Abstract
In T cells, the Tec kinases IL-2–inducible T cell kinase (ITK) and resting lymphocyte kinase (RLK) are activated by TCR stimulation and are required for optimal downstream signaling. Studies of CD4+ T cells from Itk−/− and Itk−/−Rlk−/− mice have indicated differential roles of ITK and RLK in Th1, Th2, and Th17 differentiation and cytokine production. However, these findings are confounded by the complex T cell developmental defects in these mice. In this study, we examine the consequences of ITK and RLK inhibition using a highly selective and potent small molecule covalent inhibitor PRN694. In vitro Th polarization experiments indicate that PRN694 is a potent inhibitor of Th1 and Th17 differentiation and cytokine production. Using a T cell adoptive transfer model of colitis, we find that in vivo administration of PRN694 markedly reduces disease progression, T cell infiltration into the intestinal lamina propria, and IFN-γ production by colitogenic CD4+ T cells. Consistent with these findings, Th1 and Th17 cells differentiated in the presence of PRN694 show reduced P-selectin binding and impaired migration to CXCL11 and CCL20, respectively. Taken together, these data indicate that ITK plus RLK inhibition may have therapeutic potential in Th1-mediated inflammatory diseases.
Introduction
The Tec family tyrosine kinases play a key role in Ag receptor–mediated signaling pathways in lymphocytes. Among these kinase family members, T cells express IL-2–inducible kinase (ITK), resting lymphocyte kinase (RLK), and tyrosine kinase expressed in hepatocellular carcinoma (1). Although each of these kinases is expressed in mature naive T cells, ITK is the most predominant. Based on mRNA analysis, RLK is expressed at 3- to 10-fold lower levels than ITK, and Tec is 30- to 100-fold reduced compared with ITK (2, 3). Following TCR stimulation, ITK is activated and directly phosphorylates phospholipase C (PLC)γ1. Activated PLCγ1 hydrolyzes phosphatidylinositol 4,5-biphosphate to produce inositol triphosphate and diacylglycerol, secondary messengers that lead to Ca2+ influx and MAPK and protein kinase C activation (4). As a consequence, Itk−/− T cells have significant defects in T cell activation and differentiation (5–8). For RLK, some evidence supports a role in TCR signaling, as Itk−/−Rlk−/− double-deficient T cells are more impaired than those lacking only ITK (5, 9). Nonetheless, based on present data, the precise functions of RLK and tyrosine kinase expressed in hepatocellular carcinoma in T cell activation are unclear.
To elucidate the role of Tec kinases in TCR signaling, several studies have addressed the impact of a deficiency in ITK, or ITK plus RLK, in CD4+ Th cell differentiation and function. Initial studies showed that Itk−/− mice exhibited impaired Th2 differentiation and Th2-biased responses to parasitic infection, with little effect on protective Th1 responses to intracellular protozoans (2, 10). These data were further supported by controlled in vitro studies that demonstrated that naive Itk−/− CD4+ T cells were defective in Th2 but not Th1 differentiation, in part due to the fact that differentiated Th2 cells fail to express any RLK protein, as do Th1 cells (2). Additionally, ITK and RLK functions in Th cells are at least partially redundant, as RLK overexpression in Itk−/− mice was able to restore Th2 responses in animal models of allergic asthma and schistosome egg–induced lung granuloma formation (11). Nonetheless, it has been difficult to distinguish which phenotypes observed in these mice are due to the functions of ITK and/or RLK in mature naive CD4+ T cells, and which are the consequence of altered T cell development generating an abnormal cytokine environment in the Itk−/− or Itk−/−Rlk−/− mice.
More recently, studies by Schwartzberg and colleagues (12, 13) have indicated an additional role for ITK in Th17 differentiation. Specifically, Itk−/− T cells showed reduced IL-17A production and increased Foxp3 expression following in vitro polarization. Additionally, Itk−/− T cells provided enhanced regulatory T cell (Treg)–mediated protection in an adoptive transfer model of colitis owing to their increased potential to upregulate Foxp3 (13), although another study found that Itk−/− Tregs were unable to protect against T cell–mediated colitis (14). Despite some disparities between studies, in general, these findings have provided impetus for the development of small-molecule ITK kinase inhibitors, with the intent of using them as treatments for atopic diseases, as well as for their potential as an immunosuppressant to block graft rejection or autoimmunity.
The complex phenotype of Itk−/− mice, including defects in T cell development, activation, differentiation, and effector function, has made it difficult to precisely assess the function of ITK in each lineage of T cells and at different stages of an immune response. It has also been challenging to distinguish functions of ITK in T cell activation and differentiation from effects due to altered T cell development in Itk−/− mice. A more direct strategy to address ITK and/or RLK function in T cells is to use a selective small-molecule inhibitor of these Tec kinases. PRN694 is a small molecule that forms an irreversible covalent bond with C442 in ITK or C350 in RLK, and it has recently been shown to selectively inhibit ITK and RLK in T cells (15). To date, the inhibitory effects of PRN694 on CD4+ Th cell differentiation and effector function have not been tested.
In this study, we examined the effects of PRN694 on CD4+ T cell differentiation and function in vitro and in vivo. Surprisingly, we found that PRN694 showed potent inhibitory effects on Th1 differentiation and IFN-γ production as well as on Th17 differentiation and IL-17A production, with decreased potency on Th2 differentiation. To test the relevance of this inhibitory activity in vivo, we used the T cell adoptive transfer model of colitis, an inflammatory condition mediated by IFN-γ–producing Th1 cells (16, 17). Consistent with our in vitro data, PRN694 administration ameliorated colitis disease progression and markedly inhibited colonic inflammation. Taken together, these data indicate that simultaneous inhibition of ITK and RLK by a small-molecule inhibitor may be an effective treatment for Th1-biased inflammatory diseases.
Materials and Methods
Mice
C57BL/6 wild-type (WT), C57BL/6 Rag2−/−, C57BL/6 OT-II Rag1−/− TCR transgenic (Tg), and B10.A 5C.C7 Rag2−/− TCR Tg mice were purchased from Taconic Biosciences and housed in specific pathogen-free conditions at the University of Massachusetts Medical School in accordance with Institutional Animal Care and Use Committee guidelines. CD-1 mice were purchased from Charles River Laboratories; all housing and procedures with these mice were in accordance with the guidelines approved by Principia Biopharma’s Institutional Animal Care and Use Committee.
Th cell polarization and CFSE dilution assay
Naive CD4+ T cells (3.0 × 105) from B10.A 5C.C7 Rag2−/− TCR Tg or C57BL/6 OT-II Rag1−/− TCR Tg mice were isolated using CD4 (L3T4) microbeads (Miltenyi Biotec). Isolated cells were plated in 12- or 24-well plates and activated by plate-bound anti-CD3 (1.0 μg/ml) and anti-CD28 (4.0 μg/ml) (BD Biosciences) for 72 h in the presence of the following conditions: Th0 (anti-CD3/CD28 with no cytokines), Th1 (IL-12, 10 ng/ml plus anti–IL-4, 10 μg/ml), Th2 (IL-4, 10 ng/ml plus anti–IFN-γ, 10 μg/ml), Th17 (IL-6, 20 ng/ml, TGF-β, 5.0 ng/ml, IL-1β, 20 ng/ml plus anti–IFN-γ, anti–IL-4, and anti–IL-2, each at 10 μg/ml) (all anti-cytokine Abs were purchased from R&D Systems and BD Biosciences). For CFSE dilution assay, CD4+ T cells isolated by magnetic separation were resuspended in 1.0 ml 1× PBS with 0.1% BSA at 5.0 × 106 cells/ml. Then, cells were stained with CFSE at a final concentration of 2.0 μM at 37°C for 10 min. After incubation, stained cells were quenched by adding 5 vol ice-cold RPMI 1640 complete medium (10% FBS) for 5 min on ice. Cells were washed three times with complete medium and then plated for Th polarization for 72 h.
Cytokine and transcription factor analysis
T cells were stimulated with PMA (50 ng/ml) and ionomycin (1.0 μg/ml) for 5 h with protein transport inhibitors, GolgiStop and GolgiPlug (BD Biosciences), each at 1.0 μg/ml. All cells were first stained with Abs to CD4 and CD44 (BD Biosciences) and a Live/Dead fixable aqua dead cell stain kit (Life Technologies). Stained cells were then fixed and permeabilized by using a BD Cytofix/Cytoperm kit (BD Biosciences). For cytokine staining, cells were stained with Abs to IFN-γ, IL-4, IL-17A, or IL-2 (BD Biosciences). For transcription factor staining, cells were fixed and permeabilized using a Foxp3/transcription factor staining buffer set (eBioscience) and stained with Abs to T-bet, GATA-3, retinoic acid–related orphan receptor γt, or Foxp3 (BD Biosciences). Cells were analyzed on an LSR II flow cytometer (BD Biosciences), and data were analyzed with FlowJo (Tree Star).
Measurements of cytokine production by human PBMCs and in-mouse plasma
Human PBMCs isolated from whole blood were incubated for 1 h with and without PRN694 and stimulated with plate-bound anti-CD3 (2.5 μg/ml) and soluble anti-CD28 (1.0 μg/ml) (BD Biosciences) for 18 h. Supernatants were collected, frozen, and analyzed using the human InflammationMAP v.1.0 biomarker panel (Myriad RBM). For the IL-2 inhibition dose response, PBMCs were incubated for 1 h with a concentration range of PRN694 and stimulated either with anti-CD3/CD28 or 10 μM thapsigargin (Sigma-Aldrich) for 18 h. Supernatants were analyzed for IL-2 using the AlphaLISA IL-2 kit (PerkinElmer). For cytokine inhibition in vivo, triplicate CD-1 mice were administered a 20 mg/kg i.p. dose of PRN694 followed either 1 or 6 h later by administration of 10 μg/mouse anti-CD3 (R&D Systems). Plasma was collected 2 h after anti-CD3 injection and analyzed for cytokines using the RodentMAP v.3.0 biomarker panel (Myriad RBM).
Mouse adoptive transfer colitis model and PRN694 administration
CD4+CD45RBhi T cells were sorted from spleens of C57BL/6 WT mice, and 4.0 × 105 cells were injected i.p. into Rag2−/− hosts. Recipient mice were weighed daily and sacrificed 7 wk posttransfer, at which time colons were removed for length measurement, histologic analysis, and lymphocyte isolation, and lymphoid organs were harvested for flow cytometry analysis. Vehicle (5% ethanol/95% Captex 355 EP/NF, ABITEC) or PRN694 was orally administered according to the following regimen: once a day (40 mg/kg) for weeks 0–2 and 4–7, and twice a day (20 mg/kg) for weeks 2–4.
Histologic examination of colon
Colon specimens obtained from vehicle-treated or PRN694-treated hosts were fixed in 10% buffered formalin and stained with H&E. To assess the severity of colitis, histologic scores of the proximal, middle, and distal colon were examined.
In vivo PRN694 target engagement assay and assessments of toxicity
Naive C57BL/6 mice were dosed with vehicle or PRN694 (40 mg/kg). After 2 or 6 h of oral administration, the dosed mice were sacrificed and CD4+ T cells were isolated from the spleen by using an EasySep mouse CD4+ T cell isolation kit (StemCell Technologies). Then, isolated CD4+ T cells were stimulated with anti-CD3 (10 μg/ml) and anti-CD28 (5.0 μg/ml) for 5 min at 37°C. After stimulation, CD4+ T cells (4.0 × 106) were lysed by CelLytic M buffer (Sigma-Aldrich) containing proteinase and phosphatase inhibitor. The expression of phosphorylated PLCγ1 and total PLCγ1 was examined by Western blot using anti–p-PLCγ1 (Tyr783) mAb and anti–PLCγ-1 mAb (D9H10) (Cell Signaling Technology) at 1:1000 dilution. The secondary Ab was Alexa Fluor 647–conjugated anti-rabbit IgG (H+L) (Invitrogen) at 1:1000 dilution, and the signal was evaluated using a Typhoon biomolecular imager (GE Healthcare Life Sciences) with signal quantification using ImageQuant TL 7.0 software (GE Healthcare Life Sciences). Following long-term in vivo administration, we noted that the mice treated with PRN694 gained weight comparable to control untreated Rag2−/− controls and showed no behavioral indications of drug-mediated toxicity. Furthermore, to address whether PRN694 might be generally cytotoxic, we examined the inhibition of proliferation of the HCT116 colorectal carcinoma cell line in the presence or absence of PRN694 and found an IC50 of 10 μM in this assay. Because this is a concentration higher than the maximum concentration of PRN694 in plasma following a 40 mg/kg oral dose, we concluded that PRN694 had no apparent toxicity during the 7-wk dosing time frame.
Isolation of intraepithelial lymphocytes and lamina propria lymphocytes from the colon
Colons were opened longitudinally and then cut into 1- to 2-cm pieces. After incubation with EDTA and DTT in HBSS at 37°C for 20 min with vigorous vortexing, the cell suspension was passed through a 70-μm strainer, and the flow-through containing intraepithelial lymphocytes (IELs) was collected by centrifugation. For lamina propria (LP) lymphocytes, the remaining tissues were digested with collagenase D (Roche), Dispase II (Roche), and DNase I (Roche) at 37°C for 20 min and then vortexed vigorously. Final cell suspensions were isolated using 40/80% Percoll (GE Healthcare Life Sciences) density gradients, and the viability of extracted cells was tested using trypan blue.
P-selectin glycoprotein ligand-1 binding and cell migration assays
Naive CD4+ T cells were isolated with CD4 (L3T4) microbeads (Miltenyi Biotec) and then cultured in the presence or absence of PRN694 (25 or 50 nM) for 72 h in each Th polarization condition as described above. For P-selectin glycoprotein ligand 1 binding assay, cultured and polarized CD4+ T cells were first treated with 2.4G2 Fc Block (anti-CD16/CD32) for 10 min at 4°C and then washed with FACS buffer (1× PBS containing 5.0% FBS). Recombinant P-selectin–human IgG fusion protein (BD Biosciences) was added to the cells (1:200 dilution) at 4°C for 30 min. Cells were washed and then stained with allophycocyanin-conjugated anti-human IgG (Jackson ImmunoResearch Laboratories) and surface marker Abs. The cell migration assay was performed by using HTS Transwell-96 well permeable supports with 3.0-μm pores (Corning). Cultured CD4+ T cells were washed twice and resuspended at 2.5 × 106 cells/ml with serum-free RPMI 1640 containing 0.1% BSA. Lower chambers were loaded with 200 μl diluted chemokines (CXCL11, 100 ng/ml; CCL20, 100 ng/ml; CCL25, 300 ng/ml) (PeproTech and R&D Systems). Twenty-five microliters cell suspension (5.0 × 105 cells) was loaded in the upper chamber of a polycarbonate filter. Cell migration was performed at 37°C, 5% CO2 for 3 h, and nonmigrated cells on the upper chamber were rinsed off with 1× PBS containing 0.1% BSA. After centrifugation (1500 rpm for 10 min), the upper chamber was removed and the migrated cells were resuspended in 1× PBS with 0.1% BSA. The numbers of migrated cells were counted by the flow cytometer. To calculate the chemotaxis index, the numbers of cells migrated in response to each chemokine were divided by the numbers of spontaneously migrated cells.
Statistical analysis
Data are represented as mean ± SEM. Statistically significant differences were determined by a Student t test.
Results
Inhibition of ITK and RLK kinases potently inhibits CD4+ Th cell differentiation
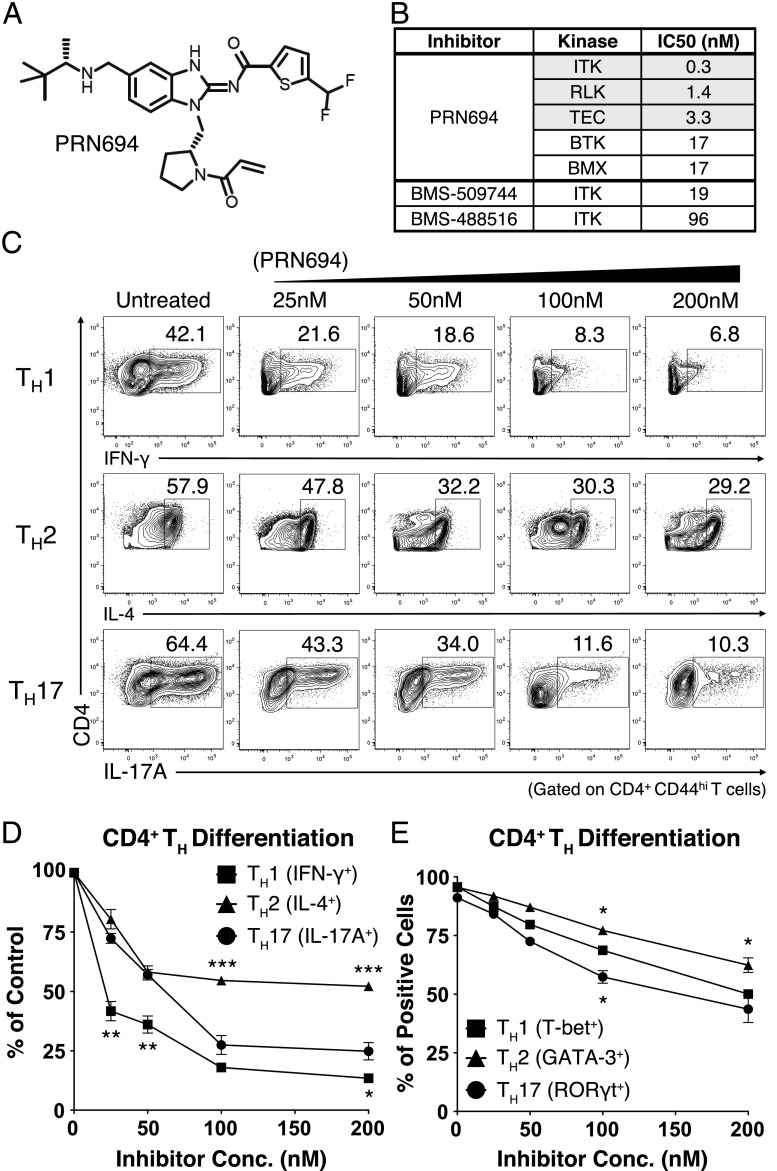
To determine the consequences of Tec kinase inhibition on Th differentiation, we used a recently reported small-molecule covalent inhibitor of ITK and RLK, PRN694 (Fig. 1A) (15). Based on in vitro kinase assays, PRN694 is a potent inhibitor of all three Tec kinases expressed in T cells, demonstrates less potency toward the other Tec kinases BTK and BMX (Fig. 1B), and shows excellent kinome-wide selectivity (15). As reported, PRN694 binds covalently to a conserved cysteine residue in the ATP binding sites of ITK and RLK (ITK C442, RLK C350) and is highly selective for these two kinases in T cells (15). Unlike its binding to ITK and RLK, PRN694 does not appear to bind covalently to BTK and hence displays a limited duration of BTK inhibition biochemically and in cells (15). Furthermore, PRN694 is 63- to 320-fold more potent in inhibiting ITK than previously described ITK inhibitors, for example, BMS-509744 and BMS-488516 (Fig. 1B) (18–22).
FIGURE 1.
PRN694 inhibits CD4+ Th cell differentiation. (A) Chemical structure of PRN694. (B) The selectivity and potency of PRN694 were examined by in vitro kinase assay. The table shows the IC50 values for the kinases indicated; for the complete dataset, see Zhong et al. (15). (C and D) Purified naive mouse splenic CD4+ T cells were stimulated in Th-polarizing conditions in the presence of PRN694 at the indicated concentrations for 72 h. CD4+ T cells were then restimulated with PMA and ionomycin for 5 h and analyzed by intracellular cytokine staining. The untreated controls were cultured in the presence of DMSO. The percentages of cytokine-producing CD4+ T cells (gated on CD4+CD44hi cells) are shown (C). Compilation of data from three independent experiments indicating the inhibitory effect of PRN694 on the cytokine production are shown. For each cytokine, the data were normalized to the percentage of cytokine-producing cells in the absence of inhibitor (D). (E) Data from three experiments were compiled, and for each Th transcription factor, the data were normalized to the percentage of positive cells in the absence of inhibitor. *p < 0.05, **p < 0.01, ***p < 0.001.
To assess the potential effect of PRN694 on CD4+ Th cell differentiation, we used naive CD4+ T cells from B10.A 5C.C7 Rag2−/− TCR Tg mice and stimulated these cells under Th1-, Th2-, or Th17-polarizing conditions. Cells were cultured for 3 d in the presence of increasing doses of PRN694 and then analyzed for cytokine production after restimulation with PMA and ionomycin for 5 h. Among these conditions, PRN694 inhibited Th1 differentiation more potently than was observed for Th2 and Th17 differentiation. Whereas a low concentration of PRN694 (25 nM) inhibited IFN-γ production by Th1 cells by >50%, 50–100 nM PRN694 was required to achieve a similar level of inhibition of IL-17A production by Th17 cells (Fig. 1C, 1D). Furthermore, none of the doses tested achieved 50% inhibition of IL-4 production by Th2 cells. The effects of PRN694 on cytokine production did not correlate with effects on cell proliferation (Supplemental Fig. 1A, 1B), indicating that the inhibition of IFN-γ production by Th1 cells was not due to reduced proliferation of these cells in the presence of PRN694.
We also examined the expression of Th cell lineage-determining transcription factors. Overall, the expression levels of all transcription factors tested (T-bet, GATA-3, and retinoic acid–related orphan receptor γt) were decreased upon the inhibitor treatment; however, the modest differences observed between the three Th cell lineages did not correlate precisely with diminished cytokine production (Fig. 1E). Collectively, these data indicate that PRN694 specifically targets ITK and RLK kinase activity and also impairs CD4+ Th cell differentiation and cytokine production, with a range of potency as follows: Th1 > Th17 > Th2.
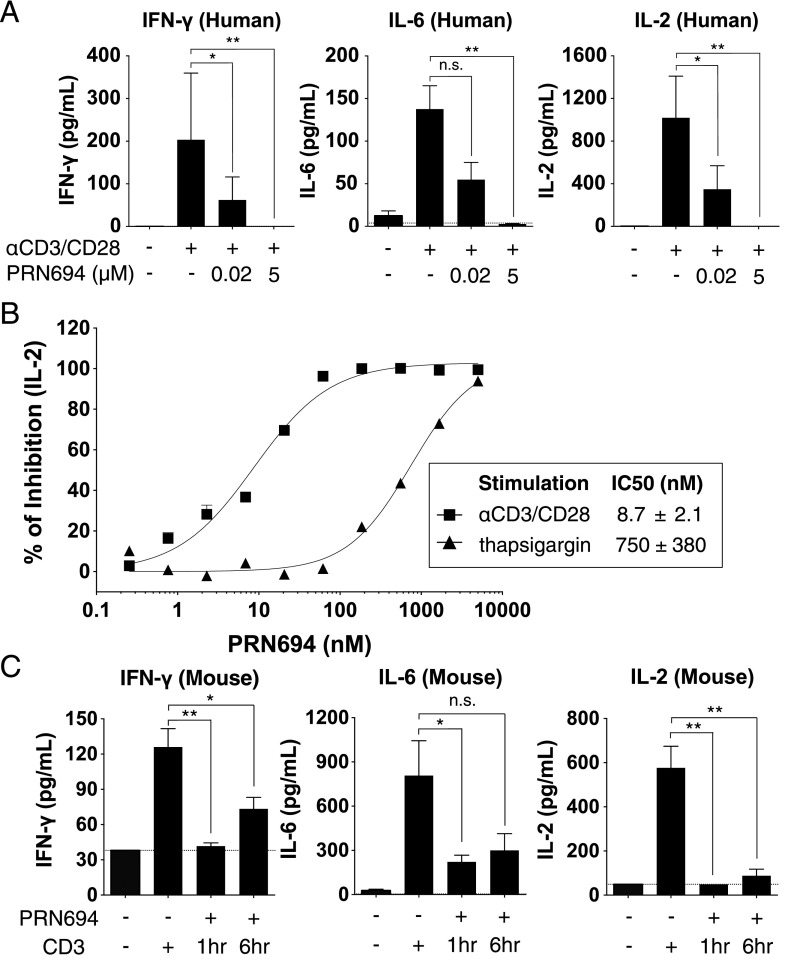
PRN694 inhibits TCR-induced cytokine production from human PBMCs and in mouse plasma
To further study the effect of dual ITK/RLK inhibition on T cell function, human PBMCs were stimulated with anti-CD3/CD28 in both the absence and presence of PRN694, and the cytokines produced after 18 h were measured using a biomarker panel. We used both a moderate (20 nM) and high (5.0 μM) concentration of PRN694 and focused on cytokines that were robustly induced in this short-term assay, including IFN-γ, IL-6, and IL-2. The increase in each of these cytokines induced by anti-CD3/CD28 stimulation was completely blocked by 5.0 μM PRN694 and was strongly inhibited by 20 nM PRN694 (Fig. 2A, Supplemental Table I). To quantify the potency of inhibition of IL-2 production, a 12-concentration dose range of PRN694 was tested. These data indicated an IC50 value of 8.7 ± 2.1 nM (Fig. 2B). To confirm that the inhibition of IL-2 production was due to selective ITK and RLK inhibition, we bypassed the requirement for these kinases by stimulating PBMCs with the calcium pump inhibitor thapsigargin, and we observed that the ability of PRN694 to inhibit IL-2 production was strongly impaired (Fig. 2B). To determine whether PRN694 inhibited these cytokine responses in vivo, mice were injected with a 20 mg/kg i.p. dose of PRN694, followed by injection of anti-CD3 either 1 or 6 h later to stimulate cytokine production. Plasma was collected 2 h after anti-CD3 injection, and cytokines were analyzed using a cytokine biomarker panel (Fig. 2C). The robust increase in IFN-γ, IL-6, and IL-2 induced by anti-CD3 stimulation was inhibited by PRN694 at both time points (Fig. 2C, Supplemental Table II).
FIGURE 2.
PRN694 inhibits TCR-induced cytokine production from human PBMCs and in mouse plasma. (A) Human PBMCs were stimulated in vitro with anti-CD3/CD28 for 18 h in the absence or presence of PRN694 at 20 nM or 5.0 μM, and levels of IFN-γ, IL-6, and IL-2 in the supernatants were measured as part of the human InflammationMAP v.1.0 biomarker panel. A complete list of all biomarkers of the panel is found in Supplemental Table I. Data are shown as means ± SEM (n = 3/group). (B) A dose response of PRN694 was used to assess the potency of inhibition of IL-2 production by PBMCs. Cells were stimulated with anti-CD3/CD28 (▪) or thapsigargin (▴). IL-2 was quantified using an AlphaLISA IL-2 immunoassay. (C) Mice were injected i.p. with PRN694 (20 mg/kg) followed by anti-CD3 (10 μg/mouse) 1 or 6 h later. Two hours after anti-CD3 injection, plasma was collected and cytokines were measured as part of the RodentMAP v.3.0 biomarker panel. A complete list of all biomarkers of the panel is found in Supplemental Table II. Dotted lines indicate the detection limit for each cytokine in the assay. Data are shown as means ± SEM (n = 3/group). *p < 0.05, **p < 0.01.
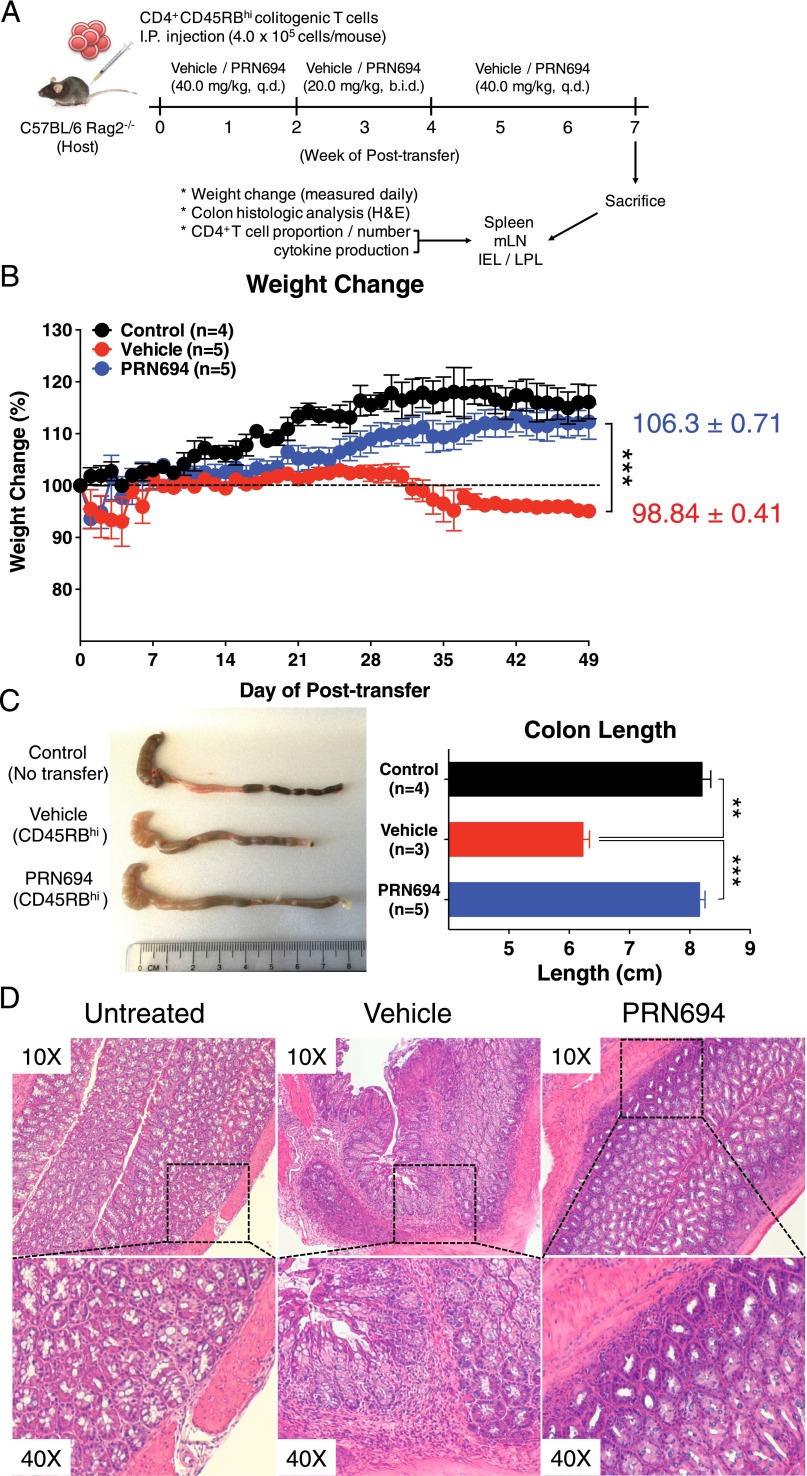
PRN694 treatment in vivo ameliorates symptoms in the CD4+CD45RBhi T cell transfer model of colitis
Owing to the potent inhibitory effect of PRN694 on IFN-γ production by Th1 cells in in vitro culture experiments, we considered whether PRN694 might function in vivo to suppress a Th1/IFN-γ–mediated disease. To test this hypothesis, we adoptively transferred WT colitogenic CD4+CD45RBhi T cells from C57BL/6 mice into RAG2-deficient hosts and monitored the mice for weight loss as a surrogate for disease progression. Previous studies have demonstrated that the colitis induced in this model is due to Th1-mediated inflammation, with little involvement of Th2 or Th17 effector responses (16, 23, 24). In addition to the transferred cells, mice received vehicle alone or PRN694 by oral gavage. Based on studies of ITK target occupancy in thymocytes following in vivo administration of PRN694 in mice (15), we dosed mice daily and, for a 2-wk period (weeks 2–4), twice daily (Fig. 3A). As expected, recipient mice receiving vehicle alone failed to gain weight and progressively lost weight, beginning 4 wk after T cell transfer. In contrast, PRN694-treated mice exhibited no weight loss and remained similar to control RAG2-deficient mice that did not receive a colitogenic T cell transfer (Fig. 3B). Consistent with these data, analysis of colon length at 7 wk posttransfer indicated that the reduced colon length seen in the vehicle-treated mice was prevented in the PRN694-treated mice (Fig. 3C). Additionally, histological analysis of the colonic epithelium revealed lymphocytic infiltration in the colon of the vehicle-treated mice (Fig. 3D), in contrast to the reduced inflammation seen in the colon of PRN694-treated mice. To further confirm target engagement of PRN694 with ITK in vivo, splenic CD4+ T cells were isolated from vehicle-dosed or PRN694-dosed mice (2 or 6 h posttreatment) and then stimulated with anti-CD3/CD28 mAb to examine PLCγ1 phosphorylation. As shown, the phosphorylation of PLCγ1 was completely blocked at both time points following PRN694 administration (Supplemental Fig. 1C). Taken together, these data demonstrate that PRN694 reduces T cell–mediated colonic inflammation by inhibiting ITK/RLK.
FIGURE 3.
PRN694 administration ameliorates colitis disease progression. (A–D) Colitogenic CD4+CD45RBhi splenic T cells (4.0 × 105 cells/mouse) from C57BL/6 WT mice were injected i.p. into C57BL/6 Rag2−/− hosts. Recipients were treated with vehicle (red, n = 5) or PRN694 (blue, n = 5) with the indicated regimen (A), and disease progression of dosed recipients and untreated (no CD4+CD45RBhi T cell transfer) Rag2−/− controls (black, n = 4) was monitored by weight change (B). Data are shown as means ± SEM. At 7 wk posttransfer, untreated Rag2−/− control, vehicle-dosed, and PRN694-dosed mice were sacrificed for the measurement of colon length (C) and histologic analysis using H&E staining (D). Data were compiled from two independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001.
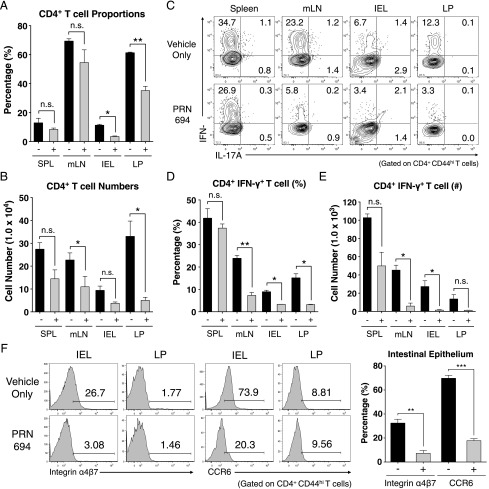
PRN694 impairs IFN-γ–mediated Th1 responses and prevents T cell migration to the inflamed colon
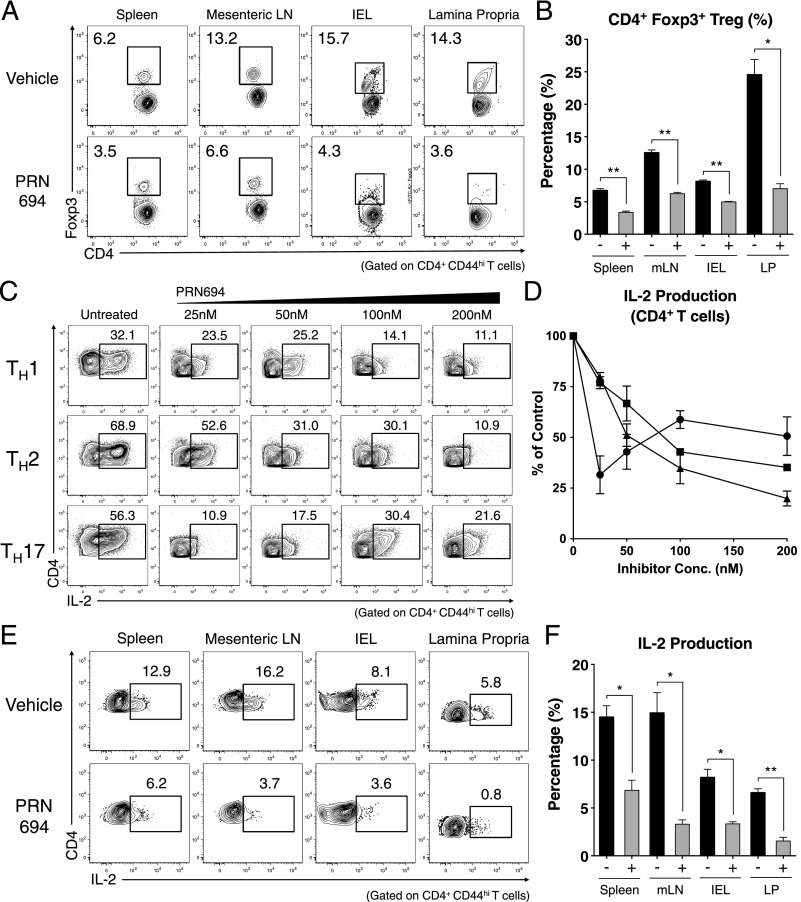
To determine the basis of PRN694-mediated inhibition of colitis, we analyzed the T cell populations in the spleen, mesenteric lymph node (mLN), intestinal epithelium (intraepithelial lymphocytes [IELs]), and LP of recipient Rag2−/− mice at 7 wk posttransfer. Assessment of CD4+ T cell proportions did not reveal any significant differences in the spleens or mLNs between the vehicle- or PRN694-treated mice, although a modest but significant difference in absolute numbers of CD4+ T cells was observed in mLNs (Fig. 4A, 4B). However, CD4+ T cell proportions in intestinal epithelium and LP of PRN694-treated mice were significantly less than those seen in the vehicle-treated mice, along with a statistically significant difference in absolute numbers in the LP (Fig. 4A, 4B). To test whether PRN694 exerted any inhibitory effect on T cell function, we examined cytokine production from transferred colitogenic T cells in several different organs after a brief in vitro stimulation. As expected, CD4+ T cells showed robust IFN-γ production in all analyzed sites of vehicle-treated mice, whereas no IL-17A production was observed (Fig. 4C). Interestingly, PRN694 administration markedly reduced the proportions of CD4+ T cells producing IFN-γ in mLNs, IELs, and LP, although this effect was less prominent in the spleen (Fig. 4C, 4D). There was also a large decrease in the numbers of IFN-γ–producing CD4+ T cells in all organs following PRN694 treatment, although the suppression did not reach significance in the LP (Fig. 4E). Consistent with our in vitro studies, the reduced production of IFN-γ by T cells in the PRN694-treated mice could not be accounted for simply by defects in expression of the Th1 transcription factor, T-bet (Supplemental Fig. 2A, 2B).
FIGURE 4.
PRN694 treatment reduces colitogenic T cell proportions and IFN-γ production in the intestinal epithelium. (A and B) The proportions and absolute numbers of CD4+ T cells in spleen, mLNs, intestinal epithelium, and colon LP are shown. (C–E) Isolated lymphocytes from various sites were stimulated with PMA and ionomycin and analyzed for IL-17A and IFN-γ production. Dot plots show gated CD4+CD44hi T cells (C). The proportions (D) and numbers (E) of IFN-γ–producing CD4+ T cells are shown. Data are compiled from two independent experiments. (F) Histograms show the expression of gut-homing receptors, integrin α4β7 (LPAM-1) and CCR6, on colonic IELs and LP CD4+CD44hi T cells. Numbers indicate the percentages of cells in each region. Graph shows a compilation of data from three to five mice in each group. *p < 0.05, **p < 0.01, ***p < 0.001.
Owing to the reduced proportion of CD4+ T cells in the inflamed intestines of PRN694-treated mice, we also investigated expression of gut-homing receptors, integrin α4β7 (LPAM-1) and CCR6, on colitogenic T cells (25–27). Consistent with the proportions of gut-infiltrating CD4+ T cells, expression of both integrin α4β7 and CCR6 were reduced on intestinal epithelial CD4+ T cells when PRN694 was administered (Fig. 4F). In contrast, we did not detect any significant level of expression of these receptors on LP-isolated CD4+ T cells from either group of mice (Fig. 4F). Taken together, these data strongly suggest that PRN694 attenuates in vivo Th1-biased intestinal inflammation through the inhibition of IFN-γ production and the expression of intestine-homing surface receptors.
PRN694 impairs induced Treg differentiation and IL-2 production by transferred colitigenic CD4+ T cells
A recent study has shown that Itk−/− CD4+ T cells have an increased propensity to upregulate Foxp3 and differentiate into induced Tregs (iTregs) when stimulated under Th17-polarizing conditions (13). We therefore considered whether the inhibitory effect of PRN694 on colitis might be due to enhanced differentiation of Foxp3+ iTregs in the inhibitor-treated mice. Instead, analysis of T cells at 7 wk posttransfer indicated that PRN694 significantly reduced the proportions of CD4+ T cells expressing Foxp3 compared with controls, a result consistent with the findings of Huang et al. (14). This was evident in all organs examined (Fig. 5A, 5B). Examination of surface markers commonly found on most Tregs, including CTLA-4, PD-1, and GITR, also showed reduced expression on Foxp3+CD4+ T cells in PRN694-treated mice (Supplemental Fig. 2C–E).
FIGURE 5.
PRN694 treatment decreases Treg frequencies and IL-2 production from CD4+ T cells in vitro and in vivo. (A and B) CD4+ T cells from spleen, mLNs, IELs, and LP from vehicle-treated or PRN694-treated mice at 7 wk posttransfer were examined for Foxp3 expression. (A) Dot plots show CD4 versus Foxp3 staining, with numbers indicating the percentages of CD4+Foxp3+ Tregs, and (B) the graph shows a compilation of data from three to five mice in each group. (C and D) Naive CD4+ T cells were stimulated in Th-polarizing conditions with PRN694 at the indicated concentrations for 72 h. Cells were then restimulated with PMA and ionomycin for 5 h and then analyzed for IL-2 production. (C) The percentages of IL-2–producing CD4+ T cells (gated on CD4+CD44hi) are shown. (D) Compilation of data from three independent experiments is shown, with the data for each Th subset normalized to the percentage of positive cells in the absence of inhibitor. (E and F) Isolated lymphocytes from vehicle-treated or PRN694-treated mice at 7 wk posttransfer were stimulated with PMA and ionomycin and analyzed for IL-2 production. Plots show gated CD4+CD44hi T cells, and the graph shows a compilation of data from three to five mice in each group. *p < 0.05, **p < 0.01.
Because upregulation of Foxp3 and iTreg differentiation are dependent on IL-2 (28–31), we examined the effects of PRN694 on IL-2 production by in vitro polarized CD4+ T cells. As shown, PRN694 inhibited IL-2 production by all three lineages of Th cells (Fig. 5C, 5D). Consistent with these data, T cells from the adoptive transfer colitis studies also showed reduced proportions of cells capable of producing IL-2 following treatment with PRN694 (Fig. 5E, 5F). These data indicate that reduced disease progression in mice treated with PRN694 is not a consequence of enhanced differentiation of Foxp3+ iTreg cells, but rather a result of inhibition of differentiation and activation of the IFN-γ–producing Th1 effector cell population.
PRN694 inhibits P-selectin binding activity and chemokine-induced migration of polarized Th1 and Th17 cells
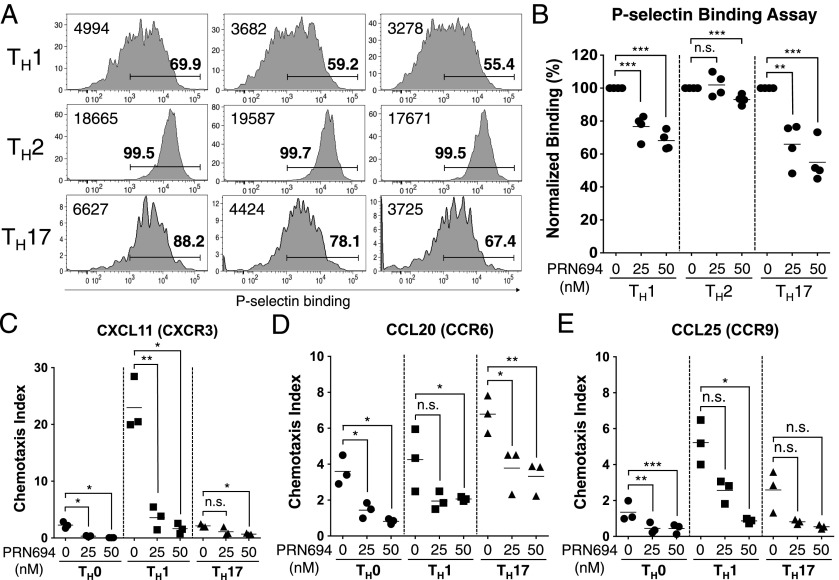
Our results in the adoptive transfer colitis studies showed reduced numbers of CD4+ T cells present in the colonic epithelium of PRN694-treated mice compared with controls. As previous studies using naive T cells from Itk−/− and Itk−/−Rlk−/− mice had shown a role for these kinases in CCL19- and CXCL12-induced migration (7, 32), we considered whether CD4+ effector T cells might also require ITK and RLK for efficient migration into tissues. Using CD4+ T cells polarized in vitro to Th1, Th2, or Th17 lineages in the presence or absence of PRN694, we first examined binding to P-selectin, a ligand expressed on activated endothelium (33). Both Th1 and Th17 cells showed reduced P-selectin binding following differentiation in the presence of both doses of PRN694 tested; in contrast, Th2 cells showed only a modest reduction, and this reduction was only visible in the higher dose of inhibitor (Fig. 6A, 6B).
FIGURE 6.
PRN694 leads to impaired P-selectin binding and inhibits chemokine-induced CD4+ T cell migration. (A–E) Isolated naive CD4+ T cells were cultured in the presence or absence of PRN694 (25 or 50 nM) for 72 h in each Th polarization condition. (A) Histograms show polarized CD4+ T cells stained with recombinant P-selectin–human IgG fusion protein as an assay for P-selectin glycoprotein ligand 1 binding. Bold numbers above region indicate the percentages of cells staining positively for P-selectin–Ig based on comparison with an isotype control stain; numbers at the left of each histogram indicate the mean fluorescence intensity of P-selectin–Ig staining in each panel. (B) Graph shows a compilation of data from two independent experiments. The data were normalized to the percentage of P-selectin–binding cells among each Th-polarizing condition in the absence of inhibitor. (C–E) Cultured CD4+ T cells (5.0 × 105 cells) were plated in the upper chamber of HTS Transwell-96 well permeable supports (3.0-μm pore); bottom chambers were loaded with diluted CXCL11 (100 ng/ml) (C), CCL20 (100 ng/ml) (D), or CCL25 (300 ng/ml) (E). After 3 h at 37°C, the numbers of migrated cells were counted by the flow cytometer. The chemotaxis index was calculated by dividing the number of cells that migrated in response to each chemokine by the number of spontaneously migrated cells. *p < 0.05, **p < 0.01, ***p < 0.001.
As a second approach, we tested migration of CD4+ effector T cells to chemokines implicated in trafficking to the gastrointestinal epithelium. We focused on responses to CXCL11, a ligand for CXCR3 commonly found on effector Th1 cells (34), CCL20, a ligand for CCR6 found on Th17 cells (35, 36), and CCL25, a ligand for CCR9 (37, 38), which is expressed on gut-resident T cells. Using a Transwell migration assay, we found that PRN694 inhibited Th1 migration to CXCL11, with little migration observed by other subsets to this chemokine. CCL20 induced a modest degree of migration of Th17 cells, and this migration was reduced ∼2-fold by PRN694. Th0 and Th1 cells showed fewer cells migrating in response to CCL20, and these responses were further reduced by PRN694. Overall, few cells of any lineage migrated in response to CCL25, with significant inhibition seen only in Th1 and Th0 cells. Polarized Th2 cells showed no migration to any of the chemokines tested (not shown). Overall, these data confirm that PRN694 impacts differentiating Th1 and Th17 cells to reduce their ability to bind to activated endothelium and to migrate to inflammatory chemokines.
Discussion
In this study, we show that inhibition of ITK and RLK by PRN694 leads to a significant impairment in CD4+ Th cell differentiation and to protection from Th1-mediated colitis. Although PRN694 treatment inhibited the differentiation of all CD4+ Th subsets in vitro, the most potent inhibitory effect we observed was on Th1 differentiation. These findings are consistent with a recent study using an allele-sensitive mutant of ITK coupled with a selective inhibitor, which also revealed an important role for ITK kinase activity in Th1 differentiation and cytokine production (39). Our data with PRN694 are also consistent with previous studies showing that Itk−/−Rlk−/− mice mounted a normal protective type II cytokine response against Schistosoma mansoni, but were highly susceptible to the intracellular protozoan Toxoplasma gondii (5, 6). Furthermore, these results strongly suggest that a dual inhibitor of ITK and RLK would likely have a different effect on Th2-skewed inflammation such as asthma or atopic diseases compared with an inhibitor that was selective for ITK alone.
Also consistent with previous studies using Itk−/− T cells (12), we observed that PRN694 was a potent inhibitor of Th17 differentiation and IL-17A production in vitro. Nonetheless, it is unlikely that inhibition of Th17 responses can account for the effects of PRN694 in ameliorating disease in our colitis experiments. Although some studies have shown a role for Th17 cells in colitis (40–42), these effects were seen either when colitogenic BALB/c T cells were transferred to syngeneic hosts (41) or when the cecal bacterial Ag-specific C3H/HeJBir CD4+ T cell lines were transferred to C3H.SCID hosts and then restimulated for 10 d in the presence of bacterial Ag-pulsed dendritic cells and with or without IL-23 prior to analysis (42). In contrast, a study using a protocol most similar to our own found that colitis was actually enhanced when Th17 responses were eliminated (40), and the transfer of naive CD4+ T cells from Il17a−/− mice still induced a severe colitis (41). Furthermore, several other reports suggest that Th17 responses arise early on during colitis disease progression and that in vitro polarized Th17 cells are eventually converted into Th1 cells after adoptive transfer to lymphopenic hosts (43, 44). Using this CD4+CD45RBhi adoptive transfer model of T cell–mediated colitis, we were unable to detect IL-17A production from colitogenic T cells in mice that were treated with vehicle alone, suggesting that Th17 responses were not contributing to disease progression in our studies. Thus, we were unable to assess the efficacy of PRN694 to inhibit IL-17A production in vivo in this disease model.
In the present study, we cannot rule out an effect of PRN694 on the Tec kinase family member Tec. Previous studies have indicated that Tec mRNA expression is negligible in primary T cells examined ex vivo, including memory phenotype cells (45) (http://www.immgen.org), and that only modest increases in Tec protein expression (<2-fold) are observed even after Th1 polarization in vitro (46). Furthermore, studies of T cell responses in Tec−/− mice have failed to reveal any significant defects due to the absence of Tec. Therefore, it seems unlikely that the inhibitory effects of PRN694 observed in our studies are due to its effect on Tec activity in T cells.
Interestingly, we observed reduced differentiation of Foxp3+ iTreg cells in PRN694-treated mice compared with controls. In contrast, a previous study showed increased differentiation of Itk−/− naive CD4+ cells into Foxp3+ iTreg cells in a similar adoptive transfer model of colitis (13). The difference in these two sets of findings could be due to a number of factors. First, the numbers of cells transferred and the time points analyzed were each different in the two studies. More importantly, there may be an unappreciated difference in the starting populations of CD4+CD45RBhiCD25− T cells isolated from WT mice, as in our studies, compared with those isolated from Itk−/− mice. Additionally, PRN694 is a potent inhibitor of both ITK and RLK, and this dual inhibition may lead to a different outcome than the single genetic deficiency in Itk. It is also important to consider that a small-molecule kinase inhibitor might have a different effect than a genetic deficiency that causes a complete absence of ITK protein, as the former situation would not abolish any kinase-independent functions of ITK. Finally, it is important to consider the possibility that mice housed in two different environments have significant differences in their microbiota, and that these differences may impact iTreg differentiation in vivo.
Our studies indicate that PRN694 treatment reduces the numbers of colitogenic CD4+ T cells in the intestinal epithelium at 7 wk posttransfer. It is possible that one component of this decrease is reduced proliferation of the T cells in the presence of PRN694, although our in vitro studies examining proliferation suggest that this explanation is unlikely. Instead, we propose that impaired T cell migration to the gastrointestinal tract is contributing to this deficit. We find that PRN694 treatment during Th cell polarization leads to reduced P-selectin binding on Th1 and Th17 cells, an alteration that could impact T cell rolling on the activated endothelium prior to extravasation into the gastrointestinal tract. This possibility is consistent with our previous studies showing impaired tissue homing and transendothelial migration of Itk−/− T cells in an autoimmune disease model (47). Furthermore, we observed reduced expression of integrin α4β7 and CCR6, as well as reduced in vitro chemotaxis to inflammatory chemokines, by PRN694-treated CD4+ T cells. Taken together, these data provide strong support for the conclusion that ITK/RLK inhibition leads to impaired T cell migration into the gastrointestinal tissue.
Overall, we show that PRN694 inhibits CD4+ effector T cell differentiation and cytokine production, particularly for Th1 and Th17 lineage cells. Together with the inhibitory effect of PRN694 on T cell migration and on colitis disease progression, the data presented in the present study provide strong support for the development of a combined ITK/RLK inhibitor as a potential therapeutic strategy for Th1-mediated, and possibly Th17-mediated, inflammatory diseases.
Supplementary Material
Acknowledgments
We thank all members of the Berg Laboratory, especially Regina Whitehead and Sharlene Hubbard, for technical assistance. We also thank the University of Massachusetts Medical School’s Department of Animal Medicine for the maintenance of mouse colonies and Flow Cytometry Core Facility for assistance in cell sorting.
This work was supported by National Institutes of Health Grants AI084987 and AI083505 (both to L.J.B.).
The online version of this article contains supplemental material.
- IEL
- IEL, intraepithelial lymphocyte
- ITK
- IL-2–inducible T cell kinase
- iTreg
- induced regulatory T cell
- LP
- lamina propria
- mLN
- mesenteric lymph node
- PLC
- phospholipase C
- RLK
- resting lymphocyte kinase
- Tg
- transgenic
- Treg
- regulatory T cell
- WT
- wild-type.
Disclosures
H.H.-D., T.D.O., J.O.F., R.J.H., and J.M.B. are members of a biotechnology company developing molecules for inflammatory and autoimmune diseases. The other authors have no financial conflicts of interest.
References
- 1.Berg L. J., Finkelstein L. D., Lucas J. A., Schwartzberg P. L. 2005. Tec family kinases in T lymphocyte development and function. Annu. Rev. Immunol. 23: 549–600. [DOI] [PubMed] [Google Scholar]
- 2.Miller A. T., Wilcox H. M., Lai Z., Berg L. J. 2004. Signaling through Itk promotes T helper 2 differentiation via negative regulation of T-bet. Immunity 21: 67–80. [DOI] [PubMed] [Google Scholar]
- 3.Colgan J., Asmal M., Neagu M., Yu B., Schneidkraut J., Lee Y., Sokolskaja E., Andreotti A., Luban J. 2004. Cyclophilin A regulates TCR signal strength in CD4+ T cells via a proline-directed conformational switch in Itk. Immunity 21: 189–201. [DOI] [PubMed] [Google Scholar]
- 4.Andreotti A. H., Schwartzberg P. L., Joseph R. E., Berg L. J. 2010. T-cell signaling regulated by the Tec family kinase, Itk. Cold Spring Harb. Perspect. Biol. 2: a002287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schaeffer E. M., Debnath J., Yap G., McVicar D., Liao X. C., Littman D. R., Sher A., Varmus H. E., Lenardo M. J., Schwartzberg P. L. 1999. Requirement for Tec kinases Rlk and Itk in T cell receptor signaling and immunity. Science 284: 638–641. [DOI] [PubMed] [Google Scholar]
- 6.Schaeffer E. M., Yap G. S., Lewis C. M., Czar M. J., McVicar D. W., Cheever A. W., Sher A., Schwartzberg P. L. 2001. Mutation of Tec family kinases alters T helper cell differentiation. Nat. Immunol. 2: 1183–1188. [DOI] [PubMed] [Google Scholar]
- 7.Takesono A., Horai R., Mandai M., Dombroski D., Schwartzberg P. L. 2004. Requirement for Tec kinases in chemokine-induced migration and activation of Cdc42 and Rac. Curr. Biol. 14: 917–922. [DOI] [PubMed] [Google Scholar]
- 8.Ellmeier W., Jung S., Sunshine M. J., Hatam F., Xu Y., Baltimore D., Mano H., Littman D. R. 2000. Severe B cell deficiency in mice lacking the tec kinase family members Tec and Btk. J. Exp. Med. 192: 1611–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schaeffer E. M., Broussard C., Debnath J., Anderson S., McVicar D. W., Schwartzberg P. L. 2000. Tec family kinases modulate thresholds for thymocyte development and selection. J. Exp. Med. 192: 987–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fowell D. J., Shinkai K., Liao X. C., Beebe A. M., Coffman R. L., Littman D. R., Locksley R. M. 1999. Impaired NFATc translocation and failure of Th2 development in Itk-deficient CD4+ T cells. Immunity 11: 399–409. [DOI] [PubMed] [Google Scholar]
- 11.Sahu N., Venegas A. M., Jankovic D., Mitzner W., Gomez-Rodriguez J., Cannons J. L., Sommers C., Love P., Sher A., Schwartzberg P. L., August A. 2008. Selective expression rather than specific function of Txk and Itk regulate Th1 and Th2 responses. J. Immunol. 181: 6125–6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gomez-Rodriguez J., Sahu N., Handon R., Davidson T. S., Anderson S. M., Kirby M. R., August A., Schwartzberg P. L. 2009. Differential expression of interleukin-17A and -17F is coupled to T cell receptor signaling via inducible T cell kinase. Immunity 31: 587–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gomez-Rodriguez J., Wohlfert E. A., Handon R., Meylan F., Wu J. Z., Anderson S. M., Kirby M. R., Belkaid Y., Schwartzberg P. L. 2014. Itk-mediated integration of T cell receptor and cytokine signaling regulates the balance between Th17 and regulatory T cells. J. Exp. Med. 211: 529–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang W., Jeong A.-R., Kannan A. K., Huang L., August A. 2014. IL-2-inducible T cell kinase tunes T regulatory cell development and is required for suppressive function. J. Immunol. 193: 2267–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhong Y., Dong S., Strattan E., Ren L., Butchar J. P., Thornton K., Mishra A., Porcu P., Bradshaw J. M., Bisconte A., et al. 2015. Targeting interleukin-2-inducible T-cell kinase (ITK) and resting lymphocyte kinase (RLK) using a novel covalent inhibitor PRN694. J. Biol. Chem. 290: 5960–5978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Powrie F., Leach M. W., Mauze S., Menon S., Caddle L. B., Coffman R. L. 1994. Inhibition of Th1 responses prevents inflammatory bowel disease in scid mice reconstituted with CD45RBhi CD4+ T cells. Immunity 1: 553–562. [DOI] [PubMed] [Google Scholar]
- 17. Read, S., and F. Powrie. 2001. Induction of inflammatory bowel disease in immunodeficient mice by depletion of regulatory T cells. Curr. Protoc. Immunol. Chapter 15: Unit 15.13. doi:10.1002/0471142735.im1513s30. [DOI] [PubMed]
- 18.Lin T.-A., McIntyre K. W., Das J., Liu C., O’Day K. D., Penhallow B., Hung C.-Y., Whitney G. S., Shuster D. J., Yang X., et al. 2004. Selective Itk inhibitors block T-cell activation and murine lung inflammation. Biochemistry 43: 11056–11062. [DOI] [PubMed] [Google Scholar]
- 19.Das J., Furch J. A., Liu C., Moquin R. V., Lin J., Spergel S. H., McIntyre K. W., Shuster D. J., O’Day K. D., Penhallow B., et al. 2006. Discovery and SAR of 2-amino-5-(thioaryl)thiazoles as potent and selective Itk inhibitors. Bioorg. Med. Chem. Lett. 16: 3706–3712. [DOI] [PubMed] [Google Scholar]
- 20.Snow R. J., Abeywardane A., Campbell S., Lord J., Kashem M. A., Khine H. H., King J., Kowalski J. A., Pullen S. S., Roma T., et al. 2007. Hit-to-lead studies on benzimidazole inhibitors of ITK: discovery of a novel class of kinase inhibitors. Bioorg. Med. Chem. Lett. 17: 3660–3665. [DOI] [PubMed] [Google Scholar]
- 21.Riether D., Zindell R., Kowalski J. A., Cook B. N., Bentzien J., Lombaert S. D., Thomson D., Kugler S. Z., Jr., Skow D., Martin L. S., et al. 2009. 5-Aminomethylbenzimidazoles as potent ITK antagonists. Bioorg. Med. Chem. Lett. 19: 1588–1591. [DOI] [PubMed] [Google Scholar]
- 22.Sahu N., August A. 2009. ITK inhibitors in inflammation and immune-mediated disorders. Curr. Top. Med. Chem. 9: 690–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neurath M. F., Weigmann B., Finotto S., Glickman J., Nieuwenhuis E., Iijima H., Mizoguchi A., Mizoguchi E., Mudter J., Galle P. R., et al. 2002. The transcription factor T-bet regulates mucosal T cell activation in experimental colitis and Crohn’s disease. J. Exp. Med. 195: 1129–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Strober W., Fuss I. J. 2011. Proinflammatory cytokines in the pathogenesis of inflammatory bowel diseases. Gastroenterology 140: 1756–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petrovic A., Alpdogan O., Willis L. M., Eng J. M., Greenberg A. S., Kappel B. J., Liu C., Murphy G. J., Heller G., van den Brink M. R. M. 2004. LPAM (α4β7 integrin) is an important homing integrin on alloreactive T cells in the development of intestinal graft-versus-host disease. Blood 103: 1542–1547. [DOI] [PubMed] [Google Scholar]
- 26.Varona R., Cadenas V., Gómez L., Martínez-A C., Márquez G. 2005. CCR6 regulates CD4+ T-cell-mediated acute graft-versus-host disease responses. Blood 106: 18–26. [DOI] [PubMed] [Google Scholar]
- 27.Monteiro P., Gosselin A., Wacleche V. S., El-Far M., Said E. A., Kared H., Grandvaux N., Boulassel M.-R., Routy J.-P., Ancuta P. 2011. Memory CCR6+CD4+ T cells are preferential targets for productive HIV type 1 infection regardless of their expression of integrin β7. J. Immunol. 186: 4618–4630. [DOI] [PubMed] [Google Scholar]
- 28.Boyman O., Sprent J. 2012. The role of interleukin-2 during homeostasis and activation of the immune system. Nat. Rev. Immunol. 12: 180–190. [DOI] [PubMed] [Google Scholar]
- 29.Malek T. R., Castro I. 2010. Interleukin-2 receptor signaling: at the interface between tolerance and immunity. Immunity 33: 153–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu A., Zhu L., Altman N. H., Malek T. R. 2009. A low interleukin-2 receptor signaling threshold supports the development and homeostasis of T regulatory cells. Immunity 30: 204–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fontenot J. D., Rasmussen J. P., Gavin M. A., Rudensky A. Y. 2005. A function for interleukin 2 in Foxp3-expressing regulatory T cells. Nat. Immunol. 6: 1142–1151. [DOI] [PubMed] [Google Scholar]
- 32.Fischer A. M., Mercer J. C., Iyer A., Ragin M. J., August A. 2004. Regulation of CXC chemokine receptor 4-mediated migration by the Tec family tyrosine kinase ITK. J. Biol. Chem. 279: 29816–29820. [DOI] [PubMed] [Google Scholar]
- 33.Ley K., Kansas G. S. 2004. Selectins in T-cell recruitment to non-lymphoid tissues and sites of inflammation. Nat. Rev. Immunol. 4: 325–335. [DOI] [PubMed] [Google Scholar]
- 34.Qin S., Rottman J. B., Myers P., Kassam N., Weinblatt M., Loetscher M., Koch A. E., Moser B., Mackay C. R. 1998. The chemokine receptors CXCR3 and CCR5 mark subsets of T cells associated with certain inflammatory reactions. J. Clin. Invest. 101: 746–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Esplugues E., Huber S., Gagliani N., Hauser A. E., Town T., Wan Y. Y., O’Connor W., Jr., Rongvaux A., Van Rooijen N., Haberman A. M., et al. 2011. Control of TH17 cells occurs in the small intestine. Nature 475: 514–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang C., Kang S. G., Lee J., Sun Z., Kim C. H. 2009. The roles of CCR6 in migration of Th17 cells and regulation of effector T-cell balance in the gut. Mucosal Immunol. 2: 173–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stenstad H., Ericsson A., Johansson-Lindbom B., Svensson M., Marsal J., Mack M., Picarella D., Soler D., Márquez G., Briskin M., Agace W. W. 2006. Gut-associated lymphoid tissue-primed CD4+ T cells display CCR9-dependent and -independent homing to the small intestine. Blood 107: 3447–3454. [DOI] [PubMed] [Google Scholar]
- 38.Cassani B., Villablanca E. J., Quintana F. J., Love P. E., Lacy-Hulbert A., Blaner W. S., Sparwasser T., Snapper S. B., Weiner H. L., Mora J. R. 2011. Gut-tropic T cells that express integrin α4β7 and CCR9 are required for induction of oral immune tolerance in mice. Gastroenterology 141: 2109–2118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kannan A., Lee Y., Qi Q., Huang W., Jeong A.-R., Ohnigian S., August A. 2015. Allele-sensitive mutant, Itkas, reveals that Itk kinase activity is required for Th1, Th2, Th17, and iNKT-cell cytokine production. Eur. J. Immunol. 45: 2276–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.O’Connor W., Jr., Kamanaka M., Booth C. J., Town T., Nakae S., Iwakura Y., Kolls J. K., Flavell R. A. 2009. A protective function for interleukin 17A in T cell-mediated intestinal inflammation. Nat. Immunol. 10: 603–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Noguchi D., Wakita D., Tajima M., Ashino S., Iwakura Y., Zhang Y., Chamoto K., Kitamura H., Nishimura T. 2007. Blocking of IL-6 signaling pathway prevents CD4+ T cell-mediated colitis in a Th17-independent manner. Int. Immunol. 19: 1431–1440. [DOI] [PubMed] [Google Scholar]
- 42.Elson C. O., Cong Y., Weaver C. T., Schoeb T. R., McClanahan T. K., Fick R. B., Kastelein R. A. 2007. Monoclonal anti-interleukin 23 reverses active colitis in a T cell-mediated model in mice. Gastroenterology 132: 2359–2370. [DOI] [PubMed] [Google Scholar]
- 43.Feng T., Qin H., Wang L., Benveniste E. N., Elson C. O., Cong Y. 2011. Th17 cells induce colitis and promote Th1 cell responses through IL-17 induction of innate IL-12 and IL-23 production. J. Immunol. 186: 6313–6318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Harbour S. N., Maynard C. L., Zindl C. L., Schoeb T. R., Weaver C. T. 2015. Th17 cells give rise to Th1 cells that are required for the pathogenesis of colitis. Proc. Natl. Acad. Sci. USA 112: 7061–7066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Felices M., Yin C. C., Kosaka Y., Kang J., Berg L. J. 2009. Tec kinase Itk in γδT cells is pivotal for controlling IgE production in vivo. Proc. Natl. Acad. Sci. USA 106: 8308–8313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tomlinson M. G., Kane L. P., Su J., Kadlecek T. A., Mollenauer M. N., Weiss A. 2004. Expression and function of Tec, Itk, and Btk in lymphocytes: evidence for a unique role for Tec. Mol. Cell. Biol. 24: 2455–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jain N., Miu B., Jiang J.-K., McKinstry K. K., Prince A., Swain S. L., Greiner D. L., Thomas C. J., Sanderson M. J., Berg L. J., Kang J. 2013. CD28 and ITK signals regulate autoreactive T cell trafficking. Nat. Med. 19: 1632–1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.