Abstract
Objectives
The tumor suppressor LKB1 has recently been shown to be involved in the regulation of microtubule dynamics, thus cancer cells with inactivated LKB1 may have developed a means to overcome dysregulated microtubule functions, making them intrinsically resistant to microtubule targeting agents. Here, we generated isogenic LKB1-wild type and mutant non-small cell lung cancer (NSCLC) cell lines to evaluate the role of LKB1 in paclitaxel resistance.
Materials and methods
SRB, flow cytometry and immunoblotting were used to assess cell proliferation and apoptosis in NSCLC cell lines after paclitaxel treatment. Expression of LKB1 was restored in LKB1-null cells by retrovirus infection and was reduced in LKB1-wild type cells by shRNA knock down.
Results and Conclusion
The restoration of LKB1 in LKB1-null cells failed to promote paclitaxel-induced apoptosis in both p53-wild type and p53-mutant backgrounds, indicating that LKB1 was not required for paclitaxel-induced apoptosis. Interestingly, the re-establishment of LKB1 expression led to the up-regulation of class III beta-tubulin and MDR1 in EKVX cells. The up-regulation of MDR1 protein and transcripts in EKVX cells was specifically associated with the expression of wild-type LKB1 and mainly responsible for the increased cellular resistance to paclitaxel. However, the presence of LKB1 protein was not required to maintain this increased MDR1 expression even though there was no genetic amplification or promoter de-methylation of the ABCB1 locus in EKVX-LKB1-WT cells. These data suggest that LKB1 does not promote paclitaxel-induced apoptosis in most NSCLC cell lines. In contrast, in some NSCLC, the presence of LKB1 may facilitate increases in either MDR1 or class III beta-tubulin expression which can lead to paclitaxel resistance.
Keywords: microtubule targeting agents, paclitaxel resistance, tumor suppressor, promoter methylation
1. Introduction
Lung cancer is the leading cause of cancer death in the United States, with more people dying of lung cancer than prostate, breast, and colon cancers combined (1). The majority of patients suffer from non-small cell lung cancer (NSCLC), and various anti-microtubule agents, such as paclitaxel, have been used as a component of chemotherapy against this disease. Even though paclitaxel is an effective chemotherapeutic agent against ovarian, breast, and gastric cancer, the response rate in NSCLC is only 30–40% (2) suggesting that many NSCLC tumors have intrinsic resistance to anti-microtubule agents. The best known paclitaxel resistance mechanisms are the over-expression of the MDR1 efflux pump, the expression of class III β-tubulin, and mutations in β-tubulin (3). However, some NSCLC cells have intrinsic resistance to paclitaxel in the absence of these alterations, and it is possible that other mechanisms are also involved (3).
LKB1 is the third most frequently mutated gene in smoking-related NSCLC (4, 5). It encodes a serine/threonine kinase that is involved in the regulation of many cellular processes, including energy metabolism, cell polarity and cell mobility (6). Most LKB1 mutations found in NSCLC inactivate its kinase activity, and some of its down-stream substrates, such as microtubule/MAP-affinity regulating kinases (MARKs), are known to regulate the function of microtubules. For example, tau has been implicated in microtubule stability (7), and LKB1 was found to regulate tau phosphorylation through its downstream target MARK2 (8, 9). In addition, LKB1 was recently found to promote microtubule destabilization in myoblasts (10). Therefore, LKB1 may play a role in the regulation of microtubule dynamics. Furthermore, a 2001 study indicated that the over-expression of a dominant negative LKB1-K78M mutant in LKB1-wild type HT1080 cells led to paclitaxel and vincristine resistance, and LKB1 was found to mediate p53-dependent apoptosis in this cell line (11). This result suggests that LKB1 may also be a mediator of paclitaxel-induced apoptosis, and the inactivation of LKB1 in some NSCLC cells may be partially responsible for their intrinsic resistance to microtubule targeting agents.
In this study, we generated various isogenic NSCLC cancer cell lines by either expressing wild-type (WT) LKB1 in LKB1-null NSCLC cell lines or depleting the endogenous LKB1 in LKB1-wild type NSCLC cell lines. The establishment of these isogenic cell lines allowed us to determine whether LKB1 is a mediator of paclitaxel-induced apoptosis in NSCLC cells.
2. Materials and methods
2.1 Cell culture
NSCLC cell lines A549, EKVX, H1299, H157, and H460 were purchased from the American Type Culture Collection (ATCC); all cells were maintained in RPMI1640 media. The identities of these cells were verified by genotyping service at Emory. Phoenix A and HEK293T (ATCC) cells were used as virus packaging cell lines, and were cultivated in DMEM media. Both media contained 10% fetal bovine serum (FBS) (Invitrogen) and 100 units/mL of Pen-Strep (Invitrogen).
2.2 Reagents
AICAR, paclitaxel, puromycin, doxycycline, PSC833 were purchased from Sigma-Aldrich. Paclitaxel and PSC833 were dissolved in DMSO; AICAR, puromycin, and doxcycline were dissolved in water. Antibodies against LKB1, phospho-AMPKα, total-AMPKα, phospho-ACC, caspase3, PARP, MDR1 and β-tubulin were purchased from Cell Signaling Technology. Class III β-tubulin (TUBB3) antibody was purchased from Millipore. Antibody against phosphor-tau(S262) was purchased from Invitrogen. β-Actin antibody was purchased from Sigma-Aldrich.
2.3 Small interfering RNA (siRNA) knockdown
LKB1 siRNAs were purchased from Sigma-Aldrich. The sequences of these siRNAs are provided in Table 1. Cells were grown to 40~60% confluence in 6-well plates, and each well was transfected with 100 pmol of indicated siRNA per 2ml of medium using Lipofectamine 2000 as previously described (12).
Table 1.
| siRNA/primer sequence (5′ -> 3′) | Product size (bp) | |
|---|---|---|
| LKB1 siRNA | ||
| siRNA_1 | GGACUGACGUGUAGAACAATT | |
| siRNA_2 | GCUCUUACGGCAAGGUGAA | |
| mRNA (cDNA) | ||
| ABCB1 forward | GGAAGACATGACCAGGTATGC | 93 |
| ABCB1 reverse | GCCAGGCACCAAAATGAAACC | |
| TUBB3 forward | GCCTCTTCTCACAAGTACGTG | 130 |
| TUBB3 reverse | CCCCACTCTGACCAAAGATGAA | |
| β-Actin forward | CGTCACCAACTGGGACGACA | 130 |
| β-Actin reverse | CTTCTCGCGGTTGGCCTTGG | |
| Genomic copy number | ||
| ABCB1 forward | TGCTGGTCCTGAAGTTGATCTGTGAAC | 248 |
| ABCB1 reverse | ACATTAGGCAGTGACTCGATGAAAGGCA | |
| GAPDH forward | CCCCACACACATGCACTTACC | 97 |
| GAPDH reverse | CCTAGTCCCAGGGCTTTGATT | |
| Bisulfate sequencing | ||
| BS forward | GGAAGTTAGAATATTTTTTTTGGAAAT | 223 |
| BS reverse | ACCTCTACTTCTTTAAACTTAAAAAAACC | |
2.4 Re-expression of LKB1 in LKB1-mutant cells
Re-expression of LKB1 was achieved using pBABE retrovirus and the Phoenix A cell line as host, pBABE vectors (from Addgene) encoding LKB1-WT or LKB1-K78M (with pBABE-puro as control vector) were transfected as described above, and viral supernatants were collected after 48 hrs. These viruses were used to infect A549, H157, H460 and EKVX cells, which were selected with puromycin for two weeks. Stably-infected cell line pools were used for subsequent experiments.
2.5 Tet-inducible LKB1 shRNA knockdown
TRIPZ lentiviruses containing vector control or doxycycline-inducible shRNAs against LKB1 (Open Biosystem, RHS4696-99705612, RHS4696-101314769, and RHS4696-101321040) were produced in the HEK293T cell line using psPAX2 and pVSVG as packaging plasmids (gifts from Dr. Trent Spencer’s lab) (13). Viral supernatants were collected after 64hrs to infect H1299 cells. Infected cells were selected with 2 μg/ml puromycin for 2 weeks to obtain stably infected pools. shRNA expression was induced by 1 μg/ml doxycycline treatment for 72 hrs.
2.6 Immunoblotting analysis
30 μg of total cell lysates were separated by 8 or 12% SDS-PAGE gels, blotted onto PVDF membranes (Bio-Rad), and probed with the indicated antibodies. β-Actin was used as a loading control.
2.7 Immunofluorescence analysis
Fixed-cell immunofluorescence was performed as described previously (14). Tubulin was visualized by anti-α-tubulin antibody and Alexa Fluor 488 secondary antibody (Invitrogen). DNA was visualized by DAPI staining (350 nM, 10 mins). Mitotic index is defined as the percentage of mitotic figures. Cells were fixed 24 hrs after 100 nM paclitaxel treatment and stained with DAPI to visualize nuclei. At least 200 cells were counted in each field.
2.8 Proliferation assay
Cells were plated at a density of 5000~6000 per well in 96-well plates, and treated with indicated drugs after 20 hours. Cells were fixed at indicated time with 10% trichloroacetic acid (TCA), and SRB assay was carried out as previously described (15).
2.9 Cell cycle analysis
Cells were seeded in 6-well plates, subjected to the desired treatment, and then trypsinized and collected. Cells were washed twice with PBS, then fixed with cold (−20°C) 70% ethanol for 30 mins. After removal of ethanol, the cells were stained with PI/RNASE staining kit (BD Pharmingen, San Jose, CA) for 30 mins at RT in the dark, and then analyzed by FACS system (Becton Dickinson, San Jose, CA). At least a total of 10,000 gated cells were acquired for each analysis. Results were analyzed by FlowJo version 7 software.
2.10 Flow cytometry apoptosis analysis
Annexin V-PE apoptosis binding assay (BD Pharmingen, San Jose, CA) was used as recommended by the manufacturer and was carried out by FACS analysis (BD, San Jose, CA); 10,000 cells were acquired for each analysis. Results were analyzed by FlowJo version 7 software as previously described (15).
2.11 Quantitative real-time PCR analysis
qPCR was carried out as previously described (16). β-Actin was used as the control for transcript quantitation, and GAPDH was used for genomic DNA copy number control. DNA and RNA isolated from EKVX-puro cells were used as the calibrator when measuring ABCB1 genomic copy number and transcript levels. RNA isolated from EKVX or H1299-vector treated cells was used as calibrator for measuring TUBB3 Expression. Primers sequences are shown in Table 1.
2.12 Methylation analysis
Bisulfite modification of genomic DNA followed by PCR amplification was carried out as described previously (17). Sequences of primers for bisulfate sequencing are listed in Table 1.
3. Results
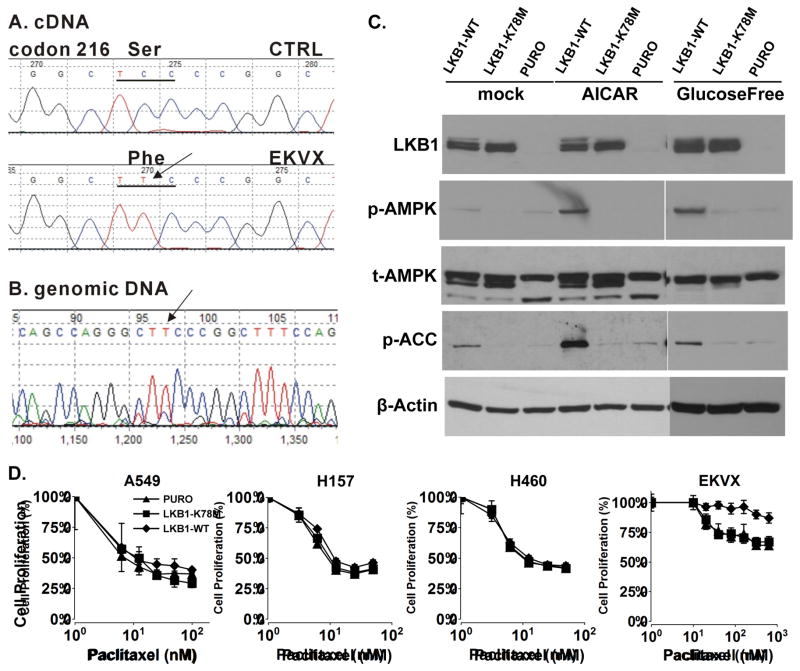
We chose to restore LKB1 expression in four LKB1-null NSCLC cell lines, A549, H460, H157 and EKVX. Both A549 and H460 cells contain a Q37 nonsense mutation, and we previously reported a homozygous LKB1 deletion in H157 cells (18). Here, we discovered that EKVX cells contains a Ser to Phe missense mutation at codon 216 in the kinase domain of LKB1 (Figure 1A). Genomic sequencing analysis revealed that the EVKX cell line contained a homozygous mutation in the LKB1 gene (Figure 1B). Functionally, this mutation leads to the suppression of endogenous LKB1 expression, and energetic stress conditions, such as glucose-free or AICAR treatment, failed to activate the phosphorylation of AMPK at Thr172, which is known to be a target phosphorylation site for LKB1 kinase (Figure 1C, lanes 6 and 9). Therefore, EVKX is an LKB1-defective NSCLC lung cancer cell line.
Fig. 1.
EKVX cells contain LKB1-inactivation mutation. (A) Sequence trace of LKB1 cDNA isolated from EKVX cell line. (B) Sequence trace of LKB1 genomic DNA isolated from EKVX cell line. (C) AICAR and glucose-free treatment induced phosphorylation alterations in LKB1 downstream targets. Isogenic EKVX cells were treated with 1mM AICAR for 5hrs, or glucose-free media for 2hrs, and cell lysates were analyzed on immunoblots with indicated antibodies. Each experiment was repeated three times. (D) A549, H157, H460 and EKVX isogenic cell lines were treated with varying dose of paclitaxel for 48hrs, and cell proliferation was analyzed by SRB assay. Each experiment was carried out in quadruplicates, and repeated three times.
Because of the potential role of p53 in paclitaxel-induced apoptosis, we also designed our study to evaluate the role of p53 in this process. A549 and H460 cells contain wild-type p53, while H157 and EKVX contain mutant p53 (p53-null and E204-to-nonsense). We restored LKB1 expression in these cells using retrovirus containing either a wild-type LKB1 or a kinase-dead LKB1-K78M mutant. For example, the expression of a wild-type LKB1 restored the ability of EKVX cells to phosphorylate AMPK at Thr172 under glucose-free conditions for 2 hours or after AICAR treatment for 5 hours (Figure 1C, lanes 4 and 7). The restoration of AMPK kinase activity was also supported by the phosphorylation of ACC at Ser79. In contrast, the introduction of the LKB1-K78M mutant or the empty virus failed to restore the phosphorylation of AMPK or ACC under energetic stress conditions (Figure 1C, lanes 5 and 8). Similar data were obtained for the A549, H460 and H157 isogenic cell line panel (data not shown).
We next used a cell proliferation assay to determine whether the restoration of LKB1 function in LKB1-null NSCLC cell lines renders them more sensitive to paclitaxel treatment (Figure 1D). Our results indicated that the restoration of LKB1 function in LKB1-null NSCLC cells did not promote paclitaxel-induced growth suppression in either p53 wild-type or p53-mutant background. We also compared caspase-3 and PARP cleavage in these cells but failed to observe any increases in the cleaved proteins after the restoration of wild-type LKB1 expression (data not shown). Therefore, LKB1 is not a mediator of paclitaxel-induced cell killing in the NSCLC cell lines tested in this study.
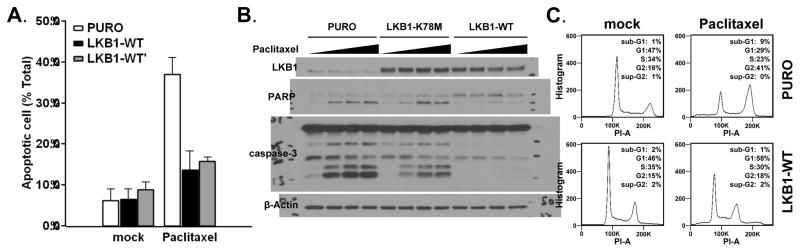
One surprising finding in the LKB1-WT expressing EKVX cell line was its ability to resist paclitaxel treatment (Figure 1D). Flow-cytometry analysis with Annexin-V and 7AAD revealed significantly fewer apoptotic cells after paclitaxel treatment in EKVX-LKB1-WT cells (Figure 2A). The suppression of apoptosis in EKVX-LKB1-WT cells was also supported by immunoblotting analysis of caspase-3 and PARP-cleavage (Figure 2B). Paclitaxel treatment usually results in extensive G2/M phase cell cycle arrest, which was also absent in EKVX-LKB1-WT cells (Figure 2C). These data indicate that EKVX-LKB1-WT cells are resistant to paclitaxel-induced apoptosis. To rule out any experimental artifact, we carried out an independent selection process by introducing wild-type LKB1 into the parental EKVX cell line. This independent restoration of wild type LKB1 in the EKVX cell line (EKVX-LKB1-WT′) again led to resistance to paclitaxel, indicating that the re-expression of LKB1 preferentially promoted resistance to paclitaxel specifically in the EKVX cell line (Figure 2A).
Fig. 2.
Re-expression of LKB1 in EKVX leads to resistance to paclitaxel. (A) EKVX isogenic cell lines were treated with 80nM paclitaxel for 48hrs then subjected to Annexin V-7AAD flow cytometry analysis. (B) EKVX isogenic cells were treated with varying doses (0, 10, 20, 40nM) of paclitaxel for 48hrs, and cell lysates were analyzed by immunoblot for apoptosis bio-markers caspase-3 and PARP. (C) Cell cycle analysis of the EKVX isogenic cells treated with 50nM paclitaxel for 24hrs. All experiments described in this figure were repeated three times.
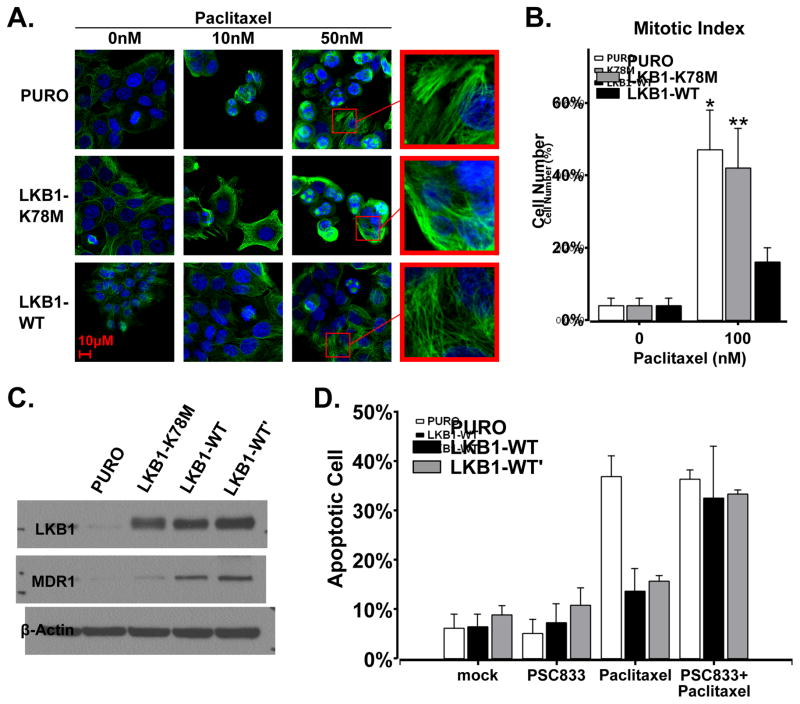
To explore the mechanism of paclitaxel resistance in EKVX-LKB1-WT cells, we assessed tubulin polymerization status by immunofluorescent analysis of tubulin. While paclitaxel treatment led to microtubule polymerization in cells with control vector, it failed to polymerize microtubules in EKVX-LKB1-WT cells (Figure 3A). Mitotic index analysis also revealed a significant decrease in the number of EKVX-LKB1-WT cells that entered mitosis (Figure 3B). These data suggested that paclitaxel maybe actively exported by EKVX-LKB1 cells, and immunoblotting analysis revealed significant up-regulation of MDR1 protein expression in both EKVX-LKB1-WT and EKVX-LKB1-WT′ cells (Figure 3C). To determine whether MDR1 was mainly responsible for paclitaxel resistance of EVKX-LKB1-WT cells, we used an MDR1-specific inhibitor PSC833, and found that pre-treatment with 4 μM PSC833 was sufficient to restore paclitaxel-induced apoptosis in EKVX-LKB1-WT and WT′ cells (Figure 3D). These findings indicate that the over-expression of MDR1 is the molecular basis of paclitaxel resistance in EKVX-LKB1-WT cells.
Fig. 3.
Elevated MDR1 expression is responsible for paclitaxel-resistance of EKVX cells expressing a wild-type LKB1. (A) Immunofluorescence assay of microtubule morphological changes after treatment with 0, 10, 50nM paclitaxel for 48hrs in EKVX isogenic cell lines. Original magnification 100×, green signaling for α-tubulin, and blue for nuclei. This experiment was repeated twice. (B) Mitotic index of EKVX isogenic cells after treated with 100nM taxol at 24 hrs (two-tail t-test, *P<0.01, **P<0.01 vs EKVX-LKB1-WT group). (C) Immunoblotting analysis of MDR1 protein expression in EKVX isogenic cell lines. (D) Annexin V-7AAD flow cytometry analysis of EKVX isogenic cell lines after treated with 4μM PSC833, 80nM paclitaxel, or the combination of these two drugs for 48 hrs. Experiments in panel B-C were repeated three times.
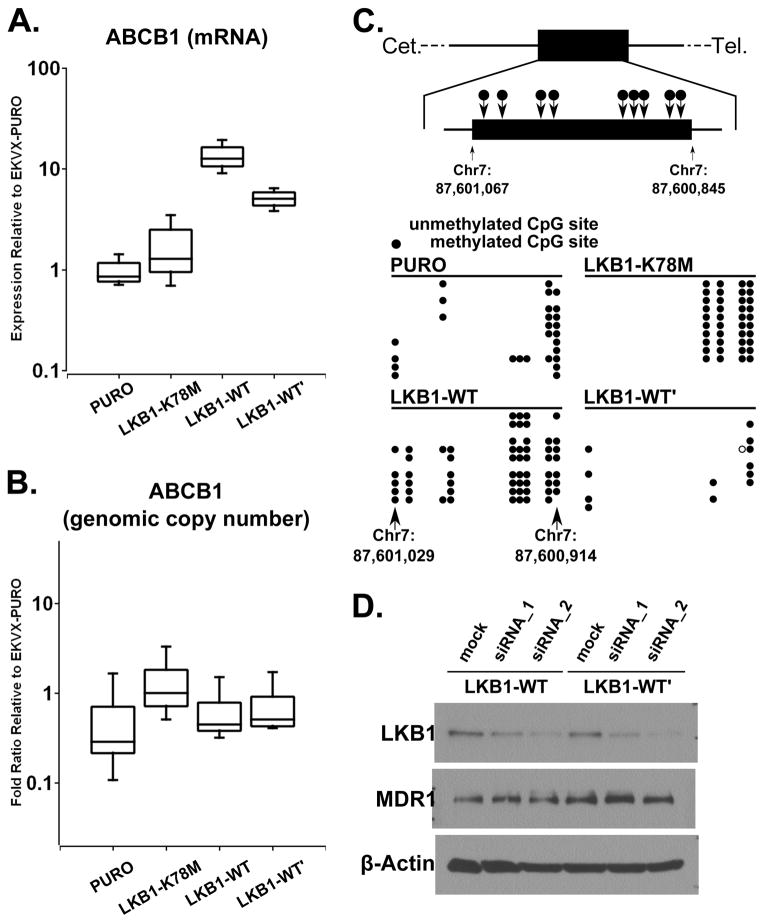
MDR1 is encoded by the ABCB1 gene, and quantitative real-time PCR analysis indicated that mRNA for ABCB1 was up-regulated 10-fold in EKVX-LKB1-WT cells and 7-fold in EKVX-LKB1-WT′ cells (Figure 4A). This up-regulated ABCB1 mRNA expression was not due to genomic amplification as the genomic copy number of the ABCB1 gene was not amplified in either cell line (Figure 4B). We also assessed the ABCB1 promoter methylation patterns in the EKVX cell line after LKB1 expression. ABCB1 has a unique CpG island which contains 13 CpG repeats and is present in exon 2 of ABCB1. Bisulfite sequencing revealed an increase in CpG island methylation of the ABCB1 promoter in EKVX-LKB1-WT cells but not in EKVX-LKB1-WT′ cells (Figure 4C). Therefore, the promoter of ABCB1 was not demethylated after LKB1 expression, and the up-regulation of ABCB1 mRNA is unlikely due to CpG island demethylation. Furthermore, the depletion of LKB1 in EKVX-LKB1-WT or EKVX-LKB1-WT′ cells by RNAi did not down-regulate MDR1 expression (Figure 4D). In combination, these data indicate that the presence of LKB1 is required to initiate the over-expression of MDR1 in the EKVX cell line, but not to maintain the elevated MDR1 expression.
Fig. 4.
Genetic and epigenetic analyses of ABCB1 loci. (A) Real-time qPCR of the ABCB1 (MDR1) transcript level in EKVX isogenic cells. (B) Quantitative PCR of the ABCB1 genomic copy number variation in EKVX isogenic cells. (C) Bisulfate sequencing of the ABCB1 promoter methylation changes in EKVX isogenic cells. (D) Immunoblotting of MDR1 after knocking down LKB1 with siRNAs in EKVX-LKB1-WT and EKVX-LKB1-WT′ cells. These experiments were repeated three times.
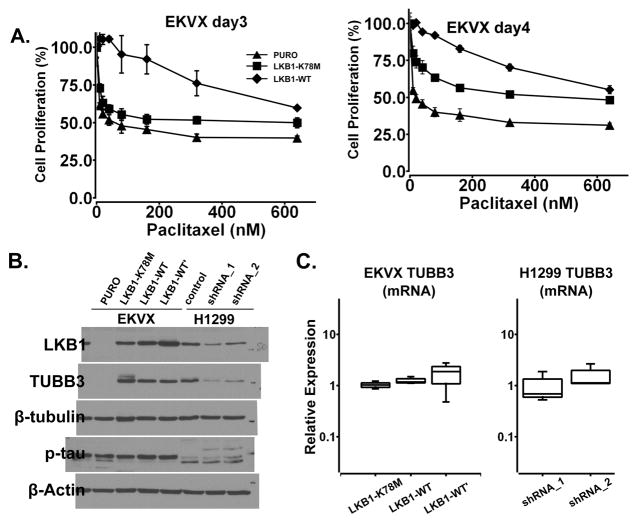
We initially designed this studying using kinase-dead LKB1-K78M mutant as a negative control. The over-expression of LKB1-K78M mutant was not associated with MDR1 over-expression (Figure 3C, lane 2), and paclitaxel was still capable of polymerizing microtubules (Figure 3A), promoting mitotic arrest (Figure 3B) and inducing caspase-3 and PARP cleavage (Figure 2B, lanes 6–8). Nevertheless, even though paclitaxel had similar growth inhibitory effort on vector-treated cells and LKB1-K78M expressing cells at 48 hours (Figure 1D), we consistently observed a small but statistical significant increase of cell proliferation in EVKX-K78M cells at 72 and 96 hours (Figure 5A). These findings led us to explore other mechanisms that have previously been associated with microtubule resistance, such as the phosphorylation of tau or the altered expression class III β-tubulin. Tau phosphorylation was not altered with LKB1 expression in EKVX cells, but immunoblotting analysis revealed an up-regulation of class III β-tubulin in both EKVX-K78M and EKVX-LKB1-WT cells (Figure 5B, lanes 2~4), indicating that the kinase activity of LKB1 may not be required for the up-regulation of class III β-tubulin. Interestingly, a down-regulation of class III β-tubulin was also observed in H1299 cells when LKB1 was depleted by shRNA (Figure 5B, lanes 6 and 7). In both cases, the alteration of class III β-tubulin protein expression was not associated with a change in their transcripts level (Figure 5C), indicating that the presence of wild type or LKB1-K78M LKB1 protein may regulate the expression of this protein at post-transcription level. It is also important to point that this associated was only observed in a subset of NSCLC cell lines because we failed to observe similar alteration in A549, H460 or H157 cells (data not shown).
Fig. 5.
The expression of LKB1 protein correlates with class III β-tubulin expression in EKVX and H1299 cells. (A) EKVX isogenic cell lines were treated with varying dose of paclitaxel at 72 and 96hrs in proliferation assay. (B) Immunoblotting analysis of class III β-tubulin and phosphorylated tau in EKVX isogenic cell lines and H1299 doxycycline-inducible LKB1-shRNA cell lines. (C) Real-time qPCR of the TUBB3 transcript level in EKVX isogenic cells and H1299 doxycycline-inducible LKB1-shRNA cell lines. These experiments were repeated three times.
4. Discussion
Early genetic evidence from Caenorhabditis elegans and Drosophila indicated that LKB1 plays an important role in cell polarity (19, 20), 6 of which have been reported to regulate microtubule dynamics (21, 22). The direct involvement of LKB1 in microtubule dynamics was demonstrated in MEF cells and myoblasts (9, 10). Because LKB1 is inactivated in a significant portion of NSCLC, the regulation of microtubule dynamics may be disrupted in these cancer cells. Consequently, these cancer cells may already adapted to dysregulated microtubule function, making them more resistant to microtubule targeting agents.
The first major finding of this study is that the restoration of LKB1 function in LKB1-null NSCLC cells was not sufficient to promote paclitaxel-induced cell killing. This finding differs from that of a previous report in an osteosarcoma cell line, where LKB1 was found to be a mediator of p53-dependent cell death (11). We carried out our studies in two p53-wild type and two p53-mutant NSCLC cell lines but the restoration of LKB1 function failed to decrease cell proliferation. It is important to point out that the apoptotic machinery is still intact in these cells, because energetic stress, such as that induced by phenformin treatment, is capable of inducing caspase-3 and PARP cleavage in our cell lines (data not shown), which is similar to previous observations by others (23). Therefore, either LKB1 does not play a significant role in paclitaxel-induced apoptosis in these NSCLC cell lines or this signaling pathway has additional defects. In either case, the restoration of LKB1 function alone was not sufficient to restore paclitaxel sensitivity in the NSCLC cell lines tested in this study. We also knocked-down LKB1 expression in H1299 and H1792 NSCLC cell lines. However, these cell lines are p53-mutant and we did not observe a difference in paclitaxel-sensitivity, as expected (data not shown). Our data are also consistent with drug sensitivity data from The Genomics of Drug Sensitivity in Cancer (GDSC), where the mutation status of LKB1 was not found to be correlated with paclitaxel-sensitivity in 700 cancer cell lines (24).
The surprising finding of this work is that the restoration of LKB1 function in EKVX cells leads to decreased cellular sensitivity to paclitaxel, which is in contrast to our initial hypothesis. The major contributing factor for paclitaxel resistance in EVKX-LKB1-WT cells is the up-regulation of MDR1 expression, since the pre-treatment of EKVX-LKB1-WT cells with an MDR1 inhibitor was sufficient to restore paclitaxel sensitivity. This up-regulation is specific for the selection of LKB1-WT protein as it is not observed in vector or LKB1-K78M mutant expressing cells. Interestingly, the continuous presence of LKB1 is not required to maintain up-regulated MDR1 expression, indicating a permanent change in the regulation of MDR1 expression. Our data indicate that this up-regulation is due to the increase of MDR1 transcription, which is not related to genomic amplification or the demethylation of CpG islands in the promoter. Hence, EKVX-LKB1-WT cells may acquire a mean to stabilize MDR1 transcripts to promote paclitaxel resistance.
Another interesting finding in this study is the potential protein expression association between class III β-tubulin and LKB1. The overexpression of class III β-tubulin was reported to be associated with paclitaxel-resistance in ovarian cancer cell lines (25) and in tumors from ovarian cancer patients (26). Recently, a combined meta-analysis of ten previous studies indicated that low class III β-tubulin expression level is correlated with anti-tubulin therapy in NSCLC (27). In our cell line model, the over-expression of both wild-type and LKB1-K78M mutant in EKVX cells was associated with an increase in class III β-tubulin expression. In addition, the depletion of LKB1 expression in H1299 cells also led to decreased class III β-tubulin expression. Hence, in some NSCLC cell lines, LKB1 may regulate the expression of class III β-tubulin in a kinase-independent manner. It is important to note that this correlation was only observed in a subset of NSCLC cell lines, so the expression of class III β-tubulin may be regulated by other factors as well. Nevertheless, it will be interesting to determine in the future whether LKB1 depletion or non-sense mutations, but not missense mutations, are associated with decreased class III β-tubulin expression in clinical specimens.
Highlights.
LKB1 is not a mediator of paclitaxel-induced apoptosis in non-small cell lung cancers.
Restoration of wild-type LKB1 in LKB1-mutant EKVX cells leads to the overexpression of MDR1 and paclitaxel resistance.
The expression of LKB1 correlates with class III β-tubulin expression in EKVX and H1299 cells.
Acknowledgments
This study was supported by PERPG 2011B09040056 (M. Li), NIH grant R01-CA140571 (W. Zhou), P01 CA116676 (F.R. Khuri), China Scholarship Council (F. Liu), and Anise McDaniel Brock Scholar fund (W. Zhou). This research project was also supported in part by the Emory University Integrated Cellular Imaging Microscopy Core of the Winship Cancer Institute comprehensive cancer center grant, P30CA138292. W. Z. and F.R. K. are Georgia Cancer Coalition Distinguished Cancer Scholars. W. Z. is an Anise McDaniel Brock Scholar and an American Cancer Society Research Scholar. We would like to thank Dr. Anthea Hammond for editing this manuscript.
Footnotes
Conflict of interest
The authors declare that they have no conflict of interest.
Authors’ Contributions
Conception and design: K. Mao, M. Li and W. Zhou.
Development of methodology: K. Mao, F. Liu, X. Liu.
Acquisition of data: K. Mao, F. Liu, X. Liu.
Analysis and interpretation of data; K. Mao, A. Marcus, F. Khuri, M. Li and W. Zhou.
Writing, review, and/or revision of the manuscript: K. Mao, A. Marcus, F. Khuri, M. Li and W. Zhou.
Study supervision: M. Li, W. Zhou
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 2.Pujol JL, Barlesi F, Daures JP. Should chemotherapy combinations for advanced non-small cell lung cancer be platinum-based? A meta-analysis of phase III randomized trials. Lung Cancer. 2006;51(3):335–345. doi: 10.1016/j.lungcan.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 3.Zhou J, Giannakakou P. Targeting microtubules for cancer chemotherapy. Current medicinal chemistry Anti-cancer agents. 2005;5(1):65–71. doi: 10.2174/1568011053352569. [DOI] [PubMed] [Google Scholar]
- 4.Ding L, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455(7216):1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gill RK, et al. Frequent homozygous deletion of the LKB1/STK11 gene in non-small cell lung cancer. Oncogene. 2011;30(35):3784–3791. doi: 10.1038/onc.2011.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marcus AI, Zhou W. LKB1 regulated pathways in lung cancer invasion and metastasis. J Thorac Oncol. 2010;5(12):1883–1886. doi: 10.1097/JTO.0b013e3181fbc28a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dehmelt L, Halpain S. The MAP2/Tau family of microtubule-associated proteins. Genome biology. 2005;6(1):204. doi: 10.1186/gb-2004-6-1-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thies E, Mandelkow EM. Missorting of tau in neurons causes degeneration of synapses that can be rescued by the kinase MARK2/Par-1. J Neurosci. 2007;27(11):2896–2907. doi: 10.1523/JNEUROSCI.4674-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kojima Y, et al. Suppression of tubulin polymerization by the LKB1-microtubule-associated protein/microtubule affinity-regulating kinase signaling. J Biol Chem. 2007;282(32):23532–23540. doi: 10.1074/jbc.M700590200. [DOI] [PubMed] [Google Scholar]
- 10.Mian I, et al. LKB1 destabilizes microtubules in myoblasts and contributes to myoblast differentiation. PLoS One. 2012;7(2):e31583. doi: 10.1371/journal.pone.0031583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karuman P, et al. The Peutz-Jegher gene product LKB1 is a mediator of p53-dependent cell death. Mol Cell. 2001;7(6):1307–1319. doi: 10.1016/s1097-2765(01)00258-1. [DOI] [PubMed] [Google Scholar]
- 12.Zhong D, et al. The glycolytic inhibitor 2-deoxyglucose activates multiple prosurvival pathways through IGF1R. J Biol Chem. 2009;284(35):23225–23233. doi: 10.1074/jbc.M109.005280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Johnston JM, Denning G, Doering CB, Spencer HT. Generation of an optimized lentiviral vector encoding a high-expression factor VIII transgene for gene therapy of hemophilia A. Gene Ther. 2013;20(6):607–615. doi: 10.1038/gt.2012.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Thaiparambil JT, Eggers CM, Marcus AI. AMPK regulates mitotic spindle orientation through phosphorylation of myosin regulatory light chain. Mol Cell Biol. 2012;32(16):3203–3217. doi: 10.1128/MCB.00418-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhong D, et al. LKB1 is Necessary for Akt-mediated Phosphorylation of Pro-apoptotic Proteins. Cancer Res. 2008;68(18):7270–7277. doi: 10.1158/0008-5472.CAN-08-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhong D, et al. Homozygous deletion of SMAD4 in breast cancer cell lines and invasive ductal carcinomas. Cancer Biol Ther. 2006;5(6):601–607. doi: 10.4161/cbt.5.6.2660. [DOI] [PubMed] [Google Scholar]
- 17.Guo L, et al. Sox7 is an independent checkpoint for beta-catenin function in prostate and colon epithelial cells. Mol Cancer Res. 2008;6(9):1421–1430. doi: 10.1158/1541-7786.MCR-07-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhong D, et al. LKB1 mutation in large cell carcinoma of the lung. Lung Cancer. 2006;53(3):285–294. doi: 10.1016/j.lungcan.2006.05.018. [DOI] [PubMed] [Google Scholar]
- 19.Kemphues KJ, Priess JR, Morton DG, Cheng NS. Identification of genes required for cytoplasmic localization in early C. elegans embryos. Cell. 1988;52(3):311–320. doi: 10.1016/s0092-8674(88)80024-2. [DOI] [PubMed] [Google Scholar]
- 20.Martin SG, St Johnston D. A role for Drosophila LKB1 in anterior-posterior axis formation and epithelial polarity. Nature. 2003;421(6921):379–384. doi: 10.1038/nature01296. [DOI] [PubMed] [Google Scholar]
- 21.Kishi M, Pan YA, Crump JG, Sanes JR. Mammalian SAD kinases are required for neuronal polarization. Science. 2005;307(5711):929–932. doi: 10.1126/science.1107403. [DOI] [PubMed] [Google Scholar]
- 22.Timm T, Matenia D, Li XY, Griesshaber B, Mandelkow EM. Signaling from MARK to tau: regulation, cytoskeletal crosstalk, and pathological phosphorylation. Neuro-degenerative diseases. 2006;3(4–5):207–217. doi: 10.1159/000095258. [DOI] [PubMed] [Google Scholar]
- 23.Shackelford DB, et al. LKB1 inactivation dictates therapeutic response of non-small cell lung cancer to the metabolism drug phenformin. Cancer Cell. 2013;23(2):143–158. doi: 10.1016/j.ccr.2012.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang W, et al. Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res. 2013;41(Database issue):D955–961. doi: 10.1093/nar/gks1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kavallaris M, et al. Taxol-resistant epithelial ovarian tumors are associated with altered expression of specific beta-tubulin isotypes. J Clin Invest. 1997;100(5):1282–1293. doi: 10.1172/JCI119642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mozzetti S, et al. Class III beta-tubulin overexpression is a prominent mechanism of paclitaxel resistance in ovarian cancer patients. Clin Cancer Res. 2005;11(1):298–305. [PubMed] [Google Scholar]
- 27.Zhang HL, et al. Association between class III beta-tubulin expression and response to paclitaxel/vinorebine-based chemotherapy for non-small cell lung cancer: a meta-analysis. Lung Cancer. 2012;77(1):9–15. doi: 10.1016/j.lungcan.2012.01.005. [DOI] [PubMed] [Google Scholar]