Abstract
Avian-origin H7N9 influenza is a novel influenza A virus (IAV) that emerged in humans in China in 2013. Using immunoinformatics tools, we identified several H7N9 T cell epitopes with T cell receptor (TCR)-facing residues identical to those of multiple epitopes from human proteins. We hypothesized that host tolerance to these peptides may impair T helper response and contribute to the low titer, weak hemagglutination inhibiting (HI) antibody responses and diminished seroconversion rates that have been observed in human H7N9 infections and vaccine trials. We found that the magnitude of human T effector responses to individual H7N9 peptides was inversely correlated with the peptide's resemblance to self. Furthermore, a promiscuous T cell epitope from the hemagglutinin (HA) protein suppressed responses to other H7N9 peptides when co-administered in vitro. Along with other highly ‘human-like’ peptides from H7N9, this peptide was also shown to expand FoxP3+ regulatory T cells (Tregs). Thus, H7N9 may be camouflaged from effective human immune response by T cell epitope sequences that avert or regulate effector T cell responses through host tolerance.
Keywords: cross-conservation, H7N9 influenza, immune evasion, immunoinformatics, JanusMatrix, regulatory T cells (Tregs), tolerance, vaccine
Abbreviations
- IAV
influenza A virus
- TCR
T cell receptor
- HI
hemagglutination inhibiting
- A(H1N1)pdm09
pandemic H1N1
- H7N9
H7N9 avian influenza
- HA
hemagglutinin
- NA
neuraminidase
- EBV
Epstein Barr Virus
- HSV
herpes simplex virus
- HIV
human immunodeficiency virus
- HCV
hepatitis C virus
- HLA
human leukocyte antigen
- Tregs
T regulatory cells
- PBMC
peripheral blood mononuclear cells
- ICS
immunogenic consensus sequences
- TRF
time resolved fluorescence
- DPBS
Dulbecco's phosphate-buffered saline
- ELISpot
enzyme-linked immunospot
- SI
stimulation index
- LAIV
live attenuated influenza vaccine
Introduction
The discordant immunogenicity of vaccines developed for 2 distinct emerging influenza A viruses (IAV), 2009 pandemic H1N1 (A(H1N1)pdm09) and H7N9 avian influenza (H7N9), provides an opportunity to evaluate the role of T cells in the development of effective humoral immune response. For example, although A(H1N1)pdm09 was highly transmissible1,2 and spread to more than 200 countries within 12 months of emergence due to the lack of pre-existing antibodies,3,4 morbidity and mortality due to the A(H1N1)pdm09 influenza were lower than expected, presumably due to pre-existing T cell responses among individuals exposed to or vaccinated with seasonal A(H1N1) strains.5 H7N9's emergence in China in 20136 was associated with much higher lethality. Due to concerns about its lethality and pandemic potential, H7N9 vaccines were prioritized for production.7
In stark contrast with A(H1N1)pdm09, unadjuvanted H7N9 vaccines were poorly antigenic and vaccination with unadjuvanted H7N9 hemagglutinin (HA) resulted in hemagglutination inhibition (HI) seroconversion rates of only 6% and 15.6% in Phase I clinical trials8,9 (as compared to 89% for similar unadjuvanted A(H1N1)pdm09 subunit vaccines10). Even when 2 doses of H7N9 vaccine were administered with adjuvant, only 59% of subjects seroconverted in a recent Phase II clinical trial.11 The development of neutralizing antibodies to H7N9 is also delayed in H7N9-infected humans, when compared to the typical immune response to other IAV infections, and IgG avidity to H7N9 HA is significantly lower.12 In clinical trials of other H7 subtypes, an attenuated H7N1 vaccine elicited low HI titers,13 and an inactivated subunit H7N7 vaccine was poorly immunogenic.14 H7 HA appears to be uniquely non-antigenic: human antibody response to a related H7 HA in the H7N7 outbreak in 2003 in the Netherlands was also diminished in HI titer.15 Taken together, these studies suggest that adaptive immune responses to H7N9 infection may be diminished and delayed, even in the context of natural infection.
Limited T helper response could explain the attenuation of humoral immunity to H7N9. CD4+ T cells provide help to B cells, supporting isotype conversion and affinity maturation16,17; thus, these clinical results suggest that T cell help was limited or abrogated. We previously reported that there are fewer CD4+ T helper epitopes in the H7N9 sequences than in other IAV.18 Similar patterns of epitope deletion have been observed in chronic (‘hit-and-stay’) viruses that have adapted to the human host, such as Epstein-Barr virus (EBV) and herpes simplex virus (HSV), but not in acute (‘hit-and-run’) viruses.19 Immune escape mediated by epitope deletion is a well-established mechanism of viral pathogenesis for human immunodeficiency virus (HIV) and hepatitis C virus (HCV),20,21 but this escape mechanism has not been previously described for influenza.
Another means by which H7N9 may minimize host response is to adopt ‘immune camouflage’, a new mechanism of immune escape identified by our group. T cell epitopes derived from pathogens that have high T cell receptor (TCR) ‘cross-conservation’ with human sequences can be identified using JanusMatrix, an algorithm that compares TCR-facing patterns of CD4+ T cell epitopes.22 Commensal viruses contain a significantly higher number of these JanusMatrix-defined ‘human-like’ T cell epitopes than viruses that do not establish chronic infections in humans.19
For example, we have identified an epitope in HCV that is highly cross-conserved with self and significantly expands T regulatory cells (Tregs) in vitro. T cells that respond to this peptide exhibit markers that are characteristic of Tregs and actively suppress bystander effector T cell responses in vitro.23 The striking difference observed between commensal viruses, which appear to have many such epitopes, and acute-disease, pathogenic viruses, suggests that immune camouflage may be an important method by which certain human pathogens escape adaptive immune response.24
When evaluating the potential immunogenicity of H7N9 using our computational tools, we identified several H7N9 CD4+ T cell epitopes that are more cross-conserved with human sequences than were similar epitopes found in other influenza strains. Notably, an H7 HA sequence that corresponds in sequence location to an immunodominant epitope of A(H3N2) and A(H1N1) bears mutations at TCR-facing positions that increase its resemblance to self antigens in the context of human leukocyte antigen (HLA)-DR presentation. We considered the possibility that these human-like H7N9 epitopes might reduce H7N9 vaccine efficacy and contribute to lower titer, lower affinity antibody development.25 We therefore performed in vitro T cell assays using peripheral blood mononuclear cells (PBMC) from naïve human donors, examining the phenotype and function of cells responding to H7N9 class II-restricted T cell epitopes that are cross-conserved with the human genome, and compared responses to these peptides with responses to corresponding peptides derived from human proteins and to less cross-conserved peptides in H7N9. As described here, we confirm that highly cross-conserved epitopes contained in H7N9 protein sequences exhibited low immunogenicity and stimulated functional Tregs, a finding that has significant implications for H7N9 vaccines and viral immunopathogenesis.
Results
Genome analysis and epitope prediction
In previously published work, we analyzed 4 human H7N9 influenza sequences (A/Hangzhou/1/2013, A/Anhui/1/2013, A/Shanghai/1/2013, and A/Shanghai/2/2013) from GISAID (http://platform.gisaid.org/) for HLA class II-restricted epitopes, and constructed immunogenic consensus sequences (ICS) to enable broad HLA and strain coverage.18 For the present study, we selected 15 representative ICS from the original publication with varying degrees of cross-conservation with self. An additional 4 published influenza A epitopes from A(H1N1), A(H3N2), and A(H5N1) and 5 peptides from human proteins were selected to serve as positive controls and human ‘analogs’ of the H7N9 peptides, respectively. The human analog peptides were among those identified by JanusMatrix as likely targets of mimicry by selected H7N9 peptides.
A quantitative measure of human genome cross-conservation called ‘JanusMatrix Delta’ was calculated for all peptides; scoring is described in detail in the Methods. A higher JanusMatrix Delta indicates a greater number of TCR matches with autologous (human) peptides, which themselves share HLA restrictions with the query peptide. JanusMatrix Delta values for the peptides used in this study ranged from 0 to 37.89. A complete list of peptides, along with their sequence similarity to corresponding peptides in circulating IAV strains and cross-conservation with the human genome, is provided in Table 1.
Table 1.
Selected ICS peptides from H7N9 influenza and controls from circulating IAV strains or human proteins
| % Similarity with IAV |
Cross-conservation with Human Sequences |
||||||
|---|---|---|---|---|---|---|---|
| Peptide Description | Peptide Name | Peptide Sequence | Source Protein | A/California/7/2009 (H1N1) | A/Victoria/361/2011 (H3N2) | JanusMatrix Delta | # of Matches |
| Immunodominant HA peptides from circulating IAV strains | IAV-1 | PKYVKSTKLRLATG | HA | 100% | 85% | 6.70 | 10 |
| IAV-2 | PRYVKQSTLKLATG | HA | 85% | 100% | 8.45 | 13 | |
| IAV-3 | PRYVKQNTLKLATG | HA | — | 97% | 6.57 | 7 | |
| IAV-4 | PKYVKSNRLVLATG | HA | 89% | — | 9.37 | 11 | |
| H7N9 ICS peptides | H7N9-1 | RIDFHWLMLNPNDTVTFS | HA | — | — | 0.00 | 0 |
| H7N9-2a | YAEMKWLLSNTDNAAFPQ | HA | — | — | 6.37 | 8 | |
| H7N9-3 | KGILGFVFTLTVPSERGLQ | M1 | 100% | 100% | 6.77 | 10 | |
| H7N9-4 | QPEWFRNVLSIAPIMFSNK | PB1 | 97% | 99% | 11.50 | 14 | |
| H7N9-5 | GFTFKRTSGSSVKRE | PB2 | 93% | 93% | 12.05 | 17 | |
| H7N9-6 | RRDQKSLRGRSSTLGLDI | NS1 | 94% | 94% | 12.33 | 15 | |
| H7N9-7 | NYLLTWKQVLAELQDIE | PA | 96% | 97% | 13.00 | 14 | |
| H7N9-8 | DKLYERVKRQLRENAEED | HA | — | 83% | 13.22 | 24 | |
| H7N9-9 | AVKLYKKLKREMTFHGA | M1 | 97% | 95% | 13.68 | 35 | |
| H7N9-10 | AANIIGILHLILWILDRL | M2 | 96% | 100% | 15.36 | 16 | |
| H7N9-11 | SRKLLLIVQALRDNLEPG | PA | 100% | 98% | 16.99 | 51 | |
| H7N9-12 | QITFMQALQLLLEVE | NEP | 100% | 93% | 17.22 | 18 | |
| H7N9-13 | PRYVKQRSLLLATG | HA | — | 89% | 21.85 | 24 | |
| H7N9-14Ab | IVYWKQWLSLKNLTQ | PB1-F2 | — | — | 25.87 | 24 | |
| H7N9-14Bb | WKQWLSLKNLTQGSL | PB1-F2 | — | — | 26.05 | 25 | |
| Autologous peptides sharing identical TCR contact residues with selected H7N9 ICS peptides | 5-HUMAN-A | LSGLKRASASSLRSI | Rho GTPase-activating protein 42 | — | — | 27.97 | 36 |
| 5-HUMAN-B | RGILKRNSSSSSTDS | Synaptotagmin-like protein 2 | — | — | 37.89 | 38 | |
| 12-HUMAN | VFHFMQSLALLMSPV | ectopic p granules protein 5 homolog | — | — | 22.71 | 14 | |
| 14A-HUMAN | EEDLKQLLALKGSSY | mitochondrial NAD kinase 2 | — | — | 32.21 | 46 | |
| 14B-HUMAN | NLELLSLKRLTLTTS | Hyccin | — | — | 26.76 | 51 | |
Column 1: groups assigned to peptides based on their immunological characteristics. Column 2: peptide names. H7N9 ICS peptide names are ordered by JanusMatrix Delta. Human analog peptides are numbered according to their corresponding H7N9 peptide. Column 3: peptide sequence. Column 4: source protein of each peptide, either from IAV or the human proteome. Column 5: percentage of similarity with IAV. Similarity to circulating strains of IAV was determined by comparing each peptide to its corresponding sequence in either of the 2 IAV strains from the 2012/13 TIV. There was no conservation with Influenza B strain Wisconsin/1/2010 for any peptide. Any percentage that was lower than 80% was represented by ‘−’. Column 6: cross-conservation of each peptide with the human genome represented by JanusMatrix Delta and the number of matches found in the human database.aSimilar to avian H7.bSimilar to Trivalent Inactivated Influenza Vaccine and Live Attenuated Influenza Vaccine backbone strains.
So as to visualize the peptide relationships identified by JanusMatrix, we used Cytoscape to provide a qualitative analysis of the predicted cross-reactivity between each peptide and the human genome.26 Figure 1 shows Cytoscape networks for each of the peptides included in this study.
Figure 1.
Three categories of peptides analyzed in JanusMatrix for human sequence cross-conservation. Cross-conservation with the human genome, visualized in Cytoscape networks, is shown. Each peptide is represented by a green diamond. HLA-binding 9-mer frames contained within the source peptide are depicted as gray squares. For each 9-mer frame, human 9-mers with similar HLA binding affinities and identical TCR-facing residues are shown as dark blue triangles, and the human proteins from which they are derived are shown as light blue circles. Accordingly, peptides with limited cross-conservation to the human genome have sparse networks surrounding the central node, and peptides with high cross-conservation to the human genome have more complex, interconnected networks with many branches.
Class II HLA binding assay
HLA class II binding affinity assays were performed to validate the computational predictions. All 24 peptides were evaluated for binding affinity in competition assays for 5 common HLA DRB1 alleles: HLA DRB1*0101, DRB1*0301, DRB1*0401, DRB1*0701, and DRB1*0801 (Fig. S1). Of all the peptide-HLA binding interactions assayed, 50% displayed strong binding affinity (estimated IC50<1μM), 13% showed moderate binding (1μM < estimated IC50<10μM), 11% showed weak binding affinity (10μM < estimated IC50<100μM) and 11% exhibited no significant affinity (estimated IC50>100μM) to the target allele. In 18 cases, the data were not sufficient to establish binding affinity.
The concordance of computational predictions and binding assay results was evaluated by classifying peptide-HLA binding pairs as either true positive (TP), false positive (FP), true negative (TN), or false negative (FN). For a given HLA allele, an EpiMatrix Z-score ≥ 1.64 indicates that the peptide is in the top 5% of predicted binders and is considered a ‘hit’. The overall predictive success rate was 85%, excluding indeterminate measurements. The correlation between prediction and binding was 82% for DRB*0101, 75% for DRB1*0301, 95% for DRB1*0401, 73% for DRB1*0701, and 100% for DRB1*0801 (Fig. 2). These correlations fall in the range of previously published results for IAV peptides predicted using EpiMatrix.27,28
Figure 2.
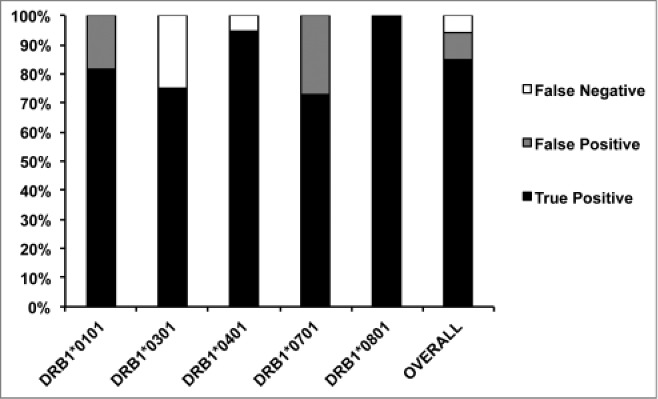
Comparison of immunoinformatic predictions and HLA binding in vitro. The HLA class II binding result for each peptide was compared to its EpiMatrix Z-score for the corresponding HLA allele. True positive (black) reflected correctly predicted HLA-binding peptide results. False positive (gray) reflected incorrectly predicted HLA-binding peptide results. False negative (white) reflected incorrectly predicted non-binding peptide results. No true negatives, which reflected correctly predicted non-binding peptide results, were found. The correspondence between predicted binding and observed binding was high, and consistent with previous observations.
HLA-DR blocking assay
To identify whether the peptides were presented by HLA-DR, we examined the effect of an anti-HLA-DR antibody on the epitope-specific T cell responses by IFNγ enzyme-linked immunospot (ELISpot) in 3 healthy donors. For ten of the peptides (IAV-1, H7N9-2, -4, -7, -9, -12, -13, -14A, 5-HUMAN-A, and -B), peptide-specific spot formation was 100% inhibited by blocking HLA-DR, indicating that these peptides are restricted by HLA-DR (Table 2).
Table 2.
Inhibition of peptide-specific responses by HLA-DR blocking antibody
| Peptide Name | % Inhibition by HLA-DR blocking Ab |
|---|---|
| IAV-1 | 100% |
| IAV-2 | 73% |
| IAV-3 | 78% |
| IAV-4 | 96% |
| H7N9-1 | 46% |
| H7N9-2 | 100% |
| H7N9-3 | 97% |
| H7N9-4 | 100% |
| H7N9-5 | N/A |
| H7N9-6 | N/A |
| H7N9-7 | 100% |
| H7N9-8 | 89% |
| H7N9-9 | 100% |
| H7N9-10 | 68% |
| H7N9-11 | increased response |
| H7N9-12 | 100% |
| H7N9-13 | 100% |
| H7N9-14A | 100% |
| H7N9-14B | N/A |
| 5_HUMAN-A | 100% |
| 5-HUMAN-B | 100% |
| 12-HUMAN | N/A |
| 14A-HUMAN | N/A |
PBMC were incubated with H7N9 peptides and controls in the presence or absence of an anti-HLA-DR blocking antibody as described in Methods. Most peptide-specific responses were inhibited by the addition of the antibody, suggesting the peptides were indeed presented by HLA-DR molecules. However, as the inhibition was not always complete, and in one case (H7N9-11) the response increased in the presence of the blocking antibody, the possibility of presentation by other class II alleles (DP, DQ) and/or class I HLA cannot be ruled out. N/A: response in absence of blocking Ab was below assay background.
Seven of the predicted peptides (IAV-2, -3, -4, H7N9-1, -3, -8, and -10) induced T cell responses that were only partially inhibited by blocking HLA-DR. This may be due to presentation by another HLA molecule such as HLA-DP, HLA-DQ, or class I HLA in addition to (or instead of) HLA-DR. Indeed, several of the peptides in this category contained class I HLA binding motifs that were identified by EpiMatrix (data not shown). In the case of peptide H7N9-11, response was absent except when HLA-DR was blocked, suggesting that other HLA alleles may present this peptide.
T cell reactivity of individual peptides
PBMC from 18 individual healthy donors were stimulated in culture with individual peptides over 8 days, and human IFNγ production was measured in response to restimulation with individual peptides in ELISpot assays (Fig. 3A). ELISpot assays were performed following a cell expansion phase because ex vivo responses to the peptides did not rise significantly above background at 24–48 hours, suggesting that epitope-specific T cell frequencies were too low to detect without expanding precursor populations (data not shown). The data were analyzed by calculating the stimulation index (SI) of each response. Although the variability among donor responses was high, when the average ELISpot SI was plotted against the JanusMatrix Delta value for each peptide, a significant negative correlation (p < 0.05) was observed, as shown in Figure 3B. A regression model relating averaged SI to JanusMatrix Delta was significant (F significance 0.02) but underpowered (Power 0.65). By considering all 432 data points generated from this 24-peptide screen of 18 subjects, JanusMatrix Delta was identified as a significant and well-powered predictor of variation in SI (F significance 0.004, Power 0.85).
Figure 3.
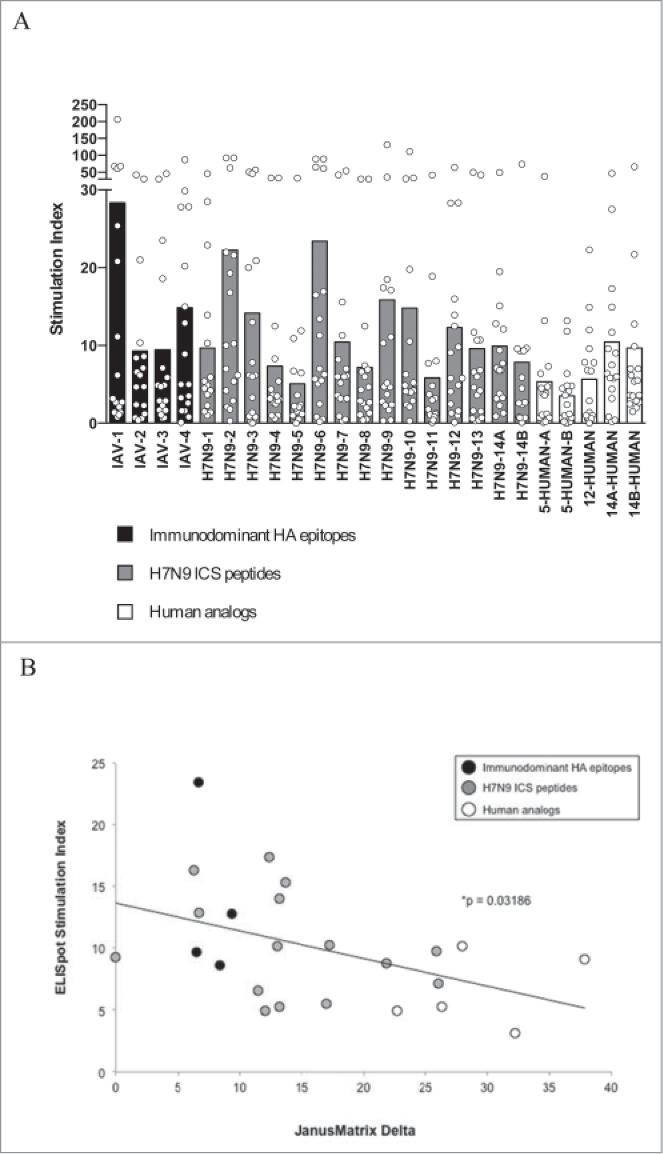
Human IFNγ responses to individual H7N9 peptides and controls. (A) The chart depicts the individual (circles) and average (bars) SI across donors (n = 18). The H7N9 peptides are arranged on the chart according to the degree of predicted cross-conservation with peptides from the human genome, as measured by JanusMatrix Delta. (B) The average response to each peptide across 18 healthy donors, as measured by SI, was significantly negatively correlated with the JanusMatrix Delta, which is a measure of cross-conservation with self. More specifically, peptides presenting the same amino acids to T cells as human protein sequences were significantly less immunogenic in vitro.
T cell reactivity of pooled peptides
So as to relate the observation described above more specifically to H7N9 infection and/or vaccination, we performed the same experiment with 2 peptide pools (Fig. 4). The first pool was comprised of H7N9 ICS peptides with JanusMatrix Delta values between 10 and 20 (H7N9-4 to -12), and the second pool contained peptides with JanusMatrix Delta values higher than 20 (H7N9-13 to -14B). Because there were more peptides in the first pool than the second, the pool concentrations were equalized to achieve the same total per unit of volume. In 4 of the 5 tested individuals, the SI values of the second pool, consisting of the most human-like H7N9 peptides, were lower than the first pool. The other individual responded with the same SI values to the 2 peptide pools. In summary, the mean SI of the second pool was significantly lower than that of the first pool (p < 0.05).
Figure 4.
Human IFNγ responses to pooled H7N9 peptides. Human IFNγ responses to pools composed of H7N9 peptides with different levels of human genome cross-conservation were measured in PBMC from healthy donors (n=5). The SI upon re-stimulation with the same peptides in ELISpot was significantly lower (*p < 0.05) in the PBMC that received the more human-like peptides. Lines connect data collected from the same donor.
These results reflect the responses of 5 donors. As there may have been significant variation between subjects due to HLA differences, additional studies were performed. For example, we also pooled peptides in groups according to their predicted immunological properties, their similarity to circulating IAV, cross-conservation with human sequences, or status as self antigens, and the results were consistent with those observed for the individual peptides in the pools (Fig. S3).
Treg phenotyping
Peptides were also tested individually for their ability to expand Tregs in healthy donor PBMC. All three peptides with JanusMatrix Delta values greater than 20 induced the expansion of significantly higher proportions of CD3+CD4+FoxP3+T cells in vitro (n = 3) (Fig. 5, p < 0.05) than culture medium. We observed similar trends in the frequency of CD25+FoxP3+ and CD39+FoxP3+ Tregs in the same assays (Fig. S5), although only the increase in CD39+FoxP3+frequency was statistically significant. Pooled influenza A epitopes that are similar to circulating IAV did not induce a similar expansion of CD25+FoxP3+ and CD39+FoxP3+ T cells in vitro (n = 9) (Fig. S4).
Figure 5.

Treg cell expansion. Human-like peptides from H7N9 induced Treg expansion. (A) The gating strategy was based on live CD3+lymphocytes, then analyzed for CD4 vs FoxP3. (B) Representative results for a single subject are shown in the dot plots, with the averages for 3 subjects shown in the chart below. *p <0.05.
Bystander suppression
We performed bystander suppression experiments to determine whether a peptide with known HLA promiscuity and a human TCR signature could exert a regulatory effect on adjacent inflammatory responses, as may occur in natural infection or vaccination. Normal subject PBMC were stimulated with a pool of H7N9 ICS peptides (including all except H7N9-1, -2, -9, and -13) in the presence or absence of peptide H7N9-13, the H7 homolog of the seasonal influenza HA immunodominant epitope. This peptide had a JanusMatrix Delta value of 21.85. To ensure that the pool was not diluted by the addition of H7N9-13, the concentrations were adjusted so that both cultures had the same absolute concentration of the pooled peptides, with the addition of H7N9-13 being the only variable. Coincubation with H7N9-13 significantly suppressed T cell response to the pooled peptides in 6 of the 7 tested individuals (Fig. 6A, p < 0.01).
Figure 6.
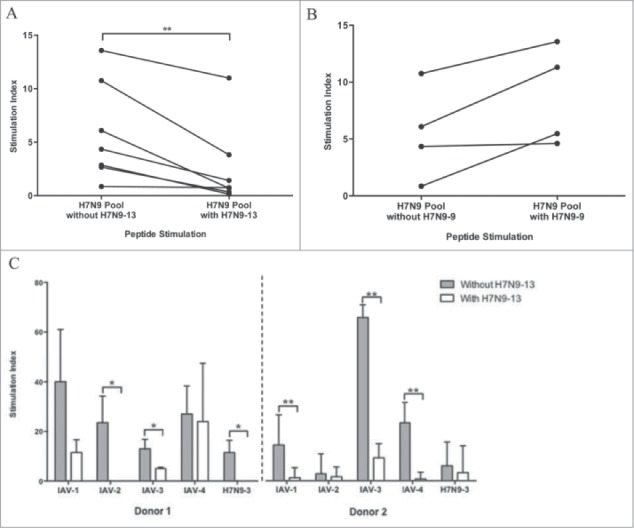
Suppressive activity of the H7 homolog of the seasonal influenza immunodominant HA epitope. Peptide H7N9-13, the H7N9 variant of an immunodominant HA epitope, was associated with a reduction in T cell response when co-administered with other peptides. Healthy donor ELISpot responses to a pool of H7N9 peptides were significantly decreased in the presence of H7N9-13 (n = 7) (A), but not H7N9-9, a less human-like peptide (n = 4) (B). H7N9-13 was able to suppress responses to other immunodominant HA peptides from circulating IAV strains (n = 2) (C). Lines connect data collected from the same donor. *p < 0.05. **p < 0.01.
In contrast, coincubation with peptide H7N9-9, which is not as cross-conserved with the human genome as H7N9-13, did not suppress T cell responses to the pool of H7N9 epitopes in all 4 individuals tested (Fig. 6B), suggesting that the immunosuppressive activity of H7N9-13 is peptide-specific.
To confirm this effect using peptides from other IAV strains, the same experiment was performed using individual or pooled peptides from the HA of circulating IAV strains (IAV-1 through -4) or an H7N9 peptide from M1 with high similarity to the sequence of circulating IAV strains (H7N9-3). Using PBMC from 2 individual donors, peptide H7N9-13 significantly suppressed T cell response to IAV-3 when co-cultured with H7N9-3 as compared to responses to IAV-3 in the absence of H7N9-3; similar reductions in T cell responses were observed to peptide IAV-1 in the first donor and IAV-2 in the second when H7N9-3 was present (Fig. 6C, all p < 0.05). Reduced responses to the pool of IAV peptides (1–4) were also observed in the presence of H7N9-13, -14A and -14B, which also had high JanusMatrix Delta scores (>20), though these reductions were not statistically significant (Fig. S6).
Discussion
Influenza vaccines can call upon memory T cells to generate protective immunity and stimulate antibody response in the absence of adjuvants; thus, usually only one vaccination is required to generate protective immunity to seasonal influenza strains. Traditionally, vaccines developed for new influenza subtypes such as A(H1N1)pdm09 and H7N9 (for which cross-reactive humoral immunity is presumed to be absent), have been adjuvanted to improve immunogenicity, and 2 immunizations are recommended to generate new memory T helper cells to the novel virus. However, pre-existing heterotypic T cell memory specific for epitopes contained in the new flu strain may obviate the need for adjuvants and effective antibody titers may develop following a single dose, as was observed for A(H1N1)pdm09.29 Thus, the T cell epitope content of vaccine immunogens appears to be a critical contributor to protective immunity. Adjuvanting poorly antigenic vaccines can amplify low-level T cell responses, augmenting the contribution of the few epitopes that are present to improve protective antibody titers.
While T cell epitopes that recall pre-existing immunity may help protect against multiple viral subtypes as was observed for A(H1N1)pdm09 influenza,30 epitopes that resemble host sequences may be detrimental to immunity. Using the JanusMatrix tool, we identified epitopes in H7N9 that are cross-conserved with multiple predicted HLA ligands from human proteins. Based on our previous discovery of human-like Treg epitopes in HCV,23 we hypothesized that similarly cross-conserved epitopes in H7N9 might be responsible for the attenuation of adaptive immunity to H7N9. As exposed donor blood was not available, we evaluated the responses of H7N9-naïve subject PBMC to H7N9 influenza T cell epitope peptides. Even though the response of individual peptide varied in different subjects (Fig. 3A), it was inversely correlated with their degree of cross-conservation with the human genome on their TCR face (Fig. 3B). The high variability in antigen-specific IFNγ release that is observed in ELISpot assays using whole PBMC cultures may be attributable to multiple factors including (i) efficiency of antigen presentation, (ii) binding affinity of a peptide for an individual's HLA type, (iii) availability of T cells with TCRs that recognize the peptide-HLA complex, and (iv) competition between T cells for domains on the antigen-presenting cells. A further source of variability is that the peptides tested comprise multiple 9-mers with potential to bind HLA. It is the sum of IFNγ responses to the individual 9-mers, each of which is governed by the factors described above, that is measured in the ELISpot assay, which further compounds variability. Despite the high potential for inconsistency, a clear inverse correlation emerges with 65% power in an “averaged data” model and 85% power in an “all data” construct. Future studies that measure responses to individual 9-mers would help reduce variability and strengthen the correlation.
Hypothesizing that attenuation of response could be due to Tregs that recognize the H7 epitopes, we discovered that Tregs expand in vitro in co-cultures with the human-like H7 epitopes (Fig. 5). We also confirmed the functionality of these expanded Tregs in bystander suppression assays (Fig. 6). While the exact origin of the Treg cells that respond to the human-like H7N9 epitopes remains to be defined (thymic-derived natural Tregs or induced peripheral Tregs), the implications for vaccine development are clear. If, in the context of natural infection or unadjuvanted vaccination using H7N9 HA, Treg responses are induced by these epitopes, humoral immune responses may be diminished and delayed, as has been reported in H7N9 infection.12
Although exploration of the impact of Treg epitopes and Tregs on humoral immune responses in the context of infection is relatively new,31 we have described the importance of Treg-activating epitopes derived from IgG (now known as ‘Tregitopes’) in the context of anti-drug antibodies to protein therapeutics.32-34 In a retrospective analysis of published viral epitopes in a large epitope database, greater human cross-conservation was associated with absent or Treg responses.19 Taken together, these findings demonstrate that certain human pathogens may evolve to contain T cell epitopes in their proteomes that resemble important human Treg epitopes, a phenomenon we have called ‘immune camouflage’.24
The T cell epitope profile of H7N9 (few effector T cell epitopes and many cross-conserved epitopes) is much closer to these ‘hit-and-stay’ viruses than viruses that ‘hit-and-run’ as described in our previous publication on host pathogens.19 Although human-to-human transmission of H7N9 is rare, the virus has been noted to have a ‘mammalian signature’.35,36 Cases of human-to-human transmission case have been reported.37 Perhaps human-to-human transmission of H7N9 occurs more frequently than suspected,38 as it is harder to detect due to low titers of antibody. The discovery of human-like epitopes in the H7N9 proteome raises an important question about the origin and evolution of H7N9 and the duration of its circulation in human beings or other mammals.
Building on our previous work, this report provides additional in vitro evidence that T cells responding to epitopes that are cross-conserved between pathogens and their hosts may be immunomodulatory; a discovery that deserves further investigation in other human pathogens.
The discovery of these T cell epitopes (and others like them) was possible using a new tool, JanusMatrix. JanusMatrix is a homology analysis tool that considers aspects of antigen recognition that are not captured by raw sequence alignment. At the level of the TCR-HLA-II-peptide interaction, there is evidence to support the designation of amino acids in positions 2, 3, 5, 7, and 8 as ‘TCR-facing’39; these are used to identify homologous epitopes in sets of peptides predicted to be restricted by the same HLA. The same authors also describe a role for several positions in the class II HLA ligand that lie outside of the central binding groove, notably at the N-terminus. Currently, the JanusMatrix tool does not integrate the contributions of these distal residues. Further studies are needed to determine whether these residues would improve the discovery of cross-conserved epitopes with the JanusMatrix tool.
For this study, we limited the JanusMatrix evaluation of ‘analogs’ to human-origin peptides that were cross-conserved with selected H7N9 ICS peptides, and did not evaluate the influence of TCR cross-conservation with epitopes derived from other human pathogens, or from human commensals. Others have been pioneers in that regard, establishing evidence for immune modulation (termed ‘heterologous immunity’) in a range of viral infections; these studies have focused on class I HLA-restricted epitopes.40,41 Epitopes that are cross-conserved with the human microbiome have also been described, and may contribute to T cell reactivity.42
Cross-reactivity with human pathogens or commensals may explain in part why one of the H7N9 ICS peptides, H7N9-2, was more immunogenic in vitro, in H7N9-unexposed donor PBMC than H7N9-1, despite their lack of similarity to circulating strains of IAV (Fig. 3A). We applied JanusMatrix to retrospectively analyze these peptides, but instead of searching the human genome for matches, we searched databases of bacterial and viral pathogens.43,44 JanusMatrix identified 168 HLA-binding sequences from the databases that present the same amino acids to the TCR as peptide H7N9-2. By contrast, only 7 such sequences were found corresponding to peptide H7N9-1, which was less immunogenic in vitro. Pre-existing memory T cells, expanded in the course of past infections, may have contributed to the response observed to H7N9-2.
With so many possible avenues for cross-reactivity at the TCR face of like-HLA-binding peptides, the calculation of a fully predictive metric for T cell epitope response remains an elusive goal. Previously, we identified a ratio of human genome to human microbiome cross-conservation that was associated with a regulatory, rather than effector, T cell response.22 Here, we used the JanusMatrix Delta score, which was significantly inversely correlated with the magnitude of effector T cell response (Fig. 3B). Additional prospective studies and retrospective analyses need to be performed to better define how JanusMatrix Delta or other measures of ‘human-ness’ can be used to predict the phenotype of T cells responding to specific epitopes.
Experiments performed on H7N9-infected cells would be instructive for the elaboration of these important relationships, but we did not have access to H7N9-exposed subjects in Providence, RI. Instead, we examined T cell responses in restimulation assays using PBMC from unexposed donors. These in vitro studies of naïve donors are still relevant, since the responses observed may be representative of responses that might be generated following vaccination or infection of H7N9-unexposed human subjects.45
Finally, we limited our studies to a manageable number of possible T cell epitopes from H7N9 and did not complete a comprehensive analysis of all H7N9 cross-conserved epitopes. Nonetheless, these studies serve as a benchmark against which future in vitro studies can be conducted for H7N9 and other influenza strains. Future studies should also examine the impact of these immunomodulatory H7N9 epitopes on CD8+ T cell responses.46,47 We are now evaluating whether removal or modification of the human-like epitopes that trigger Tregs will improve vaccine efficacy in vivo.
In conclusion, H7N9 influenza T cell epitopes that have a high degree of cross-conservation with the human host can expand Tregs in vitro and reduce IFNγ secretion in PBMC when co-incubated with other H7N9 peptides, in contrast to epitopes that are less cross-conserved with self. When this study is considered in the context of previous work by our group and other laboratories,23,25 cross-conservation of T helper epitopes with epitope sequences in the human proteome appears to be an important modulator of immune response to viral pathogens.
Modulation of T and B cell responses by these human-like epitopes probably reduces the titer and affinity of neutralizing antibodies to H7N9 HA, in vaccination and infection. In addition to posing a barrier to the success of conventional approaches currently being used to develop H7N9 vaccines, ‘immune camouflage’ can be added to the list of mechanisms by which human pathogens may escape from or modulate human immune defense.
Materials and Methods
Peptide similarity to circulating IAV and cross-conservation with human genome
The similarity of H7N9 peptides to other IAV strains was identified as previously published.18 Cross-conservation with the human genome was evaluated using JanusMatrix.22 In this analysis, we translated the UniProt reviewed human genome database48 as the source of human sequences for comparison.
H7N9 ICS peptides were generated as previously described.18 Given a peptide containing multiple HLA-binding 9-mer frames, JanusMatrix divides each such frame into T cell receptor-facing residues (positions 2, 3, 5, 7, and 8) and HLA-binding residues (positions 1, 4, 6, and 9).39 Subsequently, JanusMatrix searches for potentially cross-conserved epitopes (100% TCR-facing identity and predicted to bind at least one of the same HLA supertypes) in the human genome database. Finally, a differential (Delta) score is calculated by applying a user-defined deduction to each EpiMatrix hit in the source peptide for each TCR-matched 9-mer found in the human genome (set for the purpose of the current study at 10% of the human 9-mer's Z-score). After deduction, the hits in the source peptide are summed and used to calculate a JanusMatrix-adjusted Cluster Score. The difference between a peptide's original EpiMatrix Cluster Score and its JanusMatrix-adjusted Cluster Score is called the JanusMatrix Delta. A higher JanusMatrix Delta value implies that the original potential for immunogenicity may be discounted by greater cross-conservation with the human genome. The calculation is illustrated below:
Grouping peptides by predicted immunological properties
To compare immune responses to IAV epitopes in vitro using human PBMC, IAV peptides that could elicit several types of possible immune responses were selected in this study. The first group consisted of peptides representing variants of the immunodominant and highly conserved HA epitope, from IAV strains other than H7: A(H1N1), A(H3N2) and A(H5N1).49-52
The second group of peptides was selected from a list of ICS peptides derived from the H7N9 antigens (H7N9 ICS peptides) identified in a previous publication.18 A subset of the 101 ICS generated by the EpiAssembler algorithm were selected for this study on the basis of promiscuous HLA binding potential, lack of cysteines and hydrophobic domains known to result in difficulties with peptide synthesis, and predicted TCR/HLA matches with the human genome using the JanusMatrix algorithm described above. The H7N9 ICS peptides are ordered by their JanusMatrix Delta scores. In some of the assays described, this set of peptides was further separated into pools according to their degree of cross-conservation with the human genome.
For those H7N9 peptides with the most extensive human cross-conservation, one or 2 peptides from human sequences with which the corresponding H7N9 peptide shared TCR-facing residues were selected for synthesis. These human ‘analog’ peptides were numbered by the H7N9 peptide with which they share TCR-facing amino acids. For example, 12-HUMAN is the human analog of peptide H7N9-12.
Peptide synthesis
Synthetic peptides were manufactured using 9-fluoronylmethoxycarbonyl (Fmoc) chemistry by 21st Century Biochemicals (Marlboro, MA). Peptide purity was >80% as ascertained by analytical reversed phase HPLC. Peptide mass was confirmed by tandem mass spectrometry.
Class II HLA binding assays
Class II HLA binding assays were performed to screen predicted epitope sequences for binding to multiple HLA alleles as previously described.53 Briefly, non-biotinylated test peptides over 3 concentrations (1, 10, and 100 µM) were used to compete for binding against a biotinylated standard peptide (25 nM) to soluble class II molecules (Benaroya Institute, Seattle, WA). The reaction was incubated at 37°C for 24 hours to reach equilibrium. Class II HLA-peptide complexes were then captured on 96-well plates coated with pan anti-HLA-DR antibodies (L243, anti-HLA-DRA, BioXCell). The microwell plates were then washed to remove excess peptide and incubated with Europium-labeled streptavidin (Perkin-Elmer) for one hour at room temperature. Europium activation buffer (Perkin-Elmer) was added to develop the plates for 15-20 minutes at room temperature before the plates were read on a Time Resolved Fluorescence (TRF) plate reader. Assays were performed in triplicate. Binding assays were performed for all 24 peptides, for 5 alleles: DRB1*0101, DRB1*0301, DRB1*0401, DRB1*0701, and DRB1*0801, a selection of HLA class II alleles that provides a broad representation of class II HLA allele binding pockets. 54
PBMC isolation
Leukocyte reduction filters were obtained from de-identified healthy donors (Rhode Island Blood Center, RI) and buffy coats were obtained from age-identified healthy donors (Research Blood Components, MA). All studies using human blood were performed in accordance with NIH regulations and with the approval of the URI institutional review board.
All leukocyte reduction filters and buffy coats were obtained and processed on the same day as the blood was drawn. Fresh PBMC were isolated from leukocyte reduction filters or buffy coats by Ficoll density gradient centrifugation as follows: leukocyte reduction filters were back-flushed by Hank's Balanced Salt Solution (HBSS, Cellgro, Manassas, VA) with 2.5% sucrose and 5mM EDTA (pH = 7.2). Buffy coats were removed by a syringe and diluted in Dulbecco's Phosphate-Buffered Saline (DPBS, Thermo Fisher Scientific, Waltham, MA). Blood from filters or buffy coats was underlaid with Ficoll (Histopaque 1077, Sigma-Aldrich, St. Louis, MO) before centrifugation to isolate mononuclear cells. PBMC were transferred to separate tubes and washed twice in DPBS. PBMC were then resuspended in cell culture medium: RPMI 1640 (Cellgro) with 10% human AB serum (Valley Biomedical, Winchester, VA), 1% L-glutamine (Life Technologies, Carlsbad, CA) and 0.1% Gentamycin (Cellgro).
PBMC culture
Freshly isolated PBMC were cultured with individual peptides (10 μg/ml) or pools of peptides (10 μg/ml) over 8 days at 37°C under a 5% CO2 atmosphere to expand antigen-specific T cells. Prior to placing the peptides in culture, they were dissolved in DMSO and further diluted in culture medium. The maximum concentration of DMSO per peptide per well was 0.2%. In wells of a 48-well cell culture plate, 2 × 106 cells in 150 μl of culture medium were stimulated with 150 µl each individual peptide or pool. Positive control wells received PHA (Thermo Fisher Scientific, Waltham, MA) at 1 μg/ml or CEFT peptide pool (CTL, Shaker Heights, OH) at 10 μg/ml. Negative control wells only received culture medium with 0.2% DMSO. At days 3 and 6, cells were supplemented with 10 ng/ml of IL-2 (BD PharMingen, San Diego, CA) by half media replacement. At day 8, PBMC were collected and washed in preparation for antigen re-stimulation to measure cytokine secretion by ELISpot assay. For HLA-DR blocking experiments, PBMC from the same donor were cultured in the presence or absence of 5 μg/ml purified NA/LE mouse anti-human HLA-DR antibody (BD PharMingen, San Diego, CA).
ELISpot Assay
Human IFNγ ELISpot assays were performed using a Mabtech (Cincinnati, OH) IFNγ ELISpot Kit according to the manufacturer's protocol. Briefly, cells from the 8-day expansion period were transferred at 1×105/well or 1.5×105/well to ELISpot plates that were pre-coated with anti-human IFNγ capture antibody, and re-stimulated with corresponding peptides at 10 µg/ml. Positive control wells were stimulated with PHA at 1 µg/ml or CEFT at 10 µg/ml. Negative controls only received culture medium with DMSO at the same concentration as would be present in peptide-stimulated cultures (0.2%). All stimulations and controls were administered in triplicate wells. ELISpot plates were incubated for 24 hours at 37°C under a 5% CO2 atmosphere, washed and incubated with a secondary HRP-labeled anti-IFNγ detection antibody, and developed by the addition of TMB substrate. Raw spot counts were recorded using an ImmunoSpot reader (CTL S5 UV Analyzer). Responses were considered positive if the number of spots was greater than 50 over background per million PBMC and at least twice the background. The ELISpot SI was determined by dividing the average number of spots in each peptide triplicate by the average number of spots in the negative control wells.
Multicolor flow cytometry
2 × 106 PBMC were stimulated with individual or pooled peptides at 10 μg/ml, or culture medium with 0.2% DMSO (negative control), in the presence of anti-CD49d and anti-CD28 antibody at 0.5 μg/ml (BD PharMingen) over 8 days. IL-2 (10 ng/ml) was added at days 3 and 6. At day 8, PBMC were re-stimulated with the corresponding peptides at 10 μg/ml or negative control for 24 hours. At day 9, cells were collected and washed in preparation for flow cytometric analysis.
Re-stimulated PBMC were first incubated with fixable viability stain 450 (BD Horizon) for 15 minutes at room temperature. Afterwards, cells were stained with fluorochrome-conjugated anti-human monoclonal antibodies against T cell surface antigens (Alexa Fluor 700 anti-CD3, PerCP-Cy5.5 anti-CD4, APC anti-CD25, and FITC anti-CD39, BD PharMingen) for 30 minutes at 4°C. Cells were then fixed and permeabilized by using FoxP3 Fixation/Permeabilization solution (FoxP3/Transcription Factor Staining Buffer Set, eBioscience, San Diego, CA) for 30 minutes at room temperature before being stained with PE-conjugated anti-human FoxP3 antibody (clone 259D/C7, BD PharMingen) for at least 30 minutes at room temperature. Cells were washed with FoxP3 Permeabilization Buffer (eBioscience, San Diego, CA) and acquired by flow cytometry using a Beckton-Dickinson LSR-II flow cytometer. Data were analyzed in FlowJo software (Treestar, Ashland, OR).
Statistical analysis
Tests to determine p-value and statistical significance were performed using Graphpad Prism or Microsoft Excel. When correlating JanusMatrix Delta with SI, the Pearson function was used to determine the R value and a regression model of variance to retrospectively determine study power. Student's t-test was used to calculate statistical significance between paired or unpaired T cell reactivity values.
Disclosure of Potential Conflicts of Interest
Anne S. De Groot and William D. Martin are founders and majority owners of EpiVax, Inc. a biotechnology company that provides access to immunoinformatics tools and designs vaccines for commercial clients. Leonard Moise holds options in EpiVax, Inc. These authors acknowledge that there is a potential conflict of interest related to their relationship with EpiVax and attest that the work contained in this research report is free of any bias that might be associated with the commercial goals of the company.
Acknowledgments
The authors thank Dr. Alan Rothman (University of Rhode Island), Dr. Loren Fast (Rhode Island Hospital), Dr. Yoshimasa Takahashi and Dr. Manabu Ato (National Institute of Infectious Diseases, Japan) for helpful discussions.
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
Funding
This work was supported in part by NIH Grant AI082642.
References
- 1.Cumulative number of confirmed human death cases of pandemic influenza A/(H1N1) reported to WHO. Geneva, SWITZERLAND: 2010. [Google Scholar]
- 2.Dawood FS, Iuliano AD, Reed C, Meltzer MI, Shay DK, Cheng PY, Bandaranayake D, Breiman RF, Brooks WA, Buchy P, et al.. Estimated global mortality associated with the first 12 months of 2009 pandemic influenza A H1N1 virus circulation: a modelling study. Lancet Infect Dis 2012; 12:687-95; PMID:22738893; http://dx.doi.org/ 10.1016/S1473-3099(12)70121-4 [DOI] [PubMed] [Google Scholar]
- 3.Katz J, Hancock K, Veguilla V, Zhong W, Lu XH, Sun H, Butler E, Dong L, Liu F, Li ZN, et al.. Serum cross-reactive antibody response to a novel influenza A (H1N1) virus after vaccination with seasonal influenza vaccine. MMWR Morb Mortal Wkly Rep 2009; 58:521-24; PMID:19478718 [PubMed] [Google Scholar]
- 4.Hancock K, Veguilla V, Lu X, Zhong W, Butler EN, Sun H, Liu F, Dong L, DeVos JR, Gargiullo PM, et al.. Cross-reactive antibody responses to the 2009 pandemic H1N1 influenza virus. N Engl J Med 2009; 361:1945-52; PMID:19745214; http://dx.doi.org/ 10.1056/NEJMoa0906453 [DOI] [PubMed] [Google Scholar]
- 5.Schanen BC, De Groot AS, Moise L, Ardito M, McClaine E, Martin W, Wittman V, Warren WL, Drake DR 3rd. Coupling sensitive in vitro and in silico techniques to assess cross-reactive CD4 (+) T cells against the swine-origin H1N1 influenza virus. Vaccine 2011; 29:3299-309; PMID:21349362; http://dx.doi.org/ 10.1016/j.vaccine.2011.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao R, Cao B, Hu Y, Feng Z, Wang D, Hu W, Chen J, Jie Z, Qiu H, Xu K, et al.. Human infection with a novel avian-origin influenza A (H7N9) virus. N Engl J Med 2013; 368:1888-97; PMID:23577628; http://dx.doi.org/ 10.1056/NEJMoa1304459 [DOI] [PubMed] [Google Scholar]
- 7.NIH begins testing H7N9 avian influenza vaccine candidate, possible role for adjuvants to be examined. 2013; (Press Release, National Institutes of Health, Bethesda, Maryland: ). http://www.nih.gov/news/health/sep2013/niaid-18.htm [Google Scholar]
- 8.Novartis announces positive clinical trial results for novel H7N9 vaccine. Media Release Novartise 2013; http://www.novartis.com/newsroom/media-releases/en/2013/1743124.shtml [Google Scholar]
- 9.Fries LF, Smith GE, Glenn GM. A Recombinant Viruslike Particle Influenza A (H7N9) Vaccine. N Engl J Med 2013; 369:2564-6; PMID:24224560; http://dx.doi.org/ 10.1056/NEJMc1313186 [DOI] [PubMed] [Google Scholar]
- 10.Griffin MR, Monto AS, Belongia EA, Treanor JJ, Chen Q, Chen J, Talbot HK, Ohmit SE, Coleman LA, Lofthus G, et al.. Effectiveness of non-adjuvanted pandemic influenza A vaccines for preventing pandemic influenza acute respiratory illness visits in 4 U.S. communities. PLoS One 2011; 6:e23085; PMID:21857999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mulligan MJ, Bernstein DI, Winokur P, Rupp R, Anderson E, Rouphael N, Dickey M, Stapleton JT, Edupuganti S, Spearman P, et al.. Serological responses to an avian influenza A/H7N9 vaccine mixed at the point-of-use with MF59 adjuvant: a randomized clinical trial. JAMA 2014; 14:1409-19; http://dx.doi.org/ 10.1001/jama.2014.12854 [DOI] [PubMed] [Google Scholar]
- 12.Guo L, Zhang X, Ren L, Yu X, Chen L, Zhou H, Gao X, Teng Z, Li J, Hu J, et al.. Human antibody responses to avian influenza A (H7N9) virus. Emerg Infect Dis 2013; 20:192-200; http://dx.doi.org/ 10.3201/eid2002.131094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cox RJ, Madhun AS, Hauge S, Sjursen H, Major D, Kuhne M, Höschler K, Saville M, Vogel FR, Barclay W, Donatelli I, Zambon M, Wood J, Haaheim LR. A phase I clinical trial of a PER.C6 cell grown influenza H7 virus vaccine. Vaccine 2009; 27:1889-97; PMID:19368768; http://dx.doi.org/ 10.1016/j.vaccine.2009.01.116 [DOI] [PubMed] [Google Scholar]
- 14.Couch RB, Patel SM, Wade-Bowers CL, Niño D. A randomized clinical trial of an inactivated avian influenza A (H7N7) vaccine. PLoS One 2012; 7:e49704; PMID:23239968; http://dx.doi.org/ 10.1371/journal.pone.0049704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meijer A, Bosman A, van de Kamp EE, Wilbrink B, Du Ry van Beest Holle M, Koopmans M. Measurement of antibodies to avian influenza virus A (H7N7) in humans by hemagglutination inhibition test. J Virol Methods 2006; 132:113-20; PMID:16271401; http://dx.doi.org/ 10.1016/j.jviromet.2005.10.001 [DOI] [PubMed] [Google Scholar]
- 16.Leddon SA, Sant A. The peptide specificity of the endogenous T follicular helper cell repertoire generated after protein immunization. PLoS One 2012; 7:e46952; PMID:23077537; http://dx.doi.org/ 10.1371/journal.pone.0046952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nayak JL, Fitzgerald TF, Richards KA, Yang H, Treanor JJ, Sant AJ. CD4+ T-cell expansion predicts neutralizing antibody responses to monovalent, inactivated 2009 pandemic influenza A (H1N1) virus subtype H1N1 vaccine. J Infect Dis 2013; 207:297-305; PMID:23148285; http://dx.doi.org/ 10.1093/infdis/jis684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Groot AS, Ardito M, Terry F, Levitz L, Ross T, Moise L, Martin W. Low immunogenicity predicted for emerging avian-origin H7N9: implication for influenza vaccine design. Hum Vaccin Immunother 2013; 9:950-6; PMID:23807079; http://dx.doi.org/ 10.4161/hv.24939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He L, De Groot AS, Gutierrez AH, Martin WD, Moise L, Bailey-Kellogg C. Integrated assessment of predicted MHC binding and cross-conservation with self reveals patterns of viral camouflage. BMC Bioinformatics 2014; 15:S1; PMID:25104221; http://dx.doi.org/ 10.1186/1471-2105-15-S4-S1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vider-Shalit T, Sarid R, Maman K, Tsaban L, Levi R, Louzoun Y. Viruses selectively mutate their CD8+ T-cell epitopes—a large-scale immunomic analysis. Bioinformatics 2009; 25:i39-i44; PMID:19478014; http://dx.doi.org/ 10.1093/bioinformatics/btp221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neumann-Haefelin C, Frick DN, Wang JJ, Pybus OG, Salloum S, Narula GS, Eckart A, Biezynski A, Eiermann T, Klenerman P, et al.. Analysis of the Evolutionary Forces in an Immunodominant CD8 Epitope in Hepatitis C Virus at a Population Level. J Virol 2008; 82:3438-51; PMID:18216107; http://dx.doi.org/ 10.1128/JVI.01700-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moise L, Gutierrez AH, Bailey-Kellogg C, Terry F, Leng Q, Abdel Hady KM, VerBerkmoes NC, Sztein MB, Losikoff PT, Martin WD, et al.. The two-faced T cell epitope: examining the host-microbe interface with JanusMatrix. Hum Vaccin Immunother 2013; 9:1577-86; PMID:23584251; http://dx.doi.org/ 10.4161/hv.24615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Losikoff PT, Mishra S, Terry F, Gutierrez A, Ardito MT, Fast L, Nevola M, Martin WD, Bailey-Kellogg C, De Groot AS, et al.. HCV Epitope, homologous to multiple human protein sequences, induces a regulatory T cell response in infected patients. J Hepatol 2015; 62(1):48-55. [DOI] [PubMed] [Google Scholar]
- 24.Moise L, Liu R, Gutierrez AH, Tassone R, Bailey-Kellogg C, Martin WD, De Groot A. Immune Camouflage: Relevance to Vaccine Design and Human Immunology. Hum Vaccin. Immunother 2014; 1:e36134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.De Groot AS, Moise L, Liu R, Gutierrez AH, Terry F, Koita OA, Ross TM, Martin W. Cross-conservation of T-cell epitopes: now even more relevant to (H7N9) influenza vaccine design. Hum. Vaccin Immunother 2014; 10:256-62; http://dx.doi.org/ 10.4161/hv.28135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 2003; 13:2498-504; PMID:14597658; http://dx.doi.org/ 10.1101/gr.1239303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moise L, Terry F, Ardito M, Tassone R, Latimer H, Boyle C, Martin WD, De Groot AS. Universal H1N1 influenza vaccine development: identification of consensus class II hemagglutinin and neuraminidase epitopes derived from strains circulating between 1980 and 2011. Hum Vaccin Immunother 2013; 9:1598-1607; PMID:23846304; http://dx.doi.org/ 10.4161/hv.25598 [DOI] [PubMed] [Google Scholar]
- 28.De Groot AS, Martin W. Reducing risk, improving outcomes: bioengineering less immunogenic protein therapeutics. Clin Immunol 2009; 131:189-201; PMID:19269256; http://dx.doi.org/ 10.1016/j.clim.2009.01.009 [DOI] [PubMed] [Google Scholar]
- 29.Greenberg ME, Lai MH, Hartel GF, Wichems CH, Gittleson C, Jillian B, Gail D, Wilson H, Connie L, Diane W, et al.. Response to a monovalent 2009 influenza A (H1N1) vaccine. N Engl J Med 2009; 361:2405-13; PMID:19745216; http://dx.doi.org/ 10.1056/NEJMoa0907413 [DOI] [PubMed] [Google Scholar]
- 30.Laurie KL, Carolan LA, Middleton D, Lowther S, Kelso A, Barr IG. Multiple infections with seasonal influenza A virus induced cross-protective immunity agasint A(H1N1)pandemic influenza virus in a ferret model. J Infect Dis 2010; 202:1011-20; PMID:20715930; http://dx.doi.org/ 10.1086/656188 [DOI] [PubMed] [Google Scholar]
- 31.Wang SM, Tsai MH, Lei HY, Wang JR, Liu CC. The regulatory T cells in anti-influenza antibody response post influenza vaccination. Hum Vaccin Immunother 2012; 8:1243-1249; PMID:22894960; http://dx.doi.org/ 10.4161/hv.21117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cousens L, De Groot AS, Najafian N, Martin WD. Tregitopes: an immunomodulation powerhouse. Hum Immunol 2014; 75:1139-46; PMID:25454619; http://dx.doi.org/ 10.1016/j.humimm.2014.10.012 [DOI] [PubMed] [Google Scholar]
- 33.De Groot AS, Terry F, Cousens L, Martin W. Beyond Humanization and De-immunization: tolerization as a method for reducing the immunogenicity of biologics. Expert Rev Clin Pharmacol 2013; 6:651-62; PMID:24164613; http://dx.doi.org/ 10.1586/17512433.2013.835698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jawa V, Cousens LP, Awwad M, Wakshull E, Kropshofer H, De Groot AS. T-cell dependent immunogenicity of protein therapeutics: preclinical assessment and mitigation. Clin Immunol 2013; 149:534-55; PMID:24263283; http://dx.doi.org/ 10.1016/j.clim.2013.09.006 [DOI] [PubMed] [Google Scholar]
- 35.Background and summary of human infection with avian influenza A (H7N9) virus – as of 31 January 2014 World Health Organization, Geneva, Switzerland: 2014; (http://www.who.int/influenza/human_animal_interface/latest_update_h7n9/en/) [Google Scholar]
- 36.Kageyama T, Fujisaki S, Takashita E, Xu H, Yamada S, Uchida Y, Neumann G, Saito T, Kawaoka Y, Tashiro M. Genetic analysis of novel avian A (H7N9) influenza viruses isolated from patients in China, February to April 2013. Euro Surveill 2013; 18:20453; PMID:23594575 [PMC free article] [PubMed] [Google Scholar]
- 37.Gao HN, Yao HP, Liang WF, Wu XX, Wu HB, Wu NP, Yang SG, Zhang Q, Su KK, Guo J, et al.. Viral genome and antiviral drug sensitivity analysis of two patients from a family cluster caused by the influenza A (H7N9) virus in Zhejiang, China. Int J Infect Dis 2014; 29C:254-8; http://dx.doi.org/ 10.1016/j.ijid.2014.10.029 [DOI] [PubMed] [Google Scholar]
- 38.Kucharski A, Mills H, Pinsent A, Fraser C, Van Kerkhove M, Donnelly CA, Riley S. Distinguishing between reservoir exposure and human-to-human transmission for emerging pathogens using case onset data. PLoS Curr 2014; 6: pii: ecurrents.outbreaks.e1473d9bfc99d080ca242139a06c455f; PMID:24619563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Patarroyo ME, Cifuentes G, Salazar LM, Espejo F, Alba MP, Bermúdez A. Based on HLA-DR beta1* allele binding specificities, striking differences in distance and TCR Contacting Residue Orientation can be observed in modified protection-inducing malarial synthetic peptides. Curr Med Chem 2005; 12:2849-65; PMID:16305475; http://dx.doi.org/ 10.2174/092986705774454733 [DOI] [PubMed] [Google Scholar]
- 40.Clute SC, Watkin LB, Cornberg M, Naumov YN, Sullivan JL, Luzuriaga K, Welsh RM, Selin LK. Cross-reactive influenza virus-specific CD8+ T cells contribute to lymphoproliferation in Epstein-Barr virus-associated infectious mononucleosis. J Clin Invest 2005; 115: 3602-12; PMID:16308574; http://dx.doi.org/ 10.1172/JCI25078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Welsh RM, Che JW, Brehm MA, Selin LK. Heterologous immunity between viruses. Immunol Rev 2010; 235:244-66; PMID:20536568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su LF, Kidd BA, Han A, Kotzin JJ, Davis MM. Virus-specific CD4 (+) memory-phenotype T cells are abundant in unexposed adults. Immunity 2013; 38:373-83; PMID:23395677; http://dx.doi.org/ 10.1016/j.immuni.2012.10.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Calis JJ, de Boer RJ, Keşmir C. Degenerate T-cell recognition of peptides on MHC molecules creates large holes in the T-cell repertoire. PLoS Comput Biol 2012; 8:e1002412; PMID:22396638; http://dx.doi.org/ 10.1371/journal.pcbi.1002412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hertz T, Nolan D, James I, John M, Gaudieri S, Phillips E, Huang JC, Riadi G, Mallal S, Jojic N. Mapping the landscape of host-pathogen coevolution: HLA class I binding and its relationship with evolutionary conservation in human and viral proteins. J Virol 2011; 85:1310-21; PMID:21084470; http://dx.doi.org/ 10.1128/JVI.01966-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wullner D, Zhou L, Bramhall E, Kuck A, Goletz TJ, Swanson S, Chirmule N, Jawa V. Considerations for optimization and validation of an in vitro PBMC derived T cell assay for immunogenicity prediction of biotherapeutics. Clin Immunol 2010; 137:5-14; PMID:20708973; http://dx.doi.org/ 10.1016/j.clim.2010.06.018 [DOI] [PubMed] [Google Scholar]
- 46.Epstein SL, Lo CY, Misplon JA, Bennink JR. Mechanism of protective immunity against influenza virus infection in mice without antibodies. J Immunol 1998; 160:322-7; PMID:9551987 [PubMed] [Google Scholar]
- 47.Price GE, Soboleski MR, Lo CY, Misplon JA, Pappas C, Houser KV, Tumpey TM, Epstein SL. Vaccination focusing immunity on conserved antigens protects mice and ferrets against virulent H1N1 and H5N1 influenza A viruses. Vaccine 2009; 27:6512-21; PMID:19729082; http://dx.doi.org/ 10.1016/j.vaccine.2009.08.053 [DOI] [PubMed] [Google Scholar]
- 48.The UniProt Consortium . Reorganizing the protein space at the Universal Protein Resource (UniProt). Nucleic Acids Res 2012; 40:D71-5; PMID:22102590; http://dx.doi.org/ 10.1093/nar/gkr981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yang J, James E, Gates TJ, DeLong JH, LaFond RE, Malhotra U, Kwok WW. CD4+ T cells recognize unique and conserved 2009 H1N1 influenza hemagglutinin epitopes after natural infection and vaccination. Int Immunol 2013; 25:447-57; PMID:23524391; http://dx.doi.org/ 10.1093/intimm/dxt005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Markovic-Plese S, Hemmer B, Zhao Y, Simon R, Pinilla C, Martin R. High level of cross-reactivity in influenza virus hemagglutinin-specific CD4+ T-cell response: implications for the initiation of autoimmune response in multiple sclerosis. J Neuroimmunol 2005; 169:31-8; PMID:16150497; http://dx.doi.org/ 10.1016/j.jneuroim.2005.07.014 [DOI] [PubMed] [Google Scholar]
- 51.Roti M, Yang J, Berger D, Huston L, James EA, Kwok WW. Healthy human subjects have CD4+ T cells directed against H5N1 influenza virus. J Immunol 2008; 180:1758-68; PMID:18209073; http://dx.doi.org/ 10.4049/jimmunol.180.3.1758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sun J, Li R, Guo J, Jia Y, Sun X, Liu Y, Li Y, Huang F, Lu L, Li Z. Superior molecularly altered influenza virus hemagglutinin peptide 308-317 inhibits collagen-induced arthritis by inducing CD4+ Treg cell expansion. Arthritis Rheum 2012; 64:2158-68; PMID:22231228; http://dx.doi.org/ 10.1002/art.34372 [DOI] [PubMed] [Google Scholar]
- 53.Reijonen H, Kwok WW: Use of HLA class II tetram-ers in tracking antigen-specific T cells and map-ping T-cell epitopes. Methods 2003, 29:282-288; PMID:12725793 [DOI] [PubMed] [Google Scholar]
- 54.Southwood S, Sidney J, Kondo A, del Guercio MF, Appella E, Hoffman S, Kubo RT, Chesnut RW, Grey HM, Sette A. Several common HLA-DR types share largely overlapping peptide binding repertoires. J Immunol 1998; 160:3363-73; PMID:9531296 [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.