Abstract
Sex chromosomes are subject to unique evolutionary forces that cause suppression of recombination, leading to sequence degeneration and the formation of heteromorphic chromosome pairs (i.e., XY or ZW). Although progress has been made in characterizing the outcomes of these evolutionary processes on vertebrate sex chromosomes, it is still unclear how recombination suppression and sequence divergence typically occur and how gene dosage imbalances are resolved in the heterogametic sex. The threespine stickleback fish (Gasterosteus aculeatus) is a powerful model system to explore vertebrate sex chromosome evolution, as it possesses an XY sex chromosome pair at relatively early stages of differentiation. Using a combination of whole-genome and transcriptome sequencing, we characterized sequence evolution and gene expression across the sex chromosomes. We uncovered two distinct evolutionary strata that correspond with known structural rearrangements on the Y chromosome. In the oldest stratum, only a handful of genes remain, and these genes are under strong purifying selection. By comparing sex-linked gene expression with expression of autosomal orthologs in an outgroup, we show that dosage compensation has not evolved in threespine sticklebacks through upregulation of the X chromosome in males. Instead, in the oldest stratum, the genes that still possess a Y chromosome allele are enriched for genes predicted to be dosage sensitive in mammals and yeast. Our results suggest that dosage imbalances may have been avoided at haploinsufficient genes by retaining function of the Y chromosome allele through strong purifying selection.
Keywords: threespine stickleback, sex chromosome evolution, dosage compensation
Introduction
Heteromorphic sex chromosomes (i.e., XY or ZW) have repeatedly evolved from autosomal ancestors across a diverse array of taxa and exhibit evolutionary patterns distinct from the remainder of the genome. Theoretical work as well as empirical studies in vertebrates, invertebrates, plants, and fungi (Ellegren 2011; Bachtrog 2013) suggest that the independent evolution of heteromorphic sex chromosomes in diverse taxa follows a similar trajectory. First, suppression of recombination is a hallmark of heteromorphic sex chromosomes, and is thought to occur as a result of selection for linkage between a sex-determination locus and genes with sexually antagonistic alleles (Bull 1983; Rice 1987a; Charlesworth 1991, 1996). Suppression can either occur through inversions or changes in genetic modifiers controlling recombination rate (Charlesworth et al. 2005). Once recombination is suppressed, the efficacy of natural selection is reduced, and deleterious mutations can quickly accumulate on the Y or W chromosome (Charlesworth 1978; Rice 1987b; Bachtrog 2013).
These evolutionary milestones have been studied in a variety of sex chromosome systems. In humans, the Y chromosome evolved over the last 180 My through at least four independent steps of recombination suppression, creating regions of different ages (termed “evolutionary strata”) (Lahn and Page 1999; Skaletsky et al. 2003; Hughes et al. 2012; Bellott et al. 2014; Cortez et al. 2014). At least in the younger strata, inversions seem to have suppressed recombination between the X and Y chromosomes (Lemaitre et al. 2009). Inversions have also been correlated with evolutionary strata on the papaya sex chromosomes (∼7 My old) (Wang et al. 2012). However, the relationship between evolutionary strata and inversions is less clear in other systems that span a variety of ages from the young sex chromosomes of the plant Silene latifolia (5–10 My old) (Filatov and Charlesworth 2002; Filatov 2005; Bergero et al. 2008, 2013; Rautenberg et al. 2010; Chibalina and Filatov 2011) to the old sex chromosomes of birds (140 My old) (Cortez et al. 2014; Wright et al. 2014). Furthermore, suppression of recombination does not occur in many sex chromosome systems, and even ancient sex chromosomes can remain homomorphic (Bachtrog et al. 2014). Thus, the evolutionary forces and molecular mechanisms that lead to the suppression of recombination on sex chromosomes are not completely understood.
As a result of suppressed recombination, many genes are eventually lost from the Y chromosome. This is exemplified by mammalian Y chromosomes, in which only a small fraction of genes remain on the Y chromosome, compared with their X-linked gametologs (Hughes et al. 2012; Bellott et al. 2014; Cortez et al. 2014). In response to sequence degeneration and gene loss, different mechanisms have evolved across taxa to restore gene dosage balance in the heterogametic sex. In some old systems, like Drosophila melanogaster (Gelbart and Kuroda 2009; Larschan et al. 2011) and Caenorhabditis elegans (Ercan et al. 2007), chromosome-wide dosage compensation mechanisms have evolved. However, recent work has revealed that chromosome-wide dosage compensation has not evolved in many other systems with heteromorphic sex chromosomes of different ages (Mank and Ellegren 2009; Mank 2013). Rather, in many systems dosage compensation can operate locally throughout the chromosome to specifically restore balance at dosage-sensitive genes (Mank and Ellegren 2009; Mank 2013). In eutherian mammals, dosage has been maintained at some haploinsufficient genes through local upregulation of the X chromosome (Lin et al. 2012; Pessia et al. 2012). At other genes, dosage imbalances have been avoided entirely by preserving the Y chromosome allele through strong selection (Bellott et al. 2014; Cortez et al. 2014). These recent findings have raised a number of new questions about the selective forces that shape degeneration and gene loss across the Y chromosome and whether dosage compensation evolves more often at particular types of genes, or even at all.
The threespine stickleback, Gasterosteus aculeatus, is a powerful vertebrate model system to further explore sex chromosome evolution. Gasterosteus aculeatus has an XY sex chromosome system that evolved sometime since the species arose at least approximately 13–16 Ma (Bell et al. 2009; Kawahara et al. 2009; Ross et al. 2009; Aldenhoven et al. 2010). In this time, the Y chromosome has structurally differentiated from the X chromosome through a series of at least three pericentric inversions and an apparent approximately 6 Mb deletion (Ross and Peichel 2008). Recombination has been suppressed between the X and Y chromosome across the region containing the inversions and deletion (Ross and Peichel 2008), resulting in elevated sequence divergence for the handful of loci that were studied (Peichel et al. 2004). However, it remains unknown whether there are evolutionary strata correlated with the chromosomal rearrangements that have occurred on the Y chromosome. Similar to other young sex chromosome systems, female-biased expression ratios across the sex chromosomes suggest that there are incomplete levels of dosage compensation within the nonrecombining region (Leder et al. 2010). However, it remains unknown whether dosage compensation occurs locally to restore ancestral gene expression levels.
Here, we used a combination of high-throughput DNA-sequencing (DNA-seq) and RNA-sequencing (RNA-seq) across a collection of male and female fish to explore sequence evolution and dosage compensation on the X and Y chromosomes of the threespine stickleback. We analyzed sequence divergence at genes across the nonrecombining region of the X and Y chromosomes to search for evolutionary strata that correlate with known chromosomal rearrangements and to determine if certain classes of genes maintain functional Y chromosome alleles despite rapid chromosome-wide degeneration. To directly test whether dosage compensation occurs, it is necessary to compare gene expression of the X chromosome with expression of orthologous genes in the ancestor (the proto-X chromosome) (Julien et al. 2012; Lin et al. 2012; Mank 2013; Vicoso, Emerson, et al. 2013). Therefore, we explored whether local dosage compensation has evolved at individual genes by comparing gene expression on the threespine stickleback sex chromosomes with their autosomal orthologs in a closely related outgroup species, the ninespine stickleback (Pungitius pungitius). This species does not possess the threespine stickleback XY sex chromosome system and diverged approximately 13–16 Ma from the threespine stickleback (Bell et al. 2009; Kawahara et al. 2009; Ross et al. 2009; Aldenhoven et al. 2010). Our results highlight the power in merging patterns of molecular evolution with allele-specific gene expression in males to understand the evolution of heteromorphic sex chromosome pairs.
Results and Discussion
Protein-Coding Divergence between the X and Y Chromosomes Reveals Two Evolutionary Strata
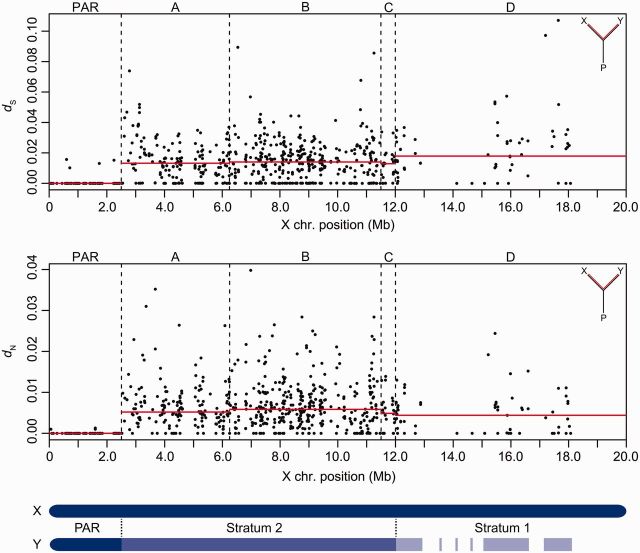
Divergence between the X and Y chromosomes was significantly higher among genes in the nonrecombining region than among genes within the recombining pseudoautosomal region (PAR). This pattern was evident for both synonymous site divergence (dS) (median XY nonrecombining: 0.0139, N = 657 genes; median XY recombining PAR: 0.0000, N = 87 genes; Mann–Whitney U test, P < 0.001) (fig. 1) and nonsynonymous site divergence (dN) (median XY nonrecombining: 0.0056, N = 657 genes; median XY recombining PAR: 0.0000, N = 87 genes; Mann–Whitney U test, P < 0.001) (fig. 1). In addition, divergence between the X and Y chromosomes showed evidence of significantly less purifying selection (i.e., higher dN/dS) in genes in the nonrecombining region than in genes in the recombining PAR of the sex chromosomes (median dN/dS XY nonrecombining: 0.2832, N = 562 genes; median dN/dS XY recombining PAR: 0.0000, N = 84 genes; Mann–Whitney U test, P < 0.001).
Fig. 1.
Protein-coding divergence between the X and Y chromosomes. Protein-coding divergence was quantified in a pairwise fashion between the X and Y chromosomes to estimate synonymous site divergence (dS) and nonsynonymous site divergence (dN). dS and dN are shown for each gene, organized by position in megabases (Mb) on the X chromosome (chromosome 19) assembly. Median divergence across each region is indicated by the red line. A schematic representation of the evolutionary strata as defined by synonymous divergence is shown below the divergence plots. The PAR (N = 87 genes), the three pericentric inversions (A: N = 180 genes; B: N = 378 genes; C: N = 24 genes), and the distal region, where a large proportion of the Y chromosome has either degenerated or been deleted (D: N = 75 genes), are indicated.
We investigated whether there were differences in sequence divergence among the three major cytogenetically characterized pericentric inversions (here referred to as A, B, and C) of the G. aculeatus Y chromosome (Ross and Peichel 2008). This difference in dS would support a model where inversions suppressed recombination in a stepwise fashion across the G. aculeatus Y chromosome. Despite the presence of pericentric inversions, we detected no significant differences in median nonsynonymous and synonymous site divergence (dN and dS) (fig. 1) or in median dN/dS ratio between the X and Y among the three pericentric inversions (median dS: A: 0.0132, N = 180 genes; B: 0.0140, N = 378 genes; C: 0.0129, N = 24 genes; median dN: A: 0.0052, N = 180 genes; B: 0.0059, N = 378 genes; C: 0.0049, N = 24 genes; median dN/dS: A: 0.3195, N = 151 genes; B: 0.2991, N = 316 genes; C: 0.3303, N = 21 genes; P>0.05 in all comparisons using Mann–Whitney U test). Similar values of dS suggest that recombination ceased at a comparable time across the three regions. These results are consistent with several scenarios. In one scenario, a single large inversion could have suppressed recombination simultaneously across the region. If two nested inversions then formed within the prior inversion (Ross and Peichel 2008), delineating regions A, B, and C, the nested inversions would have no differential effect on dS because recombination was already suppressed across the region. A second plausible scenario is that inversions A, B, and C were not nested within an initial large inversion and instead occurred sequentially, shutting down recombination in a stepwise manner across the Y chromosome. If these inversions occurred close enough together temporally, dS may not be discernable between the strata. Indeed, simulations indicate that the ages must be considerably different to generate detectable strata in the values of dS (Chibalina and Filatov 2011). Third, it is possible that other genetic modifiers evolved to suppress recombination (Charlesworth et al. 2005) and that the pericentric inversions did not play a role. Finally, intrachromosomal rearrangements have been documented on Y chromosomes between closely related species (Hughes et al. 2010) and even within species (Knebel et al. 2011; Lange et al. 2013). Thus, it is possible that the lack of differentiation between the inversions is because the inversions are polymorphic within the Japanese Pacific Ocean G. aculeatus population used in both this study and the previous cytogenetic study (Ross and Peichel 2008). Regardless of the reason, because we found equal dS across A, B, and C, we combined these regions into a single region for subsequent analyses.
In addition to the pericentric inversions, previous cytogenetic evidence indicated that the distal approximately 6 Mb of the X chromosome is largely missing from the Y chromosome (Ross and Peichel 2008). Next-generation sequencing and microarrays from multiple worldwide freshwater and marine populations have also revealed a similar pattern, with read coverage or hybridization in this region nearly half of that in females (Leder et al. 2010; Roesti et al. 2013; Yoshida et al. 2014; Schultheiss et al. 2015). This indicates that the region is largely degenerated and/or deleted, and that these changes are likely fixed within the species. Surprisingly, we found a number of Y-linked transcripts that aligned to this region of the X, revealing that some genes from this region still exist on the Y chromosome (fig. 1). Together, these results raise the intriguing possibility that this region is an older evolutionary stratum. To test this possibility, we estimated divergence in the remaining genes of this distal region (D). Median dS between X and Y in region D was significantly higher than in region ABC (D: 0.0179, N = 75 genes; ABC: 0.0135, N = 582 genes; Mann–Whitney U test, P = 0.021) (fig. 1), suggesting that recombination first ceased between the X and Y chromosomes in region D. Interestingly, a previous study did not report a significant difference in dS between these regions (Schultheiss et al. 2015). However, the previous study was likely underpowered to detect a difference in divergence as dS was estimated among a smaller pool of genes in the oldest stratum (N = 38 in Schultheiss et al. 2015; N = 75 in this study).
Median dN in region D (0.0044, N = 75 genes) was significantly lower than in region ABC (0.0057, N = 582 genes) (Mann–Whitney U test, P = 0.010) (fig. 1). Consequently, genes within region D have been under stronger purifying selection (i.e., lower dN/dS) than genes in the rest of the nonrecombining region since the divergence of the X and Y chromosomes from their autosomal ancestor (D: 0.1500, N = 74 genes; ABC: 0.3085, N = 488 genes; Mann–Whitney U test, P = 0.019). Similar patterns were observed in the oldest regions of the S. latifolia sex chromosomes, where high dS was correlated with low dN/dS ratios (Chibalina and Filatov 2011). After large-scale loss of coding and intergenic regions, the only genes that will remain in old strata are those that are critically important for biological functions. This has been observed in mammals, where a highly conserved set of genes involved in transcription and translation independently survived deletion across Y chromosome lineages (Bellott et al. 2014; Cortez et al. 2014). As in threespine sticklebacks, this conserved set of genes had significantly lower dN/dS ratios than the remainder of genes on the X chromosome (Bellott et al. 2014). Hereafter, we refer to the older evolutionary stratum (D) as stratum 1 and the younger region (ABC) as stratum 2, to be consistent with previous literature on sex chromosome evolution (fig. 1) (Lahn and Page 1999; Ross et al. 2005; Bellott et al. 2014; Cortez et al. 2014). Note that this nomenclature differs from previous studies of the threespine stickleback sex chromosomes (Roesti et al. 2013; Schultheiss et al. 2015).
Stratum 1 may have evolved structurally under a variety of scenarios. One possibility is that the distal end of the Y chromosome was not subject to a single large deletion. Instead, most of the genes and intergenic regions could have degraded over a long period of time, leaving little homologous sequence for aligning next-generation sequencing reads (Roesti et al. 2013; Yoshida et al. 2014; Schultheiss et al. 2015) or for hybridizing cytogenetic probes (Ross and Peichel 2008). Under this model, if most of the genes and intergenic regions have simply degraded over time, the remaining genes under purifying selection would be clustered together. Alternatively, biologically important genes from the old stratum could have duplicated and translocated elsewhere on the Y chromosome over time. Following these gene duplication events, the ancestral copies of stratum 1 would be deleted from the Y chromosome in a single event. This scenario is less parsimonious as it requires multiple translocation steps; however, extensive Y chromosome rearrangements have been documented in mammals (Hughes et al. 2010). Under this model, the remaining genes of stratum 1 would be scattered in different regions of the Y chromosome and would not be syntenic with the X chromosome. Ultimately, the complete sequencing of the Y chromosome will reveal the order and location of all transcripts from stratum 1, allowing us to distinguish these models.
Lineage-Specific Sequence Divergence of the X and Y Chromosomes
Substitutions occur at different rates along the X and Y chromosome lineages for several reasons, including higher mutation rates in males (Shimmin et al. 1993; Li et al. 2002; Ellegren 2007; Wilson Sayres and Makova 2011), Y-specific sequence degeneration (Charlesworth and Charlesworth 2000; Bachtrog 2013), and the faster-X effect (X-linked divergence occurs at a higher rate than divergence on the autosomes) (Charlesworth et al. 1987; Vicoso and Charlesworth 2006; Mank et al. 2010; Meisel and Connallon 2013). To separate lineage-specific evolution of the X and Y chromosome of G. aculeatus, we used the sequence of a P. pungitius female as an outgroup. This species has independently evolved an XY sex chromosome system from chromosome 12 (the G. aculeatus XY sex chromosome system evolved from chromosome 19) and so the sex-linked transcripts of G. aculeatus are autosomal in P. pungitius (Ross et al. 2009; Shapiro et al. 2009). There were no significant differences in dS among the X and Y lineages in any regions of the sex chromosomes (fig. 2A) (Kruskal–Wallis test, P = 0.227), indicating an absence of a biased mutation rate in male threespine sticklebacks. However, we found different levels of dN among regions of the sex chromosomes (fig. 2B) (Kruskal–Wallis test, P < 0.001). dN was higher in stratum 2 of the Y chromosome as compared with the autosomes, although this difference was not significant when corrected for multiple comparisons (autosomes, N = 27,858 genes; stratum 2, N = 490 genes; post hoc Mann–Whitney U test, P = 0.016; corrected for multiple comparisons using Holm’s method, P = 0.097; Holm 1979). dN was significantly higher in nonfunctional stratum 2 genes that contain frameshifts or nonsense mutations on the Y chromosome as compared with the autosomes (autosomes, N = 27,858 genes; stratum 2, N = 92 genes; post hoc Mann–Whitney U test, P < 0.001) (fig. 2B). Similarly, the dN/dS ratio was significantly higher in nonfunctional Y chromosome genes of stratum 2 than autosomes (autosomes, N = 27,328 genes; stratum 2, N = 91 genes; Kruskal–Wallis test, P < 0.001; post hoc Mann–Whitney U test, P = 0.006) (supplementary fig. S1, Supplementary Material online), consistent with a loss of selective constraint on Y chromosome genes as they degenerate.
Fig. 2.
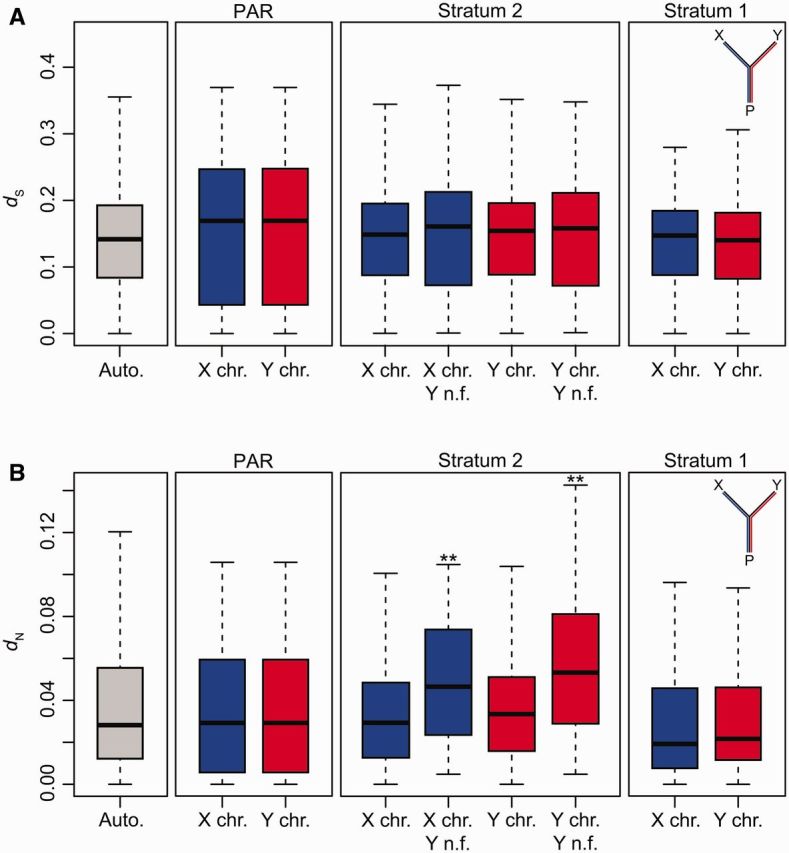
Lineage-specific divergence of the X and Y chromosomes. The X chromosome, Y chromosome, and autosomes of Gasterosteus aculeatus were aligned to the ninespine stickleback (Pungitius pungitius) to quantify (A) synonymous site divergence (dS) and (B) nonsynonymous site divergence (dN) between G. aculeatus and P. pungitius orthologs (autosomes: N = 27,858 genes; PAR: N = 87 genes; stratum 2: N = 490 genes; stratum 2 non-functional (n.f.): N = 92 genes; stratum 1: N = 75 genes). n.f. genes are those that include frameshifts or nonsense mutations on the Y chromosome. Significant differences among groups were determined using a Kruskal–Wallis test. Groups significantly different from the autosomes using a post hoc Mann–Whitney U test (corrected for multiple comparisons) are indicated with asterisks (**P < 0.05). Whiskers are 1.5 × the interquartile range. Outliers are not shown.
Synonymous divergence between the sex chromosomes should be similar to dS between P. pungitius and G. aculeatus autosomal orthologs if the sex chromosomes evolved immediately after the species split (∼13–16 Ma) (Bell et al. 2009; Aldenhoven et al. 2010). Instead, we found that XY dS was significantly lower than autosomal ds between the species in both strata (autosomes: 0.1417, N = 27,858 genes; stratum 1: 0.0179, N = 75 genes; stratum 2: 0.0135, N = 582 genes; P < 0.001 in both comparisons using Mann–Whitney U test), indicating that both strata are considerably younger than the split between G. aculeatus and P. pungitius. It is possible that we were unable to recover all evolutionary strata with next-generation sequencing. Smaller, ancient strata on the Y chromosome may lack adequate sequence coverage because of difficulty mapping divergent reads to the X chromosome. A Sanger sequenced assembly of the G. aculateus Y chromosome will be necessary to reveal the complete set of evolutionary strata and to accurately estimate the age of the sex chromosomes.
Recessive male beneficial mutations are predicted to accumulate more rapidly in regions of the X chromosome that are hemizygous in males due to the faster-X effect (Charlesworth et al. 1987; Vicoso and Charlesworth 2006; Mank et al. 2010; Meisel and Connallon 2013). To explore whether genes on the X chromosome also evolve faster in the threespine stickleback, we examined whether dN/dS ratios (between the G. aculeatus X chromosome and P. pungitius orthologs) were higher in hemizygous X chromosome genes from stratum 1 and stratum 2, and in genes that had nonfunctional alleles on the Y chromosome (i.e., functionally hemizygous) from stratum 2. When compared with autosomes (median dN/dS: 0.1798, N = 27,328 genes), the median dN/dS ratio was higher for X chromosome genes of stratum 2 that were hemizygous (median dN/dS: 0.2530, N = 148 genes; Mann–Whitney one-tailed U test, P = 0.006) or that had nonfunctional alleles on the Y chromosome (median dN/dS: 0.2712, N = 91 genes; Mann–Whitney one-tailed U test, P = 0.002), and for hemizygous genes in stratum 1 (median dN/dS: 0.1981, N = 591 genes; Mann–Whitney one-tailed U test, P = 0.048). Our results complement previous studies on relatively old sex chromosome systems that have demonstrated the faster-X effect (Baines and Harr 2007; Mank et al. 2007; Baines et al. 2008; Meisel and Connallon 2013). Previous work on the threespine stickleback did not detect elevated dN/dS ratios on the X chromosome (Yoshida et al. 2014). However, the study was underpowered to detect elevated rates of evolution on the X chromosome. First, the authors did not focus on hemizygous regions of the sex chromosomes, where the effect should be strongest. In addition, the analysis did not use an autosomal outgroup for the sex chromosomes, effectively analyzing dN/dS over a shorter time period. Here, we use an autosomal outgroup and show an elevated rate of evolution in the hemizygous regions of two differently aged strata, indicating that selection can act rapidly to increase beneficial mutations on the X chromosome.
We used the outgroup to show that insertions, deletions, and nonsense mutations were also distributed differently across the X and Y chromosomes. Within stratum 2, there were significantly more indels on the Y chromosome than the X chromosome (table 1) (χ2 = 75.06, df = 1, P < 0.001). Across the genome, deletions are primarily driven by replication errors, rather than recombination (Kvikstad et al. 2007, 2009). Therefore, in nonrecombining regions of the genome, such as the Y chromosome, deletions should be favored because mutations would primarily be driven by replication errors (Wilson Sayres and Makova 2011). Consistent with this prediction, we detected a higher frequency of deletions on the G. aculeatus Y chromosome than insertions (χ2 = 9, df = 1, P = 0.003) (table 1). Frameshift indels were nonexistent on the X chromosome; however, both frameshifts and nonframeshift indels occurred at similar frequencies on the Y chromosome (χ2 = 0.1011, df = 1, P = 0.751), indicating a lack of selection to purge deleterious frameshift indels from the Y chromosome. Within stratum 1, indels only occurred on the Y chromosome and were at a significantly lower frequency than stratum 2 (stratum 1, N = 75 genes; stratum 2, N = 582 genes; permutation test with 10,000 random samples, P = 0.010), indicating that purifying selection is also acting to purge deleterious insertions and deletions from the remaining genes of stratum 1.
Table 1.
Insertions and deletions across the sex chromosomes.
| X Chromosome Deletion | X Chromosome Insertion | Y Chromosome Deletion | Y Chromosome Insertion | |
|---|---|---|---|---|
| Stratum 2 | ||||
| Nonframeshift | 0 | 5 | 31 | 12 |
| Frameshift | 0 | 0 | 32 | 14 |
| Stratum 1, X and Y alleles | ||||
| Nonframeshift | 0 | 0 | 4 | 0 |
| Frameshift | 0 | 0 | 0 | 0 |
Accumulation of Pseudogenes on the Threespine Stickleback Y Chromosome
Pseudogenes have been quantified as a measure of degeneration on Y and W chromosomes in other systems. We defined pseudogenes on the G. aculeatus Y chromosome as the genes that contain frameshift or nonsense mutations along with the genes that are hemizygous based on the DNA read depth threshold (see Methods). This is likely an overestimate, as hemizygous genes in our analysis combine two types of gene loss. Lower read depth can result from poor sequencing read alignments to highly divergent pseudogenes, or lower read depth can reflect true deletions. Still, these results allow for comparison of the threespine stickleback Y chromosome to other sex chromosome systems. By our estimates, 8.9% of genes were pseudogenes, whereas 53.5% of genes were hemizygous in males. Thus, 62.4% of genes are predicted to be nonfunctional pseudogenes on the G. aculeatus Y chromosome.
Lack of Global Dosage Compensation in Threespine Sticklebacks
In light of this extensive Y chromosome degeneration, we wanted to test whether dosage compensation has evolved in threespine sticklebacks. Thus, we compared gene expression of the X chromosome with orthologous genes in the ninespine stickleback, P. pungitius. In these analyses, we assume that the expression levels in the outgroup reflect the ancestral levels of expression on the proto-X chromosome (Julien et al. 2012; Lin et al. 2012; Mank 2013; Vicoso, Emerson, et al. 2013). Using RNA-seq data from brain tissues of males and females, we first compared the expression of autosomal genes in threespine stickleback relative to their expression in ninespine stickleback. As expected, autosomal gene expression in both males and females was highly correlated with the expression of orthologous genes in the outgroup (Spearman’s rank correlation; male rho = 0.888, N = 24,308 genes, P < 0.001; female rho = 0.876, N = 24,308 genes, P < 0.001) (fig. 3A). Median autosomal expression ratios were also nearly identical between males and females (Mann–Whitney U test, N = 24,308 genes, P = 0.926) (fig. 3B). Across the sex chromosomes, gene expression significantly deviated from ancestral levels, with both region-specific and sex-specific effects. In the youngest regions, the PAR and stratum 2, females had expression ratios that were significantly greater than ancestral expression (one-sample Mann–Whitney U test, mu = 0; PAR: N = 64 genes, P = 0.039; stratum 2: N = 496 genes, P = 0.009), whereas expression levels in males were indistinguishable from ancestral levels (one-sample Mann–Whitney U test, mu = 0; PAR: N = 64 genes, P = 0.075; stratum 2: N = 496 genes, P = 0.291) (fig. 3B). Different patterns were observed in the older stratum 1. Males had gene expression levels significantly lower than the ancestor (one-sample Mann–Whitney U test, mu = 0; stratum 1 X and Y alleles: N = 65 genes, P < 0.001; stratum 1 X hemizygous: N = 522 genes, P < 0.001), and females had expression levels that matched ancestral levels (one-sample Mann–Whitney U test, mu = 0; stratum 1 X and Y alleles: N = 65 genes, P = 0.092; stratum 1 X hemizygous: N = 522 genes, P = 0.520) (fig. 3B).
Fig. 3.
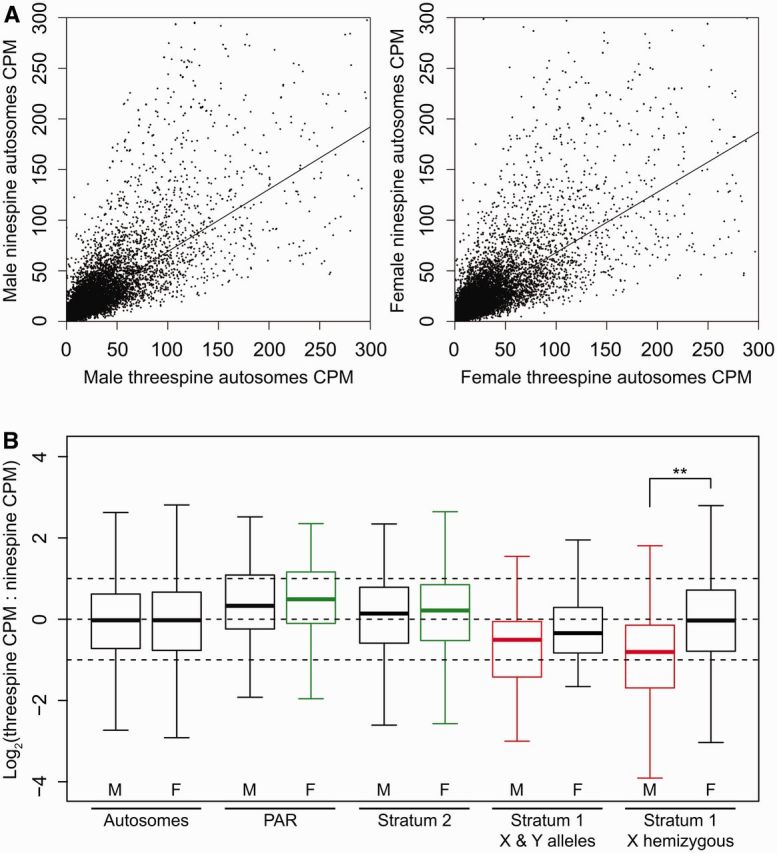
Threespine stickleback gene expression compared with outgroup gene expression levels. (A) Gene expression of autosomal genes in the threespine stickleback (Gasterosteus aculeatus) is highly correlated with gene expression of orthologous genes in the ninespine stickleback (Pungitius pungitius) (males: N = 24,308 genes; females: N = 24,308 genes; CPM: counts per million). Linear regression lines are shown. (B) Gene expression levels in male and female threespine stickleback and male and female ninespine stickleback were normalized to their respective median autosomal gene expression. Median threespine stickleback:ninespine stickleback expression ratios were measured across the autosomes and the threespine stickleback sex chromosomes. Medians that are significantly greater than ancestral levels are indicated in green, medians that are significantly less than ancestral levels are indicated in red, and medians that do not differ from ancestral levels are indicated in black (Mann–Whitney U test, mu = 0, autosomes and the sex chromosomes were corrected for multiple comparisons separately). Within each region (autosomes: N = 24,308 genes; PAR: N = 64 genes; stratum 2: N = 496 genes; stratum 1 X and Y alleles: N = 65 genes; stratum 1 X hemizygous: N = 522 genes), significant differences between males and females are marked with asterisks (pairwise Mann–Whitney U test, corrected for multiple comparisons, **P < 0.05). “X & Y alleles” refer to the genes within stratum 1 that retain coding sequence on the Y chromosome. Whiskers are 1.5 × the interquartile range. Outliers are not shown.
Stratum 2 and the PAR exhibited strong patterns of feminization. Feminization of the X chromosome has been observed in other XY systems (Bachtrog 2006; Vicoso and Charlesworth 2006; Dean and Mank 2014) and can result from two mechanisms. Because the X chromosome is transmitted more frequently through the female germline, female-beneficial traits are selected for more efficiently than male beneficial traits (Rice 1984; Bachtrog 2006; Dean and Mank 2014). In addition, genes can evolve sex-biased expression in recombining regions of sex chromosomes to resolve conflict from sexually antagonistic mutations in the absence of recombination suppression (Rice 1984; Scotti and Delph 2006; Otto et al. 2011; Vicoso, Kaiser, et al. 2013). In this case, genes would be upregulated in the sex they benefit. Consistent with these patterns, we observed significant upregulation of genes only in females in the youngest regions of the sex chromosomes. Within the PAR, feminization could be a mechanism to resolve sexual conflict. In stratum 2, female gene expression was still higher than ancestral expression levels, but lower than female expression levels in the PAR (Mann–Whitney U test, N = 65 genes, P = 0.047) (fig. 3B). In this stratum, feminization could reflect its recent history as a recombining region where sexually antagonistic genes once needed to resolve sexual conflict by evolving sex-biased expression. Alternatively, female-beneficial genes may have accumulated after recombination ceased. Feminization in stratum 2 may also reflect a combination of these two mechanisms.
Reduced gene expression in males within older stratum 1 might reflect a lack of global dosage compensation. To more closely assess whether dosage compensation may be operating in males, we compared male and female gene expression ratios in each of the regions across the sex chromosomes. In the PAR and stratum 2, the expression in males and females did not differ significantly (Mann–Whitney U test; PAR: N = 65 genes, P = 0.569; stratum 2: N = 496 genes, P = 0.168) (fig. 3B). In stratum 1, there were two different patterns. Among the genes of stratum 1 that retained a Y chromosome allele, male gene expression levels were lower than females, but this reduction was not statistically significant (Mann–Whitney U test, N = 65 genes, P = 0.183), suggesting some maintenance of gene expression in males. In the hemizygous genes, males had a significantly lower median gene expression than females (Mann–Whitney U test, N = 522 genes, P < 0.001), matching female-biased expression patterns previously observed in this region (Leder et al. 2010; Schultheiss et al. 2015). Lower gene expression in hemizygous genes in males argues against a global mechanism for dosage compensation. Our results contrast previous work which postulated that dosage compensation was evolving in stratum 1 on the basis of sex chromosome to autosome expression ratios of hemizygous genes with no comparison with ancestral expression levels (Schultheiss et al. 2015). By comparing the expression of sex-linked genes with their autosomal orthologs in the outgroup, we find no evidence for upregulation of hemizygous genes in either males or females, indicating a general lack of dosage compensation across stratum 1.
X-Biased Gene Expression in Males
Males had similar median gene expression ratios to females in regions of the sex chromosomes that still possessed a Y chromosome allele (stratum 2 and stratum 1 X and Y alleles). However, simply assessing relative expression levels in males and females alone does not reveal whether similar expression ratios in these regions are the result of increased expression from the X chromosome and/or maintenance of transcript expression from the Y chromosome. To distinguish these possibilities, we examined allele-specific expression to quantify whether Y chromosome alleles matched the expression of X chromosome alleles. In both stratum 2 and stratum 1, X chromosome alleles were expressed at significantly higher levels than Y chromosome alleles (one-sample Mann–Whitney U test, mu = 0; stratum 1: N = 52 genes, P = 0.003; stratum 2: N = 414 genes, P < 0.001), indicating either some loss of Y chromosome expression or an upregulation of the X chromosome (i.e., dosage compensation) (fig. 4). To distinguish the two possibilities, we examined whether male or female gene expression (normalized to ancestral proto-X expression) changed in response to decreasing expression of the Y chromosome allele. If X-biased gene expression is caused by dosage compensation, overall male gene expression should remain equal to female expression as transcription from the Y chromosome decreases. On the other hand, if X-biased gene expression is mainly due to downregulation of Y chromosome genes without dosage compensation, male gene expression should decrease as gene expression is lost from the Y chromosome. In both strata, among genes where X chromosome expression more closely matched Y chromosome expression, male and female median gene expression was more similar (Mann–Whitney U test; stratum 1: N = 8 genes, P = 0.557; stratum 2, N = 65 genes, P = 0.557) (fig. 5). In stratum 2, when X-biased expression increases, male gene expression decreases compared with females (Mann–Whitney U test, N = 67 genes, P < 0.001), suggesting no dosage compensation (fig. 5A). In stratum 1, male gene expression was reduced to a similar magnitude among the most X-biased genes, but this result was not significant (Mann–Whitney U test, N = 8 genes, P = 0.482) (fig. 5B). The lack of significance was likely due to the small sample size of stratum 1 (upper quartile of X-biased genes in stratum 1, N = 8; upper quartile of X-biased genes in stratum 2, N = 67). Unlike in S. latifolia where local dosage compensation may be evolving (Muyle et al. 2012), our results suggest that X-biased gene expression in G. aculeatus is primarily driven by a decay of transcription from the Y chromosome. This argues that most genes that have degenerated across the sex chromosomes do not have strong haploinsufficiency phenotypes that would require a fine-tuning of dosage in the heterogametic sex.
Fig. 4.
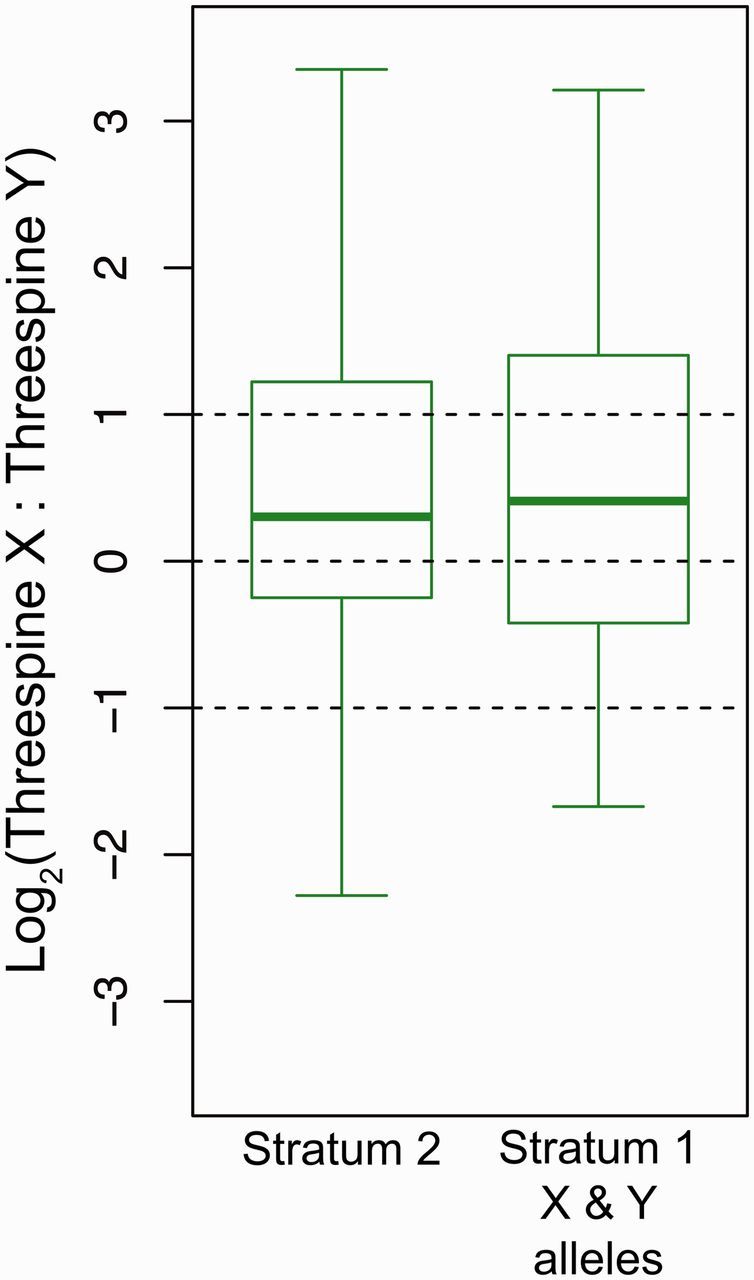
X-biased gene expression across the sex chromosomes. X chromosome and Y chromosome SNPs were identified to quantify allele-specific gene expression on the sex chromosomes. Genes are expressed at higher levels on the X chromosome as compared with the Y chromosome in both stratum 2 (N = 414 genes) and among the genes that retain coding sequence on the Y chromosome in stratum 1 (N = 52 genes). Medians that are significantly greater than an equal X to Y expression ratio are shown in green (one-sample Mann–Whitney U test, mu = 0, corrected for multiple comparisons). Whiskers are 1.5× the interquartile range. Outliers are not shown.
Fig. 5.
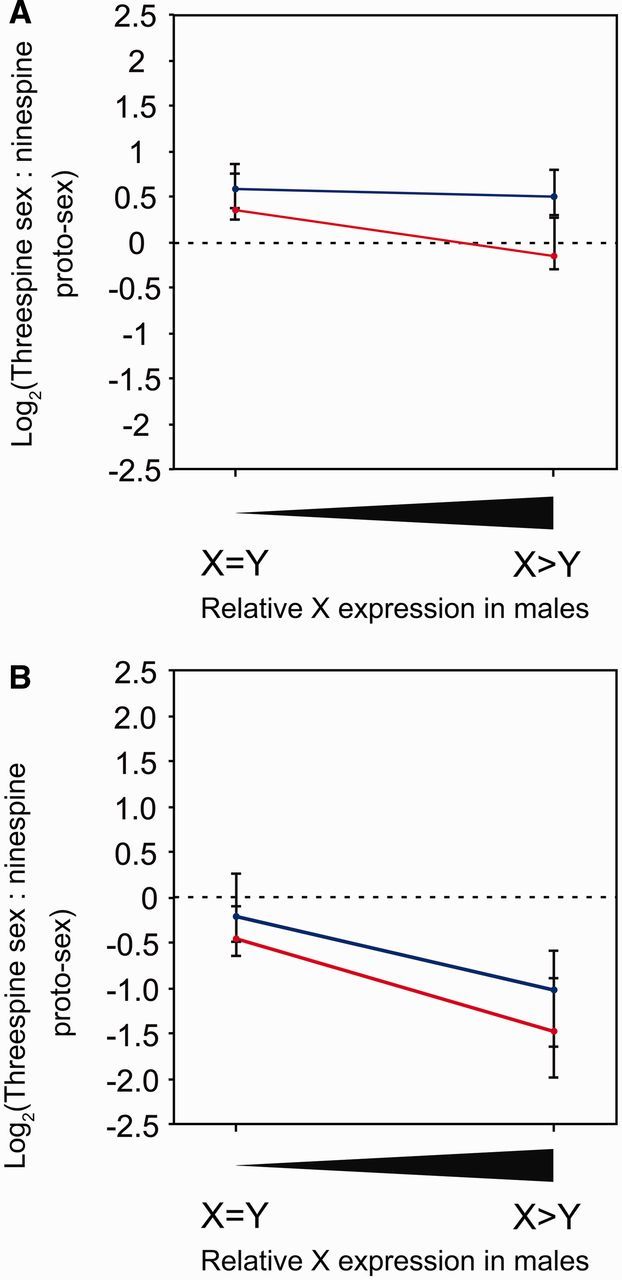
X-biased gene expression in males is driven by lower transcription from the Y chromosome. Median male (red) and female (blue) gene expression was measured in the upper and lower quartiles of X-biased expression among genes in (A) stratum 2 (upper quartile: N = 67 genes; lower quartile: N = 65 genes) and (B) stratum 1 (upper quartile: N = 8 genes; lower quartile: N = 8 genes). In the lower quartile of genes, X chromosome expression is nearly equal to Y chromosome expression, whereas in the upper quartile of genes, X chromosome expression is much higher than Y chromosome expression. Y-biased genes were omitted from the analysis. In males, median gene expression decreases as X-biased gene expression increases, indicating Y degeneration instead of dosage compensation in both strata. Error bars are 95% confidence intervals from bootstrapping.
Retained Genes Are Enriched for Dosage-Sensitive Functions
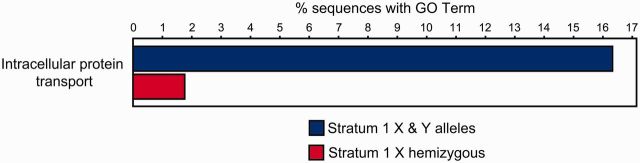
However, some genes were retained under purifying selection in stratum 1, so we explored whether they were enriched for certain classes of genes. Using the X chromosome hemizygous genes in stratum 1 as the reference pool, there was a significant overenrichment of genes predicted to have a role in intracellular protein transport among the genes that retained a Y chromosome allele (stratum 1 X and Y alleles, N = 49 total genes, 8 intracellular protein transport genes; stratum 1 X hemizygous genes, N = 398 total genes, 7 intracellular protein transport genes; Fisher’s exact test, P = 0.026, false discovery rate [FDR] corrected) (fig. 6). We conducted permutation tests to explore whether stratum 1 had an unusually high complement of intracellular protein transport genes by randomly drawing stratum 1-sized regions across the autosomes. We did not detect a significantly higher density of these genes in stratum 1 than on the autosomes (8 Mb regions, 10,000 random permutations, P = 0.070). Therefore, our results do not reflect ancestral clustering of genes with a function in intracellular transport and instead likely reflect the outcome of strong purifying selection to maintain protein-coding function of important housekeeping genes on the Y chromosome. A core set of housekeeping genes are also conserved across a wide range of highly degenerate mammalian Y chromosomes (Bellott et al. 2014; Cortez et al. 2014) preserved under purifying selection because of dosage constraints. Interestingly, known dosage-sensitive genes in the yeast genome are enriched for intracellular transport (Makanae et al. 2013), indicating that this function is particularly sensitive to dosage imbalances.
Fig. 6.
The remaining genes of stratum 1 are enriched for intracellular protein transport. The remaining genes of stratum 1 (blue; stratum 1 X & Y alleles, N = 49 total genes, 8 intracellular protein transport genes) are significantly enriched for genes involved in intracellular protein transport as compared with genes across the hemizygous genes of stratum 1 (red; stratum 1 X hemizygous, N = 398 total genes, 7 intracellular protein transport genes) (Fisher’s exact test, P = 0.026, FDR corrected).
Genes involved in protein complexes should also be sensitive to dosage imbalances, because altering the dosage of a single member will imbalance the stoichiometry of the entire protein complex (Papp et al. 2003; Pessia et al. 2012; Makanae et al. 2013). We searched for orthologs of mammalian protein complex genes (identified using the Comprehensive Resource of Mammalian Protein Complexes [CORUM] database) within the remaining genes of stratum 1. We found a highly significant enrichment of genes involved in protein complexes in the remaining genes of stratum 1, compared with the hemizygous genes of stratum 1 (stratum 1 X and Y alleles, N = 65 total genes, 21 protein complex genes; stratum 1 X hemizygous genes, N = 522 total genes, 58 protein complex genes; Fisher’s exact test, P < 0.001). It is possible that this enrichment could be an artifact resulting from ancestral clustering of protein complex genes within stratum 1. To investigate this, we conducted the same permutation tests as above to determine whether stratum 1 had a significantly higher density of protein complex genes than the autosomes. Unlike the intracellular protein transport genes, stratum 1 did contain a higher number of protein complex genes than most of the autosomal regions (8 Mb regions, 10,000 random permutations, P = 0.050), indicating some ancestral clustering. However, ancestral clustering would not necessarily result in enrichment of protein complex genes preferentially within regions that retained Y chromosome alleles. If gene loss within stratum 1 was a stochastic process, there should not be a difference in the number of protein complex genes between the hemizygous genes and the genes that still possess a Y chromosome allele. Instead, our results suggest that selection differentially favored the retention of Y chromosome alleles for many of the protein complex genes. Consistent with this, protein complex genes that retained a Y chromosome allele interacted with a higher number of proteins than protein complex genes that are hemizygous (median number of interacting proteins of remaining genes in stratum 1: 25, N = 21; median number of interacting proteins of hemizygous genes in stratum 1: 9, N = 58; Mann–Whitney U test, P = 0.001). Proteins with the highest number of interactions in complexes were found to be the most dosage sensitive in mammals (Pessia et al. 2012). Thus, our data suggest that the genetic architecture of the threespine stickleback Y chromosome has been shaped by dosage sensitivity.
Conclusions
Our results provide a detailed characterization of Y chromosome evolution in the threespine stickleback. Despite extensive sequence divergence and gene loss, we did not find clear evidence of local dosage compensation across a majority of genes. Instead, X-biased gene expression in males was largely due to reduced expression of the Y chromosome allele. Dosage balance was likely maintained at some haploinsufficient genes by preserving function of the Y chromosome allele in the face of Y chromosome degeneration.
Several closely related species of sticklebacks will help clarify the early stages of sequence divergence and dosage compensation. There is a young neo-Y sex chromosome system in the Japan Sea lineage of the threespine stickleback (Higuchi and Goto 1996; Kitano et al. 2007, 2009). The neo-Y sex chromosome formed only 1.5–2 million generations ago, but genes on this chromosome are already diverging from the neo-X (Yoshida et al. 2014). Furthermore, the fourspine stickleback (Apeltes quadracus) will be useful to compare male and female heterogametic systems. The fourspine stickleback ZZ/ZW system (Chen and Reisman 1970; Ross et al. 2009; Urton et al. 2011) is approximately of the same age as the threespine stickleback XX/XY system, enabling a direct comparison of whether the Y or W chromosomes degrade faster (Naurin et al. 2010). Thus, the stickleback family (Gasterosteidae) offers a rich resource to explore sequence divergence and dosage compensation across the early stages of sex chromosome evolution in vertebrates.
Materials and Methods
DNA and RNA Sequencing
All research on live animals was approved by the Fred Hutchinson Cancer Research Center Institutional Animal Care and Use Committee (protocol 1575) and the National Institute of Genetics, Japan (protocol 25-15). Genomic DNA was isolated from four male and four female adults, which were the laboratory-reared progeny of wild-caught fish collected in the Bekanbeushi River, Akkeshi, Japan, from the Japanese Pacific Ocean population of G. aculeatus (Kitano et al. 2007, 2009), and from a single female P. pungitius collected from a tidepool in Biwase, Japan (Ishikawa et al. 2013). Whole caudal fin clips were digested overnight in 0.33 µg/µl proteinase-K at 55°. Genomic DNA was extracted using the DNeasy Blood & Tissue Kit (Qiagen), following manufacturer recommended protocols. For G. aculeatus, paired-end reads were sequenced for 50 cycles using the Illumina Genome Analyzer IIx (GAIIx) system by the Genomics Shared Resource at The Fred Hutchinson Cancer Research Center. For P. pungitius, paired-end reads were sequenced for 100 cycles on a HiSeq2000 by the Takara Bio Dragon Genomics Center (Mie, Japan). Each individual was sequenced to at least 10 × coverage on average, with one Pacific Ocean male sequenced to 40 × coverage.
Total RNA was isolated from the whole brain tissue of nine male and nine female G. aculeatus individuals, which were siblings of the Japanese Pacific Ocean fish used for DNA sequencing. Total RNA was also isolated from the whole brain tissue of three male and three female P. pungitius individuals, which were laboratory-reared fish raised from a single male and female collected from the tidepool in Biwase, Japan. Brain tissue was homogenized in TRIzol reagent (Invitrogen) and RNA was extracted following manufacturer recommended protocols. For G. aculeatus, each RNA sample replicate was composed of a pool of three different individuals of each sex. There were a total of three male replicates and three female replicates. Single-end reads were sequenced for 50 cycles using the Illumina Genome Analyzer IIx (GAIIx) system by the Genomics Shared Resource at The Fred Hutchinson Cancer Research Center. For P. pungitius, RNA was sequenced from individual brains of each male and female, rather than pooled samples, also resulting in three male replicates and three female replicates. For P. pungitius, RNA libraries were constructed with the TruSeq RNA Sample Preparation Kit (Illumina). Paired-end reads were sequenced for 150 cycles on the rapid run mode of HiSeq2500 by the Riken Genesis (Yokohama, Japan). For both the RNA and DNA sequencing, the reads were cleaned of adapters and barcodes, and only reads that passed the default Illumina CASAVA chastity threshold (≥0.6) were used in downstream analyses.
Sequence Assembly and Transcriptome Annotation
Paired-end DNA sequencing reads were aligned using Bowtie 2 (v. 2.0.2) (Langmead and Salzberg 2012) to the G. aculeatus reference genome, which was generated from a single individual female from a lake population in Alaska (BROADS1) (Jones et al. 2012). Physical positions of the X chromosome (i.e., chromosome 19) were based on a revised version of the X chromosome assembly (Ross and Peichel 2008). The PAR boundary and putative deleted region (region D) boundary were set at 2.50 and 12.00 Mb, respectively (Roesti et al. 2013). Boundaries for the pericentric inversions (regions A, B, and C) were set as the midpoints between Stn187/Stn 235 (6.25 Mb) and Idh/Stn194 (11.50 Mb) (Ross and Peichel 2008). For the sequencing reads from the Japanese Pacific Ocean G. aculeatus fish, the following parameters were used: -D 15 -R 2 -N 0 -L 22 -i S,1,1.15–rdg 5,3 –rfg 5,3 –mp 6,2. These default parameters produced an average alignment rate of 90.8% of female reads and 89.5% of male reads. For the sequencing reads from the P. pungitius fish, less stringent alignment parameters were used to compensate for greater sequence divergence between the two species (Bruneaux et al. 2013) (-D 20 -R 3 -N 1 -L 20 -i S,1,0.50 –rdg 3,2 –rfg 3,2 –mp 3). The less stringent parameters resulted in an alignment rate of 46.4% of the female P. pungitius reads to the G. aculeatus reference genome (only 12.2% of the reads aligned using the default parameters above).
RNA sequencing reads were aligned to the G. aculeatus reference genome and assembled using a combination of TopHat (v. 2.0.6) (Kim et al. 2013) and Cufflinks (v. 2.2.1) (Roberts et al. 2011). For G. aculeatus RNA sequencing reads, the following parameters were used: –b2-D 15 –b2-R 2 –b2-N 0 –b2-L 20 –b2-i S,1,1.25 –b2-rdg 5,3 –b2-rfg 5,3 –b2-mp 6,2 -N 2 –max-insertion-length 3 –max-deletion-length 3 –read-gap-length 2 –read-edit-dist 2. These default parameters produced an average alignment rate of 86.6% of female reads and 83.4% of male reads. Similar to the DNA alignments, less stringent TopHat parameters were used to increase the alignment rate of P. pungitius RNA sequencing reads to the reference genome (–b2-D 20 –b2-R 3 –b2-N 1 –b2-L 20 –b2-i S,1,0.50 –b2-rdg 3,2 –b2-rfg 3,2 –b2-mp 3,1 -N 36 –max-insertion-length 12 –max-deletion-length 12 –read-gap-length 24 –read-edit-dist 50). With these parameters, 70.3% of female reads and 63.6% of male reads aligned to the reference genome (only 2.3% of female reads aligned using the default parameters).
To define the total G. aculeatus transcriptome, only female RNA sequencing reads were used. Male reads were not included in the transcript annotations, as transcripts on the Y chromosome harbor a large number of single nucleotide polymorphisms (SNPs) and insertions/deletions (indels), complicating transcript assembly when included with the X chromosome. All three pools of female RNA reads were combined and aligned to the G. aculeatus reference genome with TopHat and assembled with Cufflinks, both using default parameters. The total set of transcripts used for analyses included all Cufflinks transcripts (1,108 transcripts from 848 genes) as well as all predicted transcripts in the Ensembl (release 69) database (846 transcripts from 661 genes), allowing for overlapping transcripts between the two annotation methods (1,954 combined transcripts from 1,509 combined genes).
Molecular Evolution of the X and Y Chromosomes
SNP and indel variants were called using the Genome Analysis Toolkit (GATK v. 2.2-16) (McKenna et al. 2010; DePristo et al. 2011). First, reads were locally realigned around indels in each male and female G. aculeatus DNA sample (using the RealignerTargetCreator and IndelRealigner tools). Variants were then called simultaneously across the four male samples and simultaneously across the four female samples. To maximize the number of variants detected from the Y chromosome, variants were called less stringently with UnifiedGenotyper (–genotype_likelihoods_model BOTH -stand_call_conf 4 –stand_emit conf 0 -dcov 200). Variants were called in the single female P. pungitius DNA sample using the same parameters for UnifiedGenotyper (–genotype_likelihoods_model BOTH -stand_call_conf 4 –stand_emit conf 0 -dcov 200). To reduce the number of false positives in G. aculeatus, variants were only considered if the SNP or indel was heterozygous in all four males and homozygous in all four females. The allele that was homozygous in females was treated as the X chromosome allele, whereas the alternate allele in males was treated as the Y chromosome allele. Custom Perl scripts were used to filter the SNPs and indels and to call the X and Y chromosome alleles (available from Dryad Digital Repository).
Synonymous (dS) and nonsynonymous (dN) site divergence was quantified in protein-coding regions of the transcripts defined above; the protein-coding regions were defined as the longest open reading frame (ORF) among all six reading frames. Divergence was only estimated in ORFs greater than 200 bp in length. The X chromosome, Y chromosome, and P. pungitius DNA sequences of each ORF were constructed from the GATK variants and G. aculeatus reference genome by substituting in the X chromosome, Y chromosome, and P. pungitius SNP and indel variants from the filtered UnifiedGenotyper output into the G. aculeatus reference genome. Nucleotide positions where the P. pungitius read depth was less than two were considered missing and were coded as “N” in the analysis. Divergence was not calculated where more than 10% of the ORF sequence was missing from P. pungitius. In the G. aculeatus data, coding sequences with no variants can reflect no divergence within the region, but can also be due to a lack of Y chromosome reads aligning to the region. Therefore, average read depth across all four males in the coding region was used to determine if a region was missing Y chromosome sequence, that is, was hemizygous. The minimum read depth was empirically determined from coding regions within the recombining PAR (average read depth: 59.20) and from coding regions in the nonrecombining region where Y chromosome variants were detected (region ABC average read depth: 55.74; region D average read depth: 45.18). Based on these thresholds, coding regions with average read depths below 45 were considered hemizygous in males.
dN, dS, and dN/dS were quantified in a pairwise manner between the G. aculeatus X and Y chromosomes, between the G. aculeatus X chromosome and P. pungitius orthologs, and between the G. aculeatus Y chromosome and P. pungitius orthologs, using the codeml module of PAML (phylogenetic analysis by maximum likelihood) (runmode = −2) (Yang 2007) after removal of stop codons and codons that included gaps or ambiguity characters. Pungitius pungitius has an independently evolved XY sex chromosome system and so the sex-linked transcripts of G. aculeatus are autosomal in P. pungitius (Ross et al. 2009; Shapiro et al. 2009). To estimate autosomal divergence, dN, dS, and dN/dS were also calculated between the autosomal transcripts of G. aculeatus and orthologous transcripts of P. pungitius, using the same procedure. All Mann–Whitney U tests were corrected for multiple pairwise comparisons using the method of Holm (Holm 1979). If a gene had multiple transcripts, dN, dS, and dN/dS were averaged across transcripts to provide single gene estimates. Genes with dN/dS = 99 were omitted in calculations of median dN/dS as these values represent infinity (Yang 2007). Divergence between the X and Y chromosome may be underestimated from the stringent SNP filtering scheme. To determine if this was the case, dS was estimated across 18 Sanger sequenced genes from the Y chromosome (identified within bacterial artificial chromosome library inserts) (Peichel et al. 2004). There was a marginal reduction in median dS between the sex chromosomes when Y chromosome sequence was reconstructed from the next-generation sequencing data (0.015) as compared with complete Sanger sequenced Y chromosome genes (0.020). However, this difference was not significant (Mann–Whitney U test, P = 0.052), suggesting that the next-generation sequencing analysis closely approximates the true level of sequence divergence. Perl scripts were written to find the longest ORF, construct the sequence alignments, tally frameshifts and nonsense mutations, and parse PAML output (available from Dryad Digital Repository).
Expression Levels of Genes in Males and Females
Transcript expression levels were measured independently in the three male and three female G. aculeatus RNA sample pools as well as the three male and three female individual P. pungitius RNA samples. Each male and female sample was mapped separately to the G. aculeatus reference genome using TopHat (as previously described). For each sample, the Python script, htseq-count (HTSeq software package v. 0.6.1) (http://www-huber.embl.de/users/anders/HTSeq, last accessed March 31, 2015), was used to count the total number of RNA reads that mapped to each gene annotation (-m = union, –stranded = no). Ambiguous reads that mapped to more than one location in the genome were not considered in the counts. Genes were removed from the analysis if they had a read count of zero in any of the 12 samples. This stringent filtering removed all genes with potentially interesting sex- or species-limited expression patterns. However, these genes are uninformative in our tests for dosage compensation, which rely on comparisons of logarithms of gene expression ratios. Genes with a DNA read depth below 45 on the autosomes, within the PAR of the sex chromosomes, and within stratum 2 of the sex chromosomes were not considered (see above). Reads were normalized across the three male G. aculeatus replicates, the three female G. aculeatus replicates, the three male P. pungitius replicates, and the three female P. pungitius replicates by calculating scaling factors with the trimmed mean of M-values method implemented in the Bioconductor package, edgeR, which minimizes the log-fold changes of gene expression between samples (Robinson et al. 2010). To calculate a single gene expression value for female G. aculeatus, male G. aculeatus, female P. pungitius, and male P. pungitius, the normalized read counts per million (Robinson et al. 2010) were averaged across the three replicates in each of these groups.
Allele-Specific Transcript Expression in Gasterosteus aculeatus Males
Male RNA reads were combined across the three sample replicates. Ambiguous reads that mapped to more than one location in the genome were not considered in the counts. The X and Y SNP variants discovered by the UnifiedGenotyper module of GATK (see above) were used to identify X and Y RNA SNP variants within each ORF. The number of X and Y RNA alleles present at each SNP within an ORF were summed to derive the total X:Y RNA expression ratio. Because transcripts expressed at low levels have a higher chance of being skewed stochastically toward either allele, only SNPs with a minimum total RNA read depth of four were considered. Short sequencing reads are subject to mapping biases (Dohm et al. 2008; Bullard et al. 2010), which could differentially affect the X chromosome or Y chromosome read counts. To correct for mapping biases, the X:Y RNA expression ratio was normalized to the X:Y DNA ratio. To have an accurate estimate of X and Y chromosome allele counts in the DNA sequences, SNPs were only used if there was a minimum read depth of six in each of the four male DNA sequences. It is important to note that transcripts were only considered if they contained at least one variant between the X and Y chromosomes within the DNA samples; thus, the status of hemizygous genes that no longer have a recognizable homolog on the Y chromosome could not be investigated in this analysis. If a gene had multiple transcripts, the X and Y chromosome read counts were averaged across transcripts to provide single gene estimates. A custom Perl script was written to compute the normalized X:Y RNA expression ratios (available from Dryad Digital Repository).
Functional Annotation of Genes
The total pool of genes were functionally annotated using the Blast2GO software package (v. 2.6.6) (Götz et al. 2008). If there were multiple transcripts per gene, genes consisted of the union of all transcripts. The remaining genes within region D (i.e., genes that had X and Y alleles) were tested for overenrichment of functional classes using the Enrichment Analysis tool and a Fisher’s exact test. The hemizygous genes of region D (i.e., genes with only an X chromosome allele) were used as the reference set. A functional class was considered over-enriched if the FDR-adjusted P value was less than 0.05.
Threespine stickleback genes involved in protein complexes were identified by searching for orthologs to proteins in the CORUM database using a translated BLAST search (BLASTX) and default parameters. If there were multiple transcripts per gene that matched orthologs, only the transcript with the highest bit score was used. The remaining genes of region D (i.e., genes that had X and Y alleles) were tested for an enrichment of genes involved in protein complexes using a Fisher’s exact test. The hemizygous genes of region D (i.e., genes with only an X chromosome allele) were used as the reference comparison.
Supplementary Material
Supplementary figure S1 is available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank Matt Fitzgibbon and the Genomics Shared Resource at The Fred Hutchinson Cancer Research Center for help with sequence analysis, Kohta Yoshida for advice on assembling the Pungitius pungitius sequence, Doris Bachtrog for discussion, and Judith Mank and Mark Kirkpatrick for discussion and comments on a previous version of the manuscript. The threespine stickleback (Gasterosteus aculeatus) sequences are deposited in the National Center for Biotechnology Information Short Sequence Read Archive (PRJNA277770). The ninespine stickleback (Pungitius pungitius) sequences are deposited in DNA Data Bank of Japan (DRA001085 and DRA002524). This work was funded by the Fred Hutchinson Cancer Research Center (C.L.P.) and the MEXT Grant-in-Aid for Scientific Research on Innovative Areas (23113007 and 23113001) (J.K.). M.A.W. was a Howard Hughes Medical Institute Fellow of the Life Sciences Research Foundation.
References
- Aldenhoven JT, Miller MA, Corneli PS, Shapiro MD. Phylogeography of ninespine sticklebacks (Pungitius pungitius) in North America: glacial refugia and the origins of adaptive traits. Mol Ecol. 2010;19:4061–4076. doi: 10.1111/j.1365-294X.2010.04801.x. [DOI] [PubMed] [Google Scholar]
- Bachtrog D. A dynamic view of sex chromosome evolution. Curr Opin Genet Dev. 2006;16:578–585. doi: 10.1016/j.gde.2006.10.007. [DOI] [PubMed] [Google Scholar]
- Bachtrog D. Y-chromosome evolution: emerging insights into processes of Y-chromosome degeneration. Nat Rev Genet. 2013;14:113–124. doi: 10.1038/nrg3366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachtrog D, Mank JE, Peichel CL, Kirkpatrick M, Otto SP, Ashman TL, Hahn MW, Kitano J, Mayrose I, Ming R, et al. Sex determination: why so many ways of doing it? PLoS Biol. 2014;12:e1001899. doi: 10.1371/journal.pbio.1001899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines JF, Harr B. Reduced X-linked diversity in derived populations of house mice. Genetics. 2007;175:1911–1921. doi: 10.1534/genetics.106.069419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baines JF, Sawyer SA, Hartl DL, Parsch J. Effects of X-linkage and sex-biased gene expression on the rate of adaptive protein evolution in Drosophila. Mol Biol Evol. 2008;25:1639–1650. doi: 10.1093/molbev/msn111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell MA, Stewart JD, Park PJ. The world’s oldest fossil threespine stickleback fish. Copeia. 2009;2:256–265. [Google Scholar]
- Bellott DW, Hughes JF, Skaletsky H, Brown LG, Pyntikova T, Cho TJ, Koutseva N, Zaghlul S, Graves T, Rock S, et al. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature. 2014;508:494–499. doi: 10.1038/nature13206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergero R, Charlesworth D, Filatov DA. Defining regions and rearrangements of the Silene latifolia Y chromosome. Genetics. 2008;178:2045–2053. doi: 10.1534/genetics.107.084566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergero R, Qiu S, Forrest A, Borthwick H, Charlesworth D. Expansion of the pseudo-autosomal region and ongoing recombination suppression in the Silene latifolia sex chromosomes. Genetics. 2013;194:673–686. doi: 10.1534/genetics.113.150755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruneaux M, Johnston SE, Herczeg G, Merilä J, Primmer CR, Vasemägi A. Molecular evolutionary and population genomic analysis of the nine-spined stickleback using a modified restriction-site-associated DNA tag approach. Mol Ecol. 2013;22:565–582. doi: 10.1111/j.1365-294X.2012.05749.x. [DOI] [PubMed] [Google Scholar]
- Bull JJ. Evolution of sex determining mechanisms. Menlo Park (CA): Benjamin-Cummings Publishing Company; 1983. [Google Scholar]
- Bullard JH, Purdom E, Hansen KD, Dudoit S. Evaluation of statistical methods for normalization and differential expression in mRNA-Seq experiments. BMC Bioinformatics. 2010;11:94. doi: 10.1186/1471-2105-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. Model for evolution of Y chromosomes and dosage compensation. Proc Natl Acad Sci U S A. 1978;75:5618–5622. doi: 10.1073/pnas.75.11.5618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B. The evolution of sex chromosomes. Science. 1991;251:1030–1033. doi: 10.1126/science.1998119. [DOI] [PubMed] [Google Scholar]
- Charlesworth B. The evolution of chromosomal sex determination and dosage compensation. Curr Biol. 1996;6:149–162. doi: 10.1016/s0960-9822(02)00448-7. [DOI] [PubMed] [Google Scholar]
- Charlesworth B, Charlesworth D. The degeneration of Y chromosomes. Philos Trans R Soc Lond B Biol Sci. 2000;355:1563–1572. doi: 10.1098/rstb.2000.0717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Coyne JA, Barton N. The relative rates of evolution of sex chromosomes and autosomes. Am Nat. 1987;130:113–146. [Google Scholar]
- Charlesworth D, Charlesworth B, Marais G. Steps in the evolution of heteromorphic sex chromosomes. Heredity. 2005;95:118–128. doi: 10.1038/sj.hdy.6800697. [DOI] [PubMed] [Google Scholar]
- Chen TR, Reisman HM. A comparative chromosome study of the North American species of sticklebacks (Teleostei: Gasterosteidae) Cytogenetics. 1970;9:321–332. doi: 10.1159/000130102. [DOI] [PubMed] [Google Scholar]
- Chibalina MV, Filatov DA. Plant Y chromosome degeneration is retarded by haploid purifying selection. Curr Biol. 2011;21:1475–1479. doi: 10.1016/j.cub.2011.07.045. [DOI] [PubMed] [Google Scholar]
- Cortez D, Marin R, Toledo-Flores D, Froidevaux L, Liechti A, Waters PD, Grützner F, Kaessmann H. Origins and functional evolution of Y chromosomes across mammals. Nature. 2014;508:488–493. doi: 10.1038/nature13151. [DOI] [PubMed] [Google Scholar]
- Dean R, Mank JE. The role of sex chromosomes in sexual dimorphism: discordance between molecular and phenotypic data. J Evol Biol. 2014;27:1443–1453. doi: 10.1111/jeb.12345. [DOI] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohm JC, Lottaz C, Borodina T, Himmelbauer H. Substantial biases in ultra-short read data sets from high-throughput DNA sequencing. Nucleic Acids Res. 2008;36:e105. doi: 10.1093/nar/gkn425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H. Characteristics, causes and evolutionary consequences of male-biased mutation. Proc Biol Sci. 2007;274:1–10. doi: 10.1098/rspb.2006.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellegren H. Sex-chromosome evolution: recent progress and the influence of male and female heterogamety. Nat Rev Genet. 2011;12:157–166. doi: 10.1038/nrg2948. [DOI] [PubMed] [Google Scholar]
- Ercan S, Giresi PG, Whittle CM, Zhang X, Green RD, Lieb JD. X chromosome repression by localization of the C. elegans dosage compensation machinery to sites of transcription initiation. Nat Genet. 2007;39:403–408. doi: 10.1038/ng1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filatov DA. Evolutionary history of Silene latifolia sex chromosomes revealed by genetic mapping of four genes. Genetics. 2005;170:975–979. doi: 10.1534/genetics.104.037069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filatov DA, Charlesworth D. Substitution rates in the X- and Y-linked genes of the plants, Silene latifolia and S. dioica. Mol Biol Evol. 2002;19:898–907. doi: 10.1093/oxfordjournals.molbev.a004147. [DOI] [PubMed] [Google Scholar]
- Gelbart ME, Kuroda MI. Drosophila dosage compensation: a complex voyage to the X chromosome. Development. 2009;136:1399–1410. doi: 10.1242/dev.029645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz S, García-Gómez JM, Terol J, Williams TD, Nagaraj SH, Nueda MJ, Robles M, Talón M, Dopazo J, Conesa A. High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 2008;36:3420–3435. doi: 10.1093/nar/gkn176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higuchi M, Goto A. Genetic evidence supporting the existence of two distinct species in the genus Gasterosteus around Japan. Environ Biol Fishes. 1996;47:1–16. [Google Scholar]
- Holm S. A simple sequentially rejective multiple test procedure. Scand J Stat. 1979;6:65–70. [Google Scholar]
- Hughes JF, Skaletsky H, Brown LG, Pyntikova T, Graves T, Fulton RS, Dugan S, Ding Y, Buhay CJ, Kremitzki C, et al. Strict evolutionary conservation followed rapid gene loss on human and rhesus Y chromosomes. Nature. 2012;483:82–86. doi: 10.1038/nature10843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes JF, Skaletsky H, Brown LG, Pyntikova T, Graves T, Fulton RS, Dugan S, Ding Y, Buhay CJ, Kremitzki C, et al. Chimpanzee and human Y chromosomes are remarkably divergent in structure and gene content. Nature. 2010;463:536–539. doi: 10.1038/nature08700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa A, Takeuchi N, Kusakabe M, Kume M, Mori S, Takahashi H, Kitano J. Speciation in ninespine stickleback: reproductive isolation and phenotypic divergence among cryptic species of Japanese ninespine stickleback. J Evol Biol. 2013;26:1417–1430. doi: 10.1111/jeb.12146. [DOI] [PubMed] [Google Scholar]
- Jones FC, Grabherr MG, Chan YF, Russell P, Mauceli E, Johnson J, Swofford R, Pirun M, Zody MC, White S, et al. The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 2012;484:55–61. doi: 10.1038/nature10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Julien P, Brawand D, Soumillon M, Necsulea A, Liechti A, Schütz F, Daish T, Grützner F, Kaessmann H. Mechanisms and evolutionary patterns of mammalian and avian dosage compensation. PLoS Biol. 2012;10:e1001328. doi: 10.1371/journal.pbio.1001328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara R, Miya M, Mabuchi K, Near TJ, Nishida M. Stickleback phylogenies resolved: evidence from mitochondrial genomes and 11 nuclear genes. Mol Phylogenet Evol. 2009;50:401–404. doi: 10.1016/j.ympev.2008.10.014. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitano J, Mori S, Peichel CL. Phenotypic divergence and reproductive isolation between sympatric forms of Japanese threespine sticklebacks. Biol J Linn Soc. 2007;91:671–685. [Google Scholar]
- Kitano J, Ross JA, Mori S, Kume M, Jones FC, Chan YF, Absher DM, Grimwood J, Schmutz J, Myers RM, et al. A role for a neo-sex chromosome in stickleback speciation. Nature. 2009;461:1079–1083. doi: 10.1038/nature08441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knebel S, Pasantes JJ, Thi DAD, Schaller F, Schempp W. Heterogeneity of pericentric inversions of the human Y chromosome. Cytogenet Genome Res. 2011;132:219–226. doi: 10.1159/000322080. [DOI] [PubMed] [Google Scholar]
- Kvikstad EM, Chiaromonte F, Makova KD. Ride the wavelet: a multiscale analysis of genomic contexts flanking small insertions and deletions. Genome Res. 2009;19:1153–1164. doi: 10.1101/gr.088922.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvikstad EM, Tyekucheva S, Chiaromonte F, Makova KD. A macaque’s-eye view of human insertions and deletions: differences in mechanisms. PLoS Comp Biol. 2007;3:1772–1782. doi: 10.1371/journal.pcbi.0030176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lahn BT, Page DC. Four evolutionary strata on the human X chromosome. Science. 1999;286:964–967. doi: 10.1126/science.286.5441.964. [DOI] [PubMed] [Google Scholar]
- Lange J, Noordam MJ, van Daalen SKM, Skaletsky H, Clark BA, Macville MV, Page DC, Repping S. Intrachromosomal homologous recombination between inverted amplicons on opposing Y-chromosome arms. Genomics. 2013;102:257–264. doi: 10.1016/j.ygeno.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larschan E, Bishop EP, Kharchenko PV, Core LJ, Lis JT, Park PJ, Kuroda MI. X chromosome dosage compensation via enhanced transcriptional elongation in Drosophila. Nature. 2011;471:115–118. doi: 10.1038/nature09757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leder EH, Cano JM, Leinonen T, O’Hara RB, Nikinmaa M, Primmer CR, Merilä J. Female-biased expression on the X chromosome as a key step in sex chromosome evolution in threespine sticklebacks. Mol Biol Evol. 2010;27:1495–1503. doi: 10.1093/molbev/msq031. [DOI] [PubMed] [Google Scholar]
- Lemaitre C, Braga MDV, Gautier C, Sagot MF, Tannier E, Marais GAB. Footprints of inversions at present and past pseudoautosomal boundaries in human sex chromosomes. Genome Biol Evol. 2009;1:56–66. doi: 10.1093/gbe/evp006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li WH, Yi S, Makova K. Male-driven evolution. Curr Opin Genet Dev. 2002;12:650–656. doi: 10.1016/s0959-437x(02)00354-4. [DOI] [PubMed] [Google Scholar]
- Lin F, Xing K, Zhang J, He X. Expression reduction in mammalian X chromosome evolution refutes Ohno’s hypothesis of dosage compensation. Proc Natl Acad Sci U S A. 2012;109:11752–11757. doi: 10.1073/pnas.1201816109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makanae K, Kintaka R, Makino T, Kitano H, Moriya H. Identification of dosage-sensitive genes in Saccharomyces cerevisiae using the genetic tug-of-war method. Genome Res. 2013;23:300–311. doi: 10.1101/gr.146662.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE. Sex chromosome dosage compensation: definitely not for everyone. Trends Genet. 2013;29:677–683. doi: 10.1016/j.tig.2013.07.005. [DOI] [PubMed] [Google Scholar]
- Mank JE, Axelsson E, Ellegren H. Fast-X on the Z: rapid evolution of sex-linked genes in birds. Genome Res. 2007;17:618–624. doi: 10.1101/gr.6031907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mank JE, Ellegren H. All dosage compensation is local: gene-by-gene regulation of sex-biased expression on the chicken Z chromosome. Heredity. 2009;102:312–320. doi: 10.1038/hdy.2008.116. [DOI] [PubMed] [Google Scholar]
- Mank JE, Vicoso B, Berlin S, Charlesworth B. Effective population size and the Faster-X effect: empirical results and their interpretation. Evolution. 2010;64:663–674. doi: 10.1111/j.1558-5646.2009.00853.x. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meisel RP, Connallon T. The faster-X effect: integrating theory and data. Trends Genet. 2013;29:537–544. doi: 10.1016/j.tig.2013.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muyle A, Zemp N, Deschamps C, Mousset S, Widmer A, Marais GAB. Rapid de novo evolution of X chromosome dosage compensation in Silene latifolia, a plant with young sex chromosomes. PLoS Biol. 2012;10:e1001308. doi: 10.1371/journal.pbio.1001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naurin S, Hansson B, Bensch S, Hasselquist D. Why does dosage compensation differ between XY and ZW taxa? Trends Genet. 2010;26:15–20. doi: 10.1016/j.tig.2009.11.006. [DOI] [PubMed] [Google Scholar]
- Otto SP, Pannell JR, Peichel CL, Ashman TL, Charlesworth D, Chippindale AK, Delph LF, Guerrero RF, Scarpino SV, McAllister BF. About PAR: the distinct evolutionary dynamics of the pseudoautosomal region. Trends Genet. 2011;27:358–367. doi: 10.1016/j.tig.2011.05.001. [DOI] [PubMed] [Google Scholar]
- Papp B, Pál C, Hurst LD. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424:194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- Peichel CL, Ross JA, Matson CK, Dickson M, Grimwood J, Schmutz J, Myers RM, Mori S, Schluter D, Kingsley DM. The master sex-determination locus in threespine sticklebacks is on a nascent Y chromosome. Curr Biol. 2004;14:1416–1424. doi: 10.1016/j.cub.2004.08.030. [DOI] [PubMed] [Google Scholar]
- Pessia E, Makino T, Bailly-Bechet M, McLysaght A, Marais GAB. Mammalian X chromosome inactivation evolved as a dosage-compensation mechanism for dosage-sensitive genes on the X chromosome. Proc Natl Acad Sci U S A. 2012;109:5346–5351. doi: 10.1073/pnas.1116763109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautenberg A, Hathaway L, Oxelman B, Prentice HC. Geographic and phylogenetic patterns in Silene section Melandrium (Caryophyllaceae) as inferred from chloroplast and nuclear DNA sequences. Mol Phylogenet Evol. 2010;57:978–991. doi: 10.1016/j.ympev.2010.08.003. [DOI] [PubMed] [Google Scholar]
- Rice WR. Sex chromosomes and the evolution of sexual dimorphism. Evolution. 1984;38:735–742. doi: 10.1111/j.1558-5646.1984.tb00346.x. [DOI] [PubMed] [Google Scholar]
- Rice WR. The accumulation of sexually antagonistic genes as a selective agent promoting the evolution of reduced recombination between primitive sex chromosomes. Evolution. 1987a;41:911–914. doi: 10.1111/j.1558-5646.1987.tb05864.x. [DOI] [PubMed] [Google Scholar]
- Rice WR. Genetic hitchhiking and the evolution of reduced genetic activity of the Y sex chromosome. Genetics. 1987b;116:161–167. doi: 10.1093/genetics/116.1.161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts A, Pimentel H, Trapnell C, Pachter L. Identification of novel transcripts in annotated genomes using RNA-Seq. Bioinformatics. 2011;27:2325–2329. doi: 10.1093/bioinformatics/btr355. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesti M, Moser D, Berner D. Recombination in the threespine stickleback genome-patterns and consequences. Mol Ecol. 2013;22:3014–3027. doi: 10.1111/mec.12322. [DOI] [PubMed] [Google Scholar]
- Ross JA, Peichel CL. Molecular cytogenetic evidence of rearrangements on the Y chromosome of the threespine stickleback fish. Genetics. 2008;179:2173–2182. doi: 10.1534/genetics.108.088559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross JA, Urton JR, Boland J, Shapiro MD, Peichel CL. Turnover of sex chromosomes in the stickleback fishes (Gasterosteidae) PLoS Genet. 2009;5:e1000391. doi: 10.1371/journal.pgen.1000391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross MT, Grafham DV, Coffey AJ, Scherer S, McLay K, Muzny D, Platzer M, Howell GR, Burrows C, Bird CP, et al. The DNA sequence of the human X chromosome. Nature. 2005;434:325–337. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultheiss R, Viitaniemi HM, Leder EH. Spatial dynamics of evolving dosage compensation in a young sex chromosome system. Genome Biol Evol. 2015;7:581–590. doi: 10.1093/gbe/evv013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti I, Delph LF. Selective trade-offs and sex-chromosome evolution in Silene latifolia. Evolution. 2006;60:1793–1800. [PubMed] [Google Scholar]
- Shapiro MD, Summers BR, Balabhadra S, Aldenhoven JT, Miller AL, Cunningham CB, Bell MA, Kingsley DM. The genetic architecture of skeletal convergence and sex determination in ninespine sticklebacks. Curr Biol. 2009;19:1140–1145. doi: 10.1016/j.cub.2009.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimmin LC, Chang BH, Li WH. Male-driven evolution of DNA sequences. Nature. 1993;362:745–747. doi: 10.1038/362745a0. [DOI] [PubMed] [Google Scholar]
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntikova T, Ali J, Bieri T, et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–837. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- Urton JR, McCann SR, Peichel CL. Karyotype differentiation between two stickleback species (Gasterosteidae) Cytogenet Genome Res. 2011;135:150–159. doi: 10.1159/000331232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Charlesworth B. Evolution on the X chromosome: unusual patterns and processes. Nat Rev Genet. 2006;7:645–653. doi: 10.1038/nrg1914. [DOI] [PubMed] [Google Scholar]
- Vicoso B, Emerson JJ, Zektser Y, Mahajan S, Bachtrog D. Comparative sex chromosome genomics in snakes: differentiation, evolutionary strata, and lack of global dosage compensation. PLoS Biol. 2013;11:e1001643. doi: 10.1371/journal.pbio.1001643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicoso B, Kaiser VB, Bachtrog D. Sex-biased gene expression at homomorphic sex chromosomes in emus and its implication for sex chromosome evolution. Proc Natl Acad Sci U S A. 2013;110:6453–6458. doi: 10.1073/pnas.1217027110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Na JK, Yu Q, Gschwend AR, Han J, Zeng F, Aryal R, VanBuren R, Murray JE, Zhang W, et al. Sequencing papaya X and Yh chromosomes reveals molecular basis of incipient sex chromosome evolution. Proc Natl Acad Sci U S A. 2012;109:13710–13715. doi: 10.1073/pnas.1207833109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson Sayres MA, Makova KD. Genome analyses substantiate male mutation bias in many species. Bioessays. 2011;33:938–945. doi: 10.1002/bies.201100091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright AE, Harrison PW, Montgomery SH, Pointer MA, Mank JE. Independent stratum formation on the avian sex chromosomes reveals inter-chromosomal gene conversion and predominance of purifying selection on the W chromosome. Evolution. 2014;68:3281–3295. doi: 10.1111/evo.12493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yoshida K, Makino T, Yamaguchi K, Shigenobu S, Hasebe M, Kawata M, Kume M, Mori S, Peichel CL, Toyoda A, et al. Sex chromosome turnover contributes to genomic divergence between incipient stickleback species. PLoS Genet. 2014;10:e1004223. doi: 10.1371/journal.pgen.1004223. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.