Abstract
Objectives
Combination therapy is an important option in the fight against Gram-negative ‘superbugs’. This study systematically investigated bacterial killing and the emergence of polymyxin resistance with polymyxin B and chloramphenicol combinations used against New Delhi metallo-β-lactamase (NDM)-producing MDR Klebsiella pneumoniae.
Methods
Four NDM-producing K. pneumoniae strains were employed. The presence of genes conferring resistance to chloramphenicol was examined by PCR. Time–kill studies (inocula ∼106 cfu/mL) were conducted using various clinically achievable concentrations of each antibiotic (range: polymyxin B, 0.5–2 mg/L; chloramphenicol, 4–32 mg/L), with real-time population analysis profiles documented at baseline and 24 h. The microbiological response was examined using the log change method and pharmacodynamic modelling in conjunction with scanning electron microscopy (SEM).
Results
Multiple genes coding for efflux pumps involved in chloramphenicol resistance were present in all strains. Polymyxin B monotherapy at all concentrations produced rapid bacterial killing followed by rapid regrowth with the emergence of polymyxin resistance; chloramphenicol monotherapy was largely ineffective. Combination therapy significantly delayed regrowth, with synergy observed in 25 out of 28 cases at both 6 and 24 h; at 24 h, no viable bacterial cells were detected in 15 out of 28 cases with various combinations across all strains. No polymyxin-resistant bacteria were detected with combination therapy. These results were supported by pharmacodynamic modelling. SEM revealed significant morphological changes following treatment with polymyxin B both alone and in combination.
Conclusions
The combination of polymyxin B and chloramphenicol used against NDM-producing MDR K. pneumoniae substantially enhanced bacterial killing and suppressed the emergence of polymyxin resistance.
Keywords: synergy, New Delhi metallo-β-lactamase, K. pneumoniae
Introduction
Since the first report of New Delhi metallo-β-lactamase (NDM)-producing Klebsiella pneumoniae in December 2009, a major international crisis has arisen due to the rapid spread of NDM-producing Enterobacteriaceae.1,2 NDM-1 is a novel metallo-β-lactamase (28 kDa) encoded by the blaNDM-1 gene that can be disseminated via horizontal gene transfer (with a frequency of ∼10−4–10−5).1–3 NDM-1 catalyses hydrolysis of the β-lactam ring and confers resistance to a wide range of β-lactams including cephalosporins, penicillins and carbapenems.2 Typically, NDM-producing Enterobacteriaceae isolates are MDR and cannot be treated with commonly used antibiotics such as aminoglycosides and fluoroquinolones.2 The intransigence to therapy of NDM-producing pathogens leads to prolonged illness, increased morbidity and mortality and higher economic costs.4,5 A recent report by the US CDC classified carbapenem-resistant Enterobacteriaceae (CRE) as an urgent threat to the antibiotic armamentarium.6 CRE cause ∼9300 healthcare-associated infections in the USA per year, with the majority of these being due to Klebsiella spp.6 Facing dwindling treatment options, clinicians have increasingly turned to polymyxins, which retain significant activity against NDM-producing pathogens, including K. pneumoniae.1,7
Two polymyxins are used clinically: colistin (polymyxin E; administered intravenously as its inactive prodrug colistin methanesulphonate) and polymyxin B.8,9 Polymyxins, which entered clinical use in 1959, target the negatively charged lipid A moiety of LPS in the outer membrane and as such are active only against Gram-negative bacteria.10 Although polymyxins are active against NDM-producing K. pneumoniae, recent studies have shown that polymyxin resistance emerges rapidly in NDM-producing organisms that are treated with polymyxin alone.2,7,11 Combination therapy has been proposed as a strategy to increase antimicrobial activity and reduce the emergence of resistance to polymyxins.12,13 Given the role of polymyxins as a treatment option of ‘last resort’ for infections caused by NDM-producing pathogens, investigating combinations including polymyxin is critical to maintaining its efficacy in the clinic. Chloramphenicol, which was introduced into clinics in 1949,14 has a broad spectrum of activity that includes K. pneumoniae.7,15 Unlike polymyxins, it is bacteriostatic and inhibits bacterial protein synthesis.16 The aim of this study was to investigate, using clinically relevant concentrations of each antibiotic, the antibacterial activity and emergence of polymyxin resistance that were associated with polymyxin B monotherapy and combination therapy with chloramphenicol used against NDM-producing MDR K. pneumoniae.
Materials and methods
Bacterial strains and MIC measurements
Four strains of NDM-producing K. pneumoniae were examined in this study: a reference strain, ATCC BAA-2146 (ATCC, Manassas, VA, USA), and three K. pneumoniae clinical isolates (1, S01 and 129).17–19 The strains are described in detail in Table 1. All the strains were MDR, which was defined as resistance to at least one antimicrobial agent from three or more antimicrobial categories.20 The MICs of both polymyxin B (Sigma-Aldrich, Castle Hill, Australia; Batch number BCBD1065V) and chloramphenicol (Sigma-Aldrich; Batch number 02111LB) were determined for all strains in two replicates on separate days using broth microdilution in CAMHB [Ca2+ at 22.5 mg/L and Mg2+ at 11.25 mg/L (Oxoid, Hampshire, UK)].21 Susceptibility and resistance to chloramphenicol were defined as MICs of ≤8 mg/L and >8 mg/L, respectively, as per the EUCAST guidelines on Enterobacteriaceae.22 Although no breakpoints for polymyxin B have currently been established for the Enterobacteriaceae, susceptibility and resistance breakpoints to colistin have been set at ≤2 mg/L and >2 mg/L, respectively.22 Given the comparable activity of colistin and polymyxin B,23 the colistin breakpoints were applied to polymyxin B for the purposes of this study.
Table 1.
MICs for and carbapenemase typing and genotyping of NDM-producing K. pneumoniae strains used in this study
| Straina | MIC (mg/L)b |
Carbapenemase |
Presence of genes coding for efflux pumps related to chloramphenicol resistance |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PMB | CHL | blaNDM | blaCTX-M | blaCMY-2 | blaSHV | blaTEM | acrA | eefB | emrA | mdfA | oqxA | oqxB | |
| ATCC BAA-2146 | 0.5 | 256 | + | + | + | + | + | + | + | + | + | + | + |
| 1 | 0.5 | 4 | + | − | + | + | − | + | − | + | + | − | − |
| S01 | 0.5 | 16 | + | + | − | + | + | + | + | + | + | + | + |
| 129 | 0.5 | 64 | + | + | − | + | − | + | + | + | + | + | + |
PMB, polymyxin B; CHL, chloramphenicol.
aAll the strains were MDR, defined as resistance to at least one antimicrobial agent from three or more antimicrobial categories.20
bEUCAST breakpoints (S, susceptible; R, resistant) for chloramphenicol were S ≤8 mg/L and R >8 mg/L; for polymyxin B, the EUCAST breakpoints for colistin of S ≤2 mg/L and R >2 mg/L were applied.22
Genotyping of NDM-producing K. pneumoniae strains
The presence of β-lactamase genes in these strains was previously investigated using PCR.17,24 β-Lactamases of Ambler classes A (ESBLs), B (metallo-β-lactamases) and C (extended-spectrum cephalosporinases) were examined and are presented in Table 1. The reference strain and all three NDM-producing MDR K. pneumoniae clinical isolates contained blaNDM genes.
Bacterial genomic DNA was extracted using a DNeasy® Blood & Tissue Kit (Qiagen, USA) according to the manufacturer's instructions. The genotyping of known chloramphenicol resistance genes was carried out using standard PCR using GoTaq® Green Master Mix (M712) (Promega, Madison, USA) with the primers shown in Table S1 (available as Supplementary data at JAC Online). PCR amplifications were carried out using a T100 Thermal cycler (Bio-Rad, USA) and analysed by agarose gel electrophoresis. The published sequence for K. pneumoniae ATCC BAA-2146 was used for primer design and as a positive control.25
Population analysis profiles (PAPs)
PAPs were used to determine heteroresistance to polymyxins in the K. pneumoniae strains that were examined.26 Each K. pneumoniae strain was cultured in CAMHB to an inoculum of ∼109 cfu/mL before viable counting on Mueller–Hinton agar plates containing polymyxin B (0.5, 1, 2, 4 and 8 mg/L). The plates were incubated for 24 h at 35°C. The limit of detection was 20 cfu/mL (equivalent to one colony per plate). PAPs for polymyxin B were also constructed following 24 h of polymyxin B treatment in the time–kill studies (see below).
Time–kill studies
Static time–kill studies26 were used to examine bacterial killing and the emergence of polymyxin resistance in the absence (growth controls) and presence of polymyxin B and chloramphenicol monotherapy and combination therapy against the four K. pneumoniae strains. Polymyxin B monotherapy was investigated at 0.5, 1 and 2 mg/L. Chloramphenicol monotherapy of 8 and 16 mg/L against all the strains was examined, and concentrations of 4 and 32 mg/L were examined against strains with an MIC <64 mg/L and ≥64 mg/L, respectively. A total of 10 combination regimens were examined across the four strains, as follows: for isolates 1 and S01 (chloramphenicol MICs <64 mg/L), polymyxin B 0.5 mg/L plus chloramphenicol 16 mg/L, and polymyxin B 1 and 2 mg/L plus chloramphenicol 4, 8 or 16 mg/L; for ATCC BAA-2146 and isolate 129 (chloramphenicol MICs ≥64 mg/L), polymyxin B 0.5 mg/L plus chloramphenicol 32 mg/L, and polymyxin B 1 and 2 mg/L plus chloramphenicol 8, 16 or 32 mg/L. The concentrations of both polymyxin B and chloramphenicol investigated represent clinically achievable concentrations of unbound (free) plasma that can be achieved in patients.27–29 Viable counting was conducted at 0, 0.5, 1, 2, 4, 6 and 24 h, with PAPs (see above) documented at 24 h for all experiments involving polymyxin B (including combination regimens).
Quantification of antibacterial activity
The antibacterial activity of antibiotic monotherapies and combination therapies was quantified using two methods. The first was the change in log10 cfu/mL (Δcfu) from 0 h to 6 and 24 h. Activity was defined as a Δcfu value ≤−1, while synergy was defined as a Δcfu value ≥2 log10 cfu/mL lower than the most active monotherapy. Additive combinations were defined as those that achieved a Δcfu value that was between >1 and <2 log10 cfu/mL lower than the most active monotherapy.26
The activity of monotherapies and combination therapies was also assessed using a recently developed rate–area–shape model (equation 1).30
| (1) |
The extent of bacterial killing is described by parameter A and the extent of bacterial regrowth by parameter B. Kd describes the rate of bacterial killing and Kr the rate of regrowth, while parameter C characterizes the time delay of bacterial regrowth. These parameters were then used to calculate the time to 2 log10 killing (T2LK; equation 2) and the time to 3 log10 regrowth (T3LR; equation 3), both of which were employed as metrics to assess the activity of each therapy.
| (2) |
| (3) |
To account for the sampling schedule and the duration of the time–kill study, the T2LK was constrained to ≥6.59 min and the T3LR to ≤24 h. A pooled analysis over all the concentrations was performed via analysis of covariance (ANCOVA) to differentiate between the activity and regrowth of polymyxin B and chloramphenicol monotherapy and combination therapy.
Scanning electron microscopy (SEM) of cells treated with antibiotics
A single colony of K. pneumoniae isolate 1 was used to prepare an overnight culture from which 20 mL log-phase cultures (at ∼108 cfu/mL) were obtained. Sterile stock solutions of polymyxin B and chloramphenicol were added to achieve concentrations of 2 mg/L (4 × MIC) polymyxin B and 32 mg/L (8 × MIC) chloramphenicol. The tubes were incubated at 37°C in a shaking water bath for 1 h and then centrifuged at 3220 g for 10 min. The bacterial cells were fixed with 2.5% glutaraldehyde before being washed and resuspended three times in PBS. The bacterial cultures were incubated on polyethylenimine-coated coverslips (22 mm × 22 mm) for 1 h and immersed for a further hour in 2.5% glutaraldehyde in PBS before rinsing in PBS for 10 min for three times. Dehydration was then performed using increasing concentrations of ethanol in water (10%, 30%, 50%, 70%, 90% and 100%) for 10 min in each step. The coverslips were dried in a Balzers critical point dryer (Balzers, Liechtenstein, Germany) prior to mounting on 25 mm aluminium stubs with double-sided carbon tabs. Silver liquid was applied to the edges of each coverslip, and these were then dried and gold-coated in an Edwards S150B sputter coater (Edwards High Vacuum, Crawley, West Sussex, UK). The cells were imaged with a Philips XL30 field-emission scanning electron microscope (Philips, Eindhoven, The Netherlands) at a voltage of 2 kV.
Results
Chloramphenicol resistance genotyping, MIC measurements and polymyxin heteroresistance
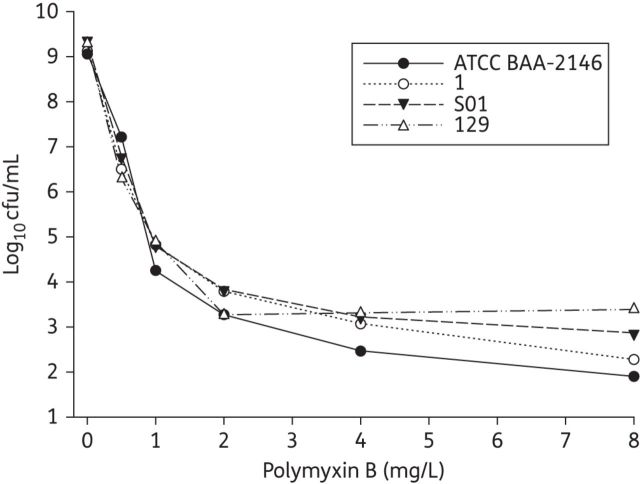
All the strains carried chloramphenicol resistance genes encoding the efflux pumps AcrA, EmrA and MdfA, as shown in Table 1. All the strains except K. pneumoniae 1 also carried the eefB and oqxAB genes. The MICs of both polymyxin B and chloramphenicol are shown in Table 1. All the strains were polymyxin heteroresistant at baseline, with the proportion of resistant bacterial cells on plates containing polymyxin B at 4 mg/L ranging from 2.4 × 10−7 to 1.1 × 10−6 (Figure 1).
Figure 1.
Baseline polymyxin B PAPs of the reference strain and all clinical isolates at an initial inoculum of ∼109 cfu/mL. The y-axis starts from the limit of detection.
Microbiological response and emergence of polymyxin resistance
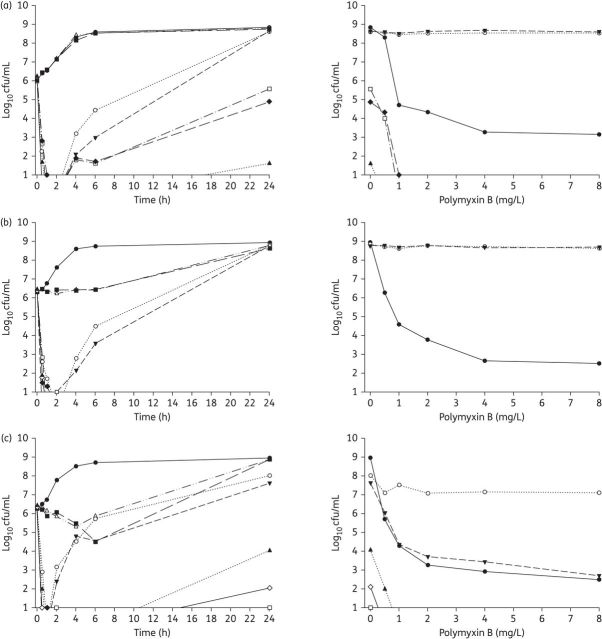
The time–kill curves for selected regimens of polymyxin B and chloramphenicol alone and in combination are shown in Figure 2. The data were well explained (median R2 = 0.98 for all regimens) by the rate–area–shape model. The estimated parameters for the model (Table 2 and Table S2) were then used to calculate the T2LK and T3LR for each therapy examined. The growth rates were derived from the Kr values for the control groups.
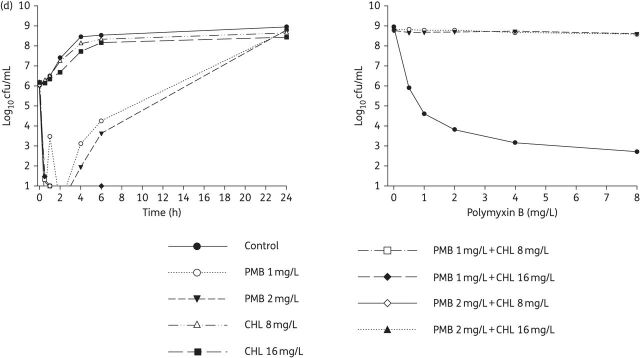
Figure 2.
Left: Time–kill curves with various clinically relevant concentrations of polymyxin B and chloramphenicol alone and in combination with an inoculum of ∼106 cfu/mL. Right: PAPs after 24 h of exposure to polymyxin B monotherapy, polymyxin B/chloramphenicol combination therapy or neither antibiotic (control). (a) ATCC BAA-2146 (chloramphenicol resistant, MDR). (b) Isolate 1 (chloramphenicol susceptible, MDR). (c) Isolate S01 (chloramphenicol resistant, MDR). (d) Isolate 129 (chloramphenicol resistant, MDR). The y-axis starts from the limit of detection. For isolates 1 and 129, only three time–kill curves are seen in the PAPs as there were no viable counts above the limit of detection for the two combination regimens. Symbols not seen at certain times are below the limit of detection. PMB, polymyxin B; CHL, chloramphenicol.
Table 2.
Parameter estimates of the model based on the time–kill studies with polymyxin B and chloramphenicol against NDM-producing K. pneumoniae ATCC BAA-2146, with a growth rate of 0.32 h−1
| Parameter | PMB 0.5 mg/L | PMB 1 mg/L | PMB 2 mg/L | CHL 8 mg/L | CHL 16 mg/L | CHL 32 mg/L | PMB 0.5 mg/L + CHL 32 mg/L | PMB 1 mg/L + CHL 8 mg/L | PMB 1 mg/L + CHL 16 mg/L | PMB 1 mg/L + CHL 32 mg/L | PMB 2 mg/L + CHL 8 mg/L | PMB 2 mg/L + CHL 16 mg/L | PMB 2 mg/L + CHL 32 mg/L |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A (log10 cfu/mL) | 4.70 | 5.19 | 5.23 | 0.00 | 0.00 | 2.70 | 3.90 | 5.97 | 5.80 | 4.98 | 5.94 | 6.31 | 5.59 |
| B (log10 cfu/mL) | 8.54 | 8.76 | 8.77 | 9.08 | 8.96 | 8.51 | 6.96 | 5.63 | 4.92 | 6.88 | 0.00 | 1.93 | 4.20 |
| C (h) | 5.09 | 6.78 | 8.80 | −2.19 | −2.39 | 1.22 | 2.45 | 8.69 | 7.63 | 5.09 | 5.73 | 19.15 | 5.60 |
| Kd (h−1) | 1.50 | 3.06 | 3.06 | 0.00 | 0.97 | 0.18 | 1.99 | 2.52 | 2.09 | 1.58 | 3.06 | 2.89 | 3.06 |
PMB, polymyxin B; CHL, chloramphenicol.
Parameter A describes the extent of bacterial killing and parameter B describes the extent of bacterial regrowth. Kd describes the rate of bacterial killing, while parameter C characterizes the time delay of bacterial regrowth.
Polymyxin monotherapy
Polymyxin B monotherapy produced extensive bacterial killing within 1 h against all strains, with 3–4 log10 cfu/mL killing at 0.5 mg/L (Table S3) and ≥5 log10 cfu/mL killing at 1 and 2 mg/L (Figure 2). Bacterial killing with polymyxin was very rapid, as shown by an estimated T2LK for polymyxin B, pooled across all the strains and concentrations examined, of 12.9 ± 3.99 min (mean ± SD). Despite good initial killing, significant bacterial regrowth was observed at all concentrations of polymyxin B examined; within 6 h, a regrowth of >3 log10 cfu/mL was observed for all strains, and by 24 h the total bacterial populations approached those observed in the control experiments (Figure 2). In line with these findings, the model-derived T3LR, pooled for all the strains and concentrations that were examined, was 3.87 ± 2.13 h. The PAPs of the bacterial regrowth at 24 h (Figure 2, right-hand panels) revealed a large proportion of highly polymyxin-resistant cells (∼100% were able to grow on plates containing 8 mg/L polymyxin B) in all but one polymyxin monotherapy time–kill study (2 mg/L polymyxin B against K. pneumoniae S01) (Figure 2).
Chloramphenicol monotherapy
In the time–kill studies examining chloramphenicol alone, modest bacterial killing (up to ∼1.5 log10 cfu/mL at 6 h with 16 mg/L) was achieved only against K. pneumoniae S01 (Figure 2). In this strain, the rate of bacterial killing for chloramphenicol was much slower than that for polymyxin B, as illustrated by T2LK values for chloramphenicol of 178 min at 8 mg/L and 416 min at 16 mg/L (compared with T2LK values for polymyxin B of 13.0 min at 1 mg/L and 12.7 min at 2 mg/L). No bacterial killing was observed for any of the remaining strains; however, an inhibition of bacterial growth (0–6 h) was observed against strain 1. Significant bacterial regrowth occurred across all the strains and chloramphenicol concentrations, with no significant differences in the total bacterial counts observed between the treated and control experiments at 24 h (Figure 2).
Combination therapy
The combination of chloramphenicol and polymyxin B increased the extent of bacterial killing observed in the first hour following the initiation of therapy by ∼1–2 log10 cfu/mL (Figure 2). However, the rate of bacterial killing, as measured by the T2LK (10.6 ± 4.37 min) assessed jointly across the combinations, did not significantly improve compared with polymyxin monotherapy (P = 0.283, ANCOVA). Notably, combination therapy significantly delayed bacterial regrowth compared with polymyxin monotherapy. Bacterial regrowth, pooled for all strains and characterized by the model-estimated T3LR, for combination therapy was markedly delayed compared with that for polymyxin monotherapy (T3LRcombination 19.5 h versus T3LRmonotherapy 3.89 h; P < 0.01). These findings were mirrored in the Δcfu values calculated at 6 and 24 h, where synergy was observed in 25 out of 28 cases at each timepoint (Table S3). The emergence of polymyxin-resistant bacteria was also suppressed with combination therapy at all the concentrations examined, with no colonies detected on polymyxin-containing plates following 24 h of treatment (Figure 2, right-hand panels). Importantly, at 24 h no viable bacterial cells were detected in 15 out of 28 (54%) cases with the various combinations across all strains (Table S3).
Morphological changes
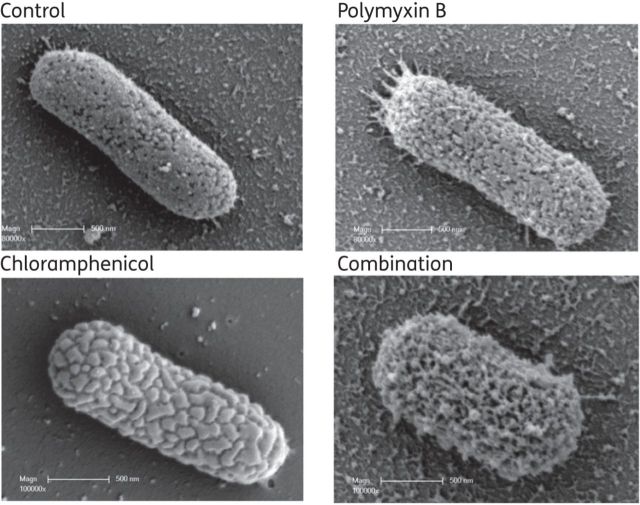
An analysis of K. pneumoniae isolate 1 by SEM revealed morphological changes in the cell surface after treatment with each antibiotic both alone and in combination (Figure 3). The cell surface in the untreated (control) group was relatively smooth, while the cells treated with chloramphenicol alone showed uneven surface bulges. Treatment with polymyxin B alone produced significant changes to the outer membrane, with numerous pits and protrusions. The combination treatment caused more severe cell surface damage that resembled the combined changes observed with the individual treatments.
Figure 3.
SEM images of K. pneumoniae 1 in the absence of drug therapy (control) or in the presence of polymyxin B at 2 mg/L, chloramphenicol at 32 mg/L or the combination of these. Scale bar = 500 nm.
Discussion
As NDM-producing K. pneumoniae continues to spread globally, a lack of new antibiotics active against MDR Gram-negative bacteria in the drug development pipeline means that polymyxins will be an essential component of our antibiotic armamentarium for many years to come.2,7 However, the regrowth of a variety of Gram-negative organisms including K. pneumoniae with polymyxin monotherapy (both polymyxin B and colistin) has been well established both in vivo31 and in vitro,26,32–34 even at concentrations far exceeding those that can be safely achieved clinically. The amplification of pre-existent polymyxin-resistant subpopulations within heteroresistant strains is known to contribute to the regrowth.32 Adaptive resistance and the induction of genetic mutations within two-component systems involved in modifications of LPSs may also contribute.35,36 Combined with known suboptimal exposures to polymyxins in critically ill patients12,27 and dose-limiting nephrotoxicity,12 polymyxin combination therapy has been suggested as a possible way not only to increase bacterial killing, but also to reduce the emergence of polymyxin resistance.12,13,37 To date, studies utilizing time–kill methodology to examine polymyxin combinations against NDM-producing Enterobacteriaceae are virtually absent from the literature. Furthermore, the emergence of polymyxin resistance has never been examined.38
In the present study, we systematically investigated the effectiveness of polymyxin B alone and in combination with chloramphenicol against polymyxin-susceptible NDM-producing MDR K. pneumoniae. Similarly to polymyxins, chloramphenicol fell out of favour with clinicians due to the potential adverse events, which include aplastic anaemia.39 Chloramphenicol was chosen because it has a broad spectrum of activity against MDR organisms, including a significant subset of NDM-producing Enterobacteriaceae (up to 25% in a recent study).7,39,40 In addition, chloramphenicol lacks the nephrotoxicity and exerts its antibacterial effect via a different mode of action from that of polymyxins. The concentrations of polymyxin B employed were chosen to reflect clinically achievable unbound plasma concentrations in critically ill patients following intravenous administration of the currently recommended doses.27,37 For chloramphenicol, pharmacokinetic data for critically ill patients are limited. In hospitalized patients, total serum concentration of 5–20 mg/L are recommended.29,41 Given that chloramphenicol is ∼40% unbound in plasma, clinically desirable unbound chloramphenicol concentrations would be ∼2.3–9.4 mg/L, although free chloramphenicol concentrations of 16–32 mg/L (which might be undesirable due to the potential side effects) have been reported.29 The concentrations employed in the current study (4, 8, 16 and 32 mg/L) thus span the full range of clinically achievable plasma concentrations of unbound chloramphenicol.
Genes encoding multiple key efflux pumps involved in chloramphenicol resistance were present in all four strains. Three strains contained all the efflux pumps examined (i.e. AcrA, EefB, OqxAB, EmrA and MdfA) and were resistant to chloramphenicol, as indicated by the MIC results (Table 1); the other isolate (K. pneumoniae 1) contained only three efflux pumps and was susceptible to chloramphenicol (MIC 4 mg/L). As monotherapy, chloramphenicol merely delayed the growth of strains 1 and S01, and was completely ineffective against both strains with the highest MICs (ATCC BAA-2146 and 129); as genes for all the efflux pumps were present in isolate S01, other underlying mechanisms of resistance and/or other efflux pumps may be present in ATCC BAA-2146 and isolate 129. Nevertheless, all polymyxin B/chloramphenicol combinations substantially enhanced bacterial killing against all the strains irrespective of the chloramphenicol MIC (Figure 2 and Table S3). Importantly, although both polymyxin B monotherapy and the combinations produced rapid and extensive bacterial killing within the first 1–2 h following antibiotic exposure, in all cases the subsequent rapid regrowth observed with polymyxin B monotherapy was either substantially reduced by combination therapy or, in 54% of cases, entirely eliminated (Figure 2 and Table S3). That even low concentrations of polymyxin B (e.g. 0.5 mg/L) can prevent or minimize regrowth is an important finding given that polymyxin-induced nephrotoxicity is the dose-limiting adverse effect. Additionally, and in stark contrast to polymyxin B monotherapy, no emergence of resistance to polymyxin B across 24 h was observed with any combination even when regrowth occurred.
After treatment with polymyxin B alone, the formation of projections and blebs on the surface of K. pneumoniae 1 cells indicated a pronounced disruption of the structural integrity of the bacterial outer membrane (Figure 3). These observations are consistent with its mode of action, which is centred on the bacterial membrane and membrane-anchored lipid A.42 These changes in the outer membrane have been reported after treatment with polymyxins against other bacteria including Escherichia coli,43,44 Salmonella typhimurium43,45 and Pseudomonas aeruginosa.44,46 While previous investigations using other bacterial species such as E. coli and P. aeruginosa have not shown significant morphological changes following treatment with chloramphenicol as monotherapy,46 morphological changes in the form of convoluted surfaces were observed in the present study. The combination of polymyxin B and chloramphenicol caused even more dense projections, morphologically a combination of the changes caused by polymyxin B and chloramphenicol alone (Figure 3). Our SEM results also support the synergistic killing of polymyxin B with chloramphenicol.
Two mechanisms, namely subpopulation synergy and mechanistic synergy, have been proposed to explain the enhanced pharmacodynamic effects seen with polymyxin combination therapy.47 Subpopulation synergy involves the killing of a subpopulation resistant to one drug in combination with the second drug and vice versa, whereas mechanistic synergy involves two drugs in a combination acting on different cellular pathways in the same organism to increase the rate and extent of killing of the other drug. We have demonstrated, for the first time, the phenomenon of polymyxin heteroresistance in strains of NDM-producing K. pneumoniae (Figure 1); as has been previously observed, the presence of highly polymyxin-resistant cells is likely to be a key contributor to the regrowth that was observed in our study following polymyxin monotherapy (Figure 2). Interestingly, for isolate 129, no bacterial killing was observed following exposure to chloramphenicol monotherapy at 32 mg/L, with the bacterial density reaching that of the control at 24 h. However, a reduction of ∼2 log10 cfu/mL lower than the initial inoculum was achieved with the same regimen against a polymyxin-resistant subpopulation taken from the same isolate (data not shown). This suggests that at least part of the improvement in bacterial killing observed by the addition of chloramphenicol to polymyxin B may be due to chloramphenicol acting on the polymyxin-resistant subpopulation that was present in the heteroresistant strains.
In summary, this study is the first to show the existence of polymyxin heteroresistance within NDM-producing K. pneumoniae and to demonstrate that polymyxin B in combination with chloramphenicol at clinically relevant concentrations significantly enhances bacterial killing. Notably, the combination suppressed the emergence of polymyxin-resistant subpopulations over a 24 h period. Further studies are warranted to investigate the effect of chloramphenicol resistance genes (e.g. the chloramphenicol acetyltransferase gene) on synergistic killing with polymyxins and to optimize combination therapy against NDM-producing ‘superbugs’ using animal infection models. As this information becomes available, its translation into the clinic should prolong the efficacy of these important last-line antibiotics.
Funding
This study was partially funded by the Australian National Health and Medical Research Council (NHMRC APP1046561). J. L., R. L. N. and T. V. are supported by research grants from the National Institute of Allergy and Infectious Diseases of the National Institutes of Health [R01 A1098771 (J. L., R. L. N. and T. V.) and R01 AI111965 (J. L. and T. V.)]. M. A. C. is an Australian NHMRC Principal Research Fellow. T. V. is an Australian NHMRC Industry Career Development Research Fellow. J. L. is an Australian NHMRC Senior Research Fellow.
Transparency declarations
None to declare.
Disclaimer
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Allergy and Infectious Diseases or the National Institutes of Health.
Supplementary data
Tables S1–S3 are available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/).
Acknowledgements
Part of this study was presented at the Fifty-second Interscience Conference on Antimicrobial Agents and Chemotherapy, San Francisco, CA, USA, 2012 (Abstract E-795).
We are grateful to Dr Simon Crawford (University of Melbourne, Parkville) for his technical support with the SEM experiment.
References
- 1.Yong D, Toleman MA, Giske CG, et al. Characterization of a new metallo-β-lactamase gene, blaNDM-1, and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Chemother 2009; 53: 5046–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumarasamy KK, Toleman MA, Walsh TR, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis 2010; 10: 597–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Potron A, Poirel L, Nordmann P. Plasmid-mediated transfer of the blaNDM-1 gene in Gram-negative rods. FEMS Microbiol Lett 2011; 324: 111–6. [DOI] [PubMed] [Google Scholar]
- 4.Boucher HW, Talbot GH, Bradley JS, et al. Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 2009; 48: 1–12. [DOI] [PubMed] [Google Scholar]
- 5.ECDC. Updated Risk Assessment on the Spread of NDM and its Variants Within Europe. Stockholm: ECDC, 2011. http://ecdc.europa.eu/en/publications/Publications/1111_TER_Risk-assessment-NDM.pdf. [Google Scholar]
- 6.CDC. Antibiotic Resistance Threats in the United States, 2013. http://www.cdc.gov/drugresistance/pdf/ar-threats-2013-508.pdf.
- 7.Livermore DM, Warner M, Mushtaq S, et al. What remains against carbapenem-resistant Enterobacteriaceae? Evaluation of chloramphenicol, ciprofloxacin, colistin, fosfomycin, minocycline, nitrofurantoin, temocillin and tigecycline. Int J Antimicrob Agents 2011; 37: 415–9. [DOI] [PubMed] [Google Scholar]
- 8.Velkov T, Roberts KD, Nation RL, et al. Pharmacology of polymyxins: new insights into an ‘old’ class of antibiotics. Future Microbiol 2013; 8: 711–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bergen PJ, Li J, Rayner CR, et al. Colistin methanesulfonate is an inactive prodrug of colistin against Pseudomonas aeruginosa. Antimicrob Agents Chemother 2006; 50: 1953–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li J, Nation RL, Milne RW, et al. Evaluation of colistin as an agent against multi-resistant Gram-negative bacteria. Int J Antimicrob Agents 2005; 25: 11–25. [DOI] [PubMed] [Google Scholar]
- 11.Arpin C, Noury P, Boraud D, et al. NDM-1-producing Klebsiella pneumoniae resistant to colistin in a French community patient without history of foreign travel. Antimicrob Agents Chemother 2012; 56: 3432–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garonzik SM, Li J, Thamlikitkul V, et al. Population pharmacokinetics of colistin methanesulfonate and formed colistin in critically ill patients from a multicenter study provide dosing suggestions for various categories of patients. Antimicrob Agents Chemother 2011; 55: 3284–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rahal JJ. Novel antibiotic combinations against infections with almost completely resistant Pseudomonas aeruginosa and Acinetobacter species. Clin Infect Dis 2006; 43 Suppl 2: S95–9. [DOI] [PubMed] [Google Scholar]
- 14.Laferriere CI, Marks MI. Chloramphenicol: properties and clinical use. Pediatr Infect Dis 1982; 1: 257–64. [PubMed] [Google Scholar]
- 15.Kawakami M, Nagai Y, Shimizu S, et al. Anti-microbial effect of combinations of colistin methanesulfonate and chloramphenicol. I. In vitro effect. J Antibiot (Tokyo) 1971; 24: 884–91. [DOI] [PubMed] [Google Scholar]
- 16.Brock TD. Chloramphenicol. Bacteriol Rev 1961; 25: 32–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sidjabat H, Nimmo GR, Walsh TR, et al. Carbapenem resistance in Klebsiella pneumoniae due to the New Delhi metallo-β-lactamase. Clin Infect Dis 2011; 52: 481–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Netikul T, Sidjabat HE, Paterson DL, et al. Characterization of an IncN2-type blaNDM-1-carrying plasmid in Escherichia coli ST131 and Klebsiella pneumoniae ST11 and ST15 isolates in Thailand. J Antimicrob Chemother 2014; 69: 3161–3. [DOI] [PubMed] [Google Scholar]
- 19.Day KM, Salman M, Kazi B, et al. Prevalence of NDM-1 carbapenemase in patients with diarrhoea in Pakistan and evaluation of two chromogenic culture media. J Appl Microbiol 2013; 114: 1810–6. [DOI] [PubMed] [Google Scholar]
- 20.Magiorakos AP, Srinivasan A, Carey RB, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 2012; 18: 268–81. [DOI] [PubMed] [Google Scholar]
- 21.Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: Twenty-third Informational Supplement M100-S23. CLSI, Wayne, PA, USA, 2013. [Google Scholar]
- 22.EUCAST. Breakpoint Tables for Interpretation of MICs and Zone Diameters, Version 4.0. 2014. http://www.eucast.org/fileadmin/src/media/PDFs/EUCAST_files/Breakpoint_tables/Breakpoint_table_v_4.0.pdf.
- 23.Gales AC, Jones RN, Sader HS. Contemporary activity of colistin and polymyxin B against a worldwide collection of Gram-negative pathogens: results from the SENTRY Antimicrobial Surveillance Program (2006–09). J Antimicrob Chemother 2011; 66: 2070–4. [DOI] [PubMed] [Google Scholar]
- 24.Rogers BA, Sidjabat HE, Silvey A, et al. Treatment options for New Delhi metallo-β-lactamase-harboring enterobacteriaceae. Microb Drug Resist 2013; 19: 100–3. [DOI] [PubMed] [Google Scholar]
- 25.Hudson CM, Bent ZW, Meagher RJ, et al. Resistance determinants and mobile genetic elements of an NDM-1-encoding Klebsiella pneumoniae strain. PLoS One 2014; 9: e99209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bergen PJ, Forrest A, Bulitta JB, et al. Clinically relevant plasma concentrations of colistin in combination with imipenem enhance pharmacodynamic activity against multidrug-resistant Pseudomonas aeruginosa at multiple inocula. Antimicrob Agents Chemother 2011; 55: 5134–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zavascki AP, Goldani LZ, Cao G, et al. Pharmacokinetics of intravenous polymyxin B in critically ill patients. Clin Infect Dis 2008; 47: 1298–304. [DOI] [PubMed] [Google Scholar]
- 28.Ambrose PJ. Clinical pharmacokinetics of chloramphenicol and chloramphenicol succinate. Clin Pharmacokinet 1984; 9: 222–38. [DOI] [PubMed] [Google Scholar]
- 29.Koup JR, Lau AH, Brodsky B, et al. Chloramphenicol pharmacokinetics in hospitalized patients. Antimicrob Agents Chemother 1979; 15: 651–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheah SE, Li J, Nation RL, et al. A novel rate-area-shape modeling approach to quantify bacterial killing and regrowth for in vitro static time–kill studies. Antimicrob Agents Chemother 2015; 59: 381–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ketthireddy S, et al. In vivo pharmacodynamics of colistin against Pseudomonas aeruginosa in thighs of neutropenic mice. In: Abstracts of the Forty-seventh Interscience Conference on Antimicrobial Agents and Chemotherapy, Chicago, IL, USA, 2007. Abstract A-4, p. 1 American Society for Microbiology, Washington, DC, USA. [Google Scholar]
- 32.Poudyal A, Howden BP, Bell JM, et al. In vitro pharmacodynamics of colistin against multidrug-resistant Klebsiella pneumoniae. J Antimicrob Chemother 2008; 62: 1311–8. [DOI] [PubMed] [Google Scholar]
- 33.Deris ZZ, Yu HH, Davis K, et al. The combination of colistin and doripenem is synergistic against Klebsiella pneumoniae at multiple inocula and suppresses colistin resistance in an in vitro pharmacokinetic/pharmacodynamic model. Antimicrob Agents Chemother 2012; 56: 5103–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bergen PJ, Tsuji BT, Bulitta JB, et al. Synergistic killing of multidrug-resistant Pseudomonas aeruginosa at multiple inocula by colistin combined with doripenem in an in vitro pharmacokinetic/pharmacodynamic model. Antimicrob Agents Chemother 2011; 55: 5685–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tam VH, Schilling AN, Vo G, et al. Pharmacodynamics of polymyxin B against Pseudomonas aeruginosa. Antimicrob Agents Chemother 2005; 49: 3624–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Choi MJ, Ko KS. Mutant prevention concentrations of colistin for Acinetobacter baumannii, Pseudomonas aeruginosa and Klebsiella pneumoniae clinical isolates. J Antimicrob Chemother 2014; 69: 275–7. [DOI] [PubMed] [Google Scholar]
- 37.Sandri AM, Landersdorfer CB, Jacob J, et al. Population pharmacokinetics of intravenous polymyxin B in critically ill patients: implications for selection of dosage regimens. Clin Infect Dis 2013; 57: 524–31. [DOI] [PubMed] [Google Scholar]
- 38.Tangden T, Hickman RA, Forsberg P, et al. Evaluation of double- and triple-antibiotic combinations for VIM- and NDM-producing Klebsiella pneumoniae by in vitro time–kill experiments. Antimicrob Agents Chemother 2014; 58: 1757–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Civljak R, Giannella M, Di Bella S, et al. Could chloramphenicol be used against ESKAPE pathogens? A review of in vitro data in the literature from the 21st century. Expert Rev Anti Infect Ther 2014; 12: 249–64. [DOI] [PubMed] [Google Scholar]
- 40.Wareham DW, Wilson P. Chloramphenicol in the 21st century. Hosp Med 2002; 63: 157–61. [DOI] [PubMed] [Google Scholar]
- 41.Burke JT, Wargin WA, Sherertz RJ, et al. Pharmacokinetics of intravenous chloramphenicol sodium succinate in adult patients with normal renal and hepatic function. J Pharmacokinet Biopharm 1982; 10: 601–14. [DOI] [PubMed] [Google Scholar]
- 42.Velkov T, Thompson PE, Nation RL, et al. Structure–activity relationships of polymyxin antibiotics. J Med Chem 2010; 53: 1898–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schindler PR, Teuber M. Action of polymyxin B on bacterial membranes: morphological changes in the cytoplasm and in the outer membrane of Salmonella typhimurium and Escherichia coli B. Antimicrob Agents Chemother 1975; 8: 95–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Koike M, Iida K, Matsuo T. Electron microscopic studies on mode of action of polymyxin. J Bacteriol 1969; 97: 448–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lounatmaa K, Makela PH, Sarvas M. Effect of polymyxin on the ultrastructure of the outer membrane of wild-type and polymyxin-resistant strain of Salmonella. J Bacteriol 1976; 127: 1400–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Waisbren SJ, Hurley DJ, Waisbren BA. Morphological expressions of antibiotic synergism against Pseudomonas aeruginosa as observed by scanning electron microscopy. Antimicrob Agents Chemother 1980; 18: 969–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bulitta JB LJ, Poudyal A, Yu HH, et al. Quantifying synergy of colistin combinations against MDR gram negatives by mechanism-based models. In: Abstracts of the Forty-ninth Interscience Conference on Antimicrobial Agents and Chemotherapy, San Francisco, CA, USA, 2009. Abstract A1-573, p. 41 American Society for Microbiology, Washington, DC, USA. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.