 % SDS-PAGE gel, the biotinylated sample was brought to a final volume of 1 ml with water. Both 1 ml (nonbiotinylated and biotinylated) samples were boiled for 10 min and stored in the − 20 °C freezer. Samples were run on an SDS-PAGE gel and detected using anti-Pal sera in an immunoblot, as described above. To serve as a control, the same samples were also detected using anti-RNA polymerase B or anti-alkaline phosphatase. To quantify the nonbiotinylated and biotinylated proteins, the chemiluminescence of the immunoblot was detected using a ChemiDoc XRS system (Bio-Rad) and band volumes were calculated using Quantity One 1-D Analysis software (Bio-Rad).
Biotinylation experiment: NHS-LC-LC-biotin
Biotinylation of surface proteins using NHS-LC-LC-biotin was performed as described in the literature (Cowles et al., 2011), with some modifications. Briefly, E. coli bacteria were cultured as described above until the OD600 reached approximately 0.8. Cell culture (10 ml) was gently pelleted, washed with PBS twice, and resuspended in 1 ml of PBS. A 25 mg ml− 1 stock of NHS-LC-LC-biotin was prepared in DMSO (Pierce) and added to a final concentration of 2 % v/v (20 μl per 1 ml of cell suspension). After a 20 min incubation at room temperature (rocking), the NHS-LC-LC-biotin was capped using 500 μl of the quenching solution from a Cell Surface Protein Isolation kit (Pierce). The cells were washed several times with TBS, and resuspended to a final volume of 10 ml of TBS. Cells were lysed via sonication (15 s on, 45 s off, for 20 cycles) in the presence of 30 μl of Triton X-100. After the cells were pelleted (15 000 g), the supernatant was concentrated in an Amicon Ultra-15 Centrifugal Filter unit (10 kDa MWCO) to a final volume of 500 μl. The supernatant was added to a NeutrAvidin Agarose column and allowed to incubate with 500 μl of NeutrAvidin Agarose beads (washed and spun dry prior to addition of the supernatant) for 60 min (as described above). The nonbiotinylated proteins were eluted off the column, added to 500 μl of 2 × SDS sample buffer, and boiled for 10 min. The NeutrAvidin beads and bound biotinylated proteins were washed four times with Wash buffer (provided in the Pierce Cell Surface Protein Isolation kit). Finally, 500 ml of 2 × SDS sample buffer was added to the beads and the mixture was boiled for 10 min to release the biotinylated proteins. The samples were brought to their final volume using TBS. Samples were separated and quantified as described in the other biotinylation experiment.
Results
Pal is expressed on the surface of Escherichia coli
The periplasmic population of Pal has been well documented (Godlewska et al., 2009). Therefore, our focus was to determine if Pal was surface exposed. To indirectly visualize surface Pal, we used confocal microscopy to detect Pal after monoclonal antibody (Valentine et al., 2006) and AlexaFluor 488-conjugated secondary antibody labelling. Surface Pal was observed and its expression was ‘all or nothing’; that is, E. coli (XL1-Blue) cells displayed either no visible surface Pal or were highly illuminated, as seen in Fig. 1(a). Confocal images demonstrated that only a few E. coli cells exhibited fluorescent staining (we estimate between 2.5–7 % of the total cells), suggesting that the cells with exposed Pal were scarce in a given population. The fluorescently labelled cells appeared to be of normal morphology and size compared to non-fluorescent E. coli cells.
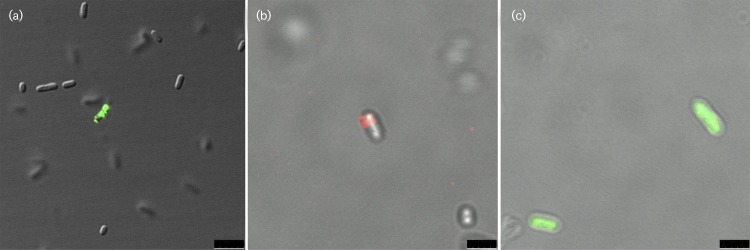
Fig. 1. Confocal images of E. coli stained with anti-Pal and vancomycin. (a) This pseudo-colour confocal image shows that only a small percentage of whole E. coli cells are stained with anti-Pal and Alexa Fluor 488 (green). Scale bar, 5 μm. (b) This merged pseudo-colour confocal image shows an E. coli cell stained with Pal and Alexa Fluor 647 (red), but not BODIPY FL vancomycin (green). Scale bar, 2.5 μm. (c) This pseudo-colour confocal image shows that BODIPY FL vancomycin (green) readily stains E. coli cells that were permeabilized by pretreatment with 0.1% Triton X-100.Scale bar, 2.5 μm.
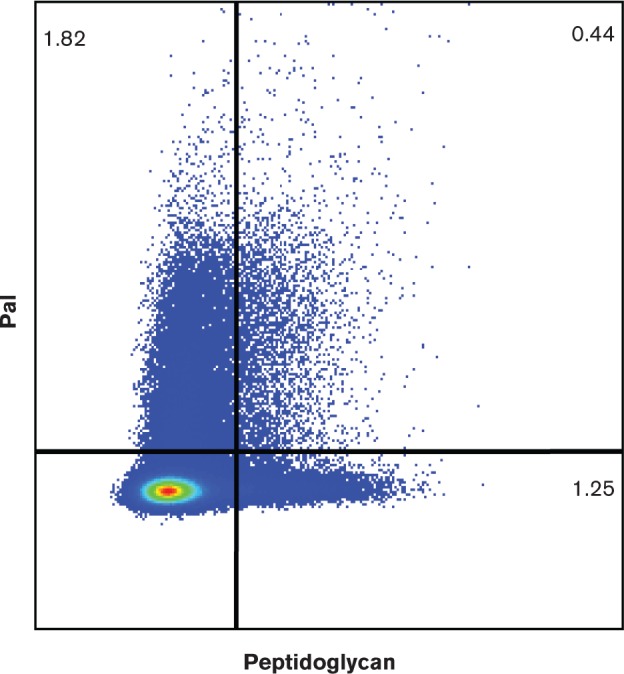
To ensure that the cells that exhibited surface Pal (as seen by confocal microscopy) were not the result of fragile or perforated bacterial membranes, similar experiments were performed with E. coli (XL1-Blue) cells using green fluorescently labelled vancomycin (BODIPY FL vancomycin) which targets the peptidoglycan of Gram-positive bacteria. Since vancomycin is too large to diffuse through the major porins of the outer membrane in Gram-negative cells (Cascales et al., 2000), sensitivity to vancomycin could only occur if the outer membranes of the E. coli cells were fragile or easily penetrable. We did visualize Pal-labelled cells that did not exhibit vancomycin labelling (Fig. 1b). As a control, we observed that the same concentration of vancomycin was able to readily label E. coli cells that were pre-treated with 0.1 % Triton X-100 detergent (Fig. 1c). We analysed the same Pal-labelled cell samples with vancomycin using flow cytometry, and observed that 1.82 % of the cells were labelled with anti-Pal only, 1.25 % of cells were labelled with vancomycin only, and 0.44 % of cells were labelled with both anti-Pal and vancomycin (Fig. 2; controls in Fig. S1 and S2, available in the online Supplementary Material).
Fig. 2. Flow cytometric detection of E. coli cells stained with anti-Pal and vancomycin. Flow cytometry data demonstrate that a small percentage of E. coli cells are stained with BODIPY FL vancomycin (peptidoglycan) only (1.25%), a small percentage with Alexa Fluor 647 (Pal) only (1.82%), and an even smaller percentage of cells with both stains (0.44%). The percentages of labeled bacteria (from total bacteria) are noted in the quadrants.
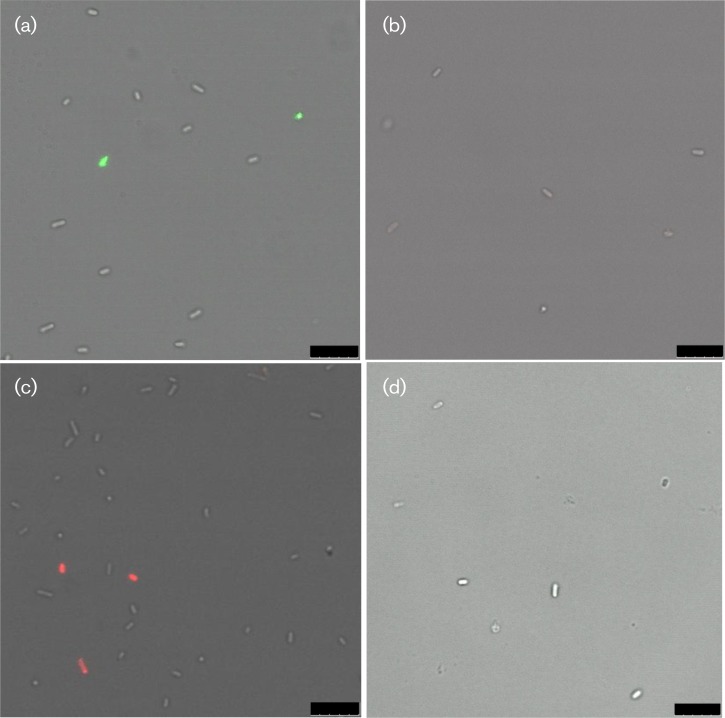
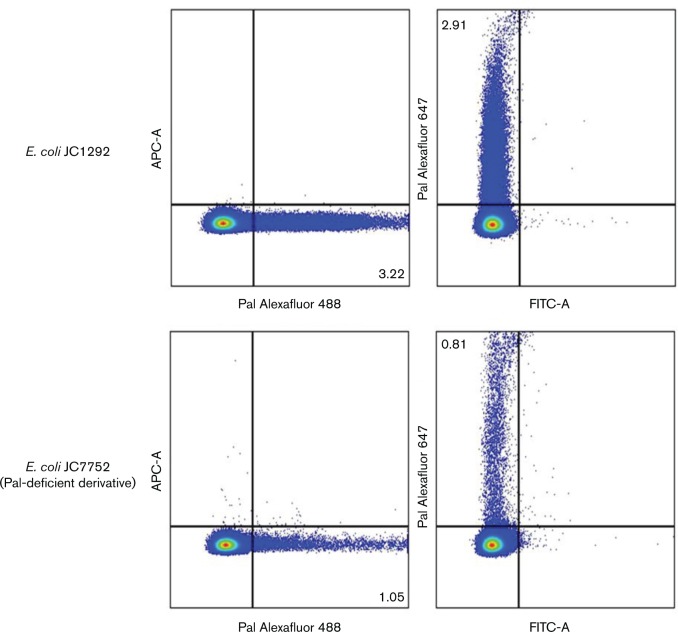
As an additional control, we performed labelling experiments using the Pal deletion strain JC7752 and compared them with experiments using the parent E. coli strain JC1292 (Hellman et al., 2000). The parent strain exhibited labelling with Pal and secondary antibody conjugated to Alexa Fluor 488 (green) or Alexa Fluor 647 (red), while the Pal deletion strain showed low Pal labelling above background (Fig. 3). Fig. 3 contains only single snapshots of the cells, but these snapshots are representative of what we saw throughout the entire sample. We performed flow cytometry on the same samples (parent strain: with anti-Pal and 488 or 647; Pal deletion strain: with anti-Pal and 488 or 647) (Fig. 4; controls in Fig. S3). The Pal deletion strain consistently showed some Pal labelling, but labelling was only approximately three times the background level. In contrast, the parent strain showed labelling approximately 23 times the background level.
Fig. 3. Confocal images comparing the Pal deletion strain with WT E. coli. Pseudo-colour confocal images show that the E. coli Pal deletion strain does not fluoresce when incubated with anti-Pal and Alexa Fluor 488 (b) or Alexa Fluor 647 (d). (a) and (c) show the E. coli parent strain fluorescing under the same incubation conditions, respectively. Bars, 10 μm.
Fig. 4. Flow cytometric detection of Pal in a Pal deletion strain and WT E. coli. E. coli JC1292 cells (top) and JC7752 (Pal-deficient derivative) cells (bottom). The left panels are cells stained with Alexa Fluor 488 (Pal) and the right panels are stained with Alexa Fluor 647 (Pal). These data were representative of the three replicates that were collected for the experiment. The percentages of labeled bacteria (from total bacteria) are noted in the quadrants.
The majority of Pal is periplasmic
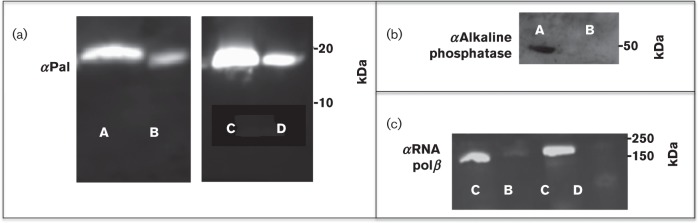
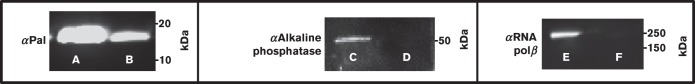
As determined with confocal microscopy and flow cytometry, Pal appeared to be on the surface of only a limited number of E. coli cells, in an ‘all or nothing’ manner. We estimated using confocal microscopy that between 2.5 % and 7 % (mean average of 5 %) of the total cells had surface Pal; not surprisingly, our flow cytometry experiments yielded similar values (approximately 3 % of total cells were labelled with anti-Pal). To quantify the average surface versus periplasmic Pal populations using an alternate method, we employed a biotinylation technique previously described in the literature (Cowles et al., 2011). The hydrophobic, primary amine-reactive biotinylation reagent, NHS-LC-LC-biotin, was shown to be impermeable to the E. coli outer membrane due to its hydrophobicity and relatively high molecular mass (Cowles et al., 2011). We employed the same reagent to label surface Pal in E. coli with biotin, thus allowing us to quantify internal and surface Pal populations. Monoclonal anti-Pal Western blots of the two populations showed that the majority of Pal was internally localized, while a much smaller population was surface exposed [E. coli K-12 RS218 cells: Fig. 5a (A and B) and Table 1]. Several other E. coli strains were also tested using the same methodology (Fig. S4 and Table 1), all yielding similar results with internal Pal population around 72 % and surface Pal around 28 %.
Fig. 5. Biotinylation of surface proteins in E. coli. (a) Representative immunoblots of Pal from E. coli K1 RS218: nonbiotinylated Pal (A) and Sulfo-NHS-SS-biotinylated Pal (B); nonbiotinylated Pal (C) and NHS-LC-LC-biotinylated Pal (D). (b) The immunoblot for alkaline phosphatase from E. coli K1 RS218 shows that all of the protein is nonbiotinylated (A) and no biotinylated protein (by NHS-LC-LC-biotin) was detected (B). (c) The immunoblot for RNA polymerase β from E. coli K1 RS218 shows that only a very small percentage of RNA polymerase β is biotinylated with NHS-LC-LC-biotin (14 %) (B) or Sulfo-NHS-SS-biotin (5 %) (D), while most of RNA polymerase β is nonbiotinylated (A, C).
Table 1. Biotinylation of Pal in several E. coli strains.
Average biotinylated (B) and nonbiotinylated (NB) Pal populations, using Sulfo-NHS-SS-biotin and NHS-LC-LC-biotin with different E. coli strains, suggest that most of the Pal populations are nonbiotinylated.
| E. coli strain | Biotin | NB (% population) | B (% population) |
| RS218 | Sulfo-NHS-SS-biotin | 74 | 26 |
| RS218 | NHS-LC-LC-biotin | 74 | 26 |
| XL1-Blue | NHS-LC-LC-biotin | 72 | 28 |
| Top10 | NHS-LC-LC-biotin | 69 | 31 |
| JM109 | NHS-LC-LC-biotin | 72 | 28 |
| JC1292 | NHS-LC-LC-biotin | 73 | 27 |
To confirm the NHS-LC-LC-biotin results, we used a second biotinylation reagent, Sulfo-NHS-SS-biotin (Lei et al., 2011) to quantify internal and surface Pal. The Sulfo-NHS-SS-biotin labelling results were similar to those with NHS-LC-LC-biotin. Internal Pal was present as 74 % of the total population, and surface Pal was present as 26 % of the population [Fig. 5a (C and D) and Table 1].
To ascertain that the biotin experiments were not a result of a compromised membrane, we used antibodies to two non-exposed proteins in E. coli to test the leakiness of the biotin reagents (Fig. 5b, c). We could not detect any labelling using our periplasmic control antibody (anti-alkaline phosphatase) (Fig. 5b), and the cytoplasmic control antibody (anti-RNA polymerase β) was only minimally leaky (NHS-LC-LC: 14 %, Sulfo: 5 %) (Fig. 5c), indicating that the quantification of the surface Pal populations using both Sulfo-NHS-SS-biotin and NHS-LC-LC-biotin were, at most, overestimates by 14 % and 5 %, respectively.
OmpA does not play a role in Pal surface exposure
OmpA is a large transmembrane protein with a periplasmic domain that has been shown to undergo a temperature-sensitive transition (Zakharian & Reusch, 2005) where it combines with the transmembrane region to form a larger β-barrel pore at elevated temperatures. The structure of the OmpA periplasmic domain was recently solved (Park et al., 2012) and, as suggested by its sequence homology to Pal (29 % sequence identity), its 3D structure is similar to the structure of Pal. We, therefore, wanted to assess whether or not OmpA played an integral important role in ‘flipping’ Pal to the cell surface. We biotinylated surface Pal using the NHS-LC-LC-biotin protocol on an E. coli ompA knockout mutant. The results of this experiment showed that Pal is surface exposed in the OmpA knockout mutant cells at the same level as in the WT E. coli parent strain (K1 RS218): 71 % internal Pal and 29 % surface Pal (Fig. 6). Neither biotinylated RNA polymerase β, nor biotinylated alkaline phosphatase, were detected in the same samples.
Fig. 6. Biotinylation of surface proteins in an OmpA deletion strain of E. coli. Immunoblots from the same biotinylation experiment with E. coli OmpA deletion cells: nonbiotinylated (A) and biotinylated Pal (B); nonbiotinylated (C) and biotinylated alkaline phosphatase (D); nonbiotinylated (E) and biotinylated RNA polymerase β (F).
Discussion
Since its discovery in 1981 (Mizuno, 1981), Pal has been well studied (Godlewska et al., 2009); mostly notably for its interactions with other bacterial proteins (Bouveret et al., 1995; Cascales et al., 2000, 2002; Clavel et al., 1998), its proposed role in maintaining the integrity of the outer membrane of E. coli (Cascales et al., 2002) and its other physiological interaction with the peptidoglycan layer (Lazzaroni & Portalier, 1992). TolB and peptidoglycan compete for the same binding site on Pal, which happens to be on the opposite end (structurally) of the N-terminal lipid anchor (Abergel et al., 1999; Bouveret et al., 1995; Cascales & Lloubès, 2004; Parsons et al., 2006), thus making the interactions of Pal with TolB and peptidoglycan mutually exclusive. These interactions all point to the necessity for Pal to be localized in the periplasm of the cell. This study describes, for the first time, a subpopulation of Pal that is localized to the cell surface of E. coli.
The discovery that the homologue of Pal in NTHi, P6, is expressed both on the cell surface and in the periplasm (Michel et al., 2013) led us to the hypothesis that Pal might also be dual oriented. As corroborative evidence, it was also suggested that both Lpp and Pal epitopes were exposed on the surface of E. coli that had been incubated in human serum (Hellman et al., 1997). Here we have shown with flow cytometry and confocal microscopy that Pal is exposed on the surface of uncompromised E. coli cells. We also demonstrate that Pal is surface exposed in an ‘all or nothing’ manner, with a few cells seemingly covered with surface Pal and most other cells with undetectable levels of surface Pal. Biotinylation of surface Pal allowed us to separate the ‘inside’ and ‘outside’ Pal populations to estimate that less than 25 % of the total Pal population is surface exposed (taking into account leakiness, we estimate that between 10–20 % of Pal may be surface exposed).
The biotinylation experiment yields slightly higher percentages of surface Pal compared to estimates using confocal microscopy and flow cytometry. This discrepancy is likely to be the result of our different labelling techniques. For the biotinylation experiment, our biotin reagents label any/all exposed lysine residues on Pal (six lysine residues available on the Pal structure). In contrast, the confocal and flow experiments rely on monoclonal antibody binding to a specific epitope on Pal. The monoclonal antibody interaction can be hindered by crowding on the cell surface, thus yielding lower estimates for surface Pal.
OmpA is a large transmembrane protein that exhibits a unique temperature-sensitive folding transition where, at elevated temperatures, its periplasmic C-terminal tail is incorporated into the membrane-spanning domain thus forming a large ‘megapore’ (Zakharian & Reusch, 2005). Since the periplasmic tail of OmpA is similar in both sequence and structure to Pal (Park et al., 2012), we considered the possibility that Pal might interact with the periplasmic domain of OmpA such that it could be ‘flipped’ to the surface during the OmpA temperature transition. Our experimental data, however, showed that surface Pal was biotinylated in the OmpA knockout (in E. coli), suggesting that OmpA is not required for surface exposure of Pal.
We know that lipoproteins are transported (as dictated by their signal sequences) to the outer membrane via the Lol transport system (Narita et al., 2004; Narita & Tokuda, 2010). Therefore, we consider only two ways for Pal to be inserted into the outer membrane. Either the Lol system inserts Pal into both the inner and outer leaflets of the outer membrane, or Lol inserts Pal into the inner leaflet of the outer membrane and then Pal ‘flips’ to the cell surface sporadically or in response to a signal/event. Since it would be energetically unfavourable for Pal to flip to the surface on its own, the latter hypothesis would require a lipoprotein ‘flippase’ to accomplish the task.
In summary, we determined that less than 25 % of Pal is surface exposed, and that Pal surface exposure occurs in an ‘all or nothing’ manner. Although currently we have no hypotheses as to how or why Pal is dual oriented, with this report we add Pal to Lpp and NTHi P6 as dual oriented lipoproteins in Gram-negative bacteria. It has also been shown that Pal is released from bacteria in experimental models of sepsis, and that a proportion of released Pal was tightly associated with LPS, OmpA and Lpp (Hellman et al., 2000, 2002). A second population of released Pal, however, was not associated with LPS, OmpA or Lpp (Hellman et al., 2000). Furthermore, serum-released Pal was preferentially bound by IgG in J5 antiserum, which has been shown to protect in sepsis (Liang et al., 2005). Pal has also been reported to cause inflammation and death in LPS-hyporesponsive mice, and Pal-deficient E. coli mutants are less virulent than their WT progenitors. Taken together, these reports suggest that Pal may be a bacterial mediator of Gram-negative sepsis (Hellman et al., 2002). We speculate that serum-released Pal (perhaps more specifically the unassociated population of released Pal) may be derived from bacteria with surface expressed Pal.
Acknowledgements
This study was supported by the Camille and Henry Dreyfus Special Grant Program in the Chemical Sciences, NIH NIDCD RO1 08671 (to M. E. P.) and Rochester Institute of Technology. We thank Cheryl Hanzlik, Steven Wilbert and Evan Darling, technicians at the Confocal Microscopy Lab at Rochester Institute of Technology, for assistance and use of the microscope to obtain images. The confocal lab is funded, in part, by a Major Research Instrumentation Grant from NSF (1126629). We thank Dr Kwang Sik Kim (Johns Hopkins Children's Center) for gifts of E. coli K1 RS218 and OmpA deletion cells, Dr Jean-Claude Lazzaroni (University of Lyon) for E. coli K-12 JC1292 and JC7752 (Pal deletion) cells, Dr André Hudson (Rochester Institute of Technology) for use of his E. coli strains, and Dr. Martin Pavelka and Dr. Michelle Dziejman (University of Rochester Medical Center) for thoughtful discussions.
Abbreviations:
- Pal
peptidoglycan associated lipoprotein.
References
- 1. Abergel C., Bouveret E., Claverie J. M., Brown K., Rigal A., Lazdunski C., Bénédetti H. (1999). Structure of the Escherichia coli TolB protein determined by MAD methods at 1.95 A resolution. Structure 7, 1291–1300 10.1016/S0969-2126(00)80062-3 [DOI] [PubMed] [Google Scholar]
- 2. Bernadac A., Gavioli M., Lazzaroni J. C., Raina S., Lloubès R. (1998). Escherichia coli tol-pal mutants form outer membrane vesicles. J Bacteriol 180, 4872–4878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bouveret E., Derouiche R., Rigal A., Lloubès R., Lazdunski C., Bénédetti H. (1995). Peptidoglycan-associated lipoprotein-TolB interaction. A possible key to explaining the formation of contact sites between the inner and outer membranes of Escherichia coli . J Biol Chem 270, 11071–11077 10.1074/jbc.270.19.11071 [DOI] [PubMed] [Google Scholar]
- 4. Bouveret E., Bénédetti H., Rigal A., Loret E., Lazdunski C. (1999). In vitro characterization of peptidoglycan-associated lipoprotein (PAL)-peptidoglycan and PAL-TolB interactions. J Bacteriol 181, 6306–6311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Braun V., Bosch V. (1972). Sequence of the murein-lipoprotein and the attachment site of the lipid. Eur J Biochem 28, 51–69 10.1111/j.1432-1033.1972.tb01883.x [DOI] [PubMed] [Google Scholar]
- 6. Braun V., Rehn K. (1969). Chemical characterization, spatial distribution and function of a lipoprotein (murein-lipoprotein) of the E. coli cell wall. The specific effect of trypsin on the membrane structure. Eur J Biochem 10, 426–438 10.1111/j.1432-1033.1969.tb00707.x [DOI] [PubMed] [Google Scholar]
- 7. Cascales E., Lloubès R. (2004). Deletion analyses of the peptidoglycan-associated lipoprotein Pal reveals three independent binding sequences including a TolA box. Mol Microbiol 51, 873–885 10.1046/j.1365-2958.2003.03881.x [DOI] [PubMed] [Google Scholar]
- 8. Cascales E., Gavioli M., Sturgis J. N., Lloubès R. (2000). Proton motive force drives the interaction of the inner membrane TolA and outer membrane pal proteins in Escherichia coli . Mol Microbiol 38, 904–915 10.1046/j.1365-2958.2000.02190.x [DOI] [PubMed] [Google Scholar]
- 9. Cascales E., Bernadac A., Gavioli M., Lazzaroni J. -C., Lloubès R. (2002). Pal lipoprotein of Escherichia coli plays a major role in outer membrane integrity. J Bacteriol 184, 754–759 10.1128/JB.184.3.754-759.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Clavel T., Germon P., Vianney A., Portalier R., Lazzaroni J. C. (1998). TolB protein of Escherichia coli K-12 interacts with the outer membrane peptidoglycan-associated proteins Pal, Lpp and OmpA. Mol Microbiol 29, 359–367 10.1046/j.1365-2958.1998.00945.x [DOI] [PubMed] [Google Scholar]
- 11. Cowles C. E., Li Y., Semmelhack M. F., Cristea I. M., Silhavy T. J. (2011). The free and bound forms of Lpp occupy distinct subcellular locations in Escherichia coli . Mol Microbiol 79, 1168–1181 10.1111/j.1365-2958.2011.07539.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Deprez C., Lloubès R., Gavioli M., Marion D., Guerlesquin F., Blanchard L. (2005). Solution structure of the E. coli TolA C-terminal domain reveals conformational changes upon binding to the phage g3p N-terminal domain. J Mol Biol 346, 1047–1057 10.1016/j.jmb.2004.12.028 [DOI] [PubMed] [Google Scholar]
- 13. Gerding M. A., Ogata Y., Pecora N. D., Niki H., de Boer P. A. (2007). The trans-envelope Tol-Pal complex is part of the cell division machinery and required for proper outer-membrane invagination during cell constriction in E. coli . Mol Microbiol 63, 1008–1025 10.1111/j.1365-2958.2006.05571.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Godlewska R., Wiśniewska K., Pietras Z., Jagusztyn-Krynicka E. K. (2009). Peptidoglycan-associated lipoprotein (Pal) of Gram-negative bacteria: function, structure, role in pathogenesis and potential application in immunoprophylaxis. FEMS Microbiol Lett 298, 1–11 10.1111/j.1574-6968.2009.01659.x [DOI] [PubMed] [Google Scholar]
- 15. Hellman J., Zanzot E. M., Loiselle P. M., Amato S. F., Black K. M., Ge Y., Kurnick J. T., Warren H. S. (1997). Antiserum against Escherichia coli J5 contains antibodies reactive with outer membrane proteins of heterologous gram-negative bacteria. J Infect Dis 176, 1260–1268 10.1086/514121 [DOI] [PubMed] [Google Scholar]
- 16. Hellman J., Loiselle P. M., Tehan M. M., Allaire J. E., Boyle L. A., Kurnick J. T., Andrews D. M. (2000). Outer membrane protein A, peptidoglycan-associated lipoprotein, and murein lipoprotein are released by Escherichia coli bacteria into serum. Infect Immun 68, 2566–2572 10.1128/IAI.68.5.2566-2572.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hellman J., Roberts J. D., Jr, Tehan M. M., Allaire J. E., Warren H. S. (2002). Bacterial peptidoglycan-associated lipoprotein is released into the bloodstream in gram-negative sepsis and causes inflammation and death in mice. J Biol Chem 277, 14274–14280 10.1074/jbc.M109696200 [DOI] [PubMed] [Google Scholar]
- 18. Lazzaroni J. C., Portalier R. (1992). The excC gene of Escherichia coli K-12 required for cell envelope integrity encodes the peptidoglycan-associated lipoprotein (PAL). Mol Microbiol 6, 735–742 10.1111/j.1365-2958.1992.tb01523.x [DOI] [PubMed] [Google Scholar]
- 19. Leduc M., Ishidate K., Shakibai N., Rothfield L. (1992). Interactions of Escherichia coli membrane lipoproteins with the murein sacculus. J Bacteriol 174, 7982–7988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lei H., Velez G., Kazlauskas A. (2011). Pathological signaling via platelet-derived growth factor receptor alpha involves chronic activation of Akt and suppression of p53. Mol Cell Biol 31, 1788–1799 10.1128/MCB.01321-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Liang M. D., Bagchi A., Warren H. S., Tehan M. M., Trigilio J. A., Beasley-Topliffe L. K., Tesini B. L., Lazzaroni J. C., Fenton M. J., Hellman J. (2005). Bacterial peptidoglycan-associated lipoprotein: a naturally occurring toll-like receptor 2 agonist that is shed into serum and has synergy with lipopolysaccharide. J Infect Dis 191, 939–948 10.1086/427815 [DOI] [PubMed] [Google Scholar]
- 22. Michel L. V., Snyder J., Schmidt R., Milillo J., Grimaldi K., Kalmeta B., Khan M. N., Sharma S., Wright L. K., Pichichero M. E. (2013). Dual orientation of the outer membrane lipoprotein P6 of nontypeable Haemophilus influenzae . J Bacteriol 195, 3252–3259 10.1128/JB.00185-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Mizuno T. (1981). A novel peptidoglycan-associated lipoprotein (PAL) found in the outer membrane of Proteus mirabilis and other Gram-negative bacteria. J Biochem 89, 1039–1049 [PubMed] [Google Scholar]
- 24. Narita S., Tokuda H. (2010). Protein secretion. In Methods in Molecular Biology. vol. 619, Chapter 7. Edited by A. Economou. New York, NY: Springer Science+Business Media, LLC. [Google Scholar]
- 25. Narita S., Matsuyama S., Tokuda H. (2004). Lipoprotein trafficking in Escherichia coli . Arch Microbiol 182, 1–6 10.1007/s00203-004-0682-4 [DOI] [PubMed] [Google Scholar]
- 26. Park J. S., Lee W. C., Yeo K. J., Ryu K. -S., Kumarasiri M., Hesek D., Lee M., Mobashery S., Song J. H., other authors (2012). Mechanism of anchoring of OmpA protein to the cell wall peptidoglycan of the gram-negative bacterial outer membrane. FASEB J 26, 219–228 10.1096/fj.11-188425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Parsons L. M., Lin F., Orban J. (2006). Peptidoglycan recognition by Pal, an outer membrane lipoprotein. Biochemistry 45, 2122–2128 10.1021/bi052227i [DOI] [PubMed] [Google Scholar]
- 28. Valentine C. H., Hellman J., Beasley-Topliffe L. K., Bagchi A., Warren H. S. (2006). Passive immunization to outer membrane proteins MLP and PAL does not protect mice from sepsis. Mol Med 12, 252–258 10.2119/2006-00065.Valentine [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Vollmer W., Bertsche U. (2008). Murein (peptidoglycan) structure, architecture and biosynthesis in Escherichia coli . Biochim Biophys Acta 1778, 1714–1734 10.1016/j.bbamem.2007.06.007 [DOI] [PubMed] [Google Scholar]
- 30. Wang Y., Kim K. S. (2002). Role of OmpA and IbeB in Escherichia coli K1 invasion of brain microvascular endothelial cells in vitro and in vivo . Pediatr Res 51, 559–563 10.1203/00006450-200205000-00003 [DOI] [PubMed] [Google Scholar]
- 31. Yeh Y. -C., Comolli L. R., Downing K. H., Shapiro L., McAdams H. H. (2010). The caulobacter Tol-Pal complex is essential for outer membrane integrity and the positioning of a polar localization factor. J Bacteriol 192, 4847–4858 10.1128/JB.00607-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zakharian E., Reusch R. N. (2005). Kinetics of folding of Escherichia coli OmpA from narrow to large pore conformation in a planar bilayer. Biochemistry 44, 6701–6707 10.1021/bi047278e [DOI] [PubMed] [Google Scholar]