Abstract
Nicotine dependence (ND) is a heterogeneous phenotype with complex genetic influences that may vary across ethnicities. The use of intermediate phenotypes may clarify genetic influences and reveal specific etiological pathways. Prior work in European Americans has found that the four Primary Dependence Motives (PDM) subscales (Automaticity, Craving, Loss of Control, and Tolerance) of the Wisconsin Inventory of Smoking Motives represent core features of nicotine dependence and are promising intermediate phenotypes for understanding genetic pathways to ND. However, no studies have examined PDM as an intermediate phenotype in African American smokers, an ethnic population that displays unique patterns of smoking and genetic variation. In the current study, 268 African American daily smokers completed a phenotypic assessment and provided a sample of DNA. Associations among haplotypes in the NCAM1-TTC12-ANKK1-DRD2 gene cluster, a dopamine-related gene region associated with ND, PDM intermediate phenotypes, and ND were examined. Dopamine-related genetic variation in the DBH and COMT genes was also considered on an exploratory basis. Mediational analysis was used to test the indirect pathway from genetic variation to smoking motives to nicotine dependence. NCAM1-TTC12-ANKK1-DRD2 region variation was significantly associated with the Automaticity subscale and, further, Automaticity significantly mediated associations among NCAM1-TTC12-ANKK1-DRD2 cluster variants and ND. DBH was also significantly associated with Automaticity, Craving, and Tolerance; Automaticity and Tolerance also served as mediators of the DBH-ND relationship. These results suggest that PDM, Automaticity in particular, may be a viable intermediate phenotype for understanding dopamine-related genetic influences on ND in African American smokers. Findings support a model in which putatively dopaminergic variants exert influence on ND through an effect on patterns of automatic routinized smoking.
Keywords: nicotine, intermediate phenotype, dopamine, haplotype, SNP, motives, African American
INTRODUCTION
Although African Americans (AAs) initiate smoking later than European Americans (EAs), AA smokers display a greater persistence of smoking into mid-adulthood, lower cessation rates, and higher mortality rates from tobacco-associated diseases (Kandel et al., 2011, Safford et al., 2012). Genetic factors contribute substantially to nicotine dependence (ND; Goldman et al., 2005, MacKillop et al., 2010), with the preponderance of molecular genetic evidence coming from studies focusing on EAs (Munafò et al., 2004, Munafò et al., 2009). Although many non-genetic factors contribute to observed EA/AA differences in smoking (e.g. Primack et al., 2007), examinations of genetic influences in AA smokers are critical due to the distinct phenotypic and genotypic patterns in this population. In addition to the potential for unique etiological influences within AA smokers, AA smokers represent an ethnic subgroup known for its greater polymorphic variation and associated shorter haplotypes (Gabriel et al., 2002). Thus, investigating genetic influences on smoking phenotypes in AAs may broadly inform etiological studies of smoking by providing data that narrows genetic findings from large regions of interest to more specific susceptibility loci.
NCAM1-TTC12-ANKK1-DRD2 variants became a focus of smoking molecular genetics due to the critical role of dopamine D2 receptors (DRD2) in nicotine pharmacodynamics (Benowitz, 2010). This chromosome 11q23 gene cluster has been associated with ND in studies using both genome-wide and candidate approaches (Bergen et al., 2009, Ducci et al., 2011, Laucht et al., 2008, Morley et al., 2006, Saccone et al., 2007), including a handful in AAs (David et al., 2010, Gelernter et al., 2006, Huang et al., 2009). Although meta-analyses support a role for DRD2 in smoking risk (Li et al., 2004, Munafò et al., 2004), heterogeneity of effects exists across studies and meta-analytic reports call for additional studies using non-EA participants. Due to their role in dopamine pathway, also of interest are catechol-O-methyltransferase (COMT; located on chromosome 22q11) (Akil et al., 2003), involved in dopamine degradation, and dopamine beta hydroxylase (DBH; located on chromosome 9q34), involved in converting dopamine to norepinephrine (Cubells and Zabetian, 2004). Although effects of variation of these genes on smoking have been inconsistent (Han et al., 2008, McKinney et al., 2000, Shiels et al., 2008, Ton et al., 2007), studies in AAs are limited and there is suggestion of ethnic-specificity of effects (Beuten et al., 2006, Colilla et al., 2005).
Further, genetic influences may be clarified by using intermediate/mechanistic phenotypes that are putatively more narrow and more proximal to the differences in genetic variation (NCI, 2009, MacKillop and Munafò, 2013). This strategy is intended to reveal larger effect sizes, clarify mechanisms of genetic risk and/or protection, and identify a more homogeneous group of smokers who may share a particular genetically-mediated vulnerability to ND. Preliminary work in EAs has shown the four primary dependence motives (PDM) subscales of the Wisconsin Inventory of Smoking Motives (WISDM-68) Automaticity, Craving, Loss of Control, and Tolerance (Piasecki 2010 and Piper et al 2008) are viable intermediate phenotypes that can explicate genetic mechanisms of dependence. For example, neuronal cholinergic receptor (CHRNA5-A3-B5) haplotypes were associated with PDM subscales in early onset smokers (Baker et al., 2009). Our recent work in EAs has shown that the PDM subscales are a mediator of the association with NCAM1-TTC12-ANKK1-DRD2 haplotype variation and ND, and thus support their role as viable intermediate phenotypes that can explicate pathways between genetic risk and dependence (Bidwell et al., 2015). This work suggested that, rather than PDM and FTND being alternative manifestations of the clinical ND phenotype without evidence an indirect effect, the PDM motivational intermediate phenotypes serve as a mediator along an etiological pathway that explains the association between these risk loci and ND in EAs. Within the context of the aforementioned differences in smoking topography of AAs and unique genetic variability, it is important to attempt to replicate our previous findings on EAs to AAs. In this way, studies that employ formal mediation analyses can connect established genotype–ND relationships empirically as credible mechanisms by which genetic variation exerts influence on clinical dependence phenotypes across ethnic groups.
Current Study
Thus, given the need for studies that examine the intersection of smoking intermediate phenotypes, biologically-implicated candidate genes, and nicotine dependence in AAs, we examined WISDM motivational profiles as intermediate phenotypes for ND in an AA sample. Based on limited prior work on the WISDM in AAs, we did not expect phenotypic motivational differences based on race. Modeling our approach after our prior work in EAs (Bidwell et al., 2015), we tested DRD2-ANKK1-TTC12-NCAM1 haplotypes in association with both clinical dependence and PDM phenotypes. Single nucleotide variation in COMT and DBH were also considered on an exploratory basis. We then used mediation to evaluate mechanistic pathways testing whether PDM subscales were significant mediators of the genotype-ND relationship.
MATERIALS & METHODS
Sample Description
Participants (N=268; 57% [N=153] males) were recruited via newspaper, Internet, and flyer advertising as part of a larger study of behavioral economics and smoking (MacKillop et al., 2012). Inclusion criteria were ≥18 years old, ≥5 cigarettes/day, and ≥8th grade education. The full sample consisted of 1,124 participants enrolled across three sites: Providence, RI; Athens, GA; and Aiken, SC. All study procedures were approved by the Brown University and University of Georgia Institutional Review Boards. The current study examines data from the subset of 268 African American (AA) daily smokers who provided both DNA and self-report smoking data. As expected, FTND was significantly higher in AA than the 734 EA participants (t = 5.13, p <.0001; e.g. Luo et al., 2008), but there were no significant differences on PDM subscales by ethnicity. Of the AA sample, 74% (n = 198) stated a preference for menthol cigarettes, compared to 29% (n = 212) of the EA sample (χ2 = 168.1, p <.0001). Demographic, smoking, and alcohol use characteristics are listed in Table 1.
Table 1.
Sample demographics, smoking, and other substance use (N = 268)
| Sample characteristics | Mean | SD |
|---|---|---|
| Age | 37.3 | 12.6 |
| Years education | 11.8 | 2.0 |
| Number of daily cigarettes | 16.9 | 12.9 |
| Age smoked first cigarette | 14.8 | 3.9 |
| AUDIT Total | 9.0 | 8.0 |
Note. AUDIT = Alcohol Use Disorders Test
Phenotypic Measures
Fagerström Test of Nicotine Dependence (FTND; Heatherton et al., 1991)
The FTND measures the severity of ND using six-items. Two of the items are reversed scored and higher FTND reflects greater dependence on nicotine.
WISDM PDM
The 68-item WISDM-68 (Piper et al., 2004) was used to assess the four primary dependence motives (PDM; Automaticity, Craving, Locus of Control, and Tolerance) reflecting heavy, pervasive smoking (Piasecki et al., 2010, Piper et al., 2008). An index score for each of the four PDM was created by averaging the items within each subscale: Automaticity (e.g. “I often smoke without thinking about it.”), Craving (e.g. “I frequently crave cigarettes.”), Loss of Control (LOC) (e.g. “Cigarettes control me.”), and Tolerance (e.g. “I can only go a couple hours between cigarettes.”). The means and distributions for each subscale are listed in Table 2.
Table 2.
Means, standard deviations, and bivariate correlations for smoking phenotypes.
| Correlations among smoking measures (r) |
||||||
|---|---|---|---|---|---|---|
| Mean | SD | FTND | Automaticity | Craving | Locus of Control | |
| FTND | 4.9 | 2.1 | - | |||
| WISDM-Automaticity | 4.0 | 1.6 | 0.40 | - | ||
| WISDM-Craving | 4.1 | 1.6 | 0.39 | 0.69 | - | |
| WISDM-Locus of Control | 3.5 | 1.6 | 0.41 | 0.63 | 0.70 | - |
| WISDM-Tolerance | 4.4 | 1.6 | 0.55 | 0.69 | 0.74 | 0.62 |
Note. FTND= Fagerström Test of Nicotine Dependence; WISDM = Wisconsin Inventory of Smoking Motives; all correlations significant at p <.001.
Marker Information and Haplotype Derivation
Genotyping and SNP selection
This study examined 13 markers across the NCAM1-TTC12-ANKK1-DRD2 candidate gene region and two exploratory markers in the COMT and DBH genes. HapMap was used to determine the tag SNPs required to capture >80% of the variance within the NCAM1-TTC12-ANKK1-DRD2 gene cluster and these tag SNPs were augmented with loci that had been implicated in prior studies of this region and nicotine dependence (e.g. Gelernter et al., 2006). Ethanol precipitation was used to extract DNA from collected saliva samples. Samples were genotyped using a MassEXTEND Sequenom assay based on the annealing of an oligonucleotide primer adjacent to the SNP of interest. The assay was performed in multiplex with 20 reactions in a single well; 20% of all samples were randomly run in duplicate resulting in a genotyping error rate of 0.02%. Primer sequences are available in Supplementary Table 1. Genotypes were determined by investigators blinded to phenotypic data.
Frequencies of genotypes/alleles and Hardy-Weinberg Equilibrium (HWE) p-values for each marker are listed in Table 3. One marker (rs4938012) was excluded from subsequent analyses due to greater than 15% missing genotypes and another was excluded due to HW failure (rs1799732).
Table 3.
Genotype and minor allele frequencies of dopamine-related loci.
| Polymorphism | Genotypes N (%) | Allele Frequency | HWE p-value | |||
|---|---|---|---|---|---|---|
| NCAM1-TTC12-ANKK1-DRD2 region loci | ||||||
| NCAM1 | ||||||
| rs618114 Frequency |
GG 156 (58.2) |
GA / AG 85 (31.7) |
AA 8 (3.0) |
G 0.81 |
A 0.19 |
0.44 |
| TTC12 | ||||||
| rs2303380 Frequency |
GG 24 (9.0) |
GA / AG 109 (40.7) |
AA 124 (46.3) |
G 0.29 |
A 0.71 |
0.99 |
| rs2282511 Frequency |
CC 155 (57.8) |
CA / AC 78 (29.1) |
AA 21 (7.8) |
C 0.78 |
A 0.22 |
0.03 |
| ANKK1 | ||||||
| rs877138 Frequency |
GG 18 (6.7) |
GA / AG 77 (28.7) |
AA 164 (61.2) |
G 0.21 |
A 0.79 |
0.06 |
| rs17115439 Frequency |
TT 99 (36.9) |
TC / CT 107 (39.9) |
CC 51 (19.0) |
T 0.61 |
C 0.39 |
0.04 |
| rs4938013 Frequency |
CC 165 (61.6) |
CA / AC 78 (29.1) |
AA 18 (6.7) |
C 0.79 |
A 0.21 |
0.05 |
| rs4938015 Frequency |
TT 101 (37.7) |
TC / CT 107 (39.9) |
CC 51 (19.0) |
T 0.61 |
C 0.39 |
0.02 |
| rs11604671 Frequency |
GG 203 (75.7) |
GA / AG 51 (19.0) |
AA 51 (19.0) |
G 0.90 |
A 0.10 |
0.33 |
| DRD2 | ||||||
| rs1079597 Frequency |
GG 190 (70.9) |
GA / AG 63 (23.5) |
AA 8 (3.0) |
G 0.85 |
A 0.15 |
0.31 |
| rs6277 Frequency |
TT 3 (1.1) |
TC / CT 51 (19.0) |
CC 207 (77.2) |
T 0.11 |
C 0.89 |
1 |
| rs1800497 Frequency |
TT 34 (12.7) |
TC / CT 112 (41.8) |
CC 111 (41.4) |
T 0.34 |
C 0.66 |
0.24 |
| rs1799732 Frequency |
Del/Del 56 (20.9) |
C.Del/Del.C 86 (32.1) |
CC 94 (35.1) |
Del 0.37 |
C 0.63 |
3.59 E-05 |
| Exploratory loci | ||||||
| COMT | ||||||
| rs4680 Frequency |
GG 121 (45.1) |
GA / AG 110 (41.0) |
AA 25 (9.3) |
G 0.70 |
A 0.30 |
.71 |
| DBH | ||||||
| rs1611115 Frequency |
TT 5 (1.9) |
TC / CT 76 (28.4) |
CC 178 (66.4) |
T 0.16 |
C 0.84 |
.39 |
Note. Table shows the genotypes and frequencies for each marker (in order of chromosomal location). All loci are single nucleotide polymorphisms except for rs1799732 which is an InDel polymorphism. Proportions do not sum to 100 to allow for the accurate depiction of missingness in the data.
Haplotype derivation
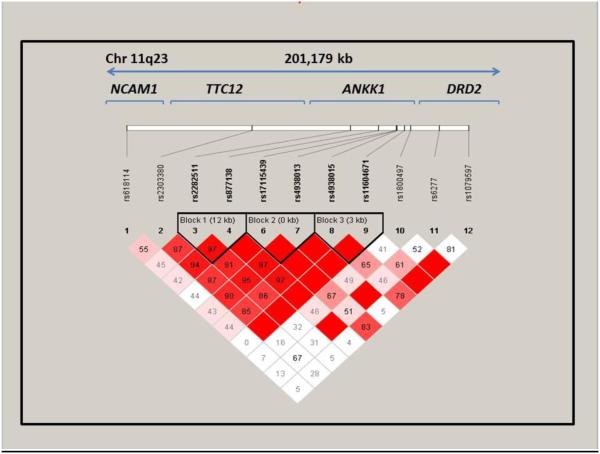
Haploview was used to visualize haplotype blocks (i.e., the combinations of SNP/InDel markers that are statistically associated) using all available polymorphic data (Barrett, 2009, Barrett et al., 2005). As expected, greater variation with more, but shorter, haplotype blocks were observed than in our prior paper of EAs (Bidwell et al., 2015). Three haplotype blocks were observed (Figure 1): Block 1 was based on rs2282511 (TTC12) and rs877138 (ANKK1); Block 2 on rs17115439 (ANKK1), rs4938013 (ANKK1); and Block 3 on rs4938015 (ANKK1); rs11604671 (ANKK1).
Figure 1.
Marker-to-marker D’ values for the NCAM1-TTC12 -ANKK1-DRD2 region polymorphisms. D’ varies between 0 and 1 describes the extent of linkage disequilibrium, a measure of interdependency between genetic loci. A value of 0 for D' suggests that the examined polymorphisms are independent of one another, while a value of 1 suggests that the polymorphisms provide redundant information. Numbers in the boxes are shown as Haploview output.as whole numbers, but reflect D' correlations that do not exceed 1 (e.g., 91 = .91); an empty box with no numerical value represents D' of 1.
Haplotype blocks were then confirmed and extracted using PHASE [Version 2.1] (Stephens and Donnelly, 2003, Stephens and Scheet, 2005, Stephens et al., 2001), with the requirement that the probability of a phased haplotype had to be greater than or equal to 0.80 (Oroszi et al., 2009). Diplotypes (i.e., combination of haplotypes across the pair of homologous chromosomes) were then constructed for use in the regression analyses. Table 4 describes the haplotypes and their frequencies in the sample population. Because of the limited and inconsistent literature indicating a putative risk allele at our candidate loci, haplotype, and thus diplotype, scores were created assuming an additive effect for each of the identified haplotypes (Lu et al., 2006, Pajewski et al., 2011). This additive scoring scheme where an individual may possess 0, 1, or 2 copies of each haplotype observed was utilized for every haplotype that was more frequent than .20 in the study sample. Less frequent haplotypes were excluded from further analysis.
Table 4.
NCAM1-TTC12-ANKK1-DRD2 region haplotypes and frequencies.
| Block 1 | Population Frequency |
N(%) of Carrying 0, 1, or 2 Copies | |||
|
|
|||||
| rs2282511 | rs877138 | 0 | 1 | 2 | |
|
| |||||
| C | A | 0.77 | 22 (8.2) | 78 (29.1) | 159 (59.3) |
| A | G | 0.21 | 164 (61.2) | 78 (29.1) | 17 (6.3) |
|
Block 2
|
|||||
| rs17115439 | rs4938013 | ||||
|
| |||||
| T | C | 0.38 | 108 (40.3) | 105 (39.2) | 43 (16.0) |
| T | A | 0.21 | 162 (60.4) | 76 (28.4) | 18 (6.7) |
| C | C | 0.41 | 98 (36.6) | 107 (39.9) | 51 (19.0) |
|
Block 3
|
|||||
| rs4938015 | rs11604671 | ||||
|
| |||||
| T | G | 0.59 | 50 (18.7) | 104 (38.8) | 101 (37.7) |
| C | G | 0.31 | 133 (49.6) | 93 (34.7) | 29 (10.8) |
Note. Table shows haplotype blocks and estimated population frequencies for haplotypes extracted from participants at a probability of at least 80% using PHASE. Proportions do not sum to 100 to allow for the accurate depiction of missingness in the data.
Statistical Analyses
Analyses were executed in PLINK v1.07 (Purcell, http://pngu.mgh.harvard.edu/purcell/plink/, Purcell et al., 2007) and SPSS 19.0 (IBM, Released 2010.). Smoking phenotypes were initially examined for outliers (using standard scores, criterion Z=3.29) and for distribution normality with no observed violations (Tabachnick and Fidell, 2001). Bivariate correlations among smoking phenotypes were examined. Univariate linear regressions were used to test the main effects of genetic variation on smoking phenotypes, using the observed haplotypes as the primary tests and following up with test of the individual loci that did not fall into haplotype blocks. Separate models were run for each smoking measure. Finally, where initial regression analyses determined a significant main effect of a genetic variant on a PDM subscale, we subsequently used selected mediation to test the extent to which the genetic effect (IV) exerts its influence on FTND (DV) through the PDM (mediator). Testing of indirect effects was not conditional on the presence of a statistically significant direct effect from the genotype/haplotype to FTND (i.e., the direct A→C path) because it is not required for the indirect test and several scenarios can give rise to a nonsignificant A→C relationship (for a review, see MacKinnon & Fairchild, 2009). Mediation was tested using the products of the coefficients method in SPSS 19.0 (Preacher and Hayes, 2008). Bias corrected 95% confidence intervals of the indirect effect were estimated using bootstrapping methods (Preacher and Hayes, 2008). Given the relatively large number of phenotypes tested in this preliminary investigation, we adopted a significance threshold p < .01 for all analyses; p values of < .05 were considered significant at trend level.
RESULTS
Correlations among Smoking Measures
As expected, the smoking motives were each significantly correlated with each other and FTND (all p’s < .001; Table 2).
Main Effects of Genetic Variation on Smoking Phenotypes
Table 5 presents the results of linear regression models testing association among the genetic variants (3 NCAM1-TTC12-ANKK1-DRD2 haplotype blocks and 7 individual polymorphisms) and smoking phenotypes (FTND and PDM). The direction of the regression coefficient represents the effect of each extra minor allele (i.e., a negative regression coefficient means that the minor allele decreases risk/phenotype mean). For Block 1, the CA haplotype was significantly associated with higher FTND (R2 = .02, p < .01) and higher Automaticity (R2 = .02, p < .01). In the same block, the AG haplotype was associated at trend level with lower FTND (R2 = .02, p < .05) and lower Automaticity (R2 = .02, p < .05). For Block 2, the TA haplotype was associated at trend level with lower FTND (R2 = .02, p < .05) and lower Automaticity (R2 = .02, p < .05). None of the Block 3 haplotypes were associated with the smoking phenotypes. None of the NCAM1-TTC12-ANKK1-DRD2 loci were significantly associated with FTND or PDM phenotypes, but two were associated at trend level with lower Automaticity, including loci from TTC12 [rs2303380 (G)] and DRD2 [rs1079597 (A)] (p’s < .05). In addition, the T allele of rs1611115 in DBH was associated with higher levels of each of the three of the four PDM intermediate phenotypes, Automaticity, Craving, and Tolerance (p’s < .01) and at trend level with FTND and the LOC subscale (p’s < .05). Each of the significantly associated haplotype or loci variant accounted for 2-3% of the variance in the phenotype (R2‘s range from .018 to .032).
Table 5.
Unstandardized estimates from main effects models predicting FTND and WISDM intermediate phenotypes from genetic variants
|
|
WISDM Subscales |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FTND |
Automaticity |
Craving |
Locus of Control |
Tolerance |
||||||||
| β | SE | β | SE | β | SE | β | SE | β | SE | |||
| Haplotype Block 1 [rs2282511 (TTC12), rs877138 (ANKK1)] | ||||||||||||
| CA | 0.49* | 0.20 | 0.38* | 0.01 | 0.22 | 0.16 | 0.23 | 0.15 | 0.22 | 0.15 | ||
| AG | −0.50† | 0.21 | −0.34† | 0.16 | −0.23 | 0.17 | −0.23 | 0.16 | −0.20 | 0.16 | ||
| Haplotype Block 2 [rs17115439 (ANKK1) and rs4938013 (ANKK1) | ||||||||||||
| TC | 0.23 | 0.18 | 0.07 | 0.14 | 0.02 | 0.14 | 0.10 | 0.14 | −0.03 | 0.14 | ||
| TA | −0.45† | 0.21 | −0.34† | 0.16 | −0.20 | 0.17 | −0.22 | 0.16 | −0.19 | 0.16 | ||
| CC | 0.10 | 0.18 | 0.17 | 0.13 | 0.12 | 0.14 | 0.07 | 0.13 | 0.16 | 0.13 | ||
| Haplotype Block 3 [rs4938015 (ANKK1) and rs11604671 (ANKK1)] | ||||||||||||
| TG | −0.16 | 0.18 | −0.19 | 0.13 | −0.14 | 0.14 | −0.06 | 0.13 | −0.18 | 0.13 | ||
| CG | 0.09 | 0.19 | 0.17 | 0.15 | 0.14 | 0.15 | 0.01 | 0.14 | 0.20 | 0.14 | ||
| Polymorphism | Gene | MA | ||||||||||
| rs618114 | NCAM1 | A | −0.07 | 0.00 | −0.16 | 0.18 | 0.03 | 0.19 | 0.04 | 0.18 | −0.07 | 0.18 |
| rs2303380 | TTC12 | G | −0.30 | 0.01 | −0.32† | 0.15 | −0.16 | 0.16 | −0.05 | 0.15 | −0.15 | 0.15 |
| rs1079597 | DRD2 | A | −0.07 | 0.00 | −0.39† | 0.19 | −0.25 | 0.20 | −0.13 | 0.19 | −0.29 | 0.18 |
| rs6277 | DRD2 | T | 0.02 | 0.00 | −0.12 | 0.22 | −0.18 | 0.23 | −0.12 | 0.22 | −0.04 | 0.22 |
| rs1800497 | DRD2 | T | 0.30 | 0.01 | −0.19 | 0.14 | −0.18 | 0.15 | −0.04 | 0.14 | −0.23 | 0.14 |
| rs4680 | COMT | A | −0.02 | 0.20 | −0.17 | 0.15 | −0.13 | 0.16 | −0.04 | 0.15 | −0.07 | 0.15 |
| rs1611115 | DBH | T | 0.59† | 0.25 | 0.55* | 0.19 | 0.52* | 0.20 | 0.41† | 0.19 | 0.49* | 0.19 |
Note. FTND=Fagerstrom Test of Nicotine Dependence; WISDM = Wisconsin Inventory of Smoking Motives; COMT = Catechol-O-methyltransferase gene; DBH = Dopamine beta-hydroxylase gene; ANKK1 = Ankyrin repeat and kinase domain containing 1 gene; DRD2 = Dopamine D2 receptor gene; NCAM1 = Neural cell adhesion molecule 1 gene; TTC12 = Tetratricopeptide Repeat Domain 12 gene. Block 1 = rs2282511 (TTC12), rs877138 (ANKK1); Block 2 = rs17115439 (ANKK1) and rs4938013 (ANKK1); Block 3 rs4938015 (ANKK1) and rs11604671 (ANKK1).
p < .01,
p < .05.
Indirect Effects of Genetic Variation through Smoking Motives
Table 6 reports the results of the indirect effects for the selected mediation tests. When mediation was formally tested, nearly all of the genetic variants that were significantly associated with a PDM subscale exerted a significant indirect effect on FTND through the requisite PDM intermediate phenotype. Specifically, significant mediation was found for rs1611115 of DBH on higher FTND through increasing scores on both Automaticity and Tolerance (p’s <.01) and a trend level mediation effect on higher FTND was found for Craving (p = .02). With regard to NCAM1-TTC12-ANKK1-DRD2 gene cluster, there was a significant indirect effect in the direction of increased risk of the Block 1 AC haplotype on FTND through Automaticity.
Table 6.
Tests of the indirect effects of the genotypes/haplotypes associated with smoking motives in relation to nicotine dependence.
| Genetic Variant | Mediator | β | 95% BC CI | Z | p |
|---|---|---|---|---|---|
| Polymorphism | |||||
| rs1611115 | Automaticity | 0.29 | 0.10-0.51 | 2.71 | <.01 |
| rs1611115 | Craving | 0.24 | 0.06-0.46 | 2.40 | 0.02 |
| rs1611115 | Tolerance | 0.36 | 0.08-0.64 | 2.51 | 0.01 |
| Haplotype | |||||
| Block 1 CA | Automaticity | 0.21 | 0.06- 0.39 | 2.45 | 0.01 |
Note. BC CI = Bias Corrected Confidence Intervals. Block 1 = rs2282511 (TTC12) and rs877138 (ANKK1).
DISCUSSION
Albeit preliminary, these data provide novel support for associations among smoking motives and common genetic variation within the NCAM1-TTC12-ANKK1-DRD2 gene-cluster and the DBH gene in an AA sample. With regard to the NCAM1-TTC12-ANKK1-DRD2 gene-cluster, analyses revealed significant associations between variation in haplotype block 1 (consisting of rs2282511 in TTC12 and rs877138 in ANKK1) and Automaticity motives. Trend level findings were present for block 2 (consisting of rs17115439 in ANKK1 and rs4938013 in ANKK1) with FTND and Automaticity as well as two additional independent SNPs (rs2303380 in TTC12 and rs1079597 in DRD2) with Automaticity. Each of these NCAM1-TTC12-ANKK1-DRD2 cluster variants demonstrated significant association with Automaticity in our prior study in a large sample of EAs. In contrast, block 3 (consisting of rs4938015 and rs11604671 both in ANKK1) was not associated with any smoking phenotype in the current study, whereas rs4938015 had been significantly associated with Automaticity in our prior study of EA smokers (significant both at an independent SNP level and as part of a five SNP haplotype block). Thus, the strategy of combining haplotype with individual loci analyses sheds light on the potential role of specific individual variants, while simultaneously accounting for LD and genetic background across the region. Further, examining associations and variation across the region specifically in a subsample of AA smokers revealed shorter haplotype blocks and more specific associations that may allow for narrowing of the susceptibility loci in the region. DBH and COMT SNPs were considered on an exploratory basis. The T allele at rs161115 of the DBH gene was significantly associated with three of the four PDM subscales, Automaticity, Craving, and Tolerance, and at trend level with ND and the Locus of Control subscale. There were no associations with any smoking phenotypes and COMT.
Mediation analyses supported a significant indirect effect via PDM of both NCAM1-TTC12-ANKK1-DRD2 cluster (Automaticity subscale) and DBH (Automaticity and Tolerance subscales) genetic effects on ND. Findings of a significant indirect effect via PDM is suggestive that, rather than these smoking motives being an alternative manifestation of the clinical ND phenotype, PDM serve as a mechanism along the causal pathway from genetic risk to clinical dependence. Thus, DBH variation, linked to ND via the correlated Automaticity, Craving, and Tolerance subscales, may increase the likelihood that a person will become dependent via increased craving, tolerance, and automatic smoking. NCAM1-TTC12-ANKK1-DRD2 variants showed a particular risk pathway related to Automaticity, potentially exerting their effect through a more narrow pattern of automatic smoking behaviors that can be elicited with little awareness. These findings are consistent with our data in EA smokers demonstrating associations among NCAM1-TTC12-ANKK1-DRD2 variation and the PDM intermediate phenotypes, as well as a mechanistic role for PDM phenotypes in risk for ND (Bidwell et al., 2015). Taken together, findings support the use of smoking motives as viable intermediate phenotypes for ND in AA populations and suggest that genetic studies of the complex ND phenotype may benefit from focusing on more discrete smoking behaviors like motivational profiles.
Our results are consistent with prior AA studies associating variation in the NCAM1-TTC12-ANKK1-DRD2 gene cluster with ND (David et al., 2010, Gelernter et al., 2006, Huang et al., 2009). Several of the SNPs suggested here to be part of a pathway to ND via Automaticity motives have been associated with ND in prior studies using AA samples, including rs2303380 (TTC12), rs2282511 (TTC12), rs877138 (ANKK1) and rs4938013 (ANKK1) (David et al., 2010, Gelernter et al., 2006). DRD2’s rs1079597 has previously been associated with ND and the severity of self-reported smoking withdrawal (Gelernter et al., 2006, Robinson et al., 2007); however, these prior associations were present in primarily EA or pooled samples. Thus, across studies using both EA and AA samples data are broadly supportive that a substantive susceptibility locus for ND is located in this region, although the specific location or structure of this vulnerability may differ across ethnicities.
As far as we know, this is the first report of an association with rs161115 DBH SNP and smoking phenotypes in an exclusively AA sample. Prior studies demonstrating associations among DBH variants and smoking phenotypes have almost exclusively focused on EA samples (e.g. Tobacco and Genetics Consortium, 2010, Freire et al., 2006, Johnstone et al., 2002, Siedlinski et al., 2011). In our study, the low activity T allele at this locus was associated significantly or at trend level with increased levels of FTND and each of the four PDM; further, Automaticity and Tolerance intermediate phenotypes were a risk mechanism linking the T allele to increased ND severity. Given these associations and the associations among the specific PDM, the most cogent interpretation appears to be that this SNP confers risk via higher PDM for smoking, not a particular subset of motives. This putatively functional variant, located upstream of the DBH translational start site, accounts for between 1/3 to 1/2 of the variation in plasma-DBH activity in both EA and AA samples (Zabetian et al., 2001). Although in that study genotypic relationships with DBH enzyme activity values were broadly similar across ethnicities, some profile differences between EA and AA subsamples were reported, particularly in the prevalence of very low activity individuals. Thus, studies should continue to examine the role of functional DBH variants in ND and test intermediate phenotypic mechanisms in samples stratified by ethnicity in order to understand if risk mechanisms are operating similarly across various populations.
Our results are consistent with some prior work indicating a lack of association among smoking cessation phenotypes and COMT in AA smokers (Colilla et al., 2005). However, these findings are inconsistent with prior family-based work supporting an association among the Val/Met COMT polymorphism and FTND in AAs (Beuten et al., 2006) and laboratory work indicating associations with withdrawal severity in a pooled EA/AA sample (Herman et al., 2013). The mixed literature suggests that results may be specific to particular phenotypic features of ND and may vary depending on the ethnic make-up of the sample. Despite these inconsistencies, given the prominent role of COMT in dopamine metabolism, it remains an attractive candidate for molecular genetic studies of smoking.
A strength of our study is the focus on a restricted sample of AA smokers. When there are phenotypic and genotypic differences by ethnicity, the use of mixed samples can increase the rate of false discoveries and/or reduce power (Clayton et al., 2005). Further, AA subpopulations tend have greater polymorphic variation, resulting in shorter haplotypes which may aid (as available samples get larger) in narrowing genetic findings from large regions of interest and/or long haplotype blocks to specific susceptibility loci. As with all studies using a candidate based approach, our findings need to be substantiated through replication with more and larger samples. This is especially the case given that, due to the relatively small sample size and the number of SNPs examined, some of the associations reported here would not survive stringent type I error rate correction. In addition, the genotype and analytic approach focused on characterizing common variation in the NCAM1-TTC12-ANKK1-DRD2 gene region. However, there is additional genotypic variance within this region that was not measured (e.g. both common and rare variants) and additional mechanisms involved that may act in concert with putatively dopamine-related variants in order to influence risk for smoking.
In sum, this investigation, using individual genotype, haplotype, and mediation analyses, extends the literature and preliminarily implicates NCAM1-TTC12-ANKK1-DRD2 gene cluster variants in risk for ND in AAs. Our findings, which are consistent with our prior study of EAs, suggest that the effects of genetic variation within the NCAM1-TTC12-ANKK1-DRD2 gene region on ND may exert itself through a heavy and pervasive smoking ritual that can be elicited with little awareness. In addition, it was found that a variant in DBH appeared to increase risk of greater ND by way of PDM subscales more broadly. Together, these findings suggest that smoking motives, particularly Automaticity, are viable intermediate phenotypes for understanding genetic contributions to ND in AA smokers that may shed light on the genetic pathways to dependence and clinical outcomes.
Supplementary Material
Highlights.
Dopaminergic (DA) genetic variation is important in nicotine dependence (ND) etiology
DA genetic risk is understudied in African American (AA) smokers
This study examined DA genes and nicotine dependence (ND) phenotypes in AA smokers
DA genes and haplotypes were associated with both ND and automatic smoking behaviors
DA genes may increase ND risk through effects on automatic, habitual smoking behaviors
ACKNOWLEDGEMENTS
Funding was provided by the following grants: SAPRP 65626 from the Robert Wood Johnson Foundation and K23 AA016936 from NIH to James MacKillop; K23 DA033302 from NIDA to L. Cinnamon Bidwell; a Research Career Development Award from the Medical Research Service of the Department of Veteran Affairs, 1S10RR023457-01A1 and Shared equipment grants (ShEEP) from the Medical Research Service of the Department of Veteran Affairs to John McGeary; and K01 AA021113 from NIAAA to Rohan Palmer; and R01 DA023134 from NIDA to Valerie Knopik. Dr. MacKillop is the holder of the Peter Boris Chair in Addictions Research, which partially supported his role.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have no conflicts of interest to report. The views expressed in this article are those of the authors and do not necessarily reflect the position or policy of the National Institutes of Health or Department of Veterans Affairs.
Bibliography and References Cited
- Akil M, Kolachana BS, Rothmond DA, Hyde TM, Weinberger DR, Kleinman JE. Catechol-O-methyltransferase genotype and dopamine regulation in the human brain. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2003;23:2008–13. doi: 10.1523/JNEUROSCI.23-06-02008.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker TB, Weiss RB, Bolt D, von Niederhausern A, Fiore MC, Dunn DM, et al. Human neuronal acetylcholine receptor A5-A3-B4 haplotypes are associated with multiple nicotine dependence phenotypes. Nicotine & tobacco research : official journal of the Society for Research on Nicotine and Tobacco. 2009;11:785–96. doi: 10.1093/ntr/ntp064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JC. Haploview: Visualization and analysis of SNP genotype data. Cold Spring Harb Protoc. 2009;2009 doi: 10.1101/pdb.ip71. pdb ip71. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Benowitz NL. Nicotine addiction. The New England journal of medicine. 2010;362:2295–303. doi: 10.1056/NEJMra0809890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergen AW, Conti DV, Van Den Berg D, Lee W, Liu J, Li D, et al. Dopamine genes and nicotine dependence in treatment-seeking and community smokers. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2009;34:2252–64. doi: 10.1038/npp.2009.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beuten J, Payne TJ, Ma JZ, Li MD. Significant association of catechol-O-methyltransferase (COMT) haplotypes with nicotine dependence in male and female smokers of two ethnic populations. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2006;31:675–84. doi: 10.1038/sj.npp.1300997. [DOI] [PubMed] [Google Scholar]
- Bidwell LC, McGeary JE, Gray JC, Palmer RH, Knopik VS, MacKillop J. NCAM1-TTC12-ANKK1-DRD2 variants and smoking motives as intermediate phenotypes for nicotine dependence. Psychopharmacology. 2015;232:1177–86. doi: 10.1007/s00213-014-3748-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton DG, Walker NM, Smyth DJ, Pask R, Cooper JD, Maier LM, et al. Population structure, differential bias and genomic control in a large-scale, case-control association study. Nature genetics. 2005;37:1243–6. doi: 10.1038/ng1653. [DOI] [PubMed] [Google Scholar]
- Colilla S, Lerman C, Shields PG, Jepson C, Rukstalis M, Berlin J, et al. Association of catechol-O-methyltransferase with smoking cessation in two independent studies of women. Pharmacogenetics and genomics. 2005;15:393–8. doi: 10.1097/01213011-200506000-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium TTaG Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nature genetics. 2010;42:441–7. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubells JF, Zabetian CP. Human genetics of plasma dopamine beta-hydroxylase activity: applications to research in psychiatry and neurology. Psychopharmacology. 2004;174:463–76. doi: 10.1007/s00213-004-1840-8. [DOI] [PubMed] [Google Scholar]
- David SP, Mezuk B, Zandi PP, Strong D, Anthony JC, Niaura R, et al. Sex differences in TTC12/ANKK1 haplotype associations with daily tobacco smoking in Black and White Americans. Nicotine & tobacco research : official journal of the Society for Research on Nicotine and Tobacco. 2010;12:251–62. doi: 10.1093/ntr/ntp201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducci F, Kaakinen M, Pouta A, Hartikainen AL, Veijola J, Isohanni M, et al. TTC12-ANKK1-DRD2 and CHRNA5-CHRNA3-CHRNB4 influence different pathways leading to smoking behavior from adolescence to mid-adulthood. Biological psychiatry. 2011;69:650–60. doi: 10.1016/j.biopsych.2010.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freire MT, Marques FZ, Hutz MH, Bau CH. Polymorphisms in the DBH and DRD2 gene regions and smoking behavior. European archives of psychiatry and clinical neuroscience. 2006;256:93–7. doi: 10.1007/s00406-005-0610-x. [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, et al. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Gelernter J, Yu Y, Weiss R, Brady K, Panhuysen C, Yang BZ, et al. Haplotype spanning TTC12 and ANKK1, flanked by the DRD2 and NCAM1 loci, is strongly associated to nicotine dependence in two distinct American populations. Human molecular genetics. 2006;15:3498–507. doi: 10.1093/hmg/ddl426. [DOI] [PubMed] [Google Scholar]
- Goldman D, Oroszi G, Ducci F. The genetics of addictions: uncovering the genes. Nature reviews Genetics. 2005;6:521–32. doi: 10.1038/nrg1635. [DOI] [PubMed] [Google Scholar]
- Han DH, Joe KH, Na C, Lee YS. Effect of genetic polymorphisms on smoking cessation: a trial of bupropion in Korean male smokers. Psychiatric genetics. 2008;18:11–6. doi: 10.1097/YPG.0b013e3282df0939. [DOI] [PubMed] [Google Scholar]
- Heatherton TF, Kozlowski LT, Frecker RC, Fagerstrom KO. The Fagerstrom Test for Nicotine Dependence: a revision of the Fagerstrom Tolerance Questionnaire. British journal of addiction. 1991;86:1119–27. doi: 10.1111/j.1360-0443.1991.tb01879.x. [DOI] [PubMed] [Google Scholar]
- Herman AI, Jatlow PI, Gelernter J, Listman JB, Sofuoglu M. COMT Val158Met modulates subjective responses to intravenous nicotine and cognitive performance in abstinent smokers. The pharmacogenomics journal. 2013;13:490–7. doi: 10.1038/tpj.2013.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Payne TJ, Ma JZ, Beuten J, Dupont RT, Inohara N, et al. Significant association of ANKK1 and detection of a functional polymorphism with nicotine dependence in an African-American sample. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2009;34:319–30. doi: 10.1038/npp.2008.37. [DOI] [PubMed] [Google Scholar]
- IBM . IBM SPSS Statistics for Windows, Version 19.0. IBM Corp; Armonk, NY: 2010. Released. [Google Scholar]
- Johnstone EC, Clark TG, Griffiths SE, Murphy MF, Walton RT. Polymorphisms in dopamine metabolic enzymes and tobacco consumption in smokers: seeking confirmation of the association in a follow-up study. Pharmacogenetics. 2002;12:585–7. doi: 10.1097/00008571-200210000-00012. [DOI] [PubMed] [Google Scholar]
- Kandel D, Schaffran C, Hu MC, Thomas Y. Age-related differences in cigarette smoking among whites and African-Americans: evidence for the crossover hypothesis. Drug and alcohol dependence. 2011;118:280–7. doi: 10.1016/j.drugalcdep.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laucht M, Becker K, Frank J, Schmidt MH, Esser G, Treutlein J, et al. Genetic variation in dopamine pathways differentially associated with smoking progression in adolescence. Journal of the American Academy of Child and Adolescent Psychiatry. 2008;47:673–81. doi: 10.1097/CHI.0b013e31816bff77. [DOI] [PubMed] [Google Scholar]
- Li MD, Ma JZ, Beuten J. Progress in searching for susceptibility loci and genes for smoking-related behaviour. Clinical genetics. 2004;66:382–92. doi: 10.1111/j.1399-0004.2004.00302.x. [DOI] [PubMed] [Google Scholar]
- Lu J, Wei Q, Bondy ML, Li D, Brewster A, Shete S, et al. Polymorphisms and haplotypes of the NBS1 gene are associated with risk of sporadic breast cancer in non-Hispanic white women <or=55 years. Carcinogenesis. 2006;27:2209–16. doi: 10.1093/carcin/bgl077. [DOI] [PubMed] [Google Scholar]
- Luo Z1, Alvarado GF, Hatsukami DK, Johnson EO, Bierut LJ, Breslau N. Race differences in nicotine dependence in the Collaborative Genetic study of Nicotine Dependence (COGEND) Nicotine and Tobacco Research. 2008;10(7):1223–30. doi: 10.1080/14622200802163266. [DOI] [PubMed] [Google Scholar]
- MacKillop J, Few LR, Murphy JG, Wier LM, Acker J, Murphy C, et al. High-resolution behavioral economic analysis of cigarette demand to inform tax policy. Addiction. 2012;107:2191–200. doi: 10.1111/j.1360-0443.2012.03991.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacKillop J, Munafò M. Genetic Influences on Addiction: An Intermediate Phenotype Approach. MIT Press; Cambridge, MA: 2013. [Google Scholar]
- MacKillop J, Obasi E, Amlung MT, McGeary JE, Knopik VS. The Role of Genetics in Nicotine Dependence: Mapping the Pathways from Genome to Syndrome. Current cardiovascular risk reports. 2010;4:446–53. doi: 10.1007/s12170-010-0132-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinney EF, Walton RT, Yudkin P, Fuller A, Haldar NA, Mant D, et al. Association between polymorphisms in dopamine metabolic enzymes and tobacco consumption in smokers. Pharmacogenetics. 2000;10:483–91. doi: 10.1097/00008571-200008000-00001. [DOI] [PubMed] [Google Scholar]
- Morley KI, Medland SE, Ferreira MA, Lynskey MT, Montgomery GW, Heath AC, et al. A possible smoking susceptibility locus on chromosome 11p12: evidence from sex-limitation linkage analyses in a sample of Australian twin families. Behavioral Genetics. 2006;36:87–99. doi: 10.1007/s10519-005-9004-0. [DOI] [PubMed] [Google Scholar]
- Munafò M, Clark T, Johnstone E, Murphy M, Walton R. The genetic basis for smoking behavior: a systematic review and meta-analysis. Nicotine & tobacco research : official journal of the Society for Research on Nicotine and Tobacco. 2004;6:583–97. doi: 10.1080/14622200410001734030. [DOI] [PubMed] [Google Scholar]
- Munafò MR, Timpson NJ, David SP, Ebrahim S, Lawlor DA. Association of the DRD2 gene Taq1A polymorphism and smoking behavior: a meta-analysis and new data. Nicotine & tobacco research : official journal of the Society for Research on Nicotine and Tobacco. 2009;11:64–76. doi: 10.1093/ntr/ntn012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oroszi G, Anton RF, O'Malley S, Swift R, Pettinati H, Couper D, et al. OPRM1 Asn40Asp predicts response to naltrexone treatment: a haplotype-based approach. Alcoholism, clinical and experimental research. 2009;33:383–93. doi: 10.1111/j.1530-0277.2008.00846.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- National Cancer Institute (NCI) Phenotypes and Endophenotypes: Foundations for Genetic Studies of Nicotine Use and Dependence. National Cancer Institute Tobacco Control Monograph Series No. 20; Bethesda, MD: 2009. p. 654. [Google Scholar]
- Pajewski NM, Parker SD, Poland GA, Ovsyannikova IG, Song W, Zhang K, et al. The role of HLA-DR-DQ haplotypes in variable antibody responses to anthrax vaccine adsorbed. Genes & Immunity. 2011;12:457–65. doi: 10.1038/gene.2011.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piasecki TM, Piper ME, Baker TB. Refining the tobacco dependence phenotype using the Wisconsin Inventory of Smoking Dependence Motives: II. Evidence from a laboratory self-administration assay. J Abnorm Psychol. 2010;119:513–23. doi: 10.1037/a0020235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piper ME, Bolt DM, Kim SY, Japuntich SJ, Smith SS, Niederdeppe J, et al. Refining the tobacco dependence phenotype using the Wisconsin Inventory of Smoking Dependence Motives. J Abnorm Psychol. 2008;117:747–61. doi: 10.1037/a0013298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Primack BA, Bost JE, Land SR, Fine MJ. Volume of tobacco advertising in African American markets: systematic review and meta-analysis. Public Health Rep. 2007;122:607–15. doi: 10.1177/003335490712200508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S. PLINK v1.07. http://pngu.mgh.harvard.edu/purcell/plink/.
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JD, Lam CY, Minnix JA, Wetter DW, Tomlinson GE, Minna JD, et al. The DRD2 TaqI-B polymorphism and its relationship to smoking abstinence and withdrawal symptoms. The pharmacogenomics journal. 2007;7:266–74. doi: 10.1038/sj.tpj.6500427. [DOI] [PubMed] [Google Scholar]
- Saccone SF, Hinrichs AL, Saccone NL, Chase GA, Konvicka K, Madden PA, et al. Cholinergic nicotinic receptor genes implicated in a nicotine dependence association study targeting 348 candidate genes with 3713 SNPs. Human molecular genetics. 2007;16:36–49. doi: 10.1093/hmg/ddl438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safford MM, Brown TM, Muntner PM, Durant RW, Glasser S, Halanych JH, et al. Association of race and sex with risk of incident acute coronary heart disease events. JAMA : the journal of the American Medical Association. 2012;308:1768–74. doi: 10.1001/jama.2012.14306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiels MS, Huang HY, Hoffman SC, Shugart YY, Bolton JH, Platz EA, et al. A community-based study of cigarette smoking behavior in relation to variation in three genes involved in dopamine metabolism: Catechol-O-methyltransferase (COMT), dopamine beta-hydroxylase (DBH) and monoamine oxidase-A (MAO-A) Preventive medicine. 2008;47:116–22. doi: 10.1016/j.ypmed.2008.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siedlinski M, Cho MH, Bakke P, Gulsvik A, Lomas DA, Anderson W, et al. Genome-wide association study of smoking behaviours in patients with COPD. Thorax. 2011;66:894–902. doi: 10.1136/thoraxjnl-2011-200154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Donnelly P. A comparison of bayesian methods for haplotype reconstruction from population genotype data. Am J Hum Genet. 2003;73:1162–9. doi: 10.1086/379378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Scheet P. Accounting for decay of linkage disequilibrium in haplotype inference and missing-data imputation. American journal of human genetics. 2005;76:449–62. doi: 10.1086/428594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–89. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabachnick BG, Fidell LS. Using Multivariate statistics. 4th Allyn & Bacon; 2001. [Google Scholar]
- Ton TG, Rossing MA, Bowen DJ, Srinouanprachan S, Wicklund K, Farin FM. Genetic polymorphisms in dopamine-related genes and smoking cessation in women: a prospective cohort study. Behavioral and brain functions : BBF. 2007;3:22. doi: 10.1186/1744-9081-3-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabetian CP, Anderson GM, Buxbaum SG, Elston RC, Ichinose H, Nagatsu T, et al. A quantitative-trait analysis of human plasma-dopamine beta-hydroxylase activity: evidence for a major functional polymorphism at the DBH locus. American journal of human genetics. 2001;68:515–22. doi: 10.1086/318198. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.