Abstract
Genetic evidence has supported the hypothesis that schizophrenia (SZ) is a polygenic disorder caused by the disruption in function of several or many genes. The most common and reproducible cellular phenotype associated with SZ is a reduction in dendritic spines within the neocortex, suggesting alterations in dendritic architecture may cause aberrant cortical circuitry and SZ symptoms. Here, we review evidence supporting a multifactorial model of mitochondrial dysfunction in SZ etiology and discuss how these multiple paths to mitochondrial dysfunction may contribute to dendritic spine loss and/or underdevelopment in some SZ subjects. The pathophysiological role of mitochondrial dysfunction in SZ is based upon genomic analyses of both the mitochondrial genome and nuclear genes involved in mitochondrial function. Previous studies and preliminary data suggest SZ is associated with specific alleles and haplogroups of the mitochondrial genome, and also correlates with a reduction in mitochondrial copy number and an increase in synonymous and nonsynonymous substitutions of mitochondrial DNA. Mitochondrial dysfunction has also been widely implicated in SZ by genome-wide association, exome sequencing, altered gene expression, proteomics, microscopy analyses, and induced pluripotent stem cell studies. Together, these data support the hypothesis that SZ is a polygenic disorder with an enrichment of mitochondrial targets.
Key Words: Schizophrenia, Mitochondria, Dendritic spines, Polygenic disorder, Genome, Transcriptome, Proteome, Induced pluripotent stem cell, Fluorescence deconvolution tomography, Antipsychotic drug
Introduction
Schizophrenia (SZ) is a severe mental disorder in which an underlying dysfunction in synaptic transmission has been implicated across multiple brain regions [1,2,3,4]. The most notable cellular pathology observed in SZ is a reduction in dendritic spines in layer III pyramidal neurons of the neocortex, though spine alterations in other cortical layers and brain regions have also been observed [1,5,6,7]. Studies of twins, families with a higher incidence of SZ, and larger case/control populations, including recent evidence from the Psychiatric Genomics Consortium (PGC), have all suggested that SZ has a primary genetic component [8,9,10,11,12]. The genetic etiology of SZ is strikingly complex, however, given the lack of association with any one specific gene or locus in all SZ cases. Multiple studies have utilized pathway enrichment analyses to explore this complexity and have suggested the loci associated with SZ converge on specific molecular networks [e.g., postsynaptic density (PSD), cytoskeleton organization and mitochondria], supporting the hypothesis that SZ is a polygenic disorder where deficits in several or many genes coalesce to form a common cellular pathology and disease phenotype [5,12,13,14,15,16,17,18,19].
The dendritic spine pathology observed in SZ brains and the potential consequences of these defects have been extensively reviewed by Glausier and Lewis [1]. These pathological alterations in dendritic spine morphology have additionally been correlated with multiple molecular alterations, including but not limited to, a reduction in expression in specific RhoGTPase genes, a loss of microtubule-associated protein 2 (MAP2) immunoreactivity, and transcriptome alterations in the mitochondrial and ubiquitin-proteasome system pathways [5,6,7]. Interestingly, the mitochondrial network has been observed to have the most robust and significant transcriptome alterations in layer III pyramidal cells, which is where the most dramatic decrease in dendritic spine density has been observed, supporting a molecular link between signatures of mitochondrial dysfunction and dendritic spine pathophysiology in SZ [5].
Mitochondria have central roles in dendritic spine architecture and many neuronal processes, and thus affect the connections that underlie functional cortical circuitry [20,21]. More specifically, mitochondria provide nearly all of the energy for dendritic spine formation, synaptic transmission, maintenance of ionic homeostasis in synaptic terminals, synaptic vesicle recycling, and long-term potentiation [20,21,22,23,24]. In neurons, mitochondria are transported in both retrograde and anterograde directions along molecular motors, and their motility can be altered by a variety of different factors including prolonged synaptic activation, neurotransmitters, growth factors, medications, viruses, calcium level alterations, immune activation, and mutational disruption of specific genes [20,21,25,26,27,28,29,30,31,32,33]. Mitochondria have been shown to positively regulate dendritic spine morphogenesis and plasticity, but are also implicated in the negative regulation of dendritic branching during development [20,34]. Overall, there is an extensive body of evidence in the literature that intrinsically links mitochondrial copy number, localization and function with dendritic spine morphology and synaptic transmission.
This review summarizes evidence of mitochondrial dysfunction within the complex genetic etiology of SZ. Both published reports and preliminary data described here indicate the mitochondrial network may be affected in SZ through genetic aberrations of the mitochondrial genome and mitochondrial genes located on the nuclear chromosomes; downstream alterations in mitochondrial transcript expression, mitochondrial function and localization are also discussed. In order to prioritize gene candidates, we review the mitochondrial genes that have been associated with SZ multiple times by a variety of independent methods and studies, including analyses of copy number variants (CNVs), rare and de novo mutations, genome-wide associations, transcriptome alterations in SZ brains and during adolescence, and proteomics (table 1) [5,12,13,14,15,18,19,35,36,37,38,39,40]. We present a multifactorial model, where alterations in gene networks specific to PSD, cytoskeleton organization and an array of mitochondrial processes may independently (or additively) lead to dendritic spine deficits and SZ symptom onset (fig. 1). We will also address the influence of antipsychotic drugs (APDs) on mitochondrial function and describe how these effects are likely independent of the role mitochondrial dysfunction plays in SZ pathophysiology. Lastly, we highlight the positive associations between mitochondrial dysfunction and a number of adult-onset and neurological disorders, and emphasize the need to investigate drugs that specifically target mitochondrial function and/or localization as a potential therapeutic strategy for SZ patients.
Table 1.
Mitochondrial genes associated with SZ in at least two independent studies
| Symbol | SZ associations, n | Association type and references |
|---|---|---|
| MDH2 | 4 | de novo CNV [19], CNV [18], ↓ RNA (DLPFC pyramidal) [5], ↓ RNA (DG) [34] |
| NDUFS2 | 4 | GWAS (early onset) [33], ↓ RNA (DLPFC pyramidal) [5], peak expression (l5–25 years) [35], DER (teen > adult) [36] |
| ATP5A1 | 3 | rare mutation [15], ↓ RNA (PFC area 9) [13], ↑ protein (DLPFC/ACC) [37] |
| IDH3A | 3 | ↓ RNA (DLPFC pyramidal) [5], ↓ RNA (DG) [34], ↓ protein (ACC) [37] |
| NDUFA2 | 3 | GWAS (PGC) [12], ↓ RNA (DLPFC pyramidal) [5], ↑ protein (DLPFC) [37] |
| BDH1 | 3 | de novo CNV [19], CNV [18], DER (teen > adult) [36] |
| CLPX | 3 | de novo mutation [14], peak expression (15–25 years) [35], DER (teen > child) [36] |
| GOT2 | 3 | CNV [18], ↓ RNA (PFC area 9) [13], peak expression (15–25 years) [35] |
| MDH1 | 3 | ↓ RNA (PFC area 9) [13], DER (teen > child) [36], ↓ protein (DLPFC) [37] |
| QDPR | 3 | peak expression (15–25 years) [35], DER (teen > child) [36], ↓ protein (DLPFC) [37] |
| UQCRC1 | 2 | DER (teen > child) [36], ↓↑ protein (DLPFC) [37] |
| IDH3B | 2 | ↓ RNA (DLPFC pyramidal) [5], peak expression (15–25 years) [35] |
| ACO2 | 2 | rare mutation [15], ↓ protein (DLPFC) [37] |
| DLD | 2 | ↓ RNA (DLPFC pyramidal) [5], peak expression (15–25 years) [35] |
| SDHA | 2 | ↓ RNA (DG) [34], ↓ protein (ACC) [37] |
| NDUFS4 | 2 | ↓ RNA (DLPFC pyramidal) [5], ↓ RNA (DG) [34] |
| UQCRFS1 | 2 | ↓ RNA (DLPFC pyramidal) [5], ↓ RNA (DG) [34] |
| ATP5B | 2 | ↓ RNA (DLPFC pyramidal) [5], ↓ protein (DLPFC/CC) [37] |
| NDUFS1 | 2 | GWAS (early onset) [33], ↓ protein (DLPFC) [37] |
| NDUFV1 | 2 | ↓ RNA (DLPFC pyramidal) [5], peak expression (15–25 years) [35] |
| NDUFA6 | 2 | GWAS (PGC) [12], ↓ RNA (DLPFC pyramidal) [5] |
| SLC25A3 | 2 | rare mutation [15], peak expression (15–25 years) [35] |
| TUFM | 2 | CNV [18], ↓ protein (DLPFC) [37] |
| HIBADH | 2 | peak expression (15–25 years) [35], ↑ protein (DLPFC) [37] |
| NDUFB9 | 2 | ↓ RNA (DLPFC pyramidal) [5], peak expression (15–25 years) [35] |
| NDUFV2 | 2 | ↓ RNA (DLPFC pyramidal) [5], ↑ protein (DLPFC) [37] |
| PDHB | 2 | ↓ RNA (DLPFC pyramidal) [5], ↓ RNA (DG) [34] |
| HSPD1 | 2 | GWAS (PGC) [12], ↓ protein (DLPFC) [37] |
| NDUFB5 | 2 | ↓ RNA (DG) [34], DER (teen > child) [36] |
| TXNRD2 | 2 | CNV (22q11.2) [32], CNV [18] |
| PRODH | 2 | CNV (22q11.2) [32], CNV [18] |
| TIMM17A | 2 | ↓ RNA (DG) [34], ↓ RNA (PFC area 9) [13] |
| SLC25A4 | 2 | ↓ RNA (DLPFC pyramidal) [5], ↓ RNA (DG) [34] |
| MRPL40 | 2 | CNV (22q11.2) [32], CNV [18] |
| SLC25A1 | 2 | CNV (22q11.2) [32], CNV [18] |
| USMG5 | 2 | GWAS (PGC) [12], DER (teen > adult) [36] |
| NDUFA13 | 2 | GWAS (PGC) [12], peak expression (15–25 years) [35] |
| ATP5J | 2 | CNV [18], peak expression (15–25 years) [35] |
| PNKD | 2 | peak expression (15–25 years) [35], DER (teen > child) [36] |
| PPA2 | 2 | ↓ RNA (DLPFC pyramidal) [5], ↓ protein (DLPFC) [37] |
| SFXN2 | 2 | GWAS (PGC) [12], DER (teen > child) [36] |
| VDAC1 | 2 | de novo mutation [14], ↓ protein (DLPFC) [37] |
| IMMT | 2 | DER (teen > child) [36], ↓ protein (ACC) [37] |
| MRPS9 | 2 | peak expression (15–25 years) [35], DER (teen > adult) [36] |
| CYB5B | 2 | CNV [18], peak expression (15–25 years) [35] |
| HSDL1 | 2 | CNV [18], DER (teen > adult) [36] |
| PRDX1 | 2 | rare mutation [15], ↓ protein (DLPFC) [37] |
| SHMT1 | 2 | peak expression (15–25 years) [35], DER (teen > adult) [36] |
| COMTD1 | 2 | peak expression (15–25 years) [35], DER (teen > adult) [36] |
| PRDX2 | 2 | rare mutation [15], ↓ protein (DLPFC) [37] |
| VAMP1 | 2 | peak expression (15–25 years) [35], DER (teen > child) [36] |
| CERK | 2 | de novo mutation [14], DER (teen > adult) [36] |
| KYNU | 2 | rare mutation [15], peak expression (15–25 years) [35] |
| RNMTL1 | 2 | CNV [18], DER (teen > adult) [36] |
| OXR1 | 2 | peak expression (15–25 years) [35], DER (teen > child) [36] |
| COMT | 2 | CNV (22q11.2) [32], CNV [18] |
| TDRKH | 2 | de novo mutation [14], DER (teen > child) [36] |
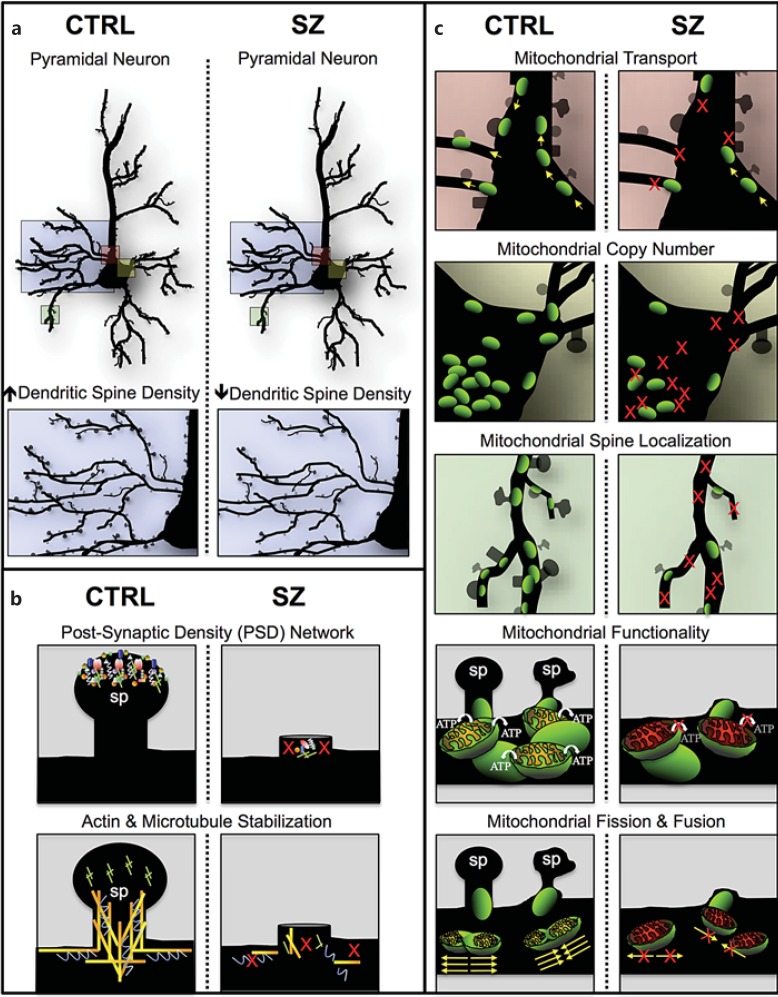
Fig. 1.
Multifactorial model of mitochondrial dysfunction in SZ. Cartoon illustration of our hypothesis of a multifactorial model where alterations in gene networks specific to PSD, cytoskeleton organization, and an array of mitochondrial processes may independently (or additively) lead to dendritic spine deficits, SZ symptom onset and polygenic disease risk. a Pyramidal neurons in control (CTRL) and SZ brains displaying a reduction in dendritic spine density in SZ pathology (blue box) and neuronal regions potentially affected by mitochondrial dysfunction (pink, yellow and green boxes). Colors refer to the online version only. b Dendritic spines (sp) in CTRL brains and spine loss in SZ brains due to polygenic variants in nonmitochondrial networks that have been implicated in SZ etiology (i.e., postsynaptic density and cytoskeleton organization). c Neuronal regions potentially affected by a variety of mitochondrial processes (i.e., transport, copy number, spine localization, functionality, and fusion/fission) and their possible role in dendritic spine loss in SZ brains compared to CTRL. We hypothesize that a signature of mitochondrial dysfunction can occur in SZ independently or additively in b and c. In b, mitochondrial deficits are a direct result of spine loss due to the polygenic burden in nonmitochondrial pathways like the PSD and cytoskeleton networks (i.e., PSD/cytoskeleton gene aberrations → spine loss ← → mitochondrial dysfunction); in c, mitochondrial deficits are a direct result of aberrations in nuclear genes with mitochondrial function and/or in the mitochondrial genome, and there is also spine loss-induced mitochondria loss (i.e., mitochondrial gene aberrations → mitochondrial dysfunction ← → spine loss); in c the mitochondria loss of function is primary, while in b the spine loss is the primary causative event.
Results and Discussion
The Mitochondrial Genome and SZ
The mitochondrial genome is a double-stranded circular molecule of approximately 16,569 nucleotides and codes for 13 protein-coding genes, in addition to a series of ribosomal and transfer RNAs [41,42,43]. Both single nucleotide polymorphisms (SNPs) within the hypervariable region of the mitochondrial genome, and mitochondrial haplogroups have been associated with SZ (or protection against SZ) in several studies [42,44,45,46,47,48]. We previously described two SNPs that nominally associate with protection against SZ - the T allele at position 16519 (rs3937033) and the C allele at position 195 (rs2857291) in individuals with European ancestry using the Genetic Association Information Network (GAIN) and Wellcome Trust Case Control Consortium 2 (WTCCC2) datasets. These specific SNP associations were specific to SZ, and were not significant for bipolar disorder (BD) or psychiatric illness in general (SZ + BD) [42]. Allelic association at position 16519 was not observed as significant in a published Santiago cohort of SZ patients; however, allele frequencies were reported in this study, so we utilized these data for a meta-analysis across three separate cohorts [49]. Specifically, analysis was performed on pooled data from (1) the GAIN-WTCCC2 cohort set, (2) the Santiago cohort, and (3) a dataset of 632 Bulgarian father-offspring pairs, resulting in a meta-analysis of 7,375 subjects (2,209 SZ vs. 5,166 controls) [19,42,49]. The results from the preliminary meta-analysis confirm the protective association of the T allele at position 16519, with a Mantel-Haenszel odds ratio (OR) of 0.83, and a 95% confidence interval range of 0.74-0.93. Two-tailed t tests demonstrated that the meta-analysis of this SNP was significant (p < 0.001). The nominal OR described in this study, as well as other reports about individuals of European and non-European ancestry, do not suggest these SNPs play a primary role in SZ pathology. However, due to the complex genetic etiology of SZ, these variants may be useful for population stratification and subsequent pathway enrichment analysis on patient cohorts with or without risk (or protective) alleles, and may thus increase statistical power in some circumstances. Another study also identified mitochondrial variants within the hypervariable region that were associated with risk or protection in multiple late-onset diseases including SZ [50]. In addition to mitochondrial allele associations with SZ, several studies have reported nominal associations with specific mitochondrial haplogroups and SZ, although other studies have additionally suggested such nominal associations are not pathogenic [44,45,48,49,51]. As such, these associations and haplogroup assignments may also be better warranted for population stratification, analysis of additional variables like disease onset and symptom severity, and enhancement of statistical power to discover variants enriched within SZ subgroups or populations with differences in mitochondrial ancestry.
There have been additional associations between the mitochondrial genome and SZ, specifically with regard to the ratio of the mitochondrial common deletion observed in brain tissue, and to alterations in peripheral mitochondrial copy number. The common deletion is a 4,977 nucleotide deletion of the mitochondrial genome occurring as a somatic, heteroplasmic event, and is most commonly observed in tissues with high metabolic rates and during the aging process [52]. Several studies have demonstrated an age-dependent increase of the mitochondrial common deletion in control subjects' brains, but this age-dependent increase was not observed in SZ [52,53,54,55,56]. Moreover, we have previously shown that pooled analysis across 10 brain regions supports a significantly lower accumulation of the common deletion in SZ compared to control subjects (p < 0.01), or compared to individuals with other psychiatric disorders [i.e., BD (p < 0.02) and major depression disorder (p < 0.001)] [52]. It should be noted that there is a substantial difference in the levels of the common deletion across brain regions, and that the differences associated with SZ were not specific to any one region but were, rather, a cumulative observation of the average brain burden. Because these results are obtained from quantitative PCR analysis, where the relative ratio of the common deletion is compared and normalized to nondeleted mitochondria, one possible explanation for the lower amount of common deletion in SZ, and the lack of correlation between deletion load and aging in these individuals, is that the number and/or stability of mitochondria organelles is compromised in SZ subjects. A lower number of mitochondria per cell, or a higher proportion of unstable mitochondria capable of appropriate fusion or fission processes, may make the natural mechanisms that lead to accumulation of large mitochondrial deletions less likely to occur in SZ brains. However, we have observed a significant increase in both synonymous and nonsynonymous single base pair substitutions, and an increased transition/transversion bias, in SZ brains compared to controls, suggesting these variants may play a role in the lack of accumulation of the common deletion in SZ or may be correlated features of the same stress response [43,44]. But we have not tested sufficient germline samples to determine whether this disease bias is exclusively due to somatic mutations or is also a reflection of inherited variants. Together, these data suggest that mitochondria in SZ may have more mutations or variants that make them more susceptible to processes like oxidative stress or replication errors that cause single-point mutations. We speculate that neurons of SZ subjects may be less tolerant of large aberrations like the accumulation of the common deletion that normally occurs during aging.
In order to preliminarily investigate whether mitochondrial copy number is reduced in SZ, and thus may be (partially or fully) responsible for the lower proportion of the common deletion observed in SZ brains, we utilized genotype data from Affymetrix SNP arrays from the GAIN case-control dataset to analyze peripheral mitochondrial copy number. Briefly, the statistical method for mitochondrial copy number assessment on genotype arrays utilizes probe intensities from a series of mitochondrial SNPs and relates this to principle component analysis data obtained from a subset (∼1,000) of autosomal SNPs to obtain standardized residual values of mitochondrial copy number for each subject [57]. After correcting for plate effects, we observed a significant reduction (p < 10-40) in peripheral mitochondrial copy number in SZ subjects compared to controls, and also a significant reduction in BD, although the effect was less dramatic (p < 10-6). In order to further correct for additional factors such as sex and age, we performed a corresponding analysis of the 632 Bulgarian family trios [19]. This analysis did not demonstrate a significant reduction in peripheral mitochondrial copy number in SZ versus controls after correcting for plate effect, age, and sex, but did show a significantly lower mitochondrial copy number in males versus females (p < 10-16), and a significantly negative correlation between age and mitochondrial copy number in males exclusively (p < 0.01). The discrepancy between these two preliminary data sets could be due to a number of factors, including lack of corrections in the GAIN data set, and/or confounding environmental factors that may influence mitochondrial copy number in either cohort, including lifestyle factors such as smoking, medications, metabolic syndromes, infections, or environmental exposures. It is encouraging, however, that one analysis resulted in a significant reduction of mitochondrial copy number in SZ, indicating that further evaluation of this trait as one possible mechanism of mitochondrial dysfunction in SZ is warranted. Another study using a polymerase chain reaction (PCR)-based quantification of mitochondrial and nuclear DNA in SZ and control brains also did not find a decrease in mitochondrial CN; however, this analysis was from a small sample size (control, n = 8; SZ, n = 6) [58]. Given our hypothesis that a multifactorial model of mitochondrial dysfunction occurs within SZ as a whole, it is also possible that mitochondrial copy number reductions will only be observed in specific SZ cohorts and that mitochondrial dysfunction is not related to copy number in all individuals but is more a reflection of (mitochondrial or nuclear) mutational load affecting mitochondrial functioning and not crude numbers of mitochondria. Future evaluations on mitochondrial copy number should perhaps focus on brain tissue evaluations, rather than those obtained from peripheral blood samples to avoid the confounding factors that may influence mitochondrial copy number in mitotically active or immune-specific cells. Moreover, mitochondrial copy number has previously been shown to have a relatively high degree of narrow-sense heritability (h2), which was described to be 0.65 in a twin study on renal carcinoma; this result was partially supported by our new analysis of the Bulgarian trio cohort using the GHOST and MERLIN software packages [56] and was likewise determined to be heritable (h2 = 0.37), although to a lesser extent than previously described [59].
Lastly, reduced expression in a majority of the protein-coding genes (10/13) residing in the mitochondrial genome has also been observed in SZ brains, but this difference was not observed in BD brains [41]. The genes found to have reduced expression in SZ include MT-ATP6, MT-ATP8, MT-CO3, MT-CYB, MT-ND1, MT-ND2, MT-ND3, MT-ND4, MT-ND5, and MT-ND6. Similar to the arguments of why a reduction in the common deletion has been observed for SZ, it is possible that the reduced expression of mitochondrial transcripts may be due to a decrease in mitochondrial copy number in SZ brains. Alternatively, this difference may also be due to differences in transcriptional regulation of the mitochondrial genome in SZ, or stability of the mitochondrial polycistronic transcript, which may precede or follow the observed reductions in dendritic spine density.
Mitochondrial Genes in the Nuclear Genome and SZ
In addition to the 13 protein-coding genes located on the mitochondrial genome, there are an estimated 1,500 protein-coding genes located in the nuclear genome involved in mitochondrial function, replication, or localization [42,60,61]. These genes have been identified as having mitochondria-specific functions through a number of methods, including computational prediction, proteomic analysis such as mass spectroscopy, localization analyses by GFP tagging, and homology with mitochondrial genes described for murine tissues, the latter of which has undergone extensive experimental validation and includes over 1,000 of the 1,500 predicted mitochondrial proteins [61]. It should be additionally noted that this list does not encompass all noncoding RNAs, such as microRNAs and long noncoding RNAs, that are likely involved in mitochondrial function or regulation of mitochondrial gene transcription, and as such the number of identified transcripts involved in some form of mitochondrial function will likely grow as functional evaluations of noncoding RNAs continues to expand these reference sets [62,63,64,65]. Nuclear-encoded mitochondrial genes have been implicated in SZ etiology and pathology in a number of published studies.
In evaluating overlap with the human MitoCarta reference set of 1,013 human mitochondrial genes [61], and review of others' published observations, we describe the association between SZ and a number of mitochondrial genes. Studies evaluated include analyses of CNVs, rare and de novo mutations, genome-wide associations, transcriptome alterations in SZ brains and during adolescence (i.e., during stages of brain development physiologically relevant to the onset of SZ symptoms), and proteomics [5,12,13,14,15,18,19,35,36,37,38,39,40]. We summarize how, together, these independent studies support a multifactorial model of mitochondrial dysfunction within the complex genetic etiology of SZ (fig. 1). The 57 mitochondrial genes associated with SZ etiology or pathology in at least 2 independent studies of unique design (and the study type and reference(s) where each gene was identified) are displayed in table 1. A complete list of the 295 mitochondrial genes identified by these various methodologies is provided online (online suppl. table S1; for all online suppl. material, see www.karger.com/doi/10.1159/000441252).
The 22q11.2 deletion syndrome is caused by a microdeletion in human chromosome 22, and results in a very high predisposition to SZ, with up to one third of individuals developing SZ or schizoaffective disorder by adolescence or early adulthood, which is a much higher incidence than the ∼1% reported for the general population at large [35]. As such, historically, 22q11.2 deletion syndrome has been one of the most well-studied models for evaluating disease-causing genes due to its pathological association with a single locus that harbors 25-40 potentially pathogenic genes depending on the size of the deletion and genomic reference. Genes with mitochondrial function within this region include PRODH, SLC25A1, MRPL40, TXNRD2, and COMT. It should be emphasized that these specific genes may have, and likely do have, multiple functions outside of their role in mitochondria; COMT, for example, has been well studied for its role in dopamine degradation, but studies employing GFP-tagging have provided evidence of mitochondrial localization as well [61,66]. Also, although SZ has been reported to have a much higher prevalence in individuals with 22q11.2 deletion syndrome, a lack of complete penetrance for these psychiatric symptoms supports a polygenic etiology that includes genes outside of this genomic region. More recently, studies have focused on genomic evaluations of larger case-control studies to identify genes contributing to SZ pathogenicity.
In addition to the 22q11.2 deletion, a number of other CNVs have been implicated in SZ etiology using genome-wide survey of several large cohorts, including one meta-analysis of 7,907 SZ cases and 10,585 controls, and a Swedish cohort study of 4,719 cases and 5,917 controls [18,19]. The meta-analysis of 34 de novo CNVs identified in SZ did not show a significant enrichment of the mitochondrial network in comparison to several control groups; however, the mitochondrial reference set used for this study only encompassed 189 genes, which was curated from protein correlation profiling in specific organelles and is considerably less robust than the predicted 1,000-1,500 proteins that have been identified in the MitoCarta reference set. Nonetheless, this study did show several mitochondrial genes hit by de novo CNV events in SZ, specifically MDH2 and BDH1 [19]. The genome-wide CNV analysis of the Swedish cohort did find a significant enrichment of the mitochondrial network in SZ, using both a smaller reference list of 193 genes that correspond to the previously described meta-analysis data set, and using 892 mitochondrial genes representative of the majority of validated murine mitochondrial genes in MitoCarta [18,61]. Specifically, for the smaller mitochondrial reference set, CNVs identified only as large deletion events (>500 kb) were significant (p < 0.01) after multiple-comparison adjustments and overlapped with 12 mitochondrial genes: ACAD8, AMACR, ATP5J, BDH1, GOT2, HSD17B4, MDH2, MLYCD, SLC25A1, TUFM, TXNRD2, and UQCRC2. For the larger MitoCarta reference set, similarly only large deletion events (>500 kb) were significant (p < 0.05) after multiple comparison adjustments and overlapped with 31 mitochondrial genes: ACAD8, ACP6, AGXT2, AIFM3, AMACR, ATP5J, BDH1, COMT, COX10, CYB5B, GOT2, HSD17B4, HSDL1, LYRM2, MDH2, MIPEP, MLYCD, MRPL39, MRPL40, MSRA, MTRF1, NT5C3, PRODH, PXMP2, RG9MTD1, RNMTL1, SLC25A1, TOMM70A, TUFM, TXNRD2, and UQCRC2 [18]. Contrary to this, genes identified by duplications (or additive analysis of deletions and duplications combined) were not significantly enriched for this pathway, suggesting mitochondrial involvement in SZ may be more related to loss of function or loss of copy number than dominant-negative or increased gene dosage effects.
A large genome-wide association study (GWAS) recently conducted by the PGC included analyses of 36,989 cases and 113,075 controls and identified 108 credible genomic loci through 128 independent associations [12]. These 108 loci encompass over 300 genes, and enrichment analysis demonstrated an overrepresentation of genes expressed in the brain, as well as those specific to glutamatergic signaling and immunity. Although not mentioned specifically, these 108 loci also overlap with the following 22 genes implicated in mitochondrial function: NDUFA6, HSPD1, PCCB, NDUFA2, SHMT2, ATPAF2, NDUFA13, IMMP2L, DDX28, EFHD1, COQ10B, USMG5, C2orf47, DUS2L, HSPA9, MARS2, SFXN2, HSPE1, IREB2, AS3MT, DRG2, and TMTC1 [12]. Statistical tests using hypergeometric distribution indicate this represents a significant enrichment (p < 0.05) of the mitochondrial network relative to all protein-coding genes. We also evaluated the 7.6-Mb region of the extended human major histocompatibility complex locus which was highly associated with SZ in the PGC GWAS study; however, we did not identify any genes within this region that are implicated in mitochondrial function [12,67]. An independent GWAS analysis of the PGC data and another cohort of Han Chinese origin evaluated nuclear-encoded core genes specific to mitochondrial complex I, and did not find a significant association of any of the SNPs with SZ in both studies; however, this report did identify a nominal association with SNPs in two genes, NDUFS1 and NDUFS2, and early-onset SZ (onset ≤18 years), supporting a possible involvement of mitochondrial dysfunction and the temporal variation of SZ symptom onset [36].
A large exome sequencing study of a Swedish cohort of 2,536 SZ cases and 2,543 controls implicated a polygenic burden of rare disruptive mutations across multiple pathways, including the PSD proteome [15]. Extended association of gene sets associated with intracellular loci or organelles demonstrated a significant (p < 0.05) enrichment of disruptive variants (non-sense, essential splice site and frameshifts) in a mitochondrial reference list of 197 genes, while other organelle compartments such as the endosomes, endoplasmic reticulum, Golgi, nucleus, and plasma membrane were not significantly enriched in this PSD gene dataset [15]. This study also demonstrated that 29% of the mitochondrion gene list overlapped with the PSD (human core) gene list. These data further support the functional link between mitochondria and dendritic spine density, and additionally suggest that associations with enrichment in the PSD network should be further analyzed for subcellular involvement of mitochondrial genes. In further evaluation of their list of genes with high ORs (>5) that were listed as those ‘prioritized as more likely to harbor large-effect alleles’, we identified 8 mitochondrial genes (including their top-ranked gene): SLC25A3, GLUD1, ACO2, ATP5A1, PRDX1, PRDX2, SUCLA2, and KYNU [15]. As such, the functional effect these genes may have on both PSD and mitochondria should be evaluated for their potentially pathological role in SZ.
Another large exome-sequencing study was performed using blood from 623 SZ family trios, and was used to determine the impact of de novo coding or canonical splice site variants within SZ probands [14]. This analysis revealed an overrepresentation of synaptic proteins, similar to the enrichment described earlier for the exome results of rare disruptive mutations. Analysis of the ∼640 validated coding de novo mutations discovered in subjects with SZ demonstrated these mutations resided in 613 unique genes, from which the following 24 were related to mitochondrial function: TDRKH, SLC25A12, ALAS1, ALDH1L1, CBR4, VDAC1, ALDH5A1, ACN9, SND1, ACO1, ACSL5, FOXRED1, BCAT1, ALDH1L2, CLPX, EARS2, COX4I1, MRPL27, LONP1, SLC25A23, GTPBP3, GTPBP5, CERK, and ACSL4 [14]. Together, these data support our multifactorial model of mitochondrial dysfunction (fig. 1) within the complex genetic etiology of SZ, suggesting nuclear genes involved in mitochondrial function may be impacted by large copy number variations [18,19,35], rare and common SNPs [12,68], rare disruptive mutations [15], and de novo mutational events [14].
RNA and Protein Alterations of Mitochondrial Genes in SZ and Adolescence
Given the genetic evidence of mitochondrial dysfunction, it is not surprising that an enrichment of differentially expressed, nuclear-encoded mitochondrial genes has been observed in SZ brains in multiple studies. One study evaluated gene signatures of layer III and layer V pyramidal cells between 36 SZ and 36 control subjects using microarrays and subsequently performed pathway enrichment analysis [5]. A striking observation was that genes involved in the mitochondrial network were significantly reduced in SZ compared to control in both layers analyzed, and that this reduction was more significant for layer III pyramidal cells, which has been the site observed to have the most robust reduction in dendritic spine density [1,5]. Additionally, a reduction in expression of genes involved in the ubiquitin-proteasome network was also observed in this study, but these differences were specific only to layer V cells [5]. Mitochondrial genes with significantly reduced expression described in this study included the following: NDUFA11, COX8A, UQCRQ, NDUFS2, NDUFA6, COX7B, SDHB, NDUFB7, NDUFV2, NDUFB3, NDUFC1, NDUFB6, NDUFV1, UQCRFS1, NDUFA2, NDUFS4, COX7C, NDUFA8, NDUFB9, ATP5B, ATP5C1, ATP5F1, ETFA, SLC25A4, COX7A1, IDH3B, IDH3A, PDHB, MDH2, DLD, ATP5L, and PPA2 [5]. Another study analyzed hippocampal dentate gyrus neurons from the granule cell layer in 22 SZ brains and 24 control subjects from two separate cohorts, and also identified enrichment of the mitochondrial network [37]. In this analysis, 13 mitochondrial (MitoCarta) genes were found to be significantly decreased in expression in subjects with SZ across both cohorts: ACAT1, UQCRFS1, IDH3A, LDHA, MDH2, NDUFB2, NDUFB5, NDUFS4, PDHB, SCO1, SLC25A4, SDHA, and TIMM17A [37]. Lastly, one study evaluating prefrontal cortex (PFC) area 9 tissue identified 10 metabolic genes that displayed significantly different expression in SZ brains [13]. These 10 genes included 6 with known mitochondrial function, including ATP5A1, GOT2, MDH1, OAT, OXCT1, and TIMM17A[13].
English et al. [40] previously performed an extensive review of the proteomics studies of SZ and other psychiatric disorders. In further support of our multifactorial model of dendritic spine loss (fig. 1), their proteomics review article summarizes multiple studies that have observed alterations in cytoskeletal, synaptic, and mitochondrial (i.e., oxidative stress and metabolism) proteome networks in SZ [40]. The supplementary data in the review by English et al. [40] covers 11 proteomic studies of both white and grey matter, and was used here to identify alterations in proteins coded by genes with mitochondrial function. From this review, we identified 42 mitochondrial genes whose protein product has been shown to have alterations in SZ brains including the following: ACAT2, ACO2, ALDH7A1, ATP5A1, ATP5B, ATP5H, CHAT, CKMT1A, CKMT1B, CS, DDAH1, FTH1, GPD1, GPX1, HAGH, HIBADH, HK1, HSPD1, IDH3A, IMMT, LAP3, PDHB, MDH1, NAGS, NDUFA2, NDUFA5, NDUFS1, NDUFS3, NDUFV2, PARK7, PDHA1, PHB, PPA2, PRDX1, PRDX2, QDPR, SDHA, TUFM, TXN, UQCRC1, VDAC1, and VDAC2 [40]. Impressively, 18 of these 42 mitochondrial genes (43%) were found to be independently associated with SZ by genomic screening and/or transcriptome analysis in the studies reviewed here (table 1).
Additionally, there have been 2 studies evaluating transcriptome alterations that occur during brain development and across multiple stages of life, which both have implications for SZ pathophysiology [38,39]. First, one study using whole-genome microarray analysis of postmortem PFC demonstrated a peak in gene expression in brains derived from subjects in adolescence or early adulthood (15-25 years old), which correlates with the onset of SZ symptoms and supports the hypothesis that this time frame represents a ‘window of vulnerability’ [38]. This study identified >3,000 probe sets, specific to 2,330 genes with a unique identifier, that significantly correlated with a template described as an expression pattern where ‘a peak or trough in expression level [was observed] between ages 15 and 25’ [38]. Of these 2,330 genes identified from their supplemental data table, we found that 128 were related to mitochondrial function based on their presence in the human MitoCarta reference set [61]. Given the length of this list of genes, their symbols are not fully provided here; however, any of these genes overlapping with other genomic/transcriptomic lists are listed in table 1. This study also emphasized enrichment of functional groups related to mitochondrial function in gene sets with a positive correlation to their template, including the electron transport chain (ETC), glycolysis and oxidative stress [38].
A second expression study with relevance to SZ development performed transcriptome sequencing on 72 PFC samples across six stages of life and discovered differentially expressed regions (DERs) that were enriched for sites related to clinical risk of SZ [39]. Analysis of the gene ontology (GO) enrichment analysis specific to the genes with the highest expression in the teen age group revealed a significant enrichment for several mitochondrial functional groups, including (but not limited to) apoptotic mitochondrial changes, release of cytochrome c from mitochondria, regulation of mitochondrial membrane permeability involved in apoptotic process, mitochondrial membrane organization, mitochondrial ATP synthesis-coupled electron transport, mitochondrial outer membrane permeabilization, response to oxidative stress, and mitochondrion organization [39]. Of note, a similar search for mitochondrial terms in genes that peaked in expression during adulthood did not find an enrichment of mitochondrial functional groups, further supporting the hypothesis that mitochondrial genes may play a specifically important role during this ‘window of vulnerability’ described for the adolescent onset of SZ symptoms. Analysis of DERs with at least a 2-fold greater expression in teens compared to adults (range 2- to 151-fold) and an adjusted mean expression value >0 for both groups identified 3,722 DERs that were annotated as proximal or overlapping with 1,597 unique gene symbols. Of these >1,500 genes, the following 36 genes were represented in the human MitoCarta reference as having mitochondrial function: HADH, SLC25A16, ME2, RNMTL1, SHMT1, MRPL42, BDH1, MPST, HSDL1, MRPS9, NDUFS2, ATAD3B, PITRM1, AIFM2, SLC25A29, MTG1, ABCB10, LYRM7, CERK, DAP3, RAB8B, USMG5, DARS2, ATP5O, TMEM11, PDK3, NDUFA7, ABHD10, MRPS30, NME6, PPM1K, C1QBP, MRPL37, COMTD1, PTRH2, and RPL34. Analysis of DERs with at least a 2-fold greater expression in teens compared to children (range 2- to 83.5-fold) and an adjusted mean expression value >0 for both groups identified 2,161 DERs that were annotated as proximal or overlapping with 899 unique gene symbols. Of these ∼900 genes, the following 25 genes were represented in the human MitoCarta reference as having mitochondrial function: HAHD, AIFM2, RAB8B, DARS2, PPOX, MPST, ABCB10, THEM5, VAMP1, NDUFA7, PNKD, TMEM11, NDUFB5, PDK3, OXR1, PITRM1, MRPS30, TDRKH, SFXN2, QDPR, STAR, CLPX, IMMT, MDH1, and UQCRC1 [39,61].
Studies like these provide an excellent framework to decipher the functional and developmental role of genes associated with SZ pathophysiology and help elucidate the role they may play in disease onset and symptom progression. In summary, we identified 295 mitochondrial genes that associated with SZ in at least 1 study (online suppl. table S1), 57 of which associated with SZ etiology or pathology in at least 2 independent studies of unique design (table 1). There were 26 genes cross-identified exclusively in the RNA and protein studies that were not represented in the genomic studies we evaluated, while the remaining 31 genes were represented in at least one genomic analysis dataset. Interestingly, 19.2% (5/26) of the genes identified exclusively at the RNA/protein level have been previously associated with Mendelian disorders of the ETC (i.e., complex I-IV deficiencies), while only 9.7% (3/31) of the genes cross-identified by genomic (DNA) analysis displayed this affiliation with Mendelian ETC disorders according to the Online Mendelian Inheritance in Man (OMIM) catalogue [69]. These data suggest that genomic alterations are less likely to be identified in SZ for major mitochondrial genes associated with the ETC as loss-of-function mutations would more likely lead to a developmental (and possibly fatal) disorder typified by deficiencies in complex I-IV; however, transcriptome and proteome alterations in these genes are heavily observed in SZ and during adolescence, suggesting other loci or aberrant processes lead to hypofunctional effects, which appear to be targeted on genes required for complex I and II activity in SZ brains.
The 57 mitochondrial genes identified across multiple studies (table 1) are associated with protein products with both synaptic and nonsynaptic enrichment. Völgyi et al. [70] performed proteomic analysis on brain synaptic and nonsynaptic mitochondria from mice and identified proteins enriched in each group. Proteins with synaptic enrichment that have shown alterations in SZ (table 1) are involved in ATP metabolism (ATB5B and ATP5A1) and the ETC (CYB5A and UQCRC1), while proteins with nonsynaptic enrichment that have shown alterations in SZ (table 1) are involved in the tricarboxylic acid cycle (ACO2, IDH3A, MDH1, and PDHB), protein transport and folding (HSPD1), and the ETC (NDUFS1, DLD, and UQCRC1)[70]. These pathways have been repeatedly implicated as mechanisms of metabolic dysfunction in SZ pathology. These data suggest that some mitochondrial SZ candidates may only show alterations in human brain tissue or neuronal cell cultures where synaptic compartments can be isolated or evaluated, while others with nonsynaptic enrichment may be more applicable to peripheral studies using blood or other nonneuronal sources. The other mitochondrial genes we describe (table 1) did not exhibit compartmental enrichment in the proteome study by Volgyi et al. [70], suggesting these genes may be equally involved in mitochondrial processes necessary for both synaptic and nonsynaptic functions.
Functional Signatures of Mitochondrial Dysfunction in SZ
In addition to the genetic associations between mitochondrial genes and SZ, and the observations related to altered expression of the mitochondrial network, functional signatures of mitochondrial dysfunction have been observed by several microscopy techniques. Alterations in mitochondrial morphology, density and number have been observed in ultrastructural studies of SZ brains using transmission electron microscopy [71,72,73,74]. Specifically, a reduction in mitochondrial density has been observed in oligodendrocytes of SZ brains; additional reductions in mitochondrial density have been observed within the caudate and putamen regions of the striatum [71,72]. More recently, a reduction in mitochondrial density specific to synaptic compartments within the striatum has been observed, and showed density reductions specific to both the putamen and caudate and more dramatic alterations in chronic paranoid SZ subjects compared to nonparanoid (undifferentiated) SZ subjects, further supporting the role of synaptic mitochondrial density and SZ symptom variation and disease onset [73,74].
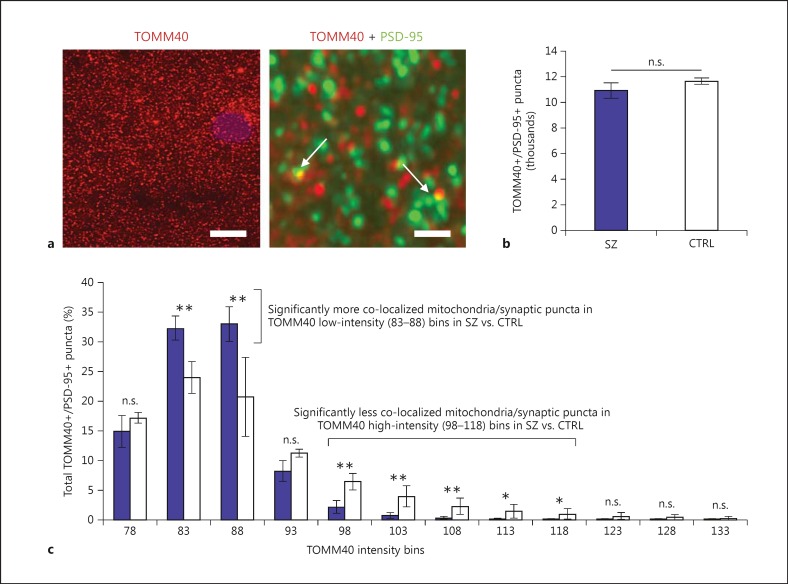
As an additional microscopy-based observation of mitochondrial dysfunction in SZ, we provide preliminary evidence here of a reduced density in (aggregated) mitochondria specifically at postsynaptic density compartments using co-localization morphometric analysis and fluorescence deconvolution tomography (FDT; fig. 2) [75,76,77]. Specifically, we utilized co-localization of TOMM40 as a mitochondrial marker (translocase of the outer mitochondria membrane 40 homolog; Millipore Ab. Cat. No. ABT296), along with PSD-95 (postsynaptic density 95 kDa; Thermo Scientific Ab. Cat. No. MA1-045) to evaluate the relationship between mitochondria and synapses in SZ brains compared to controls. We ran a preliminary comparison of PSD-95 and TOMM40 co-localization profiles in cortical layer I from postmortem dorsolateral PFC of 6 SZ subjects and 2 controls, counting an average of 11,300 co-localized synaptic and mitochondria puncta per subject (fig. 2a). Interestingly, analysis of crude co-localization counts did not demonstrate a difference between SZ and controls in this small cohort (fig. 2b); however, there was a significant difference observed between SZ and controls after binning the co-localized puncta based on fluorescence intensity of the TOMM40 signal (fig. 2c). The results of this analysis demonstrated that there was a significant reduction of ‘high intensity’ TOMM40-immunopositive mitochondria that co-localized with PSD-95 in SZ compared to controls, and a significant increase in ‘low intensity’ mitochondrial puncta that co-localized with PSD-95 in SZ compared to controls (fig. 2c). Given that the morphometric analysis by this FDT technique cannot necessarily resolve individual mitochondria, one interpretation of these results is that the differences in TOMM40 intensity signal are a reflection of mitochondrial density, or the number of mitochondria aggregated within a small space specific to the synaptic compartment. Likewise, differences in TOMM40 intensity could additionally be reflective of differences in mitochondrial size or morphology that affect the immunofluorescence signal at these co-localization sites in, or proximal to, the postsynaptic compartment. Future studies should utilize immunohistochemical imaging techniques such as FDT to evaluate the mitochondrial and spine deficits in SZ brains, and should include analyses of larger sample size, multiple brain regions involved in SZ, and additional markers such as spinophilin or MAP2 [7,78], which have been previously used to demonstrate alterations in SZ brains, but to our knowledge, have not been used in conjunction with mitochondrial markers.
Fig. 2.
FDT morphometric analysis of mitochondria and synaptic puncta in SZ brains. FDT from dorsolateral PFC layer I of SZ (n = 6) compared to controls (CTRL; n = 2) suggests synaptic puncta of SZ subjects have lower density of (aggregated) mitochondria or abnormal mitochondrial morphology. a Fluorescence images from mitochondria and synaptic puncta immunohistochemistry co-localization used for FDT analysis. Left: Low-magnification image of TOMM40 immunofluorescence (i.e., mitochondria label). Scale bar = 10 μm. Right: High-magnification image of TOMM40 (mitochondria = red) and PSD-95 (synaptic puncta = green) immunofluorescence. Colors refer to the online version only. Scale bar = 1 μm. Arrows point to co-localization of both markers used for morphometric analysis between SZ and CTRL. b Mean and standard deviation of co-localized puncta (TOMM40+/PSD-95+) across all subjects suggests no difference between SZ and CTRL (i.e., the same number of co-localized puncta were observed in both groups). c Mean and standard deviation of co-localized puncta (TOMM40+/PSD-95+) across all subjects, binned according to TOMM40 fluorescence intensity and normalized to the total number of puncta counts per subject, demonstrates significant differences between SZ and CTRL and suggests SZ synapses have lower density of (aggregated) mitochondria or abnormal mitochondrial morphology. n.s. = Not significant, * p < 0.05, ** p < 0.01.
In addition to the electron and fluorescence microscopy analyses of SZ postmortem brain tissue, functional signatures of mitochondrial dysfunction have been observed in induced pluripotent stem cell (iPSC) models derived from SZ patients and other disorders. Mitochondrial deficits have already been observed in iPSC-derived neuronal models of several human disorders, including Pearson marrow pancreas syndrome [79], Friedreich's ataxia [80], Parkinson's disease (PD) [81], and SZ [82,83]. Outside of the iPSC models generated from SZ patients thus far, the PINK1 PD model data are particularly intriguing as a cohort of individuals carrying PINK1 mutations have been shown to have a higher occurrence of SZ and schizoaffective disorder than noncarriers [84], although lack of complete penetrance for the SZ phenotype does not suggest PINK1 mutations alone are sufficient for SZ disease liability. However, this observation regarding a link between PINK1 and SZ is also interesting because a loss in PINK1 function has been shown to increase Drp1-dependent mitochondrial fragmentation [85]. Elegant work by Li et al. [20] demonstrated the Drp1- and Opa1-mediated mechanisms and described how a loss in dendritic mitochondria contents leads to a loss of spines and synapses, and showed how a gain in dendritic mitochondria has the opposite effect; together, these datasets provide a strong mechanistic link between mitochondrial dysfunction, dendritic spines, and SZ pathophysiology. The PINK1 PD iPSC model demonstrated PINK1 homozygotes are significantly more susceptible to chemical stressors than healthy controls, resulting in an increased release of lactate dehydrogenase (LD), an increased production of mitochondrial reactive oxygen species (ROS), and reduced glutathione (GSH) levels. Moreover, the authors were able to demonstrate that the iPSC-derived neural cells were more sensitive to the chemical stressors than the parental fibroblasts cells, highlighting the importance of making neural-specific cell types for such evaluations [81].
Several SZ iPSC models have demonstrated mitochondrial defects in addition to synaptic dysfunction and expression alterations in coding and noncoding RNAs. Robicsek et al. [82] evaluated 3 SZ cases versus 2 controls, and reported a significant decrease in mitochondrial membrane potential and an uneven mitochondrial distribution in all evaluated cell types of SZ versus controls, including keratinocytes, iPSCs, neural precursor cells (NPCs), dopaminergic neurons, and glutamatergic neurons. Brennand et al. [83] evaluated NPCs from 4 SZ cases and 6 controls, and reported a significant decrease in mitochondrial membrane potential, an increase in ROS, and a difference in mitochondrial size/distribution in SZ versus controls. In addition to these reports of mitochondrial deficits in SZ iPSC-derived neural cells, other SZ-iPSC studies have also revealed differences in extramitochondrial oxygen consumption and ROS [86], density of synaptic boutons [87], frequency of synaptic currents [87], depolarization-induced vesicle release [87], neuronal connectivity [88], and number of neurites [88] in SZ iPSC models compared to controls. Given that neurons differentiated from human iPSC models represent a much earlier stage of life than those present in the adolescent or adult brain, and that mitochondrial defects have also been observed in nondifferentiated NPCs, these results also suggest mitochondrial dysfunction may precede the onset of SZ symptoms and support the hypothesis that SZ is a developmental disorder.
The Role of APDs
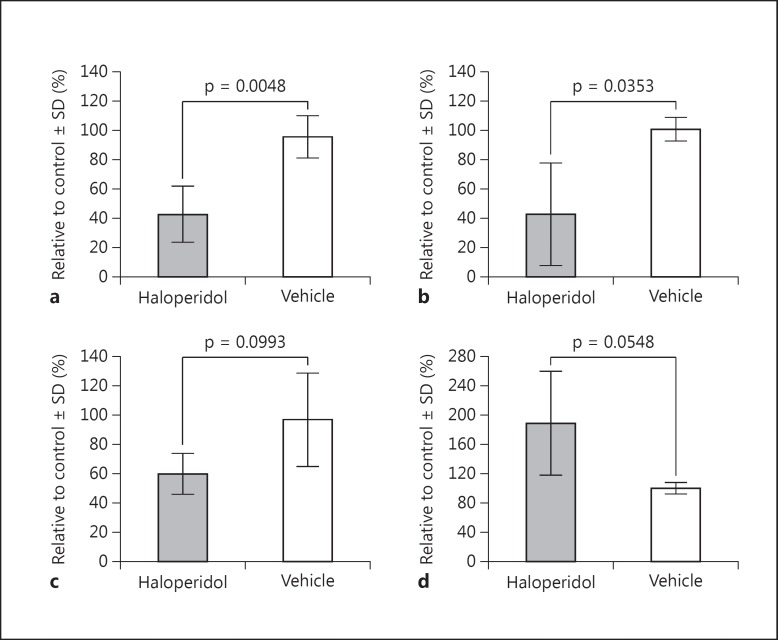
There is a long history of in vitro studies demonstrating that APDs and dopamine agonists can cause mitochondrial impairment, most often described by a hypofunctional effect on mitochondrial complex I [89,90,91,92,93,94,95]. Additionally, APD treatment in rodents has been observed to have long-term effects on mitochondrial protein in synaptosomal preparations [96]. Consistent with these reports, we have observed a significant reduction in mitochondrial function after treating lymphoblastoid cell lines (LCLs) with 30 μm haloperidol for 24-48 h (fig. 3). Specifically, we observed a significant reduction in both adjusted basal respiration (i.e., basal respiration - nonmitochondrial respiration; fig. 3a) and coupling efficiency (fig. 3b) following chronic haloperidol treatments in vitro. In addition, we also found a robust decrease in maximum respiratory capacity after drug treatment (fig. 3c), and a clear increase in ATP turnover (fig. 3d), although these latter two results were not statistically significant across our four independent experiments. It should be noted that the experiment was performed with a supratherapeutic dose of haloperidol, and utilized a nonneuronal cell line; nevertheless, these results are consistent with previously published observations of mitochondrial hypofunction following in vitro APD treatment.
Fig. 3.
Haloperidol induces hypofunctional effects on mitochondria in vitro. LCLs treated with 30 μm haloperidol for 24-48 h exhibit mitochondrial hypofunction. Results display mean and standard deviation (SD) across 4 independent experiments analyzed with the XF Cell Mito Stress Test Kit and XF Analyzer (Seahorse Bioscience), comparing 30 μm haloperidol with vehicle control treatments. a Adjusted basal respiration (i.e., basal respiration - nonmitochondrial respiration). b Coupling efficiency. c Maximum respiratory capacity. d ATP turnover. Statistical comparisons (p values) displayed are from Welch's t tests between both treatment groups.
However, despite the effects observed in vitro, several reports of haloperidol and olanzapine treatment in cynomolgus and macaque monkeys do not support a hypofunctional effect on a variety of SZ-related pathological metrics. In these studies, APD treatment did not result in significant differences in gene expression signatures of mitochondria [97], specific RhoGTPase genes involved in dendritic spine morphogenesis [6], the immunoreactivity of MAP2 [7], or dendritic spine density in general [78]. These results were obtained from gene expression analysis of brain tissue homogenate and region-specific (but not cell-specific) imaging studies, so it is possible that APD treatment effects cannot be observed in these models without isolation or imaging co-localization of specific neuronal cell populations or subtypes.
Despite the potential for APDs to have hypofunctional effects on a number of molecular pathways relevant to SZ pathology, consistent data from blood, postmortem brain tissue, and iPSC models of SZ demonstrate an enrichment of mitochondrial genetic aberrations or reductions in mitochondrial function and thus do not support the idea that mitochondrial dysfunction is exclusively due to APD treatment, but rather is intrinsic to the complex genetic etiology of SZ. The effects of APDs should, nonetheless, be investigated further as hypofunctional effects on pathways relevant to SZ pathology may exacerbate symptoms or vitiate the effect of alternative treatment strategies. Likewise, a variety of APD treatments have been linked to metabolic syndrome and, as such, investigations should continue to evaluate their effects on appetite, metabolic processes, and mitochondria [98]. The motility and function of mitochondria should additionally be evaluated in the context of immune activation, viral infections, drug exposure, etc. as these gene-environment interactions may play a role in SZ pathophysiology, disease onset, or penetrance. Maternal immune activation, for example, has been shown to alter mitochondrial proteins involved in core metabolic pathways, and may be one mechanism by which genetic susceptibility and environmental stressors collectively contribute to SZ [99].
Drug-Targeting Mitochondria in Dendritic Spines and Relevance to Other Disorders
Given the array of evidence that suggests a pathological role of mitochondrial dysfunction in SZ, it may be extremely beneficial to investigate existing therapies and develop new drugs that target mitochondrial function and/or localization. Moreover, there is a wealth of information describing mitochondrial deficits in a number of other disorders, many of which are neurological in nature, so strategies using drugs targeting mitochondria could potentially provide beneficial outcomes to many different patient groups.
A reduction in mitochondrial copy number has been associated with a general increased risk of disability and is a ‘significant predictor of all-cause mortality’ across multiple ethnic populations [57]. In addition, one study evaluated 11 major late-onset human diseases, and found significant association between mitochondrial haplogroups and 5 of the studied diseases - SZ, primary biliary cirrhosis, multiple sclerosis, ulcerative colitis, and PD [50]. Additional associations have been described for mitochondrial variants in a variety of complex human disorders [50,100]. In addition, mitochondrial genes are heavily implicated in Mendelian disorders of both childhood and adult onset, resulting in the emergence of focused exome panels and genetic evaluations where mitochondrial dysfunction is suspected to be involved [101]. A range of mitochondrial abnormalities has also been described for several neuroimmune and neuropsychiatric disorders, including BD, multiple sclerosis, PD, SZ, autism, and chronic fatigue syndrome [102]. Collectively, there is considerable evidence that mitochondrial dysfunction (in general) may not be specific to SZ pathophysiology, but rather the specific causes of mitochondrial dysfunction and the types of mitochondrial abnormalities observed may be disease-specific [102,103,104]. Using BD as an example and point of comparison, mitochondrial alterations that appear specific to SZ include the following: (1) studies evaluating peripheral blood mononuclear cells or LCLs identified significantly reduced complex I activity, increased protein oxidation, and abnormal mitochondrial networks in SZ compared to controls, but not in BD [103,104]; (2) mitochondrial ultrastructure abnormalities have been observed for both SZ and BD, though SZ-specific mitochondrial alterations are more supportive of a reduction in mitochondrial density, while BD-specific mitochondrial alterations allude to changes in mitochondrial size or cellular distribution [41,72,73,74,105,106]; (3) specific alterations in the mitochondrial genome within the brain (e.g., increased substitution rate, increased number of nonsynonymous mutations, and decreased levels of the common deletion) have been observed in SZ but not in BD [42,43,44,52], and (4) large CNVs, specifically large deletions, have been shown to confer risk for SZ and contain an enrichment of mitochondrial targets, but currently available CNV data suggest large deletions do not play a significant role in BD etiology [18,19,35,107,108]. We hypothesize that mitochondrial dysfunction will be observed with greater overlap between psychiatric disorders at the RNA and protein levels due to the downstream effects of increased oxidative stress and altered glycolytic activity, while genetic aberrations in mitochondrial genes will be observed more frequently in SZ. This supports the concept that mitochondrial dysfunction is a redundant feature of psychiatric and neurological disorder pathologies, with a more disease-specific signature of mitochondrial dysfunction occurring in SZ as it relates to genetic etiology.
These lines of evidence support the need to investigate drugs that target mitochondrial dysfunction; for SZ specifically, we hypothesize that therapies observed to modulate mitochondrial density or function in dendritic spines would be the most beneficial. Acetyl-l-carnitine, which is responsible for transporting fatty acids into mitochondria, has been shown to restore dendritic spine density in a rat model of ischemic stroke but only when administered prior to vessel occlusion [109]; likewise, acetyl-l-carnitine treatment has been shown to rescue hippocampal dendritic morphology abnormalities in a mouse model of Rett syndrome when administered daily starting at birth [110]. Given that this treatment has been shown to rescue dendritic spine abnormalities when given early or prior to disease phenotype onset in these models, this type of mitochondrial therapy may likewise be more beneficial in SZ if administered prior to dendritic spine loss and symptom onset, highlighting the need to identify at-risk individuals by genetic analysis. Latrepiridine (Dimebon), an anti-histamine, has been shown to affect mitochondrial morphology and is reported to have a beneficial effect on the cognitive decline observed in Alzheimer's Disease (AD), and displayed some effect on the negative and cognitive symptoms in a small trial of SZ patients [111,112,113]. Rosiglitazone, a proliferator-activated receptor-γ (PPARγ) agonist, has been shown to rescue dendritic spines by increasing mitochondrial numbers in a variety of studies, including an animal model of frontotemporal lobar degeneration (FTLD) and hippocampal slice cultures exposed to toxic amyloid-β oligomers [114,115]. A proliferator-activated receptor-α (PPARα) agonist, fenofibrate, demonstrated significant behavioral modifications in a neurodevelopmental rat model of SZ [116]. Agonists targeting PPARs or transcriptional co-activators of PPARs may be an especially intriguing family of drugs for SZ investigation, as their role in modulating mitochondrial biogenesis, dendritic spines and metabolic effects has been described extensively and is currently a topic of intense study [117,118,119,120,121,122,123]. Interestingly, PPARγ and PPARα agonists were also shown to reverse the behavior changes associated with an animal model of oral dyskinesia, a phenotype that is induced by chronic administration of haloperidol [124]. This leads to the additional speculation described above that APD treatments could potentially vitiate alternative treatment strategies. Consistent with this speculation, rosiglitazone was not shown to have a significant impact on cognitive performance in a small clinical trial of clozapine-treated SZ patients [125]. Future studies should include investigation of currently available and newly developed drugs that target mitochondrial function and density in dendritic spines.
Conclusions
This review summarizes evidence supporting a multifactorial model of mitochondrial dysfunction within the complex genetic etiology of SZ. There is an abundance of published data, in addition to the preliminary evidence we provide, that suggests there are multiple paths to mitochondrial dysfunction in SZ pathophysiology. Given the intrinsic link between mitochondria and dendritic spines, we hypothesize that mitochondrial dysfunction provides one route of pathology leading to the deficits in dendritic spine architecture observed in SZ brains, and ultimately leads to flawed connections in cortical circuitry and SZ symptoms.
Because of the link between mitochondria and dendritic spines, one avenue of pathophysiology is that signatures of mitochondrial dysfunction are caused by other aberrations in genes involved in spine formation and maintenance, such as the PSD system or cytoskeleton organization. If signatures of mitochondrial dysfunction were exclusively related to differences in gene expression, mitochondrial function, or copy number, this speculation would be supported. However, mitochondrial genes are redundantly associated with SZ in GWAS, suggesting it is at least equally or more likely that mitochondrial dysfunction alone may contribute to dendritic spine loss and/or underdevelopment in some, but not necessarily all, SZ subjects. The dendritic spine deficits observed in SZ could, therefore, be independently associated with mitochondrial dysfunction or be an additive phenotype resulting from alterations in the mitochondrial, cytoskeletal, and PSD networks as our model shows in figure 1. There is also evidence suggesting mitochondrial variants may contribute to disease onset variation and symptom severity in SZ, and thus may be ideal candidates for population stratification and post hoc analyses. Aberrations in mitochondrial genes have been observed in the form of large CNVs, rare disruptive mutations, and de novo mutations in a number of independent studies. While most of these events are identified by screening genomic alterations that occur with low frequency in the general population, it should also be noted that SNPs have been predicted to contribute to possibly one quarter of SZ disease liability, many of which are common with a high minor allele frequency. Likewise, genetic associations between SZ and mitochondrial variants within the hypervariable region of the mitochondrial genome and mitochondrial haplogroups support an association between mitochondrial DNA and risk of or protection against SZ. It could be worthwhile to stratify subjects by mitochondrial variants when testing for association of nuclear variants with SZ, which has not yet been reported.
Another possibility that should be addressed is that APD treatment leads to the observed signatures of mitochondrial dysfunction. In vitro evidence does suggest that APD treatment can lead to mitochondrial hypofunction; however, genetic associations in mitochondrial genes further support the argument that these effects are most likely independent of the role mitochondrial dysfunction plays in the complex genetic etiology of SZ. The role of APD treatments in mitochondrial function should, nonetheless, be investigated further as hypofunctional effects could potentially exacerbate SZ symptoms or vitiate the effect of alternative treatment strategies. High-throughput in vitro drug screens may benefit from combinatorial testing designs, where mitochondrial density in dendritic spines can be rescued in the presence of APD treatments, as clinical trials will likely continue to be performed on SZ patients who are currently on some form of APD treatment given the efficacy these drugs have on the positive symptoms.
Lastly, we suggest that drugs targeting mitochondrial function, biogenesis, copy number, or dendritic localization may be a beneficial therapeutic strategy for some SZ patients. Genetic analyses aimed to identify individuals at risk for SZ and schizoaffective disorder need to continue as drugs targeting mitochondrial function and localization may be more efficacious as a preventative strategy in high-risk groups. It is also imperative that large-scale genomic analyses continue on SZ cohorts to potentially allow subgroup identification or stratification for drug trials, as it is possible that drugs aimed to target mitochondrial function or localization will only be helpful for a portion of SZ patients carrying mitochondrial-specific risk variants, while others harboring mutations outside of the mitochondrial network may benefit more from drugs aimed to stabilize microtubules or directly affect neurotransmitter levels. We hypothesize that there are many ways dendritic spine architecture and density could be modulated in SZ pathophysiology, and that mitochondrial dysfunction occurs as one major mode of disease susceptibility. This model further supports the hypothesis that SZ is a polygenic disorder, and thus warrants continued genomic analysis of large SZ cohorts, including covariate or post hoc analyses of phenotypic subgroups or cohort stratification based on mitochondrial variants or haplogroups, in order to understand the molecular mechanisms in play during brain development, symptom onset, and disease progression. We hope that understanding mitochondrial dysfunction within SZ pathophysiology will lead to more efficacious treatments and provide relief to patients afflicted with this devastating psychiatric disorder.
Statement of Ethics
The authors have no ethical conflicts to disclose.
Supplementary Material
Supplementary data
Supplementary data
Supplementary data
Supplementary data
Erratum
In the article by Hjelm BE, Rollins B, Mamdani F et al., entitled ‘Evidence of mitochondrial dysfunction within the complex genetic etiology of schizophrenia’ [Mol Neuropsychiatry 2015;1:201–219, DOI: 10.1159/000441252], the authors would like to add two additional co-authors (marked in bold).
Brooke E. Hjelma Brandi Rollinsa Firoza Mamdania Julie C. Lauterbornb Foram N. Ashare Dan E. Arkinge George Kirovd Gary Lyncha, b Christine M. Gallb, c Adolfo Sequeiraa Marquis P. Vawtera
Departments of a Psychiatry & Human Behavior, b Anatomy & Neurobiology, and c Neurobiology & Behavior, University of California, Irvine, Calif., USA; d MRC Centre for Neuropsychiatric Genetics and Genomics, Institute of Psychological Medicine and Clinical Neurosciences, Cardiff University, Cardiff, UK; e McKusick-Nathans Institute of Genetic Medicine, Johns Hopkins University School of Medicine, Baltimore, Md., USA
References
- 1.Glausier JR, Lewis DA. Dendritic spine pathology in schizophrenia. Neuroscience. 2013;251:90–107. doi: 10.1016/j.neuroscience.2012.04.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry. 2005;10:40–68. doi: 10.1038/sj.mp.4001558. [DOI] [PubMed] [Google Scholar]
- 3.Katsel P, Davis KL, Gorman JM, Haroutunian V. Variations in differential gene expression patterns across multiple brain regions in schizophrenia. Schizophr Res. 2005;77:241–252. doi: 10.1016/j.schres.2005.03.020. [DOI] [PubMed] [Google Scholar]
- 4.Vawter MP, Barrett T, Cheadle C, Sokolov BP, Wood WH 3rd, Donovan DM, Webster M, Freed WJ, Becker KG. Application of cDNA microarrays to examine gene expression differences in schizophrenia. Brain Res Bull. 2001;55:641–650. doi: 10.1016/s0361-9230(01)00522-6. [DOI] [PubMed] [Google Scholar]
- 5.Arion D, Corradi JP, Tang S, Datta D, Boothe F, He A, Cacace AM, Zaczek R, Albright CF, Tseng G, Lewis DA. Distinctive transcriptome alterations of prefrontal pyramidal neurons in schizophrenia and schizoaffective disorder. Mol Psychiatry. doi: 10.1038/mp.2014.171. DOI: 10.1038/mp.2014.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hill JJ, Hashimoto T, Lewis DA. Molecular mechanisms contributing to dendritic spine alterations in the prefrontal cortex of subjects with schizophrenia. Mol Psychiatry. 2006;11:557–566. doi: 10.1038/sj.mp.4001792. [DOI] [PubMed] [Google Scholar]
- 7.Shelton MA, Newman JT, Gu H, Sampson AR, Fish KN, MacDonald ML, Moyer CE, DiBitetto JV, Dorph-Petersen KA, Penzes P, Lewis DA, Sweet RA. Loss of microtubule-associated protein 2 immunoreactivity linked to dendritic spine loss in schizophrenia. Biol Psychiatry. doi: 10.1016/j.biopsych.2014.12.029. DOI: 10.1016/j.biopsych.2014. 12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sullivan PF, Kendler KS, Neale MC. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch Gen Psychiatry. 2003;60:1187–1192. doi: 10.1001/archpsyc.60.12.1187. [DOI] [PubMed] [Google Scholar]
- 9.Cardno AG, Gottesman II. Twin studies of schizophrenia: from bow-and-arrow concordances to star wars Mx and functional genomics. Am J Med Genet. 2000;97:12–17. [PubMed] [Google Scholar]
- 10.Lichtenstein P, Bjork C, Hultman CM, Scolnick E, Sklar P, Sullivan PF. Recurrence risks for schizophrenia in a Swedish national cohort. Psychol Med. 2006;36:1417–1425. doi: 10.1017/S0033291706008385. [DOI] [PubMed] [Google Scholar]
- 11.van den Oord EJ, Sullivan PF, Jiang Y, Walsh D, O'Neill FA, Kendler KS, Riley BP. Identification of a high-risk haplotype for the dystrobrevin binding protein 1 (DTNBP1) gene in the Irish study of high-density schizophrenia families. Mol Psychiatry. 2003;8:499–510. doi: 10.1038/sj.mp.4001263. [DOI] [PubMed] [Google Scholar]
- 12.Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–427. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Middleton FA, Mirnics K, Pierri JN, Lewis DA, Levitt P. Gene expression profiling reveals alterations of specific metabolic pathways in schizophrenia. J Neurosci. 2002;22:2718–2729. doi: 10.1523/JNEUROSCI.22-07-02718.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fromer M, Pocklington AJ, Kavanagh DH, Williams HJ, et al. De novo mutations in schizophrenia implicate synaptic networks. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Purcell SM, Moran JL, Fromer M, Ruderfer D, et al. A polygenic burden of rare disruptive mutations in schizophrenia. Nature. 2014;506:185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Network and Pathway Analysis Subgroup of Psychiatric Genomics Consortium Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat Neurosci. 2015;18:199–209. doi: 10.1038/nn.3922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Potkin SG, Macciardi F, Guffanti G, Fallon JH, Wang Q, Turner JA, Lakatos A, Miles MF, Lander A, Vawter MP, Xie X. Identifying gene regulatory networks in schizophrenia. Neuroimage. 2010;53:839–847. doi: 10.1016/j.neuroimage.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Szatkiewicz JP, O'Dushlaine C, Chen G, Chambert K, et al. Copy number variation in schizophrenia in Sweden. Mol Psychiatry. 2014;19:762–773. doi: 10.1038/mp.2014.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirov G, Pocklington AJ, Holmans P, Ivanov D, et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry. 2012;17:142–153. doi: 10.1038/mp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li Z, Okamoto K, Hayashi Y, Sheng M. The importance of dendritic mitochondria in the morphogenesis and plasticity of spines and synapses. Cell. 2004;119:873–887. doi: 10.1016/j.cell.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 21.Sheng ZH, Cai Q. Mitochondrial transport in neurons: impact on synaptic homeostasis and neurodegeneration. Nat Rev Neurosci. 2012;13:77–93. doi: 10.1038/nrn3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vos M, Lauwers E, Verstreken P. Synaptic mitochondria in synaptic transmission and organization of vesicle pools in health and disease. Front Synaptic Neurosci. 2010;2:139. doi: 10.3389/fnsyn.2010.00139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Picard M, McEwen BS. Mitochondria impact brain function and cognition. Proc Natl Acad Sci USA. 2014;111:7–8. doi: 10.1073/pnas.1321881111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pathak D, Shields LY, Mendelsohn BA, Haddad D, Lin W, Gerencser AA, Kim H, Brand MD, Edwards RH, Nakamura K. The role of mitochondrially derived ATP in synaptic vesicle recycling. J Biol Chem. 2015;290:22325–22336. doi: 10.1074/jbc.M115.656405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chada SR, Hollenbeck PJ. Mitochondrial movement and positioning in axons: the role of growth factor signaling. J Exp Biol. 2003;206:1985–1992. doi: 10.1242/jeb.00263. [DOI] [PubMed] [Google Scholar]
- 26.Su B, Ji YS, Sun XL, Liu XH, Chen ZY. Brain-derived neurotrophic factor (BDNF)-induced mitochondrial motility arrest and presynaptic docking contribute to BDNF-enhanced synaptic transmission. J Biol Chem. 2014;289:1213–1226. doi: 10.1074/jbc.M113.526129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen S, Owens GC, Crossin KL, Edelman DB. Serotonin stimulates mitochondrial transport in hippocampal neurons. Mol Cell Neurosci. 2007;36:472–483. doi: 10.1016/j.mcn.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 28.Chen S, Owens GC, Edelman DB. Dopamine inhibits mitochondrial motility in hippocampal neurons. PLoS One. 2008;3:e2804. doi: 10.1371/journal.pone.0002804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen S, Owens GC, Makarenkova H, Edelman DB. HDAC6 regulates mitochondrial transport in hippocampal neurons. PLoS One. 2010;5:e10848. doi: 10.1371/journal.pone.0010848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kramer T, Enquist LW. Alphaherpesvirus infection disrupts mitochondrial transport in neurons. Cell Host Microbe. 2012;11:504–514. doi: 10.1016/j.chom.2012.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anand SK, Tikoo SK. Viruses as modulators of mitochondrial functions. Adv Virol. 2013;2013:738794. doi: 10.1155/2013/738794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.West AP, Shadel GS, Ghosh S. Mitochondria in innate immune responses. Nat Rev Immunol. 2011;11:389–402. doi: 10.1038/nri2975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Martin-Cofreces NB, Baixauli F, Sanchez-Madrid F. Immune synapse: conductor of orchestrated organelle movement. Trends Cell Biol. 2014;24:61–72. doi: 10.1016/j.tcb.2013.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kimura T, Murakami F. Evidence that dendritic mitochondria negatively regulate dendritic branching in pyramidal neurons in the neocortex. J Neurosci. 2014;34:6938–6951. doi: 10.1523/JNEUROSCI.5095-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karayiorgou M, Simon TJ, Gogos JA. 22q11.2 microdeletions: linking DNA structural variation to brain dysfunction and schizophrenia. Nat Rev Neurosci. 2010;11:402–416. doi: 10.1038/nrn2841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li X, Zhang W, Tang J, Tan L, Luo XJ, Chen X, Yao YG. Do nuclear-encoded core subunits of mitochondrial complex I confer genetic susceptibility to schizophrenia in Han Chinese populations? Sci Rep. 2015;5:11076. doi: 10.1038/srep11076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Altar CA, Jurata LW, Charles V, Lemire A, Liu P, Bukhman Y, Young TA, Bullard J, Yokoe H, Webster MJ, Knable MB, Brockman JA. Deficient hippocampal neuron expression of proteasome, ubiquitin, and mitochondrial genes in multiple schizophrenia cohorts. Biol Psychiatry. 2005;58:85–96. doi: 10.1016/j.biopsych.2005.03.031. [DOI] [PubMed] [Google Scholar]
- 38.Harris LW, Lockstone HE, Khaitovich P, Weickert CS, Webster MJ, Bahn S. Gene expression in the prefrontal cortex during adolescence: implications for the onset of schizophrenia. BMC Med Genomics. 2009;2:28. doi: 10.1186/1755-8794-2-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jaffe AE, Shin J, Collado-Torres L, Leek JT, Tao R, Li C, Gao Y, Jia Y, Maher BJ, Hyde TM, Kleinman JE, Weinberger DR. Developmental regulation of human cortex transcription and its clinical relevance at single base resolution. Nat Neurosci. 2015;18:154–161. doi: 10.1038/nn.3898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.English JA, Pennington K, Dunn MJ, Cotter DR. The neuroproteomics of schizophrenia. Biol Psychiatry. 2011;69:163–172. doi: 10.1016/j.biopsych.2010.06.031. [DOI] [PubMed] [Google Scholar]
- 41.Shao L, Martin MV, Watson SJ, Schatzberg A, Akil H, Myers RM, Jones EG, Bunney WE, Vawter MP. Mitochondrial involvement in psychiatric disorders. Ann Med. 2008;40:281–295. doi: 10.1080/07853890801923753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sequeira A, Martin MV, Rollins B, Moon EA, Bunney WE, Macciardi F, Lupoli S, Smith EN, Kelsoe J, Magnan CN, van Oven M, Baldi P, Wallace DC, Vawter MP. Mitochondrial mutations and polymorphisms in psychiatric disorders. Front Genet. 2012;3:103. doi: 10.3389/fgene.2012.00103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sequeira A, Rollins B, Magnan C, van Oven M, Baldi P, Myers RM, Barchas JD, Schatzberg AF, J. Watson S, Akil H, Bunney WE, Vawter MP. Mitochondrial mutations in subjects with psychiatric disorders. PLoS One. 2015;10:e0127280. doi: 10.1371/journal.pone.0127280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rollins B, Martin MV, Sequeira PA, Moon EA, Morgan LZ, Watson SJ, Schatzberg A, Akil H, Myers RM, Jones EG, Wallace DC, Bunney WE, Vawter MP. Mitochondrial variants in schizophrenia, bipolar disorder, and major depressive disorder. PLoS One. 2009;4:e4913. doi: 10.1371/journal.pone.0004913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang GX, Zhang Y, Zhang YT, Dong YS, Lv ZW, Sun M, Wu D, Wu YM. Mitochondrial haplogroups and hypervariable region polymorphisms in schizophrenia: a case-control study. Psychiatry Res. 2013;209:279–283. doi: 10.1016/j.psychres.2013.01.001. [DOI] [PubMed] [Google Scholar]
- 46.Amar S, Shamir A, Ovadia O, Blanaru M, Reshef A, Kremer I, Rietschel M, Schulze TG, Maier W, Belmaker RH, Ebstein RP, Agam G, Mishmar D. Mitochondrial DNA HV lineage increases the susceptibility to schizophrenia among Israeli Arabs. Schizophr Res. 2007;94:354–358. doi: 10.1016/j.schres.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 47.Magri C, Gardella R, Barlati SD, Valsecchi P, Sacchetti E, Barlati S. Mitochondrial DNA haplogroups and age at onset of schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2007;144B:496–501. doi: 10.1002/ajmg.b.30496. [DOI] [PubMed] [Google Scholar]
- 48.Zhang W, Tang J, Zhang AM, Peng M-S, Xie H-B, Tan L, Xu L, Zhang Y-P, Chen X, Yao Y-G. A matrilineal genetic legacy from the last glacial maximum confers susceptibility to schizophrenia in Han Chinese. J Genet Genom. 2014;41:397–407. doi: 10.1016/j.jgg.2014.05.004. [DOI] [PubMed] [Google Scholar]
- 49.Mosquera-Miguel A, Torrell H, Abasolo N, Arrojo M, Paz E, Ramos-Rios R, Agra S, Paramo M, Brenlla J, Martinez S, Vilella E, Valero J, Gutierrez-Zotes A, Martorell L, Costas J, Salas A. No evidence that major mtDNA European haplogroups confer risk to schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2012;159B:414–421. doi: 10.1002/ajmg.b.32044. [DOI] [PubMed] [Google Scholar]
- 50.Hudson G, Gomez-Duran A, Wilson IJ, Chinnery PF. Recent mitochondrial DNA mutations increase the risk of developing common late-onset human diseases. PLoS Genet. 2014;10:e1004369. doi: 10.1371/journal.pgen.1004369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Torrell H, Salas A, Abasolo N, Moren C, Garrabou G, Valero J, Alonso Y, Vilella E, Costas J, Martorell L. Mitochondrial DNA (mtDNA) variants in the European haplogroups HV, JT, and U do not have a major role in schizophrenia. Am J Med Genet B Neuropsychiatr Genet. 2014;165B:607–617. doi: 10.1002/ajmg.b.32264. [DOI] [PubMed] [Google Scholar]
- 52.Mamdani F, Rollins B, Morgan L, Sequeira PA, Vawter MP. The somatic common deletion in mitochondrial DNA is decreased in schizophrenia. Schizophr Res. 2014;159:370–375. doi: 10.1016/j.schres.2014.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Corral-Debrinski M, Horton T, Lott MT, Shoffner JM, Beal MF, Wallace DC. Mitochondrial DNA deletions in human brain: regional variability and increase with advanced age. Nat Genet. 1992;2:324–329. doi: 10.1038/ng1292-324. [DOI] [PubMed] [Google Scholar]
- 54.Cortopassi GA, Arnheim N. Detection of a specific mitochondrial DNA deletion in tissues of older humans. Nucleic Acids Res. 1990;18:6927–6933. doi: 10.1093/nar/18.23.6927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cortopassi GA, Shibata D, Soong NW, Arnheim N. A pattern of accumulation of a somatic deletion of mitochondrial DNA in aging human tissues. Proc Natl Acad Sci USA. 1992;89:7370–7374. doi: 10.1073/pnas.89.16.7370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Meissner C, Bruse P, Mohamed SA, Schulz A, Warnk H, Storm T, Oehmichen M. The 4,977 bp deletion of mitochondrial DNA in human skeletal muscle, heart and different areas of the brain: a useful biomarker or more? Exp Gerontol. 2008;43:645–652. doi: 10.1016/j.exger.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 57.Ashar FN, Moes A, Moore AZ, Grove ML, Chaves PH, Coresh J, Newman AB, Matteini AM, Bandeen-Roche K, Boerwinkle E, Walston JD, Arking DE. Association of mitochondrial DNA levels with frailty and all-cause mortality. J Mol Med (Berl) 2015;93:177–186. doi: 10.1007/s00109-014-1233-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kakiuchi C, Ishiwata M, Kametani M, Nelson C, Iwamoto K, Kato T. Quantitative analysis of mitochondrial DNA deletions in the brains of patients with bipolar disorder and schizophrenia. Int J Neuropsychopharmacol. 2005;8:515–522. doi: 10.1017/S1461145705005213. [DOI] [PubMed] [Google Scholar]
- 59.Xing J, Chen M, Wood CG, Lin J, Spitz MR, Ma J, Amos CI, Shields PG, Benowitz NL, Gu J, de Andrade M, Swan GE, Wu X. Mitochondrial DNA content: its genetic heritability and association with renal cell carcinoma. J Natl Cancer Inst. 2008;100:1104–1112. doi: 10.1093/jnci/djn213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wallace DC. The mitochondrial genome in human adaptive radiation and disease: on the road to therapeutics and performance enhancement. Gene. 2005;354:169–180. doi: 10.1016/j.gene.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 61.Pagliarini DJ, Calvo SE, Chang B, Sheth SA, Vafai SB, Ong SE, Walford GA, Sugiana C, Boneh A, Chen WK, Hill DE, Vidal M, Evans JG, Thorburn DR, Carr SA, Mootha VK. A mitochondrial protein compendium elucidates complex I disease biology. Cell. 2008;134:112–123. doi: 10.1016/j.cell.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Srinivasan H, Das S. Mitochondrial miRNA (MitomiR): a new player in cardiovascular health. Can J Physiol Pharmacol. 2015;93:855–861. doi: 10.1139/cjpp-2014-0500. [DOI] [PubMed] [Google Scholar]
- 63.Rackham O, Shearwood AM, Mercer TR, Davies SM, Mattick JS, Filipovska A. Long noncoding RNAs are generated from the mitochondrial genome and regulated by nuclear-encoded proteins. RNA. 2011;17:2085–2093. doi: 10.1261/rna.029405.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang K, Long B, Zhou LY, Liu F, Zhou QY, Liu CY, Fan YY, Li PF. CARL lncRNA inhibits anoxia-induced mitochondrial fission and apoptosis in cardiomyocytes by impairing miR-539-dependent PHB2 downregulation. Nat Commun. 2014;5:3596. doi: 10.1038/ncomms4596. [DOI] [PubMed] [Google Scholar]
- 65.Dorn GW., 2nd LIPCAR: a mitochondrial lnc in the noncoding RNA chain? Circ Res. 2014;114:1548–1550. doi: 10.1161/CIRCRESAHA.114.304028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen J, Lipska BK, Halim N, Ma QD, Matsumoto M, Melhem S, Kolachana BS, Hyde TM, Herman MM, Apud J, Egan MF, Kleinman JE, Weinberger DR. Functional analysis of genetic variation in catechol-O-methyltransferase (COMT): effects on mRNA, protein, and enzyme activity in postmortem human brain. Am J Hum Genet. 2004;75:807–821. doi: 10.1086/425589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, Khodiyar VK, Lush MJ, Povey S, Talbot CC Jr, Wright MW, Wain HM, Trowsdale J, Ziegler A, Beck S. Gene map of the extended human MHC. Nat Rev Genet. 2004;5:889–899. doi: 10.1038/nrg1489. [DOI] [PubMed] [Google Scholar]
- 68.Lee SH, DeCandia TR, Ripke S, Yang J, Sullivan PF, Goddard ME, Keller MC, Visscher PM, Wray NR. Estimating the proportion of variation in susceptibility to schizophrenia captured by common SNPs. Nat Genet. 2012;44:247–250. doi: 10.1038/ng.1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Amberger JS, Bocchini CA, Schiettecatte F, Scott AF, Hamosh A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 2015;43:D789–D798. doi: 10.1093/nar/gku1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Volgyi K, Gulyassy P, Haden K, Kis V, Badics K, Kekesi KA, Simor A, Gyorffy B, Toth EA, Lubec G, Juhasz G, Dobolyi A. Synaptic mitochondria: a brain mitochondria cluster with a specific proteome. J Proteomics. 2015;120:142–157. doi: 10.1016/j.jprot.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 71.Uranova N, Orlovskaya D, Vikhreva O, Zimina I, Kolomeets N, Vostrikov V, Rachmanova V. Electron microscopy of oligodendroglia in severe mental illness. Brain Res Bull. 2001;55:597–610. doi: 10.1016/s0361-9230(01)00528-7. [DOI] [PubMed] [Google Scholar]
- 72.Kung L, Roberts RC. Mitochondrial pathology in human schizophrenic striatum: a postmortem ultrastructural study. Synapse. 1999;31:67–75. doi: 10.1002/(SICI)1098-2396(199901)31:1<67::AID-SYN9>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- 73.Somerville SM, Conley RR, Roberts RC. Mitochondria in the striatum of subjects with schizophrenia. World J Biol Psychiatry. 2011;12:48–56. doi: 10.3109/15622975.2010.505662. [DOI] [PubMed] [Google Scholar]
- 74.Somerville SM, Conley RR, Roberts RC. Striatal mitochondria in subjects with chronic undifferentiated vs chronic paranoid schizophrenia. Synapse. 2012;66:29–41. doi: 10.1002/syn.20981. [DOI] [PubMed] [Google Scholar]
- 75.Seese RR, Babayan AH, Katz AM, Cox CD, Lauterborn JC, Lynch G, Gall CM. LTP induction translocates cortactin at distant synapses in wild-type but not Fmr1 knock-out mice. J Neurosci. 2012;32:7403–7413. doi: 10.1523/JNEUROSCI.0968-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Seese RR, Chen LY, Cox CD, Schulz D, Babayan AH, Bunney WE, Henn FA, Gall CM, Lynch G. Synaptic abnormalities in the infralimbic cortex of a model of congenital depression. J Neurosci. 2013;33:13441–13448. doi: 10.1523/JNEUROSCI.2434-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Rex CS, Chen LY, Sharma A, Liu J, Babayan AH, Gall CM, Lynch G. Different Rho GTPase-dependent signaling pathways initiate sequential steps in the consolidation of long-term potentiation. J Cell Biol. 2009;186:85–97. doi: 10.1083/jcb.200901084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Sweet RA, Henteleff RA, Zhang W, Sampson AR, Lewis DA. Reduced dendritic spine density in auditory cortex of subjects with schizophrenia. Neuropsychopharmacology. 2009;34:374–389. doi: 10.1038/npp.2008.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cherry AB, Gagne KE, McLoughlin EM, Baccei A, Gorman B, Hartung O, Miller JD, Zhang J, Zon RL, Ince TA, Neufeld EJ, Lerou PH, Fleming MD, Daley GQ, Agarwal S. Induced pluripotent stem cells with a mitochondrial DNA deletion. Stem Cells. 2013;31:1287–1297. doi: 10.1002/stem.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hick A, Wattenhofer-Donze M, Chintawar S, Tropel P, Simard JP, Vaucamps N, Gall D, Lambot L, Andre C, Reutenauer L, Rai M, Teletin M, Messaddeq N, Schiffmann SN, Viville S, Pearson CE, Pandolfo M, Puccio H. Neurons and cardiomyocytes derived from induced pluripotent stem cells as a model for mitochondrial defects in Friedreich's ataxia. Dis Model Mech. 2013;6:608–621. doi: 10.1242/dmm.010900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Cooper O, Seo H, Andrabi S, Guardia-Laguarta C, et al. Pharmacological rescue of mitochondrial deficits in iPSC-derived neural cells from patients with familial Parkinson's disease. Sci Transl Med. 2012;4 doi: 10.1126/scitranslmed.3003985. 141ra190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Robicsek O, Karry R, Petit I, Salman-Kesner N, Muller FJ, Klein E, Aberdam D, Ben-Shachar D. Abnormal neuronal differentiation and mitochondrial dysfunction in hair follicle-derived induced pluripotent stem cells of schizophrenia patients. Mol Psychiatry. 2013;18:1067–1076. doi: 10.1038/mp.2013.67. [DOI] [PubMed] [Google Scholar]
- 83.Brennand K, Savas JN, Kim Y, Tran N, Simone A, Hashimoto-Torii K, Beaumont KG, Kim HJ, Topol A, Ladran I, Abdelrahim M, Matikainen-Ankney B, Chao SH, Mrksich M, Rakic P, Fang G, Zhang B, Yates JR 3rd, Gage FH. Phenotypic differences in hiPSC NPCs derived from patients with schizophrenia. Mol Psychiatry. 2015;20:361–368. doi: 10.1038/mp.2014.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Steinlechner S, Stahlberg J, Volkel B, Djarmati A, Hagenah J, Hiller A, Hedrich K, Konig I, Klein C, Lencer R. Co-occurrence of affective and schizophrenia spectrum disorders with PINK1 mutations. J Neurol Neurosurg Psychiatry. 2007;78:532–535. doi: 10.1136/jnnp.2006.105676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lutz AK, Exner N, Fett ME, Schlehe JS, Kloos K, Lammermann K, Brunner B, Kurz-Drexler A, Vogel F, Reichert AS, Bouman L, Vogt-Weisenhorn D, Wurst W, Tatzelt J, Haass C, Winklhofer KF. Loss of parkin or PINK1 function increases Drp1-dependent mitochondrial fragmentation. J Biol Chem. 2009;284:22938–22951. doi: 10.1074/jbc.M109.035774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Paulsen Bda S, de Moraes Maciel R, Galina A, Souza da Silveira M, dos Santos Souza C, Drummond H, Nascimento Pozzatto E, Silva H Jr, Chicaybam L, Massuda R, Setti-Perdigao P, Bonamino M, Belmonte-de-Abreu PS, Castro NG, Brentani H, Rehen SK. Altered oxygen metabolism associated to neurogenesis of induced pluripotent stem cells derived from a schizophrenic patient. Cell Transplant. 2012;21:1547–1559. doi: 10.3727/096368911X600957. [DOI] [PubMed] [Google Scholar]
- 87.Wen Z, Nguyen HN, Guo Z, Lalli MA, et al. Synaptic dysregulation in a human iPS cell model of mental disorders. Nature. 2014;515:414–418. doi: 10.1038/nature13716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Brennand KJ, Simone A, Jou J, Gelboin- Burkhart C, Tran N, Sangar S, Li Y, Mu Y, Chen G, Yu D, McCarthy S, Sebat J, Gage FH. Modelling schizophrenia using human induced pluripotent stem cells. Nature. 2011;473:221–225. doi: 10.1038/nature09915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brenner-Lavie H, Klein E, Ben-Shachar D. Mitochondrial complex I as a novel target for intraneuronal DA: modulation of respiration in intact cells. Biochem Pharmacol. 2009;78:85–95. doi: 10.1016/j.bcp.2009.03.024. [DOI] [PubMed] [Google Scholar]
- 90.Rosenfeld M, Brenner-Lavie H, Ari SG, Kavushansky A, Ben-Shachar D. Perturbation in mitochondrial network dynamics and in complex I dependent cellular respiration in schizophrenia. Biol Psychiatry. 2011;69:980–988. doi: 10.1016/j.biopsych.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 91.Ben-Shachar D. Mitochondrial dysfunction in schizophrenia: a possible linkage to dopamine. J Neurochem. 2002;83:1241–1251. doi: 10.1046/j.1471-4159.2002.01263.x. [DOI] [PubMed] [Google Scholar]
- 92.Bachmann E, Zbinden G. Effect of antidepressant and neuroleptic drugs on respiratory function of rat heart mitochondria. Biochem Pharmacol. 1979;28:3519–3524. doi: 10.1016/0006-2952(79)90394-0. [DOI] [PubMed] [Google Scholar]
- 93.Sagara Y. Induction of reactive oxygen species in neurons by haloperidol. J Neurochem. 1998;71:1002–1012. doi: 10.1046/j.1471-4159.1998.71031002.x. [DOI] [PubMed] [Google Scholar]
- 94.Casademont J, Garrabou G, Miro O, Lopez S, Pons A, Bernardo M, Cardellach F. Neuroleptic treatment effect on mitochondrial electron transport chain: peripheral blood mononuclear cells analysis in psychotic patients. J Clin Psychopharmacol. 2007;27:284–288. doi: 10.1097/JCP.0b013e318054753e. [DOI] [PubMed] [Google Scholar]
- 95.Balijepalli S, Boyd MR, Ravindranath V. Inhibition of mitochondrial complex I by haloperidol: the role of thiol oxidation. Neuropharmacology. 1999;38:567–577. doi: 10.1016/s0028-3908(98)00215-9. [DOI] [PubMed] [Google Scholar]
- 96.Farrelly LA, Dicker P, Wynne K, English J, Cagney G, Focking M, Cotter DR. Adolescent risperidone treatment alters protein expression associated with protein trafficking and cellular metabolism in the adult rat prefrontal cortex. Proteomics. 2014;14:1574–1578. doi: 10.1002/pmic.201300466. [DOI] [PubMed] [Google Scholar]
- 97.Martin MV, Mirnics K, Nisenbaum LK, Vawter MP. Olanzapine reversed brain gene expression changes induced by phencyclidine treatment in non-human primates. Mol Neuropsychiatry. 2015;1:82–93. doi: 10.1159/000430786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Baptista T, De Mendoza S, Beaulieu S, Bermudez A, Martinez M. The metabolic syndrome during atypical antipsychotic drug treatment: mechanisms and management. Metab Syndr Relat Disord. 2004;2:290–307. doi: 10.1089/met.2004.2.290. [DOI] [PubMed] [Google Scholar]
- 99.Farrelly L, Focking M, Piontkewitz Y, Dicker P, English J, Wynne K, Cannon M, Cagney G, Cotter DR. Maternal immune activation induces changes in myelin and metabolic proteins, some of which can be prevented with risperidone in adolescence. Dev Neurosci. 2015;37:43–55. doi: 10.1159/000368305. [DOI] [PubMed] [Google Scholar]
- 100.Pieczenik SR, Neustadt J. Mitochondrial dysfunction and molecular pathways of disease. Exp Mol Pathol. 2007;83:84–92. doi: 10.1016/j.yexmp.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 101.Falk MJ, Pierce EA, Consugar M, Xie MH, Guadalupe M, Hardy O, Rappaport EF, Wallace DC, LeProust E, Gai X. Mitochondrial disease genetic diagnostics: optimized whole-exome analysis for all MitoCarta nuclear genes and the mitochondrial genome. Discov Med. 2012;14:389–399. [PMC free article] [PubMed] [Google Scholar]
- 102.Morris G, Berk M. The many roads to mitochondrial dysfunction in neuroimmune and neuropsychiatric disorders. BMC Med. 2015;13:68. doi: 10.1186/s12916-015-0310-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rosenfeld M, Brenner-Lavie H, Ari SG, Kavushansky A, Ben-Shachar D. Perturbation in mitochondrial network dynamics and in complex I dependent cellular respiration in schizophrenia. Biol Psychiatry. 2011;69:980–988. doi: 10.1016/j.biopsych.2011.01.010. [DOI] [PubMed] [Google Scholar]
- 104.Gubert C, Stertz L, Pfaffenseller B, Panizzutti BS, Rezin GT, Massuda R, Streck EL, Gama CS, Kapczinski F, Kunz M. Mitochondrial activity and oxidative stress markers in peripheral blood mononuclear cells of patients with bipolar disorder, schizophrenia, and healthy subjects. J Psychiatr Res. 2013;47:1396–1402. doi: 10.1016/j.jpsychires.2013.06.018. [DOI] [PubMed] [Google Scholar]
- 105.Cataldo AM, McPhie DL, Lange NT, Punzell S, Elmiligy S, Ye NZ, Froimowitz MP, Hassinger LC, Menesale EB, Sargent LW, Logan DJ, Carpenter AE, Cohen BM. Abnormalities in mitochondrial structure in cells from patients with bipolar disorder. Am J Pathol. 2010;177:575–585. doi: 10.2353/ajpath.2010.081068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Clay HB, Sillivan S, Konradi C. Mitochondrial dysfunction and pathology in bipolar disorder and schizophrenia. Int J Dev Neurosci. 2011;29:311–324. doi: 10.1016/j.ijdevneu.2010.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Napoli E, Tassone F, Wong S, Angkustsiri K, Simon TJ, Song G, Giulivi C. Mitochondrial citrate transporter-dependent metabolic signature in the 22q11.2 deletion syndrome. J Biol Chem. 2015;290:23240–23253. doi: 10.1074/jbc.M115.672360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Green EK, Rees E, Walters JT, Smith KG, Forty L, Grozeva D, Moran JL, Sklar P, Ripke S, Chambert KD, Genovese G, McCarroll SA, Jones I, Jones L, Owen MJ, O'Donovan MC, Craddock N, Kirov G. Copy number variation in bipolar disorder. Mol Psychiatry. doi: 10.1038/mp.2014.174. DOI: 10.1038/mp.2014.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Kocsis K, Knapp L, Gellert L, Olah G, Kis Z, Takakuwa H, Iwamori N, Ono E, Toldi J, Farkas T. Acetyl-L-carnitine normalizes the impaired long-term potentiation and spine density in a rat model of global ischemia. Neuroscience. 2014;269:265–272. doi: 10.1016/j.neuroscience.2014.03.055. [DOI] [PubMed] [Google Scholar]
- 110.Schaevitz LR, Nicolai R, Lopez CM, D'Iddio S, Iannoni E, Berger-Sweeney JE. Acetyl-L-carnitine improves behavior and dendritic morphology in a mouse model of Rett syndrome. PLoS One. 2012;7:e51586. doi: 10.1371/journal.pone.0051586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Eckert SH, Eckmann J, Renner K, Eckert GP, Leuner K, Muller WE. Dimebon ameliorates amyloid-beta induced impairments of mitochondrial form and function. J Alzheimers Dis. 2012;31:21–32. doi: 10.3233/JAD-2012-120310. [DOI] [PubMed] [Google Scholar]
- 112.Morozova MA, Beniashvili AG, Lepilkina TA, Rupchev GE. Double-blind placebo-controlled randomized efficacy and safety trial of add-on treatment of dimebon plus risperidone in schizophrenic patients during transition from acute psychotic episode to remission. Psychiatr Danub. 2012;24:159–166. [PubMed] [Google Scholar]
- 113.Cano-Cuenca N, Solis-Garcia del Pozo JE, Jordan J. Evidence for the efficacy of latrepirdine (Dimebon) treatment for improvement of cognitive function: a meta-analysis. J Alzheimers Dis. 2014;38:155–164. doi: 10.3233/JAD-130872. [DOI] [PubMed] [Google Scholar]
- 114.Huang C, Tong J, Bi F, Wu Q, Huang B, Zhou H, Xia XG. Entorhinal cortical neurons are the primary targets of FUS mislocalization and ubiquitin aggregation in FUS transgenic rats. Hum Mol Genet. 2012;21:4602–4614. doi: 10.1093/hmg/dds299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Xu S, Liu G, Bao X, Wu J, Li S, Zheng B, Anwyl R, Wang Q. Rosiglitazone prevents amyloid-beta oligomer-induced impairment of synapse formation and plasticity via increasing dendrite and spine mitochondrial number. J Alzheimers Dis. 2014;39:239–251. doi: 10.3233/JAD-130680. [DOI] [PubMed] [Google Scholar]
- 116.Rolland B, Marche K, Cottencin O, Bordet R. The PPARα agonist fenofibrate reduces prepulse inhibition disruption in a neurodevelopmental model of schizophrenia. Schizophr Res Treatment. 2012;2012:839853. doi: 10.1155/2012/839853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Rolland B, Deguil J, Jardri R, Cottencin O, Thomas P, Bordet R. Therapeutic prospects of PPARs in psychiatric disorders: a comprehensive review. Curr Drug Targets. 2013;14:724–732. doi: 10.2174/1389450111314070002. [DOI] [PubMed] [Google Scholar]
- 118.Martinez-Gras I, Perez-Nievas BG, Garcia-Bueno B, Madrigal JL, Andres-Esteban E, Rodriguez-Jimenez R, Hoenicka J, Palomo T, Rubio G, Leza JC. The anti-inflammatory prostaglandin 15d-PGJ2 and its nuclear receptor PPARgamma are decreased in schizophrenia. Schizophr Res. 2011;128:15–22. doi: 10.1016/j.schres.2011.01.018. [DOI] [PubMed] [Google Scholar]
- 119.Chen CH, Lu ML, Kuo PH, Chen PY, Chiu CC, Kao CF, Huang MC. Gender differences in the effects of peroxisome proliferator-activated receptor gamma2 gene polymorphisms on metabolic adversity in patients with schizophrenia or schizoaffective disorder. Prog Neuropsychopharmacol Biol Psychiatry. 2011;35:239–245. doi: 10.1016/j.pnpbp.2010.11.014. [DOI] [PubMed] [Google Scholar]
- 120.Garcia-Bueno B, Perez-Nievas BG, Leza JC. Is there a role for the nuclear receptor PPARgamma in neuropsychiatric diseases? Int J Neuropsychopharmacol. 2010;13:1411–1429. doi: 10.1017/S1461145710000970. [DOI] [PubMed] [Google Scholar]
- 121.Herken H, Erdal M, Aydin N, Sengul C, Karadag F, Barlas O, Akin F. The association of olanzapine-induced weight gain with peroxisome proliferator-activated receptor-gamma2 Pro12Ala polymorphism in patients with schizophrenia. DNA Cell Biol. 2009;28:515–519. doi: 10.1089/dna.2009.0893. [DOI] [PubMed] [Google Scholar]
- 122.Cheng A, Wan R, Yang JL, Kamimura N, Son TG, Ouyang X, Luo Y, Okun E, Mattson MP. Involvement of PGC-1alpha in the formation and maintenance of neuronal dendritic spines. Nat Commun. 2012;3:1250. doi: 10.1038/ncomms2238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Cheng A, Hou Y, Mattson MP. Mitochondria and neuroplasticity. ASN Neuro. 2010;2:e00045. doi: 10.1042/AN20100019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Grover S, Kumar P, Singh K, Vikram V, Budhiraja RD. Possible beneficial effect of peroxisome proliferator-activated receptor (PPAR)-alpha and gamma agonist against a rat model of oral dyskinesia. Pharmacol Biochem Behav. 2013;111:17–23. doi: 10.1016/j.pbb.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 125.Yi Z, Fan X, Wang J, Liu D, Freudenreich O, Goff D, Henderson DC. Rosiglitazone and cognitive function in clozapine-treated patients with schizophrenia: a pilot study. Psychiatry Res. 2012;200:79–82. doi: 10.1016/j.psychres.2012.05.020. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary data
Supplementary data
Supplementary data